- 1Department of Microbial Diseases, UCL Eastman Dental Institute, University College London, London, United Kingdom
- 2Immunology Catalyst, GlaxoSmithKline plc, Stevenage, United Kingdom
Staphylococcus aureus can undergo phenotypic switching between a normal colony phenotype (NCP) and a small colony variant (SCV). The SCV phenotype confers increased antibiotic resistance and the capacity to persist within human tissues and cells, and because these cells can revert back to the NCP they cause chronic and/or recurrent infections that are very difficult to treat. A complete picture of the genetic events that can lead to phenotypic switching in S. aureus is currently lacking. We describe the selection of an SCV with a previously unreported genetic alteration leading to an ochre mutation of aroD. In addition to the known mechanisms of phenotypic switching between the SCV and the NCP we describe a previously unreported mechanism involving tRNA ochre suppressors arising. The ochre suppressor strains had wild-type growth rates and restored antibiotic sensitivity, similar to the wild-type strain. However, whilst they had increased virulence compared to the SCV parent strain, their virulence was not restored to that of the NCP parental strain. These findings establish that phenotypic switching between the NCP and SCV states can give rise to strains with different pathogenic potential.
Introduction
Staphylococcus aureus small colony variants (SCVs) are a naturally arising slow-growing subpopulation of S. aureus. They are termed SCVs because their colonies are about 1/10 the size of normal colonies on agar after 24 h of growth. S. aureus SCVs are frequently associated with persistent and recurrent infections, such as osteomyelitis, implant infections and airway infections in cystic fibrosis patients (von Eiff et al., 2006), or patients receiving long-term antibiotic treatment (Proctor et al., 1995; von Eiff et al., 1997a, 2001). This is in part because SCVs can persist within non-phagocytic host cells such as epithelial cells (Atalla et al., 2010), endothelial cells (von Eiff et al., 1997b) and bone cells (Wright and Nair, 2010). This is because SCVs have reduced toxin production and do not induce an immune response (Tan et al., 2014; Ou et al., 2016). In addition genes responsible for adhesion and biofilm formation are generally upregulated in SCVs (Proctor et al., 2006), enhancing the capacity of the bacterium to persist in implant infections, cystic fibrosis patients, and osteomyelitis.
Staphylococcus aureus SCVs isolated from patients are often found to be auxotrophic for metabolites such as heme, menadione, or thymidine (Proctor et al., 2006). The auxotrophic nature of SCVs, generally, seems to relate to the antibiotic treatment that the patient was receiving with aminoglycosides giving rise to heme and menadione auxotrophs (Balwit et al., 1994), while trimethoprim-sulfamethoxazole treatment gives rise to thymidine auxotrophs (Kriegeskorte et al., 2015). A number of studies have shown that SCVs can also be generated by exposure to very low levels of triclosan (Bayston et al., 2007; Seaman et al., 2007; Latimer et al., 2012). However, the auxotrophy of these SCVs was not defined and the likelihood of them arising in situ in clinical settings is questionable because of the levels of triclosan in hand washes and other cleaning products (Seaman et al., 2007; Latimer et al., 2012). Heme and menadione are required for the synthesis of cytochromes and menaquinone, respectively. These molecules are essential for respiration and thus SCVs defective in the biosynthesis of either of these two molecules switch to generating energy via fermentation resulting in reduced growth and bacterial numbers, they have a decreased bacterial membrane potential, and thus a decreased electrochemical gradient, which is associated with increased resistance to antimicrobials (Sendi and Proctor, 2009; Mike et al., 2013). Thymidine auxotrophs have similar phenotypes to heme and menadione auxotrophs due to decreased TCA cycle activity, which results in down-regulation of the ETC (Proctor et al., 2014).
The SCV phenotype is usually unstable and phenotypic switching back to the normal colony phenotype (NCP) occurs (Massey et al., 2001; Kahl, 2014). This phenotypic switching may explain why SCV infections recur after periods of apparent remission (Kahl, 2014). Whilst numerous studies on S. aureus SCVs have been published, there are only a few studies that show the genetic basis for phenotypic switching back to the NCP and the frequency of reversion (Vestergaard et al., 2016). It was reported that phenotypic switching from SCV to NCP restored antibiotic sensitivity and haemolytic activity to that of the wild-type (Lannergård et al., 2008; Dean et al., 2014). Given the nature of the reversions one might predict that other phenotypes, such as the capacity to form biofilms and virulence, would be restored to normal wild-type levels, though this was not examined.
In this study, we isolated a spontaneous menadione auxotrophic SCV to determine the genetic basis of SCV reversion to NCP and the consequences of this phenotypic switching on antibiotic resistance, biofilm formation, and virulence. We determined that this SCV had a point mutation in the aroD gene, which has not been previously reported. The gene encodes 3-dehydroquinate dehydratase, an enzyme participating in the shikimate pathway that catalyzes the conversion of 3-dehydroquinate to 3-dehydroshikimate, the third step in the shikimate pathway leading to the synthesis of chorismate. Chorismate is a precursor for the synthesis of a range of aromatic compounds, including aromatic amino acids and menaquinone. As expected, this SCV is auxotrophic for all aromatic amino acids and menaquinone. Two classes of revertant arose upon phenotypic switching to the NCP, those in which the aroD gene mutation was reversed and those that had a compensatory mutation in one of two tyr-tRNA genes.
Materials and Methods
Strains, Plasmids, and Oligonucleotides
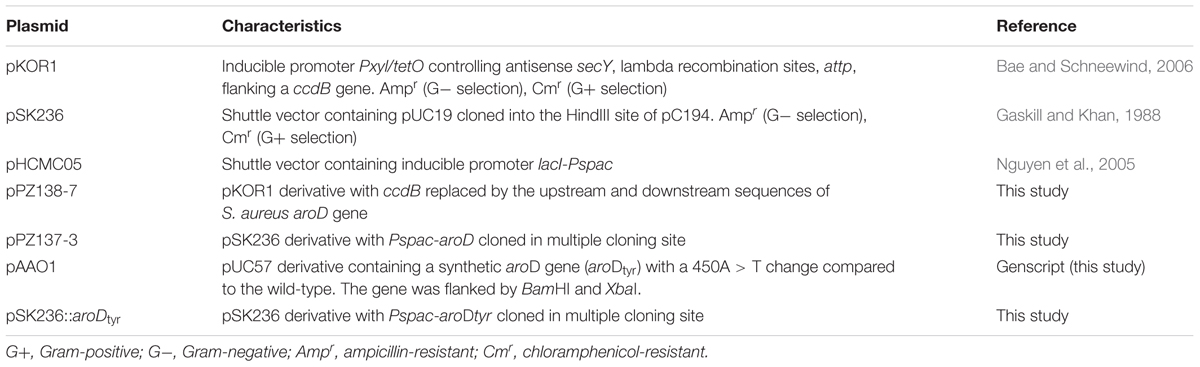
The bacterial strains and plasmids used in this study are summarized in Tables 1, 2, respectively. The oligonucleotides used are listed in Supplementary Table S1. E. coli was grown under aerobic conditions in Luria-Bertani (LB) broth or LB agar (supplemented with 10 μg/ml ampicillin to maintain plasmids when required) at 37°C. S. aureus was grown under aerobic conditions in tryptic soy broth (TSB) or tryptic soy agar (TSA) at 37°C. Appropriate antibiotics (erythromycin at 5 μg/ml or chloramphenicol at 10 μg/ml) were supplemented if the strains carried antibiotic markers. The S. aureus SCV, SCV445, was grown in the presence of 50 μg/ml kanamycin to select against phenotypic switching to the NCP.
Growth Experiments in TSB
For the characterization of S. aureus growth dynamics in TSB, a starter culture was prepared by inoculating a single colony of S. aureus into 5 ml TSB containing appropriate antibiotics and grown for 16 h. The optical density at 600 nm (OD600 nm) of the starter culture was measured using a Pharmacia Biotech Ultrospec 2000 UV/Visible Spectrophotometer (GE Healthcare Life Sciences, Buckinghamshire, United Kingdom), then diluted in TSB supplemented with appropriate antibiotics and/or indicated substance to a calculated OD600 nm of 0.05. This was then incubated at 37°C with shaking at 200 rpm. Growth was monitored by measuring the OD600 nm at 1-h intervals, or by determining colony-forming units (CFU/ml) every 3 h by spreading serial dilutions of the culture onto TSA and incubating aerobically at 37°C for 24–48 h before colonies were counted.
Growth Experiments in Chemically Defined Media (CDM)
The CDM used to characterize S. aureus growth dynamics was adapted from a described recipe (Pattee and Neveln, 1975), the constituents are listed in Supplementary Table S2. A starter culture was prepared by inoculating a single colony of S. aureus into 5 ml TSB containing appropriate antibiotics, which was then grown for 16 h. The cells were collected by centrifugation at 2500 × g for 5 min, washed once with phosphate buffered saline (PBS, pH = 7.4), and re-suspended in 5 ml of the CDM used for growth experiments. The OD600 nm of the suspension was measured, followed by diluting into CDM to a calculated OD600 nm of 0.05. The remaining steps were as described for the growth experiments in TSB.
DNA Sequencing
Genomic DNA from S. aureus strains was prepared from overnight cultures using QIAamp DNA Mini Kit (Qiagen). Overnight cultures of SCV445 were grown in TSB with 50 μg/ml of kanamycin, an aliquot of which was plated out on TSA to establish that none of the cell population had undergone phenotypic switching from SCV to NCP, before the remaining culture was used to obtain DNA. All DNA fragments for sequencing were amplified using Vent DNA polymerase (New England Biolabs). The primers used for PCR amplification and sequencing are listed in Supplementary Table S3. Sanger sequencing was performed by Genewiz UK Limited.
Construction of aroD and menD Mutants
Deletion of the aroD gene in S. aureus LS-1 was performed according to the method of Bae and Schneewind (2006) and as previously described (Wright and Nair, 2012). A deletion from nucleotide 448 bp to 717 bp was constructed by amplifying regions flanking the gene using primer pairs P194/P195 and P196/P197.
A S. aureus LS-1 menD mutant was constructed by homologous recombination. A 3.1 kb fragment containing menD disrupted by an ermC insertion was amplified from S. aureus 8325-4 menD::ermC (Bates et al., 2003) using primers P125 and P126. The amplified fragment was treated with T4 polynucleotide kinase and ligated to pKOR1 which had been digested with EcoRI and EcoRV and treated with Klenow fragment. Subsequent steps were as described previously (Wright and Nair, 2012). The insertion of ermC into menD in the chromosome of LS-1 was confirmed by PCR amplification using primers P125 and P128.
