- 1Department of Cell Biology and Molecular Genetics, Maryland Pathogen Research Institute, University of Maryland, College Park, College Park, MD, United States
- 2CosmosID Inc., College Park, MD, United States
- 3Maryland Department of Natural Resources, Annapolis, MD, United States
- 4Maryland Department of the Environment, Baltimore, MD, United States
- 5Department of Environmental Science, Louisiana State University, Baton Rouge, LA, United States
- 6Maryland Institute for Applied Environmental Health, University of Maryland, College Park, College Park, MD, United States
- 7Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, United States
- 8Center for Bioinformatics and Computational Biology, University of Maryland, College Park, College Park, MD, United States
Vibrio parahaemolyticus is the leading cause of bacterial gastroenteritis associated with seafood consumption in the United States. Here we investigated the presence of virulence factors and genetic diversity of V. parahaemolyticus isolated from water, oyster, and sediment samples from the Chesapeake Bay, Maryland. Of more than 2,350 presumptive Vibrio collected, more than half were confirmed through PCR as V. parahaemolyticus, with 10 encoding both tdh and trh and 6 encoding only trh. Potentially pathogenic V. parahaemolyticus were then serotyped with O1:KUT and O3:KUT predominant. Furthermore, pulsed-field gel electrophoresis was performed and the constructed dendrogram displayed high diversity, as did results from multiple-locus VNTR analysis. Vibrio parahaemolyticus was readily isolated from Chesapeake Bay waters but was less frequently isolated from oyster and sediment samples collected during this study. Potentially pathogenic V. parahaemolyticus was isolated in fewer numbers and the isolates displayed expansive diversity. Although characteristics of the pathogenic V. parahaemolyticus were highly variable and the percent of pathogenic V. parahaemolyticus detected was low, it is important to note that, pathogenic V. parahaemolyticus are present in the Chesapeake Bay, warranting seafood monitoring to minimize risk of disease for the public, and to reduce the economic burden of V. parahaemolyticus related illness.
Introduction
Vibrio parahaemolyticus, a halophilic Gram-negative bacterium, is both autochthonous to the marine environment and a causative agent of seafood-related illnesses (Alam et al., 2009). First reported in Japan in the 1950s, V. parahaemolyticus has now been recognized as one of the leading causes of seafood-related bacterial gastroenteritis worldwide and accounts for almost 50% of all food poisoning outbreaks in Taiwan, Japan, and Southeast Asia (Martinez-Urtaza et al., 2004; Alam et al., 2009). In the United States, V. parahaemolyticus is the leading cause of seafood-induced bacterial enteritis, typically related to consumption of raw or undercooked seafood (DePaola et al., 2003). This pathogen was first identified in 1971 in Maryland, United States after three outbreaks of 425 gastroenteritis cases, in total, were found to be associated with consumption of improperly cooked crabs (Molenda et al., 1972). Subsequently, sporadic outbreaks have occurred throughout the coastal United States (Letchumanan et al., 2014). According to the Centers for Disease Control and Prevention, infection by V. parahaemolyticus is estimated to have an annual rate of 4,500 cases per year in the United States (DePaola et al., 2003). The nationwide Cholera and Other Vibrio Illness Surveillance (COVIS) system and Foodborne Diseases Active Surveillance Network (FoodNet) have both reported an increase in vibriosis per 100,000 population from 1996 to 2010 (Newton et al., 2012). Similarly, between 2005 and 2013, there have been 326 reported cases of Vibrio related infection in Maryland and of non-cholera related Vibrio infections, 38.9% (n = 129) were traced to V. parahaemolyticus (Agarwal, 2014). Illness caused by V. parahaemolyticus can occur 3–24 h after the consumption of contaminated food and symptoms include diarrhea, nausea, vomiting, abdominal cramps, and low-grade fever (Taniguchi et al., 1985). Despite growing understanding of the occurrence and pathogenicity of V. parahaemolyticus, the burden of V. parahaemolyticus related disease has constantly increased in frequency and range since 2000 (Martinez-Urtaza et al., 2004; Caburlotto et al., 2010). Similarly, in 2013, the USDA estimates that the cost estimate for V. parahaemolyticus related disease is 43 million dollars per year (USDA, 2017).
Vibrio parahaemolyticus is both oxidative and fermentative and occurs naturally in both marine and freshwater environments where it interacts with various marine and estuarine organisms (Alam et al., 2009; Caburlotto et al., 2010). Vibrio species are known to concentrate in the gut of oysters and other filter-feeding bivalves, leading to a higher risk of infection to humans ingesting raw or undercooked seafood (Froelich et al., 2013). Although not the focus of this study, previous studies have detected the occurrence of V. parahaemolyticus in various fish species, prawn, and shrimp (Kagiko et al., 2001; Pal and Das, 2010). Prior studies have demonstrated environmental parameters most closely associated with occurrence and distribution of V. parahaemolyticus are water temperature and salinity (Kagiko et al., 2001; Caburlotto et al., 2010). When environmental conditions are favorable, increased growth of Vibrio species in the water column can lead to increased abundance in filter-feeding bivalves and mollusks. Earlier studies carried out in the Chesapeake Bay region have shown V. parahaemolyticus is rarely isolated when the water temperature is below 15°C (Kaneko and Colwell, 1973; Caburlotto et al., 2010). However, it is hypothesized that Vibrio species can persist in sediment during colder months, and can then be released back into the water column once temperatures are conducive for growth, usually in the late spring and early summer. Since V. parahaemolyticus can persist in estuarine and marine environments year-round, there is a need to determine when the risk of illness, from pathogenic V. parahaemolyticus, is highest.
Despite their abundance in estuarine and marine environments, the vast majority of V. parahaemolyticus isolated from the environment are not pathogenic, whereas the majority isolated from clinical sources are Shinoda and Miyoshi (2006). The two major and most commonly referenced virulence factors for V. parahaemolyticus are thermostable direct hemolysin (tdh) and thermostable direct hemolysin-related hemolysin (trh) (Nishibuchi and Kaper, 1985; Taniguchi et al., 1986; Kagiko et al., 2001; DePaola et al., 2003; Shinoda and Miyoshi, 2006; Alam et al., 2009; Wang et al., 2015). Both trh and tdh have similar hemolytic activity in vitro, both cause the lysis of human erythrocytes (Wang et al., 2015). The tdh gene, which codes for the Kanagawa phenomenon (KP), characterized by β-hemolysis of human erythrocytes, and is typically absent (<1%) in environmental isolates whereas more than 90% of clinical isolates are positive (Martinez-Urtaza et al., 2004; Alam et al., 2009). The KP has been regarded as an important indicator in the identification of the pathogenic and non-pathogenic V. parahaemolyticus strains (Wang et al., 2015).
Kanagawa phenomenon negative, clinical V. parahaemolyticus isolates were discovered to produce a second hemolysin, trh, which unlike tdh, is heat labile but immunologically similar to tdh (Honda and Iida, 1993). In 1997 in Calcutta, an outbreak of V. parahaemolyticus revealed the beginning of a unique serotype, O3:K6, which later became the predominant serotype for V. parahaemolyticus related outbreaks (Nair et al., 2007; Ansede-Bermejo et al., 2010). Pandemic O3:K6 strains carry the tdh but not the trh gene and are generally defined by a positive group-specific PCR (GS-PCR) based on the gene sequences of toxRS and opening reading frame, ORF8, from the f237 phage (Matsumoto et al., 2000). The ORF8 of f237 is claimed to be a specific genetic marker of the pandemic isolates of O3:K6 (Nair et al., 2007).
