- Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, Galveston, TX, United States
Mosquito-borne flaviviruses are important human pathogens that represent global threats to human health. The genomes of these positive-strand RNA viruses have been shown to be substrates of both viral and cellular methyltransferases. N7-methylation of the 5′ cap structure is essential for infection whereas 2′-O-methylation of the penultimate nucleotide is required for evasion of host innate immunity. N6-methylation of internal adenosine nucleotides has also been shown to impact flavivirus infection. Here, I summarize recent progress made in understanding roles for methylation in the flavivirus life-cycle and discuss relevant emerging hypotheses.
Introduction
The genomes of mosquito-borne flaviruses are complex, multi-functional RNA molecules that must be translated, replicated and packaged in the face of innate host defenses to accomplish the ultimate viral goal: production of infectious particles to initiate new rounds of infection. Viral genomes must interface with viral proteins and host machinery to accomplish these critical tasks. Such interactions are specified by RNA features within viral genomes, including sequences and secondary/tertiary structures, and trans-acting factors that recognize these cis-acting features (Campos et al., 2017). In addition to RNA sequence and structure, covalent modifications of individual nucleotides represent another layer of cis-acting features that have been shown to impact RNA function (Saletore et al., 2012). An RNA modification fundamental to flavivirus infection is methylation, as evidenced by the existence of virus-encoded RNA methyltransferases (MTase) (Dong et al., 2014). Moreover, a few recent studies implicate flavivirus genomes to be functionally methylated by host enzymes. In this review I summarize the current state of knowledge of flavivirus RNA methylation as well-effects of RNA methylation on flavivirus infection.
Multiple flaviviruses transmitted by arthropods represent serious human health concerns. These include yellow fever virus (YFV), West Nile virus (WNV), Zika virus (ZIKV), Japanese encephalitis virus (JEV) and the four serotypes of dengue viruses (DENV) which are the most prevalent, causing nearly 100 million symptomatic infections world-wide (Bhatt et al., 2013). These viruses, comprising part of the flavivirus genus, belong to the Flaviviridae which includes the significant blood-borne human pathogen within the hepacivirus genus, hepatitis C virus (HCV). The genomes of viruses within this family share a similar organization: each contains a single open reading frame flanked by untranslated regions (UTRs) of various sequence, length and structure. The viral UTRs contain functional RNA elements that control viral translation and RNA synthesis (Garcia-Blanco et al., 2016). Unique to members of the flavivirus genus is the presence of a so called “cap” structure at the 5′ end of the genome. As discussed in detail below, methylation of the cap structure and the adjacent penultimate nucleotide of the viral genome critically promotes virus infection by multiple mechanisms. In contrast, HCV, the most prominent member of the hepacivirus genus, is characterized by an uncapped genome that contains an internal ribosome entry site within the 5′ UTR (Tsukiyama-Kohara et al., 1992).
At the level of the individual cell all Flaviviridae use a fundamental infection strategy: (i) virus particles attach to various cellular receptors and are internalized via endocytosis, (ii) endosome acidification causes fusion between the viral envelope and endosomal membrane allowing for escape of the viral nucleocapsid into the cytoplasm, (iii) the viral RNA dissociates from capsid and engages the translational machinery to synthesize viral proteins at the cytosolic face of the endoplasmic reticulum (ER), (iv) viral proteins engage the positive-strand genome to synthesize a negative strand intermediate, (v) the negative-strand asymmetrically templates the synthesis of many genomes, (vi) some of which associate with viral structural proteins and bud into the ER to form immature viral particles that (vii) transit through the golgi apparatus where they are modified by host enzymes, and finally, (viii) mature virions are secreted into the extracellular space. Note that multiple phases of the life-cycle, including translation, RNA synthesis and virus assembly, occur concurrently on separate genomes once infection is established although completion of each process is a prerequisite for the following to occur.
