- 1Department of Biotechnology and Food Microbiology, Faculty of Food Science, Wrocław University of Environmental and Life Sciences, Wrocław, Poland
- 2Department of Fermentation and Cereals Technology, Faculty of Food Science, Wrocław University of Environmental and Life Sciences, Wrocław, Poland
- 3Department of Immunology, Pathophysiology and Veterinary Preventive Medicine, Faculty of Veterinary Medicine, Wroclaw University of Environmental and Life Sciences, Wrocław, Poland
- 4Department of Veterinary Medicine, Institute for Animal Hygiene and Environmental Health, Freie Universitaet Berlin, Berlin, Germany
Extended-spectrum β-lactamases (ESBLs) and AmpC β-lactamases are plasmid (but also chromosomally) encoded enzymes found in Enterobacteriaceae, determining resistance to a variety of important antibiotics including penicillins, cephalosporins, and monobactams. In recent decades, the prevalence of ESBL/AmpC-producing bacteria has increased rapidly across the world. Here, we evaluate the potential use of bacteriophages in terms of a reduction of antibiotic-resistant bacteria in healthy animals. The aim of our studies was to isolate bacteriophages capable of destroying ESBL/AmpC-producing Escherichia coli isolated from livestock habitats. The efficacy of isolated phages against ESBL/AmpC E. coli strains varies, but creation of a phage cocktail with broad activity spectrum is possible. This may indicate that the role of phages may not be limited to phage therapy, but bacterial viruses may also be applied against spread of bacteria with antibiotic resistance genes in the environment. We also addressed the hypothesis, that phages, effective for therapeutic purposes may be isolated from distant places and even from different environments other than the actual location of the targeted bacteria. This may be beneficial for practical purposes, as the construction of effective phage preparations does not require access to disease outbreaks.
Introduction
Extended-spectrum β-lactamases (ESBLs) and AmpC β-lactamases are plasmid and/or chromosomally encoded enzymes found in Enterobacteriaceae, conferring resistance to different β-lactam antibiotics, including penicillins, cephalosporins, and monobactams (Rawat and Nair, 2010). ESBL/AmpC-producing organisms are often co-, or multiresistant, exhibiting resistance to other anti-microbial classes including fluoroquinolones, aminoglycosides, and trimethoprim-sulfamethoxazole (EFSA Panel on Biological Hazards [BIOHAZ], 2011; Xu et al., 2015). To date the most common ESBL types are TEM, SHV, and CTX-M, which have been divided into five groups known as CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25 (Bonnet, 2004). The most common plasmid-mediated AmpC type β-lactamase worldwide is CMY-2 (Jacoby, 2009). In veterinary medicine, ESBL-producing bacteria have been proved to cause mastitis in dairy cattle (Timofte et al., 2014), urinary tract infections (UTI) in companion animals (Keefe et al., 2010), and soft skin infections in companion animals and horses (Ewers et al., 2014). In humans, they may also cause UTI, pneumonia, or even sepsis (Peña et al., 2006; Falagas and Karageorgopoulos, 2009).
According to the European Food Safety Authority (EFSA), rapid emergence of resistance caused by ESBL in Enterobacteriaceae is a major public health concern in Europe (EFSA Panel on Biological Hazards [BIOHAZ], 2011). A report from the Infectious Diseases Society of America from 2006 listed ESBL-producing Klebsiella spp. and Escherichia coli among the six drug-resistant microbes for which new therapies are urgently needed (Pitout and Laupland, 2008).
In light of the growing threat associated with the spread of antibiotic resistance, the World Health Organization (WHO) and the European Commission (EC) have recognized the importance of studying anti-microbial resistance and highlighted the need to devise appropriate strategies for its control (Picozzi et al., 2014). Also the World Medical Association (WMA) appealed for increased research funding to develop new anti-microbial agents (Kelly et al., 2011).
Application of bacteriophages to combat bacterial diseases (phage therapy), despite its initial fast development, was significantly marginalized after the discovery of penicillin and other antibiotics (Sulakvelidze et al., 2001; Deresinski, 2009). Nowadays, a rapidly growing number of drug-resistant bacterial strains have already evolved as the most serious threats in public health. Therefore, phage therapy is recognized as one of the most attractive alternatives to classical antibiotic-based treatment and is becoming an important research objective.
Although phage therapy for the treatment of infections caused by antibiotic-resistant bacterial strains has been studied extensively (Biswas et al., 2002; Jensen et al., 2015), until now only a few studies have described the use of phages in terms of a reduction of antibiotic-resistant bacteria in healthy animals (Teng-Hern et al., 2014). In this study it was demonstrated that phages may be potentially applied to reduce the prevalence of ESBL/AmpC E. coli in healthy food-producing animals and thus may play an important role in the ecology of resistance. We also addressed the hypothesis that phages for this purpose can easily be isolated from distant places and even different environments than the targeted bacteria.
