- 1Laboratory of Microbiology, Wageningen University & Research, Wageningen, Netherlands
- 2Department of Medical Microbiology, University Medical Center Utrecht, Utrecht, Netherlands
- 3Centre for Zoonoses and Environmental Microbiology, National Institute for Public Health and the Environment, Bilthoven, Netherlands
Enterococci have emerged as important opportunistic pathogens in intensive care units (ICUs). In this study, enterococcal population size and Enterococcus isolates colonizing the intestinal tract of ICU patients receiving Selective Digestive Decontamination (SDD) were investigated. All nine patients included in the study showed substantial shifts in the enterococcal 16S rRNA gene copy number in the gut microbiota during the hospitalization period. Furthermore, 41 Enterococcus spp. strains were isolated and characterized from these patients at different time points during and after ICU hospitalization, including E. faecalis (n = 13), E. faecium (n = 23), and five isolates that could not unequivocally assigned to a specific species (E. sp. n = 5) Multi locus sequence typing revealed a high prevalence of ST 6 in E. faecalis isolates (46%) and ST 117 in E. faecium (52%). Furthermore, antibiotic resistance phenotypes, including macrolide and vancomycin resistance, as well as virulence factor-encoding genes [asa1, esp-fm, esp-fs, hyl, and cyl (B)] were investigated in all isolates. Resistance to ampicillin and tetracycline was observed in 25 (61%) and 19 (46%) isolates, respectively. Furthermore, 30 out of 41 isolates harbored the erm (B) gene, mainly present in E. faecium isolates (78%). The most prevalent virulence genes were asa1 in E. faecalis (54%) and esp (esp-fm, 74%; esp-fs, 39%). Six out of nine patients developed nosocomial enterococcal infections, however, corresponding clinical isolates were unfortunately not available for further analysis. Our results show that multiple Enterococcus species, carrying several antibiotic resistance and virulence genes, occurred simultaneously in patients receiving SDD therapy, with varying prevalence dynamics over time. Furthermore, simultaneous presence and/or replacement of E. faecium STs was observed-, reinforcing the importance of screening multiple isolates to comprehensively characterize enterococcal diversity in ICU patients.
Introduction
The genus Enterococcus encompasses indigenous commensal bacteria reported from the human and animal gut as well as the oral cavity and vagina in humans, where they have adapted to nutrient-rich, oxygen-depleted, and ecologically complex environments (Fisher and Phillips, 2009).
In the human gut, the genus Enterococcus can constitute up to 1% of the total bacterial microbiota in healthy individuals, with Enterococcus faecium and Enterococcus faecalis as most common species (Sghir et al., 2000). In contrast to their commensal role, over the past decades E. faecium and E. faecalis have also emerged as agents of nosocomial infections such as endocarditis, bacteraemia, meningitis, wound, and urinary tract infections (Guzman Prieto et al., 2016). In addition, other enterococcal species including Enterococcus durans, Enterococcus avium, Enterococcus gallinarum, Enterococcus casseliflavus, Enterococcus raffinosus, and Enterococcus hirae have sporadically been associated with infections in humans (Klein, 2003).
Most of the E. faecium and E. faecalis infections are opportunistic and are increasingly difficult to treat due to high rates of resistance to β-lactams, aminoglycosides, and vancomycin, which are mostly associated with E. faecium strains (Cattaneo et al., 2010). Similar to resistance genes, virulence genes are also frequently encoded on mobile elements and are therefore thought to disseminate frequently via intra- and interspecies horizontal gene transfer within the genus Enterococcus (Laverde Gomez et al., 2011). Both E. faecium and E. faecalis can carry a variety of genes that contribute to virulence in the immunocompromised patient. For E. faecalis these include genes encoding aggregation substance (asa1) (Galli and Wirth, 1991), cytolysin (cyl) (Jett et al., 1992), and enterococcal surface protein (esp-fs) (Vankerckhoven et al., 2004),whereas for E. faecium genes associated with virulence encode a putative hyalorunidase (hyl) (Klare et al., 2005) and enterococcal surface protein (esp-fm) (Hendrickx et al., 2013), among others.
Studies using Multi Locus Sequence Typing (MLST) have shown that there is a remarkable difference in the population structure between E. faecalis and E. faecium (Palmer et al., 2014). The phylogeny of E. faecalis did not reveal clustering of strains according to their source, e.g., human, clinical, or animal strains (Guzman Prieto et al., 2016).
In contrast, in E. faecium, high-risk clonal-complexes exist, which exhibit high levels of antibiotic resistance and are significantly associated with clinical infections in hospitalized patients (Leavis et al., 2006; Willems et al., 2012). Genome sequencing confirmed that the majority of clinical isolates form a distinct sub-population of E. faecium (Lebreton et al., 2013). Recently, Tedim et al. (2015) studied the population biology of intestinal Enterococcus isolates from hospitalized and non-hospitalized individuals in different age groups. They found that E. faecium populations differ with respect to the observed clonal lineages between hospitalized patients and community-based individuals.
Moreover, the previous identification by whole genome sequencing of E. faecium isolates of distinct hospital (A1) and commensal (B) clades suggests that a distinct evolutionary background exists between commensal and clinical isolates (Palmer et al., 2012). Likewise, Muruzábal-Lecumberri et al. (2015) reported a high prevalence of E. faecalis sequence type (ST) 6 (CC2) in patients undergoing selective decontamination of the digestive tract and indicated that a rapid detection is necessary to avoid a dissemination outside intensive care units (ICUs).
Patients in an ICU are at a high risk for developing nosocomial infections with multi-drug resistant bacteria and are often exposed to strong selective antibiotic pressure (Streit et al., 2004). Several studies have shown that the exposure of patients to broad-spectrum antibiotics, combined with prolonged hospital stay, can result in colonization by multi-drug resistant enterococci leading to nosocomial transmission and infection (Austin et al., 1999; Carmeli et al., 2002). The prophylactic therapy Selective Digestive Decontamination (SDD) aims to prevent secondary infection with opportunistic pathogens, including Enterobacteriaceae, Staphylococcus aureus and yeasts, in ICU patients and to decrease mortality (de Smet et al., 2009).
Previous studies have shown that SDD therapy can select for intestinal colonization by enterococci (Humphreys et al., 1992; de Smet et al., 2009; Benus et al., 2010). A recent meta-analysis of van der Bij et al. (2016), determined the antibiotic resistance rate of Gram-positive cocci in blood and respiratory specimens in 42 Dutch ICUs in the period from 2008 to 2013, indicating that prophylactic therapy was not associated with an increase of antibiotic resistance in Gram-positive cocci. In this study, we characterized Enterococcus isolates colonizing the intestinal tract of ICU patients receiving SDD therapy and to evaluate in more detail the genetic relatedness of E. faecalis and E. faecium isolates, using MLST and Bayesian analysis of the population structure (BAPS). Furthermore, we determined carriage of genes encoding antimicrobial resistance and virulence determinants in this population.
