- 1Center of Infection and Immunity, The First Hospital, Jilin University, Changchun, China
- 2Key Laboratory of Zoonosis, Ministry of Education, Institute of Zoonosis, College of Veterinary Medicine, Jilin University, Changchun, China
- 3Tianjin International Travel Healthcare Center, Tianjin, China
Sortase A (SrtA)-catalyzed anchorage of surface proteins in most Gram-positive bacteria is indispensable for their virulence, suggesting that this transpeptidase is a promising target for antivirulence therapy. Here, an oligopeptide, LPRDA, was identified as an effective inhibitor of SrtA via virtual screening based on the LPXTG substrate sequence, and it was found to inhibit SrtA activity in vitro and in vivo (IC50 = 10.61 μM) by competitively occupying the active site of SrtA. Further, the oligopeptide treatment had no anti-Staphylococcus aureus activity, but it provided protection against S. aureus-induced mastitis in a mouse model. These findings indicate that the oligopeptide could be used as an effective anti-infective agent for the treatment of infection caused by S. aureus or other Gram-positive bacteria via the targeting of SrtA.
Introduction
Mastitis, or inflammation of the mammary gland, is a common and devastating disease that causes serious economic losses for the dairy industry worldwide (Duarte et al., 2015). The contamination of dairy products with antibiotic residues due to antibiotic therapy seriously affects milk quality and poses public health risks (Addo et al., 2011). Furthermore, antimicrobial therapy for this disease has been increasingly challenging due to drug resistance (Virdis et al., 2010). Among the etiological agents, Staphylococcus aureus is the most important and prevalent contagious mammary pathogen, and it causes other clinical infections ranging in severity from mild to fatal, including dermonecrotic skin infection, pneumonia, sepsis, and endocarditis (de Almeida et al., 2013; Zhang et al., 2015). The remarkable ability of S. aureus to transmit and acquire resistance to currently available antibiotics further amplifies the gravity of infections, including mastitis, caused by this pathogen, especially multi-drug-resistant strains, such as methicillin-resistant S. aureus (MRSA) (Tam et al., 2007; Ciustea et al., 2012). Thus, novel antibacterial agents for the treatment of infections, including mastitis, caused by antibiotic-resistant S. aureus are urgently needed.
To successfully establish infection, S. aureus produces a vast array of virulence factors, including surface proteins and secreted toxins and enzymes that facilitate bacterial adherence, tissue invasion and destruction, and host defense evasion (Foster et al., 2014; Jusko et al., 2014). Among these processes, bacterial adhesion and invasion mediated by surface proteins (fibronectin-binding proteins, adhesin, protein A, clumping factors, and collagen-binding proteins) are the initial steps that are essential for S. aureus infection (Foster et al., 2014). Additionally, surface proteins play important roles in the evasion of host immune attack (Maresso and Schneewind, 2008). Similar to other Gram-positive pathogens, the anchorage of many virulence-associated surface proteins to the cell wall of S. aureus is catalyzed by the transpeptidase Sortase A (SrtA, encoded by the srtA gene) via a mechanism requiring a C-terminal sorting signal with an LPXTG motif (Maresso and Schneewind, 2008; Bradshaw et al., 2015). SrtA recognizes and cleaves the LPXTG recognition motif between the threonine (T) and glycine (G) residues of the surface proteins and subsequently catalyzes amide bond formation between the resulting surface protein fragment and the Gly5 cross-bridges of the cell wall peptidoglycan (Jonsson et al., 2002; Bradshaw et al., 2015). Previous studies have described a S. aureus SrtA mutant that fails to process and display surface proteins and exhibits abnormal pathogenesis of animal infections (Mazmanian et al., 2000, 2002). Furthermore, the structure of SrtA is highly conserved among Gram-positive pathogens (Suree, 2009). Therefore, SrtA inhibitors may represent new agents with novel strategies that may be used in the battle against bacterial infections.
Several studies have reported that SrtA inhibitors, including natural and synthetic compounds, significantly attenuate S. aureus virulence both in vitro and in vivo (Suree, 2009; Zhang et al., 2014). Although a phosphinic peptidomimetic inhibitor has been demonstrated to inhibit S. aureus SrtA activity (McCafferty et al., 2004), the therapeutic effect of an oligopeptide against S. aureus infection in vivo has not yet been reported. Here, we designed and evaluated several oligopeptide inhibitors of SrtA based on the LPXTG substrate sequence by virtual screening. The oligopeptide LPRDA was chosen for further study due to its relatively high inhibitory activity. The potential mechanism underlying this inhibition was characterized by molecular dynamics (MD) simulations, and the therapeutic effect of the oligopeptide inhibitor against S. aureus virulence was further determined in an animal model of mastitis. This study is the first, to our knowledge, to identify an oligopeptide inhibitor of SrtA with effects against S. aureus mastitis in vivo that does not affect cell viability.
Materials and Methods
Bacterial Strains, Growth Conditions and Reagents
The MRSA strain USA 300 was obtained from the American Type Culture Collection (ATCC, Manassas, VA, United States) and grown at 37°C in tryptic soy broth (TSB). USA 300ΔsrtA was cultured in TSB supplemented with spectinomycin. The modified oligopeptide used in this study was synthesized by Scilight Biotechnology (Beijing, China) and was dissolved in phosphate-buffered solution (PBS). To improve its pharmacokinetics and therapeutic efficacy, the oligopeptide was modified by PEG2000 modification and amide modification at the N- and C-termini, respectively.
Virtual Screening for SrtA Inhibitors
Virtual screening performance was carried out using Autodock vina software. According to previous literature (Zong et al., 2004; Suree et al., 2009), residues of C184, W194, and P197 in SrtA play the key role in the binding of substrate LPXTG with SrtA. Therefore, these residues are used as the binding sites of molecular docking. A grid box of dimensions (15 × 20 × 15) with a spacing of 1 Å was created and centered on the mass center of the ligand. The inhibitors were ranked according to the lowest energy representative from each binding sites.
Molecular modeling was used in this study for the virtual screening of potential inhibitors of SrtA (Gao et al., 2016). First, the initial structure of SrtA was obtained from the 3D X-ray structure (PDB code: 1T2P). To determine the starting structure of the ligand/SrtA complex for MD simulations, a standard docking procedure for a rigid protein and a flexible ligand was performed with AutoDock 4 (Hu et al., 2010). Subsequently, MD simulations of the complex system were performed; details of the computational biology method used have been previously described (Lv et al., 2013; Niu et al., 2013).
Recombinant SrtA Protein Expression and Purification
An Escherichia coli BL21 (DE3) strain harboring a recombinant vector encoding SrtA∆N59 was used, as described previously (Wang et al., 2015c). Cells were cultured overnight at 16°C with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG, Invitrogen, Carlsbad, CA, United States) in TSB containing ampicillin to induce SrtA expression until the OD600 reached 0.6 ∼ 0.8. Then, the cells were harvested by centrifugation at 5,500 × g for 10 min, resuspended in reaction buffer (50 mM Tris-HCl, 5 mM CaCl2, and 150 mM NaCl, pH 7.5) and lysed by sonication. After centrifugation (16,000 × g, 30 min, 4°C), the supernatant was loaded onto a self-packaged GST-affinity column and was then washed with PBS. The bound GST-tagged proteins were incubated with Precision Protease at 4°C overnight prior to elution with reaction buffer. Purified SrtA proteins were concentrated to 10 mM via centrifugal filtration (Millipore, Bedford, MA, United States) and subsequently analyzed by SDS-PAGE.
The procedures followed for expression and purification of the mutant proteins (SrtAN114A, SrtAC184A, and SrtAR197A) were identical to those followed for the wild-type protein (SrtAΔN59).
