- 1Food Animal Health Research Program, Department of Veterinary Preventive Medicine, Ohio Agricultural Research and Development Center, The Ohio State University, Wooster, OH, United States
- 2Department of Animal Hygiene, Zoonoses and Animal Ethology, Faculty of Veterinary Medicine, Suez Canal University, Ismailia, Egypt
- 3Department of Nutrition and Food Sciences, Faculty of Agricultural and Food Sciences, American University of Beirut, Beirut, Lebanon
Campylobacter jejuni is a leading cause of bacterial food poisoning in humans. Due to the rise in antibiotic-resistant Campylobacter, there exists a need to develop antibiotic-independent interventions to control infections in humans. Here, we evaluated the impact of Escherichia coli Nissle 1917 (EcN), a probiotic strain, on C. jejuni’s invasion and intracellular survival in polarized human colonic cells (HT-29). To further understand how EcN mediates its impact, the expression of 84 genes associated with tight junctions and cell adhesion was profiled in HT-29 cells after treatment with EcN and challenge with C. jejuni. The pre-treatment of polarized HT-29 cells with EcN for 4 h showed a significant effect on C. jejuni’s invasion (∼2 log reduction) of the colonic cells. Furthermore, no intracellular C. jejuni were recovered from EcN pre-treated HT-29 cells at 24 h post-infection. Other probiotic strains tested had no significant impact on C. jejuni invasion and intracellular survival. C. jejuni decreased the expression of genes associated with epithelial cells permeability and barrier function in untreated HT-29 cells. However, EcN positively affected the expression of genes that are involved in enhanced intestinal barrier function, decreased cell permeability, and increased tight junction integrity. The results suggest that EcN impedes C. jejuni invasion and subsequent intracellular survival by affecting HT-29 cells barrier function and tight junction integrity. We conclude that EcN might be a viable alternative for controlling C. jejuni infections.
Introduction
Campylobacter jejuni is a leading cause of foodborne gastroenteritis worldwide (Fullerton et al., 2007; CDC, 2013; Kaakoush et al., 2015). Campylobacter is also associated with post-infectious neurological and joint disorders, such as the Guillain-Barre’ syndrome, the Miller Fisher syndrome, and reactive arthritis (Nachamkin et al., 1998; Zia et al., 2003; CDC, 2014; Mohan, 2015; Kassem et al., 2016b). In the majority of the cases, Campylobacter infections are self-limiting and treatment mainly relies on supportive therapy, while antibiotics can be deployed in severe cases (Alemka et al., 2010). However, the emergence of antibiotic-resistant Campylobacter strains has emphasized a need to develop alternatives to treat human infections (Kassem et al., 2016a). Of particular interest is exploiting probiotic bacteria as an antibiotic-independent approach to enhance the hosts’ immunity and control infections. The application of probiotic bacteria has also received wide attention as a potential intervention to limit the use of antibiotics in food animal production. The latter is proposed to significantly reduce the impact of agricultural practices on the emergence of antibiotic-resistant foodborne pathogens that affect public health (Sahin et al., 2015).
Probiotic bacteria are defined as non-pathogenic and viable microorganisms that can confer beneficial effects on the host by maintaining gut microbial balance and homeostasis and facilitating mucosal repair in the gastrointestinal tract (Behnsen et al., 2013; Mohan, 2015). Additionally, probiotic bacteria have been shown to attenuate the impact of several enteropathogens (Corry et al., 1995; Sherman et al., 2005). For example, Escherichia coli Nissle 1917 (EcN) reduced the invasion of human intestinal epithelial cells by important bacterial pathogens, including Salmonella enterica serovar Typhimurium, Shigella flexneri, enteroinvasive E. coli, Listeria monocytogenes, and Yersinia enterocolitica (Altenhoefer et al., 2004; Kleta et al., 2006; Reissbrodt et al., 2009; Sonnenborn and Schulze, 2009). EcN is one of the widely characterized probiotic strains that demonstrate beneficial activity in both humans and animals (Kruis et al., 2004; Schultz et al., 2004; von Buenau et al., 2005; Krammer et al., 2006; Schroeder et al., 2006; Henker et al., 2008). EcN persistently colonizes its hosts (Schultz, 2008) and has been shown to (1) produce antimicrobial compounds such as bacteriocins or microcins, (2) modulate host immune responses, and (3) participate in competitive exclusion of pathogens (Helwig et al., 2006; Ukena et al., 2007; Zyrek et al., 2007; Kamada et al., 2008; Bar et al., 2009; Bickert et al., 2009; Reissbrodt et al., 2009; Trebichavsky et al., 2010; Behnsen et al., 2013). Furthermore, EcN interacts with intestinal epithelial cells to express proteins that mediate normal gut barrier functions, normalize gut permeability, and improve mucosal integrity (Zyrek et al., 2007). Consequently, EcN has a plethora of desirable probiotic properties, which can be beneficial to the overall gut health and provide protection against enteric infections.
In contrast to some other probiotic strains (Wine et al., 2009), the impact of EcN on C. jejuni’s interaction with intestinal cells has not been characterized. Consequently, we investigated the effect of EcN on C. jejuni’s invasion of human intestinal epithelial cells in vitro. Furthermore, we assessed the response of intestinal cells to C. jejuni in the presence and absence of EcN using human tight junction RT2 Profiler PCR Arrays (Qiagen, Array # PAHS-143Z), which evaluates the expression of 84 genes associated with tight junctions (Moradipoor et al., 2016). The integrity of cell to cell junctions (including tight junctions) is key to normal gut barrier functions and permeability, which affect the pathophysiology of enteric infections (Bischoff et al., 2014). This is very important, because C. jejuni was shown to impact tight junctions in intestinal epithelial monolayers, causing the redistribution of occludin (a tight junction transmembrane protein) from an intercellular to an intracellular location and potentially compromising the intestinal barrier (Chen et al., 2006). Therefore, we also used a polarized human colon cells (HT-29), which was shown to be valuable for evaluating the impact of pathogens on cell barrier permeability, transcytosis mechanisms, and cell invasion (Bras and Ketley, 1999). Furthermore, the HT-29 cells have been considered as one of the more appropriate cell types for assessing Campylobacter virulence in vitro (Haddad et al., 2010).
Materials and Methods
Bacterial Strains and Growth Conditions
Campylobacter jejuni 81-176 is a well-characterized invasive and wild type strain that has been routinely used as a “global model” in studies that characterize C. jejuni virulence and host pathogen interactions (Korlath et al., 1985; Hendrixson and DiRita, 2004; Papp-Szabo et al., 2005; Hofreuter et al., 2006). In this study, C. jejuni 81-176 was routinely cultured using Mueller-Hinton (MH) agar (Difco) with a Campylobacter selective supplement (CSS) (SR0117; Oxoid) at 42°C under microaerobic conditions (5% O2, 10% CO2, and 85% N2) (Kassem et al., 2012). E. coli strain Nissle 1917 (EcN) was cultured aerobically using Luria-Bertani (LB) broth at 37°C to achieve logarithmic growth. Other bacterial strains, Lactobacillus rhamnosus GG (LGG; ATCC 53703), Lactobacillus acidophilus NCFM (LA; ATCC 700396), and Bifidobacterium animalis subsp. Lactis (Bb-12; Christian Hansen, Ltd, Hørsholm, Denmark) were cultured using MRS (de Man, Rogosa and Sharpe) media under anaerobic condition, which was generated using the GasPakTM EZ Anaerobe Container System Sachets (BD, United States) (Kassem et al., 2012). To facilitate the growth of Bb-12, the MRS broth was supplemented with 0.05% cysteine hydrochloride. LA, LGG, and Bb-12 were grown at 37°C for 18 h (Kumar et al., 2014).
