- Graduate Institute of Biotechnology, National Chung Hsing University, Taichung, Taiwan
The genus Potexvirus is one of the eight genera belonging to the family Alphaflexiviridae according to the Virus Taxonomy 2015 released by International Committee on Taxonomy of Viruses (www.ictvonline.org/index.asp). Currently, the genus contains 35 known species including many agricultural important viruses, e.g., Potato virus X (PVX). Members of this genus are characterized by flexuous, filamentous virions of 13 nm in diameter and 470–580 nm in length. A potexvirus has a monopartite positive-strand RNA genome, encoding five open-reading frames (ORFs), with a cap structure at the 5′ end and a poly(A) tail at the 3′ end. Besides PVX, Bamboo mosaic virus (BaMV) is another potexvirus that has received intensive attention due to the wealth of knowledge on the molecular biology of the virus. In this review, we discuss the enzymatic activities associated with each of the functional domains of the BaMV replication protein, a 155-kDa polypeptide encoded by ORF1. The unique cap formation mechanism, which may be conserved across the alphavirus superfamily, is particularly addressed. The recently identified interactions between the replication protein and the plant host factors are also described.
BaMv Genome
Bamboo mosaic virus (BaMV) primarily infects members of the Bambusoideae in nature; nonetheless, it also replicates in Nicotiana benthamiana, which thereby has been used as the surrogate in laboratories. The RNA genome of BaMV contains 6366 nucleotides (nts) plus a 5′ m7GpppG (cap0) structure and a 3′ poly(A) tail (Figure 1A). It is functionally organized into a 94-nt 5′ untranslated region (UTR), five ORFs, and a 142-nt 3′ UTR (Lin et al., 1994). Two major subgenomic RNAs, co-terminal with the viral 3′ UTR, would be produced once the virus starts to replicate in host cells. The first ORF encodes a 155-kDa non-structural protein (REPBaMV) that has been thought to be essential for replication/transcription of the viral genome and the formation of the 5′ cap based on the presence of signature motifs of Sindbis virus-like methyltransferase (Rozanov et al., 1992), helicase (Habili and Symons, 1989), and RNA polymerase (Koonin and Dolja, 1993). As many positive strand RNA viruses, BaMV must encode its own enzymes for replication/transcription and 5′ cap formation because it replicates only in the cytoplasm. ORF2, 3 and 4 are overlapped, often referred to as the triple gene block (TGB), and their translated proteins, TGBp1, TGBp2, and TGBp3, respectively, are indispensable for BaMV movement in plants (Lin et al., 2004, 2006). In-depth discussions about the functions of each of the TGB proteins of PVX in the intracellular trafficking and intercellular transport can be referred in a couple of recent reviews (Verchot-Lubicz et al., 2010; Park et al., 2014). ORF5 encodes the viral coat protein (CP) that is the only structural protein required for the assembly of BaMV virions. CP also exerts a critical function in the accumulation of BaMV RNAs in protoplasts (Lee et al., 2011). It is unclear whether CP protects BaMV RNAs from being destroyed by the host defense mechanisms or if it actually participates in the viral replication process. In addition, CP of potexvirus was reported to play a role in the virus movement. For instance, White clover mosaic virus needs CP to spread efficiently in plants (Forster et al., 1992), and PVX is defective in cell-to-cell movement if it carries a C-terminally truncated CP (Fedorkin et al., 2001). Occasionally, an 836-nt satellite RNA (satBaMV) is found in association with BaMV in nature (Lin and Hsu, 1994). satBaMV contains one ORF that encodes a 20-kDa polypeptide (P20). P20 is not necessary for the replication of satBaMV; nonetheless, the accumulation rate of satBaMV in systemic leaves decreases in the absence of P20 (Lin et al., 1996).
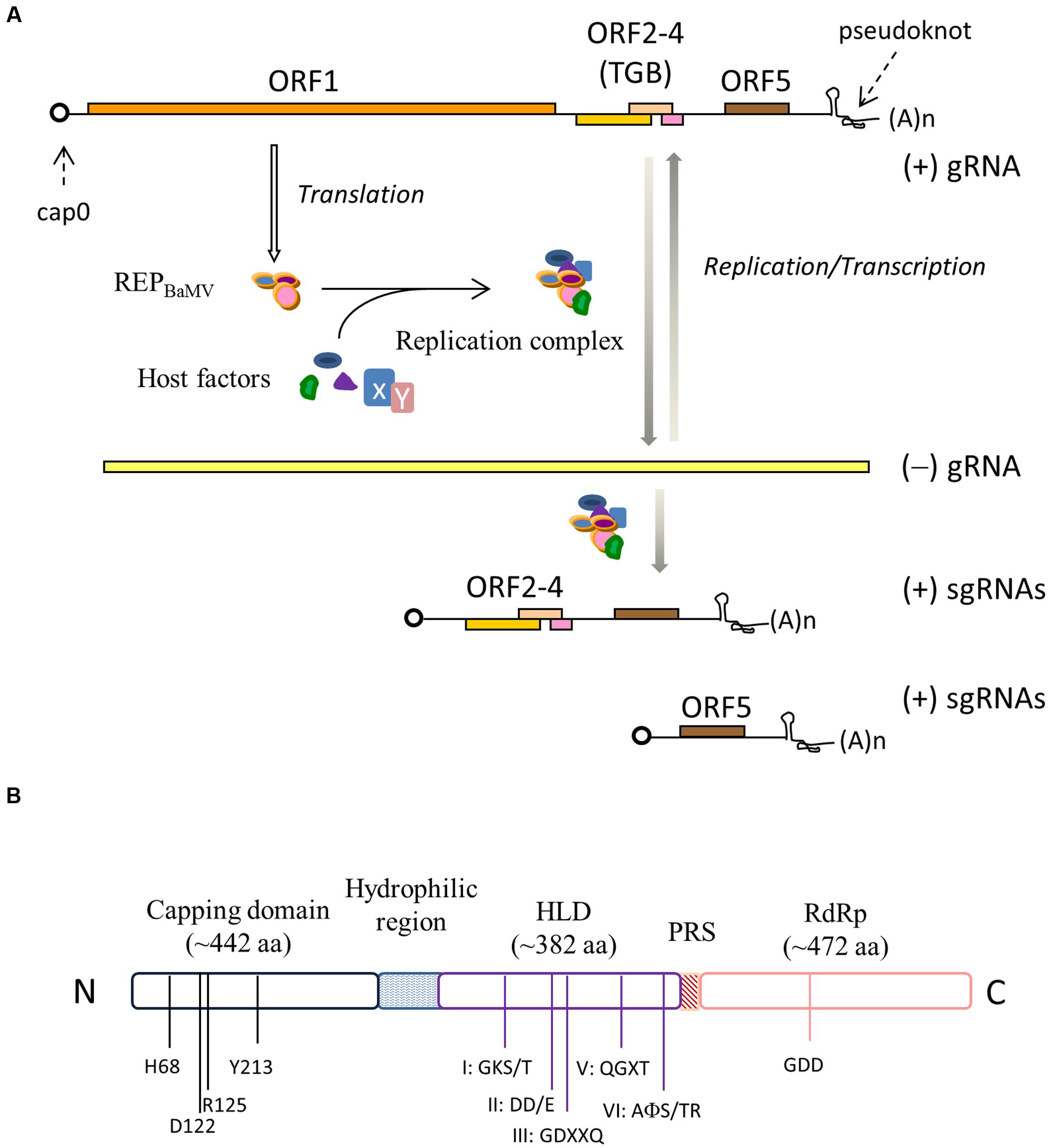
FIGURE 1. (A) The genome organization of Bamboo mosaic virus (BaMV). REPBaMV, the translation product of ORF1, associates with host factors to form the viral replication complex. Different subsets of host factors may be recruited to perform negative- and positive-strand replication and transcription. (B) Functional domains in RFPBaMV. The conserved residues in the N-terminal capping enzyme domain and the featured motifs suggestive of the helicase-like domain (HLD) and the C-terminal RdRp domain are indicated. The domains are separated by a disordered hydrophilic region and a proline rich segment (PRS).
