- College of Animal Science and Technology, Shandong Agricultural University, Tai'an, China
The current study aimed to analyze the prevalence and characterization of Salmonella enterica isolated from animals in slaughterhouses before slaughter. A total of 143 non-duplicate Salmonella were recovered from 1,000 fresh fecal swabs collected from four major pig slaughterhouses (49/600, 8.2%) and four major chicken slaughterhouses (94/400, 23.5%) between March and July 2016. Among Salmonella isolates from pigs, the predominant serovars were Salmonella Rissen (28/49, 57.1%) and Typhimurium (14/49, 28.6%), and high antimicrobial resistance rates were observed for tetracycline (44/49, 89.8%) and ampicillin (16/49, 32.7%). Class 1 integrons were detected in 10.2% (5/49) of these isolates and all contained gene cassettes aadA2 (0.65 kb). Two β-lactamase genes were detected among these isolates, and most of these isolates carried blaTEM-1 (46/49), followed by blaOXA-1(4/49). Seven STs (MLST/ST, multilocus sequence typing) were detected in these isolates, and the predominant type was ST469 (19.6%). Among Salmonella isolates from chickens, the predominant serovars were Salmonella Indiana (67/94, 71.3%) and Enteritidis (23/94, 24.5%), and high antimicrobial resistance rates were observed for nalidixic acid (89/94, 94.7%), ampicillin (88/94, 93.6%) and tetracycline (81/94, 86.2%). Class 1 integrons were detected in 23 isolates (23/94, 24.5%), which contained empty integrons (0.15 kb, n = 6) or gene cassettes drfA17-aadA5 (1.7 kb, n = 6), aadA2 (1.2 kb, n = 5), drfA16-blaPSE-1-aadA2-ereA2 (1.6 kb, n = 5) or drfA1-aadA1 (1.4 kb, n = 1). Three β-lactamase genes were detected, and all 94 isolates carried blaTEM-1, followed by blaCTX-M-55 (n = 19) and blaSPE−1 (n = 3). Five STs were found in these isolates, and the predominant type was ST17 (71.3%). Our findings indicated that Salmonella was widespread in animals at slaughter and may be transmitted from animal to fork.
Introduction
Salmonella enterica is a major global foodborne pathogen (Chiu et al., 2010; Scallan et al., 2011). More than 2,600 different serovars have been identified worldwide (Guibourdenche et al., 2010). In China, Salmonella causes an estimated 22.2% of foodborne diseases, and the majority of diseases are associated with the ingestion of contaminated meat products (Wang et al., 2007). Both pigs and chickens have been recognized as an important reservoir for antibiotic resistant Salmonella, and the resistance genes can be transferred to other bacteria via mobile genetic elements, such as plasmids and gene cassettes (Vo et al., 2006).
Agents of different antimicrobial classes, such as β-lactams or fluoroquinolones, are frequently used in clinical practice for Salmonella enterica infections. Unfortunately, Salmonella has gradually developed a high resistance rate to these antimicrobials, leading to the increase of healthcare costs and even clinical treatment failure (Cui et al., 2009; Gonzalez-Sanz et al., 2009). To date, numerous studies have been conducted to monitor antimicrobial resistance and molecular epidemiology of Salmonella isolated from pigs and chickens in slaughterhouses (Van et al., 2013; Mohamed et al., 2014).
However, little information concerning prevalence and characterization of Salmonella from animals in slaughterhouses in China is available. Shandong province, as a major breeding region, is the main producer of meat products in China. Therefore, major animal slaughterhouses in Shandong province, China were singled out as sampling sites to analyze the prevalence and characterization of Salmonella from animals in slaughterhouses.
Materials and Methods
Description of Sampling Sites
From March to July 2016, 1,000 fresh fecal swabs were collected from four major pig slaughterhouses with process capacity of 1,500–2,500 pigs per day (150 samples per pig slaughterhouse) and four major chicken slaughterhouses with process capacity of 10,000–40,000 chickens per day (100 samples per chicken slaughterhouse). The animal slaughterhouses are respectively located in Weihai, Ciyao, Zhucheng, and Yantai regions in Shandong province, China. Sampling was carried out before slaughter, and at the time animals from different farms has been mixed.