This study characterized a large number of V. parahaemolyticus isolates (1,304) collected from sampling sites in the Chesapeake Bay over a 3 year period, focusing on trh and/or tdh positive strains to determine potential pathogenicity of V. parahaemolyticus collected from the Chesapeake Bay, and to determine both genomic relatedness and environmental distribution, if any, of potentially pathogenic V. parahaemolyticus.
Materials and Methods
Sample Collection
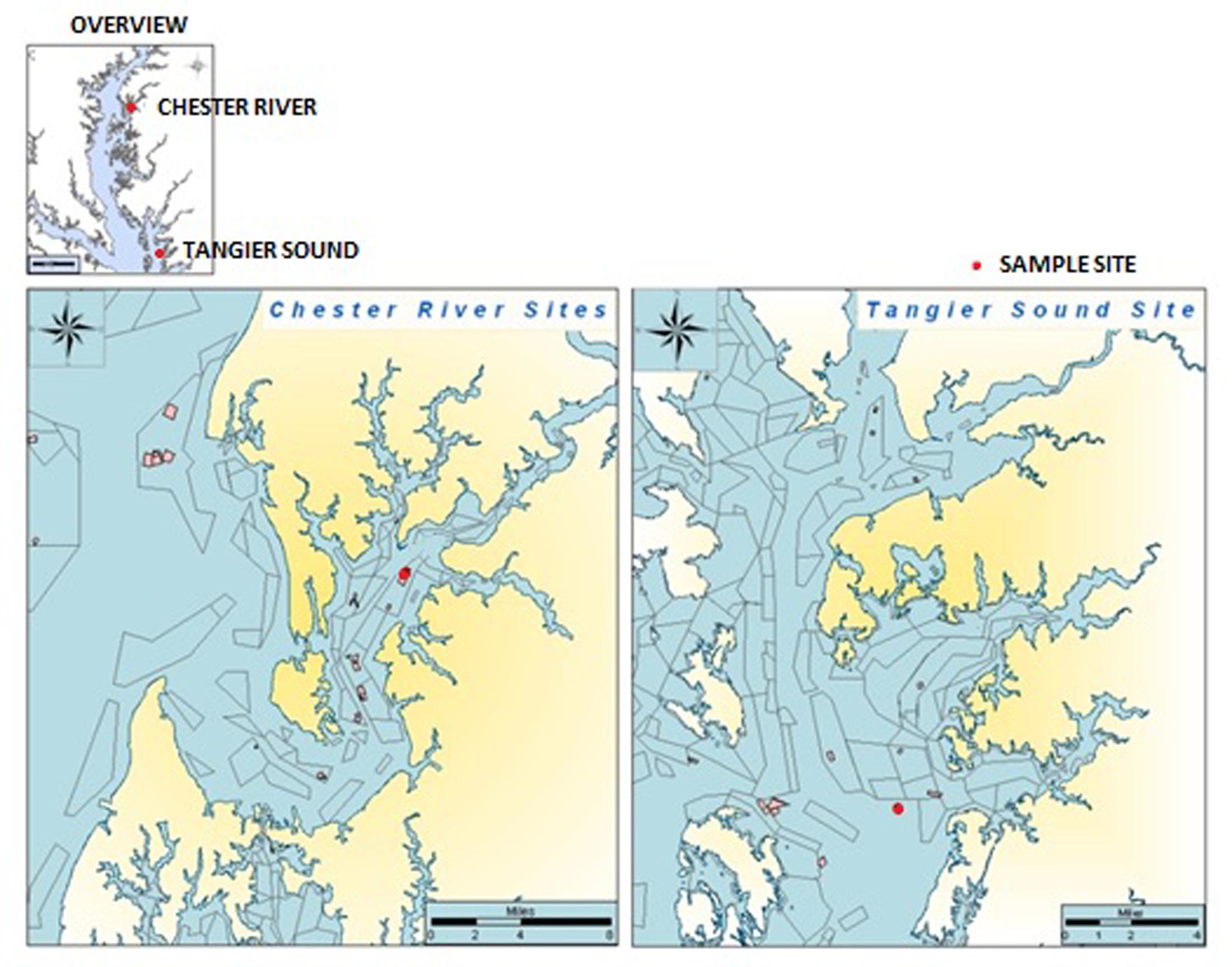
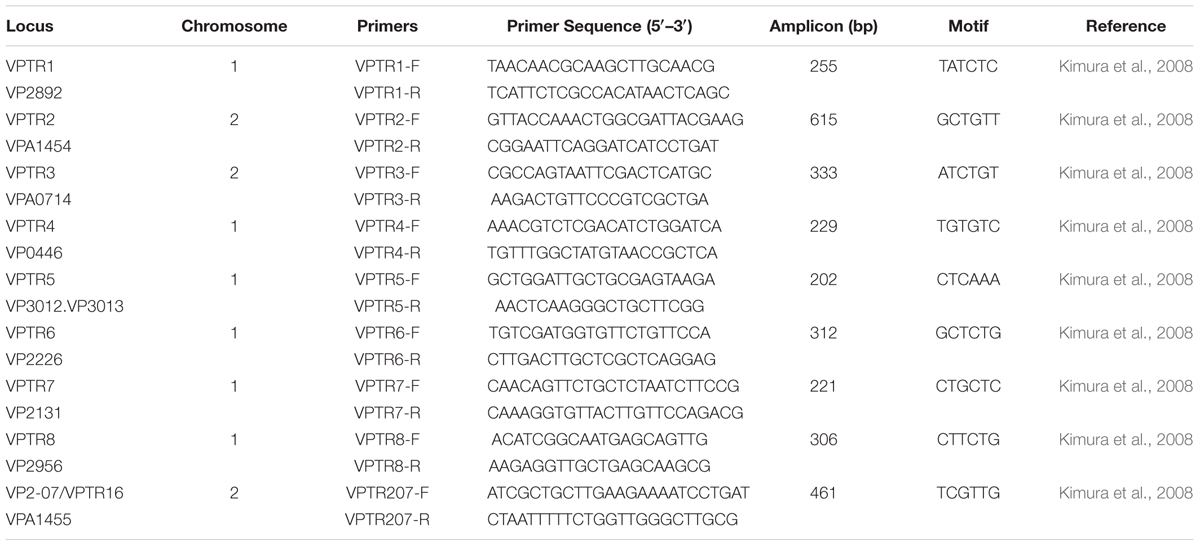
Water, oyster, and sediment samples were collected at two locations in the Chester River (39°05.09′N, 76°09.50′W) and Tangier Sound (38°10.97′N, 75°57.90′W) in the Chesapeake Bay, Maryland from June, 2009 to August, 2012 (Figure 1). During the warmer months of June through August, sampling was done twice each month and once each month during September through May. At each site, 12 liters of surface water, 20–25 oysters, and 80–100 g of sediment were collected. Oysters were collected by dredging. Samples were kept on ice during transport to the University of Maryland, College Park, MD, United States and, upon arrival, stored overnight at 15°C until processing the following morning.
