- 1Evolutionary Genomics Group, Departamento de Producción Vegetal y Microbiología, Universidad Miguel Hernández, San Juan de Alicante, Spain
- 2Institute of Hydrobiology, Department of Aquatic Microbial Ecology, Biology Center of the Academy of Sciences of the Czech Republic, České Budějovice, Czechia
- 3Cavanilles Institute of Biodiversity and Evolutionary Biology, University of Valencia, Valencia, Spain
Freshwater picocyanobacteria including Synechococcus remain poorly studied at the genomic level, compared to their marine representatives. Here, using a metagenomic assembly approach we discovered two novel Synechococcus sp. genomes from two freshwater reservoirs Tous and Lake Lanier, both sharing 96% average nucleotide identity and displaying high abundance levels in these two lakes located at similar altitudes and temperate latitudes. These new genomes have the smallest estimated size (2.2 Mb) and average intergenic spacer length (20 bp) of any previously sequenced freshwater Synechococcus, which may contribute to their success in oligotrophic freshwater systems. Fluorescent in situ hybridization confirmed that Synechococcus sp. Tous comprises small cells (0.987 ± 0.139 μm length, 0.723 ± 0.119 μm width) that amount to 90% of the picocyanobacteria in Tous. They appear together in a phylogenomic tree with Synechococcus sp. RCC307 strain, the main representative of sub-cluster 5.3 that has itself one of the smallest marine Synechococcus genomes. We detected a type II phycobilisome (PBS) gene cluster in both genomes, which suggests that they belong to a phycoerythrin-rich pink low-light ecotype. The decrease of acidic proteins and the higher content of basic transporters and membrane proteins in the novel Synechococcus genomes, compared to marine representatives, support their freshwater specialization. A sulfate Cys transporter which is absent in marine but has been identified in many freshwater cyanobacteria was also detected in Synechococcus sp. Tous. The RuBisCo subunits from this microbe are phylogenetically close to the freshwater amoeba Paulinella chromatophora symbiont, hinting to a freshwater origin of the carboxysome operon of this protist. The novel genomes enlarge the known diversity of freshwater Synechococcus and improve the overall knowledge of the relationships among members of this genus at large.
Introduction
Cyanobacteria are among the most diverse and widely distributed group of bacteria. They are the only prokaryotes capable of performing oxygenic photosynthesis, and greatly contribute to global primary production, fixing a substantial amount of carbon both in marine and freshwater environments. Among them, unicellular picocyanobacteria (Pcy) are globally distributed in almost all lakes and oceans. Typically, Pcy clades (Synechococcus, Prochlorococcus, and Cyanobium) are designated as non-bloomers (Steffen et al., 2012) and are either single or colonial rod-shaped cells ranging from 0.2 to 3 μm. However, they can form dense populations of up to several million cells per ml at the deep chlorophyll maximum (DCM) of mesotrophic stratified lakes (Camacho et al., 2003a,b) and, though less abundant, they also peak at the DCM of oceans and some seas (Partensky et al., 1999). The genera Synechococcus and Cyanobium are the dominant picocyanobacteria in freshwater systems (Callieri, 2008). However, the number of strains of marine origin with their genomes sequenced is much larger than their freshwater counterparts, providing a rather incomplete picture of the diversity of the genus. Furthermore, freshwater Synechococcus strains play a critical role in the ecological health of water bodies that are important human resources.
Similar to other cyanobacteria, Synechococcus ecotypes display differences in their accessory pigments and phycobilisomes (PBS) that make them adapted to different wavelengths of light (Camacho et al., 2000). Synechococcus PBS, responsible for light absorption and energy transfer to chlorophyll a (photosystem II) for the photosynthesis process, are also diagnostic and important for the type of light range spectrum in which they live, providing an advantage for some Synechococcus ecotypes in deep ecosystems with low light (Scanlan et al., 2009). There are three major types within this genus depending on the phycobiliprotein genes that they contain: Type I strains contain only C-phycocyanin resulting in green pigmentation; type II strains contain C-phycocyanin and phycoerythrin I and produce pink pigmentation; type III strains contain phycocyanin, phycoerythrin I and phycoerythrin II, presenting a wide range of pigmentation and some of them display chromatic adaptation (Six et al., 2007; Dufresne et al., 2008). Moreover, the morphometry and trophic state of lakes and ponds strongly influences composition, diversity and abundance of Pcy communities (Callieri, 2008; Callieri et al., 2012). Typically, deep, clear and oligotrophic/mesotrophic lakes contain mainly phycoerythrin (PE) rich cells while in shallow, turbid (humic) eutrophic lakes phycocyanin (PC) rich cells predominate (Callieri and Stockner, 2002). The success of Synechococcus in oligotrophic systems is explained by its capacity for adaptation to low-light conditions (Callieri et al., 2012). Their affinity to orthophosphate and other organic phosphorous sources apart from inorganic phosphates and their capacity for nitrogen storage in phycobilins (Camacho, 2006) enhance Synechococcus competition against algae and other bacteria (Vadstein, 2000).
Analysis of 16S rRNA genes and the internal transcribed spacer (ITS) of Synechococcus clearly suggests a polyphyletic nature (Robertson et al., 2001). These studies have revealed the existence of three marine sub-clusters: 5.1, 5.2, and 5.3 (Fuller et al., 2003; Scanlan et al., 2009; Mazard et al., 2012), and 13 clusters of non-marine Pcy (Callieri et al., 2013). Cluster 5.1 encompasses most marine clades (Rocap et al., 2003; Dufresne et al., 2008) but the less studied clusters 5.2 and 5.3 appear to be very important to understand the evolution of Synechococcus and Cyanobium and have unexpected relations to non-marine strains (Callieri et al., 2013). Recently, a new group, ‘halotolerants’ from a Mexican athalassohaline crater-lake has been found to be very close to the marine subcluster 5.3 (Synechococcus sp. RCC307), demonstrating that euryhaline and marine strains affiliate closely (Callieri et al., 2013). Phylogeny and ancestral state reconstruction approaches have shown that the earliest Pcy lineages were freshwater inhabitants, whose communities possess greater diversity than marine Pcy (Sanchez-Baracaldo et al., 2005, 2008; Blank and Sanchez-Baracaldo, 2010).
Although a large number (33) of marine Synechococcus strains have been sequenced (Scanlan et al., 2009) only a few (6) freshwater genomes are available. Among them, Synechococcus elongatus (Frenkel et al., 1950) plays now an important role as a model organism used in genetic engineering. Additional genomes, e.g., Synechococcus sp. GFB01 from a Brazilian Amazon lagoon (Guimarães et al., 2015), strains JA-3-3Ab and JA-2-3B’a(2-13) from Yellowstone (Bhaya et al., 2007) and PCC6312, PCC7502 freshwater representatives (Shih et al., 2013) were sequenced in the last few years. Metagenomic studies have suggested that Pcy (including Synechococcus) are far outcompeted by filamentous cyanobacteria (Ghai et al., 2012) in hypertrophic systems (e.g., Albufera de Valencia, Spain) but can be found in significant numbers in mesotrophic reservoirs (Oh et al., 2011; Ghai et al., 2012). Remarkably, it has been suggested that only a single abundant species of Synechococcus occurs in Lake Lanier (located in Georgia, United States), as all overlapping metagenomic reads belonging to Synechococcus were restricted to nearly 98–99% average nucleotide identity (ANI) (Oh et al., 2011).
In this manuscript we have used genome reconstruction from metagenomics (Ghai et al., 2014) to identify the most abundant Synechococcus genome obtained from a metagenomic sample of the freshwater reservoir of Tous (Valencia, Spain), an oligotrophic to slightly mesotrophic environment with high abundance of Synechococcus. We also assembled a highly similar Synechococcus relative from a Lake Lanier metagenome (Oh et al., 2011) through the same approach. This metagenomics genome reconstruction approach has provided a global view of the genomic properties and ecophysiological features of these two microbes. They represent an important and hitherto unknown species of freshwater Synechococcus with a widespread distribution.
Materials and Methods
Sampling Site Description and Physicochemical Profiles
The Tous New Dam is located in the lower course of the river Júcar, in the province of Valencia (Eastern Spain). A total catchment area of 17,821 km2 leads to the Tous dam, which is the last and one of the largest of a system of eight regulating reservoirs along the Júcar (Xúquer) river. It is an embankment-type dam with a clay core that has been rebuilt on top of an older dam that suffered severe failure in 1982 during a major flood. It has a standard maximum water surface elevation of 32.5 m corresponding to 340 × 106 m3 total water capacity, occupying an area of 9.80 km2, although its flood-regulating capacity reaches 700 × 106 m3.
Detailed vertical profiles of physical and chemical parameters were obtained with a CTD probe (SeaBird 19) equipped with specific sensors for pressure, conductivity and temperature. The probe was also fitted with sensors for oxygen (SB-43), chlorophyll-a (fluorimetric, Wetstar), phycoerythrin (fluorimetric, Seapoint), phycocyanin (fluorimetric, Cyclops-7 Turner), turbidity (Seapoint 880 nm) and c-DOM (WETStar Ex370/Em460). Sampling was performed on February 20th, 2015 when the reservoir was thermally mixed. Sampling depths were chosen on the basis of the in situ acquired physical and chemical profiles (Supplementary Figure S1). The samples were obtained from a boat, moored ∼500 m from the dam near the deepest point of the reservoir, by using a double cone fine layer sampler (Camacho, 2006). Water samples for chemical analyses were obtained from the same sample used for metagenomics analysis and a surface sample was also analyzed. These samples were filtered (e.g., for dissolved nutrient analyses) through Whatman GF/F filters, then poured into glass or PVC bottles, both previously washed with acid, and subsequently preserved at 4°C until analysis.
Major chemical variables including inorganic soluble forms of nitrogen (nitrate and ammonium) and phosphorus (soluble reactive phosphorus, SRP), as well as total nitrogen (TN) and total phosphorus (TP), were determined according to Standard Methods (American Public Health Association et al., 1915). Total organic carbon (TOC) was determined on a Shimadzu TOC-VCSN Analyser. Photosynthetic pigments were obtained by HPLC after extraction in acetone (Picazo et al., 2013).
