- Department of Biotechnology and Food Science, Norwegian University of Science and Technology, Trondheim, Norway
Aeromonas spp. are ubiquitous bacteria that have received increasing attention as human pathogens because of their widespread occurrence in food, especially seafood and vegetables. The aim of this work was to assess the species identity and phylogenetic relationship of 118 Aeromonas strains isolated from fresh retail sushi from three producers, and to characterize the isolates with respect to genetic and phenotypic virulence factors. We also evaluate the potential hazard associated with their presence in ready-to-eat seafood not subjected to heat treatment. Mesophilic Aeromonas salmonicida was most prevalent (74%), followed by A. bestiarum (9%), A. dhakensis (5%), A. caviae (5%), A. media (4%), A. hydrophila (2%), and A. piscicola (1%). All isolates were considered potentially pathogenic due to the high prevalence of genes encoding hemolysin (hlyA) (99%), aerolysin (aerA) (98%), cytotoxic enterotoxin (act) (86%), heat-labile cytotonic enterotoxin (alt) (99%), and heat-stable cytotonic enterotoxin (ast) (31%). The shiga-like toxins 1 and 2 (stx-1 and stx-2) were not detected. Moreover, there was heterogeneity in toxin gene distribution among the isolates, and the combination of act/alt/hlyA/aerA was most commonly detected (63%). β-hemolysis was species-dependent and observed in 91% of the isolates. All A. media and A. caviae strains were non-hemolytic. For isolates belonging to this group, lack of hemolysis was possibly related to the absence of the act gene. Swimming motility, linked to adhesion and host invasion, occurred in 65% of the isolates. Partial sequencing of the gyrB gene demonstrated its suitability as a genetic marker for Aeromonas species identification and for assessment of the phylogenetic relationship between the isolates. The gyrB sequence divergence within a given species ranged from 1.3 to 2.9%. A. bestiarum, A. salmonicida, and A. piscicola were the most closely related species; their sequences differed by 2.7–3.4%. The average gyrB sequence similarity between all species was 93%, demonstrating its acceptable taxonomic resolution. The presence of multiple species of potential pathogenic Aeromonas in fresh retail sushi raises new food safety issues related to the increased consumption of ready-to-eat food composed of raw ingredients.
Introduction
Aeromonas species are ubiquitous aquatic bacteria that have received increasing attention as opportunistic and primary pathogens in humans. The genus Aeromonas can be classified into two main groups. The first is the psychrophilic non-motile strains, primarily Aeromonas salmonicida, that infect fish. The second and larger group is motile, mesophilic aeromonads associated with human diseases such as gastrointestinal diseases, wound infections, and septicemia (Janda and Abbott, 2010; Parker and Shaw, 2011). Of the approximately 30 recognized species of Aeromonas (Martínez-Murcia et al., 2016), a subset of four species are more frequently implicated in human infections (Aeromonas hydrophila, A. caviae, A. veronii biovar sobria, and A. dhakensis) (Janda and Abbott, 2010; Teunis and Figueras, 2016).
In a previous study, we detected Aeromonas spp. in 71% of fresh retail ready-to-eat (RTE) sushi boxes purchased in Norwegian supermarkets (Hoel et al., 2015). These products are offered as complete meals with a selection of different sushi and are distributed cold (≤4°C) with a shelf life of 2–3 days after production. In addition to acidified rice (pH < 4.6), the sushi meals typically consist of various species of both raw fish and vegetables, which are excellent substrates for bacterial growth. The widespread occurrence of Aeromonas ssp. in fish and seafood, vegetables, meat, poultry, raw milk, and different water sources has been confirmed by others (Villari et al., 2000; Kingombe et al., 2004; Heaton and Jones, 2008; Xanthopoulos et al., 2010; Ottaviani et al., 2011; Carvalho et al., 2012; Latif-Eugenín et al., 2016).
Most aeromonads grow optimally around 28°C but can grow at a wide range of temperatures (4–42°C) (Palumbo et al., 1985a). Environmental isolates (such as food isolates) seem to be more adapted to low temperatures and can proliferate at refrigeration temperatures (Mateos et al., 1993). Growth at temperatures as low as -0.1°C has been reported for some strains (Daskalov, 2006). Species belonging to the genus Aeromonas are generally recognized as sensitive to low pH (min. pH∼6), but food isolates have demonstrated increased tolerability to low pH (Hwang, 2010).
The pathogenesis of Aeromonas-mediated infections is multi-factorial, and the role of the virulence determinants in human infections have not been conclusively established (Pablos et al., 2010). An assortment of virulence factors enables these bacteria to colonize, invade, and infect different hosts. The most reviewed virulence factors are the pore-forming hemolytic toxins, hemolysin, and aerolysin, (Howard et al., 1987; Parker and Shaw, 2011), and three different enterotoxins, including cytotoxic (Act), heat-labile cytotonic (Alt), and heat-stable cytotonic (Ast) enterotoxins. All these toxins have been linked to cases of diarrhea (Galindo et al., 2006). Furthermore, the expression of lateral or peritrichous flagella is associated with enhanced adherence and invasiveness in addition to the ability to biofilm formation (Gavín et al., 2003).
The taxonomy of the genus Aeromonas is complex and has undergone numerous changes with the implementation of new phylogenetic markers. Conventional phenotypic tests do not necessarily correspond to results achieved by genetic methods, and this is especially evident in environmental isolates of Aeromonas species (Ørmen et al., 2005; Puthucheary et al., 2012). Sequencing of the 16S rDNA has proven unsuccessful for identification of Aeromonas species due to low taxonomic resolution (Morandi et al., 2005; Nagar et al., 2013). Therefore, sequencing of the housekeeping genes gyrB (encoding the B-subunit of DNA gyrase, a type II DNA topoisomerase) (Yanez et al., 2003) and rpoD (encoding the σ70 factor conferring promoter-specific transcription initiation on RNA polymerase) (Soler et al., 2004) is now preferred for phylogenetic analysis and species identification of Aeromonas spp.
The aim of this work was to assess the species identity and phylogenetic relationship of Aeromonas strains isolated from fresh retail sushi from three producers and to characterize the isolates with respect to genetic and phenotypic virulence factors. Furthermore, we evaluate the potential hazard associated with their presence in minimally processed RTE seafood such as sushi. To the best of our knowledge, no studies have been published which concurrently assess prevalence, species distribution, and genetic characterization of several Aeromonas species in seafood intended for raw consumption. In light of the taxonomical controversies, including recent species reclassifications and the lack of definite virulence markers, studies providing updated characterization of current environmental strains are required in order to provide insight into the impact of these bacteria as foodborne pathogens.
Materials and Methods
Bacterial Strains
Mesophilic Aeromonas spp. were isolated from retail sushi and verified phenotypically according to Nordic Committee on Food Analysis [NMKL] (2004) method no. 150, as described previously (Hoel et al., 2015) and by PCR using primers targeting the Aeromonas gyrB gene (Table 1). In brief, Aeromonas spp. were isolated from sushi using starch ampicillin agar (SAA) supplemented with 10 mg/L ampicillin (Sigma–Aldrich, Oslo, Norway) and incubated at 37°C. All subsequent cultivation was carried out on tryptone soy agar (TSA) (Oxoid, Oslo, Norway) at 37°C. One hundred and three Aeromonas isolates were isolated from sushi products from three different producers: A (n = 54), B (n = 24), and C (n = 25). To increase the resolution in species diversity, 15 additional Aeromonas isolates were collected from producer A in a follow-up study. Hence, a total of 118 Aeromonas isolates were analyzed. A. hydrophila (CCUG 14551T), A. caviae (CCUG 25939T), A. veronii biovar veronii (CCUG 27821T), and A. veronii biovar sobria (CCUG 30360) were included as reference strains. Pseudomonas aeruginosa (CCUG 2080), Escherichia coli (CCUG 49263), and Staphylococcus aureus (CCUG 41582) were used as negative controls in all PCR reactions to verify the amplification specificity.