Complementation of the aroD Mutant
To complement the aroD mutant, the aroD gene from S. aureus LS-1 was amplified using primers P192 and P193. The PCR product was digested using BamHI and XbaI. The lacI-Pspac region of plasmid pHCMC05 was excised (Nguyen et al., 2005) using SacI and BamHI, and together with the aroD fragment was ligated with SacI-XbaI digested pSK236 to give plasmid pPZ137-3, which was introduced into the aroD mutant as previously described (Wright and Nair, 2012). In addition a synthetic aroD gene, with a base change at 448 of A to T, flanked by BamHI and XbaI sites was synthesized by Genscript and cloned as described above to complement the aroD mutant with a mutated aroD gene (aroDtyr) coding for a K149Y substitution.
Characterization of SCV445 Auxotrophism
Disk diffusion assays were used to determine the auxotrophic phenotype of SCV445 as previously described (Bauer et al., 1966). Heme (Fluka), menadione (Sigma), and thymidine (Fluka) were prepared at 1, 10, 100, and 1000 μg/ml, along with negative controls, and 15 μL of each substance was inoculated on to a disk and placed in the center of a TSA plate onto which SCV445 had been spread.
Susceptibility to Antimicrobial Agents
The minimum inhibitory concentration (MIC) of kanamycin against S. aureus LS-1, SCV445 and the phenotypically switched strains PZ164 and PZ165 were determined in TSB according to a protocol described previously (Wiegand et al., 2008) and following CLSI guidelines (M07-A10). Briefly, an overnight culture was prepared by growing a single colony of S. aureus in 5 ml TSB for 16 h. The culture was diluted in fresh TSB to approximately 1 × 106 CFU/ml. Aliquots of 50 μl were added to an equal volume of TSB containing antibiotics at a range of concentrations 0, 1, 2, 4, 8, 16, 32, 64, 128, 256, 512, and 1024 μg/ml in polystyrene 96-well plates (Thermo Scientific NUNC). The microtiter plate was incubated at 37°C with shaking at 200 rpm. The MIC was defined as the lowest concentration of antibiotic that resulted in no detectable bacterial growth.
Measurement of the Frequency of Phenotypic Switching
The frequency of SCV445 phenotypic switching to the NCP was measured in fluctuation tests as originally described by Luria and Delbrück (1943). A colony of SCV445 on TSA containing 50 μg/ml kanamycin was inoculated into 10 ml TSB containing 50 μg/ml kanamycin, followed by incubation at 37°C with agitation at 200 rpm overnight. The 100 μl aliquots 10-fold serial dilutions of the culture were spread on TSA and incubated at 37°C. TSA plates were visually scanned for fast-growing NCP colonies after 24 and 48 h of incubation. The mutation rate was calculated according to the formula μ = -(1/N) ln P0, where μ is the mutation rate per cell per generation, N is the total number of colonies counted, and P0 is the proportion of SCV phenotype colonies. Visually identified NCP colonies were streaked on TSA and compared to the wild-type LS-1 to confirm the normal-growth phenotype.
Biofilm Formation
Staphylococcus aureus was grown overnight in TSB, pelleted by centrifugation and resuspended to an OD600 nm of 0.02 in BHI containing 1% glucose. Bacterial suspensions were inoculated into 96 well microtiter plates (200 μl per well) and incubated statically for 24 h at 37oC. Plates were washed three times with PBS and stained with 0.1% crystal violet for 5 min. The plates were rinsed with water to remove excess dye and then dried. The dye was solubilized by adding 200 μl of 33% glacial acetic acid to each well and incubated for 10 min with shaking. Biomass was measured by reading the absorbance at 590 nm. For determination of P-values, paired Student’s t-tests were performed.
Galleria mellonella Virulence Assay
The Galleria mellonella virulence assay was performed essentially as described previously (Peleg et al., 2009) with slight modification. Galleria mellonella (obtained from Cornish Crispa Co, United Kingdom) in the larval stage and weighing 0.2–0.3 g were used to study the virulence of S. aureus. Ten randomly picked individuals were used in each experiment for each strain, and all the strains were tested in three independent experiments. S. aureus strains were cultured in TSB with appropriate antibiotics at 37°C for 16 h. The cultures were centrifuged at 2500 × g for 5 min and the supernatant was removed. Cells were washed once with PBS, then resuspended in PBS to 1.0 × 109 CFU/ml. The 10 μl of the cell suspension or PBS was injected into the hemocoel of the larvae via the last left pro-leg. Once infected, larvae were incubated at 37°C for 5 days, and the survival rate scored every 24 h. Kaplan–Meier survival analysis was performed on the data and the significance of the difference between the percentage survivals of G. mellonella infected with different strains was determined using the Log Rank test. A p-value of less than 0.05 was considered statistically significant.
Results
Isolation of a S. aureus SCV and Characterization of the Auxotrophic Phenotype
A S. aureus SCV, SCV445, was selected by growth of S. aureus LS-1 on TSB agar plates containing 50 μg/ml kanamycin. This strain exhibited typical SCV phenotypes: pinpoint colonies about 1/10 of the size of the wild-type, which were mannitol fermentation negative and had low coagulase activity and low haemolytic activity (data not shown). SCV445 also had greatly increased resistance to kanamycin compared to the wild-type. The MIC of kanamycin against the wild-type LS-1 strain was 16 μg/ml, while it increased to 256 μg/ml for SCV445. The auxotrophism of SCV445 was determined using the disk diffusion method, and it was found that menadione restored growth but hemin or thymidine did not (data not shown).
Identification of the Genetic Mutation in SCV445
To identify the genetic basis of the SCV phenotype of SCV445, genes involved in menadione biosynthesis were amplified and sequenced. These included previously identified menadione biosynthetic genes menA, menB, menC, menD, menE, menF (Nowicka and Kruk, 2010), a gerC locus (also named hepT-menG-hepS), and genes with locus tags SAOUHSC_01348 and SAOUHSC_02556. The gerC locus has been reported to be involved in menadione biosynthesis in Bacillus subtilis (Leatherbarrow et al., 1998), and SAOUHSC_01348 and SAOUHSC_02556 are homologs of a thioesterase-encoding gene that participates in menadione biosynthesis in the cyanobacterium Synechocystis sp. PCC6803 (Widhalm et al., 2009). Unexpectedly, no mutations were found in any of these genes when SCV445 was compared to the wild-type strain.
To investigate whether a mutation occurred in genes coding for enzymes involved in a pathway upstream of menadione biosynthesis, we investigated the shikimate pathway which is shown in Figure 1. The products of the aro genes are responsible for the synthesis of chorismate, which is a branching point for several metabolic pathways, including aromatic amino acid biosynthesis, folate biosynthesis and menaquinone biosynthesis. Therefore a mutation in any of the aro genes would cause a defect in the other metabolic pathways downstream of chorismate in addition to menadione auxotrophy. Therefore to test the hypothesis that there was a defect in the shikimate pathway, we measured the growth of SCV445 and the wild-type LS-1 in defined medium with or without the aromatic amino acids phenylalanine, tyrosine, and tryptophan. Surprisingly, both SCV445 and LS-1 were unable to grow in the absence of aromatic amino acids (Figure 2). To examine this further, we tested the growth of both strains in defined medium in the absence of each of the individual aromatic amino acids. The wild-type strain LS-1 was unable to grow in the absence of tyrosine, while SCV445 failed to grow in the absence of any single aromatic amino acid (Figure 3). This finding suggested the presence of a mutation in a gene involved in tyrosine synthesis in the wild-type strain LS-1, while the mutation in SCV445 conferring the SCV phenotype was likely to be in one of the aro genes.
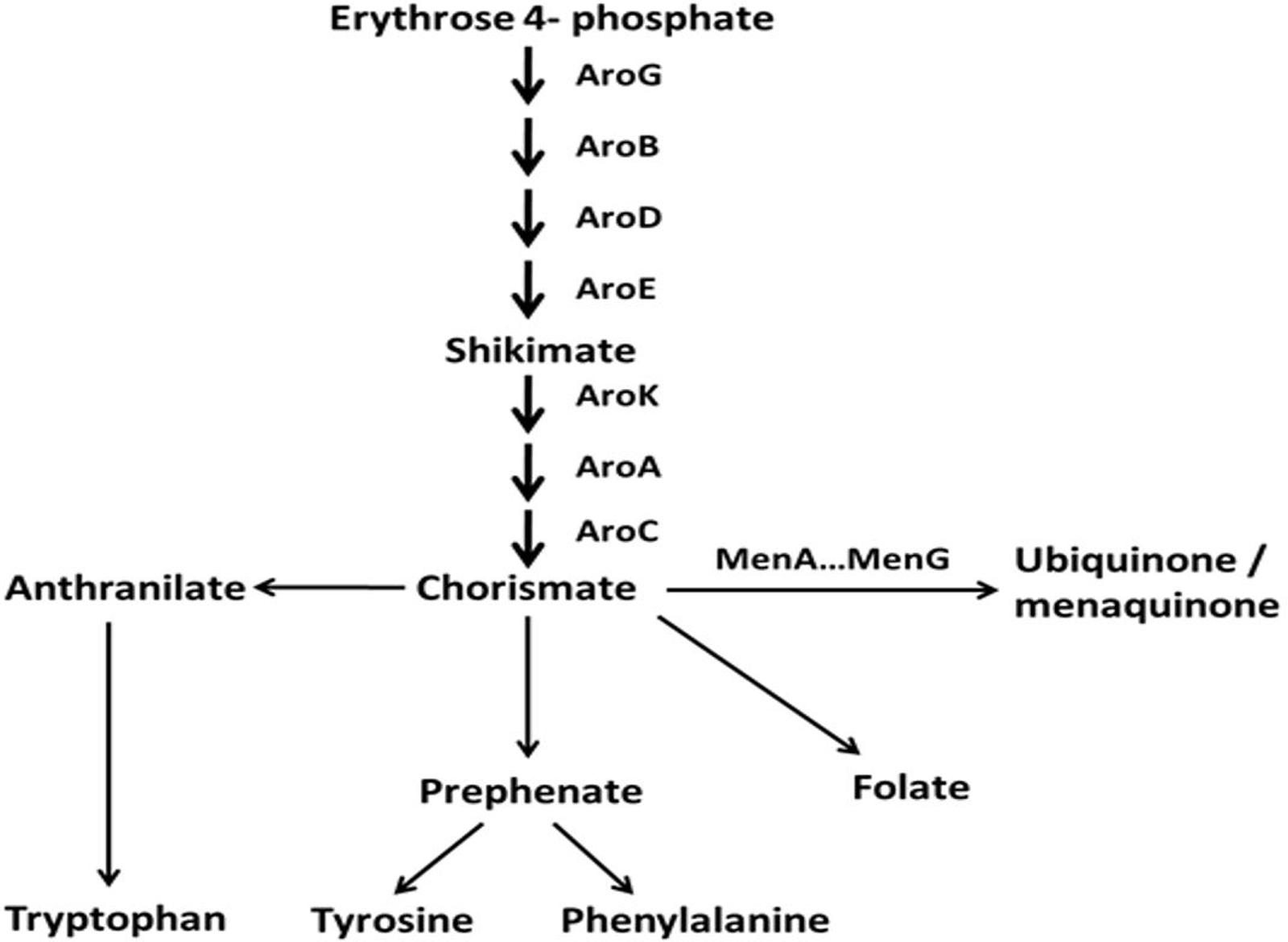
FIGURE 1. Schematic representation of the shikimate pathway and several downstream biosynthetic pathways in S. aureus. The pathway is modified from KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways: phenylalanine, tyrosine, and tryptophan biosynthesis, http://www.genome.jp/kegg/pathway/map/map00400.html.
