- 1Department of Animal Science, University of Minnesota, Saint Paul, MN, United States
- 2Bacterial Epidemiology and Antimicrobial Resistance Research Unit, USDA-ARS, Richard B. Russell Research Center, Athens, GA, United States
- 3Poultry Production and Product Safety Research Unit, USDA, Fayetteville, AR, United States
- 4Department of Poultry Science, University of Arkansas, Fayetteville, AR, United States
- 5Department of Microbiology and Molecular Genetics, University of California, Irvine, Irvine, CA, United States
- 6Department of Animal Science, University of Connecticut, Storrs, CT, United States
Background: Salmonella Enteritidis phage type 8 (PT8) is a major poultry-associated Salmonella strain implicated in foodborne outbreaks in the United States. We previously reported that two plant-derived compounds generally recognized as safe (GRAS), trans-cinnamaldehyde (TC), and eugenol (EG), significantly reduced S. Enteritidis colonization in broiler and layer chickens. To elucidate potential PT8 genes affected by TC and EG during colonization, a whole-genome microarray analysis of the bacterium treated with TC and EG was conducted.
Results: S. Enteritidis PT8 was grown in Luria-Bertani broth at 37°C to an OD600 of ~0.5. Subinhibitory concentrations (SICs; concentration that does not inhibit bacterial growth) of TC (0.01%; 0.75 mM) or EG (0.04%; 2.46 mM) were then added to the culture. S. Enteritidis PT8 RNA was extracted before and 30 min after TC or EG addition. Labeled cDNA from three replicate experiments was subsequently hybridized to a microarray of over 99% of S. Enteritidis PT4 genes, and the hybridization signals were quantified. The plant-derived compounds down-regulated (P < 0.005) expression of S. Enteritidis PT8 genes involved in flagellar motility, regulation of the Salmonella Pathogenicity Island 1, and invasion of intestinal epithelial cells. TC and EG also suppressed transcription of genes encoding multiple transport systems and outer membrane proteins. Moreover, several metabolic and biosynthetic pathways in the pathogen were down-regulated during exposure to the plant-derived compounds. Both TC and EG stimulated the transcription of heat shock genes, such as dnaK, dnaJ, ibpB, and ibpA in S. Enteritidis PT8 (P < 0.005). The results obtained from microarray were validated using a quantitative real-time PCR.
Conclusion: The plant-derived compounds TC and EG exert antimicrobial effects on S. Enteritidis PT8 by affecting multiple genes, including those associated with virulence, colonization, cell membrane composition, and transport systems.
Background
Salmonella enterica serovar Enteritidis (S. Enteritidis) is one of the most commonly isolated Salmonella serotypes from poultry (Centers for Disease Control and Prevention, 2010; Campioni et al., 2013; Gould et al., 2013), and is responsible for about one third of the reported human salmonellosis outbreaks in the United States (Gould et al., 2013). The pathogen poses a significant health concern for human health due to the contamination of poultry meat and eggs, which constitute the most common food products linked to human salmonellosis (Guard-Petter, 2001; Marcus et al., 2007). In chickens, the cecum is the most common site for Salmonella residency (Gantois et al., 2009; Kollanoor Johny et al., 2012a,b,c). The cecal colonization by the pathogen results in fecal shedding, invasion of reproductive organs, contamination of egg shells and yolks, and carcass contamination during slaughter (Keller et al., 1995; Gantois et al., 2009). Therefore, reducing S. Enteritidis in the chicken intestinal tract would reduce contamination of poultry products, minimizing human health risk (Altekruse et al., 1993). Thus, intervention strategies for controlling S. Enteritidis colonization in chickens are critical for improving the microbiological safety of poultry-derived foods. Since a multitude of sources can transmit S. Enteritidis to chickens at farms, a variety of pre-harvest approaches, especially in-feed supplementation of antimicrobials, has been explored for reducing the pathogen persistence in poultry (reviewed in Kollanoor Johny et al., 2012a,b,c).
We previously reported that two phytophenolics, namely trans-cinnamaldehyde (TC) and eugenol (EG), were effective in reducing S. Enteritidis in vitro and in broiler chickens (Kollanoor Johny et al., 2008, 2010, 2012a,b,c). Further, we observed that in-feed supplementation of TC in layers significantly reduced S. Enteritidis colonization in the internal organs of birds and egg-borne transmission of the bacterium (Upadhyaya et al., 2015). Trans-cinnamaldehyde, an aromatic aldehyde extracted from the bark of cinnamon (Cinnamomum zeylandicum), has well-known antimicrobial properties (Friedman et al., 2002, reviewed by Burt, 2004). Eugenol (EG), a compound obtained from clove (Eugenia caryophillis) oil, is also reported to be effective in killing pathogenic microorganisms (Suhr and Nielsen, 2003, reviewed by Burt, 2004). Both these compounds are classified as generally recognized as safe (GRAS) for use in foods by the United States Food and Drug Administration (USFDA) (TC–21CFR182.60; EG–21CFR582.60).
In light of our previous findings, the objective of this study was to analyze the genome-wide response of S. Enteritidis Phage Type 8 (or PT8) to a sub-inhibitory concentration of TC or EG using DNA microarrays. We sought to delineate the arsenal of genes affected by the two phytochemicals.
Results
The DNA microarray analysis revealed that about 10% of genes in the S. Enteritidis PT8 genome were significantly modulated after exposure to TC and EG. The major genes predicted to be differentially regulated in PT8 exposed to TC and EG based on the microarray data are provided in Table 1. The analysis identified 566 genes (9.7% genes on the array) with differentially expressed transcripts due to exposure to TC, and 483 genes (8.4% on the array) with differential expression as a result of EG exposure (M ± 1.5, P < 0.005; Tables 2, 3). TC significantly down-regulated 275 genes, whereas EG did so for 289 genes (P < 0.005). Many of the genes modulated are known to play a critical role in S. Enteritidis virulence in vivo based on previously published reports (Dhawi et al., 2011; Harvey et al., 2011) and are discussed here. Additional data on genes that are not discussed in depth in the manuscript can be found in the Supplementary Tables S1–S19.