Materials and Methods
Selection of Patients
The patients were selected as part of an approved study to determine the effects of antibiotic prophylactic therapy on antibiotic resistance (Buelow et al., 2014). The inclusion criteria included patients discharged from the ICU who received SDD for at least 96 h. Exclusion criteria included a hospital stay and/or antibiotic treatment prior to ICU admission and discontinuation of SDD before ICU discharge. All patients included in this study were > 18 years of age. The SDD protocol was reviewed and approved by the institutional review board of the University Medical Center Utrecht (Utrecht, The Netherlands). The SDD protocol comprised the oral application of 0.5 g of a paste containing 2% tobramycin 2% polymyxin E and 2% amphotericin B, given four times daily. In addition, a 10 ml suspension containing 80 mg tobramycin, 100 mg polymyxin E and 500 mg amphotericin B was administered through a gastric tube four times daily, and cefotaxime (4 × 1,000 mg) was given intravenously for the first 4 days after ICU admission.
Samples Collection
Fecal samples were collected at different time points during ICU hospitalization and, for five patients, after ICU discharge and cessation of SDD. All the samples were de-identified, received a sample code, and were subsequently categorized according to the collection time for subsequent analyses: ICU stay (including samples collected during ICU hospitalization for up to 40 days, n = 27), and post-ICU (samples taken after ICU discharge and discontinuation of SDD, n = 7) (Table 1). Samples were collected upon defecation and stored at 4°C for 30 min to 4 h. Two aliquot of 0.5 g of fecal material were collected, one aliquot was stored directly −80°C for fecal DNA isolation, and the other aliquot was suspended in 5 ml of 20 mM anoxic phosphate buffer (pH 7.0) with 40% glycerol, and transferred to −80°C for further analysis.
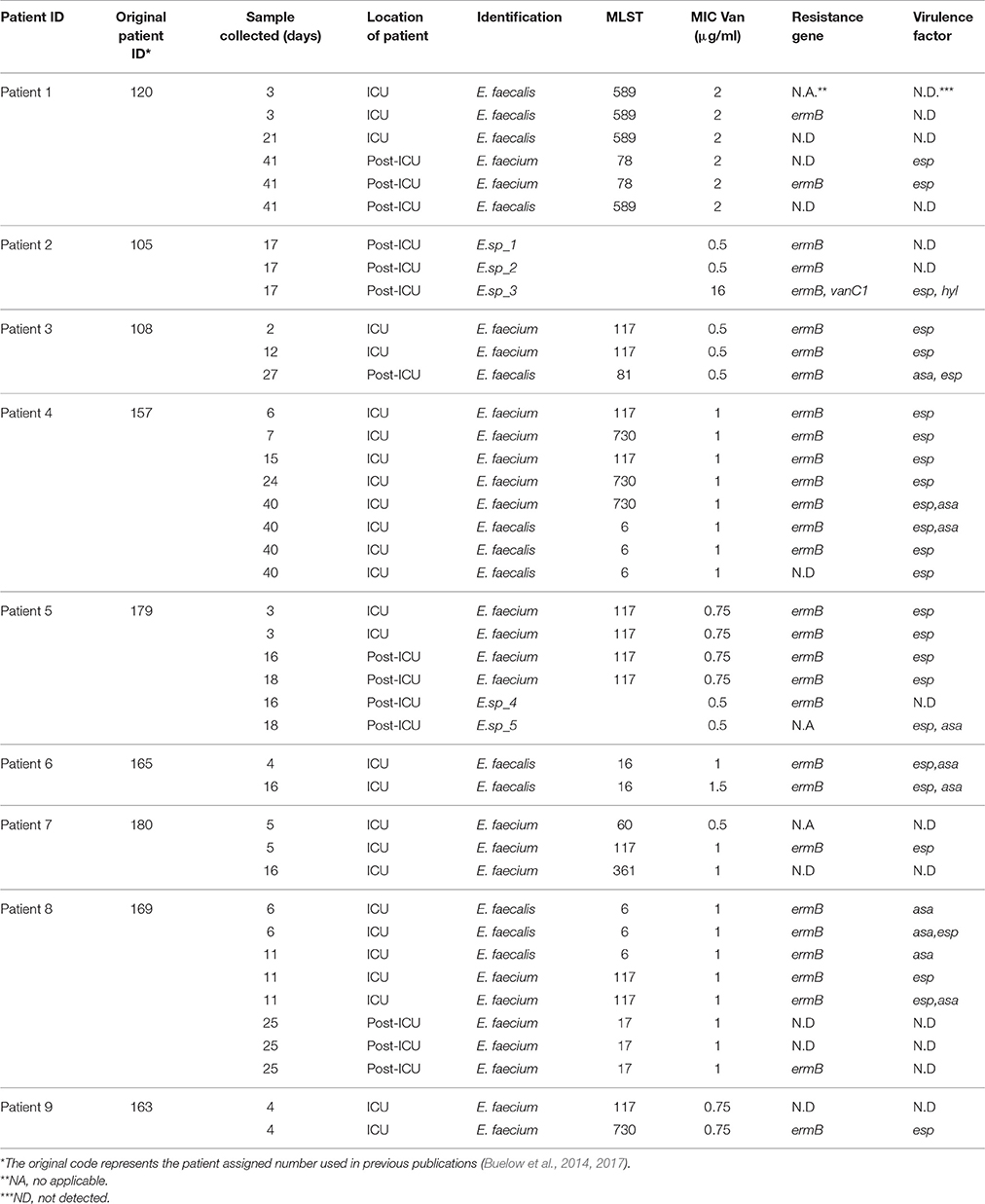
Table 1. Overview of the Enterococcus species isolated and characterized per patient during the different time points (ICU stay and Post-ICU).
Bacterial Culture Conditions and Initial Characterization
Enterococci were isolated on Bile-Esculin Agar (BEA) (Oxoid B.V., Landsmeer, The Netherlands). Colonies growing on BEA media were selected based on colony morphology (up to five colonies per plate) for phenotypic characterization (Winn et al., 2006). Haemolysis was determined by cultivation on Blood Agar supplemented with 5% sheep blood (Oxoid) after incubation at 37°C for 24 h. Isolates that showed phenotypic differences in antibiotic resistance patterns (see below) were selected for further characterization.
DNA isolation was performed using the protocol for Gram-positive bacteria of the QIAamp® DNA Mini Kit (Qiagen Benelux B.V., Venlo, The Netherlands). DNA was used for the identification of the isolates and detection of antibiotic resistance and virulence genes by Polymerase Chain Reaction (PCR) as described below. In addition, total bacterial DNA extraction was performed from 0.5 g of fecal material using the modified repeated bead beating method previously described (Salonen et al., 2010) and used for the quantification of the enterococcal population as described below.