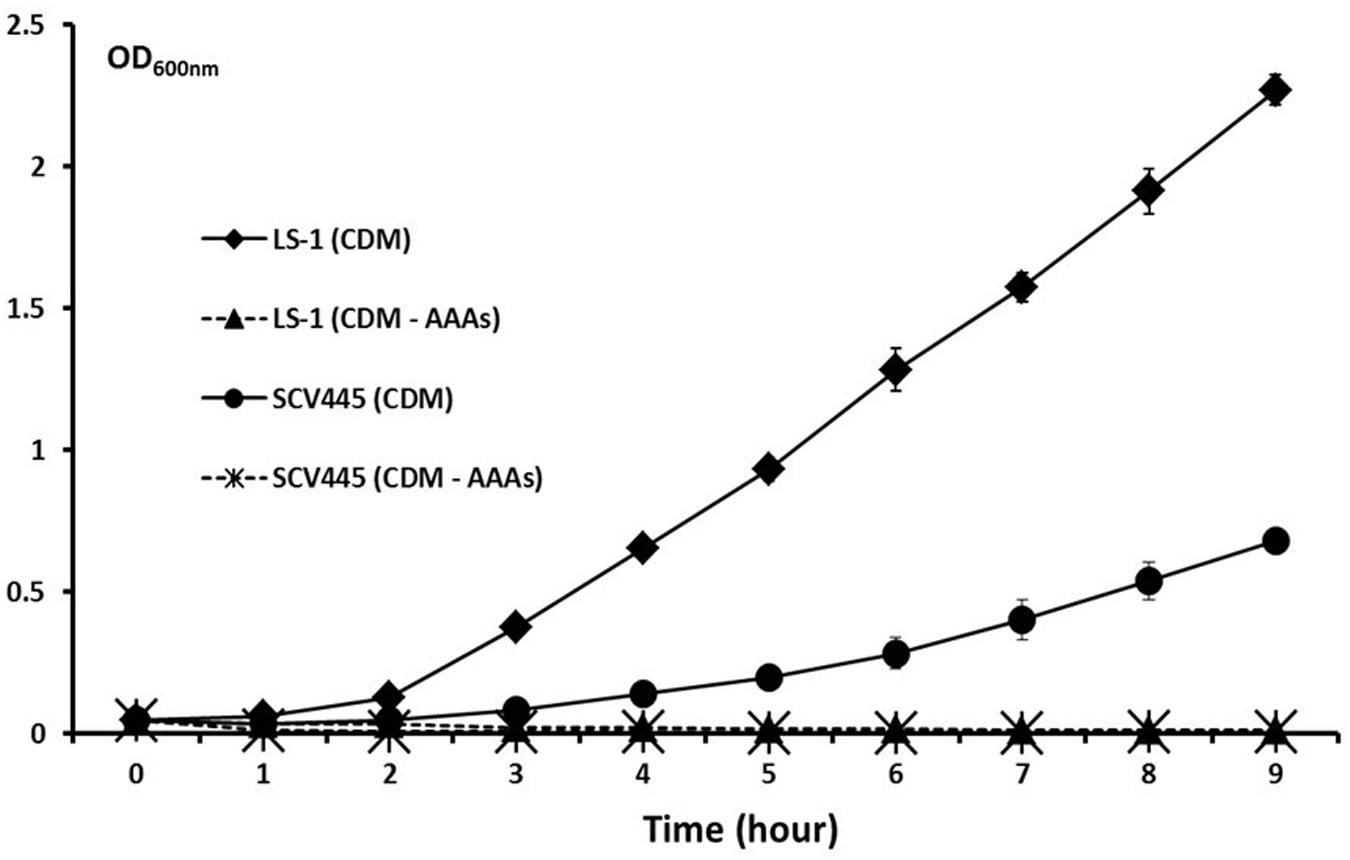
FIGURE 2. Neither the SCV SCV445 nor the parent strain LS-1 can grow in chemically defined medium lacking aromatic amino acids. Growth of S. aureus LS-1 and SCV445 in chemically defined medium (CDM). CDM without aromatic amino acids (CDM – AAAs). Data are presented as the mean of three independent experiments with error bars showing the standard deviations.
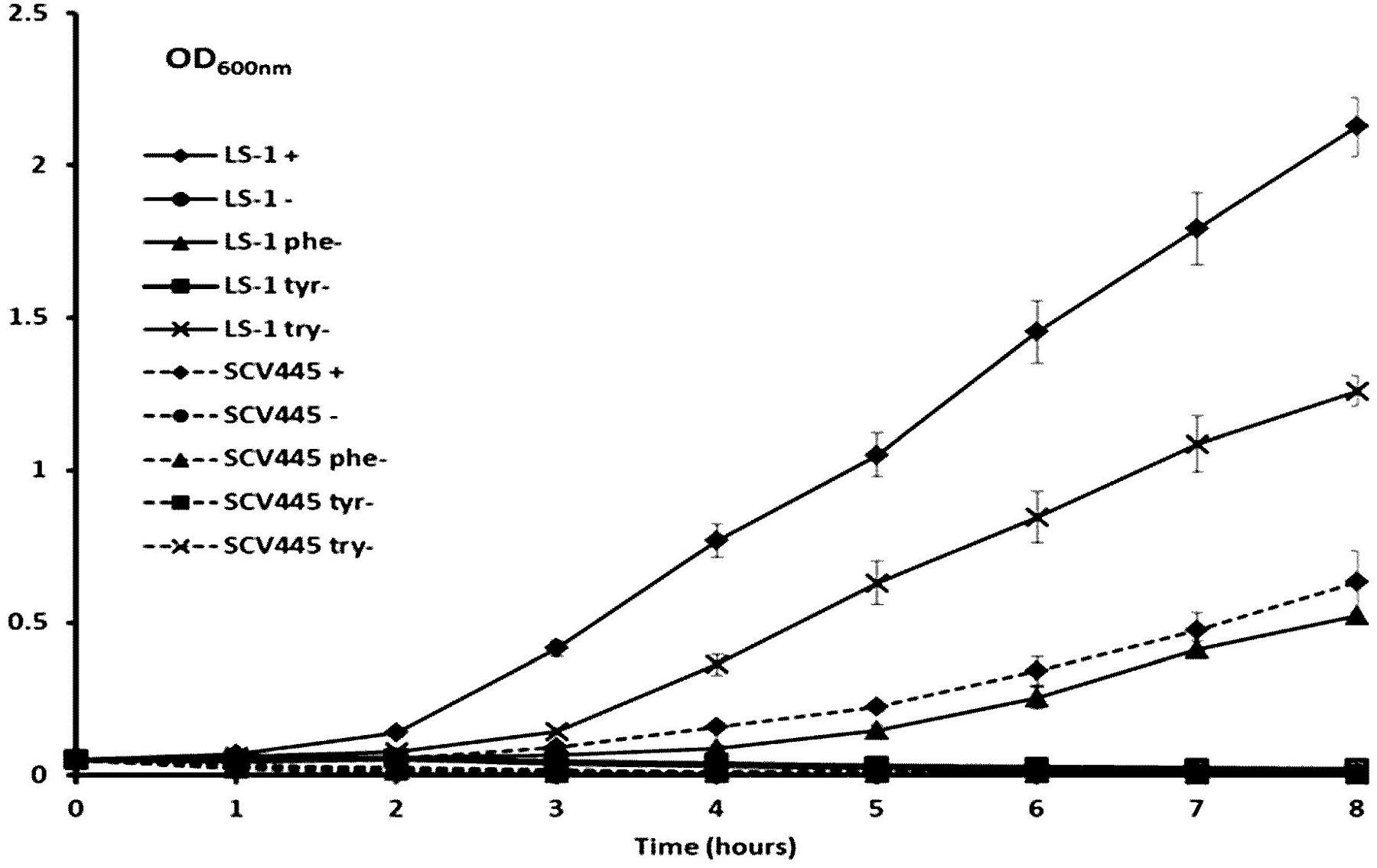
FIGURE 3. The SCV SCV445 requires all three aromatic amino acids to grow in CDM. Growth of S. aureus LS-1 and SCV445 in chemically defined medium (CDM) with or without aromatic amino acid(s). LS-1+ or SCV445+:S. aureus LS-1 or SCV445 with the three aromatic amino acids (AAAS). LS-1- or SCV445-:S. aureus LS-1 or SCV445 without the three AAAs. LS-1 phe- or SCV445 phe-: bacteria grown in CDM without phenylalanine but with the other two AAAs. LS-1 tyr- or SCV445 tyr-: bacteria grown in CDM without tyrosine but with the other two AAAs. LS-1 try- or SCV445 try-: bacteria grown in CDM without tryptophan but with the other two AAAs. Data are presented as the mean of three independent experiments with error bars showing standard deviations.
To locate the mutation in SCV445, the seven aro genes were amplified from both LS-1 and SCV445 and sequenced. Sequence alignment between S. aureus LS-1 and SCV445 showed that the aroD gene in SCV445 had a substitution of 448A > T, resulting in the 149th codon changing from AAA (lysine), to a stop codon TAA. Given that the product of aroD, 3-dehydroquinate dehydratase, is essential for the synthesis of chorismate, this mutation was likely to be the cause of both the menadione and the aromatic amino acid auxotrophy in SCV445. To confirm this, we constructed an aroD mutant of LS-1 by deleting the sequence beyond nucleotide 447 in the aroD gene and adding a TAA stop codon to the truncated gene. The LS-1 aroD mutant exhibited the same phenotypes as SCV445, menadione and aromatic amino acid auxotrophy (data not shown) and had a reduced growth compared to LS-1 (Figure 4). Expression of aroD in trans from the plasmid pPZ137-3 in the aroD deletion strain of LS-1 restored its prototrophic phenotype (data not shown) and growth rate to the same level of LS-1 (Figure 4).