Domain Organization of REPBaMv
There are apparently three functional domains in REPBaMV (Figure 1B), separated by a disordered hydrophilic region, from approximately amino acid residues 406–520, and a proline-rich segment (PRS), residues 895–910, according to a secondary structure prediction using the PHD algorithm (Rost et al., 1994). The N-terminal one-third of REPBaMV shares a few dispersedly conserved residues with the putative Sindbis-like methyltransferase domains (Rozanov et al., 1992) of a variety of plant and animal alphavirus-like viruses such as Brome mosaic virus and Semliki Forest virus (Li et al., 2001a). Sequence comparison also revealed that the central domain contains several NTP-binding motifs of RNA helicase superfamily 1 (SF1) (Kadaré and Haenni, 1997) and the C-terminal domain contains featured motifs of RNA polymerases, e.g., the catalytic GDD motif (Koonin and Dolja, 1993). Since REPBaMV is barely discernible in BaMV-infected N. benthamiana, the enzymatic activity associated with each of the domains has been investigated using the domains expressed in heterologous hosts such as Escherichia coli and Saccharomyces cerevisiae.
Capping Enzyme Domain
The enzymatic activity of the N-terminal 442 amino acids of REPBaMV was successfully characterized by using the domain expressed in S. cerevisiae (Li et al., 2001a). The recombinant domain, strongly associated with the yeast membrane, could be radiolabeled by [α-32P]GTP if S-adenosylmethionine (AdoMet) was provided in the reaction buffer. Alternatively, it could be radiolabeled by Ado[methyl-3H]Met when GTP was present. The radiolabeled moiety covalently linked to the domain was subsequently determined to be m7GMP. This [m7GMP-enzyme] adduct was thought to represent an intermediate in the pathway to form the 5′ cap. In other words, this viral domain could be a guanylyltransferase (mRNA capping enzyme) except that it is covalently modified by m7GMP rather than GMP. In addition, this N-terminal domain of REPBaMV was found capable of catalyzing a methyl transfer reaction from AdoMet to GTP, leading to the formation of m7GTP, consistent with the prediction of its function as a methyltransferase. This viral domain was therefore proposed to possess an AdoMet-dependent guanylyltransferase activity, by which the methyl group of AdoMet is transferred to GTP, leading to m7GTP formation, and then the m7GMP moiety of m7GTP was transferred to an active-site residue to form the covalent [m7GMP-enzyme] intermediate. Analogous reactions have been observed also in other members of alphavirus-like superfamily including alphavirus (Ahola and Kääriäinen, 1995), Brome mosaic virus (Ahola and Ahlquist, 1999), Semliki Forest virus (Ahola et al., 1997), Hepatitis E virus (Magden et al., 2001) and Tobacco mosaic virus (TMV) (Merits et al., 1999), suggesting that this unique mRNA capping process is conserved throughout diverse members within the superfamily in spite of the fact that only limited amino acid identities (e.g., H68, D122, R125, and Y213 in REPBaMV) are conserved.
Site-directed mutagenesis indicated that H68, D122, R125, and Y213 are essential for the BaMV capping domain to form the covalent [m7GMP-enzyme] intermediate (Huang et al., 2004). Alanine substitution for each of the conserved residues, except H68, also disabled the domain to produce m7GTP (Huang et al., 2004). Intriguingly, H68A mutant increased m7GTP production by a factor of ∼10, implying a special role of H68 in the pathway to form the covalent [m7GMP-enzyme] intermediate. The H68A mutant was thus treated as the pseudo wild type to investigate the aromatic residues important for the formation of m7GTP (Hu et al., 2011). A number of aromatic residues, including Y126, F144, F161, Y192, Y203, Y213, and W222, were found critical for AdoMet recognition. Alanine substitution for these residues, except Y213, also reduced the binding affinity to GTP. Probably, the BaMV capping domain binds AdoMet and GTP in close proximity and many of these aromatic residues participate in the binding of the two substrates simultaneously. It is noteworthy that all the indicated aromatic residues are well conserved among the capping domains of potexviruses. The primary function of Y213 is to bind AdoMet. The inability to substitute phenylalanine for Y213 suggests that the hydroxyl group on Y213 provides an essential hydrogen bond to AdoMet. Presumably, Y231 locks AdoMet in a correct spatial position so that the methyl group from the electrophilic methylsulfonium of AdoMet can be transferred to the N7 of GTP.
Peptide mapping using alkaline hydroxylamine, which specifically cleaves the asparaginyl-glycyl bond (Bornstein and Balian, 1977), indicated that the m7GMP-linking residue of the BaMV capping domain is located within the region of residues 44–76 (Lin et al., 2012). The covalent [m7GMP-enzyme] intermediate was sensitive to 0.1 N HCl but tolerant of 0.1 N NaOH (Lin et al., 2012), suggesting that the link connecting the domain and m7GMP is a phosphoamide bond (Duclos et al., 1991). Amino acids with nucleophilic side chains including lysine, arginine, asparagine, glutamine, serine, threonine, tyrosine, and cysteine were used to replace His68 (Lin et al., 2012). All the mutants, except H68C, failed to form the covalent [m7GMP-enzyme] intermediate. H68C retained a detectable activity for the covalent intermediate formation despite at considerably lower extent. The bond connecting m7GMP and the H68C mutant enzyme was moderately stable in 0.1 N HCl and 0.1 N NaOH (Lin et al., 2012), a characteristic of a phosphocysteine bond (Duclos et al., 1991). The change of the nature of the bond connecting the enzyme and m7GMP and the result of peptide mapping lead to the conclusion that His68 acts as the nucleophile to attack the α-phosphate of m7GTP, consequently leading to the formation of the covalent [m7GMP-enzyme] intermediate.
The catalytic step after formation of the [m7GMP-enzyme] intermediate was characterized by monitoring the transfer of 32P-radiolabeled m7GMP of the covalent intermediate to various RNAs (Huang et al., 2005). A RNA transcript with 5′-terminal diphosphate is a prerequisite to receive m7GMP from the covalent intermediate, and RNA led by GDP is a better substrate than that led by ADP. The putative stem-loop structure in the 5′ region of BaMV genome, nts 34–118, has a critical effect on the capping efficiency of the genomic RNA, suggesting that most of the cap formation events occur after the stem-loop sequence has been synthesized in nascent transcripts. This result also implies that the RNA polymerase domain and the capping domain of REPBaMV need to coordinate to some extent.
According to the data aforementioned and others, the cap formation pathway catalyzed by the capping domain of REPBaMV is delineated in Figure 2. (1) GTP and AdoMet bind to the capping domain of REPBaMV in proximity (Hu et al., 2011). The presence of AdoMet actually enhances the binding affinity of the domain for GTP. (2) The precise disposition of GTP and AdoMet in the domain facilitates a nucleophilic attack of the N7 of GTP on the methyl group of AdoMet, leading to the production of m7GTP and S-adenosyl-L-homocysteine (AdoHcy). (3) The nitrogen atom (not determined whether Nδ1 or N𝜀2) of His68 functions as a nucleophile attacking the α-phosphate of m7GTP, under the assistance of Mg2+, to form the covalent [m7GMP-enzyme] intermediate (Lin et al., 2012). This step is reversible because excess pyrophosphate could drive the m7GMP moiety on the covalent intermediate backward to form m7GTP (Huang et al., 2004). (4) The 5′-terminal diphosphate of nascent RNA binds to the domain in proximity to the m7GMP moiety. The 5′ β-phosphate of the RNA launches a nucleophilic attack on the phosphorus atom of m7GMP, leading to the break of the phosphohistidine bond. (5) Finally, the RNA with a 5′ cap0 structure is released from the domain.