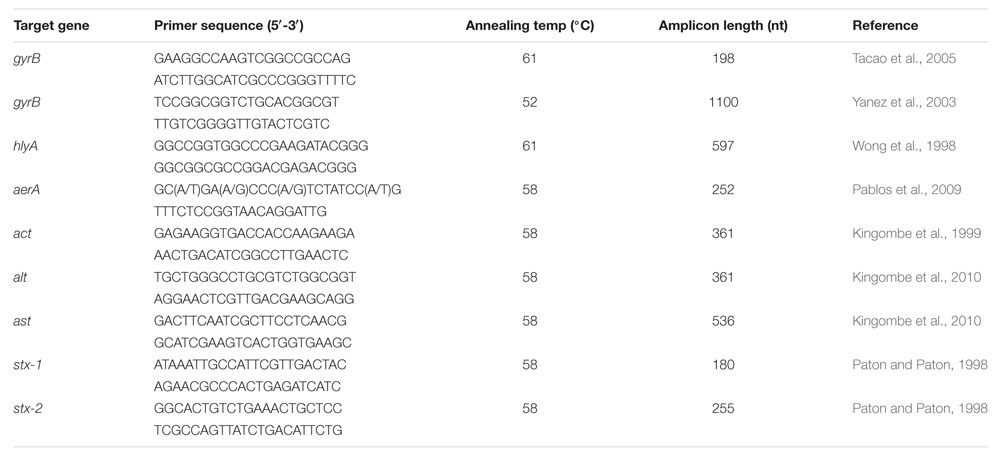
TABLE 1. PCR primers, annealing temperatures, and expected amplicon length for Aeromonas genus-specific gyrB primers and virulence-associated genes.
DNA Isolation and PCR Protocols
Total genomic DNA was extracted from overnight cultures grown in tryptone soy broth (TSB) (Oxoid) at 37°C using the protocol for Gram-negative bacteria in the DNeasy Blood and Tissue kit (Qiagen, Oslo, Norway). All PCR reactions were performed with 25 μl reactions containing 1x PCR buffer (1.5 mM MgCl2), 200 μM of each nucleotide, 0.2 μM each primer, 2.5 U Taq polymerase (Qiagen), and 50–100 ng template DNA. The PCR amplification cycles were as follows: Initial denaturation at 95°C for 15 min, 30 cycles of denaturation at 95°C for 30 s, annealing for 30 s at temperature given in Table 1, and extension at 72°C for 60 s, followed by a final extension at 72°C for 7 min. All PCR reactions were performed in duplicate in separate experiments. PCR products were visualized by electrophoresis in a 1.5% agarose gel (SeaKem, Lonza Group Ltd., Basel, Switzerland) in 1x TAE buffer. Two random PCR products of each detected virulence gene (ast, act, alt, aerA, and hlyA) were excised from the agarose gel and purified using the GeneJET Gel Extraction Kit (Thermo Scientific, Oslo, Norway) and sequenced (Eurofins Genomics, Ebersberg, Germany) to confirm the amplicon specificity.
Species Identification and Phylogenetic Analysis
An approximately 1100 nt fragment of the Aeromonas gyrB gene was amplified using primers gyrB3F/gyrB14R (Table 1). PCR products were purified using the GeneJet PCR Purification Kit (Thermo Scientific). Eurofins Genomics performed the DNA sequencing, and the resulting sequences were compared with available GenBank database sequences using the BLAST program1.
Multiple sequence alignments were performed using ClustalW, integrated in the MEGA7 software (Kumar et al., 2016). The gyrB sequences from all isolates and their corresponding type or reference sequences were included in the alignment, but identical sequences were not included in the phylogenetic analysis. The reference or type sequences for the gyrB gene for species not available as cultures at our laboratory were retrieved from NCBI: A. salmonicida strain CECT 894T (GeneBank acc. No. AY101773), A. bestiarum strain CDC 9533-76T (AJ868362), A. media strain CECT 4232T (AY101782), A. dhakensis strain MDC 2406T (HQ442711) and strain MDC 401 (EU268453), A. piscicola strain S1.2T (AY011790) and E. coli strain KCTC 2441 (EU014649).
A phylogenetic tree was constructed using the neighbor-joining method (Saitou and Nei, 1987) in the MEGA 7 software, and evolutionary distances were calculated using the Kimura two-parameter method (Kimura, 1980) with 1000 bootstrap replicates to assess tree topology robustness. To test the tree stability, we also created a phylogenetic tree using the maximum-likelihood method. The phylogenetic analysis involved 38 nucleotide sequences with a continuous stretch of 944 nt. Thirty-four partial gyrB nucleotide sequences were submitted to the GenBank database2 with accession numbers KY652231–KY652264.
Phenotypic Characterization
To test for β-hemolytic activity, one colony of each isolate was streaked onto bovine blood agar and incubated at 37°C. Hemolysis was recorded after 24 and 48 h (± 2 h) and done twice for each isolate.
Swimming (defined as flagella-directed movement in an aqueous environment) and swarming (defined as multiple, lateral, flagella-directed rapid movements on a solid surface) (Jahid et al., 2015) motility were examined using modifications of methods described by Grim et al. (2013) and Jahid et al. (2013). For swimming motility, a single colony from an exponentially growing culture was spotted into the center of a plate containing Nutrient Agar (3 g/L beef extract [Difco, Becton, Dickinson & Company, Franklin Lakes, NJ, USA], 5 g/L bacterial peptone [Oxoid], and 0.3% agar [Oxoid]). The swimming plates were incubated face up at 25°C for 24 ± 2 h. The test was performed twice for each isolate. For swarming motility, two different methods were applied. First, material from the periphery of the swimming zone was selected and streaked onto the surface of an agar plate containing LB (10 g/L bacterial peptone, 10 g/L NaCl, 5 g/L yeast extract [Oxoid], and 0.5% agar [Oxoid]). Second, a single colony from an overnight culture on TSA was streaked onto the surface of an LB plate containing 0.5% agar. In both cases, plates were incubated at 37°C for 24 and 48 h (± 2 h). Using both methods, an incubation temperature of 25°C was also tested once for all isolates. After incubation, the motility diameter was measured by examining bacterial migration through the agar from the center toward the periphery of the plate.
Statistical Analysis
A chi-square test was applied to test for differences in species distribution among the three producers and to test for variability in species distribution over time for producer A (α = 0.05) using the online resources for the textbook by Barnard et al. (2011)3.
Results
Aeromonas Species Identification
All presumptive Aeromonas isolates (n = 118) were verified as Aeromonas spp. using genus-specific primers targeting gyrB. Nucleotide sequences of gyrB amplicons were determined for 118 isolates and 4 Aeromonas reference strains. The resulting sequences were 701–1048 nt with 93% > 1030 nt. The prevalence and distribution of species from the initial sampling from producers A, B, and C, and the follow-up sampling of producer A are shown in Table 2. There were no differences in the relative occurrence of species between the three producers (p = 0.686), and mesophilic (non-typical) A. salmonicida was the most commonly isolated species from all producers. However, in the follow-up study of producer A, two more species were detected, and A. dhakensis, synonymous with A. hydrophila ssp. dhakensis/A. aquariorum, was the most prevalent. Thus, there was a significant difference in the occurrence of species over time for producer A (p < 0.001).
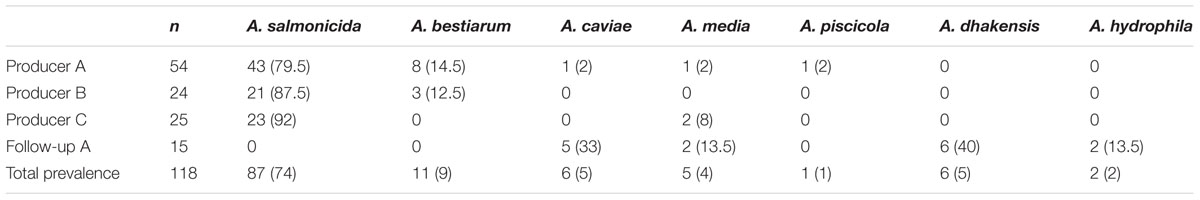
TABLE 2. Number (percentage) of Aeromonas species sampled from three sushi producers (A, B, and C), and a follow-up sampling from producer A.
Prevalence of Virulence-Associated Genes
The presence of seven virulence-associated genes (Table 3) was analyzed by PCR. At least one of these genes was present in all Aeromonas isolates. The reference strains A. hydrophila (CCUG 14551) and A. veronii biovar veronii (CCUG 27821) harbored all genes, except stx-1 and stx-2. A. veronii biovar sobria (CCUG 30360) harbored alt, ast, aerA, and hlyA, whereas only the alt gene was detected in the A. caviae reference strain (CCUG 25939). The non-Aeromonas strains (E. coli, P. aeruginosa, and S. aureus) gave negative results for all genes, except S. aureus, which exhibited a very faint band for hlyA.