Sample Processing
Details of the sample processing have been described elsewhere (Johnson et al., 2010, 2012). Briefly, water samples were shaken and three volumes (1000, 100, and 10 ml), each in triplicate, were resuspended into alkaline peptone water (10X APW, pH 8.5) (111, 11, and 1.1 ml, respectively) and incubated for 16–18 h, with shaking at 30 rpm. Water was not filtered before resuspension with APW. Oysters were rinsed and scrubbed under running water to remove debris stuck to oyster shells, shucked, and the oyster tissue was homogenized 1:1 with 1X phosphate buffer solution (1X PBS; pH 7.4) in a sterile blender for 90 s. Homogenized oyster tissue was inoculated (10 g, 1 g, 0.1 g, in triplicate) into 10X APW and incubated at 33°C for 16–18 h, with shaking at 30 rpm. Sediment samples were weighed and vortexed in equal part 1X PBS, after which 10X APW was added and the samples incubated at 33°C for 16–18 h, with shaking at 30 rpm. The following day, a loopful of pellicle from each overnight samples were collected along with a loopful of shaken overnight samples and streaked individually onto selective media, including CHROMagarTM (CHROMagar, Springfield, NJ, United States) and thiosulfate citrate bile salts sucrose agar, TCBS (Oxoid, Nepean, ON, Canada). The plates were incubated at 37°C for 16–18 h. Presumptive colonies of V. parahaemolyticus, based on growth media, were picked and streaked onto LB agar (BD Diagnostic Systems, Sparks, MD, United States) to obtain pure cultures.
DNA Extraction and PCR
Presumptive isolates of V. parahaemolyticus were inoculated into LB broth, incubated at 37°C for 16–18 h with shaking at 150 rpm. A 1.5 ml aliquot of inoculum was centrifuged for 10 min at 13G and the supernatant discarded. To each pellet, 700 μl Tris-EDTA Buffer (TE Buffer; pH 8.0) was added and mixed. Cell suspensions were boiled for 10 min at 99°C, after which the samples were cooled before centrifugation for 10 min at 13G. The supernatant was transferred to a clean, sterile tube and adjusted to concentration for PCR analysis. Multiplex PCR targeting the toxR gene (Bauer and Rørvik, 2007) was used to differentiate V. parahaemolyticus, V. vulnificus, and V. cholerae, and to confirm identification of the isolates. Subsequent PCR targeting virulence factors, tlh, trh, and tdh (Bej et al., 1999), was done for all confirmed V. parahaemolyticus strains. PCRs targeting the group-specific toxR variant, GS-PCR, and opening reading frame, ORF8, were performed (Matsumoto et al., 2000). All PCR assays were performed using Promega GoTaq Green Master Mix (Promega, Madison, WI, United States). Each reaction tube contained a total of 25 μl, including 5 μl template DNA. Thermal cycling conditions were as follows: one 10 min cycle of denaturation at 94°C, followed by 36 cycles of denaturation at 94°C for 30 s, annealing temperature for 30 s, extension at 72°C for 60 s, and final extension for 10 min at 72°C. PCR products were stored at 4°C until gel electrophoresis visualization. Sequences, amplicon size and annealing temperatures for each PCR can be found in Tables 1, 2. Positive controls included VPTX2103, VPFIHES98, VPAQ41037, and VPF11-3A. Appropriate negative controls were included in all PCR reactions.
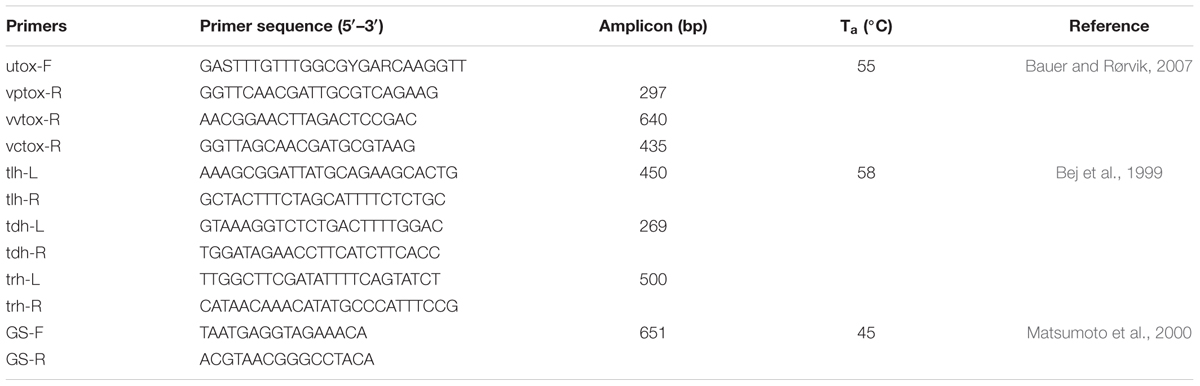
TABLE 1. List of primers, annealing temperatures (Ta), and sequences used to characterize Vibrio parahaemolyticus isolates.
Hemolysis
Cultures of V. parahaemolyticus were grown overnight on LB for 18 h at 37°C, streaked onto 5% sheep blood agar, and incubated at 37°C for 18 h. Green hemolysis was defined as α, β as clear hemolysis, and γ as no hemolysis.
Serotyping
Denken antisera kit containing 13 lipopolysaccharide (O) and 71 capsular (K) sera was used to determine serotypes of pathogenic isolates via slide agglutination. First, V. parahaemolyticus isolates were grown overnight at 37°C on 3% NaCl LB agar. Subsequently, a loopful of culture was mixed with 1 ml of 90% normal saline. Half of the cell suspension was boiled at 99°C for 2 h and used for O serotyping whereas the remaining suspension was used for K serotyping. (Denka; Seiken Corp., Tokyo, Japan).
PFGE
Pulsed-field gel electrophoresis (PFGE) of V. parahaemolyticus DNA was performed using a slight modification of the CDC Pulse-Net protocol created by the CDC (PulseNet United States, 2013), as follows.
Gel Plug Creation and Lysis
Cultures were grown for 16–18 h at 37°C on LB plates and confirmed for purity. A loopful of each broth culture was mixed with 1 ml cell suspension buffer (CSB) (100 mM Tris: 100 mM EDTA, pH 8). The concentration of cell suspension was adjusted to final absorbance of 0.9 ± 0.1 at 610 nm. Half of the cell suspension was incubated with 25 μl of 20 mg/ml Proteinase K for 10 min at room temperature. Following incubation, 500 μl of cell suspension was mixed with an equal volume of 1% SeaKem Gold agarose pre-warmed to 55–60°C. The solution was transferred to a gel plug mold, dispensed to avoid bubbles, and allowed to solidify for 5 min at 4°C. Each plug was transferred to individual 50 ml Falcon tubes. Each tube contained 5 ml cell lysis buffer (CLB) (50 mM Tris: 50 mM EDTA, 1% sarkosyl, pH 8) and 25 μl Proteinase K (20 mg/ml). Tubes containing plugs, CLBr and Proteinase K were incubated in a 54–55°C water bath with shaking at 150 rpm, for 2 h. Plugs were washed twice with 10 ml sterilized ultrapure water previously warmed to 54–55°C, with shaking, and temperature conditions as above, for 10 min. Additional washes with TE Buffer (10 mM Tris: 1 mM EDTA, pH 8) were performed a minimum of four times. Plugs were stored at 4°C with 5 ml sterile TE buffer until digestion was complete.
