- 1Infectious Disease Biology, Institute of Life Sciences, Bhubaneswar, India
- 2Bioinformatics Center, Institute of Life Sciences, Bhubaneswar, India
We examined the presence of virulence and antibiotic resistance genes, SCCmec types and determined the genomic diversity among ocular S. epidermidis isolates (patients-23, healthy controls-29). PCR determined the presence of antibiotic resistance genes, virulence genes and SCCmec types among all isolates. MLST and PFGE determined the genomic relatedness among them. All isolates of S. epidermidis showed resistance to at least one class of antibiotics of which 48 isolates were multidrug resistant and carried ARGs. Thirty-five isolates were methicillin resistant and carried mecA gene. Majority of the isolates were resistant to fluoroquinolones and showed mutation in gyrA, parC, and parE genes, however, few isolates showed additional novel mutations in parC gene. Of the MRSE strains, 17 strains carried SCCmec type IV, four type V, two type II, and two UT4. Seven strains carried novel combination of ccr complex and SCCmercury element, not reported earlier. All the S. epidermidis strains harbored icaA and icaD genes, 47 carried ACME operon, and 50 contained IS256. A noteworthy finding was the presence of ST179 among 43% of infected eye isolates an observation rarely reported among S. epidermidis. PFGE and MLST analysis showed genomic diversity among them. Statistical analysis suggests that few healthy conjunctiva isolates had characteristics similar to infected eye isolates. S. epidermidis strains carrying mecA gene are multidrug resistant, virulent and diverse irrespective of sources of isolation. IS256 cannot be used as marker to differentiate isolates of infected eye from healthy conjunctiva.
Introduction
Staphylococcus epidermidis an opportunistic pathogen and member of coagulase negative Staphylococci (CoNS) are normal inhabitant of human skin, mucosal and ocular surfaces and cause hospital acquired infections (Graham et al., 2007; Rogers et al., 2009; Le et al., 2014). This organism can cause number of ocular diseases like bacterial endophthalmitis conjunctivitis, blepharitis, and keratitis (Melo et al., 2011; Schimel et al., 2013; Bispo et al., 2014; Park et al., 2015). The predisposing risk factors associated with infections are mostly use of contact lenses, ocular surgery, and ocular inflammatory diseases (Bourcier et al., 2003; Keay et al., 2006; Park et al., 2015). Although methicillin is not used in the treatment of eye infections, increasing resistance to methicillin is known to contribute significantly to the spread and persistence of multidrug-resistant strains in a given setting (Alekshun and Levy, 2007; Asbell et al., 2008; Cavuoto et al., 2008; Lichtinger et al., 2012). The alteration in penicillin-binding protein (PBP2a) confers resistance to methicillin (Hartman and Tomasz, 1984). The mecA gene encodes for PBP2a and is present on the mobile genetic element termed as staphylococcal cassette chromosome mec (SCCmec) that carries a set of recombinase genes (ccr) (Katayama et al., 2000). A combination of class of mec gene complex and the ccr gene complex types determines the SCCmec types and diversity among staphylococci (Kondo et al., 2007; Bloemendaal et al., 2010; Martínez-Meléndez et al., 2016). Methicillin-resistant S. epidermidis (MRSE) carrying untypeable SCCmec element are often reported (Ruppé et al., 2009; McManus et al., 2015).
Resistance to fluoroquinolones occurs due to mutation(s) in the quinolone resistance determining region (QRDR) of gyrA, gyrB, parC and parE (Yamada et al., 2008). There are three accomplished mechanisms of macrolide resistance. (i) Modification of the target site, (ii) efflux of antibiotics, and (iii) drug inactivation. Plasmid-borne drug-resistance genes encoding for ABC transporter superfamily efflux pump and msrA can mediate the export of macrolide (Leclercq, 2002). Other plasmid located genes, tetK confers resistance to tetracycline and pC194, pC221, and pC223 inactivates chloramphenicol by converting it to 3- acetyl and 1,3-diacetyl derivatives (Trieu-Cuot et al., 1993; Trzcinski et al., 2000).
Pathogenicity of S. epidermidis is attributed to its ability to form biofilm comprised of polysaccharide, protein and eDNA, which confers resistance to antibiotics. During biofilm formation bacteria first, adhere onto abiotic polymeric substances mediated by polysaccharide intercellular adhesin (PIA) encoded by icaADBC locus (Schommer et al., 2011). Arginine catabolic mobile element (ACME) also play a significant role in the pathogenicity by enhancing the fitness of the bacteria by evading the host immune system (Otto, 2009).
Several molecular typing methods have been used to study epidemiology and clonal relationships of S. epidermidis; however, not a single technique alone could discriminate the bacteria because of differences in the degree of typeability, reproducibility, and discriminatory power (Tenover et al., 1994). Pulsed-Field Gel Electrophoresis (PFGE) is known as a “gold standard” and widely used for studying the epidemiological investigations (Hookey et al., 1998). Multi-Locus Sequence Typing (MLST) is currently used for long-term evolutionary research and to study population structure of S. epidermidis isolates (Bloemendaal et al., 2010; Martínez-Meléndez et al., 2016). Although S. epidermidis is rated as one of the emerging ocular pathogens and showing resistance to antibiotics; not much information is available on their molecular diversity. It is not clear how the isolates from healthy conjunctiva are related to and cause ocular infection.
We analyzed antibiotic resistance and virulence profiles of S. epidermidis isolated from infected eye and healthy conjunctiva during 2007–2011 at L.V. Prasad Eye Institute, Bhubaneswar, India. We used PFGE and MLST to determine the genomic diversity and correlated results with antibiotic resistance, SCCmec type, and virulence genes, respectively. Genetic and phenotypic data was used to determine whether isolates from healthy conjunctiva had characteristics similar to infected eye isolates.
Materials and Methods
Ethical Statement
The study was approved by Institutional Review Board (IRB) of LV Prasad Eye Institute (LEC/08/110/2009), and the data were analyzed anonymously and reported.
Bacterial Strains
Twenty-three non-duplicate S. epidermidis were isolated from patients with a variety of ocular infections at L.V. Prasad Eye Institute, Bhubaneswar, India, during 2007–2011. These isolates were from patients with keratitis (n = 13), endophthalmitis (n = 2), conjunctivitis (n = 2), marsupialization of cyst (n = 1), chronic dacryocystitis (n = 1), traumatic cataract (n = 1), canaliculitis (n = 1), blepharitis (n = 1) and graft infiltrate (n = 1). Also, 29 strains were isolated from asymptomatic healthy conjunctiva. The study was conducted following the guidelines mentioned in the Declaration of Helsinki. All the 52 isolates were identified by using biochemical tests including Gram staining, catalase production, fermentation of glucose and mannitol and ID32 STAPH strips using ATB™ NEW v.1.0.0 software on an ATB™ reader (bioMerieux, France). The amplification of S. epidermidis nuc gene confirmed the identity of isolates (Hirotaki et al., 2011). S. epidermidis strains ATCC 35984 and ATCC 12228 obtained from American Type Culture Collection (ATCC, Manassas, VA), were used as controls in this study. Staphylococcus aureus ATCC 25293 and Pseudomonas aeruginosa ATCC 27853 was used as quality control strains for antibiotic susceptibility testing.
Antibiotic Resistance and Genetic Characteristics
Antibiotic susceptibility was performed with S. epidermidis for 13 antibiotics; oxacillin, penicillin, chloramphenicol, ciprofloxacin, ofloxacin, gatifloxacin, moxifloxacin, erythromycin, clindamycin, gentamicin, tetracycline, vancomycin, and cefazolin by microbroth dilution methods according to the Clinical Laboratory Standard Institute (CLSI) guidelines (Clinical and Laboratory Standards Institute, 2015). The interpretation criteria for each antibiotic tested were those published previously as CLSI document M100-S25. Isolates showing resistance to three or more than three class of antibiotics were referred here as multidrug resistant (MDR).