Sampling, Sequencing, and Annotation
Two samples were taken for metagenomic analyses from the Tous reservoir on February 20th, 2015 at 12 and 25 m depths. Approximately 40 L of water from each sample was sequentially filtered through series of 20, 5, and a 0.22 μm-pore-size polycarbonate filters (Millipore). DNA extraction was only performed on the 0.22 μm filter for both depths as previously described (Ghai et al., 2010). Briefly, filters were treated with final concentrations of 1 mg/mL lysozyme and 0.2 mg/mL proteinase K and DNA was extracted with the Power isolation soil kit (Mo Bio). Summary statistics of sequencing was performed using Illumina HiSeq 4000 (Macrogen, South Korea), with an insert size of 350 bp, obtaining an average read length of 79.4 bp. A total of 284 and 281 million sequence reads (PE 2x100 bp) representing 28 and 27 Gb of sequence data were produced for 12 and 25 m 0.22 μm fraction, respectively. Each data set was assembled independently using the IDBA-UD assembler (Peng et al., 2012) with the following parameters: mink 70, maxk 100, step 10, pre-correction. Gene predictions on the assembled contigs were done using Prodigal in metagenomic mode (Hyatt et al., 2010), tRNAs were predicted using tRNAscan-SE (Lowe and Eddy, 1997) and ribosomal rRNA genes were identified using ssu-align (Nawrocki, 2009; Nawrocki and Eddy, 2010) and meta-rna (Huang et al., 2009). Comparisons of predicted protein sequences against NCBI NR, COG (Tatusov et al., 2001) and TIGFRAM (Haft et al., 2001) databases were performed for taxonomic binning and functional annotation. The same assembly and annotation procedure was used for contigs from the Lake Lanier (Oh et al., 2011). The samples used to reconstruct the Lanier genome were Lanier S2 (August 28, 2009) and Lanier S3 (September 7, 2009) both generated by Illumina GAII instrument. Summary statistics of the Tous and Lanier samples used for Synechococcus genome reconstruction is provided in Supplementary Table S1. Previously described assembled fosmid contigs (>10 Kb) from Amadorio were also annotated similarly and 151 cyanobacterial fosmids were selected (Ghai et al., 2014).
16S rRNA Reads Classification
A non-redundant version of the RDP database was prepared by clustering all available 16S rRNA coding sequences (approximately 2.3 million) into approximately 800,000 sequences at 90% identity level using UCLUST (Edgar, 2010). This database was used to identify candidate 16S rRNA fragments among the Illumina reads (unassembled). If a sequence matched this database at an e-value < 1e-5 it was considered a potential 16S rRNA fragment. These candidate fragments were aligned to archaeal, bacterial and eukaryal 16S/18S rRNA HMM models using ssu-align to identify true 16S/18S sequences (Nawrocki, 2009). The 16S rRNA fragments retrieved were compared to the entire RDP database and classified into a high level taxon if the sequence identity was ≥80% (BLASTN) and the alignment length was ≥90 bp. Fragments failing these thresholds were discarded.
Identification of Cyanobacterial Contigs and Genome Reconstruction
Contigs longer than 5 Kb were used for the Synechococcus sp. Tous and Synechococcus sp. Lanier genome reconstructions. Contigs were considered cyanobacterial if >60% of genes gave top BLAST hit to Cyanobacteria. More specifically, contigs with >60% of genes with top BLAST hit to Synechococcus sp. RCC307 were grouped using taxonomy, principal component analysis of tetranucleotide frequencies, GC content and coverage values in Tous and Lanier metagenomes (Oh et al., 2011). Tetranucleotide frequencies were computed using wordfreq program in the EMBOSS package (Rice et al., 2000). Principal component analysis was performed using the FACTOMINER package in R (Lê et al., 2008).
To improve the completeness and remove redundant contigs of the novel freshwater Synechococcus sp. Tous assembled genome, a second assembly step was performed. Tous metagenome reads were mapped using BWA (Li and Durbin, 2009) to Synechococcus contigs (>5 Kb contigs from 12 to 25 m metagenomes of Tous) and an additional 151 cyanobacterial contigs (>10 Kb) obtained from a fosmid library made from the metagenome of the Amadorio reservoir (Ghai et al., 2014). The mapped reads were reassembled along with the contigs using SPAdes (Bankevich et al., 2012). Finally, a composite Synechococcus genome comprising multiple highly related clones was extracted by considering contigs that grouped together based on GC%, PCA, and abundances in the Tous datasets. Similarly, for Synechococcus sp. Lanier we used Synechococcus contigs from two Lake Lanier metagenomes SRA029314.1 (S2), SRA029315.1 (S3) (Oh et al., 2011) for the reassembly and binning step to obtain the final genome. To estimate the genome size and completeness of the assembled genomes two different universal gene sets were used, one with 35 genes (Raes et al., 2007) and another with 111 genes (Albertsen et al., 2013).
Phylogenomic Tree
A total of 35 representative genomes from marine, euryhaline, and freshwater Synechococcus for each clade and sub-cluster (Scanlan et al., 2009; Shih et al., 2013) were used together with the two novel Synechococcus in a reference protein-concatenate-based phylogenomic tree. Three complete Prochlorococcus genomes were also used as outgroup. Proteins were concatenated and subsequently aligned among the different genomes. A total of 122 conserved genes were found in all genomes (based on the TIGR database) and were used to create the reference phylogeny of Synechococcus (Supplementary Table S2). The alignment was performed using Kalign (Lassmann and Sonnhammer, 2005) and trimmed using trimal (Capella-Gutiérrez et al., 2009) using default parameters. A maximum-likelihood tree was constructed using FastTree2 (Price et al., 2010) using a JTT+CAT model, a gamma approximation and 100 bootstrap replicates.
Single Gene Trees
16S rRNA Synechococcus sequences were aligned using MUSCLE (Edgar, 2004) and maximum likelihood 16S rRNA trees were constructed with MEGA7 using a gamma distribution and 100 bootstraps and considering a Tamura-Nei model (Tamura and Nei, 1993) with partial deletion, 95% of site coverage cut-off and Nearest-Neighbor-Interchange ML Heuristic Method (Kumar et al., 2016). RuBisCo small and large subunit trees were constructed using MEGA7 using a JTT model, a gamma approximation and 100 rapid bootstraps.
Metagenomic Fragment Recruitment
Recruitments were performed against several publicly available freshwater and brackish metagenomes using BLASTN (Altschul et al., 1997). A hit was considered only when it was at least 50 nucleotides long, with >95% identity and an e-value of <= 1e-5. These hits were used to compute the RPKG (reads recruited per Kb of genome per Gb of metagenome) values that provide a normalized number comparable across metagenomes from different locations. All metagenomic data sets used in this work are publicly available: Tous reservoir (SRR4198666 and SRR4198832), Amadorio reservoir (Ghai et al., 2014), Lake Lanier (Oh et al., 2011) (SRR947737, SRR948334, SRR948334) and Dexter reservoir (SRR3184716). Two southwestern Mediterranean samples from TARA oceans consortium database (Sunagawa et al., 2015) were also included in the analysis (ERR315856 and ERR315857).
Flow Cytometry Counts and Bacterioplankton Size Spectrum Determination
Water samples were fixed in situ with a paraformaldehyde:glutaraldehyde solution to a final concentration in the sample of 1%: 0.05% (w/v) (Marie et al., 1997). For flow cytometry identification and counting of the photosynthetic microbes and bacterioplankton, a Coulter Cytomics FC500 flow cytometer equipped with an argon laser (488 excitation), a red emitting diode (635 excitation), and five filters for fluorescent emission (FL1-FL5) was used. Cytometric parameter settings were as follows: FSC (550), SSC (390), FL1 (600), FL2 (670), FL3 (670), FL4 (620), and FL5 (700). Side scattering (SSC) was used for measuring size. Pcy enumeration was performed with discrimination by FL4, whereas for bacterioplankton identification, after 1h Sybr Green-I staining, discrimination was made by fluorescence in FL1 (Veldhuis and Kraay, 2000). Analyses were run for 120 s at the highest single flow rate of 128 μL min-1 of our machine. Abundance of each population was calculated according to the formula: N = (n × 1,000)/q × t, where q is the flow rate (microliter per minute), t is the length (minutes) of the data acquisition, n is the number of events counted by the flow cytometer, and N is the number of cells per milliliter. Flow rate was obtained gravimetrically considering the processed volume. Data were collected with the Beckman Coulter acquisition software for acquisition “CXP Version 2.2 Acquisition” and the analysis of the data was performed using Beckman Coulter analysis software for analysis “CXP Version 2.2 Analysis.”
Microscopy and FISH Counts
For microscopic counts of picoplankton, water samples were fixed in situ with a paraformaldehyde:glutaraldehyde solution to a final concentration in the sample of 1%: 0.05% (w/v) (Marie et al., 1997). Once in the laboratory, subsamples of 5–10 mL were filtered through 0.2 μm pore size black filters (Nuclepore, Whatman). For quantification, a section of the filter was stained with 4′,6-diamidino-2-phenylindole (DAPI) (Porter and Feig, 1980) (SIGMA) and counted with an inverted Zeiss III RS epifluorescence microscope (1250×, resolution 0.02857 lm/pixel) using a G365 exciting filter, LP420 suppression filter for blue light and G546 exciting filter, LP590 suppression filter for green light (MacIsaac and Stockner, 1993). Autotrophic cells were identified by chlorophyll autofluorescence under green light.
