- 1Center for Ecological Research, Kyoto University, Otsu, Japan
- 2Precursory Research for Embryonic Science and Technology, Japan Science and Technology Agency, Kawaguchi, Japan
- 3Graduate School of Human and Environmental Studies, Kyoto University, Kyoto, Japan
Arbuscular mycorrhizal and ectomycorrhizal symbioses are among the most important drivers of terrestrial ecosystem dynamics. Historically, the two types of symbioses have been investigated separately because arbuscular mycorrhizal and ectomycorrhizal plant species are considered to host discrete sets of fungal symbionts (i.e., arbuscular mycorrhizal and ectomycorrhizal fungi, respectively). Nonetheless, recent studies based on high-throughput DNA sequencing technologies have suggested that diverse non-mycorrhizal fungi (e.g., endophytic fungi) with broad host ranges play roles in relationships between arbuscular mycorrhizal and ectomycorrhizal plant species in forest ecosystems. By analyzing an Illumina sequencing dataset of root-associated fungi in a temperate forest in Japan, we statistically examined whether co-occurring arbuscular mycorrhizal (Chamaecyparis obtusa) and ectomycorrhizal (Pinus densiflora) plant species could share non-mycorrhizal fungal communities. Among the 919 fungal operational taxonomic units (OTUs) detected, OTUs in various taxonomic lineages were statistically designated as “generalists,” which associated commonly with both coniferous species. The list of the generalists included fungi in the genera Meliniomyces, Oidiodendron, Cladophialophora, Rhizodermea, Penicillium, and Mortierella. Meanwhile, our statistical analysis also detected fungi preferentially associated with Chamaecyparis (e.g., Pezicula) or Pinus (e.g., Neolecta). Overall, this study provides a basis for future studies on how arbuscular mycorrhizal and ectomycorrhizal plant species interactively drive community- or ecosystem-scale processes. The physiological functions of the fungi highlighted in our host-preference analysis deserve intensive investigations for understanding their roles in plant endosphere and rhizosphere.
Introduction
In terrestrial ecosystems, most plant species form intimate interactions with mycorrhizal fungi, which play essential roles in the growth and survival of their hosts (van der Heijden et al., 2008; Bever et al., 2010; Peay et al., 2016). Those fungi, for example, supply soil nitrogen and phosphorous to associated plants, thereby enhancing hosts' physiological states (Smith and Read, 2008). They are also known to reduce deleterious effects of pathogens on host plants (Marx, 1972; Azcón-Aguilar and Barea, 1997; Borowicz, 2001). Moreover, mycorrhizal fungi can contribute to physiological homeostasis of plants by increasing hosts' resistance to abiotic stress (Grover et al., 2011). Therefore, understanding and managing below-ground integrations between plants and their mycorrhizal fungal symbionts are major challenges not only in basic ecology but also in forestry and agronomy.
Among the several categories of mycorrhizal fungi, arbuscular mycorrhizal, and ectomycorrhizal fungi are major groups of below-ground fungal communities in temperate forests (Smith and Read, 2008; Peay et al., 2016). Arbuscular mycorrhizal fungi (the phylum Glomeromycota) first appeared early in the history of land plants (Remy et al., 1994) and hence they associate with plant species in diverse plant taxa (Schüβler et al., 2001). They are obligate mutualistic symbionts and hence rely entirely on carbon supply from host plants (Smith and Read, 2008). While they are abundant in root systems of herbaceous plants (Hiiesalu et al., 2014), they are hosted also by diverse tree species (Liu et al., 2015). Ectomycorrhizal fungi, which consist mainly of the phyla Ascomycota and Basidiomycota, appeared in the era of seed plant diversification (Hibbett and Matheny, 2009). In contrast to arbuscular mycorrhizal fungi, some of them may obtain carbon not only from plants but also from soil by decomposing dead organic matter (Talbot et al., 2008) (but see Lindahl and Tunlid, 2015). Ectomycorrhizal fungi play important roles in forest community dynamics because they promote the dominance of the specific plant families (e.g., Pinaceae, Fagaceae, Betulaceae, and Dipterocarpaceae; Tedersoo et al., 2010; Tedersoo and Smith, 2013) through “positive plant–soil feedbacks” (Booth, 2004; McGuire, 2007; Bennett et al., 2017). Due to the difference in their major host taxa, arbuscular mycorrhizal and ectomycorrhizal fungi have been considered to form distinct sets of symbioses with their arbuscular mycorrhizal plant and ectomycorrhizal plant hosts (Smith and Read, 2008), potentially driving discrete community ecological dynamics. As a consequence, arbuscular mycorrhizal and ectomycorrhizal symbioses have been investigated separately in most mycological studies.
Nonetheless, recent studies integrating high-throughput DNA sequencing and host–symbiont network analyses have shown that diverse non-mycorrhizal fungi with broad host ranges are associated with roots of both arbuscular mycorrhizal and ectomycorrhizal plants within terrestrial ecosystems (Toju et al., 2014a, 2015). Furthermore, mycorrhizal, endophytic, and other types of root-associated fungi have been reported to co-occur within/around a tiny segment of plant roots (Read and Haselwandter, 1981; Mandyam and Jumpponen, 2005; Nakamura et al., 2017), potentially interacting with each other positively or negatively (Toju et al., 2016b) (cf. Kennedy et al., 2009; Werner and Kiers, 2015). Interestingly, an increasing number of studies have shown that non-mycorrhizal fungi (e.g., endophytic fungi) can supply host plants with phosphorous, potentially playing physiological roles similar to those of mycorrhizal fungi (Jumpponen, 2001; Narisawa et al., 2002; Newsham, 2011; Hiruma et al., 2016; Almario et al., 2017). Thus, host plant ranges of those non-mycorrhizal fungi are of particular interest because they will provide a basis for uncovering potential sharing of soil nutrients between arbuscular mycorrhizal and ectomycorrhizal plants and its consequences on the community- or ecosystem-level dynamics (Kadowaki et al., in review). However, while an increasing number of studies have evaluated host preferences (or generality) of diverse functional groups of root-associated fungi including possible endophytes (Huang et al., 2008; Kernaghan and Patriquin, 2011; Botnen et al., 2014; Sato et al., 2015), most studies have investigated either arbuscular mycorrhizal or ectomycorrhizal plant species but not both. Consequently, we still have limited knowledge of how co-occurring arbuscular mycorrhizal and ectomycorrhizal plant species can interact with each other indirectly through below-ground webs of symbioses involving not only mycorrhizal but also diverse non-mycorrhizal fungi.
In this study, we statistically examined host preferences of not only mycorrhizal but also root-endophytic fungi in a mixed forest of arbuscular mycorrhizal and ectomycorrhizal coniferous trees in Japan. We sampled roots of Chamaecyparis obtusa (arbuscular mycorrhizal) and Pinus densiflora (ectomycorrhizal) and then revealed community compositions of the fungi associated with the two plant species based on Illumina sequencing. The dataset allowed us to classify those fungi in terms of their host preferences, highlighting endophytic fungi preferentially found from either Chamaecyparis or Pinus, and those commonly associated with both plant species. Overall, this study provides a basis for future studies examining how diverse functional groups of below-ground fungi mediate interactions between arbuscular mycorrhizal and ectomycorrhizal plant species in terrestrial ecosystems.
