Abstract
The genus Nocardiopsis is an unique actinobacterial group that widely distributed in hypersaline environments. In this study, we investigated the growth conditions, transcriptome analysis, production and accumulation of ectoine by Nocardiopsis gilva YIM 90087T under salt stress. The colony color of N. gilva YIM 90087T changed from yellow to white under salt stress conditions. Accumulation of ectoine and hydroxyectoine in cells was an efficient way to regulate osmotic pressure. The ectoine synthesis was studied by transferring the related genes (ectA, ectB, and ectC) to Escherichia coli. Transcriptomic analysis showed that the pathways of ABC transporters (ko02010) and glycine, serine, and threonine metabolism (ko00260) played a vital role under salt stress environment. The ectABC from N. gilva YIM 90087T was activated under the salt stress. Addition of exogenous ectoine and hydroxyectoine were helpful to protect N. gilva YIM 90087T from salt stress.
Introduction
The genus Nocardiopsis is an unique actinobacterial group which was first described by Meyer (1976). Members of this genus are widely distributed in hypersaline environments and produce very distinct secondary metabolites (Tsujibo et al., 2003; Sun et al., 2015, 2017; Sharma and Singh, 2016). In past few years, attempts to isolate novel Nocardiopsis strains were made and many novel species such as N. algeriensis (Bouras et al., 2015), N. mangrovei (Huang et al., 2015), N. oceani and N. nanhaiensis (Pan et al., 2015), N. ansamitocini (Zhang et al., 2016b), N. sediminis (Muangham et al., 2016), N. rhizosphaerae (Zhang et al., 2016c), and N. akesuensis (Gao et al., 2016) have been reported. At present, the genus comprises 53 species and 5 subspecies (12017). A large number of Nocardiopsis species were isolated from saline or alkaline environments and approximately two-thirds of them were halophilic or halotolerant (Bouras et al., 2015; Pan et al., 2015). To survive in hypersaline environments, Nocardiopsis uses various strategies such as reinforcement of cell walls and accumulation of various osmolytes (Ameur et al., 2011; Zhang et al., 2016a). Accumulation of osmolytes play a vital role during salt stress and help in providing osmotic balance without interfering with the essential cellular processes and the normal metabolism (Liu et al., 2017). One of the most abundant osmolytes present in nature is ectoine, which was first found in the halophilic bacterium Ectothiorhodospira halochloris (Galinski et al., 1985). Ectoines found to improve protein folding and protect biomolecules such as enzymes, nucleic acids, antibodies, and even whole cells against various stress conditions (Barth et al., 2000).
The next-generation sequencing technologies and whole genome sequencing provide a new perspective for the research (Liu et al., 2017). In the past few years, about 22 genomes of Nocardiopsis have been sequenced (22017). Comparative genomic analysis of Nocardiopsis species revealed that they retain core genome [at least 2,517 genes (approximately 43% of the genome content)] and also continuously acquire genes and expand their genomes to cope with environmental pressures (Li et al., 2013). Interestingly, the genes related to ectoine biosynthesis were found in genomes of all Nocardiopsis species.
Though extensive research was conducted to isolate novel Nocardiopsis members and analyzing their genome, yet only few reports focused on understanding mechanism for their survival under salt stress.
The objectives of the present study were: (1) compare the growth status of Nocardiopsis gilva YIM 90087T under different saline conditions. (2) Analyze intracellular and extracellular concentrations of ectoine and hydroxyectoine under different salt stress. (3) Evaluate the mRNA expressions of the genes related to ectoine biosynthesis by RNA-seq sequencing under different salt stress. (4) Study the effects of ectoine and hydroxyectoine on the growth of N. gilva YIM 90087T in presence and absence of salt stress. (5) Investigate the effects of saline conditions on ectoine biosynthesis of N. gilva YIM 90087T, by reconstructing its biosynthetic pathway and studying the process of ectoine biosynthesis in E. coli BW25113. (6) Identify the ectoine from recombinant E. coli using liquid chromatography–mass spectrometry (LC–MS).