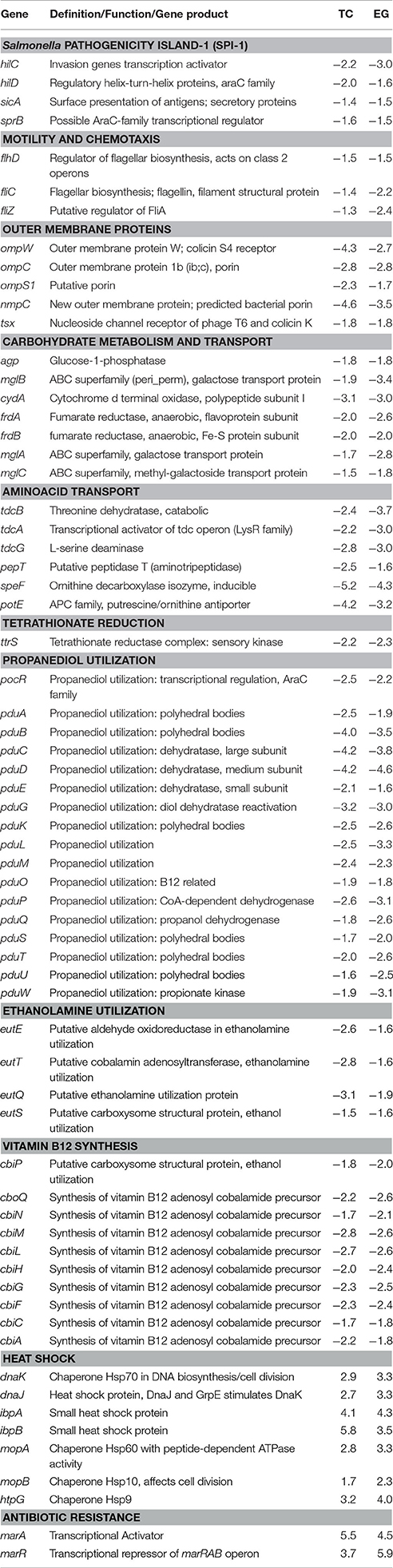
Table 1. Major genes predicted to be differentially regulated in PT8 exposed to SICs of both TC and EG (P < 0.005, M ± 1.5), based on the microarray data.
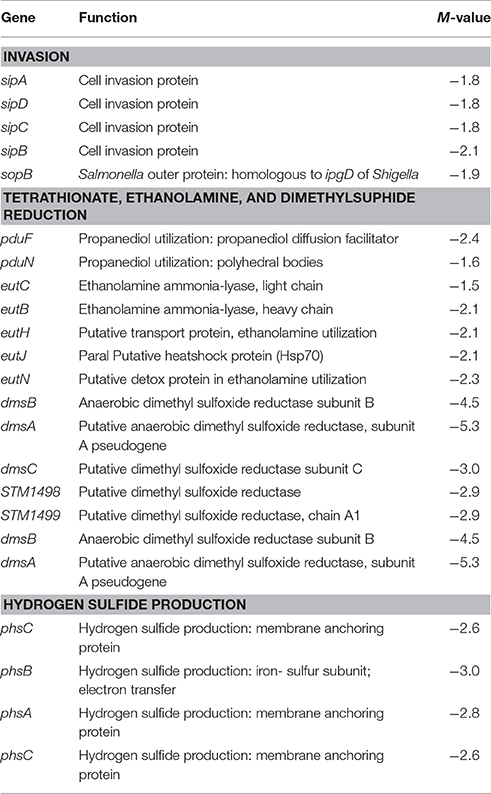
Table 2. Select genes predicted to be differentially regulated in PT8 exposed to SICs of TC but not EG (P < 0.005, M ± 1.5), based on the microarray data.
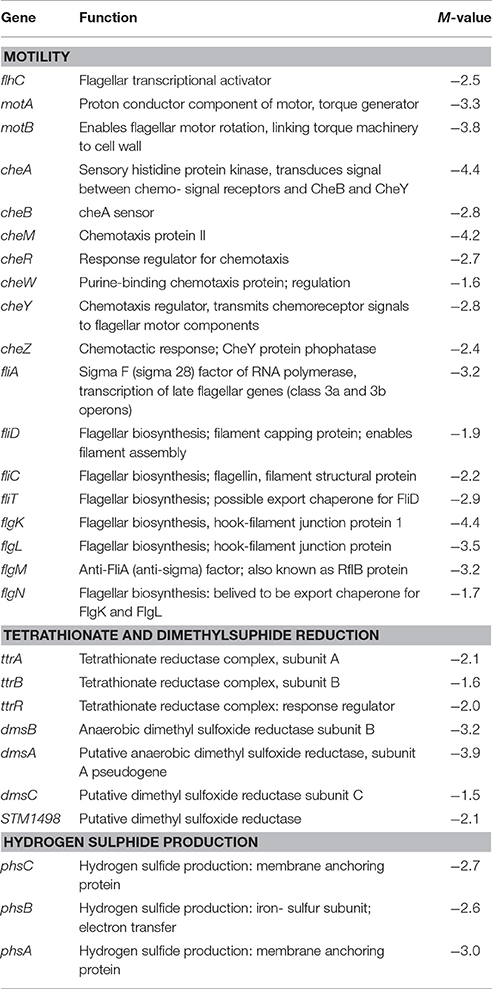
Table 3. Select genes predicted to be differentially regulated in PT8 exposed to SICs of EG, but not TC (P < 0.005, M ± 1.5), based on the microarray data.
Salmonella Pathogenicity Island-1 (SPI-1) and Type Three Secretion System Genes (T3SS)
The virulence genes associated with the major pathogenicity island, SPI-1, were affected in response to TC exposure, including major regulators, such as hilC and hilD (Table 1). Transcription of these genes was down-regulated 2.0- and 2.2-fold, respectively, by TC (P < 0.005). In addition, genes associated with cell invasion (sipABCD) and outer membrane protein syntheses (sopB) were significantly down-regulated by TC (Table 2).
Motility, Chemotaxis, and Adherence
Several genes associated with Salmonella motility, chemotaxis and adherence to host cells were down-regulated by both plant-derived compounds, although EG was significantly more effective than TC (Tables 1, 3). For example, EG significantly down-regulated the transcription of genes responsible for bacterial motility, including motA, motB, and flhC, chemotaxis genes, cheA, cheY and cheZ, and the major flagellin fliC (Table 3).