Using published methods, PCR determined the presence of blaZ, catpC221, catpC223, ermC, msrA, mphC, aac6′-aph2′, aph3′, and tetK genes encoding for penicillin, chloramphenicol, erythromycin/clindamycin, gentamicin and tetracycline in both phenotypic-resistant and -susceptible strains of S. epidermidis (Schmitz et al., 1999; Schlegelova et al., 2008; Argudin et al., 2011; Duran et al., 2012). We determined the mutations in gyrA, gyrB, parC, and parE genes by sequencing the PCR product (Yamada et al., 2008).
PCR tested all isolates for the presence of icaA, icaD, arcA, and opp3AB genes to assess the presence of ica and ACME operons (Arciola et al., 2001; Diep et al., 2008). Each strain was given a particular ACME type depending on the presence of either of the genes (Hellmark et al., 2013). Also PCR determined the presence of insertion sequence element IS256 (Gu et al., 2005).
PFGE, SCCmec Typing, and MLST
PFGE of S. epidermidis genomic DNA digested with SmaI was carried out by the protocol described for S. aureus by Centre for Disease Control and Prevention, Atlanta, USA. Banding patterns were determined by using Bionumerics software, version 7.1 (Applied Maths, Belgium) using the Dice index and un-weighted pair group method with arithmetic average (UPGMA) with 0.5% optimization and 1.5% position tolerance. Isolates showing similarity coefficient of up to 80% were considered belonging to similar pulsotype (Van Belkum et al., 2007).
Two Multiplex PCRs (MPCR-1 and MPCR-2) were performed by the method described previously using specific primers and assigned SCCmec type based on the combination of mec and ccr complexes (Kondo et al., 2007; Zong et al., 2011). Also, PCR determined the presence of SCCmercury element (Kondo et al., 2007).
Internal fragments of seven housekeeping genes, arcC, aroE, gtr, mutS, pyr, tpi, and yqil were amplified using specific primers (Thomas et al., 2007). Amplified fragments were purified using ExoSAP-IT (Affymetrix) and performed sequencing using Life Technology ABI sequencer model 3537 at the sequencing facility of Institute of Life Sciences, Bhubaneswar, India. Sequence alignments were done using MEGA 6. The sequences of the housekeeping genes were analyzed using Bionumerics software version 7.1, and a particular sequence type (ST) was assigned based on the nucleotide polymorphism. Minimum Spanning Tree (MST) was constructed using “MST for categorical data” as analysis template and accordingly assigned ST. We performed partitioning analysis according to the eBURST algorithm. Clonal complex (CC) analysis was performed according to the classification described by Miragaia et al. (2007).
Indexing Diversity
Simpson's index of diversity (SID) was calculated for all the three typing techniques (PFGE, MLST, SCCmec typing) as described by Simpson (1949).
Statistical Analysis
We performed Principal Coordinates Analysis (PCoA) and Discriminant Analysis (DA) using PAST program v2.17 (Hammer et al., 2001). We carried out the discriminant analysis using default values to confirm the hypothesis whether two groups of isolates are different. All the genotypic and phenotypic data of the strains are given in the supplementary material (Tables S1, S2).
Results
Antibiotic Resistance Pattern
Of the 52 strains, 35 strains were resistant to oxacillin and carried mecA gene (Table 1). However, all isolates were resistant to one or more than one class of antibiotics and showed 32 resistance patterns (Table S3). None of the isolates were resistant to vancomycin or cefazolin. Forty-eight strains were multidrug resistant (Table 1) comprising all isolates of MRSE from both sources, all MSSE from the infected eye and six from healthy conjunctiva (Table S3). Twenty-one isolates from the infected eye and 13 from healthy conjunctiva showed resistance to fluoroquinolones. However, two from healthy conjunctiva showed intermediate resistance to ofloxacin and ciprofloxacin (Table 2).
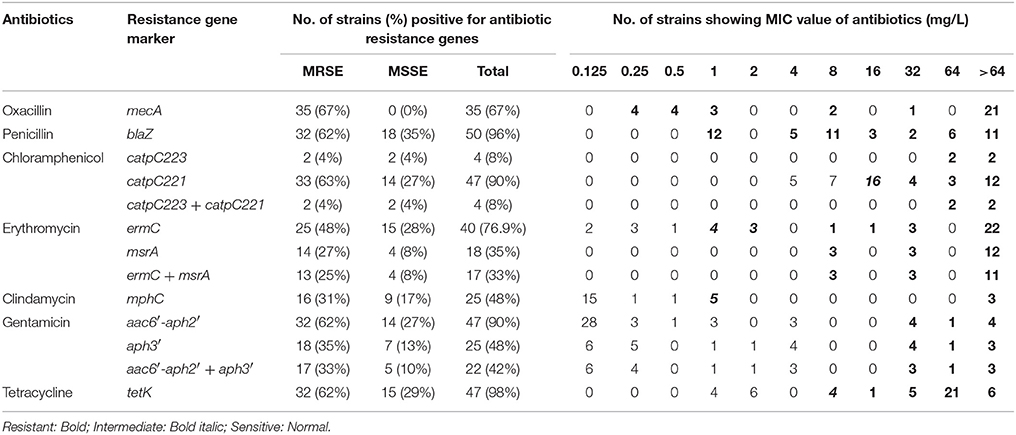
Table 1. Minimum inhibitory concentration and presence of antibiotic resistance genes of methicillin resistant and susceptible S. epidermidis isolated from the infected eye and healthy conjunctiva.
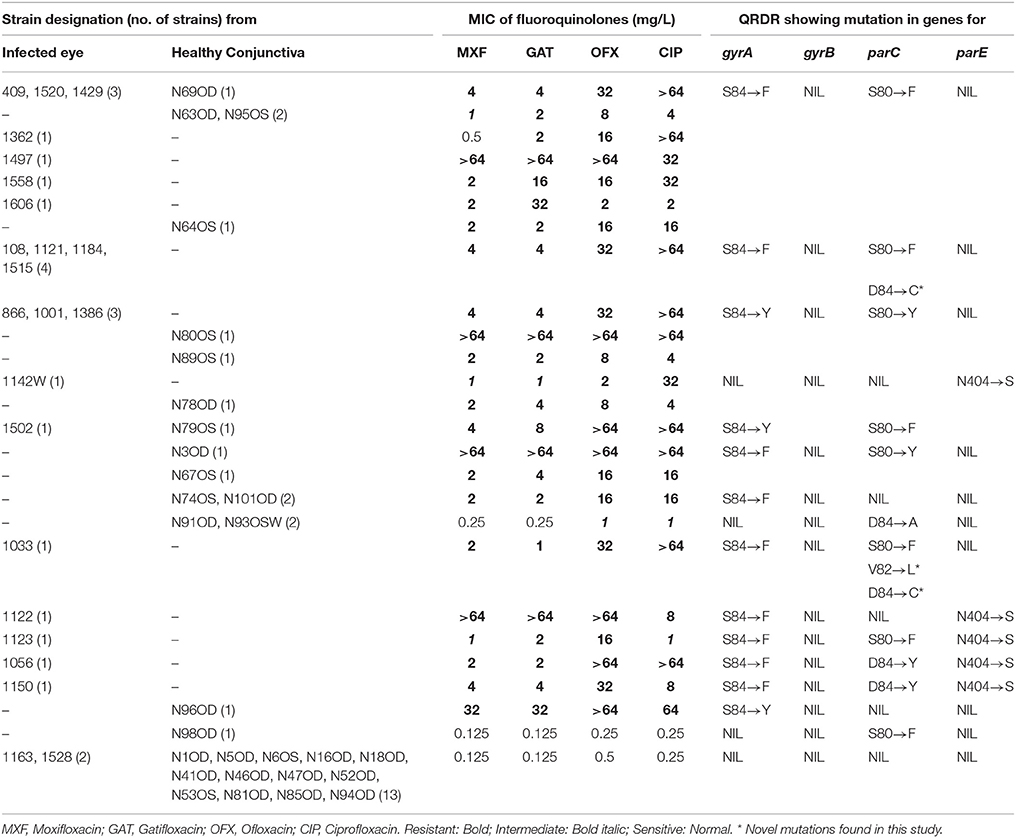
Table 2. Results of MIC obtained with fluoroquinolones and mutations identified in gyrA, gyrB, parC, and parE among S. epidermidis strains isolated from the infected eye and healthy conjunctiva.