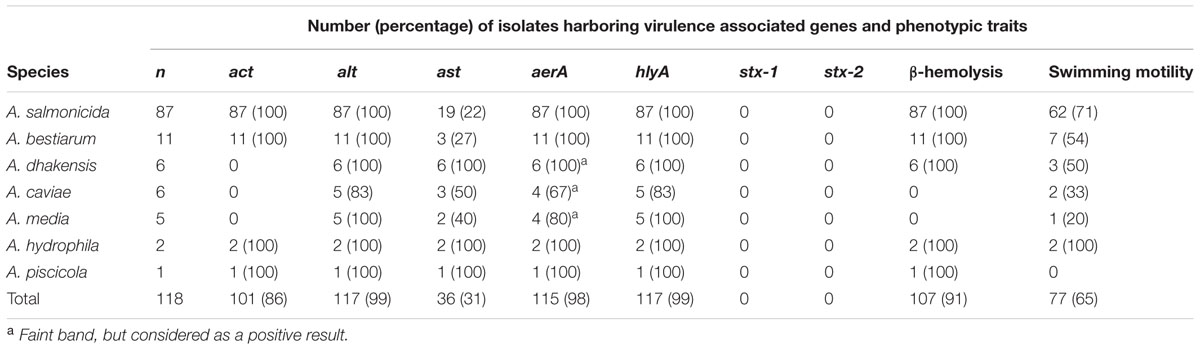
TABLE 3. Virulence associated genes, β-hemolysis on blood agar, and swimming motility in soft agar for Aeromonas spp. isolated from retail sushi.
Four different combinations of the act/alt/ast genes encoding Aeromonas enterotoxins were observed. The most frequent enterotoxin gene combination was act/alt, present in 75 isolates (64%), followed by act/alt/ast in 26 isolates (22%), and alt/ast in 11 isolates (9%). A single gene (alt) was detected in six isolates (5%), and they were all A. media and A. caviae species. A species-dependent heterogeneity in the enterotoxin gene distribution was observed (Figure 1). The most frequent enterotoxin gene combination act/alt was only observed in A. salmonicida and A. bestiarum isolates. The combination act/alt/ast was found in A. salmonicida, A. bestiarum, A. piscicola, and A. hydrophila isolates, whereas the alt/ast and alt dominated in the other species.
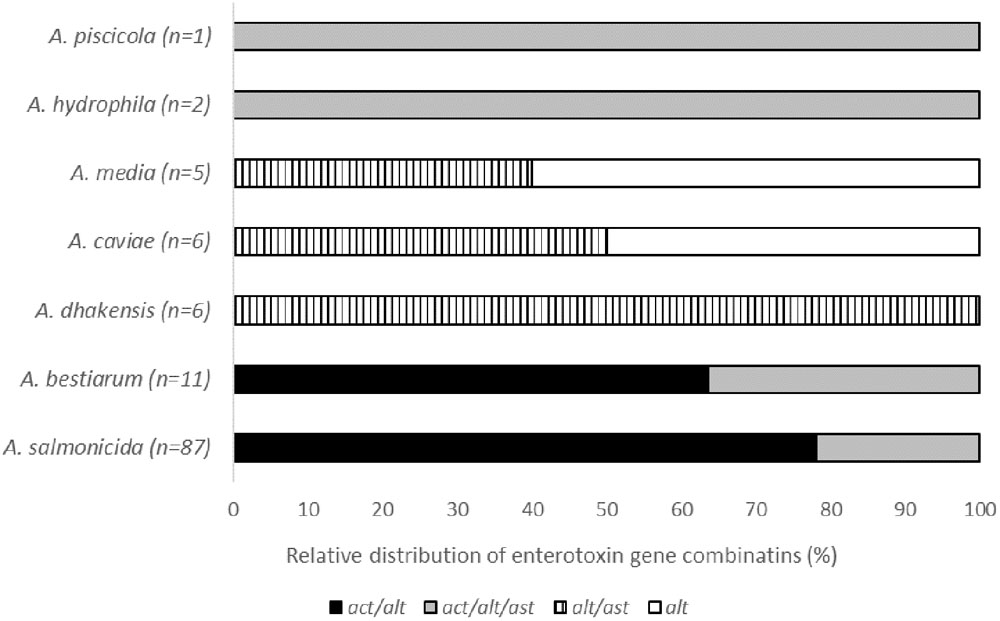
FIGURE 1. Relative distribution of the combinations of the enterotoxin genes act, alt, and ast in the different groups of Aeromonas spp.
Genes encoding the hemolytic and pore-forming toxins hemolysin and aerolysin were detected in 99 and 98% of all isolates, respectively (Table 3). However, the results for aerA were ambiguous for the isolates identified as A. caviae, A. media, and A. dhakensis. PCR yielded faint bands, regardless of DNA template concentration and attempts to optimize PCR running conditions. However, faint bands were considered positive results. All isolates gave negative results for the stx-1 and stx-2 genes, encoding shiga-like toxins 1 and 2, respectively. Sequencing of two PCR products from each gene confirmed their specificity, and the non-template controls were negative for all genes for all samples.
Hemolytic Activity and Swimming Motility
β-hemolysis was observed in 91% of the isolates, and the outcome of this test was clearly related to species identity (Table 3). The non-hemolytic isolates belonged to the species A. media and A. caviae. In fact, all A. media and A. caviae in this study were non-hemolytic, including the A. caviae reference strain.
Isolates with a circular migration zone surrounding the point of inoculation on the soft agar plates were classified as motile. In cases in which growth was limited to the inoculation point, or the diameter of the growth was ≤10 mm, the isolates were classified as non-motile under the tested conditions, and 41 isolates (35%) were assigned to this group (Table 3). Some intra-species variability was observed. All species had both motile and non-motile isolates except A. piscicola, which was represented by only one non-motile isolate. Interestingly, most of the non-motile A. salmonicida were isolated from producer C’s sushi. We were not able to induce swarming motility in any isolates or reference strains under the tested conditions.
Phylogenetic Analysis of Aeromonas gyrB Sequences
The gyrB sequences from all Aeromonas isolates and the reference strain sequences were included in the multiple sequence alignment, and a distance matrix was created for all pairs of sequences for identification and removal of duplicate sequences. The sequence similarity between all strains in the final dataset ranged from 90.1 to 99.9 % (1–93 nucleotide differences).
Percentage nucleotide substitutions were calculated for a continuous segment of 944 nt (Supplementary Table A). At the intra-species level (between isolates of a given species), the average nucleotide substitution ranged from 1.3 to 2.9% and were <2% for the majority of species (1.3% for A. bestiarum and A. piscicola, 1.4% for A. hydrophila, 1.6% for A. salmonicida, and 1.9% for A. caviae), with the exception of A. dhakensis and A. media (2.3 and 2.9%, respectively).
The inter-species sequence divergence was higher and >3.4% for all pairs of species (Table 4). The only exception was a tighter group of three species consisting of A. salmonicida, A. bestiarum, and A. piscicola with nucleotide substitutions ranging from 2.7 to 3.4% (2.7% for A. piscicola and A. bestiarum, 3.0% for A. piscicola and A. salmonicida, and 3.4% for A. bestiarum and A. salmonicida).
