- 1Microbiology, University College of Medical Sciences and Guru Teg Bahadur Hospital, New Delhi, India
- 2Dermatology, University College of Medical Sciences and Guru Teg Bahadur Hospital, Dilshad Garden, New Delhi, India
- 3Department of Minimal Access and Bariatric Surgery, Fortis Flt. Rajan Dhall Hospital, New Delhi, India
- 4Pediatrics, University College of Medical Sciences and Guru Teg Bahadur Hospital, New Delhi, India
- 5Community Medicine, University College of Medical Sciences, and Guru Teg Bahadur Hospital, Dilshad Garden, New Delhi, India
- 6Vardhman Mahavir Medical College and Sardarjung Hospital, New Delhi, India
Integrons by means of horizontal gene transfer carry multidrug resistance genes (MDR) among bacteria, including E. coli. The aim of this study was to determine the antibiotic resistance profiles and the genes associated with them, to gain insights in the distribution of phylogroups, prevalence, and characterization of class 1, 2 and 3 integrons among Enteropathogenic E. coli (EPEC) isolates, from children upto 5 years of age from Delhi and National Capital Region (NCR), India. A total of 120 E. coli isolates, including 80 from diarrheagenic E. coli (cases) and 40 from healthy isolates (controls) were recruited in this study. After isolation of E. coli, screening for EPEC was done by conventional multiplex PCR. Antibiotic suseptibility test was performed using disk diffusion method and further confirmed by minimum inhibitory concentration (MICs) by E-test. The presence and characterization of integrons and antimicrobial resistance genes were performed by PCR and DNA sequencing. Phylogeny determination was carried out by quadruplex PCR. EPEC strains were found in 64 of the 80 diarrheagenic cases, out of which 38 were MDR. In the 40 healthy controls, 23 were found to be EPEC strain, out of which only 2 were MDR. Amongst 80 diarrheagenic cases, class 1 integron were observed in 43 isolates, class 2 integron in 12 isolates and 9 isolates were found with co-existence of both. Similarly, in healthy controls; class 1 integron in 9 and class 2 integron in 7 isolates were observed with co-existence in 3 isolates. None of the isolates included class 3 integron. The dfr was the most commonly identified gene cassette within the integron-positive isolates. Phylogenetic studies showed considerable representation of phylogroup B2 in both diarrheagenic cases and healthy controls. This study reiterates the importance of class 1 integron predominantly for acquisition of antibiotic resistance genes among EPEC isolates. Furthermore, it also ascertains the possible association between multidrug resistance and presence of integrons. Approximately 91% of isolates were easily assigned to their respective phylogroups. Assessment of the relationship between antibiotic resistance and dominant phylogroups detected was also attempted. This study also highlights the increased burden of antimicrobial resistance in healthy controls.
Introduction
Childhood diarrhea is a predominant cause of child mortality worldwide, with an estimated of 2.2 million children getting affected in the developing countries alone (Ramana and Tamanna, 2012). Among the six known types of E. coli, Enteropathogenic Escherichia coli (EPEC) are one of the major causative agents of infantile diarrhea in developing countries. EPEC is classified into two subgroups; atypical EPEC (aEPEC) and typical EPEC (tEPEC) based on bundle-forming pili (bfp), present in typical E. coli (Trabulsi et al., 2002). EPEC adherence factor plasmid (pEAF) encodes bundle-forming pili which is essential for EPEC virulence, antigenicity, auto-aggregation, and localized adherence to epithelial cells (Trabulsi et al., 2002). The bfpA gene present on EAF plasmid, promotes the stabilization of bacteria by interconnecting within microcolonies. Intimin is a 94-kDa outer membrane protein encoded by eae gene, which is responsible for the adherence between bacteria and enterocyte membranes (Nataro and Kaper, 1998). The genetic determinants that produce A/E lesions are located on the locus of enterocyte effacement (LEE), a pathogenicity island that contains the genes eae encoding intimin (McDaniel et al., 1995). Attaching and effacing (A/E) lesions are the distinctive feature of EPEC infection, which are characterized by intimate attachment of the bacterium to the intestinal epithelial surface, effacement of surface-absorptive microvilli, and reorganization of actin filaments in the intestinal cell just beneath the site of attachment, which leads to the formation of pedestal-like structures (Moon et al., 1983; Croxen and Finlay, 2010). Atypical EPEC strains (lacking pEAF) are considered less virulent, as the genes encoding virulence factors are not only located on transmissible plasmids but also on transposons or bacteriophages (Trabulsi et al., 2002); nevertheless, these have not been proven to be less pathogenic (Levine et al., 1985).
The World Health Organization (WHO) has identified growing antibiotic resistance as a major problem among pediatric population. Many antibiotic resistance genes are located on plasmids, transposons, and integrons and they are greatly enhanced when they form part of a mobile gene cassette, since this provides for horizontal transfer by several mechanisms (White et al., 2001). The dihydrofolate reductase gene, dfr, plays a key role in maintaining intracellular folate homeostasis, cell growth and proliferation and is an important target for cytostatic drugs (Holger et al., 2011). Integrons commonly found in bacteria are involved in antibiotic resistance as they carry resistance genes in the form of gene cassettes. Integron-positive isolates have a greater tendency to acquire antibiotic resistance than isolates without integrons (White et al., 2001). The integrons are not mobile themselves with some exceptions (Hall and Collis, 1995) but are transported by transposons/plasmids (Barlow and Hall, 2002; Normark and Normark, 2002). There are mainly five classes of integrons known so far, each with a difference in sequence of integrase gene (Mazel, 2006). Integron carries antibiotic-resistance genes and is highly disseminated because of its close association with transposons, often embedded in conjugative plasmids (Cambray et al., 2010). Class 1 integrons were found to be more prevalent as compared to other classes of integrons (Phongpaichit et al., 2008; Rezaee et al., 2011). Most integrons carry dihydrofolate reductase gene (dfr), which confers resistance to trimethoprim and an aminoglycoside adenyltransferase gene (aadA), which confers resistance to streptomycin and spectinomycin (El-Najjar et al., 2010).
Extended spectrum β-lactamases (ESBL), AmpC β-lactamases, and carbapenemases extensively accrued by Enterobacteriaceae have now emerged as important nosocomial pathogens in India and other parts of the world (Dallenne et al., 2010; Jia et al., 2014). The ESBLs belonging to Group 2be (Bush et al., 1995) and AmpC beta-lactamases are well defined enzymes with wide range substrate specificity which could be chromosomal as well as plasmid mediated (Barlow and Hall, 2002; Babic et al., 2006). Carbapenems being stable in presence of ESBLs and AmpC enzymes are often the last choice of antibiotics in the treatment of infections due to multidrug-resistant Enterobacteriaceae isolates. Class 1 integrons contain bacterial resistance determinants to aminoglycosides as aided by aminoglycoside adenyltransferase (aadA), aminoglycoside acetyltransferases (AACs), and aminoglycoside phosphotransferases (APHs), which inactivate aminoglycosides by various identified mechanisms. The sul1 gene that encodes resistance to sulphonamides, is found exclusively on large conjugative plasmids and on class 1 integron at the 3′ end (Hall and Collis, 1995).
Based on the presence of certain genes or DNA fragments, E. coli populations are categorized into eight major phylogenetic groups namely; A, B1, B2, C, D, E, F (belonging to E. coli sensu stricto) and clade I (belonging to Escherichia clade) which can be identified using a new method of Clermont et al. (2013). These genes have been identified as chuA, encoding for heme transport protein (Torres and Payne, 1997; Bonacorsi et al., 2000); yjaA, for a hypothetical protein with unknown function (Anton et al., 2015); TSPE4.C2 as a putative lipase esterase-encoding gene (Clermont et al., 2013) with arpA, as an internal control for phylo-group F. Most of the commensal E. coli strains are mainly assigned to phylogroups A and B1 (Dobrindt, 2005; Kaper, 2005). However, for diarrheagenic E. coli (DEC), the arrangement of phylogroups is still unclear with multiple studies reporting diverse occurrences (Mosquito et al., 2015). Similarly, there is not much information available between the association of antibiotic resistance and phylogeny (Hannah et al., 2009).
The aim of this study was to determine antimicrobial resistance profiles and the genes associated with them, the prevalence and diversity of integrons, to identify the predominant phylogenetic groups, to analyze the possible relationship between presence of integrons and multidrug resistance in enteropathogenic E. coli isolates from diarrheagenic cases and healthy controls in children under 5 years of age in Delhi, India.
Materials and Methods
Study Design and Ethical Committee Approval
During the study period (July 2013–July 2014), 80 stool samples were collected from children suffering from acute diarrhea (<72 h duration) from the Out-Patient Department (OPD) of a tertiary care hospital and 40 stool samples were also collected from healthy children (neither receiving antibiotics nor suffering from any disease). This study was carried out in accordance with the recommendations of “WHO guidelines, Institutional Ethics Committee—Human Research (IEC-HR) University College of Medical Sciences, University of Delhi, New Delhi” with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The study was approved by the “Institutional Ethics Committee—Human Research (IEC-HR) University College of Medical Sciences, University of Delhi, New Delhi.”
Sample Collection and Processing
Fresh stool samples were collected in clean, leak-proof, labeled, sterile, and wide-mouthed plastic containers and were transported immediately to the laboratory and cultured. Up to five dark pink colonies (lactose fermentation) with the typical appearance of E. coli on MacConkey agar were selected and subjected to conventional biochemical tests for identifying E. coli such as gram staining (gram-negative and rod shaped bacterium), catalase test (+), oxidase test (−), glucose fermentation with production of gas, fermentation of other sugars (lactose, sucrose, maltose, and mannitol), nitrate reduction (+, reduces nitrate into nitrite), urease (−), Methyl red + Voges Proskauer (MR + and VP −), OF glucose test (glucose Fermenter), decarboxylase test [lysine (+), arginine (−) and ornithine (+/−)], indol test (+), Simon's citrate (−) and hydrogen sulfide (−) (Collee et al., 1996; Koneman et al., 1997). PCR for 16SrRNA gene was also performed, which was used as an internal quality control for E. coli confirmation (Wang et al., 2002).