Antibiotic Resistance Genes (ARG) and Gyrase Mutation
Majority of S. epidermidis isolates showing phenotypic resistance to multiple antibiotics carried ARGs (Table 1). Of the 36 fluoroquinolone resistant S. epidermidis, 32 strains comprising 20 from the infected eye and 12 from healthy conjunctiva showed mutations S84 → F/Y in gyrA gene, but none in the gyrB gene (Table 2). Similarly, 31 strains comprising 19 isolates from the infected eye and 12 from healthy conjunctiva showed a known mutation in parC gene. All isolates showing high MIC value for fluoroquionolones (especially that ≥ 64 mg/L) had mutation in either gyrA gene or gyrA and parC genes. In addition, five isolates from infected eye also showed novel mutation D84 → C and one to V82 → L in parC, not reported earlier. Of the six strains, five isolates from the infected eye and one from healthy conjunctiva showed mutation N404 → S in the parE gene (Table 2).
Virulence Profile of S. epidermidis
All the S. epidermidis strains harbored icaA and icaD genes. Of the 52 isolates, 20 from infected eye and 27 from healthy conjunctiva contained ACME operon (Table 4). Similarly 22 isolates from the infected eye and 28 from healthy conjunctiva amplified a fragment of IS256 indicating the presence of insertion sequence in S. epidermidis (Table S4, Figure 2). These observations thus suggest that virulence determinants were present in S. epidermidis strains, irrespective of sources of isolation.
SCCmec Typing
Of the MRSE strains, 25 belong to known SCCmec types of which 17 strains carried SCCmec type IV, four type V, two type II, and two carried UT4 (Table 3, Figure 1). The remaining 10 MRSE isolates showed four different combinations of mec complex and ccr complex, not reported earlier and thus do not fall within the traditional classification scheme. Of these strains, seven carried multiple ccr complexes and SCC mercury element (SCCmer). One strain from infected eye belonging to UT4 was also positive for SCCmer (Table 3, Figure 1). Although ten of the 17 MSSE isolates failed to amplify mec and ccr complex, five isolates carried ccrA2B2, and one isolate carried ccrC1 but lacked mec complex. One of the MSSE isolates carried more than one ccr type (A2B2, C1) and SCCmer (Table 3, Figure 1). Strains belonging to unreported SCCmec types (UT12 – UT14) showed a high level of oxacillin resistance (data not shown).
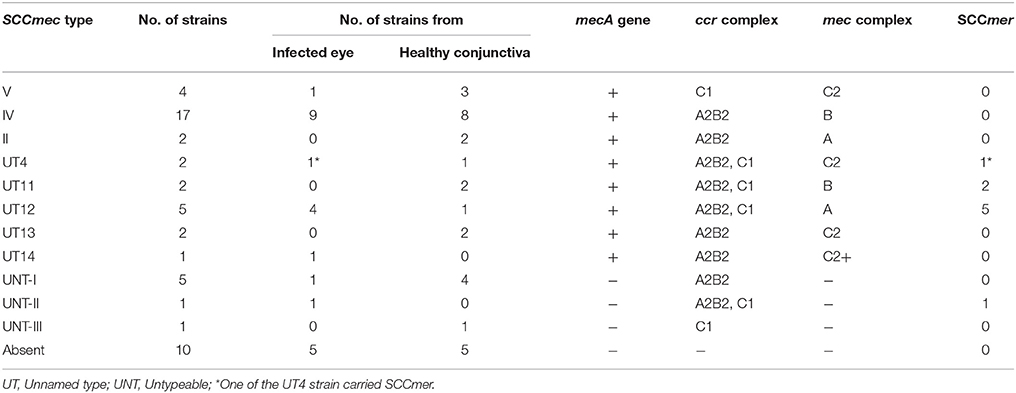
Table 3. Distribution of SCCmec types and SCCmer element among S. epidermidis strains isolated from the infected eye and healthy conjunctiva.
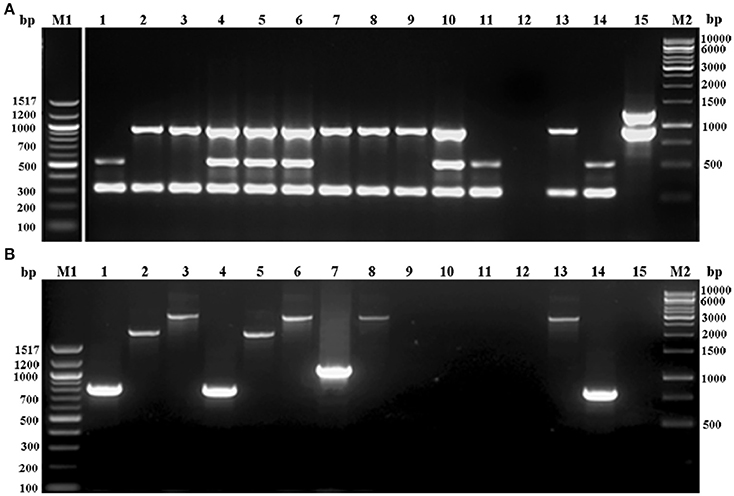
Figure 1. Agarose gel electrophoresis of multiplex PCR employed for (A) ccr complex typing (MPCR1), (B). mec complex typing (MPCR2) of S. epidermidis strains. Lane M1: 100bp ladder; lane 1: S. epidermidis strain 866; lane 2: S. epidermidis strain 1056; lane 3: S. epidermidis strain N6OS; lane 4: S. epidermidis strain 1150; lane 5: S. epidermidis strain N91OD; lane 6: S. epidermidis strain 108; lane 7: S. epidermidis strain N96OD; lane 8: S. epidermidis strain 1558; lane 9: S. epidermidis strain N89OD; lane 10: S. epidermidis strain 1386; lane 11: S. epidermidis strain N81OD; lane 12: S. epidermidis strain 1163; lane 13: S. aureus strain Mu50; lane 14: S. epidermidis strain 1295; lane 15: S. epidermidis strain 12228; lane M2, 1kb ladder.
Genomic Diversity
SmaI restriction digestion of DNA from S. epidermidis generated 14–21 bands, classifying the isolates into 35 pulsotypes, 16 subtypes, and three identical pairs (Table 4, Figure 2). Of the 52 S. epidermidis strains, 47 comprising 20 from infected eye and 27 from healthy conjunctiva harbored ACME operon of which 33 belonged to ACME type I, 11 to type II, and three to type III, respectively (Table 4). MLST analysis of the S. epidermidis strains identified 28 distinct STs of which ST179 was found in 43% of infected eye isolates followed by ST59 that was found in 13% of healthy conjunctiva isolates (Table 4). Partitioning analysis of the MST generated two CCs of which CC59 included 12 STs (ST59, ST6, ST210, ST48, ST291, ST142, ST153, ST89, ST5, ST384, ST280 and ST2) and CC197 includes two STs (ST197 and ST564). ST6 seems to be the founder of CC59 according to the eBURST algorithm. The remaining 14 STs were singletons of which ST179 comprised of 11 strains, ST69 and ST173 three strains, ST11 two strains, ST4, ST10, ST19, ST44, ST72, ST183, ST325, ST347, ST488, and ST490 one strain each (Table 4, Figure 3). A good correlation was found between high level resistance to fluoroquinolones and major sequence type, ST179.
