- 1State Key Laboratory of Microbial Technology, School of Life Science, Shandong University, Jinan, China
- 2College of Marine Life Sciences, Ocean University of China, Qingdao, China
Chaperonin GroEL (Cpn60) requires cofactor GroES (Cpn10) for protein refolding in bacteria that possess single groEL and groES genes in a bicistronic groESL operon. Among 4,861 completely-sequenced prokaryotic genomes, 884 possess duplicate groEL genes and 770 possess groEL genes with no neighboring groES. It is unclear whether stand-alone groEL requires groES in order to function and, if required, how duplicate groEL genes and unequal groES genes balance their expressions. In Myxococcus xanthus DK1622, we determined that, while duplicate groELs were alternatively deletable, the single groES that clusters with groEL1 was essential for cell survival. Either GroEL1 or GroEL2 required interactions with GroES for in vitro and in vivo functions. Deletion of groEL1 or groEL2 resulted in decreased expressions of both groEL and groES; and ectopic complementation of groEL recovered not only the groEL but also groES expressions. The addition of an extra groES gene upstream groEL2 to form a bicistronic operon had almost no influence on groES expression and the cell survival rate, whereas over-expression of groES using a self-replicating plasmid simultaneously increased the groEL expressions. The results indicated that M. xanthus DK1622 cells coordinate expressions of the duplicate groEL and single groES genes for synergistic functions of GroELs and GroES. We proposed a potential regulation mechanism for the expression coordination.
Introduction
GroEL proteins belong to the chaperonin of the chaperonin-60 family (Cpn60) that assists in protein folding, assembly, transport, and degradation (Ranson et al., 1998; Lund, 2001), and are essential for many physiological processes in cells (Houry et al., 1999). GroEL is usually a major heat-shock protein that is over-produced under non-permissive temperatures and aids in proper refolding of the proteins denatured by heat shock (Vanbogelen et al., 1987; Fayet et al., 1989; Kerner et al., 2005). The GroEL protein is a Type I chaperonin and is typically characterized by the formation of a 14-mer homopolymer arranged as two back-to-back/stacked rings, each of which comprises seven subunits (Horwich et al., 2006). In bacteria with single groEL genes, the co-chaperonin GroES, belonging to the chaperonin of the chaperonin-10 family (Cpn10) and encoded by the bicistronic groESL operon, forms a heptamer to bind to the GroEL homopolymer in the presence of ATP, forming a large central cavity that encapsulates substrate proteins and enables correct folding through multiple cycles of binding and release (Saibil and Ranson, 2002).
While most bacteria contain single copies of groEL, some possess two or more highly conserved groEL genes in their genomes (Craig et al., 1993; Kong et al., 1993; Karlin and Brocchieri, 2000; Gould et al., 2007; Jiang et al., 2008; Wang et al., 2013, 2014). In bacteria possessing single groEL genes, groES is always present upstream groEL in a bicistronic operon (groESL); in species with duplicate groEL genes, not all groELs are preceded by a groES gene (Lund, 2009). There are two possibilities for functioning manner of the stand-alone groEL: the encoded protein has evolved into a GroES-independent chaperonin or kept to function in a GroES-dependent manner. Goyal et al. suggested that, groES might be selectively lost from the operon after duplication of groESL, resulting in stand-alone groEL genes that then evolved to function in a groES-independent manner (Goyal et al., 2006). In Anabaena L-31, there are a groESL operon and a single groEL gene. Potnis et al. (2016) found that Anabaena GroEL1 formed stable higher oligomer (>12-mer) and prevented thermal aggregation of malate dehydrogenase (MDH), a substrate that requires a chaperonin for refolding following denaturation (Kumar et al., 2015). The refolding activity of Anabaena GroEL1 was lower than that of Escherichia coli GroEL, but independent of both GroES and ATP; and the presence of GroES enhanced the ATPase activity of Anabaena GroEL (Potnis et al., 2016). Qamra et al. also determined that the cpn60.2 (a GroEL homolog) proteins of Mycobacterium tuberculosis were able to polymerize into smaller oligomers, which alone retained the ability to suppress the aggregation of substrate proteins (Qamra et al., 2004). Interestingly, Kong et al. observed an independent expression of the groES and groEL genes in an operon in M. tuberculosis (Kong et al., 1993). However, it is still largely unknown whether stand-alone groEL genes require groES in order to function and, if required, how duplicate groELs and a single groES balance their expressions.
Myxobacteria are phylogenetically located in the delta division of Proteobacteria. These bacteria are distinguished by their complex multicellular behaviors, such as moving in swarms on solid surfaces, cooperative feeding on macromolecules or other microbial cells, and the development of multicellular fruiting bodies embodied with numerous adversity-resistant myxospores (Shimkets, 1990; Dworkin and Kaiser, 1993). Myxococcus xanthus DK1622 is a model strain of myxobacteria. The bacterium contains two groEL genes; groEL1 appears in a groESL operon, while groEL2 has no neighboring groES (Goldman et al., 2006; Li et al., 2010). The two groEL genes are alternatively deletable for cell growth, but function divergently in specific cellular processes: groEL1 is essential for the development, while groEL2 is required for cell predation and the biosynthesis of secondary metabolite myxovirescin (Li et al., 2010; Wang et al., 2013, 2014). The expression of groEL2 is usually less than half of groEL1 in either the growth or the developmental stage. However, in the mutant with the deletion of groEL1 (YL0301), the expression of the groEL2 gene increased more than two-fold of that in wild-type strain DK1622; whereas the expression of groEL1 in the groEL2 deletion mutant (YL0302) was similar to that in the wild-type strain DK1622 (Li et al., 2010). In response to heat shock, the expression of the groEL genes was regulated in a complex manner in groEL deletion and complementary mutants, which led to the total expressions of groEL to approach the expression level in the wild type strain (Wang et al., 2013). Seemly, there is a balance of the expressions of groEL genes in Myxococcus cells for their cellular functions. In this study, comparative genomics analysis revealed that stand-alone groEL gene often appears in bacteria possessing two or more groEL genes. Using M. xanthus DK1622 as a model, we determined that the single groES gene was essential for cell survival, and both GroEL1 and GroEL2 required GroES in order to function. The duplicate groEL genes and the single groES gene are expressed in a coordinated manner for their synergetic functions in M. xanthus cells. A self-regulatory mechanism was proposed to explain how a single groES gene simultaneously meets the requirements of two endogenous groEL genes.
Materials and Methods
Cultures, Plasmids, and Growth Conditions
The strains, plasmids and primers employed in this study are listed in Supplementary Tables 1, 2. The M. xanthus strains were cultivated in Casitone-based rich-nutrient CTT medium (Hodgkin and Kaiser, 1977) for growth assays. E. coli strains were routinely grown on Luria-Bertani (LB) agar or in LB broth. The incubation temperatures were 37°C and 30°C for E. coli cells and M. xanthus cells, respectively. When required, final concentrations of 40 μg/ml kanamycin, 100 μg/ml ampicillin, 34 μg/ml chloramphenicol, and 10 μg/ml tetracycline were added to the media.