Materials and Methods
Strains and Media
Nocardiopsis gilva YIM 90087T, a halotolerant actinobacterium, used in this study was isolated by one of our group members from hypersaline soil in Xinjiang Province, China (Li et al., 2006). E. coli DH5α, and BW25113 [F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, rph-1, Δ(rhaD-rhaB)568, hsdR514] were used as the host strains for the construction of recombinant plasmids and ectoine biosynthesis, respectively.
2YT medium composed of (L-1) 16 g tryptone and 10 g yeast extract. Luria-Bertani (LB) medium composed of (L-1) 10 g NaCl, 10 g tryptone, and 5 g yeast extract. ZYM medium composed of (L-1) 10 g tryptone, 5 g yeast extract, 5 g glycerol, 0.5 g glucose, 25 mmol Na2HPO4, 50 mmol NH4Cl, 25 mmol KH2PO4, 5 mmol Na2SO4, 2 mmol MgSO4, 50 mmol FeCl2, 0.02 mmol CaCl2, 0.01 mmol MgCl2, 0.01 mmol ZnSO4, 0.002 mmol CoCl2, 0.002 mmol CuCl2, 0.002 mmol NiCl2, 0.002 mmol Na2MoO4, 0.002 mmol Na2SeO3, 0.002 mmol H3BO3, and 0.06 mmol HCl. EB buffer consisted of 100 mM sodium aspartate, 100 mM glutamate, 100 mM glycerol, 100 mM KCl, and 100 mM sodium phosphate buffer (pH 7.0).
Effect of Salt Concentrations on Colony Morphology, Growth, and Ectoine Biosynthesis of N. gilva YIM 90087T
To investigate the effects of NaCl concentrations on the growth of N. gilva YIM 90087T, 1 ml of culture was inoculated in 100 ml of 2YT medium having NaCl concentration ranging 0–15% w/v (with an increment of 5%). The flasks were incubated at 30°C for 96 h. The growth was estimated by measuring the optical density at 600 nm (OD600). The variations of colony morphology under different NaCl stress were evaluated using 2YT agar at 30°C for 96 h. To study the effects of NaCl concentartions on exogenous ectoine and hydroxyectoine, 1 mg/ml of ectoine and hydroxyectoine were added to 2YT medium. N. gilva YIM 90087T grew in these media under different salt concentartions at 30°C for 96 h.
Molecular Methods
Genomic DNA from N. gilva YIM 90087T was extracted and purified using the Bacterial DNA Kit (Omega Bio-Tek, United States). Plasmid DNA was extracted and purified from E. coli DH5α using the Plasmid Mini Kit I (Omega Bio-Tek). Restriction endonucleases, T4 DNA ligase, and Q5 High-Fidelity DNA polymerase (New England Biolabs, United States) were used according to the manufacturer’s instructions. Standard methods were used for transformation (Chong, 2001).
NgiectA and NgiectBC were amplified from the genomic DNA of N. gilva YIM 90087T (NCBI Reference Sequence: NZ_ANBG01000167.1) by using PCR primers NgiectA-F/R and NgiectBC-F/R as follows: NgiectA-F: 5′-CATGCCATGGGCTCCCGTGATAATTCCTCCC-3′, NgiectA-R: 5′-CGGGATCCTCAGTTCCGGCCGCTGACCGCCGAC-3′; NgiectBC-F: 5′-CGGGATCCAAGGAGATATACATGGAGATCTTCGACCGCCTCG-3′, NgiectBC-R: 5′-GGAATTCCTACTCGGCCGTGGCCAGGCTCTCC-3′ (Restriction enzyme cutting site were underlined, and ribosome bind site were in italicized). The NgiectA PCR product was digested with Nco I and BamH I, while NgiectBC PCR product was digested with BamH I and EcoR I. The two DNA fragments were inserted together into the sites of Nco I and EcoR I in pBAD-HisB (Invitrogen, United States) resulting in the plasmid pBNABC. The vector pBNABC was transformed into E. coli BW25113 (named pBNABC/BW25113).