Materials and Methods
Sampling
Fieldwork was conducted in a secondary temperate forest in Sasayama, Hyogo Prefecture, Japan (35.094 °N, 135.238 °E) on June 6, 2016. Sampling in the forest was permitted by the committee of the local residents. Within the forest consisted mainly of Pinus densiflora (Pineaceae), Quercus serrata (Fagaceae), and Ilex pedunculosa (Aquifoliaceae), there were patches of planted Chamaecyparis obtuse (Cupressaceae). Along a mountain trail in the forest, we collected 2 cm segments of terminal roots at 3 cm below the soil surface at 1 m horizontal intervals, screening coniferous tree roots based on root morphology: angiosperm roots were excluded in the sampling. In total, 247 root samples were collected and delivered to the laboratory within the sampling day. The samples were stored at −80°C until DNA extraction.
DNA Extraction, PCR, and Sequencing
Each root sample was washed in 80% ethanol by sonication for 5 min. DNA extraction was then performed with a cetyltrimethylammonium bromide (CTAB) method (Sato and Murakami, 2008). We amplified the internal transcribed spacer 1 (ITS1) region of root-associated fungi using the forward primer ITS1F-KYO1 (Toju et al., 2012) fused with 3–6-mer Ns for improved Illumina sequencing quality (Lundberg et al., 2013) and the forward Illumina sequencing primer (5′- TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG- [3–6-mer Ns]–[ITS1-KYO2]−3′) and the reverse primer ITS2-KYO2 (Toju et al., 2012) fused with 3–6-mer Ns and the reverse sequencing primer (5′- GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA G [3–6-mer Ns]—[ITS2_KYO2]−3′). In the PCR, the buffer and DNA polymerase kit of KOD FX Neo (Toyobo) was used with a temperature profile of 94°C for 2 min, followed by 35 cycles at 98°C for 10 s, 50°C for 30 s, 68°C for 50 s, and a final extension at 68 °C for 5 min. The ramp rate through the thermal cycles was set to 1°C/s in order to prevent generation of chimeric sequences (Stevens et al., 2013). To add Illumina sequencing adaptors to respective samples, supplemental PCR was performed using the forward fusion primers consisting of the P5 Illumina adaptor, 8-mer indexes for sample identification (Hamady et al., 2008), and a partial sequence of the sequencing primer (5′- AAT GAT ACG GCG ACC ACC GAG ATC TAC AC—[8-mer index]—TCG TCG GCA GCG TC−3′) and the reverse fusion primers consisting of the P7 adaptor, 8-mer indexes, and a partial sequence of the sequencing primer (5′- CAA GCA GAA GAC GGC ATA CGA GAT—[8-mer index]—GTC TCG TGG GCT CGG−3′). KOD FX Neo was used with a temperature profile of 94°C for 2 min, followed by 8 cycles at 98°C for 10 s, 55°C for 30 s, 68°C for 50 s (ramp rate = 1°C/s), and a final extension at 68°C for 5 min. The PCR amplicons of the 247 root samples and a negative control sample were pooled with equal volume after a purification/equalization process with AMPureXP Kit (Beckman Coulter). The ratio of AMPure reagent to amplicons was set to 0.6 (v/v) in order to remove primer dimers (i.e., sequences shorter than 200 bp).
To discriminate Chamaecyparis and Pinus root samples, we performed, another set of PCR targeting plant chloroplast rbcL region using the rbcL_F3 and rbcL_R4 primers (Toju et al., 2013a) with the same DNA polymerase system, temperature profiles, and purification processes used in the fungal ITS analysis. The sequencing libraries of fungal ITS and plant rbcL regions were processed in an Illumina MiSeq sequencer (run center: KYOTO-HE) with the 2 × 250 cycle sequencing kit (20% PhiX spike-in).
Bioinformatics
The raw sequencing data were converted into FASTQ files using the program bcl2fastq 1.8.4 distributed by Illumina. The output FASTQ files were demultiplexed with the program Claident v0.2.2016.07.05 (Tanabe and Toju, 2013; Tanabe, 2016). Sequencing reads whose 8-mer index positions included nucleotides with low (<30) quality scores were removed in this process. Given that the quality of reverse Illumina sequences is generally much lower than that of forward sequences, only forward sequences were used after removing low-quality 3'-ends using Claident: the sequencing data are available on the DNA Data Bank of Japan (DDBJ) (DDBJ Sequence Read Archive accession; DRA006340). Noisy reads (Tanabe, 2016) were subsequently discarded and then 2,177,205 ITS and 92,013 rbcL filtered reads were obtained.
For the analysis of the ITS region, filtered reads were clustered with the program VSEARCH (Rognes et al., 2016) as implemented in Claident. Taking into account the high intraspecific ITS-sequence variation of Glomeromycota (Thiéry et al., 2016), the cut-off sequence similarity in the clustering of the fungal ITS region was set to 95%. The molecular identification of the output 1183 OTUs (Supplementary Data 1) was conducted based on the combination of the query-centric auto-k-nearest neighbor (QCauto) method (Tanabe and Toju, 2013) and the lowest common ancestor (LCA) algorithm (Huson et al., 2007) as implemented in Claident. Note that taxonomic identification results based on the combination of the QCauto search and the LCA taxonomic assignment are comparable to, or sometimes more accurate than, those with the alternative approach combining the UCLUST algorithm (Edgar, 2010) with the UNITE database (Kõljalg et al., 2013) (see Toju et al., 2016a,b for detailed comparison between the QCauto-LCA and UCLUST–UNITE approaches). The functional group of each fungal OTU was inferred using the program FUNGuild 1.1 (Nguyen et al., 2016) (Supplementary Data 2). As the FUNGuild program often output multiple guilds for a single OTU (e.g., “Ectomycorrhizal-Orchid Mycorrhizal-Root Associated Biotroph”), the output guild information was grouped into the following categories in light of Tedersoo et al. (2010) and Smith and Read (2008): i.e., arbuscular mycorrhizal, ectomycorrhizal, ericoid mycorrhizal, saprotrophic/endophytic, plant pathogenic, animal pathogenic, and unclassified fungi (see Supplementary Data 2 for details). Given that fungi in the endosphere often shift their lifestyle when host plant tissue dies (Porras-Alfaro and Bayman, 2011), we grouped potentially endophytic and saprotrophic fungal OTUs into a single category (saprotrophic/endophytic).
The Illumina sequences of the plant rbcL region were processed with a cut-off sequence similarity of 97%. Based on the taxonomic assignment results with Claident, Chamaecyparis and Pinus samples were discriminated: four samples turned out to be angiosperm roots were discarded. We then obtained a sample × fungal OTU matrix, in which a cell entry depicted the number of sequencing reads of an OTU in a sample. The cell entries whose read counts represented less than 0.1% of the total read count of each sample were removed to minimize the effects of PCR/sequencing errors (Peay et al., 2015). The filtered matrix was then rarefied to 2,000 reads per sample using the “rrarefy” function of the vegan 2.4-1 package (Oksanen et al., 2012) of R 3.3.2 (R-Core-Team, 2015). The samples with less than 2,000 reads and the 264 fungal OTUs with no read counts in the rarefied matrix were eliminated. We then obtained a matrix consisting of 208 root samples (157 Chamaecyparis and 51 Pinus samples) and 919 fungal OTUs (Supplementary Data 3).
Fungal Diversity
For all the statistical analyses below, the vegan package of R was used. We first examined relationship between the number of sequencing reads and that of detected fungal OTUs with the “rarecurve” function. Likewise, relationship between the number of root samples and that of fungal OTUs was visualized with the “specaccum” function. Root-associated fungal community compositions were then compared between Chamaecyparis and Pinus, focusing on the functional groups and order-level taxonomy of observed fungi. Difference in fungal community compositions between the two plant species was further examined by the permutational analysis of variance (Anderson, 2001) with the “adonis” function (PERMANOVA; 10,000 permutations). We also performed the permutational analysis for the multivariate homogeneity of dispersions (PERMDISP) (Anderson, 2006) with the “betadisper” function. Differentiation of fungal community structure between Chamaecyparis and Pinus was also examined by non-metric multidimensional scaling (NMDS) with the “metaMDS” function. Potential spatial autocorrelation in the fungal community data was evaluated based on a Mantel's correlogram analysis with the “mantel.correlog” function. The “Raup-Crick” metric of β-diversity (Chase et al., 2011) (“raupcrick” function) was used in the PERMANOVA, PERMDISP, NMDS, and Mantel's correlogram analyses.