For FISH detection of the specific Synechococcus sp. group isolated from Tous reservoir, water samples were fixed in situ with paraformaldehyde to 2% final concentration, then filtered within the next 2 h onto white polycarbonate filters (0.2 μm pore size, Whatman). Filter sections were stained with the different oligonucleotide probes and with DAPI, and then mounted for microscopic evaluation. We primarily used a previously described general probe “Syn405”1 (West et al., 2001; Arnosti et al., 2012) (5′-AGAGGCCTTCATCCCTCA-3′) for total Synechococcus counts. For specific Synechococcus sp. identification we designed a specific probe using the PRIMER3 tool (Rozen and Skaletsky, 1999). This new probe (“SynTo,” ‘5′-TGGCCCAGCAGAGCGCTTTC-3′’) was double labeled with the indocarbocyanine dye Cy3 and Cy5 (Thermo Scientific, Waltham, MA, United States). These probes were checked for specificity using the Probe Match RDP (Cole et al., 2009) RDP Release 11, Update 5: September 30, 2016 (3,356,809 16S rRNA database2). No specific experimental test with Synechococcus pure cultures were performed with the probe “SynTo” but RPD probe match show extremely high specificity for genus GPIIa (family II) of cyanobacteria and specifically hit to Synechococcus RCC307 (CT978603). The FISH protocol was performed as previously described (Behnam et al., 2012). Hybridization conditions for the probe were adjusted by formamide (VWR BDH Prolabo) 20-40 % series applied to different subsamples. Each filter was cut in sections to be used with different probes. For cell-wall permeabilization, each filter section was incubated in a lysozyme solution for 30 min at 37°C. The sections were washed with MQ water and 80% ethanol for 3 min. Subsequently, the filter sections were placed on glass slides and covered with 20 ml of hybridization solution, which contains 5 M NaCl, 1M Tris-HCl, 10% sodium dodecyl sulfate, and 35% formamide; next 2 μl of “SynTo” probe were added and the filter section was incubated at 46°C for 90 min in an equilibrated chamber (Eppendorf thermomixer for slides). After hybridization, the sample was transferred into a washing solution containing 1 M Tris-HCl, 0,5 M EDTA, 10% sodium dodecyl sulfate, 5 M NaCl and 10 mg/ml DAPI, then incubated twice at 46°C for 15 min. After incubation, the sections were rinsed in 80% ethanol for a few seconds and placed in MQ water for 5 min. After complete drying each section was mounted in oil for microscopic determinations. Absolute densities of hybridized Synechococcus were calculated as the product of their relative abundances on filter sections (percentage of DAPI-stained objects) and the DAPI-stained direct cell counts. A minimum of 500 DAPI and probe-stained cells were measured per sample. Images from FISH determinations were analyzed using the NIH IMAGEJ software to determine cell dimensions3.
Results and Discussion
Environmental Parameters of Tous Reservoir
At the time of sampling (February 20th, 2015) the water column of the freshwater (conductivity <400 μS⋅cm-1) Tous reservoir was mixed and consequently both samples, from 12 and 25 m depth, had mostly similar physicochemical features (Supplementary Figure S1). Water had a relatively high transparency (Secchi Disk depth of nearly 8 m) as corresponds to an oligotrophic system, with maximum chlorophyll-a concentration lower than 2 μg⋅L-1 found at medium depths (10–15 m). Phosphorus appeared as the limiting nutrient, since inorganic nitrogen, mainly nitrate, was far more abundant (Supplementary Figure S1). The determinations of abundances by flow cytometry showed that Synechococcus cells were distributed throughout the water column at moderate abundances of around 2 × 104 cells⋅mL-1, whereas heterotrophic bacteria reached around 6 × 105 cells⋅mL-1 at all tested depths.
Community Structure of the Tous Metagenome Based on 16S rRNA Reads
The taxonomic composition based on 16S rRNA coding raw read fragments found in the Tous reservoir shows that taxa are present in similar proportions to the ones found in Amadorio (10 m, 0.22 μm collected sample), the closest freshwater water body to Tous (Ghai et al., 2014). In comparison to Lake Lanier, a similar temperate reservoir (Georgia, United States) (Supplementary Figure S2), some differences could be detected such as the higher numbers of Actinobacteria (nearly 40% in Tous 12 m in contrast to the 20% found in Lanier S2 sample (taken in July at 5 m depth), Bacteroidetes (around 10–15% in Tous samples compared to <5% in Lake Lanier) and Alphaproteobacteria (20% in Tous samples vs. <10% in Lake Lanier). On the other hand, Verrucomicrobia comprise 14% of the total 16S rRNA reads in Lake Lanier samples, compared to the 7–8% in Tous and Amadorio. Gammaproteobacteria were also present at higher proportions in Lake Lanier. Cyanobacteria comprise significant relative numbers according to the 16S rRNA classification in freshwater lakes like Albufera, Lake Lanier, Mendota (summer), Trout, Vattern, Amadorio (Ghai et al., 2014) and Tous 12 and 25 m samples, ranging between 4 and 10% in these freshwater bodies. Also, there was an important increase in Lanier 16S rRNA cyanobacterial reads in the Lake Lanier S2 sample reaching 14% of the reads compared to the other sites.
Genome Reconstruction and Phylogenomics of Synechococcus sp. Assembled Genomes
By a metagenomic approach we were able to reconstruct genomes of novel freshwater Synechococcus. Genomes derived from metagenomes are always composites of several clones that coexist in aquatic environments (Rodriguez-Valera et al., 2009; Kashtan et al., 2014). However, the quality of the assembly (long contigs) and relatively easy binning, points to all the clones belonging to a single species.
A total of 1.798 Mb of assembled composite genome were retrieved for this new freshwater Synechococcus sp. (hereafter referred to as Synechococcus sp. Tous). A genome completeness of 97.14% and genome size estimation of 1.85 Mb were obtained utilizing a set of 35 essential genes (Raes et al., 2007), whereas 74.77% and 2.4 Mb were estimated using a set of 112 essential genes (Albertsen et al., 2013), respectively. A total of 1959 proteins were predicted and annotated in this genome.
Lake Lanier, a freshwater reservoir on Chattahoochee River in Georgia (United States) extends over a surface of 150 km2. It is larger than the Tous reservoir, with a maximum depth of 78 m and a maximum volume of around 1294 Hm3, but presents a much smaller catchment area than Tous (2700 km2). Given the high recruitment (at >95% sequence identity in nucleotide comparisons) of the Synechococcus sp. Tous genome in the Lake Lanier metagenomes we attempted to assemble the relative present there as well. Synechococcus sp. Lanier genome reconstruction provided 1.475 Mb in 79 contigs. A total of 1619 proteins were predicted and annotated for Synechococcus sp. Lanier. The completeness of 40 and 50.45% according to Raes et al. (2007) and Albertsen et al. (2013) was much lower, likely due to the lack of the large contigs provided by fosmid libraries that were not constructed for Tous. For example, the Lanier genome is missing 30 genes from one of the ribosomal proteins cluster and the rRNA operon.
To infer the phylogenetic relationships of the new Tous and Lanier Synechococcus sp. genomes, a phylogenomic tree consisting of a concatenation of 122 common genes (Supplementary Table S1) of marine and freshwater Synechococcus sp. was constructed, using Prochlorococcus as an outgroup (Figure 1A; for a detailed tree see Supplementary Figure S3). The resulting tree recapitulates the phylogeny of the entire Synechococcus genus with marine and freshwater representatives. A 16S rRNA tree is also provided in Supplementary Figure S4. The phylogenetically closest species to these new freshwater genomes, as expected from the similarities detected in the metagenomic contigs (see above), was the marine Synechococcus sp. RCC307, a representative of the marine sub-cluster 5.3 isolated from a 15 m depth sample from off-shore Western Mediterranean waters. Classically, the sub-cluster 5.1 has been considered exclusively marine (Six et al., 2007; Dufresne et al., 2008; Scanlan et al., 2009), while sub-cluster 5.2 contains phycocyanin-containing euryhaline strains widely distributed in coastal areas and estuaries and sub-cluster 5.3 contains marine (Synechococcus sp. RCC307) and other less studied phycoerythrin-containing strains (Scanlan et al., 2009). Our two freshwater genomes clearly associate with sub-cluster 5.3 while the freshwater Synechococcus sp. GFB01 (Guimarães et al., 2015) associates with sub-cluster 5.2, which indicates that these two sub-clusters contain strains of both marine and non-marine origin, including freshwater. Actually, a 16S rRNA study of halotolerant strains showed that they also belonged to the 5.3 sub-cluster (Callieri et al., 2013).
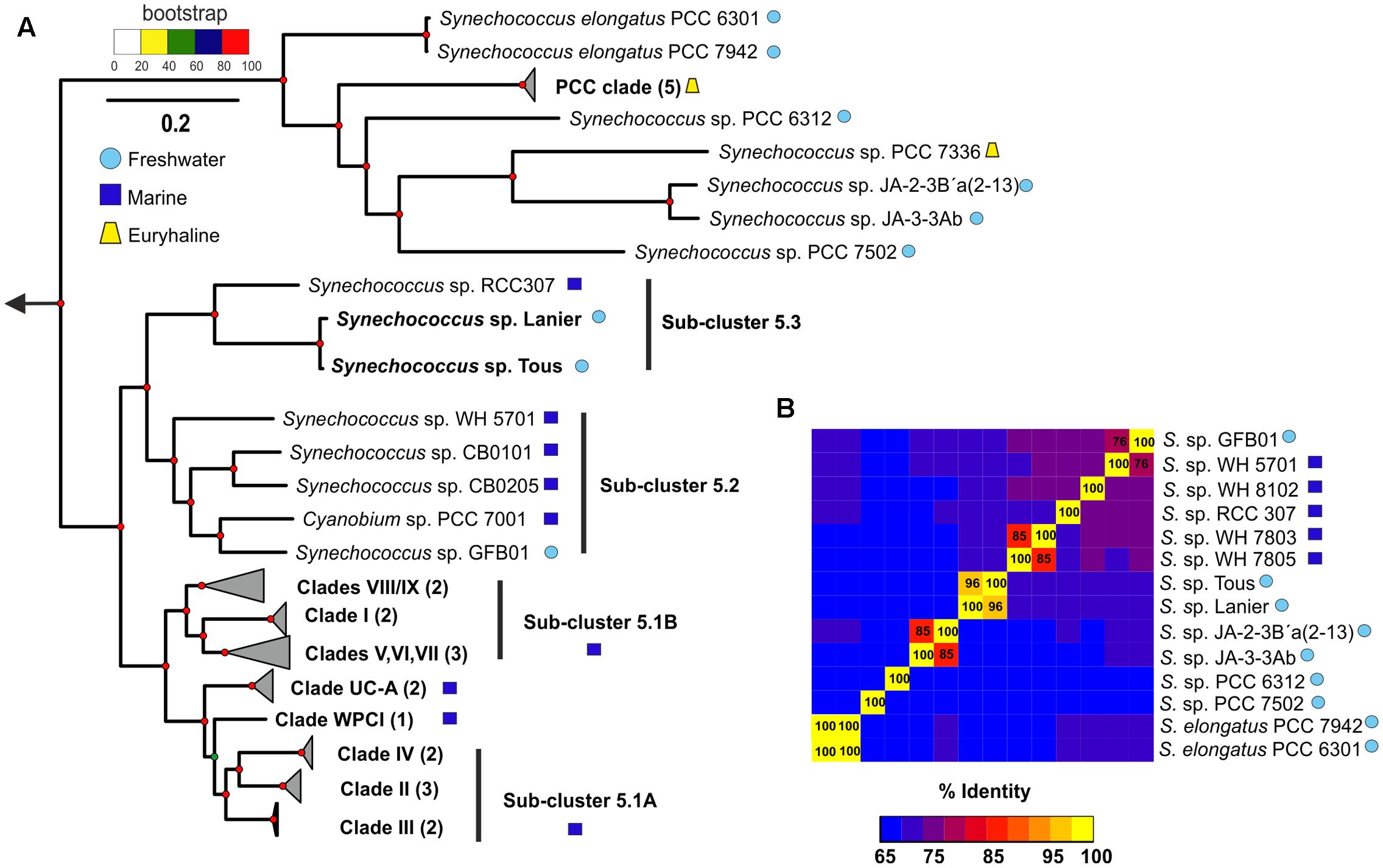
FIGURE 1. (A) Phylogenomics of the genus Synechococcus sp. One hundred twenty-two conserved genes were used to generate a maximum-likelihood phylogenetic tree with marine, freshwater and the novel Synechococcus sp. representatives. Three Prochlorococcus genomes were used as an outgroup. (B) ANI (average nucleotide identity) between the closest marine and freshwater representatives to the novel Synechococcus sp. Tous/Lanier.