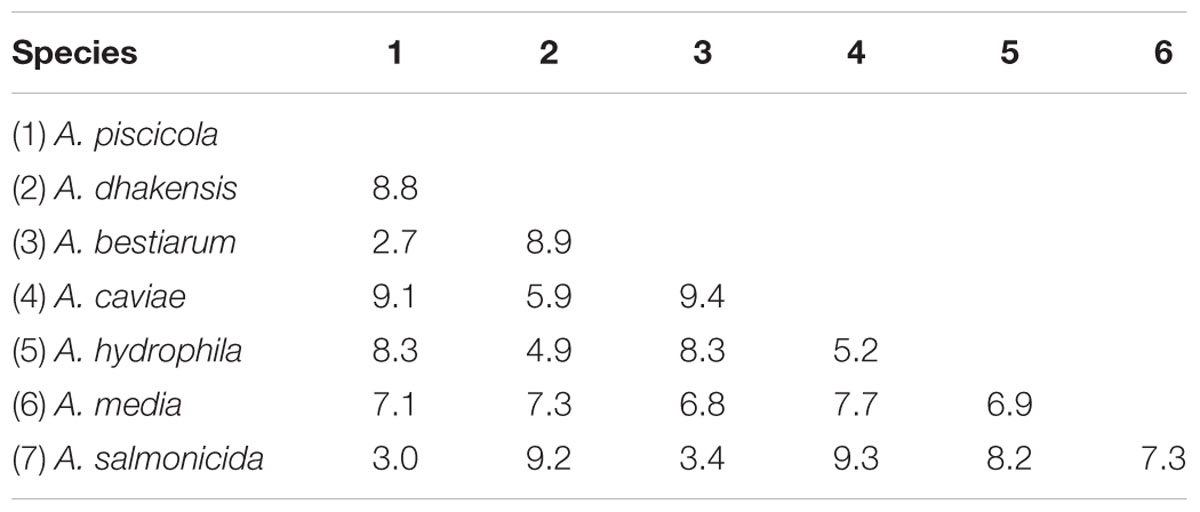
TABLE 4. Comparison of average gyrB sequence evolutionary divergence (percentage nucleotide substitutions) between the seven Aeromonas spp. isolated from retail sushi.
A neighbor-joining tree was constructed from the nucleotide alignment, and the constructed tree showed distinct clustering of all species with high bootstrap values (>75%) (Figure 2). All isolates clustered with their respective reference or type strain sequences. Moreover, the constructed phylogenetic tree demonstrated branching into two main clusters. One subsection included the species A. dhakensis, A. hydrophila, and A. caviae whereas the other main cluster included A. salmonicida, A. piscicola, A. bestiarum, and A. media. Overall, a nearly identical clustering was obtained by phylogenetic analysis using the maximum-likelihood method, confirming the robustness of the neighbor-joining tree topology (data not shown).
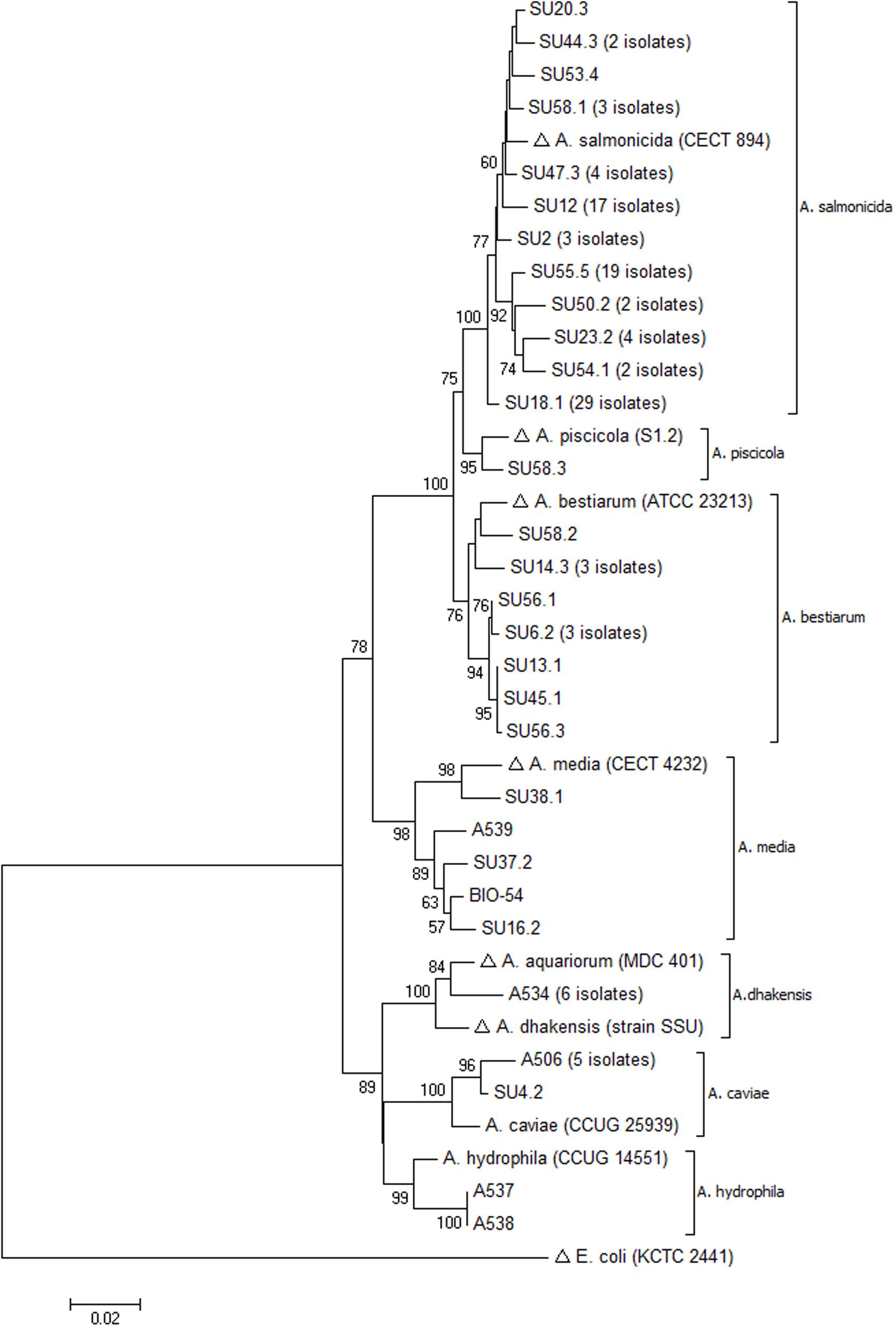
FIGURE 2. Neighbor-joining tree based on gyrB sequences showing inter-and intra-species evolutionary relationships of representative Aeromonas spp. isolated from sushi (with number of identical sequences indicated). Sequences retrieved from the GenBank database are marked with (Δ). The percentage of 1000 bootstrap replicates is shown next to the branches (only values > 50% are shown). The scale bar indicates evolutionary distance of 0.02 nucleotide substitutions per site. The analysis involved 38 sequences and a continuous stretch of 944 nt.
Discussion
In this study, Aeromonas strains isolated from retail sushi products from three producers were identified at species level and characterized genetically and phenotypically with respect to virulence traits, hemolysis and motility. We identified seven different Aeromonas species, equally distributed between the three producers, with the exception of A. hydrophila and A. dhakensis, which were isolated from one producer a year after the initial sampling. The variability of species distribution over time for producer A demonstrates the microbiological complexity of these products based on an assortment of raw ingredients. The most frequently isolated species, A. salmonicida, is by definition known as a psychrophilic, non-motile species, associated with fish disease (Austin et al., 1998). However, the A. salmonicida isolates in the present study fall into a group of so-called non-typical A. salmonicida characterized as mesophilic, sometimes motile, bacteria with no or reduced pigment production (Dallaire-Dufresne et al., 2014). Infections attributed to non-typical A. salmonicida are an increasing problem in aquaculture, mainly affecting the cleaner fish (wrasse) (Hjeltnes et al., 2015; Biering et al., 2016). A high prevalence of mesophilic A. salmonicida was also reported in studies of Italian food samples (Ottaviani et al., 2011) and in Mexican frozen fish samples (Castro-Escarpulli et al., 2003).
Phylogenetic analysis of Aeromonas spp. based on 16S rDNA sequencing has revealed that the genus is composed of a very tight group of species, some of them differing by only a few nucleotides (Martinez-Murcia et al., 1992; Kupfer et al., 2006). As an example, the inter-species similarity for the 16S rDNA sequences of A. salmonicida, A. bestiarum, and A. piscicola were reported as high as 99.8–100%, and the species were thus impossible to separate by 16S analysis (Figueras et al., 2011). Moreover, the overall mean 16S rDNA sequence similarity for Aeromonas spp. was 97.3%, highlighting the poor discriminatory power of the 16S rRNA gene (Nagar et al., 2013). The discriminatory power of some housekeeping genes was reported to be considerably higher than 16S rDNA, as demonstrated by an 89 and 92% mean sequence similarity for rpoD and gyrB, respectively (Soler et al., 2004; Nagar et al., 2013). The mean gyrB sequence similarity in the present study was 93%, and thus is in accordance with previous studies. The range of nucleotide substitutions within the tighter group of species A. salmonicida, A. bestiarum, and A. piscicola (2.7–3.4%) was on the border between the values of intra- and inter-species sequence divergence observed here, as reported previously (Yanez et al., 2003). Still, the sequence divergence between the three species were higher than the overall intra-species value (1.8%), demonstrating the power of gyrB to separate these phylogenetically close species.
