- 1State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai, China
- 2Department of Applied Biology, East China University of Science and Technology, Shanghai, China
The yigP locus is widely conserved among γ-proteobacteria. Mutation of the yigP locus impacts aerobic growth of Gram-negative bacteria. However, the underlying mechanism of how the yigP locus influences aerobic growth remains largely unknown. Here, we demonstrated that the yigP locus in Escherichia coli encodes two transcripts; the mRNA of ubiquinone biosynthesis protein, UbiJ, and the 3′ untranslated region small regulatory RNA (sRNA), EsrE. EsrE is an independent transcript that is transcribed using an internal promoter of the yigP locus. Surprisingly, we found that both the EsrE sRNA and UbiJ protein were required for Q8 biosynthesis, and were sufficient to rescue the growth defect ascribed to deletion of the yigP locus. Moreover, our data showed that EsrE targeted multiple mRNAs involved in several cellular processes including murein biosynthesis and the tricarboxylic acid cycle. Among these targets, sdhD mRNA that encodes one subunit of succinate dehydrogenase (SDH), was significantly activated. Our findings provided an insight into the important function of EsrE in bacterial adaptation to various environments, as well as coordinating different aspects of bacterial physiology.
Introduction
Gene regulation, which occurs at multiple levels including transcriptional and post-translational control, is a vital mechanism across all domains of life (Bobrovskyy and Vanderpool, 2013). One emerging class of post-transcriptional regulators in bacteria is small regulatory RNAs (sRNAs; Bobrovskyy and Vanderpool, 2013). Previous studies suggest that sRNAs are typically short transcripts that vary from 50 to 500 nucleotides and rarely contain an open reading frame (ORF; Bobrovskyy and Vanderpool, 2013). sRNAs act by perfect or limited base-pairing to target mRNAs to influence their stability or/and translation (Updegrove et al., 2015). In γ-proteobacteria, most sRNAs require the RNA chaperone, Hfq, to stabilize themselves and promote sRNA–mRNA intermolecular interaction (Chao and Vogel, 2010; Vogel and Luisi, 2011). Such interaction principally inhibits the translation and/or induces the degradation of their target mRNAs in a ribonuclease E (RNase E)-dependent manner (Masse and Gottesman, 2002; Masse et al., 2003; Morita et al., 2005; Frohlich and Vogel, 2009). However, compared to their negative effect on gene expression, the positive regulation by sRNAs remains largely unknown (Papenfort and Vanderpool, 2015). Besides the most characterized intergenic sRNAs, recent RNA-seq studies suggest bacterial sRNA-encoding genes are also located in coding regions (Lalaouna et al., 2015; Miyakoshi et al., 2015a). The 3′ untranslated region (UTR) sRNAs belong to a newly characterized non-intergenic class. They are produced by either mRNA processing or transcription from independent promoters within genes. In both cases, the 3′ UTR sRNAs share the same sequences with the 3′ regions of the coding genes (Miyakoshi et al., 2015b). Therefore, these 3′ UTR sRNAs pose a challenge for ongoing gene or locus function characterization.
All 11 ubi genes (ubiA–J and ubiX) are required for the biosynthesis of Q8 in Escherichia coli. Most of their products catalyze specific reactions in the biosynthetic process, starting from chorismate (Van Beilen and Hellingwerf, 2016). The yigP (renamed ubiJ) locus is highly conserved among almost 80 organisms, many of which are γ-proteobacteria. In E. coli, yigP is located between ubiE and ubiB loci, and may be synchronously transcribed with them under an upstream promoter of ubiE (Poon et al., 2000; Aussel et al., 2014a). UbiE is well-known for the C methylation reactions in both Q8 and menaquinone (MK8) biosynthesis (Aussel et al., 2014a). A mammalian homolog of UbiB, COQ8A (ADCK3), was recently found to display ATPase activity and interacted with lipid CoQ intermediates (Stefely et al., 2016). However, though yigP mutant strains of Salmonella and E. coli showed significant defects in Q8 biosynthesis (Aussel et al., 2014a), the underlying mechanism remains unrevealed.
Our previous studies found that the 3′ region of yigP transcribed an sRNA named EsrE with indispensable function in E. coli (Chen et al., 2012). A recent study in Salmonella found that the yigP locus encoded a protein named UbiJ required for Q8 biosynthesis. Based on evidence that a “scrambled mutation” of the 3′ region of Salmonella ubiJ had no phenotype, it was concluded that the aerobic growth-promoting function of the yigP locus was mediated by UbiJ but not the sRNA, EsrE (Aussel et al., 2014a). In this work, we provide new evidence that the yigP locus contains two genes, ubiJ and esrE. UbiJ encoded a protein, UbiJ, while esrE transcribed an sRNA, EsrE. Furthermore, our results showed that besides UbiJ, the sRNA EsrE was also critical for the aerobic growth of E. coli and required for Q8 biosynthesis. In addition, we found that EsrE might maintain cellular processes of E. coli by activating multiple mRNAs.
Materials and Methods
Bacterial Strains and Growth Conditions
The wild-type is E. coli K-12 strain JM83. Unless otherwise stated, the cells were grown at 37°C in liquid or on solid Luria-Bertani (LB) media supplemented with chloramphenicol (30 μg/mL) or ampicillin (100 μg/mL) as indicated. All strains and plasmids used here are listed in Supplementary Table S1.
Inactivation of the yigP Locus
The λ Red-mediated recombination method was used to delete different regions of the yigP locus with an insert of apramycin resistance cassette. The mutant fragments were transferred to E. coli JM83 by electroporation.
Construction of a 3 × Flag-yigP-Fused E. coli Strain
To carry out an E. coli strain that expresses a fused YigP protein with a C-terminal [Gly4Ser]3 linker plus a triple-Flag tag (DYKDHDGDYKDHDIDYKDDDDK), a pMAK705-derived temperature-sensitive plasmid, pMAK-yigP-flag, was created using an approach described previously (Bush et al., 2013). Using the pMAK-yigP-flag according to previous methods (Chen et al., 2012), the chromosomal yigP insertion strain, WT::flag, was constructed.
Quinone Extraction and Analysis
The extraction and analysis of quinones was performed as described by Ozeir et al. (2015) and Van Beilen and Hellingwerf (2016). The cells were cultured for 10 h before collection by centrifugation, washed twice with distilled water, and their wet weights determined. A total of 150 μL of 0.15 M KCl (a Q10 solution used as an internal standard, 100 μg/mL in ethyl alcohol, 0.863 μL/mg of wet weight) and 0.6 mL of methanol were added to cell pellets, and the tubes vortexed for 15 min. Then 0.4 mL of petroleum ether (boiling range, 40–60°C) was added and the tubes vortexed for 5 min. The phases were separated by centrifugation at 5000 rpm at room temperature for 5 min. The upper petroleum ether layer was transferred to a fresh tube. Petroleum ether (0.4 mL) was added to the methanol-containing tube, and the extraction was repeated once more. The petroleum ether layers were combined and dried. The lipids were resuspended in 100 μL of ethyl alcohol.
The samples were fractionated with high-performance liquid chromatography (HPLC) using a reversed-phase C18 column (Betabasic-18, 5 μm, 4.6 mm × 250 mm, Agilent Technologies). The column was equilibrated with pure ethyl alcohol as the mobile phase at a flow rate of 1 mL/min. Detection of quinones was performed using an UV/Vis absorption detector at 275 nm for ubiquinone (UQ).
Real-time qRT-PCR
Isolation of bacterial total RNA was performed with High Pure RNA Isolation Kit (Roche Diagnostics, Germany) according to the manufacturer’s instructions. Total RNA was the template for qRT-PCR reaction using Reverse Transcription M-MLV (RNase H-) kit (TaKaRa, Japan) and SYBR Green PCR Master Mix (TOYOBO, Japan). The primers for qRT-PCR are listed in Supplementary Table S2. The levels of gene expressions were normalized using the rpoD transcript data.
SDS-PAGE and Western Blot Analysis
To investigate whether the yigP locus encodes a protein, SDS-PAGE and western blotting were performed. All the strains were grown to an indicated time in LB media, and the cells were washed with phosphate-buffered saline. The samples were sonicated and total protein quantified following standard protocols. The protein was denatured at 100°C for 5 min. Total protein from each sample was loaded for SDS-PAGE, and western blotting was performed with Flag-tag primary and horseradish-peroxidase-conjugated goat anti-mouse secondary (AOGMA, United States) antibodies.