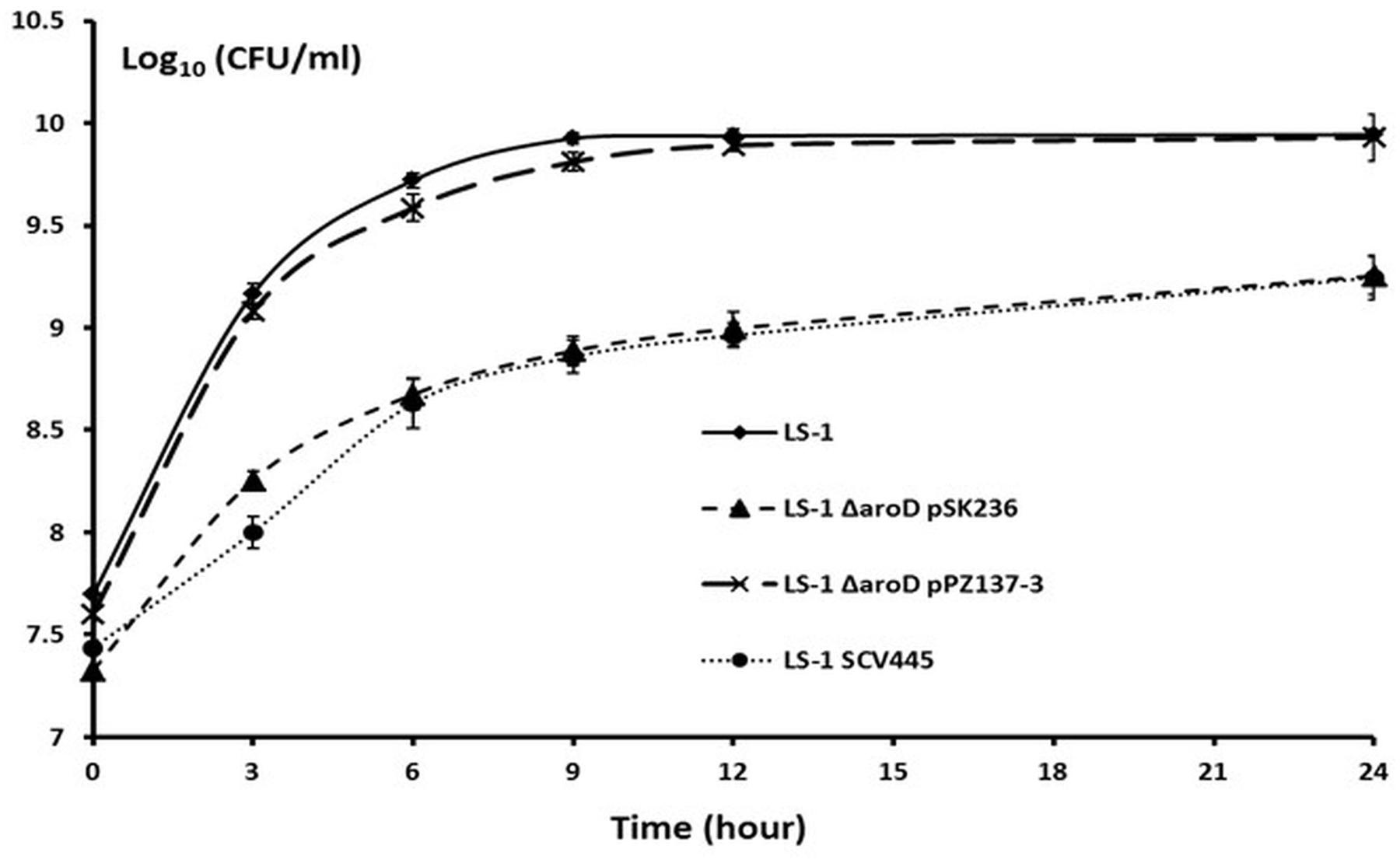
FIGURE 4. Disruption of aroD in S. aureus results in a growth defect similar to that of SCV445 and this can be complemented in trans by a plasmid borne copy of aroD. Viable counts of strains LS-1, LS-1 ΔaroD pSK236 (plasmid without an insert), LS-1 ΔaroD pPZ137-3 (pSK236::aroD) and SCV445 growing in TSB at different time points. Data are presented as the means of three independent experiments with error bars showing the standard deviations.
Disruption of aroD in S. aureus Results in a Greater Capacity to Form Biofilm
As mentioned, apart from SCVs that arose as a result of triclosan treatment (Latimer et al., 2012) the SCV phenotype in S. aureus has generally been reported to be associated with increased biofilm formation (Mitchell et al., 2010a,b). SCV445, an aroD and a menD deletion mutant of strain LS-1 all had a greater capacity to form biofilms than the LS-1 wild-type strain (Figure 5). Given the SCVs are slow growing compared to the wild-type, the increased capacity to form biofilms is remarkable, if the SCVs also grow slowly in the biofilm. Unexpectedly, a hemB deletion mutant of LS-1 had a reduced capacity to form biofilms compared to the parent strain (Figure 5).
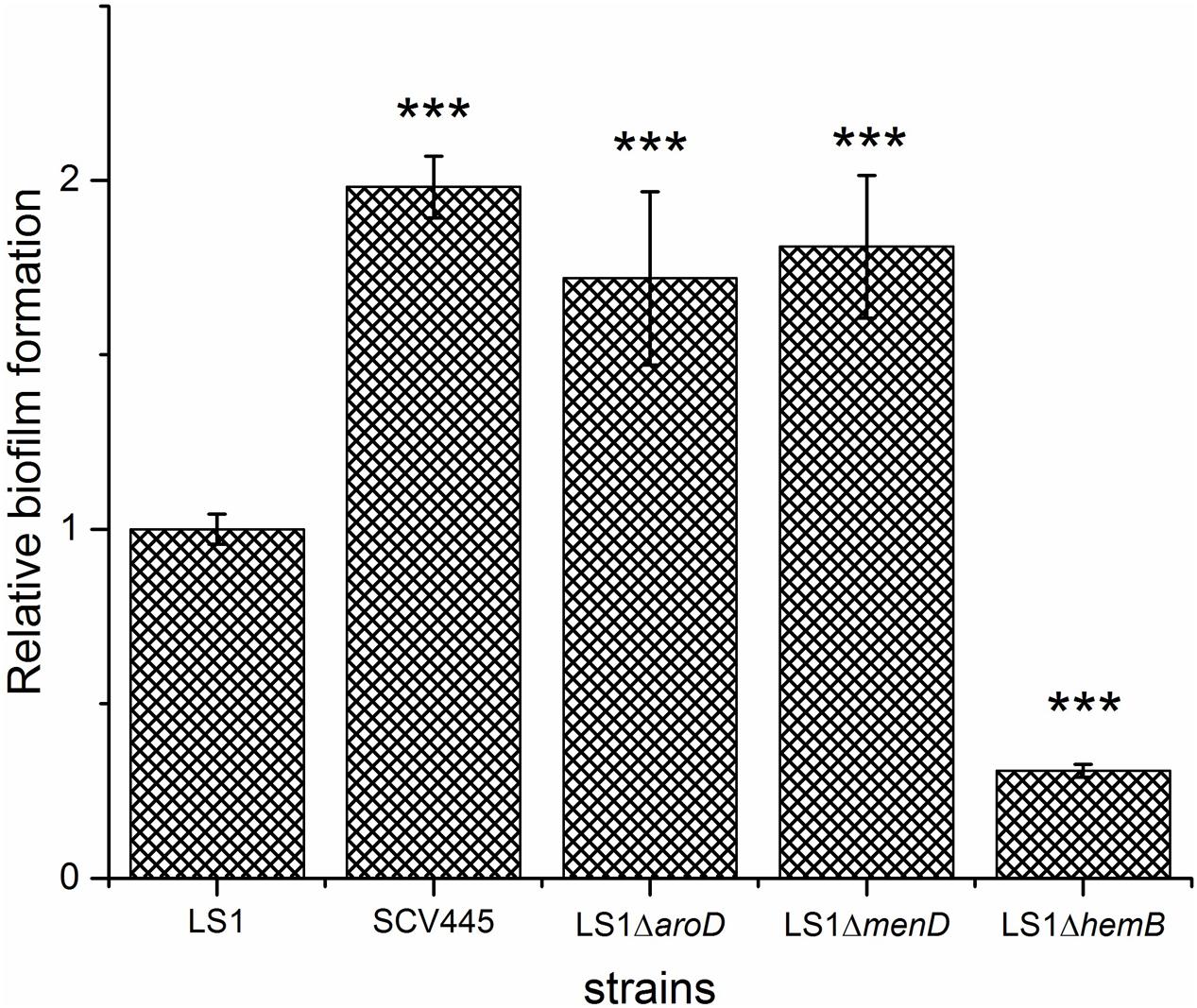
FIGURE 5. Disruption of aroD in S. aureus results in a greater capacity to form biofilm. Biofilm formation on plastic plates in BHI containing 1% glucose. Bacteria were added to microtiter wells at an OD600 nm of 0.02 and incubated for 24 h statically at 37°C. The bars represent the mean with error bars showing the standard deviation. ∗∗∗P < 0.001.
Rate of Phenotypic Switching from SCV to NCP
In three independent cultures plated out on TSA plates no NCPs were visible after 24 h of incubation, while a total of six large colonies were identified from the three cultures after 48 h of incubation, suggesting phenotypic switching had occurred. Streaking these six colonies on TSA confirmed that they were NCPs that formed normal sized colonies after 24 h incubation. In three independent experiments, the reversion rates were determined to be 2.29 × 10-8, 2.83 × 10-8, and 1.52 × 10-7 per cell per generation giving a mean rate of 6.79 × 10-8 per cell per generation.
Genetic Basis for Phenotypic Switching from SCV to NCP
The aroD gene was amplified from the six clones that had switched back to NCPs and their sequences analyzed. Four of the NCPs had the same aroD sequence as the original wild-type, whereas the other two NCPs (PZ164 and PZ165) retained the same mutation as SCV445. Furthermore there were no other mutations in the gene or the intergenic region of aroD in these clones. We reasoned that the only way these clones could have the NCP phenotype was if a functional 3-dehydroquinate dehydratase was still produced despite the presence of the aroD mutation. One possible mechanism that could account for this was that these clones may possess a tRNA gene with a mutated anticodon that recognizes the stop codon UAA and therefore allows read through translation of their aroD mRNA. There are seven tRNA molecules with a complementary anticodon that, with a single base change, could recognize an UAA stop codon. These are tRNA-tyrosine (UAU), tRNA-tyrosine (UAC), tRNA-leucine (UUA), tRNA-serine (UCA), tRNA-lysine (AAA), tRNA-glutamine (CAA), and tRNA-glutamic acid (GAA). The gene encoding each of these was amplified and sequenced. A mutation was found in the tyr-tRNA gene of both NCPs that had switched from the SCV phenotype and retained the aroD mutation. As shown in Figure 6, the tyr-tRNA mutation causes an anticodon change from AUG to AUU, which would therefore recognize the stop codon UAA. While both NCPs had the same mutation they were in different copies of the two tyr-tRNA genes in S. aureus (corresponding to locus tags SAOUHSC_T00057 and SAOUHSC_T00058 in the genome sequence of strain NCTC8325).