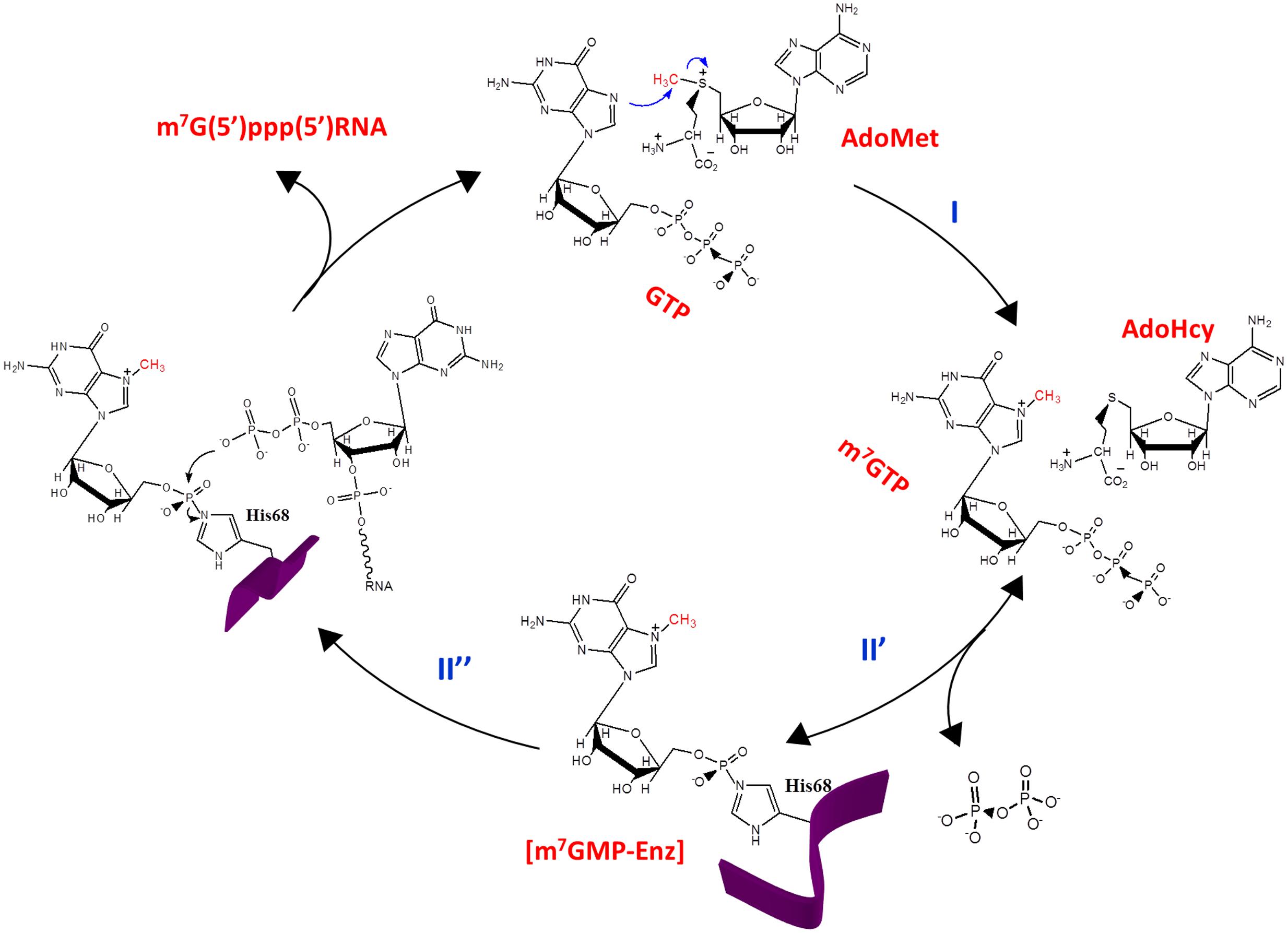
FIGURE 2. The cap0 formation pathway of BaMV. The AdoMet-dependent guanylyltransferase activity exhibited by REPBaMV is composed of activities of (I) GTP methyltransferase and (II) m7GTP:RNA guanylyltransferase, which can be further divided into two half reactions with the [m7GMP-enzyme] adduct as the intermediate.
Helicase-Like Domain (HLD)
The HLD of REPBaMV (residues 514–895) forms inclusion bodies when it is expressed in E. coli. This domain resumes soluble after denaturation and refolding processes. The purified HLD is able to remove the γ phosphate from nucleoside triphosphates as well as RNA (Li et al., 2001b); in other words, it can be a nucleoside triphosphatase (NTPase) or RNA 5′-triphosphatase (5′-TPase), depending on the substrate. Both of these reactions required the presence of divalent Mg2+ or Mn2+ cations. Mutations at any of the signature motifs I, II, III, or VI of SF1 abrogate both types of activity (Han et al., 2007). Adenylyl-imidodiphosphate (AMPPNP), a non-hydrolyzable ATP analog, is a competitive inhibitor of the RNA 5′-TPase activity. The inhibition constant Ki(AMPPNP) was determined to be 93 μM, which is close to the Km value of ATP (150 μM) for the NTPase activity (Han et al., 2007). The closeness between the values of Ki(AMPPNP) and Km(ATP) and the simultaneous inactivation of both activities by mutations at the featured motifs of helicases suggest that a common catalytic site is used for the hydrolysis of both NTP and RNA. Nonetheless, the greater value of Km(ATP) than Km(RNA), which is about 2.5 fold, suggests that more active-site residues are involved in RNA binding. The peptidyl regions employed by the HLD to bind biotinylated RNA were mapped by the reversible formaldehyde crosslinking method followed by tandem mass spectrometry (Han et al., 2009). Five peptidyl regions were identified. Regions of residues 625–645 and 696–706 encompass the helicase motif I and II, respectively; while regions of residues 585–610, 789–799, and 833–843 do not contain conserved sequences known to SF1. Compared with the well-characterized members in SF1, e.g., DNA helicase PcrA, the BaMV HLD seems to bind RNA using a different set of peptidyl regions. Mutagenesis of positively charged residues in these regions showed that some residues, e.g., K603 and R628, have a role in the virus movement (Han et al., 2009).
TGBp1 of BaMV is also a member of SF1. TGBp1 is capable of hydrolyzing NTPs but not RNA (Li et al., 2001b), implying that the RNA 5′-TPase activity embedded in the HLD of REPBaMV is not necessarily a property of all helicase proteins. The biological relevance of the RNA 5′-TPase activity was demonstrated in an in vitro assay, in which an RNA transcript would be capped at the 5′ end by the capping domain of REPBaMV only if the RNA transcript had been pretreated with the HLD (Li et al., 2001b). Taken together, the first two domains of REPBaMV work in a concerted manner to complete the formation of the 5′ cap on the nascent viral positive-strand RNAs. Besides participating in 5′ cap formation, the HLD of REPBaMV has also been proposed to act as a bona fide helicase in the replication/transcription process of BaMV. Unfortunately, convincing evidence for duplex RNA-unwinding activity is still lacking even though a great deal of time and effort has been spent. To our knowledge, no helicase activity has been reported in the HLD of any other potexviruses. Perhaps, a more sophisticated assay is needed to discern this peculiar helicase activity. It is also possible that a host protein (other than a host helicase) may be recruited as an accessory subunit of the helicase to confer unwinding activity on the viral protein.