Bioinformatics Analysis of the Occurrence of groEL and groES Genes in Prokaryotic Genomes
The two groEL genes and one groES gene in the M. xanthus DK1622 genome are MXAN_RS23765 (groEL1), MXAN_RS21695 (groEL2), and MXAN_RS23760 (groES). The NCBI accession numbers for the GroEL1, GroEL2, and GroES protein sequences of M. xanthus DK1622 are WP_011554876.1, WP_011554465.1, and WP_002640434.1, respectively. The conserved domain information of GroEL and GroES was obtained from the CDD protein family (cdd239460 for GroEL and cdd238197 for GroES of M. xanthus DK1622, respectively; Marchler-Bauer et al., 2017). Based on the sequence similarity of the conserved domains, we performed RPS-BLAST (Reverse Position-Specific BLAST), which searches against the NCBI genome database (http://www.ncbi.nlm.nih.gov/genome/), and retrieved the superset of all the GroEL and GroES protein sequences in prokaryotic genomes. The E-value for the search was set to be equal to or higher than the default cutoff of 0.01.
A whole-genome-based myxobacterial phylogenetic tree was constructed online utilizing the CVTree3 program and a composition vector approach (Zuo and Hao, 2015).
Deletion of groES
Deletion of groES was carried out in M. xanthus DK1622 using the positive-negative KG cassettes (Ueki et al., 1996). Genomic DNA from DK1622 served as the template for PCR amplification of the upstream and downstream homologous arms utilizing Pfu DNA polymerase. The arms were fused at the BamHI site to create internal deletion fragments, which were cloned into SmaI-digested pBJ113, forming the deletion plasmid pBJ-groES. The deletion plasmid was transferred via electroporation into M. xanthus DK1622 cells as previously described (Kashefi and Hartzell, 1995). Individual Km-resistant colonies were selected and inoculated onto CTT agar plates supplemented with 1% galactose for a second round of screening. Deletion mutants were identified based on their galactose resistance and kanamycin sensitivity phenotypes, as well as by PCR and sequencing verification (Li et al., 2010).
Complementary Inactivation Experiments to Knock Out groES
The groES gene was fused to the pilA promoter (1 kb), and the fused sequence was digested with XbaI and EcoRI and ligated into the site-specific integration plasmid pSWU30, which had been digested with XbaI and EcoRI, to obtain the plasmid pSWU-groES. This plasmid was transferred via electroporation into M. xanthus DK1622 cells as previously described (Kashefi and Hartzell, 1995). Individual tetracycline-resistant colonies were selected, and the selected mutant was designated YL0308. The mutant was further employed to delete the local groES gene via electroporation with pBJ-groES. The kmr and terr mutants were selected, and proper integration of the fragment was confirmed by PCR and sequencing (Lobedanz and Sogaard-Andersen, 2003).
Insertion of groES before groEL2
The groES gene, including the groES-groEL1 intergenic region, was fused to the promoter (500 bp) of groEL2, which was then fused upstream of groEL2. The fused fragments were digested with XbaI and EcoRI and ligated into the site-specific integration plasmid pSWU30, which was digested with XbaI and EcoRI to obtain the plasmid pSWU-PgroEL2+groES+groEL2. This plasmid was transferred via electroporation into groEL2 deletion mutant YL0302 cells. Individual colonies with tetracycline-resistance were selected, and the mutant was designated YL1101. All primers mentioned above are listed in Supplementary Table 2.
Construction of groES Over-Expression Mutants via the pZJY41 Plasmid
The promoter of either groEL2 or groEL1 was fused to the groES gene, and the fused fragments were digested with BamHI and EcoRI and ligated into the Myxococcus-E. coli shuttle plasmid pZJY41 (which had also been digested with BamHI and EcoRI) to obtain the plasmids pZJY-PgroEL2+groES and pZJY-PgroEL1+groES. The constructed plasmids were then transferred via electroporation into M. xanthus DK1622 cells, and the mutants were designated YL1102 and YL1103, respectively (Supplementary Table 1).
Protein Purification
The M. xanthus groEL and groES genes were separately cloned into pET22b and over–expressed in E. coli BL21(DE3) cells. The heterogeneously expressed GroELs and GroES were purified as described (Klunker et al., 2003), with some modifications. Briefly, BL21 cells were grown at 37°C in LB medium with 100 μg/ml ampicillin to an OD600 of 0.7 and induced with 1 mM IPTG. Cells were resuspended in lysis buffer (25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 2 mM dithiothreitol) and lysed via sonication. After gentle ultrasonication, the mixtures were centrifuged at 4°C for 30 min (12,000 rpm). The supernatant containing His-tagged GroES was applied to a Ni2+-NTA column and purified by Superdex 200 chromatography. His-tag sequences were not added to the GroEL proteins to maintain target protein activity. The supernatants containing GroEL were concentrated using tubes with a 100-kDa cut-off and were further purified by chromatography (Superdex 200).
ATPase Assay
An ATPase assay was performed as previously described (Figueiredo et al., 2004) with minor modifications. Briefly, 1 μM GroEL1 and GroEL2 were individually added to a reaction buffer containing 20 mM MOPS, pH 7.5, 100 mM KCl, 5 mM MgCl2, and 0.5 M ammonium sulfate with or without 2 μM GroES at 37°C. The reaction was initiated by adding 2 mM ATP and stopped with 20 mM CDTA after 10 min. Liberated inorganic phosphate was quantified by the malachite green assay (Adenosinetriphosphatase assay kit from Nanjing Jiancheng Bioengineering Institute, China) after incubation at 25°C for 30 min. The absorption was measured at 640 nm. Three independent replicates were performed for each experiment.
Refolding Assay
Twenty micromolars malate dehydrogenase (MDH) was denatured in 1 M guanidinium HCl for 60 min, and the reaction system was diluted 100-fold with buffer A (50 mM Tris-HCl, pH 7.0, 0.5 M ammonium sulfate, and 2 mM ATP). Next, 1 μM GroEL1 and GroEL2 were separately added into buffer A with or without 2 μM GroES. The concentration of added chaperonin was determined by NanoDrop 2000 Spectrophotometer. The reaction mixture was incubated at room temperature for 60 min (Figueiredo et al., 2004). Fifty millimolars CDTA was added to stop the reaction. The enzymatic activity of MDH was measured every 10 min at 340 nm using buffer B, containing 50 mM Tris-HCl, pH 7.0, and 0.5 M ammonium sulfate, 1 mg/ml bovine serum albumin, 0.22 mM NADH, 0.55 mM oxaloacetate, 1 mM CDTA. For citrate synthase (CS) refolding assay, after a similar process, the enzymatic activity of CS was determined every 10 min at 412 nm using the Citrate Synthase Activity Assay Kit (Sigma-Alorich). Three independent replicates were performed for each experiment.
Protein Pull-Down Assay
Proteins were purified as described above. Excess amount of GroEL1 (20 mg) or GroEL2 (20 mg) proteins was mixed with 1–4 mg His-tagged GroES in suspension buffer (25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 2 mM dithiothreitol, and 2 mM ATP) for 3 h. The mixtures were eluted through nickel columns. Then the columns were washed three times using washing buffer, and the bound proteins were eluted from the columns with 500 mM imidazole and detected by SDS-PAGE.
Native PAGE
Hetero-expression of the duplicate GroELs and GroES was performed using pET22b-GroEL1-NoHis, pET22b-GroEL2-NoHis, and pET28a-GroES-NoHis respectively (Supplementary Table 1), in E. coli BL21(DE3) cells. BL21 cells were treated as described as above. After gentle ultrasonic treatment, cells were centrifuged at 4°C for 30 min (12,000 rpm) for the native PAGE assay. Compared to normal sodium dodecyl sulfate (SDS)-PAGE, the protein buffer and gel for native PAGE contained no SDS or mercaptoethanol. Electrophoresis was performed in a mixture of ice and water to avoid the depolymerization of multimer proteins.