Host Preference
To explore root-associated fungi showing preference for Chamaecyparis or Pinus, we performed an analysis based on the multinomial species classification method (CLAM test; Chazdon et al., 2011) with the “clamtest” function of the R vegan package. The CLAM test has been used for exploring “generalists” and “specialists” based on comparisons between contrasting habitats or host species (Toju et al., 2013b, 2014b). Importantly, the multinomial model implemented in the test minimizes biases due to differing sampling intensities between the two habitats or host species compared (Chazdon et al., 2011). Based on a CLAM test, fungal OTUs were classified into four categories: fungal OTUs displaying statistically significant preferences for Chamaecyparis, those with significant preference for Pinus, those commonly found from both plant species, and those too rare to be evaluated statistically.
Results
Fungal Diversity
After a series of bioinformatics and rarefaction process, 815 and 412 fungal OTUs were found from Chamaecyparis and Pinus, respectively (Figure 1). The fungal community compositions differed between the two species. For example, arbuscular mycorrhizal fungi occurred almost exclusively on Chamaecyparis, while ectomycorrhizal and saprotrophic/endophytic fungi occurred on both plant species (Figure 2A). Regarding order-level taxonomy, Helotiales, Chaetothyriales, Agaricales, and Glomerales occurred frequently on Chamaecyparis (Figure 2B). In contrast, the root-associated fungal community of Pinus was characterized by Neolectales, Boletales, Russulales, and Thelephorales, although it resembled the Chamaecyparis fungal community in terms of Helotiales relative abundance (Figure 2B). The differentiation of fungal community structure between the two plant species was statistically significant (PERMANOVA; df = 1, Fmodel = 58.4, P < 0.0001) (Figure 3), although the structural difference was attributed, at least partly, to the heterogeneity of among-sample variation (PERMDISP; df = 1, F = 9.4, P = 0.003). The structure of root-associated fungal communities displayed spatial autocorrelation within 50 m and 20 m in Chamaecyparis and Pinus, respectively (Figure 4).
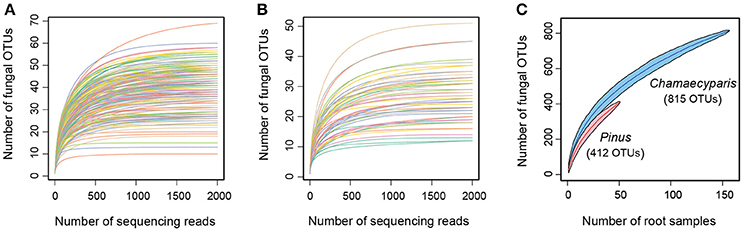
Figure 1. Fungal OTU richness. (A) Relationship between the number of sequencing reads and that of fungal OTUs (Chamaecyparis root samples). (B) Relationship between the number of sequencing reads and that of fungal OTUs (Pinus root samples). (C) Relationship between the number of root samples and that of fungal OTUs.
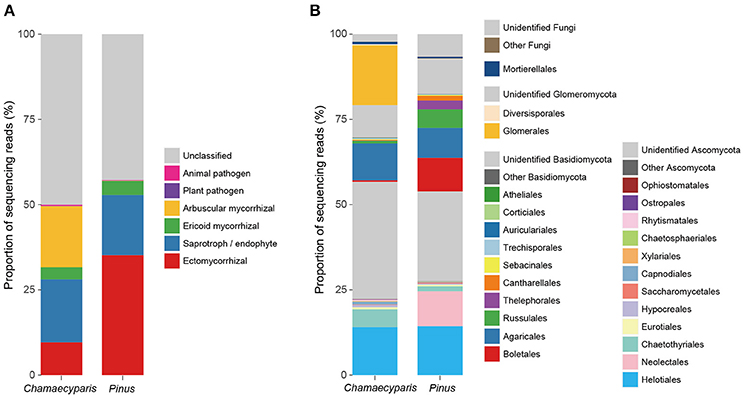
Figure 2. Compositions of the fungal community. (A) Functional groups (category). Average proportions of sequencing reads were calculated for Chamaecyparis and Pinus root samples. (B) Order-level taxonomy.
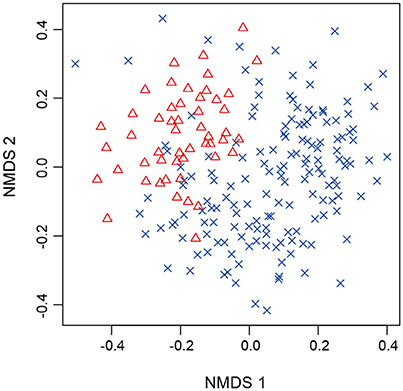
Figure 3. NMDS of root samples. Chamaecyparis (cross) and Pinus (triangle) samples were plotted on a NMDS surface (stress = 0.288).
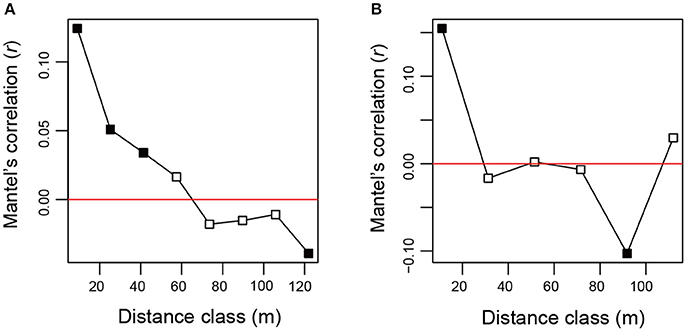
Figure 4. Spatial autocorrelation of fungal community structure. (A) Mantel's correlogram analysis of Chamaecyparis samples. A positive value indicated by filled squares represents statistically significant spatial autocorrelation at the spatial distance class (α = 0.05). (B) Mantel's correlogram analysis of Pinus samples.