The contigs obtained from the metagenomic reconstruction showed remarkable synteny with the Synechococcus sp. RCC307 genome. Therefore, we used this genome as a reference for the virtual reconstruction of the novel genomes (Figure 2A). ANI between Synechococcus sp. from Tous and Lanier was 96%, which indicates that the two assembled genomes are the same species within a novel Synechococcus sp. (Figure 1B). Despite the proximity and closeness of these two genomes to Synechococcus sp. RCC307 in the phylogenomic tree (Figure 1A) and their localized (at least) synteny (>70% similarity and 50 bp alignment length) (Figure 2A), ANI was only ca. 68% among both genomes and the marine isolate (Figure 1B). ANI was also performed against other marine and freshwater representatives, finding in all cases less than 70% among these novel species and the rest of the known Synechococcus. However, 16S rRNA showed high values over 97% similarities with several Synechococcus species confirming the lack of discriminatory power of this marker in Pcy in general. Taken together, both these assembled genomes (even considering their incompleteness) fall at the lower size range of all known Synechococcus genomes that are extremely variable in size, ranging from ca. 2 to 6 Mb) (Figures 2B,C). They have the smallest median intergenic spacer lengths of all known Synechococcus genomes so far (Figure 2C), suggesting a streamlined genome.
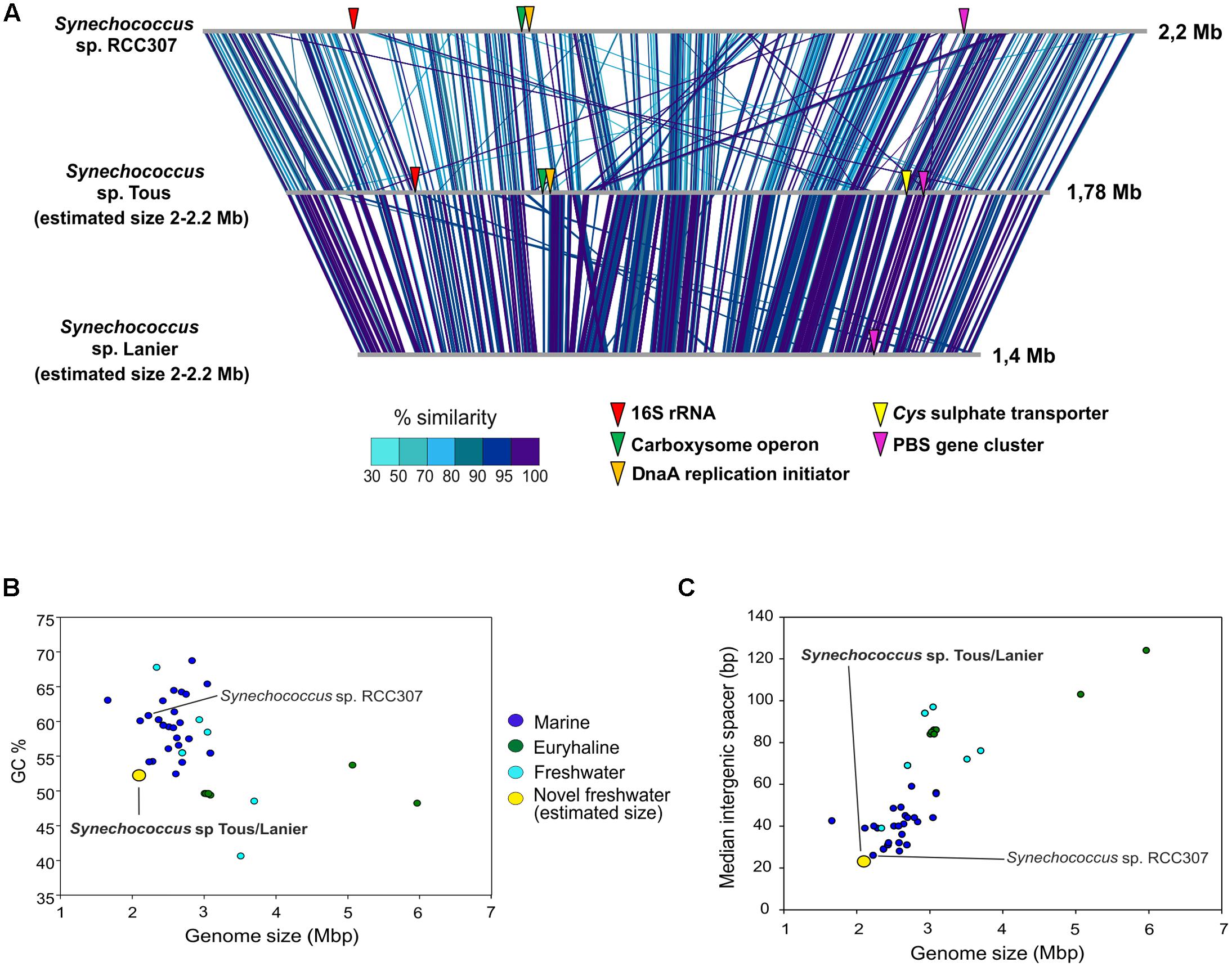
FIGURE 2. (A) Genomic comparison between marine Synechococcus sp. RCC307 and the novel freshwater assembled genomes from Tous and Lanier. Comparison made with BLASTN with 50 bp alignment length and >70% similarity. 16S rRNA, DnaA replication initiator, carboxysome operon, Cys sulfate transporter and PBS genes are indicated in the genomes. (B) Genome size (Mbp) vs. GC content of all marine, freshwater and euryhaline Synechococcus sp. sequenced genomes. (C) Genome size (Mbp) vs. median intergenic spacer (bp) of all Synechococcus sp. genomes. Estimated genome size was used for the novel freshwater Synechococcus sp.
Abundance of the Novel Synechococcus Genomes
To assess the distribution of the novel assembled Synechococcus sp. in other freshwater and brackish habitats, fragment recruitment was carried out against different metagenomes available from water bodies of different salinities (see Materials and Methods). The only available reference freshwater Synechococcus genomes, Synechococcus sp. GFB01 (Guimarães et al., 2015), S. elongatus PCC7942 (Holtman et al., 2005), Yellowstone representatives Synechococcus JA-3-3Ab and JA-2-3B’a(2-13) (Bhaya et al., 2007) and Synechococcus sp. PCC7502 and PCC6312 (Shih et al., 2013) were used for recruitment together with the phylogenetically closest marine representative Synechococcus sp. RCC307 and another marine Synechococcus sp. WH8102 (Scanlan et al., 2009). The freshwater metagenomes which provided significant recruitment values higher than 5 RPKGs for the Synechococcus sp. Tous and Lanier genomes are shown in Figure 3A. Tous and Lanier reservoirs from which the genomes were assembled showed, as might be expected, the highest number of RPKG (reads per Kb of genome per Gb of metagenome at >95% identity) (Figure 3). Both reconstructed Synechococcus spp. genomes clearly dominate Lanier and Tous metagenomes, which is reflected by the fragment recruitment identity values between 95 and 100% (Figure 3B). Both temperate reservoirs are located in very similar latitudes and altitudes (39°N and 64 m above average sea level for Tous reservoir and 34°N and 326 m for Lake Lanier) but are very distant geographically and located on different continents. Highest abundance values for both assembled genomes were found in Tous 12 m and Tous 25 m samples. In the Amadorio reservoir, that is only 100 km from Tous, 5–10 times lower values were found. Abundance in Lake Lanier samples used for assembly was comparable to that in Tous (Figure 3A). The novel genomes were also found in one metagenomic sample taken during a cyanobacterial bloom in Dexter reservoir (Oregón, United States). Only very low levels were found in Sparkling lake (Martinez-Garcia et al., 2012) and Albufera lagoon (Ghai et al., 2012). This suggests that these novel Synechococcus sp. could be widespread in many other temperate freshwater reservoirs with similar environmental parameters, being globally distributed at similar latitudes as has been previously described for marine Prochlorococcus and Synechococcus (Flombaum et al., 2013). The absence of the novel Synechococcus spp. in cold lakes with freezing cycles during the winter season, like Yellowstone, Lake Mendota, Swedish lakes (Erken, Ekoln and Vattern) or the Laurentian Great Lakes (Michigan and Ontario) also supports its temperate specialization. Low recruitment values were also found in Lake Gatun, Trout, Damariscotta, Lake Houston and the Amazon River. Interestingly, no other freshwater Synechococcus genomes recruited significant amounts from any of these metagenomes. Neither marine Synechococcus sp. RCC307 nor WH8102 were significantly detected (less than 2 RPKG) in the freshwater or brackish datasets tested. In the two marine metagenomes from the TARA oceans database, one from a 4 m sample in the southwest Mediterranean Sea (ERR315856) and another from 40 m depth from the same location (ERR315857), marine Synechococcus sp. RCC307 and WH8102 recruited significantly (20–25 RPKG) supporting their broad distribution in marine habitats, contrastingly to their absence in freshwater. The assemblies from Tous and Lanier did not recruit at all from the marine samples. These data suggest that the two assembled genomes represent a species of Synechococcus that is widely distributed in fresh water bodies at temperate latitudes and are among the abundant photosynthetic bacteria in these habitats.