Digestion and Gel Casting
Vibrio parahaemolyticus isolates were SfiI digested and Salmonella enterica ATCC BAA-664, serving as control, was XbaI digested. Plugs were cut to 2.0 mm wide slices and inserted into individual 1.5 ml Eppendorf tubes containing pre-digestion master mix consisting of 180 μl sterile ultrapure water and 20 μl 10X restriction buffer per plug. Pre-digestion of V. parahaemolyticus was done with incubation at 50°C. S. enterica was incubated at 37°C and after 10 min, the pre-digestion buffer was removed and restriction enzyme master mix added. The restriction enzyme master mix for V. parahaemolyticus contained 177 μl sterile ultrapure water, 20 μl 10X restriction buffer, 2 μl BSA (10 mg/ml), and 1 μl SfiI (40 U/μl) per plug. The restriction enzyme master mix for S. enterica contained 174 μl sterile ultrapure water, 20 μl 10X restriction buffer, 2 μl BSA (10 mg/ml), and 4 μl XbaI (10U/μl) per plug. Plugs were incubated for 4 h at 50°C (V. parahaemolyticus) or 37°C (S. enterica). Following digestion, the restricted enzyme master mix was removed and 200 μl 0.5X TBE was added to each tube and incubated at room temperature for 5 min. Plugs were loaded onto a gel comb, including control plugs of S. enterica. A 1% SeaKem Gold Agarose gel was cast in 0.5X TBE, ensuring plug slices did not move. The agarose gel and plugs were allowed to solidify for at least 30 min and inserted into an electrophoresis chamber containing 4 L freshly prepared 0.5X TBE adjusted to 14°C, with a flow rate of 1 L/min.
CHEF Mapper and Staining
The CHEF Mapper electrophoresis chamber program was set to Auto Algorithm, with a low MW of 78 kb and high MW of 396 kb. After running for 18–19 h, the gel was stained in ethidium bromide (10 mg/ml) and visualized.
Dendrogram Preparation
Restriction patterns were analyzed using BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium). The background was subtracted and the normalized before fingerprint patterns were typed.
DNA Extraction for Sequencing
Vibrio parahaemolyticus isolates were grown overnight in LB broth at 37°C for 16–18 h, with shaking at 150 rpm. A 1.5 ml aliquot of inoculum was centrifuged for 10 min at 13G and the supernatant discarded. DNA was extracted using a Qiagen MiniPrep kit, following the manufacturer’s protocol (Qiagen, Venlo, Limburg).
MLVA
Multiple-locus variable nucleotide tandem repeat (MLVA) analysis was performed for 16 of the tdh+, trh+ V. parahaemolyticus strains, employing nine primer sets belonging to both chromosomes 1 and 2. PCR conditions were identical to those described for conventional PCR. After confirmation by PCR, 25 μl PCR product was purified using DNA Clean and ConcentratorTM-5 (ZymoResearch, Irvine, CA, United States) and mailed for sequencing (Eurofins MWG Operon, Louisville, KY, United States). After sequencing, the number of repeat motifs were counted for each isolate at each individual loci. Primers and repeat motifs for each loci can be found in Table 2.
Results
Water, oyster, and sediment samples collected at sampling stations located in Tangier Sound and the Chester River in the Chesapeake Bay, between June, 2009 and August, 2012, yielded 2,350 presumptive Vibrio isolates, of which 1,304 were confirmed V. parahaemolyticus by toxR targeted multiplex PCR. The remaining isolates were mainly V. vulnificus and V. cholerae. All 1,304 V. parahaemolyticus isolates possessed the species-specific tlh gene. Of all V. parahaemolyticus isolates, 16 (1.2%) were potentially pathogenic, 10 of which (62.7%) contained both of the virulence encoding genes, tdh and trh. Six isolates (37.5%) were positive for trh. The majority of the Chesapeake Bay V. parahaemolyticus strains (83.2%) were isolated from water (whole water, plankton free water, plankton and water), followed by oyster (9.1%), and sediment (7.7%). Of the 16 potentially pathogenic V. parahaemolyticus, none were isolated from oyster, assumed because of limitations associated with relying on culture based methods.
The majority of presumptively pathogenic V. parahaemolyticus strains collected from Tangier Sound were isolated during the colder months of September, December, and January, 2009–2011. In contrast, the presumptively pathogenic V. parahaemolyticus were isolated from the Chester River during the warmer months of May, June, and August, 2009–2010, except for one strain in September, 2010 and two in December, 2009.
Serotyping was performed on all potentially pathogenic V. parahaemolyticus strains and the majority contained O1 antigen, followed by O3 and then O5. Most strains could not be typed for the K antigen using conventional kits and the most frequently occurring serotype was O1:KUT, a serovariant of O3:K6, accounted for 37.5% of strains tested, followed by O3:KUT (18.75%) (Table 3).
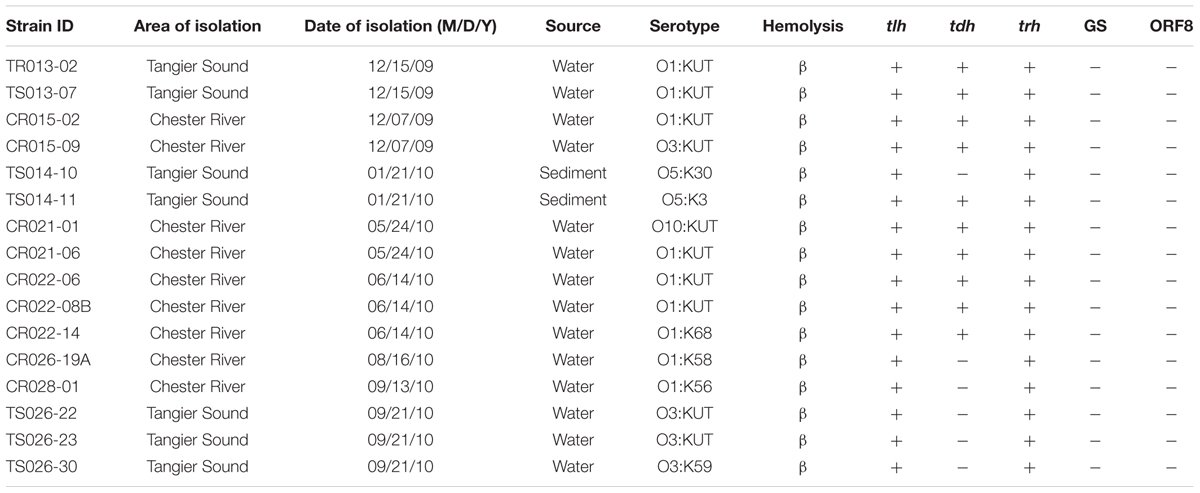
TABLE 3. Characterization of sixteen water and sediment V. parahaemolyticus isolates collected from the Chester River and Tangier Sound, Maryland.
Pulsed-field gel electrophoresis patterns of the 16 potentially pathogenic V. parahaemolyticus showed significant diversity, five falling into a cluster of related strains, but none showing similar banding patterns (Figure 2). None of the strains shared similar MLVA patterns, confirming the diversity detected by PFGE (Tables 2, 4).