Roles for Methylation At the 5′ End of the Flavivirus Genome
Cellular mRNAs are modified in the nucleus with a 7-methylguanosine (m7GpppN) cap structure attached to the first base of the transcript via a 5′-5′ triphosphate linker (Figure 1; Shatkin, 1976). This occurs early after the initiation of transcription via recruitment and sequential action of capping enzymes, including an RNA triphosphatase, guanylyltransferase and N7-guanine MTase, to the C-terminal domain of elongating RNA pol II (Phatnani and Greenleaf, 2006). The RNA triphosphatase acts to remove the γ-phosphate from the 5′ nucleotide of the nascent RNA making it available for cap addition by guanylyltransferase. Methylation of the guanosine cap at N7 completes the reaction to generate a so called “type 0” cap structure (Wei C. M. et al., 1975). Importantly, in higher eukaryotic organisms the mRNA is further modified by a separate ribose MTase at the penultimate nucleotide with a 2′-O-methyl group (Figure 1; type 1 cap) and to a lesser extent at the following nucleotide (type 2 cap) (Wei and Moss, 1975). The 5′ cap structure impacts every aspect of mRNA metabolism, including splicing, nuclear export, translation, and decay (Cowling, 2010). In contrast, 2′-O-methylation is a mark that signifies an mRNA as a “self” versus foreign molecule.
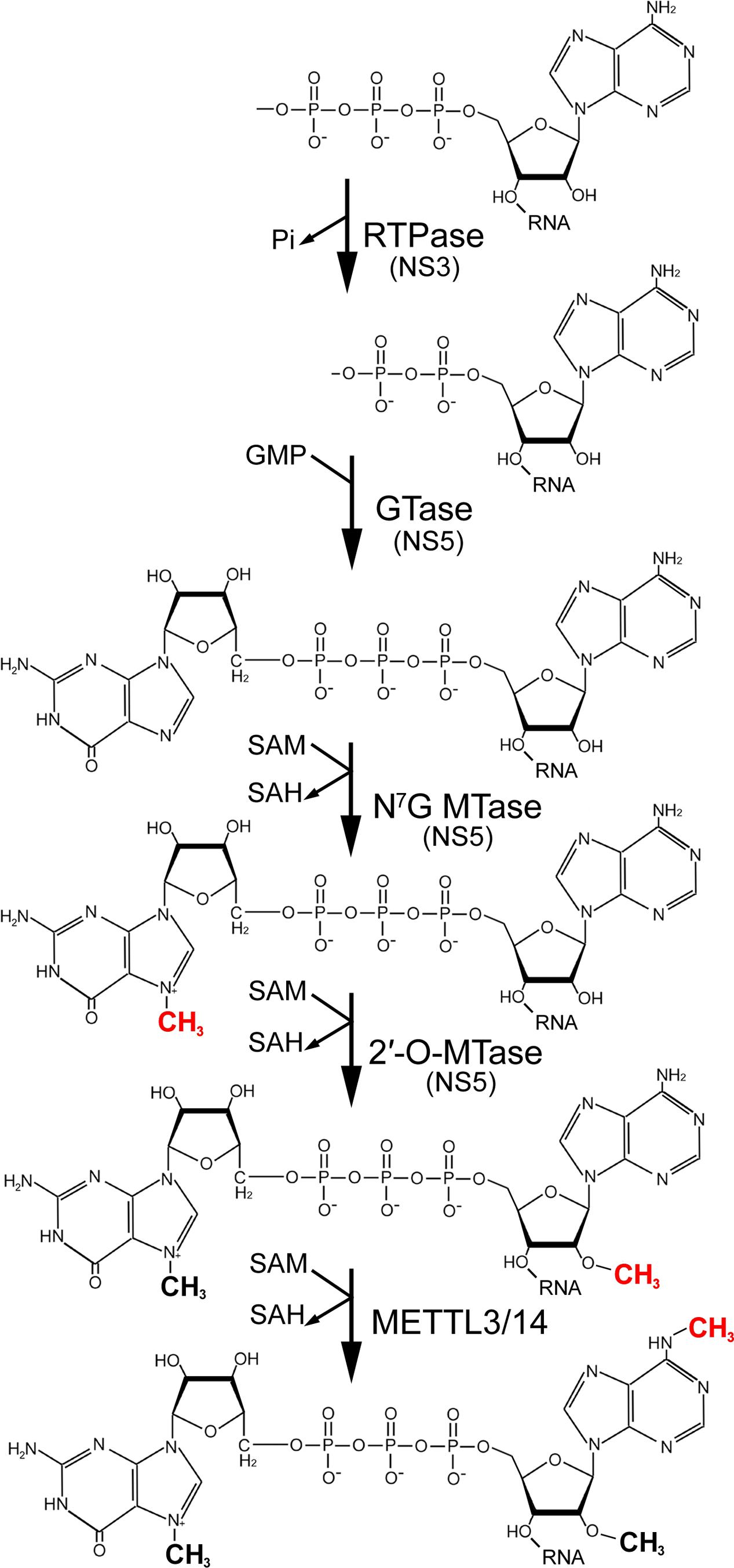
FIGURE 1. Depiction of the flavivirus RNA capping and methylation pathway. Nascent flavivirus genomes initiate with a 5′ triphosphorylated adenosine that is dephosphorylated by the RNA triphosphatase (RTPase) activity of NS3. Next, the putative NS5 guanylyltransferase (GTase) attaches guanosine monophosphate (GMP) via a 5′-5′ linkage. NS5 then methylates the guanine N7 position to form the type 0 cap using S-adenosyl methionine (SAM) as a cofactor. Methyl group donation by SAM converts it to S-adenosyl homocysteine (SAH). NS5-mediated 2′-O-methylation of the adenosine nucleotide generates the type I cap structure. Finally, hypothetical m6A methylation of flaviviral RNA at the penultimate adenosine by the METTL3/14 protein complex would result in the formation m6Am.