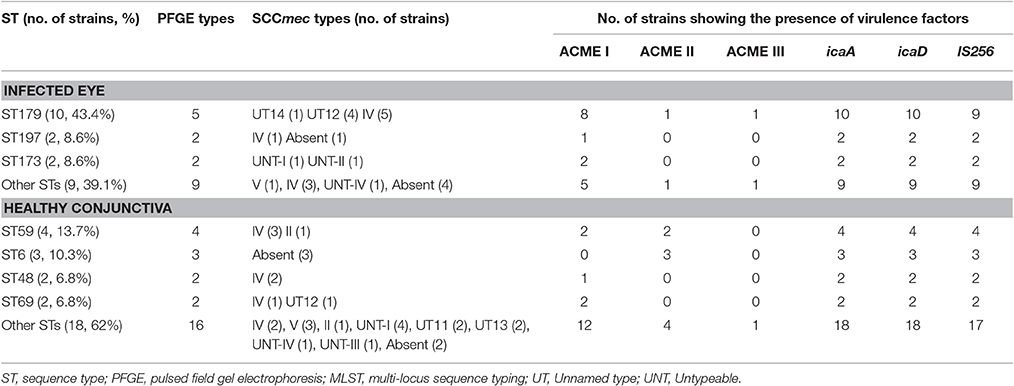
Table 4. Distribution of ST, PFGE types, SCCmec types and virulence genes among S. epidermidis isolated from the infected eye and from healthy conjunctiva.
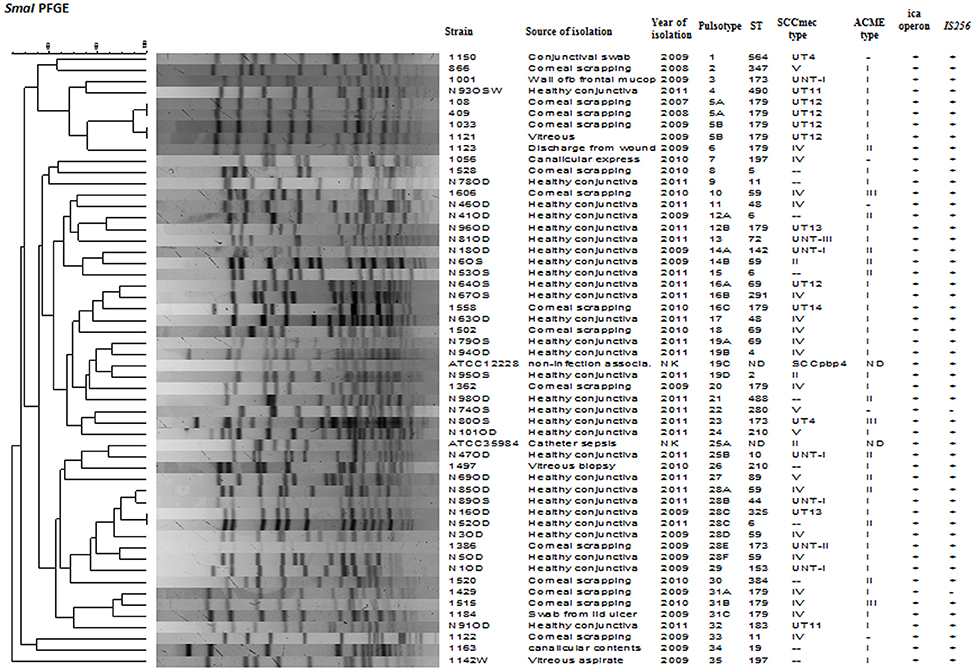
Figure 2. Dendrogram representation (Dice co-efficient) for macro-restriction banding patterns of S. epidermidis strains isolated from infected eye and healthy conjunctiva along with ATCC reference strains, generated by Pulsed-field gel electrophoresis of total chromosomal DNA digested with SmaI restriction enzyme. A correlation between Pulsotype, ST, SCCmec type, ACME, ica operon, IS256, and source of isolation is also shown. UT: Unnamed type, UNT: Untypeable, ND: not detected, NK: not known.
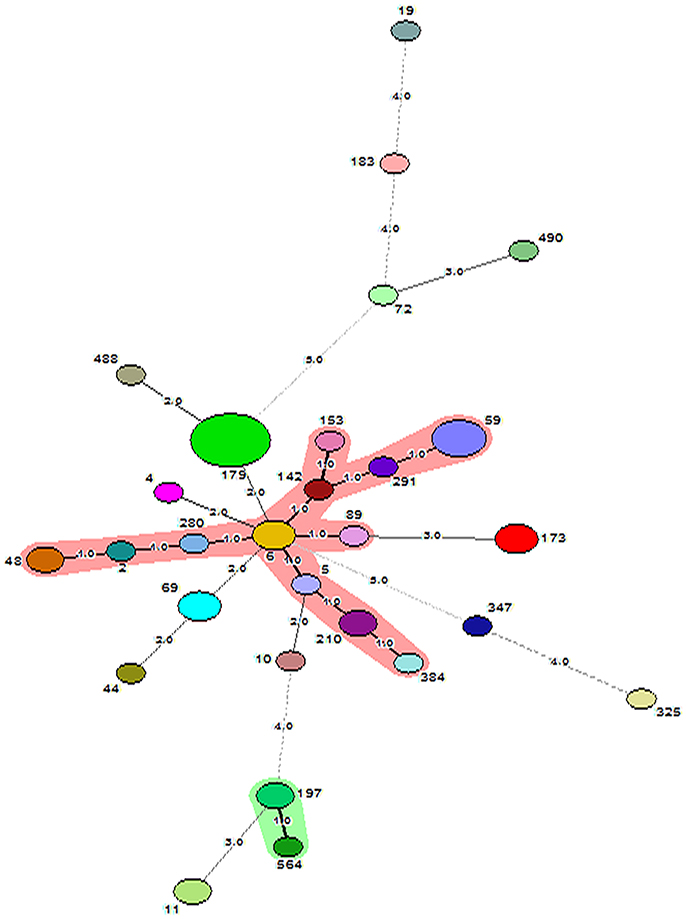
Figure 3. Minimum Spanning Tree showing relationship between different STs assigned by the analysis of MLST data. Each node represents one sequence type and the corresponding ST is given beside the node. The size of each node is directly proportional to the number of strains included under that ST. The number given on the strings connecting the nodes stands for the number of genes by which the strains under those STs differ from each other. Bold lines connect types that are identical for 6 loci, solid lines connecting types identical for ≥4 but ≤ 6 locus and dotted lines connecting STs differing from each other by ≥4 genes out of seven gene loci. CC59 and CC197 are represented as shaded area.
SID for MLST, SCCmec, and PFGE
We calculated SID for all the three typing techniques. The values derived for MLST, SCCmec typing and PFGE were 0.95, 0.85, and 0.97, suggesting the existence of genomic diversity among the isolates irrespective of sources of isolation. Overall result suggests that PFGE and MLST were more discriminatory than SCCmec typing to study genomic diversity among S. epidermidis.
Statistical Analysis
PCoA can segregate isolates belonging to the healthy conjunctiva and infected eye, except for few isolates from healthy conjunctiva (axis one was used for the highest percentage of representation), with 33% of explained variance (Figure 4A). From DA graph, it was clear that, whereas isolates from infected eye grouped within more negative values, majority isolates from healthy conjunctiva grouped within positive values (Figure 4B). Predominant biomarkers were determined by calculating the coefficient of discriminant function and considered when the value was equal to 0.5 or >0.5. Whereas infected eye isolates are discriminating in the biomarker of resistance to gatifloxacin (−3.332), ofloxacin (−3.332), and tetracycline (−2.131), the healthy conjunctiva isolates were discriminating in resistance to moxifloxacin (0.64542), ciprofloxacin (0.69144), and erythromycin (1.5531).