In vivo Refolding Assay
To investigate the in vivo refolding activities of GroEL1/2 proteins with or without the presence of GroES, we constructed the plasmids pET22b-HrcA, pBAD33-GroESL1/2, and pBAD33-GroEL1/2 (Supplementary Figure 1). The compatible plasmids pET22b-HrcA and pBAD33-GroESL1/2 (or pBAD33-GroEL1/2) were transformed into E. coli Top 10F' cells (Supplementary Table 1), and transformants were selected under the double-resistance conditions of 34 μg/ml chloramphenicol and 100 μg/ml ampicillin. When the culture OD600 reached 0.7, 100 mM IPTG was added for the induction. Then, cells were harvested, resuspended in lysis buffer containing 25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 2 mM dithiothreitol, and lysed via sonication. After gentle ultrasonication, the mixtures were centrifuged at 4°C for 30 min (12,000 rpm), which divided the mixture into supernatants and sediments. The samples were tested the expression and solubility of HrcA protein by SDS-PAGE.
Quantitative Real-Time PCR Analysis
M. xanthus DK1622 cells and groEL mutants were inoculated at final concentration of 1 × 107 cells/ml in CTT medium and cultured for 36 h to the exponential growth stage, respectively. Then 0.5 ml of each of the cultures was transferred into 50 ml fresh CTT medium for further cultivation. The cultures were harvested at periodic time points, and the RNA was extracted immediately using a bacterial RNA extraction kit (Thermo Fisher) according to the manufacturer's instructions. The purified RNA extracts were reverse-transcribed to cDNA. Quantitative real-time PCR was performed in a total reaction volume of 20 μl containing 1 μl of 250 nM primers, 10 μl of SYBR Green PCR master mix, 8.5 μl of RNase-free water, and 0.5 μl of a 10-fold-diluted cDNA template. 16S rRNA gene was used as the reference. The primers used for real-time PCR are listed in Supplementary Table 2.
Heat-Shock Assays
Mid-log-phase cultures of M. xanthus cells were harvested as described above. After heat-shock treatment at 42°C for 30 min or 60 min, the cells were immediately serially diluted and plated on 1.5 and 0.3% CTT agar. After 6 days, colony-forming unit (CFU) numbers were calculated, using untreated cells as a control. The survival rate was calculated as CFU divided by the cell number before heat shock (5 × 109; Li et al., 2010).
Statistical Analysis
The difference significance was analyzed statistically by using IBM SPSS Statistics for independent-samples T-test.
Results
Stand-Alone groEL Gene Often Appears in Bacteria Possessing Duplicate groEL Genes
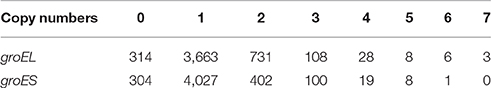
We surveyed the occurrence of groEL and groES genes by RPS-BLAST searching based on sequence similarity, and retrieved a total of 5,658 groEL and 5,253 groES genes in 4,861 completely sequenced prokaryotic genomes (Supplementary Table 3). The number of groEL genes in a genome range from zero to seven, and 93.5% of these genomes contain one or more groEL genes (Table 1). Consistent with the previous report (Williams and Fares, 2010), Mycoplasma often lack the groEL genes (67 of the 105 sequenced genomes). In addition, some Spiroplasma, Ureaplasma, and unclassified bacteria have no groEL gene. In Archaea, strains from Euryarchaeota and TACK group also lack groEL. In the genomes with single groELs, the groEL gene is always neighbored with a groES gene, forming a bicistronic groESL operon. There are 884 bacterial genomes possessing two or more groEL genes (19.5% of the 4,861 sequenced genomes). Of the total 5,658 groEL genes, 4,824 were found closely downstream a groES gene in a groESL operon, while the other 834 (14.7% of groEL genes) have no neighboring groES gene. The stand-alone groEL genes are distributed in 770 genomes; thus, some genomes contain two or more stand-alone groEL genes. This is consistent with a previous survey of 669 bacterial genomes in which the majority of groEL genes co-occurred with a neighboring groES gene, while others stood alone (Lund, 2009). Notably, of the total 5,253 groES genes, 435 stand alone with no neighboring groEL gene; these genes are distributed across 419 genomes.
There are 24 sequenced myxobacterial genomes available in the GenBank database. Among them, 19 genomes contain two groEL genes, two contain three groEL genes, and three contain single groEL genes. Figure 1 is a diagrammatic sketch of the occurrence of groEL and groES genes in myxobacteria (details referred to Supplementary Table 4). All myxobacterial genomes possess at least one complete bicistronic groESL operon. Interestingly, the loss of a neighboring groES is rather a characteristic of duplicate groEL genes in the Cystobacterineae suborder of myxobacteria, while the four sequenced genomes of Sorangium and Chondromyces in the Sorangineae suborder and the marine halophilic strain Haliangium ochraceum DSM14365 contain a second groESL operon. Of the two genomes possessing three copies of groEL, the Sorangineae strain (S. cellulosum So0157-2) has two complete groESL operons and a stand-alone groEL, while the Cystobacterineae strain (C. fuscus DSM2262) has a complete groESL operon and two stand-alone groEL genes. The three myxobacteria with single groEL genes are newly identified myxobacterial strains that are phylogenetically distant from “the classical myxobacteria.” Although normally coinciding at a high taxonomic level, the occurrence of the groES and groEL genes may vary at the level of genus or even the species. For example, while one of the two groEL genes in Cystobacter violaceus Cb vi76 neighbors groES and one does not, the C. fuscus DSM 2262 strain has three groEL genes, of which one neighbors a groES gene and two stand alone. S. cellulosum So ce56 possesses two complete groESL clusters, but S. cellulosum So0157-2 contains two groESL clusters and a third stand-alone groEL gene.

Figure 1. A phylogenomic tree constructed using 24 sequenced myxobacterial genomes. The bar equals to 0.05 of the phylogenetic distance. The occurrences of groEL and groES genes in each genome were correspondingly integrated into the tree. Blue, red, and white arrows indicate groES, groEL and flanking genes, respectively.
The groES Gene is Essential for the Cell Survival of M. xanthus DK1622
M. xanthus DK1622 possesses two groEL genes that share 79% similarity of their amino acid sequences (Goldman et al., 2006; Li et al., 2010). Based on our previous studies, a single groEL1 or groEL2 gene is capable of individually supporting the growth of DK1622 cells, but these two genes function divergently in predation and development processes (Li et al., 2010; Wang et al., 2013). Another significant characteristic of the two paralogous groEL genes is their organization differences in genome. The groEL1 gene (MXAN_RS23765) lies downstream of groES, forming a complete bicistronic groESL operon, verified using RT-PCR (referred to Supplementary Figure 2), while groEL2 (MXAN_RS21695) does not neighbor with a groES gene.
We attempted to knock out the groES gene but failed, suggesting that inactivation of the groES gene is fatal to DK1622 cells. To confirm the essentiality of groES, we complemented this gene at the Mx8 attB site in the DK1622 genome using pSWU30 to form a mutant with two copies of the groES gene (strain YL0308). We further attempted to delete the original groES from YL0308, and, as expected, obtained a viable mutant (YL0309), having an ectopically expressed groES gene and the stand-alone genes groEL1 and groEL2. The YL0309 mutant had a nearly identical growth curve to the wild-type strain DK1622 (P > 0.05 for each detected time point; Figure 2). Thus, the groES gene is essential for the cell survival of M. xanthus DK1622. The groEL1 and groEL2 genes were alternatively deletable for cell survival, which suggested that GroEL2 also required GroES in order to function. M. xanthus cells were likely able to self-regulate groEL and groES gene expression to meet cellular requirements for these proteins.