Identification and Sreotyping of Salmonella
From each animal slaughterhouse, fresh fecal swabs were randomly collected from different individual animals, and transported in an ice box to our laboratory within 6 h for further bacteriological analysis. Each swab sample was added into 50 mL buffered peptone water (BPW) and was incubated at 37°C for 16 to 18 h. After that, 0.1 mL of the BPW suspensions was sub-cultured in 10 mL subpackaged Rappaport-Vassiliadis (RV) broth at 42°C for 24 h. One loopful of each RV broth culture was then plated onto xylose lysine tergitol 4 agar plates, and was incubated at 37°C for 24 to 48 h (Yan et al., 2010). Presumptive Salmonella colonies were identified using both the VITEK system (BioMerieux, Marcy 1'Etoile, France) and polymerase chain reaction (PCR) amplification of the inherent gene invA (Malorny et al., 2003).
All Salmonella isolates were serotyped according to the Kauffmann-White scheme by slide agglutination with O and H antigen-specific sera (Tianrun Bio-Pharmaceutical, Ningbo, China) (Grimont and Weill, 2007).
Antimicrobial Susceptibility Testing
The Kirby-Bauer disk diffusion method was used in this study to examine resistance of Salmonella to 10 commonly used antibiotics, including amoxicillin/clavulanic acid (AMC, 20/10 μg), ampicillin (AMP, 10 μg), cefotaxime (CTX, 30 μg), ciprofloxacin (CIP, 5 μg), florfenicol (FFC, 30 μg), gentamicin (GEN, 10 μg), nalidixic acid (NAL, 10 μg), spectinomycin (SPT, 10 μg), tetracycline (TET, 30 μg), and sulfamethoxazole/trimethoprim (SXT, 1.25/23.75 μg). Escherichia coli (ATCC25922) was used as a quality control. The results were interpreted based on the Clinical and Laboratory Standards Institute (CLSI) standards guidelines (CLSI, 2013). Salmonella isolates resistant to more than three classes of antimicrobials were defined as multidrug resistance (MDR) isolates.
Detection of Class I Integrons and β-Lactamase-Encoding Genes
Bacterial DNA was extracted using a TIANamp bacteria DNA kit (Tiangen, Beijing, China) according to the manufacturer's instructions. The gene cassettes within the variable region of class I integrons were detected via polymerase chain reaction (PCR), using previously described primers and procedures (Kerrn et al., 2002). The PCR products were cloned into the pMD18-T vector using the pMD18-T cloning kit (Takara, Dalian, China) and submitted for sequencing (Invitrogen, Beijing, China).
PCR screening for β-lactamase-encoding genes blaTEM, blaPSE-1, blaCMY-2, blaSHV, blaDHA-1, blaOXA, and blaCTX-M was performed as previously described (Guerra et al., 2001; Chen et al., 2004; Batchelor et al., 2005; Hasman et al., 2005; Li et al., 2013). The PCR products were purified and subsequently sequenced.
MLST
The MLST analysis was performed by sequencing the fragments of seven housekeeping genes (aroC, dnaN, hemD, hisD, purE, sucA, and thrA), and the alleles and STs were assigned according to the MLST scheme at http://mlst.warwick.ac.uk/mlst/dbs/Senterica. A minimum spanning tree was created using Bionumerics software 6.5 (Applied Maths, Kortrijk, Belgium), according to the instructions (the unweighted pair group method of arithmetic averages method).
Statistical Analyses
All statistical analyses were performed using package SPSS 15.0 (SPSS Inc., Chicago, IL, USA). The chi-square test was used to compare the prevalence, multidrug resistance rate and carriage of class 1 integron of Salmonella isolated from pigs and chickens, and P < 0.05 was considered difference significant.
Results
Prevalence and Serotypes of Salmonella
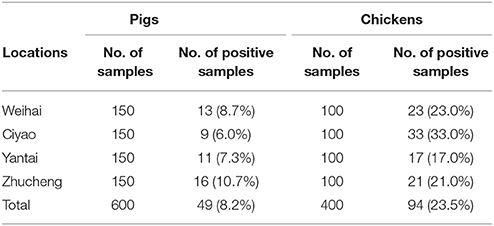
From pig slaughterhouses, 49 Salmonella isolates were recovered (49/600, 8.2%), including 13 from Weihai (13/150, 8.7%), 9 from Ciyao (9/150, 6.0%), 11 from Yantai (11/150, 10.7%), and 16 from Zhucheng (16/150, 10.7%) (Table 1). In terms of isolation rate of Salmonella, no significant difference was found between the pig slaughterhouses (P > 0.05). The 49 Salmonella belonged to 6 serovars, including Salmonella Rissen (n = 28), Typhimurium (n = 14), Grampian (n = 3), Derby (n = 2), Indiana (n = 1), and Enteritidis (n = 1). The most common serovars were Salmonella Rissen (28/49, 57.1%) and Typhimurium (14/49, 28.6%) (Table 2).
