- 1Departments of Biomedical Sciences and Public Health, Polytechnic University of Marche, Ancona, Italy
- 2Departments of Life and Environmental Sciences, Polytechnic University of Marche, Ancona, Italy
A commentary on
Nationwide Surveillance of Novel Oxazolidinone Resistance Gene optrA in Enterococcus Isolates in China from 2004 to 2014
by Cui, L., Wang, Y., Lv, Y., Wang, S., Song, Y., Li, Y. et al. (2016). Antimicrob. Agents Chemother. 60, 7490–7493. doi: 10.1128/AAC.01256-16
A distictive feature of the novel oxazolidinone-phenicol resistance gene optrA is a variability yielding an encoded OptrA protein—a 655 amino acid sequence—which is variable in turn. The issue of the OptrA variants was more regularly addressed in the early studies of the new resistance than in the following reports. It is thus with particular interest that we read the recent nationwide surveillance study by Cui et al. (2016), where a wide screening of Chinese enterococci for optrA gave the authors an opportunity to repropose the issue of the different variants of the optrA protein.
When optrA was first reported in China from a 1998 to 2014 collection of human and animal enterococci (incidence, 2.0 and 15.9%, respectively), the optrA-carrying plasmid from a human Enterococcus faecalis isolate (E349) was sequenced (accession no. KP399637) (Wang et al., 2015). The relevant optrA-encoded protein is regarded as the wild type, and is hereinafter referred to as OptrAE349. Soon after the discovery of optrA, over a thousand enterococci, randomly collected in 2010–2014, were screened for the gene: among the optrA-positive isolates (incidence, 2.9%), nine different variants of the OptrA sequence (one being identical to OptrAE349) were detected (Cai et al., 2015). Seventeen optrA-positive, unrelated isolates of E. faecalis from the aforementioned 1998–2014 collection disclosed optrA sequences consistent with no new OptrA variant (seven isolates had OptrAE349) (He et al., 2016). Finally—in China, yet again—while screening over two thousand enterococci collected in 2004–2014, Cui et al. (2016) detected among the optrA-positive isolates (incidence, 2.0%) three new OptrA variants. Thus, the different OptrA sequences so far described in Chinese enterococci total 12 (including OptrAE349).
Meanwhile, as soon as the optrA sequence became available, we detected in Italy the gene—first report of optrA outside China—in two clinically distinct but virtually identical Enterococcus faecium isolates from a collection of 81 blood enterococci (incidence, 2.5%) recovered in 2015 (Brenciani et al., 2016). One of the two E. faecium isolates (strain E35048) was investigated for molecular traits, and its optrA gene (accession no. KT892063) displayed 98% DNA identity to the wild type gene.
In the light of the later data about the diversity of OptrA variants detected in China, it's apparent that our variant (hereinafter referred to as OptrAE35048) is much more dissimilar from OptrAE349 than Chinese variants. Altogether, the reported Chinese variants differ from OptrAE349 for two, three, or six amino acid substitutions, whereas OptrAE35048 differs from OptrAE349 for 21 substitutions, 17 of which (i.e., except those at positions 3, 12, 176, and 393) undetected in Chinese isolates. OptrAE35048 adds thus as a more distant variant to the OptrA variants detected in Chinese enterococci. OptrAE349 and the currently available enterococcal OptrA variants are summarized in Table 1 together with a number of relevant properties (the optrA gene location and the species, year of isolation, source, sequence type and linezolid MIC of individual isolates, whenever available). In particular, the frequent location of the optrA gene on conjugative plasmids makes the OptrA-mediated linezolid resistance transferable, an obvious cause for concern in view of possible resistance spread (Wang et al., 2015; He et al., 2016).