DNA Extraction
‘Eluted lactose fermenting colonies on MacConkey agar were selected for DNA extraction using the commercial kit from Real Biotech Corporation, Taiwan as per manufacturer's guidelines. The extracted DNA was isolated in 100 μL of elution buffer. This DNA was used to perform conventional PCR for identification of genes associated with EPEC virulence, antibiotic resistance, class 1, 2, and 3 integrons, gene cassettes and different phylogenetic groups. Primers used for amplifying the various sequences were selected from previously published literature (Hollingshead and Vapnek, 1985; Yu and Kaper, 1992; Van de Klundert and Vliegenthart, 1993; Franke et al., 1994; Gunzburg et al., 1995; Levesque et al., 1995; Guardabassi et al., 2000; Mazel et al., 2000; Navia et al., 2003; Machado et al., 2005; Clermont et al., 2008, 2013; Kim et al., 2009; Dallenne et al., 2010; Naas et al., 2011; Manoharan et al., 2012; Lescat et al., 2013; Li et al., 2014; Table 1, Supplementary File).
Detection of EPEC Virulence
The criteria to determine typical enteropathogenic E. coli and atypical enteropathogenic E. coli were defined as follows: the presence of eae and bfpA genes for typical EPEC and presence of eae gene only depicts atypical E. coli (Kaper, 1996; Trabulsi et al., 2002). Typical EPEC strains (tEPEC) also contain the EPEC adherence factor (EAF) plasmid which carries genes encoding bundle-forming pili (BFP) (Baldini et al., 1983; Giron et al., 1991).
Conventional Multiplex PCR for Detection of Genes for EPEC Virulence, Antibiotic Resistance, Class 1, 2, and 3 Integrons, Gene Cassettes, and Phylogroups
For each set of multiplex PCR assay, 0.2 ml tubes were used, containing total volume of 25 μl of PCR mixture including, 2.5 μl buffer (10X), 1 μl dNTP's (200 uM), 1 μl MgCL2 (1.5 mM), 1 μL of each forward and reverse primers (10 μM), 5 μl of the extracted DNA and nuclease free water to make up the volume. All PCR reagents were purchased from Genei, Bangalore and amplifications were performed on an Eppendorf thermo cycler (Vidal et al., 2004). PCR amplification involved an initial denaturation step at 94°C for 10 min which was followed by 35 amplification cycles of 40 s at 94°C and 30 s at annealing temperature (as shown in Table 1, Supplementary File) and 40 s at 72°C and a final extension of 7 min at 72°C followed by a hold step at 4°C. Negative controls were PCR mixtures containing water in place of template DNA and for EPEC it was E. coli ATCC 11775, which is devoid of all the virulence genes (Janisiewicz et al., 1999). Amplified PCR products were stored at −20°C and analyzed by electrophoresis on 1.5% agarose gel (stained with ethidium bromide) at 125 volts with 15 mA current in an 18-slot apparatus for 30 min. A molecular marker of 100 bp (Fermentas) was used to determine the size of the amplicons (Sambrook et al., 1989). Uniplex PCR was also performed initially for confirmation of multiple genes.
Antibiotic Susceptibility Testing
For initial screening, antimicrobial susceptibility testing was performed with 16 antimicrobial agents (HiMedia Laboratories Mumbai, India) namely; norfloxacin (10 μg), cefotaxime (30 μg), imipenem (10 μg), meropenem (10 μg), ceftazidime (30 μg), aztreonam (30 μg), nalidixic acid (30 μg), amoxicillin (20/10 μg), gentamicin (10 μg), ciprofloxacin (5 μg), ampicillin (10 μg), amikacin (30 μg), polymixin B (300 units/disc), cefotaxime + clavulanic acid (30/10 μg), ceftriaxone (30 μg) and Piperacillin+tazobactam (100/10 μg) on Mueller-Hinton agar plates by the Kirby Bauer disc diffusion method as per CLSI guidelines (CLSI, 2012) and later minimum inhibitory concentration (MIC) by E-strips (HiMedia Laboratories Mumbai, India) was performed for confirmation. Multidrug resistant (MDR) isolates show resistance to ≥3 antibiotic classes (Magiorakos et al., 2012). Escherichia coli (ATCC) strain 25922 was included as a quality control.
Amplification and Sequencing of Gene Cassette Regions
Gene cassettes aadA1/aadA2, dfrB1, dfrB2, dfrB3, and hep 58 and hep 59 which bind to 3′ and 5′CS region of class 1 integrons were amplified using specific primers (Table 1, Supplementary File). Sequencing of the class I integrons gene cassettes (amplified by the primers hep58 and hep59) were performed commercially (Yaazh xenomics, Chennai, India). In order to increase the accuracy of the results, sequencing was performed with both forward and reverse primers and sequences were compared against GenBank database by using Basic Local Alignment Search Tool (BLAST) and submitted in the NCBI database.
Detection of Antibiotic Resistance Genes
Antibiotic resistance genes of the beta-lactamase class namely; extended spectrum beta lactamases (ESBL); TEM (Temoneira), SHV (sulfhydryl variable), CTX (cefotaxime hydrolyzing capabilities), OXA (oxacillin hydrolyzing capabilities), Metallo beta-lactamases (MBL); NDM-1 (New Delhi metallo beta lactamase), IMP (imipenem), VIM (Verona integron-encoded metallo-β-lactamase) and AmpC β-Lactamases (AmpC); ACT (AmpC type), CMY (cephamycins), DHA (Dhahran Hospital) and other antibiotic resistance genes namely; sul-1 (sulphonamides), tet-A (tetracycline) and aacC-1 (gentamicin) were determined by PCR (Table 1, Supplementary File).
Molecular Typing of Isolates
All the isolates were screened for phylogenetic groups A, B1, B2, C, D, E, F, and Clade I using quadruplex multiplex PCR as described by Clermont et al. (2013). The chuA gene was present in all strains belonging to groups B2 and D and absent from all strains belonging to groups A and B1 which separates groups B2 and D from groups A and B1. The yjaA gene allows discrimination between group B2 and group D.
Statistical Analysis
Collected Data was statistically analyzed using Statistical Package for the Social Science (SPSS; Version 20.0). Chi square test and Fisher's exact test were used to determine the statistical significance of data. P < 0.05 was considered significant.
Results
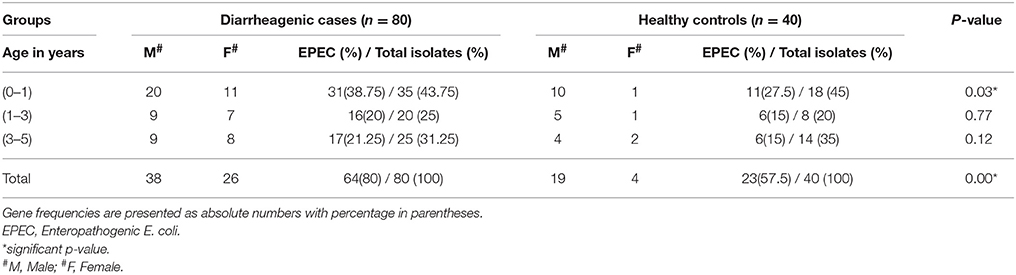
This study highlighted male prevalence in all the three age groups in diarrheagenic cases and healthy controls as shown in Table 1. The highest number of EPEC was found in the children with age up to 1 year (P < 0.05) while in the remaining two age groups, the percentage of EPEC isolates were almost similar in diarrheagenic cases (20–21.5%) and healthy controls (15% each), though not statistically significant as depicted in Table 1. The amplified PCR products of EPEC virulence for eae, bfpA and eaf genes on 1.5% agarose gel represents the uniplex PCR (Figure 1, Supplementary File), while Figure 1 shows that of multiplex PCR for all the three genes. Typical EPEC [eae+bfpA(eaf)] was found in 18.75% and 2.5% isolates (P = 0.001), atypical EPEC (eae) was found in 25% and 37.5% isolates (P = 0.673) while the total EPEC was found in 80% and 57.5% isolates (P = 0.012) of diarrheagenic cases and healthy controls respectively. 50/80 (62.5%) diarrheagenic cases (38 EPEC and 12 non-EPEC isolates) and 5/40 (12.5%) healthy controls (2 EPEC and 3 non-EPEC isolates) were multi drug resistant. The bfpA gene (usually encoded on the EAF plasmid) alone was found in 20% diarrheagenic cases and 17.5% healthy controls (P = 0.523). Three isolates were found to be positive for eaf (plasmid) gene in the absence of its virulence (bfpA) gene.
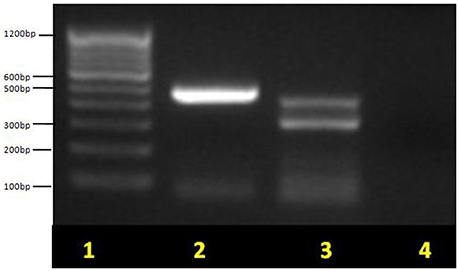
Figure 1. Multiplex PCR for EPEC virulence genes showing typical EPEC and atypical EPEC on 1.5% agarose gel. Lane 1, molecular weight marker (100 bp, Fermentas); lane 2, eae (482 bp) for atypical EPEC; lane 3, bfpA (326 bp) + eae (482 bp) for typical EPEC and lane 4, no template control (negative) control. Non-pathogenic E. coli ATCC 11775 was used as a negative control.