Yeast two-hybrid screen using a cDNA library prepared from BaMV-infected leaves of N. benthamiana identified a strong protein–protein interaction between the HLD of REPBaMV and the viral CP (Lee et al., 2011). Interacting with CP does not alter the in vitro enzymatic activity of the HLD. Mutations of A209G and N210S in CP, which diminish the CP-HLD interaction, were identified by a bacterial two-hybrid screen using a CP random mutant library generated by error-prone PCR (Lee et al., 2011). Mutant BaMV carrying A209G and/or N210S reproduces as efficiently as the wild type virus in N. benthamiana protoplasts (Lee et al., 2011). CP with the mutations retains a full activity for RNA binding, and the mutant virions exhibit similar morphologies as the wild type under transmission electron microscope (Lee et al., 2011). Nonetheless, the CP mutations do exert a profound effect on BaMV cell-to-cell movement in plants. With the A209G mutation, BaMV spreads much less effectively in leaves of N. benthamiana and Chenopodium quinoa (Lee et al., 2011). Notably, A209 of BaMV CP is well conserved among many potexviruses such as PVX and Foxtail mosaic virus (FoMV). A230G mutation in FoMV CP, analogous to BaMV A209G, also reduces the viral HLD-CP interaction and restricts the cell-to-cell movement of FoMV in C. quinoa (Lee et al., 2011). This finding suggests that the HLD-CP interaction is rather common in potexviruses; moreover, this interaction is relevant to the ability of the virus to move between cells. The critical role of the HLD-CP interaction in BaMV movement prompts us to suspect that REPBaMV is recruited into the viral movement complex, which is composed of mainly the viral RNA, TGBps, and CP. More importantly, REPBaMV may pass through plasmodesmata along with the viral RNA. With this strategy, the viral RNA can be re-replicated immediately in the newly invaded cells so that the virus has a greater chance to defeat the silencing mechanism imposed by the hosts. Involvement of the replication protein in the viral movement complex has also been proposed in TMV based on the observation that TMV requires a significantly longer time for movement from primary inoculated cells to secondary cells than is required for movement from secondary to tertiary cells (Kawakami et al., 2004).
A Distinct Pathway/Machinery for the 5′ Cap Formation
The 5′ cap0, m7G(5′)ppp(5′)Np, in eukaryotic mRNAs is a basic structural unit required for mRNA export from the nucleus, prevention of mRNA degradation by 5′-exonucleases, and recognition by eIF4F complex to initiate the translation process (Furuichi and Shatkin, 2000; Shuman, 2001). Different pathways leading to the formation of the cap structure have been reported (Figure 3). Three consecutive enzymatic reactions are responsible for the cap formation in the nucleus. First, the γ-phosphate of a nascent mRNA is removed by RNA 5′-triphosphatase. The GMP moiety of GTP is then transferred to the 5′ end of the 5′-diphosphorylated mRNA via a covalent enzyme-lysyl-GMP intermediate by GTP:mRNA guanylyltransferase. Finally, the guanine-N7 of G(5′)ppp(5′)Np cap is methylated by RNA (guanine-N7) methyltransferase to produce the cap0 structure (Mizumoto and Kaziro, 1987; Shuman, 1995). This canonical cap formation pathway also occurs in some DNA viruses, e.g., vaccinia virus (Shuman et al., 1980; Niles and Christen, 1993) and chlorella virus (Håkansson et al., 1997), and the double-stranded RNA reovirus (Luongo et al., 2000; Luongo, 2002). In the case of BaMV, the RNA 5′-triphosphatase activity embedded in the helicase-like domain of REPBaMV catalyzes the removal of γ-phosphate from the 5′ end of nascent positive-strand RNA (Li et al., 2001b). The capping domain of REPBaMV exhibits an AdoMet-dependent mRNA guanylyltransferase activity, by which the methyl group of AdoMet is transferred to the N7 of GTP, and then the m7GMP moiety is transferred from the newly formed m7GTP to the 5′ end of a 5′-diphosphorylated RNA via a covalent enzyme-histidyl-m7GMP intermediate (Li et al., 2001a,b; Huang et al., 2005; Lin et al., 2012). Plausibly, this type of cap formation pathway for BaMV also occurs across the alphavirus-like superfamily of human, animal, and plant-infection positive-strand RNA viruses. Vesicular stomatitis virus (VSV) performs another unconventional mRNA 5′ cap formation pathway (Ogino and Banerjee, 2007; Ogino et al., 2010). Besides exhibiting a RNA-dependent RNA polymerase activity, the L protein of VSV has a RNA:GDP polyribonucleotidyltransferase activity that catalyzes the transfer of the 5′-monophosphorylated viral mRNA to GDP via an enzyme-histidyl-pRNA intermediate. Two methylation reactions at the capped RNA follow to form the cap1 structure by the viral methyltransferase activity.
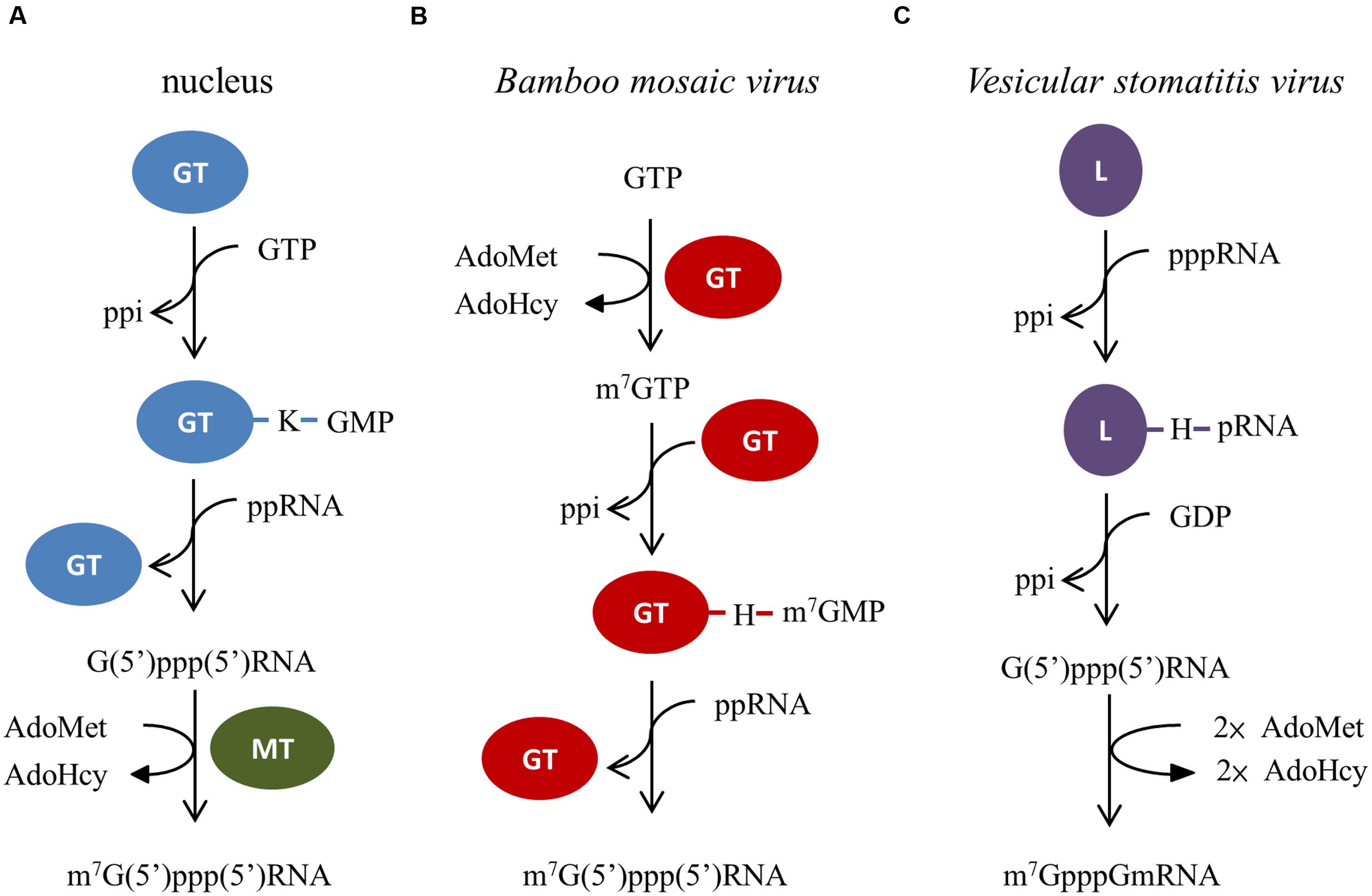
FIGURE 3. The distinct cap formation pathways among eukaryotic nucleus, Bamboo mosaic virus, and Vesicular stomatitis virus. (A) In nucleus, GTP:mRNA guanylyltransferase (GT) and RNA (guanine-N7) methyltransferase (MT) are responsible for the cap formation. (B) Only the capping enzyme domain (GT) of REPBaMV is required for Bamboo mosaic virus to form the cap structure. (C) Whereas, the L protein of Vesicular stomatitis virus possesses both the activities of RNA:GDP polyribonucleotidyltransferase and mRNA methyltransferase.