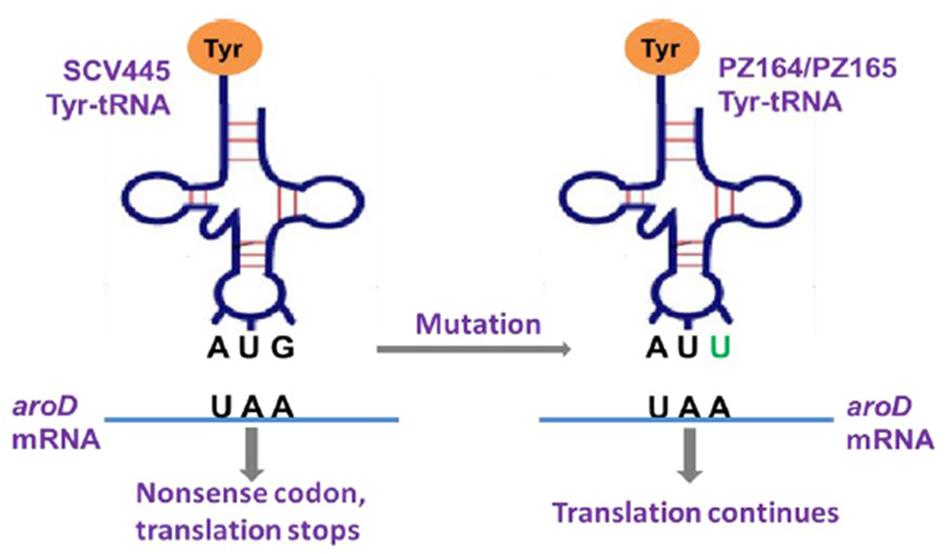
FIGURE 6. Graphic illustration of the mechanism that a tyr-tRNA mutation restores a functional AroD in PZ164 and PZ165 (Murgola, 1990).
The Two NCP tyr-tRNA Mutants Have Equivalent Planktonic Growth Characteristics to the Wild-type, Have the Same Capacity to Form Biofilms and Have Restored Aminoglycoside Sensitivity
The difference between the 3-dehydroquinate dehydratase produced by the wild-type strain and the two tyr-tRNA mutants was a K149Y substitution. Given the NCP of the two-tRNA mutants, this amino acid substitution must be tolerated, giving an enzyme that retains activity. However, the suppressor tyr-tRNAs in PZ164 and PZ165 should partially repress the translation termination of genes with TAA stop codons, which could affect global gene expression and therefore impact on other phenotypes. To test this possibility we initially examined the susceptibility to kanamycin, planktonic growth characteristics and the capacity of PZ164 and PZ165 to form biofilms. PZ164 and PZ165 had identical sensitivity to kanamycin as the wild-type LS-1 and the four NCP revertants PZ160 – PZ163 (16 μg/ml), similar growth rates to LS-1 (Figure 7) and similar capacities to form biofilms as the wild-type and revertants PZ160 – PZ163 (Figure 8). To confirm that the 3-dehydroquinate dehydratase with a K149Y substitution, which would be produced in the ochre suppressor strains, accounted for the NCP we complemented the LS-1 aroD mutant with a mutated gene encoding this change using plasmid pSK236::aroDtyr. This trans complementation restored the NCP (data not shown) and resulted in similar growth to LS-1 (Figure 9).
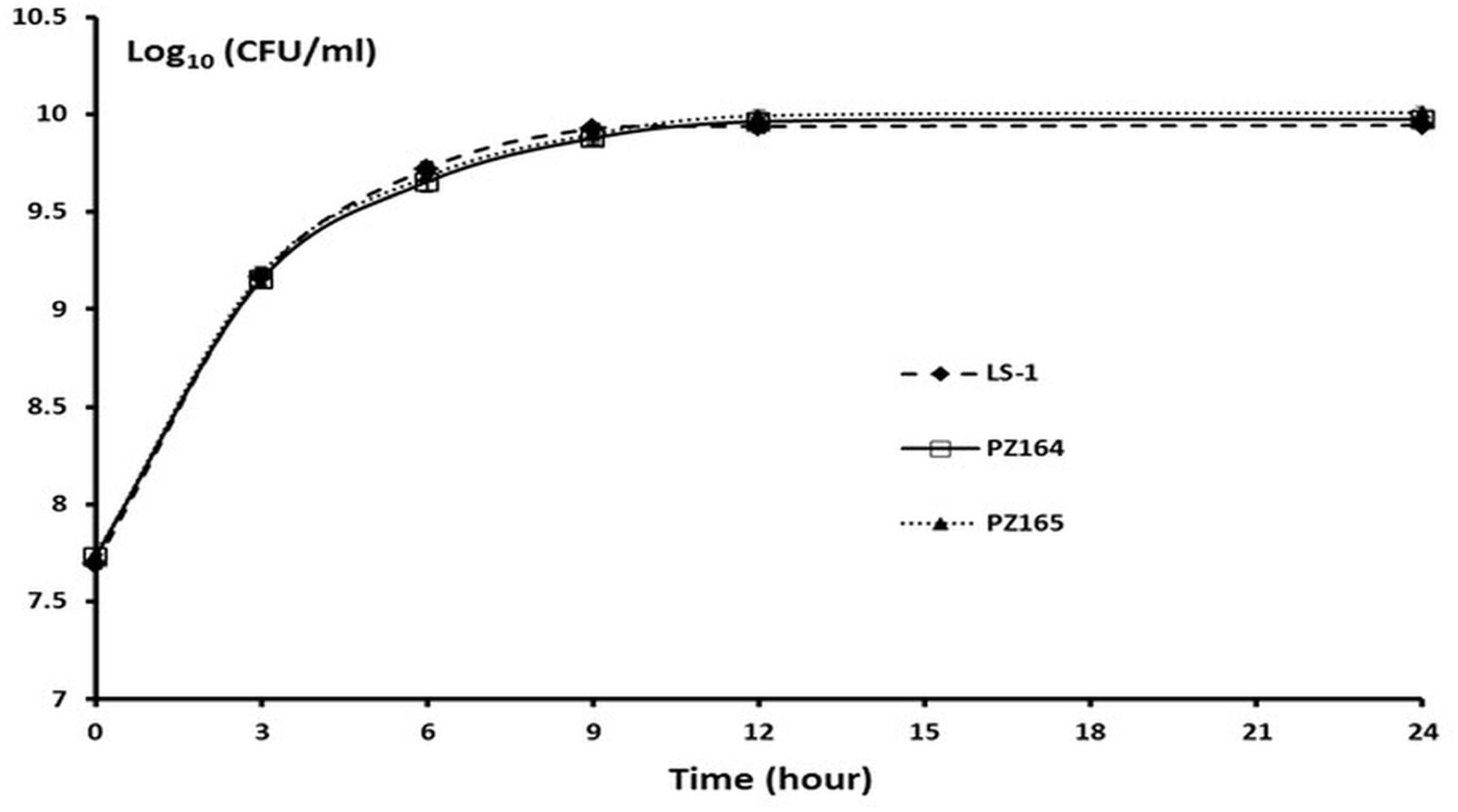
FIGURE 7. The NCP tyr-tRNA mutants grow at the same rate as the wild-type strain LS-1. Viable counts of strains LS-1, PZ164, and PZ165 growing in TSB at different time points. Data are presented as the means of three independent experiments with error bars showing the standard deviations.
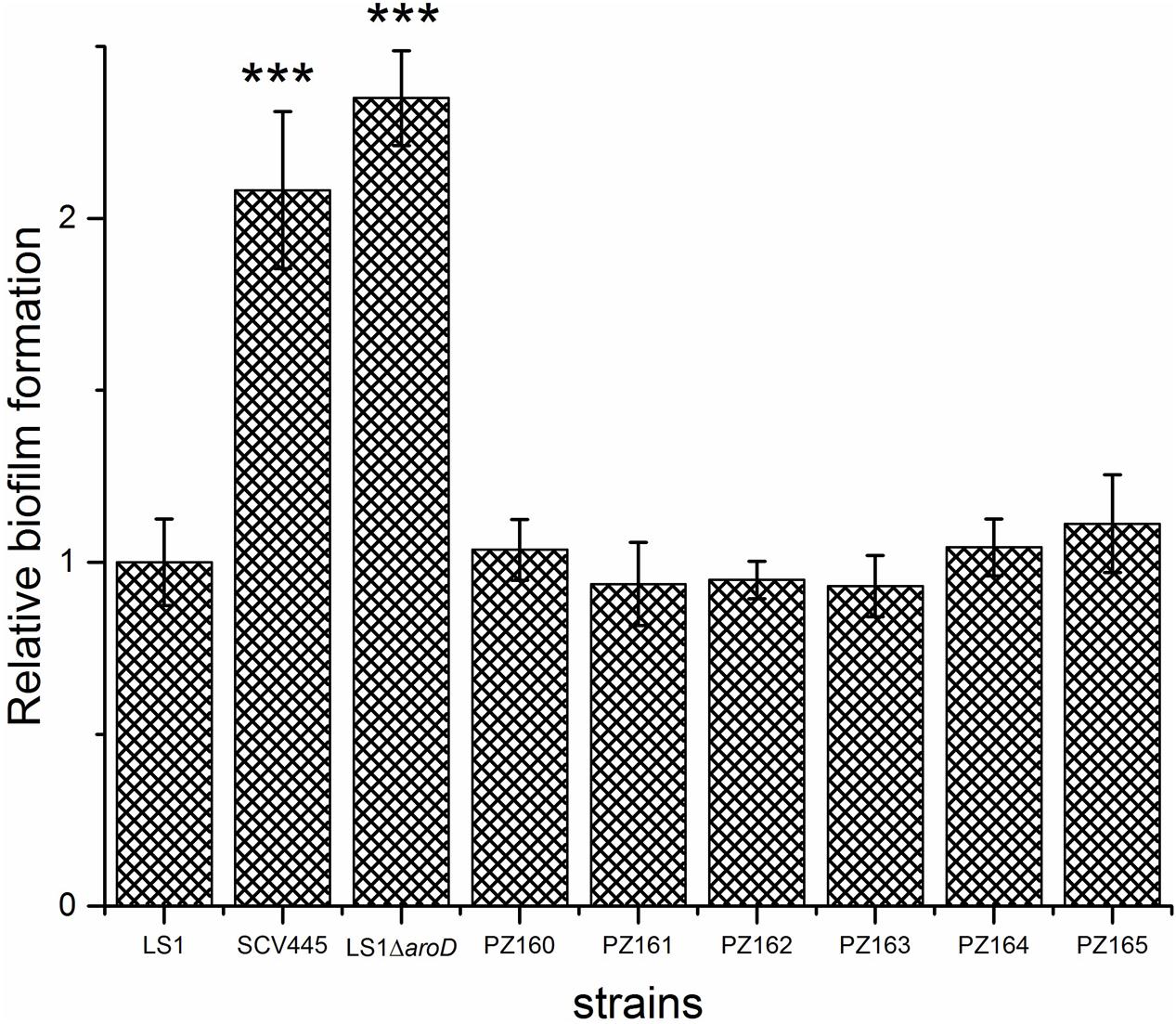
FIGURE 8. Phenotypic switching from the SCV to the NCP restores the wild-type biofilm phenotype regardless of the genetic mechanism. Biofilm formation on plastic plates in BHI containing 1% glucose. Bacteria were added to microtiter wells at an OD600 nm of 0.02 and incubated for 24 h statically at 37°C. The bars represent the mean with error bars showing the standard deviations. ∗∗∗P < 0.001.