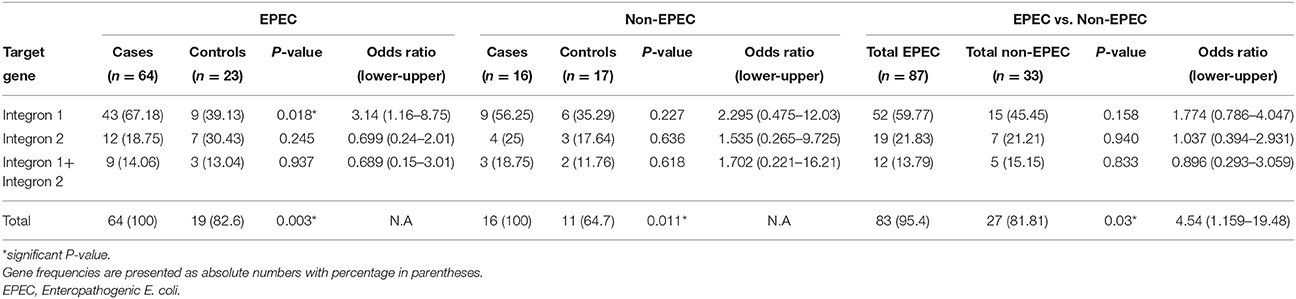
The amplified PCR products of integrase genes on 1.5% agarose gel represents the uniplex PCR for class 1 integron (160 bp) and class 2 integron (403 bp) (Figure 2, Supplementary Files). Figure 2 depicts the multiplex PCR for class 1 and class 2 integrase genes when compared with a 100 bp ladder. The distribution of integrons among EPEC and non-EPEC isolates are shown in Table 2, class 1 integron was found to be significantly (p = 0.018) associated with EPEC virulence, compared to class 2 integron, between diarrheagenic cases and healthy controls. Class 3 integron was not found in any of the isolates. No significant association was found when class 1 and 2 integrons were present together. The total number of integron-positive isolates in diarrheagenic cases and healthy controls among EPEC and non-EPEC isolates were also found to be highly significant (p = 0.003 and 0.011 respectively) as shown in Table 2. The occurrence of class 1 integron were found to be 3.14 and 2.295 times higher in EPEC and non-EPEC isolates compared to class 2 integron or both classes of integrons together (Odds ratio). However; in non-EPEC isolates a remarkable difference was found between integron carrying isolates in diarrheagenic cases and healthy controls. The occurrence of integrons was found to be 4.54 times higher (Odds ratio) in total EPEC isolates as compared to total non-EPEC isolates which were significantly different (p = 0.03) as indicated in Table 2.
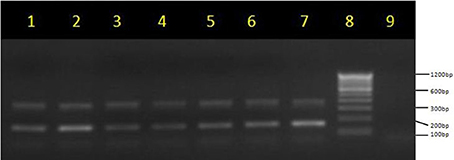
Figure 2. Multiplex PCR for integrase genes on 1.5% agarose gel. Multiplex PCR products of class 1 integrase (170 bp) and class 2 integrase genes (403 bp) lane 1–7: lane 8, molecular weight marker (100 bp, Fermentas); lanes 9, no template control (negative) control.
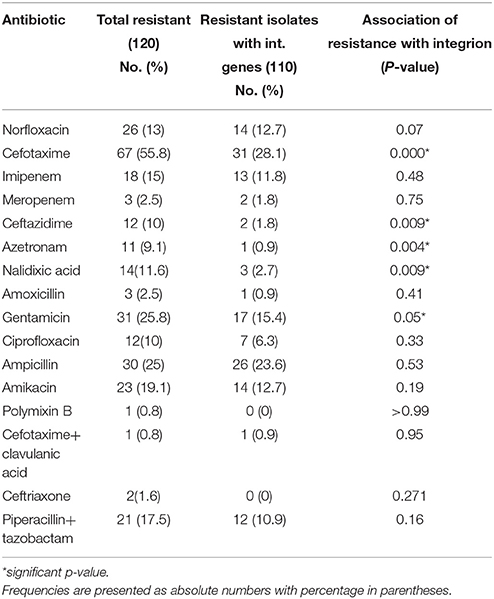
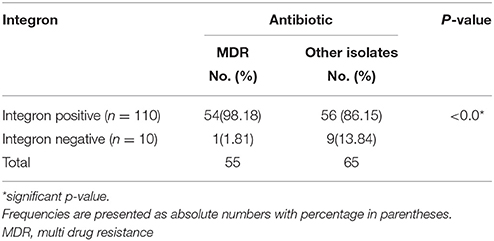
Antibiotic susceptibility testing was performed for 16 antibiotic agents as mentioned above. MIC-test was also performed for selected antibiotics by E-strip method. For resistant isolates, MIC of norfloxacin was in the range of 16–256 μg/ml, amikacin (64–256 μg/ml), cefotaxime (4–256 μg/ml), Piperacillin/tazobactam (128–256 μg/ml), gentamicin (16–256 μg/ml) and ciprofloxacin (4–256 μg/ml). Table 3 shows the relation between antibiotic resistance and presence of integrons. In our study, resistance to gentamicin, cefotaxime, ceftazidime, nalidixic acid and aztreonam was found to be significantly associated with integrons. (P < 0.05) as shown in Table 3. Table 4 depicts the relationship between the presence of integron and occurrence of multidrug resistance. A strong association was found between MDR and presence of integrons (P = 0.01).
Integron and Gene Cassettes in E. coli Isolates
PCR and sequencing analysis of class I integrons gene cassettes (hep58 and hep59) depicted different variants of dfr and aadA genes with dfrA7 at 750 bp, dfrA1/ aadA1 at 1,600 bp and dfrA12/ aadA2 at 2,000 bp (Table 2, Supplementary File). The amplicon length of the variable region of the integrons varied between 750 and 2,000 bp. Nucleotide sequences were compared against GenBank database by using BLAST. Sequence of variable region of class1 integron was submitted to NCBI and accession numbers were obtained (KY706079, KY706080, KY753816). Sequence analysis of the variable region of the class 1 integron have demonstrated the presence of nine different cassette combinations for seven different genes namely dfrA7 (9), aadA (10), dfrII (13), dfrA7+ aadA (5), dfrII+ aadA (7), dfrA7+ dfrA1/ aadA1 (2), dfrA1+aadA1 (3), dfrA12+aadA2 (10), and dfrA 7+dfrA12+aadA2 (2). Most of the gene cassettes found within the variable region of class 1 integrons in our isolates corresponded to different variants of dfrA and aadA genes. The dfr gene cassette (dfrA1, -A7, -A12, -B1, -B2, and -B3) was found to be rampant as compared to aadA gene cassette. Among the integron-positive isolates, 61.8% isolates carried gene cassettes with aadA1/2 (40.9%), dfrII (17.2%), and dfrA (31.8%).
Detection of Genes Encoding Resistance to Beta-Lactamases, Tetracycline, Gentamicin, and Sulfonamide
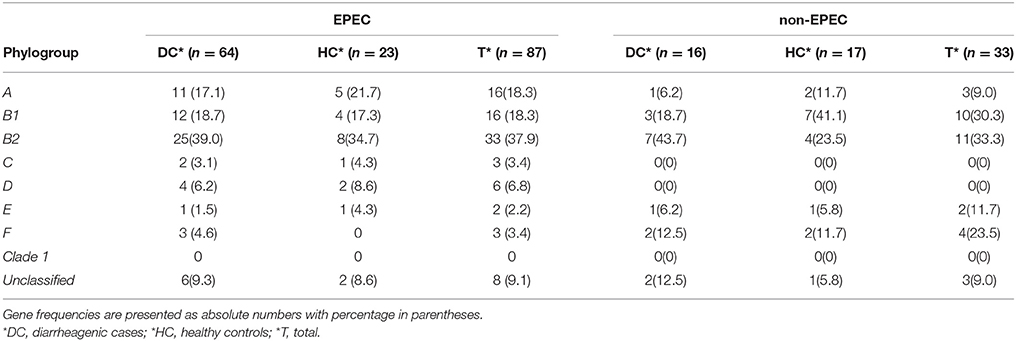
Table 5 shows the distribution of antibiotic resistance genes detected in the EPEC and non-EPEC isolates among diarrheagenic cases and healthy controls. Resistance was more commonly seen in typical EPEC of diarrheagenic cases. High frequency of resistance was observed against ampicillin, sulfonamides and tetracycline. Frequency of antibiotic resistance genes detected were compared as follows (% distribution among EPEC/ % distribution among non-EPEC): TEM (35.6/57.5), SHV (31/36.3), CTX (19.5/18.1), OXA (16/18.1), NDM-1 (12.6/3), IMP (11.5/9), VIM (17.2/12.1), ACT (11.5/15.1), DHA (4.5/6), CMY (4.5/18.1), sul 1 (49.4/42.4), tet A (21.8/15.1) and aacC 1 (18.3/21.2). In healthy controls representing EPEC, 6/23 (26%) isolates were positive for NDM and VIM each, IMP gene was seen in 5/23 (21.7%) while AmpC encoding ACT and DHA gene were positive in 4/23 (17.4%) and 1/23 (4.3%) isolates respectively. In healthy controls representing non-EPEC, only 1/17 (5.8%) isolate was positive for VIM and IMP each, 2 (11.7%) isolates were positive for ACT and DHA genes each while CMY gene was positive in 3/17 (17.6%) isolates. Among total EPEC (87) and total non-EPEC (33) isolates, carriage of resistance genes; TEM was found in 31(35.6%) and 19 (57.5%) isolates, while CMY was found in 4 (4.5%) and 6 (18.1%) isolates respectively, which were statistically significant (p < 0.05) as shown in Table 5.