Host Preference
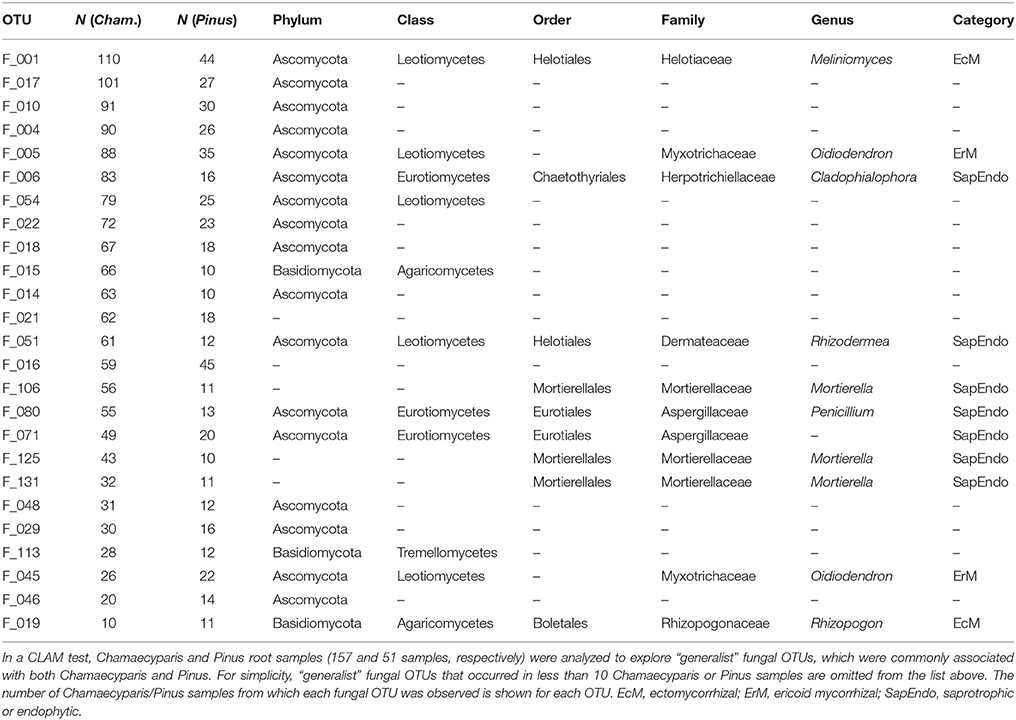
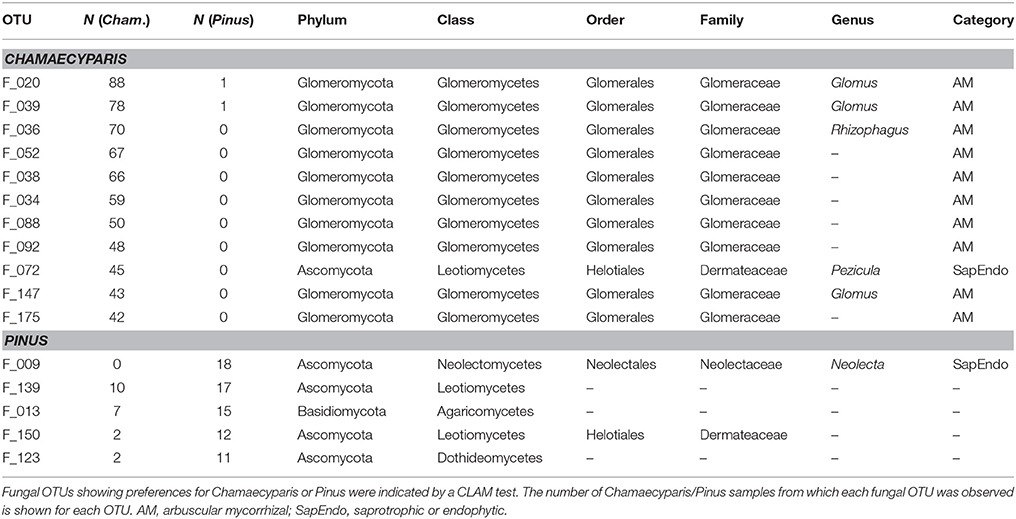
In the CLAM test (Figure 5), fungal OTUs in various taxonomic lineages were classified as “generalists,” meaning fungi commonly found from both plant species (Table 1). Among them, a Meliniomyces fungus in the order Helotiales appeared in 70.1% (110/157) and 86.3% (44/51) of Chamaecyparis and Pinus samples, respectively (Table 1). The Meliniomyces OTU was allied to M. variabilis (NCBI accession: HM190129), which were reported as saprotrophic, endophytic, and ericoid mycorrhizal but not ectomycorrhizal (Vrålstad et al., 2002a,b; Hambleton and Sigler, 2005): note that all Meliniomyces fungal OTUs were automatically designated as “ectomycorrhizal” by the FUNGuild program used in this study. The ascomycetous genera Oidiodendron, Cladophialophora, Rhizodermea, and Penicillium were also commonly found from the two coniferous species (Table 1). The statistical test also highlighted fungal OTUs showing host preferences for Chamaecyparis or Pinus (Table 2). A fungus in the genus Pezicula (Helotiales) and 10 glomeromycete OTUs were classified as OTUs associated preferentially with Chamaecyparis. In contrast, Neolecta sp. (Neolectales), Dermataceae sp. (Helotiales), and three other fungi were preferentially associated with Pinus.
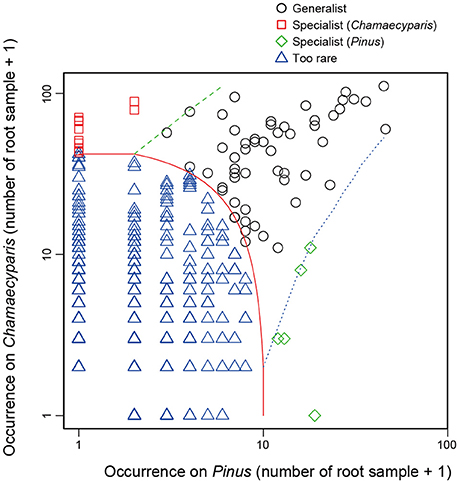
Figure 5. Screening of specialists and generalists. Fungal OTUs commonly detected from both Chamaecyparis and Pinus root samples (circle), those preferentially found from Chamaecyparis (square) or Pinus (diamond) samples, and rare fungal OTUs (triangle) were classified by a CLAM test.
Although the difference in sample size between Chamaecyparis and Pinus (157 and 51 samples, respectively) might have caused biases in the statistical analyses conducted in this study, results qualitatively similar with the abovementioned analyses were obtained in a series of supplementary analyses with equalized sample size (i.e., 51 randomly chosen Chamaecyparis samples vs. 51 Pinus samples) (Supplementary Figures 1–3; Supplementary Tables 1–2).
Discussion
Our data provided a novel opportunity to compare mycorrhizal and non-mycorrhizal fungal communities between arbuscular mycorrhizal (Chamaecyparis) and ectomycorrhizal (Pinus) plants in a temperate forest. One of the recent conceptual advances in mycology is that plant species in the wild interact not only with mycorrhizal fungi but also with diverse taxonomic/functional groups of endosphere and rhizosphere fungi (Mandyam and Jumpponen, 2005; Newsham, 2011). Those recent findings challenge the classic view that plant species differing in mycorrhizal type form discrete sets of below-ground plant–fungus interactions. Hereafter, we discuss fungi potentially mediating arbuscular mycorrhizal and ectomycorrhizal symbioses as well as those that preferentially interact with either mycorrhizal type of plant hosts.
Many of the fungi found commonly from both plant species belonged to major orders in Ascomycota, namely, Helotiales, Chaetothyriales, and Eurotiales (Figure 2B; Table 1). Among them, Meliniomyces (Helotiales) (Hambleton and Sigler, 2005) showed surprisingly high infection rates, appearing in 70 and 86% of Chamaecyparis and Pinus root samples, respectively. Although some species in Meliniomyces–Rhizoscyphus complex have been confirmed to be ectomycorrhizal in pure culture synthetic trials, the most abundant OTU detected in this study was allied to M. variabilis, which has been inferred as saprotrophic, endophytic, or ericoid mycorrhizal (Vrålstad et al., 2002a,b; Hambleton and Sigler, 2005; Tedersoo et al., 2009). Interestingly, M. variabilis obtained from a Norway spruce (Picea abies) microhabitats lacking ericaceous plants formed ericoid mycorrhizae with European blueberry (Vaccinium myrtillus) under experimental conditions, promoting the growth of the host (Vohník et al., 2013). Another Helotiales fungus frequently detected from both Chamaecyparis and Pinus roots belonged to the genus Rhizodermea. A fungus in the genus has been reported to enhance heavy-metal stress tolerance of host plants (Yamaji et al., 2016). Our analysis also detected Cladophialophora (Chaetothyriales), a lineage of so-called “dark septate endophytes” (Jumpponen and Trappe, 1998; Jumpponen, 2001). A species in the genus (C. chaetospira) has been known to enhance growth and pathogen resistance of host plants (Morita et al., 2003; Usuki and Narisawa, 2007). Penicillium (Eurotiales) fungi are also reported frequently from roots of diverse plant taxa, although they are generally considered as saprotrophic soil fungi (Watanabe, 2010) or postharvest pathogens of fruits (Agrios, 2005). However, given the repeated reports of Penicillium fungi from seemingly benign roots of diverse plant species (Cao et al., 2002; Toju et al., 2016b), some of them may play positive roles. Indeed, some Penicillium species associated with wheat are known to solubilize phosphorous in rhizosphere or endosphere (Wakelin et al., 2004). Penicillium species are also known to produce a series of antibiotics, which potentially inhibits growth of plant pathogens (Yang et al., 2008).