Site-Directed Mutagenesis
Site-directed mutagenesis for SrtAN114A, SrtAC184A, and SrtAR197A was conducted using a recombinant vector encoding SrtAΔN59 with a QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, United States). The primer pairs for these three mutations were as follows: N114A forward, 5′-GAATCACTAGATGATCAAGCGATTTCAATTGCAGGACAC-3′; N114A reverse, 5′-GTGTCCTGCAATTGAAATCGCTTGATCATCTAGTGATTC-3′; C184A forward, 5′-CAATTAACATTAATTACTGCGGATGATTACAATGAAAAG-3′; C184A reverse, 5′-CTTTTCATTGTAATCATCCGCAGTAATTAATGTTAATTG-3′; R197A forward, 5′-GGCGTTTGGGAAAAAGCGAAAATCTTTGTAGC-3′; and R197A reverse, 5′-GCTACAAAGATTTTCGCTTTTTCCCAAACGCC-3′.
SrtA Activity Inhibition Assay
The inhibitory effect of the oligopeptide against SrtA and its mutants activity was determined as previously described in 96-well black plates (PerkinElmer, Boston, MA, United States) (Wang et al., 2015c). Briefly, 100 μL of purified SrtA (4 mM) was preincubated with 100 μL of increasing concentrations of the oligopeptide (3.125–25 μM) in reaction buffer at 37°C for 20 min. The model fluorescent peptide substrate Dabcyl-QALPTTGEE-Edans (GL Biochem, Shanghai, China) (final concentration of 10 μM) was added to the reaction system, followed by incubation at 37°C for 1 h. Fluorescence intensity was measured at emission and excitation wavelengths of 350 and 520 nm, respectively. The experiments were repeated three times independently for each sample.
Anti-S. aureus Activity of Oligopeptide
The broth microdilution method was used to determine the minimum inhibitory concentration (MIC) of the oligopeptide against the MRSA strain USA 300 according to the NCCLS guideline M31-A2. For the growth curve experiment, an overnight bacterial culture was inoculated in fresh TSB (1:100) with the indicated concentrations of the oligopeptide, and the optical density (OD) at 600 nm was measured for each sample at 1-h intervals.
Construction of srtA Deletion Mutant of USA 300
The srtA gene of S. aureus USA 300 was inactivated by allelic exchange as previously described (Chen et al., 2014). Briefly, two DNA fragments were PCR amplified from the USA 300 genome using the primers Down-srtA-f (GCGGAATTCTCTTTTCATCTTTATCTTTATCGTG) and Down-srtA-r (GCGGGATCCAAATACAAAGATAGGCTCAT), as well as Up-srtA-f (GCGGTCGACCCGTAGAAAAAATATTGAATAA) and Up-srtA-r (GC GCCATGGTAAGAGATGTTAAGCCTACA). A 1.5-kb fragment including the spectinomycin resistance gene was PCR amplified using the primers Spc-f (CATGCCATGGGTTCGTGAATACATGTTATA) and Spc-r (CCGGAATTCGTTTTCTAAAATCTGAT) from a pSET2 plasmid. These three fragments were then mixed, digested with EcoRI and NcoI, and ligated at 4°C for 1 h. Using the primers Up-srtA-f and Down-srtA-r, a 2.4-kb fragment of the ligated product was PCR amplified, digested with BamHI and SalI, inserted into pBT2, and used for allele replacement as previously described. The presence of the mutation was confirmed by PCR sequencing analysis based on the known sequences of USA 300 and its SrtA mutants. The strA-knockout strain showed normal growth in TSB.
Immunofluorescence Assay
Detection of surface protein A by immunofluorescence analysis was performed as described previously (Chen et al., 2014). Briefly, S. aureus were cultured in TSB with or without the oligopeptide at 37°C for 2.5 h, pelleted by centrifugation (3,000 × g, 10 min), washed three times with PBS, blocked with 2% BSA for 1 h at 37°C, stained with fluorescein isothiocyanate (FITC)-labeled goat anti-rabbit IgG for 2 h at 37°C, and observed under a laser scanning confocal microscope (Olympus, Tokyo, Japan).
Fibronectin-Binding Assay
Ninety-six-well flat-bottom polystyrene plates were coated with 100 μL of 2 μg/mL fibronectin in PBS (fibronectin from bovine plasma; Sigma–Aldrich, St. Louis, MO, United States) per well and incubated at 4°C overnight. Each well was washed three times with PBS and blocked with 5% BSA in PBS at 37°C for 2 h. An overnight bacterial culture was inoculated in fresh TSB (1:100, OD600 nm ≈ 0.2) with the indicated concentrations of the oligopeptide and incubated until the OD600 nm reached 0.5. Then, the cells were centrifuged at 5000 × g for 10 min and resuspended in PBS to an OD600 nm of 1.0. One hundred microliters of bacterial suspension were added to each fibronectin-coated plate well as described above, and the plates were incubated at 37°C for 2 h. After removal of the cell suspension, 25% (v/v) formaldehyde was added to fix S. aureus. Then, the bacteria attached to fibronectin were stained with 100 μL of 0.4% crystal violet dye for 20 min at 37°C after being washed once with PBS. Following removal of the supernatant, adherence was quantified by measuring the OD570 nm for each well. USA 300 without the oligopeptide was used as a positive control, and PBS was used as a negative control.
Biofilm Formation Assay
An overnight bacterial culture was standardized to an OD600 nm of 0.01 in TSB supplemented with the indicated concentrations of the oligopeptide. One hundred microliters of S. aureus (USA 300 and its SrtA mutants) suspension were added to the wells of microtiter plates and incubated for 24 h at 37°C. Then, the suspension was removed, and the plates were washed twice with PBS. Adherent biofilms were fixed with 100% ethanol, stained with 0.1% (v/v) crystal violet, washed several times with PBS, and quantitatively assessed by adding 100% ethanol and measuring the OD570 nm for each well. USA 300 without the oligopeptide was used as a positive control, and PBS was used as a negative control.
Invasion Assay
J774 cells were adjusted to 4 × 105 per glass slide in 24-well plates in Dulbecco’s minimum essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and cultured in an incubator at 5% CO2 and 37°C overnight. An overnight bacterial culture was inoculated in fresh TSB (1:100, OD600 nm ≈ 0.2) with the indicated concentrations of the oligopeptide and incubated until the OD600 nm reached 1.0. Then, the bacteria were resuspended in DMEM containing 10% FBS at a concentration of 1 × 108 CFU/mL, and 1 mL of the resuspension was added to each well for infection at 37°C for 1 h. Invasion was terminated by incubation with 300 μg/mL gentamicin for 30 min. After three washes with PBS, coverslips were placed in 0.2% Triton X-100 with vigorous vortexing, and the diluted lysates were plated onto TSB agar plates to enumerate the number of bacteria associated with each sample.
Animal Experiments
BALB/c mice (from the Experimental Animal Center of Jilin University) were obtained and housed under specific pathogen-free conditions. All animal studies were approved by and conducted in accordance with the guidelines of the Animal Welfare and Research Ethics Committee of Jilin University. An overnight bacterial culture was inoculated in fresh TSB broth (1:100, OD600 nm ≈ 0.2) and incubated until the OD600 nm reached 0.8. Cells were harvested by centrifugation at 3,000 × g for 10 min, washed three times with PBS, and resuspended in PBS to a concentration of 1 × 109 CFU/mL for the mastitis infection experiment.
For establishment of a mastitis model of infection, lactating BALB/c mice, 10–12 weeks of age and weighing 30–32 g, were anesthetized using sodium pentobarbital and infected by injecting the canal glands with 50 μL of staphylococcal suspension per breast. The infected mice were administered 50 mg/kg of the oligopeptide subcutaneously at the time of infection and then at 12-h intervals. The control mice received an equal volume of PBS at the same time points. To evaluate the pathological correlates of mastitis, mammary gland tissues isolated from sacrificed mice at 48 h post-infection were fixed in 4% paraformaldehyde, stained with hematoxylin and eosin, and visualized by light microscopy. Furthermore, additional mammary gland tissues from the mice sacrificed at 48 h post-infection were homogenized in PBS containing 2% Triton X-100, after which they were appropriately diluted and inoculated onto TSB agar plates for assessment of the bacterial burden. The levels of the inflammatory cytokines IL-1β, IL-6, and TNF-α in the supernatants were measured using ELISA kits.