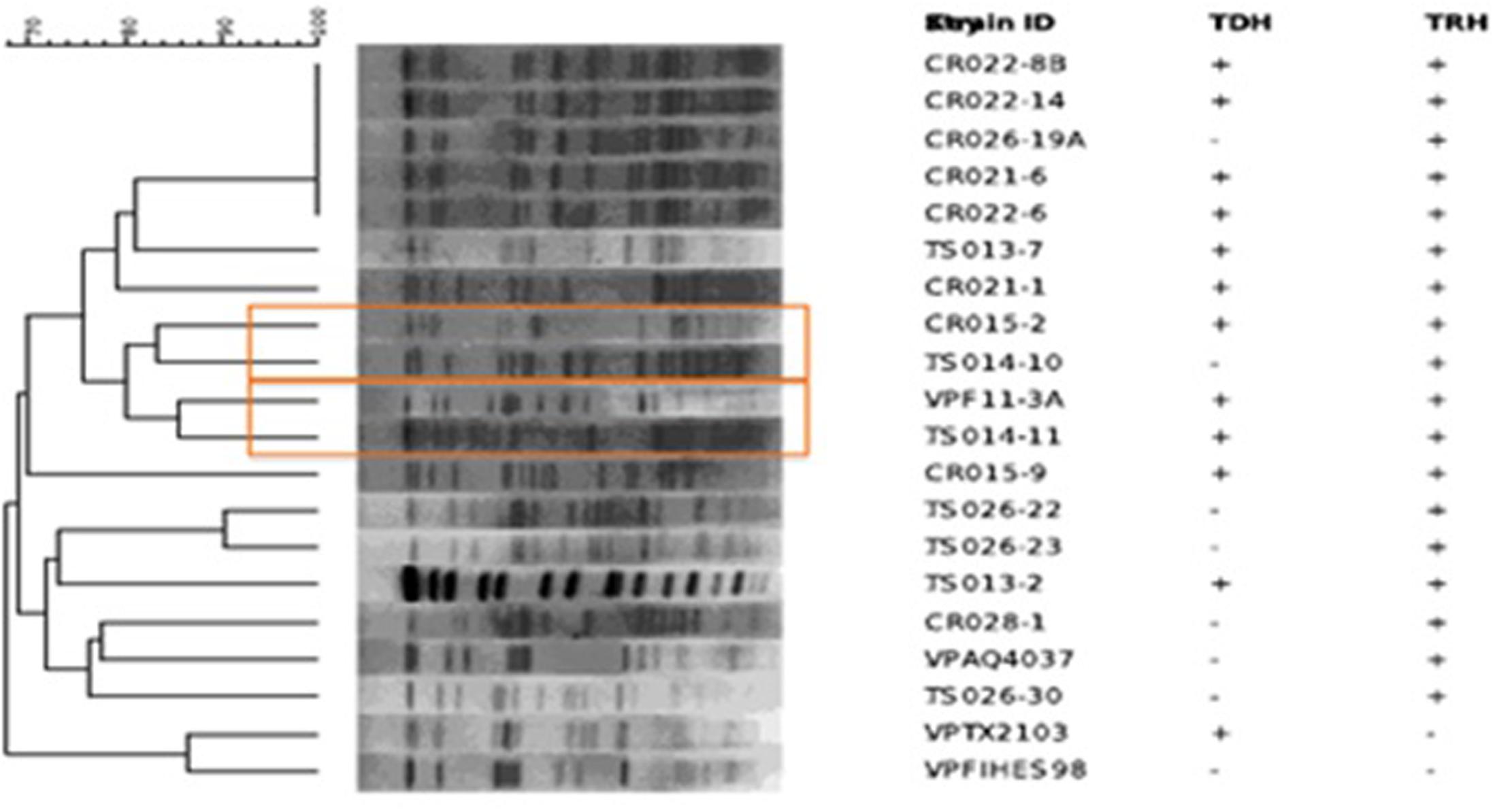
FIGURE 2. Dendrogram created with Bionumerics showing pulsed-field gel electrophoresis (PFGE) patterns of Sfi digested Vibrio parahaemolyticus isolates.
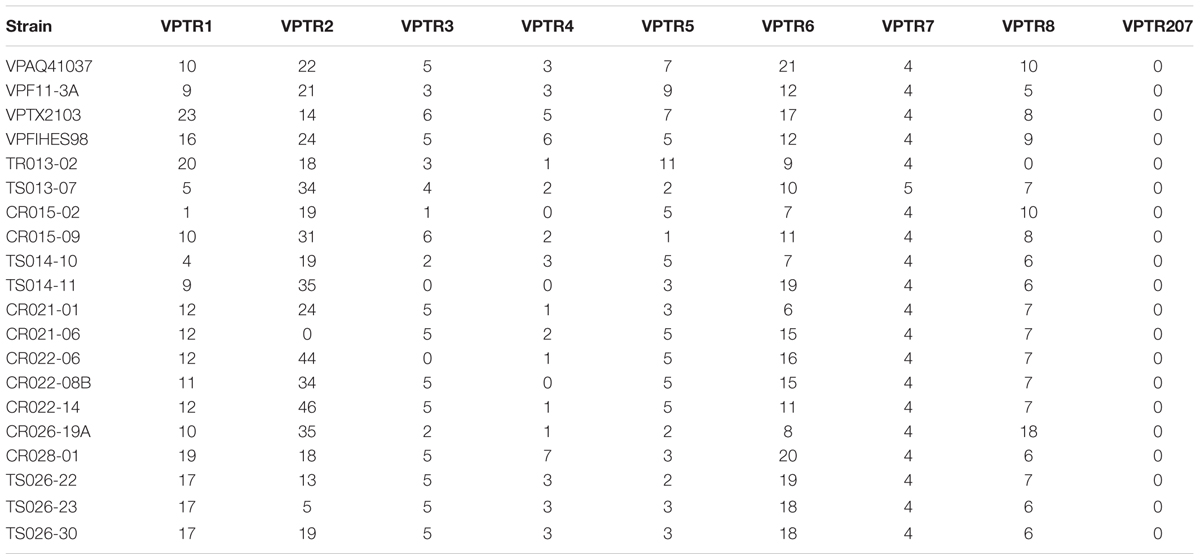
TABLE 4. Number of tandem repeats present in 16 trh and/or tdh positive V. parahaemolyticus isolates and four reference strains included in the study.
Discussion
During the course of this study, V. parahaemolyticus was collected from both locations from all sample types in large numbers. However, out of all V. parahaemolyticus strains characterized for pathogenicity, based on the presence of either trh or tdh, less than 2% were found to be potentially pathogenic. In a study performed by Parveen et al. (2008), by comparable enrichment techniques, all samples were negative for both trh and tdh positive V. parahaemolyticus. However, when real time PCR was performed on the same samples collected by Parveen et al. (2008), detection of tdh and trh positive V. parahaemolyticus increased to 20 and 40%, respectively, for oyster samples and 13 and 40% for water samples. Conversely, in a study performed by Davis et al. (2017), water samples collected from the Chesapeake Bay between 2007 and 2010, were all negative for both trh and tdh during the course of the whole study. Ultimately, the ability to detect pathogenic V. parahaemolyticus is greatly impacted by sample processing techniques.
