- 1Department of Respiratory Medicine, The Second Hospital of Jilin University, Changchun, China
- 2Department of Clinical Laboratory, The First Hospital of Jilin University, Changchun, China
- 3Key Laboratory of Zoonosis Research, Ministry of Education, College of Veterinary Medicine, Jilin University, Changchun, China
- 4Agricultural Experiment Base, Jilin University, Changchun, China
Biofilm formation, one of the most important virulence factors of pathogenic bacteria, protects bacteria against desiccation, antibiotics, phages and host immune responses. However, phage-derived depolymerases show antibiofilm activity and demonstrate great potential to treat infections caused by biofilm-forming bacteria. In this study, the Escherichia coli phage vB_EcoM_ECOO78 was isolated and characterised, and we observed its ability to lyse five out of 34 tested E. coli clinical isolates. The highest phage titre was observed at a multiplicity of infection of 10-5 and a burst size of approximately 74 plaque forming units (PFU)/infection. Electron micrographs indicated that vB_EcoM_ECOO78 belongs to the family Myoviridae. The presence of increasing halos surrounding the lysis plaques formed by vB_EcoM_ECOO78 indicated that this phage may encode a depolymerase. Based on a sequencing analysis, the complete genome of vB_EcoM_ECOO78 was found to be 41,289 bp in size, with a GC content of 53.07%. Additionally, vB_EcoM_ECOO78 has 56 predicted open reading frames, 51 (91.07%) of which are assumed to be functional. A BLAST analysis indicated that ORF42 of vB_EcoM_ECOO78 (Dpo42) has low identity with other reported phage-associated depolymerases. Dpo42 was expressed and purified as a soluble protein using E. coli BL21. The biofilm formation ability of E. coli isolates and the antibiofilm activity of Dpo42 were tested by performing spot assays and using a 96-well micro-titre plate method. Dpo42 degraded the capsular polysaccharides surrounding E. coli and exhibited dose-dependent biofilm-formation prevention activity. Based on these results, Dpo42 appears to be a novel phage-derived depolymerase that represents a new potential strategy for preventing E. coli biofilm formation.
Introduction
Extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli is a common Gram-negative pathogen in China (Zhang et al., 2016) that causes community- and hospital-acquired infections. The number of ESBL-positive E. coli strains has significantly increased in recent years (Canton et al., 2008). One of the most important virulence factors of E. coli is the ability to form biofilms. There are three necessary steps involved in biofilm formation: initial adhesion, early development of biofilm and maturation of the developed biofilm (Sharma et al., 2016). The structure and composition of bacterial biofilms were first described in Zobell (1943), and vary between species or strains based on biofilm age and environmental conditions (Harper et al., 2014). Usually, biofilms are microbial communities attached to a surface and encased in an extracellular matrix mainly composed by extracellular polymeric substances (EPSs), but also by proteins, nucleic acids, lipids, water and mineral ions (Costerton, 1995; O’Toole et al., 2000; Flemming and Wingender, 2010). The EPSs can be capsular polysaccharides or biofilm matrix polysaccharides (Latka et al., 2017). Three major EPSs have been reported, including β-1,6-N-acetyl-D-glucosamine polymer (PGA), cellulose and colanic acid, that comprise the E. coli biofilm matrix (Sharma et al., 2016). For E. coli, more than 80 distinct serotypes have been established depending on their capsular polysaccharides (Whitfield and Roberts, 1999). The ability of bacteria to form biofilms represents a serious medical challenge given the increasing prevalence of ESBL-positive E. coli strains. Therefore, there is a need to identify and develop new therapeutic strategies to eradicate E. coli biofilms (Stewart and Costerton, 2001).
Phages are viruses that can specifically infect bacteria. Therefore, in recent years, lytic phages have been reconsidered for the treatment of multidrug-resistant bacteria infections and several clinical trials have proved a great potential exists (Summers, 2001; Chanishvili, 2012; Oliveira et al., 2015). Further more, phage-derived proteins (such as lysins and holins) have been developed as antibacterial agents in vitro and in vivo (Nelson et al., 2001; Fischetti, 2005). Although numerous studies have investigated phages and lysins, little is known about phage depolymerases. Phage depolymerases are enzymes encoded by phages that specifically degrade bacterial EPSs (Latka et al., 2017). The existence of depolymerase is commonly identified by the development of an increasing halo surrounding each lysis plaque after prolonged incubation (Cornelissen et al., 2012; Harper et al., 2014; Pires et al., 2016). Additionally, phage-derived depolymerases potentially control the formation of E. coli biofilms (Stewart et al., 1995). A recent review reported on the putative depolymerases present in phages infecting different bacterial genera (Pires et al., 2016); among these phages, only a few E. coli phages produced depolymerases. In fact, we have discovered that more and more putative depolymerases can be found.
The activities of depolymerases extracted from phage lysates against serotype K30 capsular E. coli polysaccharide have been described (McCallum et al., 1989). The bacteriophage K1F tail protein is a depolymerase (endo-N-acylneuraminidase) that degrades the α-(2, 8)-linked polysialic acid chains of E. coli K1 serotype (Petter and Vimr, 1993). In 2004, bacteriophage-derived endosialidase E (endoE) was expressed and purified, and was observed to selectively degrade the polysialic acid capsule on the surface of E. coli K1 strains (Mushtaq et al., 2004). Besides these, other bacterial phage-derived depolymerases have also been studied, including Enterobacter agglomerans phage depolymerase (Hughes et al., 1998a), Erwinia amylovora phage depolymerase (DpoL1) (Born et al., 2014), Acinetobacter baumannii phage depolymerase ORF40ΦAB6 (Lai et al., 2016), Staphylococcus epidermidis phage depolymerase Dpo7 (Gutierrez et al., 2015) and Klebsiella phage depolymerase depoKP36 (Majkowska-Skrobek et al., 2016).
In this study, an E. coli phage was isolated and a novel phage-derived depolymerase, Dpo42, was characterised. Dpo42 could degrade the capsular polysaccharides surrounding E. coli and exhibited biofilm-formation prevention activity in vitro.
Materials and Methods
Bacterial Strains and Drug Resistance
The E. coli ATCC 25922 strain was purchased from the American Type Culture Collection (ATCC). Drug-resistant E. coli isolates were isolated from patients at the First Hospital of Jilin University (Changchun, Jilin province, China). The antibiotic susceptibility of the E. coli isolates was evaluated using the VITEK® 2 Compact system (bioMérieux, Marcy-l’Étoile, France). Glycerol bacterial stocks were stored at -80°C and transferred to lysogeny broth (LB) medium (BioCorp, Warszawa, Poland) before recovery at 37°C. Tryptic Soy Broth (TSB) medium (bioMérieux, Marcy-l’Étoile, France) was used to culture bacterial biofilms. All strains were tested to determine the host range of the phage and to obtain the depolymerase antibiofilm activity.
Isolation and Purification of Phage Particles
Sewage samples were obtained from the First Hospital of Jilin University in Changchun. Phage isolation and purification were performed as described previously, with minor modifications (Lin et al., 2010). Each 100-mL water sample was filtered using a sterile 0.22 μm Filter (Millex-GP Filter Unit; Millipore, Bedford, MA, United States; LOT R6MA05262) to remove any impurities and microorganisms. Then, the filtered solution was utilised to prepare LB liquid medium (tryptone 1%, yeast extract 0.5%, NaCl 1%) instead of purified water. Host E. coli O78-3 cells were cultured to the logarithmic phase, added to LB liquid medium and mixed. The culture was centrifuged at 4°C and 3,762 × g for 15 min after incubation at 37°C for 18 h. The supernatant was then filtered using a sterile 0.22 μm philtre. Phages were purified using the double-layer agar plate method. A total of 100 μL of overnight-cultured E. coli O78-3 and phage were added to melted semi-solid medium (tryptone 1%, yeast extract 0.5%, NaCl 1%, agar 0.7%) (45°C) and immediately mixed, then poured onto plates containing LB solid medium (tryptone 1%, yeast extract 0.5%, NaCl 1%, agar 1.5%). The plates were cultured at 37°C for 12 h. Phages were purified until the plaques were uniform in size. Phage lysates were centrifuged at 4°C at 3,762 × g for 30 min. The purified phages were stored in Suspended Medium (SM) buffer (gelatine 0.01%, NaCl 100 mmol/L, Tris-HCl 50 mmol/L, and MgSO4 10 mmol/L) at 4°C. A total of 1 μg/mL RNase A and DNase I (Sigma–Aldrich, St. Louis, MO, United States) was added into the phage supernatant, and then the mixture was incubated for 30 min at room temperature. To separate bacterial fragments and phage particles, 1 mol/L NaCl was added to the supernatant on ice and incubated for 1 h. The supernatant was then centrifuged at 4°C and 10,451 × g for 10 min. To precipitate the phage particles, 10% (w/v) PEG 8000 was added to the supernatant and gently mixed on ice, followed by incubation for 3 h. Samples were centrifuged at 10,451 × g for 10 min, resuspended in SM buffer, extracted with chloroform to remove bacterial debris, and centrifuged at 4°C and 3,762 × g for 10 min.
Transmission Electron Microscopy
Phages were concentrated via CsCl density gradient (1.32, 1.45, 1.5, and 1.7 g/mL) centrifugation at 4°C and 35,000 × g for 3 h (Russell, 2001). The purified phages were dropped onto a 200-mesh copper film on a slide and negatively stained with phosphotungstic acid (2% w/v). A transmission electron microscope (JEOL JEM-1200EXII; Japan Electronics and Optics Laboratory, Tokyo, Japan) was used to observe the samples at an acceleration voltage of 80 kV.
