- APC Microbiome Institute and School of Microbiology, University College Cork, Cork, Ireland
Bacteria evolved over millennia in the presence of the vital micronutrient iron. Iron is involved in numerous processes within the cell and is essential for nearly all living organisms. The importance of iron to the survival of bacteria is obvious from the large variety of mechanisms by which iron may be acquired from the environment. Random mutagenesis and global gene expression profiling led to the identification of a number of genes, which are essential for Bifidobacterium breve UCC2003 survival under iron-restrictive conditions. These genes encode, among others, Fe-S cluster-associated proteins, a possible ferric iron reductase, a number of cell wall-associated proteins, and various DNA replication and repair proteins. In addition, our study identified several presumed iron uptake systems which were shown to be essential for B. breve UCC2003 growth under conditions of either ferric and/or ferrous iron chelation. Of these, two gene clusters encoding putative iron-uptake systems, bfeUO and sifABCDE, were further characterised, indicating that sifABCDE is involved in ferrous iron transport, while the bfeUO-encoded transport system imports both ferrous and ferric iron. Transcription studies showed that bfeUO and sifABCDE constitute two separate transcriptional units that are induced upon dipyridyl-mediated iron limitation. In the anaerobic gastrointestinal environment ferrous iron is presumed to be of most relevance, though a mutation in the sifABCDE cluster does not affect B. breve UCC2003's ability to colonise the gut of a murine model.
Introduction
Iron is an essential nutrient for nearly all forms of life, having co-evolved with biological systems for billions of years; it is a key element in many redox reactions and is involved in various cellular processes such as DNA replication, nitrogen fixation, nucleotide biosynthesis and the production of various metabolites (Ilbert and Bonnefoy, 2013). The prevalence and importance of iron in biological systems is most likely due to the abundance of Fe2+ during the initial evolution of living organisms (Crichton and Pierre, 2001; Ilbert and Bonnefoy, 2013). The ionic forms of iron found in nature represent two interchangeable redox states: ferrous iron (Fe2+) and its more oxidized form ferric iron (Fe3+). These attributes make iron a versatile prosthetic component in many proteins as a biocatalyst or electron carrier (Andrews et al., 2003). Under aerobic neutral pH conditions iron is present in its oxidized, but essentially insoluble trivalent, ferric form, while under anaerobic, neutral pH conditions iron is found in its reduced, divalent ferrous state, the latter being more biologically accessible due to its greater solubility (Kortman et al., 2014).
Iron speciation, which relates to the changing concentration of varying forms of an ion as the pH of the solution changes, and iron availability are two factors which are important for the accessibility of iron within the gastrointestinal environment. Iron availability can vary greatly along the length of the gastrointestinal tract due to its tendency to complex with other molecules and because of its ability to exist in various oxidation states depending on its surrounding environment (Ilbert and Bonnefoy, 2013). The low pH in the stomach favors the solubility of both ferric and ferrous iron, whereas the subsequent rise in pH upon entry into the small intestine results in a decrease in the solubility of ferric iron. Iron may then complex with food components or host/microbiota-derived compounds such as citrate, ascorbate, mucin, certain amino acids, or lactate (Andrews et al., 2003). Certain insoluble forms of ferric iron, for example when complexed with phosphate, carbonate or oxides, are not readily available to the microbiota and may require reduction or removal from these complexes by siderophore activity (Kortman et al., 2014). Soluble forms of ferric iron may be reduced to ferrous iron upon entry into the cytoplasm, while ferrous citrate or ferrous ascorbate may be taken up directly by (certain components of) the microbiota. Finally, lactate and short chain fatty acid (SCFA) production by the microbiota may also cause a modest, possibly localised drop in the pH in the colon resulting in an increase in iron solubility (den Besten et al., 2013).
The human gut microbiota typically includes clostridia, eubacteria, and species of the genera Bacteroides and Bifidobacterium (Picard et al., 2005). Of particular interest, the genus Bifidobacterium belongs to the Bifidobacteriaceae family, which in turn belongs to the Actinobacteria phylum. Henri Tissier was the first to isolate a Bifidobacterium species in 1899 from the faeces of a healthy, breast-fed infant, and due to his pioneering work and subsequent research of many others, members of this genus are today considered to represent health-promoting or probiotic bacteria (Sanders, 2008).
Competitive sequestration/withholding of iron has long been known in both Gram-positive and -negative bacteria as a defense mechanism, conferring a competitive advantage to certain commensals, such as bifidobacteria which thrive in low iron environments (Collins, 2008; Dostal et al., 2015). Bacteria have developed a wide range of mechanisms for iron sequestration, including a variety of systems for the uptake of ferrous iron, ferric iron, heme, hemephores (which scavenge heme from various hemoproteins), and siderophores (selective iron chelators which have a high affinity for ferric iron). Iron sequestration via siderophore-mediated and/or direct iron uptake has been reported in the Bifidobacterium genus, with Bifidobacterium breve exhibiting the highest siderophore activity and Bifidobacterium kashiwanohense exhibiting the highest iron uptake (Hood and Skaar, 2012; Vazquez-Gutierrez et al., 2015). Furthermore, bifidobacterial carbohydrate fermentation typically acidifies its surrounding environment, favouring the solubility of ferric and ferrous iron in a localised fashion (Vernazza et al., 2006). Interestingly, certain species of Bifidobacterium have also been found to bind ferric iron to their cell wall and membranes, thereby limiting its availability to other bacteria (Kot and Bezkorovainy, 1999). Bifidobacteria, like many other bacteria, are known to import iron (ions) across the cytoplasmic membrane by means of ABC-type transporters (Brown and Holden, 2002). In the case of B. breve, this leads to a cytoplasmic iron concentration (100/200 μM) which is about 14–16 times higher than its surrounding environment (Kot et al., 1993; Brown and Holden, 2002). Global gene expression analysis of B. breve UCC2003 under iron-limiting conditions has also revealed a number of iron-responsive genes, including bfeUOB (Cronin et al., 2012). The bfeUOB gene cluster is homologous to the Escherichia coli efeUOB genes, which are known to encode an iron transport system. The promoter of bfeUOB was found to be inducible under iron-limiting conditions and was utilised to create an inducible promoter system in bifidobacteria. A subsequent study found that insertional mutants created in the bfeUO genes and adjacent genes (with locus tags Bbr_0223-Bbr_0227) caused a decreased ability of B. breve to confer protection against Salmonella infection in the C. elegans model and to survive iron limitation (Christiaen et al., 2014).
In the current study random mutagenis was combined with global transcriptional profiling and led to the identification of a number of genes which are vital for B. breve UCC2003 survival under ferrous and/or feric iron limitiation. The identified genes are predicted to represent a variety of iron-related functions, such as a ferric iron reductase, Fe-S cluster associated proteins, a number of cell wall associated proteins, several DNA replication/repair proteins and various transport systems.
Materials and Methods
Bacterial Strains and Culture Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. B. breve UCC2003 was routinely grown at 37°C in either de Man Rogosa and Sharpe medium (MRS medium; Difco, BD, Le Pont de Claix, France), modified de Man Rogosa and Sharpe (mMRS) medium made from first principles (Man et al., 1960), or reinforced clostridial medium (RCM; Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) supplemented with 0.05% cysteine-HCl. Iron-limiting experiments were carried out in filtered RCM (fRCM) prepared in de-mineralised water. Maximal tolerable concentration (MTC) of dipyridyl, ciclopirox olamine, and phenanthroline were 275, 100, and 80 μM respectively, as determined by Christiaen et al. (2014). Bifidobacterial cultures were incubated anaerobically in a modular, atmosphere-controlled system (Davidson and Hardy, Belfast, Ireland). Where appropriate growth medium was supplemented with tetracycline (Tet; 10 μg ml−1), chloramphenicol (Cm; 5 μg ml−1 for L. lactis and E. coli, 2.5 μg ml−1 for B. breve), ampicillin (Amp; 100 μg ml−1), erythromycin (Em; 100 μg ml−1) or kanamycin (Kan; 50 μg ml−1) for plasmid selection and maintenance.
Phenotypic Screening and Monitoring of Dipyridyl-Sensitive Growth
Phenotypic screening and determination of transposon insertion sites was carried out as described previously (Ruiz et al., 2013) with the following adjustments: B. breve UCC2003 transposon mutants were sub-cultured twice in RCM supplemented with tetracycline and spotted onto Q-Trays (Molecular Devices, Berkshire, United Kingdom) containing RCA or RCA supplemented with the iron chelator dipyridyl at a concentration of either 250 or 275 μM. These two different concentrations of dipyridyl were chosen as they did not impair growth of the WT strain and to reduce the number of false positives identified in subsequent confirmatory analysis. Transposon mutants which grew on RCA, but failed to grow or showed poor growth on RCA supplemented with (either concentration of) dipyridyl were then selected for further analysis. The phenotype of such dipyridyl-sensitive mutants was confirmed by monitoring growth in fRCM and fRCM containing dipyridyl (at a final concentration of 250 or 275 μM). Such mutants were incubated anaerobically at 37°C, with OD600 readings taken at 24 h following inoculation. Mutants which were found to be sensitive to growth on dipyridyl were further tested for growth on fRCM in the presence of ciclopirox olamine (100 μM) and phenanthroline (80 μM).
Nucleotide Sequence Analysis
Sequence data was obtained from the Artemis-mediated (Rutherford et al., 2000) genome annotations of the B. breve UCC2003 genome sequence (O'Connell Motherway et al., 2011). Data base searches were carried out using non-redundant sequences accessible at the National Centre for Biotechnology Information internet site (https://www.ncbi.nlm.nih.gov/) utilising the basic alignment search tool (Blast). Sequence analysis was performed by using the Seqbuilder and Seqman programs of the DNASTAR software package (DNASTAR, Madison, WI). Protein functions were assigned with the use of the basic protein alignment search tool BlastP and homology detection and structure prediction by HMM-HMM comparison; HHpred (Altschul et al., 1990, 1997). Rho-independent terminators were identified utilising ARNold (Gautheret and Lambert, 2001). Bacterial promoters were predicted with BPROM (http://softberry.com).