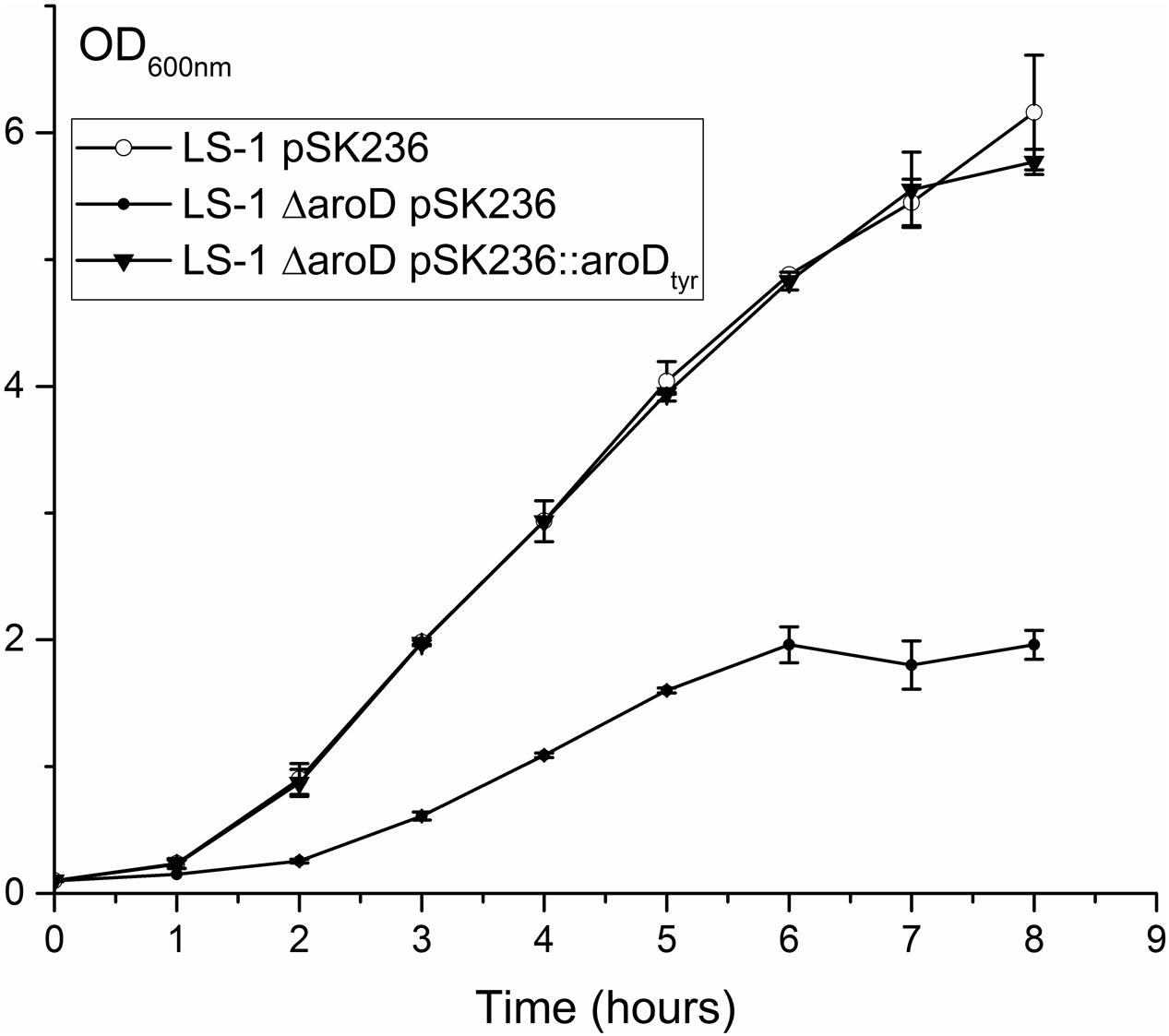
FIGURE 9. Disruption of aroD in S. aureus results in a growth defect similar to that of SCV445 and this can be complemented in trans by a plasmid borne copy of a synthetic aroD gene coding for a K149Y substitution. Growth curves (OD600 nm) of strains LS-1, LS-1 aroD pSK236 (plasmid without an insert) and LS-1 aroD containing pSK236::aroDtyr growing in TSB at different time points. Data are presented as the means of three independent experiments with error bars showing the standard deviations.
The SCV Strains and the Two NCP tyr-tRNA Mutants Are Attenuated in a Galleria mellonella Infection Model
The virulence potential of the wild-type LS-1, the SCV, SCV445, the aroD mutants and the two tyr-tRNA mutants (PZ164 and PZ165) was evaluated using a G. mellonella infection model. Figure 10 shows a Kaplan–Meier plot for survival of infected G. mellonella over time. Compared to the parental strain LS-1, SCV445 and the defined aroD mutant showed significantly reduced virulence (P < 0.001) and there was no significant difference between SCV445 and LS-1 aroD mutant (P = 0.171). Complementation with a functional aroD gene using plasmid pPZ137-3 restored the virulence of the defined mutant to the same levels as the wild-type. The Kaplan–Meier plot shown in Figure 11 demonstrates that compared to the parental strain LS-1 both tyr-tRNA mutants have significantly attenuated virulence (P < 0.001).
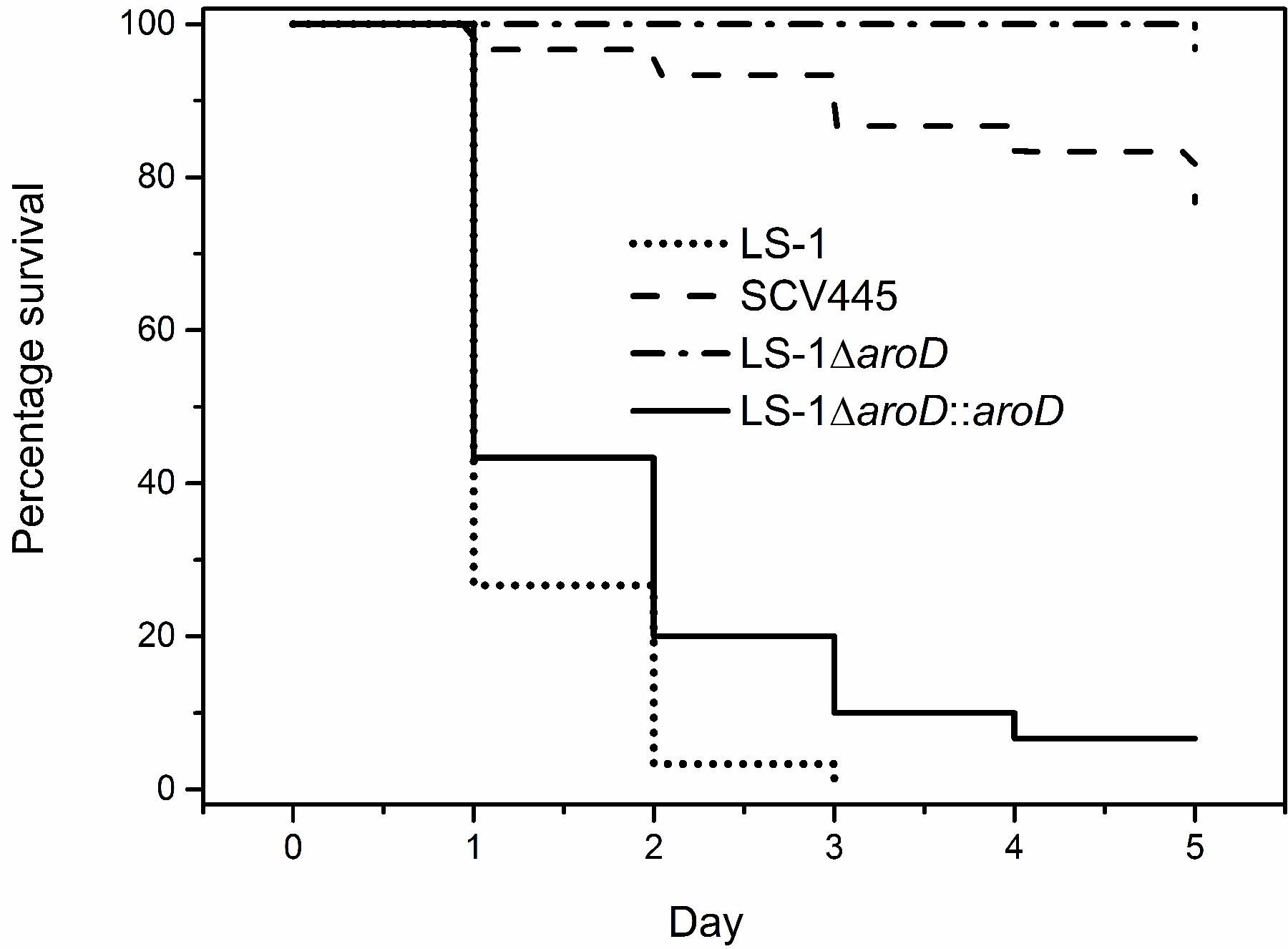
FIGURE 10. The SCV strains are attenuated for virulence in Galleria mellonella. Percentage survival of G. mellonella infected with S. aureus strains and monitored over 5 days. G. mellonella were infected with either the wild-type strain LS-1, or the SCV (SCV445), or an aroD mutant of LS-1 (LS-1ΔaroD) or an aroD mutant of LS-1 complemented with aroD in trans (LS-1ΔaroD::aroD). Data are presented as the percentages of 30 worms, which were assayed in groups of 10 on three different days.