The phylogenetic analysis based on partial gyrB sequences resulted in neighbor-joining and maximum-likelihood trees with robust and well-separated clusters of species. Of particular interest, all A. salmonicida and A. bestiarum isolates were separated into two robust clusters with a 100% bootstrap value (Figure 2). Isolate SU58.3 formed a separate sub-branch between the A. salmonicida and A. bestiarum clusters. Comparison with its closest relatives in the GenBank database produced significant alignment with one sequence of A. pisciciola (type strain sequence) and several sequences of A. bestiarum. However, isolate SU58.3 clustered with the type strain sequence of A. piscicola and their sequence similarity was 98.9%.
The taxonomical controversy of the genus Aeromonas was further demonstrated by the recent reclassification of A. hydrophila subsp. dhakensis (Huys et al., 2002) and A. aquariorum (Martinez-Murcia et al., 2008) as A. dhakensis (Beaz-Hidalgo et al., 2013). A nucleotide database search with the gyrB sequences from the six A. dhakensis isolates included in our study resulted mainly in alignments with sequences annotated as A. hydrophila. In fact, it is estimated that 30% of the genomes deposited in the GenBank database under the name A. hydrophila do not belong to this species (Teunis and Figueras, 2016). However, the phylogenetic analysis of the obtained gyrB sequences demonstrated that the six isolates clustered strongly with known sequences of A. aquariorum/A. dhakensis (Figure 2), and they were clearly separated from the A. hydrophila cluster. For decades, A. dhakensis has been mistaken for A. hydrophila, which might have contributed to an overestimation of the clinical significance of A. hydrophila. Increasing evidence indicates that A. dhakensis is widely distributed in the environment and must be recognized as a potent human pathogen (Chen et al., 2016).
The Aeromonas isolates in the present study were considered potentially pathogenic due to the high frequency of virulence-related genes. Aeromonas pathogenicity is associated with numerous virulence factors, but there is no definite link between the presence of specific toxin genes and clinical presentation. The lack of classification as a true pathogen is also related to the low number of registered outbreaks of foodborne disease linked to Aeromonas spp. Nevertheless, several studies have established an epidemiological link between the sources of infection and clinical isolates of Aeromonas spp. (Khajanchi et al., 2010; Pablos et al., 2011; Teunis and Figueras, 2016). As for other opportunistic pathogens, infections by potentially pathogenic Aeromonas may not always lead to disease due to host responses and the infectious bacterial dose. However, studies of clinical isolates from patients with various infections have brought more attention to some virulence factors, including the enterotoxins, hemolysins, and their secretion systems, and the clinical Aeromonas isolates have been reported to harbor a variety of toxin gene profiles (Rasmussen-Ivey et al., 2016). Furthermore, several studies have effaced the differences between clinical and environmental strains with respect to pathogenicity. The prevalence of toxin genes in isolates from food, environmental, and clinical sources was compared by Ottaviani et al. (2011), and no differences between these groups were detected. In agreement with previous studies of clinical and environmental isolates (Sen and Rodgers, 2004; Khajanchi et al., 2010), we detected multiple virulence-related genes in all sushi isolates. Moreover, we found heterogeneity in the distribution of toxin genes among the isolates, also within species. In our study, the combination of act/alt/hlyA/aerA was the most common, observed in 63% of the isolates. Interestingly, we observed variability in the combination of enterotoxin genes between species, but some of the species were represented by only one or a few isolates. In accordance with previous studies, we demonstrated lack of or reduced prevalence of the classical virulence genes in A. caviae (Kingombe et al., 1999; Carvalho et al., 2012). A significant proportion of clinical A. caviae strains that lacked all the toxin genes or harbored only the aerA gene alone were reported (Ottaviani et al., 2011). However, A. caviae is considered as one of the most potent human pathogen belonging to the genus Aeromonas and is frequently associated with infections (Pablos et al., 2010; Puthucheary et al., 2012). This suggests that for this species, there is a possible involvement of other virulence factors, and it highlights the relevant role of the aerolysin toxin in pathogenesis (Aguilera-Arreola et al., 2007; Ottaviani et al., 2011).
The act gene encoding a cytotoxic enterotoxin has been shown to be highly prevalent in strains from drinking water (70%) and seafood (75%) (Sen and Rodgers, 2004; Yano et al., 2015). For this gene, we observed a species-dependent prevalence consisting of a 100% prevalence in A. salmonicida, A. bestiarum, A. hydrophila, and A. piscicola (represented by only one isolate). In contrast, the act gene was not detected in any A. dhakensis, A. caviae, or A. media isolates. Act and aerolysin are pore-forming toxins, and the hemolytic, cytotoxic, and enterotoxic activities of these proteins have been previously demonstrated (Xu et al., 1998; Buckley and Howard, 1999; Sha et al., 2002). The vast majority of Aeromonas isolated from sushi in our study harbored both the aerA and hlyA genes, but not all of these isolates displayed β-hemolysis on blood agar. Shared characteristics of the non-hemolytic isolates were the absence of act, the faint aerA band observed on the agarose gel, and the presence of hlyA. For these isolates, a link between the absence of the act gene and the observed lack of hemolysis is suggested. In contrast, all A. dhakensis (act negative) isolates in the present study displayed strong hemolysis, suggesting that hemolysis cannot be attributed to a single factor. Wong et al. (1998) proposed a two-toxin model for aerA and hlyA, in which both genes must be knocked out to reduce virulence. The vast majority of Aeromonas isolated from sushi in our study harbored both the aerA and hlyA genes, but not all of these isolates displayed β-hemolysis on blood agar. Thus, the lack of observed hemolysis might be a result of altered gene expression and/or posttranslational processes not analyzed here.
Regardless of geographical origin, the alt and ast genes seem to be less prevalent, and their impact on pathogenesis has not been completely recognized. In the present study, the proportion of isolates harboring these genes were higher than reported previously (Di Pinto et al., 2012; Senderovich et al., 2012; Yano et al., 2015). It has been hypothesized that the species A. caviae and A. veronii lack the ast gene (Puthucheary et al., 2012). However, three out of six A. caviae isolates from the present study harbored this gene. Moreover, we detected ast in the A. veronii biovar veronii reference strain. Keeping in mind the heterogeneous distribution of various virulence genes in strains from various sources and geographical origin and the possibility of horizontal gene transfer, it is unlikely that the lack of specific genes can be attributed to certain species.
Human Aeromonas-mediated infections have occasionally been associated with hemolytic uremic syndrome (HUS), which can lead to acute renal failure and death. HUS is mainly associated with shiga-like toxins (Stx1 and/or Stx2) produced from enterohemorrhagic E. coli strains (EHEC) (Figueras et al., 2007). These toxins are encoded by stx genes in the genome of a lysogenic bacteriophage (Stx phage) of EHEC and represent a horizontal transfer mechanism (Palma-Martínez et al., 2016). The stx-1 and stx-2 genes have been detected in clinical and environmental isolates of A. hydrophila, A. caviae, and A. veronii biovar sobria (Snowden et al., 2006; Alperi and Figueras, 2010; Palma-Martínez et al., 2016). None of our isolates was positive for the stx-1/-2 genes, and to our knowledge, these genes have not been detected in foodborne isolates.
Bacterial motility, enabled by a lateral or polar flagella, have a number of biological functions in pathogens such as chemotaxis, adhesion, and invasion as seen in E. coli, P. aeruginosa, and Clostridium difficile (Haiko and Westerlund-Wikström, 2013; Qin et al., 2016). A recent study demonstrated that flagellar motility is necessary for A. hydrophila to adhere to the host mucus (Qin et al., 2016). We were able to demonstrate motility on soft agar plates in a significant proportion of A. salmonicida (71%) and A. hydrophila (100%) isolates, and less in the other species. However, it must be noted that perturbations in the motility assays such as inoculum state, incubation temperature, and brand of agar can give false negative results (Grim et al., 2013).