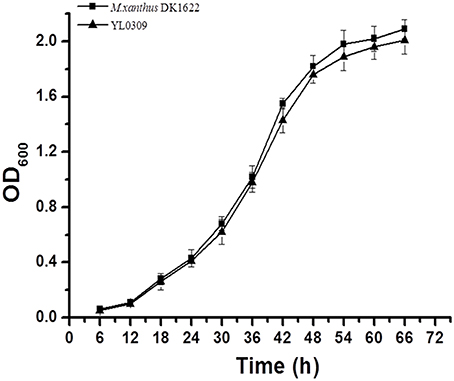
Figure 2. Growth curves of the wild-type strain DK1622 and the groES-deletion mutant YL0309 complemented with an ectopic groES gene. Error bars represent standard deviation of three time repeats.
GroES is Able to Bind to Either GroEL1 or GroEL2 and Is Required for GroEL Refolding Activities In vitro
To ascertain the relationship between the two GroELs and the single GroES protein of M. xanthus DK1622, we hetero-expressed these genes in E. coli cells and purified the target proteins. The in vitro binding activities of GroES and GroEL1/GroEL2 were assayed using two methods. We designed GroES with a His-tag at the C-terminus, which made the protein to be able to bind to a nickel column. The GroEL proteins had no His-tag and were unable to bind to the nickel column by themselves. When added to a GroES-containing column, either GroEL1 or GroEL2 was able to retain on the column (Figure 3A). As the concentration of the column-bound GroES increased, the recovery of GroEL proteins by the column also increased. Furthermore, GroEL is an ATPase, and binding to GroES reduces the ATPase activity of GroEL proteins (Martin et al., 1991; Viitanen et al., 1991). After the addition of GroES, the ATPase activities of the GroEL1 and GroEL2 proteins decreased to a similar extent (P < 0.001 and 0.001 < P < 0.01 for GroEL1 and GroEL2, respectively; Figure 3B). The above results confirmed the binding activities of both GroEL1 and GroEL2 to GroES in vitro. Based on the results in Figure 3B, GroEL1 showed higher in vitro ATPase activity than GroEL2 in the absence or presence of GroES, suggesting their disparate refolding activities in cells.
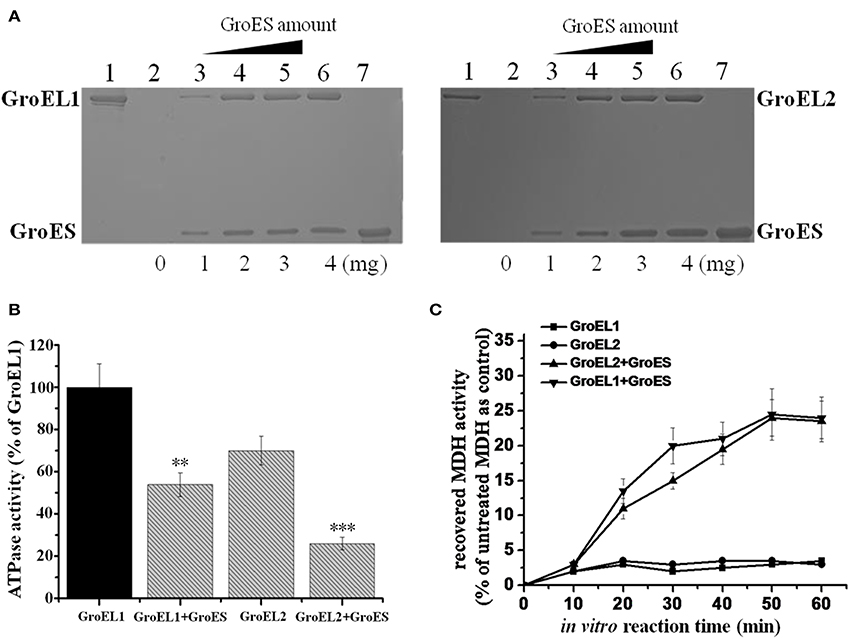
Figure 3. In vitro binding and refolding activities of GroELs and GroES from M. xanthus DK1622. (A) GroEL1 (left) and GroEL2 (right) proteins bound by the His-tag GroES-containing nickel column. Bound GroEL-GroES complexes were eluted from the column with 500 mM imidazole and assayed by SDS-PAGE. Line 1, GroEL1 and GroEL2 proteins that passed through the His-tag GroES column; lines 2–6, increased His-tag GroES bound by the nickel column (0-4 mg) with the same amounts of GroEL proteins for each line; line 7, GroES control with no GroEL. (B) ATPase activities of GroELs in the absence or presence of GroES. For statistical analysis, ***P < 0.001 and **P < 0.01, respectively. (C) MDH renaturation activities of GroELs in the absence or presence of GroES. Error bars in the pictures represent standard deviation of three time repeats.
We assayed the in vitro refolding activities of the two GroELs with the malate dehydrogenase (MDH) protein. In the presence of GroEL1 or GroEL2, but without GroES, denatured MDH exhibited slight renaturation. When GroES was added, the MDH-folding activity of GroEL1 had a slightly higher rate than that of GroEL2 after 30 min (P < 0.05), but the final refolding activities were nearly identical (~25% re-natured MDH) for both GroEL1 and GroEL2 (Figure 3C). In addition, we also performed the refolding assay on citrate synthase (CS), which showed similar result as the MDH refolding (Supplementary Figure 3).
GroES Forms Polymer Complexes with Either GroEL1 or GroEL2 and Aids GroEL Refolding Functions In vivo
We estimated the in vivo polymerization of the Myxococcus GroEL1 and GroEL2 proteins in E. coli cells in the presence or absence of the GroES. As the native-PAGE shown, the alone-expressed GroEL1 and GroEL2 proteins had a molecular weight (Mw) of ~800 kDa, respectively (the lanes 3–4 and 6–7 in Figure 4A). The molecular weight is about 14-times of that of the GroEL proteins (the Mws of GroEL1 and GroEL2 are 57.9 and 58.1 kDa, respectively, and their 14-mer polymers are 810.6 and 813.4 kDa, respectively). Similarly, the only-expressed GroES proteins formed a 7-mer complex (lanes 1–2 in Figure 4B; the Mw of GroES is 10.7 kDa, and the Mws of 7-mer GroES are 74.9 kDa). Co-polymers of GroEL and GroES were also observed in E. coli cells containing co-expressed groEL1/groEL2 and groES genes of M. xanthus DK1622. The GroEL1-GroES and GroEL2-GroES complexes formed retarded bands with a similar molecular size (lanes 1–2 and 8–9 in Figure 4A; the Mws of 7-mer GroES plus 14-mer GroEL1 or 14-mer GroEL2 are 885.5 or 888.3 kDa, respectively). These results suggested that the individual GroEL1 and GroEL2 proteins both existed as 14-mer homopolymers, each of which was able to bind the 7-mer of GroES in vivo.