Identification and Classification of Isolates
The complete bacterial 16S ribosomal RNA (rRNA) gene was amplified from genomic DNA using T7prom-Bact-27-F and Uni-1492-R primers as described previously (Rajilić-Stojanović et al., 2009). The amplified fragments were selected for partial sequence analysis of the 16S rRNA gene (~800 bp) using the 16S-1392R primer 5′- ACGGGCGGTGTGTRC -3′ (GATC Biotech, Cologne, Germany).
Partial 16S rRNA gene sequences of isolates assigned to other Enterococcus species obtained in this study were deposited at GenBank under accession numbers KX577731, KX577732, KX577733, KX577734.
Quantification of the Enterococcal Population
The enterococcal population present in the fecal material was quantified by qPCR with 16S rRNA gene targeted enterococci-specific primers described by Matsuda et al. (2009). A standard curve was made from bacterial genomic DNA (E. faecium E5), using 10-fold dilution series (1.0 × 101–1.0 × 109 DNA copies μl−1). The standard curve had a correlation coefficient of R = 0.97–0.99 and an efficiency of amplification between 97.2 and 103.1%.
The qPCR was performed with the SYBR-Green PCR kit (Roche Applied Science). The total reaction volume was 10 μl: 5 μl SYBR-Green I, 0.2 μl of each forward and reverse primers, 1.6 μl of sterilized nuclease free water and 3 μl extracted DNA (1 ng/μl). The amplification program included an initial denaturation step at 94°C for 5 min, followed by 40 cycles of denaturation for 20 s at 94°C, annealing for 20 s at 60°C and extension for 50 s at 720C. A melting curve analysis was performed by slowly heating the sample from 60 to 95°C (0.5°C per 15 s) with simultaneous monitoring of fluorescence. All reactions were performed in duplicates in a 384- well PCR plate sealed with optical sealing tape (Bio-Rad) on a iQ5 real-time PCR detection system.
One way-ANOVA was used for statistical analysis to indicate significant differences of enterococcal abundance during and after SDD therapy, with Bonferoni correction (p < 0.05) for multiple testing.
Antimicrobial Susceptibility
Vancomycin resistance of enterococci was screened on Mueller-Hinton Agar (MHA) (Oxoid) containing 6 μg/ml vancomycin. Colonies growing on this medium were tested by E-test (Biomerieux) to determine the minimal inhibitory concentration (MIC) of vancomycin, following CLSI guidelines (CLSI, 2013). In addition, the disk diffusion method was used to test for susceptibility to ampicillin (10 μg) and tetracycline (30 μg) (CLSI, 2013).
Detection of Antibiotic Resistance- and Virulence Factor-Encoding Genes
Antibiotic resistance genes were detected using a multiplex PCR for the vancomycin-resistance genes van (A), van (B), and van (C) (van (C1)–van (C2)/van (C3)) (Depardieu et al., 2004), and a single PCR for erm (A), erm (B), erm (C), and mef (A)/mef (E) genes (Zou et al., 2011). PCR products of mef (A) and mef (E) genes were discriminated by BamHI restriction analysis, as only mef (A) carries a single restriction site, giving rise to fragments of 284 and 64 bp as described previously (Klaassen and Moutin, 2005).
Genes coding for virulence factors, i.e., enterococcal surface protein (esp-fm, esp-fs), aggregation substance (asa1), cytolysin (cyl (B)), and hyalorunidase (hyl), were selected for detection by PCR as described previously (Vankerckhoven et al., 2004; Hällgren et al., 2009).
E. faecalis ATCC29212, E. faecium E5 and E. faecalis E507 (Department of Medical Microbiology, Utrecht Medical Centre, UMC, The Netherlands) and E. gallinarum HSIEG1 (van den Bogert et al., 2013) (Laboratory of Microbiology, Wageningen University, The Netherlands) were used as positive controls for the detection of antibiotic resistance and virulence factor encoding genes. Amplicons were visualized by agarose gel electrophoresis.
Clonal Relatedness and Analysis of Population Structure
In order to establish the clonal relationship of Enterococcus isolates, we applied the MLST schemes proposed by Ruiz-Garbajosa et al. (2006) and Homan et al. (2002) or E. faecalis and E. faecium, respectively.
Sequences were compared with published alleles, and sequence types (STs) were assigned using the MLST database (http://pubmlst.org/efaecium/ and http://pubmlst.org/efaecalis/). BAPS groups were determined as previously described (Willems et al., 2012). E. faecalis and E. faecium MLST products were stored in the MLST database (http://pubmlst.org/efaecium/ and http://pubmlst.org/efaecalis).
Results
Identification of Intestinal Enterococcus Isolates from ICU Patients Receiving SDD
A total of nine ICU hospitalized patients that underwent SDD prophylactic antibiotic therapy were followed. From these patients, 34 fecal samples were collected. Besides SDD, additional antibiotics were administrated to seven patients for the control of infections and/or as an agent to accelerate gastric motility during ICU stay (Pilot, 1994; Galligan et al., 2005). Enterococci were isolated from 23 out of 34 fecal samples; the number of isolates per patient ranged from 2 (patient 6, 9) to 8 (patient 4, 8) (Table 1). Thirty six isolates were classified to the enterococcal species level by 16S rRNA gene sequencing and MLST products. The most commonly found species were E. faecium (23 isolates) and E. faecalis (13 isolates). The remaining five enterococcal isolates, all of which were isolated only during the post-ICU phase (Figure 1, Table 1), could not be unequivocally identified to the species level by 16S rRNA gene sequencing. Six patients receiving SDD developed nosocomial enterococcal infections during ICU stay (Figure 1), including one pleural infection caused by E. faecium, six urinary tract infections (two episodes in a single patient) caused by E. faecalis (five cases) and E. faecium (one case), and one central line catheter associated infection caused by E. faecalis (two episodes in a single patient). Unfortunately, however, the corresponding clinical isolates were not available for further analysis.