We also detected Mortierella and Oidiodendron fungi as common symbionts of Chamaecyparis and Pinus roots (Table 1). Fungi in the genus Mortierella are often isolated from soil and root systems in various types of habitats (Watanabe, 2010). Although they are generally regarded as saprotrophs, some of them potentially promote plant growth by suppressing root-knot nematodes or phytopathogens such as Rhizoctonia and Cercospora (Eroshin and Dedyukhina, 2002; AL-Shammari et al., 2013). Fungi in the genus Oidiodendron (anamorph of Myxotrichum) are also reported from diverse soil environments, while the genus include ericoid mycorrhizal fungi, O. maius and O. griseum (Couture et al., 1983; Douglas et al., 1989; Rice and Currah, 2005; Vohník et al., 2005). Oidiodendron fungi were also reported from roots of non-ericaceous plants such as Betula, Picea, and Abies trees in a boreal forest (Kernaghan and Patriquin, 2011).
While there were ectomycorrhizal fungi frequently detected from both Chamaecyparis and Pinus roots (Rhizopogon), no arbuscular mycorrhizal fungi were designated as “host generalists” in our study (Table 1). This pattern is of particular interest in light of previous studies reporting asymmetry in host–symbiont associations between ectomycorrhizal and arbuscular mycorrhizal symbioses (Plattner and Hall, 1995; Dickie et al., 2001). For example, colonization of ectomycorrhizal fungi might be deleterious to non-ectomycorrhizal plants as reported in a herbaceous plant species, whose roots suffered from severe necrosis after infection of the truffle fungus, Tuber melanosporum (Plattner and Hall, 1995) (see also Booth, 2004). Thus, in the forest studied in this study, the presence of Pinus and its ectomycorrhizal fungi may have negative impacts on Chamaecyparis, although possibilities that those ectomycorrhizal fungi play positive or neutral roles in Chamaecyparis root systems deserve further investigations. Arbuscular mycorrhizal fungi have been also reported to interact with non-typical host plant species. For example, an oak species (Quercus rubra) is known to host not only ectomycorrhizal but also arbuscular mycorrhizal fungi in the vicinity of arbuscular mycorrhizal plants (Dickie et al., 2001). The nearly complete absence of arbuscular mycorrhizal fungi in Pinus roots in our study (Figure 2A) highlights context dependency in such host–symbiont associations that span conventional categories of mycorrhizal symbioses.
The statistical analysis conducted in this study also allowed us to explore fungal species preferentially associated with either Chamaecyparis or Pinus. As expected, many arbuscular mycorrhizal fungi were found almost exclusively from Chamaecyparis. Meanwhile, a Helotiales fungus in the genus Pezicula (anamorph, Cryptosporiopsis; Verkley, 1999) (Chen et al., 2016) showed a strong preference for Chamaecyparis. Given that fungi in the genus produce secondary metabolites (e.g., mullein and echinocandin) that inhibit growth of plant pathogens (Noble et al., 1991; Schulz et al., 1995; Wang et al., 2014), Chamaecyparis hosts may be benefited by the presence of the endophytic fungi. Among the fungi preferentially associated with Pinus, an ascomycete fungus in the genus Neolecta (Neolectales) displayed the strongest host preference. Although their functions remain unknown, Neolecta fungi are known to associate with plant roots (Redhead, 1979; Landvik et al., 2003): an observation of co-occurrence of a Neolecta fungus and an ectomycorrhizal fungus in root tips (Redhead, 1979) is intriguing in postulating their functions. Although these results on potential host preferences are of particular ecological interest, it should be acknowledged that this study did not take into account possible spatial heterogeneity of edaphic factors (e.g., soil pH and C/N ratios) within the study site: there were too many sampling positions to perform detailed chemical analyses. In the dataset, we observed spatial autocorrelations in the occurrences of Chamaecyparis/Pinus root samples (Supplementary Figure 4) and fungal community structure (Figure 4). To evaluate relative contributions of host preference and spatial environmental heterogeneity, more sophisticated statistical analyses (e.g., latent variable model analyses; Warton et al., 2015) needs to be tried in future studies.
Our screening of plant-associated below-ground fungi with narrow/broad host ranges provides crucial implications for the understanding of dynamic linkage between plant and below-ground fungal communities (Klironomos, 2002; Bever et al., 2010; van der Putten et al., 2013). Previous studies on arbuscular mycorrhizal plants have shown “negative plant–soil feedbacks”, in which increases of host-specific soil microbes result in decline of the host plant populations (Bever, 2002; Kardol et al., 2007; Mangan et al., 2010). In contrast, positive feedbacks leading to monodominance have been suspected for interactions between ectomycorrhizal plants and their ectomycorrhizal fungi (Booth, 2004; McGuire, 2007; Bennett et al., 2017). While most of those previous studies focused on plant–soil feedbacks operating in interactions involving a single plant species and their mycorrhizal (and pathogenic) fungi, arbuscular mycorrhizal, and ectomycorrhizal fungi often coexist within a forest (Dickie et al., 2001; Toju et al., 2014a), potentially driving feedbacks across different mycorrhizal types (Kadowaki et al., in review). In this respect, the observed asymmetry in infection patterns of arbuscular mycorrhizal and ectomycorrhizal fungi (Figure 2A) helps us postulate possible directionality in such across-mycorrhizal-type dynamics of plant and below-ground fungal communities.
Also intriguingly, this study identified a number of endophytic fungi associated with both arbuscular mycorrhizal and ectomycorrhizal plants and those specific to either mycorrhizal type of plant species (Tables 1, 2; Figure 5). Given the prevalence of endophytic fungi and their functional effects on host plant growth (Jumpponen and Trappe, 1998; Jumpponen, 2001; Newsham, 2011), understanding of plant–soil feedbacks would be never complete without taking into account the entire association networks involving not only mycorrhizal but also non-mycorrhizal fungi. Among endophytic fungal taxa potentially playing pivotal roles in such plant–soil feedbacks, the ascomycete order Helotiales (Tedersoo et al., 2009; Almario et al., 2017; Nakamura et al., 2017) is of particular interest because they included not only OTUs specific to either arbuscular mycorrhizal or ectomycorrhizal plant species but also generalist OTUs associated with both categories of host plants. Experimental studies testing the roles of host-specific and generalist endophytic fungi are awaited to build frameworks for describing and forecasting forest community dynamics.
Author Contributions
HT designed the work. HT and HS performed fieldwork. HT conducted molecular experiment. HT wrote the manuscript with HS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Takashi Kitaba, Tomio, Morimoto, and Mieko Kiyono for their support in fieldwork and reviewers for their helpful comments. This work was financially supported by JSPS KAKENHI Grant (26711026 & 15KT0032) and JST PRESTO (JPMJPR16Q6) to HT.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00433/full#supplementary-material
References
Almario, J., Jeena, G., Wunder, J., Langen, G., Zuccaro, A., Coupland, G., et al. (2017). Root-associated fungal microbiota of nonmycorrhizal Arabis alpina and its contribution to plant phosphorus nutrition. Proc. Natl. Acad. Sci. U.S.A. 114, E9403–E9412. doi: 10.1073/pnas.1710455114
AL-Shammari, T. A., Bahkali, A. H., Elgorban, A. M., El-Kahky, M. T., and Al-Sum, B. A. (2013). The use of Trichoderma longibrachiatum and Mortierella alpina against root-knot nematode, Meloidogyne javanica on tomato. J. Pure Appl. Microbiol. 7, 199–207.