Flaviviruses do not have access to the nuclear m7G-capping machinery and instead have evolved enzymatic activities to carry out all the necessary steps to generate capped genomes. The NS5 protein, in addition to its essential RNA-dependent RNA-polymerase function, harbors guanylyltransferase and MTase enzymes (Egloff et al., 2002; Ray et al., 2006; Issur et al., 2009). NS3 is similarly multifunctional, capable of protease, helicase and RNA triphosphatase activities (Wengler and Wengler, 1993; Bartelma and Padmanabhan, 2002). The latter of these catalyze the first step in capping: removal of the γ-phosphate from the 5′ adenosine of nascent viral RNAs. NS5 is then believed to cap the RNA and performs sequential methylation reactions to generate (i) m7GpppA (cap-0) and then (ii) m7GpppAm (cap-1) (Ray et al., 2006). The cis- and trans-determinants of putative guanylyltransferase activity have not been well-characterized but the MTase reactions are relatively well-understood. Unlike cellular MTase which is not believed to discriminate among different RNA substrates, the flavivirus MTase exhibits substrate specificity and will not efficiently methylate the cap of non-viral RNAs (Dong et al., 2007). Cap methylation by NS5 requires the second and third genome nucleotides to be GU and also the presence of a 5′ stem loop which is structurally conserved across all flaviviruses (Brinton and Dispoto, 1988). 2′-O-methylation requires the first two nucleotides (AG) and is enhanced by sequence within the first 20 residues of the genome. Notably, m7GpppA-RNA is strongly preferred over GpppA-RNA as a substrate for 2′-O-methylation, explaining the sequential order of 5′ end methylation reactions (Dong et al., 2008).
What are the functional consequences of cap and 2′-O-methylation? Mutational analyses of the NS5 MTase have identified residues that specifically ablate 2′-O-methylation, cap-methylation, or both. Interestingly, loss of 2′-O-methylation can be tolerated whereas cap methylation is essential for infection (Zhou et al., 2007; Dong et al., 2010). In considering why cap methylation is critical, it is worthwhile to consider data from Ray et al. (2006) who measured the effects of GpppA, m7GpppA and m7GpppAm caps on translation and RNA synthesis using the WNV replicon system which encodes the Renilla luciferase (RLuc) reporter. Compared to uncapped (pppA) WNV replicon RNA, the addition of GpppA strongly (∼25-fold) enhanced the accumulation of (RLuc) at 2 h post-transfection. Surprisingly, m7GpppA enhanced replicon RNA translation by only twofold compared to unmethylated GpppA cap, and m7GpppAm did not further increase RLuc expression. Unexpectedly, no differences in replicon RNA levels were detected for the differently capped replicons. By 72 h post-transfection, each of the capped replicon RNAs produced similar RLuc, indicating that methylation is not required for initial negative-strand synthesis as this is a prerequisite for synthesis of downstream positive-strand synthesis and consequent production of RLuc.
There are at least three non-mutually exclusive explanations for a cap methylation requirement by flaviviruses. First, the cap structure itself is known to protect RNA from 5′ to 3′ exonucleases such as Xrn1 (Hsu and Stevens, 1993) and likely plays a significant role in preventing viral RNA decay. However, it is not clear whether cap methylation plays a significant role in stabilizing RNA. Indeed, the human decapping protein Dcp2 cannot act on an capped RNA substrate lacking N7-methylation (Wang et al., 2002), suggesting that methylation actually enables decapping which is a prerequisite for 5′-3′ decay (Wilusz et al., 2001). Nevertheless, it is possible that cap methylation may render flaviviral genomes resistant to cellular RNases by an unknown mechanism.