Figure 3 represents the result of quadruplex PCR, for the phylogroups under study on 1.5% agarose gel. According to Clermont et al. (2013), four of the quadruplex genotypes (− − − +, − − + +, + − + + and + + + +) for arpA (400 bp), chuA (288 bp), yjaA (211 bp) and TspE4.C2 (152 bp) genes respectively have not been detected till date. In Table 6, the phylogenetic groups as determined by Clermont et al. (2013) showed the prevalence of phylogenetic group B2 in EPEC (37.9%), followed by groups B1, A, D, F, C and E in our study. The similar pattern was observed in the non-EPEC isolates with prevalence of group B2 followed by B1, F and A as shown in Table 6. All the isolates were assigned a phylogroup but eight and three of the isolates remained unclassified in EPEC and non-EPEC isolates. None of the isolates were confirmed to belong phylogroup Clade I, neither in diarrheagenic cases nor in healthy controls. However, for EPEC and non-EPEC isolates none of the phylogroups were found to be significantly associated. The absence of any significant difference between phylogroups detected in EPEC and non-EPEC isolates may be due to the small number of strains/children recruited in our study.
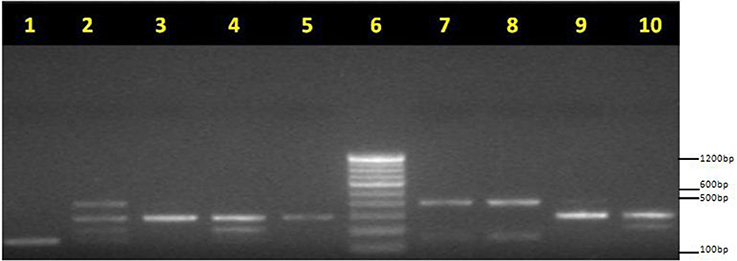
Figure 3. Phylogrouping of EPEC isolates: Quadruplex PCR profiles of new Clermont phylotyping method. arpA (400 bp), chuA (288 bp), yjaA (211 bp), and TspE4.C2 (152 bp) Lane 1, unknown (− − − +); lane 2, unknown (+ + + +); lane 3, group F (− + − −); lane 4, group B2 (− + + −); lane 5, group F (− + − −); lane 6, M: molecular weight marker (100 bp, Fermentas). lane 7, group B1 (+ − − +); lane 8, group B1 (+ − − +); lane 9, group F (− + − −); lane 10, group B2 (− + + −). PCR products were loaded on 1.5% agarose gel after electrophoresis; gels were photographed under UV light.
In Table 7, frequency of antimicrobial resistance among the prevalent phylogenetic groups in EPEC and non-EPEC isolates from diarrheagenic cases and healthy controls is shown. For EPEC isolates, high prevalence of resistance was observed against cefotaxime (71.2%) followed by gentamicin (33.3%), amikacin (26.4%), ampicillin (23%), Piperacillin/tazobactam (22.9%), norfloxacin (22.9%), nalidixic acid (16%), ciprofloxacin (13.7%), ceftazidime (13.7%), aztreonam (12.6%) and imipenem (9.1%). A significant relationship was found between antibiotic resistance (cefotaxime, gentamicin, amikacin, ampicillin and Piperacillin/tazobactam) and major phylogenetic groups detected (A, B1, and B2) in EPEC isolates. For non-EPEC isolates, ampicillin showed 30.3% resistance (P < 0.05) followed by norfloxacin (18.1%), cefotaxime (15.1%), gentamicin (6%) and Piperacillin/tazobactam (3%) as shown in Table 7.
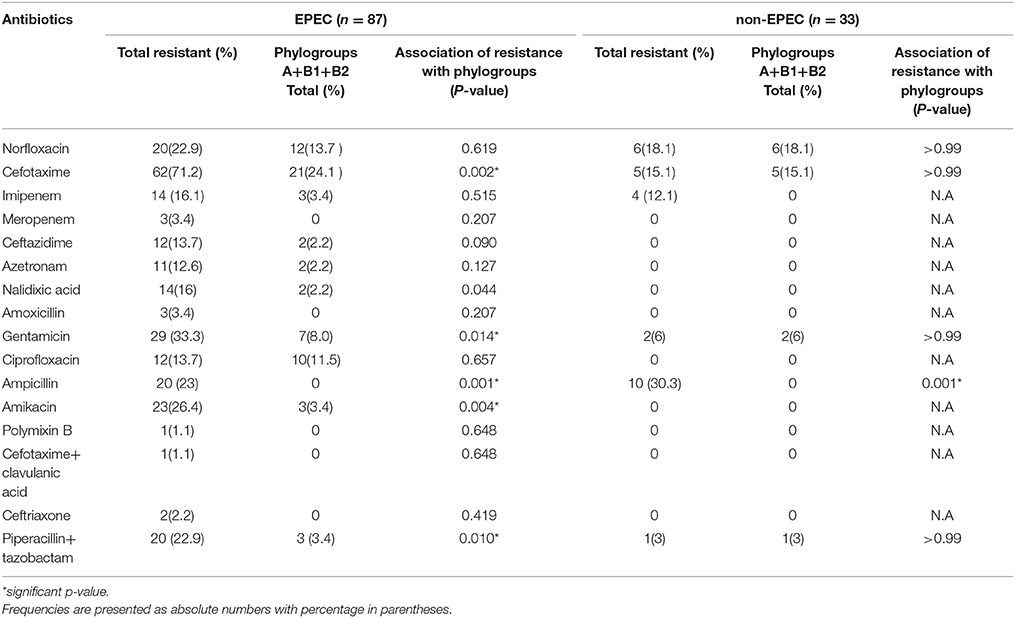
Table 7. Frequency of antimicrobial resistance among the prevalent phylogenetic groups in EPEC and non-EPEC isolates from diarrheagenic cases and healthy controls.
Discussion
EPEC is the most common cause of childhood diarrhea mainly in developing countries resulting in 30–40% deaths worldwide as they are prevalent in both community and hospital settings (Ochoa and Contreras, 2011; Ramana and Tamanna, 2012). Our study, focused on the EPEC isolated from stool samples of diarrheagenic and healthy children of up to 5 years of age for its virulence, distribution and characterization of integrons, their phylogenetic background, antibiogram and antibiotic resistance genes associated with them. EPEC strains are non-invasive and cause watery diarrhea after formation of attaching and effacing (A/E) lesions (Nataro and Kaper, 1998; Ochoa and Contreras, 2011). As indicated in Table 1, EPEC were found to be significantly present in diarrheagenic cases (80%) and healthy controls (57.5%) which is in accordance with other Indian studies [36.58% and 0% (Tilak and Mudaliar, 2012); 16 and 3% (Hegde et al., 2012)] and worldwide studies viz. 56.7 and 8.3% in Iran, 18.3 and 5.6% in Brazil, 3.95 and 0% in Hong Kong, 13.7 and 5.5% in Tunisia and 6.17 and 1.96% in Rondonia, Western Amazon region, Brazil in diarrheagenic cases and healthy controls respectively (Tsukamoto et al., 1992; Biswas et al., 1996; Alikhani et al., 2006; Orlandi et al., 2006; Ben Salem-Ben Nejma et al., 2014).
Typical EPEC isolates show lower occurrence in diarrheagenic cases and healthy controls with 18.75% and 2.5%, as compared to atypical EPEC with 25% and 37.5% isolates respectively, which is in accordance with previous reported studies (Afset et al., 2004; Ochoa et al., 2008; Nair et al., 2010). In our study, aEPEC was found predominantly in healthy controls which were similar to previous studies indicating their survival as colonizers in the intestinal mucosa (Dias et al., 2016). The higher rate of aEPEC in healthy controls also represents asymptomatic carriage rather than symptomatic infection that under favorable host conditions may become pathogenic (Afset et al., 2004; Nair et al., 2010). Since, atypical EPEC strains have not been confirmed to be nonpathogenic, or less pathogenic as compared to typical ones the only possibility is that they have adopted other virulence factors for the loss of EAF plasmid (Scaletsky et al., 2010). EPEC strains are classified into typical and atypical based on the presence of the large virulence EPEC adherence factor (EAF) plasmid (Nataro and Kaper, 1998). The EAF plasmid encodes a type IV fimbria known as the bundle-forming pilus (bfp) that mediate atypical EPEC to cause persistent diarrhea although the mechanism by which it establishes infection, is still unclear (Giron et al., 1991; Donnenberg et al., 1992; Bieber et al., 1998; Knutton et al., 1999; Afset et al., 2004).
Hall and Collis (1995) showed class 1 and 2 integrons to be prevalent in enteropathogens. Among EPEC isolates, class 1 integron was significantly associated (Table 2). Class 3 integron was not found in any isolate as reported by other studies (White et al., 2001; Yu et al., 2003; Machado et al., 2005; Skurnik et al., 2005; Su et al., 2006; Vinue et al., 2008; Muhammad et al., 2011; Rezaee et al., 2011). Contradicting to the observation, a single study in Iran showed prevalence of class 3 integron with 26.09% (Kargar et al., 2014). Our study also indicated that integrons exist in E. coli isolated from healthy controls as well. Frequency of class 1 integron in healthy controls was comparable (37.5%) to other studies with the highest prevalence of 85.5% reported from China (Su et al., 2006), followed by 49% from Estonia (Sepp et al., 2009), 29% from Spain (Vinue et al., 2008) and 21% from India (Dureja et al., 2014). Herein, after comparing the presence of integrons in cases and controls, the occurrence of class 1 integrons in healthy controls suggests a possible acquisition of resistance genes circulating in the hospital environment and may act as potential reservoir of integrons in maintaining a constant horizontal exchange of these genes (Roe et al., 2003). The integrons highlight the emergence of multidrug resistance, as usage of one antibiotic may activate the expression and transfer of a whole gene cassette emphasizing the association of class 1 integrons to MDR (Leverstein-van Hall et al., 2003; Norrby, 2005).