Materials and Methods
Environmental Samples
A total of nineteen samples were collected from six pig farms in Poland. Depending on the particular maintenance system (litter, deep litter, slatted floor), the samples were collected from bedding material (litter system) (n = 7), feces (slatted floor) – pooled or from individuals (n = 5), and feed taken from the feeders (n = 5). Additionally, two manure samples were collected from a manure lagoon.
Isolation of Bacteria from Environmental Samples
From all collected environmental samples, bacteria were isolated and subsequently used for phage isolation and host range testing. Of each environmental sample (feces/litter/feed), 5 g were introduced into 30 ml of LB broth (Sigma–Aldrich, Germany) and incubated at 37°C for 20 h. On the following day, the overnight cultures were spread on Columbia Agar with sheep blood (BioMerieux, France) to obtain single colonies. All bacteria of different phenotypes were identified using the BBL Crystal Autoreader System (BD Biosciences, USA). In addition, the generic affiliation of bacteria to which specific phages were isolated was confirmed with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis (MALDI Microflex LT and Biotyper database, Bruker Daltonics, Bremen, Germany). Subsequently, bacteriophages were isolated from the same overnight cultures by filtration through 0.22 μm syringe filters (Merck Millipore, Germany).
Isolation of Bacteriophages
The phage isolation was carried out using the modified method of Slopek et al. (1972). Each of the isolated bacterial strains was used to test for bacteriophage in each filtrate. Briefly, 5 ml of a 3-h incubated bacterial culture (LB broth) was poured out on a LB agar plate (excess culture was removed and the plate was left to dry) and 50 μl of each filtrate was introduced into each of eight sectors previously marked on the bottom of the plate. After drying of the drop, the plates were incubated at 37°C and plaque formation was monitored after 3, 4, 5, and 24 h.
Amplification of Bacteriophages
Isolated bacteriophages were amplified in the respective host bacteria. Therefore, 20 μl of phage filtrate and 20 μl of the bacterial overnight culture were added to 10 ml of LB broth and incubated at 37°C. The optical density (OD600) of the culture was measured hourly. In case of an OD fall (phages eliminated most of the bacteria), additional host bacteria were added. If the OD increased (there were more bacteria in the culture than phages), additional phage filtrate was added. After a total incubation time of 6 h, all remaining bacteria were removed by filtration through a 0.22 μm syringe filter and the phage titer was determined using the double agar method (Adams, 1959). The experiments were performed in triplicate in order to eliminate bactericidal activity of some chemical compounds which might potentially be present in the sample.
ESBL/AmpC-Producing E. coli
Extended-spectrum β-lactamases/AmpC-producing E. coli strains derived from the strain collection of the Institute for Animal Hygiene and Environmental Health, Freie Universitaet Berlin. The isolates were obtained from 7 different pig farms (n = 104) and 22 turkey farms (n = 51) in Germany. Each isolate originated from an individual sample including feces, boot swabs, manure, air or dust. ESBL/AmpC E. coli were selected using MacConkey agar (Oxoid, CM 0115, Wesel, Germany) supplemented with 1 mg/L cefotaxime, followed by species confirmation using MALDI-TOF identification (MALDI Microflex LT and Biotyper database, Bruker Daltonics, Bremen, Germany). The presence of the β-lactamase genes blaCTX, blaTEM, blaSHV and the CIT-type AmpCs (e.g., CMY-2) was confirmed by real-time PCR as described by Roschanski et al. (2014).
Bacteriolytic Activity of Isolated Bacteriophages against ESBL/AmpC E. coli Isolated from Selective Pig and Turkey Farms from Germany
One hundred four ESBL/AmpC E. coli strains originally isolated from pig farms from Germany and stored at the culture collections of the Institute for Animal Hygiene and Environmental Health, Freie Universitaet Berlin (Berlin, Germany) were chosen and tested for sensitivity to the 17 previously isolated bacteriophages. Additionally, 51 ESBL/AmpC E. coli isolated from turkey farms (from the same collection) were typed with the same phage preparations. The procedure was performed as described in the section Isolation of bacteriophages.
Activity of Phage Cocktail against ESBL/AmpC E. coli Isolated from Pig Farms
Bacteriophages selected for the cocktail were mixed in the ratio 1:1:1 (V/V/V). The final titer of each phage was 1 × 109 pfu/ml. The activity tests of the cocktail were performed as described for the tests of the monophage preparations.