The Impact of Different Probiotic Strains on Adhesion, Invasion, and Intracellular Survival of C. jejuni in HT-29 Cells
HT-29 (Human Colorectal Adenocarcinoma Cell Line; ATCC HTB-38) cells were maintained in complete Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco), 2 mM L-glutamine, 5 mM galactose, 1% antibiotic and 0.1 mM non-essential amino acids (Altenhoefer et al., 2004). The cells were incubated at 37°C in a humidified atmosphere with 5% CO2. Prior to each experiment, polarized cells were prepared after seeding 1.4 × 105 HT-29 cells into each well of a 96-well cell culture plate, which was then incubated for 3–4 days (Alemka et al., 2010). The polarized HT-29 cells were washed with- and incubated in DMEM containing no antibiotics and FBS prior to challenge with bacteria (Otte and Podolsky, 2004).
To evaluate the effect of probiotic bacteria on C. jejuni’s adhesion to HT-29 cells, the probiotic bacteria (EcN, LGG, LA, Bb-12) were grown to early exponential phase, pelleted, washed two times with Dulbecco’s phosphate-buffered saline (DPBS), and re-suspended in DMEM. Hundred microliter of each suspension (1 × 107 CFUs) was added to the wells containing the HT-29 monolayers, which were then incubated for 4 h (Altenhoefer et al., 2004). The HT-29 cells were then washed three times and infected with 1.7 × 107 CFUs of C. jejuni 2 h. After this, the infected HT-29 cells were washed three times with DBPS and the adherent C. jejuni CFUs were enumerated after lysis with 0.1% Triton X-100, serial dilution (10-fold), and spreading onto MH agar plates containing CSS.
To determine the effect of probiotics on C. jejuni’s invasion of HT-29 cells, the HT-29 cells were pre-treated with the different probiotic bacteria and C. jejuni as described above. However, following the 2 h incubation with C. jejuni, the HT-29 cells were washed three times with DPBS and treated with DMEM containing 150 μg ml-1 gentamicin for an additional 1 h. The HT-29 cells were then washed twice with DPBS, lysed with 0.1% Triton X-100 and C. jejuni CFUs were quantified as described above.
To assess C. jejuni’s intracellular survival, the HT-29 cells were treated as described above. However, after the gentamicin treatment, the HT-29 cells were washed and incubated again for 24 h in fresh DMEM containing 10 μg/ml-1 gentamicin (Altenhoefer et al., 2004; Schierack et al., 2011; Kassem et al., 2012). After this, the HT-29 cells were washed twice and lysed to quantify Campylobacter CFUs as described above. These experiments were repeated on two separate occasions and each sample was replicated four times per experiment.
We also evaluated the effect of different incubation time of EcN with HT-29 cells on C. jejuni’s adhesion, invasion, and intracellular survival. For this purpose, polarized HT-29 cells were incubated with EcN (1.7 × 107 CFUs) for 1, 2, 3, and 4 h before infection with C. jejuni as described above.
Assessment of the Impact of EcN’s Heat Killed Cells and Cell-Free Supernatant on C. jejuni’s Interaction with HT-29 Cells
Possible interactions between EcN and C. jejuni were evaluated further as follows:
(A) Treatment of HT-29 cells with heat-killed EcN:
Heat- killed EcN were prepared by heating exponentially grown cultures to 65°C for 5 min and the loss of EcN viability was confirmed by spreading the cultures on LB agar. The polarized HT-29 cells were then incubated for 4 h with heat-killed EcN (equivalent to ∼1.7 × 107 CFUs) before infection with C. jejuni as described above. Additionally, heat-killed EcN and C. jejuni mixtures (1:1 and 10:1 v/v) were pre-incubated at room temperature for 0, 1, 2, 3, and 4 h and then used to infect the polarized HT-29 cells.
(B) Treatment of HT-29 cells with EcN cell-free supernatant:
EcN-free supernatant was prepared from exponentially grown cultures, which were centrifuged at 5000 × g for 10 min. The supernatant was then collected and filtered through a sterile membrane (0.2 μm pore size) (Corning, Germany) and confirmed to be EcN free by culturing on LB agar. The EcN-free supernatant (prepared from cultures containing the equivalent of 1.7 × 107 CFUs) was used to treat the HT-29 cells prior to C. jejuni infection as described above. Additionally, cell-free supernatants were prepared from EcN cultures containing the equivalent of 1 and 10X the number of C. jejuni CFUs. These supernatants were also used to suspend C. jejuni at room temperature for 0, 1, 2, 3, and 4 h prior to infection.
In each of the experiments above, C. jejuni’s adherence, invasion, and intracellular survival were assessed by determining the number of C. jejuni CFUs ml-1 as described earlier. All experiments were repeated at least two times using four replicates of each sample per experiment.
Human Tight Junctions RT2 Profiler PCR Arrays Analysis
The expression of 84 tight junction-associated genes was determined using the human tight junctions RT2 Profiler PCR Arrays (Qiagen, Array # PAHS-143Z) (Moradipoor et al., 2016). Subsequently, polarized HT-29 cells were treated with EcN for 4 h and infected with C. jejuni for 2 h (invasion) and 24 h (intracellular survival). Untreated and unchallenged HT-29 cells and those that were challenged with EcN and C. jejuni were used as controls, respectively. Total RNA was extracted from the HT-29 cells using the TRIzol reagent (Life Technologies, United States) and the miRNeasy Mini Kit (Qiagen) and purified of DNA traces as described by the manufacturer (Qiagen). RNA quality and quantity were determined using nanodrop 2000 C spectrophotometer (Thermoscientific) and by electrophoresis in agarose gels.
Approximately, 5 μg of purified RNA were used to synthesize cDNA using the Qiagen RT2 First Strand Kit (Qiagen). As specified by the manufacturer, cDNA was added to the RT2 SYBR Green qPCR Master Mix (Qiagen) and 25 μl of the mixture were added to each well of a 96 well plate that pre-contained gene-specific primer sets (RT2 Profiler PCR Arrays) as described by the manufacturer. qRT PCR was performed using a Mastercycler® RealPlex2 (Eppendorf). The threshold cycle (Ct) values were calculated for each gene and normalized using the house-keeping genes included in the Arrays. Fold-changes in gene expression (between treated and control samples) were calculated using the ΔΔCt method (Cao et al., 2015). Significantly, affected genes were then analyzed using the Ingenuity Pathway Analysis (IPA) software1 (Kumar et al., 2014) to identify potential functions and cellular pathways that were modulated in HT-29 cells in response to EcN and C. jejuni.
Statistical Analysis
Data generated from the gentamicin protection assays (adherence, invasion, and intracellular survival) were presented as means ± standard deviations. ANOVA followed by the Tukey test was used to analyze these data and a P-value < 0.05 was used to determine statistically significant differences between means. A fold change of ±1.5 ≥ or ≤ 1.5 and a P-value ≤ 0.05 were used to determine statistically significant differences in gene expression.
Results
The Effect of Probiotic Bacteria on the Interaction of C. jejuni with HT-29 Cells
To determine the impact of EcN on the interaction of C. jejuni with polarized HT-29 cells, we incubated EcN with the HT-29 cells for 1, 2, 3, and 4 h prior to infection with C. jejuni. The treatment with EcN for 4 h resulted in the significant reduction in C. jejuni’s invasion and intracellular survival (Figure 1A). To compare the effect of EcN to other commonly known probiotic bacteria, we treated the polarized HT-29 cells with EcN, LA, LGG, and Bb-12 for 4 h then infected these cells with C. jejuni 81-176, respectively. Our results show that LA, LGG, and Bb-12 did not significantly impact the interaction of C. jejuni with the HT-29 cells. However, while EcN did not significantly impact the adherence of C. jejuni to HT-29 cells, EcN significantly reduced (P < 0.05) C. jejuni’s invasion by ∼2 logs CFU ml-1 in comparison to the control (HT-29 cells not treated with EcN). Furthermore, no intracellular C. jejuni were recovered from EcN-treated HT-29 cells in comparison to the control (Figures 1A,B).
