- Institute of Medical Mycology, Teikyo University, Tokyo, Japan
The demand for novel antibiotics to combat the global spread of multi drug-resistant pathogens continues to grow. Pathogenic bacteria and fungi that cause fatal human infections can also kill silkworms and the infected silkworms can be cured by the same antibiotics used to treat infections in the clinic. As an invertebrate model, silkworm model is characterized by its convenience, low cost, no ethical issues. The presence of conserved immune response and similar pharmacokinetics compared to mammals make silkworm infection model suitable to examine the therapeutic effectiveness of antimicrobial agents. Based on this, we utilized silkworm bacterial infection model to screen the therapeutic effectiveness of various microbial culture broths and successfully identified a therapeutically effective novel antibiotic, lysocin E, which has a novel mode of action of binding to menaquinone, thus leading to membrane damage and bactericidal activity. The similar approach to screen potential antibiotics resulted in the identification of other therapeutically effective novel antibiotics, such as nosokomycin and ASP2397 (VL-2397). In this regard, we propose that the silkworm antibiotic screening model is very effective for identifying novel antibiotics. In this review, we summarize the advantages of the silkworm model and propose that the utilization of silkworm infection model will facilitate the discovery of novel therapeutically effective antimicrobial agents.
Introduction
The conventional approach of antibiotic discovery includes purification from culture supernatants of soil bacteria by monitoring in vitro antimicrobial activity. This approach makes it difficult to identify novel antibiotics, however, due to the frequent isolation of overlapping chemical entities. In addition, only a small fraction of the antibiotics isolated using this approach exert therapeutic activity in animal models, which further limits the discovery of the therapeutically effective antibiotic. The lack of therapeutic activity by most of the compounds is due to their toxicity and poor pharmacokinetic properties. Thus, the conventional approach of antibiotic screening clearly requires remodeling. To overcome the problems associated with conventional screening methods, we used the silkworm as an animal model to evaluate the therapeutic effects of candidate samples. In this review, we discuss the advantages of the silkworm model for antimicrobial drug development.
Advantages of the Silkworm as an Animal Model
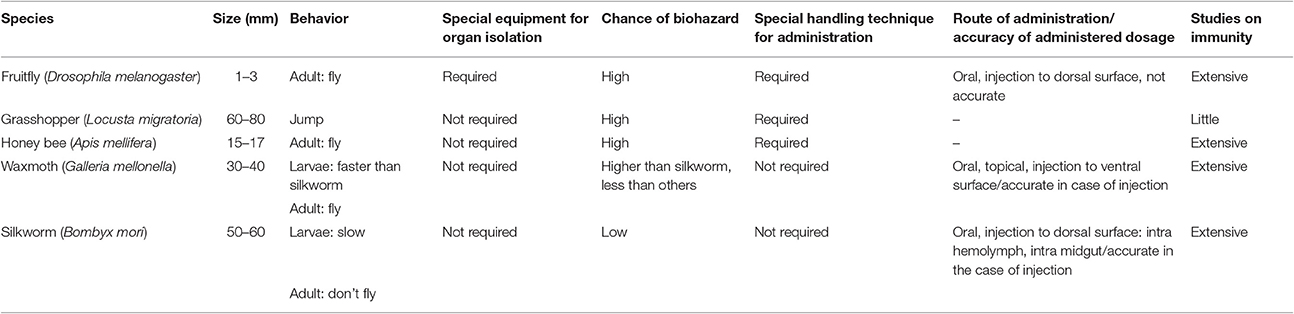
Silkworms, the larvae of the domesticated moth Bombyx mori, have been used for silk production for more than 5,000 years. The silk industry originally started in China, distributed toward several parts of Asia and the West, and has contributed greatly to the economic development of countries along the Silk Road. While B. mori continues to be a major player in sericulture throughout the world, it has also gained importance for biotechnology as a bioreactor for the production of recombinant proteins and silk-based biomaterials (Altman et al., 2003; Kato et al., 2010). From an anatomic point of view, B. mori harbors most of the organs and systems present in mammals, leading scientists to use B. mori as an excellent model organism to elucidate various processes in life sciences, which has been facilitated by the availability of its complete genome sequence and the development of technologies for genetic manipulation. As an animal model, B. mori has clear advantages over other organisms (Table 1).
Ethical Issues
Over the last several years, animal welfare concerns have forced scientists to reduce the number of vertebrates, especially mammals, used as experimental animal models, and alternative animal models that do not require approval by the ethics committee have been sought. Given that silkworms have been used in the silk industry for centuries and their application for research does not require ethical clearance, their use as experimental animal models is both less complicated and less costly compared to vertebrates.