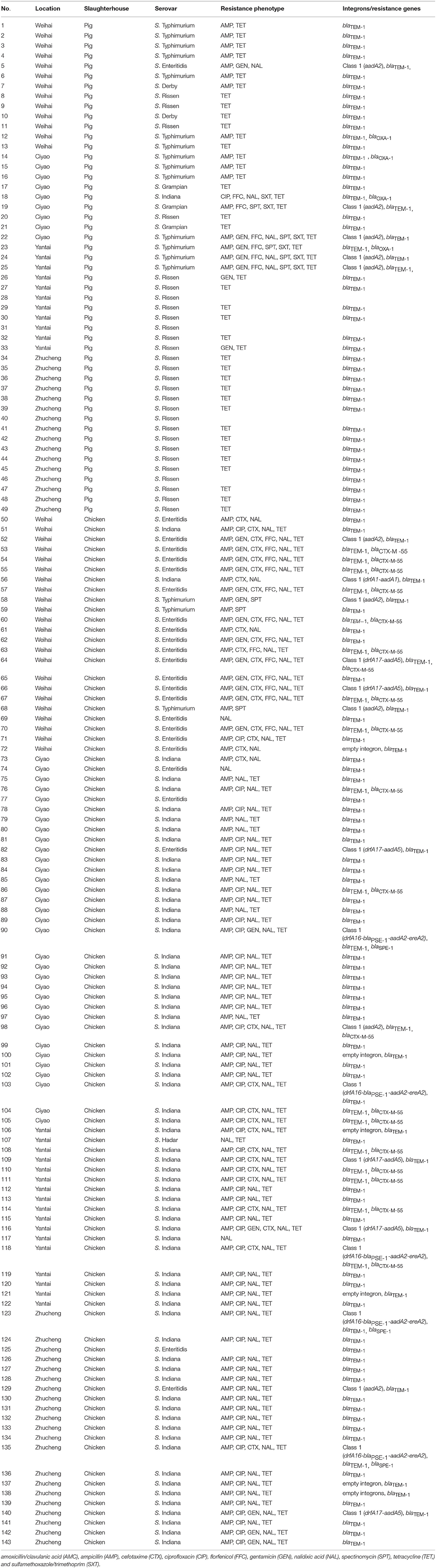
Table 2. Resistance phenotype, incidence of class 1 integron, and resistance gens in Salmonella isolated from animals in slaughterhouses.
From chicken slaughterhouses, 94 Salmonella isolates were recovered (94/400, 23.5%), including 23 from Weihai (23/100, 23.0%), 33 from Ciyao (33/100, 33.0%), 17 from Yantai (17/100, 17.0%), and 21 from Zhucheng (21/150, 21.0%) (Table 1). In terms of isolation rate of Salmonella, no significant difference was found between the chicken slaughterhouses (P > 0.05). These 94 Salmonella isolates belonged to 4 serovars, including Salmonella Indiana (n = 67), Enteritidis (n = 23), Typhimurium (n = 3), and Hadar (n = 1). The dominant serovars were Salmonella Indiana (67/94, 71.3%) and Enteritidis (23/94, 24.5%) (Table 2).
Antimicrobial Susceptibility Testing
All 49 isolates from pig slaughterhouses were susceptible to amoxicillin/clavulanic acid and cefotaxime. But most isolates were resistance to tetracycline (44/49, 89.8%) and ampicillin (16/49, 32.7%). In addition, 7 isolates (7/49, 14.3%) exhibited MDR (Table 2). In addition, 4 isolates were susceptible to all antibiotics used in this study.
All 94 isolates from chicken slaughterhouses were susceptible to amoxicillin/clavulanic acid and sulfamethoxazole/trimethoprim. But most isolates were resistant to nalidixic acid (89/94, 94.7%), ampicillin (87/94, 92.6%), and tetracycline (81/94, 86.2%). Eighty-six isolates (86/94, 91.5%) exhibited MDR (Table 2). Of note, MDR rate of Salmonella from chickens was higher than that from pigs (P < 0.05). In addition, 2 isolates were susceptible to all antibiotics used in this study.