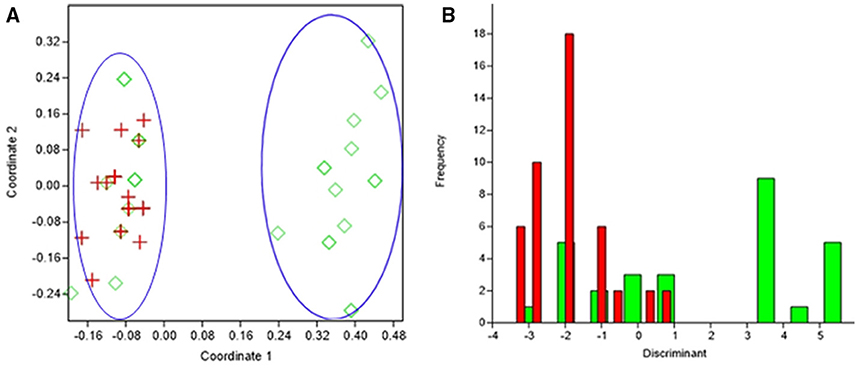
Figure 4. Results obtained by statistical analysis using phenotypic and genotypic data. (A) Principal Coordinates Analysis (PCoA) of S. epidermidis from infected eye, (IE, red), and isolates from healthy conjunctiva (HC, green). (B) Discriminant analysis of the isolates of S. epidermidis from infected eye, (IE, red), and isolates from healthy conjunctiva (HC, green). Negative values belong to IE isolates and positive value to HC isolates.
Discussion
S. epidermidis has been reported causing indwelling medical device infections, such as prosthetic valve endocarditis, and intra-cardiac abscesses leading to high mortality (Otto, 2009). The incidence of S. epidermidis in ocular disease is more frequent than any other species including S. aureus (Duggirala et al., 2007; Flores-Páez et al., 2015). The frequency of methicillin resistance among disease-associated isolates was higher than commensal strains (Duggirala et al., 2007). Several studies have shown that MDR is more frequent in MRSE than MSSE (Cherifi et al., 2013, 2014; Deplano et al., 2016). Also, strains from the infected eye were more resistant to non-beta-lactam antibiotics than those from the healthy conjunctiva (Duggirala et al., 2007; Cherifi et al., 2013, 2014). This study also showed that S. epidermidis isolates are multidrug resistant, irrespective of methicillin-resistance and sources of isolation. Similar to other workers 70% of isolates comprising isolates from healthy conjunctivae and the infected eye carried catpC221 or catpC223 or both gene(s) encoding for chloramphenicol resistance (Cherifi et al., 2013; Bispo et al., 2014). A good correlation was found between the presence of catp223 and/or catp221 gene(s) and chloramphenicol resistance. The catpC221 gene encoding for the resistance to chloramphenicol was reported previously in CoNS, especially in S. pseudintermedius of veterinary origin (Perreten et al., 2010; Kern and Perreten, 2013). A small percentage of S. epidermidis showed resistance to clindamycin and gentamicin a finding similar to those workers who reported resistance to these antibiotics among isolates from ocular and blood stream infections (Kern and Perreten, 2013; Bispo et al., 2014). In contrast, a high percentage of MLSB resistance gene ermC encoding for macrolide-lincosamide-streptograminB resistance was present among S. epidermidis isolates comprising 12 from the infected eye and 24 from the healthy conjunctiva (Argudin et al., 2011). Interestingly, most frequent ARG found in this study was tetK, though many of the isolates carrying this gene were phenotypically sensitive to tetracycline. Like Argudin et al. (2011) the bifunctional aminoglycoside resistance-conferring gene aac6′-aph2″ was present in majority strains of which only 34% were phenotypically resistant to gentamicin. The commensal isolates were reported showing multidrug resistance (Li et al., 2009). Also, in this study, isolates from the healthy conjunctiva showed resistance to chloramphenicol, fluoroquinolones, erythromycin and tetracycline. These observations thus suggest that even the commensal strains can acquire resistance without being exposed to the antibiotics, therefore, acts as reservoirs for ARGs.
Fluoroquinolones are the preferred antibiotics for the treatment of staphylococcal ocular infections. Use of inappropriate dosage and overexposure of the bacteria to the antibiotic had facilitated the emergence of resistance to this drug. Mutation(s) in the gyrA, gyrB, parC, and parE genes is the primary mechanism of fluoroquinolone resistance. Like other workers, we also did not find any mutation in gyrB among S. epidermidis (Yamada et al., 2008). These workers have shown mutations in QRDR of gyrA, gyrB, parC, and parE genes in ~50% of S. epidermidis isolates from the human conjunctiva. However, the results of the present study showed an increase in mutation in QRDR up to 70% until 2011. We like other workers also found a mutation in gyrA (Ser84 → F/Y) in S. epidermidis strains that showed a significant association with high level of resistance to fluoroquinolones (Yamada et al., 2008; Betanzos-Cabrera et al., 2009; Bispo et al., 2013). In addition to the reported mutation in parC (Ser80 → F/Y and D → A), we found two novel mutations (D84 → C and V82 → L). These mutations do not alter the level of MIC significantly for any of the tested fluoroquinolones. Although mutation in gyrA gene confers resistance to fluoroquinolone, mutations in parC or parE do not alter the level of fluoroquinolones resistance. We found high MIC value for ciprofloxacin (32 mg/L) in one of the strain 1142W that showed mutation only in parE gene but its role in high level of quninolone resistance is not known (Yamada et al., 2008). We also did not find any association of high level of MIC to ciprofloxacin in a strain that had mutation in only parE gene. It is likely that other mechanism such as efflux of quinolone may be responsible for quinolone resistance leading to high level of ciprofloxacin resistance in this strain (Betanzos-Cabrera et al., 2009).
Several studies showed that ica operon is more prevalent among S. epidermidis isolated from a variety of infections than the commensals (Cherifi et al., 2013; Du et al., 2013). In contrast, all isolates of the present study carried ica operon irrespective of sources of isolation. These findings are similar to those workers who also reported the presence of ica operon among the isolates from both infected eye and healthy conjunctiva (Duggirala et al., 2007; Cherifi et al., 2013; Flores-Páez et al., 2015). Thus, this study support the proposal of eliminating detection of ica gene as a factor for differentiating commensal from invasive isolates (Wisplinghoff et al., 2003). Similarly, ACME operon was present in 47 S. epidermidis strains. This finding is in contrast to those workers who reported less number of S. epidermidis showing the presence of ACME operon (Cherifi et al., 2013; Du et al., 2013; Deplano et al., 2016). These authors also demonstrated that ACME was found more commonly in commensals than those isolated from infections (Du et al., 2013). In contrast, we did not find much difference in the distribution of ACME operon among S. epidermidis with regard to sources of isolation.
The majority of isolates from the infected eye and healthy conjunctiva were methicillin-resistant and carried the mecA gene. We did not find the presence of mecC (a variant of mecA) in any of the S. epidermidis isolates. These findings thus suggest that there is no significant difference of oxacillin resistance and the presence of mecA gene between sources of isolation. MRSE isolates were mostly multidrug resistant and belong to SCCmec type IV. These findings are similar to those workers who also reported the presence of SCCmec type IV in S. epidermidis (Wisplinghoff et al., 2003; Duggirala et al., 2007; Cherifi et al., 2014). The findings of this study thus suggest that there is mobility of SCCmec cassette among S. epidermidis, similar to that reported in S. aureus (Miragaia et al., 2007). Also, nine S. epidermidis isolates comprising six from the infected eye and three from healthy conjunctiva showed amplification of mercury resistance and J-region suggesting the presence of SCCmercury operon. To the best of our knowledge, this is the first report of the presence of SCCmercury among ocular isolates of S. epidermidis, although this element was reported earlier in S. aureus strains (Kondo et al., 2007).