DNA Manipulations
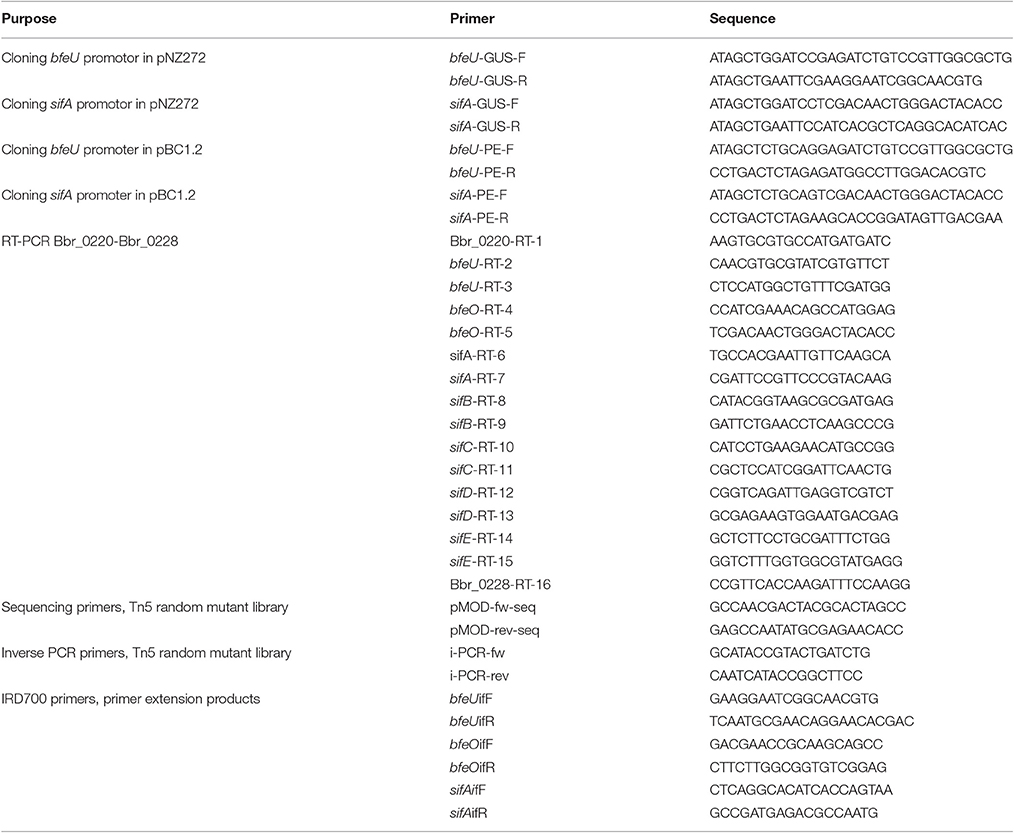
DNA manipulations were carried out as previously reported (Russell, 2001). Restriction enzymes and T4 DNA ligase were obtained from Roche Diagnostics, and were used according to the manufacturer's instructions. PCRs were performed using Extensor Long Range PCR Enzyme master mix (Thermo Scientific). Synthetic oligonucleotides were synthesized by Eurofins (Ebersberg, Germany) and are listed in Table 2. PCR products were purified by the use of a High-Pure PCR product purification kit (Roche). Plasmid DNA was introduced into E. coli and B. breve by electroporation and large-scale preparation of chromosomal DNA from B. breve was performed as described previously (O'Riordan and Fitzgerald, 1999). Plasmid DNA was obtained from B. breve and E. coli using the Roche High Pure plasmid isolation kit (Roche Diagnostics, Basel, Switzerland). An initial lysis step was performed using 30 mg ml−1 of lysozyme for 30 min at 37°C as part of the plasmid purification protocol for B. breve.
Transcriptome Analysis
B. breve UCC2003's transcriptome response to iron limitation was tested by subjecting exponentially growing cells to a high concentration (700 μM) of dipyridyl for 30 min. This concentration was chosen in order to rapidly and substantially reduce iron availability, thus allowing the monitoring of an immediate transcriptional response of B. breve UCC2003 to iron limitation. Cells were prepared as follows; an overnight culture of B. breve UCC2003 in RCM was inoculated at 1% into filtered RCM broth and incubated until an OD600 of 0.5 was reached. Cells were then exposed to 700 μM dipyridyl for 30 min. B. breve UCC2003 controls were treated in the same manner except for the addition of dipyridyl. Following dipyridyl exposure as outlined above (or in the absence of dipyridyl for controls), cells were harvested by centrifugation at 10,000 rpm for 2 min at room temperature and immediately frozen at −80°C prior to RNA isolation. DNA microarrays containing oligonucleotide primers representing each of the annotated genes on the genome of B. breve UCC2003 were designed by and obtained from Agilent Technologies (Palo Alto, CA, USA). Cell disruption, RNA isolation, RNA quality control, and cDNA synthesis and labeling were performed as described previously (Zomer et al., 2009). The labeled cDNA was hybridized using the Agilent Gene Expression hybridization kit (part number 5188–5242) as described in the Agilent Two-ColorMicroarrayBased Gene Expression Analysis v4.0 manual (G4140-90050). Following hybridization, the microarrays were washed in accordance with Agilent's standard procedures and scanned using an Agilent DNA microarray scanner (model G2565A). The generated scans were converted to data files with Agilent's Feature Extraction software (v9.5). The DNA microarray data were processed as previously described (García de la Nava et al., 2003; van Hijum et al., 2003, 2005). Differential expression tests were performed with the Cyber-T implementation of a variant of the t-test (Long et al., 2001). A gene was considered to exhibit a significantly different expression level relative to the control when p < 0.001 and an expression ratio of > 1.7 or < 0.25. The microarray data obtained in this study have been deposited in NCBI's Gene Expression Omnibus database and is accessible through GEO series accession number GSE92758.
Real Time-PCR
Primers were chosen to amplify upstream regions of all genes from Bbr_0220-Bbr_0227 (Table 1). B. breve UCC2003 was prepared for RNA extraction as follows: cells were incubated until an OD600 of 0.5 was achieved and were then exposed to 700 μM dipyridyl for 30 min, cells were harvested by centrifugation at 10,000 rpm for 2 min at room temperature and immediately frozen at −80°C. Cell disruption, RNA isolation and cDNA synthesis were performed as previously described (Gueimonde et al., 2009). Two microgram of RNA was treated with 2 units of DNase, RNase free (Roche) for 1 h at 37°C. cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen). Absence of chromosomal DNA contamination was checked by real-time PCR. RT-PCR experiments were carried out using the cDNA-containing sample as a template and employing primers as above and Extensor Hi-Fidelity PCR Master Mix (Thermo Scientific).
Beta-Glucoronidase Assay
The potential promoter-containing regions of bfeU and sifA were amplified by PCR using primer combinations bfeU-GUS-F, bfeU-GUS-R, and sifA-GUS-F, sifA-GUS-R (Table 2), which contain EcoRI and BamH1 restriction sites at their 5′ ends. Amplicons were digested with EcoRI and BamH1, and cloned upstream of the promoterless gusA gene present in the similarly restricted pNZ272 reporter vector (Platteeuw et al., 1994). Ligation mixtures were introduced by electroporation in Lactococcus lactis NZ9000 competent cells. The resulting plasmids, pNZ272-bfeU-IR and pNZ272-sifA-IR, once verified by restriction and sequence analysis, were then introduced into B. breve UCC2003 by electroporation (Table 1). GusA activity assays in B. breve UCC2003 were carried out in triplicate by independent assay as previously described (Alvarez-Martin et al., 2012), with the following modifications: cells were grown in fRCM to an OD600 of ~0.3–0.4, at which transcription was induced by the addition of 700 μM dipyridyl for 20 min. Two hundred microliters of cell culture were used in the assay. GusA activity was expressed in Miller Units and calculated using the following equation: 1000*((O.D.420 − (1.75*OD550))/(t × v × O.D.600)), where t is reaction time in minutes, v is cell volume in [ml] and OD420,OD550, and OD600 are absorbance values at 420, 550, and 600 nm.
Primer Extensions
The upstream (intergenic) regions of bfeU and sifA were PCR amplified using B. breve UCC2003 genomic DNA as the template and oligonucleotide primer combinations listed in Table 3. These PCR products were then individually ligated into pBC1.2, using PstI and XbaI restriction sites and transformed into E. coli XL1 BLUE cells by electroporation. E. coli XL1 BLUE transformants containing the recombinant pBC1.2 constructs were selected for on LB agar with appropriate antibiotics. The integrity of the constructs was then confirmed by restriction and sequence analysis, and plasmid preparations of resulting recombinant plasmids, designated pBC1.2-bfeU-IR and pBC1.2-sifA-IR (names correspond to the gene downstream of the promoter in the UCC2003 genome, see Table 1), were introduced by electroporation into B. breve UCC2003 with selection on RCA supplemented with the appropriate antibiotic.
B. breve UCC2003-pBC1.2-bfeU-IR and B. breve UCC2003-pBC1.2-sifA-IR were prepared as follows for RNA extraction; Cells were incubated until an OD600 of 0.5 was achieved and were then exposed to 700 μM dipyridyl for 30 min, cells were harvested by centrifugation at 10,000 rpm for 2 min at room temperature and immediately frozen at −80°C. RNA extraction was carried out as previously described (Zomer et al., 2009).