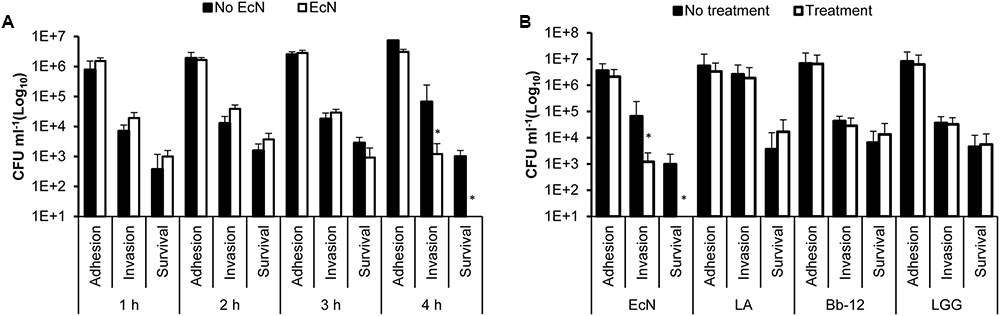
FIGURE 1. Campylobacter jejuni’s adhesion to, invasion of and intracellular survival in HT-29 cells pretreated with (A) EcN for 1, 2, 3, and 4 h; (B) EcN, Lactobacillus acidophilus NCFM (LA), Bifidobacterium animalis subsp. Lactis (Bb-12) and Lactobacillus rhamnosus GG (LGG) for 4 h. ∗Indicates statistically significant differences (P < 0.05) in C. jejuni CFU numbers between probiotic-treated and non-treated HT-29 cells. The experiments were repeated at least two times and samples were processed in four replicates in each experiment.
The Impact of EcN Cell Free Supernatant and Heat-Killed Cells on the Interaction of C. jejuni with HT-29 Cells
The pretreatment of HT-29 cells with EcN cell-free supernatant and heat-killed EcN cells for 4 h prior to infection with C. jejuni showed no significant impact (P > 0.05) on C. jejuni’s interaction with HT-29 cells (Figure 2). Additionally, pre-incubation of C. jejuni at room temperature with EcN cell-free supernatants and with heat killed EcN cells for 0, 1, 2, 3, and 4 h prior to infection of HT-29 cells also showed no significant impact on C. jejuni’s interaction with HT-29 cells (Figures 3A–D). Direct spreading of aliquots form the pre-incubated mixtures on MH agar plates showed no change in C. jejuni CFUs, which further confirmed the insignificant impact of EcN’s cell-free supernatants and heat killed cells on C. jejuni viability (data not shown).
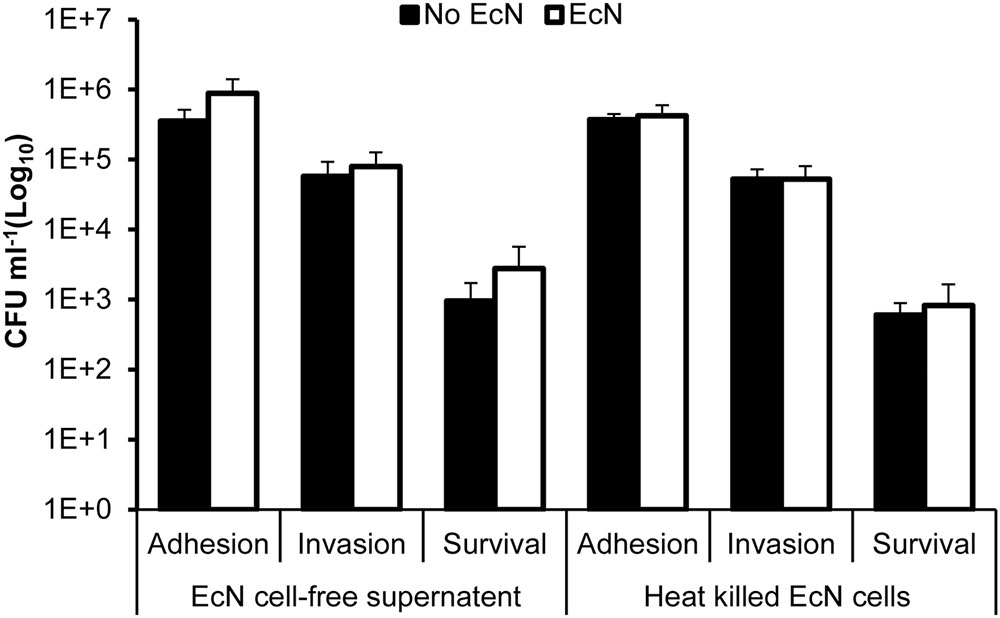
FIGURE 2. Campylobacter jejuni’s adhesion to, invasion of and intracellular survival in HT-29 cells pretreated with EcN free supernatant and heat killed cells for 4 h. Heat-killed EcN were generated by heating exponentially growing cultures to 65°C for 5 min and checking the loss of viability by culturing on LB agar plates. EcN-free supernatant was prepared from exponentially growing cultures, which were centrifuged and filtered through a sterile membrane. The experiments were repeated at least two times and samples were processed in four replicates in each experiment.
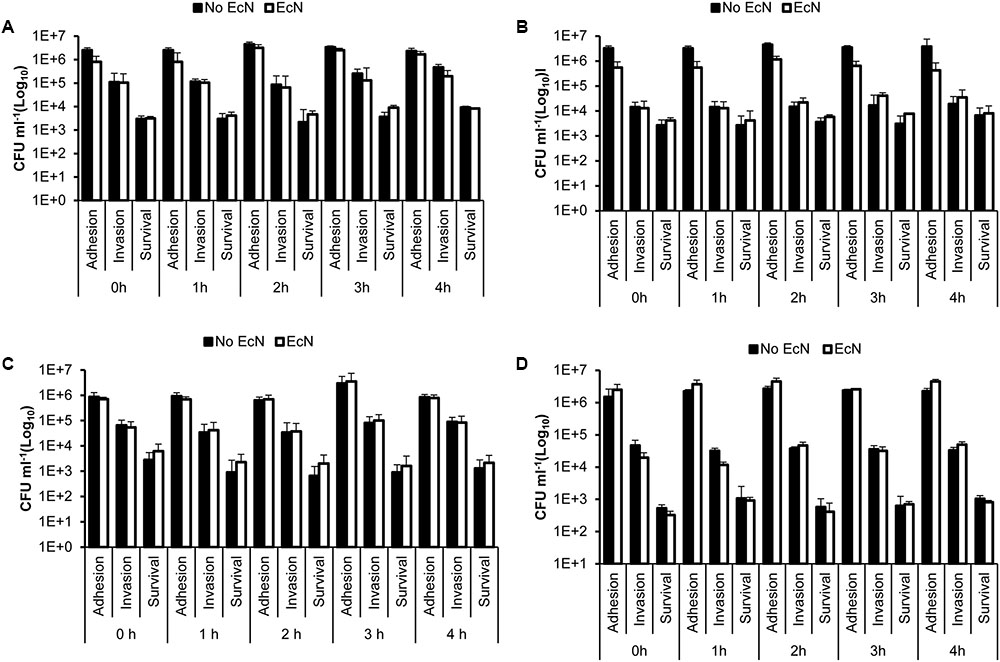
FIGURE 3. Campylobacter jejuni’s adhesion to, invasion of and intracellular survival in HT-29 cells following co-incubation of C. jejuni with EcN free supernatant and heat killed cells prior to infection of HT-29 cells. (A) Heat-killed EcN and C. jejuni 81-176 were co-incubated (1:1) for 0, 1, 2, 3, and 4 h at room temperature before the HT-29 cells were infected with the mixture for 2 h. (B) Heat-killed EcN and C. jejuni 81-176 were co-incubated (10:1) for 0, 1, 2, 3, and 4 h at room temperature before the HT-29 cells were infected with the mixture for 2 h. (C) EcN-cell free supernatant and C. jejuni 81-176 were co-incubated for 0, 1, 2, 3, and 4 h at room temperature before the HT-29 cells were infected with the mixture for 2 h. The cell-free supernatant was prepared from cultures containing the same number of EcN CFUs as C. jejuni (1X). (D) EcN-cell free supernatant and C. jejuni 81-176 were co-incubated for 0, 1, 2, 3, and 4 h at room temperature before the HT-29 cells were infected with the mixture for 2 h. The cell-free supernatant was prepared from cultures containing 10X more EcN CFUs than C. jejuni (10X). The experiments were repeated at least two times and samples were processed in four replicates in each experiment.