Host Spectrum Determination
The host range of vB_EcoM_ECOO78 was determined by performing the spot test and double-layer agar plate test (Barrow et al., 1998). For the double-layer agar plate test, dilutions of phages and the tested E. coli strains were added to melted semi-solid medium (45°C) and immediately mixed, then poured onto LB solid medium plates. To perform the spot test, 200 μL of overnight-cultured E. coli were coated on LB solid medium plates and allowed to dry. Then, 10 μL of phage suspension was dropped onto each plate, and the plates were cultured at 37°C for an additional 10 h to observe the formation of clear spots.
Determination of Multiplicity of Infection (MOI)
The MOI was detected as described by Gu et al. (2011). Briefly, E. coli O78-3 host cells were cultured to logarithmic phase, and the phage was inoculated into liquid LB medium containing the host bacteria at MOIs of 10, 1, 10-1, 10-2, 10-3, 10-4, 10-5, or 10-6. The bacteria and phage mixtures were cultured at 37°C for 12 h. Phage titres in the filtrates were measured. The experiment was repeated three times.
One-Step Growth Curve Assay
A one-step growth curve was constructed as described previously by Gu et al. (2011), with minor modifications. E. coli O78-3 host cells were grown to the mid-logarithmic phase. Phages were added at an MOI of 10-5, followed by incubation for 15 min at 4°C. The mixture was centrifuged at 4°C and 15,049 × g for 30 s, and the pellet was suspended in 10 mL of fresh LB liquid medium. The samples were then incubated at 37°C at 180 rpm. Samples were taken every 5 min for the first 30 min, and at 40, 60, 80, 90, 100, 120, and 150 min to test phage titration. Tests were conducted three times. The burst size was defined as the ratio of the mean number of phage in final rise phase to the mean number of phage in the latent period.
DNA Sequencing and Analyses
The method used to extract the vB_EcoM_ECOO78 genome has been reported previously (Russell, 2001). Briefly, the concentrated phage was treated with DNase I (final concentration, 10 μg/mL) and RNase A (final concentration, 5 μg/mL) in SM buffer at 37°C for 1 h. Subsequently, ethylenediaminetetraacetic acid (EDTA) was added (pH 8.0, final concentration 25 mmol/mL). Finally, the phage genomic DNA was extracted using a Viral DNA Kit (Omega Bio-Tek®).
Whole-genome sequencing was performed by the Suzhou GENEWIZ Biotechnology Co., Ltd. using an Illumina HiSeq 2500 sequencer and SOAPdenovo v2.01 stitching to obtain the complete genome sequence of vB_EcoM_ECOO78. The identification of coding regions in the vB_EcoM_ECOO78 genome was based on the application of ORF Finder1. Open reading frames (ORFs) were automatically predicted using Glimmer2 and Genemark (Besemer and Borodovsky, 1999). The genome map of vB_EcoM_ECOO78 was drawn using CLC Main Workbench version 7.7.3 (CLC Bio-Qiagen, Aarhus, Denmark). To improve genome annotations/predictions, all proteins encoded by ORFs in the genome sequence of vB_EcoM_ECOO78 were subjected to a BLASTP search against the phage protein database at the National Center for Biotechnology Information3 and Pfam4. PSI-BLAST5 was used to predict the genes encoding the depolymerase. Phyre26 was used to predict the structure of the depolymerase(Kelley et al., 2015).
Expression and Purification of the Phage Depolymerase
ORF42, a putative phage depolymerase, was amplified from the purified phage vB_EcoM_ECOO78 by PCR using the primers Dpo42PF (5′-CGCGGATCCGTGGAACTGAAAACAT-3′) and Dpo42PR (5′-CCGCTCGAGTTAAATCAAGCCATAAATAGC-3′). Underlined nucleotides indicate the recognition sequences for BamHI and XhoI. The PCR fragment was excised with BamHI and XhoI and inserted into the pET-28a expression vector (Novagen, Madison, WI, United States). The constructed plasmid was used to transform E. coli BL21 cells. The ORF42 protein was expressed under 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) induction at 16°C overnight, purified from the soluble fraction using a Ni-nitrilotriacetic acid (NTA) column (Genscript, Nanjing, China) according to the manufacturer’s instructions, and then analyzed using sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) (Gu et al., 2014). Lysis buffer was composed of 50 mM Na2HPO4, 0.3 M NaCl, pH 8.0; the wash buffer was composed of 50 mM Na2HPO4, 0.3 M NaCl, pH 8.0 with 50 mM imidazole; the elution buffer was composed of 50 mM Na2HPO4, 0.3 M NaCl, pH 8.0 with 250 mM imidazole. Subsequently, the purified Dpo42 was diluted in sterile phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.2).
Biofilm Assay
The biofilm formation of E. coli isolates was evaluated using the 96-well micro-titre plate method as described previously (O’Toole et al., 1999), with some modifications. Briefly, E. coli isolates were inoculated into 3 mL of sterile TSB medium and grown for 16 h at 37°C. A 1:100 dilution of each E. coli culture was created in sterile TSB medium, and 200 μL of each diluted culture was added to six wells in untreated 96-well micro-titre plates (Nest; NEST Biotech Co, China). A total of 200 μL of fresh TSB medium was used as a negative control. The plates were incubated at 37°C for 72 h without agitation. Non-adherent cells were removed by pipetting out the culture and washing the wells twice with 200 μL of sterile PBS. Then, 200 μL of methanol was added to each well, and the plates were incubated for 30 min at room temperature. The methanol was removed, and the plates were dried at room temperature. Subsequently, 200 μL of 1% w/v crystal violet solution was added to each well, and the plates were incubated for 30 min at room temperature. Wells containing the biofilm total biomass were washed gently with sterile deionized water. The plates were left to dry at room temperature. Thereafter, 200 μL of 33% v/v glacial acetic acid was added to each well to solubilize the bound crystal violet from the stained E. coli biofilms. The optical density (OD) was measured at 590 nm using a Synergy 2 multi-mode microplate reader (BioTek, United States). The isolates were classified as having strong (4 × ODc ≤ OD), moderate (2 × ODc < OD ≤ 4 × ODc), weak (ODc < OD ≤ 2 × ODc) or non-biofilm formation (OD ≤ ODc) based on a previous report (Stepanovic et al., 2000). ODc is a cut-off of the OD measurement obtained in the negative control three times. The experiments were performed in triplicate, and the results are expressed as the means ± SDs.
Activity of Dpo42 against Biofilms
A spot test was performed to observe Dpo42 activity as described previously (Verma et al., 2009). In brief, LB agar was overlaid with 0.7% top agar inoculated with 200 μL of fresh log-phase bacterial culture. After drying, the purified Dpo42 (1 μg) was spotted onto the double-layer agar plate. Serial dilutions of Dpo42 (50, 5, 2.5, 1.25, 0.625, 0.3125, 0.156, and 0.078 ng) were also dropped onto host strain double-layer agar plates. The elution buffer diluted in PBS was used as a control. After overnight incubation at 37°C, plates were observed for the formation of semi-clear spots.
The lytic activity of Dpo42 (0.25 μg/μL) toward E. coli isolates was tested at 37°C (Gu et al., 2014). Cell counts were performed at 0 min, 30 min, 1, 2, and 4 h. The elution buffer diluted in PBS was used as a negative control.
Escherichia coli isolates E9 and HXM were used to test the preventative effects of Dpo42 on biofilm formation as described previously (O’Toole et al., 1999), with some modifications. Briefly, E. coli E9 and HXM were cultured to logarithmic phase, centrifuged, resuspended in equal amounts of TSB medium, mixed and (100 μL) added to each well of a 96-well micro-titre plate. The wells were then divided into four groups. The elution buffer diluted in PBS, 10 μg/well Dpo42, 25 μg/well Dpo42 or 50 μg/well Dpo42 at the volume of 100 μL was added to different groups; 200 μL of sterile TSB medium was used as a negative control. Each group contained triplicate samples. The plate was covered, and bacteria were permitted to adhere and grow at 37°C for 24, 72, and 96 h without agitation. After incubation, the residual biofilms were stained, and the OD590 was measured.
The effects of Dpo42 on 24 and 96 h old biofilms were also detected. E. coli E9 and HXM were cultured in the 96-well micro-titre plate to form 24 h old biofilm and 96 h old biofilm. The biofilms were treated with 10 μg/well Dpo42, 25 μg/well Dpo42, 50 μg/well Dpo42 or elution buffer diluted in PBS for 12 h. As a control, 200 μL of sterile TSB medium was used. Each group contained triplicate samples. After incubation, the residual biofilms were stained, and the OD590 was measured.
Capsule Staining
The capsule staining of E. coli E9 was negatively stained with Maneval’s solution and analyzed by microscopy (Cornelissen et al., 2011). Cells grown in the absence and presence of Dpo42 were transferred to 10 μL of 1% aqueous Congo red solution (Sigma–Aldrich, St. Louis, MO, United States) and mixed. The mixture was spread across a glass slide to form a thin film and then air dried. Another 10 μL of Maneval’s solution (3.33% phenol, 4.44% glacial acetic acid, 2.67% ferric chloride, 0.02% acid fuchsin; Sigma–Aldrich) was dropped across the glass slide. Capsules were negatively stained and appeared white underneath the microscope (100×, oil; Olympus CX-41; Olympus America, Center Valley, PA 18034-0610, United States).