The 5′ ends of the RNA transcripts were determined by annealing 1 pmol of an IRD700-labeled synthetic oligonucleotide to 20 μg of RNA, as previously described (Ventura et al., 2004). The following IRD700 labeled primer pairs were used: bfeUifF and bfeUifR, sifAifF and sifAifR (Table 3). Corresponding sequence ladders of the promoter regions of bfeU and sifA were produced using the same primer as in the primer extension reaction and employing a DNA cycle-sequencing kit (Jena Bioscience, Germany) and were run alongside the primer extension products to allow precise alignment of the transcriptional start site (TSS) with the corresponding DNA sequence. Separation was achieved on a 6.5% Li-Cor Matrix KB Plus acrylamide gel. Signal detection and image capture were performed employing a Li-Cor sequencing instrument (Li-Cor Biosciences).
Murine Colonisation Experiments
Experiments with mice were approved by the University College Cork Animal Experimentation Ethics Committee and experimental procedures were conducted under license from the Irish Government (license number B100/3729). Seven-week-old female, BALB/c mice were housed in individually vented cages (Animal Care Systems) under a strict 12 h light cycle. Mice (n = 7 per group) were fed a standard polysaccharide-rich mouse chow diet and water ad libitum. Mice were inoculated by oral gavage 109 cfu of B. breve UCC2003PK1, B. breve UCC2003-bfeU and B. breve UCC2003-sifA (see Table 1 for corresponding descriptions of strains). Faecal pellets were collected at intervals during 18 days to enumerate bacteria. Eighteen days after inoculation, mice were sacrificed and their intestinal tracts immediately dissected.
Results
Phenotypic Screening for Dipyridyl-Sensitive Mutants
A library containing ~20,000 B. breve UCC2003 random transposon mutants, previously constructed by Ruiz et al. (Ruiz et al., 2013), was screened in order to identify mutants with increased sensitivity to the ferrous iron chelator dipyridyl. The dipyridyl-sensitive phenotype was then validated for nearly 250 mutants by sub-cultivation in fRCM broth supplemented with dipyridyl (at a concentration of 250 or 275 μM), resulting in a final collection of 33 verified, dipyridyl-sensitive mutants. These 33 dipyridyl-sensitive mutants, were also analysed by growth profile analysis for their sensitivity two other iron chelators, ciclopirox olamine, a ferric iron chelator, and phenanthroline, an iron chelator which binds both ferrous and ferric iron (Bayer et al., 1964). The use of these three iron chelators allowed us to assess the cause of the growth defect of the obtained mutants as will be discussed below.
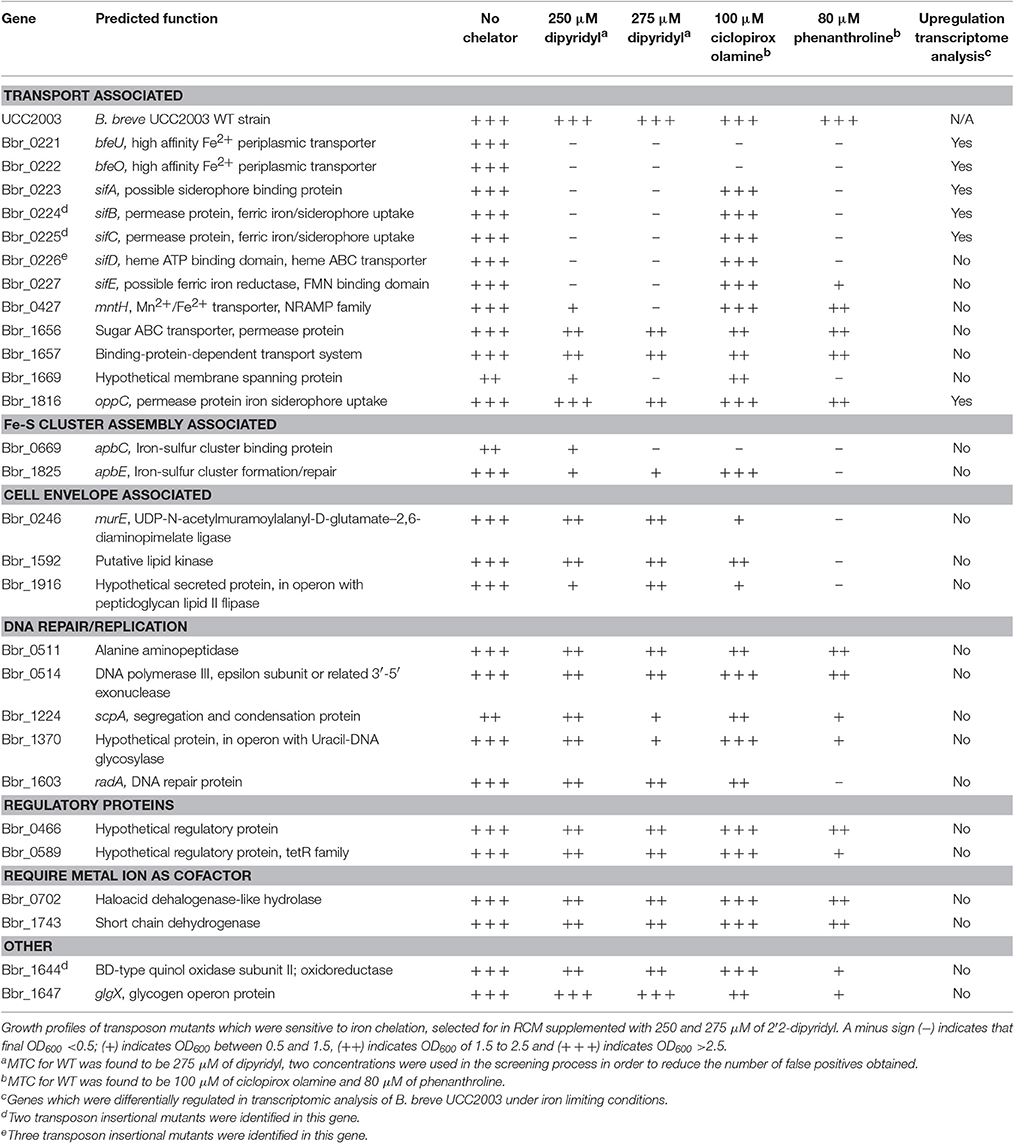
The transposon insertion site of validated dipyridyl-sensitive mutants was then determined for these 33 mutants using an approach that was described previously (Ruiz et al., 2013). A list of these mutants can be found in Table 3 and Figure 1; some mutants had been isolated more than once, resulting in a final collection of 28 genes which were identified as important for survival under (ferrous) iron limiting conditions. Based on the predicted functions of the interrupted genes, the identified dipyridyl-sensitive mutants can be divided into seven functional groups (see Table 3): (i) transport associated, (ii) Fe-S cluster assembly associated, (iii) cell envelope associated, (iv) DNA repair/replication, (v) regulatory proteins, (vi) proteins which require a metal co-factor and (vii) other.
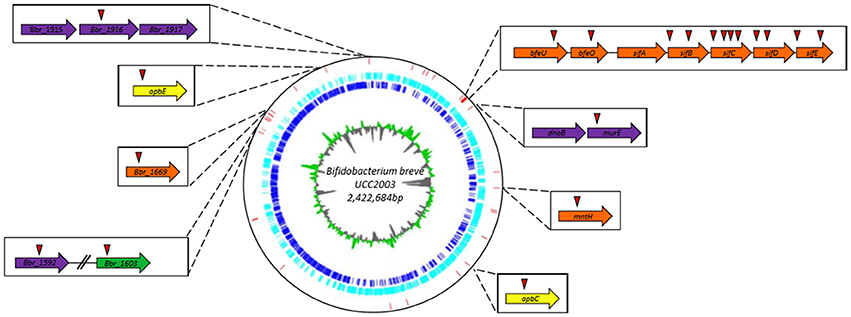
Figure 1. Circular genome map of B. breve UCC2003. The innermost circle illustrates GC skew, shown in green on the forward strand and in gray on the reverse strand. The dark blue circle displays the ORF on the forward strand, while the light blue circle representing all the ORF on the reverse strand. The red vertical bars indicate the transposon insertion sites of dipyridyl-sensitive mutants identified in the screening of the random mutant library. Genes in which transposon mutations were shown to cause complete growth impairment in the presence of either dipyridyl, phenanthroline or ciplopirox olamine are indicated in boxes together with their names or locus tags with the position(s) of the transposon insertion indicated by a red triangle.
A number of genes encoding (based on BlastP and HHpred analysis) predicted transport systems were identified in the screening, namely Bbr_0427 (designated here as mntH), specifying a homolog of a divalent cation transporter in E. coli. The mutant identified in the mntH gene was found to be sensitive to dipyridyl and phenanthroline, yet was not sensitive to ferric iron chelation with ciclopirox olamine (at the concentration used for this chelator). A transposon insertion was also identified in Bbr_1816 (oppC), encoding a permease protein of an iron siderophore ABC transport system. Similar to the mutant in mntH, the strain carrying a mutation in oppC was shown to be sensitive to iron chelation with dipyridyl and phenanthroline, but not with ciclopirox olamine, indicating that both strains are specifically affected in ferrous iron uptake or metabolism.