Results
Occurrence of E. coli and E. coli-Specific Bacteriophages in Pig Environment Sampled in Polish Pig Farms
A total of 75 bacterial strains were isolated from the 19 collected samples including: Proteus spp. (22), Escherichia ssp. (out of 23 strains 20 were E. coli), Aeromonas ssp. (5), Staphylococcus ssp. (4), Providencia ssp. (3), Klebsiella ssp. (3), Morganella ssp. (2), Citrobacter ssp. (2), Vibrio ssp. (2), Shigella ssp. (2), Serratia ssp. (2), Kytococcus ssp. (1), Corynebacterium ssp. (1), Acinetobacter ssp. (1), Aerococcus ssp. (1), Bacillus ssp. (1). A total of 19 filtrates, one per sample, with potential bacteriophages were prepared and the sensitivity of all isolated bacterial strains tested against each bacteriophage. Twenty-two different bacteriophages, all specific to E. coli, were isolated. Three rounds of successive plaque purifications were performed to isolate each phage. Seventeen phages were successfully amplified for further studies.
Activity of Isolated Bacteriophages against ESBL/AmpC E. coli Isolated from Pig Farms
The activity of 17 bacteriophages isolated from Polish pig farms was tested against 104 ESBL/AmpC E. coli strains isolated from German pig farms. The results showed that all phages were active against the tested E. coli strains and they lysed from 1.9% up to 57.7% of all strains (Figure 1). 82.7% of all bacterial isolates were sensitive to at least one of the phages, which means that 17.3% were not sensitive to any of the phage preparations.
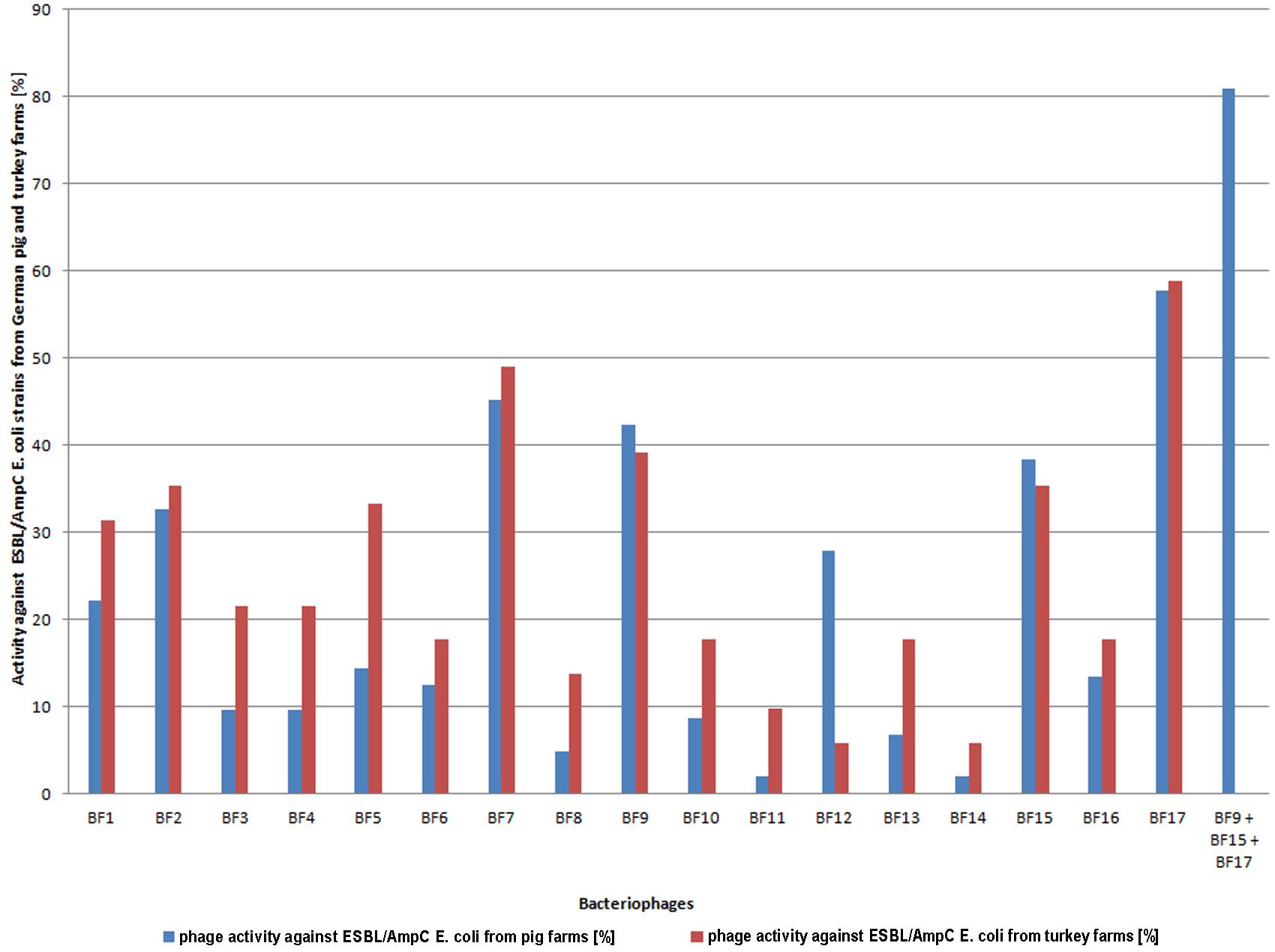
FIGURE 1. Activity of bacteriophages isolated from Polish pig farms against ESBL/AmpC Escherichia coli isolated from German pig and turkey farms. Blue and red columns mean activity (%) of bacteriophages isolated from Polish pig farms against ESBL/AmpC E. coli isolated from German pig and turkey farms, respectively, and the last column means activity (%) of a cocktail (BF9 + BF15 + BF17) against ESBL/AmpC E. coli isolated from German pig farms.