We cannot deny the fact that EPEC isolates become predominant under the antibiotic influence, and it facilitates their selective proliferation and colonization in gastrointestinal tracts of children. Such resistance with slow reversal rate has higher chances of persistence in the bacterial population (Andersson and Hughes, 2010). Higher resistance to some antimicrobial agents (gentamicin, cefotaxime, ceftazidime, nalidixic acid, and aztreonam) among integron-positive isolates (91.6%) indicated the involvement of resistance genes in the same mobile elements that carry integrons as shown in Table 3. Suspected localization of resistance genes in the conserved or variable region of integrons or the involvement of resistance genes in the same mobile elements that carry integrons may confer higher resistance. Among EPEC isolates, (40/87) 45.97 percent isolates, showed MDR phenotype with higher antibiotic resistance to cefotaxime followed by gentamicin, ampicillin, norfloxacin, amikacin, Piperacillin+tazobactam, imipenem, nalidixic acid, ciprofloxacin, as reported by others (Saravanan and Raveendaran, 2013; Lin et al., 2014; Wang et al., 2015). Such high cefotaxime-resistant (CTX-R) population of enterobacteriaceae, constitute a reservoir for transmission that may remain unidentified in hospitals which do not implement active surveillance testing (Tarchouna et al., 2014). In our study, less than half of the isolates were multidrug resistant (45.83%) which has also been reported by other workers (Elsharkawy et al., 2013). Class 1 integron was predominant among EPEC isolates, and significantly associated with MDR (P < 0.05) as shown in Table 4. There are studies which also reported association between MDR and integrons (Bakhshi et al., 2014; Kargar et al., 2014; Malek et al., 2015). Occurrence of gene cassettes in our study was different in terms of number and gene combinations as reported by many other researchers (White et al., 2001). The dfr gene cassette (dfrA1, -A7, -A12, -B1, -B2, and -B3) was found to be prevalent as compared to the aadA gene cassette, (Table 2, Supplementary File) which is in agreement with Malek et al. (2015).
Acquisition of tetracycline, sulphonamide and gentamicin resistance genes are also of great concern (Table 5). Tetracycline resistance gene tet(A) and class 1 integrons usually share the same conjugative plasmid (Sunde and Norstrom, 2006), while the acquisition of gentamicin resistance genes are yet to be established. The prevalence of sul1 gene in our integron-positive healthy isolates was higher by 47.5% to previously reported study (Infante et al., 2005). High prevalence of sulphonamide resistance in healthy controls suggests that commensal strains could also represent an important reservoir of these resistance determinants. A significant association between resistance to aminoglycosides tested (gentamicin) and the presence of integron was also explainable because many aminoglycoside resistance genes have been reported within integron structures, including aadA and aacA1 (Martinez-Freijo et al., 1999).
Escherichia coli have the potential to spread outside the hospital environment also as described by Banerjee and Johnson (2014). In our study (Table 5), production of enzymes encoding beta-lactamases genes in healthy children suggests that children may be silently harboring MDR strains with prolonged colonization, which means that they could serve as potential reservoirs for these strains in the community (Zerr et al., 2014; Yaffee et al., 2016). NDM gene is located on transmissible plasmid along with other antibiotic resistance genes, which make their spreading easy (Kumarasamy et al., 2010). The VIM encoding integron structure most likely has been acquired during hospital stay and then colonized in patients before leaving (Riccio et al., 2001). The emergence of the CMY gene has been reported among diverse genera of the Enterobacteriaceae (Qin et al., 2008). The observed increase in the proportion of beta-lactamase producers among healthy controls is probably multi-factorial, resulting from the clonal spread of virulent strains, dissemination of conjugative plasmids as well as increased selection pressure (Baudry et al., 2009). There are many other factors for their dissemination like overcrowding, availability of antibiotics, low level of hygiene and weak hospital antibiotic policies (Nordmann et al., 2011).
The phylogenetic analyses as described in Table 6 showed majority of EPEC isolates belonging to one of four major phylogenetic groups (A, B1, B2, or D) in diarrheagenic cases as well as in healthy controls. In diarrheagenic cases, EPEC isolates depicted a strong association with phylogroup B2 which is in accordance with other previously reported study, followed by B1, A, D, F, C, and E (Chakraborty et al., 2015). Six (diarrheagenic cases) and two (healthy control) isolates remained unclassified, which could either be due to their rarity of occurrence or a high frequency of loss of gene leading to low existence of these phylogroups (Touchon et al., 2009). In EPEC isolates from healthy controls, B2 (34.7%) remained the predominant phylogroup, similar to other previously reported studies followed by A, B1, D, C, or E, whereas Group F was not observed in any isolate (Bailey et al., 2010; Wang et al., 2013; Rana et al., 2016). Previous studies have indicated that phylogroups B2 and D possess more virulence properties, compared to other phylogroups. Similar pattern of occurrence of phylogenetic groups in both diarrheagenic cases and healthy controls is suggestive of a homogeneous group existing in the pediatric population with an onward circulation and transfer of the similar set of virulence genes (Nguyen et al., 2010).
Table 7 shows the antibiotic resistance of all the phylogroups revealed that group B2 displayed maximum number of antibiotic resistance genes with nearly similar number of isolates in tEPEC and aEPEC category while the isolates in phylogroups A and B1 were mainly found to be linked with aEPEC (Wang et al., 2013). We found cefotaxime resistance to be significantly associated with group B2 compared to others (Soto et al., 2007; Jakobsen et al., 2010). Many virulence genes after implanting themselves into pathogenicity islands move along with integrons into the environment via horizontal gene transfer. The preponderance of strains in different phylogroups may vary due to the intensity of antibiotic resistant strains, virulence of the strains, geographical site and the site of infection (Duriez et al., 2001; Bukh et al., 2009).
A strong association between the presence of integrons and different phylogroups was found in our study (p = 0.004) which is also in agreement with a previous study (Mabbett et al., 2009). For EPEC isolates, we found a strong association between class 1 integron with phylogenetic group A, B1 and B2 (P < 0.05) while for non-EPEC isolates class 1 integron was significantly associated with only phylogenetic group A. Phylogenetic group B2 was significantly associated with class 2 integron and when both the integrons are present together for EPEC isolates (P < 0.05).
Essentially, our study conveys a strong link between diarrheagenic E. coli populations and establishment of virulence and antibiotic resistance. The epidemiological groups' existance in children determines the circumstances of persistence of the pathogen in the community. Although, quadruplex PCR has limitations of not detecting all clades, it can still be utilized to demonstrate the role of integrons in multidrug resistance within diarrheagenic and commensal E. coli strains. Since, integrons are common among MDR isolates, hence; they can be used as a marker for the identification of MDR isolates. Isolates carrying integrons representing multidrug resistance may be explored as a useful therapeutic target for future research using specific silencing strategies and combination antimicrobial therapy. Moreover, occurrence of resistance in healthy controls indicates that antimicrobial resistant isolates in the gut are widely circulating in the community and hence continued surveillance of antibiotic resistance will provide crucial information in developing locally appropriate guidelines for efficacious treatment of diarrheagenic E. coli in developing countries like India.
Author Contributions
TS and SD conceived and designed the study, analyzed/interpreted results. TS and AS collected the samples. TS and SW collected the data. AS, TS, and SW performed experiments and analyzed the data. TS, SW, and DS carried out the literature search. TS and SW gave technical support and conceptual advice. VR, SD, and TS wrote the manuscript. VR, KM, and DS participated in design and supervision of the study and revised the final version of the manuscript. KM performed statistical analysis also. AS and KM performed the manuscript editing. SD and TS supervised the study and revised the final version of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Council of Scientific and Industrial Research (CSIR) Library Avenue, Pusa Road, New Delhi 110012. Project 08/532 (0007)/2011-EMR-I.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank all the children and their parents for providing samples. We also like to thank Mr. Praveen Kumar SIngh (UGC, JRF) for his technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00561/full#supplementary-material
References
Afset, J. E., Bevanger, L., Romundstad, P., and Bergh, K. (2004). Association of atypical enteropathogenic Escherichia coli (EPEC) with prolonged diarrhoea. J. Med. Microbiol. 53, 1137–1144. doi: 10.1099/jmm.0.45719-0
Alikhani, M. Y., Mirsalehian, A., and Aslani, M. M. (2006). Detection of typical and atypical enteropathogenic Escherichia coli (EPEC) in Iranian children with and without diarrhoea. J. Med. Microbiol. 55, 1159–1163. doi: 10.1099/jmm.0.46539-0
Andersson, D., and Hughes, D. (2010). Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8, 260–271. doi: 10.1038/nrmicro2319
Anton, B. P., Mongodin, E. F., Agrawal, S., Fomenkov, A., Byrd, D. R., Roberts, R. J., et al. (2015). Complete genome sequence of ER2796, a DNA methyltransferase-deficient strain of Escherichia coli K-12. PLoS ONE 10:e0127446. doi: 10.1371/journal.pone.0127446
Babic, M., Hujer, A. M., and Bonomo, R. A. (2006). What's new in antibiotic resistance? Focus on beta-lactamases. Drug Resist. Updat. 9, 142–156. doi: 10.1016/j.drup.2006.05.005
Bailey, J. K., Pinyon, J. L., Anantham, S., and Hall, R. M. (2010). Distribution of human commensal Escherichia coli phylogenetic groups. J. Clin. Microbiol. 48, 3455–3456. doi: 10.1128/JCM.00760-10
Bakhshi, B., Eftekhari, N., and Pourshafie, M. R. (2014). Genetic elements associated with antimicrobial resistance among intestinal bacteria. Jundishapur J. Microbiol. 7:e9924. doi: 10.5812/jjm.9924
Baldini, M. M., Kaper, J. B., Levine, M. M., Candy, D. C., and Moon, H. W. (1983). Plasmid-mediated adhesion in enteropathogenic Escherichia coli. J. Pediatr Gastroenterol. Nutr. 2, 534–539. doi: 10.1097/00005176-198302030-00023
Banerjee, R., and Johnson, J. R. (2014). A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob. Agents Chemother. 58, 4997–5004. doi: 10.1128/AAC.02824-14
Barlow, M., and Hall, B. G. (2002). Origin and evolution of the AmpC β-Lactamases of Citrobacter freundii. Antimicrob. Agents Chemother. 46, 1190–1198. doi: 10.1128/AAC.46.5
Baudry, P. J., Mataseje, L., Zhanel, G. G., Hoban, D. J., and Mulvey, M. R. (2009). Characterization of plasmids encoding CMY-2 AmpC b-lactamases from Escherichia coli in Canadian intensive care units. Diagn. Microbiol. Infect. Dis. 65, 379–383. doi: 10.1016/j.diagmicrobio.2009.08.011
Ben Salem-Ben Nejma, I., Hassine Zaafrane, M., Hassine, F., Sdiri-Loulizi, K., Ben Said, M., Aouni, M., et al. (2014). Etiology of acute diarrhea in tunisian children with emphasis on diarrheagenic Escherichia coli: prevalence and identification of E. coli virulence markers. Iran. J. Public Health 43, 947–960.