Most modern approaches for controlling levels of food contamination by microorganisms are effective against Aeromonas spp. (Daskalov, 2006). However, the increased consumption of raw or mildly processed RTE food poses some new safety concerns because the food is intended for consumption without further preparation such as washing or heat treatment. We have previously reported relatively high numbers of Aeromonas spp. (>log 4 CFU/g) in retail sushi before its stated expiration date. The risk of acute gastrointestinal disease associated with concentrations of Aeromonas spp. found in different water and food matrices was recently simulated by Teunis and Figueras (2016). Of particular concern, the concentration of Aeromonas spp. in the salad identified as the source of a substantial Chinese outbreak was estimated to be in the range of log 2.3–3.0 CFU/g. Several studies have reported the proliferation of Aeromonas spp. at low temperatures (Palumbo et al., 1985b; Palumbo and Buchanan, 1988; Mano et al., 2000; Papageorgiou et al., 2006). However, most studies were done in laboratory cultures or in products not comparable to sushi. Little information is available on the growth rates of Aeromonas spp. in sushi during refrigerated storage. Moreover, fluctuations in temperature are likely to occur during production, distribution, and display in stores. We have seen that the growth rates of other bacterial groups increased in sushi during storage at 8°C compared with storage at 4°C (submitted manuscript). Furthermore, analysis of sushi ingredients revealed that Aeromonas spp. can be introduced into the product through both raw vegetables and fish (Hoel et al., 2015). In the present study, we isolated Aeromonas spp. from a homogenized product containing two species of fish and vegetables (spring onion). Considering the number of species detected, and the fact that all isolates were mesophilic, these bacteria probably originated from more than one ingredient.
Conclusion
Our results demonstrate that potentially pathogenic Aeromonas species are widespread in retail sushi products. The isolates were assigned to seven different species according to gyrB sequencing. All isolates harbored several virulence genes, and a significant proportion were hemolytic and motile, thus showing virulence properties comparable to those in clinical strains (Wu et al., 2007; Puthucheary et al., 2012; Senderovich et al., 2012). The results highlight the potential microbiological hazard linked to the presence of Aeromonas spp. in sushi and in other perishable RTE food not subjected to heat treatment. Because of the multi-factorial virulence, a rapid discrimination between pathogenic and non-pathogenic strains remains a challenge for the genus Aeromonas.
Author Contributions
SH performed the experiments, phylogenetic analyses, and wrote the manuscript. OV did the statistical analysis, contributed to experimental planning and critically commented and revised the manuscript. AJ contributed to experimental planning and critically commented and revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00931/full#supplementary-material
Footnotes
- ^ http://blast.ncbi.nlm.nih.gov/
- ^ https://www.ncbi.nlm.nih.gov/genbank/
- ^ http://www.pearsoned.co.uk/HigherEducation/Resources/BarnardAskingQuestionsinBiology4e/
References
Aguilera-Arreola, M. G., Hernandez-Rodriguez, C., Zuniga, G., Figueras, M. J., Garduno, R. A., and Castro-Escarpulli, G. (2007). Virulence potential and genetic diversity of Aeromonas caviae, Aeromonas veronii, and Aeromonas hydrophila clinical isolates from Mexico and Spain: a comparative study. Can. J. Microbiol. 53, 877–887. doi: 10.1139/w07-051
Alperi, A., and Figueras, M. J. (2010). Human isolates of Aeromonas possess Shiga toxin genes (stx1 and stx2) highly similar to the most virulent gene variants of Escherichia coli. Clin. Microbiol. Infect. 16, 1563–1567. doi: 10.1111/j.1469-0691.2010.03203.x
Austin, B., Austin, D. A., Dalsgaard, I., Gudmundsdottir, B. K., Hoie, S., Thornton, J. M., et al. (1998). Characterization of atypical Aeromonas salmonicida by different methods. Syst. Appl. Microbiol. 21, 50–64. doi: 10.1016/S0723-2020(98)80008-8
Barnard, C. J., Gilbert, F., and McGregor, P. (2011). Asking Question in Biology: A Guide to Hypothesis Testing, Experimental Design and Presentation in Practical Work and Research Projects, 4th Edn. Harlow: Pearson.
Beaz-Hidalgo, R., Martinez-Murcia, A., and Figueras, M. J. (2013). Reclassification of Aeromonas hydrophila subsp. dhakensis Huys et al., 2002 and Aeromonas aquariorum Martinez-Murcia et al., 2008 as Aeromonas dhakensis sp. nov. comb nov. and emendation of the species Aeromonas hydrophila. Syst. Appl. Microbiol. 36, 171–176. doi: 10.1016/j.syapm.2012.12.007
Biering, E., Vaagnes,Ø., Krossøy, B., Gulla, S., and Colquhoun, D. J. (2016). Challenge models for atypical Aeromonas salmonicida and Vibrio anguillarum in farmed Ballan wrasse (Labrus bergylta) and preliminary testing of a trial vaccine against atypical Aeromonas salmonicida. J. Fish Dis. 39, 1257–1261. doi: 10.1111/jfd.12450
Buckley, J. T., and Howard, S. P. (1999). The cytotoxic enterotoxin of Aeromonas hydrophila is aerolysin. Infect. Immun. 67, 466–467.
Carvalho, M. J., Martínez-Murcia, A., Esteves, A. C., Correia, A., and Saavedra, M. J. (2012). Phylogenetic diversity, antibiotic resistance and virulence traits of Aeromonas spp. from untreated waters for human consumption. Int. J. Food Microbiol. 159, 230–239. doi: 10.1016/j.ijfoodmicro.2012.09.008
Castro-Escarpulli, G., Figueras, M. J., Aguilera-Arreola, G., Soler, L., Fernandez-Rendon, E., Aparicio, G. O., et al. (2003). Characterisation of Aeromonas spp. isolated from frozen fish intended for human consumption in Mexico. Int. J. Food Microbiol. 84, 41–49. doi: 10.1016/S0168-1605(02)00393-8
Chen, P.-L., Lamy, B., and Ko, W.-C. (2016). Aeromonas dhakensis, an increasingly recognized human pathogen. Front. Microbiol. 7:793. doi: 10.3389/fmicb.2016.00793
Dallaire-Dufresne, S., Tanaka, K. H., Trudel, M. V., Lafaille, A., and Charette, S. J. (2014). Virulence, genomic features, and plasticity of Aeromonas salmonicida subsp. salmonicida, the causative agent of fish furunculosis. Vet. Microbiol. 169, 1–7. doi: 10.1016/j.vetmic.2013.06.025
Daskalov, H. (2006). The importance of Aeromonas hydrophila in food safety. Food Control 17, 474–483. doi: 10.1016/j.foodcont.2005.02.009
Di Pinto, A., Terio, V., Pinto, P., and Tantillo, G. (2012). Detection of potentially pathogenic Aeromonas isolates from ready-to-eat seafood products by PCR analysis. Int. J. Food Sci. Technol. 47, 269–273. doi: 10.1111/j.1365-2621.2011.02835.x
Figueras, M., Aldea, M., Fernandez, N., Aspiroz, C., Alperi, A., and Guarro, J. (2007). Aeromonas hemolytic uremic syndrome. A case and a review of the literature. Diagn. Micrbiol. Infect. Dis. 58, 231–234. doi: 10.1016/j.diagmicrobio.2006.11.023
Figueras, M. J., Alperi, A., Beaz-Hidalgo, R., Stackebrandt, E., Brambilla, E., Monera, A., et al. (2011). Aeromonas rivuli sp. nov., isolated from the upstream region of a karst water rivulet. Int. J. Syst. Evol. Microbiol. 61, 242–248. doi: 10.1099/Ijs.0.016139-0
Galindo, C. L., Sha, J., Fadl, A. A., Pillai, L. L., and Chopra, A. K. (2006). Host immune responses to Aeromonas virulence factors. Curr. Immunol. Rev. 2, 13–26. doi: 10.2174/157339506775471910
Gavín, R., Merino, S., Altarriba, M., Canals, R., Shaw, J. G., and Tomás, J. M. (2003). Lateral flagella are required for increased cell adherence, invasion and biofilm formation by Aeromonas spp. FEMS Microbiol. Lett. 224, 77–83. doi: 10.1016/s0378-1097(03)00418-x
Grim, C. J., Kozlova, E. V., Sha, J., Fitts, E. C., van Lier, C. J., Kirtley, M. L., et al. (2013). Characterization of Aeromonas hydrophila wound pathotypes by comparative genomic and functional analyses of virulence genes. MBio 4:e00064-13 doi: 10.1128/mBio.00064-13
Haiko, J., and Westerlund-Wikström, B. (2013). The role of the bacterial flagellum in adhesion and virulence. Biology 2, 1242–1267. doi: 10.3390/biology2041242
Heaton, J. C., and Jones, K. (2008). Microbial contamination of fruit and vegetables and the behaviour of enteropathogens in the phyllosphere: a review. J. Appl. Microbiol. 104, 613–626. doi: 10.1111/j.1365-2672.2007.03587.x
Hjeltnes, B., Walde, C., Bang Jensen, B., and Haukaas, A. (2015). The Fish Health Report 2015. Oslo: The Norwegian Veterinary Institute.