The Impact of EcN on the Expression of Tight Junction-Associated Genes in HT-29 Cells
RT2 Profiler PCR Arrays were used to evaluate the impact of EcN on tight junctions-associated gene expression in HT-29 cells. Collectively, the three treatments (EcN, C. jejuni, and EcN + C. jejuni) affected 76 out of 84 genes included in the arrays (Supplementary Table S1). The detailed description of differentially expressed genes with the fold change values under different treatments is included in Supplementary Table S2. IPA analysis identified two canonical pathways that were significantly modulated by EcN treatment and C. jejuni infection. The two major affected canonical pathways were (1) tight junctions and other cell–cell junction signaling (TCS) and (2) cell adhesion and extravasation signaling (CAS).
Impact on TCS Associated Genes
At 2 h post-infection with C. jejuni (invasion), EcN treatment alone caused significant alteration in the expression of 33 genes (27 up-regulated and 6 down-regulated) in TCS (Figure 4A). Interestingly, nine up-regulated genes and three down-regulated genes (encoding CDK4, CTNNB1, and JAM2) were uniquely affected by treatment with EcN alone. C. jejuni infection affected the expression of 12 genes (eight up-regulated and four down-regulated), one gene encoding MPDZ (an up-regulated junction associated protein) was uniquely associated with C. jejuni (Figure 4A). EcN + C. jejuni affected 26 genes (18 up-regulated and 8 down-regulated), 8 of which were uniquely associated with this treatment. Notably, in EcN + C. jejuni, the expression of the CLDN15 encoding gene was up-regulated in comparison to infection with C. jejuni (Figure 4A).
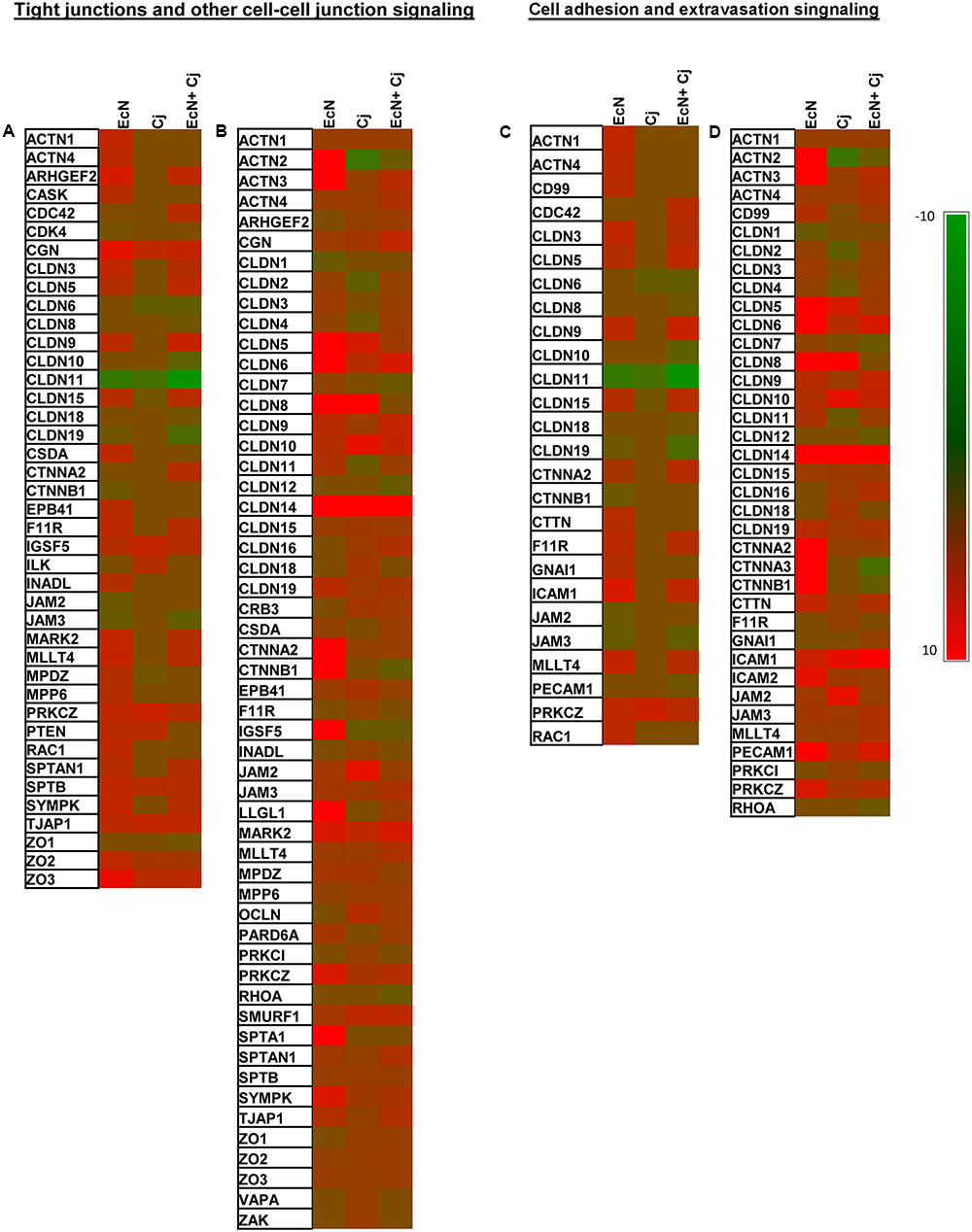
FIGURE 4. Expression of tight junction associated genes in HT-29 cells with and without EcN and C. jejuni. Expression of genes associated with two significantly modulated canonical pathways (1) tight junction and other cell–cell junction signaling (TCS) at 2 h (A) and 24 h (B) C. jejuni post-infection and (2) cell adhesion and extravasation signaling (CAS) at 2 h (C) and 24 h (D) C. jejuni post-infection. The human tight junction RT2 Profiler PCR arrays were used to assess the expression of 84 genes in EcN-pretreated cells at 2 and 24 h post-C. jejuni infection. Significant change in gene expression was determined by comparison to untreated cells (no EcN and no C. jejuni). A fold change of ±1.5 ≥ or ≤ 1.5 and a P ≤ 0.05 were used to determine significant differences in gene expression.
After 24 h post-infection with C. jejuni (intracellular survival), the expression of 40 genes was up-regulated in HT-29 treated with EcN, and one of these genes (encoding SPTA1) was uniquely associated with EcN (Figure 4B). Only the expression of one gene (encoding CLDN1) was down-regulated by EcN. C. jejuni infection altered the expression of 44 genes (39 up-regulated and 5 down-regulated). Notably, the down-regulated genes (encoding CLDN2, CLDN4, CLDN11, ACTN2, and IGSF5) associated with the C. jejuni treatment were up-regulated in the presence of EcN alone (Figure 4B). Additionally, six up-regulated genes (encoding ZAK, F11R, PRKC1, INDAL, VAPA, and CLDN18) were uniquely associated with C. jejuni infection. EcN + C. jejuni affected the expression of 45 genes (39 up-regulated and 6 down-regulated) (Figure 4B). The expression of two genes was uniquely impacted in the EcN + C. jejuni treatment and included genes that encoded CLDN12 (claudin), and RHOA (G-protein signaling). In addition, in EcN + C. jejuni, the expression of the CLDN2, CLDN4, and CLDN11 encoding genes was down-regulated in comparison to infection with C. jejuni (Figure 4B).