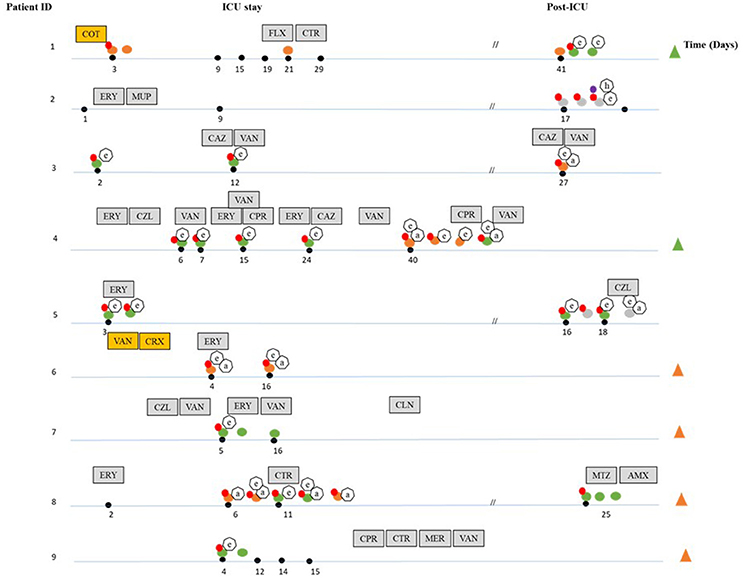
Figure 1. Overview of the dynamics of colonization by Enterococcus species and carriage of antibiotic resistance and virulence genes during and after ICU hospitalization. The black dots indicate days where fecal samples were taken during hospitalization. Discontinuation lines (//) indicate samples collected during the post-ICU period. The different species isolated are indicated by differently colored dots: orange (E. faecalis), green (E. faecium), dark gray (E.sp). Isolates not connected to a black dot were obtained from the sample closest to the left. The presence of antibiotic resistance genes is indicated by red (ermB) and purple dots (vanC1). Virulence factors are shown in heptagonal shapes (a-asa1), (e-esp-fm and esp-fs), (h-hyl). Patients that developed nosocomial infections during ICU stay with E. faecalis and E. faecium are indicated by green (E. faecium) and orange (E. faecalis) triangles. Gray boxes indicate systemic antibiotics given under clinical indications at the specific time point indicated (ERY, erythromycin; VAN, vancomycin; CPR, ciprofloxacin; CTR, ceftriaxone; CAZ, ceftazidime; CZL, cefazoline; MER, meropenem; FLX, flucoxacilin; AMX, amoxacillin; CLN, clindamycin; MTZ, metronidazole; MUP, mupirosin), yellow boxes indicate systemic antibiotics given under clinical indications during the entire ICU stay (CRX, cefuroxime; COT, cotrimoxazole).
Quantification of the Enterococcal Population
The quantification of the enterococcal population based on enterococcal 16S rRNA gene-targeted qPCR showed a significant increase in time from ICU stay to post ICU (6.0 × 10 6 vs. 2.0 × 10 7 Log DNA copies/g of feces; p < 0.05) in seven out of 9 patients. In the patients, substantial shifts in the absolute enterococcal 16S rRNA gene copy number were observed during the hospitalization period (Figure 2).
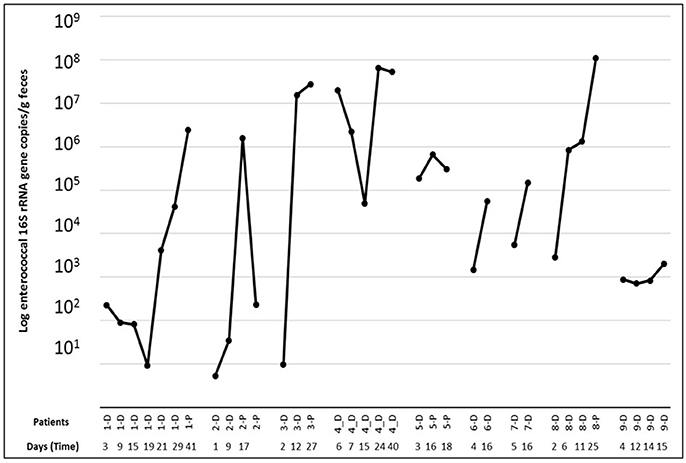
Figure 2. 16S rRNA gene-targeted qPCR based quantification of the enterococcal population present per patient in the samples taken at different time points [during ICU (D) and Post-ICU (P)].
Antimicrobial Susceptibility
All isolates were vancomycin-susceptible (MIC 0.5–2 μg/ml), except for a single vancomycin resistant (MIC 16 μg/ml) isolate closely related to E. gallinarum (E.sp_3) (99% nucleotide identity).
Ampicillin resistance was detected in 25 out of 41 isolates, with the highest prevalence of resistant strains being found amongst E. faecium isolates[21 (91%) E. faecium, three (23%) E. faecalis, and one (20%) E.sp (E.sp_5)]. Resistance to tetracycline was detected in 19 out of 41 isolates [9 E. faecalis, seven E. faecium, and three E. spp. (E.sp_1, E.sp_2, E.sp_4)], the majority of which was obtained during ICU stay and in one patient during the first 72 h after admission.
Detection of Antibiotic Resistance- and Virulence Factor-Encoding Genes
The presence of four of the genes that encode macrolide-lincosamide-streptogramin (MLS) resistance, namely erm (A), erm (B), erm (C), and mef (A)/mef (E) genes genes encoding the Major Facilitator Superfamily (MFS) transporter conferring resistance to erythromycin, were assayed by PCR-based detection. Our results revealed the presence of the erm (B) gene in 30 out of 41 enterococci isolates that were obtained during the entire study period. No other MLSB resistance genes or MFS transporter genes were detected. From the group of vancomycin resistance genes tested, the van (C1) gene was identified in the single isolate that was also found vancomycin resistant (Figure 1, Table 1).
Three out of the four targeted genes encoding enterococcal virulence factors were detected. The asa1 gene was frequently present in E. faecalis isolates (n = 7/13), whereas the esp gene was more often found in E. faecium isolates (n = 17/23). The esp gene was also present in two E. sp. isolates. Finally, the hyl gene was detected post-ICU in a single isolate of E. faecium and in a single isolate closely related to E. gallinarum (99% nucleotide identity). All isolates displayed alpha-haemolysis in blood agar. The cyl (B) gene, which would cause a beta-hemolytic phenotype (Semedo et al., 2003; Abriouel et al., 2008), was not detected in any of the isolates (Figure 1).
Clonal Relatedness and Analysis of Population Structure
Using MLST, we established the clonal relationship of all E. faecium and E. faecalis isolates obtained in this study. In total, we identified six different STs among the E. faecium isolates (Figure 3, Table 2). Further analysis of their population structure revealed that these STs belonged to three BAPS (sub) groups, which were previously associated with hospitalized patients (Willems et al., 2012). The majority of the STs belonged to BAPS group 2.1a (18 isolates), and 15 of them were resistant to ampicillin (ST117 n = 12, ST78 n = 2, and ST730 n = 1). Other sub-groups observed included BAPS 1.2 (2 isolates) as well as BAPS 3.3a2 (3 isolates). In four patients, we identified two or more different STs in the same patient during hospitalization (Figure 3).