A role in stimulating viral translation initiation is a plausible explanation for a cap methylation requirement by flaviviruses. The canonical mRNA cap-binding protein, eIF4E, is believed to be essential for translation of most cellular mRNAs through indirect recruitment of the 40S ribosomal subunit and associated initiation factors (Hershey et al., 2012). EIF4E strongly discriminates between m7Gppp and Gppp, and early studies by Shatkin and colleagues demonstrated that methylation enhanced cap-dependent translation (Both et al., 1975; Muthukrishnan et al., 1975). There are, however, a few clues that flavivirus translation initiation may occur via a non-canonical mechanism. First, as noted above cap methylation conferred a relatively small translational advantage to WNV replicon RNAs (Ray et al., 2006). Second, depletion of eIF4E by RNA interference was reported to not affect DENV replication or protein synthesis: whereas DENV translation was reduced by ∼10%, cellular translation was reduced by 60% due to eIF4E knockdown (Edgil et al., 2006). These observations suggest that flaviviruses may use a non-canonical pathway of translation initiation that depends minimally on the presence of a 5′- m7Gppp.
Finally, it is hypothetically possible that cap methylation protects viral genomes from recognition by factor(s) that sense unmethylated cap structures as invading nucleic acid. No such factor has yet been identified but there are many well-defined “pattern recognition receptors” whose tasks are to detect invading non-self nucleic acids and directly or indirectly, through innate immune pathways, antagonize infection (Wu and Chen, 2014). A pertinent example of these factors is the IFIT family of proteins discussed below.
What are the functional consequences of 2′-O-methylation at the penultimate nucleotide? As noted above, loss of this methylation event does not cause virus lethality in contrast to cap methylation. Key insights into this question were made by Diamond and colleagues who observed that a 2′-O-methylation-deficient NS5 mutant (E218A) WNV lacking m7GpppAm was attenuated in immunocompetent mice and primary cells, whereas animals and cells lacking the type I interferon (IFN) receptor (IFNAR-/-) were fully susceptible to infection (Daffis et al., 2010). These authors went on to show that IFN-inducible proteins of the IFIT family (murine IFIT1 and IFIT2) disproportionately restricted WNV lacking 2′-O-methylation compared to WT virus. Some IFIT proteins have been described to inhibit translation via binding and interfering with the function of eIF3 (Hui et al., 2003), a complex of initiation factors that recruit the 40S ribosomal subunit to mRNA via interaction with eIF4G during initiation of translation (Hershey et al., 2012). More recently, several groups have identified human IFIT1 as a protein that binds directly to cap-0 and blocks translation (Kumar et al., 2014; Abbas et al., 2017), presumably by hindering access of eIF4E to the cap structure. This translational suppression coincides with an accelerated innate immune response that compromises infection (Schmid et al., 2015; Chang et al., 2016). Taken together, these reports strongly suggest that 2′-O-methylation of the cap is an epigenetic RNA modification that allows cells to differentiate self versus non-self RNAs via IFIT proteins. Clearly flaviviruses, and indeed many other types of viruses, have evolved mechanisms to evade IFIT-mediated restriction by encoding their own 2′-O-MTases.
Roles for Internal Adenosine Methylation In Infection By Flaviviruses and HCV
It has been recognized for several decades that a prominent modification to cellular mRNA across many diverse organisms is the methylation of adenosine at the N6 position (Desrosiers et al., 1974; Perry and Kelley, 1974; Zhao et al., 2016). This occurs at internal mRNA positions (m6A) and also at the penultimate nucleotide of transcripts that initiate with A (Figure 1). The latter is referred to as m6Am as it is also methylated at the 2′-hydroxyl (Wei C. et al., 1975). In the past few years research on m6A has greatly expanded and multiple studies have addressed roles of m6A in virus infection. Several methyltransferase and demethylase enzymes have been identified as well as proteins that can recognize methyl groups in RNA (Zhao et al., 2016; Meyer and Jaffrey, 2017). These factors are referred to as “writers,” “erasers,” and “readers” of m6A. A key recent innovation is the use of m6A-specific antibodies in RNA-immunoprecipitation to allow transcriptome-wide mapping of m6A locations in RNA molecules (Dominissini et al., 2012; Meyer et al., 2012; Linder et al., 2015). This has enabled identification and functional analysis of m6A sites by mutation of the low complexity consensus motif DRACH (D = G/A/U; R = G/A; H = C/A/U).