Outer Membrane Proteins (OMPs)
Both plant-derived compounds significantly down-regulated genes encoding OMPs, namely ompW, ompC, ompS1, and nmpC (Table 1). The nmpC gene, encoding a predicted bacterial porin, was down-regulated by 4.6- and 3.5-fold by TC and EG, respectively. Similarly, ompW, a colicin S4 receptor gene, was also reduced in transcription by 4.3- and 2.7-fold, respectively, by TC and EG. However, some of the major OMP genes, such as ompA, ompR, and ompX, were not affected by both compounds.
Metabolism and Biosynthetic Pathway Genes
As revealed by microarray analysis, several genes responsible for energy production and conversion in Salmonella were down-regulated by TC and EG. For example, the majority of the genes regulating the carbohydrate metabolism and transport, amino acid transport, and carbon compound degradation (Table 1, Supplementary Table S11) were found to be down-regulated by TC and EG. There were indications of down-regulation of mixed metabolism genes, including genes related to the glucose metabolism and genes associated with C4- (e.g., fumarate), non-glucose C6- (e.g., gluconate, fucose), and C9- (sialic acid) carbohydrate metabolisms. In addition, melA, melB, and melR, genes regulated by the melibiose operon, and mannose-specific PTS (phosphotransferase) family genes manXYZ were down-regulated by both plant-derived compounds (Table 1). A complete list of these genes is found in Supplementary Tables S8–S18.
A major set of genes involved in tetrathionate reduction (ttr), propanediol utilization (pdu), ethanolamine utilization (eut), and dimethyl sulfide reduction (dms) were down regulated by TC and EG (Tables 1–3). Moreover, the cbi locus genes associated with vitamin B12 synthesis (Table 1) were down-regulated by TC and EG. Furthermore, genes responsible for H2S production, phsABC, were significantly down-regulated by both compounds (Tables 2, 3).
Up-Regulated Genes
Besides the down-regulation of several genes indicated above, a few critical genes were up-regulated in response to TC and EG exposure (Table 1, Supplementary Table S19). The greatest increase in transcription was observed with heat shock proteins. The genes encoding two small heat shock proteins, ibpA and ibpB, were up-regulated by TC by 4.1- and 5.8-fold, respectively, and 4.3- and 3.5-fold, respectively, by EG. In addition, dnaJ, and the Hsp70 chaperone gene dnaK were up-regulated on exposure to both compounds. The microarray analysis revealed increased transcription of acrA and acrB, the acridine efflux pump genes, by EG (Table 1). Similarly, multiple antibiotic resistance protein (mar) genes were also up-regulated by TC and EG. However, marR, encoding the transcriptional repressor of the marRAB operon, was also found to be up-regulated by both plant -derived compounds (P < 0.005), probably as a compensatory mechanism.
Confirmation with RT-qPCR
Nine genes with significantly different transcription levels following TC or EG exposure as identified by microarray analysis were further analyzed by RT-qPCR. Regarding exposure to TC, microarray and RT-qPCR results were in agreement for all tested genes but sipA and sopB (Figure 1). Microarray data revealed that sipA and sopB were down-regulated by 1.8- and 2.0-fold, respectively, but in RT-qPCR analysis, a slight up-regulation of both genes on exposure to TC was observed. Regarding exposure to EG, hilA, hilD, flhD, ttrS, ompC, sipA, and sopB were found significantly down-regulated in microarray analysis and in RT-qPCR results (Figure 2).
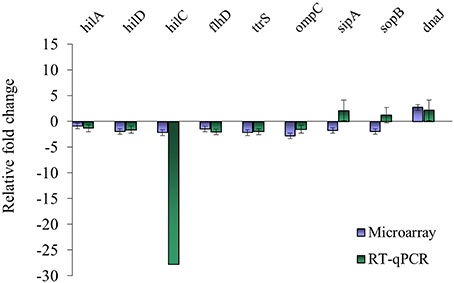
Figure 1. Comparison of microarray and RT-qPCR data on select genes after exposure of S. Enteritidis PT8 to SICs of TC. Data are from three replicates.
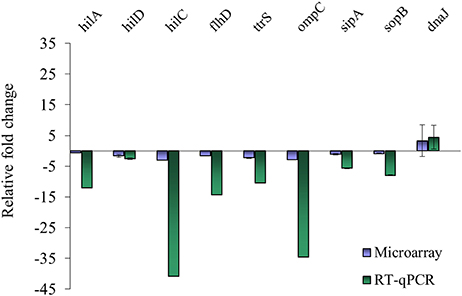
Figure 2. Comparison of microarray and RT-qPCR data on select genes after exposure of S. Enteritidis PT8 to SICs of EG. Data are from three replicates.
Discussion
Salmonella Enteritidis is a leading cause of foodborne salmonellosis worldwide. The increase in the number of outbreaks over the last few decades, where poultry and poultry products were implicated, has led to several pre- and post-harvest strategies to control the pathogen. We previously reported that supplementation of TC and EG in broilers (Kollanoor Johny et al., 2012a,b) and TC in layers (Upadhyaya et al., 2015) significantly reduced S. Enteritidis colonization in birds. We also observed that these compounds decreased Salmonella motility and invasion of avian epithelial cells in vitro (Kollanoor Johny et al., 2012b). Since analysis of transcriptional profiles of bacteria exposed to an antimicrobial molecule can yield new information regarding affected pathways and mechanisms of inhibition (Wilson et al., 1999; Gmuende et al., 2001), we conducted a microarray analysis of S. Enteritidis PT8 exposed to TC and EG. The goal was to delineate the genes affected by these plant-derived compounds, to gain insight into the molecular mechanism of how these compounds may cause reduced S. Enteritidis colonization in chickens.