β-Galactosidase Assays
Cells were cultured overnight and then subcultured at 1:100 in fresh LB. After growing for 5 h to an optical density (OD)600 of 2, equivalent cell densities were collected, pelleted, and permeabilized by ultrasonication. Ortho-nitrophenyl-β-D-galactopyranoside hydrolysis was determined in triplicate to measure β-galactosidase activity as described previously (Wang et al., 2012).
Results
The yigP Locus Encodes Two Products: EsrE and UbiJ
The ubiE, yigP, and ubiB loci in E. coli are considered an operon (Poon et al., 2000) essential for Q8 biosynthesis (Aussel et al., 2014b). However, how yigP contributes to this process is still unclear. In our previous study, we found that the 3′ region of yigP encoded a small RNA called EsrE that might have essential functions in E. coli (Chen et al., 2012). Recently, the yigP locus was reported to encode a protein called UbiJ that is required for Q8 biosynthesis and aerobic growth in Salmonella and E. coli (Aussel et al., 2014a). To further determine whether yigP locus has two transcripts, we designed two pairs of primers, yigP-S1/2 and yigP-X1/2 targeting its 5′ and 3′ regions, respectively, for transcript analysis (Figure 1A). Because ubiE, yigP, and ubiB loci are considered to form an operon (Poon et al., 2000), ubiE and ubiB mRNAs were also examined (Figure 1B). As shown by qRT-PCR, ubiJ mRNA was expressed throughout the lifetime of the bacteria (Figure 1B). In contrast, esrE expression was only detected during the late exponential growth phase (approximately 5 h) as indicated by the finding that the transcript from the 3′ region was more abundant than the transcript from the 5′ region at 5 h (Figure 1B). These observations suggested that UbiJ was the constitutively expressed product of the yigP locus, while EsrE was transiently generated during the transition to the stationary phase.
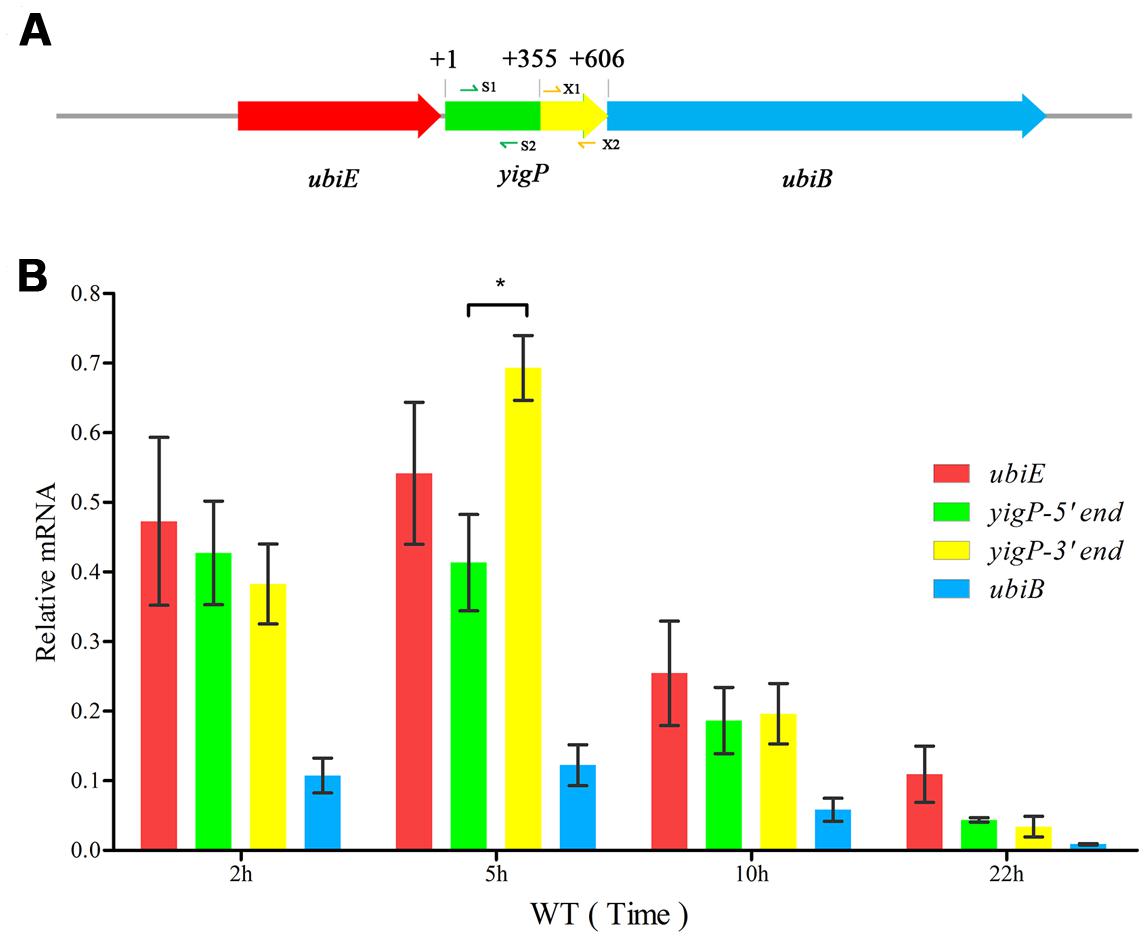
FIGURE 1. The yigP locus transcribes two products. (A) Schematic representationof the ubiE, yigP, and ubiB loci in Escherichia coli. S1/2 and X1/2 represent primer pairs used to analyze yigP expression (yigP-S1/2 and yigP-X1/2). (B) qRT-PCR analysis of the yigP locus transcripts in wild type strain at different time points. Bacteria were grown in triplicate in Luria-Bertani (LB) liquid culture, and equivalent cell densities were collected. The reactions were performed in duplicate, and the relative expression changes were calculated using the 2ΔΔCT method with the constitutively expressed rpoD gene serving as the endogenous control. ∗P < 0.05.
In order to determine the contribution of the two transcripts to aerobic growth, we constructed three deletion mutants containing different regions of the E. coli yigP locus (Figure 2A). All mutants were successfully obtained. As shown in Supplementary Figure S1, the colonies of the full region (ΔyigP) and 3′ region (ΔyigP-3′end) mutant strains that grew on LB plates under aerobic conditions were much smaller than the colonies of the 5′ region mutant strain (ΔyigP-5′end) (Supplementary Figure S1). These mutant strains and the wild type strain were then cultured in LB under aerobic conditions. Consistent with our previous results, ΔyigP and ΔyigP-3′end strains exhibited significant growth defects compared to the wild type strain. In contrast, ΔyigP-5′end strain grew just as well as the wild type strain (Figure 2B). Because the yigP locus was truncated in ΔyigP-5′end strain to prevent UbiJ translation in vivo, our results suggest EsrE was the product of the yigP locus that maintained the aerobic growth of E. coli.