Ten distinct mutants were identified, carrying Tn5 insertions in a gene cluster represented by locus tags Bbr_0221–Bbr_0227. The products of Bbr_0221-Bbr_0222 (bfeUO) are predicted to represent a high affinity ferrous iron uptake system which is similar to proteins encoded by (efeUO) of the efeUOB gene cluster found in E. coli (Grosse et al., 2006). Functional annotation analysis revealed that BfeU which contains nine transmembrane domains and encodes a conserved hypothetical membrane spanning protein with an iron permease FTR1 family domain. While BfeO encodes a conserved hypothetical protein, containing a hydrophobic N-terminus suggesting it is a membrane-associated protein that functions in metal ion binding. Interestingly, BfeO has homology to a human lactoferrin transporter of Treponema pallidum based on HHpred analysis. The deduced proteins of Bbr_0223-Bbr_0227, designated here as sifABCDE, are predicted to specify a ferrous iron/siderophore uptake system, based on similarities to previously characterised ferrous iron uptake/aerobactin siderophore uptake systems encoded by E. coli 1520, plasmid pIP1206 and Bacillis subtilis 168 (Cronin et al., 2012; Fukushima et al., 2013). Functional annotation analysis revealed that SifA is a conserved hypothetical membrane-spanning protein containing eight transmembrane domains and is homologous to a cation antiporter. SifB and SifC are ABC transporter permeases with FtsX-like family domains, while SifD is an ATP-binding subunit protein with homology to a ferric ion import ATP-binding subunit. SifE contains a hydrophobic N-terminus indicating that it is membrane associated, while it also contains an FMN-binding domain which may be associated with oxidoreductase activity. Interestingly, SifE is homologous to IsdH, a membrane bound protein which is involved in removing heme from hemoglobin in Staphylococcus lugdunensis. Mutations in the bfeUO gene cluster were shown to cause severely impaired growth in the presence of all three iron chelators, indicating that this predicted iron uptake system is important for the uptake of both ferrous and ferric iron. In contrast, mutations in the sifABCDE gene cluster were shown to negatively affect growth in medium supplemented with either dipyridyl or phenanthroline, whereas normal growth (compared to growth of UCC2003 under these conditions) was observed in the presence of ciclopirox olamine, indicating that this system is specifically responsible for the transport of ferrous iron (Table 3).
Furthermore, mutations identified in two genes involved in iron-sulfur (Fe-S) cluster formation, repair and docking were identified as causing severe growth impairment under iron limitation by dipyridyl. Fe-S clusters act as cofactors for many cellular proteins which have prominent roles in many cellular processes including but not limited to respiration, central metabolism, gene regulation, DNA repair and replication (Kiley and Beinert, 2003; Andreini et al., 2016). The first transposon mutant identified in a gene Bbr_1825 (designated here as apbE); apbE is homologous to apbE of Salmonella enterica, the product of this gene is known to play a role in the assembly and repair of Fe-S clusters (Skovran and Downs, 2003; Py and Barras, 2010). The second transposon insertion was identified was in a gene Bbr_0669 (designated as apbC), encoding a protein with similarity to a P-loop NTPase, which binds and transfers Fe-S clusters to cytosolic apo-proteins (Hausmann et al., 2005; Boyd et al., 2008). This apbC::Tn5 mutant was severely impaired in growth in the presence of ferrous and ferric iron chelators, while the apbE::Tn5 mutant was impaired in growth only in the presence of ferrous iron chelators.
A number of genes encoding cell envelope-associated proteins were identified in the phenotypic screen, namely: Bbr_0246, encoding a putative UDP-N-acetylmuramoylalanyl-D-glutamate–2,6-diaminopimelate ligase; Bbr_1592, encoding a putative lipid kinase; and Bbr_1916, which encodes a hypothetical secreted protein, and which is located in an operon with a gene specifying a predicted peptidoglycan lipid II flippase. These insertion mutants were impaired in growth in the presence of all iron chelators tested. Two genes with a predicted regulatory function were identified: Bbr_0589, a gene specifying a hypothetical TetR regulatory protein and Bbr_0466. The corresponding mutants were sensitive to ferrous iron but not ferric iron chelation. Two transposon insertions were identified in genes Bbr_1743 encoding a short chain dehydrogenase, and Bbr_0702 encoding a haloacid dehalogenase-like hydrolase; the products of these genes are reputed to require iron as a cofactor.
The remaining dipyridyl-sensitive mutants which were selected from the Tn5 library screening efforts, whose involvement in iron metabolism or requirement for iron is not immediately clear from similarity searches, are listed in Table 3.
Transcriptomic Response of B. breve UCC2003 to Iron Limitation
To gain additional insight into the genes important for B. breve UCC2003 survival under iron limiting conditions and to complement the results obtained through the phenotypic screening, microarray analysis was carried out. The transcriptomic response of B. breve UCC2003 upon iron limitation was assessed by subjecting cells in the exponential growth phase to a short but high concentration of dipyridyl (700 μM) for 30 min (Results are summarized in Table 4, complete microarray results can be found in Supplementary Tables S1, S2). Microarray analysis was carried out in this way in order to identify the immediate transcriptional response to acute and severe iron limitation. Genes with a significantly different transcription level relative to the control when grown under iron limiting conditions, were identified in this manner (p < 0.001 and an expression ratio of >1.7 or < 0.25). Based on these criteria a total of 57 genes were found to be differentially regulated under iron-limiting conditions.
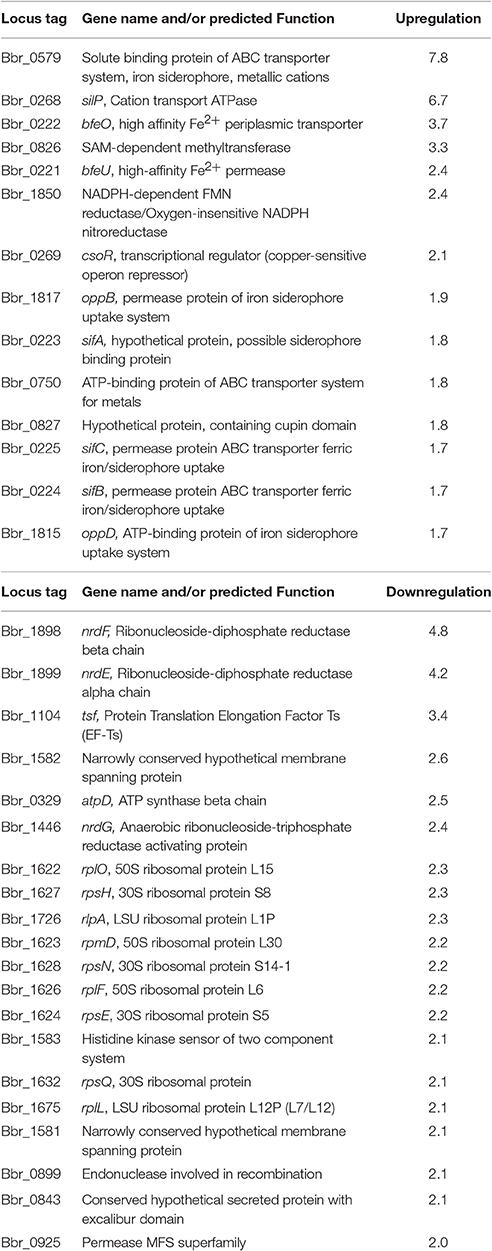
One noteworthy feature of the B. breve UCC2003 transcriptome response to iron limitation was the transcriptional activation of most genes of the bfeUO and sifABCDE clusters, as discussed above. Furthermore, the gene cluster represented by locus tags Bbr_1817–Bbr_1814, predicted to encode an iron siderophore ABC transport system, was also upregulated under the imposed iron limiting conditions. Mutations in (some of the genes of) the bfeUO/sifABCDE and Bbr_1817–Bbr_1814 gene clusters cause a dipyridyl-sensitive phenotype (see above) and were the only genes to be identified using both of these approaches (i.e., phenotypic screening and microarray analysis).
Other genes encoding putative metal uptake systems were shown to be transcriptionally upregulated under the imposed iron-limiting conditions, including a solute binding protein of an ABC-type transport system (Bbr_0579). Based on BlastP and HHpred analysis this protein displays similarity to solute binding proteins for metal cations found in many species including E. coli and Streptococcus pneumoniae (Yatsunyk et al., 2008; Begg et al., 2015). The gene associated with locus tag Bbr_0268 (silP) displayed a 7.6-fold transcriptional increase under iron-limiting conditions. The protein product of silP contains an ATPase domain and is predicted to be involved in the transport of iron-carrying compounds and copper ions. Bbr_0269 (csoR) which was shown to be upregulated 2.1-fold under microarray analysis encodes a predicted copper-sensitive operon repressor.
Also of interest was the upregulation of Bbr_1850, a NADPH-dependent FMN reductase. Studies in Pseudomonas putida have found that these reductases have ferric iron reductase activity and are believed to play a role in NADH-dependent ferric/flavin reduction under iron stress conditions (Yeom et al., 2009). It is therefore possible that B. breve UCC2003 is utilising this reductase to convert cytosolic ferric iron or ferric iron bound to siderophore complexes into its more biologically available ferrous form for further use within the cell.
Additionally, a number of iron-dependent genes were downregulated in the microarrays, for example ribonucleoside-diphosphate reductase system (Bbr_1898 and Bbr_1899) and many other genes involved in purine and pyrimidine metabolism (Table 3). The ribonucleoside-diphosphate reductase system is essential for de novo synthesis of deoxy ribonucleotides, the precursors of DNA synthesis and consequently cell division.
Transcriptional Analysis of the bfeUO/sifABCDE Gene Cluster
Phenotypic screening of the random mutant library and microarray analysis showed that the bfeUO/sifABCDE (bfe/sif) cluster is transcriptionally induced and essential for normal growth of B. breve UCC2003 under iron-limiting conditions. A previous study on the bfe gene cluster of B. breve UCC2003 identified one promoter-containing region upstream of bfeU (Bbr_0221), which was used to create an inducible expression system for Bifidobacterium (Cronin et al., 2012). Due to the size of the intergenic region between the bfeO/sifA genes and the presence of a predicted promoter based on BPROM analysis we reasoned that bfe/sif region may encompass two transcriptional units. In order to assess this notion, RT-PCR was carried out across various sections of the bfe/sif gene cluster. Total RNA was extracted from logarithmically growing B. breve UCC2003 cultured in fRCM supplemented with dipyridyl (i.e., growth under iron-limiting conditions so as to ensure/maximize transcription of the bfe/sif gene cluster), and then used to generate cDNA by reverse transcription (see Materials and Methods). The resulting cDNA was used as a template for the amplification of the intergenic regions of bfe/sif gene cluster using various primer sets listed in Table 2. RT-PCR analysis indicated that the bfe/sif gene cluster indeed encompasses two transcriptional units represented by the bfeUO operon and the sifABCDE operon (Figure 2). Consistent with this analysis was the identification of a potential rho-independent transcriptional terminator sequence located downstream of bfeO, while the sifABCDE operon is followed by a gene, corresponding to locus tag Bbr_0228, which is oriented in the opposite direction (Figure 2).