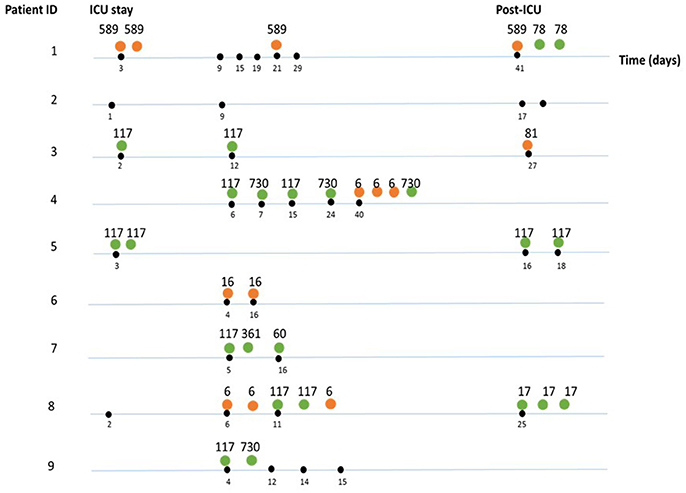
Figure 3. Sequence types (ST) identified per sample, per patient during and after SDD therapy. The differently colored dots indicate the species: orange (E. faecalis), green (E. faecium). Numbers indicate the sequence types. Black dots indicate the time point (days) where samples were taken during hospitalization. If multiple strains were isolated and characterized, these are shown to the right of each indicated timepoint.
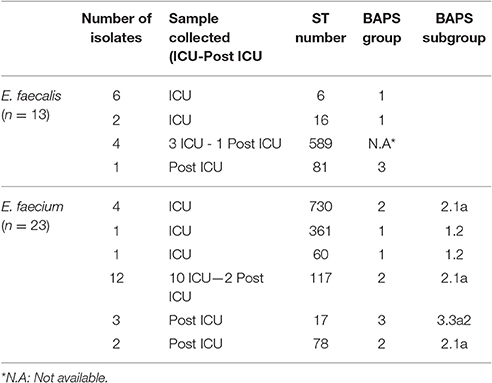
Table 2. Sequence type (ST) and BAPS analysis of Enterococcus faecalis and Enterococcus faecium isolates.
Among the E. faecalis isolates, we identified three STs (ST6, ST81, and ST16), which were previously detected among hospitalized patients (Willems et al., 2012), as well as a new ST (ST589), represented by four isolates (Figure 3, Table 2). All isolates belonging to ST589 were susceptible to ampicillin, and were obtained from a single patient from samples taken throughout the study. Three out of these four ST589 isolates carried the ermB gene.
From the group of E. faecalis isolates belonging to ST6 (n = 6), three carried ermB, asa, and esp genes and were susceptible to ampicillin, whereas the other three isolates displayed resistance to ampicillin. BAPS cluster analysis subdivided the E. faecalis isolates into BAPS groups 1 (eight isolates) and 3 (one isolate) (Table 2). In contrast to the situation in the E. faecium isolates, we neither detected the simultaneous presence of multiple E. faecalis STs nor clonal replacement over time within individual patients.
Discussion
In the present study we characterized a group of Enterococcus species isolated from fecal samples of ICU patients receiving SDD therapy. We observed a pool of diverse enterococcal species, being E. faecium and E. faecalis the most prevalent species, both previously identified as important human pathogens associated with nosocomial infections (Cattaneo et al., 2010).
In three patients, these two species were detected in samples collected during the first 72 h, which could suggest that these patients were colonized with the recovered strains before ICU admission. This is in line with previous studies, as recently reviewed by Guzman Prieto and co-authors, showing that enterococci are present in healthy humans as well as in the environment, and that the abundance of resistance genes and mobile elements rapidly increases and facilitates colonization and subsequent infection in hospitalized patients (Guzman Prieto et al., 2016).
The clinical isolates causing nosocomial infections in the patients included in our study were not available for further analysis, which limited the possibility to clarify whether the infections derived from the isolates colonizing the patient in that period, and whether the isolates obtained from fecal material correspond to a nosocomial acquisition or selection of strains that were present in the gut microbiota of these patients prior to hospitalization. Other enterococcal isolates could not be identified to the species level (n = 5), although it should be noted that these were found only post-ICU. One possible explanation could be that due to the cessation of the SDD therapy, and thus aleviation of the corresponding antibiotic selective pressure during post-ICU stay, other species than E. faecalis and E. faecium were able to colonize the gut. From these isolates, three isolates were closely related to E. gallinarum and E. avium species. Both species have been identified in fecal samples of animals and healthy humans (Layton et al., 2010; Silva et al., 2011), and infrequently linked to human enterococcal infections (Tan et al., 2010; Varun et al., 2016). We cannot exclude that these other species were present in fecal samples taken at earlier time points at very low population size and thus evaded detection with the methods used in this study.
We were furthermore able to isolate more than one enterococcal species per sample in five out of nine patients. This highlights the importance of analyzing multiple colonies per culture to adequately sample the diversity of the enterococcal population. Moreover, qPCR analysis indicated an increase in the abundance of enterococci in seven out of nine patients. This shift could be due to the administration of antibiotics and to changes in the gut microbiota composition due to the antibiotic selective pressure. This is in line with previous studies based on qPCR analyses that showed that the enterococcal population increased in hospitalized patients receiving antibiotics compared to hospitalized patients without antibiotics and healthy volunteers (Bartosch et al., 2004).
Colonization by ampicillin-resistant Enterococcus faecium (ARE) is frequently associated with previous exposure to selective antibiotics, and ampicillin resistance is a specific trait for nosocomial isolates (de Regt et al., 2012).
In our study the highest prevalence of ampicillin resistance were found in E. faecium isolates compared to non-ampicillin resistant-E. faecium isolates. Similar results were reported by Ruiz-Garbajosa P., et al. (2012); indicating that this increased population most significantly contributes to the transmission and spread of enterococcal resistance in the ICU.
In our study, vancomycin resistance was not detected among E. faecalis and E. faecium isolates. This is in line with the previously reported prevalence (<1% for both E. faecium and E. faecalis) of vancomycin-resistance among enterococci in clinical infections in the Netherlands, as shown in the European Antimicrobial Resistance Surveillance System (EARSS) (ecdc.europa.eu/en/activities/surveillance/EARS-Net). The only vancomycin-resistant isolate was identified as being closely related to E. gallinarum (vancomycin MIC of 16 μg/ml), which carried the van (C1) gene that is naturally present in this species (Toye et al., 1997).
Resistance to tetracycline was detected in 46% of all 41 isolates (n = 19) and predominantly in E. faecalis isolates, which is in accordance with previous studies (Templer et al., 2008). Moreover, the presence of the macrolide resistance gene erm (B) was detected in 30 of these isolates (73%). We can not exclude, however, that other erythromycin resistance genes were present in these isolates. Although no vancomycin resistant E. faecalis and E. faecium populations were observed in the current study, our findings highlight the importance to perform a periodic surveillance during SDD therapy in ICU patients, in order to detect resistant Enterococcus spp. strains and prevent their dissemination as a preventive infection control measure.