Activity of Phage Cocktail against ESBL E. coli Isolated from Pig Farms
Based on the above results, a cocktail of three phages was prepared and tested against ESBL E. coli. The cocktail included phages BF17 and BF15, which showed the highest bactericidal activity. Phages BF7 and BF9 showed a similar activity against the tested E. coli strains (45.2 and 42.3%, respectively); however, to cover a broader spectrum of bacteria, for the cocktail we chose phage BF9 even though it showed slightly lower activity than phage BF7. The phages from hypothetical cocktail were characterized in terms of their morphology, genetics and phage-bacteria kinetics (data not shown). The selected cocktail lysed 80.8% of all tested strains.
To confirm the activity of the phage cocktail, nine ESBL E. coli strains with different sensitivity to the individual phages were chosen. The obtained results (Table 1) confirmed that the tested phage cocktail might potentially be useful against ESBL E. coli.
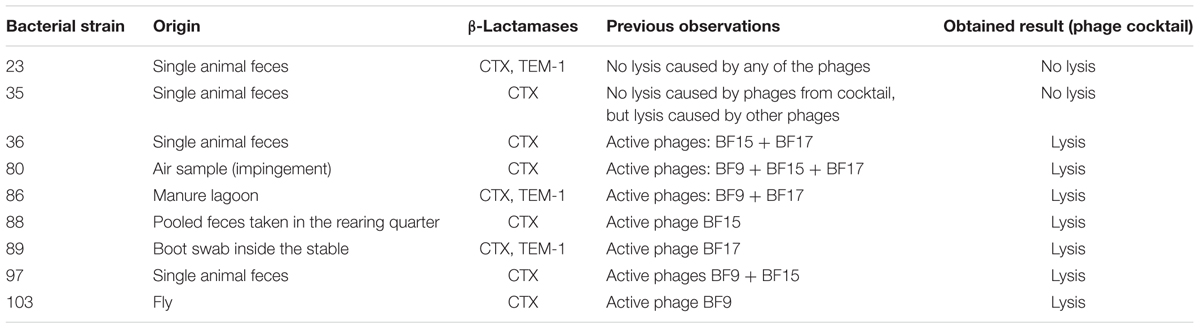
TABLE 1. Activity of bacteriophage cocktail (BF9 + BF15 + BF17) against selected Extended-spectrum β-lactamase (ESBL) Escherichia coli strains.
Activity of Isolated Bacteriophages against ESBL/AmpC E. coli Isolated from Turkey Farms
To investigate whether the 17 phages may also be active against bacterial strains isolated from different animal farm environments, additional host range determination using 51 randomly selected ESBL/AmpC E. coli isolated from German turkey farms was performed. Interestingly, almost all phages (14/17) were more active against ESBL/AmpC E. coli isolated from turkey farms than ESBL/AmpC E. coli isolated from pig farms. Eleven out of the 51 strains were not sensitive to any of the phages, which means that the complete pool of phages lysed 78.4% of all tested strains (Figure 1).