Phylogenetic Tree Analysis
The terminase large subunit is one of the conserved phage proteins (Feiss and Rao, 2012). Thus, a phylogenetic tree analysis of vB_EcoM_ECOO78 was performed based on terminase large subunit sequences. The evolutionary history of the taxa was drawn using the neighbour-joining (NJ) method (Saitou and Nei, 1987) to construct a bootstrap consensus tree from 1,000 replicates. Evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are indicated as the units for the number of amino acid substitutions per site. Evolutionary analyses were carried out using MEGA7 (Kumar et al., 2016).
Phylogenetic relationships were determined based on the protein sequences for Dpo42 and 10 other phage depolymerases. A molecular phylogenetic analysis was performed using the maximum-likelihood method. Additionally, the evolutionary history was inferred using the maximum-likelihood method based on the Poisson correction model (Zuckerkandl and Pauling, 1965). The tree with the highest log likelihood (-7059.4930) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying the NJ and BioNJ algorithms to a matrix of pairwise distances estimated using a Jones–Taylor–Thornton (JTT) model, then selecting the topology with superior log likelihood values. Evolutionary analyses were carried out using MEGA7 (Kumar et al., 2016).
Statistical Analysis
One-way analysis of variance (ANOVA) was performed for the normally distributed data. The qualitative data were analyzed using the chi-square and two-sided Fisher’s exact tests. P-values < 0.05 were considered statistically significant. All tests were performed using SPSS version 19.0 (SPSS 111 Inc., Chicago, IL, United States).
Results
General Microbiological Characteristics of vB_EcoM_ECOO78
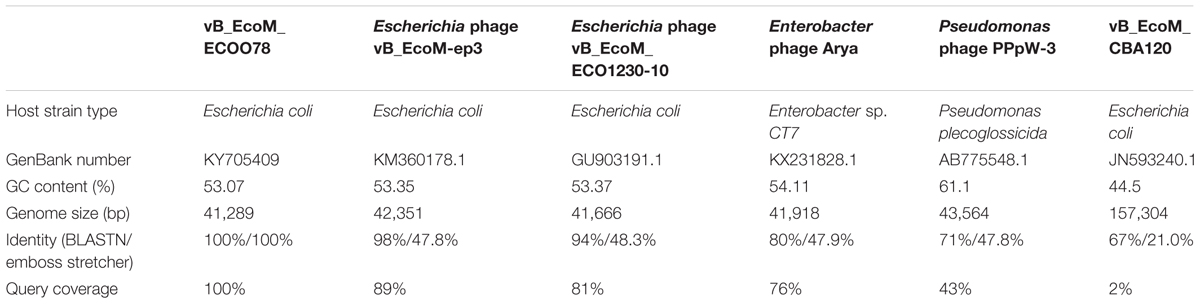
The clinical isolates were resistant to a variety of antibiotics, such as quinolones (67.6%), β-lactam antibiotics (76.5%) and aminoglycosides (58.8%) (Supplementary Table S1). The lytic phage vB_EcoM_ECOO78 was isolated successfully from sewage samples by using E. coli O78-3 as the host bacterium. vB_EcoM_ECOO78 lysed five out of the 34 tested clinical isolates of pathogenic E. coli, including E. coli O78-3 (the host strain) (Table 1). After prolonged incubation, the areas surrounding the phage plaques developed haloes that increased in size over the course of 96 h (Figure 1). The phage had an isometrically hexagonal head of 47 ± 2 nm in diameter. The head was separated from the tail sheath by a collar. The tail was contractile and approximately 149 ± 3 nm long. Based on these morphological characteristics, this phage is a member of the family Myoviridae (International Committee on Taxonomy of Viruses) (Figure 2A). The highest phage titre was observed at an MOI of 10-5 (Figure 2B). In the one-step growth curve analysis for the assessment of the infection process, vB_EcoM_ECOO78 had a latent period of approximately 60 min and a burst size of approximately 74 plaque forming units (PFU)/infection (Figure 2C).

FIGURE 1. The appearance of plaques formed by phage vB_EcoM_ECOO78 on E. coli O78-3 biofilms. Pure phage plaques appeared after 12 h, and the haloes surrounding the phage plaques grew in size after 48 and 96 h.
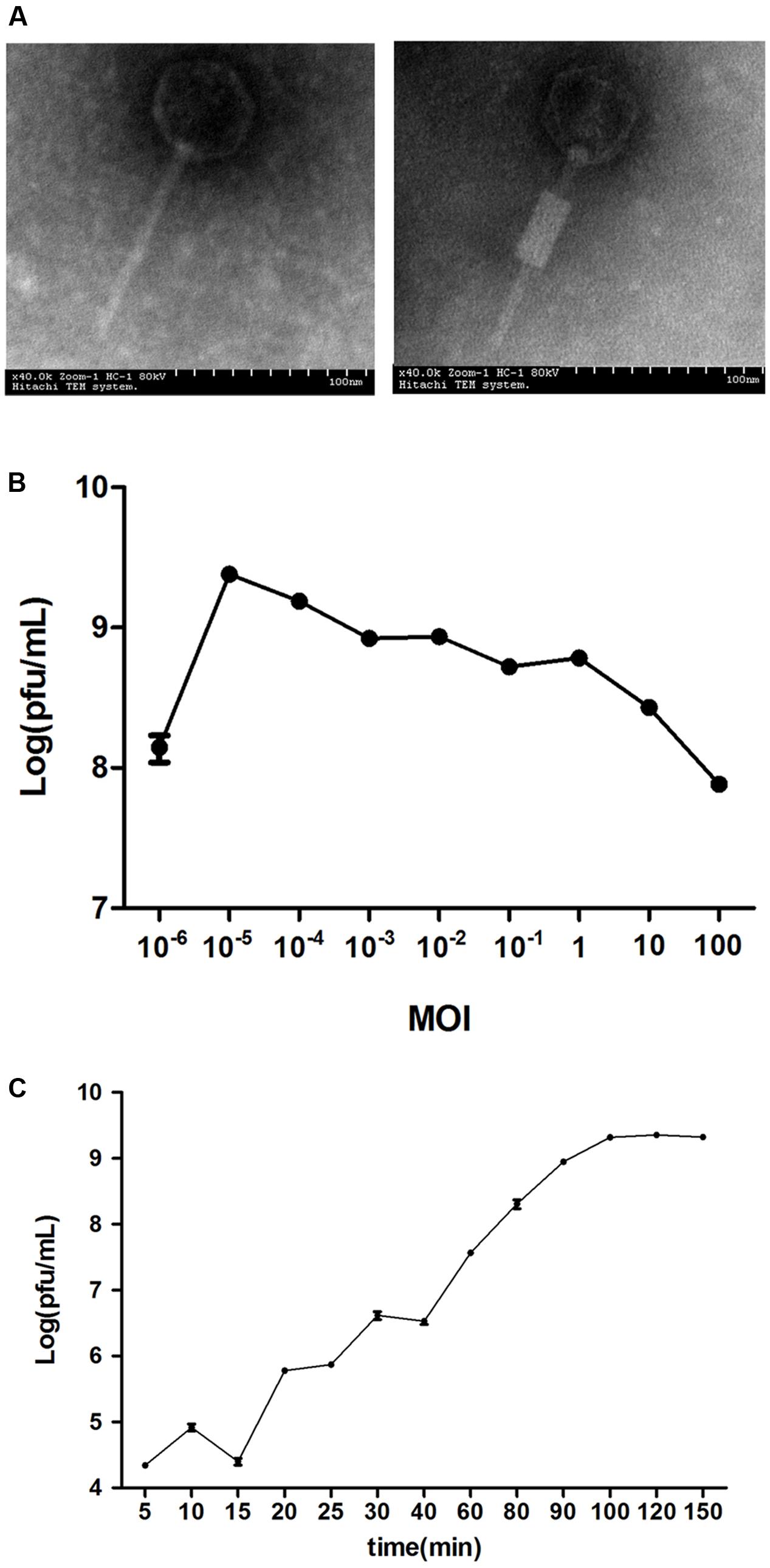
FIGURE 2. The general characteristics of vB_EcoM_ECOO78. (A) The morphology of phage vB_EcoM_ECOO78, as revealed by transmission electron microscope (scale bar: 100 nm). (B) Determination of the multiplicity of infection for this bacteriophage. The x-axis indicates different MOIs, and the y-axis indicates phage titre. Each dot on the graph represents a mean titre. (C) One-step growth curve for vB_EcoM_ECOO78. vB_EcoM_ECOO78 was added at an MOI of 10-5, and culture samples were harvested at regular intervals. The x-axis indicates the time post-infection, and the y-axis indicates phage titres. Each dot on the graph represents a mean titre.
General Features of the vB_EcoM_ECOO78 Genome
The genome sequence of vB_EcoM_ECOO78 was deposited in GenBank under accession number KY705409. The complete genome of the bacteriophage vB_EcoM_ECOO78 was 41,289 bp with the following nucleotide composition: A (21.57%), T (25.35%), G (25.74%) and C (27.33%). The GC content was 53.07%, and the genome contained both structural and non-structural genes. Based on a comparative analysis of the complete genome, five phages showed similarity to vB_EcoM_ECOO78, specifically vB_EcoM-ep3 (Lv et al., 2015), vB_EcoM_ECO1230-10 (Santos and Bicalho, 2011), Enterobacter phage Arya (unpublished), Pseudomonas phage PPpW-3 (Kawato et al., 2015) and vB_EcoM_CBA120 (unpublished) (Figure 3A and Table 2). vB_EcoM_ECOO78 contained 56 predicted ORFs, 51 (91.07%) of which were assumed to be functional. The coordinates and best matches for the sequences are shown in Supplementary Table S2. The remaining five ORFs showed no sequence identity to any of the sequences in the database. As seen from the phylogenetic trees reconstructed based on the terminase large subunit (Figure 3B), vB_EcoM_ECOO78 has higher homology with vB_EcoMep3, vB_EcoM_ECO1230-10, Pseudomonas phage PPpW-3 and Enterobacter phage Arya at the same evolutionary level. Shigella phage SfIV, Salmonella phage E1, Salmonella phage IME207, Stenotrophomonas phage Smp131, Enterobacteria phage Bp4, Escherichia phage EC1-UPM, Escherichia phage PhaxI, Escherichia phage vB EcoM CBA120, Vibrio phage vB VpaM MAR and Vibrio harveyi bacteriophage VHML are more distantly related.
