- 1Department of Biotechnology and Laboratory Science in Medicine, School of Biomedical Science and Engineering, National Yang Ming University, Taipei, Taiwan
- 2Center of Infectious Disease and Signaling Research, National Cheng Kung University, Tainan, Taiwan
- 3Department of Microbiology and Immunology, College of Medicine, National Cheng Kung University, Tainan, Taiwan
- 4Department of Internal Medicine, College of Medicine, National Cheng Kung University Hospital, National Cheng Kung University, Tainan, Taiwan
- 5Department of Internal Medicine, Tainan Hospital, Ministry of Health & Welfare, Tainan, Taiwan
CsrA has been shown to positively control the expression of flagella-related genes, including flaA and flaB, through regulating expression of an alternative sigma factor RpoN in Helicobacter pylori J99. Here, we aimed to characterize the CsrA regulatory system by comparative transcriptomic analysis carried out with RNA-seq on strain J99 and a csrA mutant. Fifty-three genes in the csrA mutant were found to be differentially expressed compared with the wild-type. Among CsrA-regulated genes, jhp0106, with unclear function, was found located downstream of flaB in the J99 genome. We hypothesized that flaB-jhp0106 is in an operon under the control of RpoN binding to the flaB promoter. The RT-qPCR results showed the expression of jhp0106 was decreased 76 and 92% in the csrA and rpoN mutants, respectively, compared to the wild-type. Moreover, mutations of the RpoN binding site in the flaB promoter region resulted in decreased expression of flaB and jhp0106 and deficient motility. Three-dimensional structure modeling results suggested that Jhp0106 was a glycosyltransferase. The role of jhp0106 in H. pylori was further investigated by constructing the jhp0106 mutant and revertant strains. A soft-agar motility assay and transmission electron microscope were used to determine the motility and flagellar structure of examined strains, and the results showed the loss of motility and flagellar structure in jhp0106 mutant J99. In conclusion, we found jhp0106, under the control of the CsrA/RpoN regulatory system, plays a critical role in H. pylori flagella formation.
Introduction
Helicobacter pylori is a highly prevalent human pathogen that colonizes roughly 50% of the world's population. Persistent infection with H. pylori increases the risk of developing gastroduodenal diseases, including chronic gastritis, gastric and duodenal ulcer, and gastric adenocarcinoma (Parsonnet et al., 1991; Graham et al., 1993; Ahmad et al., 2003). Motility of H. pylori mediated by flagella has been shown to be critical for the cells to establish initial colonization and achieve dense colonization and severe pathological outcomes in patients (Eaton et al., 1996; Ottemann and Lowenthal, 2002; Kao et al., 2012a, 2016). Despite intensive research in the role of motility in H. pylori pathogenesis, the complex regulatory network that modulates the expression of flagellar genes in H. pylori is still not fully understood.
H. pylori has five to seven polar, sheathed flagella, which are composed of three main structures: the basal body, hook and filament (Lertsethtakarn et al., 2011). Flagellar related genes are divided into three classes, governed by the housekeeping sigma factor σ80 (RpoD, regulating class I genes), the alternative sigma factors σ54 (RpoN, regulating class II genes), and σ28 (FliA, regulating class III genes) (Niehus et al., 2004; Kao et al., 2016). The flagellar filament consists of two flagellin proteins, FlaA (the major constituent) and FlaB (Kostrzynska et al., 1991; Suerbaum et al., 1993). H. pylori flagellin proteins are synthesized, then post-translationally modified intracellularly by glycosylation with a nine carbon pseudaminic acid sugar derivative that resembles sialic acid (Schirm et al., 2003; Logan, 2006). The enzymes of the pseudaminic acid biosynthetic pathway in H. pylori, in order, are PseB, PseC, PseH, PseG, and PseI (Schirm et al., 2003; Menard et al., 2014), and the glycosylation process is essential for assembly of functional flagellar filaments and consequent bacterial motility (Schoenhofen et al., 2006).
CsrA was identified as a post-transcriptional regulator of glycogen biosynthesis, motility, biofilm formation and bacterial virulence in E. coli, acting as an RNA binding protein on its target mRNA and thus affecting its stabilization or translation (Romeo et al., 1993, 2013; Liu et al., 1995; Liu and Romeo, 1997; Wang et al., 2005; Jonas et al., 2008). In H. pylori strain J99, CsrA regulates flagella formation by controlling RpoN expression, and it thereby affects bacterial motility (Kao et al., 2014). Although the decrease of FlaA/FlaB partially explains the non-flagellated phenotype of the csrA mutant observed by transmission electron microscopy (TEM) (Kao et al., 2014), other regulators or mechanisms may be involved in the CsrA regulatory system. In this study, we aimed to characterize the CsrA regulatory system by comparative transcriptomic analysis carried out with RNA-seq on H. pylori strain J99 and a csrA mutant. We demonstrated that Jhp1006, a putative glycosyltransferase involved in H. pylori J99 flagella formation and motility, is under the control of CsrA/RpoN.
Materials and Methods
Bacterial Strains and Growth Conditions
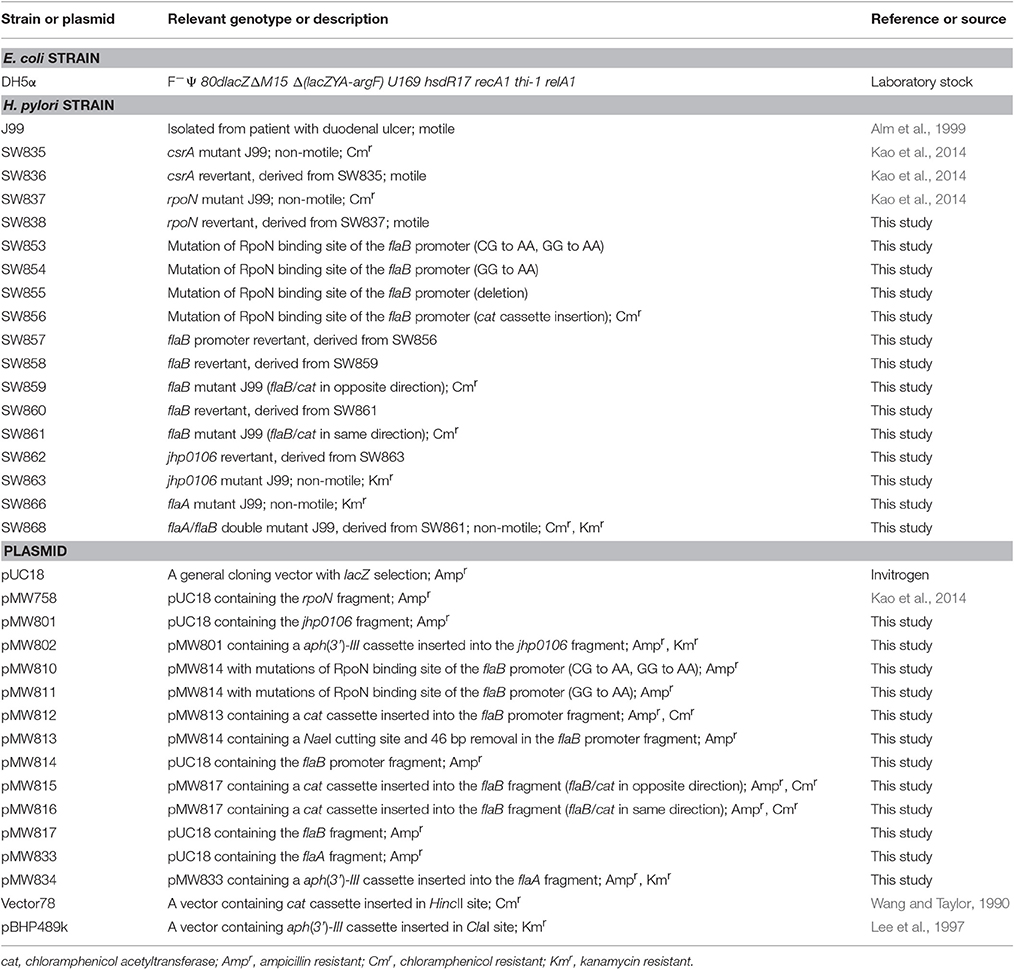
The bacterial strains and plasmids used in this study are described in Table 1. H. pylori cells were grown on CDC anaerobic blood agar (BBL, Microbiology Systems, Cockeysville, MD) or in Brucella broth containing 10% (v/v) horse serum (Gibco BRL, Life Technologies, Rockville, MD) at 37°C in microaerophilic conditions (5% O2, 10% CO2 and 85% N2). E. coli was grown on Luria-Bertani (LB) (BD Biosciences, San Jose, CA) agar or in broth. Bacteria harboring antibiotic resistance determinants were grown in the presence of the appropriate antibiotics at the following concentrations: ampicillin (Amp, 100 μg ml−1); chloramphenicol (Cm, 25 μg ml−1 for E. coli, 10 μg ml−1 for H. pylori); kanamycin (Km, 50 μg ml−1 for E. coli, 10 μg ml−1 for H. pylori). All strains were stored at −80°C in Brain-Heart Infusion (BHI) broth (H. pylori) or LB broth (E. coli) containing 20% (v/v) glycerol until testing.