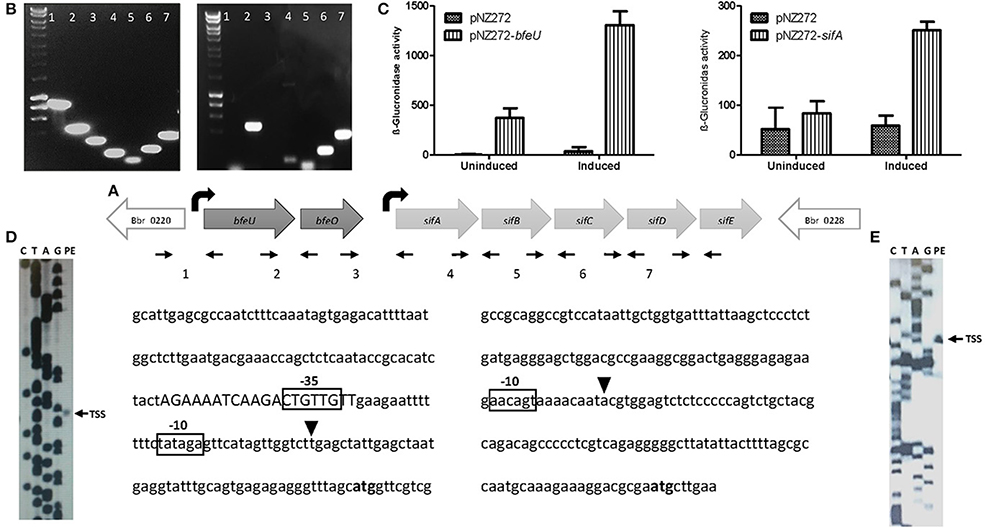
Figure 2. Transcriptional analysis of the bfeUO/sifABCDE gene cluster: (A) Schematic representation of the bfeUO/sifABCDE gene cluster, curved black arrows represent the predicted transcriptional start sites present, straight small arrows indicate positions of primers used. Intergenic region between genes bfeU to sifE, are labeled as follows; 1 (Bbr_0220/bfeU), 2 (bfeU/bfeO), 3 (bfeO/sifA), 4 (sifA/sifB), 5 (sifB/sifC), 6 (sifC/sifD), and 7 (sifD/sifE). (B) RT-PCR of the intergenic regions from Bbr_0220 to Bbr_0228, intergenic regions are labeled as in (A). (C) β-glucuronidase assay of pNZ272+bfeU, pNZ272+sifA and pNZ272 (negative control). β-glucuronidase activity is expressed in Miller units. (D) Primer extension (PE) analysis of the transcriptional start site of bfeU. (E) PE analysis of transcriptional start site of sifA. In both (D,E) the start codon (ATG) is indicated in bold, the transcriptional start site (TSS) is indicated by a black triangle, the proposed −10 and −35 motifs are boxed and a possible operator sequence is displayed in uppercase.
The RT-PCR results indicated that transcription of the bfeUO and sifABCDE operons initiates upstream of the bfeU and sifA genes, respectively. A previous study had indicated that the bfeU promoter region was inducible under iron-limiting conditions, and in order to validate this and to investigate whether the promoter region upstream of sifA is also inducible under iron limiting conditions, plasmids pNZ-bfeU and pNZ-sifA were constructed, in which the upstream regions of bfeU and sifA were cloned in front of the promoter-less gusA gene in the promoter-probe vector pNZ272 (see Materials and Methods). Plasmids pNZ-bfeU and pNZ-sifA, as well as pNZ272 (negative control) were individually introduced into B. breve UCC2003, logarithmically growing cells of these three strains that either had or had not been subjected to iron limitation by means of dipyridyl addition were then utilised for β-glucuronidase (the product of gusA) activity assays. Consistent with our earlier study, we clearly demonstrate that each of the cloned fragments in pNZ-bfeU and pNZ-sifA elicit promoter activity and that these promoters are inducible by iron limitation (Figure 2, Cronin et al., 2012).
Next, given that the bfeUO/sifABCDE gene cluster encompasses two transcriptional units and that these units elicit promoter activity inducible by iron limitation, the transcriptional initiation sites were identified by primer extension analysis. This analysis identified a single TSS upstream of bfeU and sifA. The TSS of the bfeUO operon was shown to be located 48 bp upstream of the predicted bfeU start codon, and a putative promoter sequence was identified nine base pairs upstream of this TSS with putative −10 (CATAGT) and −35 (TTGAAG) elements which are similar to vegetative promoter sequences found in B. breve (Pokusaeva et al., 2010; O'Connell et al., 2014; Egan et al., 2015, 2016). The transcriptional start site of the sifABCDE operon was determined to be located 93 bp upstream of the sifA start codon, and a putative promoter sequence was identified eight base pairs upstream of this TSS with a putative −10 (AACAGT), though without any obvious −35 sequence (Figure 2). Our previous studies identified a putative regulatory motif (AAAATCAAGACTGTTGTT) upstream of bfeUO, which is also present upstream of a number of genes involved in iron utilization. The location of this binding operator sequence in relation to the bfeUO promoter region suggests that this as yet unidentified transcription factor may act as a repressor (Figure 2, Cronin et al., 2012).
The Presence of bfeUO or sifABCDE Gene Clusters Is Not Required for Colonisation of the Healthy Murine GIT
Murine colonisation experiments were carried out to assess if bfeUO, a high affinity ferrous iron uptake cluster, or sifABCDE, a predicted ferrous iron/siderophore uptake system, play a role in gut colonisation. In order to analyse this the gut colonisation capacity of B. breve UCC2003, B. breve UCC2003-bfeU, B. breve UCC2003-sifA was tested in BALB/c mice. In conventional BALB/c mice with a resident microbiota (i.e., in a competitive environment), WT B. breve UCC2003, bfeU and the sifA insertion mutants were all able to colonise the gastrointestinal tract, as was shown by plating of fecal samples (1 × 105 CFU/g feces retrieved 15 days after last administration; Figure 3). On the basis of these results it appears that the functionality of bfeUO and sifABCDE does not have any obvious impact on the ability of B. breve UCC2003 to colonise the gut of healthy BALB/c mice with a resident microbiota (Figure 3). These results are in agreement with previous work (Christiaen et al., 2014) who found that B. breve UCC2003-bfeU and B. breve UCC2003-sifA insertion mutant were able to colonise the gastrointestinal tract of nematodes as efficiently as the wild type strain.
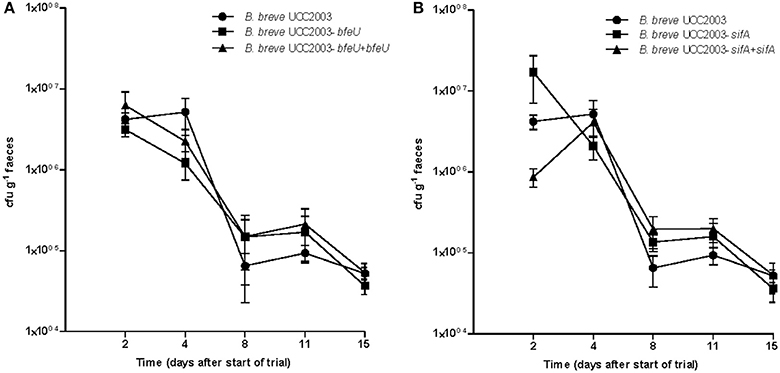
Figure 3. Murine colonisation trial: (A) illustrates the cfu g−1 faeces of B. breve UCC2003 (circle) and B. breve UCC2003-bfeU (square) and B. breve UCC2003-bfeU+pBC1.2+bfeU (triangle), while (B) illustrates cfu g−1 faeces of B. breve UCC2003 (circle), B. breve UCC2003-sifA (square), and B. breve UCC2003-sifA+pBC1.2+sifA (triangle) administered individually, administration started at day 0 and was continued for 3 consecutive days. Data shown are mean ± SEM. (n = 6).
Distribution of the bfeUO and sifABCDE Iron Uptake Systems across the Bifidobacterium Genus
The importance of bfeUO and sifABCDE gene clusters under iron limitation has been clearly displayed through transcriptomic and phenotypic analysis, while transcriptional analysis clearly demonstrates that this gene cluster is composed of two transcriptional units bfeUO and sifABCDE. In order to gain more information about distribution and conservation of these gene clusters in the Bifidobacterium genus analysis utilising balstP was carried out. This was achieved by comparing the degree of protein similarity of the BfeUO and SifABCDE proteins found in B. breve UCC2003 with deduced protein complements of representative bifidobacterial species. The results are displayed in Table 5 as a heat map, which illustrates the protein similarity, as well as the genetic organisation and presence/absence of BfeUO- and SifABCDE-encoding genes across a number of bifidobacterial species. From this table it is obvious that the bfeUO and sifABCDE gene clusters are highly conserved across the bifidobacterial genus, and that these cluster are present in the genomes of 35 of the 47 bifidobacterial species tested. Interestingly the 12 bifidobacterial species which do not contain these gene clusters are members of three distinct branch points of a phylogenetic tree constructed by Milani et al. (2014). The members of these branch points are furthest away from the root of the evolutionary phylogenetic tree, which represents the Bifidobacterium genus ancestor (Milani et al., 2014). Furthermore, the five genes of the sifABCDE cluster are, when present, always co-located on the bifidobacterial genomes. The only exception to this was SifE, which encodes a predicted ferric reductase protein, and which appears to be absent from the genomes of B. bohemicum and B. bombi, bifidobacterial species that originate from the hind-gut of a bumblebee.