Discussion
The potential therapeutic applications of phages as one of the main alternatives to classical treatment have been widely explored. Bacteriophages have already been successfully applied in treatment of antibiotic-resistant bacterial strains, including methicillin-resistant Staphylococcus aureus (Jensen et al., 2015) and vancomycin-resistant Enterococcus sp. (Biswas et al., 2002). In addition, the effectiveness of phage ø9882, isolated from hospital sewage, against a broad range of ESBL-producing E. coli clinical isolates has been shown in mice. It is worth emphasizing that the studies of Wang et al. (2006) provided a framework to evaluate the therapeutic efficacy of phages against fatal ESBL E. coli infections in humans (Wang et al., 2006). However, our research shows that the role of phages may not be limited to phage therapy, but bacterial viruses may also be applied against spread of bacteria with antibiotic resistance genes in the environment.
In the first part of the present study, a total of 17 bacteriophages specific to E. coli were isolated from Polish pig farms. Phages were isolated only with bacteria simultaneously isolated from the same environment. In the second stage of the study, the bactericidal activity of phages isolated from Polish farms was evaluated against unrelated bacterial isolates (ESBL/AmpC E. coli isolated from German pig farms). All phages were found to be active against some of the tested strains, but they lysed these strains with different efficacy.
Possible activity of the isolated phages against ESBL/AmpC E. coli from animal species other than pigs was also investigated. However, it is worth noting that the tested strains isolated from pig and turkey farms were not compared genetically, and thus it is theoretically possible that the same bacteria were isolated from both environments. Interestingly, almost all phages showed higher or similar activity against the strains originating from turkey compared to the pig isolates.
In the past, the focus of most phage therapy studies lay in finding phages against specific pathogens. This was performed by on-demand isolation methods in which targeted bacterial strains were added to the culture to enrich potential bacteriophages. This seems to be highly effective, since it was successfully done with phages against Pseudomonas aeruginosa and Salmonella but also against ESBL E. coli and Klebsiella (Mattila et al., 2015). In this study, we addressed the hypothesis that phages, isolated from distant places and even deriving from different farm environments than the targeted bacteria, may also be suitable for an efficient polyphage therapy. This is of great importance, as it proves that the construction of effective phage preparations does not require access to disease outbreaks.
Interestingly, the relationship between the presence of phages and sensitive bacteria has already been observed at the stage of phage isolation. In an initial spot tests we selected 22 potential bacteriophages for purification (17 phages were successfully amplified for further studies). As we analyzed the origin of these 22 phages and their hosts, 18 phages were active against bacteria from different farms. Furthermore, to compare the effectiveness of the on-demand method with isolation of phages from distant locations and environments, we additionally prepared phage filtrate obtained using this technique (Supplementary Figure 1). To create this preparation, we used the same 51 E. coli strains from turkey farms, which were used for activity tests of phages from pig farms. Phage filtrate obtained using the on-demand method lysed 80.4% of all tested strains. To compare, three of 17 phages from Polish pig farms (BF5 + BF15 + BF17) lysed this pool of bacteria with only slightly lower efficacy of 78%. It is noteworthy that we did not separate individual phages from the preparation obtained using the on-demand method, but while testing the activity of phages against bacteria we used the whole filtrate, which may hypothetically contain several different phages. We may assume that after isolation of individual phages it would not be possible to create a 3-phage cocktail with similar efficiency as the one from pig farms.
The results of this study confirmed previous observations of Jensen et al. (2015), who demonstrated that phages active against human methicillin-resistant and methicillin-susceptible S. aureus may be easily isolated from chicken sources, which are not necessarily predicted to be a good source of phages against these bacteria. Notably, in contrast to our studies, these phages were isolated using the on-demand method. Gutiérrez et al. (2016) found no clear association of infectivity of phages against S. aureus strains based on the geographic region of host or phage isolation. However, the authors postulated a possible association between phage infectivity and the environment. This was also mentioned by Vos et al. (2009), who suggested that phages may adapt to become more effective against bacteria living closely.
Although person-to-person spread is recognized as the main method of dissemination of ESBL/AmpC bacteria in the community, the prevalence of ESBLs nowadays has increased significantly due to E. coli isolates from food-producing animals (Carattoli, 2008; Geser et al., 2012; Samanta et al., 2015). Furthermore, the ESBL/AmpC-producing E. coli in animal farms could influence public health through environmental pollution (Li et al., 2015). Fecal emission and the airborne route might be possible ways of ESBL/AmpC emission into the environment (Yuan et al., 2010; Friese et al., 2013; Laube et al., 2014). Moreover, vectors such as flies (Usui et al., 2013) or small rodents (Kozak et al., 2009) may also spread anti-microbial resistance. The landform may also boost the dissemination of bacteria, as they may be spread with the rainfall from farms situated in the hills. In addition, if antibiotics are used in the farms, residues can enter the waterways and be transferred to distant locations, creating an antibiotic resistance problem in new locations. As bacteriophages may be transmitted in the environment via similar pathways, the spread of phages and host bacteria may have a wide range. This may be one explanation why active phages may be isolated from distant locations and different environments than targeted bacteria.