Characteristics of Class 1 Integrons and β-Lactamase-Encoding Genes
Among the 49 isolates recovered from pigs, class 1 integrons were found in 5 isolates (5/49, 10.2%), including 4 Salmonella Typhimurium and 1 Enteritidis. The 5 isolates only contained the single resistance gene cassette aadA2 (0.65 kb). Two β-lactamase genes were detected among the isolates, most of the isolates carried blaSPE-1 (n = 46) and blaOXA-1(n = 4) (Table 2).
Among the 94 isolates recovered from chicken, class 1 integrons were found in 23 isolates (23/94, 24.5%), including 16 Salmonella Indiana, 5 Enteritidis and 2 Typhimurium. Of these isolates, 5 groups of resistance gene cassettes were detected: empty integrons (0.15 kb, n = 6), drfA17-aadA5 (1.6 kb, n = 6), aadA2 (1.2 kb, n = 5), drfA16-blaPSE-1-aadA2-ereA2 (1.7 kb, n = 5), and drfA1-aadA1 (1.4 kb, n = 1). Three β-lactamase genes were detected among these isolates. Most of the isolates carried blaTEM-1 (n = 94), followed by blaCTX-M-55 (n = 19) and blaSPE-1(n = 3) (Table 2).
MLST
One hundred and forty-three Salmonella isolates were divided into 9 STs, including 7 STs from pigs (ST11, ST17, ST19, ST34, ST40, ST358, and ST469), and 5 STs from chickens (ST11, ST17, ST19, ST33, and ST3172). The STs identified in the present study showed the following correlations with Salmonella serovars: ST11 with Salmonella Enteritidis, ST17 with Indiana, and ST469 with Rissen.
BioNumerics software version 6.5 was used to generate a minimum-spanning tree based on all the sources of STs (Figure 1). The dominant ST was ST17 (68/143, 47.6%), with most of isolates from chickens and only one from pigs, followed by ST469 (28/143, 19.6%), with all isolates from pigs. ST34 and ST19 belonged to one clone complex and had the same serovars of Salmonella Typhimurium. ST11 and ST3172 belonged to one clone complex, and had the same serovars of Salmonella Enteritidis.
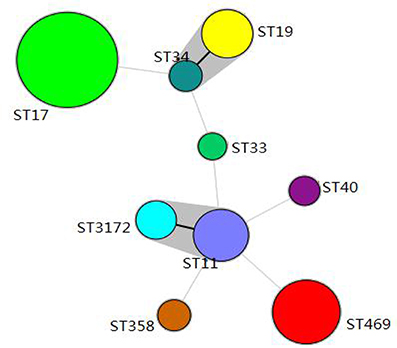
Figure 1. Minimum-spanning tree analysis of the Salmonella isolated from pigs and chickens in slaughterhouses. Each circle represents one ST, and the area of the circle corresponds to the number of isolates, the gray region indicates that ST19 and ST34 belong to a clonal complex, ST11, and ST3172 belong to a clonal complex.
Discussion
In this study, Salmonella isolation rate from pigs (8.2%) was much lower than that (71.8%) in Jiangsu province, China (Cai et al., 2016), and the most common serotype in pigs was Salmonella Rissen, which in consistent with the result from the retail pork products in Thailand (Prapas et al., 2016). However, this finding was different from that reported in EU in which Salmonella Typhimurium was the most common serotype (European Food Safety Authority, 2014). Of note, Salmonella Rissen isolates from pigs only showed resistant to tetracycline (85.7%), which may be associated with the fact that the antimicrobial is frequently used in pig farms in China (Bai et al., 2015).
The Salmonella isolation rate from chickens (23.5%) was similar to the result reported for frozen chicken meat in Shandong province (26.3%), China (Cui et al., 2016). However, the result in this study was much lower than that (45.2%) from chickens in Henan province, China (Bai et al., 2015) and was higher than that (4.5%) from large-scale chicken farms in Shanghai, China (Liu et al., 2010). The difference of these isolation rates may be related with collection seasons, culture methods, and local environments. In the present study, the most common serotypes identified in chickens were Salmonella Indiana and Enteritidis, consistent with findings reported in Henan, China (Bai et al., 2015). However, this finding differed from the result reported in Sichuan province, China, in which Salmonella Derby and Typhimurium were the most common serotypes (Li et al., 2013). Additionally, Salmonella Kentucky and Enteritidis were the most common serotypes in the USA (National Antimicrobial Resistance Monitoring System, 2011), and Salmonella Typhimurium in the EU (European Food Safety Authority, 2014). This difference may be associated with geographical regions. In the present study, Salmonella Indiana showed a high MDR rate (61/68, 89.7%), similar with the result conducted in China (Lu et al., 2011), which demonstrated that most of Salmonella Indiana showed MDR, and these bacteria were not only resistant to streptomycin and tetracycline but also were resistant to chloramphenicol, fluoroquinolones and cephalosporin antibiotics.