To date m6A mapping and some functional analyses have been performed on multiple viruses including influenza A virus (Courtney et al., 2017), human immunodeficiency virus (Kennedy et al., 2016; Lichinchi et al., 2016a; Tirumuru et al., 2016), HCV (Gokhale et al., 2016), YFV (Gokhale et al., 2016), DENV (Gokhale et al., 2016), WNV (Gokhale et al., 2016), and ZIKV (Gokhale et al., 2016; Lichinchi et al., 2016b). Relevant to this discussion are the studies conducted on Flaviviridae by Gokhale et al. (2016) and Lichinchi et al. (2016b) who characterized functional roles of m6A in HCV and ZIKV infection, respectively. To determine the effects of m6A on infection, Gokhale et al. (2016) depleted the key methylase (METTL3 plus its co-factor METTL14) and demethylases (FTO and ALKBH5) by RNA interference and assayed effects on HCV infection. Intriguingly, knockdown of METTL3/14 enhanced infection while FTO depletion correspondingly reduced infection. These results are consistent with an antiviral role for m6A in the HCV life-cycle. Notably, depletion of these enzymes had no effect on HCV translation or RNA synthesis, suggesting a role for m6A in opposing a late stage of infection such as assembly or egress of infectious virus. Consistent with this idea, several known cytosolic reader proteins (YTHDF1-3) suppressed viral titers, co-immunoprecipitated HCV RNA and localized to lipid droplets which are known sites of HCV assembly (Miyanari et al., 2007). Silent mutation of four m6A sites within the envelope coding region enhanced infection, providing further evidence for a restrictive role of m6A in HCV infection. Gokhale et al. (2016) went on to map m6A in the genomes of multiple mosquito-transmitted flaviviruses, including DENV, YFV, WNV, and two divergent strains of ZIKV. Of note, this analysis revealed abundant m6A within the NS5 coding regions of these viruses.
In their companion article to the Gokhale et al. (2016) study, Lichinchi et al. (2016b) and colleagues mapped locations m6A on ZIKV RNA and investigated the roles of readers, writers and erasers in infection. Depletion of METTL3 or METTL14 enhanced ZIKV infection in 293T cells whereas ALKBH5 and, to a lesser extent, FTO knockdown reduced infection. Moreover, YTDHF1/2 expression negatively correlated with ZIKV RNA levels released from infected cells, suggesting antagonism of ZIKV infection by these reader proteins and YTHDF2 in particular. The authors speculated that YTHDF2 may bind to and destabilize ZIKV RNA. Finally, Lichinchi et al. (2016b) reported that ZIKV infection alters the host m6A methylome, implying that gene expression changes caused by infection may be partly due to altered m6A patterns on cellular mRNA.
Many open questions remain in the nascent field addressing roles for m6A in flavivirus infection. What is the mechanism by which m6A inhibits ZIKV infection? Are similar effects and modes of action operating during infection with other flaviviruses? Does m6A impact cellular innate immune responses? How does infection impact functionality of the machinery that regulates the m6A methylome? Does m6A control flavivirus infection in vivo? In the context of HCV, how do YTHDF proteins suppress late stages of the virus life-cycle?
Lichinchi et al. (2016b) observed elevated titer and viral RNA in supernatants of cells with reduced levels of METTL3/METTL14 which implies that m6A modification opposes virus infection. It should be noted that the overall effects on ZIKV production may be considered mild (< ∼2-fold), indicating that m6A acts as a moderate restriction factor for infection in vitro. Nevertheless, the relatively small effects could reflect a viral strategy that is able to counter, in part, otherwise potent restriction by m6A and trans-acting reader proteins. One hypothesis is that the subgenomic flaviviral RNA (sfRNA), a highly stable RNA fragment produced from decay of virus genomes (Pijlman et al., 2008), could act to buffer the negative impact of m6A by sequestering YTHDF reader proteins. It would be of interest, for example, to test whether strains of DENV that produce different amounts of sfRNA would be differentially susceptible to inhibition by m6A (Manokaran et al., 2015). Of course, this is only one hypothesis and there remains much work to be done to gain a thorough understanding of how flavivirus infections are affected by m6A.