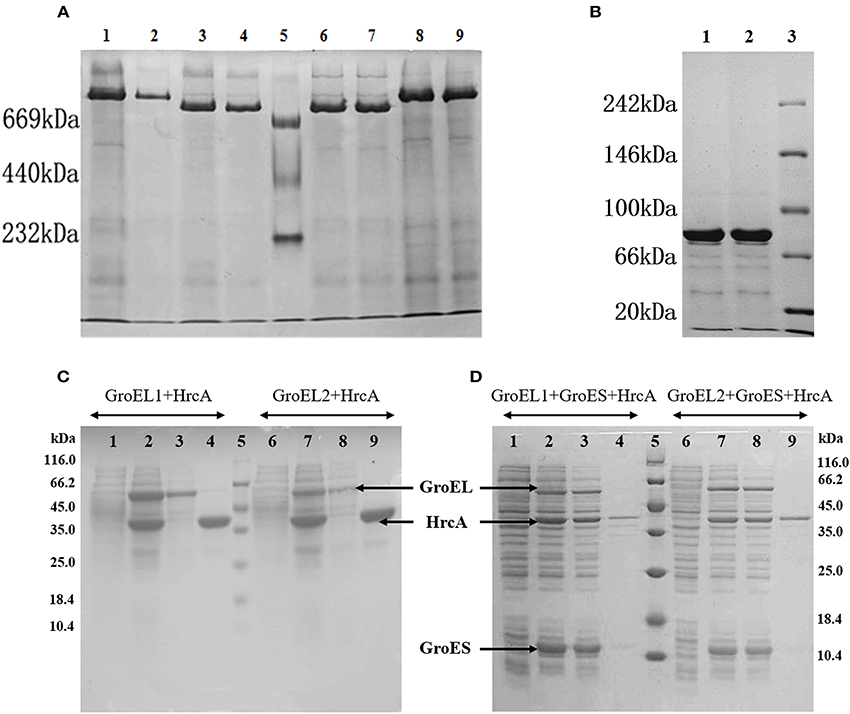
Figure 4. In vivo forms and refolding activities of GroELs and GroES proteins from M. xanthus DK1622 in E. coli cells. (A,B) Native-PAGE analysis of extracts from E. coli cells expressing various arrays of DK1622 groEL and groES genes. (A) Lanes 1–2, co-expression of GroEL1 and GroES; lanes 3–4, expression of GroEL1 alone; lane 5, native protein markers; lanes 6–7, expression of GroEL2 alone; lanes 8–9, co-expression of GroEL2 and GroES. (B) Lanes 1–2, expression of GroES alone; lane 3, native protein markers. Each sample was repeated twice in gel for reliability of the Native-PAGE results. (C,D) SDS-PAGE analysis of co-expression of hrcA with groEL1 (left) or groEL2 (right) in the absence and presence of groES in E. coli cells, respectively. Lane 1, 6, before induction with IPTG; lane 2, 7, after induction; lane 3, 8, supernatants; lane 4, 9, sediments; lane 5, Mw markers. Ten microlitres were loaded for each lane.
HrcA is a negative regulatory protein that is able to bind to the CIRCE (Controlling Inverted Repeat of Chaperone Expression) regions in front of the groESL operons and thus reduce the expression of groEL and groES genes (Wilson et al., 2005). The HrcA protein relies on GroEL to complete refolding and is a potential GroEL substrate. We assayed the in vivo refolding functions of the two GroEL proteins with or without GroES by expressing the groEL, groES and hrcA genes from M. xanthus in different combinations in E. coli cells. SDS-PAGE result indicated that in the absence of the DK1622 groES gene, HrcA primarily existed in the form of inclusion bodies in cells when co-expressed with only the groEL1 or groEL2 gene from M. xanthus (Figure 4C). However, when the DK1622 groES gene was further introduced into in the E. coli cells containing the hrcA and groEL1/groEL2 genes from M. xanthus, the HrcA protein was then mostly soluble (Figure 4D). These results suggested that both GroEL1 and GroEL2 proteins from M. xanthus were able to aid the refolding of the HrcA proteins in the presence of the M. xanthus GroES in E. coli cells.
M. xanthus Cells Coordinate Expressions of Single groES and Duplicate groELs
The above results indicated that either GroEL1 or GroEL2 requires GroES in order to function in M. xanthus. The two groELs and the single groES had to coordinate their expressions for synergy. Our previous studies showed that the two groELs had a balance of their expressions in M. xanthus cells (Li et al., 2010; Wang et al., 2013). In this study, we further assayed expressions of the groES gene in the wild type strain DK1622, the groEL1/2 deletion mutants (YL0301 and YL0302) and their corresponding complementary strains (YL0901 and YL0902) by using quantitative PCR. In the wild type strain DK1622, groES gene expression increased and reached its maximum during the first 24 h of incubation in CTT medium. Then, the expression decreased and reached its lowest level after ~42–48 h of incubation (Figure 5). There was a slight increase in groES expression in the late stationary growth stage. This groES expression curve was highly consistent with that of the groEL genes in DK1622 (Li et al., 2010). Deletion of the groEL2 gene, which has no neighboring groES gene, resulted in the decreased expression of the groES gene at each time point. This result is similar to those obtained following the deletion of the groEL1 gene (Figure 5). When groEL1 or groEL2 with its own promoter was complemented ectopically at the attB site in the genome of the groEL1 deletion mutant YL0301 (YL0901 and YL0902), the mutants recovered the total groEL expression levels comparable to that in M. xanthus DK1622 (Wang et al., 2013). Interestingly, the groES expression levels in YL0901 and YL0902 were also recovered to that in the wild type strain (Figure 5). These results suggested that there was a self-regulation mechanism to balance the expressions of the groEL and groES genes for their synergetic functions in M. xanthus cells.
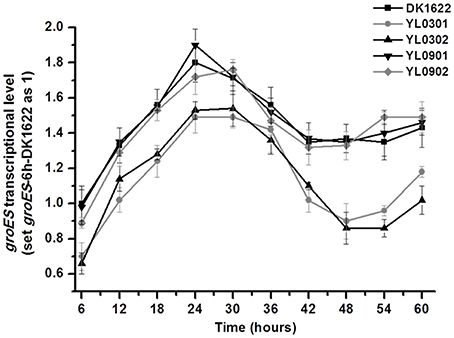
Figure 5. Quantitative PCR analysis of groES gene expression levels in groEL mutants (YL0301 and YL0302), complementary mutants (YL0901 and YL0902), and the wild-type strain DK1622. groES expression in DK1622 after 6 h incubation was set to one. Error bars represent standard deviation of three time repeats. DK1622, wild-type strain; YL0301, groEL1-deletion mutant; YL0302, groEL2-deletion mutant; YL0901, YL0301 complemented with groEL1; YL0902, YL0301 complemented with groEL2.
Over-Expression of groES Simultaneously Raises Both the groEL1 and groEL2 Expressions
In M. xanthus DK1622, the groES and groEL1 genes are in an operon under the same promoter, while groEL2 stands with a different promoter (Supplementary Figure 4 demonstrates their construction in genome). To confirm the possible coordinated expression and self-regulation of isolated groEL and groES genes, we further introduced an additional groES gene into DK1622 using the shuttle plasmid pZJY41 under the groEL2 or groEL1 promoter (referred to Supplementary Figure 4), forming the mutants YL1102 and YL1103, respectively. Compared with that in the wild-type strain DK1622, the expressions of groES markedly increased in both the YL1102 and YL1103 mutants (P < 0.001) and the groEL1 promoter is stronger than the groEL2 promoter (Figure 6). Interestingly, the expressions of either the groEL1 or the groEL2 genes also increased correspondingly (P < 0.001). The groES expression levels were comparable to the total expression changes of groEL1 and groEL2 in the two mutants. The results further demonstrated that the expression levels of the groEL and groES genes appeared to be self-regulated in M. xanthus, even when distributed in different places in the genome.