Bieber, D., Ramer, S. W., Wu, C. Y., Murray, W. J., Tobe, T., Fernandez, R., et al. (1998). Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280, 2114–2118. doi: 10.1126/science.280.5372.2114
Biswas, R., Nelson, E. A., Lewindon, P. J., Lyon, D. J., Sullivan, P. B., Elion, J., et al. (1996). Molecular epidemiology of Escherichia coli diarrhea in children in Hong Kong. J. Clin. Microbiol. 34, 3233–3234.
Bonacorsi, S. P., Clermont, P. O., Tinsley, C., Le Gall, I., Beaudoin, J. C., Elion, J., et al. (2000). Identification of regions of the Escherichia coli chromosome specific for neonatal meningitis-associated strains. Infect. Immun. 68, 2096–2101. doi: 10.1128/IAI.68.4
Bukh, A. S., Schonheyder, H. C., Emmersen, J. M. G., Sogaard, M., Bastholm, S., and Roslev, P. (2009). Escherichia coli phylogenetic groups are associated with site of infection and level of antibiotic resistance in community-acquired bacteraemia: a 10 year population-based study in Denmark. J. Antimicrob. Chemother. 64, 163–168. doi: 10.1093/jac/dkp156
Bush, K., Jacoby, G. A., and Medeiros, A. A. (1995). A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39, 1211–1233.
Cambray, G., Guerout, A. M., and Mazel, D. (2010). Integrons. Annu. Rev. Genet. 44, 141–166, doi: 10.1146/annurev-genet-102209-163504
Chakraborty, A., Saralaya, V., Adhikari, P., Shenoy, S., Baliga, S., and Hegde, A. (2015). Characterization of Escherichia coli phylogenetic groups associated with extraintestinal infections in south indian population. Ann. Med. Health Sci. Res. 5, 241–246. doi: 10.4103/2141-9248.160192
Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013). The clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
Clermont, O., Lescat, M., O'Brien, C. L., Gordon, D. M., Tenaillon, O., and Denamur, E. (2008). Evidence for a human specific Escherichia coli clone. Environ. Microbiol. 10, 1000–1006. doi: 10.1111/j.1462-2920.2007.01520.x
CLSI (2012). Performance Standards for Antimicrobial Disc Susceptibility Tests; Approved Standard, 11th Edn., Wayne, PA: CLSI; Document M02–A11
Collee, J. G., Duguid, J. P., Fraser, A. G., Marmion, B. P., and Simmons, A. B. P., (1996). Mackie and McCartney Practical Medical Microbiology, 14th Edn., New York, NY: Churchill Livingstone. 131–138.
Croxen, M. A., and Finlay, B. B. (2010). Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8, 26–38. doi: 10.1038/nrmicro2265
Dallenne, C., Da Costa, A., Decré, D., Favier, C., and Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important b-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi: 10.1093/jac/dkp498
Dias, R. C., Dos Santos, B. C., Dos Santos, L. F., Vieira, M. A., Yamatogi, R. S., Mondelli, A. L., et al. (2016). Diarrheagenic Escherichia coli pathotypes investigation revealed atypical enteropathogenic E. coli as putative emerging diarrheal agents in children living in Botucatu, São Paulo State, Brazil. APMIS 124, 299–308. doi: 10.1111/apm.12501
Dobrindt, U. (2005). Patho-genomics of Escherichia coli. Int. J. Med. Microbiol. 295, 357–371. doi: 10.1016/j.ijmm.2005.07.009
Donnenberg, M. S., Girón, J. A., Nataro, J. P., and Kaper, J. B. (1992). A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol. Microbiol. 6, 3427–3437.
Dureja, C., Mahajan, S., and Raychaudhuri, S. (2014). Phylogenetic distribution and prevalence of genes encoding class I integrons and CTX-M-15 extended- spectrum b-lactamases in Escherichia coli isolates from healthy humans in Chandigarh, India. PLoS ONE 9:e112551. doi: 10.1371/journal.pone.0112551
Duriez, P., Clermont, O., Bonacorsi, S., Bingen, E., Chaventre, A., Elion, J., et al. (2001). Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 147, 1671–1676. doi: 10.1099/00221287-147-6-1671
El-Najjar, N. G., Farah, M. J., Hashwa, F. A., and Tokajian, S. T. (2010). Antibiotic resistance patterns and sequencing of class I integron from uropathogenic Escherichia coli in Lebanon. Lett. Appl. Microbiol. 51, 456–461. doi: 10.1111/j.1472-765X.2010.02926.x
Elsharkawy, A., Elkady, L., Mansour, S., and Esmaeel, N. (2013). Detection of extended-spectrum and plasmid-mediated AmpC b-lactamases in nosocomial Klebsiella isolates. J. Microbiol. Infect. Dis. 3, 24–30. doi: 10.5799/ahinjs.02.2013.01.0074
Franke, J., Franke, S., Schmidt, H., Schwarzkopf, A., Beutin, L., and Helge, K. (1994). Nucleotide Sequence Analysis of Enteropathogenic Escherichia coli (EPEC) adherence factor probe and development of PCR for rapid detection of EPEC harboring virulence plasmids. J. Clin. Microbiol. 32, 2460–2463.
Giron, J. A., Ho, A. S., and Schoolnik, G. K. (1991). An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254, 710–713. doi: 10.1126/science.1683004
Guardabassi, L., Dijkshoorn, L., Collard, J. M., Olsen, J. E., and Dalsgaard, A. (2000). Distribution and in-vitro transfer of tetracycline resistance determinants in clinical and aquatic Acinetobacter strains. J. Med. Microbiol. 49, 929–936. doi: 10.1099/0022-1317-49-10-929
Gunzburg, S. T., Tornieporth, N. G., and Riley, L. W. (1995). Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J. Clin. Microbiol. 33, 1375–1377.
Hall, R. M., and Collis, C. M. (1995). Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15, 593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x
Hannah, E. L., Johnson, J. R., Angulo, F., Haddadin, B., Williamson, J., and Samore, M. H. (2009). Molecular analysis of antimicrobial-susceptible and -resistant Escherichia coli from retail meats and human stool and clinical specimens in a rural community setting. Foodborne Pathog. Dis. 6, 285–295. doi: 10.1089/fpd.2008.0176
Hegde, A., Ballal, M., and Shenoy, S. (2012). Detection of diarrheagenic Escherichia coli by multiplex PCR. Indian J. Med. Microbiol. 30, 279–284. doi: 10.4103/0255-0857.99485
Holger, C., Smith, D. E. C., Blom, H., Blau, N., Bode, H., Holzmann, K., et al. (2011). Dihydrofolate reductase deficiency due to a homozygous DHFR mutation causes megaloblastic anemia and cerebral folate deficiency leading to severe neurologic disease. Am. Soc. Hum. Genet. 88, 226–231. doi: 10.1016/j.ajhg.2011.01.007
Hollingshead, S., and Vapnek, D. (1985). Nucleotide sequence analysis of the gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid 13, 17–30.
Infante, B., Grape, M., Larsson, M., Kristiansson, C., Pallecchi, L., Rossolini, G. M., et al. (2005). Acquired sulphonamide resistance genes in faecal Escherichia coli from healthy children in Bolivia and Peru. Int. J. Antimicrob. Agents 25, 308–312. doi: 10.1016/j.ijantimicag.2004.12.004
Jakobsen, L., Spangholm, D. J., Pedersen, K., Jensen, L. B., Emborg, H. D., Agerso, Y., et al. (2010). Broiler chickens, broiler chicken meat, pigs and pork as sources of ExPEC related virulence genes and resistance in Escherichia coli isolates from community dwelling humans and UTI patients. Int. J. Food Microbiol. 142, 264–272. doi: 10.1016/j.ijfoodmicro.2010.06.025
Janisiewicz, W. J., Conway, W. S., Brown, M. W., Sapers, G. M., Fratamico, P., and Buchanan, R. L. (1999). Fate of Escherichia coli O157:H7 on fresh-cut apple tissue and its potential for transmission by fruit flies. Appl. Environ. Microbiol. 65, 1–5.
