Abstract
Actinobacteria are prolific producers of thousands of biologically active natural compounds with diverse activities. More than half of these bioactive compounds have been isolated from members belonging to actinobacteria. Recently, rare actinobacteria existing at different environmental settings such as high altitudes, volcanic areas, and marine environment have attracted attention. It has been speculated that physiological or biochemical pressures under such harsh environmental conditions can lead to the production of diversified natural compounds. Hence, marine environment has been focused for the discovery of novel natural products with biological potency. Many novel and promising bioactive compounds with versatile medicinal, industrial, or agricultural uses have been isolated and characterized. The natural compounds cannot be directly used as drug or other purposes, so they are structurally modified and diversified to ameliorate their biological or chemical properties. Versatile synthetic biological tools, metabolic engineering techniques, and chemical synthesis platform can be used to assist such structural modification. This review summarizes the latest studies on marine rare actinobacteria and their natural products with focus on recent approaches for structural and functional diversification of such microbial chemicals for attaining better applications.
Introduction
Actinobacteria are Gram-positive bacteria with high GC contents in DNA. They have characteristics presence of intracellular proteasomes, and spores if present are exospores (Cavalier-Smith, 2002). The order Actinomycetales under phylum Actinobacteria includes major producer strains of diverse bioactive compounds. Actinomycetales includes 11 suborders viz. Actinomycineae, Actinopolysporineae, Catenulisporineae, Corynebacterineae, Glycomycineae, Jiangellineae, Micromonosporineae, Propionibacterineae, Pseudonocardineae, Streptomycineae, and Streptosporangineae (http://www.bacterio.net/-classifphyla.html). The genus Streptomyces under sub-order Streptomycineae have been characterized as most important producer of bioactive microbial metabolites (Berdy, 2005). Recently, previously underexplored genera are reported as important resources of diverse bioactive metabolites (Tiwari and Gupta, 2013). These so called rare-actinobacteria are commonly categorized as strains other than Streptomyces (Berdy, 2005) or actinobacteria strains with less frequency of isolation under normal parameters (Lazzarini et al., 2001; Baltz, 2006).
The un-explored and under-explored habitats including marine ecosystems are believed to be rich sources of such rare actinobacteria, with tremendous potential to produce interestingly new compounds (Hong et al., 2009). These marine actinobacteria with potential of producing bioactive compounds have attracted major attention to search for unique compounds with pharmaceutical and biotechnological applications (Bull and Stach, 2007; Subramani and Aalbersberg, 2013; Azman et al., 2015). Recently, there are reports on the discovery of rare actinobacteria from wide range of terrestrial and aquatic locations, including deep seas (Goodfellow et al., 2012). Reports on the analysis of geographical origins of the marine rare actinobacteria, with special focus on the isolation of specific compounds, and precise bioactivities are predominant indications of increasing global interest on the natural compounds from marine rare actinobacteria (Blunt et al., 2007).
Isolation and characterization of marine rare actinobacteria
Generally, for uncovering the marine rare actinobacteria, isolation efforts have been focused on rare locations as deep-sea sediments to obtain new marine diversities (Fenical and Jensen, 2006). The specialized sampling techniques using sophisticated equipment (Fenical and Jensen, 2006), remotely operated vehicles (Pathom-Aree et al., 2006) and even human (Bredholdt et al., 2007), have provided easy access to unprecedented microbial diversity. However, marine rare actinobacteria are usually difficult to culture compared to their terrestrial counterparts mostly due to their special growth requirements (Zotchev, 2012) or unknown culture conditions. It has been observed that hardly <2% of bacterial cells can form colonies by conventional plate cultivation. A large number of them belong to “viable but not culturable” (VBNC) strains (Bernard et al., 2000). Recently, strategies such as mimicking the natural environment in terms of pH, oxygen gradient, nutrient compositions, etc is employed. With these improvements, some previously VBNC species can now be grown with more efficiency (Kaeberlein et al., 2002; Zengler et al., 2002; Vartoukian et al., 2010; Stewart, 2012).
Moreover, the laborious microscopic techniques are being replaced with techniques utilizing recent advances in genomics, proteomics, and bioinformatics for identification and characterization of microbial diversity in robust manner (Rastogi and Sani, 2011). The genomic analysis by genetic fingerprinting (Nübel et al., 1999), DNA-DNA hybridization techniques (Pinhassi et al., 1997), and the construction of metagenomic library and sequencing (Kisand et al., 2012) have been employed for identifying and characterizing the diversity within marine samples. The development of next generation sequencing (NGS) (Webster et al., 2010) and nanopore sequencing (Deamer et al., 2016) has made the process robust and less time consuming. The analysis of RNA expression and regulation using metatranscriptomics (Ogura et al., 2011) or determination of protein profile by metaproteomics (Slattery et al., 2012) can be directly linked to available genome in the database. The coupled metagenomics and metatranscriptomic analysis was successfully used for determining the microbial communities in deep sea water of the North Pacific Ocean (Wu J. et al., 2013). Thus, the combination of both culture dependent (grow and isolate) and culture independent (analysis of nucleic acids and proteins) approaches have revolutionized the characterization and isolation of diverse marine organisms including rare actinobacteria (Hirayama et al., 2007; Zeng et al., 2012).
Discovery of bioactive compounds from marine rare actinobacteria
Actinobacteria including Streptomyces contribute for approximately half of the characterized bioactive compounds up to date (Berdy, 2005). However, the chances of discovery of novel bioactive molecules from Streptomyces has significantly declined (Fenical et al., 1999), presumably due to easy chances of genetic exchange between species during evolution (Freel et al., 2011). Therefore, special attention is given to isolation, screening, and culturing of rare actinobacteria from rare environmental locations as marine sources. The list below summarizes some of the representative compounds isolated from diverse marine rare actinobacteria during last 10 years (Table 1A).
Table 1
| A. Examples of bioactive compounds isolated from various marine rare actinobacteria | ||||
|---|---|---|---|---|
| Compound name | Isolation source | Bacterial source | Biological activities | References |
| INDEPENDENT ISOLATES | ||||
| Pseudonocardians | Deep-sea sediment of South China Sea | Pseudonocardia sp. SCSIO 01299 | Antibacterial and cytotoxic | Li et al., 2011 |
| Caerulomycins | Marine sediments from the seashore of Weihai, China | Actinoalloteichus cyanogriseus WH1-2216-6 | Cytotoxic, antibacterial | Fu et al., 2011 |
| Marinacarbolines, | Marine sediment sample from South China Sea | Marinactinospora thermotolerans SCSIO 00652 | Antimalarial | Huang et al., 2011 |
| Salinosporamides (Commercial name Marizomib) | Deep sea-water of Bahamas Islands, Bahamas | Salinispora tropica (strain CNB-392) | Cytotoxic | Feling et al., 2003; Williams et al., 2005 |
| Abyssomicins | Sediment sample from the Sea of Japan, Japan | Verrucosispora sp. AB-18-032 | Antibacterial | Bister et al., 2004; Riedlinger et al., 2004 |
| Marinomycins | Sediment sample offshore of La Jolla, USA | Marinispora strain CNQ-140 | Cytotoxic | Kwon et al., 2006 |
| Levantilides | Deep-sea sediment Eastern Mediterranean Sea | Micromonospora M71-A77 | Cytotoxic | Gärtner et al., 2011 |
| Salinoquinones | Deep sea-water of Bahamas Islands, Bahamas | Salinispora arenicola CNS-325. | Cytotoxic | Murphy et al., 2010 |
| Neomaclafungin | Marine sediment from Usa bay, Kochi Prefecture, Japan. | Actinoalloteichus sp. NPS702 | Antifungal | Sato et al., 2012 |
| Marthiapeptide A | Deep-sea sediment of the South China Sea | Marinactinospora thermotolerans SCSIO 00652 | Antibacterial, Cytotoxic | Zhou et al., 2012 |
| Lucentamycins | Sediment sample from Bahamas island, Bahamas | Nocardiopsis lucentensis (strain CNR-712) | Cytotoxic | Cho et al., 2007 |
| Juvenimicin C | Sediment collected off the coast of Palau, USA | Micromonospora sp (CNJ-878) | Cancer chemo preventive | Carlson et al., 2013 |
| Levantilide C | Shallow coastal waters near the island of Chiloe, Chile. | Micromonospora strain FIM07-0019 | Antiproliferative | Fei et al., 2013 |
