- 1Advanced Scientific Research Leaders Development Unit, Graduate School of Medicine, Gunma University, Gunma, Japan
- 2Laboratory of Bacterial Drug Resistance, Gunma University, Graduate School of Medicine, Gunma, Japan
- 3Department of Bacteriology, Gunma University, Graduate School of Medicine, Gunma, Japan
Bacterial infections to anaerobic site are often hard to be treated because the activity of most of antimicrobials decreases under anaerobic conditions. However, fosfomycin rather provides a greater activity under anaerobic conditions than aerobic conditions. Previously, we found that expression of glpT and uhpT, fosfomycin symporters in enterohaemorrhagic Escherichia coli (EHEC) was upregulated by FNR, a global regulator during the anaerobiosis of the bacterium, which led to increased uptake and susceptibility to this drug. In this study, we showed that expression of glpT and uhpT is induced by CRP-cAMP, the regulator complex under both aerobic and anaerobic conditions. The activity of CRP-cAMP in EHEC was elevated under anaerobic conditions because levels of both CRP and cAMP were higher in the cells when grown anaerobically than those when grown aerobically. Results of expression study using mutants indicated that CRP-cAMP is indispensable for expression of glpT but not uhpT—whereas that of uhpT requires UhpA that is the response regulator composing of two-component system with the sensor kinase, UhpB. The CRP-cAMP protein bound to a region that overlaps RNA polymerase binding site for glpT and region upstream of UhpA binding site for uhpT. FNR bound to a region further upstream of CRP-cAMP binding site on region upstream of the glpT gene. These combined results suggested that increased antibacterial activity of fosfomycin to EHEC under anaerobic conditions is due to activation of FNR and increment of CRP-cAMP activity. Then, FNR enhances the expression of glpT activated by CRP-cAMP while CRP-cAMP and FNR cooperatively aids the action of UhpA to express uhpT to maximum level.
Introduction
Fosfomycin is used for treatment of infectious diseases caused by bacteria including E. coli, Pseudomonas aeruginosa and Staphylococcus aureus. The antibiotic is effective against multidrug-resistant (MDR) pathogens such as extended-spectrum β-lactamase (ESBL) producers and quinolone resistant strains because there is no cross resistance issue to other commonly used antibiotics (Michalopoulos et al., 2011; Dinh et al., 2012). In addition to this benefit, fosfomycin provides a higher antimicrobial activity under anaerobic conditions than aerobic conditions whereas many other antibiotics such as fluoroquinolones and aminoglycosides are less effective under anaerobic conditions (Inouye et al., 1989; Bryant et al., 1992; Morrissey and Smith, 1994; Grif et al., 2001; Kohanski et al., 2010). Bacteria often encounter non-oxygenic or micro-oxygenic situations during infections. For E. coli, available oxygen is depleted when they are in an enteric site and growing together with other members in a microbial complex as biofilm.
Fosfomycin is transported to the cells via GlpT and UhpT, glycerol-3-phosphate and glucose-6-phosphate symporters, respectively, then antagonizes the binding of phosphoenolpyruvate (PEP) to MurA that transfers PEP to the 3′-hydroxyl group of UDP-N-acetylglucosamine in the initial step for bacterial cell wall biosynthesis (Kadner and Winkler, 1973; Argast et al., 1978; Bush, 2012). E. coli strains that are resistant to fosfomycin have been isolated in clinical settings, and these strains have mutations in uhpA and cyaA genes. These genes encode the response regulator composing of two-component system with the cognate sensor kinase, UhpB activated by glucose-6-phosphate and adenylate cyclase that synthesizes cyclic AMP (cAMP), respectively (Nilsson et al., 2003; Takahata et al., 2010). Some laboratory studies in E. coli showed that expression of the glpT gene is activated by the complex of cAMP and its receptor termed cAMP receptor protein (CRP), while that of the uhpT gene is activated by both the CRP and cAMP complex (designated CRP-cAMP) and UhpA (Schumacher and Bussmann, 1978; Merkel et al., 1995). Hence, susceptibility to fosfomycin can be affected by expression of the genes encoding GlpT and UhpT. The effect of these regulatory elements on expression of glpT and uhpT has been investigated in E. coli grown under aerobic conditions because facultative anaerobe is often grown under aerobic conditions in laboratory studies (Schumacher and Bussmann, 1978; Merkel et al., 1995). For E. coli species including EHEC, available oxygen is depleted at enteric sites where they reside or cause infection. Therefore, studying on regulation of glpT and uhpT genes in E. coli grown under anaerobic conditions aids us to more precisely understand efficacy of fosfomycin acting on this bacterium at the infection sites.
We have been interested in studying on how antimicrobial activity of fosfomycin is enhanced under anaerobic conditions. Previously, we found that expression of glpT and uhpT genes in EHEC are activated by FNR (Fumarate Nitrate Reduction) in anaerobic culture, which led to increased susceptibility to fosfomycin (Kurabayashi et al., 2015a). FNR is a transcriptional regulator, and the protein is activated only during the anaerobiosis (Green et al., 1996). Thus, increased susceptibility of fosfomycin under anaerobic conditions is due to elevated productions of GlpT and UhpT transporters via activation of FNR.
In this study, we aim to get further insights into how expression of glpT and uhpT is activated in anaerobically-grown EHEC and how fosfomycin susceptibility is elevated. In addition to FNR that we worked on in our previous study, we investigated roles of CRP, cAMP and UhpA on susceptibility of EHEC to fosfomycin and induction of glpT and uhpT in anaerobic culture. Then, relationship among these regulatory elements was also studied.
