- 1Institute for Food Safety and Hygiene, University of Zurich, Zürich, Switzerland
- 2Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration, Laurel, MD, United States
Objectives: The aim of this study was to characterize a collection of 520 Salmonella enterica serovar Infantis strains isolated from food (poultry meat), human infections and environmental sources from the years 2010, 2013 and 2015 in Switzerland.
Methods: We performed antimicrobial susceptibility testing and pulsed-field gel electrophoresis (PFGE) analysis on all 520 S. Infantis isolates, and whole genome sequencing (WGS) on 32 selected isolates.
Results: The majority (74.8%) of the isolates was multidrug resistant (MDR). PFGE analysis revealed that 270 (51.9%) isolates shared an identity of 90%. All isolates subjected to WGS belonged to sequence type (ST) 32 or a double-locus variant thereof (one isolate). Seven (21.9%) of the sequenced isolates were phylogenetically related to the broiler-associated clone B that emerged in Hungary and subsequently spread within and outside of Europe. In addition, three isolates harboring blaCTX-M-65 on a predicted large (∼320 kb) plasmid grouped in a distinct cluster.
Conclusion: This study documents the presence of the Hungarian clone B and related clones in food and human isolates between 2010 and 2015, and the emergence of a blaCTX-M-65 harboring MDR S. serovar Infantis lineage.
Introduction
Non-typhoidal Salmonella enterica (NTS) are one of the most important etiological agents of foodborne diarrheal diseases in humans worldwide and cause an estimated 80.3 million foodborne illnesses a year (Majowicz et al., 2010). Although most cases of salmonellosis are self-limiting episodes of gastro-enteritis, severe cases of infection, including bacteremia and meningitis require antimicrobial treatment. Ciprofloxacin is a common first-line antimicrobial for treating salmonellosis but because fluoroquinolones are not used for treating children, β-lactams (ampicillin or third-generation cephalosporins) are of equal importance (Medalla et al., 2016). Multidrug-resistant (MDR) NTS is associated with higher morbidity and mortality outcomes compared to drug-susceptible strains and is a major public health concern (Mølbak, 2005). Salmonella enterica subsp. enterica serovar Infantis (S. serovar Infantis) has emerged as the fourth most common serovar causing human salmonellosis in Europe, with 1,846 cases reported by the EU/EEA countries in 2014 (European Food Safety Authority [EFSA] and European Centre for Disease Prevention [ECDC], 2016). Poultry, especially from layer and broiler farms, as well as pigs are the main animal reservoirs for S. serovar Infantis (Nógrády et al., 2012). This serovar is also dominant in broiler meat, accounting for 35.9% of all Salmonella isolates reported from EU countries in 2014 (European Food Safety Authority [EFSA] and European Centre for Disease Prevention [ECDC], 2016). Over the last few years, antimicrobial resistance has emerged in S. serovar Infantis isolates from human and animal sources in various European countries and consequently, this serovar, together with S. Kentucky, contributes significantly to the numbers of MDR Salmonella in Europe (Nógrády et al., 2008; Dionisi et al., 2011). Closely related MDR clones of S. serovar Infantis have disseminated among broiler populations and associated animal growing environments, ultimately being disseminated into the food chain and then into humans in European countries such as Hungary, Poland and Austria (Nógrády et al., 2012). Isolates belonging to these clones are characterized by their resistance to nalidixic acid, sulfamethoxazole, streptomycin and tetracycline (NaSSuT). Recently, resistance to third generation cephalosporins has emerged in S. serovar Infantis isolates in Italy, due to the circulation of an extended-spectrum β-lactamase (ESBL) producing, MDR clone with additional reduced susceptibility to ciprofloxacin (Franco et al., 2015). The spread of MDR S. serovar Infantis clones throughout the food production system (mainly poultry and poultry meat) and in humans is highly worrisome and warrants improved understanding of its epidemiology. In Switzerland, S. serovar Infantis ranks among the top five of Salmonella serovars registered by the National Centre for Enteropathogenic Bacteria and Listeria (NENT). However, currently no data on antimicrobial resistance patterns or clonal relationships of the isolates exist, despite its clinical importance.
The aim of this study was to characterize a collection of 520 S. serovar Infantis strains isolated from food (poultry meat), human infections and environmental sources from the years 2010, 2013 and 2015 in Switzerland (i) by determining their phenotypic antibiotic resistance profiles using the disk diffusion method and (ii) by assessing genotypic characteristics and clonal relatedness using molecular methods including pulsed-field gel electrophoresis (PFGE), PCR, and whole genome sequencing (WGS).
Materials and Methods
Bacterial Strains
A total of 520 non-duplicate S. serovar Infantis isolates from human infections (n = 84), poultry meat (n = 418) and other sources (n = 18) were collected during 2010, 2013 and 2015 at the National Centre for Enteropathogenic Bacteria and Listeria (NENT), Switzerland. The isolates had been forwarded by hospitals, diagnostic laboratories or surveillance programs of retail markets and food or feed producing facilities for final species-level identification according to the White-Kauffmann-Le Minor scheme (Grimont and Weill, 2008).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed using the disk-diffusion method and the antibiotics ampicillin (AM), amoxicillin-clavulanic acid (AMC), cefotaxime (CTX), nalidixic acid (Na), ciprofloxacin (CIP), gentamicin (GM), kanamycin (K), streptomycin (S), sulfamethoxazole (Su), trimethoprim (TMP), tetracycline (T), and chloramphenicol (C) (Becton Dickinson, Heidelberg, Germany). Results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) performance standards (CLSI, 2016). For sulfamethoxazole, for which breakpoints are not listed separately from trimethoprim, an inhibition zone of ≤10 mm was interpreted as resistant. Isolates displaying resistance to three or more classes of antimicrobials (counting β-lactams as one class) were defined as multidrug-resistant (MDR). Synergistic effects between AMC and CTX were regarded as an indication of the presence of an ESBL producer (Kaur et al., 2013).