A diverse repertoire of Salmonella virulence genes was down-regulated in response to treatment with TC and EG, including, critically, the genes involved in SPI-1 regulation. These genes are necessary for Salmonella uptake into non-phagocytic epithelial cells (McCormick, 2004). A direct control of SPI-1 genes is exerted by the activation by hilA-the major regulator of the island, resulting in the invasion of host tissue. Ellermeier and Slauch (2007) reported that other regulators, such as HilC and HilD work in conjunction with another regulator RtsA to express SPI-1. HilD, an AraC/XylS family member, binds directly to several sites within the hilA promoter and derepresses gene expression, which in turn results in the expression of other associated virulence genes. Both plant-derived compounds down-regulated the major regulators hilA, hilD, and hilC, although a greater effect was observed on hilC (Table 1, Supplementary Table S1). The T3SS encoded by SPI-1 is the pathogenic machinery to inject secretory proteins into host cells that help in the internalization of Salmonella (Kubori et al., 2000; Galan, 2001). The effector proteins are transported into the host cells with the help of three translocation proteins, sipB, sipC and sipD (Collazo and Galan, 1997; Galan, 2001). As revealed in the microarray data, TC down-regulated the effector protein genes (Supplementary Table S3).
Flagellar motility and chemotaxis are the two critical features by which bacteria survive under a wide variety of environments (Soutourina and Bertin, 2003), including the intestinal lumen and ceca. Flagellar motility is also required for colonization by Salmonella (Ciacci-Woolwine et al., 1998), and is considered a virulence factor in the bacterium (Josenhans and Suerbaum, 2002). In the regulation of flagella, the flhDC master operon plays a key role (Kutsukake et al., 1990; Soutourina and Bertin, 2003). Among other genes, the master operon is necessary for transcriptional activation of the fliA gene, encoding σ28, a sigma factor associated with the transcription of late flagellar genes, such as fliC (filament structural protein). It was interesting to note that that these associated genes were all down-regulated by EG (Table 1). Apart from the flhCD-regulated genes (early flagellar operon), the middle and late operon genes, such as flgKLMN, fliZ, motAB, and cheAWYZ, were also down-regulated by EG (Table 3, Supplementary Table S5). In contrast, TC did not exert strong effects on these genes.
Porins are outer membrane aqueous channels used in the diffusion of various compounds across the membrane that can also aid in adapting the bacteria to various conditions and confer antibacterial drug resistance (Gil et al., 2009). In the current study, TC and EG down-regulated porins, such as ompW, ompC, ompS1, and nmpC by 2-fold. Recently, Gil et al. (2009) reported that ompW expression in Salmonella was activated in response to oxidative stress. Both plant-derived compounds down-regulated ompW by 4.3- and 2.7-fold, respectively. Methner et al. (2004) reported that ompC deletion mutants of S. Enteritidis resulted in decreased virulence and colonization in chickens in conjunction with deletions of rpoS or phoP. Our results revealed that ompC was down-regulated by 2.8-fold by both compounds (Table 1). Porin ompS1, down-regulated by TC and EG in our study, is regulated by the EnvZ/OmpR two-component signal transduction system (Florez-Valdez et al., 2003). Rodriguez-Morales et al. (2006) reported that ompS1 is required for virulence in S. Typhimurium in mice.
It was recently reported that during colonization in the gut Salmonella makes use of a wide range of carbon compounds, including D- glucose, melibiose, L-ascorbate, and other unusual compounds, such as sialic acid, ethanolamine, and propanediol resulting from the degradation of host mucosa (Dhawi et al., 2011). We observed that TC and EG down-regulated some of the glucose metabolism genes (agp, pykA), and TC reduced the transcription of melABR genes (Supplementary Table S8). Moreover, both plant-derived compounds were found to apparently reduce the bacterial capacity to utilize propanediol and ethanolamine as an energy source or tetrathionate as a terminal electron acceptor (Table 1), suggesting that TC and EG may interrupt the pathways critical for Salmonella survival and successful competition with the host microflora. During colonization of the host intestine, Salmonella can use energy and nutrient sources unavailable to commensal bacteria, including propanediol and ethanolamine. These compounds cannot be used for energy without a terminal electron acceptor, and the host flora mainly consists of mostly fermentative bacteria unable to perform this electron transport. Salmonella, however, is uniquely capable of using these compounds by employing tetrathionate as a terminal electron acceptor. When tetrathionate is used as an electron acceptor, it is reduced to thiosulfate, which is then regenerated via oxidation by the reactive oxygen compounds released by the host in response to invasion by Salmonella. Salmonella can therefore outcompete the host flora, resulting in inflammatory diarrhea, with Salmonella cells as major bacterial component of the excreta thus accomplishing transmission to the next host organism.
Harvey et al. (2011) reported that within the lumen of chicks, the degradation of 1, 2-propanediol occurs at a much higher rate, requiring cobalamin synthesis. The pdu operon required for propanediol utilization works in conjunction with cob and cbi operons involved in vitamin B12 synthesis. Our results revealed that TC and EG significantly down-regulated pdu and cbi operons (Table 1). It was previously observed that Salmonella could utilize ethanolamine, which is derived from host cells and membranes, as a sole source of carbon, nitrogen, and energy (Garsin, 2010). Catabolism of ethanolamine is under the control of the eut operon that primarily consists of 17 clustered genes (Kofoid et al., 1999) involved in the conversion of the compound to acetyl-CoA for energy (Harvey et al., 2011). The plant compound TC was found to down-regulate these genes (Supplementary Table S14). An anaerobic environment may exist in the chicken cecum (Harvey et al., 2011), and oxygen may not be present as a terminal electron acceptor. In that case, other electron acceptors are required, and ethanolamine and 1, 2-propanediol could be utilized in that function by Salmonella in the cecum (Price-Carter et al., 2001). Tetrathionate may be present in the cecum of chicks due to the presence of yolk sac, which is rich in sulfur. Tetrathionate is reduced to thiosulphide and H2S, involving proteins encoded by ttr and phs (Harvey et al., 2011). Our results indicated that TC down-regulated the transcription of ttr genes (Supplementary Table S12), whereas both plant-derived compounds reduced phsABC transcription (Supplementary Table S18). Overall, TC and EG may disrupt Salmonella's ability to use a unique energy source, possibly reducing its ability to compete successfully with the host's microflora.
Some heat shock genes and genes conferring antibiotic resistance were up-regulated by TC and EG. While the mar (multiple antibiotic resistance) locus genes were up-regulated by both TC and EG, the efflux pump genes acrAB were only up-regulated by EG. The MarA protein positively regulates transcription of acrAB, and it was therefore unexpected that the latter genes were not up-regulated after exposure to TC. However, the transcriptional repressor marR was also found to be up-regulated by both TC and EG, potentially indicating a compensatory mechanism on reducing marAB-related resistance. Heat shock proteins dnaJ, dnaK, ibpA, ibpB, and htpG were found to be up-regulated by both plant-derived compounds (P < 0.005). However, clpPX-mediated heat shock protection mechanisms were least affected (Supplementary Table S19).