Another possibility is that phages become lysogenic during transmission in the environment and they revert back to lytic ones at distant places. They may originate from farms other than the one from which they were isolated.
Higher activity of phages from distant farms and environments than in the case of co-isolates may be due to the occurrence in different environments and the lack of direct contact that would result in the development of resistance to phages, as observed in phage-treated campylobacteriosis (Loc Carrillo et al., 2005). The phenomenon of bacterial phage resistance is, however, complex and probably depends on the phage-bacteria configuration and time points of interactions occurring between bacterial viruses and their hosts (Fischer et al., 2013). The data regarding phage resistance are scarce and contradictory; thus detailed studies to explain this phenomenon need to be conducted.
Although the results of this study indicate the relation between the geographic distance and type of environment from which active phages are isolated, they are not sufficient for the conclusion that a particular source is better for the isolation of effective phages. Studies including the use of samples from several different environments and locations would be more reliable, especially considering the contradictory data in the literature.
The present study indicated that bacterial viruses have a potential for prevention or treatment of E. coli with ESBL genes. Moreover, they may be good candidates in control strategies against the widespread presence of ESBL/AmpC E. coli in healthy food-producing animals and hence the reduction of these bacteria in the entire food chain. Phages for this purpose may be easily isolated from the environment, but the question whether distant locations and different environments are better sources of active phages remains open. Although phages are considered one of the least known entities on our planet (Grose et al., 2014) and further studies regarding their biology need to be conducted, they undoubtedly could provide a novel approach to the prevention and treatment of infections caused by drug-resistant bacteria.
Author Contributions
AS devised the experiments, acquired and processed laboratory data and wrote the manuscript, PS, MK-B, and GS performed the experiments related to isolation of bacteriophages, AR collected the environmental samples for isolation of bacteriophages, AF and NR designed the studies related to phage activity against ESBL/AmpC E. coli, JM confirmed the generic affiliation of bacteria with MALDI TOF mass spectrometry analysis, UR designed the studies and examined the manuscript. All authors participated in writing the manuscript and approved its final version.
Funding
Project supported by Wroclaw Centre of Biotechnology, program The Leading National Research Centre (KNOW) for years 2014–2018.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We thank Mr. Richard Ashcroft, for his help in preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00530/full#supplementary-material
References
Adams, M. H. (ed.). (1959). “Enumeration of bacteriophage particles,” in The Bacteriophages, (New York, NY: Interscience Publishers), 27–30.
Biswas, B., Adhya, S., Washart, P., Paul, B., Trostel, A. N., Powell, B., et al. (2002). Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect. Immun. 70, 204–210. doi: 10.1128/IAI.70.1.204-210.2002
Bonnet, R. (2004). Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48, 1–14. doi: 10.1128/AAC.48.1.1-14.2004
Carattoli, A. (2008). Animal reservoirs for extended spectrum β-lactamase producers. Clin. Microbiol. Infect. 14, 117–123. doi: 10.1111/j.1469-0691.2007.01851.x
Deresinski, S. (2009). Bacteriophage therapy: exploiting smaller fleas. Clin. Infect. Dis. 48, 1096–1101. doi: 10.1086/597405
EFSA Panel on Biological Hazards [BIOHAZ] (2011). Scientific opinion on the public health risks of bacterial strains producing extended-spectrum β-lactamases and/or AmpC β-lactamases in food and food-producing animals. EFSA J. 9, 2322–2417. doi: 10.2903/j.efsa.2011.2322
Ewers, C., Stamm, I., Pfeifer, Y., Wieler, L. H., Kopp, P. A., Schønning, K., et al. (2014). Clonal spread of highly successful ST15-CTX-M-15 Klebsiella pneumoniae in companion animals and horses. J. Antimicrob. Chemother. 69, 2676–2680. doi: 10.1093/jac/dku217
Falagas, M. E., and Karageorgopoulos, D. E. (2009). Extended-spectrum beta-lactamase-producing organisms. J. Hosp. Infect. 73, 345–354. doi: 10.1016/j.jhin.2009.02.021
Fischer, S., Kittler, S., Klein, G., and Glünder, G. (2013). Impact of a single phage and a phage cocktail application in broilers on reduction of Campylobacter jejuni and development of resistance. PLoS ONE 8:e78543. doi: 10.1371/journal.pone.0078543
Friese, A., Schulz, J., Laube, H., von Salviati, C., Hartung, J., and Roesler, U. (2013). Faecal occurrence and emissions of livestock-associated methicillin-resistant Staphylococcus aureus (laMRSA) and ESbl/AmpC-producing E. coli from animal farms in Germany. Berl. Munch. Tierarztl. Wochenschr. 126, 175–180.