Detection and Characterization of Extended-Spectrum β-Lactamase (bla) Genes
Putative ESBL producers were grown on BrillianceTMESBL agar (Oxoid, Hampshire, United Kingdom). The presence of blaESBLs was confirmed by PCR by screening for blaTEM, blaSHV, and blaCTX-M alleles belonging to CTX-M groups 1, 2, 8, 9, and 25 as described previously (Woodford et al., 2006; Geser et al., 2012; Zurfluh et al., 2015). Synthesis of primers and DNA custom sequencing was carried out by Microsynth (Balgach, Switzerland) and nucleotide sequences were analyzed with CLC Main Workbench 6.6.1. For database searches the BLASTN program of NCBI1 was used.
Pulsed-Field Gel Electrophoresis
Pulsed-field gel electrophoresis was performed according to the PulseNet protocol of the Centers for Disease Control and Prevention (CDC)2, using the restriction enzyme XbaI (Roche, Mannheim, Germany) and Salmonella serovar Braenderup strain H9812 (ATCC BAA 664) as the reference strain. Restricted DNA was separated in a 1% agarose gel (BioRad, Cressier, Switzerland) at 12°C for 20 h at 6 V/cm under linear ramp with switch times from 2 to 64 s and 120° included angle using a CHEF-DR III system (BIO-RAD, Munich, Germany). Gels were stained with ethidium bromide and visualized under UV light with Gel Doc (BIO-RAD, Munich, Germany). GelCompar II software (Applied Maths NV, Sint-Martens-Latem, Belgium) was used for analysis. Pairwise similarities between the XbaI PFGE patterns were calculated by the DICE similarity coefficient. Clustering was based on the unweighted pair-group method with averages, setting tolerance at 1% and optimization at 0.5%.
Whole Genome Sequencing
Whole genome sequencing was performed with a representative subset of 32 isolates selected with regard to their PFGE pattern, source, year of isolation or presence of blaCTX-M-65. DNA extraction was performed with the Wizard® Genomic DNA Purification Kit according to the manufacturers protocol (Promega AG, Dübendorf, Switzerland). Sequencing was performed on a MiSeq sequencer (Illumina, San Diego, CA, United States), utilizing a 600 cycle Nextera XT library kit. Trimmed Fastq data sets were de novo assembled using the recommended workflow on CLC Genomics Work bench version 8.0 (CLC bio, Aarhus, Denmark). Genomic contigs were annotated using the RAST annotation server (Aziz et al., 2008). The assembled sequences were uploaded to the http://www.genomicepidemiology.org/ server. Sequences of the seven housekeeping genes (aroC, dnaN, hemD, hisD, purE, sucA and thrA) were analyzed to identify multilocus sequence types (MLST)3, antibiotic resistance genes4, and plasmid replicon types5, using each website’s algorithms and databases. Routine processing of genome datasets was carried out by in-house perl scripts (available upon request). A local customized database of Salmonella chromosomal and plasmid genomes from NCBI was created and used for annotation, plasmid analysis, and homology detection with BLAST suite (Altschul et al., 1990). Phylogenetic analysis was conducted using a multiloci analysis based on a published core gene dataset (Leekitcharoenphon et al., 2012). Alleles in 2780 core gene loci across all the genomes were identified with Salmonella enterica subsp. enterica serovar Typhimurium LT2 genome as the reference. Of these loci, 1,500 core gene loci were randomly chosen and alleles were concatenated. The data matrix of alleles was subject to multiple alignment and phylogenetic analysis using tools available in MEGA7 suite (Kumar et al., 2016). The phylogenetic tree was built using the Neighbor-Joining algorithm in the MEGA 7 suite with 9,562 positions across 50 genomes for the final analysis.
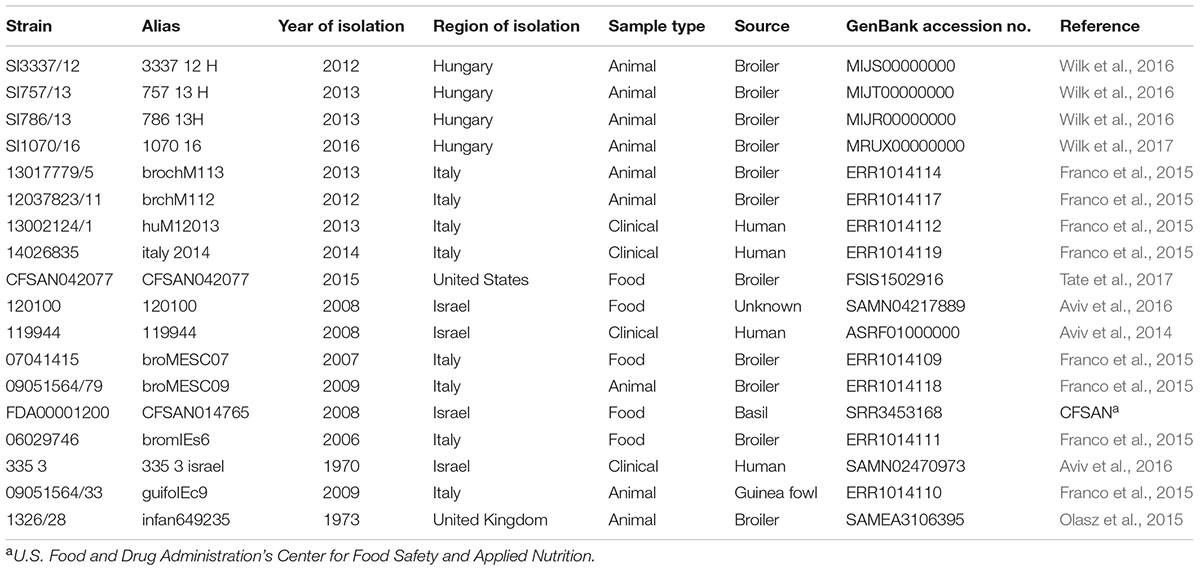
For genome comparison, whole genome draft sequences of Salmonella enterica serovar Infantis strains from public databases were retrieved, including four recent strains of the prevalent Hungarian clone B of S. serovar Infantis from Hungary (Wilk et al., 2016, 2017), five blaESBL harboring, and eight non-blaESBL harboring strains from Italy, Israel and the United States. (Aviv et al., 2014; Franco et al., 2015), and one fully susceptible strain from the United Kingdom (Olasz et al., 2015). An overview of the strains and their GenBank accession numbers is given in Table 1. For plasmid comparison, annotations were obtained from RAST (Aziz et al., 2008), and compared with pCSAM042077 (Tate et al., 2017) on the SEED server for additional verification (Overbeek et al., 2013).