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Anderson, M. J. (2006). Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62, 245–253. doi: 10.1111/j.1541-0420.2005.00440.x
Azcón-Aguilar, C., and Barea, J. (1997). Arbuscular mycorrhizas and biological control of soil-borne plant pathogens–an overview of the mechanisms involved. Mycorrhiza 6, 457–464. doi: 10.1007/s005720050147
Bennett, J. A., Maherali, H., Reinhart, K. O., Lekberg, Y., Hart, M. M., and Klironomos, J. (2017). Plant-soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science 355, 181–184. doi: 10.1126/science.aai8212
Bever, J. D. (2002). Negative feedback within a mutualism: host–specific growth of mycorrhizal fungi reduces plant benefit. Proc. R. Soc. Lond. Ser. B 269, 2595–2601. doi: 10.1098/rspb.2002.2162
Bever, J. D., Dickie, I. A., Facelli, E., Facelli, J. M., Klironomos, J., Moora, M., et al. (2010). Rooting theories of plant community ecology in microbial interactions. Trends Ecol. Evol. 25, 468–478. doi: 10.1016/j.tree.2010.05.004
Booth, M. G. (2004). Mycorrhizal networks mediate overstorey-understorey competition in a temperate forest. Ecol. Lett. 7, 538–546. doi: 10.1111/j.1461-0248.2004.00605.x
Borowicz, V. A. (2001). Do arbuscular mycorrhizal fungi alter plant–pathogen relations? Ecology 82, 3057–3068. doi: 10.2307/2679834
Botnen, S., Vik, U., Carlsen, T., Eidesen, P. B., Davey, M. L., and Kauserud, H. (2014). Low host specificity of root-associated fungi at an Arctic site. Mol. Ecol. 23, 975–985. doi: 10.1111/mec.12646
Cao, L., You, J., and Zhou, S. (2002). Endophytic fungi from Musa acuminata leaves and roots in South China. World J. Microbiol. Biotechnol. 18, 169–171. doi: 10.1023/A:1014491528811
Chase, J. M., Kraft, N. J., Smith, K. G., Vellend, M., and Inouye, B. D. (2011). Using null models to disentangle variation in community dissimilarity from variation in α-diversity. Ecosphere 2, 1–11. doi: 10.1890/ES10-00117.1
Chazdon, R. L., Chao, A., Colwell, R. K., Lin, S.-Y., Norden, N., Letcher, S. G., et al. (2011). A novel statistical method for classifying habitat generalists and specialists. Ecology 92, 1332–1343. doi: 10.1890/10-1345.1
Chen, C., Verkley, G. J., Sun, G., Groenewald, J. Z., and Crous, P. W. (2016). Redefining common endophytes and plant pathogens in Neofabraea, Pezicula, and related genera. Fungal Biol. 120, 1291–1322. doi: 10.1016/j.funbio.2015.09.013
Couture, M., Fortin, J., and Dalpe, Y. (1983). Oidiodendron griseum Robak: An endophyte of ericoid mycorrhiza in Vaccinium spp. New Phytol. 95, 375–380. doi: 10.1111/j.1469-8137.1983.tb03505.x
Dickie, I. A., Koide, R. T., and Fayish, A. C. (2001). Vesicular–arbuscular mycorrhizal infection of Quercus rubra seedlings. New Phytol. 151, 257–264. doi: 10.1046/j.1469-8137.2001.00148.x
Douglas, G., Heslin, M., and Reid, C. (1989). Isolation of Oidiodendron maius from Rhododendron and ultrastructural characterization of synthesized mycorrhizas. Can. J. Bot. 67, 2206–2212. doi: 10.1139/b89-280
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Eroshin, V., and Dedyukhina, E. (2002). Effect of lipids from Mortierella hygrophila on plant resistance to phytopathogens. World J. Microbiol. Biotechnol. 18, 165–167. doi: 10.1023/A:1014429527591
Grover, M., Ali, S. Z., Sandhya, V., Rasul, A., and Venkateswarlu, B. (2011). Role of microorganisms in adaptation of agriculture crops to abiotic stresses. World J. Microbiol. Biotechnol. 27, 1231–1240. doi: 10.1007/s11274-010-0572-7
Hamady, M., Walker, J. J., Harris, J. K., Gold, N. J., and Knight, R. (2008). Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5, 235–237. doi: 10.1038/nmeth.1184
Hambleton, S., and Sigler, L. (2005). Meliniomyces, a new anamorph genus for root-associated fungi with phylogenetic affinities to Rhizoscyphus ericae (= Hymenoscyphus ericae), Leotiomycetes. Stud. Mycol. 53, 1–27. doi: 10.3114/sim.53.1.1
Hibbett, D. S., and Matheny, P. B. (2009). The relative ages of ectomycorrhizal mushrooms and their plant hosts estimated using Bayesian relaxed molecular clock analyses. BMC Biol. 7:13. doi: 10.1186/1741-7007-7-13
Hiiesalu, I., Pärtel, M., Davison, J., Gerhold, P., Metsis, M., Moora, M., et al. (2014). Species richness of arbuscular mycorrhizal fungi: associations with grassland plant richness and biomass. New Phytol. 203, 233–244. doi: 10.1111/nph.12765
Hiruma, K., Gerlach, N., Sacristán, S., Nakano, R. T., Hacquard, S., Kracher, B., et al. (2016). Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165, 464–474. doi: 10.1016/j.cell.2016.02.028
Huang, W., Cai, Y., Hyde, K., Corke, H., and Sun, M. (2008). Biodiversity of endophytic fungi associated with 29 traditional Chinese medicinal plants. Fungal Divers. 33, 61–75.