| Nocapyrones | Sediment sample, Ulleung Basin, Eastern sea, Korea | Nocardiopsis sp. | Reduced the pro-inflammatory factor | Kim et al., 2013 |
| Nocardiamides | Sediment sample from La Jolla Canyon, San Diego, California, USA. | Nocardiopsis sp. CNX037 | Low antibacterial activity | Wu Z. C. et al., 2013 |
| Cyanogramides | Marine sediments from the seashore of Weihai, China | Actinoalloteichus cyanogriseus WH1-2216-6 | Multidrug-resistance (MDR) reversing activity | Fu et al., 2014 |
| Taromycin | Marine sediment sample from La Jolla Submarine Canyon, San Diego, California, USA. | Saccharomonospora sp. CNQ-490 | Antibacterial | Yamanaka et al., 2014 |
| Lodopyridone | Marine sediment sample from La Jolla Submarine Canyon, San Diego, California, USA. | Saccharomonospora CNQ490 | Modest cytotoxic activity | Maloney et al., 2009 |
| Lynamicins | Marine sediment off the coast of San Diego, California, USA | Marinispora NPS12745 | Antibacterial | McArthur et al., 2008 |
| Saccharothrixones | Sediment sample from Heishijiao Bay, Dalian, China | Saccharothrix sp. 10-10 | Cytotoxic | Gan et al., 2015 |
| Saliniketals | Sediment sample from Island of Guam, USA | Salinispora arenicola CNR-005 | Prevention of carcinogenesis | Williams et al., 2007a |
| Arenicolides | Sediment sample from Island of Guam, USA | Salinispora arenicola CNR-005 | Moderate cytotoxicity | Williams et al., 2007b |
| Lagumycin B, Dehydrorabelomycin, Phenanthroviridone, WS-5995 A | Sediment sample from Cát Bà Peninsula, East Sea Vietnam | Micromonospora sp. | Cytotoxic | Mullowney et al., 2015 |
| Dermacozines, Phenazine derivatives | Sediment sample from Mariana Trench | Dermacoccus abyssi sp. nov., strains MT1.1 and MT1.2 | Cytotoxic and anti-oxidant | Abdel-Mageed et al., 2010 |
| Fijiolides | Sediment sample from the Beqa Lagoon, Fiji | Nocardiopsis CNS-653 | Inhibitor of TNF-α-induced NFκB activation | Nam et al., 2010 |
| Fluostatin | Sediment sample from South China Sea | Micromonospora rosaria SCSIO N160 | Antimicrobial | Zhang et al., 2012 |
| Retimycin | Deep sea-water of Bahamas Islands, Bahamas | S. arenicola strain CNT-005. | Cytotoxic | Duncan et al., 2015 |
| Sioxanthin | Deep sea-water of Bahamas Islands, Bahamas | Salinispora tropica CNB-440 | Siderophore | Richter et al., 2015 |
| Lobosamides | Sediment sample from Point Lobos, Monterey Bay, California, USA. | Micromonospora sp. RL09-050-HVF-A | Antitryposomal | Schulze et al., 2015a |
| Salinipostins | Sediment sample from Keawekaheka Bay, Hawai, USA | Salinispora sp. RL08-036-SPS-B | Antimalarial | Schulze et al., 2015b |
| Isomethoxyneihumicin | Sediment sample at Chichijima, Ogasawara, Japan | Nocardiopsis alba KM6-1 | Cytotoxic | Fukuda et al., 2016 |
| Nocarimidazoles | Sediment sample off the coast of southern California, USA | Nocardiopsis sp. CNQ115 | Weak antibacterila | Leutou et al., 2015 |
| Cyclomarine Cyclomarazine | Marine sediment from Palau, Republic of Palau | S. arenicola CNS-205 | Anti-inflammatory | Schultz et al., 2008 |
| ISOLATES IN SYMBIOTIC ASSOCIATION | ||||
| JBIR-65 | Symbiont to unidentified marine sponge from Ishigaki Island, Okinawa Prefecture, Japan | Actinomadura sp. SpB081030SC-15 | Anti-oxidant | Takagi et al., 2010 |
| Nocapyrones | Symbiont to sponge Halichondria panacea from Baltic Sea, Germany | Nocardiopsis sp. HB383 | Weak cytotoxic | Schneemann et al., 2010 |
| Arenjimycin | Symbiont to ascidian Ecteinascidia | Salinispora arenicola | Antimicrobial and ytotoxic | Asolkar et al., 2010 |
| Turbinate from Sweetings Cay, Grand Bahama Island, USA | ||||
| Bendigoles | Symbiont to sponge Suberites japonicas from unspecified source | Alctinomadura sp. SBMs009 | Antimicrobial and cytotoxic | Simmons et al., 2011 |
| Thiocoraline | Symbiont to sponge Chondrilla caribensis from Florida Keys, USA | Verrucosispora sp. | Cytotoxic | Wyche et al., 2011 |
| Peptidolipins | Symbiont to ascidian Trididemnum orbiculatum from Florida Keys, USA | Nocardia sp. | Antibacterial | Wyche et al., 2012 |
| Anthracyclinones | Symbiont to tunicate Eudistoma vannamei from Taìba Beach, Ceará, Brazil | Micromonospora sp. | Cytotoxic | Sousa et al., 2012 |
| Halomadurone | Symbiont to ascidian Ecteinascidia turbinata, from Florida Keys, USA | Actinomadura sp. | Active against neurodegenerative diseases | Wyche et al., 2013 |
| Solwaric acids | Symbiont to ascidian, Trididemnum orbiculatum from Florida Keys, USA | Solwaraspora sp. | Antibacterial | Ellis et al., 2014 |
| Forazoline A | Symbiont to ascidian, Ecteinascidia turbinate from Florida Keys | Actinomadura sp. WMMB-499 | Antifungal | Wyche et al., 2014 |
| Rifamycins | Symbiont to sponge, Pseudoceratina clavata. From Great Barrier Reef, Australia | Salinispora sp. strain M403 | Antibacterial | Kim et al., 2006 |
| Saccharothrixmicines | Symbiont to marine mollusk Anadara broughtoni from Sea of Japan | Saccharothrix espanaensis An 113 | Antibacterial, Antifungal | Kalinovskaya et al., 2010 |
| B. Approaches used for production and structural/functional diversification of bioactive compounds derived from marine rare actinobacteria | ||||
| Compound name | Genus | Particulars | Biological activity | References |
| Retimycin | Salinospora | MS/MS spectrum pattern based genome mining | Cytotoxic, Antibacterial | Duncan et al., 2015 |
| Thiolactomycin | Salinospora | Antibiotic resistance gene based genome mining, heterologous expression | Bacterial fatty acid synthase inhibitor | Tang et al., 2015 |
| Lomaiviticin | Salinospora | Bioactivity guided genome mining | Cytotoxic | Kersten et al., 2013 |
| Salinosporamide K | Salinospora | Genome mining, metabolomics and transcriptomics | Cytotoxic | Eustáquio et al., 2011 |
| Taromycin | Saccharomonospora | BCG Genome mining, heterologous expression | Antibacterial | Yamanaka et al., 2014 |
| Enterocin | Salinispora | BCG Genome mining, heterologous expression | Antibacterial | Bonet et al., 2014 |
| Fluostatins | Micromonospora | Heterologous expression | Antibacterial | Yang et al., 2015 |
| Thiocoraline | Micromonospora | Heterologous expression | Cytotoxic | Lombó et al., 2006 |
| Bromosalinosporamide | Salinospora | Precursor directed biosynthesis | Cytotoxic | Lam et al., 2007 |
| Salinosporamide A | Salinospora | Precursor pathway modulation | Cytotoxic | Lechner et al., 2011 |
| Salinosporamide X1, Salinosporamide X2 | Salinospora | Combinatorial biosynthesis | Cytotoxic | McGlinchey et al., 2008 |
| Salinosporamide X3 | Salinospora | Mutasynthesis | Cytotoxic | Nett et al., 2009 |
| Salinosporamide X4 | ||||
| Salinosporamide X5 | ||||
| Salinosporamide X6 | ||||
| Salinosporamide X7 | ||||
| Fluorosalinosporamide | Salinospora | Mutasynthesis | Cytotoxic | Eustáquio and Moore, 2008 |
| Salinosporamides analogs | Salinospora | Chemobiosynthesis | Cytotoxic | Liu et al., 2009 |
| Salinosporamide A | Salinospora | Total chemical synthesis | Cytotoxic | Reddy et al., 2004; Endo and Danishefsky, 2005; Kaiya et al., 2011; Logan et al., 2014 |
| Homosalinosporamide | Salinospora | Total chemical synthesis | Cytotoxic | Nguyen et al., 2010 |
| Salinosporamides analogs | Salinospora | Chemobiosynthesis | Cytotoxic | Liu et al., 2009 |
| Salinosporamide E | Salinospora | Semi-synthesis | Cytotoxic | Macherla et al., 2005 |
| Bromosalinospramide | ||||
| Iodosalinosporamide, Azidosalinosporamide, Hydroxysalinosporamide | ||||
| Methylsalinosporamide | Salinospora | Semi-synthesis | Cytotoxic | Manam et al., 2008 |
| Tosylsalinosporamide | ||||
| Dansylsalinosporamide | ||||
| Hydroxysalinosporamide | ||||
| Flurosalinosporamide | ||||
Overview of achievements in study of bioactive molecules derived from marine rare actinobacteria.
Reinvigorating natural product discovery from marine rare actinobacteria
Though isolation and cultivation of marine rare actinobacteria is difficult, the development of novel and facile bacterial cultivation platforms such as hollow-fiber membrane chamber (HFMC) and iChip for in situ cultivation of previously unculturable microbial species have expanded the scope of natural product discovery (Aoi et al., 2009; Nichols et al., 2010). By utilizing rationally designed iChip platform, Ling et al. (2015) has successfully isolated previously uncultivable soil bacteria Eleftheria terrae and characterized its bioactive molecule (Ling et al., 2015).
It is assumed that strain divergence (phylogenetic or ecological) can have great impact on metabolism and biosynthetic pathway and result in novel chemistry and bioactivities, so research is focused on previously unexplored strains (Monciardini et al., 2014). However, it is unrealistic to assume that every unexplored strain can provide bioactive compounds (Donadio et al., 2010). Hence, systematic approaches need to be employed for utilizing the true potential of natural products from marine rare actinobacteria. Some of the key foundations can be categorized as:
-
Identification of target strains/molecules,
-
Systematic enrichment of production,
-
Explicit modification for functional/structural diversity.