Determination of Binding Affinities of Ligands with Proteins
In this study, the fluorescence quenching method was used to measure the binding constants (KA) of ligands to proteins. A 280-nm excitation wavelength with 5-nm band pass and a 345-nm emission wavelength with 10-nm band pass were used for the measurements. Details of the measurement procedures have been described previously (Bandyopadhyay et al., 2002; Jurasekova et al., 2009).
Statistics
The data are presented as the mean ± SD (n ≥ 3). Student’s t-test for paired sample was used to determine statistical significance using SPSS 13.0. P ≤ 0.05 was considered to be statistically significant.
Results
Deletion of srtA in USA 300
To further analyze the role of SrtA in S. aureus virulence, we first inactivated the srtA gene in the S. aureus USA 300 strain by homologous recombination (Figure 1A), as confirmed by PCR sequencing analysis of chromosomal DNA from the S. aureus USA 300 strain and its SrtA mutants (Figure 1B).
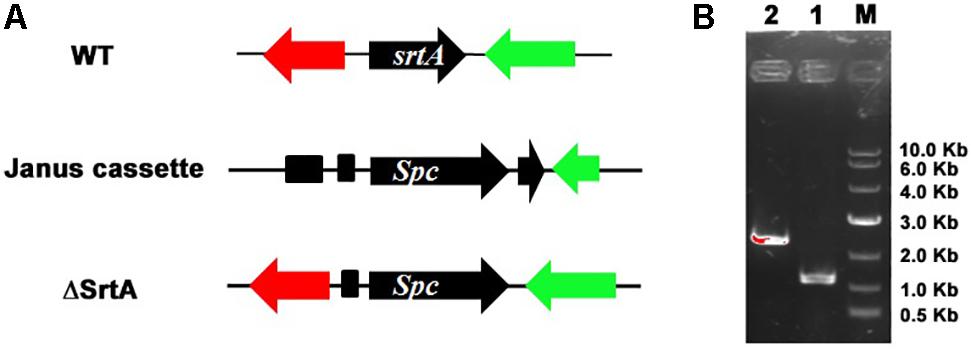
FIGURE 1. Mutation of srtA in Staphylococcus aureus USA 300. (A) The intact srtA gene in S. aureus USA 300 was disrupted by recombination deletion with a Janus cassette. (B) The srtA mutation was confirmed by PCR based on the chromosomal DNA sequences in S. aureus USA 300 and its SrtA mutant strain ΔSrtA. PCR products for the parent strain USA 300 (lane 1) and its mutant strain ΔSrtA (lane 2).
Screening and Validation of SrtA Inhibitors
The docking of a series of ligands, based on the LPXTG substrate sequence, into the SrtA active site was carried out, and their binding affinities were estimated using the Autodock vina package for virtual screening of the SrtA inhibitors. As shown in Table 1, the binding affinity between the oligopeptide LPRDA (Figure 2A) and SrtA was higher than those of all other oligopeptides tested, indicating that this oligopeptide may have the strongest inhibitory activity against SrtA. Thus, this oligopeptide was chosen for further validation of the inhibitory activity in the following experiments. A fluorescence assay was employed to assess the putative inhibition of SrtA activity by the oligopeptide based on the model peptide substrate Dabcyl-QALPTTGEE-Edans. Consistent with the computational screening results, the catalytic activity of the reaction mixture exposed to the oligopeptide was significantly decreased (Figure 2B). Importantly, the concentration required for 50% inhibition (IC50) was 10.61 μM under our experimental conditions (Figure 2B), indicating that this oligopeptide is a relatively highly efficient inhibitor of S. aureus SrtA compared with the inhibitors tested in previous studies (Cegelski et al., 2002; Zhang et al., 2014). Additionally, the MIC of the tested oligopeptide for MRSA USA 300 was greater than 200 μM, and the growth of S. aureus was not visibly affected in the presence of the oligopeptide at concentrations required to inhibit SrtA catalytic activity (3.125–25 μM) (Figure 2C). Taken together, these results indicate that the oligopeptide LPRDA is an effective SrtA inhibitor without anti-S. aureus activity, suggesting that it may be a promising antivirulence candidate for anti-infective therapy.
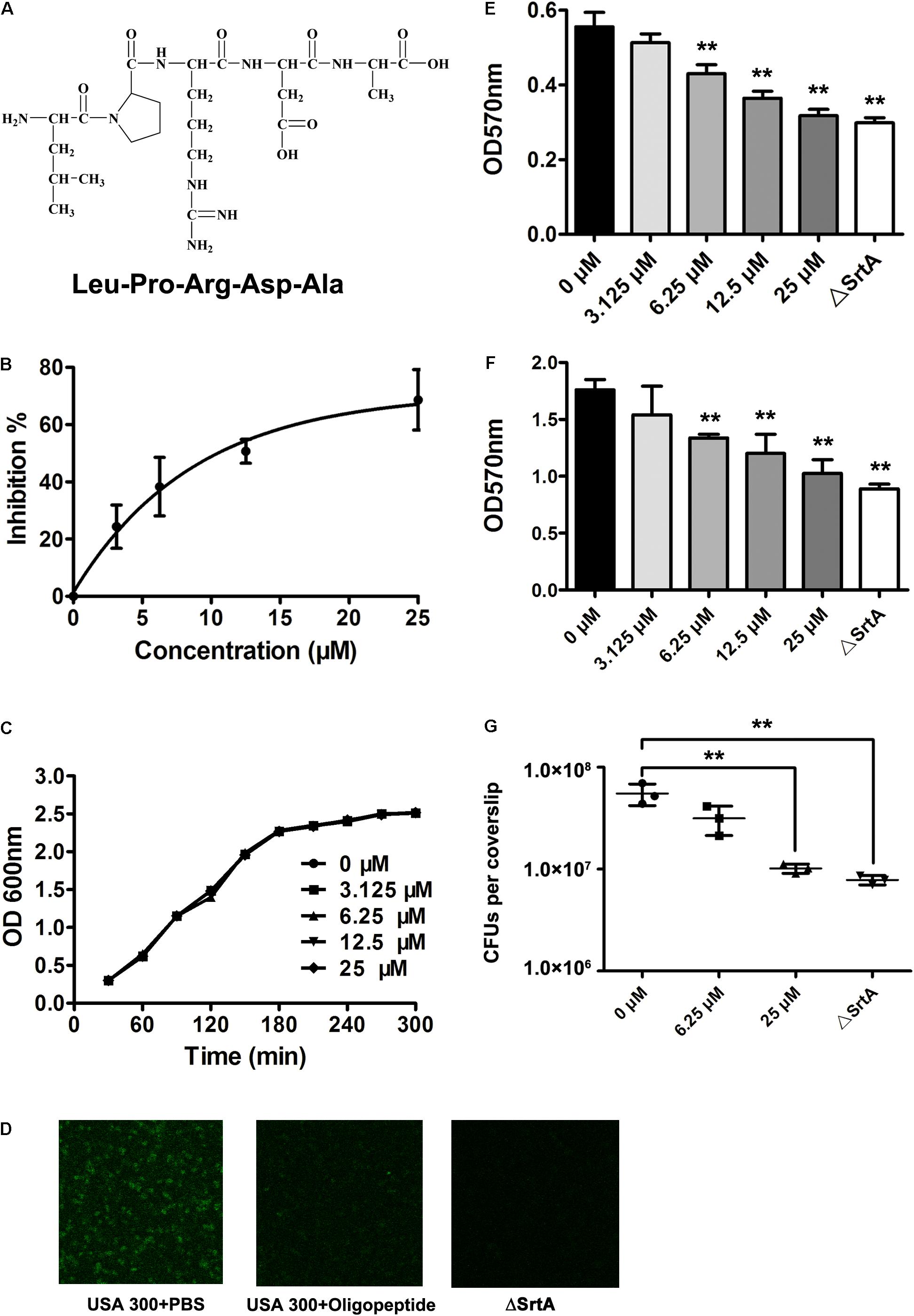
FIGURE 2. Inhibition of SrtA activity both in vitro and in vivo by the oligopeptide. (A) The molecular structure of the oligopeptide, LPRDA. (B) The inhibitory effect of the oligopeptide on SrtA activity. The fluorescent peptide substrate Dabcyl-QALPTTGEE-Edans was mixed with purified SrtA preincubated with various concentrations of the oligopeptide, and the fluorescence intensity of the reaction system was determined at emission and excitation wavelengths of 350 and 520 nm, respectively (n = 3). (C) The growth of S. aureus USA 300 cultured in TSB in the presence of increasing concentrations of the oligopeptide was monitored by measuring the OD at 600 nm every 60 min. Oligopeptide treatment inhibits the binding of S. aureus to fibronectin (D) and the formation of S. aureus biofilm (E) (n = 3) (∗p < 0.05, ∗∗p < 0.01). (F) Inhibition of S. aureus invasion into J774 cells by the oligopeptide. J774 cells were infected with S. aureus at 37°C for 1 h, and the total colony-forming units (CFU) for lysed cells was evaluated (n = 3) (∗p < 0.05, ∗∗p < 0.01). (G) The presence of protein A on the cell envelopes of S. aureus treated with or without the oligopeptide and its SrtA mutant was viewed under a confocal laser scanning microscope following staining with an FITC-conjugated antibody. Results are reported as mean ± SEM.