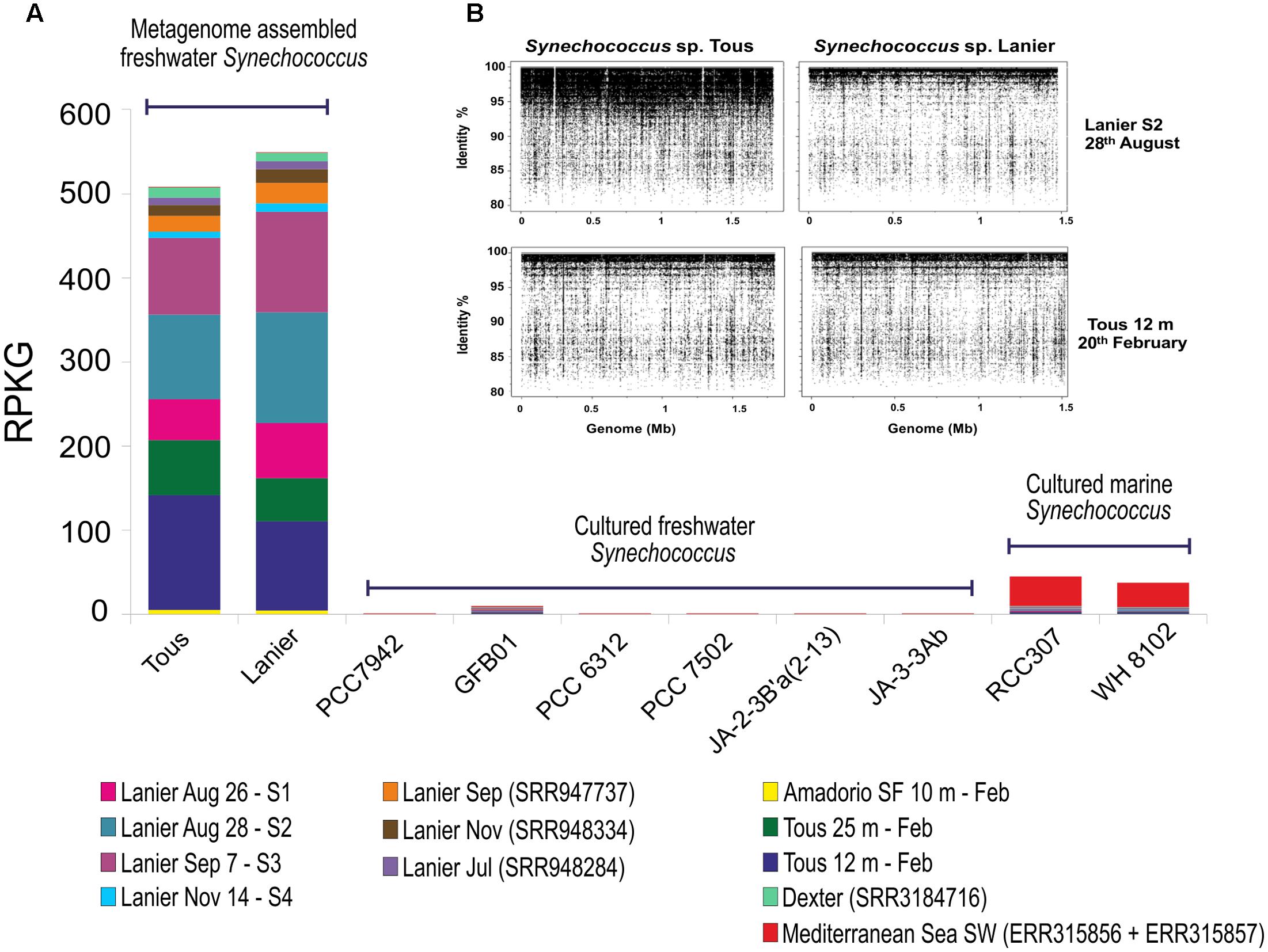
FIGURE 3. (A) Fragment recruitment of novel and reference Synechococcus sp. Genome abundance (expressed as RPKG, reads per Kb of genome per Gb of metagenome) along different freshwater metagenomes and two marine metagenomes. Only datasets with >5 RPKG values for the novel freshwater Synechococcus sp. were included in this analysis. Only hits with ≥95% identity, ≥50 bp alignment length were considered. Sep, September; Nov, November; Feb, February; Jul, July; Aug, August; SF, small fraction. (B) Metagenomic recruitment of Synechococcus sp. Tous and Lanier assembled genomes against Lanier S2 sample (5 m August 28th, 2011) (I) and Tous 12 m February 2014 (II) metagenomes. A 95% identity cut-off and 50 bp read length on each metagenome were used as restrictive parameters.
The abundance of members of the species represented by the assembled genomes in the Tous reservoir was confirmed by FISH (Figure 4). A total of 87.8% of the cells that hybridized to the general Synechococcus probe Syn405 also did with the specific probe for Synechococcus sp. Tous “SynTo.” Cells had rather small sizes (0.987 ± 0.139 μm lengths and 0.723 ± 0.119 μm width). From the general description of the genome provided below and the microscopic image we propose the name Ca. Synechococcus lacustris for the microbes identified by these two genomes and microscopically detected by FISH.
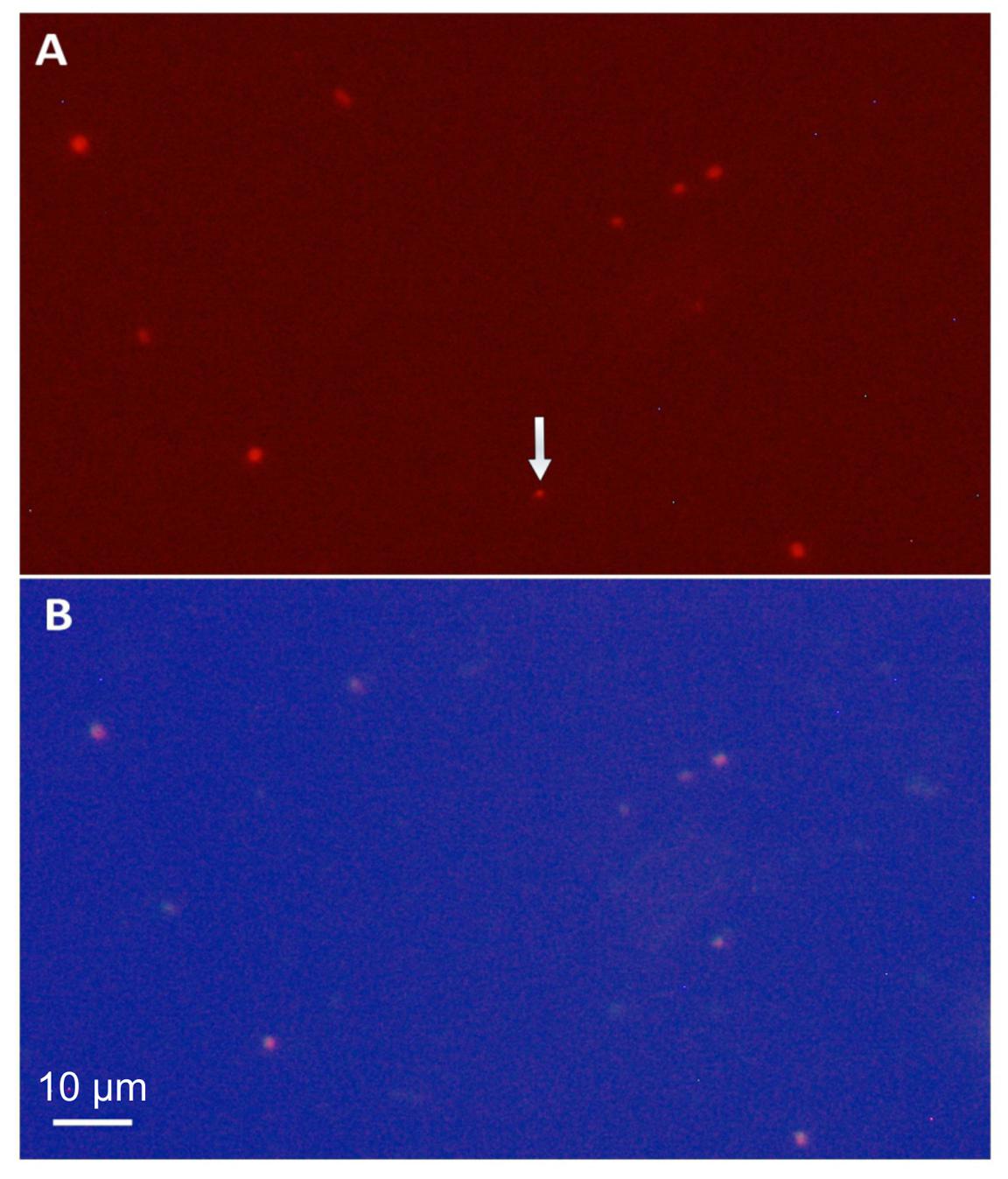
FIGURE 4. Photomicrographs (1250×) of a 12 m depth sample from Tous reservoir. (A) Phycoerythrin fluorescence under a green filter showing all picocyanobacteria in the sample. (B) FISH microphotograph of the same microscopic field stained with the Cy3, Cy5 fluorescence labeled rRNA-targeted probe specific for Synechococcus sp. Tous (“SynTo”). Note that all picocyanobacteria except that marked by the arrow are targeted by the specific “SynTous” probe. Scale bars represent 10 μm.
PBS Gene Cluster of the Novel Freshwater Synechococcus spp.
As seen in Figure 5, the novel Synechococcus sp. Tous/Lanier genome presents a highly similar PBS gene cluster structure to Synechococcus sp. WH7805, isolated in the Sargasso Sea (Six et al., 2007). The lack of PEII subunit in WH7805 and the new genomes points toward the novel Synechococcus spp. having type II pink pigmentation, being ecotypes adapted to low light ranges, typically found, for example, in deep marine oligotrophic waters (Scanlan et al., 2009; Callieri et al., 2012). Freshwater Synechococcus sp. GFB01 presents five PC subunits, one more as compared to the type I marine homolog WH5701. The only differences observable in the comparison of type II pigment containing strains WH7805 and the novel freshwater species is the lack of the low molecular weight tyrosine-phosphatase and two hypothetical proteins, which could be unique characteristics in the freshwater type II pigmentation strains. In any case, for each pigment type the structure of the PBS gene cluster is maintained in both freshwater and marine genomes.