Cell Line and Cell Culture
The human gastric carcinoma cell line AGS (purchased from ATCC; American Type Culture Collection, Manassas, VA, USA) was grown in Ham's F-12 medium (Invitrogen Life Technologies, Rockville, MD) supplemented with 10% (v/v) fetal bovine serum (Gibco BRL) in an atmosphere consisting of 5% CO2 at 37°C. The human gastric epithelial immortalized GES-1 cells (a gift from Prof. Wei-Lun Chang, National Cheng Kung University Hospital) were grown in RPMI 1640 medium (Invitrogen Life Technologies) supplemented with 10% (v/v) fetal bovine serum in an atmosphere consisting of 5% CO2 at 37°C.
DNA Techniques
Mini Qiagen columns and a QiaAmp DNA extraction kit (Qiagen, Valencia, CA, USA) were used for plasmid and chromosomal DNA extraction. PCR was carried out according to the manufacturer's instruction using Taq polymerase (Promega, Madison, WI, USA).
RNA-Seq Library Preparation and Sequencing
H. pylori cells were grown on CDC plates in microaerophilic conditions for 36 h, then transferred to 100 ml Brucella broth containing 10% (v/v) horse serum at an optical density (OD) of 0.2 and incubated with shaking (150 rpm, to reduce cell aggregation) for 18 h in microaerophilic conditions. Mini Qiagen columns and a Qiagen RNAeasy mini kit (Qiagen, Valencia, CA) were used for RNA extraction. Ribosomal RNA was depleted according to the manufacturer's instruction using Bacteria Minus™ Transcriptome Isolation Kit (Invitrogen Life Technologies). The Applied Biosystems SOLiD™ Total RNA-Seq kit was used to generate the cDNA template library. The SOLiD™ EZ Bead system (Invitrogen Life Technologies) was used to perform emulsion clonal bead amplification to generate bead templates for SOLiD™ platform sequencing. Samples were sequenced on the 5500XL SOLiD™ platform. The 50-base short read sequences produced by the 5500XL SOLiD™ sequencer were first run through SOLiD Accuracy Enhancement Tool (SAET) to improve color call accuracy, then were mapped in color space using SOLiD™ LifeScope™ software version 2.5 using default parameters against the H. pylori J99 reference genome (NCBI accession number, NC_000921). The BAM file from LifeScope™ was performed the analysis of gene expression with Partek software package. The gene expression from each sample was then tested for statistical differences using one-way ANOVA at 5% confidence level. The complete set of RNA-seq files has been deposited in Gene expression omnibus (GEO), NCBI (https://www.ncbi.nlm.nih.gov/geo/, accession number GSE95006). The fold change of each gene was measured as the mean of three independent experiments. Gene expression of the csrA mutant compare to wild-type J99 with a fold change > 1.5-fold was selected and confirmed by RT-qPCR.
Preparation of cDNA from H. pylori
H. pylori cells were grown on CDC plates in microaerophilic conditions for 36 h, then transferred to 100 ml Brucella broth containing 10% (v/v) horse serum at an OD of 0.2 and incubated with shaking (150 rpm) for 18 h in microaerophilic conditions. RNA extraction and reverse transcription PCR were described previously (Kao et al., 2012b). Thirty microliters of culture media were centrifuged at 1,000 × g for 5 min at 4°C and then washed with ice-cold 0.2 M sodium acetate buffer (pH 5.5) twice. The bacterial pellet was then re-suspended in 600 μl acetate buffer (20 mM sodium acetate, 1 mM EDTA, and 0.5% (w/v) SDS), and 600 μl acid-phenol (pH 4.5) was added to isolate bacterial RNA. The sample was incubated at 65°C for 10 min, and centrifuged at 12,000 × g for 10 min to collect the supernatant. After isopropanol precipitation, the sample was treated with DNase I (Promega) at 37°C for 2 h. Finally, phenol/chloroform was used to extract total RNA, and the sample was dissolved in diethylpyrocarbonate (DEPC)-treated deionized water and stored at −80°C until used. The RNA was quantified at an absorbance of 260 nm. Random hexamers (Mission biotech, Taiwan) and MMLV reverse transcriptase (Promega) were used to generate cDNA from 1 μg of total RNA, and the cDNA was stored at −20°C until testing.
Real-Time Quantitative RT-PCR (RT-qPCR)
The primers used for RT-qPCR are listed in Table S1. RNA quantification was carried out by RT-qPCR with a KAPA PROBE FAST Universal 2 x qPCR Master Mix (KAPA Biosystems Inc., Woburn, MA) specifically adapted for one-step RT-qPCR in glass capillaries using a Light Cycler instrument (Roche Diagnostics, Indianapolis, IN). Cycling conditions were as follows: activation of the polymerase for 10 min at 95°C, followed by 40 cycles of denaturation at 95°C for 20 s, annealing at 60°C for 1 min, and elongation at 72°C for 15 s. Fluorescence was detected at the end of each extension step, and the Cp values were calculated by the LightCycler 1.5 software.
Construction of the Mutants and Revertants
The primers used in this study are listed in Table S1. In order to construct a revertant strain from rpoN mutant J99 (SW837), the plasmid containing the rpoN fragment, pMW758 (Kao et al., 2014), was transformed into SW837 to generate a revertant strain containing the wild-type rpoN. In brief, after natural transformation (Haas et al., 1993), H. pylori was grown on CDC anaerobic blood agar without antibiotic for 3 days. Colonies were picked and subcultured on CDC anaerobic blood agar without antibiotic and Brucella agar plate containing 10% (v/v) horse serum and Cm (10 μg ml−1) at the same time and incubated at 37°C in microaerophilic conditions for 3 days. Colonies that lost the ability to grow on a Cm-containing plate (that only grew on CDC anaerobic blood agar) were considered as the revertant strain and were verified by PCR-sequencing and motility assay.
For flaB mutant construction, the 1,175 bp flaB fragment obtained from H. pylori J99 genomic DNA and PCR with flaB-Mut-1 and flaB-Mut-2 primers, was digested by PstI and KpnI and ligated to the plasmid pUC18 to generate plasmid pMW817. A chloramphenicol acetyltransferase cassette (cat cassette) containing 806 bp was obtained from plasmid vector 78, cut with HincII and inserted into plasmid pMW817 digested with HincII. Two plasmids were designated as plasmid pMW815 (flaB/cat in opposite transcriptional orientation) and pMW816 (flaB/cat in same transcriptional orientation), respectively, and transformed into J99. Cm (10 μg ml−1) was used to select for the flaB mutants, SW859 and SW861, generated from chromosomal flaB double cross-over with plasmids pMW815 and pMW816, respectively. Plasmid pMW817 was then transformed into SW859 and SW861 to generate a revertant strains SW858 and SW860, respectively.
For jhp0106 mutant construction, the 1,004 bp jhp0106 fragment, obtained from J99 genomic DNA and PCR with jhp0106-Mut-1 and jhp0106-Mut-2 primers, was digested with EcoRI and ligated into the plasmid pUC18 to generate plasmid pMW801. A kanamycin resistance cassette (aph(3′)-III cassette) was obtained from plasmid vector pBHP489K, cut with ClaI and inserted into the plasmid pMW801 digested with BsmI. This plasmid was designated as plasmid pMW802 and transformed into J99. Km (10 μg ml−1) was used to select for the jhp0106 mutant, SW863. In order to construct a jhp0106 revertant from SW863, the plasmid pMW801, was transformed into SW863 to generate SW862 containing wild-type jhp0106.
For flaA and flaA/flaB mutants construction, the 1,283 bp flaA fragment obtained from H. pylori J99 genomic DNA and PCR with FlaA-Mut-1 and FlaA-Mut-2 primers, was digested by PstI and KpnI and ligated to the plasmid pUC18 to generate plasmid pMW833. A aph(3′)-III cassette obtained from plasmid vector pBHP489K was inserted into plasmid pMW833 digested with AfeI. This plasmid was designated as plasmid pMW834 and transformed into J99 and SW861 (flaB mutant). Km (10 μg ml−1) was used to select for the flaA mutant (SW866) and flaA/flaB double mutant (SW868).
Construction of the flaB Promoter Mutants and Revertant
The primers used for construction of the flaB promoter mutants and revertant are listed in Table S1. The 1,001 bp flaB promoter fragment obtained from H. pylori J99 genomic DNA and PCR with flaB-ProMut-1 and flaB-ProMut-2 primers, was digested by HincII and EcoRV and ligated to the plasmid pUC18 to generate plasmid pMW814. Primers flaB-ProMut-3 and flaB-ProMut-4 were used to amplify pMW814 to generate plasmid pMW813 containing a 46 bp partial deletion of the flaB promoter and carrying a NaeI site in the flaB promoter fragment. A cat cassette obtained from plasmid vector 78 was inserted into plasmid pMW813 digested with NaeI. This plasmid was designated as plasmid pMW812 and transformed into J99. Cm was used to select for the flaB promoter-cat mutant, SW856. In order to construct flaB promoter mutants with GG or CG/GG nucleotide mutations, primer pairs flaB-ProMut-7/flaB-ProMut-8 and flaB-ProMut-9/flaB-ProMut-10 were used to amplify pMW814 to generate plasmid pMW811 and pMW810, respectively. pMW814, pMW813, pMW811, and pMW810 were transformed into SW856 to generate SW857 (revertant), SW855 (deletion), SW853 (CG to AA/GG to AA), and SW854 (GG to AA), respectively.