Moreover, we observed that the pool of diverse enterococcal species identified in this study also harbored a variety of virulence genes that could contribute to infections in immunocompromised patients. The esp gene was the most prevalent virulence determinant detected throughout the study period followed by the asa1 gene detected mainly during ICU stay, including two E. faecium and seven E. faecalis isolates, and one isolate closely related to E. avium. Similar results were previously reported (Billström et al., 2008; Hällgren et al., 2009; Sharifi et al., 2013). In addition, we detected the presence of the hyl gene in one E. faecium isolate and one isolate closely related to E. gallinarum only post-ICU. It should be noted, however, that the hyl gene has been identified not only in E. faecium and E. faecalis, but also in E. casseliflavus, E. mundtii, and E. durans isolated from food-stuffs (Trivedi et al., 2011), showing that the hyl gene can be present in a variety of Enterococcus spp. Furthermore, we cannot exclude that isolates obtained here contain other virulence genes that were not targeted in the present study.
Finally, the clonal relationship and population structure (BAPs groups) found in E. faecium and E. faecalis isolates indicated that the vast majority of our E. faecium isolates clustered in subgroups 2.1a and 3.3a2, representing separate hospital lineages that belong to clade A1 that contains most nosocomial E. faecium isolates (Willems et al., 2012). These BAPS groups are infrequent in healthy individuals (de Regt et al., 2012) suggesting that these isolates have been acquired during hospitalization.
Most E. faecalis isolates (62%) clustered in BAPS group 1, of which the majority belonged to ST 6 that was previously found in both hospitalized and non-hospitalized patients (Willems et al., 2012; Tedim et al., 2015).
In our study we observed the simultaneous presence of STs and clonal replacement over time among E. faecium isolates during ICU stay, whereas this was not the case for E. faecalis. Based on the current data, however, it is unclear whether the clonal replacement observed in E. faecium isolates was due to nosocomial strains or populations that were previously present in lower abundances. Future studies would be needed to test this hypothesis.
The prevalence of Enterococcus in ICU hospitalized patients, combined with the carriage of antibiotic resistance and virulence genes, described in this study, underlines the importance of this group of organisms as a potential cause of nosocomial infections in critically ill patients. Particular attention needs to be given to ICU patients during SDD therapy, with specific focus on the increased colonization by enterococci, even in non-endemic countries, considering that in SDD therapy, the enterococcal population is not a target group. SDD has been shown to not only decrease mortality and morbidity, but also to induce changes in the composition of the gut microbiota of patients (Benus et al., 2010; Buelow et al., 2014). Here we showed that it may increase the prevalence and extent of colonization by enterococci and provide insights into the diversity of the enterococcal strains that colonize these patients.
Author Contributions
TdJBG and PP: designed and performed the experiments, analyzed, and interpreted the data and wrote the paper. RW, JT and WvS: performed the MLST analysis and revised the work critically for intellectual content. HS and MWJvP: supervised the project, substantial contribution to revising it critically, and final approval of the version to be published.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by The Netherlands Organization for Health Research and Development ZonMw (Priority Medicine Antimicrobial Resistance; grant 205100015) and by the European Union Seventh Framework Programme (FP7-HEALTH-2011-single-stage) “Evolution and Transfer of Antibiotic Resistance” (EvoTAR) under grant agreement number 282004. We are grateful to Tom van den Bogert and Ana Sofia Tedim Pedrosa for their advice and suggestions.
References
Abriouel, H., Omar, N. B., Molinos, A. C., López, R. L., Grande, M. J., Martínez-Viedma, P., et al. (2008). Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. Int. J. Food Microbiol. 123, 38–49. doi: 10.1016/j.ijfoodmicro.2007.11.067
Austin, D. J., Bonten, M. J., Weinstein, R. A., Slaughter, S., and Anderson, R. M. (1999). Vancomycin-resistant enterococci in intensive-care hospital settings: transmission dynamics, persistence, and the impact of infection control programs. Proc. Natl. Acad. Sci. U.S.A. 96, 6908–6913. doi: 10.1073/pnas.96.12.6908
Bartosch, S., Fite, A., Macfarlane, G. T., and McMurdo, M. E. T. (2004). Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 70, 3575–3581. doi: 10.1128/AEM.70.6.3575-3581.2004
Benus, R. F., Harmsen, H. J., Welling, G. W., Spanjersberg, R., Zijlstra, J. G., Degener, J. E., et al. (2010). Impact of digestive and oropharyngeal decontamination on the intestinal microbiota in ICU patients. Intensive Care Med. 36, 1394–1402. doi: 10.1007/s00134-010-1826-4
Billström, H., Sullivan, A., and Lund, B. (2008). Cross-transmission of clinical Enterococcus faecium in relation to esp and antibiotic resistance. J. Appl. Microbiol. 105, 2115–2122. doi: 10.1111/j.1365-2672.2008.03983.x
Buelow, E., Bello Gonzz, T. d. J., Fuentes, S., de Steenhuijsen Piters, W. A. A., Lahti, L., Bayjanov, J. R., et al. (2017). Comparative gut microbiota and resistome profiling of intensive care patients receiving selective digestive tract decontamination and healthy subjects. Microbiome 5:88. doi: 10.1186/s40168-017-0309-z
Buelow, E., Gonzalez, T. B., Versluis, D., Oostdijk, E. A., Ogilvie, L. A., van Mourik, M. S., et al. (2014). Effects of selective digestive decontamination (SDD) on the gut resistome. J. Antimicrob. Chemother. 69, 2215–2223. doi: 10.1093/jac/dku092
Carmeli, Y., Eliopoulos, G. M., and Samore, M. H. (2002). Antecedent treatment with different antibiotic agents as a risk factor for vancomycin-resistant Enterococcus. Emerg. Infect. Dis. 8, 802–807. doi: 10.3201/eid0805.010418
Cattaneo, C., Casari, S., Bracchi, F., Signorini, L., Ravizzola, G., Borlenghi, E., et al. (2010). Recent increase in enterococci, viridans streptococci, Pseudomonas spp. and multiresistant strains among haematological patients, with a negative impact on outcome. Results of a 3-year surveillance study at a single institution. Scand. J. Infect. Dis. 42, 324–332. doi: 10.3109/00365540903496569
CLSI (2013). Performance Standards for Antimicrobial Susceptibility Testing. 22nd informational supplement. Wayne, PA: CLSI.
de Regt, M. J., van Schaik, W., van Luit-Asbroek, M., Dekker, H. A., van Duijkeren, E., Koning, C. J., et al. (2012). Hospital and community ampicillin-resistant Enterococcus faecium are evolutionarily closely linked but have diversified through niche adaptation. PLoS ONE 7:e30319. doi: 10.1371/journal.pone.0030319.