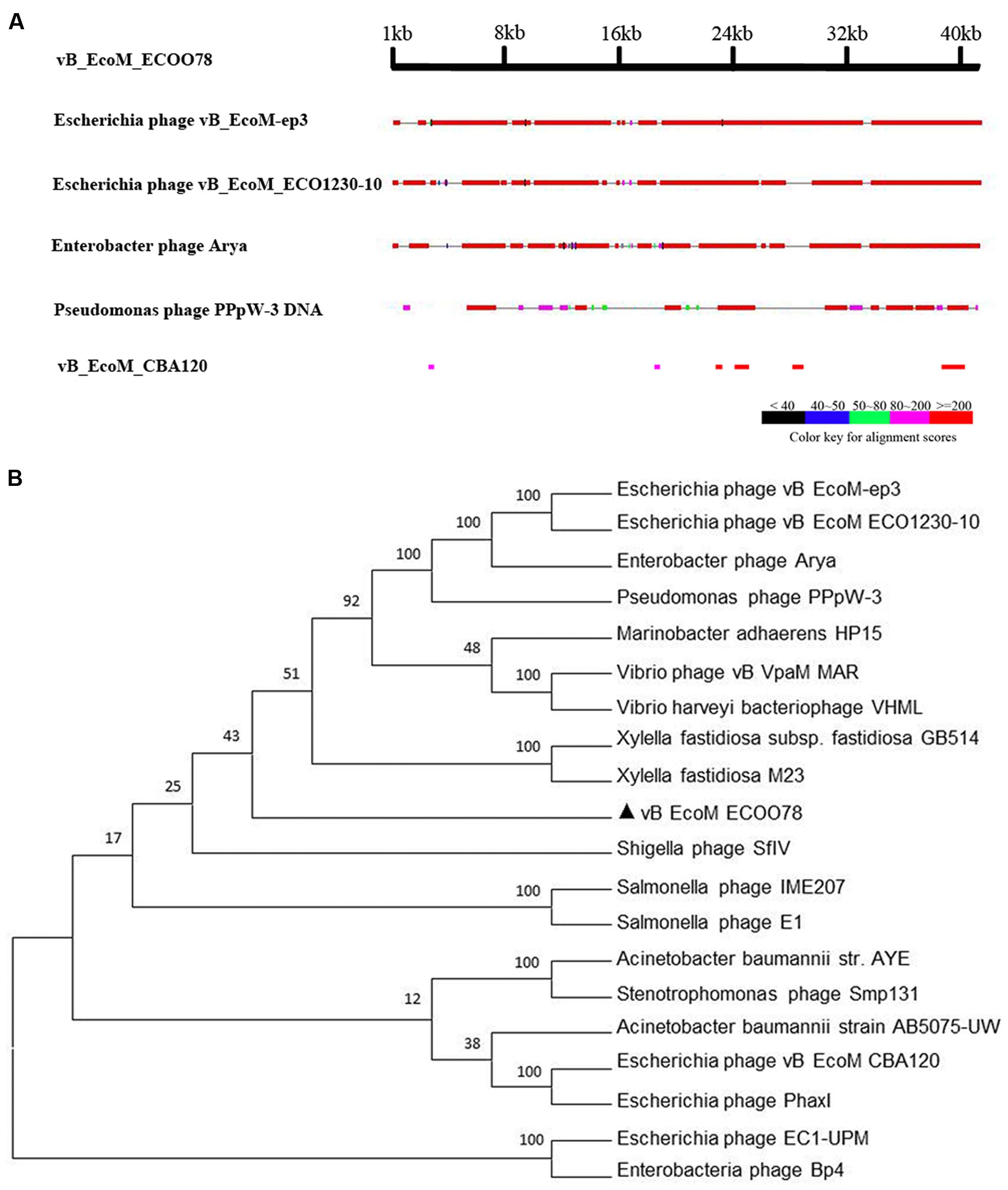
FIGURE 3. (A) A general comparison of the genome of vB_EcoM_ECOO78 with other phages. (B) Phylogenetic tree analysis of vB_EcoM_ECOO78 based on terminase large subunit sequences. The numbers next to the branches are bootstrap value and represent confidence (%).
Approximately 90.2% of ORFs (46 ORFs) started with an ATG codon, 7.8% (4 ORFs) used GTG and only one ORF started with TTG. TGA and TAA were stop codons for 27 (52.9%) and 22 (43.1%) ORFs, respectively. Only ORF48 and ORF53 had TAG as a stop codon. The vB_EcoM_ECOO78 genome is modular (Figure 4). Based on bioinformatics predictions, vB_EcoM_ECOO78 encoded a cluster of proteins involved in DNA replication and phage metabolism, including ORF09 (DNA methylase), ORF10 (primase), ORF22 (replicative DNA helicase), ORF26 (single-stranded DNA-binding protein), ORF29 (ATPase), ORF31 (transcriptional regulator) and ORF33 (putative transcriptional regulator). The lysin (ORF03) and holin (ORF05) sequences are important for phage lysis and had ORF sizes of 489 and 324 bp, respectively. The DNA packaging proteins included ORF55 (large terminase subunit) and ORF56 (putative small terminase subunit). The vB_EcoM_ECOO78 genome also encoded proteins associated with tail-baseplate-head morphogenesis, such as ORF34 (tail protein), ORF35 (putative phage tail protein), ORF36 (putative phage tail protein), ORF38 (putative phage tail protein), ORF39 (tail protein), ORF41 (tail protein), ORF43 (tail protein), ORF44 (tail protein), ORF45 (tail protein), ORF52 (major capsid protein) and ORF53 (capsid protein).
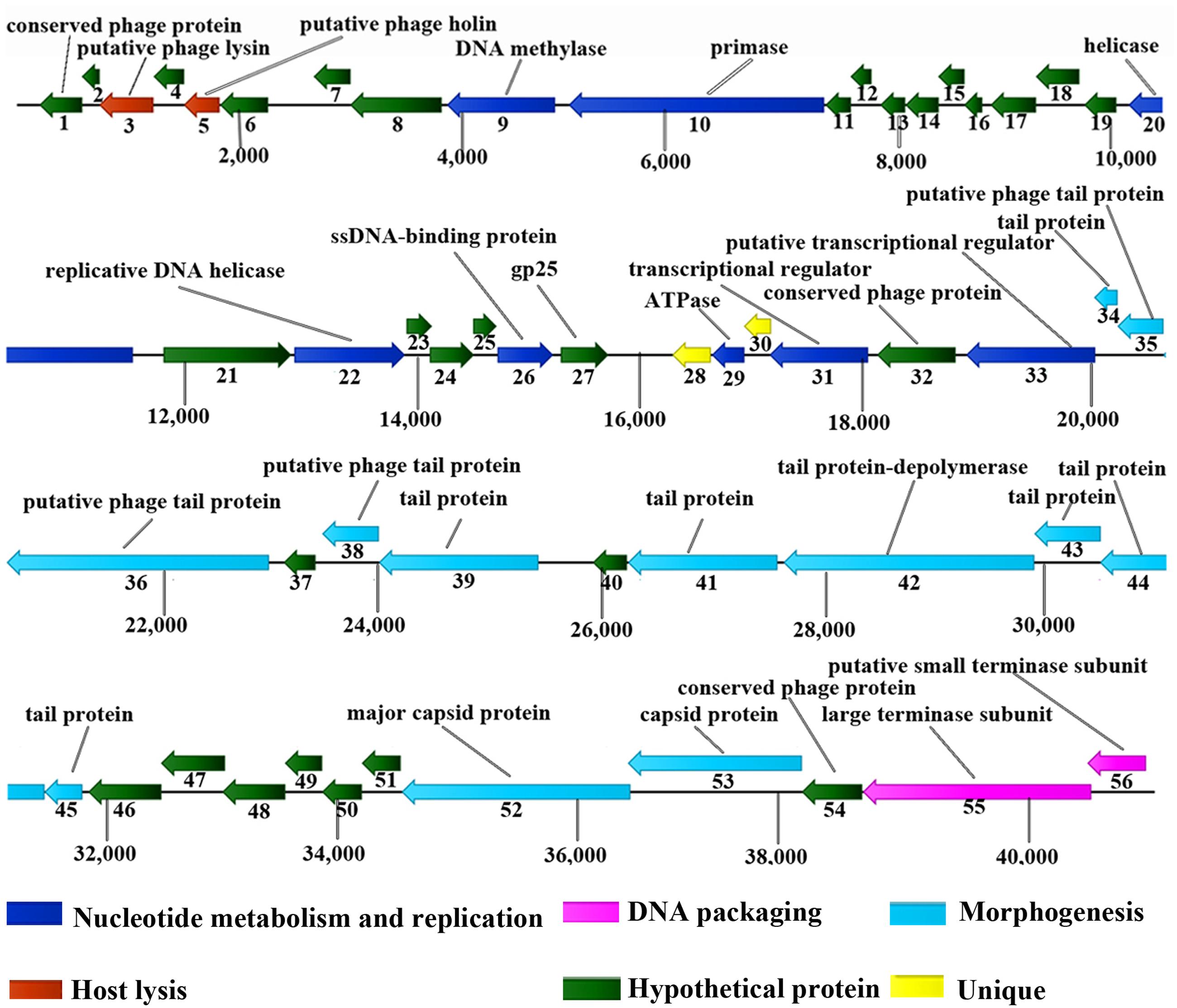
FIGURE 4. Genomic map of vB_EcoM_ECOO78. Fifty-six open ORFs are presented as arrows; the direction of each arrow represents the direction of transcription. Proposed modules are based on hypothetical functions predicted from bioinformatic analysis.