Similar to previous studies performed in the United States (DePaola et al., 2003), in this study, all 16 tdh+ and/or trh+ were negative for pandemic markers GS and ORF-8 by PCR, indicating V. parahaemolyticus isolates collected during this study are different from pandemic O3:K6 strains (Table 3). V. parahaemolyticus strains negative for GS-PCR are also negative for ORF-8, the marker for the filamentous phage, presumed associated with pandemic genotypes O3:K6 (Bhuiyan et al., 2002; Alam et al., 2009). The majority of the Chesapeake Bay V. parahaemolyticus strains (83.2%) were isolated from water (whole water, plankton free water, plankton and water), followed by oyster (9.1%) and sediment (7.7%). Of the 16 potentially pathogenic V. parahaemolyticus, none were isolated from oyster, assumed because of limitations associated with relying on culture based methods and the ability of V. parahaemolyticus to undergo a VBNC state (viable but non-culturable) (Mizunoe et al., 2000).
The majority of presumptively pathogenic V. parahaemolyticus strains collected from Tangier Sound were isolated during the colder months of September, December, and January, 2009–2011. In contrast, the presumptively pathogenic V. parahaemolyticus were isolated from the Chester River during the warmer months of May, June, and August, 2009–2010, except for one strain in September, 2010 and two in December, 2009 (Table 3). Interestingly, all presumptively pathogenic V. parahaemolyticus were isolated during the first year of the study, with the last of the isolates collected in September 2010, suggesting environmental factors determining temporal changes in the occurrence of these strains of V. parahaemolyticus. However, more importantly and likely, is the fact that environmental strains of pathogenic V. parahaemolyticus are very difficult to isolate (Mizunoe et al., 2000). In a similar study conducted in India, pathogenic V. parahaemolyticus were isolated from 59% of samples after enrichment for 18 h, but the same samples yielded strains negative for tdh when conventional methods followed by PCR were employed (Deepanjali et al., 2005). A study carried out in Japan found 41.5% seawater and 8.5% organic matter samples were positive for tdh and trh when MPN followed by PCR was done but the tdh and/or trh positive strains could not be isolated (Alam et al., 2003). Similarly, in a study performed in the Chesapeake Bay, detection of tdh positive V. parahaemolyticus, was not detected in water samples via direct plating and 55% of the time using enrichment methods (Parveen et al., 2008). Thus, it is concluded that potentially pathogenic strains of V. parahaemolyticus are present in the Chesapeake Bay, but isolation and culture of these strains can remain a challenge.
Vibrio parahaemolyticus associated with disease outbreaks is multi-serogroup, with at least 13 O and 71 K serogroups having been reported (Alam et al., 2009). Commercial kits manufactured in Japan are commonly used to distinguish serogroups (Martinez-Urtaza et al., 2004) and the one most frequently isolated from clinical cases is O3:K6, shown to be the causative agent of a massive outbreak of diarrhea cases in Kolkata, India, in 1996, and later identified in other parts of the world, including Asia, Africa, Europe, Latin America, and the United States (Nair et al., 2007). Results of previous studies have shown that the V. parahaemolyticus O3:K6 serogroup contains the O3:K6 specific filamentous phage f237 and GS sequences of the toxRS operon in addition to ORF-8. These are used as markers to distinguish O3:K6 from other serogroups (Alam et al., 2009). Serotypes O1:KUT, O1:K25, O1:K41, and O4:K68 have been shown to be serovariants of O3:K6 (Martinez-Urtaza et al., 2004). The majority of potentially pathogenic V. parahaemolyticus strains contained O1 antigen, followed by O3 and then O5. Most strains could not be typed for the K antigen using conventional kits and the most frequently occurring serotype was O1:KUT, a serovariant of O3:K6, accounted for 37.5% of strains tested, followed by O3:KUT (18.75%).
In addition to serotyping, a variety of fingerprinting techniques, including PFGE and MLVA, have been used to profile V. parahaemolyticus. Although PFGE is not a new method, few studies have employed PFGE to analyze the diversity of environmental isolates of V. parahaemolyticus, especially with respect to geographic distribution. Previous studies employing PFGE have been done in Japan, Bangladesh, Taiwan, and China (Wong et al., 1996; Suffredini et al., 2011; Wang et al., 2017). Only recently have environmental strains of V. parahaemolyticus from more than one European country been characterized using PFGE (Suffredini et al., 2011). Furthermore, very few V. parahaemolyticus isolates have been in the United States, and specifically within the Chesapeake Bay have been subjected to PFGE analysis, only one study in Texas utilized PFGE analysis for V. parahaemolyticus diversity (Daniels et al., 2000). In this study, a dendrogram constructed using PFGE patterns showed significant diversity among the 16 strains of V. parahaemolyticus isolated in this study, a conclusion also drawn from results of MLVA (Figure 2). Of the 16 potentially pathogenic V. parahaemolyticus environmental strains typed by PFGE, none had identical banding patterns, not surprisingly given past studies showing high genetic diversity among V. parahaemolyticus strains (Alam et al., 2003, 2009). The lack of duplicate banding patterns amongst these strains is important as PFGE is used to determine the ancestry of bacterial strains. Of the 16 strains, only five fell into a cluster of related strains. Interestingly, those five strains had been isolated from the Chester River on three separate days (the first on May 24, 2010, three on June 14, 2010, and the fifth on August 16, 2010). Given the high diversity among all Chesapeake Bay V. parahaemolyticus isolates observed in this study, it is intriguing to note that these five strains formed a related cluster despite having been isolated over a 4 month period. Of the five strains, four were tdh+ and trh+ and the strain last to be isolated, on June 14, 2010, was tdh-, an interesting observation since significant strain divergence was observed among those carrying trh compared to the tdh+ strains. MLVA is a fingerprinting technique to distinguish bacterial strains with little to no genetic variation (Lüdeke et al., 2015). The amplification of polymorphisms are determined in several Variable-Number Tandem-Repeat (VNTR) loci. These VNTRs are highly polymorphic and can be used to differentiate bacterial strains based on the length of repeat regions. None of the V. parahaemolyticus isolates in this study shared similar MLVA patterns, confirming the diversity detected by PFGE (Tables 2, 4). Repeats in the VPTR207 locus were not detected in any of the strains and the least variability of repeat regions was observed at locus VPTR7 of chromosome 1, with most strains carrying four or five repeats. Ultimately, strains of potentially pathogenic V. parahaemolyticus are extremely diverse in regard to location, time and sample type.
Although the occurrence of trh and/or tdh V. parahaemolyticus isolates were relatively uncommon, there are other putative virulence factors that could cause pathogenicity. Previous studies have shown that environmental isolates of V. parahaemolyticus that lacking tdh and/or trh were able to produce putative virulence factors, such as extracellular proteases, biofilm, siderophore, and remained cytotoxic (Mahoney et al., 2010). Ultimately, cytotoxicity and enterotoxicity of pathogenic V. parahaemolyticus cannot be entirely explained by tdh and trh, indicating unknown virulence factor may play a role in pathogenicity (Raghunath, 2014).