Huson, D. H., Auch, A. F., Qi, J., and Schuster, S. C. (2007). MEGAN analysis of metagenomic data. Genome Res. 17, 377–386. doi: 10.1101/gr.5969107
Jumpponen, A. (2001). Dark septate endophytes-are they mycorrhizal? Mycorrhiza 11, 207–211. doi: 10.1007/s005720100112
Jumpponen, A., and Trappe, J. M. (1998). Dark septate endophytes: a review of facultative biotrophic root-colonizing fungi. New Phytol. 140, 295–310. doi: 10.1046/j.1469-8137.1998.00265.x
Kardol, P., Cornips, N. J., van Kempen, M. M., Bakx-Schotman, J., and van der Putten, W. H. (2007). Microbe-mediated plant–soil feedback causes historical contingency effects in plant community assembly. Ecol. Monogr. 77, 147–162. doi: 10.1890/06-0502
Kennedy, P. G., Peay, K. G., and Bruns, T. D. (2009). Root tip competition among ectomycorrhizal fungi: are priority effects a rule or an exception? Ecology 90, 2098–2107. doi: 10.1890/08-1291.1
Kernaghan, G., and Patriquin, G. (2011). Host associations between fungal root endophytes and boreal trees. Microbial Ecol. 62, 460–473. doi: 10.1007/s00248-011-9851-6
Klironomos, J. N. (2002). Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417, 67–70. doi: 10.1038/417067a
Kõljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F., Bahram, M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481
Landvik, S., Schumacher, T. K., Eriksson, O. E., and Moss, S. T. (2003). Morphology and ultrastructure of Neolecta species. Mycol. Res. 107, 1021–1031. doi: 10.1017/S0953756203008219
Lindahl, B. D., and Tunlid, A. (2015). Ectomycorrhizal fungi–potential organic matter decomposers, yet not saprotrophs. New Phytol. 205, 1443–1447. doi: 10.1111/nph.13201
Liu, B., Li, H., Zhu, B., Koide, R. T., Eissenstat, D. M., and Guo, D. (2015). Complementarity in nutrient foraging strategies of absorptive fine roots and arbuscular mycorrhizal fungi across 14 coexisting subtropical tree species. New Phytol. 208, 125–136. doi: 10.1111/nph.13434
Lundberg, D. S., Yourstone, S., Mieczkowski, P., Jones, C. D., and Dangl, J. L. (2013). Practical innovations for high-throughput amplicon sequencing. Nat. Methods 10, 999–1002. doi: 10.1038/nmeth.2634
Mandyam, K., and Jumpponen, A. (2005). Seeking the elusive function of the root-colonising dark septate endophytic fungi. Stud. Mycol. 53, 173–189. doi: 10.3114/sim.53.1.173
Mangan, S. A., Schnitzer, S. A., Herre, E. A., Mack, K. M., Valencia, M. C., Sanchez, E. I., et al. (2010). Negative plant-soil feedback predicts tree-species relative abundance in a tropical forest. Nature 466, 752–755. doi: 10.1038/nature09273
Marx, D. H. (1972). Ectomycorrhizae as biological deterrents to pathogenic root infections. Ann. Rev. Phytopathol. 10, 429–454. doi: 10.1146/annurev.py.10.090172.002241
McGuire, K. L. (2007). Common ectomycorrhizal networks may maintain monodominance in a tropical rain forest. Ecology 88, 567–574. doi: 10.1890/05-1173
Morita, S., Azuma, M., Aoba, T., Satou, H., Narisawa, K., and Hashiba, T. (2003). Induced systemic resistance of Chinese cabbage to bacterial leaf spot and Alternaria leaf spot by the root endophytic fungus, Heteroconium chaetospira. J. Gen. Plant Pathol. 69, 71–75. doi: 10.1007/s10327-002-0005-z
Nakamura, N., Tanaka, E., Tanaka, C., and Takeuchi-Kaneko, Y. (2017). Localization of helotialean fungi on ectomycorrhizae of Castanopsis cuspidata visualized by in situ hybridization. Mycorrhiza 28, 17–28. doi: 10.1007/s00572-017-0803-y
Narisawa, K., Kawamata, H., Currah, R. S., and Hashiba, T. (2002). Suppression of Verticillium wilt in eggplant by some fungal root endophytes. Eur. J. Plant Pathol. 108, 103–109. doi: 10.1023/A:1015080311041
Newsham, K. K. (2011). A meta-analysis of plant responses to dark septate root endophytes. New Phytol. 190, 783–793. doi: 10.1111/j.1469-8137.2010.03611.x
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Noble, H. M., Langley, D., Sidebottom, P., Lane, S., and Fisher, P. (1991). An echinocandin from an endophytic Cryptosporiopsis sp. and Pezicula sp. in Pinus sylvestris and Fagus sylvatica. Mycol. Res. 95, 1439–1440. doi: 10.1016/S0953-7562(09)80401-2
Oksanen, J., Blanachet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O'Hara, R. B., et al. (2012). Vegan: Community Ecology Package. R Package Version 2.0-3 Available online at: http://CRAN.R-project.org/package=vegan
Peay, K. G., Kennedy, P. G., and Talbot, J. M. (2016). Dimensions of biodiversity in the Earth mycobiome. Nat. Rev. Microbiol. 14, 434–447. doi: 10.1038/nrmicro.2016.59
Peay, K. G., Russo, S. E., McGuire, K. L., Lim, Z., Chan, J. P., Tan, S., et al. (2015). Lack of host specificity leads to independent assortment of dipterocarps and ectomycorrhizal fungi across a soil fertility gradient. Ecol. Lett. 18, 807–816. doi: 10.1111/ele.12459
Plattner, I., and Hall, I. (1995). Parasitism of non-host plants by the mycorrhizal fungus Tuber melanosporum. Mycol. Res. 99, 1367–1370. doi: 10.1016/S0953-7562(09)81223-9
Porras-Alfaro, A., and Bayman, P. (2011). Hidden fungi, emergent properties: endophytes and microbiomes. Ann. Rev. Phytopathol. 49, 291–315. doi: 10.1146/annurev-phyto-080508-081831
R-Core-Team (2015). R 3.4.1: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: http://www.R-project.org/
Read, D., and Haselwandter, K. (1981). Observations on the mycorrhizal status of some alpine plant communities. New Phytol. 88, 341–352. doi: 10.1111/j.1469-8137.1981.tb01729.x
Redhead, S. A. (1979). Mycological observations: 1, on Cristulariella; 2, on Valdensinia; 3, on Neolecta. Mycologia 71, 1248–1253. doi: 10.2307/3759112
Remy, W., Taylor, T. N., Hass, H., and Kerp, H. (1994). Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. U.S.A. 91, 11841–11843. doi: 10.1073/pnas.91.25.11841
Rice, A. V., and Currah, R. S. (2005). Oidiodendron: A survey of the named species and related anamorphs of Myxotrichum. Stud. Mycol. 53, 83–120. doi: 10.3114/sim.53.1.83
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Sato, H., and Murakami, N. (2008). Reproductive isolation among cryptic species in the ectomycorrhizal genus Strobilomyces: population-level CAPS marker-based genetic analysis. Mol. Phylogenet. Evol. 48, 326–334. doi: 10.1016/j.ympev.2008.01.033
Sato, H., Tanabe, A. S., and Toju, H. (2015). Contrasting diversity and host association of ectomycorrhizal basidiomycetes versus root-associated ascomycetes in a dipterocarp rainforest. PLoS ONE 10:e0125550. doi: 10.1371/journal.pone.0125550
Schulz, B., Sucker, J., Aust, H., Krohn, K., Ludewig, K., Jones, P., et al. (1995). Biologically active secondary metabolites of endophytic Pezicula species. Mycol. Res. 99, 1007–1015. doi: 10.1016/S0953-7562(09)80766-1
Schüβler, A., Schwarzott, D., and Walker, C. (2001). A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol. Res. 105, 1413–1421. doi: 10.1017/S0953756201005196
Stevens, J. L., Jackson, R. L., and Olson, J. B. (2013). Slowing PCR ramp speed reduces chimera formation from environmental samples. J. Microbiol. Methods 93, 203–205. doi: 10.1016/j.mimet.2013.03.013