A novel combination of ccr A2B2 and mecC2, designated as UT13, was present among S. epidermidis strains and was not included in the earlier classification scheme (Zong et al., 2011). We also found amplification of more than one ccr complex among few isolates designated as UT11 and UT12 indicating the presence of composite SCCmec types. Although the presence of more than one type of ccr complex was reported earlier in S. epidermidis, 17 MSSE isolates from this study were untypeable because none of the strains amplified either one or both of the complexes (Kondo et al., 2007; Cherifi et al., 2014). Also, we did not find the presence of ΨSCCmec complex designated as a pseudo-SCCmec element, carrying mec complex but lacking ccr complex, in any of the strains. The heterogeneity of isolates including those from healthy conjunctiva indicates that S. epidermidis can act as a reservoir and play a role in the emergence of new types of SCCmec elements. It can happen due to acquisition or loss of SCCmec element which is frequent in CoNS (Zong et al., 2011).
MLST analysis revealed that ST179 is the most common ST found in 43% of infected eye isolates of S. epidermidis. This ST179 was reported earlier in single isolate each from an ocular strain in Brazil in 2009 and Iran in 2016 (Cherifi et al., 2013; Bispo et al., 2014). The second most frequent ST was ST59 and found in one strains from infected eye and three from healthy conjunctiva. This ST59 along with other STs was reported in strains isolated from blood culture, ocular infection and the healthy conjunctiva and suggested their association with the eye disease (Mertens and Ghebremedhin, 2013; Bispo et al., 2014; Soroush et al., 2016). There is good correlation between types of STs, ST179, and ST59 with source of isolation. These findings are in contrast to those workers who reported that lineage ST2 is most frequent in ocular infection and lineage ST5 was abundant in healthy conjunctiva isolates showing a substantial difference between isolates from ocular disease and healthy conjunctiva (Flores-Páez et al., 2015). However, a study conducted on clinical isolates in US hospital on bone and joint infection showed association of ST5 lineage with disease (Mendes et al., 2012; Cremniter et al., 2013). High level of fluoroquinolone resistance was significantly associated with sequence type ST179 of S. epidermidis. This observation is similar to those workers who also reported association of major clones of HA-MRSA with high-level resistance to fluoroquinolones (Horváth et al., 2012; Knight et al., 2012; Holden et al., 2013).
PFGE is the most appreciated method used for short-term epidemiological investigation. In this study, PFGE revealed that isolates from both infected eye and the healthy conjunctiva were genetically diverse. It can be seen that out of 35 pulsotypes; the infected eye isolates showed 16 pulsotypes, and the healthy conjunctiva isolates showed 17 pulsotypes. Only two pulsotypes was common among members of both the groups. Several studies also reported diversity among S. epidermidis (Cherifi et al., 2013; Martínez-Meléndez et al., 2016). Though strains from the infected eye and healthy conjunctiva clustered separately, there was high diversity within the members of the same group. This finding is in agreement with those workers who reported polyclonality among S. epidermidis isolated from the same host (Ueta et al., 2007).
It was clear from statistical analysis, that there is no specific discriminant marker to differentiate isolates of eye infection from healthy conjunctiva. However, the resistance to fluoroquinolones, erythromycin, and tetracycline can discriminate infected eye strains from the healthy conjunctiva isolates. This study thus suggests that few healthy conjunctiva isolates had characteristics of infected eye isolates.
We conclude that S. epidermidis strains are multidrug resistant, virulent and carried mecA gene, irrespective of sources of isolation. Although majority strains carried known types of SCCmec, seven strains carried unreported SCCmec types including SCCmercury. To the best of our knowledge, this is the first report of the presence of SCCmercury among ocular isolates of S. epidermidis. Few strains besides possessing known mutation in gyrA, parC, and parE, had additional mutation in parC gene. IS256 cannot be used as a marker to differentiate isolates from infected eye and healthy conjunctiva. Statistical analysis suggests that few healthy conjunctiva isolates had characteristics similar to infected eye isolates. A noteworthy finding was the presence of ST179 in 43% of infected eye isolates an observation rarely reported among S. epidermidis. PFGE and MLST analysis indicates genomic diversity among them irrespective of sources of isolation.
Author Contributions
SJ, SP, and DVS designed the research; SJ and SP participated in most of the experiment; SJ, and SP analyzed the data; KN performed the statistical analysis; SJ drafted the manuscript; SJ, SP, KN, and DVS revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the Department of Science and Technology, New Delhi grant no. SR/SO/HS-117 to DVS and fund contributed by the Department of Biotechnology, New Delhi to the Institute of Life Sciences, Bhubaneswar. SJ and SP are grateful to Department of Science and Technology, New Delhi and Indian Council of Medical Research, New Delhi, respectively for providing Senior Research Fellowship. Authors thank Dr. Savitri Sharma of Jhaveri Microbiology Centre, LV Prasad Eye Institute, Hyderabad, India, for critically reading the manuscript and providing S. epidermidis isolates used in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01430/full#supplementary-material
References
Alekshun, M. N., and Levy, S. B. (2007). Molecular mechanisms of antibacterial multidrug resistance. Cell 128, 1037–1050. doi: 10.1016/j.cell.2007.03.004
Arciola, C. R., Baldassarri, L., and Montanaro, L. (2001). Presence of icaA and icaD genes and slime production in a collection of Staphylococcal strains from catheter-associated infections. J. Clin. Microbiol. 39, 2151–2156. doi: 10.1128/JCM.39.6.2151-2156.2001
Argudin, M., Tenhagen, B.-A., Fetsch, A., Sachsenröder, J., Käsbohrer, A., Schroeter, A., et al. (2011). Virulence and resistance determinants in German Staphylococcus aureus ST398 isolates from non-human origin. Appl. Environ. Microbiol. 77, 3052–3060. doi: 10.1128/AEM.02260-10
Asbell, P. A., Sahm, D. F., Shaw, M., Draghi, D. C., and Brown, N. P. (2008). Increasing prevalence of methicillin resistance in serious ocular infections caused by Staphylococcus aureus in the United States: 2000 to 2005. J. Cataract. Refract. Surg. 34, 814–818. doi: 10.1016/j.jcrs.2008.01.016
Betanzos-Cabrera, G., Juárez-Verdayes, M. A., González-González, G., Cancino-Díaz, M. E., and Cancino-Díaz, J. C. (2009). Gatifloxacin, moxifloxacin, and balofloxacin resistance due to mutations in the gyrA and parC genes of Staphylococcus epidermidis strains isolated from patients with endophthalmitis, corneal ulcers and conjunctivitis. Ophthalmic Res. 42, 43–48. doi: 10.1159/000219684
Bispo, P. J., Alfonso, E. C., Flynn, H. W., and Miller, D. (2013). Emerging 8-methoxyfluoroquinolone resistance among methicillin-susceptible Staphylococcus epidermidis isolates recovered from patients with endophthalmitis. J. Clin. Microbiol. 51, 2959–2963. doi: 10.1128/JCM.00846-13
Bispo, P. J., Hofling-Lima, A. L., and Pignatari, A. C. (2014). Characterization of ocular methicillin-resistant Staphylococcus epidermidis isolates belonging predominantly to clonal complex 2 subcluster II. J. Clin. Microbiol. 52, 1412–1417. doi: 10.1128/JCM.03098-13
Bloemendaal, A. L., Brouwer, E. C., and Fluit, A. C. (2010). Methicillin resistance transfer from Staphylocccus epidermidis to methicillin-susceptible Staphylococcus aureus in a patient during antibiotic therapy. PLoS ONE 5:e11841. doi: 10.1371/journal.pone.0011841
Bourcier, T., Thomas, F., Borderie, V., Chaumeil, C., and Laroche, L. (2003). Bacterial keratitis: predisposing factors, clinical and microbiological review of 300 cases. Br. J. Ophthalmol. 87, 834–838. doi: 10.1136/bjo.87.7.834
Cavuoto, K., Zutshi, D., Karp, C. L., Miller, D., and Feuer, W. (2008). Update on bacterial conjunctivitis in South Florida. Ophthalmology 115, 51–56. doi: 10.1016/j.ophtha.2007.03.076
Cherifi, S., Byl, B., Deplano, A., Nagant, C., Nonhoff, C., Denis, O., et al. (2014). Genetic characteristics and antimicrobial resistance of Staphylococcus epidermidis isolates from patients with catheter-related bloodstream infections and from colonized healthcare workers in a Belgian hospital. Ann. Clin. Microbiol. Antimicrob. 13:20. doi: 10.1186/1476-0711-13-20
Cherifi, S., Byl, B., Deplano, A., Nonhoff, C., Denis, O., and Hallin, M. (2013). Comparative epidemiology of Staphylococcus epidermidis isolates from patients with catheter-related bacteremia and from healthy volunteers. J. Clin. Microbiol. 51, 1541–1547. doi: 10.1128/JCM.03378-12
Clinical and Laboratory Standards Institute, C. (2015). Performance Standards for Antimicrobial Susceptibility Testing: Seventeenth Informational Supplement, M100-S25. Wayne, PA: CLSI.