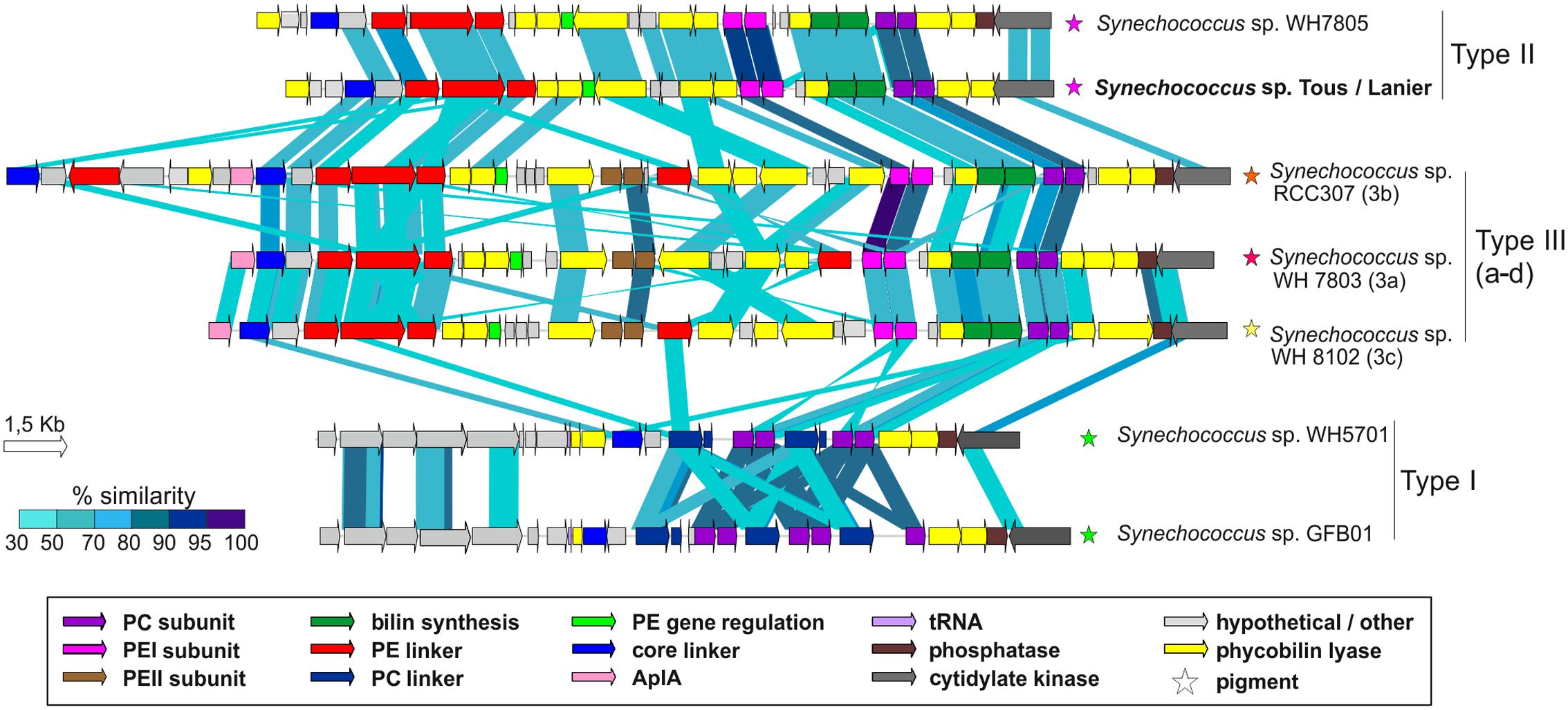
FIGURE 5. Structure and similarity of the PBS gene cluster among marine and Tous/Lanier freshwater Synechococcus sp. Comparison made with TBLASTX with >30% similarity hits and 150 bp alignment lengths. PC, Phycocyanin; PE, Phycoerythrin; AplA, Allophycoyanin-like protein; Phycobilin lyases, CpeY, CpeZ, CpeF, CpeS, CpeT, CpeU, RpcG, RpcE, RpcE, RpcF, RpcT; Bilin synthesis, pebA, pebB.
Features of the Synechococcus sp. Assembled Genomes That Support Their Freshwater Specialization
RuBisCo large and small subunits found in the Synechococcus sp. Tous assembled genome were phylogenetically close to those found in Paulinella chromatophora (Supplementary Figures S5, S6), a freshwater photosynthetic amoeba with chloroplasts originating from a photosynthetic symbiont related to Synechococcus (Marin et al., 2007). In addition, 30S ribosomal proteins and some photosynthetic proteins like photosystem I subunit VII and photosystem II D2 protein gave very high similarities (over 98%) to their homologs in the amoeba chloroplasts. That a freshwater amoeba receives an endosymbiont from a freshwater Synechococcus makes eco-evolutionary sense (Blank and Sanchez-Baracaldo, 2010).
As expected, the ntcA transcriptional regulator and nitrogen regulatory p-II protein common cyanobacterial genes were found in both Lanier and Tous genomes. Both Synechococcus sp. are able to obtain urea from the environment since the urtABCDE transport system and urease cluster ureABCDEFGH were detected in both assembled genomes. Glutamine synthetases (GlnA and GlnIII), which are missing in Synechococcus sp. RCC307 (but are present in other marine strains), were also found in both freshwater Synechococcus spp. Ammonia is the preferred N source in Pcy, as it can be incorporated easily to the amino acid glutamine and later on glutamic acid (Scanlan et al., 2009). Two amt-family ammonia channel proteins with small similarities (<60%) to other marine Synechococcus, but closer to the freshwater cyanobacteria Cyanobium sp. CACIAM14 (at 89% similarity), Cyanobium gracile PCC6307 or Synechococcus sp. GFB01, were detected in both genomes.
A common freshwater transporter Cys (specific for sulfate transport) which is absent in marine but present in freshwater cyanobacteria (see below) was found in the genomes. Synechococcus contain two sulfate permeases known as sulP (Price et al., 2004) in their genomes to acquire sulfur. A different sulfate ABC transport system has been described for some freshwater cyanobacteria like S. elongatus PCC7942. It consists of three Cys genes (CysW, CysT, and CysA) which form the membrane components and the sbpA gene, that codes for a periplasmic binding protein (Laudenbach and Grossman, 1991). S. elongatus PCC7942 contains a gene cluster with two subunits of each of these four genes; mutation of CysT, CysW or CysA genes in this strain led to no sulfate uptake and no growth when the latter was used as the sole sulfur source (Laudenbach and Grossman, 1991). Supplementary Figure S7 shows the comparisons made between Synechococcus sp. from Tous and the freshwater Cyanobium sp. CACIAM 14, Cyanobium gracile PCC6307, Synechococcus sp. GFB01 and the marine representative RCC307 showing a marked synteny among the freshwater ones and absence in the marine representative. This sulfate transport system is also preserved among many other freshwater Pcy, even filamentous genera like Tolypothrix, Fischerella, or Scytonema. The absence of this transporter in marine Synechococcus, could be explained by the high sulfate concentrations in the ocean which do not require the presence of these high affinity ABC sulfate transporters.
Freshwater cyanobacteria generally appear to have a requirement for zinc detoxification while some marine Synechococcus and Prochlorococcus do not seem to have it (Blindauer, 2008). Among the different transport systems detected in Synechococcus sp. Tous genome, we found ZnuC, ZnuB zinc/manganese and ZupT (IPR023498) zinc transporters. Surprisingly, the latter ZupT transporter was not found in marine or other freshwater cyanobacteria. Furthermore, this gene had highest similarities (<65 of relative abundance) to some found in green-sulfur photosynthetic or sulfur-reducing bacteria like Chlorobium, Desulfobacter, or Desulfuromonas.
Previous studies comparing the isoelectric point proteome of halophilic, marine and freshwater bacteria have shown that salinity adaptation positively correlates with a protein shift toward acidity in halophilic bacteria (Bardavid and Oren, 2012). Instead, a typical bimodal pattern is observed in marine and more pronouncedly in freshwater microbes, the latter presenting a lower peak in acidic proteins and a higher peak in basic proteins compared to marine representatives. We performed a whole proteome isoelectric point profile comparison between marine and freshwater Synechococcus (Supplementary Figure S8A). As expected, while marine Synechococcus showed a high percentage of acidic proteins, which presumably helps them adapt to their saline environment, improving the hydration sphere of the proteins, they also displayed a very low fraction of basic proteins. In contrast, freshwater Synechococcus sp. Tous, Lanier and GFB01 exhibited a lower peak of acidic proteins and a higher peak of basic amino acids. Freshwater S. elongatus PCC7942 showed both peaks of acid and basic proteins, consistent with its ability to tolerate different pH and salinity ranges (Billini et al., 2008).
We also compared the isoelectric points (pI) for all type of annotated transporters with transmembrane domains like ABC-type, permeases, efflux systems and also membrane proteins expecting that these exposed proteins would have the effect of salt amplified. Indeed, (Supplementary Figure S8B), marine Synechococcus showed a higher percentage of acidic transporters and membrane proteins and less basic transporters compared to freshwater genomes. Differences were even sharper when we considered exclusively those transporters and membrane proteins with pI ranging from 3.5 to 6.5 and from 8.5 to 12.5 (Supplementary Figure S8C). These pI plots further support that the novel Synechococcus sp. assembled genomes are specialized to live in freshwater.
Other mechanisms marine microbes use to deal with salinity adaptation rely on the active transport of nutrients in exchange of sodium, via co-transport symporters and translocation systems (Ventosa et al., 1998; López-Pérez et al., 2013). As expected, S. elongatus PCC7942, Synechococcus sp. GFB01 and the novel freshwater genomes did not contain any of these transporters. Sodium/proton antiporters were detected in both marine and freshwater strains, but especially in freshwater S. elongatus PCC7942 there are several copies of these transporters, as previously described (Billini et al., 2008). The accumulation of potassium as pH regulator through KefB and Trk transporters and the presence of sodium/proton antiporters, which couple with the maintenance of pH homeostasis in cells, have been described previously as strategies of salt adaptation (Mongodin et al., 2005). Either one or both potassium transporters were detected in both marine and freshwater Synechococcus. On the other hand, 11, 9, and 16 sodium dependent transporters of C-4 dicarboxylates osmoprotectants like glycine betaine and the amino acids proline or alanine were found in the marine Synechococcus sp. RCC307, WH8102 and WH7805, respectively (Scanlan et al., 2009), but none in the freshwater genomes. Sodium dependent transporters for sulfate, calcium, bicarbonate or bile acids were also found in marine genomes but not in the freshwater ones.