Rearing System
The utilization of silkworms in the silk industry has facilitated its domestication. The proper methods of feeding and maintaining silkworms are well-established. The development of artificial diet in the 1960s replaced the seasonally available mulberry leaves and allowed year-round utilization of silkworms. The feeding of artificial diet not only facilitated breeding and rearing but also contributed to a uniform quality of silkworms. Uniformity in quality is important for robust and reproducible results in studies performed using model animals. The widespread distribution of the silk industry makes it easy to obtain fertilized eggs, thus dramatically reducing the time and labor required to care for silkworms. As fifth-instar day 2 larvae are utilized for infection assays, the total silkworm rearing time required from hatching of the eggs to using the larvae for infection assays is <3 weeks, which is far shorter than the time required for mammals. Utilization of aseptic procedures during rearing allows the generation of germ-free larvae. Furthermore, larvae molt four times; thus making it easy to distinguish each instar stage and reducing individual genetic differences.
Cost Issue
A silkworm rearing room generally includes an incubator and a safety cabinet. Thus, silkworm rearing does not require large and expensive equipment, making it much less expensive than other animal models. Furthermore, in contrast to mammalian models, large numbers of larvae can be reared in a single cage, which significantly reduces the cost of maintaining silkworms. Based on our experience, the cost of using silkworms is 1% that of the cost of utilizing mice.
Handling
In general, experiments utilizing silkworms require less time than those utilizing mammalian models. Further, silkworm larvae have a large enough body size for easy handling by human hands, and samples can be injected with the same syringe type used for medical purposes in humans (Figure 1A). Samples can be injected into the hemolymph, which corresponds to intravenous injection in mammals, and the midgut, corresponding to oral administration. Moreover, silkworm organs such as the midgut can be isolated for experimental use. Silkworms are easy to work with; they do not have sharp horns/hair, teeth, or claws that can sting, and do not bite. Moreover, the adult moths cannot fly and the movement of silkworm larvae is slow and weak, making them easy to handle for injecting samples. This weak and slow movement also makes it difficult for silkworms to escape from their cages, allowing us to perform infection assays for level 2 pathogens. As silkworms have been very highly domesticated through the long history of the silk industry, they cannot survive or reproduce in a natural environment. This further adds to the safety of using silkworms for injection of pathogenic organisms and reduces the biohazard potential.
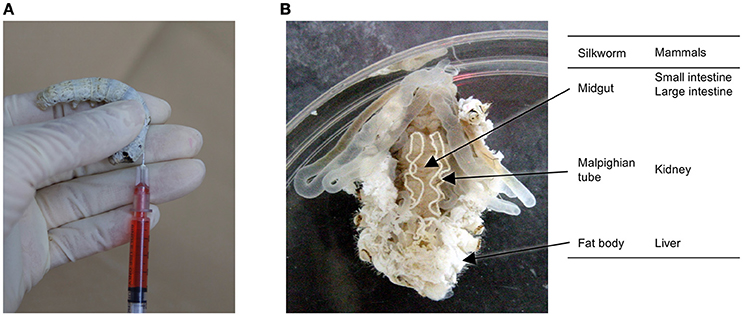
Figure 1. Silkworm and its organs. (A) Manual injection of a sample into the silkworm hemolymph using a 1-ml disposable syringe, and (B) comparison of silkworm organs involved in drug absorption, metabolism, and elimination between silkworms and mammals.
Gene Editing
Attempts to decode the B. mori genome were performed in 2004 and 2008 (Xia et al., 2004; The International Silkworm Genome Consortium, 2008). The availability of the genome sequences facilitated the development of basic genetic and molecular genetic tools and markers. Over the last decade, the development of genetic technologies for B. mori has greatly advanced. Genetic modification may be achieved by transposon-based technology, such as transgene integration or expression; RNA interference-based gene silencing; gene- and enhancer-trap methods and genome editing technology utilizing zinc finger nucleases; transcription activator-like effector nucleases (TALENs); and CRISPR/Cas9 (Xu and O'Brochta, 2015). Furthermore, Dr. Sezutsu's group in the National Agriculture and Food Research Organization, Japan, recently succeeded in utilizing these various techniques to edit the silkworm genome (Inagaki et al., 2015; Sakurai et al., 2015; Takasu et al., 2016). With these recent developments, the silkworm now has a sophisticated genetic modification system and can thus be used to establish disease models and humanized models to screen candidate compounds and analyze physiologic processes.