Cremniter, J., Sivadon-Tardy, V., Caulliez, C., Bauer, T., Porcher, R., Lortat-Jacob, A., et al. (2013). Genetic analysis of glycopeptide-resistant Staphylococcus epidermidis strains from bone and joint infections. J. Clin. Microbiol. 51, 1014–1019. doi: 10.1128/JCM.02608-12
Deplano, A., Vandendriessche, S., Nonhoff, C., Dodémont, M., Roisin, S., and Denis, O. (2016). National surveillance of Staphylococcus epidermidis recovered from bloodstream infections in Belgian hospitals. J. Antimicrob. Chemother. 71, 1815–1819. doi: 10.1093/jac/dkw086
Diep, B. A., Stone, G. G., Basuino, L., Graber, C. J., Miller, A., des Etages, S. A., et al. (2008). The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 197, 1523–1530. doi: 10.1086/587907
Du, X., Zhu, Y., Song, Y., Li, T., Luo, T., Sun, G., et al. (2013). Molecular analysis of Staphylococcus epidermidis strains isolated from community and hospital environments in China. PLoS ONE 8:e62742. doi: 10.1371/journal.pone.0062742
Duggirala, A., Kenchappa, P., Sharma, S., Peeters, J. K., Ahmed, N., Garg, P., et al. (2007). High-resolution genome profiling differentiated Staphylococcus epidermidis isolated from patients with ocular infections and normal individuals. Invest. Ophthalmol. Vis. Sci. 48, 3239–3245. doi: 10.1167/iovs.06-1365
Duran, N., Ozer, B., Duran, G. G., Onlen, Y., and Demir, C. (2012). Antibiotic resistance genes & susceptibility patterns in staphylococci. Ind. J. Med. Res. 135, 389–396.
Flores-Páez, L. A., Zenteno, J. C., Alcántar-Curiel, M. D., Vargas-Mendoza, C. F., Rodríguez-Martínez, S., Cancino-Diaz, M. E., et al. (2015). Molecular and phenotypic characterization of Staphylococcus epidermidis isolates from healthy conjunctiva and a comparative analysis with isolates from ocular infection. PLoS ONE 10:e0135964. doi: 10.1371/journal.pone.0135964
Graham, J. E., Moore, J. E., Jiru, X., Moore, J. E., Goodall, E. A., Dooley, J. S., et al. (2007). Ocular pathogen or commensal: a PCR-based study of surface bacterial flora in normal and dry eyes. Invest. Ophthalmol. Vis. Sci. 48, 5616–5623. doi: 10.1167/iovs.07-0588
Gu, J., Li, H., Li, M., Vuong, C., Otto, M., and Wen, Y. (2005). Bacterial insertion sequence IS256 as a potential molecular marker to discriminate invasive strains from commensal strains of Staphylococcus epidermidis. J. Hosp. Infect. 61, 342–348. doi: 10.1016/j.jhin.2005.04.017
Hammer, Ø., Harper, D., and Ryan, P. (2001). Paleontological statistics software: package for education and data analysis. Palaeontol. Electron. 4, 1–9. Available online at: http://palaeo-electronica.org/2001_1/past/past.pdf
Hartman, B. J., and Tomasz, A. (1984). Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158, 513–516.
Hellmark, B., Berglund, C., Nilsdotter-Augustinsson, Å., Unemo, M., and Söderquist, B. (2013). Staphylococcal cassette chromosome mec (SCCmec) and arginine catabolic mobile element (ACME) in Staphylococcus epidermidis isolated from prosthetic joint infections. Eur. J. Clin. Microbiol. Infect. Dis. 32, 691–697. doi: 10.1007/s10096-012-1796-2
Hirotaki, S., Sasaki, T., Kuwahara-Arai, K., and Hiramatsu, K. (2011). Rapid and accurate identification of human-associated staphylococci by use of multiplex PCR. J. Clin. Microbiol. 49, 3627–3631. doi: 10.1128/JCM.00488-11
Holden, M. T., Hsu, L. Y., Kurt, K., Weinert, L. A., Mather, A. E., Harris, S. R., et al. (2013). A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 23, 653–664. doi: 10.1101/gr.147710.112
Hookey, J. V., Richardson, J. F., and Cookson, B. D. (1998). Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J. Clin. Microbiol. 36, 1083–1089.
Horváth, A., Dobay, O., Kardos, S., Ghidán, Á., Tóth, Á., Pászti, J., et al. (2012). Varying fitness cost associated with resistance to fluoroquinolones governs clonal dynamic of methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 31, 2029–2036. doi: 10.1007/s10096-011-1536-z
Katayama, Y., Ito, T., and Hiramatsu, K. (2000). A new class of genetic element, Staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44, 1549–1555. doi: 10.1128/AAC.44.6.1549-1555.2000
Keay, L., Edwards, K., Naduvilath, T., Taylor, H. R., Snibson, G. R., Forde, K., et al. (2006). Microbial keratitis: predisposing factors and morbidity. Ophthalmology 113, 109–116. doi: 10.1016/j.ophtha.2005.08.013
Kern, A., and Perreten, V. (2013). Clinical and molecular features of methicillin-resistant, coagulase-negative staphylococci of pets and horses. J. Antimicrob. Chemother. 68, 1256–1266. doi: 10.1093/jac/dkt020
Knight, G. M., Budd, E. L., Whitney, L., Thornley, A., Al-Ghusein, H., Planche, T., et al. (2012). Shift in dominant hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) clones over time. J. Antimicrob. Chemother. 67, 2514–2522. doi: 10.1093/jac/dks245
Kondo, Y., Ito, T., Ma, X. X., Watanabe, S., Kreiswirth, B. N., Etienne, J., et al. (2007). Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51, 264–274. doi: 10.1128/AAC.00165-06
Le, K. Y., Dastgheyb, S., Ho, T. V., and Otto, M. (2014). Molecular determinants of staphylococcal biofilm dispersal and structuring. Front. Cell. Infect. Microbiol. 4:167. doi: 10.3389/fcimb.2014.00167
Leclercq, R. (2002). Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34, 482–492. doi: 10.1086/324626
Li, M., Wang, X., Gao, Q., and Lu, Y. (2009). Molecular characterization of Staphylococcus epidermidis strains isolated from a teaching hospital in Shanghai, China. J. Med. Microbiol. 58, 456–461. doi: 10.1099/jmm.0.007567-0
Lichtinger, A., Yeung, S. N., Kim, P., Amiran, M. D., Iovieno, A., Elbaz, U., et al. (2012). Shifting trends in bacterial keratitis in Toronto: an 11-year review. Ophthalmology 119, 1785–1790. doi: 10.1016/j.ophtha.2012.03.031
Martínez-Meléndez, A., Morfín-Otero, R., Villarreal-Trevi-o, L., Camacho-Ortíz, A., González-González, G., Llaca-Díaz, J., et al. (2016). Molecular epidemiology of coagulase-negative bloodstream isolates: detection of Staphylococcus epidermidis ST2, ST7 and linezolid-resistant ST23. Braz. J. Infect. Dis. 20, 419–428. doi: 10.1016/j.bjid.2016.05.007
McManus, B. A., Coleman, D. C., Deasy, E. C., Brennan, G. I., O'Connell, B., Monecke, S., et al. (2015). Comparative genotypes, staphylococcal cassette chromosome mec (SCCmec) genes and antimicrobial resistance amongst Staphylococcus epidermidis and Staphylococcus haemolyticus isolates from infections in humans and companion animals. PLoS ONE 10:e0138079. doi: 10.1371/journal.pone.0138079