The BaMV enzymes performing the catalytic steps in the pathway are also unique from the viewpoint of protein structures. Apparently, the BaMV RNA 5′-triphosphatase activity has emerged from the helicase motif-containing domain. By contrast, the RNA 5′-triphosphatases of yeast and DNA viruses, e.g., vaccinia virus and baculovirus, belong to a metal-dependent phosphohydrolase family (Lima et al., 1999), while those of animals and plants are classified into a cysteine phosphatase superfamily (Changela et al., 2001). Moreover, the capping domain of REPBaMV does not share similarity in amino acid sequence with either GTP:mRNA guanylyltransferase or RNA (guanine-N7) methyltransferase of eukaryotic cells and DNA viruses. With the limited genome size, BaMV has evolved an efficient capping enzyme, with merely 442 amino acids, to accomplish the work of forming the 5′ cap.
RNA-Dependent RNA Polymerase Domain
The C-terminal domain of REPBaMV had been thought to be the key component of the viral replication complex, due to the presence of the hallmark signature of polymerase S/TGX3TX3NS/TX22GDD (Koonin and Dolja, 1993). This domain (residues 893–1364), expressed in E. coli with a thioredoxin tag fused at the N terminus, exhibits an in vitro RNA polymerase activity, preferentially taking the 3′-terminal fragments of both positive and negative strands of BaMV as templates (Li et al., 1998). Mutational analysis confirmed the essential role of the GDD motif in the catalysis of polymerization reaction. Structure mapping based on selective RNA hydrolysis using a variety of ribonucleases and chemicals suggested that the 3′ UTR of BaMV folds into four consecutive stem-loop domains (A–D), followed by a tertiary pseudoknot structure (Cheng and Tsai, 1999). The hexanucleotide ACC/UUAA, conserved in the 3′ UTR of potexviruses, is situated in the apical loop of the D domain. A competition binding assay suggested that the E. coli-expressed BaMV polymerase domain binds independently to the D domain and the poly(A) tail (Huang et al., 2001). A footprinting assay further defined the D loop as the primary region protected by the polymerase domain of REPBaMV against chemical cleavages (Huang et al., 2001). Similarly, the 3′-terminal fragment (77 nts) of the BaMV negative-strand RNA was mapped to contain a 5′stem-loop, followed by a spacer and the 3′-CUUUU sequence (Lin et al., 2005). Reducing the number of uridylate in the 3′-CUUUU to less than three or changing the penultimate U to other nucleotides is deleterious to BaMV accumulation in plants (Chen et al., 2010). UV-crosslinking and competition assay indicated that the E. coli-expressed BaMV polymerase domain also binds to the 3′-terminal fragment of the negative-strand RNA through a specific interaction particularly with the 5′stem-loop (Chen et al., 2010). In summary, the polymerase domain of REPBaMV recognizes the specific sequence and structural feature formed on the 3′-terminal region of both the positive and negative strands of BaMV, enabling the viral RNA replication to be initiated at the precise positions. Without these specific protein–RNA interactions, the replication of the viral RNAs would be incorrect or even impossible.
Subcellular Localization of REPBaMv
In general, the replication complexes of plant positive-strand RNA viruses, which consist of the viral replication proteins, the viral genomic RNAs, and co-opted host factors, are embedded in membrane-enclosed micro-compartments derived from various cellular organelles (Novoa et al., 2005; Miller and Krijnse-Locker, 2008). Virus replication within the microenvironments should benefit the viral RNAs from being destroyed by the host defense mechanisms. For instances, Brome mosaic virus and Red clover necrotic mosaic virus recruit the membrane derived from endoplasmic reticulum to constitute their replication complexes (Noueiry and Ahlquist, 2003; Turner et al., 2004), while Flock house virus and Tomato bushy stunt virus employ the membrane of mitochondria and peroxisome, respectively, for replication complex assembly (Miller et al., 2001; McCartney et al., 2005). REPBaMV is also a membrane-associated protein. In fact, the membrane fraction P30 of BaMV-infected leaves, the pellet of cell extract after 30000 × g centrifugation, exhibits an in vitro BaMV RNA-dependent RNA polymerase activity; therefore, the P30 has been used in analysis of the cis-acting RNA elements required for the viral genome replication (Chen et al., 2003, 2005, 2010; Lin et al., 2005). To locate the subcellular organelle where BaMV replicates, a genetically modified BaMV positive-strand RNA that contains a phage MS2 CP-recognized sequence was inoculated into N. benthamiana leaves that had been infiltrated with Agrobacterium tumefaciens carrying the NLS-GFP-MS2 fusion protein-encoding gene (Cheng et al., 2013). The viral RNA was found located in chloroplasts according to the green fluorescent imaging of the infected cells under confocal microscope. Therefore, BaMV was proposed to replicate in chloroplasts although REPBaMV per se was invisible in the virus-infected leaves under microscope due to the low expression amount.
The chloroplast is a common target of a large number of plant viruses belonging to a variety of genera. Subcellular localization of the virus-encoded proteins in the chloroplast may constitute a basis for the viral pathogenesis or/and is critical for the viral propagation (Zhao et al., 2016). Besides BaMV, Alternanthera mosaic virus (AltMV) and PVX are two other potexviruses that have been demonstrated to be associated with chloroplasts in their infection processes. The TGB3 of AltMV preferentially accumulates around the chloroplast membrane and disruption of TGB3 targeting to chloroplast impairs cell-to-cell movement of the virus (Lim et al., 2010). Furthermore, AltMV TGB3 strongly interacts with the photosystem II oxygen-evolving complex protein PsbO and this interaction correlates with chloroplast vesiculation and veinal necrosis caused by TGB3 over-expression (Jang et al., 2013). In the case of PVX, the viral CP interacts with the transit peptide of plastocyanin, a protein involved in photosynthesis, and silencing of plastocyanin prior to PVX infection reduces CP accumulation in chloroplasts and ameliorates symptom severity in host plants (Qiao et al., 2009).