Talbot, J., Allison, S., and Treseder, K. (2008). Decomposers in disguise: mycorrhizal fungi as regulators of soil C dynamics in ecosystems under global change. Func. Ecol. 22, 955–963. doi: 10.1111/j.1365-2435.2008.01402.x
Tanabe, A. S. (2016). Claident v0.2.2016.07.05, a Software Distributed by Author. Available online at: http://www.fifthdimension.jp/
Tanabe, A. S., and Toju, H. (2013). Two new computational methods for universal DNA barcoding: a benchmark using barcode sequences of bacteria, archaea, animals, fungi, and land plants. PLoS ONE 8:e76910. doi: 10.1371/journal.pone.0076910
Tedersoo, L., May, T. W., and Smith, M. E. (2010). Ectomycorrhizal lifestyle in fungi: global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza 20, 217–263. doi: 10.1007/s00572-009-0274-x
Tedersoo, L., Pärtel, K., Jairus, T., Gates, G., Põldmaa, K., and Tamm, H. (2009). Ascomycetes associated with ectomycorrhizas: molecular diversity and ecology with particular reference to the Helotiales. Environ. Microbiol. 11, 3166–3178. doi: 10.1111/j.1462-2920.2009.02020.x
Tedersoo, L., and Smith, M. E. (2013). Lineages of ectomycorrhizal fungi revisited: foraging strategies and novel lineages revealed by sequences from belowground. Fungal Biol. Rev. 27, 83–99. doi: 10.1016/j.fbr.2013.09.001
Thiéry, O., Vasar, M., Jairus, T., Davison, J., Roux, C., Kivistik, P. A., et al. (2016). Sequence variation in nuclear ribosomal small subunit, internal transcribed spacer and large subunit regions of Rhizophagus irregularis and Gigaspora margarita is high and isolate-dependent. Mol. Ecol. 25, 2816–2832. doi: 10.1111/mec.13655
Toju, H., Guimarães, P. R. Jr., Olesen, J. M., and Thompson, J. N. (2014a). Assembly of complex plant–fungus networks. Nat. Commun. 5:5273. doi: 10.1038/ncomms6273
Toju, H., Guimarães, P. R. Jr., Olesen, J. M., and Thompson, J. N. (2015). Below-ground plant–fungus network topology is not congruent with above-ground plant–animal network topology. Sci. Adv. 1:e1500291. doi: 10.1126/sciadv.1500291
Toju, H., Sato, H., and Tanabe, A. S. (2014b). Diversity and spatial structure of belowground plant–fungal symbiosis in a mixed subtropical forest of ectomycorrhizal and arbuscular mycorrhizal plants. PLoS ONE 9:e86566. doi: 10.1371/journal.pone.0086566
Toju, H., Tanabe, A., and Ishii, H. (2016a). Ericaceous plant–fungus network in a harsh alpine–subalpine environment. Mol. Ecol. 25, 3242–3257. doi: 10.1111/mec.13680
Toju, H., Tanabe, A. S., Yamamoto, S., and Sato, H. (2012). High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 7:e40863. doi: 10.1371/journal.pone.0040863
Toju, H., Yamamoto, S., Sato, H., and Tanabe, A. S. (2013a). Sharing of diverse mycorrhizal and root-endophytic fungi among plant species in an oak-dominated cool–temperate forest. PLoS ONE 8:e78248. doi: 10.1371/journal.pone.0078248
Toju, H., Yamamoto, S., Sato, H., Tanabe, A. S., Gilbert, G. S., and Kadowaki, K. (2013b). Community composition of root-associated fungi in a Quercus-dominated temperate forest: “codominance” of mycorrhizal and root-endophytic fungi. Ecol. Evol. 3, 1281–1293. doi: 10.1002/ece3.546
Toju, H., Yamamoto, S., Tanabe, A. S., Hayakawa, T., and Ishii, H. S. (2016b). Network modules and hubs in plant–root fungal biome. J. R. Soc. Interface 13:20151097. doi: 10.1098/rsif.2015.1097
Usuki, F., and Narisawa, K. (2007). A mutualistic symbiosis between a dark septate endophytic fungus, Heteroconium chaetospira, and a nonmycorrhizal plant, Chinese cabbage. Mycologia 99, 175–184. doi: 10.1080/15572536.2007.11832577
van der Heijden, M. G., Bardgett, R. D., and van Straalen, N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
van der Putten, W. H., Bardgett, R. D., Bever, J. D., Bezemer, T. M., Casper, B. B., Fukami, T., et al. (2013). Plant–soil feedbacks: the past, the present and future challenges. J. Ecol. 101, 265–276. doi: 10.1111/1365-2745.12054
Vohník, M., Albrechtová, J., and Vosátka, M. (2005). The inoculation with Oidiodendron maius and Phialocephala fortinii alters phosphorus and nitrogen uptake, foliar C: N ratio and root biomass distribution in Rhododendron cv. Azurro. Symbiosis 40, 87–96.
Vohník, M., Mrnka, L., Lukešová, T., Bruzone, M. C., Kohout, P., and Fehrer, J. (2013). The cultivable endophytic community of Norway spruce ectomycorrhizas from microhabitats lacking ericaceous hosts is dominated by ericoid mycorrhizal Meliniomyces variabilis. Fungal Ecol. 6, 281–292. doi: 10.1016/j.funeco.2013.03.006
Vrålstad, T., Myhre, E., and Schumacher, T. (2002a). Molecular diversity and phylogenetic affinities of symbiotic root-associated ascomycetes of the Helotiales in burnt and metal polluted habitats. New Phytol. 155, 131–148. doi: 10.1046/j.1469-8137.2002.00444.x
Vrålstad, T., Schumacher, T., and Taylor, A. F. (2002b). Mycorrhizal synthesis between fungal strains of the Hymenoscyphus ericae aggregate and potential ectomycorrhizal and ericoid hosts. New Phytol. 153, 143–152. doi: 10.1046/j.0028-646X.2001.00290.x
Wakelin, S. A., Warren, R. A., Harvey, P. R., and Ryder, M. H. (2004). Phosphate solubilization by Penicillium spp. closely associated with wheat roots. Biol. Fertil. Soils 40, 36–43. doi: 10.1007/s00374-004-0750-6
Wang, J., Wang, G., Zhang, Y., Zheng, B., Zhang, C., and Wang, L. (2014). Isolation and identification of an endophytic fungus Pezicula sp. in Forsythia viridissima and its secondary metabolites. World J. Microbiol. Biotechnol. 30, 2639–2644. doi: 10.1007/s11274-014-1686-0
Warton, D. I., Blanchet, F. G., O'Hara, R. B., Ovaskainen, O., Taskinen, S., Walker, S. C., et al. (2015). So many variables: joint modeling in community ecology. Trends Ecol. Evol. 30, 766–779. doi: 10.1016/j.tree.2015.09.007
Watanabe, T. (2010). Pictorial Atlas of Soil and Seed Fungi: Morphologies of Cultured Fungi and Key to Species Boca Raton, FL: CRC press.
Werner, G. D., and Kiers, E. T. (2015). Order of arrival structures arbuscular mycorrhizal colonization of plants. New Phytol. 205, 1515–1524. doi: 10.1111/nph.13092
Yamaji, K., Watanabe, Y., Masuya, H., Shigeto, A., Yui, H., and Haruma, T. (2016). Root fungal endophytes enhance heavy-metal stress tolerance of Clethra barbinervis growing naturally at mining sites via growth enhancement, promotion of nutrient uptake and decrease of heavy-metal concentration. PLoS ONE 11:e0169089. doi: 10.1371/journal.pone.0169089
Keywords: arbuscular mycorrhizal fungi, Chaetothyriales, dark septate endophytes, ectomycorrhizal fungi, ecological communities, Helotiales, host specificity, Illumina
Citation: Toju H and Sato H (2018) Root-Associated Fungi Shared Between Arbuscular Mycorrhizal and Ectomycorrhizal Conifers in a Temperate Forest. Front. Microbiol. 9:433. doi: 10.3389/fmicb.2018.00433
Received: 16 December 2017; Accepted: 26 February 2018;
Published: 12 March 2018.
Edited by:
Stéphane Hacquard, Max Planck Institute for Plant Breeding Research (MPG), GermanyReviewed by:
Ravin Poudel, University of Florida, United StatesKezia Goldmann, Helmholtz-Zentrum für Umweltforschung (UFZ), Germany
Copyright © 2018 Toju and Sato. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hirokazu Toju, dG9qdS5oaXJva2F6dS40Y0BreW90by11LmFjLmpw
 Hirokazu Toju
Hirokazu Toju Hirotoshi Sato3
Hirotoshi Sato3