Impact on CAS Associated Genes
At 2 h post-infection with C. jejuni (invasion), EcN treatment alone caused significant alteration in the expression of 19 genes (14 up-regulated and 5 down-regulated) associated with CAS (Figure 4C). Of these, six up-regulated genes and two down-regulated genes were uniquely observed in treatment with EcN alone. C. jejuni infection affected four genes (one up-regulated and three down-regulated). Notably, one of the down-regulated genes (encoding the claudin, CLND15) was up-regulated in the EcN + C. jejuni treatment (Figure 4C). Additionally, the expression of 18 genes (10 up-regulated and 8 down-regulated) was affected in EcN + C. jejuni and change in the expression of 6 (two up-regulated and four down-regulated) of these genes was uniquely associated with this treatment. The latter genes encoded CDC42, CLDN8, CLDN10, CLDN18, CTNNA2, and PECAM1 (Figure 4C).
After 24 h post-infection with C. jejuni (intracellular survival), the expression of 29 genes was up-regulated in HT-29 treated cells with EcN, while one gene encoding CLDN1 was down-regulated (Figure 4D). Changes in one gene (encoding CLDN1) were uniquely observed in the EcN treatment (Figure 4D). In comparison to untreated controls, EcN treatment caused the up-regulation of genes encoding ACTN2, and ACTN3 by 42.5, and 87 folds, respectively (Figure 4D). C. jejuni infection altered the expression of 28 genes (24 up-regulated and 4 down-regulated), and the impact on expression of three of the up-regulated genes (encoding CLDN18, F11R, and PRKC1) was only observed in this treatment (Figure 4D). EcN + C. jejuni affected the expression of 32 genes (26 up-regulated and 6 down-regulated) and the expression of two of the down-regulated genes (encoding CLDN12, and RHOA) was uniquely associated with this treatment. In addition, in EcN + C. jejuni, the expression of the CLDN2, CLDN4, and CLDN11 encoding genes was up-regulated in comparison to infection with C. jejuni (Figure 4D).
Discussion
In light of the increase in antibiotic resistant C. jejuni (Kassem et al., 2016a), there is a need to proactively devise alternative approaches to control the proliferation of this pathogen and cognate infections. Several studies have recently investigated the effect of probiotic bacteria on C. jejuni infections in cell lines (in vitro) and in animal models such as chickens, a primary host and source of this bacterium (Baffoni et al., 2017; Ekmekciu et al., 2017; Johnson et al., 2017). Lactobacillus, Bacillus, and Enterococcus have been among the most commonly researched probiotic bacteria against C. jejuni (Johnson et al., 2017). For example, Chaveerach et al. (2004) showed that probiotic bacteria, Lactobacilli and Bifidobacteria, had a negative effect on the growth of different strains of C. jejuni and C. coli. However, previous studies also suggest probiotic bacteria did not always significantly affect C. jejuni colonization and that the desirable antagonistic impacts appeared to vary according to the probiotic strain. For example, Lactobacillus helveticus R0052 reduced C. jejuni’s invasion of human epithelial colon cells (T84), while L. rhamnosus strain R0011 did not affect the invasion of these cells (Wine et al., 2009). Despite their promise in laboratory trials, orally administered strains such as Lactobacillus acidophilus, Bacillus subtilis, and Enterococcus faecium did not significantly reduce C. jejuni in broiler chickens (Thomrongsuwannakij et al., 2016). Taken together, the aforementioned observations highlight a need to thoroughly evaluate different probiotic bacteria/strains to assess (1) their impact on C. jejuni, and (2) the mechanisms that govern the desirable antagonistic effect on this pathogen. To address this need, we evaluated the potential use of the probiotic strain, EcN, to control C. jejuni in vitro. This was motivated by previous observations that showed negative impact of EcN on the invasion of human intestinal epithelial cells by several important pathogens, including Salmonella enterica serovar Typhimurium and Listeria monocytogenes (Altenhoefer et al., 2004; Kleta et al., 2006; Reissbrodt et al., 2009; Sonnenborn and Schulze, 2009). Furthermore, the probiotic properties of EcN and its use in the treatment of various diseases of the digestive tract of humans have been strongly established (Altenhoefer et al., 2004).
In our experimental system, we found that only EcN negatively impacted C. jejuni’s invasion of and intracellular survival in polarized HT-29 cells (Figure 1) and in another human intestinal epithelial cell line (Caco-2) (Supplementary Figure S1). In comparison, the probiotic bacteria strains, LA, Bb-12, and LGG did not significantly affect C. jejuni’s interaction with the HT-29 cells (Figure 1B). EcN cell-free supernatant and heat-killed EcN did not affect C. jejuni’s interaction with intestinal cells (Figures 2, 3). This suggested that the antagonistic impact of EcN (1) might require live EcN to inhibit C. jejuni, and (2) might not significantly include metabolites secreted into the culture supernatant. This is not surprising, because it is known that viable probiotic bacteria and their cell-free supernatant differ in their ability to protect against pathogens (Campana et al., 2017). Furthermore, the optimal impact of EcN was observed after incubating this probiotic bacterium with intestinal cells for 4 h before infection with C. jejuni (Figure 1A). This suggested that the contact time between the viable probiotic bacterium and intestinal cells was crucial. Notably, in a previous report, it was suggested that EcN on the surface of the intestinal cells impedes the ability of potential pathogens to exert their impact (Boudeau et al., 2003). This might be attributed to the ability of live probiotic bacteria to compete with pathogens for nutrients and for binding to intestinal cell receptors. Additionally, we observed that EcN and C. jejuni did not significantly co-aggregate in co-cultures (data not shown). Taken together, this suggests that EcN might be forming a physical barrier between the pathogen and intestinal cells and/ or inducing intestinal cell properties that might resist infection. The latter is plausible based on reports that probiotic bacteria can increase tight junction integrity and enhance intestinal barrier function and permeability to resist bacterial invasion (Putaala et al., 2008). Furthermore, this corroborated previous studies that reported that EcN’s probiotic activity might be mediated via enhancing the intestinal barrier through the up-regulation of tight junction-associated proteins (Schulze and Downward, 2001; Ukena et al., 2007).
Based on our analysis above, we evaluated the expression of 84 genes associated with tight- and other cell to cell junctions to further analyze how EcN mediates its impact (Figure 4). Notably, several probiotic strains appear to affect the expression of occludins and cingulin (Anderson et al., 2010; Miyauchi et al., 2012; Patel et al., 2012). This is important, because C. jejuni infection can increase the epithelial cells permeability and induce epithelial barrier disruption, which in turn might facilitate the invasion of the gut (Wine et al., 2008). Therefore, EcN’s impact on epithelial cell junctions might (1) reduce C. jejuni’s impact on epithelial cells and (2) prevent the pathogen from possibly entering via the paracellular pathway to cause further damage to the cells (Ulluwishewa et al., 2011). In our study, EcN differentially impacted the expression of tight junction associated genes in HT-29 cells at 2 h (44 genes) and 24 h (55 genes), including genes encoding occludins (ZO-2 and ZO-3) and spectrins (Supplementary Table S1 and Figure 4). In several other studies, EcN increased the expression of ZO-2 and caused the redistribution of this protein, which lead to restoration of a disrupted epithelial barrier (Bischoff et al., 2014), while spectrins have been implicated in the stabilization and remodeling of epithelial junctions (Naydenov and Ivanov, 2011). Furthermore, in our study, EcN affected the expression of genes encoding claudins, including claudin 3, 5, 9, 15 at 2 h and claudin 2–14, 15, and 19 at 24 h (Figure 4). Interestingly, claudin 3, 4, 5, and 8 are associated with enhancing tight junctions and reducing space between two neighboring cells and decreasing paracellular permeability (Ulluwishewa et al., 2011). Furthermore, the EcN-associated differential expression of genes encoding actinins, catenins/cadherins, spectrins, and other adhesion molecules (Figure 4) might contribute to intestinal homeostasis by affecting adherens junctions, which are cell to cell anchoring structures that contribute to organization of the epithelium (Harris and Tepass, 2010). Taken together, we suggest that the presence of EcN appears to stimulate the expression of genes that that enhance cell to cell junction and intestinal barrier integrity. This “priming” effect might increase the resistance of the HT-29 cells to infection.