Results
Antimicrobial Susceptibility Testing
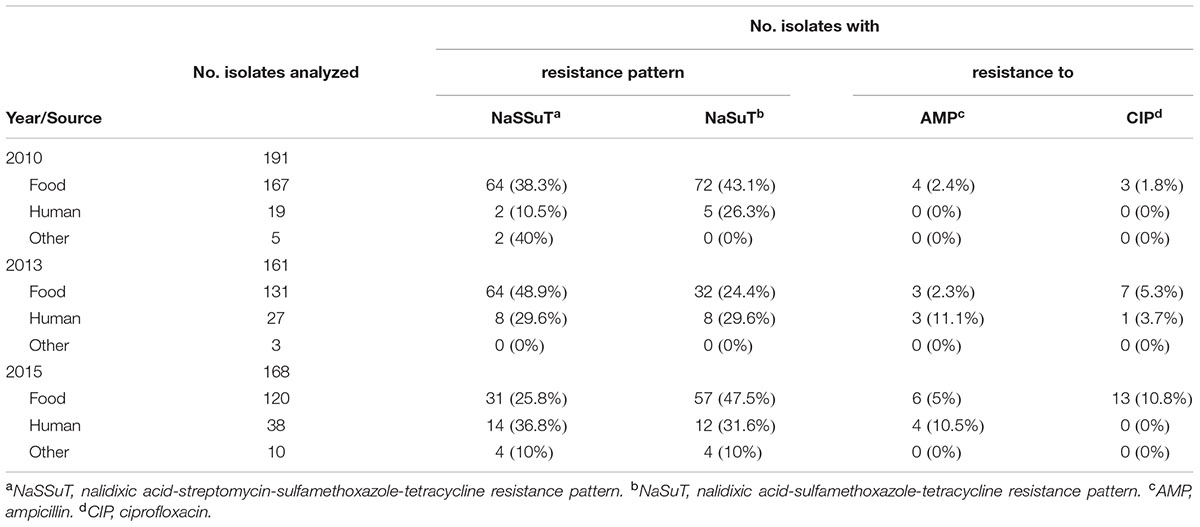
The distribution of resistance phenotypes among the S. serovar Infantis isolates is summarized in the Table 2 and shown in detail in technical Supplementary Tables S1–S3.
Multidrug resistant was detected in 389 (74.8%) of the isolates. Thereof, the majority (379/520, 72.9% of all isolates) showed a combined resistance pattern to nalidixic acid, sulfamethoxazole and tetracycline, with (NaSSuT) or without streptomycin (NaSuT). The NaSSut pattern was detected in 189 (36.3% of all isolates) and the NaSuT pattern in 190 (36.5% of all isolates). Resistance to ampicillin and ciprofloxacin was verified in 20 (3.8%) and 24 (4.6%) of the isolates, respectively, and showed a rising prevalence between 2010 and 2015 for both antimicrobials, as illustrated in Figure 1. A total of 496 (95.4% of all isolates) tested within the intermediate range for ciprofloxacin, according to CLSI breakpoints. An ESBL phenotype and growth on BrillianceTMESBL agar was recorded for three strains (0.6% of all isolates), whereof two originated from humans (isolates 21-13 and 125-15) and one (isolate 144-13) from food (Table 3 and Supplementary Tables S2, S3).
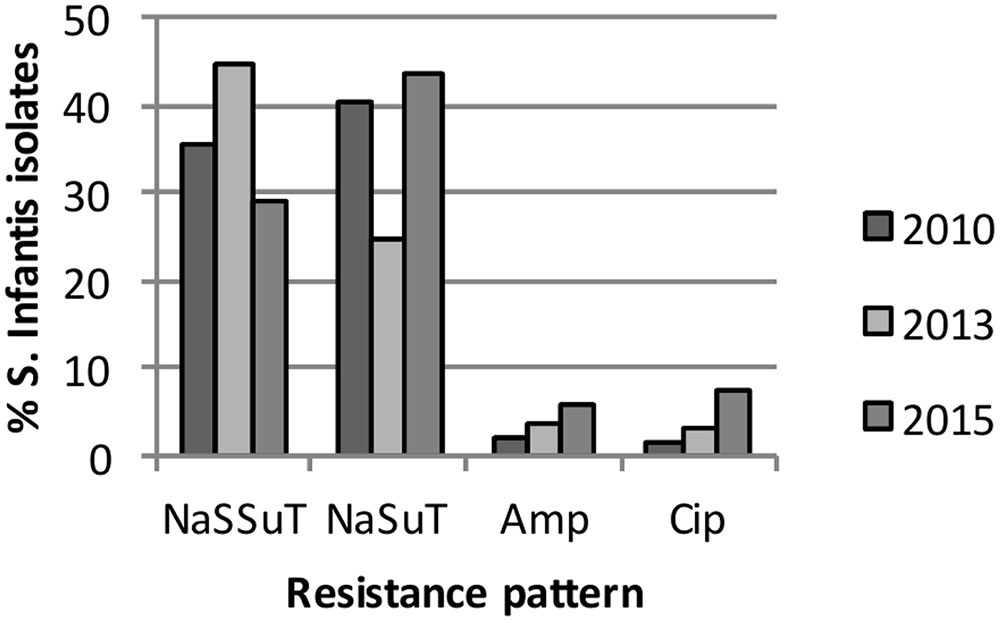
FIGURE 1. Percentage of S. serovar Infantis isolates with the nalidixic acid-sulfamethoxazole-tetracycline (NaSuT), or the nalidixic acid-streptomycin-sulfamethoxazole-tetracycline resistance pattern (NaSSuT) resistance pattern and resistance to ampicillin and ciprofloxacin in 2010, 2013 and 2015.