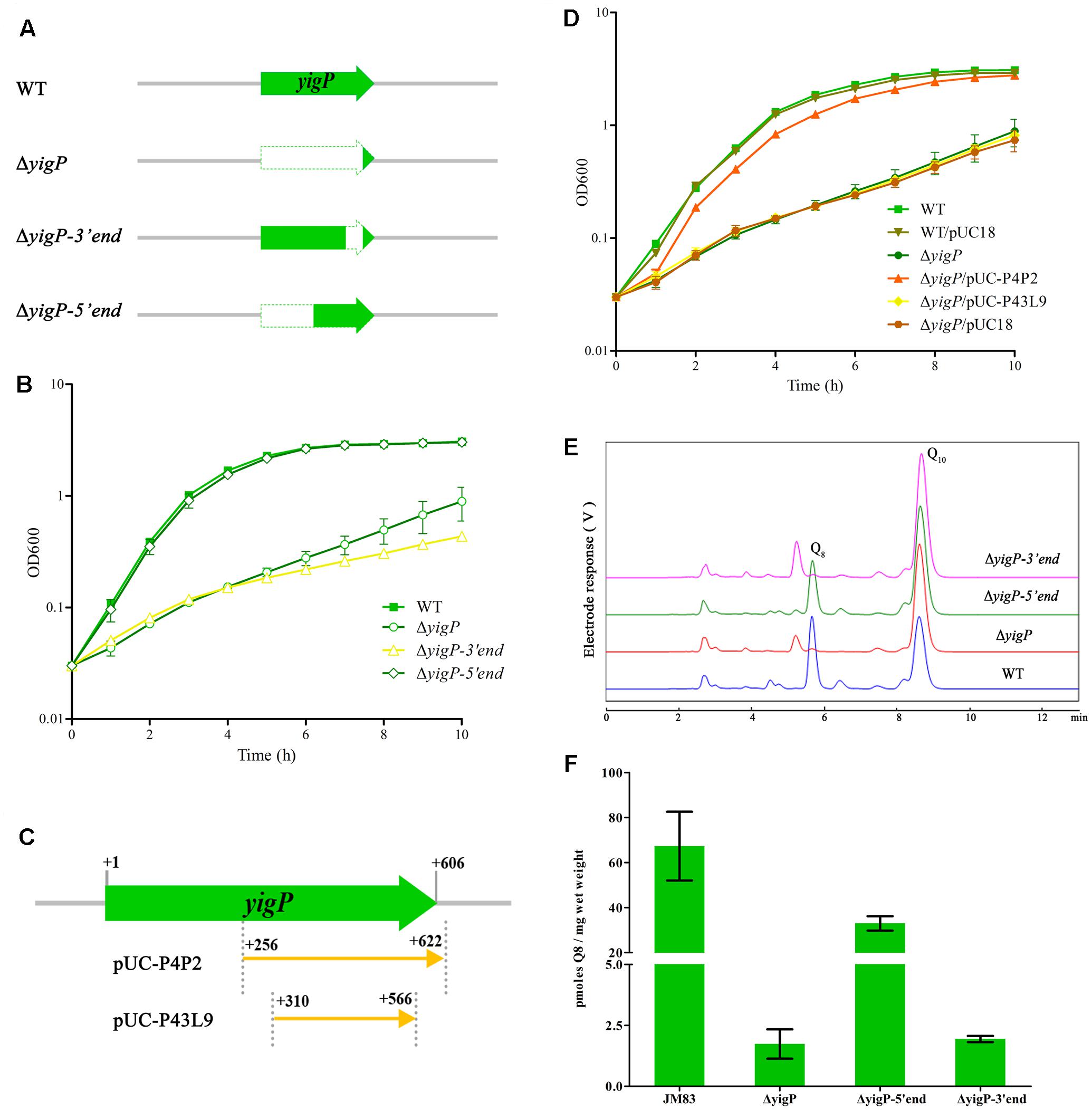
FIGURE 2. Functional regions of the yigP locus. (A) Schematic representation of the different yigP locus mutants used in this study. ΔyigP represents the 572 bp yigP deletion mutant, ΔyigP-3′end represents the mutant in which 153 bp of the 3′ end of the yigP locus was deleted, and ΔyigP-5′end represents the mutant in which 283 bp of the 5′ end of the yigP locus was deleted. (B) The results of a representative experiment are shown. Wild type (filled squares), ΔyigP (open circles), ΔyigP-3′end (open triangles), and ΔyigP-5′end (open rhombuses) strains were grown overnight and then diluted to an OD600 of 0.03 in LB medium at 37°C. Growth was monitored at 600 nm. (C) Schematic representation of the yigP plasmids used in this work. pUC-P4P2 and P43L9 represent the plasmids carrying the 367 and 267 nucleotides, respectively, located at the 3′ end of yigP. (D) Wild type (filled squares), wild type transformed with pUC18 (filled inverted triangles), ΔyigP (filled circles) and ΔyigP strains transformed with pUC-P4P2 (filled triangles), P43L9 (filled rhombuses), and pUC18 (filled hexagons) were grown overnight and then diluted to an OD600 of 0.03 in LB. Growth was monitored at 600 nm. The experiment was performed at least three times, and identical patterns were obtained. (E) High-performance liquid chromatography (HPLC) separation of the strains described for panel (A) with the eluate analyzed with A275. The identified quinones, Q8 and Q10, are indicated. (F) Quantification of cellular Q8 content (n = 3) of the same cells as in panel (A) in picomoles per milligram of wet weight. Error bars represent standard deviation.
To further determine the functional region of the yigP locus, we transformed two plasmids containing different truncated fragments (Chen et al., 2012) in proximity to the esrE-coding region into the ΔyigP strain (Figure 2C). The plasmid carrying the 367 bp whole esrE-coding sequence fragment partially (P4P2) restored the growth defect of ΔyigP strain, whereas the 267 bp partial esrE-coding sequence fragment (P43L9) failed to do so (Figure 2D). Because the PlacZ promoter located in pUC18 had been removed, the experiment also revealed that the 3′ region fragment (P4P2) had its own promoter. Hence, the EsrE transcript from an independent promoter located within the yigP locus contributed to the growth of E. coli.
In addition, the yigP locus was previously shown to be essential for Q8 biosynthesis, and the amount of Q8 was tightly related to the aerobic growth of E. coli (Aussel et al., 2014a). We therefore measured the cellular Q8 content of the three mutant (Figure 2A) and the wild type strains by HPLC analysis (Figure 2E) (Ozeir et al., 2015; Van Beilen and Hellingwerf, 2016). Unlike the growth phenotype (Figure 2B), the Q8 content in ΔyigP-5′end strain was 50% of the wild type strain (33 and 67 pmol, respectively) (Figure 2F). Reasoning that ΔyigP-5′end strain had no UbiJ protein expression in vivo, EsrE was responsible for the half amount of Q8, which might be enough to maintain normal aerobic growth of E. coli (Figure 2B; Aussel et al., 2014a,b). Meanwhile, the ΔyigP and ΔyigP-3′end strains had tiny amounts of Q8 (Figure 2F), which was in agreement with previous evidence that the yigP locus was required for the synthesis of Q8. Taken together, these results indicated that both EsrE and UbiJ from the yigP locus were required for Q8 production in E. coli.
EsrE Is Required for Aerobic Growth of E. coli as an sRNA
We investigated whether the 3′ region of the yigP locus encodes a polypeptide or only EsrE as sRNA. A 3 × Flag plus Linker coding sequence (111 bp) was chromosomally fused via homologous recombination to the 3′ end of the yigP locus in wild type strain using the temperature-sensitive plasmid pMAK-yigP-flag, yielding the WT::flag strain (Figure 3A). The bacterial cells were harvested from four growth periods, lysed, and the expression levels of the Flag-fused proteins examined by immunoblotting. As expected, a 26 kDa band of UbiJ::Linker-3 × Flag was detected (Figure 3B), which coincided with the expression pattern of its mRNA (Figure 1B). This finding verified that the yigP locus translated the 23 kDa full-length product UbiJ.