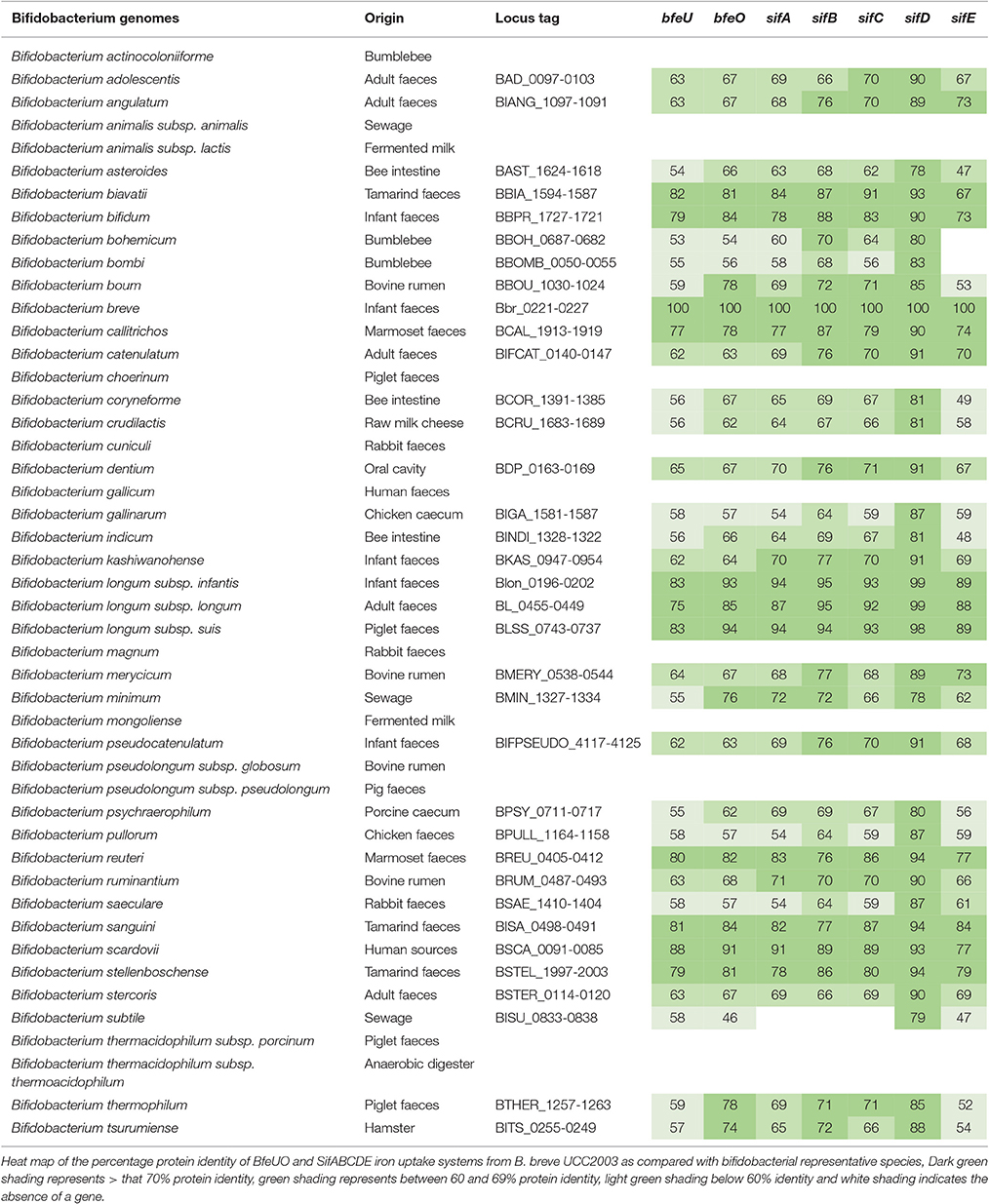
Table 5. Heat map representing the distribution and conservation of homologs of the BfeUO and SifABCDE iron uptake systems across the Bifidobacterium genus.
Discussion
Bifidobacteria are believed to be able to propagate under low iron conditions and are thought to be efficient scavengers of iron, a notion being supported by the finding that certain bifidobacterial species are found in greater numbers under low luminal iron conditions as compared with normal or high luminal iron conditions. Much of the relevant literature to date focuses on the ability of bifidobacteria to internalise and sequester iron, and to proliferate in either high or low concentrations of iron (Bezkorovainy and Solberg, 1989; Kot et al., 1993; Kim et al., 2002; Vazquez-Gutierrez et al., 2015). The current study was aimed at uncovering the molecular mechanisms and systems responsible for iron uptake and metabolism in B. breve UCC2003, as a prototypical representative of its genus.
Severe iron limitation has a profound effect on B. breve UCC2003, as illustrated by the plethora of genes whose transcription is altered under such iron restrictive conditions and by the number of genes which were identified in the screening of the random mutant library under iron limiting conditions. This response includes the upregulation of Bbr_1850, a gene encoding a predicted NADPH-dependent FMN reductase. A similar reductase was identified to be involved in iron metabolism in Pseudomonas putida, and it has been speculated that such reductases may be needed in higher amounts to reduce ferric iron during iron limitation (Yeom et al., 2009; Takeda et al., 2010). Among the B. breve UCC2003 genes that were shown to be essential for growth under iron limiting conditions, we identified apbC and apbE, which are predicted to encode proteins required for Fe-S cluster formation, repair and docking. Mutations in homologs of apbC or apbE in S. enterica result in cellular deficiencies which are reversed by the addition of ferric chloride, which suggests that iron addition compensates for such mutations by increasing repair of oxygen-labile Fe-S clusters (Skovran and Downs, 2003). Therefore, in the case of B. breve UCC2003 it is possible that under iron limiting conditions mutation of apbC or apbE causes an inability for the cell to construct, repair or to load these Fe-S clusters into apo-proteins which are essential to many cellular process. This phenomenon, although not studied in full in this paper, would be of interest for future study.
Screening of the random mutant library and transcriptome analysis led to identification of several genes encoding factors for the transportation of cations and or siderophore complexes. From the current study it appears that there are two key uptake systems important for bifidobacterial survival under the imposed, in vitro iron-limiting conditions tested: BfeUO, a predicted high affinity ferrous/ferric iron uptake system and SifABCDE, a predicted Fe2+/siderophore uptake system, which specifically transports ferrous iron. BfeUO is similar to EfeUOB, a Fe2+ uptake system identified in E. coli and to a system present in Bacillus subtilis, which is responsible for ferrous and ferric iron transport depending on the extracellular conditions and the oxidant supply (Cao et al., 2007; Gaballa et al., 2008; Castells-Roca et al., 2011). This system was previously identified as BfeUO which was sensitive to both ferrous and ferric iron chelation, is highly conserved across the Bifidobacterium genus (Table 5) and is possibly the primary/predominant system responsible for the uptake of iron in B. breve UCC2003 under the in vitro iron limiting conditions tested here (Cronin et al., 2012).
SifABCDE represents a putative ferrous iron/siderophore uptake system, which was found to be important for B. breve UCC2003 survival when availability of ferrous iron is limiting. A relatively small number of siderophore uptake systems have been characterised in Gram-positive bacteria, however, of those characterised there are a number of common features, including a siderophore binding protein (possible SifA), ABC transporter permeases (SifB/C), ATPase (SifD), and a ferric iron reductase (SifE) (Fukushima et al., 2013, 2014). The identified mutants within the bfeUO gene cluster were sensitive to both ferrous and ferric iron chelation, while mutants located within the sifABCDE only exhibited sensitivity to ferrous iron chelation. This phenotype indicates that the sifABCDE cluster is specifically responsible for the uptake of free/complexed ferrous iron. Transcriptional analysis of bfeUO and sifABCDE via RT-PCR, and primer extension analysis found that the gene cluster is organised into two transcriptional units, which are subject to transcriptional induction upon iron limitation with dipyridyl. These results indicate that the expression of both the BfeUO and SifABCDE uptake systems is induced under ferrous iron limitation, while phenotypic analysis indicates that that the sifABCDE uptake system is specifically responsible for the uptake of ferrous iron, the form of iron that is expected to more prevalent in the anaerobic GIT.
A recent study carried out by Vazquez-Gutierrez et al. identified a number of iron uptake clusters utilising a proteomic approach, this analysis identified three ferrous iron transporters in the secretome of B. kashiwanohense PV20-2: AH68_00590 and AH68_00595 which are homologous to bfeUO, respectively, AH68_00600 which is homologous to sifA (Vazquez-Gutierrez et al., 2017). The presence of BfeUO and SifA homologs in these three bifidobacterial strains illustrate the importance of these two iron uptake system in sequestering this vital micronutrient. From the phylogenetic analysis illustrated in Table 5 it is also evident that these iron uptake systems are widely distributed and are highly conserved across the Bifidobacterium genus. Furthermore, the identification of a number of other potential cation uptake systems such as MntH and that encoded by Bbr_1817–1814 illustrates the diversity and possible complementarity of iron sequestration mechanisms within the Bifidobacterium genus.