Hoel, S., Mehli, L., Bruheim, T., Vadstein, O., and Jakobsen, A. N. (2015). Assessment of microbiological quality of retail fresh sushi from selected sources in Norway. J. Food Prot. 78, 977–982. doi: 10.4315/0362-028X.JFP-14-480
Howard, S. P., Garland, W. J., Green, M. J., and Buckley, J. T. (1987). Nucleotide sequence of the gene for the hole-forming toxin aerolysin of Aeromonas hydrophila. J. Bacteriol. 169, 2869–2871. doi: 10.1128/jb.169.6.2869-2871.1987
Huys, G., Kampfer, P., Albert, M. J., Kuhn, I., Denys, R., and Swings, J. (2002). Aeromonas hydrophila subsp. dhakensis subsp. nov., isolated from children with diarrhoea in Bangladesh, and extended description of Aeromonas hydrophila subsp. hydrophila (Chester 1901) Stanier 1943 (approved lists 1980). Int. J. Syst. Evol. Microbiol. 52(Pt 3), 705–712. doi: 10.1099/00207713-52-3-705
Hwang, C.-A. (2010). “Delicatessen salads,” in Ready-to-eat Foods - Microbial Concerns and Control Measures, eds A. Hwang and L. Huang (Boca Raton, FL: CRC Press), 61–74. doi: 10.1201/EBK1420068627-c2
Jahid, I. K., Lee, N.-Y., Kim, A., and Ha, S.-D. (2013). Influence of glucose concentrations on biofilm formation, motility, exoprotease production, and quorum sensing in Aeromonas hydrophila. J. Food Prot. 76, 239–247. doi: 10.4315/0362-028x.jfp-12-321
Jahid, I. K., Mizan, M. F., Ha, A. J., and Ha, S. D. (2015). Effect of salinity and incubation time of planktonic cells on biofilm formation, motility, exoprotease production, and quorum sensing of Aeromonas hydrophila. Food Microbiol. 49, 142–151. doi: 10.1016/j.fm.2015.01.016
Janda, J. M., and Abbott, S. L. (2010). The Genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73. doi: 10.1128/cmr.00039-09
Khajanchi, B. K., Fadl, A. A., Borchardt, M. A., Berg, R. L., Horneman, A. J., Stemper, M. E., et al. (2010). Distribution of virulence factors and molecular fingerprinting of Aeromonas species isolates from water and clinical samples: suggestive evidence of water-to-human transmission. Appl. Environ. Microbiol. 76, 2313–2325. doi: 10.1128/Aem.02535-09
Kimura, M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kingombe, C. I. B., D’Aoust, J.-Y., Huys, G., Hofmann, L., Rao, M., and Kwan, J. (2010). Multiplex PCR method for detection of three Aeromonas enterotoxin genes. Appl. Environ. Microbiol. 76, 425–433. doi: 10.1128/aem.01357-09
Kingombe, C. I. B., Huys, G., Howald, D., Luthi, E., Swings, J., and Jemmi, T. (2004). The usefulness of molecular techniques to assess the presence of Aeromonas spp. harboring virulence markers in foods. Int. J. Food Microbiol. 94, 113–121. doi: 10.1016/S0168-1605(03)00105-3
Kingombe, C. I. B., Huys, G., Tonolla, M., Albert, M. J., Swings, J., Peduzzi, R., et al. (1999). PCR detection, characterization, and distribution of virulence genes in Aeromonas spp. Appl. Environ. Microbiol. 65, 5293–5302.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kupfer, M., Kuhnert, P., Korczak, B. M., Peduzzi, R., and Demarta, A. (2006). Genetic relationships of Aeromonas strains inferred from 16S rRNA, gyrB and rpoB gene sequences. Int. J. Syst. Evol. Microbiol. 56, 2743–2751. doi: 10.1099/ijs.0.63650-0
Latif-Eugenín, F., Beaz-Hidalgo, R., and Figueras, M. J. (2016). First record of the rare species Aeromonas schubertii from mussels: phenotypic and genetic reevaluation of the species and a review of the literature. Arch. Microbiol. 198, 333–345. doi: 10.1007/s00203-016-1189-5
Mano, S. B., Ordonez, J. A., and De Fernando, G. D. G. (2000). Growth/survival of natural flora and Aeromonas hydrophila on refrigerated uncooked pork and turkey packaged in modified atmospheres. Food Microbiol. 17, 657–669. doi: 10.1006/fmic.2000.0358
Martínez-Murcia, A., Beaz-Hidalgo, R., Navarro, A., Carvalho, M. J., Aravena-Román, M., Correia, A., et al. (2016). Aeromonas lusitana sp. nov., isolated from untreated water and vegetables. Curr. Microbiol. 72, 795–803. doi: 10.1007/s00284-016-0997-9
Martinez-Murcia, A. J., Benlloch, S., and Collins, M. D. (1992). Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: lack of congruence with results of DNA-DNA hybridizations. Int. J. Syst. Bacteriol. 42, 412–421. doi: 10.1099/00207713-42-3-412
Martinez-Murcia, A. J., Saavedra, M. J., Mota, V. R., Maier, T., Stackebrandt, E., and Cousin, S. (2008). Aeromonas aquariorum sp nov., isolated from aquaria of ornamental fish. Int. J. Syst. Evol. Microbiol. 58, 1169–1175. doi: 10.1099/ijs.0.65352-0
Mateos, D., Anguita, J., Naharro, G., and Paniagua, C. (1993). Influence of growth temperature on the production of extracellular virulence factors and pathogenicity of environmental and human strains of Aeromonas hydrophila. J. Appl. Bacteriol. 74, 111–118. doi: 10.1111/j.1365-2672.1993.tb03003.x
Morandi, A., Zhaxybayeva, O., Gogarten, J. P., and Graf, J. (2005). Evolutionary and diagnostic implications of intragenomic heterogeneity in the 16S rRNA gene in Aeromonas strains. J. Bacteriol. 187, 6561–6564. doi: 10.1128/JB.187.18.6561-6564.2005
Nagar, V., Shashidhar, R., and Bandekar, J. R. (2013). Characterization of Aeromonas strains isolated from Indian foods using rpoD gene sequencing and whole cell protein analysis. World J. Microbiol. Biotechnol. 29, 745–752. doi: 10.1007/s11274-012-1212-1
Nordic Committee on Food Analysis [NMKL] (2004). Mesophilic Aeromonas species. Quantification in Foods and Feeds. Oslo: NKML.