Some limitations of this study include the use of nutrient broth for the gene expression studies as opposed to growth within a characterized chicken-derived intestinal cell line, although broth cultures have been previously used for similar experiments (Cao et al., 2011; Jessica et al., 2013; Upadhyay et al., 2013). To our knowledge, immortalized chicken intestinal cell lines are unavailable, and developing and maintaining a primary chicken intestinal cell line has its challenges, including the high variability in confluency across different propagations. In a separate study, we conducted invasion assays using the only available permanent avian cancer cell line (Budgerigar Abdominal Tumor Cells; BATCs) and found that these two phytophenolics significantly reduced Salmonella invasion without affecting the viability of cells (Kollanoor Johny et al., 2012b). However, being a permanent cell line obtained from a different avian species (parakeets), a direct correlation of these results to a chicken-derived cell environment is not guaranteed. However, the selected phytophenolics reduced the cecal colonization of S. Enteritidis in broiler chickens in at least 5 independent in vivo experiments (Kollanoor Johny et al., 2012a,b), lending support to the notion that TC and EG directly affect S. Enteritidis virulence genes. However, this hypothesis needs to be further validated.
Conclusions
Genes required for S. Enteritidis PT8 virulence and colonization, including those conferring flagella-associated motility, those enabling invasion of epithelial cells, genes encoding T3SSs, genes regulating the synthesis of effector proteins delivered by T3SS, and genes encoding other surface virulence structures and OMPs were found to be down-regulated after exposure to subinhibitory concentrations of TC or EG. Moreover, the exposure to both phytochemicals brought about transcriptional changes of S. Enteritidis PT8 genes involved in the degradation of carbon compounds, those related to amino acid-, carbohydrate-, and lipid-transport, those associated with secondary metabolism, and genes encoding the biosynthesis of molybdopterin and vitamin B12. The genes regulating pathways associated with tetrathionate reduction, propanediol, and ethanolamine utilization, and H2S production were also affected. Finally, a few genes regulating antibiotic resistance and heat shock were up-regulated by the plant-derived compounds, including transcriptional activators and repressors, with as yet undefined phenotypic net results.
Methods
Bacterial Strain and Growth Conditions
S. Enteritidis PT8, obtained from the Connecticut Veterinary Diagnostic Medical Laboratory, was used for the experiments. This strain was used along with three other strains in our in vitro invasion and motility assays, and in vivo chicken experiments (Kollanoor Johny et al., 2008, 2010, 2012a,b,c). The strain was selected based on RT-qPCR analysis of virulence gene transcription (hilA, hilD, motA, flhC, and invF) of different phage types (PT-8, -13, and -13b) in response to subinhibitory concentrations of TC and EG (Kollanoor Johny et al., 2012b).
PT8 was grown overnight in Luria-Bertani (LB) broth (Difco) at 37°C with shaking at 150 rpm. For the transcription profiling experiments, 10 ml of an overnight culture was spun at 3,800 × g for 15 min, and the resulting pellet was resuspended in 10 ml phosphate buffered saline (PBS; pH 7.0). A 0.5 ml aliquot of the suspension was added to 500 ml of LB broth and grown with vigorous shaking at 37°C until it reached a late-log phase OD600 of 0.5. Assays were performed using 3 independent biological replicates for TC and EG, separately. The SIC of TC (0.01% vol/vol; 0.75 mM) or EG (0.04% vol/vol; 2.46 mM) (Kollanoor Johny et al., 2012b) was added to the culture and the mixture was vortexed for 1 min. The control flask was vortexed for 1 min as well. Exactly 100 ml were drawn from the broth before addition of TC or EG (time 0), and 30 min (time 30) after addition of the plant-derived compounds. The samples were drawn separately into 250 ml centrifuge tubes containing 16 ml of ice-cold ethanol/phenol stop solution (5% citrate-buffered phenol v/v), to halt the degradation of mRNA. The cells were spun at 8,000 × g for 2 min at 4°C, and the resulting pellet was stored at −80°C until use.
Total RNA Extraction
Total RNA from S. Enteritidis PT8 was extracted as previously described (Frye et al., 2006). The frozen pellet was lysed by resuspending in a final volume of a fresh solution of 10 ml 0.5 mg/ml lysozyme in TE (pH 8.0). A volume of 1 ml of 10% SDS was added, and the mixture was placed in a water bath at 64°C for 2 min. After incubation, 11 ml of 1 M sodium acetate solution (pH 5.2) was added to the tubes and mixed. An equal volume of phenol was added and mixed, followed by incubation at 64°C for 6 min. The tubes were then placed on ice to chill before they were spun at 10,000 × g for 10 min at 4°C. The aqueous layer was transferred to a fresh tube containing an equal volume of chloroform, mixed, and spun at 10,000 rpm for 5 min at 4°C. The aqueous layer from the samples was then transferred into fresh tubes, 1/10 volume of 3 M sodium acetate (pH 5.2) was added, and one volume of cold isopropanol. The tubes were incubated at −80°C for 20 min before they were spun at 14,000 × g for 25 min at 4°C. Isopropanol was carefully removed and the resulting pellet was washed with 40 ml 80% cold ethanol. The pellets were spun at 14,000 × g for 5 min at 4°C. Ethanol was carefully removed and the pellet was air dried. The pellet was resuspended in 2 ml of RNase-free DEPC-treated water. The sample was split into two and each was provided with 500 U of RNase inhibitor (Ribolock®, Fermentas), 250 U of DNase (Fermentas), 20 μl of 1 M Tris (pH 8.3) and 10 μl of 1 M MgCl, followed by mixing and incubation at 37°C for 30 min. The RNA sample was then extracted once each with phenol and phenol–chloroform and twice with chloroform. A 1/10th volume of 3 M sodium acetate at pH 5.2 was added, and the RNA was precipitated with isopropanol, washed with 80% ethanol and resuspended in nuclease-free water (Frye et al., 2006). The RNA concentration and purity were determined using a Nanodrop (Thermofisher Corp.).