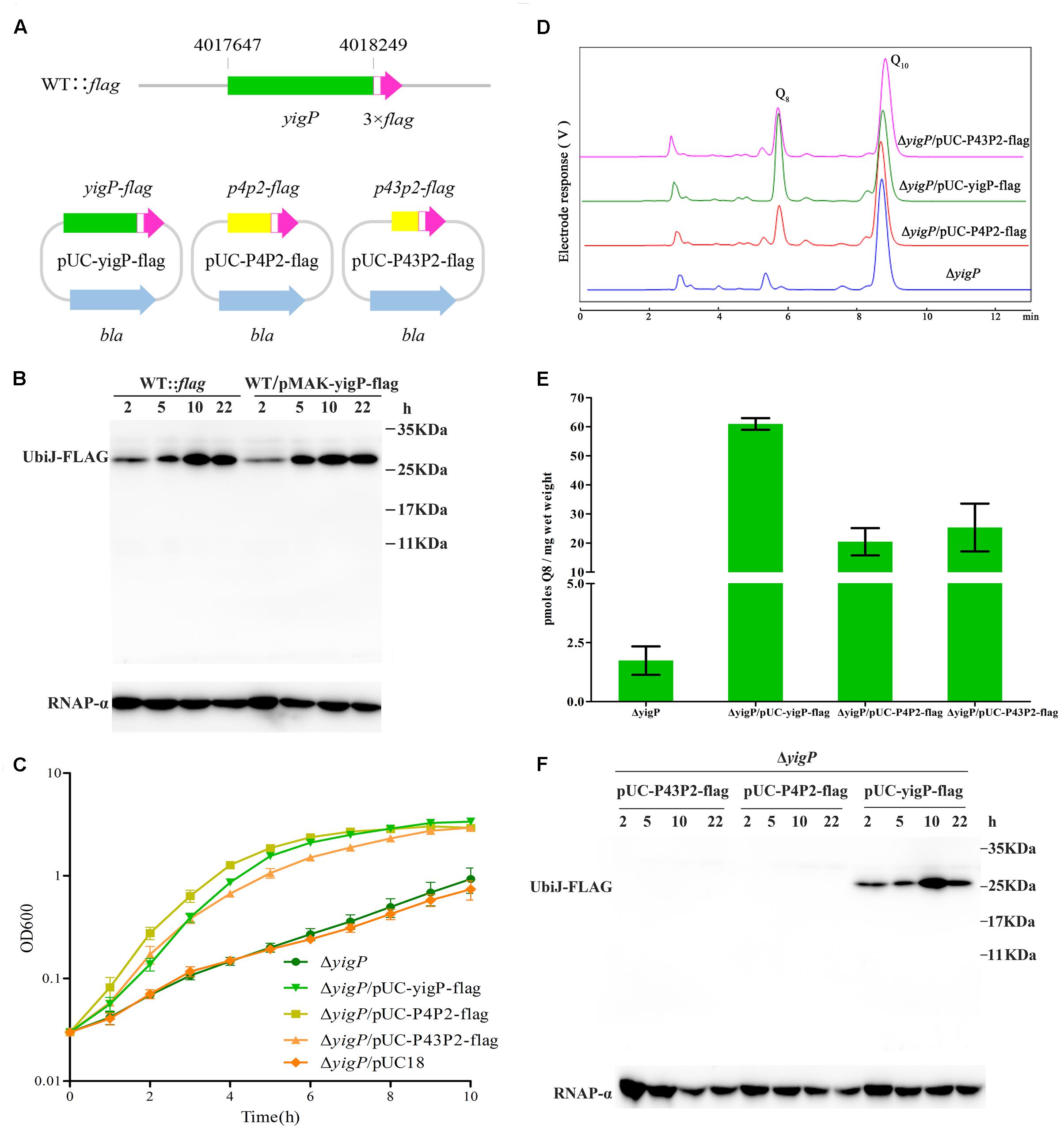
FIGURE 3. The yigP locus encodes EsrE and a 23-kDa protein, UbiJ, both of which have important effects on the growth of E. coli. (A) Schematic representation of mutant strain, which Linker plus 3 × Flag tag sequences were introduced before the stop codon of the yigP locus in wild type bacteria, named WT::flag. In addition, Linker plus 3 × Flag tag sequences were introduced before the stop codon of different yigP fragments in plasmid pUC18 to yield pUC-yigP-flag, pUC-P4P2-flag, and pUC-P43P2-flag. YigP means the full-length fragment (including 241 bp sequences upstream yigP locus, considered as the promoter region of ubiJ), P4P2 (367 bp) indicates larger region of esrE-coding sequence than P43P2, while P43P2 (313 bp) contains the minimal region of esrE-coding sequence. (B) Western blot analysis of WT::flag and wild type bacteria with pMAK-yigP-flag. The yigP variants were separated by 15% SDS-PAGE, blotted onto PVDF membranes and hybridized with a Flag tag antibody or RNAP-α antibody. RNAP-α means RNA polymerase α subunit protein used as a loading control. (C) ΔyigP (filled circles), ΔyigP strains transformed with pUC-yigP-flag (filled inverted triangles), pUC-P4P2-flag (filled squares), pUC-P43P2-flag (filled triangles), and pUC18 (filled rhombuses) were grown overnight and then diluted to an OD600 of 0.03 in LB under aerobic conditions. Growth was monitored at 600 nm. The experiment was performed at least three times, and identical patterns were obtained. (D) HPLC separation of the strains described for panel (C), except ΔyigP/pUC18 with the eluate analyzed with A275. The identified quinones, Q8 and Q10, are indicated. (E) Quantification of cellular Q8 content (n = 3) of the same cells as in panel (C), except ΔyigP/pUC18 in picomoles per milligram of wet weight. Error bars represent standard deviation. (F) Western blot analysis of pUC-yigP-flag, pUC-P4P2-flag, and pUC-P43P2-flag in ΔyigP strain. The yigP variants were separated by 15% SDS-PAGE, blotted onto PVDF membranes and hybridized with a Flag tag antibody or RNAP-α antibody. RNAP-α means RNA polymerase α subunit protein used as a loading control.
Meanwhile, the full-length fragment (including 241 bp sequences upstream of the yigP locus, considered as the promoter region of ubiJ) and 3′ region P4P2 (367 bp) or P43P2 (313 bp) fragment-fused 3 × Flag plus Linker coding sequence were inserted into pUC18 plasmid without foreign promoter to yield pUC-yigP-flag and pUC-P4P2-flag or pUC-P43P2-flag. These plasmids were then transformed into ΔyigP strain. All plasmids rescued the impaired aerobic growth of ΔyigP strain (Figure 3C). Moreover, by measuring Q8 production of the three strains (Figure 3D), we found that the ΔyigP strain with pUC-yigP-flag could recover Q8 amounts to the level of the wild type strain (Figure 3E). However, the Q8 content in the other two strains only accounted for half of that in the wild type strain (Figure 3E), and pUC18 did not affect Q8 production in ΔyigP strain (Supplementary Figure S2A). This result indicated that both EsrE and UbiJ were important for aerobic growth and Q8 biosynthesis. Similarly, only pUC-yigP-flag encoded a 26 kDa protein, while both pUC-P4P2-flag and pUC-P43P2-flag did not present any band by immunoblotting (Figure 3F). Meanwhile, all three plasmids transcribed esrE as determined by qRT-PCR (Supplementary Figure S2B). These results suggested that the function of the 3′ region fragment relied on an sRNA instead of a polypeptide. Collectively, our results showed that there were two products encoded by different regions of the yigP locus: one was the RNA, EsrE, and the other was the 23 kDa protein, UbiJ.
To further examine the role of EsrE, we employed four different P43P2 frameshift mutant fragments (Figure 4A). The ΔyigP strain was transformed with four plasmids, named pT-002, pT-003, pT-004, and pT-005 (Chen et al., 2012) that contained these mutant fragments. All plasmids significantly complemented the impaired aerobic growth of ΔyigP strain when cultured in LB (Figure 4B). It is worth mentioning that the ΔyigP strain with pT-005 was much smaller than the others when grown on LB plates for up to 24 h, but then reached similar size after 40 h of culture (Figure 4C). This indicated that the deletion of site 005 (adenine) might have partly affected the rescue capability of the P43P2 fragment. Meanwhile, we also checked the cellular Q8 content of the four strains. All four strains harbored enough amounts of Q8 (Figure 4D) to recover the growth defect. Overall, because the four mutation sites could affect all potential ORFs, their rescue capabilities provided strong evidence that all the potential ORFs were immune to frameshift mutations, even though these mutant fragments could not recover the growth defect to the same level as the P43P2 fragment (Supplementary Figure S3). Taken together, these data showed that the esrE gene located in the 3′ region of the yigP locus encoded a 3′ UTR sRNA required for E. coli aerobic growth and biosynthesis of Q8.