de Smet, A. M., Kluytmans, J. A., Cooper, B. S., Mascini, E. M., Benus, R. F., van der Werf, T. S., et al. (2009). Decontamination of the digestive tract and oropharynx in ICU patients. N. Engl. J. Med. 360, 20–31. doi: 10.1056/NEJMoa0800394
Depardieu, F., Perichon, B., and Courvalin, P. (2004). Detection of the van Alphabet and identification of Enterococci and Staphylococci at the species level by multiplex PCR. J. Clin. Microbiol. 42, 5857–5860. doi: 10.1128/JCM.42.12.5857-5860.2004
Fisher, K., and Phillips, C. (2009). The ecology, epidemiology and virulence of Enterococcus. Microbiology 155, 1749–1757. doi: 10.1099/mic.0.026385-0
Galli, D., and Wirth, R. (1991). Comparative analysis of Enterococcus faecalis sex pheromone plasmids identifies a single homologous DNA region which codes for aggregation substance. J. Bacteriol. 173, 3029–3033. doi: 10.1128/jb.173.9.3029-3033.1991
Galligan, J. J., and Vanner, S. (2005). Basic and clinical pharmacology of new motility promoting agents. Neurogastroenterol. Motil. 17, 643–653. doi: 10.1111/j.1365-2982.2005.00675.x
Guzman Prieto, A. M., van Schaik, W., Rogers, M. R. C., Coque, T. M., Baquero, F., Corander, J., et al. (2016). Global emergence and dissemination of enterococci as nosocomial pathogens: attack to the clones? Front. Microbiol. 7:788. doi: 10.3389/fmicb.2016.00788
Hällgren, A., Claesson, C., Saeedi, B., Monstein, H. J., Hanberger, H., and Nilsson, L. E. (2009). Molecular detection of aggregation substance, enterococcal surface protein, and cytolysin genes and in vitro adhesion to urinary catheters of Enterococcus faecalis and E. faecium of clinical origin. Int. J.Med. Microbiol. 299, 323–332. doi: 10.1016/j.ijmm.2008.10.001
Hendrickx, A. P., van Schaik, W., and Willems, R. J. (2013). The cell wall architecture of Enterococcus faecium: from resistance to pathogenesis. Future Microbiol. 8, 993–1010. doi: 10.2217/fmb.13.66
Homan, W. L., Tribe, D., Poznanski, S., Li, M., Hogg, G., Spalburg, E., et al. (2002). Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40, 1963–1971. doi: 10.1128/JCM.40.6.1963-1971.2002
Humphreys, H., Winter, R., and Pick, A. (1992). The effect of selective decontamination of the digestive tract on gastrointestinal enterococcal colonization in ITU patients. Intensive Care Med. 18, 459–463. doi: 10.1007/BF01708581
Jett, B. D., Jensen, H. G., Nordquist, R. E., and Gilmore, M. S. (1992). Contribution of the pADl-encoded cytolysin to the severity of experimental Enterococcus faecalis endophthalmitis. Infect. Immun. 60, 2445–2452.
Klaassen, C. H. W., and Moutin, J. W. (2005). Molecular detection of the macrolide efflux gene: to discriminate or not to discriminate between mef (A) and mef (E). Antimicrob. Agents Chemother. 49, 1271–1278. doi: 10.1128/AAC.49.4.1271-1278.2005
Klare, I., Konstabel, C., Mueller-Bertling, S., Werner, G., Strommenger, B., Kettlitz, C., et al. (2005). Spread of ampicillin/vancomycin-resistant Enterococcus faecium of the epidemic-virulent clonal complex-17 carrying the genes esp and hyl in German hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 24, 815–825. doi: 10.1007/s10096-005-0056-0
Klein, G. (2003). Taxonomy, ecology and antibiotic resistance of enterococci from food and the gastro-intestinal tract. Int. J. Food Microbiol. 88, 123–131. doi: 10.1016/S0168-1605(03)00175-2
Laverde Gomez, J. A., Hendrickx, A. P., Willems, R. J., Top, J., Sava, I., Huebner, J., et al. (2011). Intra- and interspecies genomic transfer of the Enterococcus faecalis pathogenicity island. PLoS ONE 6:e16720. doi: 10.1371/journal.pone.0016720
Layton, B. A., Walters, S. P., Lam, L. H., and Boehm, A. B. (2010). Enterococcus species distribution among human and animal hosts using multiplex PCR. J. Appl. Microbiol. 109, 539–547. doi: 10.1111/j.1365-2672.2010.04675.x.
Leavis, H. L., Bonten, M. J., and Willems, R. J. (2006). Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr. Opin. Microbiol. 9, 454–460. doi: 10.1016/j.mib.2006.07.001
Lebreton, F., van Schaik, W., McGuire, A. M., Godfrey, P., Griggs, A., Mazumdar, V., et al. (2013). Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. MBio 20, 1–10. doi: 10.1128/mBio.00534-13
Matsuda, K., Tsuji, H., Asahara, Y., Matsumoto, K., Takada, T., and Nomoto, K. (2009). Establishment of an analytical sustem for the human fecal microbota, based on reverse transcription-quantitative pcr targeting of multicopy rRNA molecules. Appl. Environ. Microbiol. 75, 1961–1969. doi: 10.1128/AEM.01843-08
Muruzábal-Lecumberri, I., Girbau, C., Canut, A., Alonso, R., and Fernández-Astorga, A. (2015). Spread of an Enterococcus faecalis sequence type 6 (CC2) clone in patients undergoing selective decontamination of the digestive tract. APMIS 123, 245–251. doi: 10.1111/apm.12336
Palmer, K. L., Godfrey, P., Griggs, A., Kos, V. N., Zucker, J., Desjardins, C., et al. (2012). Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. MBio 3:e00318-11. doi: 10.1128/mBio.00318-11
Palmer, K. L., van Schaik, W., Willems, R. J. L., Gilmore, M. S., and Enterococcal Genomics (2014). Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston, MA: Massachusetts Eye and Ear Infirmary.