The total RNA from YIM 90087T (culture of mid-log growth phase) was extracted using RNAprep Pure Cell/Bacteria Kit (TIANGEN, CN). RNA concentration was measured by UVS-99 (ActGene, United States). RNA quality was assessed by electrophoresis on a denaturing agarose gel. The samples of transcription were sequenced and analyzed by Biomarker Technologies, China.
Ectoine Biosynthesis in Recombinant E. coli
Ectoine biosynthesis was performed using the method of whole-cell catalysis in E. coli pBNABC/BW25113 (He et al., 2015). To obtain ectoine producer, E. coli pBNABC/BW25113 was grown in ZYM medium with ampicillin by adding 2 g/L L-arabinose as an inducer at 30°C, 200 rpm for 16 h. The induced cells were harvested by centrifugation (6000 × g, 20 min) and washed with 0.9% NaCl buffer twice. The cells were re-suspended with 50 ml of EB buffer (OD600 nm = 10). The catalysts mixtures were shaken in a 250-ml flask at 30°C, 200 rpm for 96 h.
HPLC and LC–MS Analysis of Ectoine and Hydroxyectoine
To identify ectoine and hydroxyectoine, 1 ml of cells suspension was harvested by centrifugation and extracted according to the method of Bligh and Dyer (1959). The intracellular and extracellular concentrations of ectoine and hydroxyectoine were measured by isocratic HPLC (Agilent 1260 series, Hewlett-Packard) following the method of He et al. (2015). The ectoine from recombinant E. coli was identified using Agilent 1260/6460 high-performance liquid chromatography/triple quadrupole mass spectrometer (Agilent, United States) using ESI source in positive ionization (He et al., 2015).
Results
Effect of Salt Concentrations on Colony Morphology and Growth of N. gilva YIM 90087T
The colony color of N. gilva YIM 90087T changed from yellow to white with the increase of salt concentration (Figure 1A). N. gilva YIM 90087T had the optimum growth at 5% NaCl concentration and the growth rate declined with the increase or decrease of the NaCl concentration (Figure 1B).
FIGURE 1
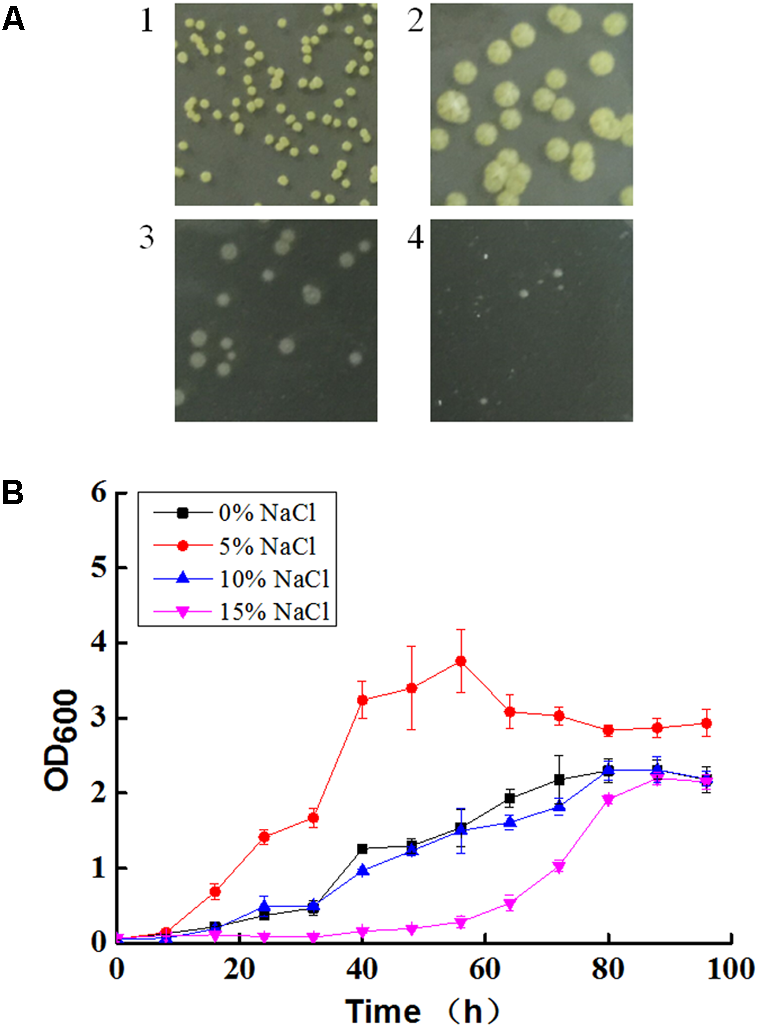
Colony morphology (A) and growth curve (B) of Nocardiopsis gilva YIM 90087T with varying concentrations of NaCl. A1: without NaCl; A2: 5% NaCl; A3: 10% NaCl; A4: 15% NaCl.