Materials and Methods
Bacterial Strains and Culture Conditions
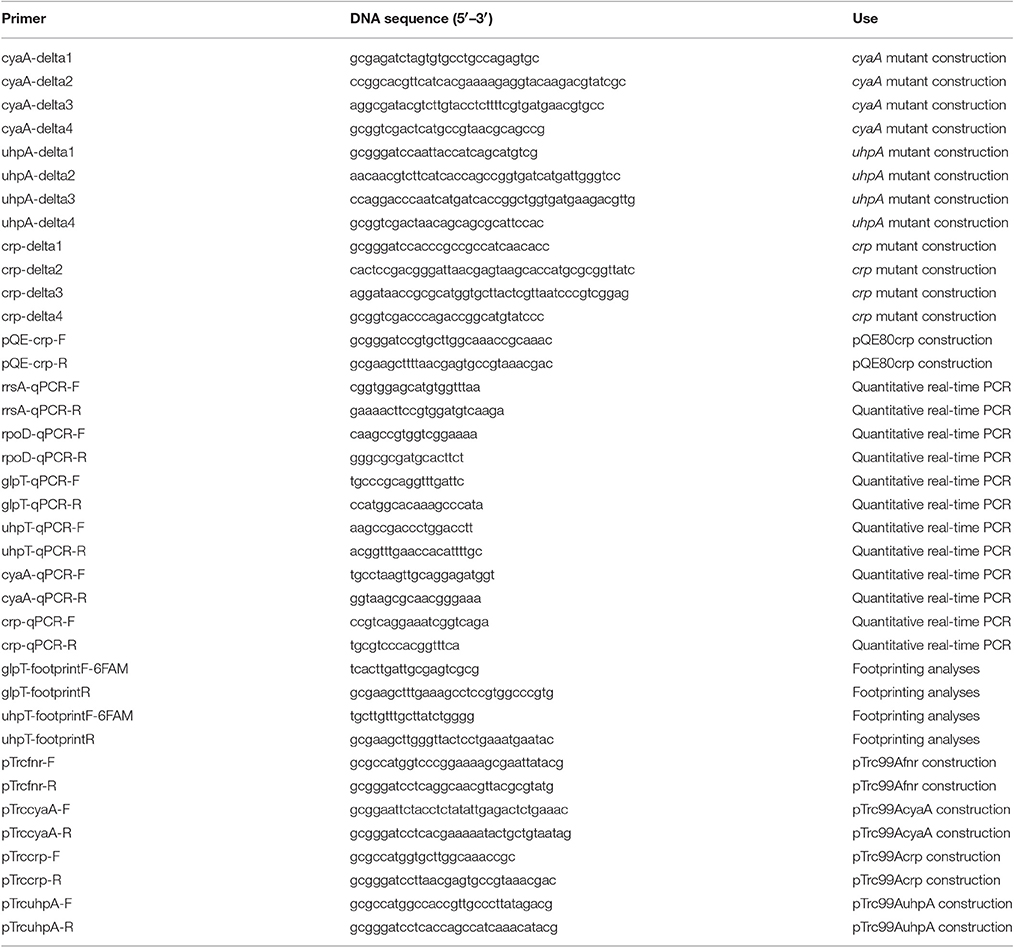
The bacterial strains and plasmids used in this study are listed in Table 1. All bacteria were grown in LB (Luria-Bertani) medium (Nacalai tesque, Kyoto, Japan). For aerobic culture, EHEC strains were grown in loosely capped glass tube with shaking at 160 rpm. For anaerobic culture, we grew EHEC in a sealed container with carbon dioxide gas generators, AnaeroPack-Anaero (Mitsubishi Gas Chemical Co., Inc., Tokyo, Japan) for MIC assays. When we need to monitor the cell growth by measuring absorbance at 600 nm for RNA extraction and cyclic AMP assays, we used Anaerobic Hungate culture tubes equipped with a rubber stopper and a screw cap (Chemglass Life Sciences, Vineland, NJ). For marker selection and maintenance of plasmids, antibiotics were added to growth media at the following concentrations; 150 μg/ml ampicillin, 25 μg/ml kanamycin and 30 μg/ml chloramphenicol.
Antibiotics and Reagents
We obtained antibiotics and reagents used in this study except ampicillin, kanamycin, chloramphenicol and isopropyl β-D-1-thiogalactopyranoside (IPTG) from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). Ampicillin, kanamycin, chloramphenicol and IPTG were purchased from Nacalai tesque (Kyoto, Japan).
Cloning and Mutant Constructions
In-frame deletions of crp, cyaA, and uhpA were constructed by sequence overlap extension PCR according to a strategy described previously (Link et al., 1997), with primer pairs, delta1/delta2 and delta3/delta4 primers for each gene as described in Table 2. The upstream flanking DNA included 450 bp and the first three amino acid codons. The downstream flanking DNA included the last two amino acid codons, the stop codon, and 450 bp of DNA. These deletion constructs were ligated into BamHI and SalI-digested temperature sensitive vector pKO3 (Link et al., 1997) and introduced into HH-H7-008, the parent strain (Hirakawa et al., 2009). We selected sucrose-resistant/chloramphenicol-sensitive colonies at 30°C and confirmed the resulting mutant strains using PCR analysis and DNA sequencing.
To construct His6-CRP expression plasmid pQE80crp, the crp gene was amplified with the primer pair shown in Table 2. The product was digested with BamHI and HindIII and ligated into similarly digested pQE80 plasmid.
To construct pTrc99Afnr, pTrc99AcyaA, pTrc99Acrp, or pTrc99AuhpA for complementation tests, we amplified the fnr, cyaA, crp or uhpA gene, and digested with NcoI and BamHI for pTrc99Afnr, pTrc99Acrp, and pTrc99AuhpA or EcoRI and BamHI for pTrc99AcyaA, then ligated into similarly digested pTrc99A plasmid. All constructs were confirmed by DNA sequencing.
Drug Susceptibility Assays
To test susceptibility of EHEC to fosfomycin, MIC assays were performed by a serial agar dilution method with the standard method of the Clinical and Laboratory Standards Institute (CLSI) (Clinical Laboratory Standards Institute, 2011). To conduct MIC assays under anaerobic conditions, we grew EHEC strains without shaking in a sealed container with AnaeroPack-Anaero for 20 h. After taking the cultures out the anaerobic container, five thousand cells were immediately inoculated on Mueller-Hinton agar (EIKEN Chemical Co., Ltd., Tochigi, Japan) containing fosfomycin, and the agar plates were incubated in the sealed container with fresh AnaeroPack-Anaero gas generator pack at 37°C. The MICs were determined as the lowest concentration at which growth was inhibited.
Promoter Assays
EHEC strains carrying pNN-glpT-P or pNN-uhpT-P, the LacZ reporter plasmid, were aerobically or anaerobically grown at 37°C in LB medium. To measure lacZ expression from pNN-uhpT-P, we added 25 μg/mL of glucose-6-phosphate into the medium because the promoter activity of uhpT gene was too low to be detected when glucose-6-phosphate is absent. β-Galactosidase activities from lacZ expression in cell lysates were determined as Miller's method (Miller, 1992).
RNA Extraction and Quantitative Real-Time PCR Analyses
Bacteria were grown to mid-logarithmic growth phase (OD600 ~0.4) in LB medium. Total RNA extraction and cDNA synthesis were performed using SV Total-RNA Isolation System and GoScript™ Reverse Transcription System as described by the manufacturer (Promega Corp., Madison, WI). Real-time PCR included 2.5 ng cDNA and 200 nM primers in SYBR Select Master Mix (Applied Biosystems, Foster City, CA) and were run on an ABI Prism 7900HT Fast Real Time PCR System. Constitutively expressed rrsA and rpoD genes that respectively encode 16S ribosomal RNA and RNA polymerase sigma 70 factor were used as an internal control (Kurabayashi et al., 2014). Primers are listed in Table 2. Amplification plot and melting curve data are available upon request.
Cyclic-AMP (cAMP) Assays
Intracellular cAMP levels of EHEC grown under aerobic or anaerobic conditions were determined by using Cyclic AMP EIA Kit (Cayman Chemical, Ann Arbor, MI). Bacteria were grown in 20 ml of LB medium to late logarithmic phase and harvested. The cell pellets were once washed in phosphate buffered saline (PBS) and suspended in 0.5 ml of EIA buffer supplied in the kit, then lysed by sonication. The lysate was centrifuged and the amount of cAMP in the resulting supernatant was quantified using cAMP AChE Tracer, cAMP EIA Antiserum and Ellman's reagent according to the manufacturer protocol. cAMP levels in fraction of the cell lysate were represented as pmol per cells producing 1 μg of protein.