Soft-Agar Motility Assay
The motility assay was described previously (Kao et al., 2014). Bacterial colonies were applied to one spot in the motility agar plate containing Brucella broth, 0.3% (w/v) Bacto agar, and supplemented with 10% (v/v) horse serum. The plates were incubated at 37°C under microaerophilic conditions for 7 days, and the motility was assessed by the diameter of migration of bacteria through the agar, from the inoculated center toward the periphery of the plate. The motility of each strain was measured as the mean of three independent experiments.
Reconstruction of Three-Dimensional Model of Jhp0106 Protein
Complete amino acid sequence of H. pylori J99 Jhp0106 (Accession number: WP_001028953) was downloaded from NCBI database (http://www.ncbi.nlm.nih.gov/protein/). A three-dimensional structural modeling was carried out on the SWISS-MODEL Workspace server (http://swissmodel.expasy.org/) (Arnold et al., 2006). The structure representation figures are generated by the program PyMOL (http://www.pymol.org).
Transmission Electron Microscope
A grid covered with a carbon-coated parlodion film (300 mesh copper grid) was floated onto a 20 μl sample drop and left for 2 min for adsorption of the sample to the grid. The grid was then removed from the drop and floated on a drop of 1% (w/v) phosphotungstic acid (Sigma-Aldrich) and left for 1 min. Excess stain was removed by touching the edge of the grid to a piece of Whatman filter paper. All samples for electron microscopy were examined in a Hitachi H-7650 transmission electron microscope (Hitachi, Tokyo, Japan).
Bacterial Adhesion and IL-8 Production Assay
The assay was performed according to a previous study with modification (Kwok et al., 2002). AGS and GES-1 cells (1 × 106/well) were grown overnight in 6-well culture dishes to approximately 80% confluence. H. pylori cells were added to the wells at a multiplicity of infection (MOI) of 100 without centrifugation and were incubated for either 30 min for the adhesion assay or 8 h for the cellular IL-8 production assay. For the adhesion assay, each dish with AGS-H. pylori coculture was washed three times with prewarmed phosphate-buffered saline (PBS) buffer to remove unbound bacteria. Adhered H. pylori were quantified by lysing the cells for 15 min with 0.1% (w/v) saponin-containing PBS buffer, followed by serial dilution and spreading on Brucella agar plates containing 10% (v/v) horse serum. The number of adhered bacteria was measured by the plate counts after 3 days incubation. For the cellular IL-8 production assay, the culture supernatants were collected and stored at −20°C until assayed. IL-8 concentration in the supernatant was determined by standard ELISA with commercially available assay kits according to the manufacturer's procedures (Arigo Biolaboratories Corp., Taiwan).
Statistics
The Student's t-test and paired t-tests were applied as appropriate for the parametric differences. ANOVA was used for comparing groups of more than two strains. All tests of significance were two-tailed with a p value < 0.05 taken as significant.
Results
CsrA Acts as a Global Positive Regulator in Strain J99
To characterize the CsrA regulatory system, RNA-seq analysis on the wild-type J99 and its respective csrA mutant, SW835, was carried out. Three independent biological replicates were sequenced for each strain. The raw sequence output of the two strain transcriptomes included ~14 and ~15 million reads of the wild-type and csrA mutant, respectively. Approximately 80% of the reads were perfectly aligned to the J99 reference genome. Based on the genomic alignment, our analysis determined the expression of 1,559 genes in each strain.
In this study, genes found to be differentially expressed compared to the wild-type J99 (a > 1.5-fold change) by RNA-seq data were taken into consideration. In the csrA mutant, 53 chromosomally encoded genes were found to be differentially expressed compared to the wild-type J99 (Table 2). Of these genes, 94% (50 genes) were expressed at a lower level in the csrA mutant compared to the wild-type, with only 6% (3 genes) having higher expression in the csrA mutant (Table 2). These results indicate that CsrA acts primarily as a global positive regulator. RT-qPCR was further employed to validate the expression of 53 CsrA regulated genes in the wild-type and csrA mutant (Table 2). In agreement with the RNA-seq data, RT-qPCR confirmed the expression of genes differentially expressed in the csrA mutant compared to the wild-type J99 with a > 2-fold change (except jhp1296). Three genes, jhp1334, jhp1169 and jhp1132, originally identified with a >1.5-fold change by RNA-seq, showed inconsistent results between the RNA-seq and RT-qPCR methods (change <1.5-fold by RT-qPCR) (Table 2). These transcriptomic analyses also indicated that most of CsrA regulated genes (18 genes) were classified as genes encoding components involved in flagella formation, chemotaxis and motility, followed by genes with unknown function (15 genes) (Figure S1).
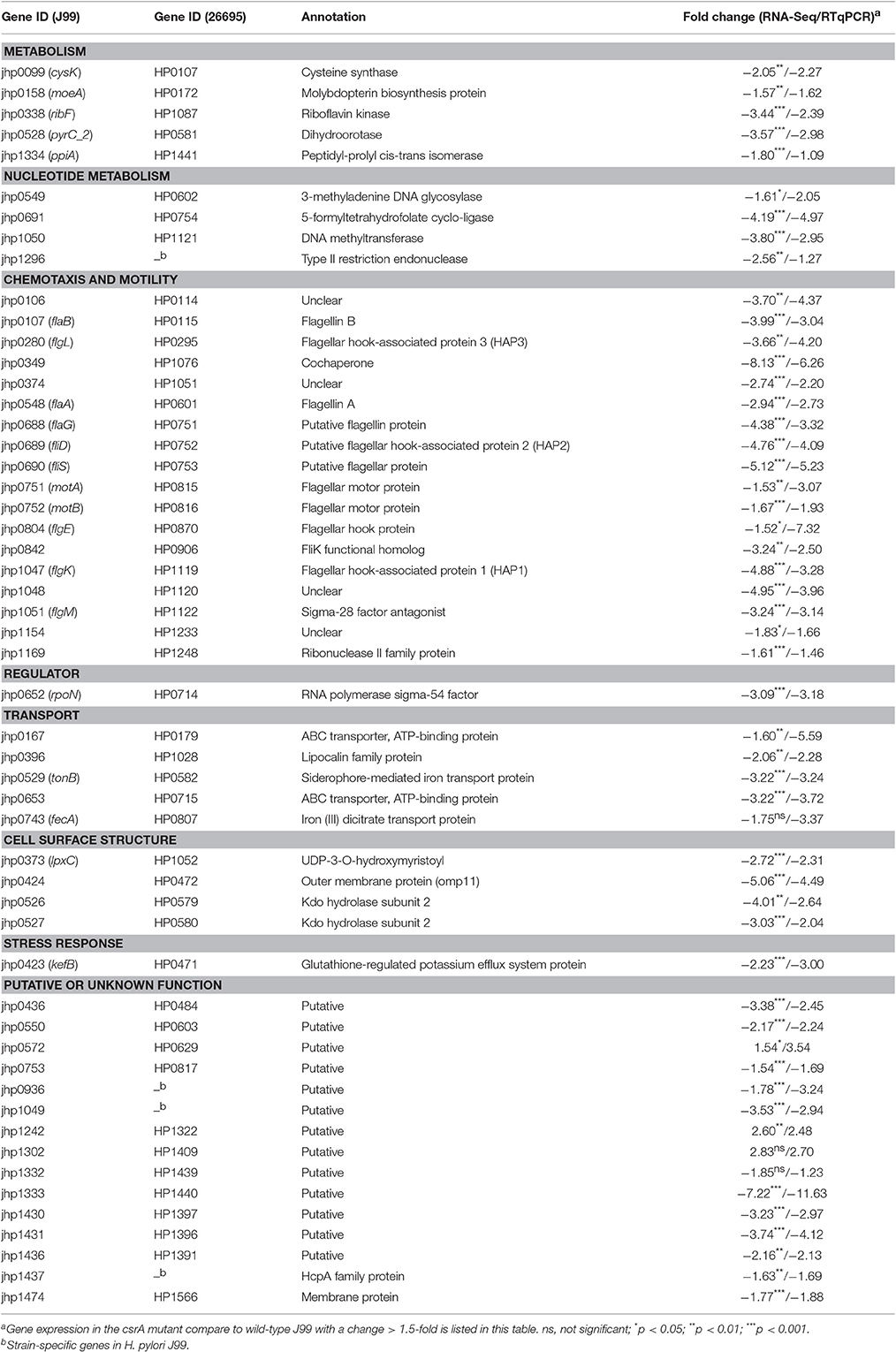
Table 2. Gene ID, annotation and function of genes regulated by the CsrA regulatory system identified by RNA-seq analysis.
CsrA/RpoN Regulates flaB-jhp0106 Expression
The CsrA-regulated jhp0106 gene was of interest, as it encodes a protein suggested to be involved in H. pylori strain 1016 flagellin glycosylation and motility (Schirm et al., 2003) (Table 3). The schematic diagram showing the pseudaminic acid biosynthetic pathway in H. pylori is shown in Figure S2. At present, the function of Jhp0106 in flagellin glycosylation is still unclear. We validated the transcription of pseB, pseC, pseH, pseG, pseI and jhp0106 in the wild-type, SW835 (csrA mutant) and SW836 (csrA revertant) strains by RT-qPCR. The results were consistent with the RNA-seq data. Only the expression of jhp0106 was dramatically reduced in SW835 compared to the wild-type J99 (Figure S3).