-
Identification of target strains/molecules
The accessible diversity of useful microbial molecules have almost been exhausted by traditional approaches, hence it is speculated that unstudied marine rare actinobacteria can provide reservoir of new microbial molecules (Schorn et al., 2016). Recently, direct connection of genomic information to biomolecule can be attained in culture independent approach as introducing environment (eDNA) into a suitable expression host (metagenomic libraries) (Handelsman, 2004). But, compound rediscovery due to similar strain replications is a major limitation of this approach. To maximize the capacity to mine metagenomes for attaining biomolecules with novel activities, there is requisite for parallel developments in techniques for bioactivity screening, isolation and separation methods, and analytical chemistry (Trindade et al., 2015). Robust techniques for analytical characterization of compounds (Figure 1A) based on UV absorbance, high pressure liquid chromatography (HPLC), mass spectrometry, and nuclear magnetic resonance (NMR) analysis can be used to scrutinize the discovery of new compounds (Liu et al., 2012). The techniques utilizing coupling of biochemical analytical methods with genome information such as, in glycogenomics (Kersten et al., 2013), peptidogenomics (Medema et al., 2014), and metabolomics (Maansson et al., 2016) are recent advances facilitating easy access to diverse biomolecules. The results of such analytical analysis can be subsequently compared against databases repositories, such as MarinLit, ChemSpider, Pubchem, etc., to avoid already known compounds (Forner et al., 2013). Hence, robust analytical facilities and comparison with reference databases can assist on characterization of diverse chemical structures.
The prime focus in drug discovery is identification of new bioactive chemical or discovery of previously unreported biological activity with known chemical structure. High throughput screening (HTS) can provide easy means for evaluating desired bioactivities against an array natural products (Monciardini et al., 2014). The robust screening strategies ranging from the classic whole cell assays to more sophisticated antisense based assay have been reviewed elsewhere (Silver and Bostian, 1990; Singh et al., 2011; Farha and Brown, 2016). Recently, the integrative approach of metabolite profiling, bioactivity studies and taxonomic studies have been utilized for characterizing different marine actinobacteria and biological properties of metabolites produced by them (Betancur et al., 2017). Such integrative approaches can be fascinating tool for directly assessing bioactivities at preliminary stages of study.
The next focus in drug discovery is understanding the biogenesis of bioactive molecule in producer strains. The rapid development of genome sequencing methods has revolutionized such studies by unveiling information about the whole genome architecture (Figure 1B). The challenge now is mining the data and connect the predicted biosynthetic gene clusters (BGC) to bioactive molecules. A plethora of in silico tools are available for determining the nature of gene clusters (Weber and Kim, 2016). The classic genome mining approach (focusing on unique biosynthetic enzyme) has transitioned to the concept of comparative genome mining (complete BGC to next BGC comparison) and culture independent-metagenome mining (Ziemert et al., 2016). Due to its efficacy in studying BGCs, the genome mining concept has been expanded to different marine rare actinobacteria for getting insight on biosynthesis mechanisms of different secondary metabolites. The analysis of genome sequence of Micromonospora sp. RV43, Rubrobacter sp. RV113, and Nocardiopsis sp. RV163 isolated from Mediterranean sponges revealed presence of numerous gene clusters of different secondary metabolites (Horn et al., 2015). The 5.2 Mb genome of marine rare actinobacteria, Salinispora tropica CNB-440 (Udwary et al., 2007) was interpreted using bioinformatics revealing at least 19 novel secondary metabolite BCGs. Later, diverse compounds have been characterized from S. tropica, including anticancer agent salinosporamide A, lymphocyte kinase inhibitor lymphostin, DNA-cleaving agent calicheamicin, novel lysin-primed polyene macrolactam polyketide, and various siderophores (Kersten et al., 2013). Biosynthetic analysis of the draft genome of Saccharomonospora sp. CNQ490 has revealed 19 conspicuous BGC, indicating diverse secondary metabolic capacity (Yamanaka et al., 2014). Using precise bioinformatics tools, 75 genomes from closely related Salinospora species were compared and 124 distinct prominent BCGs were predicted which are far greater than known compound classes from these bacteria (Ziemert et al., 2014). Duncan et al. (2015) has simultaneously compared a large number of complex microbial extract in a large number of Salinispora species. This molecular networking was coupled with genome sequence data for comparative analysis of metabolite profile and BCG to develop pattern-based genome mining (PBGM) approach. Concurrently, a novel non-ribosomal peptide, retimycin A was isolated and characterized based on genome and metabolome analysis (Duncan et al., 2015). Therefore, genome mining approach has provided new avenues on discoursing novel natural products from marine rare actinobacteria.
-
Systematic enrichment of production
Generally, genome information is the starting point for pathway discovery. Various “omics” based tools have been employed for engineering pathways for secondary metabolite production in various actinobacteria (Chaudhary et al., 2013; Hwang et al., 2014). But the lack of full understanding of physiological transition stage for secondary metabolite production is a major consideration during manipulation of cellular processes using metabolic engineering (Licona-Cassani et al., 2015). Engineering primary metabolism for enhancing the pools of building blocks without compromising the growth is a major constraint in most metabolic engineering approaches (Olano et al., 2008). System biology protocols have been successfully used to study physiological parameters, leading to the discovery of the activation of NPs biosynthesis and manipulation of pathways (Licona-Cassani et al., 2015). Genome scale metabolic models are valuable for predicting organisms' phenotypes from genotypes basically by providing simulated mathematical prediction of cellular behavior under different genetic and physiological conditions (Henry et al., 2010; Ates et al., 2011). Community system biology approaches provide understanding about the complex relationship of individual members in a community and the modes of interactions they are engaged (Zengler and Palsson, 2012). The systematic application of systems biological approaches as metabolic network analysis coupled with pathway engineering or genetic engineering (Figure 2A) from a single strain to the larger community level can provide breakthrough in rational metabolic engineering approaches.
Synthetic biology is particularly focused on precise design and construction of new biological systems (metabolic pathways or genetic circuits) that are not prevalent in nature (Andrianantoandro et al., 2006). Previously, efforts in synthetic biology have been largely focused on creating and perfecting genetic devices. But the current focus is directed to customizable larger scale system engineering by assembling devices or modular organizations (Purnick and Weiss, 2009). Most often, biologically valuable natural products are produced in lower titer or are cryptic under normal laboratory conditions, whereas many rare actinobacteria are not amenable to genetic manipulation. Hence, in such cases transferring natural products biosynthesis into well-developed heterologous host is a logical approach for producing parent NPs or generating novel analogs through biosynthetic engineering (Wenzel and Müller, 2005). Direct cloning and refactoring of previously silent lipopeptide gene cluster of Saccharomonospora sp. CNQ490 have been achieved by heterologous expression in Streptomyces coelicolor to yield taromycin A by Transformation Assisted Recombination (TAR)-based genetic platform (Yamanaka et al., 2014). Besides, tuning of metabolic pathway by altering promoters (Siegl et al., 2013; Wang et al., 2013), terminators (Pulido and Jimenez, 1987), and RBS (Bai et al., 2015) and/or host manipulation by genome engineering (Siegl and Luzhetskyy, 2012; Tong et al., 2015) are providing new avenues for systemic level metabolic engineering of actinobacteria. Promoter exchange (Horbal et al., 2012) and the use of exogenous principal sigma factor (σHrdB) (Wang et al., 2014) have been utilized for increasing teicoplanin in an industrial strain of Actinoplanes teichomyceticus. Approach for constructing genetic circuit or holistic host engineering (Figure 2B) can be an effective approach for designing and synthesizing unnatural but effective molecules from marine rare actinobacteria.
-
Explicit modification for functional/structural diversity
Fundamentally, engineering or modulating the precursor pathways can lead to enhancement or diversification of natural products (Dhakal et al., 2016). Combinatorial biosynthesis exploits the shuffling of anabolic pathways by precursor directed biosynthesis, enzyme level modulations, and pathway level recombination, leading to novel natural products (Sun et al., 2015; Winn et al., 2016). The precursor-directed in-situ synthesis (PDSS) has been successfully employed for generating new congeners of saccharothriolides from Saccharothrix sp. A1506 (Lu et al., 2016). Such type of precursor modulations can be manifested chemically or biologically to generate structural diversity in compounds from marine rare actinobacteria. Mutasynthesis is another variant of modulation of anabolic pathway by generating mutant strain deficient in key aspects of biosynthetic pathway and substituting natural precursor with analog of precursor to produce new natural products (Kennedy, 2008). Mutasynthesis couples the power of chemical synthesis with molecular biology to create diverse derivatives of medicinally valuable natural products (Weissman, 2007). One such example is the production of fluorinated analog fluorosalinosporamide. It has better proteasome inhibition and cytotoxic activity than naturally produced salinosporamides isolated from various Salinispora species (Feling et al., 2003). The halogenase gene salL in Salinispora tropica has been inactivated and 5′-fluoro-5′-deoxyadenosine, a fluorinated analog of its natural precursor 5′-chloro-5′-deoxyadenosine, has been used to generate fluorosalinosporamide by chemistry mediated mutasynthesis (Eustáquio and Moore, 2008). In another approach, salL was replaced by fluorinase gene flA from Streptomyces catteleya. The mutant strain salL−flA+ produced fluorosalinosporamide in the presence of inorganic fluoride (Eustáquio et al., 2010). Moreover, combinatorial biosynthetic approach by feeding L-3-cyclohex-2′-enylalanine (CHA) residue in SalX disruption mutant of S. tropica enabled the generation of other unnatural salinosporamide derivatives such as salinosporamide X1 and salinosporamide X2, with lower activity (McGlinchey et al., 2008). But in another approach utilizing mutasynthetic approach with fine-tuned feeding of readily available amino acid precursors to SalX disruption mutant of S. tropica led to generation of many salinosporamide derivatives. Among them salinosporamide X7 exhibited equal to slightly improved cytotoxic potential than the natural counterpart (Nett et al., 2009). Hence, such approaches of precursor engineering, mutasynthesis, and combinatorial biosynthesis (Figure 3A, Table 1B) can be rationally utilized to diversify structure and perform structure-activity relationship studies of versatile molecules from various marine rare actinobacteria.