Host Proteins Associated with REPBaMv
A number of approaches have been used in the search for host proteins involved in regulation of the polymerase activity of REPBaMV. A biochemical protocol, basically involving steps of (1) UV-induced crosslinking of proteins in leaf cell extract to the 32P-radiolabeled 3′ UTR of BaMV, (2) nuclease digestion, and (3) radiolabeled protein identification using mass spectrometry, has identified several 3′ UTR-interacting proteins including chloroplast phosphoglycerate kinase (PGK), cytosolic glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and heat shock protein 90 homolog (NbHsp90). PGK promotes BaMV accumulation presumably by facilitating transport of the viral genomic RNA to chloroplasts, the plausible replication site for BaMV (Lin et al., 2007; Cheng et al., 2013). GAPDH binds to the pseudoknot poly(A) tail of BaMV and reduces the replication efficiency of the viral negative-strand RNA probably through a competition with REPBaMV for RNA binding (Prasanth et al., 2011). NbHsp90 enhances BaMV replication presumably by either promoting the maturation of REPBaMV or bridging the interaction of REPBaMV with the viral RNA (Huang et al., 2012). The physical interaction between NbHsp90 and REPBaMV was actually confirmed by a yeast two-hybrid assay. Yeast two-hybrid screen was also used to search for host proteins interacting with the polymerase domain of REPBaMV. An uncharacterized host AdoMet-dependent methyltransferase (PNbMTS1) was thus isolated from the cDNA library prepared from N. benthamiana leaves (Cheng et al., 2009). PNbMTS1 exhibits an AdoMet-dependent inhibitory effect on BaMV CP accumulation in protoplasts. By contrast, Tobacco rattle virus-induced gene silencing of PNbMTS1 increased BaMV CP and genomic RNA in N. benthamiana. Both the membrane-targeting signal peptide and the AdoMet-binding motifs are essential for PNbMTS1 to suppress BaMV accumulation. Collectively, PNbMTS1 may have a role in the plant innate defense mechanism. Nonetheless, the target of PNbMTS1 relevant to the inhibition effect is still unknown.
Recently, we found that the expression of REPBaMV in N. benthamiana could be significantly enhanced if satBaMV was co-expressed. Probably, the positive-strand RNA of satBaMV might act as a template to facilitate the folding of REPBaMV or prevent REPBaMV from being degraded by host proteases. Based on this finding, a proteomic approach was set up to find out the plant proteins differentially present in the REPBaMV-enriched P30 fraction (Lee et al., 2016). This approach includes steps of (1) transient expression of the hemagglutinin tag (HA)-fused REPBaMV and satBaMV, or satBaMV alone as the comparative control, in N. benthamiana by agroinfiltration, (2) preparation of the P30 fraction from the agroinfiltrated leaves, (3) protein solubilization using anionic detergent Sarkosyl, (4) protein precipitation using anti-HA antiserum, and (5) identification of the co-precipitated proteins by tandem mass spectrometry. Accordingly, dozens of host proteins were identified. To examine the role of the proteins in BaMV replication, each of the genes was transiently silenced in N. benthamiana. Those plants without apparent changes in phenotype were then challenged with a genetically modified BaMV that carries GFP as a reporter gene. Several potential host factors affecting BaMV replication were thus identified based on the effect of gene silencing on GFP expression. A cytoplasmic 5′→3′ exoribonuclease (NbXRN4), a ripening-related protein, S-adenosylmethionine synthetase, and a respiratory burst oxidase homolog were found capable of promoting BaMV replication. By contrast, NADP+-dependent isocitrate dehydrogenase and MAP kinase phosphatase-like protein appeared to suppress BaMV replication. The relevance between the activity of NbXRN4 and BaMV replication was further investigated. In brief, NbXRN4 benefits BaMV replication, probably by removal of the uncapped genomic and subgenomic RNAs produced erroneously during the replication/transcription process.
Perspective
Studies on replication-related proteins of plant RNA viruses have long been limited by inefficient protein expression and difficulty in protein purification. The catalytic characteristics of REPBaMV may thus not only apply to other members of the Potexvirus but also serve as references for those of other genera that also belong to the alphavirus-like superfamily. Nonetheless, the structural information at the atomic level regarding the functional domains of REPBaMV is still lacking, thanks mostly to the aggregation nature of these viral proteins. Methods that can overcome this obstacle are urgently needed. The search for host proteins, including those either boost or attenuate the enzymatic activity of REPBaMV, should be continued. More importantly, the mechanism underlying the function of host proteins should be elucidated so that the holistic and dynamic interplay between REPBaMV and its host can be understood.
Author Contributions
MM organized the contents of the text and wrote the manuscript. C-CL participated into the discussion about the text contents and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by grant MOST 103-2321-B-005-003 from the Ministry of Science and Technology, Taiwan, ROC.
References
Ahola, T., and Ahlquist, P. (1999). Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J. Virol. 73, 10061–10069.
Ahola, T., and Kääriäinen, L. (1995). Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. U.S.A. 92, 507–511. doi: 10.1073/pnas.92.2.507
Ahola, T., Laakkonen, P., Vihinen, H., and Kääriäinen, L. (1997). Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J. Virol. 71, 392–397.
Bornstein, P., and Balian, G. (1977). Cleavage at Asn-Gly bonds with hydroxylamine. Methods Enzymol. 47, 132–145. doi: 10.1016/0076-6879(77)47016-2
Changela, A., Ho, C. K., Martins, A., Shuman, S., and Mondragón, A. (2001). Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme. EMBO J. 20, 2575–2586. doi: 10.1093/emboj/20.10.2575
Chen, I.-H., Chou, W.-J., Lee, P.-Y., Hsu, Y.-H., and Tsai, C.-H. (2005). The AAUAAA motif of Bamboo mosaic virus RNA is involved in minus-strand RNA synthesis and plus-strand RNA polyadenylation. J. Virol. 79, 14555–14561. doi: 10.1128/JVI.79.23.14555-14561.2005
Chen, I.-H., Lin, J.-W., Chen, Y.-J., Wang, Z.-C., Liang, L.-F., Meng, M., et al. (2010). The 3’-terminal sequence of Bamboo mosaic virus minus-strand RNA interacts with RNA-dependent RNA polymerase and initiates plus-strand RNA synthesis. Mol. Plant Path. 11, 203–212. doi: 10.1111/j.1364-3703.2009.00597.x
Chen, I.-H., Meng, M., Hsu, Y.-H., and Tsai, C.-H. (2003). Functional analysis of the clover leaf-like structure in the 3’ untranslated region of bamboo mosaic potexvirus RNA revealed dual roles in viral RNA replication and long distance movement. Virology 315, 415–424. doi: 10.1016/S0042-6822(03)00560-9
Cheng, C. P., and Tsai, C. H. (1999). Structural and functional analysis of the 3’ untranslated region of bamboo mosaic potexvirus genomic RNA. J. Mol. Biol. 288, 555–565. doi: 10.1006/jmbi.1999.2716
Cheng, C.-W., Hsiao, Y.-Y., Wu, H.-C., Chuang, C.-M., Chen, J.-S., Tsai, C.-H., et al. (2009). Suppression of Bamboo mosaic virus accumulation by a putative methyltransferase in Nicotiana benthamiana. J. Virol. 83, 5796–5805. doi: 10.1128/JVI.02471-08
Cheng, S.-F., Huang, Y.-P., Chen, L.-H., Hsu, Y.-H., and Tsai, C.-H. (2013). Chloroplast phosphoglycerate kinase is involved in the targeting of Bamboo mosaic virus to chloroplasts in Nicotiana benthamiana plants. Plant Physiol. 163, 1598–1608. doi: 10.1104/pp.113.229666