In the current study, most Salmonella isolates showed high resistance to tetracycline, ampicillin, and nalidixic acid, similar to the report on slaughterhouses in Italy (Piras et al., 2011), suggesting that these drugs may have been widely used on animals during disease control and prevention. A high resistance rate (63.4%) of nalidixic acid was observed in Salmonella isolates, consistent with other reports (Piras et al., 2011; Siriken et al., 2015). The resistance rate to ciprofloxacin was up to 42.7%. The results may be related with the fact that fluoroquinolone antibiotics are the most common treatment for Salmonella infections. A relatively high resistance rate to cefotaxime (29.1%) was observed in this study, which may be associated with the fact that third-generation cephalosporins have become the primary drugs for the treatment of salmonellosis because of the increase in fluoroquinolone resistance. In addition, the results of the present study showed the high prevalence of multidrug resistant Salmonella isolates in chickens (91.5%), much higher than those reported in Henan province (46.0%), China (Bai et al., 2015) and in central China (34.7%) (Kuang et al., 2015). In this study, MDR isolate rate of Salmonella (91.5%) from chickens were higher than that (14.3%) from pigs, and the higher occurrence of MDR Salmonella isolates from chickens likely reflects the extensive use of antibiotics during intensive rearing. In addition, MDR Salmonella is serotype-dependent (Clemente et al., 2014): the data provided evidence that Salmonella Indiana, Typhimurium and Enteritidis were strongly associated with MDR phenotypes. However, these findings were different from a previous study showing that Salmonella Derby is commonly associated with MDR (Newell et al., 2010).
In the present study, PCR identified class 1 integrons in 19.6% of Salmonella isolates, which was similar to the 15.0% reported from retail meat products in the USA (Zhao et al., 2009) but higher than that of (2.8%) reported from milk products (Van et al., 2013). In the present study, the incidence of class 1 integrons was higher in Salmonella from chickens (24.5%) than Salmonella from pigs (10.2%) (P < 0.05). Class 1 integrons are often associated with MDR Salmonella isolates, consistent with the result of the present study. In addition, the Salmonella isolates carrying class 1 integrons included Salmonella Typhimurium, Enteritidis, and Indiana.
Four β-lactamase genes were detected among Salmonella isolates recovered from pigs and chickens: blaTEM-1, blaPSE-1, blaOXA-1, and blaCTX-M-55. Most isolates carried blaTEM-1, consistent with the report from meat and milk products in Egypt (Ashraf et al., 2014), but different from the report from animal slaughterhouses and retail meat products in Sichuan, China, which showed the dominant β-lactamase gene was blaOXA-1, followed by blaTEM-1, blaPSE-1, and blaCMY-2 (Li et al., 2013). The fact that 46 Salmonella from pigs carried blaTEM-1 whereas only 16 were resistant to ampicillin, and only 88 out of 94 Salmonella carrying blaTEM-1 from chickens showed resistant to ampicillin may be associated with the expression status of blaTEM-1 genes and is needed to be further studied.
In addition, blaCMY-2 encodes resistance to third-generation cephalosporins, an important class of antibiotics used to treat complicated cases of salmonellosis (Gonzalez-Sanz et al., 2009). The incidence of blaCMY-2-positive Salmonella in China was low and was only reported in Shanxi and Sichuan (Yang et al., 2010; Li et al., 2013).
The MLST results revealed 9 STs identified in Salmonella from pigs and chickens. ST19 and ST34 have continually been reported to cause human salmonellosis in recent years, and these bacteria belong to the same serotype, Salmonella Typhimurium (Cai et al., 2016), and this circumstance was also true for Salmonella Enteritidis, represented by ST11 and ST3172. These findings suggested that serovars and STs were tightly coupled (Sukhnanand et al., 2005). ST358 is rare in China and corresponds to Salmonella Grampian, which causes an unusual increase in human cases of Salmonella Grampian infections (Horvath et al., 2013). This observation indicates that Salmonella could spread from animals to humans via pork and chicken products (Osman et al., 2014).