Pilot, M. A. (1994). Macrolides in roles beyond antibiotic therapy. Br. J. Surg. 81, 1423–1429. doi: 10.1002/bjs.1800811006
Rajilić-Stojanović, M., Heilig, H., Molenaar, D., Kajander, K., Surakka, A., Smidt, H., et al. (2009). Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ. Microbiol. 11, 1736–1751. doi: 10.1111/j.1462-2920.2009.01900.x
Ruiz-Garbajosa, P., Bonten, M. J., Robinson, D. A., Top, J., Nallapareddy, S. R., Torres, C., et al. (2006). Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 44, 2220–2228. doi: 10.1128/JCM.02596-05
Ruiz-Garbajosa, P., de Regt, M., Bonten, M., Baquero, F., Coque, T. M., Cantón, R., et al. (2012). High-density fecal Enterococcus faecium colonization in hospitalized patients is associated with the presence of the polyclonal subcluster CC17. Eur. J. Clin. Microbiol. Infect. Dis. 31, 519–522. doi: 10.1007/s10096-011-1342-7
Salonen, A., Nikkila, J., Jalanka-Tuovinen, J., Immonen, O., Rajilić-Stojanović, M., Kekkonen, RA., et al. (2010). Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J. Microbiol. Methods 81, 127–134. doi: 10.1016/j.mimet.2010.02.007
Semedo, T., Almeida Santos, M., Martins, P., Silva Lopes, M. F., Figueiredo Marques, J. J., Tenreiro, R., et al. (2003). Comparative study using type strains and clinical and food isolates to examine hemolytic activity and occurrence of the cyl operon in enterococci. J. Clin. Microbiol. 41, 2569–2576. doi: 10.1128/JCM.41.6.2569-2576.2003
Sghir, A., Gramet, G., Suau, A., Violaine, R., Pochart, P., and Dore, J. (2000). Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 66, 2263–2266. doi: 10.1128/AEM.66.5.2263-2266.2000
Sharifi, Y., Hasani, A., Ghotaslou, R., Naghili, B., Aghazadeh, M., Milani, M., et al. (2013). Virulence and antimicrobial resistance in enterococci isolated from urinary tract infections. Adv. Pharm. Bull. 3, 197–201. doi: 10.5681/apb.2013.032
Silva, N., Igrejas, G., and Goncalves, A. (2011). Commensal gut bacteria: distribution of Enterococcus species and prevalence of Escherichia coli pylogenetic groups in animals and humans in Portugal. Ann. Microbiol. 62, 449–459. doi: 10.1007/s13213-011-0308-4
Streit, J. M., Jones, R. N., Sader, H. S., and Fritsche, T. R. (2004). Assesment of pathogen occurrences and resistance profiles among infected patients in the intensive care unit: report from the SENTRY Antimicrobial Surveillance Program (North America, 2001). Int. J. Antimicrob. Agents 24, 111–118. doi: 10.1016/j.ijantimicag.2003.12.019
Tan, C. K., Lai, C. C., Wang, J. Y., Lin, S. H., Liao, C. H., Huang, Y. T., et al. (2010). Bacteremia caused by non-faecalis and non-faecium Enterococcus species at a Medical center in Taiwan, 2000 to 2008. J. Infect. 61, 34–43. doi: 10.1016/j.jinf.2010.04.007
Tedim, A. P., Ruiz-Garbajosa, P., Corander, J., Rodríguez, C. M., Cantón, R., Willems, R. J., et al. (2015). Population biology of intestinal Enterococcus isolates from hospitalized and nonhospitalized individuals in different age groups. Appl. Environ. Microbiol. 81, 1820–1831. doi: 10.1128/AEM.03661-14
Templer, S. P., Rohner, P., and Baumgartner, A. (2008). Relation of Enterococcus faecalis and Enterococcus faecium isolates from foods and clinical specimens. J. Food Prot. 71, 2100–2104. doi: 10.4315/0362-028X-71.10.2100
Toye, B., Shymanski, J., Bobrowska, M., Woods, W., and Ramotar, K. (1997). Clinical and epidemiologic significance of enterococci intrinsically resistant to vancomycin (possessing the vanC genotype). J. Clin. Microbiol. 35, 3166–3170.
Trivedi, K., Cupakova, S., and Karpiskov, R. (2011). Virulence factors and antibiotic resistance in enterococci isolated from food sutffs. Vet Med. 56, 352–357. Available online at: http://vetmed.vri.cz/?page=full_papers_from_2001
van den Bogert, B., Boekhorst, J., Smid, E. J., Zoetendal, E. G., and Kleerebezem, M. (2013). Draft genome sequence of Enterococcus sp. strain HSIEG1, isolated from the human small intestine. Genome Announc. 1, 1–2. doi: 10.1128/genomeA.01013-13
van der Bij, A. K., Frentz, D., and Bonten, M. (2016). Gram-positive cocci in Dutch ICUs with and without selective decontamination of the oropharyngeal and digestive tract: a retrospective database analysis. J. Antimicrob. Chemother. 71, 816–820. doi: 10.1093/jac/dkv396
Vankerckhoven, V., Van Autgaerden, T., VAel, C., Lammens, C., Chapelle, S., Rossi, R., et al. (2004). Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European Hospital isolates of Enterococcus faecium. J. Clin. Microbiol. 42, 4473–4479. doi: 10.1128/JCM.42.10.4473-4479.2004
Varun, G., Dinesh, K., Rajendra, K., Purva, M., and Sarman, S. (2016). Community acquired enterococcal urinary tract infections and antibiotic resistance profile in North India. J. Lab. Phys. 8, 50–54. doi: 10.4103/0974-2727.176237
Willems, R. J., Top, J., van Schaik, W., Leavis, H., Bonten, M., Sirén, J., et al. (2012). Restricted gene flow among hospital subpopulations of Enterococcus faecium. MBio 3, 1–10. doi: 10.1128/mBio.00151-12
Winn, C. W., Allen, S. D., Janda, W. M., Koneman, E. W., Procop, G. W., Schreckenberger, P. C., et al. (2006). “The gram positive cocci. Part II: Streptococci, Enterococci, and the ‘Streptococcus-like’ bacteria,” in Koneman's Color Atlas and Textbook of Diagnostic Microbiology, 6th, Edn, ed J. B. Lippincott (New York, NY: Lippincott Williams & Wilkins), 697–733.
Keywords: Enterococcus, intestinal colonization, antibiotic resistance, virulence factors, antibiotic prophylactic therapy, selective digestive decontamination
Citation: Bello Gonzalez TdJ, Pham P, Top J, Willems RJL, van Schaik W, van Passel MWJ and Smidt H (2017) Characterization of Enterococcus Isolates Colonizing the Intestinal Tract of Intensive Care Unit Patients Receiving Selective Digestive Decontamination. Front. Microbiol. 8:1596. doi: 10.3389/fmicb.2017.01596
Received: 08 September 2016; Accepted: 07 August 2017;
Published: 28 August 2017.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Annalisa Pantosti, Istituto Superiore di Sanità, ItalyFernando Baquero, Instituto Ramón y Cajal de Investigación Sanitaria, Spain
Copyright © 2017 Bello Gonzalez, Pham, Top, Willems, van Schaik, van Passel and Smidt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hauke Smidt, aGF1a2Uuc21pZHRAd3VyLm5s
†Present Address: Phu Pham, Laboratory of Molecular Virology, Department of Infection Biology, Faculty of Medicine, University of Tsukuba, Tsukuba, Japan
 Teresita d. J. Bello Gonzalez
Teresita d. J. Bello Gonzalez Phu Pham
Phu Pham Janetta Top2
Janetta Top2 Rob J. L. Willems
Rob J. L. Willems Willem van Schaik
Willem van Schaik Mark W. J. van Passel
Mark W. J. van Passel Hauke Smidt
Hauke Smidt