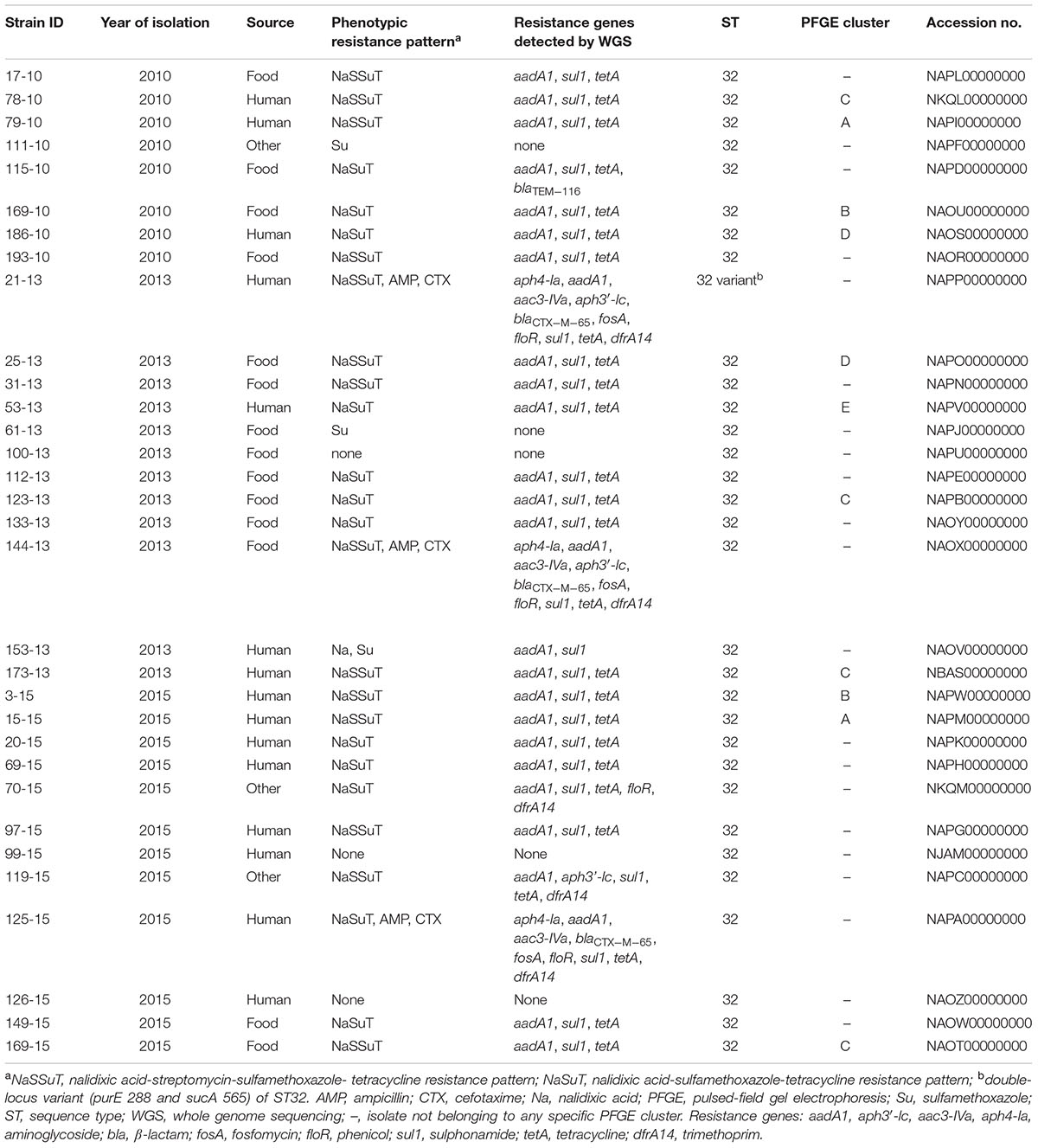
TABLE 3. Characteristics of 32 sequenced S. Infantis from food, diseased humans and other sources from 2010, 2013 and 2015 from Switzerland.
WGS
The genomic characteristics of the 32 genomes are listed in Supplementary Tables S1–S3. The average genome size, %G+C, and number of coding DNA sequences (CDS) of the isolates was 4.88 mbp (range:4.56–4.98 mbp), 52.2% (range: 52.1–52.3%), and 4812 CDS (range: 4443–4912 CDS), respectively.
Detection of Resistance Genes by PCR and by WGS
All three isolates displaying an ESBL phenotype showed the presence of the blaCTX-M-65 gene by PCR. WGS analysis using ResFinder confirmed the presence of the blaCTX-M-65 genes. All blaCTX-M-65-positive isolates carried other resistance genes aph4-la, aadA1 and aac3-IVa (aminoglycoside resistance), fosA (fosfomycin resistance), floR (phenicol resistance), sul1 (sulphonamide resistance), tetA (tetracycline resistance), and dfrA14 (trimethoprim resistance) (Table 3 and Supplementary Tables S2, S3), while strains 21-13 and 144-13 also carried the aph3′-lc gene (aminoglycoside resistance). WGS of 23 MDR, non-ESBL producing isolates revealed the presence of aadA1, sul1, and tetA resistance genes throughout. Food isolate 115-10 additionally carried the non-ESBL blaTEM-116 gene. The environmental isolate 70-15 from Israel additionally harbored floR and dfrA14 and MDR strain 119-15 isolated from animal feed also carried aph3′-lc and dfrA14 (Table 3 and Supplementary Table S3).