Overexpression and Purification of D154AFNR and CRP
N-Terminal histidine tagged D154AFNR and CRP, His6-D154AFNR, and His6-CRP were expressed in and purified from E. coli Rosetta™(DE3) (Novagen/EMD Bioscience, Philadelphia, PA), respectively. We supplied 100 μM cAMP into the buffer for purification of His6-CRP. Bacteria containing recombinant plasmid were grown at 37°C to an OD600 of 0.4 in LB, 0.5 mM IPTG was then added, and culture growth was continued for 3 h. Cells were harvested and stored at −80°C overnight. The cell pellet was suspended in lysis buffer (20 mM Tris [pH 7.9], 500 mM NaCl and 10% glycerol) and lysed by sonication. The lysate was centrifuged and the resulting supernatant was mixed with Ni-NTA agarose (Qiagen, Valencia, CA) for 1 h. The agarose was washed with 50 mM imidazol twice and then protein was eluted with 200 mM imidazol. The protein was >95% pure as estimated by SDS-PAGE electrophoresis and Coomassie brilliant blue staining. Protein concentration was determined using the Bio-Rad protein assay (Bio-Rad, Hercules, CA). The protein was diluted into gel-shift/footprinting assay buffer (20 mM Tris [pH 7.5], 50 mM KCl, 1 mM dithiothreitol, 10% glycerol and 200 μM cAMP) to a concentration of 4 pmol/μl, and stored at 4°C.
Gel-Shift Assays
Gel-shift assays were performed as previously described (Kurabayashi et al., 2015a). We used DNA probes containing the 300 bp region upstream of the glpT and uhpT gene, respectively. We also used a DNA fragment from Pseudomonas aeruginosa rhlR gene as a non-specific control probe. The probe DNA fragments (0.30 pmol) were mixed with purified CRP and/or D154AFNR in a 10 μl reaction mixture containing gel-shift/footprinting assay buffer described above. After incubation for 20 min at room temperature, samples were separated by electrophoresis on a 5% polyacrylamide Tris-glycine/EDTA (10 mM Tris [pH 8.0], 380 mM glycine and 1 mM EDTA) gel in Tris-glycine/EDTA buffer at 4°C. DNA bands in the gel were stained by 10,000-fold-diluted SYBR Green I nucleic acid stain (Lonza, Walkersville, MD), and visualized under UV light at 300 nm.
DNase I Footprinting
DNase I footprinting was performed using a previously described non-radiochemical capillary electrophoresis method on an ABI PRISM genetic analyzer equipped with an ABI PRISM GeneScan (Hirakawa et al., 2005). The 6-carboxyfluorescein (6-FAM)-labeled (5′), 300-bp DNA fragments (starting at 300 bp upstream of the glpT start codon and ending 1 bp upstream of the glpT start codon for glpT probe and starting at 300 bp upstream of the uhpT start codon and ending 1 bp upstream of the uhpT start codon for uhpT probe, respectively) were generated by PCR amplification with primer pairs a 6-FAM labeled forward primer (glpT-footprintF-6FAM) and an unlabeled reverse primer (glpT-footprintR) for glpT probe and a 6-FAM labeled forward primer (uhpT-footprintF-6FAM) and an unlabeled reverse primer (uhpT-footprintR) for uhpT probe. Each DNA fragment (0.45 pmol) was mixed with purified protein (20 pmol) in a 50 μl reaction mixture containing the same buffer as above. After incubation for 20 min at room temperature, DNase I (0.3 units, Promega Corp., Madison, WI) was added. After incubation for 60 s at room temperature, samples were purified for electrophoresis on the ABI PRISM genetic analyzer apparatus.
Statistical Analysis
P-value in each assay was determined by unpaired t-test and two-way ANOVA with the GraphPad Prism version6.00.
Results
Deletion of crp or cyaA Decreases Expression of Both glpT and uhpT in Anaerobically-Grown EHEC While Deletion of uhpA Only Decreases Expression of uhpT, but Not glpT
cAMP-bound CRP (CRP-cAMP) activates expression of both glpT and uhpT genes while UhpA activates expression of uhpT but not glpT. The role of these activators on expression of glpT and uhpT has been demonstrated in aerobically grown E. coli laboratory strains (Schumacher and Bussmann, 1978; Merkel et al., 1995). To investigate the effect of these regulators on expression of glpT and uhpT in EHEC grown under anaerobic conditions, we constructed crp, which encodes the regulatory protein, cyaA, which encodes cAMP synthase and uhpA deletion mutants in EHEC O157:H7 background, and measured promoter activities of glpT and uhpT via lacZ expression from the reporter plasmids pNN-glpT-P and pNN-uhpT-P in these mutant strains when aerobically or anaerobically grown. Previously, we found that expression of glpT and uhpT is activated by FNR during anaerobic culture (Kurabayashi et al., 2015a). Initially, we showed that levels of lacZ expression from both pNN-glpT-P and pNN-uhpT-P in the wild-type parent when grown under anaerobic conditions were indeed higher than those when grown under aerobic conditions, and expression levels from these two genes under anaerobic conditions were decreased by deletion of fnr (Figures 1A,B). We note that contribution of fnr to uhpT expression under anaerobic conditions is partial because significant level of lacZ expression from pNN-uhpT-P in the fnr mutant was still observed. This observation implies that there is yet another unknown factor that elevates expression of uhpT during EHEC anaerobiosis.
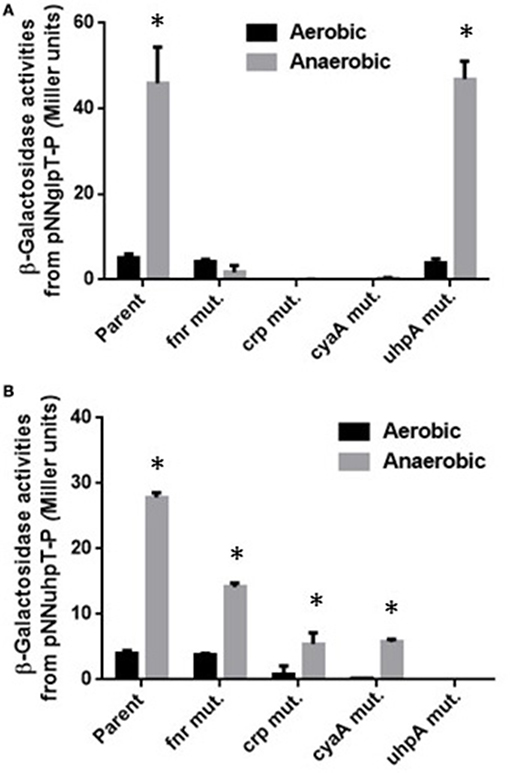
Figure 1. β-Galactosidase activities of EHEC wild-type parent and its derivative mutants containing lacZ reporter plasmids grown under aerobic or anaerobic conditions. β-Galactosidase activities from lacZ expression in these strains correspond to glpT (A) or uhpT (B) promoter activities, then were described as Miller units. Data plotted are the means from three independent experiments; error bars indicate the standard deviations, *P < 0.01. Asterisks denote significance for values of β-galactosidase activity in bacterial cultures under anaerobic conditions relative to those under aerobic conditions.