Conclusion
The scarcity of sequenced freshwater Synechococcus sp. genomes complicates an update of the current classification of the genus, particularly sub-clusters 5.2 and the less studied 5.3, both containing marine, euryhaline and freshwater species. In this manuscript we describe two freshwater genomes that belong to sub-cluster 5.3. ANI between the novel freshwater species and the rest of the marine or freshwater genomes never reached 70%, which confirms the novelty of the new freshwater genomes. Their bona fide freshwater adaptation is clear from several lines of evidence, from metagenomic recruitment to the pI of the proteome or the presence/absence of specific transporters.
The massive predominance of this single species in these two lakes located in two different continents and more than 7000 km apart indicates a potentially widespread distribution in temperate latitudes. However, more metagenomics latitudinal gradients in distant lakes are required to establish their global distribution and habitat range. Interestingly, both freshwater Synechococcus spp. described here were not found in cold lakes with annual ice-in and ice-out freezing cycles regardless of their habitat type. Neither eutrophic reservoirs or oligo-mesotrophic Laurentian Great Lakes nor Swedish Lakes (Vattern and Erken) seem to be appropriate environments for the novel Synechococcus sp. This distribution supports their specialization in temperate oligo-mesotrophic environments, which is the case for both Tous reservoir and Lake Lanier. It is noteworthy that the genomes described here are the smallest freshwater Synechococcus discovered thus far (2.2 Mb of estimated genome size), which display a certain degree of streamlining as shown by their short mean intergenic spacer size (20 bp). These reduced and compact genomes would be an advantage in oligotrophic waters such as those in the seasonally stratified temperate lakes, decreasing cell size and increasing volume to surface ratio. Attempts to isolate microbes belonging to Ca. S. lacustris are underway.
Data Accessibility
Tous reservoir metagenomic datasets have been deposited in the NCBI SRA database with BioProject number PRJNA342151 (SRR4198666 and SRR4198832). The assembled genomes Synechococcus sp. Lanier and Synechococcus sp. Tous have been deposited in the NCBI under Biosample identifiers SAMN05915837 and SAMN05915836, respectively. The cyanobacterial fosmids from Amadorio reservoir have been deposited to NCBI to the bioproject number PRJNA238866.
Author Contributions
FR-V, AC, and PC-Y conceived this work. RG, AP and AC performed the sample collection and filtration. JH-M performed the DNA extraction. Analysis was carried out by PC-Y, A-BM-C, AP, and JH-M. AP and AC analyzed the sample properties. Manuscript was written by PC-Y, AC, RG, and FR-V. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by projects “MEDIMAX”- BFPU2013-48007-P, Acciones de dinamización “REDES DE EXCELENCIA” CONSOLIDER- CGL2015-71523-REDC from the Spanish Ministerio de Economía y Competitividad and PROMETEO II/2014/012 “AQUAMET” from Generalitat Valenciana. JHM was supported by a Ph.D. fellowship from the Spanish Ministerio de Economía y Competitividad. RG was supported by the Grant Agency of the Czech Republic by the research grant 17-04828S.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01151/full#supplementary-material
Footnotes
- ^ http://probebase.csb.univie.ac.at/pb_report/probe/766
- ^ https://rdp.cme.msu.edu/index.jsp
- ^ http://rsb.info.nih.gov/ij/index.html
References
Albertsen, M., Hugenholtz, P., Skarshewski, A., Nielsen, K. L., Tyson, G. W., and Nielsen, P. H. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 31, 533–538. doi: 10.1038/nbt.2579
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
American Public Health Association, American Water Works Association, World Powerlifting Congress Federation, and Water Environmental Federation (1915). Standard Methods for the Examination of Water and Wastewater. Washington, DC: American Public Health Association.
Arnosti, C., Fuchs, B. M., Amann, R., and Passow, U. (2012). Contrasting extracellular enzyme activities of particle-associated bacteria from distinct provinces of the North Atlantic Ocean. Front. Microbiol. 3:425. doi: 10.3389/fmicb.2012.00425
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bardavid, R. E., and Oren, A. (2012). Acid-shifted isoelectric point profiles of the proteins in a hypersaline microbial mat: an adaptation to life at high salt concentrations? Extremophiles 16, 787–792. doi: 10.1007/s00792-012-0476-6
Behnam, F., Vilcinskas, A., Wagner, M., and Stoecker, K. (2012). A straightforward DOPE (double labeling of oligonucleotide probes)-FISH (fluorescence in situ hybridization) method for simultaneous multicolor detection of six microbial populations. Appl. Environ. Microbiol. 78, 5138–5142. doi: 10.1128/AEM.00977-12
Bhaya, D., Grossman, A. R., Steunou, A.-S., Khuri, N., Cohan, F. M., Hamamura, N., et al. (2007). Population level functional diversity in a microbial community revealed by comparative genomic and metagenomic analyses. ISME J. 1, 703–713. doi: 10.1038/ismej.2007.46
Billini, M., Stamatakis, K., and Sophianopoulou, V. (2008). Two members of a network of putative Na+/H+ antiporters are involved in salt and pH tolerance of the freshwater cyanobacterium Synechococcus elongatus. J. Bacteriol. 190, 6318–6329. doi: 10.1128/JB.00696-08
Blank, C., and Sanchez-Baracaldo, P. (2010). Timing of morphological and ecological innovations in the cyanobacteria–a key to understanding the rise in atmospheric oxygen. Geobiology 8, 1–23. doi: 10.1111/j.1472-4669.2009.00220.x
Blindauer, C. A. (2008). Zinc-handling in cyanobacteria: an update. Chem. Biodivers. 5, 1990–2013. doi: 10.1002/cbdv.200890183
Callieri, C. (2008). Picophytoplankton in freshwater ecosystems: the importance of small-sized phototrophs. Freshw. Rev. 1, 1–28. doi: 10.1608/FRJ-1.1.1
Callieri, C., Coci, M., Corno, G., Macek, M., Modenutti, B., Balseiro, E., et al. (2013). Phylogenetic diversity of nonmarine picocyanobacteria. FEMS Microbiol. Ecol. 85, 293–301. doi: 10.1111/1574-6941.12118
Callieri, C., Cronberg, G., and Stockner, J. G. (2012). ”Freshwater picocyanobacteria: single cells, microcolonies and colonial forms,” in Ecology of Cyanobacteria II: Their Diversity in Time and Space, 2nd Edn, ed. B. A. Whitton (Berlin: Springer), 229–269.
Callieri, C., and Stockner, J. G. (2002). Freshwater autotrophic picoplankton: a review. J. Limnol. 61, 1–14. doi: 10.4081/jlimnol.2002.1
Camacho, A. (2006). On the occurrence and ecological features of deep chlorophyll maxima (DCM) in Spanish stratified lakes. Limnetica 25, 453–478.
Camacho, A., Miracle, M. R., and Vicente, E. (2003a). Which factors determine the abundance and distribution of picocyanobacteria in inland waters? A comparison among different types of lakes and ponds. Arch. Hydrobiol. 157, 321–338. doi: 10.1127/0003-9136/2003/0157-0321
Camacho, A., Picazo, A., Miracle, M. R., and Vicente, E. (2003b). Spatial distribution and temporal dynamics of picocyanobacteria in a meromictic karstic lake. Algol. Stud. 109, 171–184. doi: 10.1127/1864-1318/2003/0109-0171
Camacho, A., Vicente, E., and Miracle, M. R. (2000). Spatio-temporal distribution and growth dynamics of phototrophic sulfur bacteria populations in the sulfide-rich Lake Arcas. Aquat. Sci. 62, 334–349. doi: 10.1007/PL00001339
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Cole, J. R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37(suppl. 1), D141-D145. doi: 10.1093/nar/gkn879
Dufresne, A., Ostrowski, M., Scanlan, D. J., Garczarek, L., Mazard, S., Palenik, B. P., et al. (2008). Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 9:R90. doi: 10.1186/gb-2008-9-5-r90
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Flombaum, P., Gallegos, J. L., Gordillo, R. A., Rincón, J., Zabala, L. L., Jiao, N., et al. (2013). Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 110, 9824–9829. doi: 10.1073/pnas.1307701110
Frenkel, A., Gaffron, H., and Battley, E. H. (1950). Photosynthesis and photoreduction by the blue green alga, Synechococcus elongatus, Näg. Biol. Bull. 99, 157–162. doi: 10.2307/1538735
Fuller, N. J., Marie, D., Partensky, F., Vaulot, D., Post, A. F., and Scanlan, D. J. (2003). Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl. Environ. Microbiol. 69, 2430–2443. doi: 10.1128/AEM.69.5.2430-2443.2003
Ghai, R., Hernandez, C. M., Picazo, A., Mizuno, C. M., Ininbergs, K., Díez, B., et al. (2012). Metagenomes of Mediterranean coastal lagoons. Sci. Rep. 2:490 doi: 10.1038/srep00490
Ghai, R., Martin-Cuadrado, A.-B., Molto, A. G., Heredia, I. G., Cabrera, R., Martin, J., et al. (2010). Metagenome of the Mediterranean deep chlorophyll maximum studied by direct and fosmid library 454 pyrosequencing. ISME J. 4, 1154–1166. doi: 10.1038/ismej.2010.44
Ghai, R., Mizuno, C. M., Picazo, A., Camacho, A., and Rodriguez-Valera, F. (2014). Key roles for freshwater Actinobacteria revealed by deep metagenomic sequencing. Mol. Ecol. 23, 6073–6090. doi: 10.1111/mec.12985
Guimarães, P. I., Leão, T. F., de Melo, A. G. C., Ramos, R. T. J., Silva, A., Fiore, M. F., et al. (2015). Draft genome sequence of the picocyanobacterium Synechococcus sp. strain GFB01, isolated from a freshwater lagoon in the Brazilian Amazon. Genome Announc. 3:e00876-15. doi: 10.1128/genomeA.00876-15
Haft, D. H., Loftus, B. J., Richardson, D. L., Yang, F., Eisen, J. A., Paulsen, I. T., et al. (2001). TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res. 29, 41–43. doi: 10.1093/nar/29.1.41
Holtman, C. K., Chen, Y., Sandoval, P., Gonzales, A., Nalty, M. S., Thomas, T. L., et al. (2005). High-throughput functional analysis of the Synechococcus elongatus PCC 7942 genome. DNA Res. 12, 103–115. doi: 10.1093/dnares/12.2.103
Huang, Y., Gilna, P., and Li, W. (2009). Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 25, 1338–1340. doi: 10.1093/bioinformatics/btp161
Hyatt, D., Chen, G.-L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Kashtan, N., Roggensack, S. E., Rodrigue, S., Thompson, J. W., Biller, S. J., Coe, A., et al. (2014). Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344, 416–420. doi: 10.1126/science.1248575
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lassmann, T., and Sonnhammer, E. L. (2005). Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6:298. doi: 10.1186/1471-2105-6-298
Laudenbach, D. E., and Grossman, A. R. (1991). Characterization and mutagenesis of sulfur-regulated genes in a cyanobacterium: evidence for function in sulfate transport. J. Bacteriol. 173, 2739–2750. doi: 10.1128/jb.173.9.2739-2750.1991
Lê, S., Josse, J., and Husson, F. (2008). FactoMineR: an R package for multivariate analysis. J. Stat. Softw. 25, 1–18. doi: 10.18637/jss.v025.i01
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
López-Pérez, M., Ghai, R., Leon, M. J., Rodríguez-Olmos,Á., Copa-Patiño, J. L., Soliveri, J., et al. (2013). Genomes of “Spiribacter”, a streamlined, successful halophilic bacterium. BMC Genomics 14:787. doi: 10.1186/1471-2164-14-787
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.0955
MacIsaac, E., and Stockner, J. G. (1993). “Enumeration of phototrophic picoplankton by autofluorescence microscopy,” in Handbook of Methods in Aquatic Microbial Ecology, eds B. Sherr and E. Sherr (Boca Raton, FL: CRC Press), 187–197.