Ideally, monitoring Vibrio species in water, sediment, and oysters should provide a good estimate of the actual occurrence of pathogenic V. parahaemolyticus relative to total Vibrio spp., if sufficient sampling is done. However, the requirement for an intensive monitoring regimen, coupled with the difficulty in isolating pathogenic V. parahaemolyticus, and related pathogens, cause environmental surveillance to remain a serious challenge. However, once patterns for the presence of pathogenic V. parahaemolyticus in relation to various environmental parameters, such as temperatures and salinity, are coupled, an effective monitoring program can be provided to guard the public from Vibrio related disease and infection. Not only can active monitoring of Vibrio safeguard the public from disease and infection, ultimately, monitoring of Vibrio can have economic benefits, as USDA estimates that V. parahaemolyticus related disease cost an estimated 23 million dollars per year (USDA, 2017).
In summary, Chesapeake Bay strains of V. parahaemolyticus can carry indicators of pathogenicity and are highly diverse, however, they represent a low proportion of the total population of V. parahaemolyticus in the Chesapeake Bay. These conclusions are in concordance with those reported globally. However, because potentially pathogenic V. parahaemolyticus can be readily isolated from the Chesapeake Bay waters, a monitoring program that include V. parahaemolyticus would be a wise public health program to help reduce the incidence of V. parahaemolyticus related illness.
Author Contributions
AC, NH, BH, ET, MT, and KB were involved in sample collection, processing and/or lab experiments. AH and RC designed the study in the Chesapeake Bay and supervised the project while CJ was a collaborator in the larger project. All authors reviewed and edited the manuscript written by AC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was primarily supported by NSF grant EF-0813285/EF-0813066 as part of the joint NSF-NIH Ecology of Infectious Diseases program. The authors are grateful to the Department of Natural Resources (David White) and the Maryland Department of the Environment (Sarah Harvey, Rusty McKay, and Steve Hiner) for their exceptionally generous support during the study, notably in providing logistical support for sample collection. This work would not have been possible without their assistance.
References
Agarwal, S. (2014). Vibrio Infections Surveillance in Maryland 2005-2013 [1-29]. Available at: https://phpa.health.maryland.gov/OEHFP/EH/SiteAssets/SitePages/harmful-algae-blooms/Vibrio%20Presentation%20SA%20final%20(1).pdf
Alam, M., Chowdhury, W. B., Bhuiyan, N. A., Islam, A., Hasan, N. A., Nair, G. B., et al. (2009). Serogroup, virulence, and genetic traits of Vibrio parahaemolyticus in the estuarine ecosystem of Bangladesh. Appl. Environ. Microbiol. 75, 6268–6274. doi: 10.1128/AEM.00266-09
Alam, M. J., Miyoshi, S., and Shinoda, S. (2003). Studies on pathogenic Vibrio parahaemolyticus during a warm weather season in the Seto Inland Sea, Japan. Environ. Microbiol. 5, 706–710. doi: 10.1046/j.1462-2920.2003.00458.x
Ansede-Bermejo, J., Gavilan, R. G., Triñanes, J., Espejo, R. T., and Martinez-Urtaza, J. (2010). Origins and colonization history of pandemic Vibrio parahaemolyticus in South America. Mol. Ecol. 19, 3924–3937.
Bauer, A., and Rørvik, L. M. (2007). A novel multiplex PCR for the identification of Vibrio parahaemolyticus, Vibrio cholerae and Vibrio vulnificus. Lett. Appl. Microbiol. 45, 371–375. doi: 10.1111/j.1472-765X.2007.02195.x
Bej, A. K., Patterson, D. P., Brasher, C. W., Vickery, M. C., Jones, D. D., and Kaysner, C. A. (1999). Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 36, 215–225. doi: 10.1016/S0167-7012(99)00037-8
Bhuiyan, N. A., Ansaruzzaman, M., Kamruzzaman, M., Alam, K., Chowdhury, N. R., Nishibuchi, M., et al. (2002). Prevalence of the pandemic genotype of Vibrio parahaemolyticus in Dhaka, Bangladesh, and significance of its distribution across different serotypes. J. Clin. Microbiol. 40, 284–286. doi: 10.1128/JCM.40.1.284-286.2002
Caburlotto, G., Haley, B. J., Lleò, M. M., Huq, A., and Colwell, R. R. (2010). Serodiversity and ecological distribution of Vibrio parahaemolyticus in the Venetian Lagoon, Northeast Italy. Environ. Microbiol. 2, 151–157. doi: 10.1111/j.1758-2229.2009.00123.x
Daniels, N. A., Ray, B., Easton, A., Marano, N., Kahn, E., McShan, A. L. II, et al. (2000). Emergence of a new Vibrio parahaemolyticus serotype in raw oysters: a prevention quandary. JAMA 284, 1541–1545. doi: 10.1001/jama.284.12.1541
Davis, B. J. K., Jacobs, J. M., Davis, M. F., Schwab, K., DePaolo, A., and Curriero, F. C. (2017). Environmental determinants of Vibrio parahaemolyticus in the Chesapeake Bay. Appl. Environ. Microbiol. doi: 10.1128/AEM.01147-17 [Epub ahead of print].
Deepanjali, A., Kumar, H. S., and Karunasagar, I. (2005). Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the Southwest Coast of India. Appl. Environ. Microbiol. 71, 3575–3580. doi: 10.1128/AEM.71.7.3575-3580.2005
DePaola, A., Nordstorm, J. L., Bowers, J. C., Wells, J. G., and Cook, D. W. (2003). Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl. Environ. Microbiol. 69, 1521–1526. doi: 10.1128/AEM.69.3.1521-1526.2003
Froelich, B., Ayrapetyan, M., and Oliver, J. D. (2013). Vibrio vulnificus integration into marine aggregates and subsequent uptake by the oyster, Crassostrea virginica. Appl. Environ. Microbiol. 79, 1454–1458. doi: 10.1128/AEM.03095-12
Honda, T., and Iida, T. (1993). The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct haemolysin and related haemolysins. Rev. Med. Microbiol. 4, 106–113. doi: 10.1097/00013542-199304000-00006
Johnson, C. N., Bowers, J. C., Griffitt, K. J., Molina, V., Clostio, R. W., Pei, S., et al. (2012). Ecology of Vibrio parahaemolyticus and Vibrio vulnificus in the coastal and estuarine waters of Louisiana, Maryland, Mississippi, and Washington (United States). Appl. Environ. Microbiol. 78, 7249–7257. doi: 10.1128/AEM.01296-12
Johnson, C. N., Flowers, A. R., Noriea, N. F. III, Zimmerman, A. M., Bowers, J. C., DePaola, A., et al. (2010). Relationships between environmental factors and pathogenic Vibrios in the Northern Gulf of Mexico. Appl. Environ. Microbiol. 76, 7076–7084. doi: 10.1128/AEM.00697-10
Kagiko, M. M., Damiano, W. A., and Kayihura, M. M. (2001). Characterisation of Vibrio parahaemolyticus isolated from fish in Kenya. East Afr. Med. J. 78, 124–127. doi: 10.4314/eamj.v78i3.9076
Kaneko, T., and Colwell, R. R. (1973). Ecology of Vibrio parahaemolyticus in Chesapeake Bay. J. Bacteriol. 113, 24–32.