The genes close to jhp0106 in H. pylori J99 are shown in Figure 1A. jhp0106 was located immediately downstream of the flaB gene, which has been shown to be regulated by RpoN. The gene order and orientation of flaB-jhp0106 are conserved in H. pylori genomes. Therefore, we propose genes flaB and jhp0106 are in an operon controlled by RpoN binding to the flaB promoter. To validate this hypothesis, RT-qPCR was carried out to determine the expression of rpoN and jhp0106 in the wild-type, SW835 (csrA mutant), SW836 (csrA revertant), SW837 (rpoN mutant), and SW838 (rpoN revertant) strains. The results showed that the expression of rpoN was reduced in SW835 (Figure 1B), consistent with our previous study (Kao et al., 2014). Moreover, expression of jhp0106 mRNA was decreased to 24 and 8% in SW835 and SW837, respectively (p < 0.001). These results indicated that CsrA and RpoN positively regulate jhp0106 expression.
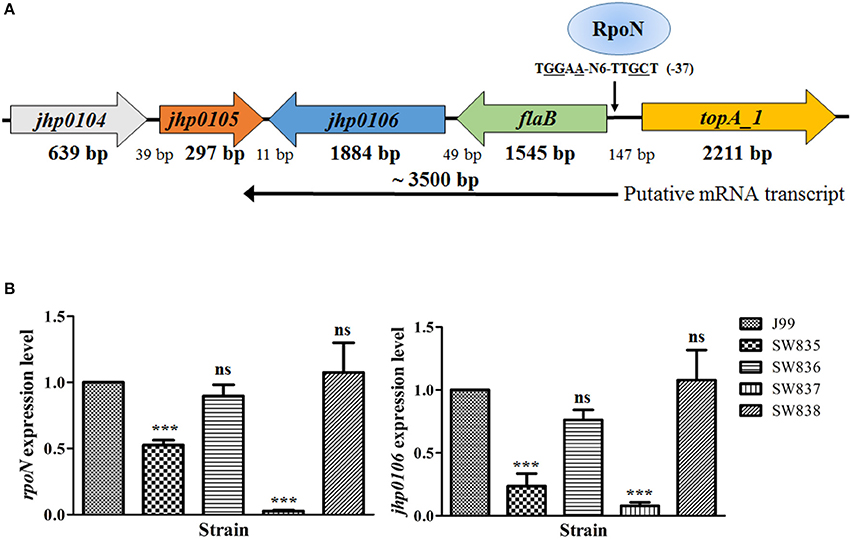
Figure 1. Reduced expression of jhp0106 in strains SW835 and SW837. (A) Schematic diagram showing the genetic loci of genes close to the putative flaB-jhp0106 operon. A predicted RpoN binding site located upstream of the flaB ORF (−37 bp) is indicated. (B) RT-qPCR was used for quantifying the mRNA levels of rpoN (left panel) and jhp0106 (right panel) in examined strains. Results are representative of 3 independent experiments (means ± SD). ***p < 0.001, ns, not significant (vs. wild-type J99). SW835, csrA mutant; SW836, csrA revertant; SW837, rpoN mutant; SW838, rpoN revertant.
RT-PCR with different primer pairs was performed to confirm the co-transcription of flaB-jhp0106, and the results showed that 1.4- and 1.8-Kb transcripts were observed using primer pairs jhp0106-1/flaB-4 and jhp0106-3/flaB-4 for RT-PCR, respectively (Figure 2A). In addition, the expression of the flaB-jhp0106 transcript was reduced in SW837 (Figure 2A). The predicted RpoN binding site of the flaB promoter was examined by construction of flaB promoter mutants, as shown in Figure 2B. The mRNA levels of flaB and jhp0106 were dramatically reduced in SW837, SW853, SW854, SW855, and SW856, as determined by RT-qPCR (Figure 2C). We further investigated whether the RpoN binding site of the flaB promoter was conserved among different H. pylori strains, and the results showed that the RpoN binding sequence was identical among 17 examined strains (Figure S4). Our results indicated the transcription of jhp0106 is positively controlled by RpoN bound to the flaB promoter. However, these results could not exclude the possibility that the jhp0106 gene had its own promoter. Therefore, northern blotting was performed to determine the size and number of jhp0106 transcripts. Although several different fragments of the jhp0106 gene were used to serve as probes, the signal was still too weak to be detectable (data not shown).
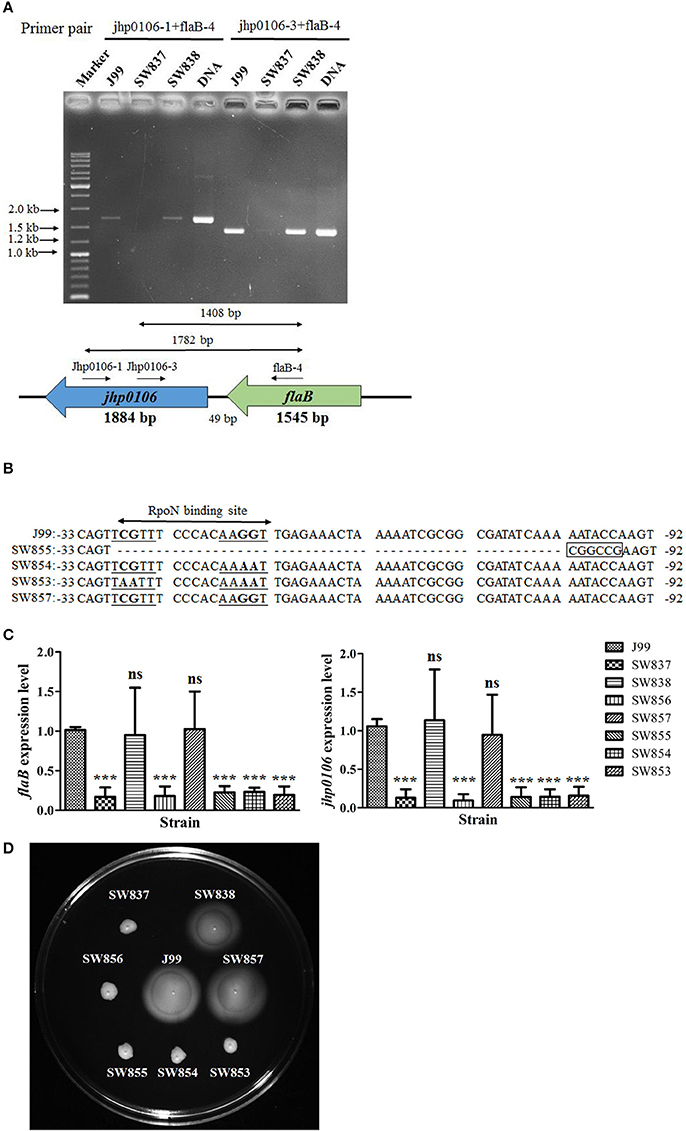
Figure 2. Characterization of the flaB-jhp0106 operon. (A) The expression of the flaB-jhp0106 co-transcript in J99, SW837, and SW838 was determine by RT-PCR. The primer pairs and predicted PCR product size are described in the lower panel. J99 DNA was considered as a positive control. Marker, GeneRuler™ DNA ladder (Fermentas). (B) Sequence alignment of the flaB promoter region (−33 to −92 bp, upstream of the flaB ORF) of wild-type, SW855, SW854, SW853, and SW857 strains. (C) mRNA levels of flaB and jhp0106 in 8 examined strains were measured by RT-qPCR. Results are representative of 3 independent experiments (means ± SD). ***p < 0.001, ns, not significant (vs. wild-type J99). (D) The motility of 8 tested strains was determined by soft-agar motility assay plates. SW837 was used as a negative control (non-motile phenotype). SW837, rpoN mutant; SW838, rpoN revertant.
In order to clarify the role of the flaB-jhp0106 operon in H. pylori J99 motility, soft-agar analysis was used to determine the motility of flaB promoter mutants (Figure 2D). SW837 was used as a non-motile negative control. The results showed that strains SW853, SW854, SW855, and SW856 exhibited deficient motility (Figure 2D). These results indicated that the flaB-jhp0106 operon was controlled by RpoN, and that it plays a critical role in H. pylori motility.
Jhp0106, but Not FlaB, Plays a Critical Role in H. pylori Motility
Josenhans et al. reported that when the flaB gene was disrupted in H. pylori strain N6, the motility decreased by 30 to 40% (Josenhans et al., 1995). To validate this observation in strain J99, we constructed flaB mutants SW861 and SW859 (Figure 3A). A cat cassette was inserted in the same (SW861) or opposite (SW859) direction to flaB transcription in wild-type J99 (Figure 3A). Strain SW861 showed decreased motility, and the quantified results revealed 83.8% motility ability compared with wild-type J99 (p < 0.05) (Figure 3B). In contrast, strain SW859 showed a non-motile phenotype, compared with the wild-type J99 (Figure 3B). To evaluate whether the deficient phenotype of SW859 was caused by the insertion of the cat cassette into the flaB gene, which interfered with downstream jhp0106 expression, RT-qPCR was performed to determine the expression of jhp0106 in the examined strains. The results showed that the transcription level of jhp0106 was decreased to 13% in SW859, but increased to 242% in SW861, compared to the wild-type (Figure 3C). The increase of jhp0106 expression in SW861 resulted from a cat cassette inserted into flaB in same transcriptional orientation. Thus triggering downstream jhp0106 transcription due to the leakage of the transcriptional terminator of the cat cassette. These results indicated that jhp0106 in flaB-jhp0106 operon, but not flaB, plays a critical role in H. pylori motility.