Analysis of Predicted Genes Encoding an EPS Depolymerase
PSI-BLAST indicated that ORF42 demonstrated homology to certain depolymerases, implying that ORF42 might have a function related to EPS synthesis. However, this ORF showed low identity (≤31.0%) to other phage depolymerases (Supplementary Table S3). Dpo42 contained 747 amino acids with a molecular weight of 78.58 KDa and a theoretical pI of 4.77. The secondary structure of Dpo42 was predicted using Phyre2 (Supplementary Figure S1) and included nine α-helices, 55 β-strands and disordered regions. Unfortunately, although the model confidence is 98.0%, due to the low coverage (9%) and identity (45%) with other homologous proteins (including C4ojpC), the modeling failed. As shown in the protein-level phylogenetic tree (Figure 5), Dpo42 (black triangle) represents a different evolutionary offshoot branch from the 10 other similar phage depolymerases.
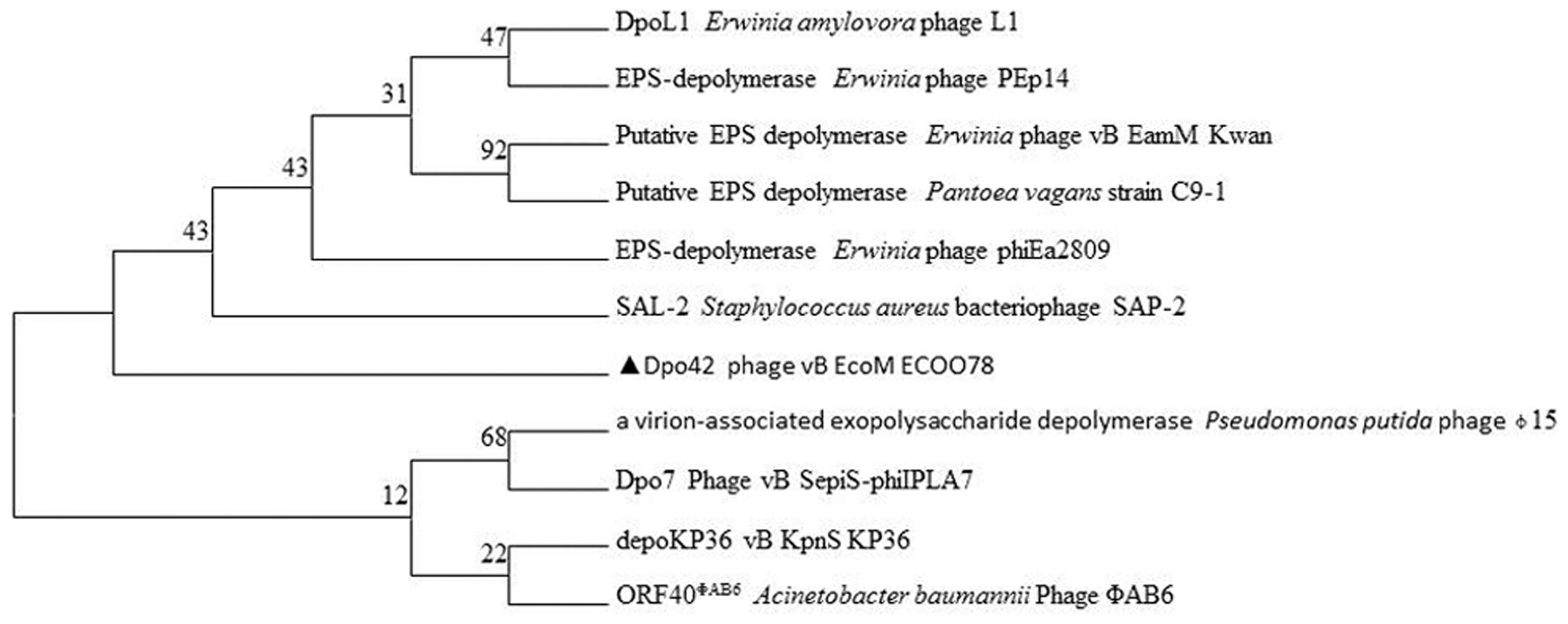
FIGURE 5. Phylogenetic relationships based on the protein sequences for Dpo42 and 10 other phage depolymerases.
Dpo42 Antibiofilm Activity
The predicted size of Dpo42 was ∼78.58 KDa, and the protein was expressed and purified (1 μg/μL) from supernatant liquid (Figure 6A). Though Dpo42 didn’t show a lytic activity against the E. coli (data not shown), Dpo42 prevented the biofilm formation of 15 clinical E. coli isolates (Table 1). In spot tests, purified Dpo42 (0.1 μg) prevented the biofilm formation of E. coli O78-3, and progressive semi-clear spot formation was observed on the plate. Spot size constantly increased after prolonged incubation until 96 h (Supplementary Figure S2). We subsequently dropped serial dilutions of Dpo42 onto the host strain (Figure 6B). As the concentration of Dpo42 decreased, the sizes of the semi-clear circles on the plate also decreased. When the concentration of Dpo42 decreased to 0.156 ng, the size of the semi-clear spot resembled that of the negative control.
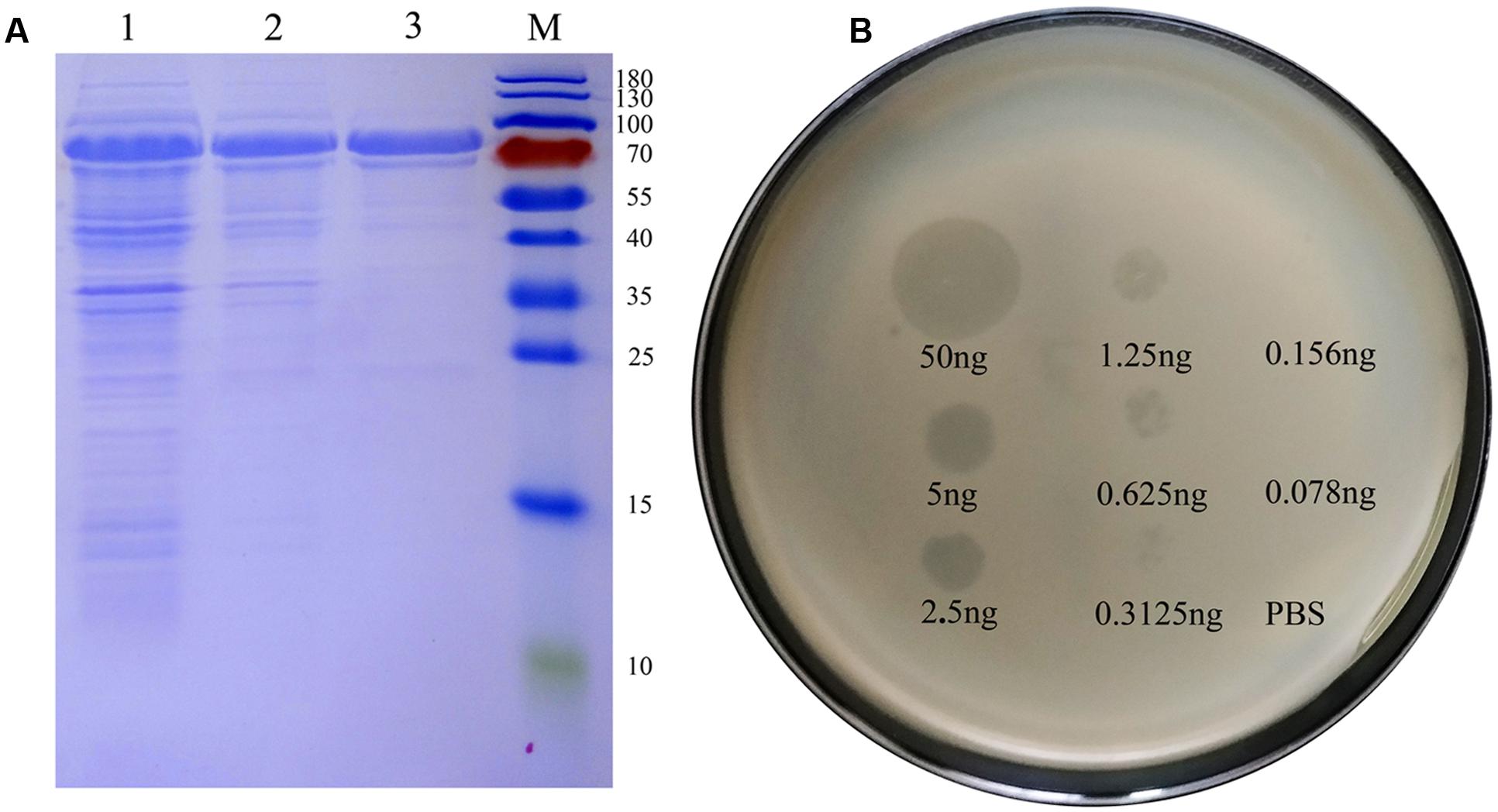
FIGURE 6. Dpo42 overexpression and activity. (A) An SDS-PAGE analysis of purified Dpo42. Lanes: M, protein markers; 1, the induced E. coli BL21 cells; 2, supernatant of the induced E. coli BL21 cells; 3, purified Dpo42. (B) Dpo42 activity against its natural substrate. Serial dilutions of Dpo42 were dropped on the host strain. Elution buffer diluted in PBS was used as a control.