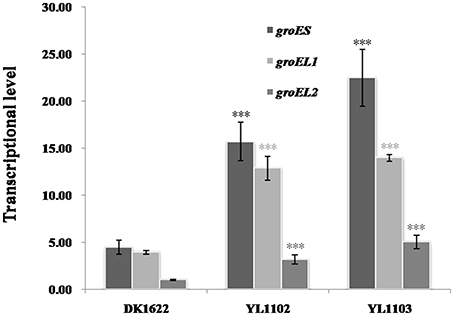
Figure 6. groES, groEL1, and groEL2 transcription levels in the wild-type strain DK1622 and the groES over–expression mutants YL1102 and YL1103. groEL2 expression in DK1622 was set to one. Error bars represent standard deviation of three time repeats. For statistical analysis, ***P < 0.001. DK1622, wild-type strain; YL1102, over-expression mutant inserted the plasmid pZJY-PgroEL2+groES; YL1103, over-expression mutant inserted the plasmid pZJY-PgroEL1+groES.
Artificial groESL2 Operon has Almost No Effect on groES and groEL Expressions
Stand-alone groELs are quite common in bacteria. In myxobacteria, the second groEL gene exists alone in the Cystobacterineae suborder but forms a complete groESL operon in the Sorangineae suborder (Figure 1). Based on our results, the stand-alone groEL gene functioned in a groES-dependent manner, at least in M. xanthus DK1622. We thus sought to determine whether an artificial groESL2 operon was able to change the coordinated expression of groEL and groES genes in M. xanthus. We inserted a second copy of the groES gene in front of groEL2 in DK1622, following the native groEL2 promoter (Supplementary Figure 4) to form a complete bicistronic groESL2 operon (mutant YL1101). The co-expression of groEL2 and the inserted groES in the artificial groESL operon was verified by RT-PCR amplification (Supplementary Figure 2). However, the total expression levels of groES, as well as that of groELs, in this mutant were still similar to those of DK1622 (P > 0.05); whereas the groES and groEL1 expressions in the groEL2 deletion mutant (YL0302) were both significantly decreased (P < 0.001; Figure 7A). The co-chaperonin GroES, in combination with the chaperonin GroEL, helps bacteria resist heat-shock conditions (Moparthi et al., 2013). We further assayed the heat-shock response of the M. xanthus cells containing two complete groESL operons. The survival rate of the YL1101 mutant after heat-shock treatment showed almost no differences from that of the wild-type strain DK1622 (P > 0.05); whereas the groEL2 deletion mutant decreased the survival rate upon heat shock (P < 0.001; Figure 7B). The results demonstrated that the single groES gene was molecularly equivalent for the expressions and functions of the duplicate groEL genes in M. xanthus cells.
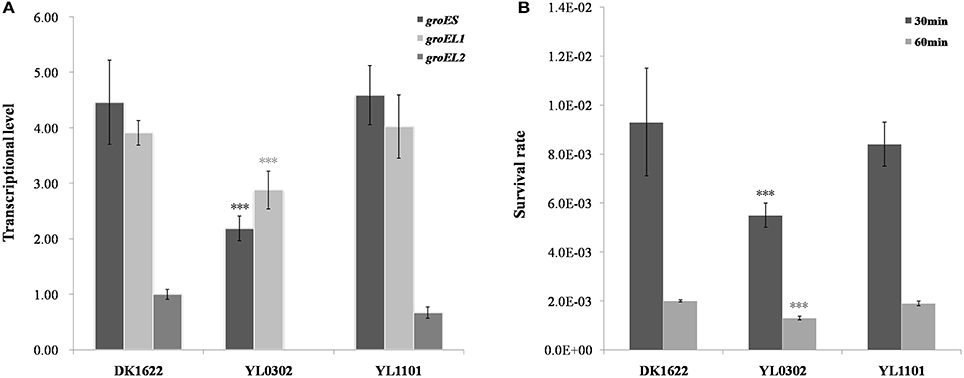
Figure 7. Comparison of the wild-type strain DK1622 and the mutant containing an artificial groESL2 operon (YL1101). (A) groES and groEL transcriptional levels after 24 h of incubation in CTT liquid growth medium, with the groEL2-deletion mutant YL0302 as a control. groEL2 expression in DK1622 was set to one. (B) Survival rates of DK1622 and YL1101 after heat-shock treatment at 42°C for 30 or 60 min, with the groEL2-deletion mutant YL0302 as a control. Error bars represent standard deviation of three time repeats. For statistical analysis, ***P < 0.001.
Discussion
Classically, the genes encoding the chaperonin GroEL and its co-factor GroES are clustered in a bicistronic groESL operon in bacterial genomes. In bacteria possessing single groEL genes, groES is always present upstream groEL to form an operon, thus probably ensuring balanced expression for their synergetic functions. However, in genomes with two or more groEL genes, the duplicated groEL genes have been observed to stand alone with no neighboring groES gene. The occurrence of stand-alone groEL genes, which are often encountered in bacterial genomes, appeared to be characteristic of certain taxonomic units, such as the Cystobacterineae suborder of myxobacteria. Based on the genetic and biochemical analyses presented in this study, the single groES gene was indispensable for M. xanthus cells, and the stand-alone groEL gene still required groES in order to function. Generally, each of the GroEL1 and GroEL2 proteins was able to form a 14-mer complex, which further bound to a 7-mer polymer of the GroES protein in vivo, consistent with that in the bacteria possessing single groEL and groES genes (Weissman et al., 1995; Liu et al., 2009). In the absence of GroES, the DK1622 GroEL proteins were unable to refold denatured proteins correctly in vitro and in vivo. Thus, similar to GroEL1, GroEL2 depends on the presence of GroES to carry out its functions, even though the groEL2 gene has lost its neighboring groES gene. However, as shown by our bioinformatics analysis, the presence of groEL and groES genes varies widely across prokaryotic genomes (Table 1). Although we determined that both GroEL proteins require the presence of GroES to carry out refolding in M. xanthus DK1622, it is unknown whether there are GroES-independent GroELs.
The results present in this paper showed that the M. xanthus cells are able to self-regulate expressions of the groEL and groES genes to meet the commensurable requirements for these proteins. For example, the deletion of groEL1 or groEL2 decreased not only groEL expression but also groES expression, while ectopic complementation of groEL recovered both groEL and groES expression in M. xanthus cells. Notably, the addition of an extra groES gene in front of groEL2 to form an artificial groESL2 operon had almost no effect on the expressions of groES and groELs, but the over-expression of groES using the self-replicating plasmid pZJY41 increased groEL expressions considerably. This analysis indicated that the duplicate groEL and single groES genes have evolved to express in a coordinated manner for their synergistic functions in M. xanthus cells. The dependence of stand-alone groEL genes on non-neighbored groES is easily understandable in the context of the functional mechanisms of GroEL and its co-factor GroES. However, it is puzzled how the single groES gene coordinate its expression to meet the requirements of two endogenous groEL genes.