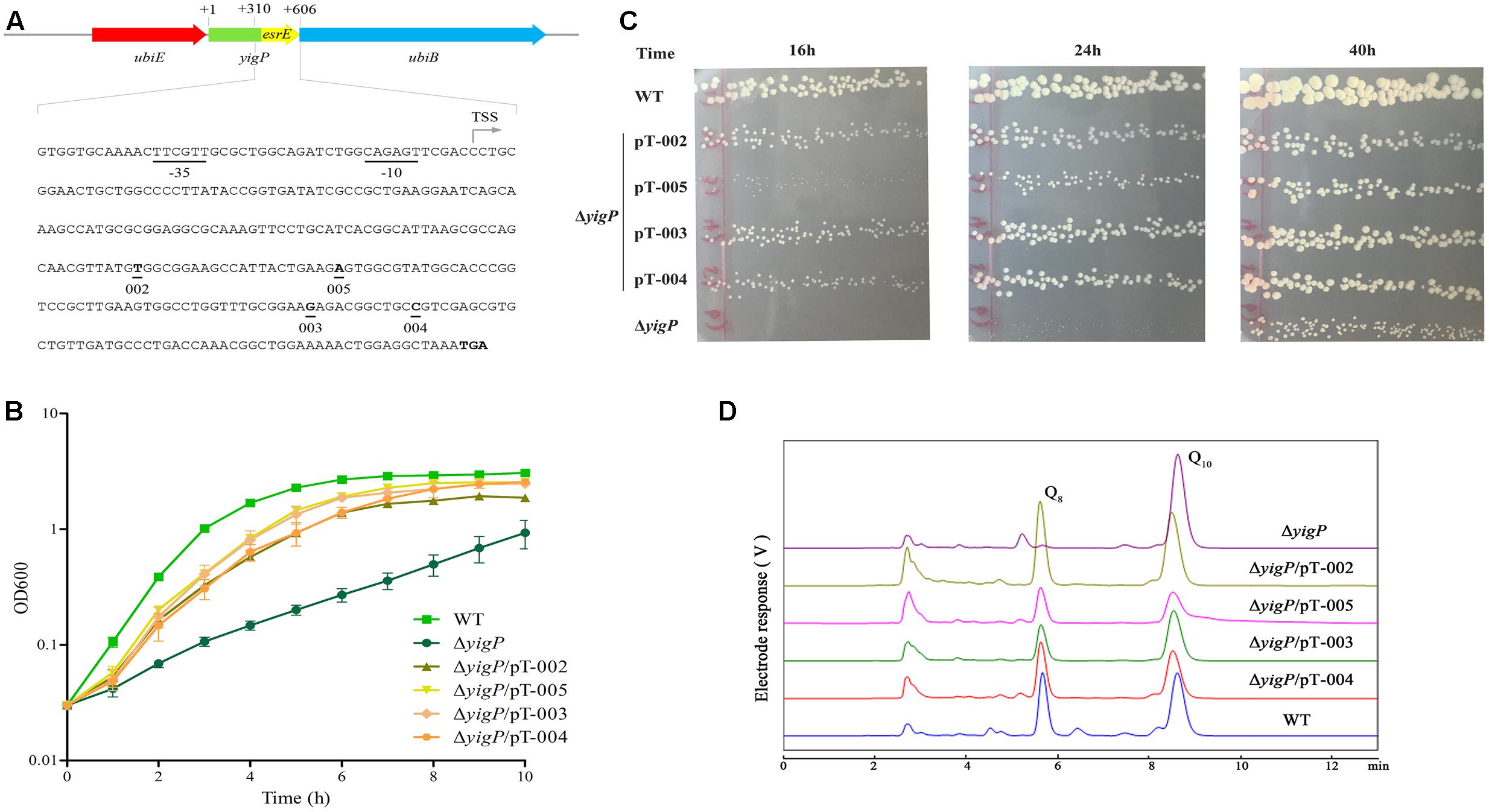
FIGURE 4. EsrE is a 3′ UTR sRNA. (A) Schematic representation and sequence analysis of EsrE. The bent arrow marks the transcription start site (TSS), and the four frame-shift mutation sites in this study are indicated. (B) Wild type (filled squares), ΔyigP (filled circles), and ΔyigP strains transformed with pT-002 (filled triangles), pT-003 (filled rhombuses), pT-004 (filled hexagons), and pT-005 (filled inverted triangles) were grown overnight and then diluted to an OD600 of 0.03 in LB. Growth was monitored at 600 nm. The experiment was performed at least three times, and identical patterns were obtained. (C) Wild type, ΔyigP, and ΔyigP strains transformed with pT-002, pT-003, pT-004, and pT-005 were grown on LB plates for 16, 24, and 40 h at 37°C. (D) HPLC separation of the strains described for panel (B) with the eluate analyzed with A275. The identified quinones, Q8 and Q10, are indicated.
EsrE Targets Multiple mRNAs In Vivo
To uncover the physiological role of EsrE, we tried to search its regulons. Because most sRNAs target mRNAs to regulate their expression levels via perfect or limited base-pairing, the potential regulons of EsrE were screened with two comparative prediction algorithms for sRNA targets, CopraRNA (Wright et al., 2014) and TargetRNA (Kery et al., 2014). Twenty-four candidates were chosen for validation in vivo (Supplementary Table S3).
To further verify these candidates, translational fusions were generated by integrating the 5′ UTRs in-frame with the first codon of gfp or lacZ (the fluorescence of some GFP fusions are not high enough for detection) and downstream of the heterologous PLtetO promoter (Supplementary Table S3) (Urban and Vogel, 2007). Wild type cells were then transformed with these fusion plasmids. GFP fluorescence or β-galactosidase activity was monitored in the presence of EsrE overexpression or control plasmids. Among all the candidates, four showed significant differences from their controls. Three genes, including murE (UDP-N-acetylmuramoylalanyl-D-glutamate-2,6-diaminopimelate ligase), murF (D-alanyl-D-alanine-adding enzyme), and sdhD [succinate dehydrogenase (SDH), membrane protein SdhD] were activated, whereas another gene, argI (ornithine carbamoyltransferase chain I) was repressed by EsrE, as shown by the activated or repressed GFP fluorescence or β-galactosidase activities (Figures 5A,B) (Keseler et al., 2013). It has been reported that MurE and MurF were essential for cell growth by catalyzing the final two reactions of the biosynthesis of UDP-N-acetylmuramoyl-pentapeptide, the monomeric unit of peptidoglycan (Keseler et al., 2013). The strongest signal difference was detected between the strains that expressed the 5′ UTR of sdhD. The β-galactosidase activity in EsrE overexpressing strain was almost threefold that of its control. The sdhD gene encodes the membrane subunit of SDH, the primary dehydrogenase involved in aerobic electron transfer in E. coli (Keseler et al., 2013). Collectively, these findings implied that EsrE targeted multiple regulons to perform its function in E. coli.
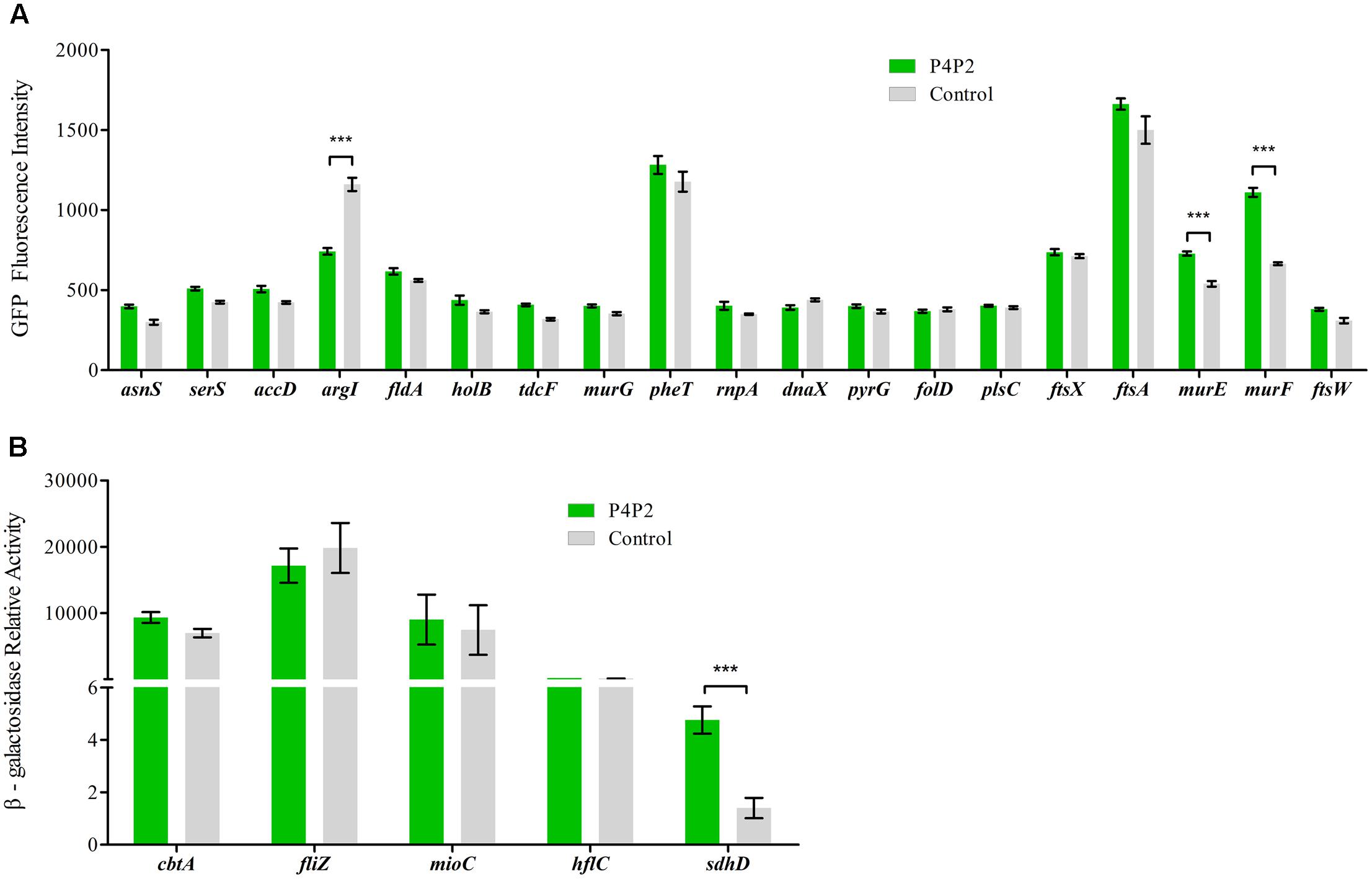
FIGURE 5. Experimental validation of EsrE target predictions. (A) Fluorescence of wild type bacteria carrying the indicated gfp fusions and EsrE plasmid (pT-P4P2) or control plasmid (pT-NS). The bacteria were grown in triplicate in LB liquid culture and measured at the indicated growth phase (at 5 h, about OD600 of 2). The fluorescence values were given in arbitrary units and corrected for basal fluorescence of a wild type strain harboring plasmid pXG-0. (B) Relative activity of β-galactosidase in wild type bacteria carrying the indicated lacZ fusions and EsrE (pT-P4P2) or control (pT-NS) plasmid. The bacteria were grown in triplicate in LB liquid culture and were collected at the indicated growth phase (at 5 h, about OD600 of 2). β-Galactosidase assays were then performed according to a protocol described previously (Wang et al., 2012). ∗∗∗P < 0.001.