The enzymatic activity of Dpo42 was further tested against clinical E. coli strains using 96-well micro-titre plates. Compared with the negative control, the E9 and HXM strains formed strong biofilms (4 × ODc ≤ OD) at 24, 72, and 96 h. By contrast, three groups containing Dpo42 only formed weak biofilms (ODc < OD ≤ 2 × ODc). The OD590 values of three groups containing Dpo42 were much lower than that of the positive control (P < 0.001; Figure 7). Moreover, the OD590 of the 50 μg of Dpo42 group was lower than that of the 10 μg of Dpo42 group. It indicated that Dpo42 can prevent the formation of the biofilm and exhibited dose-dependent activity during 96 h. Additionally, 24 and 96 h old biofilms were treated with different concentrations of Dpo42 (10 μg/well, 25 μg/well, 50 μg/well). The strong biofilms (4 × ODc ≤ OD) formed at 24 and 96 h by E9 and HXM strains couldn’t be destroyed by Dpo42. The OD590 values of three groups treated with Dpo42 were much higher than that of the negative control (P < 0.001; Figure 8). Thus, Dpo42 can only prevent the formation of biofilm, but can not remove it.
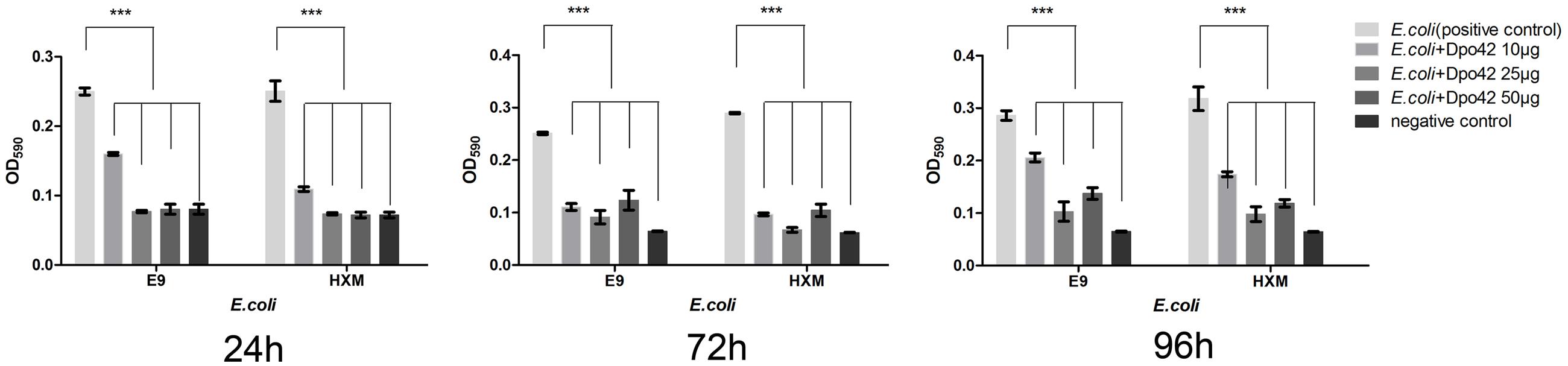
FIGURE 7. The preventative effect of Dpo42 on biofilm formation. Different doses of Dpo42 or elution buffer diluted in PBS were added to the wells of a 96-well micro-titre plate that contained E. coli E9 or HXM strains. The biofilm formation of different groups was detected at 24, 72, and 96 h. ∗∗∗P < 0.001. Error bars = ± SD; n = 3.
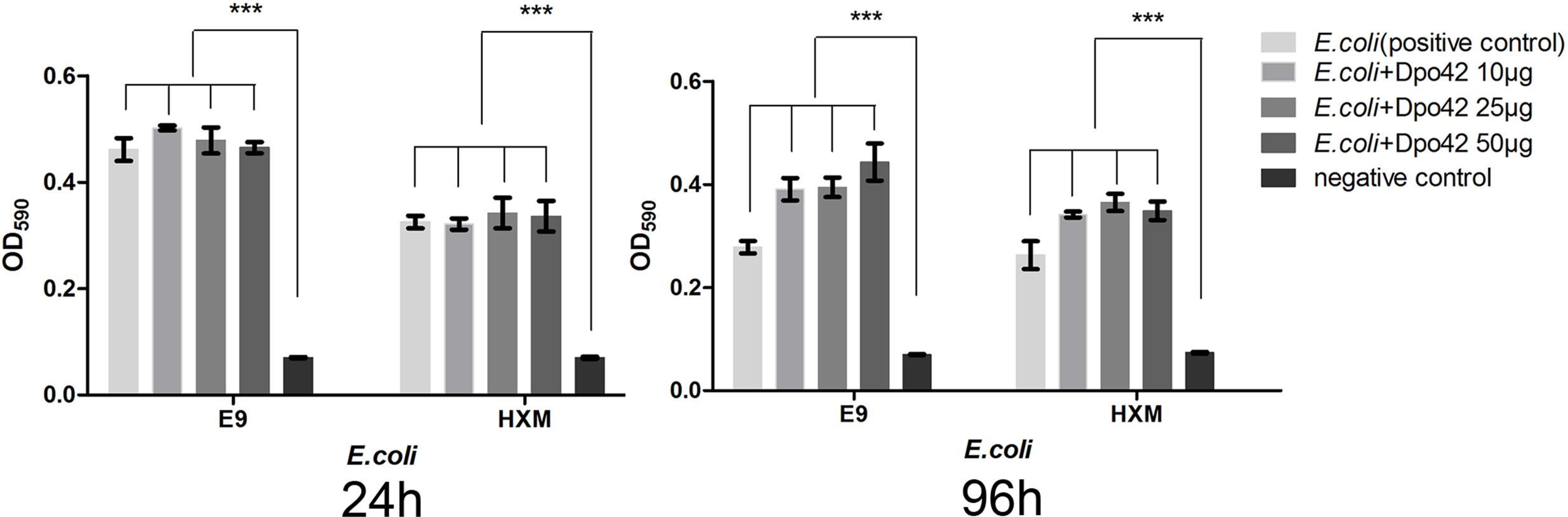
FIGURE 8. The biofilm removing activity of Dpo42. The E9 and HXM strains formed strong biofilms at 24 or 96 h. The biofilms were treated with different doses of Dpo42. ∗∗∗P < 0.001. Error bars = ± SD; n = 3.
Staining assays indicated that E. coli adhered to each other and were encircled by a white capsule. By contrast, cells were separated and almost lost the white capsule after treatment with Dpo42 (Figure 9). Thus, the target of Dpo42 belongs to the capsular polysaccharides.
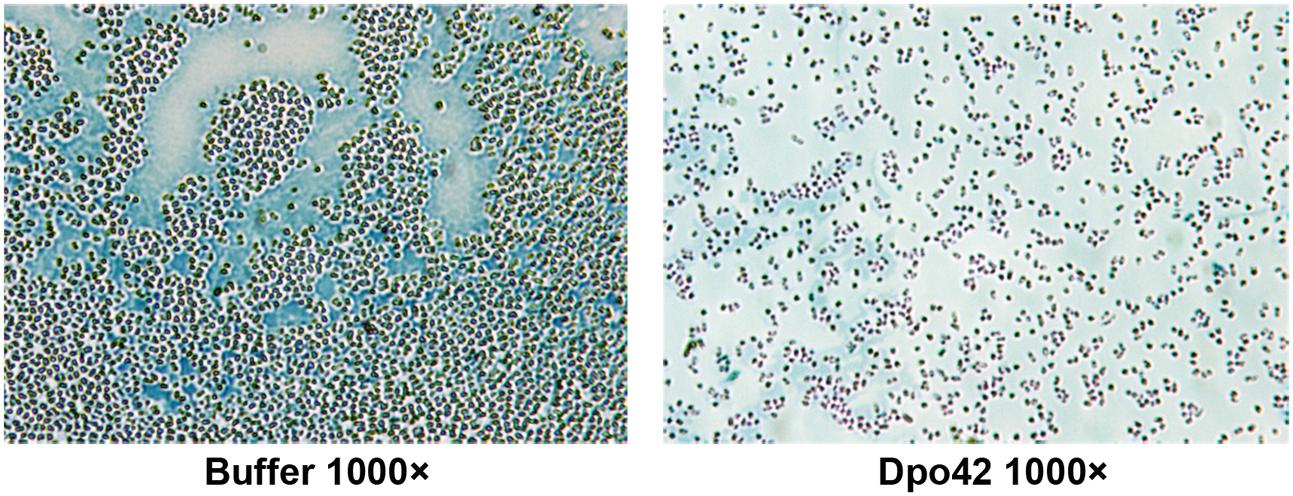
FIGURE 9. Capsule staining of E. coli E9 in the absence and presence of Dpo42. Capsules were negatively stained with Maneval’s solution and appeared white underneath the microscope.