Common Pharmacokinetics between Silkworm and Mammals
Silkworm organs, such as the gut, fat body, and malpighian tubule, correspond to the intestine, liver, and kidney, respectively, in mammals, and are involved in the metabolism and excretion of external compounds (Figure 1B). Pharmacokinetic parameters are very important from the view of therapeutic activity. Even compounds that exhibit good activity in vitro will not be effective in vivo if they have poor pharmacokinetic parameters. We demonstrated that the half-lives of model compounds in silkworms are similar to those in mammals (Hamamoto et al., 2009). We also found that a model compound is metabolized in silkworms by the cytochrome P450 enzyme, follows the metabolic pathway via the conjugation reaction, and exhibits the similar pharmacokinetics as in mammals (Hamamoto et al., 2009). We also demonstrated that the general non-specific transport of molecules through paracellular routes is comparable between the mammalian intestine and the silkworm midgut (Hamamoto et al., 2004, 2005). In addition, the doses of cytotoxic chemicals that are lethal in 50% of the animals (LD50) are similar between silkworms and mammals, indicating that the toxicity of compounds listed in the OECD guidelines can be evaluated using silkworms (Usui et al., 2016). Based on this, we have used silkworms to optimize antimicrobial agents for less acute toxicity and succeeded to increase the median lethal dose (LD50) from 100 to 230 μg/g larvae (Paudel et al., 2013).
Host–Pathogen Interaction and Immune Response in Silkworm
Pathogens respond and adapt to the signals appearing in the host environment to successfully infect the host and in most cases, the host's response is critical to determine the degree of pathogenicity. An ideal animal model should provide a response similar to that of humans, during infections. Although insects like silkworm lack acquired immunity, innate immunity is widely conserved among mammals and insects (Hoffmann and Reichhart, 2002). Immune response by silkworms includes humoral response such as: production of various proteins like phenoloxidase, antimicrobial peptides, lysozymes, lectins, serine proteases, etc.; and cellular response such as hemocyte-mediated phagocytosis, encapsulation, nodule formation, etc. We previously reported the activation of innate immunity in silkworm during pathogen invasion and found that a cytokine-like paralytic peptide induced humoral and cellular responses and played an important role in silkworm immunity. Further, we found that the paralytic peptide promoted the engulfment of bacteria and induced nitric oxide (NO) production, that is required for both p38 and MAPK activation (Ishii et al., 2013, 2015a).
All the features of silkworms mentioned above suggest that they are a suitable alternative evaluation system for determining the therapeutic effectiveness of candidate drugs. In addition, the availability of a large number of silkworms with low cost and no ethical issues allows us to screen novel antibiotics for their therapeutic effectiveness.
Advantage of Screening Novel Antibiotics Using the Silkworm Infection Model
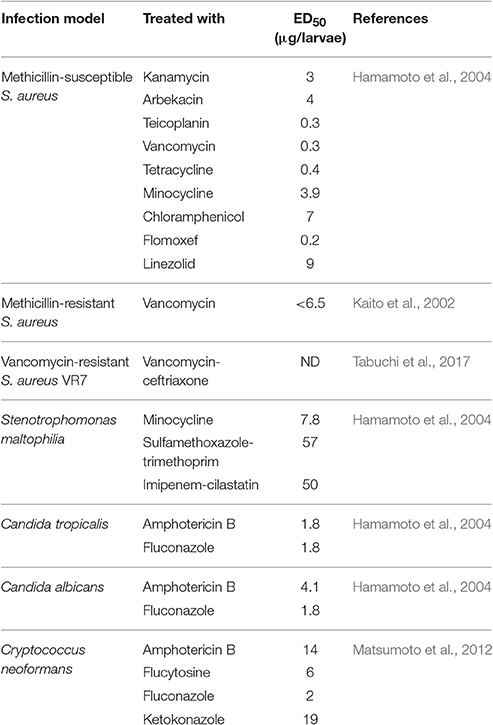
We have previously shown that pathogenic bacteria and fungi that cause fatal infections in human beings such as methicillin sensitive and resistant Staphylococcus aureus; Pseudomonas aeruginosa can also kill silkworms. Furthermore, these infections were cured with the same antibiotics used clinically to cure these infections (Kaito et al., 2002; Hamamoto et al., 2004). Since then, silkworms have been utilized to study pathogenic bacterial toxins (Hossain et al., 2006); evaluate the target specificity of antibacterial agents (Kurokawa et al., 2009); identify novel S. aureus virulence genes (Kaito et al., 2005; Miyazaki et al., 2012); and identify novel probiotic bacteria that promote survival during P. aeruginosa infection (Nishida et al., 2016). Additionally, infection model of multiple pathogenic microorganisms Stenotrophomonas maltophilia (Hamamoto et al., 2004); Vibrio vulnificus (Yamamoto et al., 2016); Vibrio cholera (Kaito et al., 2002); Candida tropicalis (Hamamoto et al., 2004); Candida albicans (Hamamoto et al., 2004); Aspergillus fumigatus (Nakamura et al., 2017a); Cryptococcus neoformans (Matsumoto et al., 2012); and laboratory generated vancomycin-resistant S. aureus VR7 (Tabuchi et al., 2017) have been developed and the success of the antimicrobial to rescue the silkworms from the effect of pathogens have been reported (Table 2). These findings suggest that silkworm infection models can be utilized to mimic the infections caused by various bacterial and fungal pathogens.