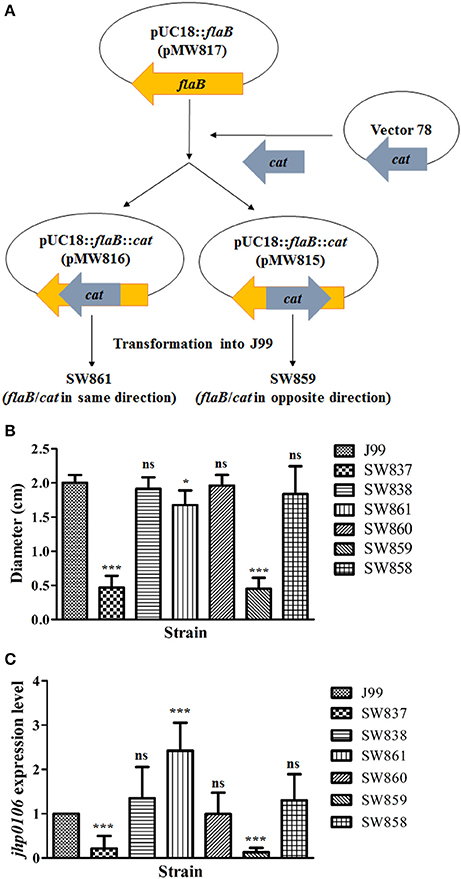
Figure 3. Characterization of the influence of flaB and jhp0106 on H. pylori motility. (A) Schematic diagram showing the construction strategy of the flaB mutants, SW861 and SW859. (B) The quantified motility diameter of 7 examined strains. SW837 was used as a negative control (non-motile phenotype). (C) The mRNA level of jhp0106 was determined by RT-qPCR. Results are representative of 3 independent experiments (means ± SD). *p < 0.05, ***p < 0.001, ns, not significant (vs. wild-type J99). SW837, rpoN mutant; SW838, rpoN revertant.
Jhp0106, a Putative Glycosyltransferase
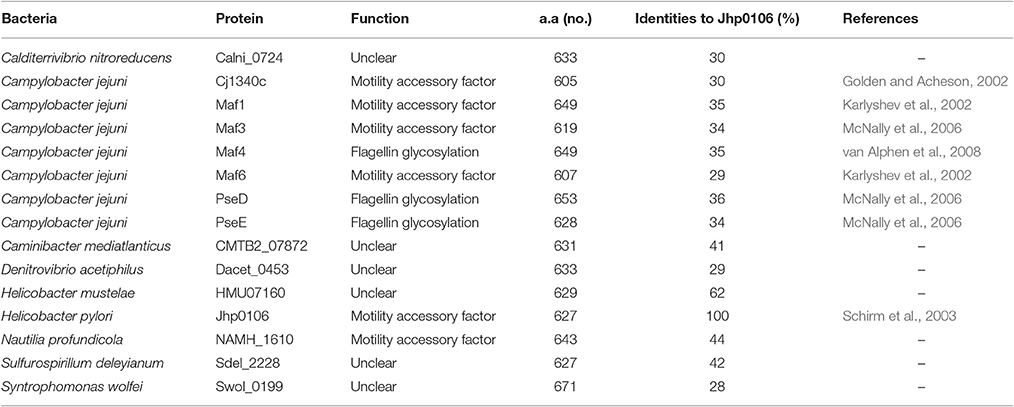
The NCBI protein database was used to search for proteins showing high similarity to Jhp0106, and the results are shown in Table 3. Among them, HMU07160 (62%) showed the highest identity to Jhp0106, followed by WS2199 (55%), NAMH_1610 (44%), Sdel_2228 (42%), and CMTB2_07872 (41%) (Table 3). The phylogenic tree based on the homologous full-length sequence of 16 proteins was also displayed in Figure S5. Previous studies reported that Maf4, PseD and PseE were involved in flagellin glycosylation in C. jejuni (McNally et al., 2006; van Alphen et al., 2008), and they also displayed high identity to Jhp0106 (Table 3).
The attempt to determine the crystal structure of Jhp0106 was unsuccessful, in spite of extensive efforts to crystallize the recombinant Jhp0106 protein from an E. coli expression system. Therefore, we performed computational modeling of the Jhp0106 protein to gain structural insights into Jhp0106 function. A 3D-model for Jhp0106 of 227 amino acid residues in length (from Asp231 to Phe457) predicted by the SWISS-MODEL server is shown in Figure 4A. The overall folding of the Jhp0106 structure is similar to an alpha-2,3/8-sialyltransferase CstII from C. jejuni in complex with a substrate analog, CMP-3FNeuAc (PDB: 1R07) (Figure 4B) (Chiu et al., 2004), suggesting Jhp0106 is a glycotransferase.
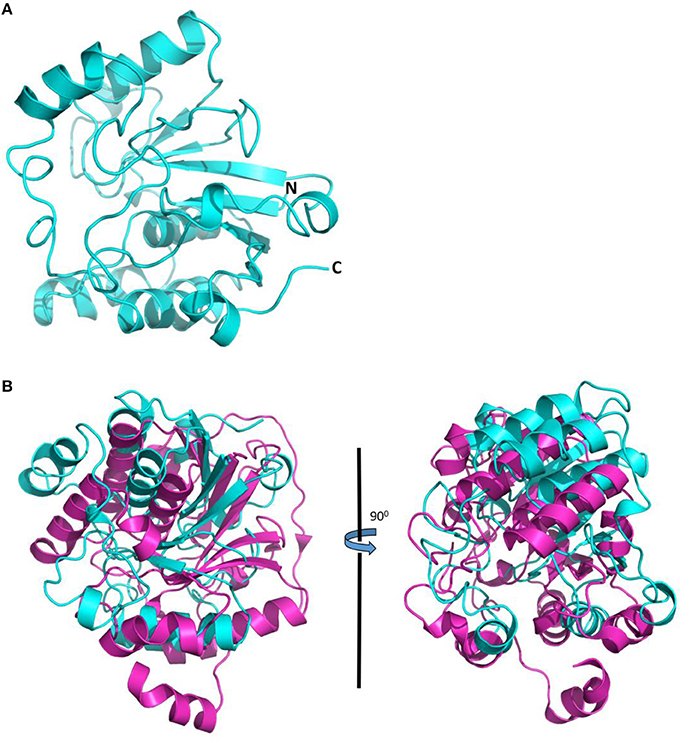
Figure 4. Computational structure model of Jhp0106 from H. pylori J99. (A) The structural model of Jhp0106 composed of 227 amino acids, by the SWISS-MODEL server. (B) Structural superimposition of Jhp0106 (cyan) with the crystal structure of sialyltransferase CstII in complex with CMP-3FNeuAc (PDB 1R07) (magenta).
The protein interaction networks for Jhp0106 showed that the protein interacts with flagella structure proteins (FlaA, FlaB, FlaG, FlaL, FliD), pseudaminic acid synthase (NeuB), CagDelta protein (Jhp0417), hypothetical proteins (PdP, Jhp0578), and a septum formation inhibitor (MinC) (Figure S6). These results suggested that Jhp0106 functions as a glycosyltransferase and is involved in the transfer of pseudaminic acid to flagellin.
Jhp0106 Is Involved in Flagella Formation
The roles of Jhp0106 in H. pylori flagella formation, adhesion and cellular cytokine induction were examined by constructing jhp0106 mutant (SW863) and revertant J99 (SW862) strains. The genetic loci of jhp0105 and flaB are close to jhp0106 in the J99 genome (Figure 1A). In order to rule out the possibility of polar effects in SW862 and SW863, the expression of jhp0105, jhp0106 and flaB were determined by RT-PCR (Figure S7). The data revealed that the mRNA expression levels of jhp0105 and flaB in strains SW863 and SW862 were similar to J99, and the transcription of jhp0106 was only disrupted in SW863 (Figure S7). In addition, the 72 h growth curves of SW863 and SW862 were similar to that of the wild-type J99 (Figure S8).
The deficient motility of SW863 is shown in Figure 5A. The bacterial shape and flagellar structure of J99, SW837, SW838, SW862, and SW863 were examined by TEM with negative staining. No flagellar structure was detected in SW837 and SW863, whereas the characteristic multiple polar, sheathed flagella were abundant on J99, SW838 and SW862 (Figure 5B). In addition, there was no dramatic difference in shape between the strains examined. These observations demonstrated that flagella formation was severely defective in the jhp0106 mutant.