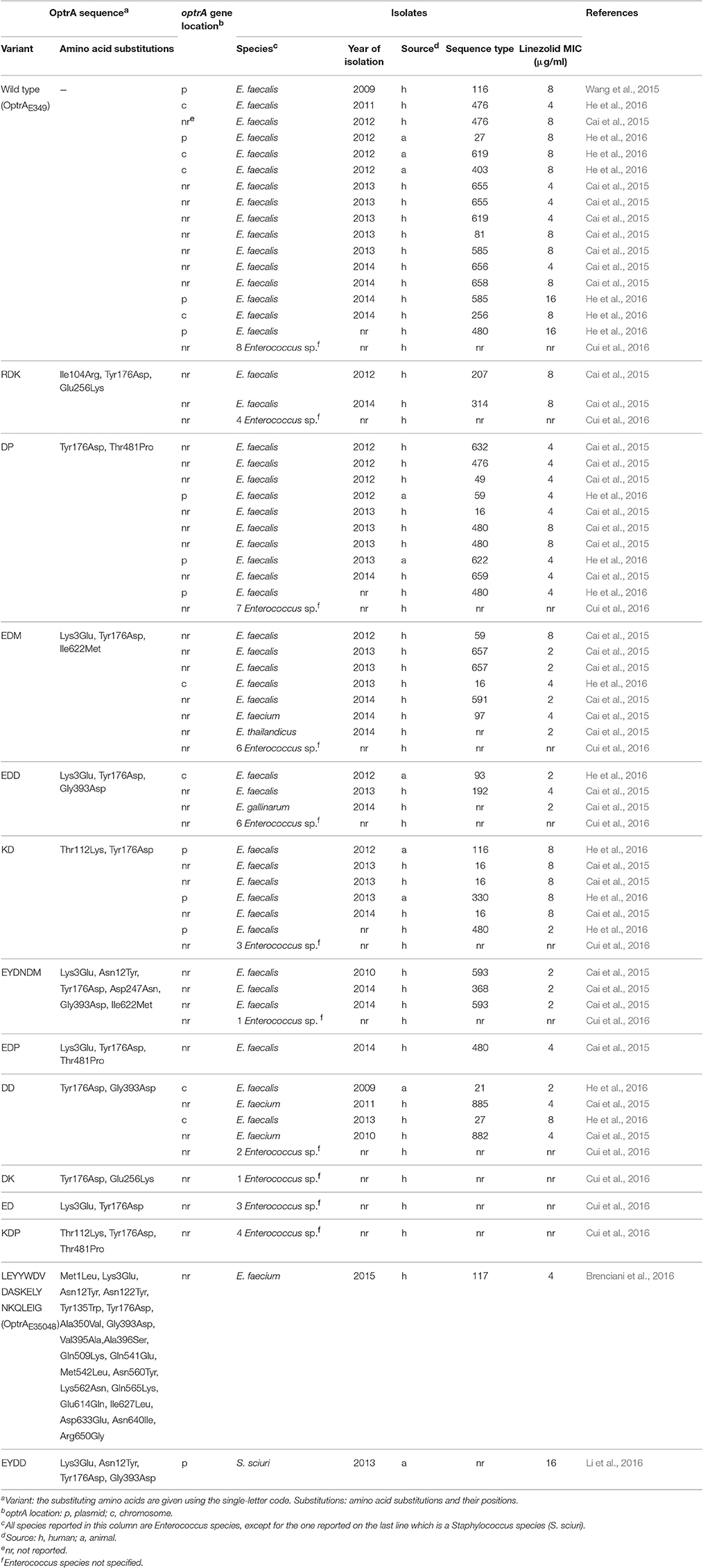
Table 1. Enterococcus isolates (plus one Staphylococcus isolate) where 14 different OptrA sequences (the E. faecalis wild type, 12 enterococcal variants, and 1 S. sciuri variant) have so far been documented.
While recently investigating three optrA-positive E. faecalis isolates of poultry origin in Colombia, Cavaco et al. (2017) deduced that two carried an optrA gene identical to one already detected in China, whereas the third isolate bore an optrA gene with a different nucleotide sequence that was defined as “more closely related” to the one we had described in Italy.
Worryingly, the optrA gene has been found in China not only in enterococci, but also in staphylococci, specifically in a Staphylococcus sciuri strain of swine origin (Li et al., 2016): optrA and its promoter region exhibited 99.1% nucleotide sequence identity to the corresponding region on the wild type E. faecalis plasmid pE349. The 655 amino acid OptrA sequence from S. sciuri is another variant exhibiting 99.4% identity to OptrAE349 (Table 1).
It's self-evident that optrA is not a conserved gene. The related variability of OptrA proteins appears to be a fitting example of that evolvability of clinical resistance by the antibiotic's effect which has been the subject of a recent reflection by Baquero et al. (2013). Given the importance of oxazolidinones as last resort antibiotics for the treatment of serious infections caused by Gram-positive pathogens, it would be important to clarify how the different amino acid substitutions affect OptrA-mediated resistance. However, irrespective of the variant, the linezolid MICs for the optrA-positive enterococci listed in Table 1 display limited variability (2–16 μg/ml), the highest MIC value in the range being shared by the optrA-positive strain of S. sciuri. Remarkably, our optrA-positive E. faecium (Brenciani et al., 2016), in spite of no less than 21 amino acid substitutions, exhibits the same linezolid MIC (4 μg/ml) as several Chinese isolates with other OptrA variants, suggesting that the number of amino acid substitutions has little influence on the level of linezolid resistance.
On the other hand, the linezolid resistance breakpoint is still an unsettled issue: indeed, an enterococcus with a linezolid MIC of 4 μg/ml is regarded as “intermediate” according to Clinical Laboratory Standards Institute (2017) and “susceptible” according to European Committee on Antimicrobial Susceptibility Testing (2017). The latter Committee, in particular, sets for enterococci a linezolid epidemiological cut-off of 4 μg/ml, and has increased the susceptible clinical breakpoint of linezolid to ≤4 μg/ml to avoid dividing wild type MIC distributions (European Committee on Antimicrobial Susceptibility Testing, 2017). In spite of the low linezolid MICs for several optrA-positive isolates, it's well established that in the clinical setting, as well as with other antibiotics, resistance levels may increase in patients with risk factors such as previous linezolid therapy, prolonged exposure to linezolid, and intensive care unit stay (Endimiani et al., 2011; Mendes et al., 2014).