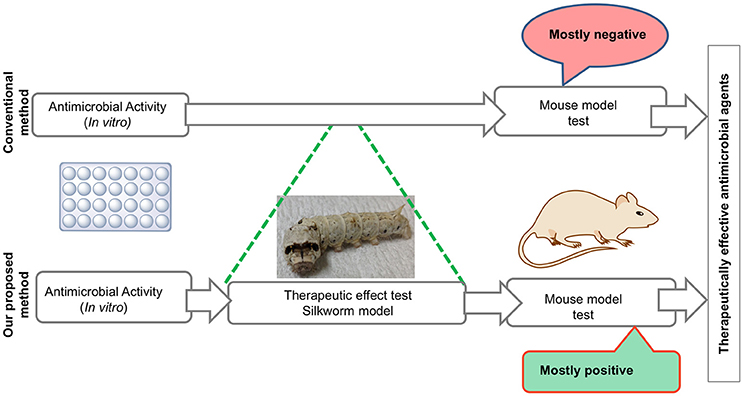
We also demonstrated that the dose required to cure 50% of fatal infections (ED50) in silkworms is similar to that in mice, suggesting that the pharmacokinetics of these antibiotics are similar between silkworms and mammals (Hamamoto et al., 2004). This shared common feature in the pharmacokinetic parameters of clinically applied antibiotics between mammals and silkworms led us to utilize the silkworm infection model to screen for antimicrobial agents. Given that most of the antibiotics currently used clinically were derived from natural products isolated from microorganisms and these microorganisms serve as a repertoire of novel antibiotics, the utilization of silkworms has a further advantage—the possibility of injecting a crude extract. The injection of a crude extract from natural products, such as plant extracts and supernatant of soil bacteria, is prohibited in mammals by the experimental guidelines for animal welfare due to the lack of a rational basis for the experiment. Soil bacteria can produce several anti-bacterial agents, which are often effective only in in vitro conditions and do not show therapeutic efficacy. Purification based on in vitro activity alone may miss to identify therapeutically effective compounds. Furthermore, our previous experience of selecting the hits from a chemical library using minimum inhibitory concentrations (MIC) as an indicator showed that these compounds had no therapeutic activity (Paudel et al., 2012, 2013). Therefore, we chose to evaluate the therapeutic activity using the silkworm infection model to purify therapeutically effective compounds. By doing so, we can easily eliminate the compounds that are only effective in vitro and focus on therapeutically effective compounds, even when their production level is lower than that of compounds that are only effective in vitro, which reduces time, effort, resources, and cost to obtain a therapeutically effective compound (Figure 2). These features of the silkworm infection model facilitate the identification of novel, therapeutically effective antibiotics from natural products.
Development of Novel Antimicrobial Agents Using the Silkworm Infection Model
We developed the silkworm infection model by evaluating the effects of various pathogenic bacteria and fungi. Further, antibiotics and antifungal agents clinically used against those bacteria and fungi effectively cured infections in silkworms. By utilizing the silkworm infection model, we and collaborators successfully identified novel antimicrobial agents—lysocin E (Hamamoto et al., 2015), nosokomycins (Uchida et al., 2010), and ASP2397 (Nakamura et al., 2017a). The details of the discovery of lysocin E and ASP2397 are presented in this review.
Identification of Novel Antibiotics Lysocin E from Soil Bacteria
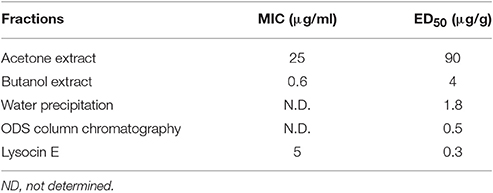
We collected soil samples from various places within Japan and isolated bacteria from these samples. We screened the supernatants of the soil bacteria for in vitro antimicrobial activity against methicillin-resistant Staphylococcus aureus as a primary screening. Of the 14,651 supernatants, 2,794 (19%) had in vitro inhibitory activity against S. aureus. Surprisingly, only 23 of the 2,794 culture supernatants had therapeutic activity in the silkworm infection model. This low hit rate for the therapeutic effectiveness of the culture supernatants indicates that the silkworm infection model is highly effective for excluding antibiotics that do not have therapeutic effectiveness. We purified antibiotics from the culture supernatant of a Lysobacter, which has recently attracted high interest due to their capacity to produce antibiotics and other natural products (Panthee et al., 2016). Purification based on the therapeutic activity observed in silkworms infected with S. aureus from the culture broth of Lysobacter sp. RH2180-5 led to the identification of a therapeutically active novel antibiotic, lysocin E (Hamamoto et al., 2015) (Table 3, Figure 3A). The detailed purification process showed that the in vitro activity was higher for the partially purified butanol extract than purified lysocin E. This indicated that the culture broth contained both therapeutically active and inactive antimicrobial components and our success in finding a therapeutically active antibiotic relied on utilization of the silkworm screening system.