The advent of combinatorial synthetic chemistry has created huge excitement in the pharmaceutical industry by generating libraries of millions of compounds which could be screened by HTS (Butler, 2004). The total synthesis of complex natural products offers greater potential for direct access to bioactive molecule from marine sources. However, large scale production of complex natural product remains elusive due to low yields and high cost (Yeung and Paterson, 2005). Recent achievement as total synthesis of natural products in absence of protecting groups can lead to development of superior molecules with greater flexibility (Young and Baran, 2009). The generation of microbial chemicals by total enzymatic synthesis has been used as alternative to total chemical synthesis (Cheng et al., 2007). There have been ample of examples illustrating improvement in physical and biological properties of natural products (including many marine natural products) by chemical modifications, semisynthesis, mutasynthesis, and chemobiosynthesis (Hamann, 2003; Kennedy, 2008) mediated by biological and chemical techniques. Bioinspired total synthesis of salinosporamides and structurally related derivatives have provided access to novel functionalities of tremendously effective molecule (Nguyen et al., 2010; Chen et al., 2012). Suitable integration of synthetic chemistry (Figure 3B, Table 1B) with biological production system can be utilized for generating structurally and functionally diverse analogs/derivatives of target molecule. One of the successful example illustrating application of synthetic chemistry in marine natural products is rationalized for structural/functional diversification of salinosporamides (Baran et al., 2007; Potts and Lam, 2010). The synergy between genome sequencing, mass spectroscopy based analysis and bio-inspired synthesis have been utilized for studying biosynthetic mechanism and structural diversification of nocardioazine B from Nocardiopsis sp. CMB-M0232 (Alqahtani et al., 2015). Hence, it is no doubt that rational integration of biological processes and chemical techniques (Dhakal and Sohng, 2015, 2017) can provide new foundations for drug discoveries from marine rare actinobacteria.
Figure 1
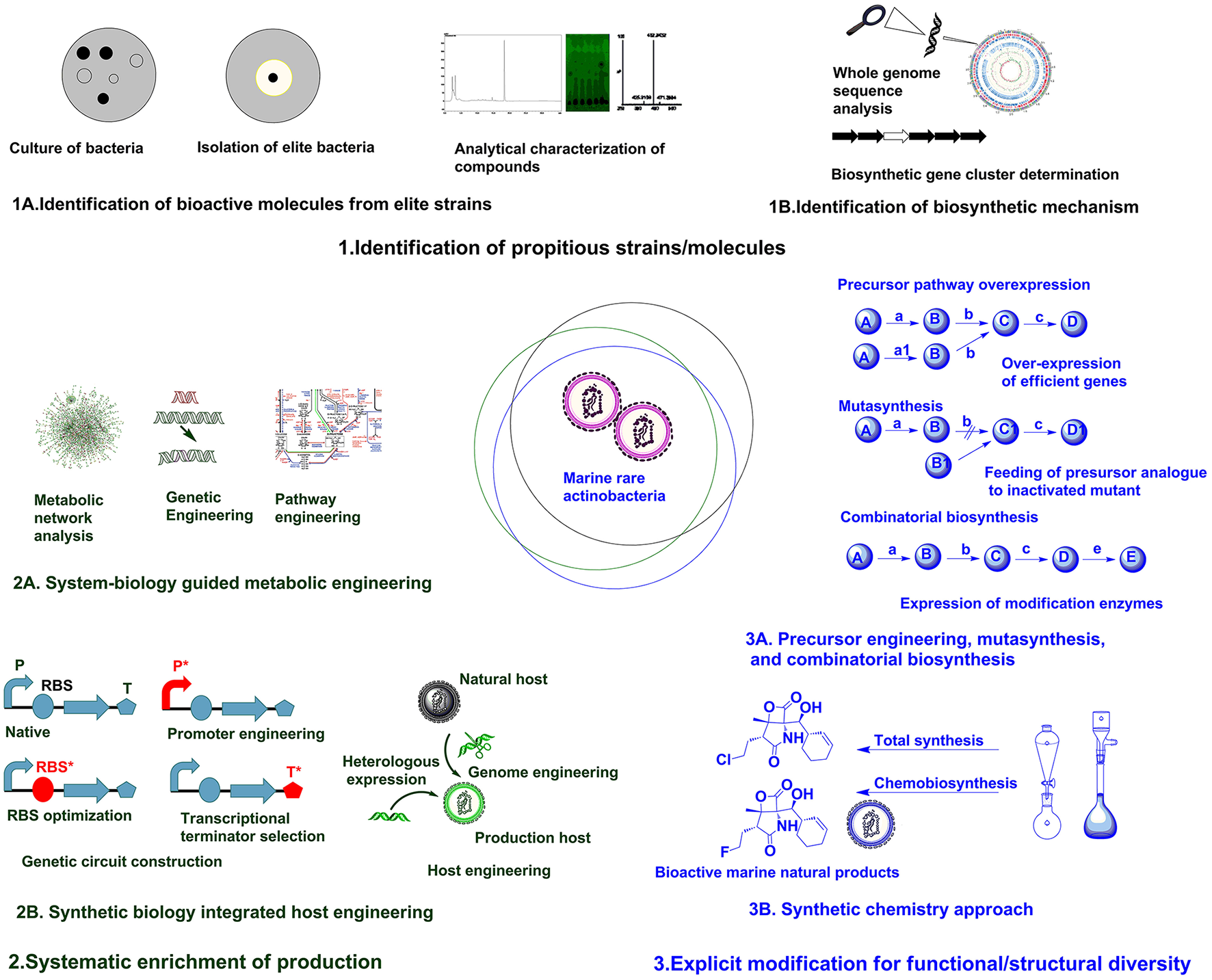
Different approaches for enhancing natural product discovery from marine rare actinobacteria.
Future outlook
As evident from examples above, the innovative methods for procurement of bioactive molecules from potent strains, efficient production and/or modifications by biological and chemical methods can assist in harnessing the full potential of biomolecules derived from marine rare actinobacteria. Further, tuning of structural and functional properties based on structure activity relationship studies can lead to development of superior analogs. But the prime focus should be on application of cutting edge translational research, such as transferring the achievements of discovery or synthesis of such biomolecule to the industrial bench-tops and clinics. The successful collaboration between biologists/chemists in academics and/or pharmaceutical companies can open new avenues for development of highly effective drugs. Salinosporamide A (Marizomib) has been a significant representation of compound derived from marine rare actinobacteria leading to phase trials. It is no doubt that exploration of new candidate strains with sophisticated techniques will certainly unravel tremendous opportunities to identify novel natural products and improve their applicability by structural/functional diversifications.
Statements
Author contributions
DD, ARP, BS, and JS made substantial, direct, and intellectual contribution to the work, and approved it for publication with full consent.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (NRF-2017R1A2A2A05000939).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Abdel-Mageed W. M. Milne B. F. Wagner M. Schumacher M. Sandor P. Pathom-aree W. et al . (2010). Dermacozines, a new phenazine family from deep-sea dermacocci isolated from a Mariana Trench sediment. Org. Biomol. Chem.8, 2352–2362. 10.1039/c001445a
2
Alqahtani N. Porwal S. K. James E. D. Bis D. M. Karty J. A. Lane A. L. et al . (2015). Synergism between genome sequencing, tandem mass spectrometry and bio-inspired synthesis reveals insights into nocardioazine B biogenesis. Org. Biomol. Chem.13, 7177–7192. 10.1039/C5OB00537J
3
Andrianantoandro E. Basu S. Karig D. K. Weiss R. (2006). Synthetic biology: new engineering rules for an emerging discipline. Mol. Syst. Biol.2:2006.0028. 10.1038/msb4100073
4
Aoi Y. Kinoshita T. Hata T. Ohta H. Obokata H. Tsuneda S. (2009). Hollow-fiber membrane chamber as a device for in situ environmental cultivation. Appl. Environ. Microbiol.75, 3826–3833. 10.1128/AEM.02542-08
5
Asolkar R. N. Kirkland T. N. Jensen P. R. Fenical W. (2010). Arenimycin, an antibiotic effective against rifampin-and methicillin-resistant Staphylococcus aureus from the marine actinomycete Salinispora arenicola. J. Antibiot.63, 37–39. 10.1038/ja.2009.114
6
Ates Ö. Oner E. T. Arga K. Y. (2011). Genome-scale reconstruction of metabolic network for a halophilic extremophile, Chromohalobacter salexigens DSM 3043. BMC Syst. Biol.5:12. 10.1186/1752-0509-5-12
7
Azman A. S. Othman I. Velu S. S. Chan K. G. Lee L. H. (2015). Mangrove rare actinobacteria: taxonomy, natural compound, and discovery of bioactivity. Front. Microbiol.6:856. 10.3389/fmicb.2015.00856
8
Bai C. Zhang Y. Zhao X. Hu Y. Xiang S. Miao J. et al . (2015). Exploiting a precise design of universal synthetic modular regulatory elements to unlock the microbial natural products in Streptomyces. Proc. Natl. Acad. Sci. U.S.A.112, 2181–12186. 10.1073/pnas.1511027112
9
Baltz R. H. (2006). Marcel Faber Roundtable: is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration?J. Ind. Microbiol. Biotechnol.33, 507–513. 10.1007/s10295-005-0077-9
10
Baran P. S. Maimone T. J. Richter J. M. (2007). Total synthesis of marine natural products without using protecting groups. Nature446, 404–408. 10.1038/nature05569
11
Berdy J. (2005). Bioactive microbial metabolites. J. Antibiot.58, 1–26. 10.1038/ja.2005.1
12