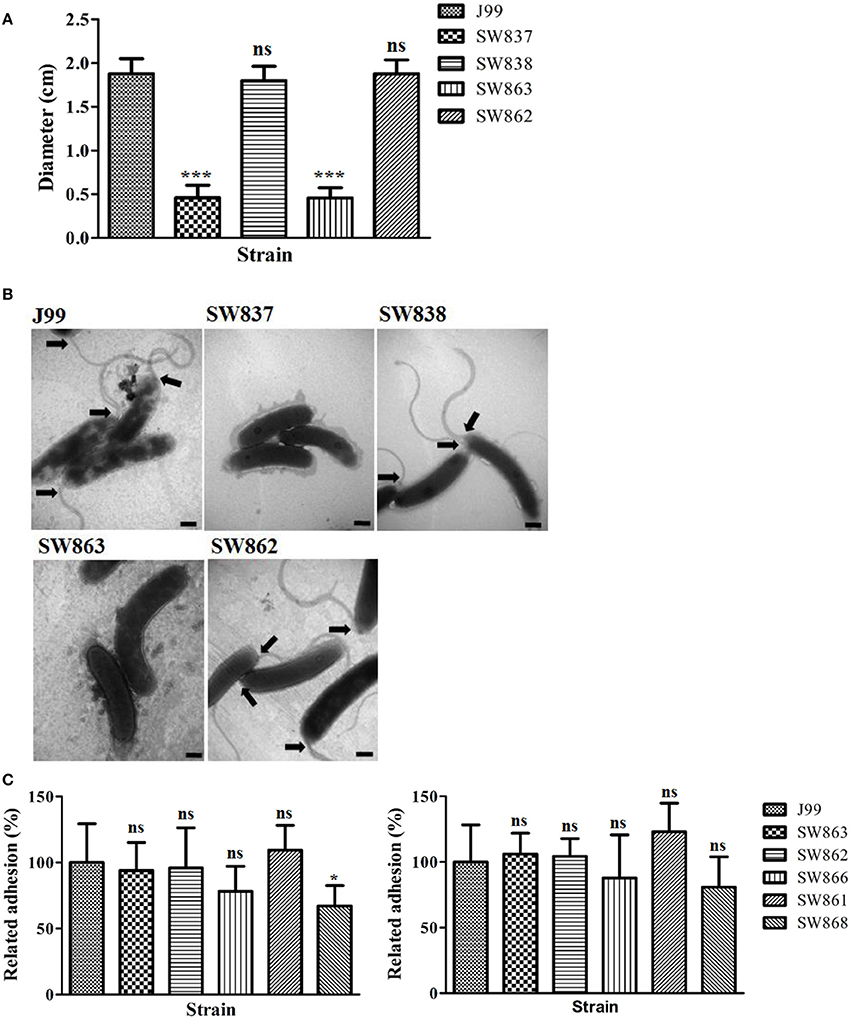
Figure 5. Motility, flagellar structure, and adhesion ability of strains J99, SW837, SW838, SW863, and SW862. (A) The quantified motility diameter of tested strains. SW837 was used as a negative control (non-motile phenotype). (B) Transmission electron micrographs of negatively stained H. pylori. Black arrowheads indicate the presence of full-length flagella at the cell poles. Scale bars represent 500 nm. (C) The adhesion of H. pylori to AGS cells (left panel) and GES-1 cells (right panel) with MOI 100 of the tested strains. Results are representative of 3 independent experiments (means ± SD). *p < 0.05, ***p < 0.001, ns, not significant (vs. wild-type J99). SW837, rpoN mutant; SW838, rpoN revertant; SW863, jhp0106 mutant; SW862, jhp0106 revertant; SW866, flaA mutant; SW861, flaB mutant; SW868, flaA/flaB mutant.
To determine the role of flagella structure and Jhp0106 in H. pylori pathogenesis, the adhesion rate of the bacteria and the IL-8 production in H. pylori-infected AGS and GES-1 cells were determined (Figure 5C and Figure S9). The results showed that only SW868 (flaA/flaB double mutant) had lower adhesion to AGS cells, compared with J99 (p < 0.05) (Figure 5C). No difference in IL-8 production was observed between cells infected with the six examined strains, as determined by an ELISA assay (p > 0.05) (Figure S9).
Characterization of Jhp0106 in Clinical Isolates
Many studies suggest that genetic diversity in H. pylori virulence factors such as sabA, babA, cagA, and vacA genes is high among isolates from different geographic regions, and may be associated with different pathological outcomes (van Doorn et al., 1998; Yamaoka et al., 2006). As a result, we evaluated the prevalence of the jhp0106 gene in 95 isolates from patients with different diseases by PCR, including 38 gastritis strains, 21 duodenal ulcer strains, 17 gastric ulcer strains, 18 gastric cancer strains, and 1 MALToma strain. The results showed that all tested isolates contained the jhp0106 gene. To determine whether Jhp0106 is a critical factor in H. pylori motility among different strains, we constructed rpoN and jhp0106 mutants of 14 clinical isolates. The results of soft-agar motility assay showed that all rpoN mutants and jhp0106 mutants were non-motile (Figure 6).
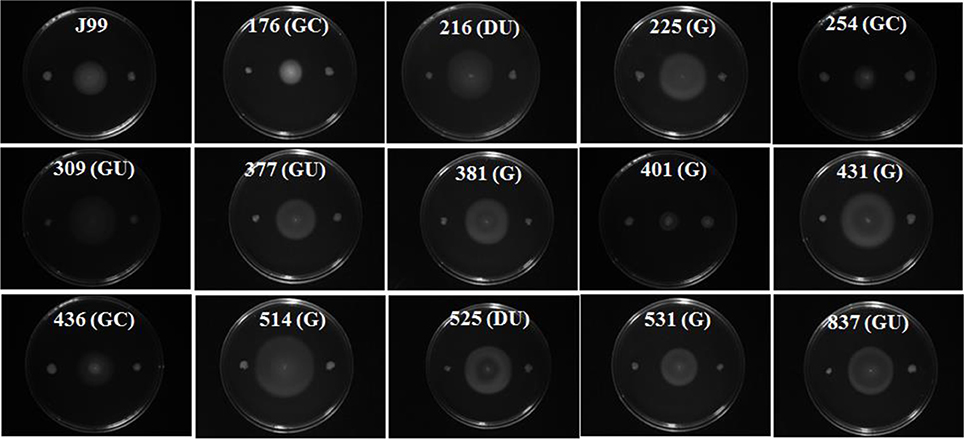
Figure 6. rpoN and jhp0106 are critical for the motility of 15 H. pylori strains. Wild-type, rpoN mutant, and jhp0106 mutant strains were inoculated in the middle, left, and right of the soft-agar plate, respectively. G, gastritis; GU, gastric ulcer; DU, duodenal ulcer; GC, gastric cancer.
Discussion
The present study aimed to reveal the CsrA regulatory system by using RNA-seq, and to identify CsrA target genes involved in H. pylori motility. We showed that CsrA regulated the level of the flaB-jhp0106 transcript in J99 by controlling the expression of the alternative sigma factor RpoN. In addition, Jhp0106 was characterized as a putative glycosyltransferase involved in flagellin glycosylation and flagella formation.
Edwards et al. identified > 700 transcripts that bind to CsrA in E. coli, indicating that CsrA affects expression of ~15% of the genes in E. coli (Edwards et al., 2011). In this study, we revealed that 53 genes (~4%) in a csrA mutant were found to be differentially expressed, compared to the wild-type J99 (Table 2). Like CsrA in E. coli and Clostridium acetobutylicum (Edwards et al., 2011; Tan et al., 2015), RNA-seq analysis showed that CsrA in H. pylori was closely involved in regulating multiple pathways including metabolism, iron uptake, flagella assembly, and oligopeptide transport (Table 2). However, the molecular mechanism through which CsrA regulates target gene expression in strain J99 remained to be clarified. Moreover, 15 CsrA-regulated genes of unknown function are worth investigating further (Table 2).
CsrA has been shown to control target gene expression by diverse mechanisms in several organisms (Romeo et al., 2013). For example, CsrA is mainly known for its post-transcriptional role in mRNA stability (Liu et al., 1995; Wei et al., 2001; Baker et al., 2002; Wang et al., 2005; Esquerre et al., 2016). In other cases, CsrA binds to multiple sites in the untranslated leader and/or initially translated region of target transcripts, and bound CsrA thus repressed translation by competing with ribosome binding to the Shine-Dalgarno sequence, but did not affect the level of targeted mRNA (Dubey et al., 2003; Ren et al., 2014). This may explain why only 4% of genes in J99 were identified to be regulated by CsrA, while 15% of genes were CsrA-regulated in E. coli (Edwards et al., 2011). As a result, it is worth carrying out a comparative proteomic analysis to evaluate any CsrA post-transcriptionally regulated genes in H. pylori.
Barnard et al. showed that the morphology of the csrA mutant N6 strain was similar to the wild-type strain, with a unipolar bundle of four or five flagella (Barnard et al., 2004). mRNA levels of flaA and flaB were elevated in the csrA mutant compared to the N6 strain. In addition, mutation of csrA in the N6 strain resulted in the increased expression of neutrophil activating protein (napA), ferric uptake regulator (fur), hspR, and groESL (Barnard et al., 2004). However, this is in contrast to our results. The expression of flaA and flaB at the mRNA level was reduced in the csrA mutant J99 (Table 2). napA, fur, hspR, and groESL transcripts were not changed in the csrA mutant J99. These results raised the possibility that the CsrA regulatory system was strain-specific, due to the high variation of the H. pylori genome.
Douillard et al. indicated that the hp0256 mutant has lower motility, significantly weaker adhesion, and induces weaker IL-8 secretion in AGS cells compared to the wild-type CCUG17874 strain (Douillard et al., 2010). In our previous study, we showed that in patients infected with higher-motility strains, the bacterial density, inflammatory score, and rate of atrophy were higher than those of patients infected with lower motility strains (Kao et al., 2012a), suggesting that H. pylori motility may be positively correlated with bacterial adhesion and il-8 gene expression in H. pylori-infected AGS cells. However, the jhp0106 mutation caused a non-flagellated phenotype in strain J99, and there was no decrease of adhesion or IL-8 production in AGS and GES-1 cells infected with SW863, compared to the wild-type (Figure 5, Figure S9). In contrast, the flaA/flaB double mutant (SW868), showing deficient motility, had weaker adhesion to AGS cells (67% compared to the wild-type) (Figure 5C). These results indicated that the motility of J99 has a minor role in cell adhesion. However, whether Jhp0106 modulates the composition/or modification of surface proteins in J99 and thus affects adhesion to AGS-1 cells is still unclear.