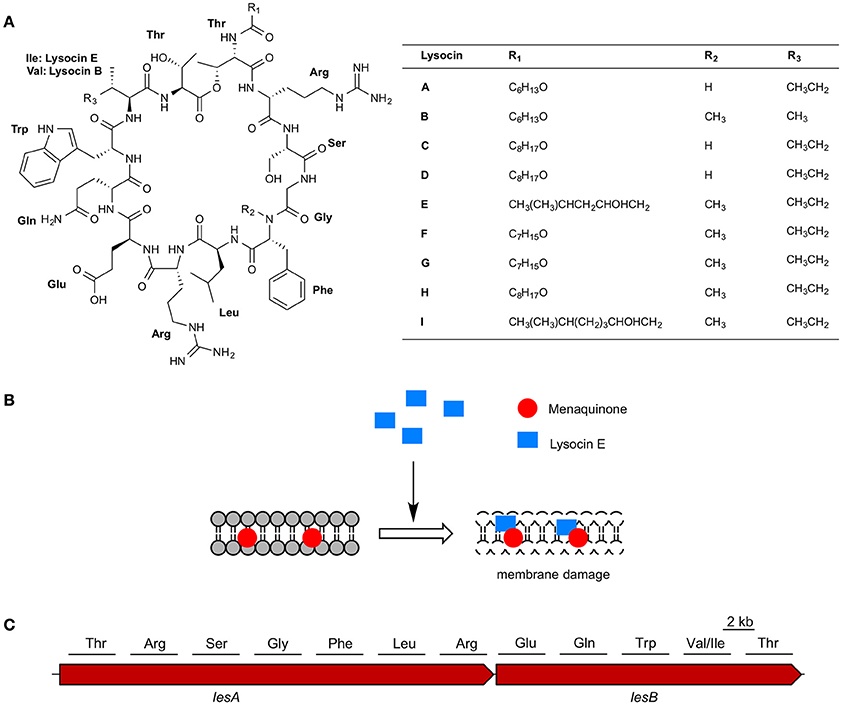
Figure 3. Lysocins, mode of action, and biosynthesis. (A) Chemical structure of lysocins A–I and (B) schematic representation of membrane damage by lysocin E (C) non-ribosomal peptide synthetases involved in lysocin biosynthesis and amino acids activated by the 12 modules in LesA and LesB.
Lysocin E is a cyclic peptide that exhibits antimicrobial activity against various Staphylococci, Bacillus subtilis, Bacillus cereus, and Listeria monocytogenes with MICs ranging from 1 to 4 μg/ml (Table 4). Based on the genetic and biochemical analysis, we identified menaquinone, a membrane molecule important for the bacterial electron transport chain, as the lysocin E target (Figure 3B). Lysocin E is the first antibiotic discovered that targets menaquinone (Hamamoto et al., 2015; Paudel et al., 2016). Lysocin E also has potent therapeutic activity in a mouse infection model of methicillin-resistant S. aureus (ED50: 0.5 mg/kg). Lysocin E did not exhibit acute toxicity in mice at a dose up to 400 mg/kg. The chemical structure of lysocin E comprises 12 amino acids arranged linearly by two large multimodular non-ribosomal peptide synthetases named lysocin E synthetase (LesA and LesB) that have a total of 12 modules and 43 domains in a 1.7-MDa core peptide (Figure 3C) (Panthee et al., 2017). The potent therapeutic activity and low toxicity of lysocin E in animal infection models suggest its high potential for clinical application and the identification of its gene cluster opened avenues for further derivatization of the structure with enhanced activity.

Table 4. MIC of lysocin E against various microorganisms (Hamamoto et al., 2015).
Identification of a Novel Antifungal Compound, ASP2397 (VL-2397)
To identify antifungal compounds, the clinical fungal isolate A. fumigatus FP1305 was used as a test strain. Astellas group screened culture broths of 310 fungal strains for in vitro activity against A. fumigatus FP1305, followed by testing the ability of the broths to cure silkworms infected with FP1305. The culture broth of Acremonium persicinum MF-347833 had therapeutic activity in the silkworm infection model. To purify the antifungal compound, they first attempted to purify the activity based on the in-vitro antifungal activity, which failed to identify some of the therapeutically active fractions. This finding led to the speculation that the culture broth of A. persicinum MF-347833 contained a mixture of antifungal compounds with therapeutic activity or without therapeutic activity. They next utilized the silkworm infection assay-guided purification method to identify therapeutically active antifungal compounds and successfully identified ASP2397 (Nakamura et al., 2017a,b) (Figure 4). ASP2397 has an MIC of 0.2 μg/ml against A. fumigatus FP1305 and is also effective against azole-resistant A. fumigatus (Arendrup et al., 2016), displays therapeutic activity in mice infected with A. fumigatus, and has no cytotoxicity toward mammalian cells at a concentration up to 50 μg/ml, indicating its potential as a therapeutic agent. The planar structure of ASP2397 indicates that it is a metal ion chelator that harbors aluminum, indicating the importance of metal chelation for its therapeutic activity. The fact that ASP2397 was not identified by the conventional approach highlights the utility of the silkworm model for identifying therapeutically active drug molecules.