Bernard L. Schäfer H. Joux F. Courties C. Muyzer G. Lebaron P. (2000). Genetic diversity of total, active and culturable marine bacteria in coastal seawater. AME23, 1–11. 10.3354/ame023001
13
Betancur L. A. Naranjo-Gaybor S. J. Vinchira-Villarraga D. M. Moreno-Sarmiento N. C. Maldonado L. A. Suarez-Moreno Z. R. et al . (2017). Marine Actinobacteria as a source of compounds for phytopathogen control: an integrative metabolic-profiling/bioactivity and taxonomical approach. PLoS ONE12:e0170148. 10.1371/journal.pone.0170148
14
Bister B. Bischoff D. Ströbele M. Riedlinger J. Reicke A. Wolter F. et al . (2004). Abyssomicin C-A polycyclic antibiotic from a marine Verrucosispora Strain as an inhibitor of the p-aminobenzoic acid/tetrahydrofolate biosynthesis pathway. Angew. Chem. Int. Ed. Engl.43, 2574–2576. 10.1002/anie.200353160
15
Blunt J. W. Copp B. R. Hu W. P. Munro M. H. Northcote P. T. Prinsep M. R. (2007). Marine natural products. Nat. Prod. Rep.24, 31–86. 10.1039/b603047p
16
Bonet B. Teufel R. Cruüsemann M. Ziemert N. Moore B. S. (2014). Direct capture and heterologous expression of Salinispora natural product genes for the biosynthesis of enterocin. J. Nat. Prod.78, 539–542. 10.1021/np500664q
17
Bredholdt H. Galatenko O. A. Engelhardt K. Fjærvik E. Terekhova L. P. Zotchev S. B. (2007). Rare actinomycete bacteria from the shallow water sediments of the Trondheim fjord, Norway: isolation, diversity and biological activity. Environ. Microbiol.9, 2756–2764. 10.1111/j.1462-2920.2007.01387.x
18
Bull A. T. Stach J. E. (2007). Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol.15, 491–499. 10.1016/j.tim.2007.10.004
19
Butler M. S. (2004). The role of natural product chemistry in drug discovery. J. Nat. Prod.67, 2141–2153. 10.1021/np040106y
20
Carlson S. Marler L. Nam S. J. Santarsiero B. D. Pezzuto J. M. Murphy B. T. (2013). Potential chemopreventive activity of a new macrolide antibiotic from a marine-derived Micromonospora sp. Mar. Drugs.11, 1152–1161. 10.3390/md11041152
21
Cavalier-Smith T. (2002). The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int. J. Syst. Evol. Microbiol.52, 7–76. 10.1099/00207713-52-1-7
22
Chaudhary A. K. Dhakal D. Sohng J. K. (2013). An insight into the “-omics” based engineering of streptomycetes for secondary metabolite overproduction. Biomed. Res. Int.2013:968518. 10.1155/2013/968518
23
Chen Z. H. Wang B. L. Kale A. J. Moore B. S. Wang R. W. Qing F. L. (2012). Coupling of sterically hindered aldehyde with fluorinated synthons: stereoselective synthesis of fluorinated analogues of salinosporamide A. J. Fluor. Chem.136, 12–19. 10.1016/j.jfluchem.2012.01.003
24
Cheng Q. Xiang L. Izumikawa M. Meluzzi D. Moore B. S. (2007). Enzymatic total synthesis of enterocin polyketides. Nat. Chem. Biol.3, 557–558. 10.1038/nchembio.2007.22
25
Cho J. Y. Williams P. G. Kwon H. C. Jensen P. R. Fenical W. (2007). Lucentamycins, A. D., cytotoxic peptides from the marine-derived actinomycete Nocardiopsis lucentensis. J. Nat. Prod.70, 321–1328. 10.1021/np070101b
26
Deamer D. Akeson M. Branton D. (2016). Three decades of nanopore sequencing. Nat. Biotechnol.34, 518–524. 10.1038/nbt.3423
27
Dhakal D. Sohng J. K. (2015). Commentary: toward a new focus in antibiotic and drug discovery from the Streptomyces arsenal. Front. Microbiol.6:727. 10.3389/fmicb.2015.00727
28
Dhakal D. Sohng J. K. (2017). Coalition of biology and chemistry for ameliorating antimicrobial drug discovery. Front. Microbiol.8:734. 10.3389/fmicb.2017.00734
29
Dhakal D. Chaudhary A. K. Yi J. S. Pokhrel A. R. Shrestha B. Parajuli P. et al . (2016). Enhanced production of nargenicin A1 and creation of a novel derivative using a synthetic biology platform. Appl. Microbiol. Biotechnol.100, 9917–9931. 10.1007/s00253-016-7705-3
30
Donadio S. Maffioli S. Monciardini P. Sosio M. Jabes D. (2010). Antibiotic discovery in the twenty-first century: current trends and future perspectives. J. Antibiot.63, 423–430. 10.1038/ja.2010.62
31
Duncan K. R. Crüsemann M. Lechner A. Sarkar A. Li J. Ziemert N. et al . (2015). Molecular networking and pattern-based genome mining improves discovery of biosynthetic gene clusters and their products from Salinispora species. Chem. Biol.22, 460–471. 10.1016/j.chembiol.2015.03.010
32
Ellis G. A. Wyche T. P. Fry C. G. Braun D. R. Bugni T. S. (2014). Solwaric acids A and B, antibacterial aromatic acids from a marine Solwaraspora sp. Mar. Drugs12, 1013–1022. 10.3390/md12021013
33
Endo A. Danishefsky S. J. (2005). Total synthesis of salinosporamide A. J. Am. Chem. Soc.127, 8298–8299. 10.1021/ja0522783
34
Eustáquio A. S. Moore B. S. (2008). Mutasynthesis of fluorosalinosporamide, a potent and reversible inhibitor of the proteasome. Angew. Chem. Int. Ed. Engl.47, 3936–3938. 10.1002/anie.200800177
35
Eustáquio A. S. Nam S. J. Penn K. Lechner A. Wilson M. C. Fenical W. et al . (2011). The discovery of salinosporamide K from the marine bacterium “Salinispora pacifica” by genome mining gives insight into pathway evolution. Chembiochem12, 61–64. 10.1002/cbic.201000564
36
Eustáquio A. S. O'Hagan D. Moore B. S. (2010). Engineering fluorometabolite production: fluorinase expression in Salinispora tropica yields fluorosalinosporamide. J. Nat. Prod.73, 378–382. 10.1021/np900719u
37
Farha M. A. Brown E. D. (2016). Strategies for target identification of antimicrobial natural products. Nat. Prod. Rep.33, 668–680. 10.1039/C5NP00127G
38
Fei P. Chuan-xi W. Yang X. Hong-lei J. Lu-jie C. Uribe P. et al . (2013). A new 20-membered macrolide produced by a marine-derived Micromonospora strain. Nat. Prod. Res.27, 1366–1371. 10.1080/14786419.2012.740038
39
Feling R. H. Buchanan G. O. Mincer T. J. Kauffman C. A. Jensen P. R. Fenical W. (2003). Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. Engl.42, 355–357. 10.1002/anie.200390115
40
Fenical W. Jensen P. R. (2006). Developing a new resource for drug discovery: marine actinomycete bacteria. Nat. Chem. Biol.2, 666–673. 10.1038/nchembio841
41
Fenical W. Baden D. Burg M. de-Goyet C. V. Grimes J. D. Katz M. et al . (1999). Marine derived pharmaceuticals and related bioactive compounds, in From Monsoons to Microbes: Understanding the Ocean's Role in Human Health, ed FenicalW. (Washington, DC: National Academies Press), 71–86.