Ørmen,Ø., Granum, P. E., Lassen, J., and Figueras, M. J. (2005). Lack of agreement between biochemical and genetic identification of Aeromonas spp. APMIS 113, 203–207. doi: 10.1111/j.1600-0463.2005.apm1130308.x
Ottaviani, D., Parlani, C., Citterio, B., Masini, L., Leoni, F., Canonico, C., et al. (2011). Putative virulence properties of Aeromonas strains isolated from food, environmental and clinical sources in Italy: a comparative study. Int. J. Food Microbiol. 144, 538–545. doi: 10.1016/j.ijfoodmicro.2010.11.020
Pablos, M., Huys, G., Cnockaert, M., Rodriguez-Calleja, J. M., Otero, A., and Garcia-Lopez, M. L. (2011). Identification and epidemiological relationships of Aeromonas isolates from patients with diarrhea, drinking water and foods. Int. J. Food Microbiol. 147, 203–210. doi: 10.1016/j.ijfoodmicro.2011.04.006
Pablos, M., Remacha, M. A., Rodriguez-Calleja, J. M., Santos, J. A., Otero, A., and Garcia-Lopez, M. L. (2010). Identity, virulence genes, and clonal relatedness of Aeromonas isolates from patients with diarrhea and drinking water. Eur. J. Clin. Microbiol. Infect. Dis. 29, 1163–1172. doi: 10.1007/s10096-010-0982-3
Pablos, M., Rodriguez-Calleja, J. M., Santos, J. A., Otero, A., and Garcia-Lopez, M. L. (2009). Occurrence of motile Aeromonas in municipal drinking water and distribution of genes encoding virulence factors. Int. J. Food Microbiol. 135, 158–164. doi: 10.1016/j.ijfoodmicro.2009.08.020
Palma-Martínez, I., Guerrero-Mandujano, A., Ruiz-Ruiz, M. J., Hernández-Cortez, C., Molina-López, J., Bocanegra-García, V., et al. (2016). Active shiga-like toxin produced by some Aeromonas spp., isolated in Mexico City. Front. Microbiol. 7:1522. doi: 10.3389/fmicb.2016.01522
Palumbo, S. A., and Buchanan, R. L. (1988). Factors affecting growth or survival of Aeromonas hydrophila in foods. J. Food Saf. 9, 37–51. doi: 10.1111/j.1745-4565.1988.tb00506.x
Palumbo, S. A., Maxino, F., Williams, A. C., Buchanan, R. L., and Thayer, D. W. (1985a). Starch-ampicillin agar for the quantitative detection of Aeromonas hydrophila. Appl. Environ. Microbiol. 50, 1027–1030.
Palumbo, S. A., Morgan, D. R., and Buchanan, R. L. (1985b). Influence of temperature, NaCI, and pH on the growth of Aeromonas hydrophila. J. Food Sci. 50, 1417–1421.
Papageorgiou, D. K., Melas, D. S., Abrahim, A., and Angelidis, A. S. (2006). Growth of Aeromonas hydrophila in the whey cheeses Myzithra, Anthotyros, and Manouri during storage at 4 and 12 degrees C. J. Food Prot. 69, 308–314. doi: 10.4315/0362-028X-69.2.308
Parker, J. L., and Shaw, J. G. (2011). Aeromonas spp. clinical microbiology and disease. J. Infect. 62, 109–118. doi: 10.1016/j.jinf.2010.12.003
Paton, A. W., and Paton, J. C. (1998). Detection and characterization of shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36, 598–602.
Puthucheary, S. D., Puah, S. M., and Chua, K. H. (2012). Molecular characterization of clinical isolates of Aeromonas species from Malaysia. PLoS ONE 7:e30205. doi: 10.1371/journal.pone.0030205
Qin, Y., Lin, G., Chen, W., Xu, X., and Yan, Q. (2016). Flagellar motility is necessary for Aeromonas hydrophila adhesion. Microb. Pathog. 98, 160–166. doi: 10.1016/j.micpath.2016.07.006
Rasmussen-Ivey, C. R., Figueras, M. J., McGarey, D., and Liles, M. R. (2016). Virulence factors of Aeromonas hydrophila: in the wake of reclassification. Front. Microbiol. 7:1337. doi: 10.3389/fmicb.2016.01337
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sen, K., and Rodgers, M. (2004). Distribution of six virulence factors in Aeromonas species isolated from US drinking water utilities: a PCR identification. J. Appl. Microbiol. 97, 1077–1086. doi: 10.1111/j.1365-2672.2004.02398.x
Senderovich, Y., Ken-Dror, S., Vainblat, I., Blau, D., Izhaki, I., and Halpern, M. (2012). A molecular study on the prevalence and virulence potential of Aeromonas spp. recovered from patients suffering from diarrhea in Israel. PLoS ONE 7:e30070. doi: 10.1371/journal.pone.0030070
Sha, J., Kozlova, E. V., and Chopra, A. K. (2002). Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect. Immun. 70, 1924–1935. doi: 10.1128/IAI.70.4.1924-1935.2002
Snowden, L., Wernbacher, L., Stenzel, D., Tucker, J., McKay, D., O’Brien, M., et al. (2006). Prevalence of environmental Aeromonas in South East Queensland, Australia: a study of their interactions with human monolayer Caco-2 cells. J. Appl. Microbiol. 101, 964–975. doi: 10.1111/j.1365-2672.2006.02919.x
Soler, L., Yáñez, M. A., Chacon, M. R., Aguilera-Arreola, M. G., Catalán, V., Figueras, M. J., et al. (2004). Phylogenetic analysis of the genus Aeromonas based on two housekeeping genes. Int. J. Syst. Evol. Microbiol. 54, 1511–1519. doi: 10.1099/ijs.0.03048-0
Tacao, M., Moura, A., Alves, A., Henriques, I., Saavedra, M. J., and Correia, A. (2005). Evaluation of 16S rDNA- and gyrB-DGGE for typing members of the genus Aeromonas. FEMS Microbiol. Lett. 246, 11–18. doi: 10.1016/j.femsle.2005.03.033
Teunis, P., and Figueras, M. J. (2016). Reassessment of the enteropathogenicity of mesophilic Aeromonas species. Front. Microbiol. 7:1395. doi: 10.3389/fmicb.2016.01395
Villari, P., Crispino, M., Montuori, P., and Stanzione, S. (2000). Prevalence and molecular characterization of Aeromonas spp. in ready-to-eat foods in Italy. J. Food Prot. 63, 1754–1757. doi: 10.4315/0362-028X-63.12.1754
Wong, C. Y. F., Heuzenroeder, M. W., and Flower, R. L. P. (1998). Inactivation of two haemolytic toxin genes in Aeromonas hydrophila attenuates virulence in a suckling mouse model. Microbiology 144, 291–298. doi: 10.1099/00221287-144-2-291
Wu, C. J., Wu, J. J., Yan, J. J., Lee, H. C., Lee, N. Y., Chang, C. M., et al. (2007). Clinical significance and distribution of putative virulence markers of 116 consecutive clinical Aeromonas isolates in southern Taiwan. J. Infect. 54, 151–158. doi: 10.1016/j.jinf.2006.04.002
Xanthopoulos, V., Tzanetakis, N., and Litopoulou-Tzanetaki, E. (2010). Occurrence and characterization of Aeromonas hydrophila and Yersinia enterocolitica in minimally processed fresh vegetable salads. Food Control 21, 393–398. doi: 10.1016/j.foodcont.2009.06.021
Xu, X. J., Ferguson, M. R., Popov, V. L., Houston, C. W., Peterson, J. W., and Chopra, A. K. (1998). Role of a cytotoxic enterotoxin in Aeromonas-mediated infections: development of transposon and isogenic mutants. Infect. Immun. 66, 3501–3509.
Yanez, M. A., Catalan, V., Apraiz, D., Figueras, M. J., and Martinez-Murcia, A. J. (2003). Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int. J. Syst. Evol. Microbiol. 53, 875–883. doi: 10.1099/ijs.0.02443-0
Keywords: Aeromonas spp., virulence factors, gyrB, sushi, ready-to-eat food, food safety
Citation: Hoel S, Vadstein O and Jakobsen AN (2017) Species Distribution and Prevalence of Putative Virulence Factors in Mesophilic Aeromonas spp. Isolated from Fresh Retail Sushi. Front. Microbiol. 8:931. doi: 10.3389/fmicb.2017.00931
Received: 10 March 2017; Accepted: 08 May 2017;
Published: 24 May 2017.
Edited by:
Mehdi Razzaghi-Abyaneh, Pasteur Institute of Iran, IranReviewed by:
Hikmate Abriouel, Universidad de Jaén, SpainJesus L. Romalde, Universidade de Santiago de Compostela, Spain
Copyright © 2017 Hoel, Vadstein and Jakobsen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sunniva Hoel, c3Vubml2YS5ob2VsQG50bnUubm8=
 Sunniva Hoel
Sunniva Hoel Olav Vadstein
Olav Vadstein Anita N. Jakobsen
Anita N. Jakobsen