EsrE Is Required for Succinate Dehydrogenase Activity
SdhD is a membrane protein in the four-subunit enzyme complex of SDH (Keseler et al., 2013). SDH is an essential component of the tricarboxylic acid (TCA) cycle. It catalyzes the oxidation of succinate to fumarate and donates electrons to the electron transport chain (ETC; Cecchini, 2003). As described above, the expression level of sdhD was upregulated by EsrE. To further verify the relationship between EsrE and SDH, SDH activity was measured in wild type and ΔyigP strains at four different growth times. In the wild type strain, SDH activity doubled at 7.5 h, reached its peak at 10 h, and then plateaued (Figure 6A). In contrast, SDH activity of ΔyigP strain increased slowly during growth and reached its peak at 22 h (Figure 6A). The deletion of EsrE caused 40–50% decrease in SDH activity at 5, 7.5, and 10 h. The decrease could be rescued by EsrE in trans expressed by pUC-P4P2-flag (Figure 6A). The rapid increase of SDH activities from the later exponential phase to the stationary growth phase in wild type strain may be due to increasing amounts of EsrE, because its expression level boosted during the transition to the stationary phase (Figure 1B). The results indicated EsrE improved the activity of the SDH complex from the later exponential phase.
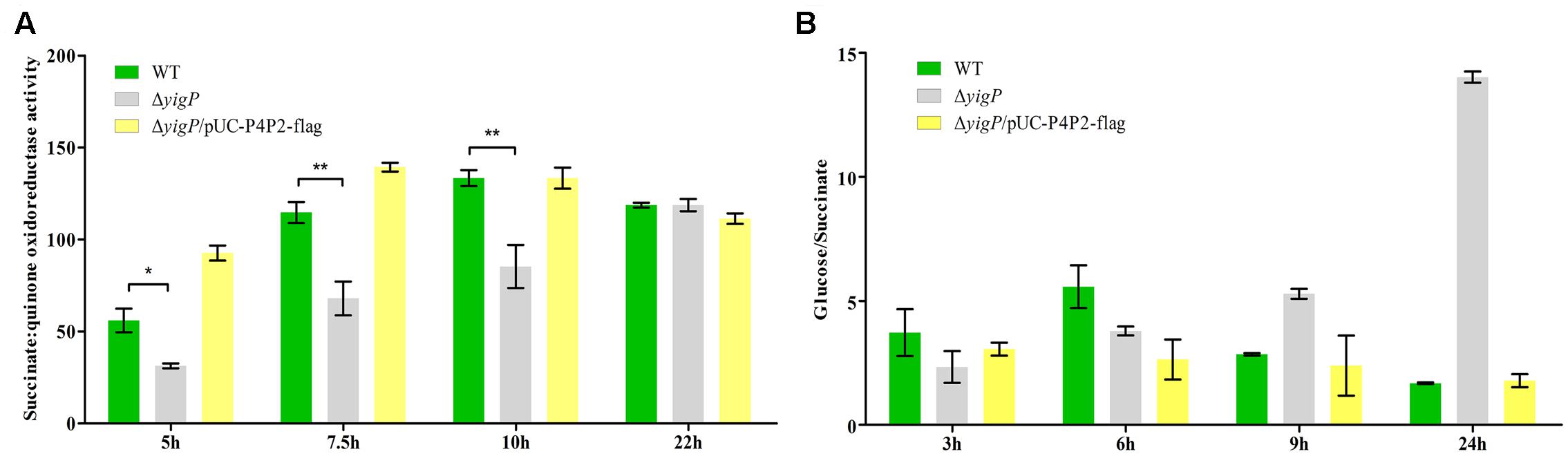
FIGURE 6. EsrE is required for succinate dehydrogenase activity. (A) Succinate dehydrogenase (SDH) activity assays were carried out on wild type, ΔyigP, and ΔyigP strains transformed with pUC-P4P2-flag at four different growth phases (5, 7.5, 10, and 22 h). Equivalent cell densities were collected, pelleted, and permeabilized by ultrasonication. The measurement of SDH activity was performed in triplicate using the Succinate Dehydrogenase assay kit (Nanjing Jiancheng Bioengineering Institute, China). The experiment was performed at least three times, and identical patterns were obtained. (B) Wild type, ΔyigP, and ΔyigP strains transformed with pUC-P4P2-flag were grown overnight in LB and then diluted to an OD600 of 0.03 in 100 mL of 0.2% minimal glucose or 0.4% minimal sodium succinate medium. Growth was monitored at 600 nm. The ordinate is the OD600 ratio. The experiment was performed at least three times, and identical patterns were obtained. The results of a representative experiment are shown. ∗P < 0.05, ∗∗P < 0.01.
To further investigate the influence of EsrE on SDH activity, we replaced succinate with glucose in the minimal media. Reasoning that the predominant means of energy generation is glycolysis, SDH is essential only when the carbon source is succinate (Park et al., 1995). As expected, though the EsrE deletion mutant strain showed impaired growth on minimal succinate, it grew much better than the wild type strain on the glucose media (Figure 6B and Supplementary Figure S4). Meanwhile, the complementary expression of EsrE by pUC-P4P2-flag could rescue the growth defect on minimal media with succinate (Figure 6B and Supplementary Figure S4). Collectively, these findings indicated that EsrE was required for SDH activity in E. coli.
Discussion
The yigP Locus Encodes Two Products: EsrE and UbiJ
UQ is a redox-active lipid that is widely distributed in nature. The UQ structure has a conserved aromatic ring and an isoprenoid side chain of various lengths. The length of isoprenyl chain varies among species; there are six in Saccharomyces cerevisiae, eight in E. coli, and 10 in humans (Turunen et al., 2004; Bentinger et al., 2010; Nowicka and Kruk, 2010). Therefore, UQ in E. coli is designated Q8. The Q8 biosynthetic pathway is highly conserved and requires almost 11 genes, named ubi (Meganathan, 2001; Tran and Clarke, 2007). Q8 acts as a redox carrier in the plasma membrane, plays vital roles in aerobic respiration, oxidative stress adaptation, regulation of gene expression and other pathways dependent on the proton motive force (Aussel et al., 2014b). Recently, the yigP locus was renamed ubiJ that was characterized as an important element for Q8 biosynthesis in Salmonella and E. coli under aerobic conditions (Aussel et al., 2014a). Aussel et al. (2014a) believed that the aerobic growth defect was attributed to the blockage of Q8 biosynthesis due to mutant yigP locus. No sRNA was produced from the yigP locus because their frameshift mutant fragments at the C-terminal (the last 50 amino acids) coding sequence failed to rescue the growth defect (Aussel et al., 2014a). Though our results also showed that the yigP locus was required for Q8 biosynthesis in E. coli, we found that EsrE sRNA was also required for this process because in the absence of UbiJ, Q8 could still be produced in E. coli (Figures 2–4). Based on the evidence that the UbiJ deficient (frameshift) strain showed no phenotype and EsrE could only partially rescue the defect caused by 3′ UTR deletion at the yigP locus in aerobic growth, we inferred that not all the cellular Q8 content were required for aerobic growth of E. coli. Some minimal level (threshold level) of Q8 could be enough to maintain the high growth rate. This then led us to question whether the functions of EsrE and UbiJ were tightly related, and if there were advantages of hosting esrE within ubiJ. Our data showed that besides promoting Q8 biosynthesis, EsrE was also required for SDH activity, which suggests that the function of EsrE could be tied to UbiJ, and the two cooperated with each other in the respiratory chain of E. coli. However, more investigations are needed to verify this hypothesis.