Levels of lacZ expression from pNN-glpT-P in crp and cyaA mutants were under detectable limit under both aerobic and anaerobic conditions while those from pNN-uhpT-P in these mutants was still detectable under anaerobic conditions, however approximately 5-fold lower than the wild-type parent (Figures 1A,B). Since CRP requires cAMP for its activity, it is convicting that crp and cyaA mutants showed same phenotype for glpT and uhpT expression. On the other hand, deletion of uhpA completely abolished lacZ expression from pNN-uhpT-P, but did not affect that from pNN-glpT-P (Figures 1A,B). These results suggest that CRP-cAMP activates expression of both glpT and uhpT during anaerobic cultures as aerobic cultures, and UhpA only activates that of uhpT.
Deletion of crp, cyaA, or uhpA Leads to a Decrease of Susceptibility to Fosfomycin
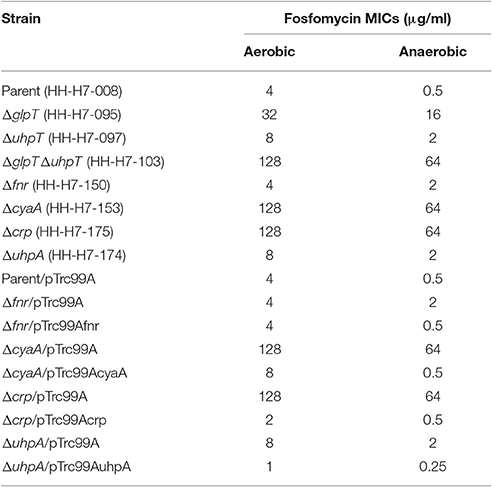
To confirm that lowered expression of glpT and uhpT in deletion of crp, cyaA, or uhpA leads to decrease of susceptibility to fosfomycin, we determined the MICs of fosfomycin for these mutants with the wild-type parent, Δfnr, ΔglpT, ΔuhpT, and ΔglpTΔuhpT strains. As observed in our previous study, the wild-type parent grown under anaerobic conditions exhibited a lower MIC than that grown under aerobic conditions, and the fnr mutant was 4-fold lower susceptible than the wild-type parent under anaerobic conditions, but no difference in the MIC between these strains was seen under aerobic conditions (Table 3). In this study, we found that deletions of crp and cyaA decreased the susceptibility to fosfomycin up to the level of ΔglpTΔuhpT strain under aerobic and anaerobic conditions whereas the susceptibility in uhpA mutant was same as that in uhpT mutant (Table 3). To confirm the contribution of these regulatory genes to glpT and uhpT-dependent susceptibility of EHEC to fosfomycin, we introduced pTrc99Afnr, pTrc99AcyaA, pTrc99Acrp, or pTrc99AuhpA plasmid into each mutant, and determined the MICs of fosfomycin. We observed that MIC phenotypes on fnr, cyaA, crp, and uhpA mutants were actually complemented by introducing exogenous gene expression plasmids, respectively (Table 3). These observations support the results observed in the promoter assay that deletions of crp and cyaA decreased expression of both glpT and uhpT, and deletion of uhpA only decreased expression of uhpT.
Anaerobically Grown EHEC Has a Higher Activity of CRP-cAMP
Activity of CRP-cAMP to regulate expression of target genes is affected by levels of both cAMP and CRP in cells (Hogema et al., 1997). These levels might be elevated during anaerobic culture, then lead to increase of glpT and uhpT expression. To test this hypothesis, we measured levels of intracellular cAMP and cyaA and crp transcripts in aerobically and anaerobically grown EHEC. We found that intracellular concentration of cAMP in EHEC when anaerobically grown was 2-fold higher than that when aerobically grown (Figure 2A). The data of qPCR analyses showed higher transcripts of cyaA and crp in EHEC grown under anaerobic conditions than aerobic conditions (Figure 2B). To confirm that increase of CRP-cAMP activity elevates expression of glpT and uhpT in anaerobic cultures, we measured levels of these genes expression in the wild-type parent grown with or without glucose under anaerobic conditions because addition of glucose as a PTS sugar reduces cAMP level associating with CRP-cAMP activity (Botsford and Harman, 1992). As expected, glucose addition reduced both expression of glpT and uhpT (Figure 2C). When the EHEC was grown with glycerol instead of glucose, we did not observe the reduction of glpT and uhpT expression levels because glycerol does not affect cAMP level (Botsford and Harman, 1992; Figure 2C). These results suggest that activity of CRP-cAMP in EHEC when grown under anaerobic conditions is higher than aerobic conditions, and leads to elevated expressions of glpT and uhpT.
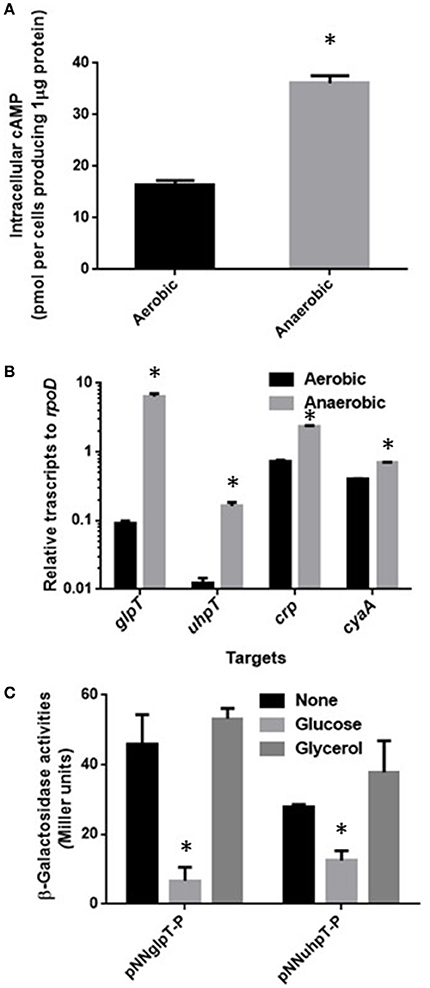
Figure 2. Activity of CRP-cAMP in the wild-type parent grown under aerobic or anaerobic conditions. Intracellular concentration of cAMP (A) and transcript levels of crp and cyaA together with glpT and uhpT (B) in the wild-type parent grown under aerobic or anaerobic conditions were compared. Intracellular concentration of cAMP is given as the amount of cAMP were represented as pmol per cells producing one μg of protein. Transcript levels of crp, cyaA, glpT, and uhpT were described as relative values to that of rpoD (housekeeping gene). Data plotted are the means of two biological replicates, error bars indicate the ranges, *P < 0.01. Asterisks denote significance for values of intracellular cAMP or mRNA level in bacterial cultures under anaerobic conditions relative to those under aerobic conditions. (C) β-Galactosidase activities from lacZ expression in the wild-type parent correspond to glpT or uhpT promoter activities were measured when grown with or without glucose under aerobic and anaerobic conditions. Data plotted are the means from three independent experiments; error bars indicate the standard deviations, *P < 0.01. Asterisks denote significance for values of β-galactosidase activity in the wild-type parent grown with glucose relative to those in the wild-type parent grown without glucose.