Discussion
In this study, vB_EcoM_ECOO78 was isolated and observed to induce the formation of increasing halos surrounding the lysis plaques. This phenomenon was presumably caused by a phage-related depolymerase (Harper et al., 2014; Pires et al., 2016). Depolymerases have multiple applications for the prevention or eradication of biofilms (Hughes et al., 1998b; Verma et al., 2010) and may be useful as therapeutic agents against bacterial pathogens (Mushtaq et al., 2004; Scorpio et al., 2008). To identify the depolymerase of vB_EcoM_ECOO78, the complete genome of this phage was sequenced and analyzed. The vB_EcoM_ECOO78 genome was modular, which is common among phages (Veesler and Cambillau, 2011). A comparative analysis indicated that five phages, specifically vB_EcoM-ep3 (Lv et al., 2015), vB_EcoM_ECO1230-10 (Santos and Bicalho, 2011), Enterobacter phage Arya (unpublished), Pseudomonas phage PPpW-3 (Kawato et al., 2015) and vB_EcoM_CBA120 (unpublished), showed similarity to vB_EcoM_ECOO78. However, there were certain genes in vB_EcoM_ECOO78 that shared no identity with these similar phages. Furthermore, three ORFs, specifically ORF03 (lysin), ORF39 (tail protein) and ORF53 (capsid protein), were related to E. coli APEC O2, Haematospirillum jordaniae and Xylella fastidiosa MUL0034, respectively. Genetic drift and recombination between different phages, as well as between vB_EcoM_ECOO78 and bacteria, were evident for certain ORFs.
Based on our bioinformatics predictions, the ORF42 of vB_EcoM_ECOO78 (Dpo42) most likely encodes a depolymerase. Interestingly, Dpo42 showed low identity (≤31.0%) to other phage depolymerases. Thus, Dpo42 is a novel depolymerase from an E. coli phage. The Klebsiella phage depolymerase depoKP36 reportedly contains a high β-sheet content. This was similar to Dpo42, which contains 55 β-sheets. Given our observation of the similarity between Dpo42 and the tail proteins of other phages, Dpo42 is likely a tail-related depolymerase.
To further characterise the activity of Dpo42, this depolymerase was expressed in E. coli. Dpo42 expression did not affect E. coli BL21. However, Dpo42 did demonstrate biofilm-formation prevention activity against several E. coli isolates in vitro. In addition, dose-dependent activity was observed when E. coli biofilms were treated with Dpo42 during 96 h. The bacterial EPS-degrading abilities of phage depolymerases have been reported for more than 70 years (Zobell, 1943; Bartell et al., 1966). To the best of our knowledge, there have only been three E. coli phage-derived depolymerases purified from E. coli phage lysates, and all were able to degrade the capsular layers on host cells (Tomlinson and Taylor, 1985; McCallum et al., 1989; Petter and Vimr, 1993). In addition, to the best of our knowledge, Dpo42 is the first genetically engineered phage-derived depolymerase with the ability to prevent E. coli biofilm formation. Notably, Dpo42 shows no similarity to the three previously reported E. coli phage-derived depolymerases. Among previously described depolymerases, only DepoKP36 has demonstrated a broad depolymerization range, hydrolysing the EPS formed Sby 8/11 Klebsiella isolates (Majkowska-Skrobek et al., 2016). In this study, Dpo42 prevented biofilm formation of 15/34 E. coli isolates and the target of Dpo42 was capsular polysaccharide.
Although most bacterial biofilms are composed of EPS, they also contain water, proteins, lipids, nucleic acids, and mineral ions (Coenye and Nelis, 2010; Flemming and Wingender, 2010). The residual proteins and nucleic acids may block depolymerases from eradicating the biofilms completely. To our knowledge, only phage-derived depolymerase Dpo7 has been proven to be effective against polysaccharidic matrixes of biofilms formed by Staphylococcus epidermidis and Staphylococcus aureus (Gutierrez et al., 2015). Dpo42 was unable to remove young or mature biofilms. We conclude that Dpo42 may have its effect on PGA, in consideration that PGA serves in the initial adhesion, which is the first step involved in biofilm formation (Sharma et al., 2016).
It may be possible to genetically modify depolymerases to amplify enzyme activity or mix different depolymerases to create a therapeutic cocktail (Yen et al., 2017). Bansal et al. (2014) evaluated the activity of depoKP36 in combination with four classes of antibiotics, specifically ciprofloxacin (a fluoroquinolone), oxytetracycline (a tetracycline), gentamicin and chloramphenicol, and observed synergistic effects for the combination of depoKP36 and gentamicin, as well as improved gentamicin activity. Thus, combination therapy employing antibiotics and a depolymerase may represent a more efficient treatment strategy for bacterial infections.
Author Contributions
ZG, JG, and JY drafted the main manuscript and performed the data analysis; ZG, JH, GY, LL, SW, LY, LZ, AG, XF, WH, JG, and JY planned and performed experiments; ZG, JG, LL, and JY were responsible for experimental design; and ZG, JY, and JG were responsible for guiding and supporting the experiments and manuscript revisions.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 31572553 and 31502103), the National Key Research and Development Program of China (No. 2017YFD0501000), the Development Program of the First Hospital of Jilin University (No. JDYY82017022), and the National Key Basic Research Program of China (No. 2013CB127205).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01460/full#supplementary-material
Footnotes
- ^ http://bioinf.ibun.unal.edu.co/servicios/sms/orf_find.html
- ^ http://ccb.jhu.edu/software/glimmer/index.shtml
- ^ https://blast.ncbi.nlm.nih.gov/
- ^ http://pfam.xfam.org/family
- ^ http://www.ebi.ac.uk/Tools/sss/psiblast
- ^ http://www.sbg.bio.ic.ac.uk/phyre2
References
Bansal, S., Harjai, K., and Chhibber, S. (2014). Depolymerase improves gentamicin efficacy during Klebsiella pneumoniae induced murine infection. BMC Infect. Dis. 14:456. doi: 10.1186/1471-2334-14456
Barrow, P., Lovell, M., and Berchieri, A. J. (1998). Use of lytic bacteriophage for control of experimental Escherichia coli septicemia and meningitis in chickens and calves. Clin. Diagn. Lab. Immunol 5, 294–298.
Bartell, P. F., Orr, T. E., and Lam, G. K. (1966). Polysaccharide depolymerase associated with bacteriophage infection. J. Bacteriol. 92, 56–62.
Besemer, J., and Borodovsky, M. (1999). Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27, 3911–3920. doi: 10.1093/nar/27.19.3911
Born, Y., Fieseler, L., Klumpp, J., Eugster, M. R., Zurfluh, K., Duffy, B., et al. (2014). The tail-associated depolymerase of Erwinia amylovora phage L1 mediates host cell adsorption and enzymatic capsule removal, which can enhance infection by other phage. Environ. Microbiol. 16, 2168–2180. doi: 10.1111/1462-2920.12212
Canton, R., Novais, A., Valverde, A., Machado, E., Peixe, L., Baquero, F., et al. (2008). Prevalence and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 14(Suppl. 1), 144–153. doi: 10.1111/j.1469-0691.2007.01850.x
Chanishvili, N. (2012). Phage therapy–history from Twort and d’Herelle through Soviet experience to current approaches. Adv. Virus Res. 83, 3–40. doi: 10.1016/B978-0-12-394438-2.00001-3
Coenye, T., and Nelis, H. J. (2010). In vitro and in vivo model systems to study microbial biofilm formation. J. Microbiol. Methods 83, 89–105. doi: 10.1016/j.mimet.2010.08.018
Cornelissen, A., Ceyssens, P. J., Krylov, V. N., Noben, J. P., Volckaert, G., and Lavigne, R. (2012). Identification of EPS-degrading activity within the tail spikes of the novel Pseudomonas putida phage AF. Virology 434, 251–256. doi: 10.1016/j.virol.2012.09.030
Cornelissen, A., Ceyssens, P. J., T’Syen, J., Van Praet, H., Noben, J. P., Shaburova, O. V., et al. (2011). The T7-related Pseudomonas putida phage phi15 displays virion-associated biofilm degradation properties. PLoS ONE 6:e18597. doi: 10.1371/journal.pone.0018597
Costerton, J. W. (1995). Overview of microbial biofilms. J. Ind. Microbiol. 15, 137–140. doi: 10.1007/BF01569816
Feiss, M., and Rao, V. B. (2012). The bacteriophage DNA packaging machine. Adv. Exp. Med. Biol. 726, 489–509. doi: 10.1007/978-1-4614-0980-9_22
Fischetti, V. A. (2005). Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 13, 491–496. doi: 10.1016/j.tim.2005.08.007
Flemming, H. C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Gu, J., Feng, Y., Feng, X., Sun, C., Lei, L., Ding, W., et al. (2014). Structural and biochemical characterization reveals LysGH15 as an unprecedented “EF-hand-like” calcium-binding phage lysin. PLoS Pathog. 10:e1004109. doi: 10.1371/journal.ppat.1004109
Gu, J., Xu, W., Lei, L., Huang, J., Feng, X., Sun, C., et al. (2011). LysGH15, a novel bacteriophage lysin, protects a murine bacteremia model efficiently against lethal methicillin-resistant Staphylococcus aureus infection. J. Clin. Microbiol. 49, 111–117. doi: 10.1128/JCM.01144-10
Gutierrez, D., Briers, Y., Rodriguez-Rubio, L., Martinez, B., Rodriguez, A., Lavigne, R., et al. (2015). Role of the pre-neck appendage protein (Dpo7) from phage vB_SepiS-phiIPLA7 as an anti-biofilm agent in Staphylococcal species. Front. Microbiol. 6:1315. doi: 10.3389/fmicb.2015.01315
Harper, D. R., Parracho, H. M. R. T., Walker, J., Sharp, R., Hughes, G., Werthén, M., et al. (2014). Bacteriophages and biofilms. Antibiotics 3, 270–284. doi: 10.3390/antibiotics3030270
Hughes, K. A., Sutherland, I. W., Clark, J., and Jones, M. V. (1998a). Bacteriophage and associated polysaccharide depolymerases–novel tools for study of bacterial biofilms. J. Appl. Microbiol. 85, 583–590.