Conclusion
Flavivirus RNA methylation critically impacts infection. Cap methylation (m7GpppA) is essential for infection, at least in part due to its role in stimulating virus translation. Methylation at the 2′-hydroxyl of the penultimate adenosine (m7GpppAm) is inessential for viability but allows the virus to escape the inhibitory actions of IFIT proteins and likely other factor(s) (Szretter et al., 2012). In contrast, internal m6A modifications are somewhat deleterious to ZIKV and HCV infections in vitro although there is much to be learned regarding roles for m6A in infection. Studies addressing viral RNA methylation are informative with respect to basic virus biology but may also allow development of approaches to control infections by pathogenic flaviruses for which there are no currently available therapeutics. Drugs specifically targeting flavivirus MTase enzymes could be potent antivirals for the treatment of patients with acute infections. Moreover, mutant flaviviruses lacking 2′-O-MTase activity have shown promise as candidate vaccine strains because they are attenuated yet induce robust immunity to heterologous infection with virulent viruses (Li et al., 2013; Züst et al., 2013). Thus, understanding the basic molecular mechanisms of flavivirus biology will hopefully lead to measures that reduce the burden caused by these viruses.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Funding
The author acknowledges Mariano Garcia-Blanco and the University of Texas Medical Branch for support.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The author thanks Gaddiel Galarza-Munoza and Rafael Campos for thoughtful comments.
References
Abbas, Y. M., Laudenbach, B. T., Martínez-Montero, S., Cencic, R., Habjan, M., Pichlmair, A., et al. (2017). Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2′-O methylations. Proc. Natl. Acad. Sci. U.S.A. 114, E2106–E2115. doi: 10.1073/pnas.1612444114
Bartelma, G., and Padmanabhan, R. (2002). Expression, purification, and characterization of the RNA 5′-triphosphatase activity of dengue virus type 2 nonstructural protein 3. Virology 299, 122–132. doi: 10.1006/viro.2002.1504
Bhatt, S., Gething, P. W., Brady, O. J., Messina, J. P., Farlow, A. W., Moyes, C. L., et al. (2013). The global distribution and burden of dengue. Nature 496, 504–507. doi: 10.1038/nature12060
Both, G. W., Banerjee, A. K., and Shatkin, A. J. (1975). Methylation-dependent translation of viral messenger RNAs in vitro. Proc. Natl. Acad. Sci. U.S.A. 72, 1189–1193. doi: 10.1073/pnas.72.3.1189
Brinton, M. A., and Dispoto, J. H. (1988). Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology 162, 290–299. doi: 10.1016/0042-6822(88)90468-0
Campos, R. K., Garcia-Blanco, M. A., and Bradrick, S. S. (2017). Roles of pro-viral host factors in mosquito-borne flavivirus infections. Curr. Top. Microbiol. Immunol. doi: 10.1007/82_2017_26 [Epub ahead of print].
Chang, D. C., Hoang, L. T., Mohamed Naim, A. N., Dong, H., Schreiber, M. J., Hibberd, M. L., et al. (2016). Evasion of early innate immune response by 2’-O-methylation of dengue genomic RNA. Virology 499, 259–266. doi: 10.1016/j.virol.2016.09.022
Courtney, D. G., Kennedy, E. M., Dumm, R. E., Bogerd, H. P., Tsai, K., Heaton, N. S., et al. (2017). Epitranscriptomic enhancement of influenza A virus gene expression and replication. Cell Host Microbe 22, 377.e–386.e. doi: 10.1016/j.chom.2017.08.004
Cowling, V. H. (2010). Regulation of mRNA cap methylation. Biochem. J. 425, 295–302. doi: 10.1042/BJ20091352
Daffis, S., Szretter, K. J., Schriewer, J., Li, J., Youn, S., Errett, J., et al. (2010). 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468, 452–456. doi: 10.1038/nature09489
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 71, 3971–3975. doi: 10.1073/pnas.71.10.3971
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Dong, H., Chang, D. C., Xie, X., Toh, Y. X., Chung, K. Y., Zou, G., et al. (2010). Biochemical and genetic characterization of dengue virus methyltransferase. Virology 405, 568–578. doi: 10.1016/j.virol.2010.06.039