Based on the computational modeling, the overall structure of Jhp0106 resembles CstII, which is reported to participate in the sialylation of lipooligosaccharide cores and thus affects the immunogenicity of C. jejuni (Guerry et al., 2000; Chiu et al., 2004). Ram et al. showed that the sialylation of gonococcal lipo-oligosaccharide enables Neisseria gonorrhoeae to bind the alternative pathway complement inhibitor, factor H, and thus provides a protective barrier to evade attack by human complement (Ram et al., 1998). However, the role of pseudaminic acid in bacterial pathogenesis remains unclear. Pseudaminic acid has been shown to have striking structural and biosynthetic similarities to sialic acid (Lewis et al., 2009). Taken together, we suggested that Jhp0106 is involved in the transfer of pseudaminic acid to flagellin FlaA/FlaB, but not in the pseudaminic acid biosynthetic pathway (Figure S2). Therefore, large-scale identification of Jhp0106 target proteins by an LC/MS-based glycoproteomic approach is worth investigating.
The current treatments for H. pylori eradication are numerous and include triple and quadruple therapy, both of which utilize two antibiotics (metronidazole, amoxicillin, tetracycline, or clarithromycin) in addition to either a proton pump inhibitor (PPI) (triple therapy), or a PPI and bismuth (quadruple therapy) (O'Connor et al., 2016). The efficacies of these therapy strategies have been severely hampered in recent years due to the rise in antibiotic resistance of H. pylori isolates worldwide. Therefore, there is an emergent need to develop alternative therapeutic strategies for the management of H. pylori infection. Currently, carbohydrate-based therapies and diagnostics in cancer research and infectious disease have received considerable attention. Menard et al. identified three inhibitors of the pseudaminic acid biosynthetic enzymes which show activity in inhibiting the flagellin proteins on the C. jejuni cell surface, by bacterial cell-based assays (Menard et al., 2014). In this study, we found the prevalence of the jhp0106 gene among 95 clinical isolates of H. pylori in Taiwan was 100%. Moreover, mutation of jhp0106 of 15 clinical strains (include J99) led to the loss of motility of all mutants (Figure 6). These results suggest that Jhp0106 is a promising target for developing an inhibitor to restrain H. pylori infection in the future.
CsrA controls flagella-related genes' expression and motility of J99 by regulating RpoN expression (Kao et al., 2014) (Figure 7). Although RpoN is the key regulator under the control of CsrA, the mechanism(s) through which CsrA modulates rpoN expression is still unclear (Figure 7). In this study, we reveal the CsrA regulatory system in H. pylori by large-scale identification of target genes using RNA-seq. Moreover, the results suggest that RpoN not only controls flagellin expression but also modulates flagella assembly by regulating the expression of the putative glycosyltransferase Jhp0106, and thus affects the post-translational modification of flagellin (Figure 7). The motility of H. pylori is a critical virulence determinant in bacterial pathogenesis, therefore, understanding the complex regulatory pathways of flagella formation could in the future lead to novel therapies against H. pylori colonization. Future work will focus on the characterization of the Jhp0106 protein, including localization, enzymatic residues, and target proteins.
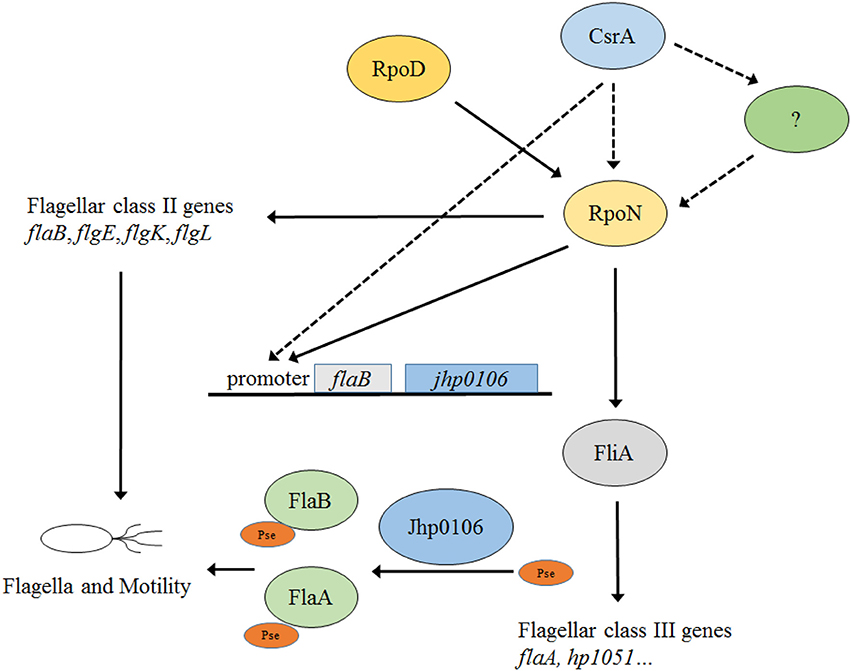
Figure 7. Current model of the CsrA/RpoN flagellar biosynthesis and motility regulatory system in H. pylori J99. Three different classes of flagellar genes are governed by the housekeeping sigma factor RpoD (class I), and the alternative sigma factors RpoN (class II) and FliA (class III). CsrA positively controls H. pylori J99 flagella formation and motility through regulating rpoN expression by an unclear mechanism(s) (shown in dotted line). In this study, the expression of jhp0106 (with a putative glycosyltransferase function) is under the control of the CsrA/RpoN system through the binding of RpoN to flaB promoter.
Author Contributions
CK, JC, and SW conceived the study, carried out experimental work and drafted the manuscript. CK, BS, and JW helped to interpret the data and draft the manuscript. All authors have read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer TC and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
We thank Robert Jonas for helpful comments on this manuscript. This study was supported by grant MOST 104-2320-B-010-046-MY3 from the Ministry of Science and Technology, Taiwan.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00483/full#supplementary-material
References
Ahmad, A., Govil, Y., and Frank, B. B. (2003). Gastric mucosa-associated lymphoid tissue lymphoma. Am. J. Gastroenterol. 98, 975–986. doi: 10.1111/j.1572-0241.2003.07424.x
Alm, R. A., Ling, L. S., Moir, D. T., King, B. L., Brown, E. D., Doig, P. C., et al. (1999). Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397, 176–180. doi: 10.1038/16495
Arnold, K., Bordoli, L., Kopp, J., and Schwede, T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201. doi: 10.1093/bioinformatics/bti770
Baker, C. S., Morozov, I., Suzuki, K., Romeo, T., and Babitzke, P. (2002). CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol. Microbiol. 44, 1599–1610. doi: 10.1046/j.1365-2958.2002.02982.x
Barnard, F. M., Loughlin, M. F., Fainberg, H. P., Messenger, M. P., Ussery, D. W., Williams, P., et al. (2004). Global regulation of virulence and the stress response by CsrA in the highly adapted human gastric pathogen Helicobacter pylori. Mol. Microbiol. 51, 15–32. doi: 10.1046/j.1365-2958.2003.03788.x
Chiu, C. P., Watts, A. G., Lairson, L. L., Gilbert, M., Lim, D., Wakarchuk, W. W., et al. (2004). Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog. Nat. Struct. Mol. Biol. 11, 163–170. doi: 10.1038/nsmb720
Douillard, F. P., Ryan, K. A., Lane, M. C., Caly, D. L., Moore, S. A., Penn, C. W., et al. (2010). The HP0256 gene product is involved in motility and cell envelope architecture of Helicobacter pylori. BMC Microbiol. 10:106. doi: 10.1186/1471-2180-10-106
Dubey, A. K., Baker, C. S., Suzuki, K., Jones, A. D., Pandit, P., Romeo, T., et al. (2003). CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J. Bacteriol. 185, 4450–4460. doi: 10.1128/JB.185.15.4450-4460.2003
Eaton, K. A., Suerbaum, S., Josenhans, C., and Krakowka, S. (1996). Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64, 2445–2448.