Jia, W., Li, G., and Wang, W. (2014). Prevalence and antimicrobial resistance of Enterococcus species: a hospital-based study in China. Int. J. Environ. Res. Public Health 11, 3424–3442. doi: 10.3390/ijerph110303424
Kaper, J. B. (2005). Pathogenic Escherichia coli. Int. J. Med. Microbiol. 295, 355–356. doi: 10.1016/j.ijmm.2005.06.008
Kargar, M., Mohammadalipour, Z., Doosti, A., Lorzadeh, S., and Japoni-Nejad, A. (2014). High prevalence of Class 1 to 3 integrons among multidrug-resistant diarrheagenic Escherichia coli in Southwest of Iran. Osong Public Health Res. Perspect. 5, 193–198. doi: 10.1016/j.phrp.2014.06.003
Kim, J., Jeon, S., Rhie, H., and Kim, S. (2009). Rapid Detection of Extended Spectrum β-Lactamase (ESBL) for Enterobacteriaceae by use of a Multiplex PCR-based Method. Infect. Chemother. 41, 181–184. doi: 10.1016/j.mimet.2010.06.006
Knutton, S., Shaw, R. K., Anantha, R. P., Donnenberg, M. S., and Zorgani, A. A. (1999). The type IV bundle forming pilus of enteropathogenic Escherichia coli undergoes dramatic alterations in structure associated with bacterial adherence, aggregation and dispersal. Mol. Microbiol. 33, 499–509.
Koneman, E. W., Allen, S. D., Janda, W. M., Schreckenberger, P. C., and Winn, W. C. Jr. (eds.). (1997). “Mycology,” in Color Atlas and Textbook of Diagnostic Microbiology, 5th Edn. (Philadelphia, PA: Lippincott Williams and Wilkins), 983–1057.
Kumarasamy, K. K., Toleman, M. A., Walsh, T. R., Bagaria, J., Butt, F., Balakrishnan, R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. doi: 10.1016/S1473-3099(10)70143-2
Lescat, M., Clermont, O., Woerther, P. L., Glodt, J., Dion, S., Skurnik, D., et al. (2013). Commensal Escherichia coli strains in Guiana reveal a high genetic diversity with host dependant population structure. Environ. Microbiol. Rep. 5, 49–57. doi: 10.1111/j.1758-2229.2012.00374.x
Leverstein-van Hall, M. A. M., Blok, H. E. T., Donders, A. R., Paauw, A., Fluit, A. C., and Verhoef, J. (2003). Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J. Infect. Dis. 187, 251–259. doi: 10.1086/345880
Levesque, C., Piche, L., Larose, C., and Roy, P. H. (1995). PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39, 185–191. doi: 10.1128/AAC.39.1.185
Levine, M. M., Nataro, J. P., Karch, H., Baldini, M. M., Kaper, J. B., Black, R. E., et al. (1985). The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J. Infect. Dis. 152, 550–559.
Li, L. M., Wang, M. Y., Yuan, X. Y., Wang, H. J., Li, Q., and Zhu, Y. M. (2014). Characterization of integrons among Escherichia coli in a region at high incidence of ESBL-EC. Pak. J. Med. Sci. 30, 177–180. doi: 10.12669/pjms.301.4079
Lin, L., Xu, X., Ren, X., and Cui, S. (2014). Prevalence and characterization of cefotaxime and ciprofloxacin co-resistant Escherichia coli isolates in retail chicken carcasses. Wei Sheng Yan Jiu. 43, 768–773.
Mabbett, A. N., Ulett, G. C., Watts, R. E., Tree, J. J., Totsika, M., Ong, C. L., et al. (2009). Virulence properties of asymptomatic bacteriuria Escherichia coli. Int. J. Med. Microbiol. 299, 53–63. doi: 10.1016/j.ijmm.2008.06.003
Machado, E., Canton, R., Baquero, F., Galán, J. C., Rollan, A., Peixe, L., et al. (2005). Integron content of extended-spectrum-b-lactamase-producing Escherichia coli strains over 12 year in a single hospital in Madrid, Spain. Antimicrob. Agents Chemother. 49, 1823–1829. doi: 10.1128/AAC.49.5.1823-1829.2005
Magiorakos, A., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Malek, M. M., Amer, F. A., Allam, A. A., El-Sokkary, R. H., Gheith, T., and Arafa, M. A. (2015). Occurrence of classes Iand II integrons in Enterobacteriaceae collected from ZagazigUniversity Hospitals, Egypt. Front. Microbiol. 6:601. doi: 10.3389/fmicb.2015.00601
Manoharan, A., Sugumar, M., Kumar, A., Jose, H., Mathai, D., and Khilnani, G. C. (2012). Phenotypic and molecular characterization of AmpC β-lactamases among Escherichia coli, Klebsiella spp. and Enterobacter spp. from five Indian Medical Centers. Indian J. Med. Res. 135, 359–364.
Martinez-Freijo, P., Fluit, A. C., Schmitz, F. J., Verhoef, J., and Jones, M. E. (1999). Many class I integrons comprise distinct stable structures occurring in different species of Enterobacteriaceae isolated from widespread geographic regions in Europe. Antimicrob. Agents Chemother. 43, 686–689.
Mazel, D. (2006). Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4, 608–620. doi: 10.1038/nrmicro1462
Mazel, D., Dychinco, B., Webb, V. A., and Davies, J. (2000). Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 44, 1568–1574.
McDaniel, T. K., Jarvis, K. G., Donnenberg, M. S., and Kaper, J. B. (1995). A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 92, 1664–1668.
Moon, H. W., Whipp, S. C., Argenzio, R. A., Levine, M. M., and Gianella, R. A. (1983). Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41, 1340–1351.
Mosquito, S., Pons, M. J., Riveros, M., Ruiz, J., and Ochoa, T. J. (2015). Diarrheagenic Escherichia coli phylogroups are associated with antibiotic resistance and duration of diarrheal episode. Sci. World J. 2015:610403. doi: 10.1155/2015/610403
Muhammad, I., Uzma, M., Yasmin, B., Mehmood, Q., and Habib, B. (2011). Prevalence of antimicrobial resistance and integrons in Escherichia coli from Punjab, Pakistan. Braz. J. Microbiol. 42, 462–466. doi: 10.1590/S1517-83822011000200008
Naas, T., Ergani, A., Carrer, A., and Nordmann, P. (2011). Real-Time PCR for detection of NDM-1 carbapenemase genes from spiked stool samples. Antimicrob. Agents Chemother. 55, 4038–4043. doi: 10.1128/AAC.01734-10
Nair, G. B., Ramamurthy, T., Bhattacharya, M. K., Krishnan, T., Ganguly, S., Saha, D. R., et al. (2010). Emerging trends in the etiology of enteric pathogens as evidenced from an active surveillance of hospitalized diarrhoeal patients in Kolkata, India. Gut Pathog. 2:4. doi: 10.1186/1757-4749-2-4
Nataro, J. P., and Kaper, J. B. (1998). Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11, 142–201.
Navia, M. M., Ruiz, J., Sanchez-Cespedes, J., and Vila, J. (2003). Detection of dihydrofolate reductase genes by PCR and RFLP. Diagn. Microb. Infect. Dis. 46, 295–298. doi: 10.1016/S0732-8893(03)00062-2
Nguyen, N. T., Ha, V., Tran, N. V., Stabler, R., Pham, D. T., Le, T. M., et al. (2010). The sudden dominance of blaCTXM harbouring plasmids in Shigella spp. circulating in Southern Vietnam. PLoS Negl. Trop. Dis. 4:e702. doi: 10.1371/journal.pntd.0000702
Nordmann, P., Poirel, L., Toleman, M. A., and Walsh, T. R. (2011). Does broadspectrum beta-lactam resistance due to NDM-1 herald the end of the antibiotic era for treatment of infections caused by Gram-negative bacteria? J. Antimicrob. Chemother. 66, 689–692. doi: 10.1093/jac/dkq520
Normark, B. H., and Normark, S. (2002). Evolution and spread of antibiotic resistance. J. Intern. Med. 252, 91–106. doi: 10.1046/j.1365-2796.2002.01026.x
Norrby, S. R. (2005). Integrons: adding another threat to the use of antibiotic therapy. Clin. Infect. Dis. 41, 10–11. doi: 10.1086/430715
Ochoa, T. J., Barletta, F., Contreras, C., and Mercado, E. (2008). New insights into the epidemiology of enteropathogenic Escherichia coli infection. Trans. R. Soc. Trop. Med. Hyg. 102, 852–856. doi: 10.1016/j.trstmh.2008.03.017
Ochoa, T. J., and Contreras, C. A. (2011). Enteropathogenic Escherichia coli infection in children. Curr. Opin. Infect. Dis. 24, 478–483. doi: 10.1097/QCO.0b013e32834a8b8b
Orlandi, P. P., Magalhães, G. F., Matos, N. B., Silva, T., Penatti, M., Nogueira P. A. et al. (2006). Etiology of diarrheal infections in children of Porto Velho (Rondonia, Western Amazon region, Brazil). Braz. J. Med. Biol. Res. 39, 507–517. doi: 10.1590/S0100-879X2006000400011
Phongpaichit, S., Wuttananupan, K., and Samasanti, W. (2008). Class 1 integrons and multidrug resistance among Escherichia coli isolates from human stools. Southeast Asian J. Trop. Med. Public Health 39, 279–287.