Conclusions
Collectively, our findings exhibit the prevalence and characteristics of Salmonella isolated from animals in slaughterhouses in Shandong province, China. In addition, this study highlights the necessity to carry out the long-term surveillance for Salmonella recovered from food-producing animals.
Author Contributions
WC and SS: conceived and designed the study. XZ and CY: performed the experiments and analyzed the data. XZ, WC, and SS: wrote and revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Taishan Scholars Program (201511023) and the National key R&D project of China (2016YFD050 1608).
References
Ashraf, M. A., Toshi, S., and Tadashi, S. (2014). Characterization of integrons and resistance genes in multidrug-resistant Salmonella enterica isolated from meat and dairy products in Egypt. Int. J. Food Microbiol. 189, 39–44. doi: 10.1016/j.ijfoodmicro.2014.07.031
Bai, L., Lan, R. T., Zhang, X. L., Cui, S. H., Xu, J., Guo, Y. C., et al. (2015). Prevalence of Salmonella isolates from chicken and pig slaughter houses and emergence of Ciprofloxacin and Cefotaxime co-resistant S. enterica Serovar Indiana in Henan, China. PLoS ONE 10:e0144532. doi: 10.1371/journal.pone.0144532
Batchelor, M., Hopkins, K., Threlfall, E. J., Clifton-Hadley, F. A., Stallwood, A. D., Davies, R. H., et al. (2005). blaCTX−M genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob. Agents Chemother. 49, 1319–1322. doi: 10.1128/AAC.49.4.1319-1322.2005
Cai, Y. Q., Tao, J., Jiao, Y., Fei, X., Zhou, L., Wang, Y., et al. (2016). Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int. J. Food Microbiol. 222, 56–64. doi: 10.1016/j.ijfoodmicro.2016.01.020
Chen, S., Zhao, S., White, D. G., Schroeder, C. M., Ran, L., Yang, H., et al. (2004). Characterization of multiple-antimicrobial-resistant Salmonella serovars isolated from retail meats. Appl. Environ. Microb. 70, 1–7. doi: 10.1128/AEM.70.1.1-7.2004
Chiu, L. H., Chiu, C. H., Horn, Y. M., Chiou, C. S., Lee, C. Y., Yeh, C. M., et al. (2010). Characterization of 13 multi-drug resistant Salmonella serovars from different broiler chickens associated with those of human isolates. BMC Microbiol. 10:86. doi: 10.1186/1471-2180-10-86
Clemente, L., Correia, I., Themudo, P., Neto, I., Canica, M., and Bernardo, F. (2014). Antimicrobial susceptibility of Salmonella enterica isolates from healthy breeder and broiler flocks in Portugal. Vet. J. 200, 276–281. doi: 10.1016/j.tvjl.2014.02.007
CLSI (2013). Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twentieth-third Informational Supplement M100-S23. Wayne, PA: Clinical and Laboratory Standards Institute.
Cui, M. Q., Xie, M. Y., Qu, Z. N., Zhao, S. J., Wang, J. W., Wang, Y., et al. (2016). Prevalence and antimicrobial resistance of Salmonella isolated from an integrated broiler chicken supply chain in Qingdao, China. Food Control. 62, 270–276. doi: 10.1016/j.foodcont.2015.10.036
Cui, S., Li, J., Sun, Z., Hu, C., Jin, S., Li, F., et al. (2009). Characterization of Salmonella enterica isolates from infants and toddlers in Wuhan, China. J. Antimicrob. Chemother. 63, 87–94. doi: 10.1093/jac/dkn452
European Food Safety Authority (2014). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2012. EFSA J. 12:3590. doi: 10.2903/j.efsa.2014.3590
Gonzalez-Sanz, R., HerreraLeon, S., de la Fuente, M., Arroyo, M., and Echeita, M. A. (2009). Emergence of extended-spectrum beta-lactamases and AmpC-type β-lactamases in human Salmonella isolated in Spain from 2001 to 2005. J. Antimicrob. Chemother. 64, 1181–1186. doi: 10.1093/jac/dkp361
Grimont, P. A. D., and Weill, F. X. (2007). Antigenic Formulae of the Salmonella Serovars, 9th Edn. Paris: WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur.