Edwards, A. N., Patterson-Fortin, L. M., Vakulskas, C. A., Mercante, J. W., Potrykus, K., Vinella, D., et al. (2011). Circuitry linking the Csr and stringent response global regulatory systems. Mol. Microbiol. 80, 156–180. doi: 10.1111/j.1365-2958.2011.07663.x
Esquerre, T., Bouvier, M., Turlan, C., Carpousis, A. J., Girbal, L., and Cocaign-Bousquet, M. (2016). The Csr system regulates genome-wide mRNA stability and transcription and thus gene expression in Escherichia coli. Sci. Rep. 6:25057. doi: 10.1038/srep25057
Golden, N. J., and Acheson, D. W. (2002). Identification of motility and autoagglutination Campylobacter jejuni mutants by random transposon mutagenesis. Infect. Immun. 70, 1761–1771. doi: 10.1128/IAI.70.4.1761-1771.2002
Graham, D. Y., Hepps, K. S., Ramirez, F. C., Lew, G. M., and Saeed, Z. A. (1993). Treatment of Helicobacter pylori reduces the rate of rebleeding in peptic ulcer disease. Scand. J. Gastroenterol. 28, 939–942. doi: 10.3109/00365529309098288
Guerry, P., Ewing, C. P., Hickey, T. E., Prendergast, M. M., and Moran, A. P. (2000). Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect. Immun. 68, 6656–6662. doi: 10.1128/IAI.68.12.6656-6662.2000
Haas, R., Meyer, T. F., and van Putten, J. P. (1993). Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol. Microbiol. 8, 753–760. doi: 10.1111/j.1365-2958.1993.tb01618.x
Jonas, K., Edwards, A. N., Simm, R., Romeo, T., Romling, U., and Melefors, O. (2008). The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol. Microbiol. 70, 236–257. doi: 10.1111/j.1365-2958.2008.06411.x
Josenhans, C., Labigne, A., and Suerbaum, S. (1995). Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J. Bacteriol. 177, 3010–3020. doi: 10.1128/jb.177.11.3010-3020.1995
Kao, C. Y., Sheu, B. S., Sheu, S. M., Yang, H. B., Chang, W. L., Cheng, H. C., et al. (2012a). Higher motility enhances bacterial density and inflammatory response in dyspeptic patients infected with Helicobacter pylori. Helicobacter 17, 411–416. doi: 10.1111/j.1523-5378.2012.00974.x
Kao, C. Y., Sheu, B. S., and Wu, J. J. (2014). CsrA regulates Helicobacter pylori J99 motility and adhesion by controlling flagella formation. Helicobacter 19, 443–454. doi: 10.1111/hel.12148
Kao, C. Y., Sheu, B. S., and Wu, J. J. (2016). Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomed. J. 39, 14–23. doi: 10.1016/j.bj.2015.06.002
Kao, C. Y., Sheu, S. M., Sheu, B. S., and Wu, J. J. (2012b). Length of thymidine homopolymeric repeats modulates promoter activity of sabA in Helicobacter pylori. Helicobacter 17, 203–209. doi: 10.1111/j.1523-5378.2012.00936.x
Karlyshev, A. V., Linton, D., Gregson, N. A., and Wren, B. W. (2002). A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology 148, 473–480. doi: 10.1099/00221287-148-2-473
Kostrzynska, M., Betts, J. D., Austin, J. W., and Trust, T. J. (1991). Identification, characterization, and spatial localization of two flagellin species in Helicobacter pylori flagella. J. Bacteriol. 173, 937–946. doi: 10.1128/jb.173.3.937-946.1991
Kwok, T., Backert, S., Schwarz, H., Berger, J., and Meyer, T. F. (2002). Specific entry of Helicobacter pylori into cultured gastric epithelial cells via a zipper-like mechanism. Infect. Immun. 70, 2108–2120. doi: 10.1128/IAI.70.4.2108-2120.2002
Lee, W. K., An, Y. S., Kim, K. H., Kim, S. H., Song, J. Y., Ryu, B. D., et al. (1997). Construction of a Helicobacter pylori-Escherichia coli shuttle vector for gene transfer in Helicobacter pylori. Appl. Environ. Microbiol. 63, 4866–4871.
Lertsethtakarn, P., Ottemann, K. M., and Hendrixson, D. R. (2011). Motility and chemotaxis in Campylobacter and Helicobacter. Annu. Rev. Microbiol. 65, 389–410. doi: 10.1146/annurev-micro-090110-102908
Lewis, A. L., Desa, N., Hansen, E. E., Knirel, Y. A., Gordon, J. I., Gagneux, P., et al. (2009). Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc. Natl. Acad. Sci. U.S.A. 106, 13552–13557. doi: 10.1073/pnas.0902431106
Liu, M. Y., and Romeo, T. (1997). The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J. Bacteriol. 179, 4639–4642. doi: 10.1128/jb.179.14.4639-4642.1997
Liu, M. Y., Yang, H., and Romeo, T. (1995). The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J. Bacteriol. 177, 2663–2672. doi: 10.1128/jb.177.10.2663-2672.1995
Logan, S. M. (2006). Flagellar glycosylation-a new component of the motility repertoire? Microbiology 152, 1249–1262. doi: 10.1099/mic.0.28735-0
McNally, D. J., Hui, J. P., Aubry, A. J., Mui, K. K., Guerry, P., Brisson, J. R., et al. (2006). Functional characterization of the flagellar glycosylation locus in Campylobacter jejuni 81-176 using a focused metabolomics approach. J. Biol. Chem. 281, 18489–18498. doi: 10.1074/jbc.M603777200
Menard, R., Schoenhofen, I. C., Tao, L., Aubry, A., Bouchard, P., Reid, C. W., et al. (2014). Small-molecule inhibitors of the pseudaminic acid biosynthetic pathway: targeting motility as a key bacterial virulence factor. Antimicrob. Agents Chemother. 58, 7430–7440. doi: 10.1128/AAC.03858-14
Niehus, E., Gressmann, H., Ye, F., Schlapbach, R., Dehio, M., Dehio, C., et al. (2004). Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52, 947–961. doi: 10.1111/j.1365-2958.2004.04006.x
O'Connor, A., Fischbach, W., Gisbert, J. P., and O'Morain, C. (2016). Treatment of Helicobacter pylori infection 2016. Helicobacter 21(Suppl. 1), 55–61. doi: 10.1111/hel.12342
Ottemann, K. M., and Lowenthal, A. C. (2002). Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect. Immun. 70, 1984–1990. doi: 10.1128/IAI.70.4.1984-1990.2002
Parsonnet, J., Friedman, G. D., Vandersteen, D. P., Chang, Y., Vogelman, J. H., Orentreich, N., et al. (1991). Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325, 1127–1131. doi: 10.1056/NEJM199110173251603
Ram, S., Sharma, A. K., Simpson, S. D., Gulati, S., McQuillen, D. P., Pangburn, M. K., et al. (1998). A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J. Exp. Med. 187, 743–752. doi: 10.1084/jem.187.5.743
Ren, B., Shen, H., Lu, Z. J., Liu, H., and Xu, Y. (2014). The phzA2-G2 transcript exhibits direct RsmA-mediated activation in Pseudomonas aeruginosa M18. PLoS ONE 9:e89653. doi: 10.1371/journal.pone.0089653
Romeo, T., Gong, M., Liu, M. Y., and Brun-Zinkernagel, A. M. (1993). Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 175, 4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993
Romeo, T., Vakulskas, C. A., and Babitzke, P. (2013). Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ. Microbiol. 15, 313–324. doi: 10.1111/j.1462-2920.2012.02794.x
Schirm, M., Soo, E. C., Aubry, A. J., Austin, J., Thibault, P., and Logan, S. M. (2003). Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 48, 1579–1592. doi: 10.1046/j.1365-2958.2003.03527.x
Schoenhofen, I. C., Lunin, V. V., Julien, J. P., Li, Y., Ajamian, E., Matte, A., et al. (2006). Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori. J. Biol. Chem. 281, 8907–8916. doi: 10.1074/jbc.M512987200
Suerbaum, S., Josenhans, C., and Labigne, A. (1993). Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J. Bacteriol. 175, 3278–3288. doi: 10.1128/jb.175.11.3278-3288.1993
Tan, Y., Liu, Z. Y., Liu, Z., Zheng, H. J., and Li, F. L. (2015). Comparative transcriptome analysis between csrA-disruption Clostridium acetobutylicum and its parent strain. Mol. Biosyst. 11, 1434–1442. doi: 10.1039/C4MB00600C
van Alphen, L. B., Wuhrer, M., Bleumink-Pluym, N. M., Hensbergen, P. J., Deelder, A. M., and van Putten, J. P. (2008). A functional Campylobacter jejuni maf4 gene results in novel glycoforms on flagellin and altered autoagglutination behaviour. Microbiology 154, 3385–3397. doi: 10.1099/mic.0.2008/019919-0
van Doorn, L. J., Figueiredo, C., Sanna, R., Plaisier, A., Schneeberger, P., de Boer, W., et al. (1998). Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology 115, 58–66. doi: 10.1016/S0016-5085(98)70365-8
Wang, X., Dubey, A. K., Suzuki, K., Baker, C. S., Babitzke, P., and Romeo, T. (2005). CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 56, 1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x
Wang, Y., and Taylor, D. E. (1990). Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94, 23–28. doi: 10.1016/0378-1119(90)90463-2
Wei, B. L., Brun-Zinkernagel, A. M., Simecka, J. W., Pruss, B. M., Babitzke, P., and Romeo, T. (2001). Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40, 245–256. doi: 10.1046/j.1365-2958.2001.02380.x
Keywords: flagella, glycosylation, motility, CsrA, jhp0106
Citation: Kao C-Y, Chen J-W, Wang S, Sheu B-S and Wu J-J (2017) The Helicobacter pylori J99 jhp0106 Gene, under the Control of the CsrA/RpoN Regulatory System, Modulates Flagella Formation and Motility. Front. Microbiol. 8:483. doi: 10.3389/fmicb.2017.00483
Received: 04 January 2017; Accepted: 08 March 2017;
Published: 28 March 2017.
Edited by:
Catherine Ayn Brissette, University of North Dakota, USAReviewed by:
Timothy Casselli, University of North Dakota, USAPablo Ivan Nikel, The Novo Nordisk Foundation Center for Biosustainability (DTU Biosustain), Denmark
Copyright © 2017 Kao, Chen, Wang, Sheu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiunn-Jong Wu, amp3dTEwMTlAeW0uZWR1LnR3
 Cheng-Yen Kao
Cheng-Yen Kao Jenn-Wei Chen
Jenn-Wei Chen Shuying Wang
Shuying Wang Bor-Shyang Sheu
Bor-Shyang Sheu Jiunn-Jong Wu
Jiunn-Jong Wu