Geser, N., Stephan, R., and Hächler, H. (2012). Occurrence and characteristics of extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet. Res. 8:21. doi: 10.1186/1746-6148-8-21
Grose, J. H., Jensen, G. L., Brunett, S. H., and Breakwell, D. P. (2014). Genomic comparison of 93 Bacillus phages reveals 12 clusters, 14 singletons and remarkable diversity. BMC Genomics 15:855. doi: 10.1186/1471-2164-15-855
Gutiérrez, D., Rodríguez-Rubio, L., García, P., Billington, C., Premarante, A., Rodríguez, A., et al. (2016). Phage sensitivity and prophage carriage in Staphylococcus aureus isolated from foods in Spain and New Zeland. Int. J. Food Microbiol. 230, 16–20. doi: 10.1016/j.ijfoodmicro.2016.04.019
Jacoby, G. A. (2009). AmpC beta-lactamases. Clin. Microbiol. Rev. 22, 161–182. doi: 10.1128/CMR.00036-08
Jensen, K. C., Hair, B. B., Wienclaw, T. M., Murdock, M. H., Hatch, J. B., Trent, A. T., et al. (2015). Isolation and host range of bacteriophage with lytic activity against methicillin-resistant Staphylococcus aureus and potential use as a fomite decontaminant. PLoS ONE 10:e0131714. doi: 10.1371/journal.pone.0131714
Keefe, A. O., Hutton, T. A., Schifferli, D. M., and Rankin, S. C. (2010). First detection of CTX-M and SHV extended-spectrum β-lactamases in Escherichia coli urinary tract isolates from dogs and cats in the United States. Antimicrob. Agents Chemother. 54, 3489–3492. doi: 10.1128/AAC.01701-09
Kelly, D., McAuliffe, O., Ross, R. P., O’Mahony, J., and Coffey, A. (2011). Development of a broad-host-range phage cocktail for biocontrol. Bioeng. Bugs 2, 31–37. doi: 10.4161/bbug.2.1.13657
Kozak, G. K., Boerlin, P., Janecko, N., Reid-Smith, R. J., and Jardine, C. (2009). Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 75, 559–566. doi: 10.1128/AEM.01821-08
Laube, H., Friese, A., von Salviati, C., Guerra, B., and Rösler, U. (2014). Transmission of ESBL/AmpC-producing Escherichia coli from broiler chicken farms to surrounding areas. Vet. Microbiol. 172, 519–527. doi: 10.1016/j.vetmic.2014.06.008
Li, S., Song, W., Zhou, Y., Tang, Y., Gao, Y., and Miao, Z. (2015). Spread of extended-spectrum beta-lactamase-producing Escherichia coli from a swine farm to the receiving river. Environ. Sci. Pollut. Res. Int. 22, 13033–13037. doi: 10.1007/s11356-015-4575-7
Loc Carrillo, C., Atterbury, R. J., el-Shibiny, A., Connerton, P. L., Dillon, E., Scott, A., et al. (2005). Bacteriophage therapy to reduce Campylobacter jejuni colonization of broiler chickens. Appl. Environ. Microbiol. 71, 6554–6653. doi: 10.1128/AEM.71.11.6554-6563.2005
Mattila, S., Ruotsalainen, P., and Jalasvuori, M. (2015). On-demand isolation of bacteriophages against drug-resistant bacteria for personalized phage therapy. Front. Microbiol. 6:1271. doi: 10.3389/fmicb.2015.01271
Peña, C., Gudiol, C., Tubau, F., Saballs, M., Pujol, M., Dominguez, M. A., et al. (2006). Risk-factors for acquisition of extended-spectrum beta-lactamase-producing Escherichia coli among hospitalised patients. Clin. Microbiol. Infect. 12, 279–284. doi: 10.1111/j.1469-0691.2005.01358.x
Picozzi, S. C. M., Casellato, S., Rossini, M., Paola, G., Tejada, M., Costa, E., et al. (2014). Extended-spectrum beta-lactamase-positive Escherichia coli causing complicated upper urinary tract infection: urologist should act in time. Urol. Ann. 6, 107–112. doi: 10.4103/0974-7796.130536
Pitout, J. D. D., and Laupland, K. B. (2008). Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8, 159–166. doi: 10.1016/S1473-3099(08)70041-0
Rawat, D., and Nair, D. (2010). Extended-spectrum β-lactamases in Gram Negative Bacteria. J. Glob. Infect. Dis. 2, 263–274. doi: 10.4103/0974-777X.68531
Roschanski, N., Fischer, J., Guerra, B., and Roesler, U. (2014). Development of a multiplex real-time PCR for the rapid detection of the predominant beta-lactamase genes CTX-M, SHV, TEM and CIT-type AmpCs in Enterobacteriaceae. PLoS ONE 9:e100956. doi: 10.1371/journal.pone.0100956
Samanta, I., Joardar, S. N., Mahanti, A., Bandyopadhyay, S., Sar, T. K., and Dutta, T. K. (2015). Approaches to characterize extended spectrum beta-lactamase/beta-lactamase producing Escherichia coli in healthy organized vis-a-vis backyard farmed pigs in India. Infect. Genet. Evol. 36, 224–230. doi: 10.1016/j.meegid.2015.09.021
Slopek, S., Durlakowa, I., Kucharewicz-Krukowska, A., Krzywy, T., Slopek, A., and Weber, B. (1972). Phage typing of Shigella flexneri. Arch. Immunol. Ther. Exp. 20, 1–60.