Previously Bifidobacterium species have been found to operate a LuxS-mediated system for gut colonisation and pathogen protection, which is linked to iron acquisition (Christiaen et al., 2014). Similar to the findings of the murine colonisation experiments carried out in this study, Christiaen et al. found that bfeU and sifA insertion mutants are able to colonise the GIT of nematodes as efficiently as the WT strain. In addition, these authors found that the bfeU and sifA mutants exhibit a significantly decreased ability to confer protection to Salmonella-infected nematodes as compared to the WT strain (Christiaen et al., 2014). Deriu et al. have elegantly demonstrated that the fecal iron concentration of S. thyphimurium infected mice is reduced to 300 mg/kg from 950 mg/kg in the absence of infection. Therefore, confirming that infection causes a reduction of iron in the gut. They also illustrate how the probiotic strain E. coli Nissle reduces growth and intestinal colonisation of S. typhimurium by competing for iron in the colonic environment (Deriu et al., 2013). Consistent with this observation, it was also demonstrated that two bifidobacterial strains; B. pseudolongum PV8-2 and B. kashiwanohense PV20-2 exhibit strain-dependent inhibitory activity on iron-dependent enteropathogens (Vazquez-Gutierrez et al., 2016). The fact that insertion mutations within these uptake systems (bfeUO and sifABCDE) do not affect the colonisation efficiency of B. breve UCC2003 in the healthy nematode or murine gut leads us to believe that iron availability in a healthy gut is sufficient for colonisation of B. breve UCC2003, B. breve UCC2003-bfeU or B. breve UCC2003-sifA, even though the latter two mutants exhibit an in vitro growth deficiency under iron-limiting conditions. Therefore, we postulate that either (sufficient) iron is available in the gut in a form which is not taken up by the bfeUO or sifABCDE, or that these systems for iron acquisition may be important for B. breve UCC2003 survival during times in which iron is more limiting, for example during GIT infection as demonstrated by Deriu et al. (2013).
Altogether this study has also helped to broaden our knowledge regarding Bifidobacterium response to iron limitation, with the identification of a variety of genes being important for B. breve UCC2003 survival under iron-limiting conditions. This study also demonstrates the effectiveness of utilising random mutagenesis as a tool for the exploration of genes involved in iron metabolism and uptake in Bifidobacterium. The characterisation of the sifABCDE and bfeUO iron uptake cluster revealed that these clusters are vitally important B. breve UCC2003 survival under ferric and ferrous iron chelation, of which the latter is the most predominant in the gastrointestinal environment. Importantly, the sifABCDE iron uptake cluster was identified as being specifically responsible for the uptake of ferrous iron, while the bfeUO iron uptake system was sensitive to both ferrous and ferric iron chelation. Taken together, this versatile response of bifidobacteria to iron limitation may be one of the factors that afford such gut commensals a competitive edge among the many microbes within the gastrointestinal tract.
Ethics Statement
Animal procedures performed in this study were approved by the UCC Animal Experimentation Ethics Committee and were regulated the Department of Health of the Irish Government. All animal housing and procedures conformed to relevant Irish and European legislation governing the use of laboratory animals for scientific experimentation.
Author Contributions
NL, Conceived and designed experiments, Carried out experiments, Analysed data, Wrote paper, Technical and scientific discussion. FB, Carried out experiments, Analyzed data, Technical and scientific discussion. PC, Carried out experiments. MO, Carried out experiments. DV, Conceived and designed experiments, Analyzed data, Wrote paper, Technical and scientific discussion.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
All authors are members of The APC Microbiome Institute, which is a research center funded by Science Foundation Ireland (SFI), through the Irish Government's National Development Plan. The authors and their work were supported by SFI (Grant Nos. 07/CE/B1368 and SFI/12/RC/2273) and HRB (Grant No. 513 PDTM/20011/9). We would like to thank Lorena Ruiz for the use of the random transposon mutant library.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00964/full#supplementary-material
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/s0022-2836(05)80360-2
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Alvarez-Martin, P., Fernandez, M., O'Connell-Motherway, M., O'Connell, K. J., Sauvageot, N., Fitzgerald, G. F., et al. (2012). A conserved two-component signal transduction system controls the response to phosphate starvation in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 78, 5258–5269. doi: 10.1128/aem.00804-12
Alvarez-Martin, P., O'Connell-Motherway, M., van Sinderen, D., and Mayo, B. (2007). Functional analysis of the pBC1 replicon from Bifidobacterium catenulatum L48. Appl. Microbiol. Biotechnol. 76, 1395–1402. doi: 10.1007/s00253-007-1115-5
Andreini, C., Banci, L., and Rosato, A. (2016). Exploiting bacterial operons to illuminate human iron-sulfur proteins. J. Proteome Res. 15, 1308–1322. doi: 10.1021/acs.jproteome.6b00045
Andrews, S. C., Robinson, A. K., and Rodriguez-Quinones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/S0168-6445(03)00055-X
Bayer, E., Fiedler, H., Hock, K. L., Otterbach, D., Schenk, G., and Voelter, W. (1964). Structure and specificity of organic chelating agents. Angew. Chem. Int. Ed. 3, 325–332. doi: 10.1002/anie.196403251
Begg, S. L., Eijkelkamp, B. A., Luo, Z., Counago, R. M., Morey, J. R., Maher, M. J., et al. (2015). Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae. Nat. Commun. 6:6418. doi: 10.1038/ncomms7418
Bezkorovainy, A., and Solberg, L. (1989). Ferrous iron uptake by Bifidobacterium breve. Biol. Trace Elem. Res. 20, 251–267.
Boyd, J. M., Pierik, A. J., Netz, D. J., Lill, R., and Downs, D. M. (2008). Bacterial ApbC can bind and effectively transfer iron-sulfur clusters. Biochemistry 47, 8195–8202. doi: 10.1021/bi800551y
Brown, J. S., and Holden, D. W. (2002). Iron acquisition by Gram-positive bacterial pathogens. Microbes Infect. 4, 1149–1156. doi: 10.1016/S1286-4579(02)01640-4
Cao, J., Woodhall, M. R., Alvarez, J., Cartron, M. L., and Andrews, S. C. (2007). EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol. Microbiol. 65, 857–875. doi: 10.1111/j.1365-2958.2007.05802.x
Castells-Roca, L., Muhlenhoff, U., Lill, R., Herrero, E., and Belli, G. (2011). The oxidative stress response in yeast cells involves changes in the stability of Aft1 regulon mRNAs. Mol. Microbiol. 81, 232–248. doi: 10.1111/j.1365-2958.2011.07689.x
Christiaen, S. E., O'Connell Motherway, M., Bottacini, F., Lanigan, N., Casey, P. G., Huys, G., et al. (2014). Autoinducer-2 plays a crucial role in gut colonization and probiotic functionality of Bifidobacterium breve UCC2003. PLoS ONE 9:e98111. doi: 10.1371/journal.pone.0098111
Collins, H. L. (2008). Withholding iron as a cellular defence mechanism - friend or foe? Eur. J. Immunol. 38, 1803–1806. doi: 10.1002/eji.200838505.
Crichton, R. R., and Pierre, J.-L. (2001). Old iron, young copper: from mars to venus. Biometals 14, 99–112. doi: 10.1023/a:1016710810701
Cronin, M., Zomer, A., Fitzgerald, G. F., and van Sinderen, D. (2012). Identification of iron-regulated genes of Bifidobacterium breve UCC2003 as a basis for controlled gene expression. Bioeng. Bugs 3, 157–167. doi: 10.4161/bbug.18985
den Besten, G., van Eunen, K., Groen, A. K., Venema, K., Reijngoud, D. J., and Bakker, B. M. (2013). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54, 2325–2340. doi: 10.1194/jlr.R036012
Deriu, E., Liu, J. Z., Pezeshki, M., Edwards, R. A., Ochoa, R. J., Contreras, H., et al. (2013). Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14, 26–37. doi: 10.1016/j.chom.2013.06.007
de Ruyter, P. G., Kuipers, O. P., and de Vos, W. M. (1996). Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62, 3662–3667.
Dostal, A., Lacroix, C., Bircher, L., Pham, V. T., Follador, R., Zimmermann, M. B., et al. (2015). Iron modulates butyrate production by a child gut microbiota in vitro. MBio 6, e01453–e01415. doi: 10.1128/mBio.01453-15
Egan, M., Jiang, H., O'Connell Motherway, M., Oscarson, S., and van Sinderen, D. (2016). Glycosulfatase-encoding gene cluster in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 82, 6611–6623. doi: 10.1128/aem.02022-16
Egan, M., O'Connell Motherway, M., and van Sinderen, D. (2015). A GntR-type transcriptional repressor controls sialic acid utilization in Bifidobacterium breve UCC2003. FEMS Microbiol. Lett. 362, 1–9. doi: 10.1093/femsle/fnu056
Fukushima, T., Allred, B. E., and Raymond, K. N. (2014). Direct evidence of iron uptake by the Gram-positive siderophore-shuttle mechanism without iron reduction. ACS Chem. Biol. 9, 2092–2100. doi: 10.1021/cb500319n
Fukushima, T., Allred, B. E., Sia, A. K., Nichiporuk, R., Andersen, U. N., and Raymond, K. N. (2013). Gram-positive siderophore-shuttle with iron-exchange from Fe-siderophore to apo-siderophore by Bacillus cereus YxeB. Proc. Natl. Acad. Sci. U.S.A. 110, 13821–13826. doi: 10.1073/pnas.1304235110
Gaballa, A., Antelmann, H., Aguilar, C., Khakh, S. K., Song, K. B., Smaldone, G. T., et al. (2008). The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 11927–11932. doi: 10.1073/pnas.0711752105
García de la Nava, J., Santaella, D. F., Alba, J. C., Carazo, J. M., Trelles, O., and Pascual-Montano, A. (2003). Engene: the processing and exploratory analysis of gene expression data. Bioinformatics 19, 657–658. doi: 10.1093/bioinformatics/btg028
Gautheret, D., and Lambert, A. (2001). Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J. Mol. Biol. 313, 1003–1011. doi: 10.1006/jmbi.2001.5102
Grosse, C., Scherer, J., Koch, D., Otto, M., Taudte, N., and Grass, G. (2006). A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol. Microbiol. 62, 120–131. doi: 10.1111/j.1365-2958.2006.05326.x
Gueimonde, M., Garrigues, C., van Sinderen, D., de los Reyes-Gavilán, C. G., and Margolles, A. (2009). Bile-inducible efflux transporter from bifidobacterium longum NCC2705, conferring bile resistance. Appl. Environ. Microbiol. 75, 3153–3160. doi: 10.1128/AEM.00172-09
Hausmann, A., Aguilar Netz, D. J., Balk, J., Pierik, A. J., Muhlenhoff, U., and Lill, R. (2005). The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc. Natl. Acad. Sci. U.S.A. 102, 3266–3271. doi: 10.1073/pnas.0406447102
Hood, M. I., and Skaar, E. P. (2012). Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10, 525–537. doi: 10.1038/nrmicro2836
Ilbert, M., and Bonnefoy, V. (2013). Insight into the evolution of the iron oxidation pathways. Biochim. Biophys. Acta 1827, 161–175. doi: 10.1016/j.bbabio.2012.10.001
Kiley, P. J., and Beinert, H. (2003). The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 6, 181–185. doi: 10.1016/S1369-5274(03)00039-0
Kim, W. S., Tanaka, T., Kumura, H., and Shimazaki, K. (2002). Lactoferrin-binding proteins in Bifidobacterium bifidum. Biochem. Cell Biol. 80, 91–94. doi: 10.1139/O01-226
Kortman, G. A., Raffatellu, M., Swinkels, D. W., and Tjalsma, H. (2014). Nutritional iron turned inside out: intestinal stress from a gut microbial perspective. FEMS Microbiol. Rev. 38, 1202–1234. doi: 10.1111/1574-6976.12086
Kot, E., and Bezkorovainy, A. (1999). Binding of ferric iron to the cell walls and membranes of Bifidobacterium thermophilum: effect of free radicals. J. Agric. Food Chem. 47, 4606–4610. doi: 10.1021/jf990474l
Kot, E., Haloftis, G., and Bezkorovainy, A. (1993). Ferrous iron uptake by bifidobacteria in the absence of glucose. Nutr. Res. 13, 1295–1303.