Oligopeptide Inhibits SrtA Activity in S. aureus
Sortase A has been shown to be a key enzyme that catalyzes the anchoring of surface proteins in most Gram-positive bacteria (Cascioferro et al., 2014). Thus, we hypothesized that treatment with the oligopeptide might inhibit the display of surface proteins on S. aureus and subsequently trigger reduced capacities for adhesion, biofilm formation and invasion of this pathogen. Consistent with previous results, the display of surface protein A was only confirmed for wild-type S. aureus, as demonstrated by its tinctorial strength, and the mutant strain was defective in displaying this surface protein (Figure 2D). However, addition of the oligopeptide significantly hindered the binding of FITC-conjugated IgG to SrtA, indicating effective inhibition of the anchoring of surface protein A by the oligopeptide treatment (Figure 2D). Adhesion and biofilm formation of S. aureus, as mediated by surface proteins, are important for prevention to be eradicated by the host. Thus, fibronectin-binding and biofilm formation assays were employed to assess the effects of the oligopeptide on these processes in S. aureus. In contrast with the weak adherence of the SrtA mutant, USA 300 exhibited strong adherence to bovine fibronectin (Figure 2E). Significantly, treatment of S. aureus with the oligopeptide resulted in decreased adherence to the fibronectin coating on the well bottom (Figure 2E). A similar phenomenon of inhibition was also observed in the biofilm formation assay (Figure 2F), in agreement with the findings of inhibition of SrtA activity and S. aureus fibronectin-binding activity by the oligopeptide. Furthermore, the invasion of S. aureus into J774 cells was examined to determine the potential inhibitory effect of the oligopeptide. As expected, the wild-type S. aureus strain exhibited a strong invasion ability compared with the SrtA mutant strain, and significant anti-invasive activity of S. aureus was observed following treatment with 25 μM oligopeptide (Figure 2G). Taken together, these results indicate that the oligopeptide treatment inhibited SrtA activity in S. aureus and subsequently hindered bacterial adhesion, biofilm formation, and invasion.
Oligopeptide Inhibits S. aureus Virulence in Animal Model of Infection
The inhibition of SrtA activity both in vitro and in S. aureus by the oligopeptide, in addition to the critical role of SrtA in S. aureus virulence, led us to reason that the SrtA inhibitor oligopeptide would also be an effective candidate for suppression of S. aureus infection. Here, the therapeutic effect of the oligopeptide against mice mastitis was evaluated. Following challenge with S. aureus, the breasts of the lactating mice were injected with the oligopeptide or PBS. To evaluate the impact of the oligopeptide treatment, histopathologic analysis of mammary gland tissues at 48 h post-infection was performed. Consistent with a previous study, almost no inflammatory responses or pathological changes were detected in the mice infected with the S. aureus SrtA mutant (Figures 3A,B), while significant inflammatory responses were observed in the mice infected with S. aureus (Figures 3A,B), with destroyed or abnormal mammary glands, dense inflammatory cells, and depletion of epithelial cells in the mammary gland tissues (Figures 3A,B). However, the oligopeptide treatment effectively alleviated the S. aureus-induced inflammatory responses and pathological changes in the infected mice, which exhibited only mild tissue injury (Figures 3A,B), demonstrating a potential prophylactic effect of the oligopeptide against S. aureus-induced mastitis. Further, the bacterial counts in the mammary gland tissues indicated a significant decrease in the bacterial burden in the oligopeptide-treated mice compared with the PBS-treated control mice (Figure 3D). Consistent with these observations, the levels of cytokines, including IL-1β, IL-6, and TNF-α, in the S. aureus-infected mice were markedly increased compared with those in the mice infected with the S. aureus SrtA mutant (Figure 3C), whereas administration of the oligopeptide significantly reduced the S. aureus-mediated increases in the cytokine levels (Figure 3C). Taken together, these results indicated that treatment with the oligopeptide LPRDA conferred effective protection against S. aureus-induced mastitis in the mouse model.
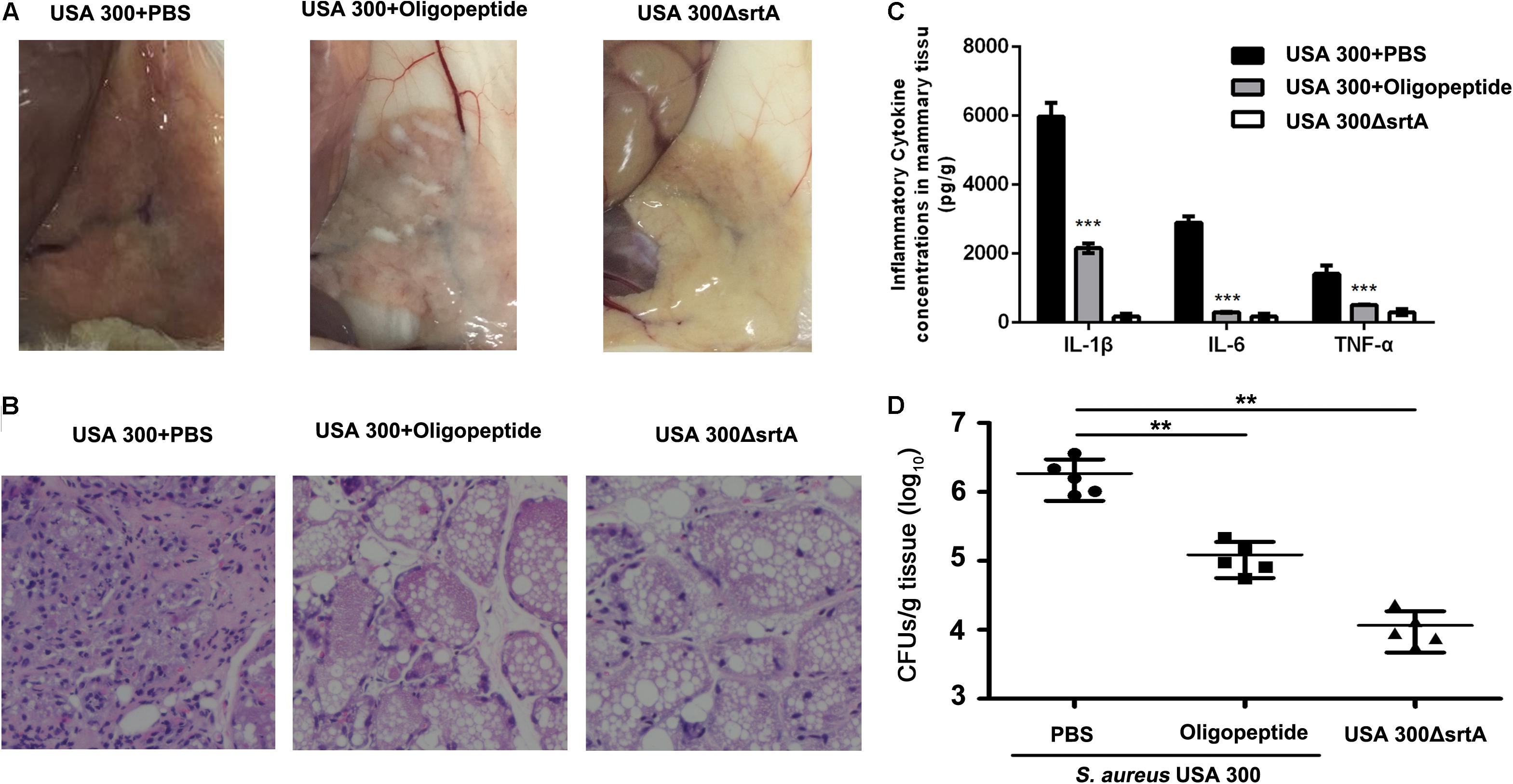
FIGURE 3. Oligopeptide provides protection against S. aureus mastitis in a mouse model. Lactating BALB/c mice were infected by injecting the canal glands with 5 × 107 CFU bacterial cells and treatment with the oligopeptide or PBS at the time of infection. Gross pathological changes (A) and histopathological analysis (B) of the mammary gland tissues at 48 h post-infection (n = 10). Significant alleviation of the pathological abnormalities was observed in the infected mice that received 50 mg/kg oligopeptide compared with the control mice treated with PBS. (C) Oligopeptide reduces the inflammatory response in infected mice. The levels of cytokines, including IL-1β, IL-6, and TNF-α, in the mammary gland tissues of infected mice were evaluated by ELISA (n = 10) (∗p < 0.05, ∗∗∗p < 0.001). (D) The effect of the oligopeptide on the bacterial burden in infected mice. Mammary gland tissues were collected, homogenized, and plated on TSB agar plates for assessment of the bacterial burden (n = 5) (∗p < 0.05, ∗∗p < 0.01). Results are reported as mean ± SEM.