In our study, EcN stimulated the expression of genes that were either not affected or were downregulated by C. jejuni (Figure 4 and Supplementary Table S1). For example, at 2 h, the expression of genes encoding claudin 3, 5, and 9 and F11R, SYMPK, ARCHGEF2, ESAM, ICMA1, MARK2, and MLLT4 was only upregulated in the EcN and the EcN + C. jejuni treatments. Similarly, at 24 h, genes encoding CD99, CLDN3, CSDA, ESAM, LLGL1, and PARD6A were only upregulated in the EcN and the EcN + C. jejuni treatments (Figure 4 and Supplementary Figure S1). Furthermore, EcN antagonized the impact of C. jejuni on the expression of certain genes in the intestinal cells. For example, genes encoding claudin 15 at 2 h and claudin 2, 4, and 11 at 24 h were down-regulated when HT-29 cells were infected with C. jejuni only. However, these genes were upregulated when the intestinal cells were pre-treated with EcN (Figure 4 and Supplementary Figure S1). These genes either directly (structure) or indirectly (signaling) affect cell junctions and adherence (Matter and Balda, 2003), which further suggest that EcN mediates its protective impact against C. jejuni by enhancing the ability of the intestinal cells to resist infection. Although it is interesting to explore the specific role of individual genes, it should also be noted that EcN’s beneficial effects should be considered in terms of the overall impact on all of the investigated genes and the associated cellular pathways.
The data presented in this study show that the pretreatment of the intestinal cells with EcN can protect against C. jejuni’s invasion and intracellular survival. It is likely that this antagonistic activity is facilitated via the probiotic EcN’s impact on HT-29 cell to cell junctions. Our data support the need for future studies that will test the effect of EcN on C. jejuni in cognate animal models such as mice and chickens. This may facilitate development of a potentially effective antibiotic-independent approach to control C. jejuni in humans and other animal reservoirs.
Author Contributions
YH, IK, and GR conceived and designed the study. YH and AK conducted the experiments. YH, IK, and GR analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
Research in the Rajashekara laboratory is funded by grants from the USDA (AFRI USDA, Grant # 2012-68003-19679) and the Ohio State University’s SEEDS program.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01588/full#supplementary-material
Footnotes
References
Alemka, A., Clyne, M., Shanahan, F., Tompkins, T., Corcionivoschi, N., and Bourke, B. (2010). Probiotic colonization of the adherent mucus layer of HT29MTXE12 cells attenuates Campylobacter jejuni virulence properties. Infect. Immun. 78, 2812–2822. doi: 10.1128/IAI.01249-09
Altenhoefer, A., Oswald, S., Sonnenborn, U., Enders, C., Schulze, J., Hacker, J., et al. (2004). The probiotic Escherichia coli strain Nissle 1917 interferes with invasion of human intestinal epithelial cells by different enteroinvasive bacterial pathogens. FEMS Immunol. Med. Microbiol. 40, 223–229. doi: 10.1016/S0928-8244(03)00368-7
Anderson, R. C., Cookson, A. L., McNabb, W. C., Park, Z., McCann, M. J., Kelly, W. J., et al. (2010). Lactobacillus plantarum MB452 enhances the function of the intestinal barrier by increasing the expression levels of genes involved in tight junction formation. BMC Microbiol. 10:316. doi: 10.1186/1471-2180-10-316
Baffoni, L., Gaggia, F., Garofolo, G., Di Serafino, G., Buglione, E., Di Giannatale, E., et al. (2017). Evidence of Campylobacter jejuni reduction in broilers with early synbiotic administration. Int. J. Food Microbiol. 251, 41–47. doi: 10.1016/j.ijfoodmicro.2017.04.001
Bar, F., Von Koschitzky, H., Roblick, U., Bruch, H. P., Schulze, L., Sonnenborn, U., et al. (2009). Cell-free supernatants of Escherichia coli Nissle 1917 modulate human colonic motility: evidence from an in vitro organ bath study. Neurogastroenterol. Motil. 21, 559–566, e16–e17. doi: 10.1111/j.1365-2982.2008.01258.x
Behnsen, J., Deriu, E., Sassone-Corsi, M., and Raffatellu, M. (2013). Probiotics: properties, examples, and specific applications. Cold Spring Harb. Perspect. Med. 3:a010074. doi: 10.1101/cshperspect.a010074
Bickert, T., Trujillo-Vargas, C. M., Duechs, M., Wohlleben, G., Polte, T., Hansen, G., et al. (2009). Probiotic Escherichia coli Nissle 1917 suppresses allergen-induced Th2 responses in the airways. Int. Arch. Allergy Immunol. 149, 219–230. doi: 10.1159/000199717
Bischoff, S. C., Barbara, G., Buurman, W., Ockhuizen, T., Schulzke, J. D., Serino, M., et al. (2014). Intestinal permeability–a new target for disease prevention and therapy. BMC Gastroenterol. 14:189. doi: 10.1186/s12876-014-0189-7
Boudeau, J., Glasser, A. L., Julien, S., Colombel, J. F., and Darfeuille-Michaud, A. (2003). Inhibitory effect of probiotic Escherichia coli strain Nissle 1917 on adhesion to and invasion of intestinal epithelial cells by adherent-invasive E. coli strains isolated from patients with Crohn’s disease. Aliment. Pharmacol. Ther. 18, 45–56. doi: 10.1046/j.1365-2036.2003.01638.x
Bras, A. M., and Ketley, J. M. (1999). Transcellular translocation of Campylobacter jejuni across human polarised epithelial monolayers. FEMS Microbiol. Lett. 179, 209–215. doi: 10.1111/j.1574-6968.1999.tb08729.x
Campana, R., van Hemert, S., and Baffone, W. (2017). Strain-specific probiotic properties of lactic acid bacteria and their interference with human intestinal pathogens invasion. Gut Pathog 9, 12. doi: 10.1186/s13099-017-0162-4
Cao, X., Lin, H., Muskhelishvili, L., Latendresse, J., Richter, P., and Heflich, R. H. (2015). Tight junction disruption by cadmium in an in vitro human airway tissue model. Respir. Res. 16, 30. doi: 10.1186/s12931-015-0191-9
CDC (2013). Trends in Foodborne Illness in the United States, 2012. Available at: https://www.cdc.gov/ncezid/dfwed/PDFs/trends-in-foodborne-Illness-1996-2012-508c.pdf
CDC (2014). Campylobacter: General Information. Available at: https://www.cdc.gov/foodsafety/diseases/campylobacter/index.html
Chaveerach, P., Lipman, L. J., and van Knapen, F. (2004). Antagonistic activities of several bacteria on in vitro growth of 10 strains of Campylobacter jejuni/coli. Int. J. Food Microbiol. 90, 43–50. doi: 10.1016/S0168-1605(03)00170-3