A New Insight into Gene Annotation: 3′ UTR-Embedded sRNAs
Ongoing studies reveal that sRNA genes are not limited to intergenic regions, as they are also detected in coding regions (Lalaouna et al., 2015; Miyakoshi et al., 2015a). These sRNAs might pose a problem for gene annotation because there can be more than one functional product from a single gene locus. One emerging class of such sRNAs is 3′ UTR sRNAs. Miyakoshi et al. (2015b) proposed that 3′ UTR sRNAs could be divided into two basic types according to their biogenesis. Type I sRNAs are transcribed from independent promoters within the coding region or the 3′ UTR of known genes. This type includes MicL (Guo et al., 2014), EsrE (described here) in E. coli, and DapZ in Salmonella enterica (Chao et al., 2012). Type II 3′ UTR sRNAs such as CpxQ (Chao and Vogel, 2016) and SorC in S. enterica (Miyakoshi et al., 2015a) strictly originate from the processing of the parental mRNAs. 3′ UTR sRNAs usually share partial sequences with their respective mRNA loci (Chao et al., 2012; Guo et al., 2014; Miyakoshi et al., 2015b; Chao and Vogel, 2016). Thus the phenotypes attributed to these loci may not depend solely on the protein. The functions of both products need to be considered. Moreover, the correlation between 3′ UTR sRNAs and their parental mRNAs are currently ambiguous since only a few examples have been studied. DapZ and MicL functions have been proven to have no relation with their parental mRNAs, while SorC and CpxQ mediate cross talk between their respective mRNAs (Chao et al., 2012; Guo et al., 2014; Miyakoshi et al., 2015a; Chao and Vogel, 2016). It seems that Type II sRNAs are more related to their parental mRNAs than Type I. Here we found that EsrE, a Type I sRNA, has four targets. Among them, three (murE, murF, and sdhD) were activated and one (argI) was repressed by EsrE (Figures 5A,B). These targets are functionally unrelated, which makes EsrE function more diversified thereby requiring more efforts to uncover. Besides, EsrE is important for cellular Q8 biosynthesis, but the corresponding targets and regulatory mechanisms are still unknown. Additionally, EsrE could activate sdhD to increase the levels of SDH, which connects the TCA cycle with the ETC. Thus, we anticipate that EsrE might have a vital role in regulating the respiratory chain of E. coli. Meanwhile, its respective mRNA, ubiJ, is also a key element of the aerobic ETC of E. coli (Aussel et al., 2014a). In general, Type I sRNA could cooperate with their parental mRNA. But still, the correlation of the parental mRNA function and targets of 3′ UTR sRNAs remain to be elucidated.
In summary, our study provided new evidence that the yigP locus had two products, the UbiJ protein and EsrE sRNA. Both products were essential for Q8 biosynthesis and further contributed to aerobic growth of E. coli (Figure 7). Our results also showed that EsrE might target multiple mRNAs, including murE, murF, sdhD, and argI, which are involved in different cellular processes. Furthermore, our data indicated EsrE was required for the activity of SDH in succinate-dependent growth of E. coli. Additionally, our characterization of EsrE indicated that sRNAs (sRNAs from UTRs) may be masked in protein-coding regions. Phenotypes generally caused by deficiencies in protein may also be due to uncovered sRNA defects.
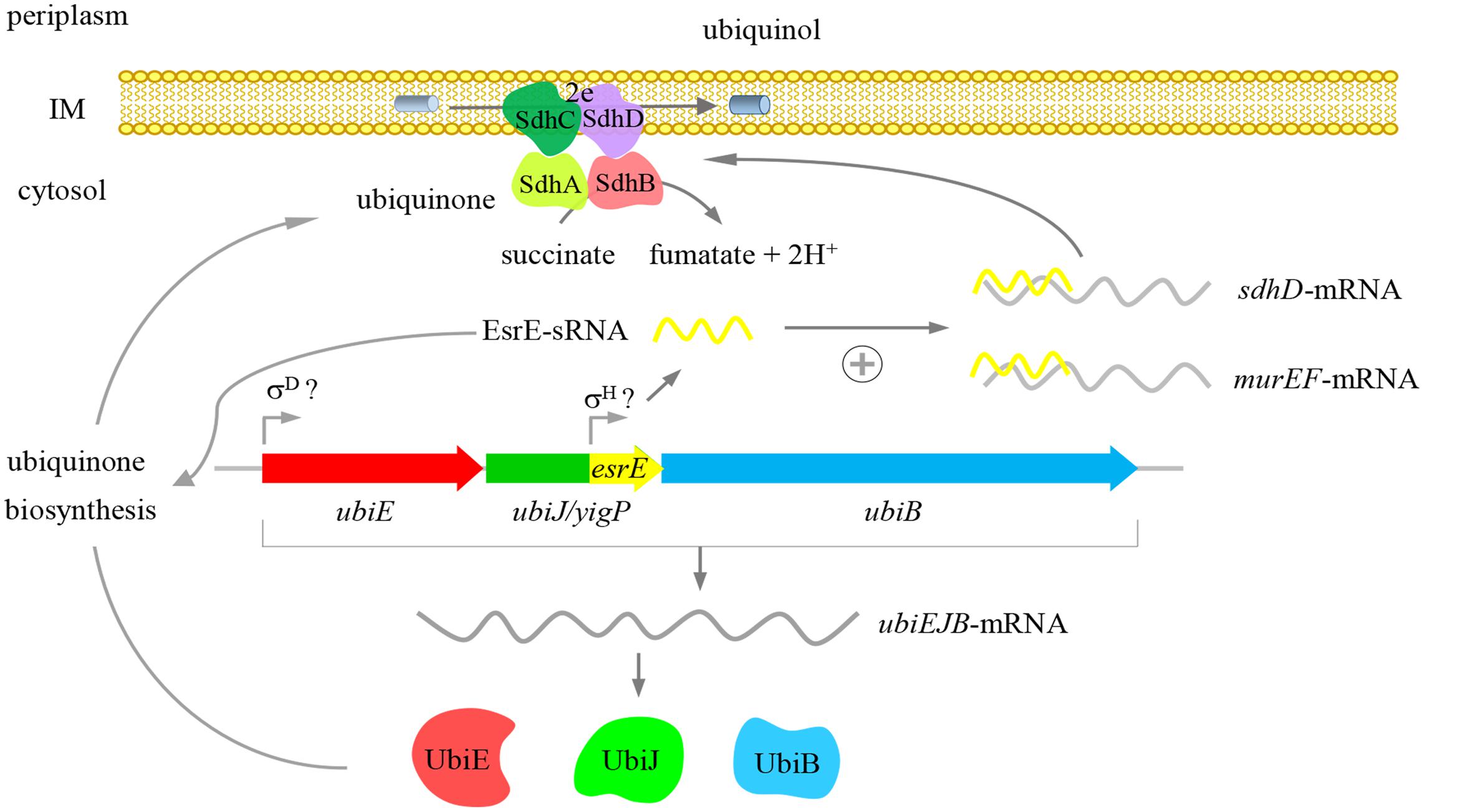
FIGURE 7. Proposed function of the yigP locus. The yigP locus may have two products, both of which are required for the aerobic growth of E. coli. One is a protein encoded by ubiJ that functions as a new factor involved in Q8 biosynthesis under aerobiosis. The other is sRNA transcribed from the 3′ region of the yigP locus and also required for Q8 biosynthesis as well as interacting with multiple targets involved in different cellular processes such as murein biosynthesis and the TCA cycle. This sRNA especially activates sdhD mRNA.
Author Contributions
HW and HZ conceived and designed the experiments. HX, XY, and QT performed the experiments. JY contributed reagents/materials/analysis tools. HX, HW, and HZ analyzed and wrote the paper.