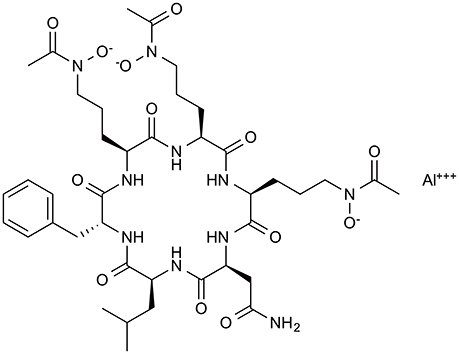
Figure 4. Chemical structure of the novel antifungal compound ASP2397 (VL-2397). ASP2397, purified based on in-vivo therapeutic effect in silkworm infection model.
Conclusion
In summary, we used silkworms as a primary screening system that has several advantages, including an established rearing system, cost effectiveness, reproducible and robust application, no ethical issues, and conserved metabolic pathways with mammals. We established silkworm infection models of various bacteria and fungi and found that silkworm infection model facilitates the discovery of therapeutically effective compounds. Furthermore, we utilized the silkworm infection model to purify compounds from natural sources and identified the antimicrobial agents with therapeutic activity that would otherwise go unidentified. As an example, ASP2397 was identified from Acremonium persicinum MF-347833 using silkworms, but was not identified by the purification approach based on in vitro activity. Similarly, the purification of lysocin E from a crude extract that had more potent in vitro activity than lysocin E itself is an example of how the silkworm screening system avoids some of the restrictions placed on mammal-based screening systems. Thus, the silkworm model is suitable for developing novel therapeutically effective antibiotics from natural products.
Author Contributions
SP wrote the manuscript, AP revised the manuscript, HH determined the outline, and KS critically revised the manuscript and approved the submission.
Conflict of Interest Statement
KS is a consultant for Genome Pharmaceutical Institute, Co. Ltd.
The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Ikuko Nakamura, Astellas Pharma Inc., for critical reading of this paper. Lysocin research in our laboratory was supported by MEXT KAKENHI (JP221S0002), Grant-in-Aid for Scientific Research on Innovative Areas (JP26102714), Grant-in-Aid for young scientists (A) (JP24689008), and Takeda Science Foundation to HH; and in part by Grant-in-Aid for scientific research (S) (JP15H05783) and Drug Discovery Support Promotion Project from Japan Agency for Medical Research and Development, AMED, to KS.
References
Altman, G. H., Diaz, F., Jakuba, C., Calabro, T., Horan, R. L., Chen, J., et al. (2003). Silk-based biomaterials. Biomaterials 24, 401–416. doi: 10.1016/S0142-9612(02)00353-8
Arendrup, M. C., Jensen, R. H., and Cuenca-Estrella, M. (2016). In vitro activity of ASP2397 against Aspergillus isolates with or without acquired azole resistance mechanisms. Antimicrob. Agents Chemother. 60, 532–536. doi: 10.1128/AAC.02336-15
Hamamoto, H., Kamura, K., Razanajatovo, I. M., Murakami, K., Santa, T., and Sekimizu, K. (2005). Effects of molecular mass and hydrophobicity on transport rates through non-specific pathways of the silkworm larva midgut. Int. J. Antimicrob. Agents 26, 38–42. doi: 10.1016/j.ijantimicag.2005.03.008
Hamamoto, H., Kurokawa, K., Kaito, C., Kamura, K., Manitra Razanajatovo, I., Kusuhara, H., et al. (2004). Quantitative evaluation of the therapeutic effects of antibiotics using silkworms infected with human pathogenic microorganisms. Antimicrob. Agents Chemother. 48, 774–779. doi: 10.1128/AAC.48.3.774-779.2004
Hamamoto, H., Tonoike, A., Narushima, K., Horie, R., and Sekimizu, K. (2009). Silkworm as a model animal to evaluate drug candidate toxicity and metabolism. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 149, 334–339. doi: 10.1016/j.cbpc.2008.08.008
Hamamoto, H., Urai, M., Ishii, K., Yasukawa, J., Paudel, A., Murai, M., et al. (2015). Lysocin E is a new antibiotic that targets menaquinone in the bacterial membrane. Nat. Chem. Biol. 11, 127–133. doi: 10.1038/nchembio.1710