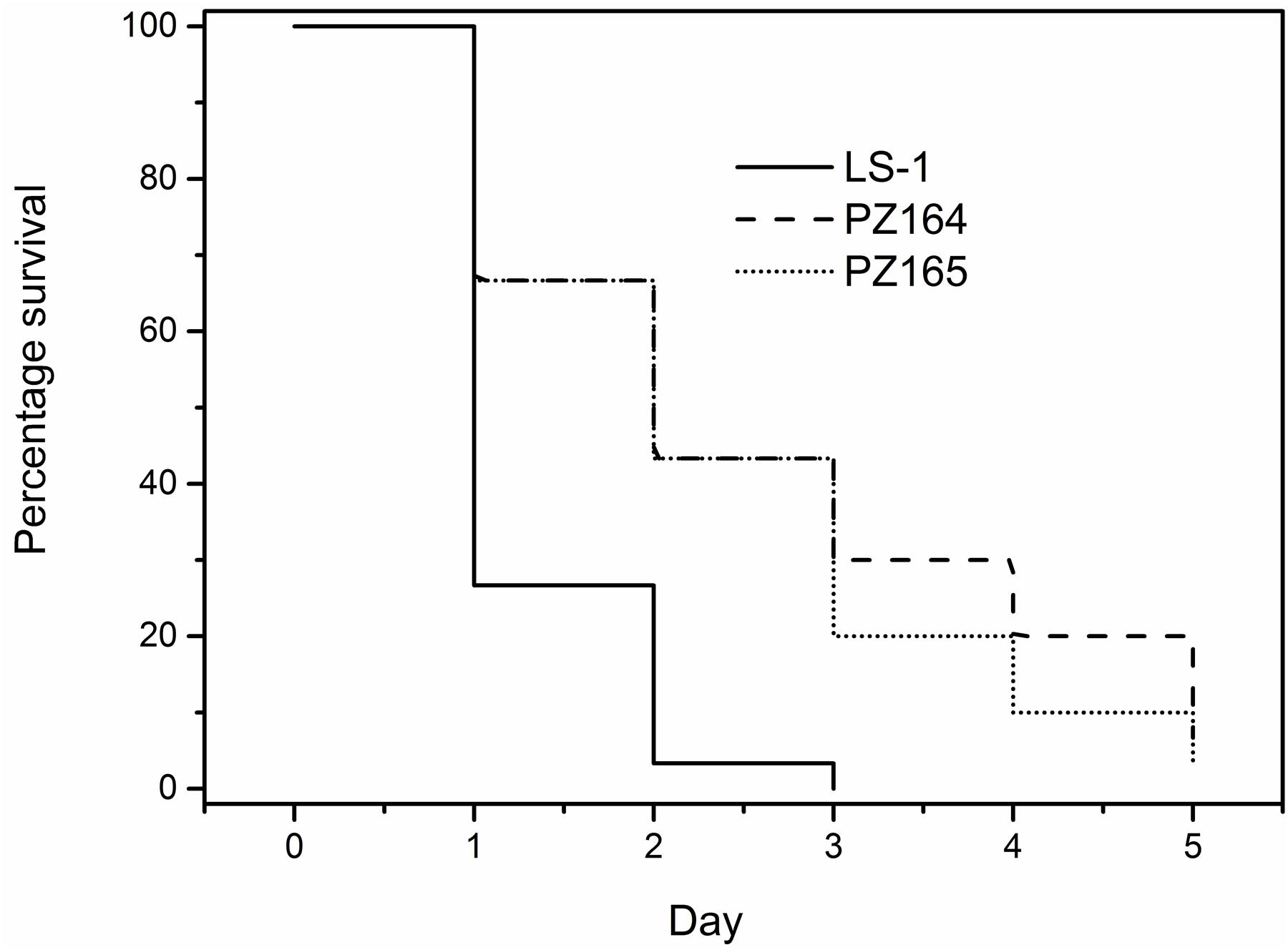
FIGURE 11. The NCP tyr-tRNA mutants are attenuated for virulence in Galleria mellonella. Percentage survival of G. mellonella infected with S. aureus strains and monitored over 5 days. G. mellonella were infected with either the wild-type LS-1, or the NCP tyr-tRNA mutants PZ164 and PZ165. Data are presented as the percentages of 30 worms, which were assayed in groups of 10 on three different days.
Discussion
Studies on S. aureus SCVs have focused on hemin or menadione auxotrophs, mainly using stable defined hemB and menD mutants (von Eiff et al., 1997b, 2006; Bates et al., 2003; Sifri et al., 2006). S. aureus SCVs are frequently isolated from patients, but the genetic basis of SCV phenotypes is not often characterized (Kahl et al., 2003; Yagci et al., 2013). Nevertheless, those SCVs that have been characterized have demonstrated that mutations in hemB or menD are not the only mutations that can give rise to the SCV phenotype. For example mutations in menB (Lannergård et al., 2008), menC, menE, menF (Dean et al., 2014), hemH (Schaaff et al., 2003), and relA, a gene that encodes guanosine polyphosphate pyrophosphohydrolase/synthetase (ppGpp synthetase) (Gao et al., 2010), give rise to a SCV phenotype. Herein, we report an S. aureus SCV with a previously undescribed genotype, an ochre mutation in the aroD gene. Although this mutation would result in a defect in the biosynthesis of a number of metabolites, including aromatic amino acids, folate and menaquinone, supplementation of growth medium with menadione alone was sufficient to reverse the SCV phenotype. The finding that a previously undescribed mutation results in menadione auxotrophy, and the SCV phenotype, suggests that there may be an under appreciation of the range of mutations that lead to this phenotype. Indeed, although the novel aroD SCV we isolated has similar growth characteristics and capacity to form biofilms as the menD SCV, and a reduced virulence potential as reported for the menD SCV (Sifri et al., 2006), it is probable that SCVs resulting from different genetic changes will have different pathogenic phenotypes. In support of this hypothesis is our finding that, while the menadione auxotrophic SCVs (due to mutation of either aroD or menD) had an increased capacity to form biofilms, the hemB SCV was less able to form biofilms than the wild-type. Therefore, knowledge of the full range of genetic changes that can lead to the SCV phenotype is necessary if we are to fully understand how these organisms persist during infection and to develop effective treatments against SCVs.
One reason why natural SCVs have not been well studied is that they are thought to be very unstable and quickly undergo phenotypic switching back to the NCP (Becker et al., 2006). However, a relatively low reversion rate of 6.79 × 10-8 per cell per generation was determined for the aroD SCV, SCV445, described in this study. In addition, a similar reversion rate of 1.8 × 10-8 per cell per generation was reported for a clinically isolated S. aureus menB mutant (Lannergård et al., 2008) and more recently for in vitro generated gentamicin resistant SCVs (Vestergaard et al., 2016). These mutation frequencies are within the range of those reported for spontaneous rifampicin resistant S. aureus mutants (between 3.5 × 10-7 and 1.10 × 10-8 per cell per generation) thus these data suggest that spontaneously formed SCVs are as stable as the NCP, and that phenotypic switching occurs as frequently as other spontaneous mutations. This is also consistent with the findings reported by Edwards (2012) that the formation of SCVs from NCP cells is a natural process during replication rather than a consequence of selective pressure, and that SCV-NCP switching occurs spontaneously during bacterial replication in the absence of selective pressure (Edwards, 2012). Given that by our reasoning SCVs are not particularly unstable, why is it frequently reported that they are? This is probably due to the fact that upon phenotypic switching of a cell from the SCV to the NCP, the NCP cell replicates much more rapidly than the SCV cells, quickly giving rise to a very high proportion of NCP cells within a culture. This may explain the higher SCV reversion rates of 1.76 × 10-6 and 1.21 × 10-6 per cell per generation observed by Dean et al. (2014), where SCVs were serially passaged in broth without selection for 3–10 days.
It has been assumed that phenotypic switching between the SCV phenotype and the NCP was due to reversion of the mutation that initially led to the SCV phenotype. Indeed until fairly recently studies on the genetic basis for phenotypic switching of S. aureus SCVs to NCP have been consistent with this assumption (Lannergård et al., 2008; Dean et al., 2014). More recently Vestergaard et al. (2016) have found new pathways that compensate for the growth fitness of gentamicin resistant SCVs, however, they did not investigate what effect these mutations had on other phenotypes such as biofilm formation and virulence (Vestergaard et al., 2016). We have found that phenotypic reversion can occur via the generation of tRNA-Tyr ochre suppressors, which has not been described previously for SCVs. These findings demonstrate that there are a number of possible genetic events that can lead to phenotypic switching to the NCP apart from the widely reported reversion of the original mutation. Such NCPs may have different phenotypes compared to the wild-type strain and the SCV from which they were derived. In support of this hypothesis is our finding that while the two NCP tyr-tRNA mutants arising from SCV445 had many of the phenotypes of the wild-type strain LS-1, they had reduced virulence in the G. mellonella infection model. Given that the reduced virulence of SCVs is associated with their ability to persist in the host, it is tempting to speculate that the tyr-tRNA NCPs might have more potential to persist in the host than the wild-type strain and therefore cause distinct clinical problems from either the wild-type NCP and the SCV.
Author Contributions
PZ, JW, and AO performed the experiments. PZ, JW, AO, and SN planned the experiments and participated in the interpretation of data and in the writing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by Arthritis Research United Kingdom project grant funding (grant number 18294). PZ received a studentship from the Eastman Foundation for Oral Research and Training (grant number 18294).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01001/full#supplementary-material
References
Atalla, H., Gyles, C., and Mallard, B. (2010). Persistence of a Staphylococcus aureus small colony variants (S. aureus SCV) within bovine mammary epithelial cells. Vet. Microbiol. 143, 319–328. doi: 10.1016/j.vetmic.2009.11.030
Bae, T., and Schneewind, O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 55, 58–63. doi: 10.1016/j.plasmid.2005.05.005
Balwit, J. M., van Langevelde, P., Vann, J. M., and Proctor, R. A. (1994). Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J. Infect. Dis. 170, 1033–1037. doi: 10.1093/infdis/170.4.1033
Bates, D. M., von Eiff, C., McNamara, P. J., Peters, G., Yeaman, M. R., Bayer, A. S., et al. (2003). Staphylococcus aureus menD and hemB mutants are as infective as the parent strains, but the menadione biosynthetic mutant persists within the kidney. J. Infect. Dis. 187, 1654–1661. doi: 10.1086/374642
Bauer, A. W., Kirby, W. M., Sherris, J. C., and Turck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496.
Bayston, R., Ashraf, W., and Smith, T. (2007). Triclosan resistance in methicillin-resistant Staphylococcus aureus expressed as small colony variants: a novel mode of evasion of susceptibility to antiseptics. J. Antimicrob Chemother. 59, 848–853. doi: 10.1093/jac/dkm031
Becker, K., Laham, N. A., Fegeler, W., Proctor, R. A., Peters, G., and von Eiff, C. (2006). Fourier-transform infrared spectroscopic analysis is a powerful tool for studying the dynamic changes in Staphylococcus aureus small-colony variants. J. Clin. Microbiol. 44, 3274–3278. doi: 10.1128/JCM.00847-06
Bremell, T., Lange, S., Svensson, L., Jennische, E., Grondahl, K., Carlsten, H., et al. (1990). Outbreak of spontaneous staphylococcal arthritis and osteitis in mice. Arthritis Rheum 33, 1739–1744. doi: 10.1002/art.1780331120
Dean, M. A., Olsen, R. J., Long, S. W., Rosato, A. E., and Musser, J. M. (2014). Identification of point mutations in clinical Staphylococcus aureus strains that produce small-colony variants auxotrophic for menadione. Infect. Immun. 82, 1600–1605. doi: 10.1128/IAI.01487-13
Edwards, A. M. (2012). Phenotype switching is a natural consequence of Staphylococcus aureus replication. J Bacteriol. 194, 5404–5412. doi: 10.1128/JB.00948-12
Gao, W., Chua, K., Davies, J. K., Newton, H. J., Seemann, T., Harrison, P. F., et al. (2010). Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog. 6:e1000944. doi: 10.1371/journal.ppat.1000944
Gaskill, M. E., and Khan, S. A. (1988). Regulation of the enterotoxin B gene in Staphylococcus aureus. J. Biol. Chem. 5, 6276–6280.