42
Forner D. Berrué F. Correa H. Duncan K. Kerr R. G. (2013). Chemical dereplication of marine actinomycetes by liquid chromatography–high resolution mass spectrometry profiling and statistical analysis. Anal. Chim. Acta805, 70–79. 10.1016/j.aca.2013.10.029
43
Freel K. C. Nam S. J. Fenical W. Jensen P. R. (2011). Evolution of secondary metabolite gene evolution in three closely related marine actinomycete species. Appl. Environ. Microbiol.20, 7261–7270. 10.1128/AEM.05943-11
44
Fu P. Kong F. Li X. Wang Y. Zhu W. (2014). Cyanogramide with a new spiro [indolinone-pyrroloimidazole] skeleton from Actinoalloteichus cyanogriseus. Org. Lett. 16, 3708–3711. 10.1021/ol501523d
45
Fu P. Wang S. Hong K. Li X. Liu P. Wang Y. et al . (2011). Cytotoxic bipyridines from the marine-derived actinomycete Actinoalloteichus cyanogriseus WH1-2216-6. J. Nat. Prod.74, 1751–1756. 10.1021/np200258h
46
Fukuda T. Takahashi M. Nagai K. Harunari E. Imada C. Tomoda H. (2016). Isomethoxyneihumicin, a new cytotoxic agent produced by marine Nocardiopsis alba KM6-1. J. Antibiot. 70, 590–594. 10.1038/ja.2016.152
47
Gan M. Liu B. Tan Y. Wang Q. Zhou H. He H. et al . (2015). Saccharothrixones A–D, tetracenomycin-type polyketides from the marine-derived actinomycete Saccharothrix sp. 10-10. J. Nat. Prod.78, 2260–2265. 10.1021/acs.jnatprod.5b00577
48
Gärtner A. Ohlendorf B. Schulz D. Zinecker H. Wiese J. Imhoff J. F. (2011). Levantilides, A., and B, 20-membered macrolides from a Micromonospora strain isolated from the mediterranean deep sea sediment. Mar. Drugs9, 98–108. 10.3390/md9010098
49
Goodfellow M. Stach J. E. Brown R. Bonda A. N. V. Jones A. L. Mexson J. et al . (2012). Verrucosispora maris sp. nov., a novel deep-sea actinomycete isolated from a marine sediment which produces abyssomicins. Antonie Van Leeuwenhoek101, 185–193. 10.1007/s10482-011-9651-5
50
Hamann M. T. (2003). Enhancing marine natural product structural diversity and bioactivity through semisynthesis and biocatalysis. Curr. Pharm. Des.9, 879–889. 10.2174/1381612033455297
51
Handelsman J. (2004). Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev.68, 669–685. 10.1128/MMBR.68.4.669-685.2004
52
Henry C. S. DeJongh M. Best A. A. Frybarger P. M. Linsay B. Stevens R. L. (2010). High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol.28, 977–982. 10.1038/nbt.1672
53
Hirayama H. Sunamura M. Takai K. Nunoura T. Noguchi T. Oida H. et al . (2007). Culture-dependent and -independent characterization of microbial communities associated with a shallow submarine hydrothermal system occurring within a coral reef off Taketomi Island, Japan. Appl. Environ. Microbiol.73, 7642–7656. 10.1128/AEM.01258-07
54
Hong K. Gao A. H. Xie Q. Y. Gao H. G. Zhuang L. Lin H. P. et al . (2009). Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs7, 24–44. 10.3390/md7010024
55
Horbal L. Zaburannyy N. Ostash B. Shulga S. Fedorenko V. (2012). Manipulating the regulatory genes for teicoplanin production in Actinoplanes teichomyceticus. World J. Microbiol. Biotechnol.28, 2095–2100. 10.1007/s11274-012-1013-6
56
Horn H. Hentschel U. Abdelmohsen U. R. (2015). Mining genomes of three marine sponge-associated actinobacterial isolates for secondary metabolism. Genome Announc.3, e01106–e01115. 10.1128/genomeA.01106-15
57
Huang H. Yao Y. He Z. Yang T. Ma J. Tian X. et al . (2011). Antimalarial β-carboline and indolactam alkaloids from Marinactinospora thermotolerans, a deep sea isolate. J. Nat. Prod.74, 2122–2127. 10.1021/np200399t
58
Hwang K. S. Kim H. U. Charusanti P. Palsson B. Ø. Lee S. Y. (2014). Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol. Adv.32, 255–268. 10.1016/j.biotechadv.2013.10.008
59
Kaeberlein T. Lewis K. Epstein S. S. (2002). Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science296, 1127–1129. 10.1126/science.1070633
60
Kaiya Y. Hasegawa J. I. Momose T. Sato T. Chida N. (2011). Total synthesis of (−)-Salinosporamide, A. Chem. Asian J.6, 209–219. 10.1002/asia.201000602
61
Kalinovskaya N. I. Kalinovsky A. I. Romanenko L. A. Dmitrenok P. S. Kuznetsova T. A. (2010). New angucyclines and antimicrobial diketopiperazines from the marine mollusk-derived actinomycete Saccharothrix espanaensis An 113. Nat. Prod. Commun. 5, 597–602.
62
Kennedy J. (2008). Mutasynthesis, chemobiosynthesis, and back to semi-synthesis: combining synthetic chemistry and biosynthetic engineering for diversifying natural products. Nat. Prod. Rep.25, 25–34. 10.1039/B707678A
63
Kersten R. D. Ziemert N. Gonzalez D. J. Duggan B. M. Nizet V. Dorrestein P. C. et al . (2013). Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules. Proc. Natl. Acad. Sci. U.S.A.110, E4407–E4416. 10.1073/pnas.1315492110
64
Kim M. C. Kwon O. W. Park J. S. Kim S. Y. Kwon H. C. (2013). Nocapyrones H–J, 3, 6-disubstituted α-pyrones from the marine actinomycete Nocardiopsis sp. KMF-001. Chem. Pharm. Bull. 61, 511–515. 10.1248/cpb.c12-00956
65
Kim T. K. Hewavitharana A. K. Shaw P. N. Fuerst J. A. (2006). Discovery of a new source of rifamycin antibiotics in marine sponge actinobacteria by phylogenetic prediction. Appl. Environ. Microbiol.72, 2118–2125. 10.1128/AEM.72.3.2118-2125.2006
66
Kisand V. Valente A. Lahm A. Tanet G. Lettieri T. (2012). Phylogenetic and functional metagenomic profiling for assessing microbial biodiversity in environmental monitoring. PLoS ONE7:e43630. 10.1371/journal.pone.0043630
67
Kwon H. C. Kauffman C. A. Jensen P. R. Fenical W. (2006). Marinomycins, A–D., antitumor-antibiotics of a new structure class from a marine actinomycete of the recently discovered genus “Marinispora”. J. Am. Chem. Soc.128, 1622–1632. 10.1021/ja0558948
68
Lam K. S. Tsueng G. McArthur K. A. Mitchell S. S. Potts B. C. Xu J. (2007). Effects of halogens on the production of salinosporamides by the obligate marine actinomycete Salinispora tropica. J. Antibiot.60, 13–19. 10.1038/ja.2007.2
69
Lazzarini A. Cavaletti L. Toppo G. Marinelli F. (2001). Rare genera of actinomycetes as potential producers of new antibiotics. Antonie Van Leeuwenhoek79, 399–405.
70
Lechner A. Eustáquio A. S. Gulder T. A. Hafner M. Moore B. S. (2011). Selective overproduction of the proteasome inhibitor salinosporamide A via precursor pathway regulation. Chem. Biol.18, 1527–1536. 10.1016/j.chembiol.2011.10.014
71
Leutou A. S. Yang I. Kang H. Seo E. K. Nam S. J. Fenical W. (2015). Nocarimidazoles, A., and B from a marine-derived actinomycete of the genus Nocardiopsis. J. Nat. Prod.78, 2846–2849. 10.1021/acs.jnatprod.5b00746
72
Li S. Tian X. Niu S. Zhang W. Chen Y. Zhang H. et al . (2011). Pseudonocardians A–C, new diazaanthraquinone derivatives from a deap-sea actinomycete Pseudonocardia sp. SCSIO 01299. Mar Drugs9, 1428–1439. 10.3390/md9081428
73
Licona-Cassani C. Cruz-Morales P. Manteca A. Barona-Gomez F. Nielsen L. K. Marcellin E. (2015). Systems biology approaches to understand natural products biosynthesis. Front. Bioeng. Biotechnol.3:19910.3389/fbioe.2015.00199
74
Ling L. L. Schneider T. Peoples A. J. Spoering A. L. Engels I. Conlon B. P. et al . (2015). A new antibiotic kills pathogens without detectable resistance. Nature517, 455–459. 10.1038/nature14098
75
Liu X. Bolla K. Ashforth E. J. Zhuo Y. Gao H. Huang P. et al . (2012). Systematics-guided bioprospecting for bioactive microbial natural products. Antonie Van Leeuwenhoek101, 55–66. 10.1007/s10482-011-9671-1
76
Liu Y. Hazzard C. Eustáquio A. S. Reynolds K. A. Moore B. S. (2009). Biosynthesis of salinosporamides from α, β-unsaturated fatty acids: implications for extending polyketide synthase diversity. J. Am. Chem. Soc.131, 10376–10377. 10.1021/ja9042824
77
Logan A. W. Sprague S. J. Foster R. W. Marx L. B. Garzya V. Hallside M. S. et al . (2014). Diastereoselective synthesis of fused lactone-pyrrolidinones; application to a formal synthesis of (−)-Salinosporamide, A. Org. Lett.16, 4078–4081. 10.1021/ol501662t
78
Lombó F. Velasco A. Castro A. De la Calle F. Braña A. F. Sánchez-Puelles J. M. et al . (2006). Deciphering the biosynthesis pathway of the antitumor thiocoraline from a marine actinomycete and its expression in two Streptomyces species. Chembiochem7, 366–376. 10.1002/cbic.200500325
79
Lu S. Nishimura S. Ito M. Kato T. Kakeya H. (2016). Precursor-directed in situ synthesis of Saccharothriolides, G., and H by the Actinomycete Saccharothrix sp. A1506. J. Antibiot.70, 718–720. 10.1038/ja.2016.153
80