Funding
This study was financially supported by National Natural Science Foundation of China (Grant Nos. 31372550, 3120026, and 31070073).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the members of our lab, specifically Dr. Bingbing Hou and Dr. Jian Mo, for the critical reading of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01658/full#supplementary-material
References
Aussel, L., Loiseau, L., Hajj Chehade, M., Pocachard, B., Fontecave, M., Pierrel, F., et al. (2014a). UbiJ, a new gene required for aerobic growth and proliferation in macrophage, is involved in coenzyme Q biosynthesis in Escherichia coli and Salmonella enterica serovar Typhimurium. J. Bacteriol. 196, 70–79. doi: 10.1128/JB.01065-13
Aussel, L., Pierrel, F., Loiseau, L., Lombard, M., Fontecave, M., and Barras, F. (2014b). Biosynthesis and physiology of coenzyme Q in bacteria. Biochim. Biophys. Acta 1837, 1004–1011. doi: 10.1016/j.bbabio.2014.01.015
Bentinger, M., Tekle, M., and Dallner, G. (2010). Coenzyme Q–biosynthesis and functions. Biochem. Biophys. Res. Commun. 396, 74–79. doi: 10.1016/j.bbrc.2010.02.147
Bobrovskyy, M., and Vanderpool, C. K. (2013). Regulation of bacterial metabolism by small RNAs using diverse mechanisms. Annu. Rev. Genet. 47, 209–232. doi: 10.1146/annurev-genet-111212-133445
Bush, M. J., Bibb, M. J., Chandra, G., Findlay, K. C., and Buttner, M. J. (2013). Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae. mBio 4:e00684-13. doi: 10.1128/mBio.00684-13
Cecchini, G. (2003). Function and structure of complex II of the respiratory chain. Annu. Rev. Biochem. 72, 77–109. doi: 10.1146/annurev.biochem.72.121801.161700
Chao, Y., Papenfort, K., Reinhardt, R., Sharma, C. M., and Vogel, J. (2012). An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 31, 4005–4019. doi: 10.1038/emboj.2012.229
Chao, Y., and Vogel, J. (2010). The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 13, 24–33. doi: 10.1016/j.mib.2010.01.001
Chao, Y., and Vogel, J. (2016). A 3′ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol. Cell 61, 352–363. doi: 10.1016/j.molcel.2015.12.023
Chen, Z., Wang, Y., Li, Y., Li, Y., Fu, N., Ye, J., et al. (2012). Esre: a novel essential non-coding RNA in Escherichia coli. FEBS Lett. 586, 1195–1200. doi: 10.1016/j.febslet.2012.03.010
Frohlich, K. S., and Vogel, J. (2009). Activation of gene expression by small RNA. Curr. Opin. Microbiol. 12, 674–682. doi: 10.1016/j.mib.2009.09.009
Guo, M. S., Updegrove, T. B., Gogol, E. B., Shabalina, S. A., Gross, C. A., and Storz, G. (2014). MicL, a new sigma(E)-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 28, 1620–1634. doi: 10.1101/gad.243485.114
Kery, M. B., Feldman, M., Livny, J., and Tjaden, B. (2014). TargetRNA2: identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res. 42, W124–W129. doi: 10.1093/nar/gku317
Keseler, I. M., Mackie, A., Peralta-Gil, M., Santos-Zavaleta, A., Gama-Castro, S., Bonavides-Martinez, C., et al. (2013). EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 41, D605–D612. doi: 10.1093/nar/gks1027
Lalaouna, D., Carrier, M. C., Semsey, S., Brouard, J. S., Wang, J., Wade, J. T., et al. (2015). A 3′ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol. Cell 58, 393–405. doi: 10.1016/j.molcel.2015.03.013
Masse, E., Escorcia, F. E., and Gottesman, S. (2003). Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17, 2374–2383. doi: 10.1101/gad.1127103
Masse, E., and Gottesman, S. (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 4620–4625. doi: 10.1073/pnas.032066599
Meganathan, R. (2001). Ubiquinone biosynthesis in microorganisms. FEMS Microbiol. Lett. 203, 131–139. doi: 10.1111/j.1574-6968.2001.tb10831.x
Miyakoshi, M., Chao, Y., and Vogel, J. (2015a). Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J. 34, 1478–1492. doi: 10.15252/embj.201490546
Miyakoshi, M., Chao, Y., and Vogel, J. (2015b). Regulatory small RNAs from the 3′ regions of bacterial mRNAs. Curr. Opin. Microbiol. 24, 132–139. doi: 10.1016/j.mib.2015.01.013
Morita, T., Maki, K., and Aiba, H. (2005). RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 19, 2176–2186. doi: 10.1101/gad.1330405
Nowicka, B., and Kruk, J. (2010). Occurrence, biosynthesis and function of isoprenoid quinones. Biochim. Biophys. Acta 1797, 1587–1605. doi: 10.1016/j.bbabio.2010.06.007
Ozeir, M., Pelosi, L., Ismail, A., Mellot-Draznieks, C., Fontecave, M., and Pierrel, F. (2015). Coq6 is responsible for the C4-deamination reaction in coenzyme Q biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 290, 24140–24151. doi: 10.1074/jbc.M115.675744
Papenfort, K., and Vanderpool, C. K. (2015). Target activation by regulatory RNAs in bacteria. FEMS Microbiol. Rev. 39, 362–378. doi: 10.1093/femsre/fuv016
Park, S. J., Tseng, C. P., and Gunsalus, R. P. (1995). Regulation of succinate dehydrogenase (sdhCDAB) operon expression in Escherichia coli in response to carbon supply and anaerobiosis: role of ArcA and Fnr. Mol. Microbiol. 15, 473–482. doi: 10.1111/j.1365-2958.1995.tb02261.x
Poon, W. W., Davis, D. E., Ha, H. T., Jonassen, T., Rather, P. N., and Clarke, C. F. (2000). Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J. Bacteriol. 182, 5139–5146. doi: 10.1128/JB.182.18.5139-5146.2000
Stefely, J. A., Licitra, F., Laredj, L., Reidenbach, A. G., Kemmerer, Z. A., Grangeray, A., et al. (2016). Cerebellar ataxia and coenzyme Q deficiency through loss of unorthodox kinase activity. Mol. Cell 63, 608–620. doi: 10.1016/j.molcel.2016.06.030
Tran, U. C., and Clarke, C. F. (2007). Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 7, S62–S71. doi: 10.1016/j.mito.2007.03.007
Turunen, M., Olsson, J., and Dallner, G. (2004). Metabolism and function of coenzyme Q. Biochim. Biophys. Acta 1660, 171–199. doi: 10.1016/j.bbamem.2003.11.012
Updegrove, T. B., Shabalina, S. A., and Storz, G. (2015). How do base-pairing small RNAs evolve? FEMS Microbiol. Rev. 39, 379–391. doi: 10.1093/femsre/fuv014
Urban, J. H., and Vogel, J. (2007). Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 35, 1018–1037. doi: 10.1093/nar/gkl1040
Van Beilen, J. W. A., and Hellingwerf, K. J. (2016). All three endogenous quinone species of Escherichia coli are involved in controlling the activity of the aerobic/anaerobic response regulator ArcA. Front. Microbiol. 7:1339. doi: 10.3389/fmicb.2016.01339
Vogel, J., and Luisi, B. F. (2011). Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578–589. doi: 10.1038/nrmicro2615
Wang, Y., Ye, J., and Zhang, H. (2012). Identification of transcriptional regulatory sequences of yigP gene in Escherichia coli. Wei Sheng Wu Xue Bao 52, 566–572.
Keywords: yigP locus, aerobic growth, Q8, EsrE sRNA, succinate dehydrogenase, E. coli
Citation: Xia H, Yang X, Tang Q, Ye J, Wu H and Zhang H (2017) EsrE–A yigP Locus-Encoded Transcript–Is a 3′ UTR sRNA Involved in the Respiratory Chain of E. coli. Front. Microbiol. 8:1658. doi: 10.3389/fmicb.2017.01658
Received: 13 May 2017; Accepted: 15 August 2017;
Published: 29 August 2017.
Edited by:
Yi-Cheng Sun, Institute of Pathogen Biology (CAMS) and Peking Union Medical College, ChinaReviewed by:
Jin-Byung Park, Ewha Womans University, South KoreaHongxia Wang, University of Alabama at Birmingham, United States
Copyright © 2017 Xia, Yang, Tang, Ye, Wu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haizhen Wu, d3VoemhAZWN1c3QuZWR1LmNu Huizhan Zhang, aHVpemh6aEBlY3VzdC5lZHUuY24=
 Hui Xia
Hui Xia Xichen Yang
Xichen Yang Qiongwei Tang
Qiongwei Tang Jiang Ye
Jiang Ye Haizhen Wu
Haizhen Wu Huizhan Zhang1,2*
Huizhan Zhang1,2*