Dong, H., Fink, K., Zust, R., Lim, S. P., Qin, C.-F., and Shi, P.-Y. (2014). Flavivirus RNA methylation. J. Gen. Virol. 95, 763–778. doi: 10.1099/vir.0.062208-0
Dong, H., Ray, D., Ren, S., Zhang, B., Puig-Basagoiti, F., Takagi, Y., et al. (2007). Distinct RNA elements confer specificity to flavivirus RNA cap methylation events. J. Virol. 81, 4412–4421. doi: 10.1128/JVI.02455-06
Dong, H., Ren, S., Li, H., and Shi, P.-Y. (2008). Separate molecules of west nile virus methyltransferase can independently catalyze the N7 and 2′-O methylations of viral RNA cap. Virology 377, 1–6. doi: 10.1016/j.virol.2008.04.026
Edgil, D., Polacek, C., and Harris, E. (2006). Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J. Virol. 80, 2976–2986. doi: 10.1128/JVI.80.6.2976-2986.2006
Egloff, M.-P., Benarroch, D., Selisko, B., Romette, J.-L., and Canard, B. (2002). An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21, 2757–2768. doi: 10.1093/emboj/21.11.2757
Garcia-Blanco, M. A., Vasudevan, S. G., Bradrick, S. S., and Nicchitta, C. (2016). Flavivirus RNA transactions from viral entry to genome replication. Antiviral Res. 134, 244–249. doi: 10.1016/j.antiviral.2016.09.010
Gokhale, N. S., McIntyre, A. B. R., McFadden, M. J., Roder, A. E., Kennedy, E. M., Gandara, J. A., et al. (2016). N6 -methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20, 654–665. doi: 10.1016/j.chom.2016.09.015
Hershey, J. W. B., Sonenberg, N., and Mathews, M. B. (2012). Principles of translational control: an overview. Cold Spring Harb. Perspect. Biol. 4:a011528. doi: 10.1101/cshperspect.a011528
Hsu, C. L., and Stevens, A. (1993). Yeast cells lacking 5′– > 3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13, 4826–4835. doi: 10.1128/MCB.13.8.4826
Hui, D. J., Bhasker, C. R., Merrick, W. C., and Sen, G. C. (2003). Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2⋅GTP⋅Met-tRNAi. J. Biol. Chem. 278, 39477–39482. doi: 10.1074/jbc.M305038200
Issur, M., Geiss, B. J., Bougie, I., Picard-Jean, F., Despins, S., Mayette, J., et al. (2009). The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA 15, 2340–2350. doi: 10.1261/rna.1609709
Kennedy, E. M., Bogerd, H. P., Kornepati, A. V. R., Kang, D., Ghoshal, D., Marshall, J. B., et al. (2016). Posttranscriptional m6 A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 19, 675–685. doi: 10.1016/j.chom.2016.04.002
Kumar, P., Sweeney, T. R., Skabkin, M. A., Skabkina, O. V., Hellen, C. U. T., and Pestova, T. V. (2014). Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5′ppp- mRNAs. Nucleic Acids Res. 42, 3228–3245. doi: 10.1093/nar/gkt1321
Li, S.-H., Dong, H., Li, X.-F., Xie, X., Zhao, H., Deng, Y.-Q., et al. (2013). Rational design of a flavivirus vaccine by abolishing viral RNA 2′-O methylation. J. Virol. 87, 5812–5819. doi: 10.1128/JVI.02806-12
Lichinchi, G., Gao, S., Saletore, Y., Gonzalez, G. M., Bansal, V., Wang, Y., et al. (2016a). Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 1, 16011. doi: 10.1038/nmicrobiol.2016.11
Lichinchi, G., Zhao, B. S., Wu, Y., Lu, Z., Qin, Y., He, C., et al. (2016b). Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe 20, 666–673. doi: 10.1016/j.chom.2016.10.002
Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., and Jaffrey, S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772. doi: 10.1038/nmeth.3453
Manokaran, G., Finol, E., Wang, C., Gunaratne, J., Bahl, J., Ong, E. Z., et al. (2015). Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350, 217–221. doi: 10.1126/science.aab3369
Meyer, K. D., and Jaffrey, S. R. (2017). Rethinking m6 a readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342. doi: 10.1146/annurev-cellbio-100616-060758
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646. doi: 10.1016/j.cell.2012.05.003
Miyanari, Y., Atsuzawa, K., Usuda, N., Watashi, K., Hishiki, T., Zayas, M., et al. (2007). The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9, 1089–1097. doi: 10.1038/ncb1631