Maansson M. Vynne N. G. Klitgaard A. Nybo J. L. Melchiorsen J. Nguyen D. D. et al . (2016). An integrated metabolomic and genomic mining workflow to uncover the biosynthetic potential of bacteria. Msystems1, e00028–e00015. 10.1128/mSystems.00028-15
81
Macherla V. R. Mitchell S. S. Manam R. R. Reed K. A. Chao T. H. Nicholson B. et al . (2005). Structure activity relationship studies of salinosporamide A (NPI-0052), a novel marine derived proteasome inhibitor. J. Med. Chem.48, 3684–3687. 10.1021/jm048995+
82
Maloney K. N. MacMillan J. B. Kauffman C. A. Jensen P. R. DiPasquale A. G. Rheingold A. L. et al . (2009). Lodopyridone, a structurally unprecedented alkaloid from a marine actinomycete. Org. Lett.11, 5422–5424. 10.1021/ol901997k
83
Manam R. R. McArthur K. A. Chao T. H. Weiss J. Ali J. A. Palombella V. J. et al . (2008). Leaving groups prolong the duration of 20S proteasome inhibition and enhance the potency of salinosporamides. J. Med. Chem.51, 6711–6724. 10.1021/jm800548b
84
McArthur K. A. Mitchell S. S. Tsueng G. Rheingold A. White D. J. Grodberg J. et al . (2008). Lynamicins A−E, chlorinated bisindole pyrrole antibiotics from a novel marine Actinomycete. J. Nat. Prod.71, 1732–1737. 10.1021/np800286d
85
McGlinchey R. P. Nett M. Eustáquio A. S. Asolkar R. N. Fenical W. Moore B. S. (2008). Engineered biosynthesis of antiprotealide and other unnatural salinosporamide proteasome inhibitors. J. Am. Chem. Soc.130:7822. 10.1021/ja8029398
86
Medema M. Paalvast Y. Nguyen D. Melnik A. Dorrestein P. Takano E. et al . (2014). Pep2Path: automated mass spectrometry-guided genome mining of peptidic natural products. PLoS Comput. Biol.10:e1003822. 10.1371/journal.pcbi.1003822
87
Monciardini P. Iorio M. Maffioli S. Sosio M. Donadio S. (2014). Discovering new bioactive molecules from microbial sources. Microbiol. Biotechnol.7, 209–220. 10.1111/1751-7915.12123
88
Mullowney M. W. Ó hAinmhire E. Tanouye U. Burdette J. E. Pham V. C. Murphy B. T. (2015). A pimarane diterpene and cytotoxic angucyclines from a marine-derived Micromonospora sp. in Vietnam's East Sea. Mar. Drugs.13, 5815–5827. 10.3390/md13095815
89
Murphy B. T. Narender T. Kauffman C. A. Woolery M. Jensen P. R. Fenical W. (2010). Saliniquinones A–F, new members of the highly cytotoxic anthraquinone-γ-pyrones from the marine actinomycete Salinispora arenicola. Aust. J. Chem.63, 929–934. 10.1071/CH10068
90
Nam S. J. Gaudêncio S. P. Kauffman C. A. Jensen P. R. Kondratyuk T. P. Marler L. E. et al . (2010). Fijiolides, A., and B, inhibitors of TNF-α-induced NFκB activation, from a marine-derived sediment bacterium of the genus Nocardiopsis. J. Nat. Prod.73, 1080–1086. 10.1021/np100087c
91
Nett M. Gulder T. A. Kale A. J. Hughes C. C. Moore B. S. (2009). Function-oriented biosynthesis of β-lactone proteasome inhibitors in Salinispora tropica. J. Med. Chem.52:6163. 10.1021/jm901098m
92
Nguyen H. Ma G. Gladysheva T. Fremgen T. Romo D. (2010). Bioinspired total synthesis and human proteasome inhibitory activity of (−)-salinosporamide A, (−)-homosalinosporamide A, and derivatives obtained via organonucleophile promoted bis-cyclizations. J. Org. Chem.76, 2–12. 10.1021/jo101638r
93
Nichols D. Cahoon N. Trakhtenberg E. M. Pham L. Mehta A. Belanger A. et al . (2010). Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol.76, 2445–2450. 10.1128/AEM.01754-09
94
Nübel U. Garcia-Pichel F. Kühl M. Muyzer G. (1999). Quantifying microbial diversity: morphotypes, 16S rRNA genes, and carotenoids of oxygenic phototrophs in microbial mats. Appl. Environ. Microbiol.65, 422–430.
95
Ogura A. Lin M. Shigenobu Y. Fujiwara A. Ikeo K. Nagai S. (2011). Effective gene collection from the metatranscriptome of marine microorganisms. BMC Genomics12:S15. 10.1186/1471-2164-12-S3-S15
96
Olano C. Lombo F. Mendez C. Salas J. A. (2008). Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab. Eng.10, 281–292. 10.1016/j.ymben.2008.07.001
97
Pathom-Aree W. Nogi Y. Sutcliffe I. C. Ward A. C. Horikoshi K. Bull A. T. et al . (2006). Dermacoccus abyssi sp. nov., a piezotolerant actinomycete isolated from the Mariana Trench. Int. J. Syst. Evol. Microbiol.56, 1233–1237. 10.1099/ijs.0.64133-0
98
Pinhassi J. Zweifel U. L. Hagstroëm A. (1997). Dominant marine bacterioplankton species found among colony-forming bacteria. Appl. Environ. Microbiol.63, 3359–3366.
99
Potts B. C. Lam K. S. (2010). Generating a generation of proteasome inhibitors: from microbial fermentation to total synthesis of salinosporamide a (marizomib) and other salinosporamides. Mar. Drugs8, 835–880. 10.3390/md8040835
100
Pulido D. Jimenez A. (1987). Optimization of gene expression in Streptomyces lividans by a transcription terminator. Nucleic Acids Res. 15, 4227–4240. 10.1093/nar/15.10.4227
101
Purnick P. E. Weiss R. (2009). The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell Biol.10, 410–422. 10.1038/nrm2698
102
Rastogi G. Sani R. (2011). Molecular techniques to assess microbial community structure, function, and dynamics in the environment, in Microbes and Microbial Technology, eds AhmadI.AhmadF.PichtelJ. (New York, NY: Springer), 29–57.
103
Reddy L. R. Saravanan P. Corey E. J. (2004). A simple stereocontrolled synthesis of salinosporamide A. J. Am. Chem. Soc.126, 6230–6231. 10.1021/ja048613p
104
Richter T. K. Hughes C. C. Moore B. S. (2015). Sioxanthin, a novel glycosylated carotenoid, reveals an unusual subclustered biosynthetic pathway. Environ. Microbiol.17, 2158–2171. 10.1111/1462-2920.12669
105
Riedlinger J. Reicke A. Zähner H. Krismer B. Bull A. T. Maldonado L. A. et al . (2004). Abyssomicins, inhibitors of the para-aminobenzoic acid pathway produced by the marine Verrucosispora strain AB-18-032. J. Antibiot. 57, 271–279. 10.7164/antibiotics.57.271
106
Sato S. Iwata F. Yamada S. Katayama M. (2012). Neomaclafungins A–I: oligomycin-class macrolides from a marine-derived actinomycete. J. Nat. Prod.75, 1974–1982. 10.1021/np300719g
107
Schneemann I. Ohlendorf B. Zinecker H. Nagel K. Wiese J. Imhoff J. F. (2010). Nocapyrones A–D, γ-pyrones from a Nocardiopsis strain isolated from the marine sponge Halichondria panicea. J. Nat. Prod.73, 1444–1447. 10.1021/np100312f
108
Schorn M. A. Alanjary M. M. Aguinaldo K. Korobeynikov A. Podell S. Patin N. et al . (2016). Sequencing rare marine actinomycete genomes reveals high density of unique natural product biosynthetic gene clusters. Microbiology162, 2075–2086. 10.1099/mic.0.000386
109
Schultz A. W. Oh D. C. Carney J. R. Williamson R. T. Udwary D. W. Jensen P. R. et al . (2008). Biosynthesis and structures of cyclomarins and cyclomarazines, prenylated cyclic peptides of marine actinobacterial origin. J. Am. Chem. Soc.130, 4507–4516. 10.1021/ja711188x
110
Schulze C. J. Donia M. S. Siqueira-Neto J. L. Ray D. Raskatov J. A. Green R. E. et al . (2015a). Genome-directed lead discovery: biosynthesis, structure elucidation, and biological evaluation of two families of polyene macrolactams against Trypanosoma brucei. ACS Chem. Biol.10, 2373–2381. 10.1021/acschembio.5b00308
111
Schulze C. J. Navarro G. Ebert D. DeRisi J. Linington R. G. (2015b). Salinipostins A–K, long-chain bicyclic phosphotriesters as a potent and selective antimalarial chemotype. J. Org. Chem.80, 1312–1320. 10.1021/jo5024409
112
Siegl T. Luzhetskyy A. (2012). Actinomycetes genome engineering approaches. Antonie Van Leeuwenhoek102, 503–516. 10.1007/s10482-012-9795-y
113
Siegl T. Tokovenko B. Myronovskyi M. Luzhetskyy A. (2013). Design, construction and characterisation of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metab. Eng.19, 98–106. 10.1016/j.ymben.2013.07.006
114
Silver L. Bostian K. (1990). Screening of natural products for antimicrobial agents. Eur. J. Clin. Microbiol. Infect. Dis.9, 455–461. 10.1007/BF01964283
115
Simmons L. Kaufmann K. Garcia R. Schwär G. Huch V. Müller R. (2011). Bendigoles D–F, bioactive sterols from the marine sponge-derived Actinomadura sp. SBMs009. Bioorg. Med. Chem.19, 6570–6575. 10.1016/j.bmc.2011.05.044
116
Singh S. B. Young K. Miesel L. (2011). Screening strategies for discovery of antibacterial natural products. Expert Rev. Anti Infect. Ther.9, 589–613. 10.1586/eri.11.81
117
Slattery M. Ankisetty S. Corrales J. Marsh-Hunkin K. E. Gochfeld D. J. Willett K. L. et al . (2012). Marine proteomics: a critical assessment of an emerging technology. J. Nat. Prod.75, 1833–1877. 10.1021/np300366a
118
Sousa T. D. S. Jimenez P. C. Ferreira E. G. Silveira E. R. Braz-Filho R. Pessoa O. D. et al . (2012). Anthracyclinones from Micromonospora sp. J. Nat. Prod.75, 489–493. 10.1021/np200795p
119