FNR is Not Responsible for Increased Activity of CRP-cAMP under Anaerobic Conditions
FNR is a transcriptional regulator that is able to induce expression of particular genes during anaerobic growth (Green et al., 1996). To investigate whether FNR induces expression of crp and cyaA genes during anaerobic growth or not, we compared levels of these genes expression between the wild-type parent and the fnr mutant when aerobically or anaerobically grown by qPCR analyses. Levels of crp and cyaA transcript in the fnr mutant when anaerobically grown were still higher than those when aerobically grown as the wild-type parent exhibited (Figure 3). Thus, FNR is not responsible for the increased activity of CRP-cAMP under anaerobic conditions.
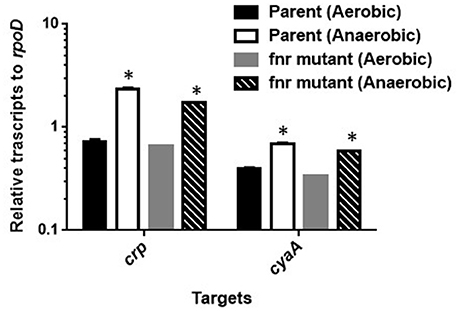
Figure 3. Transcript levels of crp and cyaA genes in the wild-type parent and the fur mutant strains grown under aerobic or anaerobic conditions. These transcript levels were described as relative values to that of rpoD (housekeeping gene). Data plotted are the means of two biological replicates, error bars indicate the ranges, *P < 0.01. Asterisks denote significance for values of mRNA level in bacterial cultures under anaerobic conditions relative to those under aerobic conditions.
CRP-cAMP and FNR Together Bind to the Region Upstream of glpT and uhpT Genes
We previously found that FNR binds to the region upstream of glpT and uhpT genes (Kurabayashi et al., 2015a). To determine whether CRP-cAMP activates expression of glpT and uhpT genes in cooperation with FNR or not, we investigated an ability of CRP-cAMP binds to the region upstream of glpT and uhpT genes in the presence of FNR by gel-shift assays using recombinant proteins. To maintain the CRP protein in active form that cAMP binds (designated CRP-cAMP), we supplied cAMP into the buffer through the process of protein purification and gel-shift assays. For the FNR protein, we used the mutant protein designated D154AFNR that aspartate at amino acid residue 154 in FNR is replaced alanine because the mutant protein is still able to form an active conformation even in the presence of oxygen and then to bind to the target DNA in the same fashion as that of wild-type FNR (Ziegelhoffer and Kiley, 1995). Consistent with results of our previous study, DNA probe bands from glpT and uhpT, but not rhlR were shifted in the presence of the D154AFNR protein (Figure 4). We also observed a retarded motility of the glpT and uhpT probes when the CRP protein is present, and the motility of these probes was further retarded in the presence of both CRP and D154AFNR (Figure 4). These results suggest that CRP-cAMP and FNR together bind to the region upstream of glpT and uhpT genes, then cooperatively activate expression of glpT and uhpT genes.
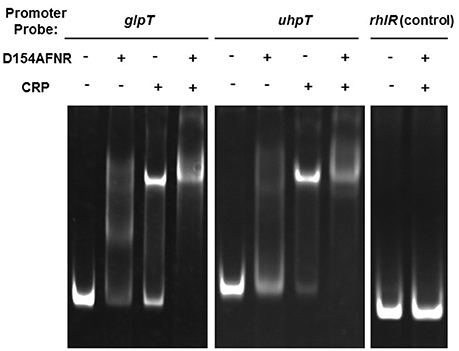
Figure 4. Gel-shift assay showing binding of CRP-cAMP and D154AFNR to the glpT and uhpT promoter. CRP and/or D154AFNR proteins (−CRP and -D154AFNR; no protein, +CRP; 1.2 pmol of the CRP protein, +D154AFNR; 10 pmol of the D154AFNR protein) were added to reaction mixtures containing 0.3 pmol of DNA probe. DNA upstream of rhlR was used as a non-binding (negative) control.
CRP-cAMP and FNR Bind to the Different Site on the Region Upstream of glpT Gene
CRP-cAMP controls target genes in two different manners depending on locations that it binds within region upstream of target genes (Zhou et al., 1994). When CRP-cAMP binds to a site which is typically distant more than 90 bp from the transcription start point of the target gene, CRP-cAMP requires a regulon specific activator protein for transcriptional activation. It has been known that the uhpT gene is in this case. CRP-cAMP binds to the inverted repeat centered at −103 position which site is located at 30 bp region upstream of the DNA site that UhpA binds, then UhpA is essential for uhpT gene activation (Figure 5D; Merkel et al., 1995; Olekhnovich et al., 1999). The data of our promoter analyses supports it because lacZ expression from pNNuhpT in the uhpA mutant was under detectable limit (Figure 1B). Thus, CRP-cAMP is an enhancer for uhpT gene activation.