Muthukrishnan, S., Both, G. W., Furuichi, Y., and Shatkin, A. J. (1975). 5′-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature 255, 33–37. doi: 10.1038/255033a0
Perry, R. P., and Kelley, D. E. (1974). Existence of methylated messenger RNA in mouse L cells. Cell 1, 37–42. doi: 10.1016/0092-8674(74)90153-6
Phatnani, H. P., and Greenleaf, A. L. (2006). Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20, 2922–2936. doi: 10.1101/gad.1477006
Pijlman, G. P., Funk, A., Kondratieva, N., Leung, J., Torres, S., van der Aa, L., et al. (2008). A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4, 579–591. doi: 10.1016/j.chom.2008.10.007
Ray, D., Shah, A., Tilgner, M., Guo, Y., Zhao, Y., Dong, H., et al. (2006). West nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 80, 8362–8370. doi: 10.1128/JVI.00814-06
Saletore, Y., Meyer, K., Korlach, J., Vilfan, I. D., Jaffrey, S., and Mason, C. E. (2012). The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 13, 175. doi: 10.1186/gb-2012-13-10-175
Schmid, B., Rinas, M., Ruggieri, A., Acosta, E. G., Bartenschlager, M., Reuter, A., et al. (2015). Live cell analysis and mathematical modeling identify determinants of attenuation of dengue virus 2′-O-methylation mutant. PLOS Pathog. 11:e1005345. doi: 10.1371/journal.ppat.1005345
Shatkin, A. J. (1976). Capping of eucaryotic mRNAs. Cell 9, 645–653. doi: 10.1016/0092-8674(76)90128-8
Szretter, K. J., Daniels, B. P., Cho, H., Gainey, M. D., Yokoyama, W. M., Gale, M., et al. (2012). 2′-O methylation of the viral mRNA cap by west nile virus evades ifit1-dependent and -independent mechanisms of host restriction in vivo. PLOS Pathog. 8:e1002698. doi: 10.1371/journal.ppat.1002698
Tirumuru, N., Zhao, B. S., Lu, W., Lu, Z., He, C., and Wu, L. (2016). N6 -methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife 5:e15528. doi: 10.7554/eLife.15528
Tsukiyama-Kohara, K., Iizuka, N., Kohara, M., and Nomoto, A. (1992). Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66, 1476–1483.
Wang, Z., Jiao, X., Carr-Schmid, A., and Kiledjian, M. (2002). The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. U.S.A. 99, 12663–12668. doi: 10.1073/pnas.192445599
Wei, C., Gershowitz, A., and Moss, B. (1975). N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 257, 251–253. doi: 10.1038/257251a0
Wei, C. M., Gershowitz, A., and Moss, B. (1975). Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 4, 379–386. doi: 10.1016/0092-8674(75)90158-0
Wei, C. M., and Moss, B. (1975). Methylated nucleotides block 5′-terminus of vaccinia virus messenger RNA. Proc. Natl. Acad. Sci. U.S.A. 72, 318–322. doi: 10.1073/pnas.72.1.318
Wengler, G., and Wengler, G. (1993). The NS 3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology 197, 265–273. doi: 10.1006/viro.1993.1587
Wilusz, C. J., Wormington, M., and Peltz, S. W. (2001). The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2, 237–246. doi: 10.1038/35067025
Wu, J., and Chen, Z. J. (2014). Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32, 461–488. doi: 10.1146/annurev-immunol-032713-120156
Zhao, B. S., Roundtree, I. A., and He, C. (2016). Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42. doi: 10.1038/nrm.2016.132
Zhou, Y., Ray, D., Zhao, Y., Dong, H., Ren, S., Li, Z., et al. (2007). Structure and function of flavivirus NS5 methyltransferase. J. Virol. 81, 3891–3903. doi: 10.1128/JVI.02704-06
Keywords: methylation, flavivirus, Zika virus, dengue virus, hepatitis C virus
Citation: Bradrick SS (2017) Causes and Consequences of Flavivirus RNA Methylation. Front. Microbiol. 8:2374. doi: 10.3389/fmicb.2017.02374
Received: 06 October 2017; Accepted: 16 November 2017;
Published: 05 December 2017.
Edited by:
Encarna Martinez-Salas, Centro de Biología Molecular Severo Ochoa (CSIC), SpainReviewed by:
Isabelle Imbert, Aix-Marseille University, FranceGraham John Belsham, Technical University of Denmark, Denmark
Copyright © 2017 Bradrick. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shelton S. Bradrick, c3NicmFkcmlAdXRtYi5lZHU=
 Shelton S. Bradrick
Shelton S. Bradrick