Qin, X., Zerr, D. M., Weissman, S. J., Englund, J. A., Denno, D. M., Klein, E. J., et al. (2008). Prevalence and mechanisms of broad-spectrum beta-lactam resistance in Enterobacteriaceae: a children's hospital experience. Antimicrob. Agents Chemother. 52, 3909–3914. doi: 10.1128/AAC.00622-08
Ramana, J., and Tamanna (2012). dbDiarrhea: the database of pathogen proteins and vaccine antigens from diarrheal pathogens. Infect. Genet. Evol. 12, 1647–1651. doi: 10.1016/j.meegid.2012.08.002
Rana, F., Siddiqui, S., Khan, A., Siddiqui, F., Noreen, Z., Bokhari, S., et al. (2016). Resistance patterns of diversified phylogroups of Escherichia coli associated with mothershaving history of preterm births in Pakistan. J. Matern. Fetal Neonatal Med. 23, 1–6. doi: 10.3109/14767058.2016.1163538
Rezaee, M. A., Sheikhalizadeh, V., and Hasani, A. (2011). Detection of integrons among multi-drug resistant (MDR) Escherichia coli strains isolated from clinical specimens in northern west of Iran. Braz. J. Microbiol. 42, 1308–1313. doi: 10.1590/S1517-838220110004000010
Riccio, M. L., Pallecchi, L., Fontana, R., and Rossolini, G. M. (2001). In70 of plasmid pAX22, a bla(VIM-1)-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrob. Agents Chemother. 45, 1249–1253. doi: 10.1128/AAC.45.4.1249-1253.2001
Roe, M. T., Vega, E., and Pillai, S. D. (2003). Antimicrobial resistance markers of class 1 and class 2 integron-bearing Escherichia coli from irrigation water and sediments. Emerg. Infect. Dis. 9, 822–826. doi: 10.3201/eid0907.030065
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual, 2nd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Saravanan, R., and Raveendaran, V. (2013). Antimicrobial resistance pattern in a tertiary care hospital: an observational study. J. Basic Clin. Pharm. 4, 56–63. doi: 10.4103/0976-0105.118797
Scaletsky, I. C., Souza, T. B., Aranda, K. R., and Okeke, I. N. (2010). Genetic elements associated with antimicrobial resistance in enteropathogenic Escherichia coli (EPEC) from Brazil. BMC Microbiol. 27:25. doi: 10.1186/1471-2180-10-25
Sepp, E., Stsepetova, J., Loivukene, K., Truusalu, K., Koljalg, S., Naaber, P., et al. (2009). The occurrence of antimicrobial resistance and class 1 integrons among commensal Escherichia coli isolates from infants and elderly persons. Ann. Clin. Microbiol. Antimicrob. 8, 8–34. doi: 10.1186/1476-0711-8-34
Skurnik, D., Le Menac'h, A., Zurakowski, D., Mazel, D., Courvalin, P., Denamur, E., et al. (2005). Integron-associated antimicrobial resistance and phylogenetic grouping of Escherichia coli isolates from healthy subjects free of recent antibiotic exposure. Antimicrob. Agents Chemother. 49, 3062–3065. doi: 10.1128/AAC.49.7.3062-3065.2005
Soto, S. M., Smithson, A., Martinez, J. A., Horcajada, J. P., Mensa, J., and Vila, J. (2007). Biofilm formation in uropathogenic E. coli. strains: relationship with prostatitis, urovirulence factors and antimicrobial resistance. J. Urol. 177, 365–368. doi: 10.1016/j.juro.2006.08.081
Su, J., Shi, L., Yang, L., Xiao, Z., Li, X., and Yamasaki, S. (2006). Analysis of integrons in clinical isolates of Escherichia coli in China during the last six years. FEMS Microbiol. Lett. 254, 75–80. doi: 10.1111/j.1574-6968.2005.00025.x
Sunde, M., and Norstrom, M. (2006). The prevalence of, associations between and conjugal transfer of antibiotic resistance genes in Escherichia coli isolated from Norwegian meat and meat products. J. Antimicrob. Chemother. 58, 741–747. doi: 10.1093/jac/dkl294
Tarchouna, M., Ferjani, A., Tilouche, S., Marzouk, M., Bouguila, J., Boughamoura, L., et al. (2014). Screening at admission for carrier prevalence of multidrug- resistant organisms in a pediatric unit. Am. J. Infect. Control. 42, 88–89. doi: 10.1016/j.ajic.2013.07.008
Tilak, G. P., and Mudaliar, J. L. (2012). Role of enteropathogenic Escherichia coli in paediatric diarrhoeas in South India. Mater. Sociomed. 24, 178–181. doi: 10.5455/msm.2012.24.178-181
Torres, A., and Payne, S. (1997). Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23, 825–833. doi: 10.1046/j.1365-2958.1997.2641628.x
Touchon, M., Hoede, C., Tenaillon, O., Barbe, V., Baeriswyl, S., Bidet, P., et al. (2009). Organised genome dynamics in Escherichia coli results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. doi: 10.1371/journal.pgen.1000344
Trabulsi, L. R., Keller, R., and Tardelli Gomes, T. A. (2002). Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8, 508–513. doi: 10.3201/eid0805.010385
Tsukamoto, T., Kimoto, T., Magalhaes, M., and Takeda, Y. (1992). Enteroadherent Escherichia coli exhibiting localized pattern of adherence among infants with diarrhoea in Brazil–incidence and prevalence of serotypes. Kansenshogaku Zasshi 66, 1538–1542.
Van de Klundert, J. A. M., and Vliegenthart, J. S. (1993). “PCR detection of genes coding for aminoglycoside-modifying enzymes,” in Diagnostic Molecular Microbiology, eds D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (Washington, DC: American Society for Microbiology), 547–552.
Vidal, R., Vidal, M., Lagos, R., Levine, M., and Prado, V. (2004). Multiplex PCR for diagnosis of enteric infections associated with diarrheagenic Escherichia coli. J. Clin. Microbiol. 42, 1787–1789. doi: 10.1128/JCM.42.4.1787-1789.2004
Vinue, L., Saenz, Y., Somalo, S., Escudero, E., Moreno, M. A., Ruiz-Larrea, F., et al. (2008). Prevalence and diversity of integrons and associated resistance genes in faecal Escherichia coli isolates of healthy humans in Spain. J. Antimicrob. Chemother. 62, 934–937. doi: 10.1093/jac/dkn331
Wang, G., Clark, C. G., and Rodgers, F. G. (2002). Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J. Clin. Microbiol. 40, 3613–3619. doi: 10.1128/JCM.40.10.3613-3619.2002
Wang, L., Wakushima, M., Aota, T., Yoshida, Y., Kita, T., Maehara, T., et al. (2013). Specific properties of enteropathogenic Escherichia coli isolates from diarrheal patients and comparison to strains from foods and fecal specimens from cattle, swine, and healthy carriers in Osaka City, Japan. Appl. Environ. Microbiol. 79, 1232–1240. doi: 10.1128/AEM.03380-12
Wang, Q., Wang, H., Yu, Y., Xu, X., Sun, Z., Lu, J., et al. (2015). Antimicrobial resistance monitoring of gram-negative bacilli isolated from 15 teaching hospitals in 2014 in China. Zhonghua Nei Ke Za Zhi 54, 837–845. doi: 10.3760/cma.j.issn.0578-1426.2015.10.005
White, P. A., McIver, C. J., and Rawlinson, W. D. (2001). Integrons and gene cassettes in the enterobacteriaceae. Antimicrob. Agents Chemother. 45, 2658–2561. doi: 10.1128/AAC.45.9.2658-2661.2001
Yaffee, A. Q., Roser, L., Daniels, K., Humbaugh, K., Brawley, R., Thoroughman, D., et al. (2016). Notes from the field: verona integron-encoded metallo-beta-lactamase-producing carbapenem-resistant Enterobacteriaceae in a neonatal and adult intensive care unit—Kentucky, 2015. MMWR Morb. Mortal. Wkly. Rep. 65, 190. doi: 10.15585/mmwr.mm6507a5
Yu, H., Lee, J., Kang, H., Ro, D. W., Chung, J. Y., Jeong, Y. S., et al. (2003). Changes in gene cassettes of class 1 integrons among Escherichia coli isolates from urine specimens collected in Korea during the last two decades. J. Clin. Microbiol. 41, 5429–5433. doi: 10.1128/JCM.41.12.5429-5433.2003
Yu, J., and Kaper, J. B. (1992). Cloning and characterization of the eae gene of enterohaemorrhagic Escherichia coli Oˆ57:H7. Mol. Microbiol. 6, 411–417. doi: 10.1111/j.1365-2958.1992.tb01484.x
Keywords: enteropathogenic Escherichia coli, phylogenetic groups, multidrug resistance, integrons, gene cassettes
Citation: Singh T, Das S, Ramachandran VG, Wani S, Shah D, Maroof KA and Sharma A (2017) Distribution of Integrons and Phylogenetic Groups among Enteropathogenic Escherichia coli Isolates from Children <5 Years of Age in Delhi, India. Front. Microbiol. 8:561. doi: 10.3389/fmicb.2017.00561
Received: 13 October 2016; Accepted: 20 March 2017;
Published: 10 April 2017.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Pramod Kumar, National Centre for Disease Control (NCDC), IndiaChew Chieng Yeo, Universiti Sultan Zainal Abidin, Malaysia
Copyright © 2017 Singh, Das, Ramachandran, Wani, Shah, Maroof and Sharma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shukla Das, c2h1a2xhZGFzXzEyM0B5YWhvby5jb20=
 Taru Singh
Taru Singh Shukla Das
Shukla Das V. G. Ramachandran2,3
V. G. Ramachandran2,3 Sayim Wani
Sayim Wani Dheeraj Shah
Dheeraj Shah Khan A. Maroof
Khan A. Maroof Aditi Sharma
Aditi Sharma