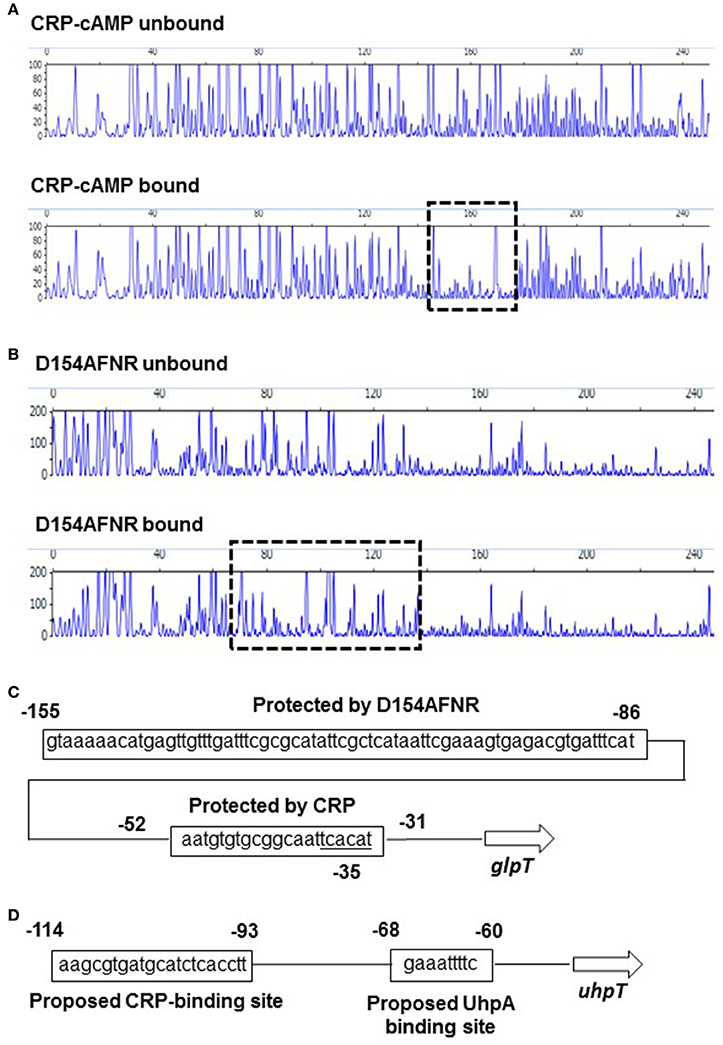
Figure 5. Sequences of the upstream glpT and uhpT region and identification of CRP-cAMP and D154AFNR binding sites. DNase I footprinting of the glpT promoter region. A 300-bp, 6-FAM-labeled DNA fragment was incubated in the presence or absence of CRP (20 pmol) containing cAMP (A) or D154AFNR (20 pmol) (B) and subjected to DNase I digestion. The fluorescence intensities of the DNA fragments (y axis) are plotted relative to their size in bases (x axis). One region (outlined by the dashed line) was protected from DNase I digestion in the presence of 20 pmol protein. (C) Sequence of the upstream glpT region. The region protected from DNase I digestion is indicated by the box. Numbers indicate relative positions to the transcript start site labeled +1. Predicted −35 box is indicated. (D) Sequence of the upstream uhpT region. The CRP and UhpA binding site reported in previous studies (Merkel et al., 1995; Olekhnovich et al., 1999) are indicated by the box. Numbers indicate relative positions to the transcript start site labeled +1.
As the other manner for CRP-cAMP, it alone activates target genes without any other transcriptional activators. In this case, CRP-cAMP binds to the DNA at near position −61.5, −72.5. −82.5, or −92.5 where CRP-cAMP and RNA polymerase bind on the same face of the DNA helix, or CRP-cAMP partly overlays RNA binding site, then it enables to form direct interaction with RNA polymerase (Straney et al., 1989; Gaston et al., 1990; Ushida and Aiba, 1990). We have shown that levels of glpT expression in crp and cyaA mutants were under detectable limit (Figure 1A). Therefore, we hypothesized that CRP-cAMP may act as an essential element for glpT gene expression interacting with RNA polymerase. To test whether this hypothesis is correct or not, we determined the DNA site which CRP-cAMP binds on the region upstream of glpT gene by DNase I footprinting analyses. We found that a 25-bp region including sequence (AATGTGTGCGGCAATTCACATT) which is similar to CRP-binding consensus motif (AA-TGTGA-[N]-TCACA-TT) was protected from DNase I digestion by CRP-cAMP (Figure 5A), suggesting that the CRP-cAMP binds to this site (Gunasekera et al., 1992). The site is located at −30 to −52 position overlaying −35 box (TCACAT) that RNA polymerase binds (Figure 5C). Thus, CRP-cAMP interacts RNA polymerase, and it acts as an essential element for glpT gene activation. In addition to CRP-cAMP binding site, we determined the DNA site which FNR binds. The D154AFNR protein protected a 70-bp region which locates at 32 bp region upstream of CRP-cAMP binding site (Figure 5B). Within this protected region, there was a sequence (TTGAT-TTCG-CGCAT) located at −123 to −136 position that is relatively similar to the proposed FNR-binding motif (TTGAT-[N]4-ATCAA) (Ziegelhoffer and Kiley, 1995; Figure 5C).
We also performed DNase I footprinting experiment to determine FNR binding site on uhpT region upstream. However, contrary to the result of gel-shift assays, we did not observe specific binding of the D154AFNR protein on the DNA probe from uhpT region upstream in footprinting experiment (data not shown).
Discussion
Susceptibility to fosfomycin is affected by expression of GlpT and UhpT transporters. CRP-cAMP is an activator of both glpT and uhpT genes while UhpA activates only the uhpT gene (Schumacher and Bussmann, 1978; Merkel et al., 1995). Among clinically isolated E. coli strains, mutations in uhpA and cyaA genes confer resistance to fosfomycin (Nilsson et al., 2003; Takahata et al., 2010). Fosfomycin is unable to be sufficiently transported into these mutant cells due to very low levels of glpT and uhpT expression. On the other hand, a mutant that overproduces MurA, the target of fosfomycin was isolated, and this strain is resistant to fosfomycin, however the mutation site remains uncharacterized (Horii et al., 1999).
Levels of glpT and uhpT are elevated when EHEC are grown under anaerobic conditions, which leads to increase of susceptibility to fosfomycin followed by increased uptake of this drug. We previously showed that levels of glpT and uhpT expression during anaerobic growth are elevated by FNR, a transcriptional regulator that is activated under anaerobic conditions (Kurabayashi et al., 2015a). In this study, we found that levels of both CRP and cAMP in EHEC were increased, and resulted in elevation of GlpT and UhpT expression (Figures 1, 2). Thus, increased susceptibility to fosfomycin under anaerobic conditions is due to elevation of GlpT and UhpT expression followed by activation of FNR and increment of CRP-cAMP amount.
According to the Clinical and Laboratory Standards Institute (CLSI), the in vivo antibacterial activity of fosfomycin against E. coli species is relatively lower than that of other commonly used antibiotics, such as β-lactams and fluoroquinolones (Clinical Laboratory Standards Institute, 2011). The antibacterial activity was evaluated in aerobic cultures. However, E. coli species including EHEC are usually in oxygen-limited situations during infections, for instance, available oxygen is depleted at enteric sites where they reside or cause infection. In many cases, E. coli grows with other members in a microbial complex as biofilm. Therefore, we suggest that in vitro test may underestimate the in vivo potency of fosfomycin utility.