Whole genome sequencing did not detect any resistance genes in six non-MDR isolates (Table 3 and Supplementary Tables S1–S3), except for human isolate 153-13 which showed intermediate resistance to streptomycin and harbored aadA1 and sul1 (Table 3 and Supplementary Table S2).
Detection of Plasmids and Replicon Types among the Isolates
By mapping WGS assemblies to the 320 kb plasmid pCFSAN42077 from blaCTX-M-65 carrying food isolate CFSAN42077 (Tate et al., 2017), contigs or regions similar to this plasmid in isolates 21-13, 144-13 and 125-15 were detected (data not shown). Specific contigs bearing the blaCTX-M-65 gene and surrounding loci were identified (Figure 2). The ∼5 kb cassette consisted of blaCTX-M-65 followed by an IS5/IS1182 family transposase and a gene encoding a TonB-dependent siderophore receptor. The 5′-end is flanked by an IS6 family transposase upstream of the fipA gene, which is truncated in pCFSAN42077. By comparison, fipA is absent in p14026835, a blaCTX-M-65 harboring plasmid from human S. Infantis isolate 14026835 from Italy (ERR1014119), as shown in Figure 2. As predicted, no replicons were detected in the genomes of these three blaCTX-M-65 harboring isolates (Franco et al., 2015).
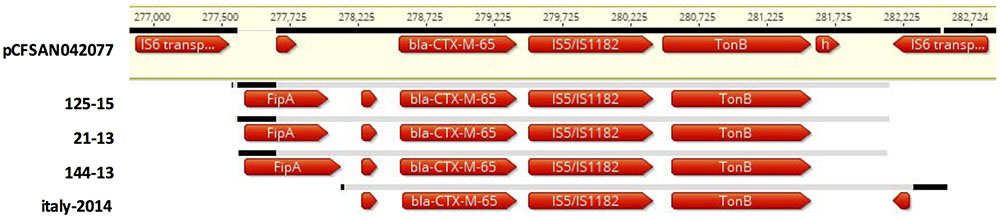
FIGURE 2. Multiple sequence alignment with pCSAM042077 from S. Infantis from food from the United States (FSIS150291), of a blaCTX-M-65 cassette on putative ∼320 kb plasmids from three S. Infantis strains from food (144-13) and diseased humans (21-13 and 125-15) from Switzerland and one S. Infantis strain from Italy (ERR1014119). The ∼5 kb cassette consists of blaCTX-M-65 followed by an IS5/IS1182 family transposase and a gene encoding a TonB-dependent siderophore receptor, and one or two hypothetical genes. The cassette is flanked on either side by the IS6 transposase. The fipA gene is of varied lengths, or absent.
For other isolates in this study, WGS analysis showed the presence of incompatibility group IncFII (p96A) plasmid (accession #JQ418521), in the fully susceptible human isolate 99-15 (N15-1280), Incl1 (accession #AP005147) in fully susceptible food isolate 100-13 (N13-1368), and pESA2 and IncFII (pCTU2) (accession nos. #CP000784 and #FN543095, respectively) in fully susceptible human isolate 126-15 (N15-1729) (data not shown).
PFGE Cluster Analysis, MLST and Phylogenetic Analysis
PFGE
Pulsed-field gel electrophoresis analysis revealed 190 distinct XbaI restriction patterns (all patterns are available upon request). Of the 520 isolates, 270 (51.9%) shared a similarity of 90%. Five clusters A–E showing a similarity of >93% and consisting of 121 (23.3% of total) isolates were detected. Isolates within a cluster had indistinguishable profiles. The clusters A–E contained 21 (4% of the total), 18 (3.5%), 27 (5.2%), 43 (8.3%) and 12 (2.3%) isolates, respectively. Isolates from sources other than human infections or food (e.g., environmental samples) were not detected within these clusters.
MLST and Phylogenetic Analysis
All 32 isolates subjected to WGS belonged to sequence type (ST) ST32, except isolate 21-13, which was a double-locus variant (purE and sucA) of ST32 (Table 3).
Genetic relatedness was investigated by mapping the 32 genomes to the genome sequences of isolates belonging to the Hungarian clone B from broilers, the blaESBL, and non-blaESBL harboring strains from Italy, Israel and United States. The resulting unrooted phylogenetic tree is shown in Figure 3. The isolates segregated into eight major clusters. Clusters determined by PFGE and by WGS did not correlate.