It is known that the HrcA protein is able to bind to the CIRCE regions in front of the groESL operons to negatively regulate expressions of the groEL and groES genes (Wilson et al., 2005). Wilson et al. reported that the GroEL protein, probably as well as the GroES protein was able to auto-regulate its own expression in Chlamydia trachomatis through direct interactions with the HrcA repressor protein (Wilson et al., 2005). Kong et al. observed an unbalanced expression of the operon-organized groES and groEL genes in M. tuberculosis (Kong et al., 1993). In Salmonella, differential expressions of bicistronic groES and groEL genes in a groESL operon were reported to be due to the mediation of an RNA thermometer in the groES-5′UTR, which regulated translation of groES, but not of groEL upon heat-shock (Cimdins et al., 2013). In addition, in E. coli cells, there is an imperfect transcriptional terminator in the intergenic region of the other major chaperonin gene operon dnaKJ, which was able to regulate the dnaK and dnaJ to express differentially (Bardwell et al., 1986). Herein, we proposed that the products of the groEL/groES genes involved in auto-regulation of expressions, and there were GroEL/GroES- dependent modulating sequences within or before the groESL1 and groEL2 operons for the coordinated expressions of the GroELs and GroES chaperonins in M. xanthus. The removal of groES from the duplicated ancient groESL2 operon is likely an important evolutionary development. Thus, controlling the expressions of duplicate groEL and single groES genes under an integrated mechanism might be easier and more efficient for cells to adapt to the changing environment. The exact regulation mechanisms for the coordinated expressions of the groES and groEL1/groEL2 genes in M. xanthus are undergoing investigation in our laboratory.
Conclusion
In general, the genes encoding the chaperonins GroEL and GroES form a groESL operon. However, stand-alone groEL genes also exist broadly in the bacteria containing multiple groEL genes. The stand-alone groELs may play functions independently or still dependently on groES. Here we prove that the stand-alone groEL2 gene strictly relies on groES to function in M. xanthus DK1622. The duplicate groEL and the single groES genes were expressed and functioned interdependently in coordination. Adding an excess groES before groEL2 had almost no influences on groES expression and the cell survival rate, and over-expression of groES increased the groEL expressions commensurably. The duplicated groEL and single groES genes thus have evolved an accurate self-regulation pattern for their cellular functions.
Author Contributions
Conceived and designed the experiments: YL, LZ, YW. Performed the experiments: LZ, YW, ZZ, JL. Analyzed the data: YL, LZ, YW, ZZ, XZ. Wrote the paper: YL, LZ, YW.
Funding
This work was financially supported by the National Natural Science Foundation of China (NSFC) (No. 31471183 & No. 31670076) and the NSFC Key Program (No. 31130004) awarded to YL.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00733/full#supplementary-material
Supplementary Figure 1. The diagrammatic sketches for the construction of the plasmids of pET22b-HrcA (A), pBAD33-GroESL1/2, and pBAD33-GroEL1/2 (B). The processing plasmids of pET22b-GroESL1/2 and pET22b-GroEL1/2 were constructed using the similar progress as pET22b-HrcA.
Supplementary Figure 2. RT-PCR detection on the operon-organization of the native groES -groEL1 locus and the artificial groES-groEL2 locus in M. xanthus. (A) Lines 1, 4, PCR amplification of the groES-groEL1 locus using the cDNA from DK1622 and YL1101 as template, respectively; Line 7, PCR amplification of the groES-groEL2 locus using YL1101 cDNA as template; Lines 2, 5, 8, positive controls using the total DNA extracted from DK1622 or YL1101 as the template; Lines 3, 6, 9, negative controls in which no reverse transcriptase was added. M, Trans 2K Plus II markers. The cDNA templates were obtained by reverse transcription of the total RNA extracted from M. xanthus DK1622 or the mutant YL1101 using random primers. The strains were grown in CTT medium at 30°C for 24 h. (B) PCR amplification verification of the existence of groES, groEL1 and groEL2 genes in the cDNA samples from DK1622 and YL1101. Lines 1, 3, PCR amplification of groES. Lines 2, 4, PCR amplification of groEL1. Line 5, PCR amplification of groEL2. M, Trans 2K Plus II markers. The primers used for amplification are listed in Supplementary Table 2.
Supplementary Figure 3. CS renaturation activities of GroELs in the absence or presence of GroES. Error bars in the pictures represent standard deviation of three time repeats.
Supplementary Figure 4. A diagrammatic sketch of the construction of the groES-groEL1 operon and the groEL2 gene in M. xanthus DK1622. The promoters that were used to construct the YL1102 and YL1103 mutants containing excess groES genes are shown in the figure. The insertion of groES in the artificial groES-groEL2 operon of YL1101 was between the groEL2-promoter and the groEL2 gene.
Supplementary Table 1. Bacterial strains and plasmids used in this study.
Supplementary Table 2. List of primers used in this study.
Supplementary Table 3. Copy numbers of groEL and groES in sequenced prokaryotic genomes.
Supplementary Table 4. The occurrence of groEL and groES genes in 24 sequenced myxobacterial genomes.
References
Bardwell, J. C., Tilly, K., Craig, E., King, J., Zylicz, M., and Georgopoulos, C. (1986). The nucleotide sequence of the Escherichia coli K12 dnaJ+ gene. A gene that encodes a heat shock protein. J. Biol. Chem. 261, 1782–1785.
Cimdins, A., Roßmanith, J., Langklotz, S., Bandow, J. E., and Narberhaus, F. (2013). Differential control of Salmonella heat shock operons by structured mRNAs. Mol. Microbiol. 89, 715–731. doi: 10.1111/mmi.12308
Craig, E. A., Gambill, B. D., and Nelson, R. J. (1993). Heat shock proteins: molecular chaperones of protein biogenesis. Microbiol. Rev. 57, 402–414.
Dworkin, M., and Kaiser, D. (1993). Myxobacteria II. Washington, DC: American Society for Microbiology.
Fayet, O., Ziegelhoffer, T., and Georgopoulos, C. (1989). The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J. Bacteriol. 171, 1379–1385. doi: 10.1128/jb.171.3.1379-1385.1989
Figueiredo, L., Klunker, D., Ang, D., Naylor, D. J., Kerner, M. J., Georgopoulos, C., et al. (2004). Functional characterization of an archaeal GroEL/GroES chaperonin system: significance of substrate encapsulation. J. Biol. Chem. 279, 1090–1099. doi: 10.1074/jbc.M310914200
Goldman, B. S., Nierman, W. C., Kaiser, D., Slater, S. C., Durkin, A. S., Eisen, J. A., et al. (2006). Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U.S.A. 103, 15200–15205. doi: 10.1073/pnas.0607335103
Gould, P. S., Burgar, H. R., and Lund, P. A. (2007). Homologous cpn60 genes in Rhizobium leguminosarum are not functionally equivalent. Cell Stress Chaperones 12, 123–131. doi: 10.1379/CSC-227R.1
Goyal, K., Qamra, R., and Mande, S. C. (2006). Multiple gene duplication and rapid evolution in the groEL gene: functional implications. J. Mol. Evol. 63, 781–787. doi: 10.1007/s00239-006-0037-7