The manner of gene activation by CRP-cAMP with FNR is different between glpT and uhpT. We suggest that CRP-cAMP probably interacts with the RNA polymerase subunit on glpT gene promoter, then acts as an essential element for glpT gene activation under both aerobic and anaerobic conditions. Results obtained in gel-shift assays, footprinting analyses and promoter assays support this idea because purified CRP-cAMP protein binds on RNA binding site and crp and cyaA mutants showed very low activity of glpT promoter even when anaerobically grown (Figures 1A, 4, 5A,C). Then, FNR further enhances the glpT transcription activated by CRP-cAMP and RNA polymerase (Figures 4, 5B,C). On the other hand, CRP-cAMP is not essential for uhpT transcription because crp and cyaA mutants are still able to express the uhpT gene at moderate level. Transcription of uhpT absolutely requires UhpA (Figure 1B). According to other studies, CRP-cAMP binds to a region that is relatively remote from the RNA polymerase binding site on the uhpT gene promoter, and then it stabilizes the complex of RNA polymerase, the promoter DNA, and UhpA (Merkel et al., 1995; Olekhnovich et al., 1999; Figure 5D). Thus, CRP-cAMP aids UhpA to activate the expression of uhpT. Results of our promoter assays agree with it. We do not still know how FNR involves in activation of uhpT by UhpA and CRP-cAMP. Deletion of fnr moderately decreases the expression of uhpT, however we did not observe a specific binding to region upstream of uhpT gene in footprinting experiment (Figure 1B and data not shown). Gel-shift assays showed that purified FNR protein bound the DNA probe from uhpT region upstream of uhpT gene with much lower affinity than that of glpT although no similar conserved consensus sequence that FNR probably binds on the region upstream was seen (Figure 4; Kurabayashi et al., 2015a). Therefore, unlikely UhpA and CRP-cAMP, the activation of uhpT gene promoter by FNR may be indirect in physiological situations. We also note that FNR does not increase activity of CRP-cAMP because there was no difference in levels of crp and cyaA transcription between the wild-type parent and fnr mutant (Figure 3). Further studies will be necessary to get insights into the molecular mechanism of uhpT gene regulation associating with FNR.
Why is expression of GlpT and UhpT elevated under anaerobic conditions? The native substrates of GlpT and UhpT transporters are glycerol-3-phosphate and glucose-6-phosphate, respectively (Kadner and Winkler, 1973; Argast et al., 1978). E. coli strains including EHEC use these compounds as carbon sources for growth (Kurabayashi et al., 2014). In addition, glycerol-3-phosphate can be utilized as an electron donor for anaerobic respiration (Varga and Weiner, 1995; Unden and Bongaerts, 1997). Therefore, elevated expression of GlpT and UhpT may reduce metabolic cost of bacteria during anaerobic growth. However, as a tradeoff of this benefit for bacteria, the susceptibility to fosfomycin is increased. As experimental observations to support this literature, constitutive reduction of GlpT and UhpT expression in mutant EHEC strains lose the biological fitness, and the strains is outcompeted by the wild-type parent strain that express GlpT and UhpT in normal level (Kurabayashi et al., 2014).
As far as we compared sequences among EHEC O157 and some other E. coli members including MG1655, laboratory-K12 strain and CFT073, uropathogenic E. coli (UPEC), sequences of glpT, uhpT and their regulatory genes such as fnr, uhpA, cyaA, and crp are highly conserved (>95% identity). Therefore, we believe that regulatory mechanism of glpT and uhpT genes associating with FNR, UhpA, CyaA, and CRP may be conserved in E. coli species although it is speculative. In addition to E. coli, some of other species are also more susceptible to fosfomycin under anaerobic conditions than aerobic conditions (Inouye et al., 1989). Several enterobacteria members genetically close to E. coli, such as Salmonella and Shigella have glpT, uhpT and their regulatory genes sharing >88% sequence identity with those in E. coli. However, typical non-enterobacterial MDR pathogens, such as P. aeruginosa and S. aureus lack some regulatory genes that probably control expression of glpT and uhpT. In addition, P. aeruginosa has no uhpT and fnr genes in chromosome (Castaneda-Garcia et al., 2009). Therefore, we presume that the regulatory system of glpT and uhpT gene expression to determine susceptibility to fosfomycin observed in EHEC could be conserved in only E. coli and genetically close members while P. aeruginosa and S. aureus could become highly susceptible to this drug under anaerobic conditions in any other unknown mechanism.
In this study, we found targets to elevate expression of glpT and uhpT transporters under anaerobic conditions, and provided insights into the molecular mechanism of increased susceptibility to fosfomycin. It will aid us to not only more precisely estimate utility and potency of fosfomycin, but also offer a method to promote fosfomycin therapy such as modeling drugs that expression of GlpT and UhpT transporters can be more activated.
Author Contributions
KK, KT, HT, and HH designed research; KK and HH performed reserch; KT, HT, and HH analyzed data; KT, HT, and HH wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was kindly supported by JSPS KAKENHI “Grant-in-Aid for Young Scientists (B)” Grant Number 25870115, JST program; “Improvement of Research Environment for Young Researchers,” Japanese Ministry of Education, Culture, Sport, Science and Technology [Gunma University Operation Grants], Sumitomo Foundation, GlaxoSmithKline (GSK) Research Grant and Research Program on Emerging and Re-emerging Infectious Diseases from Japan Agency Research and development, AMED.
References
Argast, M., Ludtke, D., Silhavy, T. J., and Boos, W. (1978). A second transport system for sn-glycerol-3-phosphate in Escherichia coli. J. Bacteriol. 136, 1070–1083.
Bryant, R. E., Fox, K., Oh, G., and Morthland, V. H. (1992). Beta-lactam enhancement of aminoglycoside activity under conditions of reduced pH and oxygen tension that may exist in infected tissues. J. Infect. Dis. 165, 676–682. doi: 10.1093/infdis/165.4.676
Bush, K. (2012). Antimicrobial agents targeting bacterial cell walls and cell membranes. Rev. Off. Int. Epizoot. 31, 43–56. doi: 10.20506/rst.31.1.2096
Castaneda-Garcia, A., Rodriguez-Rojas, A., Guelfo, J. R., and Blazquez, J. (2009). The glycerol-3-phosphate permease GlpT is the only fosfomycin transporter in Pseudomonas aeruginosa. J. Bacteriol. 191, 6968–6974. doi: 10.1128/JB.00748-09
Clinical and Laboratory Standards Institute (2011). Performance Standards for Antimicrobial Susceptibility Testing; 20th Informational Supplement. CLSI document M100-S21. Wayne, PA: Clinical and Laboratory Standards Institute.
Dinh, A., Salomon, J., Bru, J. P., and Bernard, L. (2012). Fosfomycin: efficacy against infections caused by multidrug-resistant bacteria. Scand. J. Infect. Dis. 44, 182–189. doi: 10.3109/00365548.2011.616221
Elledge, S. J., and Davis, R. W. (1989). Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev. 3, 185–197. doi: 10.1101/gad.3.2.185
Gaston, K., Bell, A., Kolb, A., Buc, H., and Busby, S. (1990). Stringent spacing requirements for transcription activation by CRP. Cell 62, 733–743. doi: 10.1016/0092-8674(90)90118-X
Green, J., Irvine, A. S., Meng, W., and Guest, J. R. (1996). FNR-DNA interactions at natural and semi-synthetic promoters. Mol. Microbiol. 19, 125–137. doi: 10.1046/j.1365-2958.1996.353884.x
Grif, K., Dierich, M. P., Pfaller, K., Miglioli, P. A., and Allerberger, F. (2001). In vitro activity of fosfomycin in combination with various antistaphylococcal substances. J. Antimicrob. Chemother. 48, 209–217. doi: 10.1093/jac/48.2.209
Gunasekera, A., Ebright, Y. W., and Ebright, R. H. (1992). DNA sequence determinants for binding of the Escherichia coli catabolite gene activator protein. J. Biol. Chem., 267, 14713–14720.