Melo, G., Bispo, P. J. M., Yu, M. C. Z., Pignatari, A., and Höfling-Lima, A. (2011). Microbial profile and antibiotic susceptibility of culture-positive bacterial endophthalmitis. Eye 25, 382–388. doi: 10.1038/eye.2010.236
Mendes, R. E., Deshpande, L. M., Costello, A. J., and Farrell, D. J. (2012). Molecular epidemiology of Staphylococcus epidermidis clinical isolates from U.S. hospitals. Antimicrob. Agents Chemother. 56, 4656–4661. doi: 10.1128/AAC.00279-12
Mertens, A., and Ghebremedhin, B. (2013). Genetic determinants and biofilm formation of clinical Staphylococcus epidermidis isolates from blood cultures and indwelling devises. Eur. J. Microbiol. Immunol. 3, 111–119. doi: 10.1556/EuJMI.3.2013.2.4
Miragaia, M., Thomas, J. C., Couto, I., Enright, M. C., and de Lencastre, H. (2007). Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol. 189, 2540–2552. doi: 10.1128/JB.01484-06
Otto, M. (2009). Staphylococcus epidermidis-the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7, 555–567. doi: 10.1038/nrmicro2182
Park, Y. M., Kwon, H. J., and Lee, J. S. (2015). Microbiological study of therapeutic soft contact lenses used in the treatment of recurrent corneal erosion syndrome. Eye Contact Lens 41, 84–86. doi: 10.1097/ICL.0000000000000068
Perreten, V., Kadlec, K., Schwarz, S., Andersson, U. G., Finn, M., Greko, C., et al. (2010). Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: an international multicentre study. J. Antimicrob. Chemother. 65, 1145–1154. doi: 10.1093/jac/dkq078
Rogers, K. L., Fey, P. D., and Rupp, M. E. (2009). Coagulase-negative staphylococcal infections. Infect. Dis. Clin. North Am. 23, 73–98. doi: 10.1016/j.idc.2008.10.001
Ruppé, E., Barbier, F., Mesli, Y., Maiga, A., Cojocaru, R., Benkhalfat, M., et al. (2009). Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob. Agents Chemother. 53, 442–449. doi: 10.1128/AAC.00724-08
Schimel, A. M., Miller, D., and Flynn, H. W. (2013). Endophthalmitis isolates and antibiotic susceptibilities: a 10-year review of culture-proven cases. Am. J. Ophthalmol. 156, e50–e52. doi: 10.1016/j.ajo.2013.01.027
Schlegelova, J., Vlkova, H., Babak, V., Holasova, M., Jaglic, Z., Stosova, T., et al. (2008). Resistance to erythromycin of Staphylococcus spp. isolates from the food chain. Vet. Med. 53, 307–314.
Schmitz, F.-J., Fluit, A. C., Gondolf, M., Beyrau, R., Lindenlauf, E., Verhoef, J., et al. (1999). The prevalence of aminoglycoside resistance and corresponding resistance genes in clinical isolates of staphylococci from 19 European hospitals. J. Antimicrob. Chemother. 43, 253–259. doi: 10.1093/jac/43.2.253
Schommer, N. N., Christner, M., Hentschke, M., Ruckdeschel, K., Aepfelbacher, M., and Rohde, H. (2011). Staphylococcus epidermidis uses distinct mechanisms of biofilm formation to interfere with phagocytosis and activation of mouse macrophage-like cells 774A. Infect. Immun. 79, 2267–2276. doi: 10.1128/IAI.01142-10
Soroush, S., Jabalameli, F., Taherikalani, M., Amirmozafari, N., Fooladi, A. A. I., Asadollahi, K., et al. (2016). Investigation of biofilm formation ability, antimicrobial resistance and the staphylococcal cassette chromosome mec patterns of methicillin resistant Staphylococcus epidermidis with different sequence types isolated from children. Microb. Pathog. 93, 126–130. doi: 10.1016/j.micpath.2016.01.018
Tenover, F. C., Arbeit, R., Archer, G., Biddle, J., Byrne, S., Goering, R., et al. (1994). Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J. Clin. Microbiol. 32, 407–415.
Thomas, J. C., Vargas, M. R., Miragaia, M., Peacock, S. J., Archer, G. L., and Enright, M. C. (2007). Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J. Clin. Microbiol. 45, 616–619. doi: 10.1128/JCM.01934-06
Trieu-Cuot, P., De Cespedes, G., Bentorcha, F., Delbos, F., Gaspar, E., and Horaud, T. (1993). Study of heterogeneity of chloramphenicol acetyltransferase (CAT) genes in streptococci and enterococci by polymerase chain reaction: characterization of a new CAT determinant. Antimicrob. Agents Chemother. 37, 2593–2598. doi: 10.1128/AAC.37.12.2593
Trzcinski, K., Cooper, B. S., Hryniewicz, W., and Dowson, C. G. (2000). Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 45, 763–770. doi: 10.1093/jac/45.6.763
Ueta, M., Iida, T., Sakamoto, M., Sotozono, C., Takahashi, J., Kojima, K., et al. (2007). Polyclonality of Staphylococcus epidermidis residing on the healthy ocular surface. J. Med. Microbiol. 56, 77–82. doi: 10.1099/jmm.0.46810-0
Van Belkum, A., Tassios, P., Dijkshoorn, L., Haeggman, S., Cookson, B., Fry, N., et al. (2007). Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13, 1–46. doi: 10.1111/j.1469-0691.2007.01786.x
Wisplinghoff, H., Rosato, A. E., Enright, M. C., Noto, M., Craig, W., and Archer, G. L. (2003). Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob. Agents Chemother. 47, 3574–3579. doi: 10.1128/AAC.47.11.3574-3579.2003
Yamada, M., Yoshida, J., Hatou, S., Yoshida, T., and Minagawa, Y. (2008). Mutations in the quinolone resistance determining region in Staphylococcus epidermidis recovered from conjunctiva and their association with susceptibility to various fluoroquinolones. Br. J. Ophthalmol. 92, 848–851. doi: 10.1136/bjo.2007.129858
Keywords: multidrug-resistant, Staphylococcus epidermidis, ACME, SCCmec typing, PFGE analysis, MLST-genotyping, diversity
Citation: Jena S, Panda S, Nayak KC and Singh DV (2017) Identification of Major Sequence Types among Multidrug-Resistant Staphylococcus epidermidis Strains Isolated from Infected Eyes and Healthy Conjunctiva. Front. Microbiol. 8:1430. doi: 10.3389/fmicb.2017.01430
Received: 15 May 2017; Accepted: 14 July 2017;
Published: 04 August 2017.
Edited by:
Johnan A. R. Kaleeba, Uniformed Services University of the Health Sciences, United StatesReviewed by:
Marquita Vernescia Gittens-St.Hilaire, University of the West Indies, BarbadosMiklos Fuzi, Semmelweis University, Hungary
Copyright © 2017 Jena, Panda, Nayak and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Durg V. Singh, ZHVyZ19zaW5naEB5YWhvby5jby5pbg==
 Smrutiti Jena
Smrutiti Jena Sasmita Panda
Sasmita Panda Kinshuk C. Nayak
Kinshuk C. Nayak Durg V. Singh
Durg V. Singh