Kahl, B. C. (2014). Small colony variants (SCVs) of Staphylococcus aureus–a bacterial survival strategy. Infect. Genet. Evol. 21, 515–522. doi: 10.1016/j.meegid.2013.05.016
Kahl, B. C., Duebbers, A., Lubritz, G., Haeberle, J., Koch, H. G., Ritzerfeld, B., et al. (2003). Population dynamics of persistent Staphylococcus aureus isolated from the airways of cystic fibrosis patients during a 6-year prospective study. J. Clin. Microbiol. 41, 4424–4427. doi: 10.1128/JCM.41.9.4424-4427.2003
Kreiswirth, B. N., Lofdahl, S., Betley, M. J., O’Reilly, M., Schlievert, P. M., Bergdoll, M. S., et al. (1983). The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 26, 709–712. doi: 10.1038/305709a0
Kriegeskorte, A., Lore, N. I., Bragonzi, A., Riva, C., Kelkenberg, M., Becker, K., et al. (2015). Thymidine-dependent Staphylococcus aureus small-colony variants are induced by trimethoprim-sulfamethoxazole (SXT) and have increased fitness during SXT challenge. Antimicrob. Agents Chemother. 59, 7265–7272. doi: 10.1128/AAC.00742-15
Lannergård, J., von Eiff, C., Sander, G., Cordes, T., Seggewiss, J., Peters, G., et al. (2008). Identification of the genetic basis for clinical menadione-auxotrophic small-colony variant isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 52, 4017–4022. doi: 10.1128/AAC.00668-08
Latimer, J., Forbes, S., and McBain, A. J. (2012). Attenuated virulence and biofilm formation in Staphylococcus aureus following sublethal exposure to triclosan. Antimicrob. Agents Chemother. 56, 3092–3100. doi: 10.1128/AAC.05904-11
Leatherbarrow, A. J., Yazdi, M. A., Curson, J. P., and Moir, A. (1998). The gerC locus of Bacillus subtilis, required for menaquinone biosynthesis, is concerned only indirectly with spore germination. Microbiology 144(Pt 8), 2125–2130. doi: 10.1099/00221287-144-8-2125
Luria, S. E., and Delbrück, M. (1943). Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28, 491–511.
Massey, R. C., Buckling, A., and Peacock, S. J. (2001). Phenotypic switching of antibiotic resistance circumvents permanent costs in Staphylococcus aureus. Curr. Biol. 11, 1810–1814. doi: 10.1016/S0960-9822(01)00507-3
Mike, L. A., Dutter, B. F., Stauff, D. L., Moore, J. L., Vitko, N. P., Aranmolate, O., et al. (2013). Activation of heme biosynthesis by a small molecule that is toxic to fermenting Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 14, 8206–8211. doi: 10.1073/pnas.1303674110
Mitchell, G., Brouillette, E., Séguin, D. L., Asselin, A. E., Jacob, C. L., and Malouin, F. (2010a). A role for sigma factor B in the emergence of Staphylococcus aureus small-colony variants and elevated biofilm production resulting from an exposure to aminoglycosides. Microb. Pathog. 48, 18–27. doi: 10.1016/j.micpath.2009.10.003
Mitchell, G., Séguin, D. L., Asselin, A. E., Déziel, E., Cantin, A. M., Frost, E. H., et al. (2010b). Staphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N-oxide. BMC Microbiol. 10:33. doi: 10.1186/1471-2180-10-33
Murgola, E. J. (1990). Suppression and the code: beyond codons and anticodons. Experientia 46, 1134–1141. doi: 10.1007/BF01936923
Nguyen, H. D., Nguyen, Q. A., Ferreira, R. C., Ferreira, L. C., Tran, L. T., and Schumann, W. (2005). Construction of plasmid-based expression vectors for Bacillus subtilis exhibiting full structural stability. Plasmid 54, 241–248. doi: 10.1016/j.plasmid.2005.05.001
Novick, R. (1967). Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33. 155–166. doi: 10.1016/0042-6822(67)90105-5
Nowicka, B., and Kruk, J. (2010). Occurrence, biosynthesis and function of isoprenoid quinones. Biochim. Biophys. Acta. 1797, 1587–1605. doi: 10.1016/j.bbabio.2010.06.007
Ou, J. J., Drilling, A. J., Cooksley, C., Bassiouni, A., Kidd, S. P., Psaltis, A. J., et al. (2016). Reduced innate immune response to a Staphylococcus aureus small colony variant compared to its wild-type parent strain. Front. Cell Infect. Microbiol. 6:187. doi: 10.3389/fcimb.2016.00187
Pattee, P. A., and Neveln, D. S. (1975). Transformation analysis of three linkage groups in Staphylococcus aureus. J. Bacteriol. 124, 201–211.
Peleg, A. Y., Monga, D., Pillai, S., Mylonakis, E., Moellering, R. C. Jr., and Eliopoulos, G. M. (2009). Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J. Infect. Dis. 15, 532–536. doi: 10.1086/596511
Proctor, R. A., Kriegeskorte, A., Kahl, B. C., Becker, K., Löffler, B., and Peters, G. (2014). Staphylococcus aureus small colony variants (SCVs): a road map for the metabolic pathways involved in persistent infections. Front. Cell Infect. Microbiol. 4:99. doi: 10.3389/fcimb.2014.00099
Proctor, R. A., van Langevelde, P., Kristjansson, M., Maslow, J. N., and Arbeit, R. D. (1995). Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 20, 95–102. doi: 10.1093/clinids/20.1.95
Proctor, R. A., von Eiff, C., Kahl, B. C., Becker, K., McNamara, P., Herrmann, M., et al. (2006). Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4, 295–305. doi: 10.1038/nrmicro1384
Schaaff, F., Bierbaum, G., Baumert, N., Bartmann, P., and Sahl, H. G. (2003). Mutations are involved in emergence of aminoglycoside-induced small colony variants of Staphylococcus aureus. Int. J. Med. Microbiol. 293, 427–435. doi: 10.1078/1438-4221-00282
Seaman, P. F., Ochs, D., and Day, M. J. (2007). Comment on: triclosan resistance in methicillin-resistant Staphylococcus aureus expressed as small colony variants: a novel mode of evasion of susceptibility to antiseptics. J. Antimicrob. Chemother. 60, 175–176. doi: 10.1093/jac/dkm118
Sendi, P., and Proctor, R. A. (2009). Staphylococcus aureus as an intracellular pathogen: the role of small colony variants. Trends Microbiol. 17, 54–58. doi: 10.1016/j.tim.2008.11.004
Sifri, C. D., Baresch-Bernal, A., Calderwood, S. B., and von Eiff, C. (2006). Virulence of Staphylococcus aureus small colony variants in the Caenorhabditis elegans infection model. Infect. Immun. 74, 1091–1096. doi: 10.1128/IAI.74.2.1091-1096.2006
Tan, N. C., Cooksley, C. M., Roscioli, E., Drilling, A. J., Douglas, R., Wormald, P. J., et al. (2014). Small-colony variants and phenotype switching of intracellular Staphylococcus aureus in chronic rhinosinusitis. Allergy 69, 1364–1371. doi: 10.1111/all.12457
Vestergaard, M., Paulander, W., Leng, B., Nielsen, J. B., Westh, H. T., and Ingmer, H. (2016). Novel pathways for ameliorating the fitness cost of gentamicin resistant small colony variants. Front. Microbiol. 7:1866. doi: 10.3389/fmicb.2016.01866
von Eiff, C., Becker, K., Metze, D., Lubritz, G., Hockmann, J., Schwarz, T., et al. (2001). Intracellular persistence of Staphylococcus aureus small-colony variants within keratinocytes: a cause for antibiotic treatment failure in a patient with darier’s disease. Clin. Infect. Dis. 32, 1643–1647. doi: 10.1086/320519
von Eiff, C., Bettin, D., Proctor, R. A., Rolauffs, B., Lindner, N., Winkelmann, W., et al. (1997a). Recovery of small colony variants of Staphylococcus aureus following gentamicin bead placement for osteomyelitis. Clin. Infect. Dis. 25, 1250–1251. doi: 10.1086/516962
von Eiff, C., Heilmann, C., Proctor, R. A., Woltz, C., Peters, G., and Götz, F. (1997b). A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 179, 4706–4712.
von Eiff, C., Peters, G., and Becker, K. (2006). The small colony variant (SCV) concept – the role of staphylococcal SCVs in persistent infections. Injury 37(Suppl. 2), S26–S33.
Widhalm, J. R., van Oostende, C., Furt, F., and Basset, G. J. (2009). A dedicated thioesterase of the hotdog-fold family is required for the biosynthesis of the naphthoquinone ring of vitamin K1. Proc. Natl. Acad. Sci. U.S.A. 106, 5599–5603. doi: 10.1073/pnas.0900738106
Wiegand, I., Hilpert, K., and Hancock, R. E. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175. doi: 10.1038/nprot.2007.521
Wright, J. A., and Nair, S. P. (2010). Interaction of staphylococci with bone. Int. J. Med. Microbiol. 300, 193–204. doi: 10.1016/j.ijmm.2009.10.003
Wright, J. A., and Nair, S. P. (2012). The lipoprotein components of the Isd and Hts transport systems are dispensable for acquisition of heme by Staphylococcus aureus. FEMS Microbiol. Lett. 329, 177–185. doi: 10.1111/j.1574-6968.2012.02519.x
Keywords: Staphylococcus aureus, small colony variants, ochre mutation, phenotypic switching, aroD
Citation: Zhang P, Wright JA, Osman AA and Nair SP (2017) An aroD Ochre Mutation Results in a Staphylococcus aureus Small Colony Variant That Can Undergo Phenotypic Switching via Two Alternative Mechanisms. Front. Microbiol. 8:1001. doi: 10.3389/fmicb.2017.01001
Received: 09 March 2017; Accepted: 18 May 2017;
Published: 31 May 2017.
Edited by:
Weihui Wu, Nankai University, ChinaReviewed by:
Joe Latimer, University of Salford, United KingdomDinesh Sriramulu, Shres Consultancy (Life Sciences), India
Copyright © 2017 Zhang, Wright, Osman and Nair. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sean P. Nair, c2Vhbi5uYWlyQHVjbC5hYy51aw==
 Ping Zhang
Ping Zhang John A. Wright
John A. Wright Ahmed A. Osman
Ahmed A. Osman Sean P. Nair
Sean P. Nair