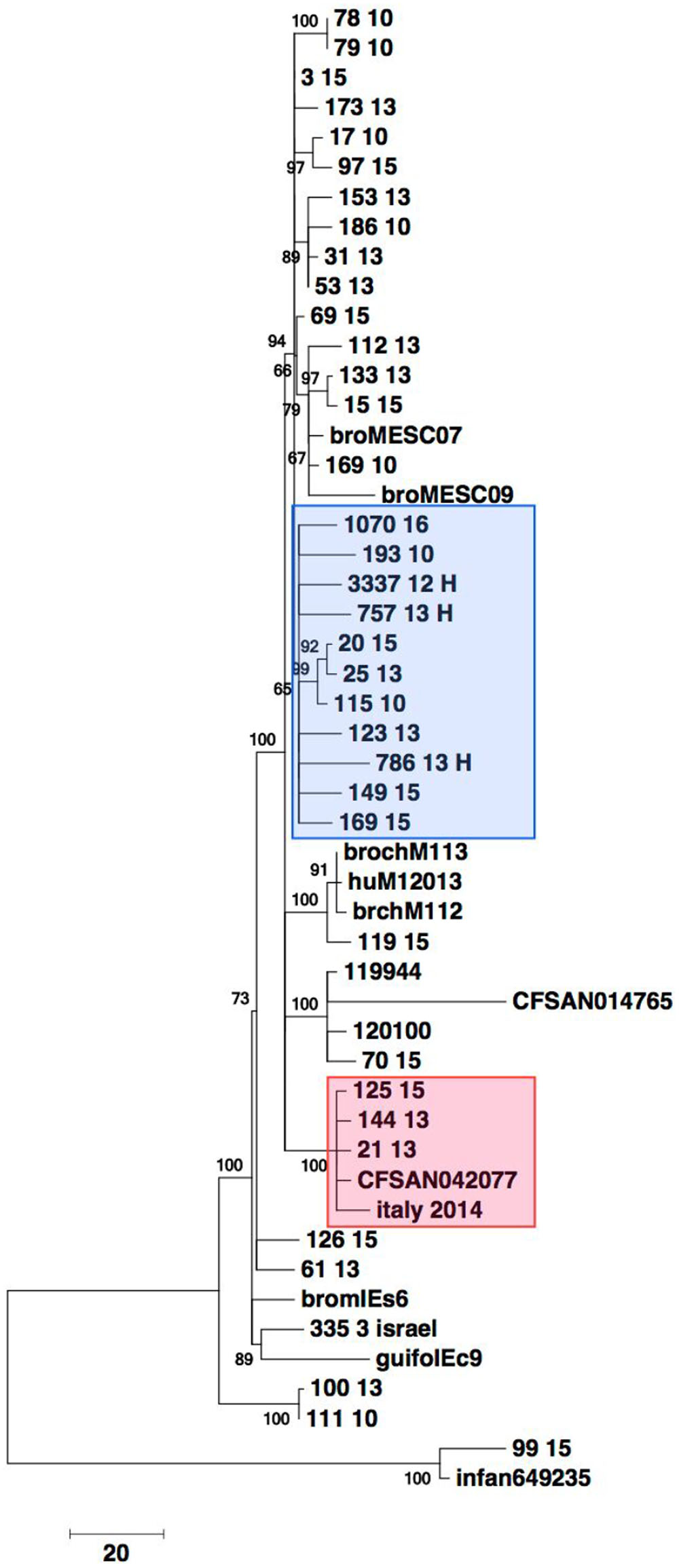
FIGURE 3. Core gene analysis based phylogeny of 32 selected S. serovar Infantis from food, diseased humans and environmental sources from 2010, 2013 and 2015 from Switzerland and of S. serovar Infantis from Hungary, Israel, Italy and United States. Alleles from 1500 randomly chosen core genes from more than 2770 conserved loci were used to build a phylogenetic tree. The data matrix of alleles was subject to multiple alignments. The phylogenetic tree was built using the Neighbor-Joining algorithm in the MEGA 7 suite with 9,562 positions across 50 genomes for the final analysis. Bar indicates 20 single nucleotide polymorphisms (SNP). Blue box: Isolates belonging to the Hungarian clone. Red box: Isolates harboring blaCTX-M-65.
Close phylogenetic relatedness with strains 3337 12 H, 757 13 H, 786 13H, 1070 16 belonging to the Hungarian clone B was detected in seven (21.9%) of the sequenced strains and included human and food isolates from 2010, 2013 and 2015 (Figure 3). The blaCTX-M-65 harboring isolates 21-13, 144-13 and 125-15 were more distantly related to the isolates that clustered with the Hungarian clone B. All three isolates were closely related to, and formed a distinct cluster with the blaCTX-M-65 harboring human isolate (italy 2014) from Italy and the food isolate CFSAN 042077 from the United States.
Four (12.5%) of the genomes (isolates 15-15, 169-10, 112-13 and 133-13, respectively) grouped in a distinct cluster with non-blaESBL, pESI-like harboring strains isolated previously from broiler meat in Italy. One environmental isolate (isolate 70-15) originating from Israel formed a cluster with three strains from Israel (pESI containing 120100 and 119944, and CFSAN014765). Two fully susceptible isolates (63-13 and 126-15) were placed in the phylogenetic tree together with the non-pESI containing strain 335 3 isolated in Israel in 1971 (Figure 3).
Accession Numbers
The accession numbers of the 32 sequenced isolates are listed in Table 3 and Supplementary Tables S1–S3. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession NAOX00000000. The version described in this paper is version NAOX00000000.1.
Discussion
S. serovar Infantis has emerged as an important disseminator of MDR in the food chain, representing a threat to human health (Nógrády et al., 2012). The majority of strains in this study exhibited either the NaSuT pattern or the characteristic NaSSuT pattern previously identified in the MDR S. Infantis clone B that emerged in broilers in farms in Hungary and subsequently spread within and outside of Europe, including to the countries of Poland, Austria, Germany, Israel and Japan (Hauser et al., 2012; Nógrády et al., 2012). WGS performed in this study detected clonality in strains deemed different by PFGE and provides evidence that the Hungarian clone B has persisted since at least 2010 in Switzerland within the food chain and is associated with human disease. Strains belonging to three other closely related clusters were less prevalent but showed similar resistance patterns and persistence within food and human isolates.
In addition, we observed the occurrence of S. serovar Infantis harboring blaCTX-M-65. This bla gene has been detected previously in one human S. serovar Infantis isolate in Great Britain and in one from Italy (Burke et al., 2014), and furthermore, in an outbreak of S. Infantis in Ecuador (Cartelle Gestal et al., 2016), as well as in food in the United States. The results presented in this study show that the blaCTX-M-65 harboring S. serovar Infantis from Switzerland belong to a unique lineage but are similar to the strains from Italy and the United States, suggesting the emergence of a blaCTX-M-65 harboring, MDR S. serovar Infantis lineage in Europe as well as in North and South America. Moreover, this study shows that this lineage has been present in food and humans at least as early as 2013 in Europe.
Whole genome sequencing of these strains indicate the presence of a ∼320 kb plasmid similar to the pESI plasmid, a megaplasmid carrying multiple resistance and virulence genes originally detected in S. serovar Infantis in Israel in 2008 (Aviv et al., 2014). In addition, resistance genes drfA14 and fosA (trimethoprim and phenicol resistance genes, respectively) which are also found on pESI-like plasmids (Franco et al., 2015), were detected in all blaCTX-M-65 harboring strains. Several other non-blaESBL harboring strains from this study had a close phylogenetic-relatedness to strains from Israel and Italy which harbor non-blaESBL pESI or pESI-like plasmids (Aviv et al., 2014), suggesting that certain S. serovar Infantis clones or lineages are acquiring these plasmids independently.