Duclos, B., Marcandier, S., and Cozzone, A. J. (1991). Chemical properties and separation of phosphoamino acids by thin-layer chromatography and/or electrophoresis. Methods Enzymol. 201, 10–21. doi: 10.1016/0076-6879(91)01004-L
Fedorkin, O. N., Solovyev, A., Yelina, N., Zamyatnin, A. Jr., Zinovkin, R., Mäkinen, K., et al. (2001). Cell-to-cell movement of Potato virus X involves distinct functions of the coat protein. J. Gen. Virol. 82, 449–458. doi: 10.1099/0022-1317-82-2-449
Forster, R. L., Beck, D. L., Guilford, P. J., Voot, D. M., Van Dolleweerd, C. J., and Andersen, M. T. (1992). The coat protein of white clover mosaic potexvirus has a role in facilitating cell-to-cell transport in plants. Virology 191, 480–484. doi: 10.1016/0042-6822(92)90215-B
Furuichi, Y., and Shatkin, A. J. (2000). Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 55, 135–184. doi: 10.1016/S0065-3527(00)55003-9
Habili, N., and Symons, R. H. (1989). Evolutionary relationship between luteoviruses and other RNA plant viruses based on sequence motifs in their putative RNA polymerase and nucleic acid helicase. Nucleic Acids Res. 17, 9543–9555. doi: 10.1093/nar/17.23.9543
Håkansson, K., Doherty, A. J., Shuman, S., and Wigley, D. B. (1997). X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell 89, 545–553. doi: 10.1016/S0092-8674(00)80236-6
Han, Y.-T., Hsu, Y.-H., Lo, C.-W., and Meng, M. (2009). Identification and functional characterization of regions that can be crosslinked to RNA in the helicase-like domain of BaMV replicase. Virology 389, 34–44. doi: 10.1016/j.virol.2009.04.013
Han, Y.-T., Tsai, C.-S., Chen, Y.-C., Lin, M.-K., Hsu, Y.-H., and Meng, M. (2007). Mutational analysis of a helicase motif-based RNA 5’-triphosphatase/NTPase from Bamboo mosaic virus. Virology 367, 41–50. doi: 10.1016/j.virol.2007.05.013
Hu, R.-H., Lin, M.-C., Hsu, Y.-H., and Meng, M. (2011). Mutational effects of the consensus aromatic residues in the mRNA capping domain of Bamboo mosaic virus on GTP methylation and virus accumulation. Virology 411, 15–24. doi: 10.1016/j.virol.2010.12.022
Huang, C.-Y., Huang, Y.-L., Meng, M., Hsu, Y.-H., and Tsai, C.-H. (2001). Sequences at the 3’ untranslated region of the bamboo mosaic potexvirus RNA interact with the viral RNA-dependent RNA polymerase. J. Virol. 75, 2818–2824. doi: 10.1128/JVI.75.6.2818-2824.2001
Huang, Y.-L., Han, Y.-T., Chang, Y.-T., Hsu, Y.-H., and Meng, M. (2004). Critical residues for GTP methylation and formation of the covalent m7GMP-enzyme intermediate in the capping enzyme domain of Bamboo mosaic virus. J. Virol. 78, 1271–1280. doi: 10.1128/JVI.78.3.1271-1280.2004
Huang, Y.-L., Hsu, Y.-H., Han, Y.-T., and Meng, M. (2005). mRNA guanylation catalyzed by the S-adenosylmethionine-dependent guanylyltransferase of Bamboo mosaic virus. J. Biol. Chem. 280, 13153–13162. doi: 10.1074/jbc.M412619200
Huang, Y.-W., Hu, C.-C., Liou, M.-R., Chang, B.-Y., Tsai, C.-H., Meng, M., et al. (2012). Hsp90 interacts specifically with viral RNA and differentially regulates replication initiation of Bamboo mosaic virus and associated satellite RNA. PLoS Pathog. 8:e1002726. doi: 10.1371/journal.ppat.1002726
Jang, C., Seo, E.-Y., Nam, J., Bae, H., Gim, Y. G., Kim, H. G., et al. (2013). Insights into Alternanthera mosaic virus TGB3 functions: Interactions with Nicotiana benthamiana PsbO correlate with chloroplast vesiculation and veinal necrosis caused by TGB3 over-expression. Front. Plant Sci. 4:5. doi: 10.3389/fpls.2013.00005
Kawakami, S., Watanabe, Y., and Beachy, R. N. (2004). Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc. Natl. Acad. Sci. U.S.A. 101, 6291–6296. doi: 10.1073/pnas.0401221101
Koonin, E. V., and Dolja, V. V. (1993). Evolution and taxonomy of positive strand RNA viruses: implication of comparative analysis of amino acid sequence. Biochem. Mol. Biol. 28, 375–430. doi: 10.3109/10409239309078440
Lee, C.-C., Ho, Y.-N., Hu, R.-H., Yen, Y.-T., Wang, Z.-C., Lee, Y.-C., et al. (2011). The interaction between Bamboo mosaic virus replication protein and coat protein is critical for virus movement in plant hosts. J. Virol. 85, 12022–12031. doi: 10.1128/JVI.05595-11
Lee, C.-C., Lin, T.-L., Lin, J.-W., Han, Y.-T., Huang, Y.-T., Hsu, Y.-H., et al. (2016). Promotion of Bamboo mosaic virus accumulation in Nicotiana benthamiana by 5′→3′ exonuclease NbXRN4. Front. Microbiol. 6:1508. doi: 10.3389/fmicb.2015.01508
Li, Y.-I., Chen, Y.-J., Hsu, Y.-H., and Meng, M. (2001a). Characterization of the AdoMet-dependent guanylyltransferase activity that is associated with the N terminus of Bamboo mosaic virus replicase. J. Virol. 75, 782–788.
Li, Y.-I., Cheng, Y.-M., Huang, Y.-L., Tsai, C.-H., Hsu, Y.-H., and Meng, M. (1998). Identification and characterization of the E. coli-expressed RNA-dependent RNA polymerase of Bamboo mosaic virus. J. Virol. 72, 10093–10099.
Li, Y.-I., Shih, T.-W., Hsu, Y.-H., Han, Y.-T., Huang, Y.-L., and Meng, M. (2001b). The helicase-like domain of plant potexvirus replicase participates in the formation of 5’ cap structure of RNA by exhibiting an RNA 5’-triphosphatase activity. J. Virol. 75, 12114–12120.
Lim, H.-S., Vaira, A. M., Bae, H., Bragg, J. N., Ruzin, S. E., Bauchan, G. R., et al. (2010). Mutation of a chloroplast-targeting signal in Alternanthera mosaic virus TGB3 impairs cell-to-cell movement and eliminates long-distance virus movement. J. Gen. Virol. 91, 2102–2115. doi: 10.1099/vir.0.019448-0
Lima, C. D., Wang, L. K., and Shuman, S. (1999). Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell 99, 533–543. doi: 10.1016/S0092-8674(00)81541-X
Lin, H.-Y., Yu, C.-Y., Hsu, Y.-H., and Meng, M. (2012). Functional analysis of the conserved histidine residue of Bamboo mosaic virus capping enzyme in the activity for the formation of the covalent enzyme-m7GMP intermediate. FEBS Lett. 586, 2326–2331. doi: 10.1016/j.febslet.2012.05.024
Lin, J.-W., Chiu, H.-N., Chen, I.-H., Chen, T.-C., Hsu, Y.-H., and Tsai, C.-H. (2005). Structural and functional analysis of the cis-acting elements required for plus-strand RNA synthesis of Bamboo mosaic virus. J. Virol. 79, 9046–9053. doi: 10.1128/JVI.79.14.9046-9053.2005
Lin, J.-W., Ding, M.-P., Hsu, Y.-H., and Tsai, C.-H. (2007). Chloroplast phosphoglycerate kinase, a gluconeogenetic enzyme, is required for efficient accumulation of Bamboo mosaic virus. Nucleic Acids Res. 35, 424–432. doi: 10.1093/nar/gkl1061
Lin, M.-K., Chang, B.-Y., Liao, J.-T., Lin, N.-S., and Hsu, Y.-H. (2004). Arg-16 and Arg-21 in the N-terminal region of the triple-gene-block protein1 of Bamboo mosaic virus are essential for virus movement. J. Gen. Viol. 85, 251–259. doi: 10.1099/vir.0.19442-0
Lin, M.-K., Hu, C.-C., Lin, N.-S., Chang, B.-Y., and Hsu, Y.-H. (2006). Movement of potexviruses requires species specific interactions among the cognate triple gene block proteins, as revealed by a trans-complementation assay based on the Bamboo mosaic virus satellite RNA-mediated expression system. J. Gen. Virol. 87, 1357–1367. doi: 10.1099/vir.0.81625-0
Lin, N.-S., and Hsu, Y.-H. (1994). A satellite RNA associated with bamboo mosaic potexvirus. Virology 202, 707–714. doi: 10.1006/viro.1994.1392
Lin, N.-S., Lee, Y.-S., Lin, B.-Y., Lee, C.-W., and Hsu, Y.-H. (1996). The open reading frame of bamboo mosaic potexvirus satellite RNA is not essential for its replication and can be replaced with a bacterial gene. Proc. Natl. Acad. Sci. U.S.A. 93, 3138–3142. doi: 10.1073/pnas.93.7.3138
Lin, N.-S., Lin, B.-Y., Lo, N.-W., Hu, C.-C., Chow, T.-Y., and Hsu, Y.-H. (1994). Nucleotide sequence of the genomic RNA of bamboo mosaic potexvirus. J. Gen. Virol. 75, 2513–2518. doi: 10.1099/0022-1317-75-9-2513
Luongo, C. L. (2002). Mutational analysis of a mammalian reovirus mRNA capping enzyme. Biochem. Biophys. Res. Commun. 291, 932–938.