Sulakvelidze, A., Alavidze, Z., and Morris, J. G. (2001). Bacteriophage therapy. Antimicrob. Agents Chemother. 45, 649–659. doi: 10.1128/AAC.45.3.649-659.2001
Teng-Hern, T. L., Kok-Gan, C., and Han, L. L. (2014). Application of bacteriophage in biocontrol of major foodborne bacterial pthogens. J. Mol. Biol. Mol. Imaging 1:9.
Timofte, D., Maciuca, I. E., Evans, N. J., Williams, H., Wattret, A., Fick, J. C., et al. (2014). Detection and molecular characterization of Escherichia coli CTX-M-15 and Klebsiella pneumoniae SHV-12 β-lactamases from bovine mastitis isolates in the United Kingdom. Antimicrob. Agents Chemother. 58, 789–794. doi: 10.1128/AAC.00752-13
Usui, M., Iwasa, T., Fukuda, A., Sato, T., Okubo, T., and Tamura, Y. (2013). The role of flies in spreading the extended-spectrum β-lactamase gene from cattle. Microb. Drug Resist. 19, 415–420. doi: 10.1089/mdr.2012.0251
Vos, M., Birkett, P. J., Birch, E., Griffiths, R. I., and Buckling, A. (2009). Local adaptation of bacteriophages to their bacterial hosts in soil. Science 325, 833. doi: 10.1126/science.1174173
Wang, J., Hu, B., Xu, M., Yan, Q., Liu, S., Zhu, X., et al. (2006). Therapeutic effectiveness of bacteriophages in the rescue of mice with extended spectrum β-lactamase-producing Escherichia coli bacteremia. Int. J. Mol. Med. 17, 347–355.
Xu, G., An, W., Wang, H., and Zhang, X. (2015). Prevalence and characteristics of extended-spectrum β-lactamase genes in Escherichia coli isolated from piglets with post-weaning diarrhea in Heilongjiang province, China. Front. Microbiol. 6:1103. doi: 10.3389/fmicb.2015.01103
Keywords: bacteriophages, antibiotic resistance, antibiotic resistance genes, ESBL/AmpC resistant E. coli, phage therapy
Citation: Skaradzińska A, Śliwka P, Kuźmińska-Bajor M, Skaradziński G, Rząsa A, Friese A, Roschanski N, Murugaiyan J and Roesler UH (2017) The Efficacy of Isolated Bacteriophages from Pig Farms against ESBL/AmpC-Producing Escherichia coli from Pig and Turkey Farms. Front. Microbiol. 8:530. doi: 10.3389/fmicb.2017.00530
Received: 07 December 2016; Accepted: 14 March 2017;
Published: 29 March 2017.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
D. Ipek Kurtboke, University of the Sunshine Coast, AustraliaPaul Hyman, Ashland University, USA
Lanying Zeng, Texas A&M University, USA
Copyright © 2017 Skaradzińska, Śliwka, Kuźmińska-Bajor, Skaradziński, Rząsa, Friese, Roschanski, Murugaiyan and Roesler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aneta Skaradzińska, YW5ldGEuc2thcmFkemluc2thQHVwd3IuZWR1LnBs
 Aneta Skaradzińska
Aneta Skaradzińska Paulina Śliwka
Paulina Śliwka Marta Kuźmińska-Bajor
Marta Kuźmińska-Bajor Grzegorz Skaradziński
Grzegorz Skaradziński Anna Rząsa
Anna Rząsa Anika Friese4
Anika Friese4 Nicole Roschanski
Nicole Roschanski Jayaseelan Murugaiyan
Jayaseelan Murugaiyan Uwe H. Roesler
Uwe H. Roesler