Microarray, Target Preparation, and Hybridization
The Salmonella whole genome microarray used in this study has been previously described (Porwollik et al., 2003) and includes 5,776 PCR products representing more than 99% of the ORFs of S. Enteritidis PT4 as well as four other serotypes of Salmonella. The microarray hybridization and analysis were done by standard methods, as previously described for Salmonella (Porwollik et al., 2003; Frye et al., 2006). Fifty microgram of RNA was transcribed into cDNA and labeled with Cy3- or Cy5-conjugated dUTP using reverse transcriptase (Superscript II®; Invitrogen) and random hexamers as primers. Unincorporated nucleotides were removed using a PCR purification kit (Qiagen) according to the manufacturer's instructions. Equal volumes of labeled probes from 0 (Cy3-labeled control) and 30 min (Cy5-labeled treatment) were mixed with an equal volume of hybridization solution (50% formamide, 10 × SSC and 0.2% SDS). Slides were prehybridized in 25% formamide, 5 × SSC and 0.1% SDS at 42°C. Probes were hybridized simultaneously to a chip containing three replicate arrays spotted onto CMT-UltraGAPS® (Corning) slides. Slides were washed in stringency-controlled buffers and dried by centrifugation before scanning.
Scanning and Data Analysis
The slides were scanned using the ScanArray 5,000 laser scanner (GSI Lumonics). The signals were recorded with scanarray 2.1 software and then quantified using Quantarray 3.0 software (Packard BioScience) (Frye et al., 2006; Ledeboer et al., 2006). Arrays were scanned for high signal intensity and low background in either of the channels, to be included for analysis. The normalization of data considered the median intensity of the spot and the local background. The data were statistically analyzed using the Partek Discovery SuiteTM v 6.2 (Partek Inc., St. Louis, MO). Mixed effects ANOVA was performed, and a significant P < 0.005 for each comparison was determined using a false discovery rate of 0.05. A total of 11 (5 treated and 6 untreated) biological replicates (separate RNA preparations) were analyzed from experiments conducted on different days, and the data have been deposited in GEO (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE102477.
Real-Time Quantitative PCR (RT-qPCR)
An RT-qPCR analysis was conducted to confirm the differential expression of the selected microarray targets. The protocol of sample preparation and collection for microarray experiment was also used for preparation of RT-qPCR samples. Total RNA was extracted from three biological replicates of PT8 cultures exposed to TC or EG using the RNeasy minikit (Qiagen) according to manufacturer's instructions. The resulting RNA was treated with RNase-free DNase. The quantitation of RNA was done by measuring the absorbance at 260 and 280 nm (Nanodrop, Biorad). Complementary DNA (cDNA) was synthesized using the Superscript II Reverse transcriptase kit (Superscript™—Invitrogen, Carlsbad, CA). The cDNA was used as the template for the amplification of Salmonella genes selected based on the results from microarray, including hilA, hilD, hilC, flhD, dnaJ, ompC, sipA, sopB, and ttrS. The specific primers for the genes, and for 16S rRNA (endogenous control) (Table 4) were designed using Primer Express software (Applied Biosystems). The primers were synthesized by Integrated DNA Technologies (Foster City, CA). Real time-qPCR was performed with the ABI PRISM 7900 sequence detection system (Applied Biosystems), using the SYBR® Green assay (Applied Biosystems) under custom thermal cycling conditions (denaturation step at 95°C for 10 min, followed by 40 cycles of a denaturation step at 95°C for 15 s, and an annealing/elongation step at 60°C for 60 s). A default melting curve stage (95°C for 15 s, 0°C for 60 s, 95°C for 15 s) was also included. The biological replicates were analyzed in duplicate and normalized against 16S rRNA gene expression. The 16S rRNA gene was used to account for the variability between the samples since the transcript levels of the endogenous control were found to be not significantly different (P > 0.05) between the control- and the TC- or EG- treated cells. The comparative Ct method (2−ΔΔCt) was used to assess the relative changes in the mRNA expression levels between the control and TC-/EG- treated PT8 (Schmittgen and Livak, 2008).
Authors Contributions
AK carried out microarray experiments, RT-qPCR assays, involved in the study design and wrote the manuscript. JF trained AK to conduct microarray analysis, preliminary interpretation of statistical analysis and corrected the manuscript. AD and DD participated in the design of the study and corrected the manuscript. SP and MM designed and constructed the microarrays, interpreted the hybridization results and performed statistical analysis of the data. KV conceived and designed the study and corrected the manuscript. All authors read and approved the manuscript.
Disclaimer
Mention of a trade name, proprietary product, or specific equipment does not constitute a guarantee or warranty by the USDA and does not imply its approval to the exclusion of other products that are suitable.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the USDA-NIFA Agriculture and Food Research Initiative Grant no. 2009-03576 awarded to KV and DD. The authors would like to thank Lari Hiott at the Bacterial Epidemiology and Antimicrobial Resistance research unit at USDA, ARS, for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01828/full#supplementary-material
Abbreviations
TC, trans-cinnamaldehyde; EG, Eugenol; SIC, subinhibitory concentration; cDNA, complementary DNA; CDC, Centers for Disease Control and Prevention; GRAS, Generally Recognized as Safe; USFDA, United States Food and Drug Administration; RT-qPCR, Real-Time Quantitative Polymerase Chain Reaction; mRNA, messenger RNA; M, Molar.
References
Altekruse, S., Koehler, J., Hickman-Brenner, F., Tauxe, R. V., and Ferris, K. (1993). A comparison of Salmonella Enteritidis phage types from egg-associated outbreaks and implicated laying flocks. Epidemiol. Infect. 110, 17–22. doi: 10.1017/S0950268800050639
Burt, S. (2004). Essential oils: their antibacterial properties and potential applications in food – a review. Int. J. Food Microbiol. 94, 223–253. doi: 10.1016/j.ijfoodmicro.2004.03.022
Campioni, F., Davis, M., Medeiros, M. I. C., Falcao, J. P., and Shah, D. H. (2013). MLVA typing reveals higher genetic homogeneity among Salmonella Enteritidis strains isolated from food, humans, and chickens in Brazil in comparison to the North American strains. Int. J. Food Microbiol. 162, 174–181. doi: 10.1016/j.ijfoodmicro.2013.01.008
Cao, B., Li, R., Xiong, S., Yao, F., Liu, X., Wang, M., et al. (2011). Use of a DNA microarray for detection and identification of bacterial pathogens associated with fishery products. Appl. Environ. Microbiol. 77, 8219–8225. doi: 10.1128/AEM.05914-11
Centers for Disease Control and Prevention (2010). Preliminary FoodNet Data on the Incidence of Infection with Pathogens Transmitted Commonly through Food—10 States, 2009. U.S. Department of Health and Human Services, Atlanta, GA: CDC.