Determination of the Molecular Mechanism of Oligopeptide Effects against SrtA
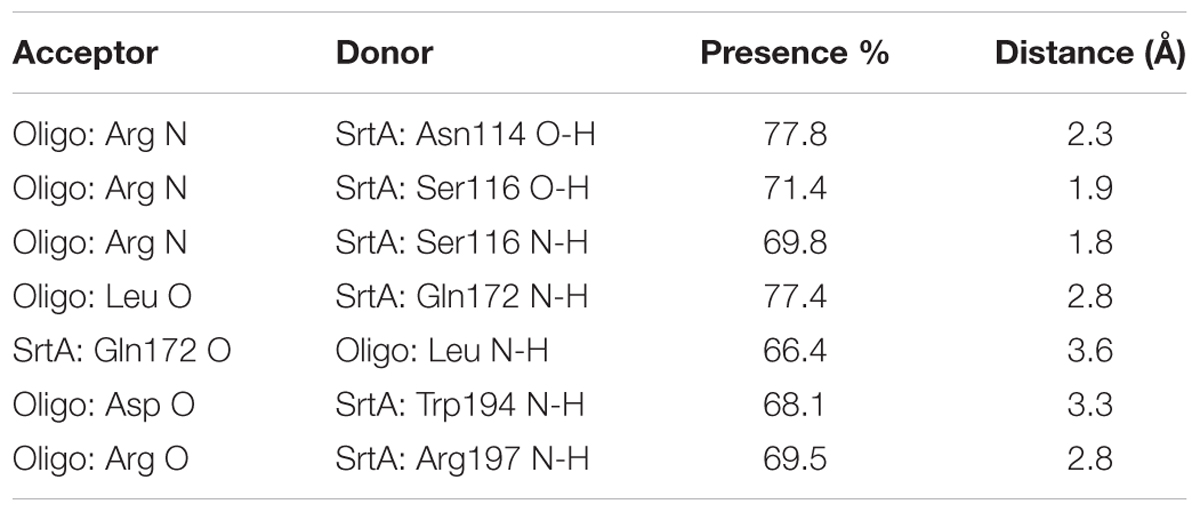
Using a computational biology method, the potential binding mode of the oligopeptide with SrtA at the active site was explored in this study. As is shown in Figure 4, the complex was found to reach equilibrium at 100 ns based on analysis of the root-mean-square deviations (RMSDs) of the backbone Cα atoms. The binding of the oligopeptide to SrtA and the binding mode are presented in Figure 5A. It was clear that the oligopeptide bound to SrtA via hydrogen bonding and hydrophobic interactions. During the time course of the simulation, the oligopeptide localized to the catalytic pocket of SrtA (residues 90–120 and 180–200). In detail, the binding model of the oligopeptide with SrtA revealed that the Arg, Leu, and Asp side chains of the oligopeptide formed seven hydrogen bonds with Asn114, Ser116, Gln172, Trp194, and Arg197, respectively. The relevant information concerning the stability of the hydrogen bonds between SrtA and oligopeptide are summarized in Table 2.
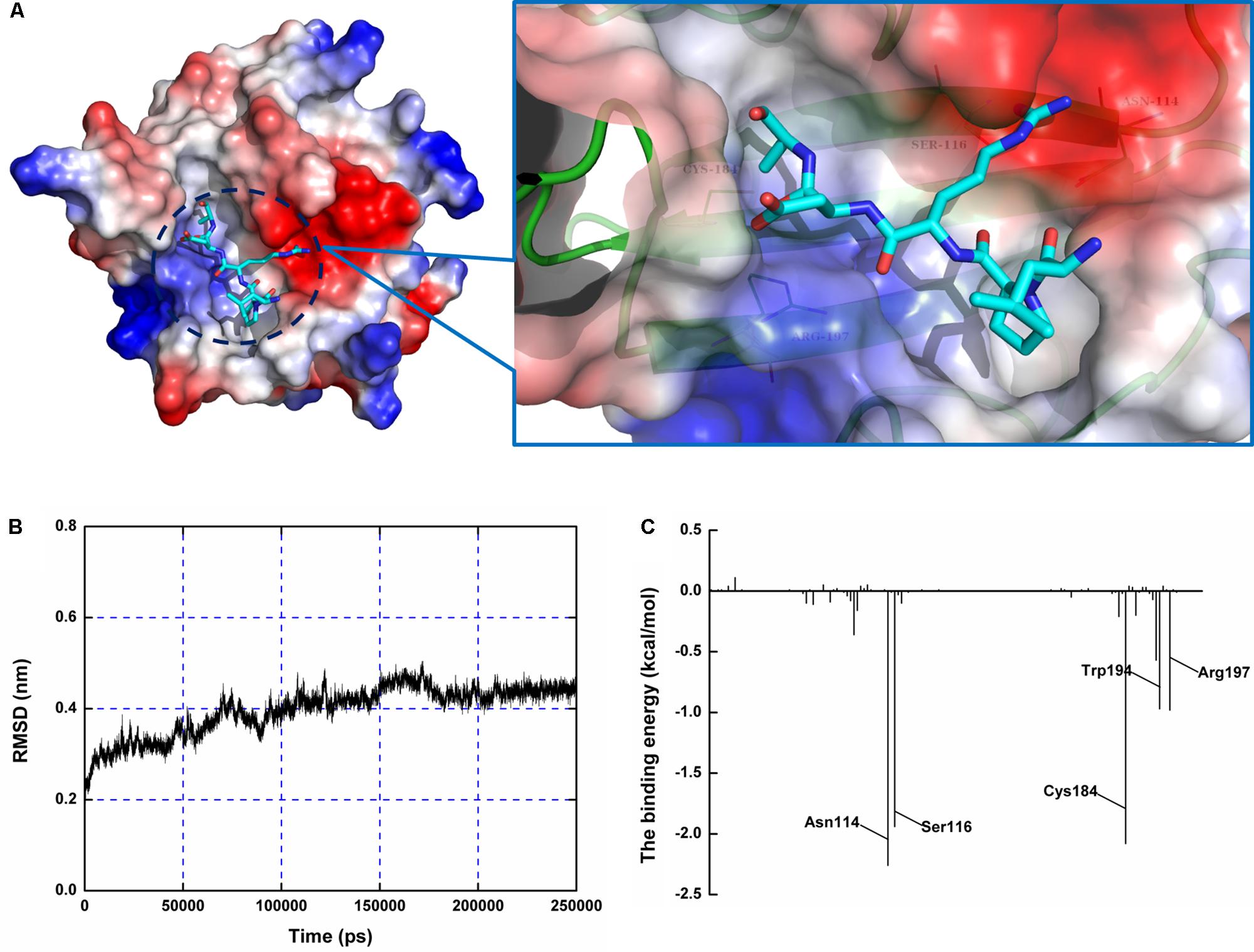
FIGURE 4. Determination of the 3D structure of the SrtA-oligopeptide complex using a molecular modeling method. (A) The structure of SrtA-oligopeptide. (B) The RMSD calculated for the backbone atoms of the protein during MD simulations of the SrtA-oligopeptide is presented. (C) Decomposition of the binding energy on a per-residue basis at the binding sites of the SrtA-oligopeptide complex.