Differential Expression Analysis
FDR < 0.01 and Fold Change ≥ 2 were used as the selection criteria to identify the differentially expressed gene (DEG). The DEGs were annotated in COG, GO, KEGG, Swissport, and NR databases. The numbers of DEGs, up-regulated and down-regulated genes, and annotated DEGs were mentioned in Table 1. COG classifications of the three DEG sets were presented in 25 COG groups (Figure 2). Amino acid transport and metabolism and carbohydrate transport and metabolism were the dominant groups in all the three DEG sets. DEGs were distributed to 34 GO groups, which constitute three domains: cellular component, molecular function, and biological process (Figure 3). The DEGs from the three sets (5%/0%, 10%/0%, and 15%/0%) were mapped to 77, 32, and 90 reference canonical pathways in KEGG database respectively. The DEGs number and enrichment analysis were mentioned in Table 2. The DEGs number and corrected P-values results indicated that DEGs in the pathways of ABC transporters (ko02010) and glycine, serine, and threonine metabolism (ko00260) play extremely significant roles under salt stress. Particularly in maximum-tolerated NaCl concentrations, expressions of ABC transporters genes of ectoine/hydroxyectoine and glycine betaine, and ectoine/hydroxyectoine synthesis genes increased evidently.
Table 1
| DEGs set |
|||
|---|---|---|---|
| 5%/0% | 10%/0% | 15%/0% | |
| DEG number | 1026 | 231 | 1578 |
| Up-regulated | 493 | 62 | 621 |
| Down-regulated | 533 | 169 | 957 |
| COG | 500 | 102 | 796 |
| GO | 408 | 75 | 668 |
| KEGG | 300 | 58 | 510 |
| Swissprot | 797 | 151 | 1219 |
| NR | 977 | 207 | 1470 |
Statistics of DEGs and annotations.
FIGURE 2
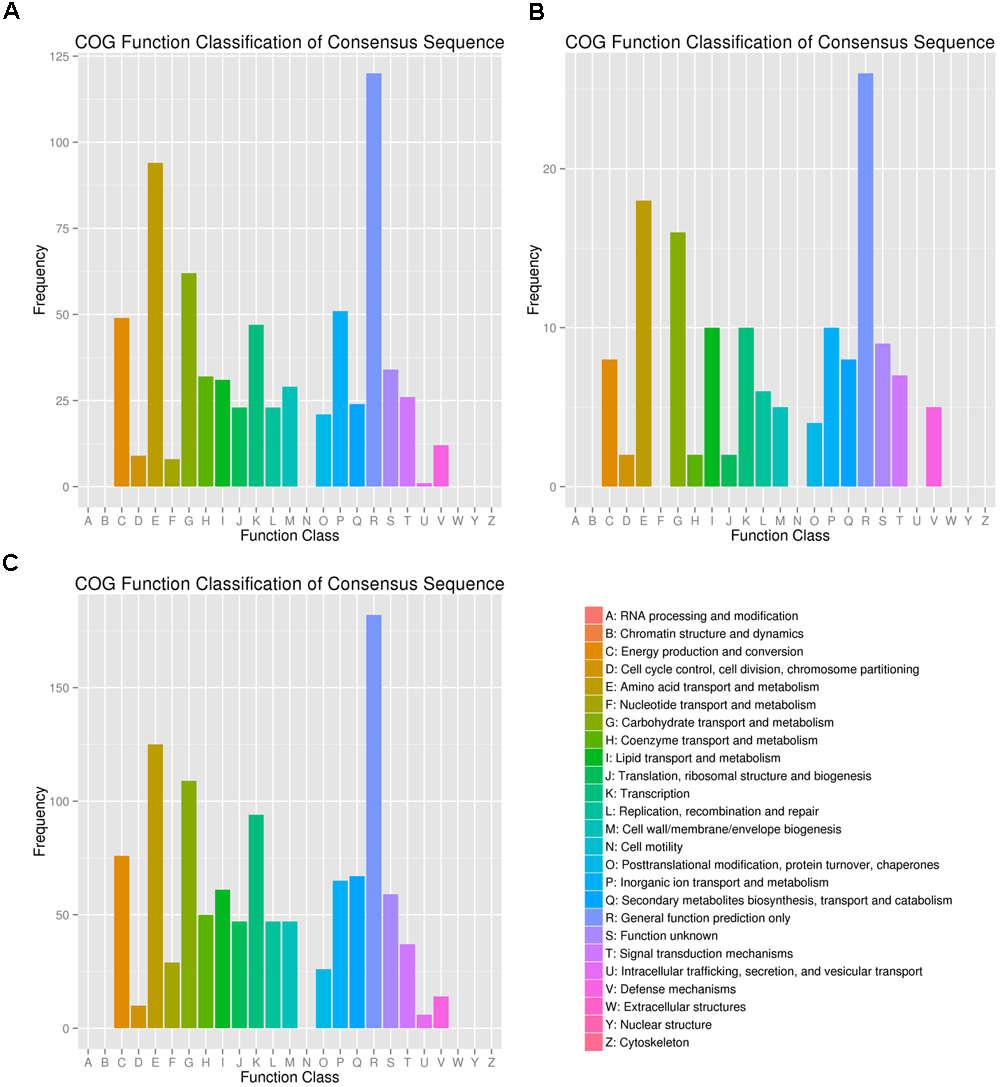
COG classifications of the three DEG sets. The DEGs from the three sets (A: 5%/0%; B: 10%/0%; C: 15%/0%) had COG classifications among the 25 groups. The capital letters on the x-axis indicates the COG groups as listed on the right of the histogram. The y-axis indicates the number of DEGs.
FIGURE 3
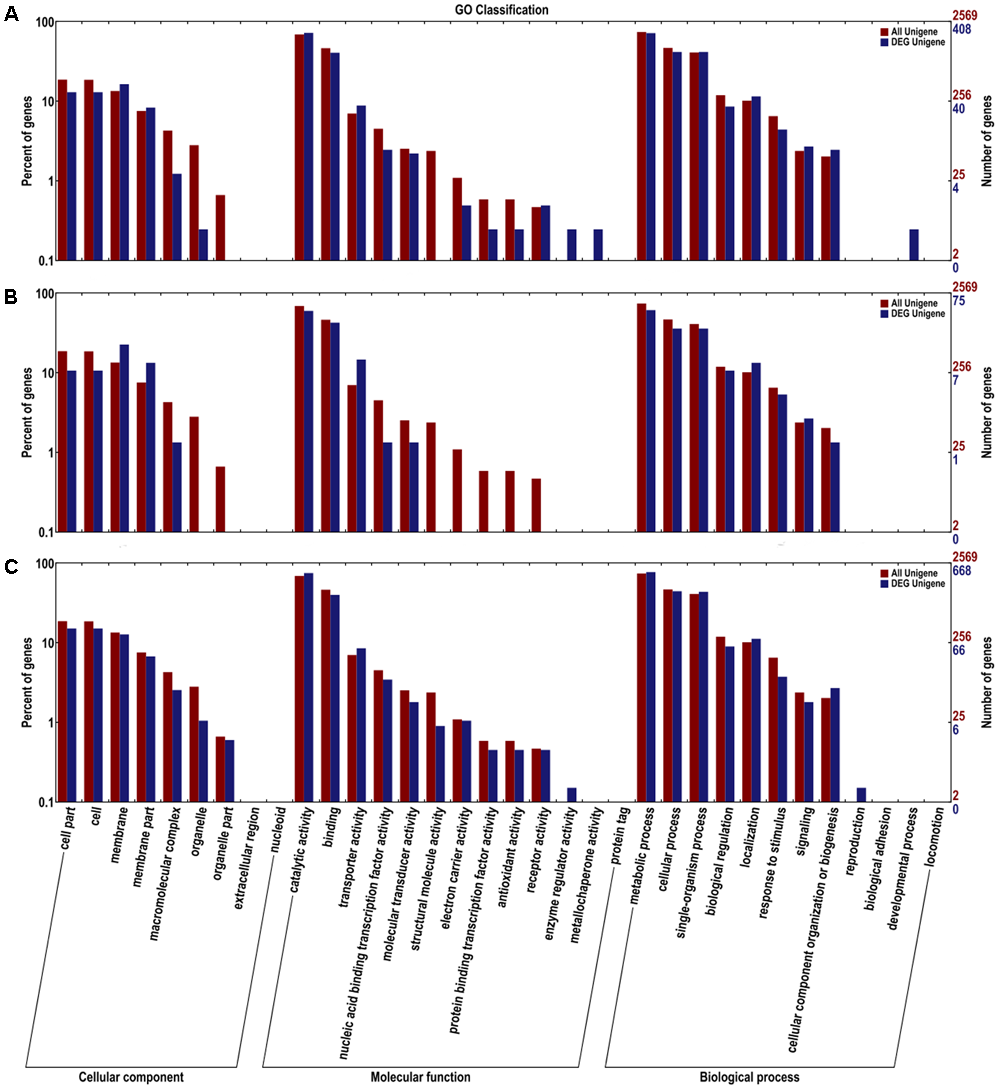
Gene ontology (GO) terms enrichments of the DEGs from the three sets. The DEGs were divided into three clusters: cellular component, molecular function, and biological process. (A) 5%/0%; (B) 10%/0%; (C) 15%/0%. The right y-axis presents the number of unigenes in the clusters, while the left y-axis presents the percentage of a specific category of unigenes in that clusters.