Ciacci-Woolwine, F., Blomfield, I. C., Richardson, S. H., and Mizel, S. B. (1998). Salmonella flagellin induces tumor necrosis factor alfa in a human promocytic cell line. Infect. Immun. 6, 1127–1134.
Collazo, C. M., and Galan, J. E. (1997). The invasion-associated type III system of S. Typhimurium directs the translocation of Sip proteins into the host cell. Mol. Microbiol. 24, 747–756. doi: 10.1046/j.1365-2958.1997.3781740.x
Dhawi, A. A., Elazomi, A. M., Jones, A. M., Lovell, A., Li, H., Emes, R. D., et al. (2011). Adaptation to the chicken intestine in Salmonella Enteritidis PT4 studied by transcriptional analysis. Vet. Microbiol. 153, 198–204. doi: 10.1016/j.vetmic.2011.07.013
Ellermeier, J. R., and Slauch, J. M. (2007). Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10, 24–29. doi: 10.1016/j.mib.2006.12.002
Florez-Valdez, M. A., Puente, J. L., and Calva, E. (2003). Negative osmoregulation of the Salmonella ompS1 porin gene independently of ompR in an hns background. J. Bacteriol. 185, 6497–6506. doi: 10.1128/JB.185.22.6497-6506.2003
Friedman, M. 1., Henika, P. R., and Mandrell, R. E. (2002). Bactericidal activities of plant essential oils and some of their isolated constituents against Campylobacter jejuni, Escherichia coli, Listeria monocytogenes, and Salmonella enterica. J. Food Prot. 65, 1545–1560. doi: 10.4315/0362-028X-65.10.1545
Frye, J. G., Jessea, T., Longb, F., Rondeaub, G., Porwollikb, S., McClelland, M., et al. (2006). DNA microarray detection of antimicrobial resistance genes in diverse bacteria. Intl. J. Antimicrob. Agents. 27, 138–151. doi: 10.1016/j.ijantimicag.2005.09.021
Galan, J. E. (2001). Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17, 53–86. doi: 10.1146/annurev.cellbio.17.1.53
Gantois, I., Ducatelle, R., Pasmans, F., Haesebrouck, F., Gast, R., Humphrey, T. J., et al. (2009). Mechanisms of egg contamination by Salmonella Enteritidis. FEMS Microbiol, Rev. 33, 718–738. doi: 10.1111/j.1574-6976.2008.00161.x
Garsin, D. A. (2010). Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat. Rev. Microbiol. 8, 290–295. doi: 10.1038/nrmicro2334
Gil, F., Hernandez-Lucas, I., Polanco, R., Pacheco, N., Collao, B., Villarreal, J. M., et al. (2009). SoxS regulates the expression of the S. Typhimurium ompW gene. Microbiology 155, 2490–2497. doi: 10.1099/mic.0.027433-0
Gmuende, H., Kuratli, K., Di Padova, K., Gray, C. P., Keck, W., and Evers, S. (2001). Gene expression changes triggeredf by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 11, 28–42. doi: 10.1101/gr.157701
Gould, H. L., Walsh, K. A., Vieira, A. R., Herman, K., Williams, I. T., Hall, A. J., et al. (2013). Surveillance for foodborne disease outbreaks–United States, 1998–2008. Morbid. Mortal. Weekly. Rep. 62, 1–34. Available online at: https://www.cdc.gov/mmwr/preview/mmwrhtml/ss6202a1.htm
Guard-Petter, J. (2001). The chicken, the egg and Salmonella Enteritidis. Environ. Microbiol. 3, 421–430. doi: 10.1046/j.1462-2920.2001.00213.x
Harvey, P. C., Watson, M., Hulme, S., Jones, M. A., Lovell, M. A., Berchieri, A. Jr., et al. (2011). Salmonella enterica serovar Typhimurium colonizing the lumen of the chicken intestine are growing slowly and up-regulate a unique set of virulence and metabolism genes. Infect. Immun. 79, 4105–4121. doi: 10.1128/IAI.01390-10
Jessica, M. A. B., Richmond, G. E., Bailey, A. M., Ivens, A., and Piddock, L. J. V. (2013). Choice of bacterial growth medium alters the transcriptome and phenotype of Salmonella enterica serovar Typhimurium. PLoS ONE 8:e63912. doi: 10.1371/journal.pone.0063912
Josenhans, C., and Suerbaum, S. (2002). The role of motility as a virulence factor in bacteria. Intl. J. Med. Microbiol. 291, 605–614. doi: 10.1078/1438-4221-00173
Keller, L. H., Benson, C. E., Krotec, K., and Eckroade, R. J. (1995). Salmonella Enteritidis colonization of the reproductive tract and forming and freshly laid eggs of chickens. Infect. Immun. 63, 2443–2449.
Kofoid, E., Rappleye, C., Stojiljkovic, I., and Roth, J. (1999). The 17-gene ethanolamine (eut) operon of Salmonella Typhimurium encodes five homologues of carboxysome cell proteins. J. Bacteriol. 181, 5317–5329.