Kimura, B., Sekine, Y., Takahashi, H., Tanaka, Y., Obata, H., Kai, A., et al. (2008). Multiple-locus variable-number of tandem-repeats analysis distinguishes Vibrio parahaemolyticus pandemic O3:K6 strains. J. Microbiol. Methods 72, 313–320. doi: 10.1016/j.mimet.2007.12.014
Letchumanan, V., Chan, K., and Lee, L. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Lüdeke, C. H., Gonzalez-Escalona, N., Fischer, M., and Jones, J. L. (2015). Examination of clinical and environmental Vibrio parahaemolyticus isolates by multi-locus sequence typing (MLST) and multiple-locus variable-number tandem-repeat analysis (MLVA). Front. Microbiol. 6:564. doi: 10.3389/fmicb.2015.00564
Mahoney, J. C., Gerding, M. J., Jones, S. H., and Whistler, C. A. (2010). Comparison of the pathogenic potentials of environmental and clinical Vibrio parahaemolyticus strains indicates a role for temperature regulation in virulence. Appl. Environ. Microbiol. 76, 7459–7465. doi: 10.1128/AEM.01450-10
Martinez-Urtaza, J., Lozano-Leon, A., DePaola, A., Ishibashi, M., Shimada, K., Nishibuchi, M., et al. (2004). Characterization of pathogenic Vibrio parahaemolyticus isolates from clinical sources in Spain and comparison with Asian and North American pandemic isolates. J. Clin. Microbiol. 42, 4672–4678. doi: 10.1128/JCM.42.10.4672-4678.2004
Matsumoto, C., Okuda, J., Ishibashi, M., Iwanaga, M., Garg, P., Rammamurthy, T., et al. (2000). Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38, 578–585.
Mizunoe, Y., Wai, S. N., Ishikawa, T., Takade, A., and Yoshida, S. (2000). Resuscitation of viable but nonculturable cells of Vibrio parahaemolyticus induced at low temperature under starvation. FEMS Microbiol. Lett. 186, 115–120. doi: 10.1111/j.1574-6968.2000.tb09091.x
Molenda, J. R., Johnson, W. G., Fishbein, M., Wentz, B., Mehlman, I. J., Dadisman, T. A., et al. (1972). Vibrio parahaemolyticus gastroenteritis in Maryland: laboratory aspects. Appl. Microbiol. 24, 444–448.
Nair, G. B., Ramamurthy, T., Bhattacharya, S. K., Dutta, B., Takeda, Y., and Sack, D. A. (2007). Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20, 39–48. doi: 10.1128/CMR.00025-06
Newton, A., Kendall, M., Vugia, D. J., Henao, O. L., and Mahon, B. E. (2012). Increasing rates of vibriosis in the United States, 1996–2010: review of surveillance data from 2 systems. Clin. Infect. Dis. 54, S391–S395. doi: 10.1093/cid/cis243
Nishibuchi, M., and Kaper, J. B. (1985). Nucleotide sequence of the thermostable direct hemolysin gene of Vibrio parahaemolyticus. J. Bacteriol. 162, 558–564.
Pal, D., and Das, N. (2010). Isolation, identification and molecular characterization of Vibrio parahaemolyticus from fish samples in Kolkata. Eur. Rev. Med. Pharmacol. Sci. 14, 545–549.
Parveen, S., Hettiarachchi, K. A., Bowers, J. C., Jones, J. L., Tamplin, M. L., McKay, R., et al. (2008). Seasonal distribution of total and pathogenic Vibrio parahaemolyticus in Chesapeake Bay oysters and waters. Int. J. Food Microbiol. 128, 354–361. doi: 10.1016/j.ijfoodmicro.2008.09.019
PulseNet United States (2013). Standard Operating Procedure for PulseNet PFGE of Vibrio cholera and Vibrio parahaemolyticus. Available at: http://www.cdc.gov/pulsenet/PDF/vibrio_pfge_protocol-508c.pdf
Raghunath, P. (2014). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 5:805. doi: 10.3389/fmicb.2014.00805
Shinoda, S., and Miyoshi, S. (2006). “Hemolysins of Vibrio cholerae and other Vibrio species,” in Comprehensive Sourcebook of Bacterial Protein Toxins, eds J. Alouf and M. Popoff (London: Academic Press).
Suffredini, E., Lopez-Joven, C., Maddalena, L., Croci, L., and Roque, A. (2011). Pulsed-Field Gel Electrophoresis and PCR characterization of environmental Vibrio parahaemolyticus strains of different origins. Appl. Environ. Microbiol. 77, 6301–6304. doi: 10.1128/AEM.00333-11
Taniguchi, H., Hirano, H., Kubomura, S., Higashi, K., and Mizuguchi, Y. (1986). Comparison of the nucleotide sequences of the genes for the thermostable direct hemolysin and the thermolabile hemolysin from Vibrio parahaemolyticus. Microb. Pathogen. 1, 425–432. doi: 10.1016/0882-4010(86)90004-5
Taniguchi, H., Ohta, H., Ogawa, M., and Mizuguchi, Y. (1985). Cloning and expression of Escherichia coli of Vibrio parahaemolyticus thermostable direct hemolysin and thermolabile hemolysin genes. J. Bacteriol. 162, 510–515.
USDA (2017). Cost Estimates of Foodborne Illnesses. Available at: https://www.ers.usda.gov/data-products/cost-estimates-of-foodborneillnesses.aspx
Wang, H., Tang, X., Su, Y. C., Chen, J., and Yan, J. (2017). Characterization of clinical Vibrio parahaemolyticus strains in Zhoushan, China, from 2013 to 2014. PLOS ONE 12:e0180335. doi: 10.1371/journal.pone.0180335
Wang, R., Zhong, Y., Gu, X., Yuan, J., Saeed, A. F., and Wang, S. (2015). The pathogenesis, detection, and prevention of Vibrio parahaemolyticus. Front. Microbiol. 6:144. doi: 10.3389/fmicb.2015.00144
Keywords: Vibrio parahaemolyticus, Chesapeake Bay, pathogenicity, virulence, environment
Citation: Chen AJ, Hasan NA, Haley BJ, Taviani E, Tarnowski M, Brohawn K, Johnson CN, Colwell RR and Huq A (2017) Characterization of Pathogenic Vibrio parahaemolyticus from the Chesapeake Bay, Maryland. Front. Microbiol. 8:2460. doi: 10.3389/fmicb.2017.02460
Received: 27 July 2017; Accepted: 27 November 2017;
Published: 15 December 2017.
Edited by:
David Rodriguez-Lazaro, University of Burgos, SpainReviewed by:
Adrian Canizalez-Roman, Autonomous University of Sinaloa, MexicoLearn-Han Lee, Monash University Malaysia, Malaysia
Copyright © 2017 Chen, Hasan, Haley, Taviani, Tarnowski, Brohawn, Johnson, Colwell and Huq. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anwar Huq, aHVxQHVtZC5lZHU= Rita R. Colwell, cmNvbHdlbGxAdW1pYWNzLnVtZC5lZHU=
†Present address: Bradd J. Haley, United States Department of Agriculture–Agricultural Research Service, Beltsville, MD, United States Elisa Taviani, Sapienza University of Rome, Rome, Italy
 Arlene J. Chen
Arlene J. Chen Nur A. Hasan
Nur A. Hasan Bradd J. Haley
Bradd J. Haley Elisa Taviani1†
Elisa Taviani1† Crystal N. Johnson
Crystal N. Johnson Rita R. Colwell
Rita R. Colwell Anwar Huq
Anwar Huq