Marie, D., Partensky, F., Jacquet, S., and Vaulot, D. (1997). Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl. Environ. Microbiol. 63, 186–193.
Marin, B., Nowack, E. C., Glöckner, G., and Melkonian, M. (2007). The ancestor of the Paulinella chromatophore obtained a carboxysomal operon by horizontal gene transfer from a Nitrococcus-like γ-proteobacterium. BMC Evol. Biol. 7:85. doi: 10.1186/1471-2148-7-85
Martinez-Garcia, M., Swan, B. K., Poulton, N. J., Gomez, M. L., Masland, D., Sieracki, M. E., et al. (2012). High-throughput single-cell sequencing identifies photoheterotrophs and chemoautotrophs in freshwater bacterioplankton. ISME J. 6, 113–123. doi: 10.1038/ismej.2011.84
Mazard, S., Ostrowski, M., Partensky, F., and Scanlan, D. J. (2012). Multi-locus sequence analysis, taxonomic resolution and biogeography of marine Synechococcus. Environ. Microbiol. 14, 372–386. doi: 10.1111/j.1462-2920.2011.02514.x
Mongodin, E. F., Nelson, K., Daugherty, S., Deboy, R., Wister, J., Khouri, H., et al. (2005). The genome of Salinibacter ruber: convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc. Natl. Acad. Sci. U.S.A. 102, 18147–18152. doi: 10.1073/pnas.0509073102
Nawrocki, E. (2009). Structural RNA Homology Search and Alignment using Covariance Models. St. Louis, MO: Washington University in St. Louis.
Nawrocki, E. P., and Eddy, S. R. (2010). SSU-Align: A Tool for Structural Alignment of SSU rRNA Sequences. Available at: http://selab.janelia.org/software.html
Oh, S., Caro-Quintero, A., Tsementzi, D., DeLeon-Rodriguez, N., Luo, C., Poretsky, R., et al. (2011). Metagenomic insights into the evolution, function, and complexity of the planktonic microbial community of Lake Lanier, a temperate freshwater ecosystem. Appl. Environ. Microbiol. 77, 6000–6011. doi: 10.1128/AEM.00107-11
Partensky, F., Blanchot, J., and Vaulot, D. (1999). Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review. Bull. Inst. Oceanogr. Monaco Numero Spec. 19, 457–476.
Peng, Y., Leung, H. C., Yiu, S.-M., and Chin, F. Y. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428. doi: 10.1093/bioinformatics/bts174
Picazo, A., Rochera, C., Vicente, E., Miracle, M. R., and Camacho, A. (2013). Spectrophotometric methods for the determination of photosynthetic pigments in stratified lakes: a critical analysis based on comparisons with HPLC determinations in a model lake. Limnetica 32, 139–158.
Porter, K., and Feig, Y. (1980). The use of DAPI for identification and enumeration of bacteria and blue-green algae. Limnol. Oceanogr. 25, 943–948. doi: 10.4319/lo.1980.25.5.0943
Price, G. D., Woodger, F. J., Badger, M. R., Howitt, S. M., and Tucker, L. (2004). Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 101, 18228–18233. doi: 10.1073/pnas.0405211101
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. doi: 10.1371/journal.pone.0009490
Raes, J., Korbel, J. O., Lercher, M. J., Von Mering, C., and Bork, P. (2007). Prediction of effective genome size in metagenomic samples. Genome Biol. 8:R10. doi: 10.1186/gb-2007-8-1-r10
Rice, P., Longden, I., and Bleasby, A. (2000). EMBOSS: the European molecular biology open software suite. Trends Genet. 16, 276–277. doi: 10.1016/S0168-9525(00)02024-2
Robertson, B. R., Tezuka, N., and Watanabe, M. M. (2001). Phylogenetic analyses of Synechococcus strains (cyanobacteria) using sequences of 16S rDNA and part of the phycocyanin operon reveal multiple evolutionary lines and reflect phycobilin content. Int. J. Syst. Evol. Microbiol. 51, 861–871. doi: 10.1099/00207713-51-3-861
Rocap, G., Larimer, F. W., Lamerdin, J., Malfatti, S., Chain, P., Ahlgren, N. A., et al. (2003). Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424, 1042–1047. doi: 10.1038/nature01947
Rodriguez-Valera, F., Martin-Cuadrado, A.-B., Rodriguez-Brito, B., Pašiæ, L., Thingstad, T. F., Rohwer, F., et al. (2009). Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7, 828–836. doi: 10.1038/nrmicro2235
Rozen, S., and Skaletsky, H. (1999). Primer3 on the WWW for general users and for biologist programmers. Bioinform. Methods Protoc. 132, 365–386. doi: 10.1385/1-59259-192-2:365
Sanchez-Baracaldo, P., Handley, B. A., and Hayes, P. K. (2008). Picocyanobacterial community structure of freshwater lakes and the Baltic Sea revealed by phylogenetic analyses and clade-specific quantitative PCR. Microbiology 154, 3347–3357. doi: 10.1099/mic.0.2008/019836-0
Sanchez-Baracaldo, P., Hayes, P., and Blank, C. (2005). Morphological and habitat evolution in the Cyanobacteria using a compartmentalization approach. Geobiology 3, 145–165. doi: 10.1111/j.1472-4669.2005.00050.x
Scanlan, D. J., Ostrowski, M., Mazard, S., Dufresne, A., Garczarek, L., Hess, W. R., et al. (2009). Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73, 249–299. doi: 10.1128/MMBR.00035-08
Shih, P. M., Wu, D., Latifi, A., Axen, S. D., Fewer, D. P., Talla, E., et al. (2013). Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, 1053–1058. doi: 10.1073/pnas.1217107110
Six, C., Thomas, J.-C., Garczarek, L., Ostrowski, M., Dufresne, A., Blot, N., et al. (2007). Diversity and evolution of phycobilisomes in marine Synechococcus spp.: a comparative genomics study. Genome Biol. 8:R259.
Steffen, M. M., Li, Z., Effler, T. C., Hauser, L. J., Boyer, G. L., and Wilhelm, S. W. (2012). Comparative metagenomics of toxic freshwater cyanobacteria bloom communities on two continents. PLoS ONE 7:e44002. doi: 10.1371/journal.pone.0044002
Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., Labadie, K., Salazar, G., et al. (2015). Structure and function of the global ocean microbiome. Science 348:1261359. doi: 10.1126/science.1261359
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526.
Tatusov, R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., Shankavaram, U. T., Rao, B. S., et al. (2001). The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29, 22–28. doi: 10.1093/nar/29.1.22
Vadstein, O. (2000). Heterotrophic, planktonic bacteria and cycling of phosphorus requirements, competitive ability and food web interactions. Adv. Microb. Ecol. 16, 115–167. doi: 10.1007/978-1-4615-4187-5_4
Veldhuis, M. J., and Kraay, G. W. (2000). Application of flow cytometry in marine phytoplankton research: current applications and future perspectives. Sci. Mar. 64, 121–134. doi: 10.3989/scimar.2000.64n2121
Ventosa, A., Nieto, J. J., and Oren, A. (1998). Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62, 504–544.
West, N. J., Schönhuber, W. A., Fuller, N. J., Amann, R. I., Rippka, R., Post, A. F., et al. (2001). Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides. Microbiology 147, 1731–1744. doi: 10.1099/00221287-147-7-1731
Keywords: Synechococcus, picocyanobacteria, freshwater reservoirs, metagenomics, abundance, smallest estimated size
Citation: Cabello-Yeves PJ, Haro-Moreno JM, Martin-Cuadrado A-B, Ghai R, Picazo A, Camacho A and Rodriguez-Valera F (2017) Novel Synechococcus Genomes Reconstructed from Freshwater Reservoirs. Front. Microbiol. 8:1151. doi: 10.3389/fmicb.2017.01151
Received: 06 March 2017; Accepted: 07 June 2017;
Published: 21 June 2017.
Edited by:
George S. Bullerjahn, Bowling Green State University, United StatesReviewed by:
Katherine McMahon, University of Wisconsin–Madison College of Engineering, United StatesMaureen Coleman, University of Chicago, United States
Copyright © 2017 Cabello-Yeves, Haro-Moreno, Martin-Cuadrado, Ghai, Picazo, Camacho and Rodriguez-Valera. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francisco Rodriguez-Valera, ZnJ2YWxlcmFAdW1oLmVz
 Pedro J. Cabello-Yeves
Pedro J. Cabello-Yeves Jose M. Haro-Moreno
Jose M. Haro-Moreno Ana-Belen Martin-Cuadrado
Ana-Belen Martin-Cuadrado Rohit Ghai
Rohit Ghai Antonio Picazo
Antonio Picazo Antonio Camacho
Antonio Camacho Francisco Rodriguez-Valera
Francisco Rodriguez-Valera