Kollanoor Johny, A., Darre, M. J., Donoghue, A. M., Donoghue, D. J., and Venkitanarayanan, K. (2010). Antibacterial effect of trans-cinnamaldehyde, eugenol, thymol and carvacrol against Salmonella Enteritidis and Campylobacter jejuni in vitro. J. Appl. Poult. Res. 19, 237–244. doi: 10.3382/japr.2010-00181
Kollanoor Johny, A., Darre, M. J., Hoagland, T. A., Schreiber, D. T., Donoghue, A. M., Donoghue, D. J., et al. (2008). Antibacterial Effect of Trans-cinnamaldehyde on Salmonella Enteritidis and Campylobacter jejuni in chicken drinking water. J. Appl. Poult. Res. 17, 490–497. doi: 10.3382/japr.2008-00051
Kollanoor Johny, A., Upadhyay, A., Upadhyaya, I., Baskaran, S. A., Moyoottu, S., Michael, D., et al. (2012a). Effect of therapeutic cinnamaldehyde and eugenol on Salmonella Enteritidis in market age broiler chicken. J. Appl. Poult. Res. 21, 816–822. doi: 10.3382/japr.2012-00540
Kollanoor Johny, A., Mattson, T., Baskaran, S. A., Amalaradjou, M. A. R., Babapoor, S., March, B., et al. (2012b). Reduction of Salmonella enterica serovar Enteritidis colonization in 20-day-old broiler chickens by the plant-derived compounds trans-cinnamaldehyde and eugenol. Appl. Environ. Microbiol. 78, 2981–2987. doi: 10.1128/AEM.07643-11
Kollanoor Johny, A., Mattson, T., Baskaran, S. A., Amalaradjou, M. A., Hoagland, T. A., Darre, M. J., et al. (2012c). Caprylic acid reduces Salmonella Enteritidis populations in various segments of digestive tract and internal organs of 3- and 6-week-old broiler chickens, therapeutically. Poult. Sci. 91, 1686–1694. doi: 10.3382/ps.2011-01716
Kubori, T., Sukhan, A., Aizawa, S. I., and Galan, J. E. (2000). Molecular characterization and assembly of the needle complex of the Salmonella Typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. U.S.A. 97, 10225–10230. doi: 10.1073/pnas.170128997
Kutsukake, K., Ohya, Y., and Iino, T. (1990). Trancriptional analysis of the flagellar regulon of S. Typhimurium. J. Bacteriol. 172, 741–747. doi: 10.1128/jb.172.2.741-747.1990
Ledeboer, N. A., Frye, J. G., McClelland, M., and Jones, B. D. (2006). Salmonella enterica serovar Typhimurium requires the Lpf, Pef, and Tafi fimbriae for biofilm formation on HEp-2 tissue culture cells and chicken intestinal epithelium. Infect. Immun. 74, 3156–3169. doi: 10.1128/IAI.01428-05
Marcus, R., Varma, J. K., Medus, C., Boothe, E. J., Anderson, B. J., Crume, T., et al. (2007). Re-assessment of risk factors for sporadic Salmonella serotype Enteritidis infections: a case-control study in five FoodNet Sites, 2002-2003. Epidemiol. Infect. 135, 84–92. doi: 10.1017/S0950268806006558
McCormick, B. A. (2004). “Invasion mechanisms of Salmonella,” in Bacterial Invasion of Host Cells, 1st Edn., ed R. J. Lamont (New York, NY: Cambridge University Press), 328.
Methner, U., Barrow, P. A., Gregorova, D., and Rychlik, I. (2004). Intestinal colonization inhibition and virulence of Salmonella phoP, rpoS and ompC deletion mutants in chickens. Vet. Microbiol. 98, 37–43. doi: 10.1016/j.vetmic.2003.10.019
Porwollik, S., Frye, J., Florea, L. D., Blackmer, F., and McClelland, M. (2003). A non-redundant microarray of genes for two related bacteria. Nucleic Acids Res. 31, 1869–1876. doi: 10.1093/nar/gkg298
Price-Carter, M., Thingey, J., Bobik, T. A., and Roth, J. R. (2001). The alternative electron acceptor tetrathionate supports B12 dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1, 2–propanediol. J. Bacteriol. 183, 2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001
Rodriguez-Morales, O., Fernandez-Mora, M., Hernandez-Lucas, I., Vazquez, A., Puente, J. L., and Calva, E. (2006). S. Typhimurium ompS1 and ompS2 mutants are attenuated for virulence in mice. Infect. Immun. 74, 1398–1402. doi: 10.1128/IAI.74.2.1398-1402.2006
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Soutourina, O. A., and Bertin, P. N. (2003). Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol. Rev. 27, 505–523. doi: 10.1016/S0168-6445(03)00064-0
Suhr, K., and Nielsen, P. V. (2003). Antifungal activity of essential oils evaluated by two different application techniques against rye bread spoilage fungi. J. Appl. Microbiol. 94, 665–674. doi: 10.1046/j.1365-2672.2003.01896.x
Upadhyay, A., Upadhyaya, I., Kollanoor-Johny, A., and Venkitanarayanan, K. (2013). Antibiofilm effect of plant derived antimicrobials on Listeria monocytogenes. Food Microbiol. 36, 79–89. doi: 10.1016/j.fm.2013.04.010
Upadhyaya, I., Upadhyay, A., Kollanoor-Johny, A., Mooyottu, S., Baskaran, S. A., Yin, H.-B., et al. (2015). In-Feed Supplementation of trans-cinnamaldehyde reduces layer-chicken egg-borne transmission of Salmonella enterica serovar enteritidis. Appl. Environ. Microbiol. 81, 2985–2994. doi: 10.1128/AEM.03809-14
Keywords: plant-derived, trans-cinnamaldehyde, eugenol, Salmonella Enteritidis PT8, microarray, antibacterial
Citation: Kollanoor Johny A, Frye JG, Donoghue A, Donoghue DJ, Porwollik S, McClelland M and Venkitanarayanan K (2017) Gene Expression Response of Salmonella enterica Serotype Enteritidis Phage Type 8 to Subinhibitory Concentrations of the Plant-Derived Compounds Trans-Cinnamaldehyde and Eugenol. Front. Microbiol. 8:1828. doi: 10.3389/fmicb.2017.01828
Received: 16 May 2017; Accepted: 06 September 2017;
Published: 26 September 2017.
Edited by:
Maria Schirone, Università di Teramo, ItalyReviewed by:
Alberto Quesada, University of Extremadura, SpainHéctor Argüello, University of Córdoba, Colombia
Copyright © 2017 Kollanoor Johny, Frye, Donoghue, Donoghue, Porwollik, McClelland and Venkitanarayanan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anup Kollanoor Johny, YW51cGpvaG5AdW1uLmVkdQ==
 Anup Kollanoor Johny
Anup Kollanoor Johny Jonathan G. Frye
Jonathan G. Frye Annie Donoghue3
Annie Donoghue3 Dan J. Donoghue
Dan J. Donoghue Kumar Venkitanarayanan
Kumar Venkitanarayanan