Chen, M. L., Ge, Z., Fox, J. G., and Schauer, D. B. (2006). Disruption of tight junctions and induction of proinflammatory cytokine responses in colonic epithelial cells by Campylobacter jejuni. Infect. Immun. 74, 6581–6589. doi: 10.1128/IAI.00958-06
Corry, J. E., Post, D. E., Colin, P., and Laisney, M. J. (1995). Culture media for the isolation of campylobacters. Int. J. Food Microbiol. 26, 43–76. doi: 10.1016/0168-1605(95)00044-K
Ekmekciu, I., von Klitzing, E., Fiebiger, U., Neumann, C., Bacher, P., Scheffold, A., et al. (2017). The probiotic compound VSL#3 modulates mucosal, peripheral, and systemic immunity following murine broad-spectrum antibiotic treatment. Front. Cell Infect. Microbiol. 7:167. doi: 10.3389/fcimb.2017.00167
Fullerton, K. E., Ingram, L. A., Jones, T. F., Anderson, B. J., McCarthy, P. V., Hurd, S., et al. (2007). Sporadic campylobacter infection in infants: a population-based surveillance case-control study. Pediatr. Infect. Dis. J. 26, 19–24. doi: 10.1097/01.inf.0000247137.43495.34
Haddad, N., Marce, C., Magras, C., and Cappelier, J.-M. (2010). An overview of methods used to clarify pathogenesis mechanisms of Campylobacter jejuni. J. Food Prot. 73, 786–802. doi: 10.4315/0362-028X-73.4.786
Harris, T. J., and Tepass, U. (2010). Adherens junctions: from molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 11, 502–514. doi: 10.1038/nrm2927
Helwig, U., Lammers, K. M., Rizzello, F., Brigidi, P., Rohleder, V., Caramelli, E., et al. (2006). Lactobacilli, bifidobacteria and E. coli nissle induce pro- and anti-inflammatory cytokines in peripheral blood mononuclear cells. World J. Gastroenterol. 12, 5978–5986. doi: 10.3748/wjg.v12.i37.5978
Hendrixson, D. R., and DiRita, V. J. (2004). Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52, 471–484. doi: 10.1111/j.1365-2958.2004.03988.x
Henker, J., Laass, M. W., Blokhin, B. M., Maydannik, V. G., Bolbot, Y. K., Elze, M., et al. (2008). Probiotic Escherichia coli Nissle 1917 versus placebo for treating diarrhea of greater than 4 days duration in infants and toddlers. Pediatr. Infect. Dis. J. 27, 494–499. doi: 10.1097/INF.0b013e318169034c
Hofreuter, D., Tsai, J., Watson, R. O., Novik, V., Altman, B., Benitez, M., et al. (2006). Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74, 4694–4707. doi: 10.1128/IAI.00210-06
Johnson, T. J., Shank, J. M., and Johnson, J. G. (2017). Current and potential treatments for reducing campylobacter colonization in animal hosts and disease in humans. Front. Microbiol. 8:487. doi: 10.3389/fmicb.2017.00487
Kaakoush, N. O., Castano-Rodriguez, N., Mitchell, H. M., and Man, S. M. (2015). Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 28, 687–720. doi: 10.1128/CMR.00006-15
Kamada, N., Maeda, K., Inoue, N., Hisamatsu, T., Okamoto, S., Hong, K. S., et al. (2008). Nonpathogenic Escherichia coli strain Nissle 1917 inhibits signal transduction in intestinal epithelial cells. Infect. Immun. 76, 214–220. doi: 10.1128/IAI.01193-07
Kassem, I. I., Helmy, Y. A., Kashoma, I. P., and Rajashekara, G. (2016a). “The emergence of antibiotic resistance on poultry farms,” in Achieving Sustainable Production of Poultry Meat: Safety, Quality and Sustainability, Vol. 1, ed. S. Ricke (Cambridge: Burleigh Dodds Science Publishing).
Kassem, I. I., Kehinde, O., Helmy, Y. A., Pina-Mimbela, R., Kumar, A., Chandrashekhar, K., et al. (2016b). “Campylobacter in poultry: the conundrums of highly adaptable and ubiquitous foodborne pathogens,” in Foodborne Diseases Case Studies of Outbreaks in the Agri-Food Industries, eds J. M. Soon, L. Manning, and C. A. Wallace (Boca Raton, FL: CRC Press), 79–112. doi: 10.1201/b19463-7
Kassem, I. I., Khatri, M., Esseili, M. A., Sanad, Y. M., Saif, Y. M., Olson, J. W., et al. (2012). Respiratory proteins contribute differentially to Campylobacter jejuni’s survival and in vitro interaction with hosts’ intestinal cells. BMC Microbiol. 12:258. doi: 10.1186/1471-2180-12-258
Kleta, S., Steinruck, H., Breves, G., Duncker, S., Laturnus, C., Wieler, L. H., et al. (2006). Detection and distribution of probiotic Escherichia coli Nissle 1917 clones in swine herds in Germany. J. Appl. Microbiol. 101, 1357–1366. doi: 10.1111/j.1365-2672.2006.03019.x
Korlath, J. A., Osterholm, M. T., Judy, L. A., Forfang, J. C., and Robinson, R. A. (1985). A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152, 592–596. doi: 10.1093/infdis/152.3.592
Krammer, H. J., Kamper, H., von Bunau, R., Zieseniss, E., Stange, C., Schlieger, F., et al. (2006). Probiotic drug therapy with E. coli strain Nissle 1917 (EcN): results of a prospective study of the records of 3,807 patients. Z. Gastroenterol. 44, 651–656. doi: 10.1055/s-2006-926909
Kruis, W., Fric, P., Pokrotnieks, J., Lukas, M., Fixa, B., Kascak, M., et al. (2004). Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53, 1617–1623. doi: 10.1136/gut.2003.037747
Kumar, A., Vlasova, A. N., Liu, Z., Chattha, K. S., Kandasamy, S., Esseili, M., et al. (2014). In vivo gut transcriptome responses to Lactobacillus rhamnosus GG and Lactobacillus acidophilus in neonatal gnotobiotic piglets. Gut Microbes 5, 152–164. doi: 10.4161/gmic.27877
Matter, K., and Balda, M. S. (2003). Signalling to and from tight junctions. Nat. Rev. Mol. Cell Biol. 4, 225–236. doi: 10.1038/nrm1055
Miyauchi, E., O’Callaghan, J., Butto, L. F., Hurley, G., Melgar, S., Tanabe, S., et al. (2012). Mechanism of protection of transepithelial barrier function by Lactobacillus salivarius: strain dependence and attenuation by bacteriocin production. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G1029–G1041. doi: 10.1152/ajpgi.00003.2012
Mohan, V. (2015). The role of probiotics in the inhibition of Campylobacter jejuni colonization and virulence attenuation. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1503–1513. doi: 10.1007/s10096-015-2392-z
Moradipoor, S., Ismail, P., Etemad, A., Wan Sulaiman, W. A., and Ahmadloo, S. (2016). Expression profiling of genes related to endothelial cells biology in patients with type 2 diabetes and patients with prediabetes. Biomed Res. Int. 2016:1845638. doi: 10.1155/2016/1845638
Nachamkin, I., Allos, B. M., and Ho, T. (1998). Campylobacter species and guillain-barre syndrome. Clin. Microbiol. Rev. 11, 555–567.