Hoffmann, J. A., and Reichhart, J.-M. (2002). Drosophila innate immunity: an evolutionary perspective. Nat. Immunol. 3, 121–126. doi: 10.1038/ni0202-121
Hossain, M. S., Hamamoto, H., Matsumoto, Y., Razanajatovo, I. M., Larranaga, J., Kaito, C., et al. (2006). Use of silkworm larvae to study pathogenic bacterial toxins. J. Biochem. 140, 439–444. doi: 10.1093/jb/mvj171
Inagaki, Y., Matsumoto, Y., Ishii, M., Uchino, K., Sezutsu, H., and Sekimizu, K. (2015). Fluorescence imaging for a noninvasive in vivo toxicity-test using a transgenic silkworm expressing green fluorescent protein. Sci. Rep. 5:11180. doi: 10.1038/srep11180
Ishii, K., Adachi, T., Hamamoto, H., Oonishi, T., Kamimura, M., Imamura, K., et al. (2013). Insect cytokine paralytic peptide activates innate immunity via nitric oxide production in the silkworm Bombyx mori. Dev. Comp. Immunol. 39, 147–153. doi: 10.1016/j.dci.2012.10.014
Ishii, K., Hamamoto, H., and Sekimizu, K. (2015a). Paralytic peptide: an insect cytokine that mediates innate immunity. Arch. Insect Biochem. Physiol. 88, 18–30. doi: 10.1002/arch.21215
Ishii, K., Tabuchi, F., Matsuo, M., Tatsuno, K., Sato, T., Okazaki, M., et al. (2015b). Phenotypic and genomic comparisons of highly vancomycin-resistant Staphylococcus aureus strains developed from multiple clinical MRSA strains by in vitro mutagenesis. Sci. Rep. 5:17092. doi: 10.1038/srep17092
Kaito, C., Akimitsu, N., Watanabe, H., and Sekimizu, K. (2002). Silkworm larvae as an animal model of bacterial infection pathogenic to humans. Microb. Pathog. 32, 183–190. doi: 10.1006/mpat.2002.0494
Kaito, C., Kurokawa, K., Matsumoto, Y., Terao, Y., Kawabata, S., Hamada, S., et al. (2005). Silkworm pathogenic bacteria infection model for identification of novel virulence genes. Mol. Microbiol. 56, 934–944. doi: 10.1111/j.1365-2958.2005.04596.x
Kato, T., Kajikawa, M., Maenaka, K., and Park, E. Y. (2010). Silkworm expression system as a platform technology in life science. Appl. Microbiol. Biotechnol. 85, 459–470. doi: 10.1007/s00253-009-2267-2
Kurokawa, K., Hamamoto, H., Matsuo, M., Nishida, S., Yamane, N., Lee, B. L., et al. (2009). Evaluation of target specificity of antibacterial agents using Staphylococcus aureus ddlA mutants and D-cycloserine in a silkworm infection model. Antimicrob. Agents Chemother. 53, 4025–4027. doi: 10.1128/AAC.00200-09
Matsumoto, Y., Miyazaki, S., Fukunaga, D. H., Shimizu, K., Kawamoto, S., and Sekimizu, K. (2012). Quantitative evaluation of cryptococcal pathogenesis and antifungal drugs using a silkworm infection model with Cryptococcus neoformans. J. Appl. Microbiol. 112, 138–146. doi: 10.1111/j.1365-2672.2011.05186.x
Miyazaki, S., Matsumoto, Y., Sekimizu, K., and Kaito, C. (2012). Evaluation of Staphylococcus aureus virulence factors using a silkworm model. FEMS Microbiol. Lett. 326, 116–124. doi: 10.1111/j.1574-6968.2011.02439.x
Nakamura, I., Kanasaki, R., Yoshikawa, K., Furukawa, S., Fujie, A., Hamamoto, H., et al. (2017a). Discovery of a new antifungal agent ASP2397 using a silkworm model of Aspergillus fumigatus infection. J. Antibiot. 70, 41–44. doi: 10.1038/ja.2016.106
Nakamura, I., Yoshimura, S., Masaki, T., Takase, S., Ohsumi, K., Hashimoto, M., et al. (2017b). ASP2397: a novel antifungal agent produced by Acremonium persicinum MF-347833. J. Antibiot. 70, 45–51. doi: 10.1038/ja.2016.107
Nishida, S., Ono, Y., and Sekimizu, K. (2016). Lactic acid bacteria activating innate immunity improve survival in bacterial infection model of silkworm. Drug Discov. Ther. 10, 49–56. doi: 10.5582/ddt.2016.01022
Panthee, S., Hamamoto, H., Paudel, A., and Sekimizu, K. (2016). Lysobacter species: a potential source of novel antibiotics. Arch. Microbiol. 198, 839–845. doi: 10.1007/s00203-016-1278-5