Hughes, K. A., Sutherland, I. W., and Jones, M. V. (1998b). Biofilm susceptibility to bacteriophage attack: the role of phage-borne polysaccharide depolymerase. Microbiology 144(Pt 11), 3039–3047. doi: 10.1099/00221287-144-11-3039
Kawato, Y., Yasuike, M., Nakamura, Y., Shigenobu, Y., Fujiwara, A., Sano, M., et al. (2015). Complete genome sequence analysis of two Pseudomonas plecoglossicida phages, potential therapeutic agents. Appl. Environ. Microbiol. 81, 874–881. doi: 10.1128/AEM.03038-14
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lai, M. J., Chang, K. C., Huang, S. W., Luo, C. H., Chiou, P. Y., Wu, C. C., et al. (2016). The tail associated protein of Acinetobacter baumannii phage PhiAB6 is the host specificity determinant possessing exopolysaccharide depolymerase activity. PLoS ONE 11:e0153361. doi: 10.1371/journal.pone.0153361
Latka, A., Maciejewska, B., Majkowska-Skrobek, G., Briers, Y., and Drulis-Kawa, Z. (2017). Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 101, 3103–3119. doi: 10.1007/s00253-017-8224-6
Lin, N. T., Chiou, P. Y., Chang, K. C., Chen, L. K., and Lai, M. J. (2010). Isolation and characterization of phi AB2: a novel bacteriophage of Acinetobacter baumannii. Res. Microbiol. 161, 308–314. doi: 10.1016/j.resmic.2010.03.007
Lv, M., Wang, S., Yan, G., Sun, C., Feng, X., Gu, J., et al. (2015). Genome sequencing and analysis of an Escherichia coli phage vB_EcoM-ep3 with a novel lysin, Lysep3. Virus Genes 50, 487–497. doi: 10.1007/s11262-015-1195-8
Majkowska-Skrobek, G., Latka, A., Berisio, R., Maciejewska, B., Squeglia, F., Romano, M., et al. (2016). Capsule-targeting depolymerase, derived from Klebsiella KP36 phage, as a tool for the development of anti-virulent strategy. Viruses 8:E324. doi: 10.3390/v8120324
McCallum, K. L., Laakso, D. H., and Whitfield, C. (1989). Use of a bacteriophage-encoded glycanase enzyme in the generation of lipopolysaccharide O side chain deficient mutants of Escherichia coli O9:K30 and Klebsiella O1:K20: role of O and K antigens in resistance to complement-mediated serum killing. Can. J. Microbiol. 35, 994–999. doi: 10.1139/m89-166
Mushtaq, N., Redpath, M. B., Luzio, J. P., and Taylor, P. W. (2004). Prevention and cure of systemic Escherichia coli K1 infection by modification of the bacterial phenotype. Antimicrob. Agents Chemother. 48, 1503–1508. doi: 10.1128/AAC.48.5.1503-1508.2004
Nelson, D., Loomis, L., and Fischetti, V. A. (2001). Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. U.S.A. 98, 4107–4112. doi: 10.1073/pnas.061038398
Oliveira, H., Sillankorva, S., Merabishvili, M., Kluskens, L. D., and Azeredo, J. (2015). Unexploited opportunities for phage therapy. Front. Pharmacol. 6:180. doi: 10.3389/fphar.2015.00180
O’Toole, G., Kaplan, H. B., and Kolter, R. (2000). Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79. doi: 10.1146/annurev.micro.54.1.49
O’Toole, G. A., Pratt, L. A., Watnick, P. I., Newman, D. K., Weaver, V. B., and Kolter, R. (1999). Genetic approaches to study of biofilms. Methods Enzymol. 310, 91–109. doi: 10.1016/S0076-6879(99)10008-9
Petter, J. G., and Vimr, E. R. (1993). Complete nucleotide sequence of the bacteriophage K1F tail gene encoding endo-N-acylneuraminidase (endo-N) and comparison to an endo-N homolog in bacteriophage PK1E. J. Bacteriol. 175, 4354–4363. doi: 10.1128/jb.175.14.4354-4363.1993
Pires, D. P., Oliveira, H., Melo, L. D., Sillankorva, S., and Azeredo, J. (2016). Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 100, 2141–2151. doi: 10.1007/s00253-015-7247-0
Russell, J. S. (2001). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press.
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Santos, T. M., and Bicalho, R. C. (2011). Complete genome sequence of vB_EcoM_ECO1230-10: a coliphage with therapeutic potential for bovine metritis. Vet. Microbiol. 148, 267–275. doi: 10.1016/j.vetmic.2010.08.034
Scorpio, A., Tobery, S. A., Ribot, W. J., and Friedlander, A. M. (2008). Treatment of experimental anthrax with recombinant capsule depolymerase. Antimicrob. Agents Chemother. 52, 1014–1020. doi: 10.1128/AAC.00741-07
Sharma, G., Sharma, S., Sharma, P., Chandola, D., Dang, S., Gupta, S., et al. (2016). Escherichia coli biofilm: development and therapeutic strategies. J. Appl. Microbiol. 121, 309–319. doi: 10.1111/jam.13078
Stepanovic, S., Vukovic, D., Dakic, I., Savic, B., and Svabic-Vlahovic, M. (2000). A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40, 175–179. doi: 10.1016/S0167-7012(00)00122-6
Stewart, P. S., and Costerton, J. W. (2001). Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138. doi: 10.1016/S0140-6736(01)05321-1
Stewart, P. S., Murga, R., Srinivasan, R., and de Beer, D. (1995). Biofilm structural heterogeneity visualized by three microscopic methods. Water Res. 29, 2006–2009. doi: 10.1016/0043-1354(94)00339-9
Summers, W. C. (2001). Bacteriophage therapy. Annu. Rev. Microbiol. 55, 437–451. doi: 10.1146/annurev.micro.55.1.437
Tomlinson, S., and Taylor, P. W. (1985). Neuraminidase associated with coliphage E that specifically depolymerizes the Escherichia coli K1 capsular polysaccharide. J. Virol. 55, 374–378.
Veesler, D., and Cambillau, C. (2011). A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol. Mol. Biol. Rev. 75, 423–433. doi: 10.1128/MMBR.00014-11
Verma, V., Harjai, K., and Chhibber, S. (2009). Characterization of a T7-like lytic bacteriophage of Klebsiella pneumoniae B5055: a potential therapeutic agent. Curr. Microbiol. 59, 274–281. doi: 10.1007/s00284-009-9430-y
Verma, V., Harjai, K., and Chhibber, S. (2010). Structural changes induced by a lytic bacteriophage make ciprofloxacin effective against older biofilm of Klebsiella pneumoniae. Biofouling 26, 729–737. doi: 10.1080/08927014.2010.511196
Whitfield, C., and Roberts, I. S. (1999). Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31, 1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x
Yen, M., Cairns, L. S., and Camilli, A. (2017). A cocktail of three virulent bacteriophages prevents Vibrio cholerae infection in animal models. Nat. Commun. 8:14187. doi: 10.1038/ncomms14187
Zhang, H., Gao, Y., and Chang, W. (2016). Comparison of extended-spectrum beta-lactamase-producing Escherichia coli isolates from drinking well water and pit latrine wastewater in a rural area of China. Biomed Res. Int. 2016:4343564. doi: 10.1155/2016/4343564
Zobell, C. E. (1943). The effect of solid surfaces upon bacterial activity. J. Bacteriol. 46, 39–56.
Keywords: bacteriophage, depolymerase, Escherichia coli, biofilm, beta-lactamase
Citation: Guo Z, Huang J, Yan G, Lei L, Wang S, Yu L, Zhou L, Gao A, Feng X, Han W, Gu J and Yang J (2017) Identification and Characterization of Dpo42, a Novel Depolymerase Derived from the Escherichia coli Phage vB_EcoM_ECOO78. Front. Microbiol. 8:1460. doi: 10.3389/fmicb.2017.01460
Received: 10 April 2017; Accepted: 20 July 2017;
Published: 02 August 2017.
Edited by:
Peter Mullany, University College London, United KingdomReviewed by:
Andrei A. Zimin, Institute of Biochemistry and Physiology of Microorganisms (RAS), RussiaDiana Gutiérrez, Instituto de Productos Lácteos de Asturias (CSIC), Spain
Ronen Nissan Hazan, Hebrew University of Jerusalem, Israel
Copyright © 2017 Guo, Huang, Yan, Lei, Wang, Yu, Zhou, Gao, Feng, Han, Gu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingmin Gu, amluZ21pbjA2MjlAMTYzLmNvbQ== Junling Yang, anVubGluZ0BqbHUuZWR1LmNu
 Zhimin Guo
Zhimin Guo Jing Huang2
Jing Huang2 Jingmin Gu
Jingmin Gu