Luongo, C. L., Reinisch, K. M., Harrison, S. C., and Nibert, M. L. (2000). Identification of the guanylyltransferase region and active site in reovirus mRNA capping protein λ2. J. Biol. Chem. 275, 2804–2810. doi: 10.1074/jbc.275.4.2804
Magden, J., Takeda, N., Li, T., Auvinen, P., Ahola, T., Miyamura, T. A., et al. (2001). Virus-specific mRNA capping enzyme encoded by Hepatitis E virus. J. Virol. 75, 6249–6255. doi: 10.1128/JVI.75.14.6249-6255.2001
McCartney, A. W., Greenwood, J. S., Fabian, M. R., White, K. A., and Mullen, R. T. (2005). Localization of the Tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17, 3513–3531. doi: 10.1105/tpc.105.036350
Merits, A., Kettunen, R., Makinen, K., Lampio, A., Auvinen, P., Kääriäinen, L., et al. (1999). Virus-specific capping of Tobacco mosaic virus RNA: methylation of GTP prior to formation of covalent complex p126-m7GMP. FEBS Lett. 455, 45–48. doi: 10.1016/S0014-5793(99)00856-X
Miller, D. J., Schwartz, M. D., and Ahlquist, P. (2001). Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 75, 11664–11676. doi: 10.1128/JVI.75.23.11664-11676.2001
Miller, S., and Krijnse-Locker, J. (2008). Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6, 363–374. doi: 10.1038/nrmicro1890
Mizumoto, K., and Kaziro, Y. (1987). Messenger RNA capping enzymes from eukaryotic cells. Prog. Nucleic Acid Res. Mol. Biol. 34, 1–28. doi: 10.1016/S0079-6603(08)60491-2
Niles, E. G., and Christen, L. (1993). Identification of the vaccinia virus mRNA guanyltransferase active site lysine. J. Biol. Chem. 268, 24986–24989.
Noueiry, A. O., and Ahlquist, P. (2003). Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 41, 77–98. doi: 10.1146/annurev.phyto.41.052002.095717
Novoa, R. R., Calderita, G., Arranz, R., Fontana, J., Granzow, H., and Risco, C. (2005). Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97, 147–172. doi: 10.1042/BC20040058
Ogino, T., and Banerjee, A. K. (2007). Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell 25, 85–97. doi: 10.1016/j.molcel.2006.11.013
Ogino, T., Yadav, S. P., and Banerjee, A. K. (2010). Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 3463–3468. doi: 10.1073/pnas.0913083107
Park, M.-R., Jeong, R.-D., and Kim, K.-H. (2014). Understanding the intracellular trafficking and intercellular transport of potexviruses in their host plants. Front. Plant Sci. 5:60. doi: 10.3389/fpls.2014.00060
Prasanth, K. R., Huang, Y.-W., Liou, M.-R., Wang, Y.-L., Hu, C.-C., Tsai, C.-H., et al. (2011). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) negatively regulates the replication of Bamboo mosaic virus and its associated satellite RNA. J. Virol. 85, 8829–8840. doi: 10.1128/JVI.00556-11
Qiao, Y., Li, H. F., Wong, S. M., and Fan, Z. F. (2009). Plastocyanin transit peptide interacts with Potato virus X coat protein, while silencing of plastocyanin reduces coat protein accumulation in chloroplasts and symptom severity in host plants. Mol. Plant Microbe. Interact. 22, 1523–1534. doi: 10.1094/MPMI-22-12-1523
Rost, B., Sander, C., and Schneider, R. (1994). PHD–an automatic mail server for protein secondary structure prediction. Comput. Appl. Biosci. 10, 53–60. doi: 10.1093/bioinformatics/10.1.53
Rozanov, M. N., Koonin, E. V., and Gorbalenya, A. E. (1992). Conservation of the putative methyltransferase domain: a hallmark of the “Sindbis-like” supergroup of positive-strand RNA viruses. J. Gen. Virol. 73, 2129–2134. doi: 10.1099/0022-1317-73-8-2129
Shuman, S. (1995). Capping enzyme in eukaryotic mRNA synthesis. Prog. Nucleic Acid Res. Mol. Biol. 50, 101–129. doi: 10.1016/S0079-6603(08)60812-0
Shuman, S. (2001). Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 66, 1–40. doi: 10.1016/S0079-6603(00)66025-7
Shuman, S., Surks, M., Furneaux, H., and Hurwitz, J. (1980). Purification and characterization of a GTP-pyrophosphate exchange activity from vaccinia virions. Association of the GTP-pyrophosphate exchange activity with vaccinia mRNA guanylyltransferase. RNA (guanine-7-)methyltransferase complex (capping enzyme). J. Biol. Chem. 255, 11588–11598.
Turner, K. A., Sit, T. L., Callaway, A. S., Allen, N. S., and Lommel, S. A. (2004). Red clover necrotic mosaic virus replication proteins accumulate at the endoplasmic reticulum. Virology 320, 276–279. doi: 10.1016/j.virol.2003.12.006
Verchot-Lubicz, J., Torrance, L., Solovyev, A. G., Morozov, S. Y., Jackson, A. O., and Gilmer, D. (2010). Varied movement strategies employed by triple gene block-encoding viruses. Mol. Plant Microbe. Interact. 23, 1231–1247. doi: 10.1094/MPMI-04-10-0086
Keywords: Bamboo mosaic virus, Potexvirus, RNA-dependent RNA polymerase, mRNA capping, virus-host interaction, positive-strand RNA virus, guanylyltransferase
Citation: Meng M and Lee C-C (2017) Function and Structural Organization of the Replication Protein of Bamboo mosaic virus. Front. Microbiol. 8:522. doi: 10.3389/fmicb.2017.00522
Received: 19 December 2016; Accepted: 13 March 2017;
Published: 28 March 2017.
Edited by:
Na-Sheng Lin, Institute of Plant and Microbial Biology (Academia Sinica), JapanReviewed by:
John Hammond, Agricultural Research Service (USDA), USASatyanarayana Tatineni, Agricultural Research Service (USDA), USA
Carmen Simón, Consejo Superior de Investigaciones Científicas (CSIC), Spain
Copyright © 2017 Meng and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Menghsiao Meng, bWhtZW5nQGRyYWdvbi5uY2h1LmVkdS50dw==
 Menghsiao Meng
Menghsiao Meng Cheng-Cheng Lee
Cheng-Cheng Lee