Our results correlate with the recent detection of a clone harboring a pESI-like plasmid and the blaCTX-M-1 gene in the broiler industry and humans in Italy (Franco et al., 2015), and suggest that some S. serovar Infantis clones harboring pESI-like plasmids may be undergoing a microevolution by acquiring blaESBL genes. While in Europe blaCTX-M-1 is prevalent within the poultry industry and has been well documented (Zurfluh et al., 2014), blaCTX-M-65 has rarely been described. By contrast, it is a prevalent blaESBL gene in animal and human E. coli strains and S. serovar Indiana isolates in China (Bai et al., 2016), from where it may have disseminated via horizontal transfer. However, further studies are needed to clarify the origins of this blaESBL gene and to characterize and compare the plasmids carrying blaESBL genes in S. serovar Infantis.
The emergence of MDR S. Infantis with resistance to third generation cephalosporins in food and in humans is of great concern, particularly since these strains show intermediate resistance to ciprofloxacin. The use of fluoroquinolones for the treatment of infections caused by such strains may be associated with unfavorable treatment outcomes and the selection of high-level ciprofloxacin resistance (Humphries et al., 2012).
This study extends our knowledge on clones of S. serovar Infantis circulating in food and causing disease in humans and provides evidence for the emergence of an MDR, ESBL-producing clone harboring blaCTX-M-65 in Switzerland. Our results highlight the necessity of strategies to reduce the prevalence of S. serovar Infantis within the food producing industry.
Author Contributions
DH, BT, and RS designed the study. KZ, DH, and DA carried out the microbiological and molecular biological tests. GG, HC, FN, and BT carried out whole genome sequencing. MN-I, RS, DH, GG, and BT analyzed and interpreted the data. MN-I drafted the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was partly supported by the Swiss Federal Office of Public Health, Division Communicable Disease.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01322/full#supplementary-material
Footnotes
- ^http://www.ncbi.nlm.nih.gov/blast/
- ^http://www.cdc.gov/pulsenet/protocols.htm
- ^https://cge.cbs.dtu.dk/services/MLST/
- ^https://cge.cbs.dtu.dk/services/ResFinder/
- ^https://cge.cbs.dtu.dk/services/PlasmidFinder/
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Aviv, G., Rahav, G., and Gal-Mor, O. (2016). Horizontal Transfer of the Salmonella enterica serovar Infantis resistance and virulence plasmid pESI to the gut microbiota of warm-blooded hosts. mBio 7:e1395-16. doi: 10.1128/mBio.01395-16
Aviv, G., Tsyba, K., Steck, N., Salmon-Divon, M., Cornelius, A., Rahav, G., et al. (2014). A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar infantis strain. Environ. Microbiol. 16, 977–994. doi: 10.1111/1462-2920.12351
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bai, L., Zhao, J., Gan, X., Gan, X., Wang, J., Zhang, X., et al. (2016). Emergence and diversity of Salmonella enterica serovar Indiana isolates with concurrent resistance to ciprofloxacin and cefotaxime from patients and food-producing animals in China. Antimicrob. Agents Chemother. 60, 3365–3371. doi: 10.1128/AAC.02849-15
Burke, L., Hopkins, K. L., Meunier, D., de Pinna, E., Fitzgerald-Hughes, D., Humphreys, H., et al. (2014). Resistance to third-generation cephalosporins in human non-typhoidal Salmonella enterica isolates from England and Wales, 2010-12. J. Antimicrob. Chemother. 69, 977–981. doi: 10.1093/jac/dkt469
Cartelle Gestal, M., Zurita, J., Paz, Y., Mino, A., Ortega-Paredes, D., and Alcocer, I. (2016). Characterization of a small outbreak of Salmonella enterica serovar Infantis that harbour CTX-M-65 in Ecuador. Braz. J. Infect. Dis. 20, 406–407. doi: 10.1016/j.bjid.2016.03.007
CLSI. (2016). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-sixth Informational Supplement M100S. Wayne, PA: Clinical and Laboratory Standards Institute.
Dionisi, A. M., Lucarelli, C., Benedetti, I., Owczarek, S., and Luzzi, I. (2011). Molecular characterisation of multidrug-resistant Salmonella enterica serotype Infantis from humans, animals and the environment in Italy. Int. J. Antimicrob. Agents 38, 384–389. doi: 10.1016/j.ijantimicag.2011.07.001
European Food Safety Authority [EFSA] and European Centre for Disease Prevention [ECDC]. (2016). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2014. EFSA J. 14, 4380. doi: 10.2903/j.efsa.2016.4380
Franco, A., Leekitcharoenphon, P., Feltrin, F., Alba, P., Cordaro, G., Iurescia, M., et al. (2015). Emergence of a clonal lineage of multidrug-resistant ESBL-producing Salmonella infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS ONE 10:e0144802. doi: 10.1371/journal.pone.0144802
Geser, N., Stephan, R., Korczak, B. M., Beutin, L., and Hächler, H. (2012). Molecular identification of extended-spectrum-β-lactamase genes from Enterobacteriaceae isolated from healthy human carriers in Switzerland. Antimicrob. Agents Chemother. 56, 1609–1612. doi: 10.1128/AAC.05539-11
Grimont, P. A. D., and Weill, F. X. (2008). Antigenic Formulae of the Salmonella Serovars. Paris: Institut Pasteur.