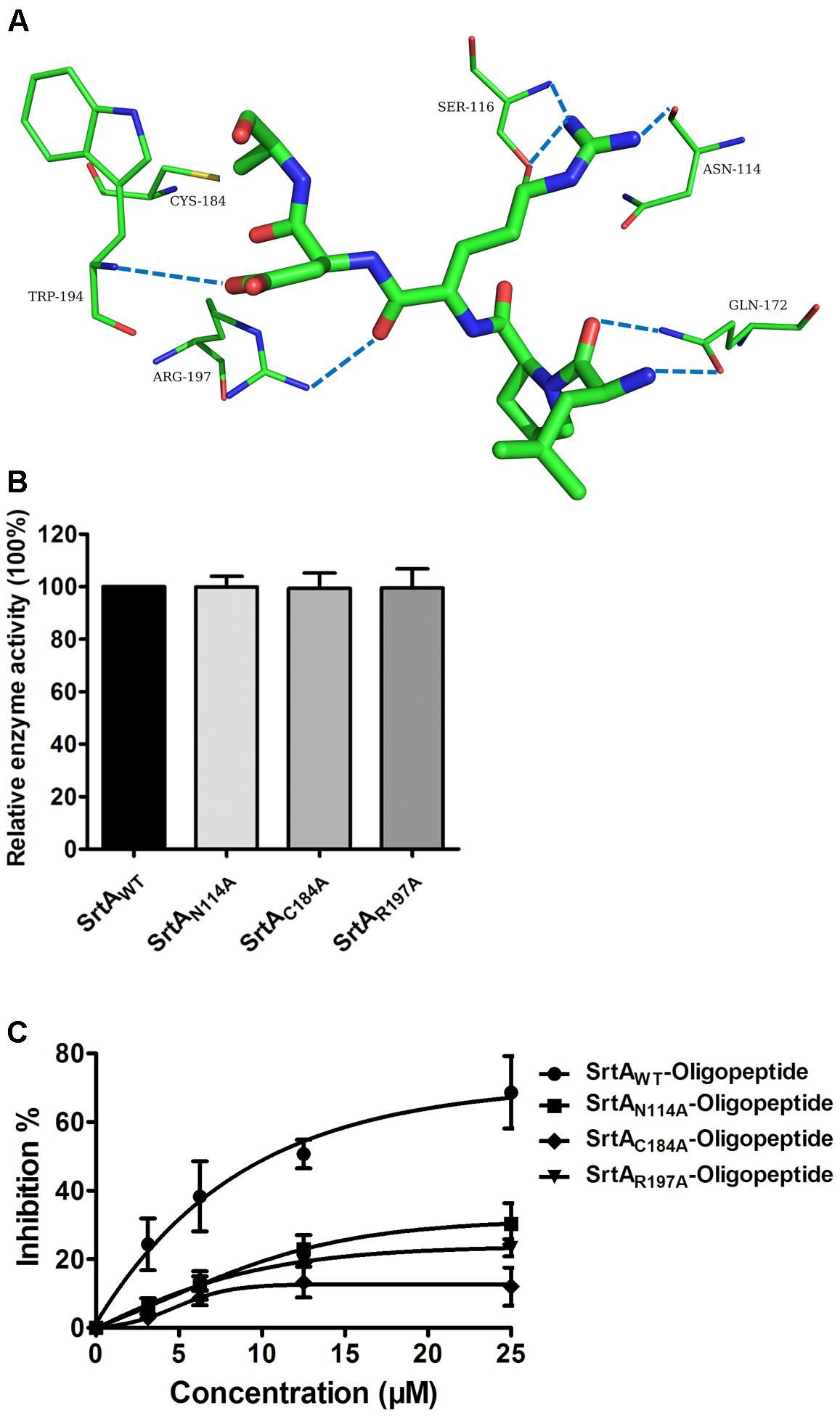
FIGURE 5. Determination of the binding sites for the oligopeptide on SrtA and its inhibitory mechanism. (A) The predicted binding mode of SrtA with the oligopeptide is shown, with labeling of key residues in the binding sites. (B) The catalytic activity of SrtA and its derivative. The activity of SrtA mutants was calculated by comparison with the wild type SrtA activity that was set to 100%. (C) Influences of the oligopeptide on the activities of SrtA and its derivatives. SrtA was incubated with various concentrations of the oligopeptide, and the fluorescent peptide substrate Dabcyl-QALPTTGEE-Edans was added for determination of SrtA protein activity, as described in Figure 1B (n = 3). Results are reported as mean ± SEM.
To examine the energy contributions from the residues of the binding sites in the SrtA-oligopeptide complex, energy decomposition was analyzed for the SrtA-oligopeptide complex system. As shown in Figure 4C, Asn114, Ser116 and Cys184 had a strong total binding energy contribution, with ΔEtotal of ≤-2.0 kcal/mol. The residues Trp194 and Arg197 also had an appreciable total binding energy contribution, with ΔEtotal of ≤-1.0 kcal/mol. These results suggest that these five residues are key residues in the sortaseA.
To confirm these results, the total binding free energy for the SrtA-oligopeptide complex and the detailed energy contributions were calculated according to the MM-PBSA approach, as summarized in Table 3. According to the results, the binding free energy, ΔGbind, of the interaction between the oligopeptide and protein decreased in the order of SrtAWT > SrtAMutants, indicating that SrtAWT exhibited the strongest binding to the oligopeptide. Further, we measured the ΔGbind and the number of binding sites between the oligopeptide and the three mutants using the fluorescence spectroscopy quenching method, and the results were highly consistent with those obtained using computational methods (Table 3). These results indicated that the information generated by the MD simulations on the SrtA-oligopeptide complex was reliable. To further confirm the molecular mechanism, we assessed the inhibitory effects of the oligopeptide against SrtA and its mutants. All the three mutants displayed the similar catalytic activity as wild-type SrtA (Figure 5B); however, the sensitivity of the SrtA mutants to oligopeptide-induced inhibition was significantly decreased compared to that of wild-type SrtA (Figure 5C), consistent with the fluorescence spectroscopy quenching assay results. Thus, these results demonstrated that binding of the oligopeptide inhibitor to the active region (residues Asn114, Ser116, Gln172, Cys184, Trp194, and Arg197) resulted in inhibition of the biology activity of SrtA.