In conclusion, we share and support many recent studies recommending routine surveillance of enterococci for the presence of the optrA gene. In addition, however, we wish for a more extensive interest in the OptrA variants and their correlation with oxazolidinone and phenicol MICs.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Pietro E. Varaldo for constructive discussion and critical reading of the manuscript.
References
Baquero, F., Tedim, A. P., and Coque, T. M. (2013). Antibiotic resistance shaping multi-level population biology of bacteria. Front. Microbiol. 4:15. doi: 10.3389/fmicb.2013.00015
Brenciani, A., Morroni, G., Vincenzi, C., Manso, E., Mingoia, M., Giovanetti, E., et al. (2016). Detection in Italy of two clinical Enterococcus faecium isolates carrying both the oxazolidinone and phenicol resistance gene optrA and a silent multiresistance gene cfr. J. Antimicrob. Chemother. 71, 1118–1119. doi: 10.1093/jac/dkv438
Cai, J., Wang, Y., Schwarz, S., Lv, H., Li, Y., Liao, K., et al. (2015). Enterococcal isolates carrying the novel oxazolidinone resistance gene optrA from hospitals in Zhejiang, Guangdong, and Henan, China, 2010-2014. Clin. Microbiol. Infect. 21, 1095.e1–1095.e4. doi: 10.1016/j.cmi.2015.08.007
Cavaco, L. M., Bernal, J. M., Zankari, E., Léon, M., Hendriksen, R. S., Perez-Gutierrez, E., et al. (2017). Detection of linezolid resistance due to the optrA gene in Enterococcus faecalis from poultry meat from the American continent (Colombia). J. Antimicrob. Chemother. 72, 678–683. doi: 10.1093/jac/dkw490
Clinical and Laboratory Standards Institute (2017). M100-S27. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Seventh Informational Supplement, Wayne, PA: Clinical and Laboratory Standards Institute.
Cui, L., Wang, Y., Lv, Y., Wang, S., Song, Y., Li, Y., et al. (2016). Nationwide surveillance of novel oxazolidinone resistance gene optrA in Enterococcus isolates in China from 2004 to 2014. Antimicrob. Agents Chemother. 60, 7490–7493. doi: 10.1128/AAC.01256-16
Endimiani, A., Blackford, M., Dasenbrook, E. C., Reed, M. D., Bajaksouszian, S., Hujer, A. M., et al. (2011). Emergence of linezolid-resistant Staphylococcus aureus after prolonged treatment of cystic fibrosis patients in Cleveland, Ohio. Antimicrob. Agents Chemother. 55, 1684–1692. doi: 10.1128/AAC.01308-10
European Committee on Antimicrobial Susceptibility Testing (2017). Available online at: http://www.eucast.org/
He, T., Shen, Y., Schwarz, S., Cai, J., Lv, Y., Li, J., et al. (2016). Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J. Antimicrob. Chemother. 71, 1466–1473. doi: 10.1093/jac/dkw016
Li, D., Wang, Y., Schwarz, S., Cai, J., Fan, R., Li, J., et al. (2016). Co-location of the oxazolidinone resistance genes optrA and cfr on a multiresistance plasmid from Staphylococcus sciuri. J. Antimicrob. Chemother. 71, 1474–1478. doi: 10.1093/jac/dkw040
Mendes, R. E., Deshpande, L. M., and Jones, R. N. (2014). Linezolid update: stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist. Updat. 17, 1–12. doi: 10.1016/j.drup.2014.04.002
Wang, Y., Lv, Y., Cai, J., Schwarz, S., Cui, L., Hu, Z., et al. (2015). A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 70, 2182–2190. doi: 10.1093/jac/dkv116
Keywords: optrA gene, OptrA protein, OptrA variants, enterococci, oxazolidinone resistance
Citation: Morroni G, Brenciani A, Simoni S, Vignaroli C, Mingoia M and Giovanetti E (2017) Commentary: Nationwide Surveillance of Novel Oxazolidinone Resistance Gene optrA in Enterococcus Isolates in China from 2004 to 2014. Front. Microbiol. 8:1631. doi: 10.3389/fmicb.2017.01631
Received: 07 June 2017; Accepted: 11 August 2017;
Published: 24 August 2017.
Edited by:
Teresa M. Coque, Instituto Ramón y Cajal de Investigación Sanitaria, SpainReviewed by:
Ana R. Freitas, University of Porto, PortugalCopyright © 2017 Morroni, Brenciani, Simoni, Vignaroli, Mingoia and Giovanetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Brenciani, YS5icmVuY2lhbmlAdW5pdnBtLml0
 Gianluca Morroni1
Gianluca Morroni1 Andrea Brenciani
Andrea Brenciani Carla Vignaroli
Carla Vignaroli Marina Mingoia
Marina Mingoia Eleonora Giovanetti
Eleonora Giovanetti