Hauser, E., Tietze, E., Helmuth, R., Junker, E., Prager, R., Schroeter, A., et al. (2012). Clonal dissemination of Salmonella enterica serovar Infantis in Germany. Foodborne Pathog. Dis. 9, 352–360. doi: 10.1089/fpd.2011.1038
Humphries, R. M., Fang, F. C., Aarestrup, F. M., and Hindler, J. A. (2012). In vitro susceptibility testing of fluoroquinolone activity against Salmonella: recent changes to CLSI standards. Clin. Infect. Dis. 55, 1107–1113. doi: 10.1093/cid/cis600
Kaur, J., Chopra, S., Sheevani, and Mahajan, G. (2013). Modified double disc synergy test to detect ESBL production in urinary isolates of Escherichia coli and Klebsiella pneumoniae. J. Clin. Diagn. Res. 7, 229–233. doi: 10.7860/JCDR/2013/4619.2734
Kumar, S. G., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Leekitcharoenphon, P., Lukjancenko, O., Friis, C., Aarestrup, F. M., and Ussery, D. W. (2012). Genomic variation in Salmonella enterica core genes for epidemiological typing. BMC Genomics 13:88. doi: 10.1186/1471-2164-13-88
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O’Brien, S. J., et al. (2010). The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
Medalla, F., Gu, W., Mahon, B. E., Judd, M., Folster, J., Griffin, P. M., et al. (2016). Estimated incidence of antimicrobial drug-resistant nontyphoidal Salmonella infections, United States, 2004-2012. Emerg. Infect. Dis. 23, 29–37. doi: 10.3201/eid2301.160771
Mølbak, K. (2005). Human health consequences of antimicrobial drug-resistant Salmonella and other foodborne pathogens. Clin. Infect. Dis. 41, 1613–1620. doi: 10.1086/497599
Nógrády, N., Kardos, G., Bistyák, A., Turcsányi, I., Mészáros, J., Galántai, Z., et al. (2008). Prevalence and characterization of Salmonella infantis isolates originating from different points of the broiler chicken-human food chain in Hungary. Int. J. Food Microbiol. 127, 162–167. doi: 10.1016/j.ijfoodmicro.2008.07.005
Nógrády, N., Király, M., Davies, R., and Nagy, B. (2012). Multidrug resistant clones of Salmonella infantis of broiler origin in Europe. Int. J. Food Microbiol. 157, 108–112. doi: 10.1016/j.ijfoodmicro.2012.04.007
Olasz, F., Nagy, T., Szabó, M., Kiss, J., Szmolka, A., Barta, E., et al. (2015). Genome sequences of three Salmonella enterica subsp. enterica serovar infantis strains from healthy broiler chicks in Hungary and in the United Kingdom. Genome Announc. 3:e1468-14. doi: 10.1128/genomeA.01468-14
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2013). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Tate, H., Folster, J. P., Hsu, C. H., Chen, J., Hoffmann, M., Li, C., et al. (2017). Comparative analysis of extended spectrum beta-lactamase CTX-M-65-producing Salmonella infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob. Agents Chemother. 61:e488-17. doi: 10.1128/AAC.00488-17
Wilk, T., Szabó, M., Szmolka, A., Kiss, J., Barta, E., Nagy, T., et al. (2016). Genome sequences of multidrug-resistant Salmonella enterica subsp. enterica serovar Infantis strains from broiler chicks in Hungary. Genome Announc. 4:e1400-16. doi: 10.1128/genomeA.01400-16
Wilk, T., Szabó, M., Szmolka, A., Kiss, J., Olasz, F., and Nagy, B. (2017). Genome sequences of Salmonella enterica subsp. enterica serovar Infantis strains from Hungary representing two peak incidence periods in three decades. Genome Announc. 5:e1735-16. doi: 10.1128/genomeA.01735-16
Woodford, N., Fagan, E. J., and Ellington, M. J. (2006). Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J. Antimicrob. Chemother. 57, 154–155.
Zurfluh, K., Nüesch-Inderbinen, M., Morach, M., Berner, A. Z., Hächler, H., and Stephan, R. (2015). Extended-spectrum ß-lactamase-producing-Enterobacteriaceae in vegetables imported from the Dominican Republic, India, Thailand and Vietnam. Appl. Environ. Microbiol. 81, 3115–3120. doi: 10.1128/AEM.00258-15
Zurfluh, K., Wang, J., Klumpp, J., Nüesch-Inderbinen, M., Fanning, S., and Stephan, R. (2014). Vertical transmission of highly similar blaCTX-M-1-harboring IncI1 plasmids in Escherichia coli with different MLST types in the poultry production pyramid. Front. Microbiol. 5:519. doi: 10.3389/fmicb.2014.00519
Keywords: Salmonella Infantis, Hungarian clone blaCTX-M-65, food, humans
Citation: Hindermann D, Gopinath G, Chase H, Negrete F, Althaus D, Zurfluh K, Tall BD, Stephan R and Nüesch-Inderbinen M (2017) Salmonella enterica serovar Infantis from Food and Human Infections, Switzerland, 2010–2015: Poultry-Related Multidrug Resistant Clones and an Emerging ESBL Producing Clonal Lineage. Front. Microbiol. 8:1322. doi: 10.3389/fmicb.2017.01322
Received: 15 May 2017; Accepted: 29 June 2017;
Published: 13 July 2017.
Edited by:
Miklos Fuzi, Semmelweis University, HungaryReviewed by:
Sebastian Guenther, Freie Universität Berlin, GermanyLiang Li, Los Angeles Biomedical Research Institute, United States
Copyright © 2017 Hindermann, Gopinath, Chase, Negrete, Althaus, Zurfluh, Tall, Stephan and Nüesch-Inderbinen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roger Stephan, c3RlcGhhbnJAZnNhZmV0eS51emguY2g=
 Denise Hindermann1
Denise Hindermann1 Gopal Gopinath
Gopal Gopinath Hannah Chase
Hannah Chase Katrin Zurfluh
Katrin Zurfluh Ben D. Tall
Ben D. Tall Roger Stephan
Roger Stephan Magdalena Nüesch-Inderbinen
Magdalena Nüesch-Inderbinen