Hirakawa, H., Inazumi, Y., Masaki, T., Hirata, T., and Yamaguchi, A. (2005). Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol. Microbiol. 55, 1113–1126. doi: 10.1111/j.1365-2958.2004.04449.x
Hirakawa, H., Kodama, T., Takumi-Kobayashi, A., Honda, T., and Yamaguchi, A. (2009). Secreted indole serves as a signal for expression of type III secretion system translocators in enterohaemorrhagic Escherichia coli O157:H7. Microbiology 155(Pt 2), 541–550. doi: 10.1099/mic.0.020420-0
Hogema, B. M., Arents, J. C., Inada, T., Aiba, H., van Dam, K., and Postma, P. W. (1997). Catabolite repression by glucose 6-phosphate, gluconate and lactose in Escherichia coli. Mol. Microbiol. 24, 857–867. doi: 10.1046/j.1365-2958.1997.3991761.x
Horii, T., Kimura, T., Sato, K., Shibayama, K., and Ohta, M. (1999). Emergence of fosfomycin-resistant isolates of Shiga-like toxin-producing Escherichia coli O26. Antimicrob. Agents Chemother. 43, 789–793.
Inouye, S., Watanabe, T., Tsuruoka, T., and Kitasato, I. (1989). An increase in the antimicrobial activity in vitro of fosfomycin under anaerobic conditions. J. Antimicrob. Chemother. 24, 657–666. doi: 10.1093/jac/24.5.657
Kadner, R. J., and Winkler, H. H. (1973). Isolation and characterization of mutations affecting the transport of hexose phosphates in Escherichia coli. J. Bacteriol. 113, 895–900.
Kohanski, M. A., Dwyer, D. J., and Collins, J. J. (2010). How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 8, 423–435. doi: 10.1038/nrmicro2333
Kurabayashi, K., Hirakawa, Y., Tanimoto, K., Tomita, H., and Hirakawa, H. (2014). Role of the CpxAR two-component signal transduction system in control of fosfomycin resistance and carbon substrate uptake. J. Bacteriol. 196, 248–256. doi: 10.1128/JB.01151-13
Kurabayashi, K., Hirakawa, Y., Tanimoto, K., Tomita, H., and Hirakawa, H. (2015a). Identification of a second two-component signal transduction system that controls fosfomycin tolerance and glycerol-3-phosphate uptake. J. Bacteriol. 197, 861–871. doi: 10.1128/JB.02491-14
Kurabayashi, K., Tanimoto, K., Fueki, S., Tomita, H., and Hirakawa, H. (2015b). Elevated expression of GlpT and UhpT via FNR activation contributes to increased fosfomycin susceptibility in Escherichia coli under anaerobic conditions. Antimicrob. Agents Chemother. 59, 6352–6360. doi: 10.1128/AAC.01176-15
Link, A. J., Phillips, D., and Church, G. M. (1997). Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179, 6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997
Merkel, T. J., Dahl, J. L., Ebright, R. H., and Kadner, R. J. (1995). Transcription activation at the Escherichia coli uhpT promoter by the catabolite gene activator protein. J. Bacteriol. 177, 1712–1718. doi: 10.1128/jb.177.7.1712-1718.1995
Michalopoulos, A. S., Livaditis, I. G., and Gougoutas, V. (2011). The revival of fosfomycin. Int. J. Infect. Dis. 15, e732–e739. doi: 10.1016/j.ijid.2011.07.007
Miller, J. H. (1992). A Short Course in Molecular Genetics. Cold Spring Harbor, NYL: Cold Spring Harbor Laboratory Press.
Morrissey, I., and Smith, J. T. (1994). The importance of oxygen in the killing of bacteria by ofloxacin and ciprofloxacin. Microbios 79, 43–53.
Nilsson, A. I., Berg, O. G., Aspevall, O., Kahlmeter, G., and Andersson, D. I. (2003). Biological costs and mechanisms of fosfomycin resistance in Escherichia coli. Antimicrob. Agents Chemother. 47, 2850–2858. doi: 10.1128/AAC.47.9.2850-2858.2003
Olekhnovich, I. N., Dahl, J. L., and Kadner, R. J. (1999). Separate contributions of UhpA and CAP to activation of transcription of the uhpT promoter of Escherichia coli. J. Mol. Biol. 292, 973–986. doi: 10.1006/jmbi.1999.3127
Schumacher, G., and Bussmann, K. (1978). Cell-free synthesis of proteins related to sn-glycerol-3-phosphate transport in Escherichia coli. J. Bacteriol. 135, 239–250.
Straney, D. C., Straney, S. B., and Crothers, D. M. (1989). Synergy between Escherichia coli CAP protein and RNA polymerase in the lac promoter open complex. J. Mol. Biol. 206, 41–57. doi: 10.1016/0022-2836(89)90522-6
Takahata, S., Ida, T., Hiraishi, T., Sakakibara, S., Maebashi, K., Terada, S., et al. (2010). Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int. J. Antimicrob. Agents 35, 333–337. doi: 10.1016/j.ijantimicag.2009.11.011
Unden, G., and Bongaerts, J. (1997). Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320, 217–234. doi: 10.1016/S0005-2728(97)00034-0
Ushida, C., and Aiba, H. (1990). Helical phase dependent action of CRP: effect of the distance between the CRP site and the -35 region on promoter activity. Nucleic Acids Res. 18, 6325–6330. doi: 10.1093/nar/18.21.6325
Varga, M. E., and Weiner, J. H. (1995). Physiological role of GlpB of anaerobic glycerol-3-phosphate dehydrogenase of Escherichia coli. Biochem. Cell Biol. 73, 147–153. doi: 10.1139/o95-018
Zhou, Y., Merkel, T. J., and Ebright, R. H. (1994). Characterization of the activating region of Escherichia coli catabolite gene activator protein (CAP). II. Role at Class I and class II CAP-dependent promoters. J. Mol. Biol. 243, 603–610. doi: 10.1016/0022-2836(94)90035-3
Keywords: drug resistance, fosfomycin, anaerobiosis, catabolite repression, transporter
Citation: Kurabayashi K, Tanimoto K, Tomita H and Hirakawa H (2017) Cooperative Actions of CRP-cAMP and FNR Increase the Fosfomycin Susceptibility of Enterohaemorrhagic Escherichia coli (EHEC) by Elevating the Expression of glpT and uhpT under Anaerobic Conditions. Front. Microbiol. 8:426. doi: 10.3389/fmicb.2017.00426
Received: 24 November 2016; Accepted: 28 February 2017;
Published: 16 March 2017.
Edited by:
Kunihiko Nishino, Osaka University, JapanReviewed by:
Yuji Morita, Aichi Gakuin University, JapanAixin Yan, University of Hong Kong, Hong Kong
Copyright © 2017 Kurabayashi, Tanimoto, Tomita and Hirakawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hidetada Hirakawa, aGlyYWthd2FAZ3VubWEtdS5hYy5qcA==
 Kumiko Kurabayashi1
Kumiko Kurabayashi1 Hidetada Hirakawa
Hidetada Hirakawa