Guerra, B., Soto, S. M., Arguelles, J. M., and Mendoza, M. C. (2001). Multidrug resistance is mediated by large plasmids carrying a class 1 integron in the emergent Salmonella enterica serotype [4,5,12:i:−]. Antimicrob. Agents Chemother. 5, 1305–1308. doi: 10.1128/AAC.45.4.1305-1308.2001
Guibourdenche, M., Roggentin, P., Mikoletit, M., Fields, P. I., Bockemuhi, J., Grimont, P., et al. (2010). Supplement 2003-2007 (No. 47) to the white-Kauffmann-le minor scheme. Res. Microbiol. 161, 26–29. doi: 10.1016/j.resmic.2009.10.002
Hasman, H., Mevius, D., Veldman, K., Olesen, I., and Aarestrup, F. M. (2005). β-Lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in the Netherlands. J. Antimicrob. Chemother. 56, 115–121. doi: 10.1093/jac/dki190
Horvath, J. K., Mengel, M., Krisztalovics, K., Nogrady, N., Paszti, J., Lenglet, A., et al. (2013). Investigation into an unusual increase of human cases of Salmonella goldcoast infection in Hungary in 2009. Euro Surveill. 18, 16–22.
Kerrn, M. B., Klemmensen, T., Frimodt-Moller, N., and Espersen, F. (2002). Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. J. Antimicrob. Chemother. 50, 513–516. doi: 10.1093/jac/dkf164
Kuang, X. H., Hao, H. H., Dai, M. H., Wang, Y. L., Ahmad, I., Liu, Z. L., et al. (2015). Serotypes and antimicrobial susceptibility of Salmonella spp. isolated from farm animals in China. Front Microbiol. 6:602. doi: 10.3389/fmicb.2015.00602
Li, R. C., Lai, J., Wang, Y., Liu, S. L., Li, Y., Liu, K. Y., et al. (2013). Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province, China. Int. J. Food Microbiol. 163, 14–18. doi: 10.1016/j.ijfoodmicro.2013.01.020
Liu, W. B., Chen, J., Huang, Y. Y., Liu, B., and Shi, X. M. (2010). Serotype, genotype, and antimicrobial susceptibility profiles of Salmonella from chicken farms in Shanghai. J. Food Protect. 73, 562–567. doi: 10.4315/0362-028X-73.3.562
Lu, Y., Wu, C. M., Wu, G. J., Zhao, H. Y., He, T., Cao, X. Y., et al. (2011). Prevalence of antimicrobial resistance among Salmonella isolates from chicken in China. Foodborne Pathog. Dis. 8, 45–53. doi: 10.1089/fpd.2010.0605
Malorny, B., Hoorfar, J., Hugas, M., Heuvelink, A., Fach, P., Ellerbroek, L., et al. (2003). Interlaboratory diagnosti acccuracy of a Salmonella specific PCR-based method. Int. J. Food Microbiol. 89, 241–249. doi: 10.1016/S0168-1605(03)00154-5
Mohamed, T., Zhao, S., White, D. G., and Parveen, S. (2014). Molecular characterization of antibiotic resistant Salmonella Typhimurium and Salmonella Kentucky isolated from pre and post-chill whole broilers carcasses. Food Microbiol. 38, 6–15. doi: 10.1016/j.fm.2013.08.002
National Antimicrobial Resistance Monitoring System (2011). Animal Arm Annual Report. Available online at: http://ars.usda.gov/SP2UserFiles/Place/60400520/NARMS/NARMS2009/NARMSAnimalArm2009.pdf
Newell, D. G., Koopmans, M., Verhoef, L., Duizer, E., Aidara-Kane, A., Sprong, H., et al. (2010). Food-borne diseases-the challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 139, 3–15. doi: 10.1016/j.ijfoodmicro.2010.01.021
Osman, K. M., Hassan, W. M., and Mohamed, R. A. (2014). The consequences of a sudden demographic change on the seroprevalence pattern, virulence genes, identification and characterisation of integron-mediated antibiotic resistance in the Salmonella enterica isolated from clinically diarrhoeic humans in Egypt. Eur. J. Clin. Microbiol. Infect. Dis. 33, 1323–1337. doi: 10.1007/s10096-014-2072-4