Hodgkin, J., and Kaiser, D. (1977). Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. U.S.A. 74, 2938–2942. doi: 10.1073/pnas.74.7.2938
Horwich, A. L., Farr, G. W., and Fenton, W. A. (2006). GroEL-GroES-mediated protein folding. Chem. Rev. 106, 1917–1930. doi: 10.1021/cr040435v
Houry, W. A., Frishman, D., Eckerskorn, C., Lottspeich, F., and Hartl, F. U. (1999). Identification of in vivo substrates of the chaperonin GroEL. Nature 402, 147–154. doi: 10.1038/45977
Jiang, D. M., Zhao, L., Zhang, C. Y., Li, J., Xia, Z. J., Wang, J., et al. (2008). Taxonomic analysis of Sorangium strains based on HSP60 and 16S rRNA gene sequences and morphology. Int. J. Syst. Evol. Microbiol. 58, 2654–2659. doi: 10.1099/ijs.0.65806-0
Karlin, S., and Brocchieri, L. (2000). Heat shock protein 60 sequence comparisons: duplications, lateral transfer, and mitochondrial evolution. Proc. Natl. Acad. Sci. U.S.A. 97, 11348–11353. doi: 10.1073/pnas.97.21.11348
Kashefi, K., and Hartzell, P. L. (1995). Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF- defect. Mol. Microbiol. 15, 483–494. doi: 10.1111/j.1365-2958.1995.tb02262.x
Kerner, M. J., Naylor, D. J., Ishihama, Y., Maier, T., Chang, H. C., Stines, A. P., et al. (2005). Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122, 209–220. doi: 10.1016/j.cell.2005.05.028
Klunker, D., Haas, B., Hirtreiter, A., Figueiredo, L., Naylor, D. J., Pfeifer, G., et al. (2003). Coexistence of group I and group II chaperonins in the archaeon Methanosarcina mazei. J. Biol. Chem. 278, 33256–33267. doi: 10.1074/jbc.M302018200
Kong, T. H., Coates, A. R., Butcher, P. D., Hickman, C. J., and Shinnick, T. M. (1993). Mycobacterium tuberculosis expresses two chaperonin-60 homologs. Proc. Natl. Acad. Sci. U.S.A. 90, 2608–2612. doi: 10.1073/pnas.90.7.2608
Kumar, C. M., Mande, S. C., and Mahajan, G. (2015). Multiple chaperonins in bacteria–novel functions and non-canonical behaviors. Cell Stress Chaperones 20, 555–574. doi: 10.1007/s12192-015-0598-8
Li, J., Wang, Y., Zhang, C. Y., Zhang, W. Y., Jiang, D. M., Wu, Z. H., et al. (2010). Myxococcus xanthus viability depends on groEL supplied by either of two genes, but the paralogs have different functions during heat shock, predation, and development. J. Bacteriol. 192, 1875–1881. doi: 10.1128/JB.01458-09
Liu, H., Kovács, E., and Lund, P. A. (2009). Characterisation of mutations in GroES that allow GroEL to function as a single ring. FEBS Lett. 583, 2365–2371. doi: 10.1016/j.febslet.2009.06.027
Lobedanz, S., and Sogaard-Andersen, L. (2003). Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus. Genes Dev. 17, 2151–2161. doi: 10.1101/gad.274203
Lund, P. A. (2001). Microbial molecular chaperones. Adv. Microb. Physiol. 44, 93–140. doi: 10.1016/S0065-2911(01)44012-4
Lund, P. A. (2009). Multiple chaperonins in bacteria–why so many? FEMS Microbiol. Rev. 33, 785–800. doi: 10.1111/j.1574-6976.2009.00178.x
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Martin, J., Langer, T., Boteva, R., Schramel, A., Horwich, A. L., and Hartl, F. U. (1991). Chaperonin-mediated protein folding at the surface of groEL through a “molten globule”-like intermediate. Nature 352, 36–42. doi: 10.1038/352036a0
Moparthi, S. B., Sjölander, D., Villebeck, L., Jonsson, B. H., Hammarström, P., and Carlsson, U. (2013). Transient conformational remodeling of folding proteins by GroES-individually and in concert with GroEL. J. Chem. Biol. 7, 1–15. doi: 10.1007/s12154-013-0106-5
Potnis, A. A., Rajaram, H., and Apte, S. K. (2016). GroEL of the nitrogen-fixing cyanobacterium Anabaena sp. strain L-31 exhibits GroES and ATP-independent refolding activity. J. Biochem. 159, 295–304. doi: 10.1093/jb/mvv100
Qamra, R., Srinivas, V., and Mande, S. C. (2004). Mycobacterium tuberculosis GroEL homologues unusually exist as lower oligomers and retain the ability to suppress aggregation of substrate proteins. J. Mol. Biol. 342, 605–617. doi: 10.1016/j.jmb.2004.07.066
Ranson, N. A., White, H. E., and Saibil, H. R. (1998). Chaperonins. Biochem. J. 333 (Pt 2), 233–242. doi: 10.1042/bj3330233
Saibil, H. R., and Ranson, N. A. (2002). The chaperonin folding machine. Trends Biochem. Sci. 27, 627–632. doi: 10.1016/S0968-0004(02)02211-9
Shimkets, L. J. (1990). Social and developmental biology of the myxobacteria. Microbiol. Rev. 54, 473–501.
Ueki, T., Inouye, S., and Inouye, M. (1996). Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183, 153–157. doi: 10.1016/S0378-1119(96)00546-X
Vanbogelen, R. A., Acton, M. A., and Neidhardt, F. C. (1987). Induction of the heat shock regulon does not produce thermotolerance in Escherichia coli. Genes Dev. 1, 525–531. doi: 10.1101/gad.1.6.525
Viitanen, P. V., Donaldson, G. K., Lorimer, G. H., Lubben, T. H., and Gatenby, A. A. (1991). Complex interactions between the chaperonin 60 molecular chaperone and dihydrofolate reductase. Biochemistry 30, 9716–9723. doi: 10.1021/bi00104a021
Wang, Y., Li, X., Zhang, W., Zhou, X., and Li, Y. Z. (2014). The groEL2 gene, but not groEL1, is required for biosynthesis of the secondary metabolite myxovirescin in Myxococcus xanthus DK1622. Microbiology 160, 488–495. doi: 10.1099/mic.0.065862-0
Wang, Y., Zhang, W. Y., Zhang, Z., Li, J., Li, Z. F., Tan, Z. G., et al. (2013). Mechanisms involved in the functional divergence of duplicated GroEL chaperonins in Myxococcus xanthus DK1622. PLoS Genet. 9:e1003306. doi: 10.1371/journal.pgen.1003306
Weissman, J. S., Hohl, C. M., Kovalenko, O., Kashi, Y., Chen, S., Braig, K., et al. (1995). Mechanism of GroEL action: productive release of polypeptide from a sequestered position under GroES. Cell 83, 577–587. doi: 10.1016/0092-8674(95)90098-5
Williams, T. A., and Fares, M. A. (2010). The effect of chaperonin buffering on protein evolution. Genome Biol. Evol. 2, 609–619. doi: 10.1093/gbe/evq045
Wilson, A. C., Wu, C. C., Yates, J. R. III., and Tan, M. (2005). Chlamydial GroEL autoregulates its own expression through direct interactions with the HrcA repressor protein. J. Bacteriol. 187, 7535–7542. doi: 10.1128/JB.187.21.7535-7542.2005
Keywords: Myxococcus xanthus, chaperonins, interdependence, duplicate groELs, single groES, coordinated expression, self-regulation
Citation: Zhuo L, Wang Y, Zhang Z, Li J, Zhang X-H and Li Y-z (2017) Myxococcus xanthus DK1622 Coordinates Expressions of the Duplicate groEL and Single groES Genes for Synergistic Functions of GroELs and GroES. Front. Microbiol. 8:733. doi: 10.3389/fmicb.2017.00733
Received: 04 February 2017; Accepted: 10 April 2017;
Published: 27 April 2017.
Edited by:
Frank T. Robb, University of Maryland, Baltimore, USAReviewed by:
Santosh Kumar C. M., University of Birmingham, UKPeter Adrian Lund, University of Birmingham, UK
Copyright © 2017 Zhuo, Wang, Zhang, Li, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yue-zhong Li, bGlsYWJAc2R1LmVkdS5jbg==
†These authors have contributed equally to this work.
 Li Zhuo1†
Li Zhuo1† Zheng Zhang
Zheng Zhang Xiao-Hua Zhang
Xiao-Hua Zhang Yue-zhong Li
Yue-zhong Li