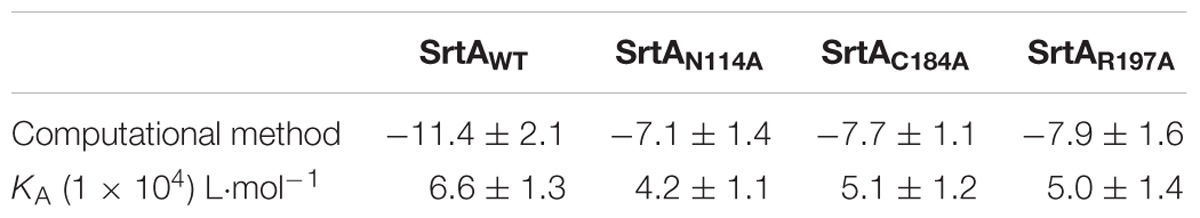
TABLE 3. The binding free energy (kcal/mol) of SrtAWT-oligopeptide, SrtAN114AT-oligopeptide, SrtAC184A-oligopeptide, and SrtAR197A-oligopeptide systems based on a computational method and the binding constant (KA) values based on fluorescence spectroscopy quenching.
Discussion
The development of drug resistance (Rasko and Sperandio, 2010) promote the alternative antivirulence strategy for the treatment of bacterial infections, especially those caused by resistant strains (Zambelloni et al., 2015). Recent research efforts have been directed at discovery of small molecules and antibodies that inhibit the expression or neutralize the functions of virulence-associated toxins (Bubeck Wardenburg and Schneewind, 2008; Wang et al., 2015a,b). However, SrtA-anchored surface proteins in the bacterial envelope are indispensable for bacterial adhesion, invasion and evasion in the host, and these processes are the first steps as well as the prerequisites for success in establishing infection (Cossart and Jonquieres, 2000; Maresso and Schneewind, 2008; Foster et al., 2014). Thus, the targeting of SrtA is more effective than that of the virulence-associated toxins (Cascioferro et al., 2014).
In the present study, virtual screening based on the LPXTG substrate sequence was used to screen for S. aureus SrtA inhibitors, among which the oligopeptide LPRDA exhibited the highest inhibitory effect against SrtA activity, with an IC50 of 10.61 μM. Importantly, the display of surface protein A by USA 300 was decreased in the presence of the oligopeptide. Subsequently, the interaction of S. aureus with fibronectin, the biofilm formation of S. aureus, and the invasion of S. aureus into host cells were all inhibited in our experimental conditions following treatment with the oligopeptide, indicating that suppression of SrtA activity by the oligopeptide in S. aureus also occurred. Furthermore, the oligopeptide had no bacteriostatic activity against S. aureus at the concentrations at which it sufficiently inhibited SrtA activity both in vitro and in vivo, suggesting that this inhibitor should be able to inhibit S. aureus virulence without exerting noticeable selective pressure. Treatment with the oligopeptide remarkably alleviated S. aureus-mediated pathological injury and inflammatory reactions in the mouse model of mastitis infection; this disease is considered the most frequent and most costly disease affecting the dairy industry. Clinical treatment failure frequently occurs with the use of antibiotics due to the development of drug resistance, and the presence of antibiotic residues in milk significantly reduces its quality and has undesirable effects on human health (Testa et al., 2007; Virdis et al., 2010; Duarte et al., 2015). Although some natural and synthetic compounds targeting virulence factors, including SrtA, have been identified as therapeutic agents for this disease (Cascioferro et al., 2014; Zhang et al., 2014; Wang et al., 2015c), the excretion of these compounds into milk also affects its quality (Addo et al., 2011). In the present study, treatment of mastitis with the oligopeptide may result in decreased drug resistance compared with that caused by the currently available drugs because of its low antibacterial activity (Zambelloni et al., 2015). Moreover, the breakdown products of the oligopeptide in the host are amino acids, which are the basic components of proteins, ensuring for production of high-quality milk, as no antibiotic residues are deposited. Therefore, the oligopeptide LPRDA may be a candidate for use in an alternative strategy for the prevention and treatment of mastitis.
Additionally, MD simulations were carried out for the SrtA-oligopeptide complex system to explore the interaction mechanism between the oligopeptide LPRDA and SrtA. This simulation revealed that the oligopeptide localized to the catalytic pocket of SrtA (residues 90–120 and 180–200), very close to the substrate binding site. Due to binding of the oligopeptide with SrtA, binding of the substrate with SrtA was blocked, leading to the loss of biological activity of SrtA. Fluorescence spectroscopy quenching assay and SrtA inhibition assay were performed to validate this inhibitory mechanism. As expected, the binding free energies of the SrtA mutants with the oligopeptide were significantly lower than that of wild-type SrtA, and the inhibitory effects of the oligopeptide against the SrtA mutants were decreased compared with that of wild-type SrtA. Our data established that inhibition of SrtA activity by the oligopeptide LPDRA via occupation of the active site of this enzyme conferred protection against S. aureus mastitis. Considering the indispensable role of SrtA in Gram-positive bacterial pathogenicity and the conserved structure of its active site (Jonsson et al., 2002; Mazmanian et al., 2000, 2002; Chen et al., 2014), the oligopeptide LPDRA should prevent a variety of bacterial infections requiring SrtA-catalyzed surface protein anchoring, especially those caused by resistant Gram-positive bacteria.
Author Contributions
XD, XN, JW, and HL conceived and designed the experiments. JW, HL, JP, JD, and XZ performed the experiments. JW and XD contributed reagents/materials/analysis tools. JW and XD wrote the paper.
Funding
This work was supported by the National Natural Science Foundation of China (Grant 31602109), the Project funded by the China Postdoctoral Science Foundation (Project No. 2017T100211) and the National Key R&D Program of China (2017YFD0501500).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Addo, K. K., Mensah, G. I., Aning, K. G., Nartey, N., Nipah, G. K., Bonsu, C., et al. (2011). Microbiological quality and antibiotic residues in informally marketed raw cow milk within the coastal savannah zone of Ghana. Trop. Med. Int. Health 16, 227–232. doi: 10.1111/j.1365-3156.2010.02666.x
Bandyopadhyay, S., Valder, C. R., Huynh, H. G., Ren, H. M., and Allison, W. S. (2002). The beta G(156)C substitution in the F-1-ATPase from the thermophilic Bacillus PS3 affects catalytic site cooperativity by destabilizing the closed conformation of the catalytic site. Biochemistry 41, 14421–14429.
Bradshaw, W. J., Davies, A. H., Chambers, C. J., Roberts, A. K., Shone, C. C., and Acharya, K. R. (2015). Molecular features of the Sortase enzyme family. FEBS J. 282, 2097–2114. doi: 10.1111/febs.13288
Bubeck Wardenburg, J., and Schneewind, O. (2008). Vaccine protection against Staphylococcus aureus pneumonia. J. Exp. Med. 205, 287–294.
Cascioferro, S., Totsika, M., and Schillaci, D. (2014). Sortase A: an ideal target for anti-virulence drug development. Microb. Pathog. 77, 105–112. doi: 10.1016/j.micpath.2014.10.007
Cegelski, L., Kim, S. J., Hing, A. W., Studelska, D. R., O’Connor, R. D., Mehta, A. K., et al. (2002). Rotational-echo double resonance characterization of the effects of vancomycin on cell wall synthesis in Staphylococcus aureus. Biochemistry 41, 13053–13058.