Piras, F., Brown, D. J., Meloni, D., Mureddu, A., and Mazzette, R. (2011). Investigation of Salmonella enterica in Sardinian slaughter pigs: prevalence, serotype and genotype characterization. Int. J. Food Microbiol. 151, 201–209. doi: 10.1016/j.ijfoodmicro.2011.08.025
Prapas, P., Kankanok, T., Tunyamai, B., Anuwat, W., Kittipat, A., Panuwat, Y., et al. (2016). Salmonella in pork retail outlets and dissemination of its pulsotypes through pig production chain in Chiang Mai and surrounding areas, Thailand. Prev. Vet. Med. 130, 99–105. doi: 10.1016/j.prevetmed.2016.06.013
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States-major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Siriken, B., Türk, H., Yildirim, T., Durupinar, B., and Erol, I. (2015). Prevalence and characterization of Salmonella isolated from chicken meat in Turkey. J. Food Sci. 80, 1044–1050. doi: 10.1111/1750-3841.12829
Sukhnanand, S., Alcaine, S., Warnick, L. D., Su, W. L., Hof, J., Craver, M. P., et al. (2005). DNA sequence-based subtyping and evolutionary analysis of selected Salmonella enterica serotypes. J. Clin. Microbiol. 43, 3688–3698. doi: 10.1128/JCM.43.8.3688-3698.2005
Van, K. J. S., Sonnier, J., Zhao, S., and Karns, J. S. (2013). Antimicrobial resistance of Salmonella enterica isolates from bulk tank milk and milk filters in the United States. J. Food Prot. 76, 18–25. doi: 10.4315/0362-028X.JFP-12-263
Vo, A. T., Van, D. E., Fluit, A. C., Heck, M. E., Verbruggen, A., Maas, H. M., et al. (2006). Distribution of Salmonella enterica serovars from humans, livestock and meat in Vietnam and the dominance of Salmonella Typhimurium phage type 90. Vet. Microbiol. 113, 153–158. doi: 10.1016/j.vetmic.2005.10.034
Wang, S. J., Duan, H. L., Zhang, W., and Li, J. (2007). Analysis of bacterial foodborne disease outbreaks in China between 1994 and 2005. FEMS Immunol. Med. Microbiol. 51, 8–13. doi: 10.1111/j.1574-695X.2007.00305.x
Yan, H., Li, L., Alam, M. J., Shinoda, S., Miyoshi, S., and Shi, L. (2010). Prevalence and antimicrobial resistance of Salmonella in retail foods in northern China. Int. J. Food Microbiol. 143, 230–234. doi: 10.1016/j.ijfoodmicro.2010.07.034
Yang, B., Qu, D., Zhang, X., Shen, J., Cui, S., Shi, Y., et al. (2010). Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shaanxi, China. Int. J. Food Microbiol. 141, 63–72. doi: 10.1016/j.ijfoodmicro.2010.04.015
Zhao, S., Blickenstaff, K., Glenn, A., Ayers, L., Friedman, S. L., Abbott, J. W., et al. (2009). β-lactam resistance in Salmonella strains isolated from retail meats in the United States by the National Antimicrobial Resistance Monitoring System between 2002 and 2006. Appl. Environ. Microbiol. 75, 7624–7630. doi: 10.1128/AEM.01158-09
Keywords: Salmonella, animal slaughterhouses, antimicrobial resistance, class 1 integrons, MLST/ST
Citation: Zhao X, Ye C, Chang W and Sun S (2017) Serotype Distribution, Antimicrobial Resistance, and Class 1 Integrons Profiles of Salmonella from Animals in Slaughterhouses in Shandong Province, China. Front. Microbiol. 8:1049. doi: 10.3389/fmicb.2017.01049
Received: 02 October 2016; Accepted: 26 May 2017;
Published: 21 June 2017.
Edited by:
Michel Stanislas Zygmunt, Institut National de la Recherche Agronomique (INRA), FranceReviewed by:
Olivier Barraud, University of Limoges, UMR Inserm 1092 and CHU Limoges, FranceKristina Kadlec, Friedrich Loeffler Institute Greifswald, Germany
Copyright © 2017 Zhao, Ye, Chang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuhong Sun, anF5YmZreWpzQDE2My5jb20=
Weishan Chang, c2R3c2NoYW5nQHNpbmEuY29t
 Xiaonan Zhao
Xiaonan Zhao Chaoqun Ye
Chaoqun Ye