Panthee, S., Hamamoto, H., Suzuki, Y., and Sekimizu, K. (2017). In silico identification of lysocin biosynthetic gene cluster from Lysobacter sp. RH2180-5. J. Antibiot. 70, 204–207. doi: 10.1038/ja.2016.102
Paudel, A., Hamamoto, H., Kobayashi, Y., Yokoshima, S., Fukuyama, T., and Sekimizu, K. (2012). Identification of novel deoxyribofuranosyl indole antimicrobial agents. J. Antibiot. 65, 53–57. doi: 10.1038/ja.2011.110
Paudel, A., Hamamoto, H., Panthee, S., and Sekimizu, K. (2016). Menaquinone as a potential target of antibacterial agents. Drug Discov. Ther. 10, 123–128. doi: 10.5582/ddt.2016.01041
Paudel, A., Kaneko, K., Watanabe, A., Shigeki, M., Motomu, K., Hamamoto, H., et al. (2013). Structure-activity relationship study of novel iminothiadiazolo-pyrimidinone antimicrobial agents. J. Antibiot. 66, 663–667. doi: 10.1038/ja.2013.69
Sakurai, T., Mitsuno, H., Mikami, A., Uchino, K., Tabuchi, M., Zhang, F., et al. (2015). Targeted disruption of a single sex pheromone receptor gene completely abolishes in vivo pheromone response in the silkmoth. Sci. Rep. 5:11001. doi: 10.1038/srep11001
Tabuchi, F., Matsumoto, Y., Ishii, M., Tatsuno, K., Okazaki, M., Sato, T., et al. (2017). Synergistic effects of vancomycin and β-lactams against vancomycin-highly resistant Staphylococcus aureus (VRSA). J. Antibiot. in press. doi: 10.1038/ja.2017.7. [Epub ahead of print].
Takasu, Y., Kobayashi, I., Tamura, T., Uchino, K., Sezutsu, H., and Zurovec, M. (2016). Precise genome editing in the silkworm Bombyx mori using TALENs and ds- and ssDNA donors – a practical approach. Insect Biochem. Mol. Biol. 78, 29–38. doi: 10.1016/j.ibmb.2016.08.006
The International Silkworm Genome Consortium (2008). The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 38, 1036–1045. doi: 10.1016/j.ibmb.2008.11.004
Uchida, R., Iwatsuki, M., Kim, Y.-P., Ohte, S., Omura, S., and Tomoda, H. (2010). Nosokomycins, new antibiotics discovered in an in vivo-mimic infection model using silkworm larvae. I: fermentation, isolation and biological properties. J. Antibiot. 63, 151–155. doi: 10.1038/ja.2010.9
Usui, K., Nishida, S., Sugita, T., Ueki, T., Matsumoto, Y., Okumura, H., et al. (2016). Acute oral toxicity test of chemical compounds in silkworms. Drug Discov. Ther. 10, 57–61. doi: 10.5582/ddt.2016.01025
Xia, Q., Zhou, Z., Lu, C., Cheng, D., Dai, F., Li, B., et al. (2004). A draft sequence for the genome of the domesticated silkworm Bombyx mori. Science 306, 1937–1940. doi: 10.1126/science.1102210
Xu, H., and O'Brochta, D. A. (2015). Advanced technologies for genetically manipulating the silkworm Bombyx mori, a model Lepidopteran insect. Proc. Biol. Sci. 282:20150487. doi: 10.1098/rspb.2015.0487
Keywords: silkworm model, pharmacokinetics, therapeutic activity, lysocin E, novel antibiotics
Citation: Panthee S, Paudel A, Hamamoto H and Sekimizu K (2017) Advantages of the Silkworm As an Animal Model for Developing Novel Antimicrobial Agents. Front. Microbiol. 8:373. doi: 10.3389/fmicb.2017.00373
Received: 17 November 2016; Accepted: 23 February 2017;
Published: 07 March 2017.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Kunihiko Nishino, Osaka University, JapanDr. Santi M. Mandal, Indian Institute of Technology Kharagpur, India
Tejas Bouklas, Stony Brook University, USA
Hiroko Tabunoki, Tokyo University of Agriculture and Technology, Japan
Copyright © 2017 Panthee, Paudel, Hamamoto and Sekimizu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kazuhisa Sekimizu, c2VraW1penVAbWFpbi50ZWlreW8tdS5hYy5qcA==
 Suresh Panthee
Suresh Panthee Atmika Paudel
Atmika Paudel Hiroshi Hamamoto
Hiroshi Hamamoto Kazuhisa Sekimizu*
Kazuhisa Sekimizu*