Chen, F. G., Liu, B. R., Wang, D. C., Wang, L., Deng, X. M., Bi, C. W., et al. (2014). Role of Sortase A in the pathogenesis of Staphylococcus aureus-induced mastitis in mice. FEMS Microbiol. Lett. 351, 95–103. doi: 10.1111/1574-6968.12354
Ciustea, M., Mootien, S., Rosato, A. E., Perez, O., Cirillo, P., Yeung, K. R., et al. (2012). Thiadiazolidinones: a new class of alanine racemase inhibitors with antimicrobial activity against methicillin-resistant Staphylococcus aureus. Biochem. Pharmacol. 83, 368–377. doi: 10.1016/j.bcp.2011.11.021
Cossart, P., and Jonquieres, R. (2000). Sortase, a universal target for therapeutic agents against Gram-positive bacteria? Proc. Natl. Acad. Sci. U.S.A. 97, 5013–5015.
de Almeida, L. M., de Almeida, M. Z., de Mendonca, C. L., and Mamizuka, E. M. (2013). Comparative analysis of agr groups and virulence genes among subclinical and clinical mastitis Staphylococcus aureus isolates from sheep flocks of the Northeast of Brazil. Braz. J. Microbiol. 44, 493–498. doi: 10.1590/S1517-83822013000200026
Duarte, C. M., Freitas, P. P., and Bexiga, R. (2015). Technological advances in bovine mastitis diagnosis: an overview. J. Vet. Diagn. 27, 665–672. doi: 10.1177/1040638715603087
Foster, T. J., Geoghegan, J. A., Ganesh, V. K., and Hook, M. (2014). Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62. doi: 10.1038/nrmicro3161
Gao, C. X., Uzelac, I., Gottfries, J., and Eriksson, L. A. (2016). Exploration of multiple Sortase A protein conformations in virtual screening. Sci. Rep. 6:20413. doi: 10.1038/srep20413
Hu, R., Barbault, F., Maurel, F., Delamar, M., and Zhang, R. (2010). Molecular dynamics simulations of 2-amino-6-arylsulphonylbenzonitriles analogues as HIV inhibitors: interaction modes and binding free energies. Chem. Biol. Drug Des. 76, 518–526. doi: 10.1111/j.1747-0285.2010.01028.x
Jonsson, I. M., Mazmanian, S. K., Schneewind, O., Verdrengh, M., Bremell, T., and Tarkowski, A. (2002). On the role of Staphylococcus aureus Sortase and Sortase-catalyzed surface protein anchoring in murine septic arthritis. J. Infect. Dis. 185, 1417–1424.
Jurasekova, Z., Marconi, G., Sanchez-Cortes, S., and Torreggiani, A. (2009). Spectroscopic and molecular modeling studies on the binding of the flavonoid luteolin and human serum albumin. Biopolymers 91, 917–927. doi: 10.1002/bip.21278
Jusko, M., Potempa, J., Kantyka, T., Bielecka, E., Miller, H. K., Kalinska, M., et al. (2014). Staphylococcal proteases aid in evasion of the human complement system. J. Innate Immun. 6, 31–46. doi: 10.1159/000351458
Lv, Z., Wang, H. S., and Niu, X. D. (2013). Molecular dynamics simulations reveal insight into key structural elements of aaptamines as Sortase inhibitors with free energy calculations. Chem. Phys. Lett. 585, 171–177.
Maresso, A. W., and Schneewind, O. (2008). Sortase as a target of anti-infective therapy. Pharmacol. Rev. 60, 128–141.
Mazmanian, S. K., Liu, G., Jensen, E. R., Lenoy, E., and Schneewind, O. (2000). Staphylococcus aureus Sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. U.S.A. 97, 5510–5515.
Mazmanian, S. K., Ton-That, H., Su, K., and Schneewind, O. (2002). An iron-regulated Sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 2293–2298.
McCafferty, D. G., Kruger, R. G., Barkallah, S., and Frankel, B. A. (2004). Inhibition of the Staphylococcus aureus Sortase transpeptidase SRTA by phosphinic peptidomimetics. Bioorg. Med. Chem. 12, 3723–3729.
Niu, X. D., Qiu, J. Z., Wang, X., Gao, X. H., Dong, J., Wang, J. F., et al. (2013). Molecular insight into the inhibition mechanism of cyrtominetin to alpha-hemolysin by molecular dynamics simulation. Eur. J. Med. Chem. 62, 320–328. doi: 10.1016/j.ejmech.2013.01.008
Rasko, D. A., and Sperandio, V. (2010). Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Dis. 9, 117–128. doi: 10.1038/nrd3013
Suree, N. (2009). Structural Studies and Drug Discovery of the Staphylococcus aureus Sortase A Enzyme. Dissertations & Theses - Gradworks. Los Angeles, CA: University of California.
Suree, N., Liew, C. K., Villareal, V. A., Thieu, W., Fadeev, E. A., Clemens, J. J., et al. (2009). The structure of the Staphylococcus aureus Sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized. J. Biol. Chem. 284, 24465–24477. doi: 10.1074/jbc.M109.022624
Tam, V. H., Louie, A., Fritsche, T. R., Deziel, M., Liu, W., Brown, D. L., et al. (2007). Impact of drug-exposure intensity and duration of therapy on the emergence of Staphylococcus aureus resistance to a quinolone antimicrobial. J. Infect. Dis. 195, 1818–1827.
Testa, C., Marogna, G., Secchi, L., Rubattu, N., Leori, G., and Calaresu, G. (2007). Antibiotics mastitis therapy: drug residue in lambs. Ital. J. Anim. Sci. 6(Suppl. 1):601.
Virdis, S., Scarano, C., Cossu, F., Spanu, V., Spanu, C., and De Santis, E. P. (2010). Antibiotic resistance in Staphylococcus aureus and coagulase negative staphylococci isolated from goats with subclinical mastitis. Vet. Med. Int. 2010:517060. doi: 10.4061/2010/517060
Wang, J. F., Qiu, J. Z., Tan, W., Zhang, Y., Wang, H. S., Zhou, X., et al. (2015a). Fisetin inhibits Listeria monocytogenes virulence by interfering with the oligomerization of listeriolysin O. J. Infect. Dis. 211, 1376–1387. doi: 10.1093/infdis/jiu520
Wang, J. F., Zhou, X., Liu, S., Li, G., Zhang, B., Deng, X. M., et al. (2015b). Novel inhibitor discovery and the conformational analysis of inhibitors of listeriolysin O via protein-ligand modeling. Sci. Rep. 5:8864. doi: 10.1038/srep08864
Wang, L., Bi, C., Cai, H., Liu, B., Zhong, X., Deng, X., et al. (2015c). The therapeutic effect of chlorogenic acid against Staphylococcus aureus infection through Sortase A inhibition. Front. Microbiol. 6:1031. doi: 10.3389/fmicb.2015.01031
Zambelloni, R., Marquez, R., and Roe, A. J. (2015). Development of antivirulence compounds: a biochemical review. Chem. Biol. Drug Des. 85, 43–55. doi: 10.1111/cbdd.12430
Zhang, J., Liu, H., Zhu, K., Gong, S., Dramsi, S., Wang, Y. T., et al. (2014). Antiinfective therapy with a small molecule inhibitor of Staphylococcus aureus Sortase. Proc. Natl. Acad. Sci. U.S.A. 111, 13517–13522.
Zhang, J. Y., Yang, F., Zhang, X. L., Jing, H. M., Ren, C. Y., Cai, C. Z., et al. (2015). Protective efficacy and mechanism of passive immunization with polyclonal antibodies in a sepsis model of Staphylococcus aureus infection. Sci. Rep. 5:15553. doi: 10.1038/srep15553
Keywords: Staphylococcus aureus, Sortase A, oligopeptide, antivirulence, mastitis
Citation: Wang J, Li H, Pan J, Dong J, Zhou X, Niu X and Deng X (2018) Oligopeptide Targeting Sortase A as Potential Anti-infective Therapy for Staphylococcus aureus. Front. Microbiol. 9:245. doi: 10.3389/fmicb.2018.00245
Received: 30 October 2017; Accepted: 31 January 2018;
Published: 14 February 2018.
Edited by:
Henrietta Venter, University of South Australia, AustraliaReviewed by:
Osmar Nascimento Silva, Universidade Católica Dom Bosco, BrazilSunil D. Saroj, Symbiosis International University, India
Copyright © 2018 Wang, Li, Pan, Dong, Zhou, Niu and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaodi Niu, bml1eGRAamx1LmVkdS5jbg== Xuming Deng, ZGVuZ3htQGpsdS5lZHUuY24=
†These authors have contributed equally to this work.
 Jianfeng Wang
Jianfeng Wang Hongen Li1,2†
Hongen Li1,2† Xuming Deng
Xuming Deng