Naydenov, N. G., and Ivanov, A. I. (2011). Spectrin-adducin membrane skeleton: A missing link between epithelial junctions and the actin cytoskeletion? Bioarchitecture 1, 186–191. doi: 10.4161/bioa.1.4.17642
Otte, J. M., and Podolsky, D. K. (2004). Functional modulation of enterocytes by gram-positive and gram-negative microorganisms. Am. J. Physiol. Gastrointest. Liver Physiol. 286, G613–G626. doi: 10.1152/ajpgi.00341.2003
Papp-Szabo, E., Kanipes, M. I., Guerry, P., and Monteiro, M. A. (2005). Cell-surface alpha-glucan in Campylobacter jejuni 81-176. Carbohydr. Res. 340, 2218–2221. doi: 10.1016/j.carres.2005.06.023
Patel, R. M., Myers, L. S., Kurundkar, A. R., Maheshwari, A., Nusrat, A., and Lin, P. W. (2012). Probiotic bacteria induce maturation of intestinal claudin 3 expression and barrier function. Am. J. Pathol. 180, 626–635. doi: 10.1016/j.ajpath.2011.10.025
Putaala, H., Salusjarvi, T., Nordstrom, M., Saarinen, M., Ouwehand, A. C., Bech Hansen, E., et al. (2008). Effect of four probiotic strains and Escherichia coli O157:H7 on tight junction integrity and cyclo-oxygenase expression. Res. Microbiol. 159, 692–698. doi: 10.1016/j.resmic.2008.08.002
Reissbrodt, R., Hammes, W. P., dal Bello, F., Prager, R., Fruth, A., Hantke, K., et al. (2009). Inhibition of growth of Shiga toxin-producing Escherichia coli by nonpathogenic Escherichia coli. FEMS Microbiol. Lett. 290, 62–69. doi: 10.1111/j.1574-6968.2008.01405.x
Sahin, O., Kassem, I., Shen, Z., Lin, J., Rajashekara, G., and Zhang, Q. (2015). Campylobacter in poultry: ecology and potential interventions. Avian Dis. 59, 185–200. doi: 10.1637/11072-032315
Schierack, P., Kleta, S., Tedin, K., Babila, J. T., Oswald, S., Oelschlaeger, T. A., et al. (2011). E. coli Nissle 1917 affects Salmonella adhesion to porcine intestinal epithelial cells. PLoS ONE 6:e14712. doi: 10.1371/journal.pone.0014712
Schroeder, B., Duncker, S., Barth, S., Bauerfeind, R., Gruber, A. D., Deppenmeier, S., et al. (2006). Preventive effects of the probiotic Escherichia coli strain Nissle 1917 on acute secretory diarrhea in a pig model of intestinal infection. Dig. Dis. Sci. 51, 724–731. doi: 10.1007/s10620-006-3198-8
Schultz, M. (2008). Clinical use of E. coli Nissle 1917 in inflammatory bowel disease. Inflamm. Bowel Dis. 14, 1012–1018. doi: 10.1002/ibd.20377
Schultz, M., Strauch, U. G., Linde, H. J., Watzl, S., Obermeier, F., Gottl, C., et al. (2004). Preventive effects of Escherichia coli strain Nissle 1917 on acute and chronic intestinal inflammation in two different murine models of colitis. Clin. Diagn. Lab. Immunol. 11, 372–378. doi: 10.1128/CDLI.11.2.372-378.2004
Schulze, A., and Downward, J. (2001). Navigating gene expression using microarrays–a technology review. Nat. Cell Biol. 3, E190–E195. doi: 10.1038/35087138
Sherman, P. M., Johnson-Henry, K. C., Yeung, H. P., Ngo, P. S., Goulet, J., and Tompkins, T. A. (2005). Probiotics reduce enterohemorrhagic Escherichia coli O157:H7- and enteropathogenic E. coli O127:H6-induced changes in polarized T84 epithelial cell monolayers by reducing bacterial adhesion and cytoskeletal rearrangements. Infect. Immun. 73, 5183–5188. doi: 10.1128/IAI.73.8.5183-5188.2005
Sonnenborn, U., and Schulze, J. (2009). The non-pathogenic Escherichia coli strain Nissle 1917 – features of a versatile probiotic. Microb. Ecol. Health Dis. 21, 122–158. doi: 10.3109/08910600903444267
Thomrongsuwannakij, T., Chuanchuen, R., and Chansiripornchai, N. (2016). Identification of competitive exclusion and its ability to protect against Campylobacter jejuni in broilers. Thai J. Vet. Med. 46, 279–286.
Trebichavsky, I., Splichal, I., Rada, V., and Splichalova, A. (2010). Modulation of natural immunity in the gut by Escherichia coli strain Nissle 1917. Nutr. Rev. 68, 459–464. doi: 10.1111/j.1753-4887.2010.00305.x
Ukena, S. N., Singh, A., Dringenberg, U., Engelhardt, R., Seidler, U., Hansen, W., et al. (2007). Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS ONE 2:e1308. doi: 10.1371/journal.pone.0001308
Ulluwishewa, D., Anderson, R. C., McNabb, W. C., Moughan, P. J., Wells, J. M., and Roy, N. C. (2011). Regulation of tight junction permeability by intestinal bacteria and dietary components. J. Nutr. 141, 769–776. doi: 10.3945/jn.110.135657
von Buenau, R., Jaekel, L., Schubotz, E., Schwarz, S., Stroff, T., and Krueger, M. (2005). Escherichia coli strain Nissle 1917: significant reduction of neonatal calf diarrhea. J. Dairy Sci. 88, 317–323. doi: 10.3168/jds.S0022-0302(05)72690-4
Wine, E., Chan, V. L., and Sherman, P. M. (2008). Campylobacter jejuni mediated disruption of polarized epithelial monolayers is cell-type specific, time dependent, and correlates with bacterial invasion. Pediatr. Res. 64, 599–604. doi: 10.1203/PDR.0b013e31818702b9
Wine, E., Gareau, M. G., Johnson-Henry, K., and Sherman, P. M. (2009). Strain-specific probiotic (Lactobacillus helveticus) inhibition of Campylobacter jejuni invasion of human intestinal epithelial cells. FEMS Microbiol. Lett. 300, 146–152. doi: 10.1111/j.1574-6968.2009.01781.x
Zia, S., Wareing, D., Sutton, C., Bolton, E., Mitchell, D., and Goodacre, J. A. (2003). Health problems following Campylobacter jejuni enteritis in a Lancashire population. Rheumatology 42, 1083–1088. doi: 10.1093/rheumatology/keg303
Zyrek, A. A., Cichon, C., Helms, S., Enders, C., Sonnenborn, U., and Schmidt, M. A. (2007). Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO-2 and PKCzeta redistribution resulting in tight junction and epithelial barrier repair. Cell Microbiol. 9, 804–816. doi: 10.1111/j.1462-5822.2006.00836.x
Keywords: probiotic, Campylobacter, E. coli Nissle 1917, HT-29 cells, adhesion, invasion, intracellular survival, tight junctions
Citation: Helmy YA, Kassem II, Kumar A and Rajashekara G (2017) In Vitro Evaluation of the Impact of the Probiotic E. coli Nissle 1917 on Campylobacter jejuni’s Invasion and Intracellular Survival in Human Colonic Cells. Front. Microbiol. 8:1588. doi: 10.3389/fmicb.2017.01588
Received: 27 June 2017; Accepted: 04 August 2017;
Published: 22 August 2017.
Edited by:
Yi-Cheng Sun, Institute of Pathogen Biology (CAMS), ChinaReviewed by:
Alessandra De Cesare, Università di Bologna, ItalyHeriberto Fernandez, Austral University of Chile, Chile
Copyright © 2017 Helmy, Kassem, Kumar and Rajashekara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gireesh Rajashekara, cmFqYXNoZWthcmEuMkBvc3UuZWR1
†Present address: Anand Kumar, B-10 Group, Biosecurity and Public Health, Bioscience Division, Los Alamos National Laboratory, Los Alamos, NM, United States
 Yosra A. Helmy
Yosra A. Helmy Issmat I. Kassem
Issmat I. Kassem Anand Kumar
Anand Kumar Gireesh Rajashekara
Gireesh Rajashekara