Stewart E. J. (2012). Growing unculturable bacteria. J. Bacteriol.194, 4151–4160. 10.1128/JB.00345-12
120
Subramani R. Aalbersberg W. (2013). Culturable rare Actinomycetes: diversity, isolation and marine natural product discovery. Appl. Microbiol. Biotechnol.97, 9291–9321. 10.1007/s00253-013-5229-7
121
Sun H. Liu Z. Zhao H. Ang E. L. (2015). Recent advances in combinatorial biosynthesis for drug discovery. Drug Des. Devel. Ther.9, 823–833. 10.2147/DDDT.S63023
122
Takagi M. Motohashi K. Khan S. T. Hashimoto J. Shin-ya K. (2010). JBIR-65, a new diterpene, isolated from a sponge-derived Actinomadura sp. SpB081030SC-15. J. Antibiot. 63, 401–403. 10.1038/ja.2010.61
123
Tang X. Li J. Millán-Aguinaaga N. Zhang J. J. O'Neill E. C. Ugalde J. A. et al . (2015). Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem. Biol.10, 2841–2849. 10.1021/acschembio.5b00658
124
Tiwari K. Gupta R. K. (2013). Diversity and isolation of rare actinomycetes: an overview. Crit. Rev. Biotechnol.39, 256–294. 10.3109/1040841x.2012.709819
125
Tong Y. Charusanti P. Zhang L. Weber T. Lee S. Y. (2015). CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth. Biol.4, 1020–1029. 10.1021/acssynbio.5b00038
126
Trindade M. van Zyl L. J. Navarro-Fernández J. Abd Elrazak A. (2015). Targeted metagenomics as a tool to tap into marine natural product diversity for the discovery and production of drug candidates. Front. Microbiol.6:890. 10.3389/fmicb.2015.00890
127
Udwary D. W. Zeigler L. Asolkar R. N. Singan V. Lapidus A. Fenical W. et al . (2007). Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. U.S.A.104, 10376–10381. 10.1073/pnas.0700962104
128
Vartoukian S. R. Palmer R. M. Wade W. G. (2010). Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol. Lett.2010, 1–7. 10.1111/j.1574-6968.2010.02000.x
129
Wang H. Yang L. Wu K. Li G. (2014). Rational selection and engineering of exogenous principal sigma factor (σ HrdB) to increase teicoplanin production in an industrial strain of Actinoplanes teichomyceticus. Microb. Cell Fact.13:10. 10.1186/1475-2859-13-10
130
Wang W. Li X. Wang J. Xiang S. Feng X. Yang K. (2013). An engineered strong promoter for streptomycetes. Appl. Environ. Microbiol.79, 4484–4492. 10.1128/AEM.00985-13
131
Weber T. Kim H. U. (2016). The secondary metabolite bioinformatics portal: Computational tools to facilitate synthetic biology of secondary metabolite production. Synth. Syst. Biotechnol.1, 69–79. 10.1016/j.synbio.2015.12.002
132
Webster N. S. Taylor M. W. Behnam F. Lücker S. Rattei T. Whalan S. et al . (2010). Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol.12, 2070–2082. 10.1111/j.1462-2920.2009.02065.x
133
Weissman K. J. (2007). Mutasynthesis–uniting chemistry and genetics for drug discovery. Trends Biotechnol.25, 139–142. 10.1016/j.tibtech.2007.02.004
134
Wenzel S. C. Müller R. (2005). Recent developments towards the heterologous expression of complex bacterial natural product biosynthetic pathways. Curr. Opin. Biotechnol.16, 594–606. 10.1016/j.copbio.2005.10.001
135
Williams P. G. Asolkar R. N. Kondratyuk T. Pezzuto J. M. Jensen P. R. Fenical W. (2007a). Saliniketals, A., and B, bicyclic polyketides from the marine actinomycete Salinispora arenicola. J. Nat. Prod.70, 83–88. 10.1021/np0604580
136
Williams P. G. Buchanan G. O. Feling R. H. Kauffman C. A. Jensen P. R. Fenical W. (2005). New cytotoxic Salinosporamides from the marine actinomycete Salinispora tropica. J. Org. Chem.70, 6196–6203. 10.1021/jo050511+
137
Williams P. G. Miller E. D. Asolkar R. N. Jensen P. R. Fenical W. (2007b). Arenicolides, A. C., 26-membered ring macrolides from the marine actinomycete Salinispora arenicola. J. Org. Chem.72, 5025–5034. 10.1021/jo061878x
138
Winn M. Fyans J. K. Zhuo Y. Micklefield J. (2016). Recent advances in engineering nonribosomal peptide assembly lines. Nat. Prod. Rep.33, 317–347. 10.1039/C5NP00099H
139
Wu J. Gao W. Johnson R. H. Zhang W. Meldrum D. R. (2013). Integrated metagenomic and metatranscriptomic analyses of microbial communities in the meso-and bathypelagic realm of North Pacific Ocean. Mar. Drugs11, 3777–3801. 10.3390/md11103777
140
Wu Z. C. Li S. Nam S. J. Liu Z. Zhang C. (2013). Nocardiamides, A., and B, two cyclohexapeptides from the marine-derived actinomycete Nocardiopsis sp. CNX037. J. Nat. Prod. 76, 694–701. 10.1021/np400009a
141
Wyche T. P. Hou Y. Braun D. Cohen H. C. Xiong M. P. Bugni T. S. (2011). First natural analogs of the cytotoxic thiodepsipeptide thiocoraline A from a marine Verrucosispora sp. J. Org. Chem.76, 6542–6547. 10.1021/jo200661n
142
Wyche T. P. Hou Y. Vazquez-Rivera E. Braun D. Bugni T. S. (2012). Peptidolipins B–F, antibacterial lipopeptides from an ascidian-derived Nocardia sp. J. Nat. Prod.75, 735–740. 10.1021/np300016r
143
Wyche T. P. Piotrowski J. S. Hou Y. Braun D. Deshpande R. McIlwain S. et al . (2014). Forazoline A: marine-derived polyketide with antifungal in vivo Efficacy. Angew. Chem. Int. Ed.126, 11767–11770. 10.1002/ange.201405990
144
Wyche T. P. Standiford M. Hou Y. Braun D. Johnson D. A. Johnson J. A. et al . (2013). Activation of the nuclear factor E2-related factor 2 pathway by novel natural products halomadurones A–D and a synthetic analogue. Mar. Drugs11, 5089–5099. 10.3390/md11125089
145
Yamanaka K. Reynolds K. A. Kersten R. D. Ryan K. S. Gonzalez D. J. Nizet V. et al . (2014). Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl. Acad. Sci. U.S.A.111, 1957–1962. 10.1073/pnas.1319584111
146
Yang C. Huang C. Zhang W. Zhu Y. Zhang C. (2015). Heterologous expression of fluostatin gene cluster leads to a bioactive heterodimer. Org. Lett.17, 5324–5327. 10.1021/acs.orglett.5b02683
147
Yeung K. S. Paterson I. (2005). Advances in the total synthesis of biologically important marine macrolides. Chem. Rev.105, 4237–4313. 10.1021/cr040614c
148
Young I. S. Baran P. S. (2009). Protecting-group-free synthesis as an opportunity for invention. Nat. Chem.1, 193–205. 10.1038/nchem.216
149
Zeng Y. Zou Y. Grebmeier J. He J. Zheng T. (2012). Culture-independent and -dependent methods to investigate the diversity of planktonic bacteria in the northern Bering Sea. Polar Biol.35, 117–129. 10.1007/s00300-011-1044-8
150
Zengler K. Palsson B. O. (2012). A road map for the development of community systems (CoSy) biology. Nat. Rev. Microbiol.10, 366–372. 10.1038/nrmicro2763
151
Zengler K. Toledo G. Rappé M. Elkins J. Mathur E. J. Short J. M. et al . (2002). Cultivating the uncultured. Proc. Natl. Acad. Sci. U.S.A.99, 15681–15686. 10.1073/pnas.252630999
152
Zhang W. Liu Z. Li S. Lu Y. Chen Y. Zhang H. et al . (2012). Fluostatins I–K from the South China Sea-derived Micromonospora rosaria SCSIO N160. J. Nat. Prod.75, 1937–1943. 10.1021/np300505y
153
Zhou X. Huang H. Chen Y. Tan J. Song Y. Zou J. et al . (2012). Marthiapeptide, A., an anti-infective and cytotoxic polythiazole cyclopeptide from a 60 L scale fermentation of the deep sea-derived Marinactinospora thermotolerans SCSIO 00652. J. Nat. Prod.75, 2251–2255. 10.1021/np300554f
154
Ziemert N. Alanjary M. Weber T. (2016). The evolution of genome mining in microbes–a review. Nat. Prod. Rep.33, 988–1005. 10.1039/c6np00025h
155
Ziemert N. Lechner A. Wietz M. Millán-Aguiñaga N. Chavarria K. L. Jensen P. R. (2014). Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc. Natl. Acad. Sci. U.S.A.111, E1130–E1139. 10.1073/pnas.1324161111
156
Zotchev S. B. (2012). Marine actinomycetes as an emerging resource for the drug development pipelines. J. Biotechnol.158, 68–175. 10.1016/j.jbiotec.2011.06.002
Summary
Keywords
marine rare actinobacteria, bacterial characterization, bioactive compounds, metagenomics, host engineering
Citation
Dhakal D, Pokhrel AR, Shrestha B and Sohng JK (2017) Marine Rare Actinobacteria: Isolation, Characterization, and Strategies for Harnessing Bioactive Compounds. Front. Microbiol. 8:1106. doi: 10.3389/fmicb.2017.01106
Received
30 December 2016
Accepted
31 May 2017
Published
15 June 2017
Volume
8 - 2017
Edited by
Jem Stach, Newcastle University, United Kingdom
Reviewed by
Polpass Arul Jose, Central Salt and Marine Chemicals Research Institute (CSIR), India; Vijay Kumar, Doon (P.G) College of Agriculture Science and Technology, India; D. Ipek Kurtboke, University of the Sunshine Coast, Australia
Updates
Copyright
© 2017 Dhakal, Pokhrel, Shrestha and Sohng.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jae Kyung Sohng sohng@sunmoon.ac.kr
This article was submitted to Antimicrobials, Resistance and Chemotherapy, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.