Law, J., Buist, G., Haandrikman, A., Kok, J., Venema, G., and Leenhouts, K. (1995). A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177, 7011–7018.
Long, A. D., Mangalam, H. J., Chan, B. Y., Tolleri, L., Hatfield, G. W., and Baldi, P. (2001). Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276, 19937–19944. doi: 10.1074/jbc.M010192200
Man, J. C. D., Rogosa, M., and Sharpe, M. E. (1960). A medium for the cultivation of lactobacilli. J. Appl. Microbiol. 23, 130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x
Milani, C., Lugli, G. A., Duranti, S., Turroni, F., Bottacini, F., Mangifesta, M., et al. (2014). Genomic encyclopedia of type strains of the genus bifidobacterium. Appl. Environ. Microbiol. 80, 6290–6302. doi: 10.1128/aem.02308-14
O'Connell, K. J., Motherway, M. O., Liedtke, A., Fitzgerald, G. F., Paul Ross, R., Stanton, C., et al. (2014). Transcription of two adjacent carbohydrate utilization gene clusters in Bifidobacterium breve UCC2003 is controlled by LacI- and repressor open reading frame kinase (ROK)-type regulators. Appl. Environ. Microbiol. 80, 3604–3614. doi: 10.1128/aem.00130-14
O'Connell Motherway, M., Zomer, A., Leahy, S. C., Reunanen, J., Bottacini, F., Claesson, M. J., et al. (2011). Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U.S.A. 108, 11217–11222. doi: 10.1073/pnas.1105380108
O'Riordan, K., and Fitzgerald, G. F. (1999). Molecular characterisation of a 5.75-kb cryptic plasmid from Bifidobacterium breve NCFB 2258 and determination of mode of replication. FEMS Microbiol. Lett. 174, 285–294.
Picard, C., Fioramonti, J., Francois, A., Robinson, T., Neant, F., and Matuchansky, C. (2005). Review article: bifidobacteria as probiotic agents – physiological effects and clinical benefits. Aliment. Pharmacol. Ther. 22, 495–512. doi: 10.1111/j.1365-2036.2005.02615.x
Platteeuw, C., Simons, G., and de Vos, W. M. (1994). Use of the Escherichia coli beta-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60, 587–593.
Pokusaeva, K., Neves, A. R., Zomer, A., O'Connell-Motherway, M., MacSharry, J., Curley, P., et al. (2010). Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb. Biotechnol. 3, 311–323. doi: 10.1111/j.1751-7915.2009.00152.x
Py, B., and Barras, F. (2010). Building Fe–S proteins: bacterial strategies. Nat Rev Micro 8, 436–446. doi: 10.1038/nrmicro2356
Ruiz, L. M., Motherway, M. O., Lanigan, N., and van Sinderen, D. (2013). Transposon mutagenesis in Bifidobacterium breve: construction and characterization of a Tn5 transposon mutant library for Bifidobacterium breve UCC2003. PLoS ONE 8:e64699. doi: 10.1371/journal.pone.0064699.
Russell, S. J. D. W. (2001). Molecular Cloning: A Laboratory Manual Cold Spring. Harbor, CA: Cold Spring Harbor Laboratory Press.
Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A., et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945. doi: 10.1093/bioinformatics/16.10.944
Sanders, M. E. (2008). Probiotics: definition, sources, selection, and uses. Clin. Infect. Dis. 46(Suppl. 2), S58–S61; discussion S144–S151. doi: 10.1086/523341
Skovran, E., and Downs, D. M. (2003). Lack of the ApbC or ApbE protein results in a defect in Fe-S cluster metabolism in Salmonella enterica serovar typhimurium. J. Bacteriol. 185, 98–106. doi: 10.1128/jb.185.1.98-106.2003
Takeda, K., Sato, J., Goto, K., Fujita, T., Watanabe, T., Abo, M., et al. (2010). Escherichia coli ferredoxin-NADP+ reductase and oxygen-insensitive nitroreductase are capable of functioning as ferric reductase and of driving the Fenton reaction. Biometals 23, 727–737. doi: 10.1007/s10534-010-9339-8
van Hijum, S. A., de Jong, A., Baerends, R. J., Karsens, H. A., Kramer, N. E., Larsen, R., et al. (2005). A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. doi: 10.1186/1471-2164-6-77
van Hijum, S. A., Garcia de la Nava, J., Trelles, O., Kok, J., and Kuipers, O. P. (2003). MicroPreP: a cDNA microarray data pre-processing framework. Appl. Bioinformatics 2, 241–244.
Vazquez-Gutierrez, P., de Wouters, T., Werder, J., Chassard, C., and Lacroix, C. (2016). High iron-sequestrating bifidobacteria inhibit enteropathogen growth and adhesion to intestinal epithelial cells in vitro. Front. Microbiol. 7:1480. doi: 10.3389/fmicb.2016.01480
Vazquez-Gutierrez, P., Lacroix, C., Jaeggi, T., Zeder, C., Zimmerman, M., and Chassard, C. (2015). Bifidobacteria strains isolated from stools of iron deficient infants can efficiently sequester iron. BMC Microbiol. 15:3. doi: 10.1186/s12866-014-0334-z
Vazquez-Gutierrez, P., Stevens, M. J. A., Gehrig, P., Barkow-Oesterreicher, S., Lacroix, C., and Chassard, C. (2017). The extracellular proteome of two Bifidobacterium species reveals different adaptation strategies to low iron conditions. BMC Genomics 18:41. doi: 10.1186/s12864-016-3472-x
Ventura, M., Canchaya, C., van Sinderen, D., Fitzgerald, G. F., and Zink, R. (2004). Bifidobacterium lactis DSM 10140: identification of the atp (atpBEFHAGDC) operon and analysis of its genetic structure, characteristics, and phylogeny. Appl. Environ. Microbiol. 70, 3110–3121. doi: 10.1128/AEM.70.5.3110-3121.2004
Vernazza, C. L., Gibson, G. R., and Rastall, R. A. (2006). Carbohydrate preference, acid tolerance and bile tolerance in five strains of Bifidobacterium. J. Appl. Microbiol. 100, 846–853. doi: 10.1111/j.1365-2672.2006.02832.x
Yatsunyk, L. A., Easton, J. A., Kim, L. R., Sugarbaker, S. A., Bennett, B., Breece, R. M., et al. (2008). Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli. J. Biol. Inorg. Chem. 13, 271–288. doi: 10.1007/s00775-007-0320-0
Yeom, J., Jeon, C. O., Madsen, E. L., and Park, W. (2009). Ferredoxin-NADP+ reductase from Pseudomonas putida functions as a ferric reductase. J. Bacteriol. 191, 1472–1479. doi: 10.1128/jb.01473-08
Keywords: ferrous, ferric, ferric reductase, siderophore, Fe-S cluster
Citation: Lanigan N, Bottacini F, Casey PG, O'Connell Motherway M and van Sinderen D (2017) Genome-Wide Search for Genes Required for Bifidobacterial Growth under Iron-Limitation. Front. Microbiol. 8:964. doi: 10.3389/fmicb.2017.00964
Received: 23 December 2016; Accepted: 15 May 2017;
Published: 31 May 2017.
Edited by:
Ivan Mijakovic, Chalmers University of Technology, SwedenReviewed by:
Christian U. Riedel, University of Ulm, GermanyFrancisco Barona-Gomez, Center for Research and Advanced Studies of the National Polytechnic Institute (CINVESTAV), Mexico
Copyright © 2017 Lanigan, Bottacini, Casey, O'Connell Motherway and van Sinderen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Douwe van Sinderen, ZC52YW5zaW5kZXJlbkB1Y2MuaWU=
 Noreen Lanigan
Noreen Lanigan Francesca Bottacini
Francesca Bottacini Pat G. Casey
Pat G. Casey Mary O'Connell Motherway
Mary O'Connell Motherway Douwe van Sinderen
Douwe van Sinderen