Table 2
| DEGs sets | Pathway number | Function | DEG number | Proportion (%) | Corrected P-value |
|---|---|---|---|---|---|
| 5%/0% | ko02010 | ABC transporters | 37 | 21.14 | 0.02 |
| ko00260 | Glycine, serine, and threonine metabolism | 14 | 8.00 | 0.2 | |
| ko00650 | Butanoate metabolism | 14 | 8.00 | 0.48 | |
| ko00230 | Purine metabolism | 12 | 6.86 | 1 | |
| ko02020 | Two-component system | 9 | 5.14 | 1 | |
| ko00280 | Valine, leucine, and isoleucine degradation | 9 | 5.14 | 1 | |
| ko00071 | Fatty acid metabolism | 9 | 5.14 | 1 | |
| ko00362 | Benzoate degradation | 9 | 5.14 | 1 | |
| ko00190 | Oxidative phosphorylation | 8 | 4.57 | 1 | |
| ko00290 | Valine, leucine, and isoleucine biosynthesis | 8 | 4.57 | 1 | |
| ko00500 | Starch and sucrose metabolism | 8 | 4.57 | 0.96 | |
| 10%/0% | ko02010 | ABC transporters | 8 | 29.63 | 0.42 |
| ko00500 | Starch and sucrose metabolism | 3 | 11.11 | 0.42 | |
| ko00260 | Glycine, serine, and threonine metabolism | 2 | 7.41 | 1 | |
| 15%/0% | ko02010 | ABC transporters | 34 | 10.4 | 1 |
| ko00260 | Glycine, serine, and threonine metabolism | 22 | 6.73 | 0.09 | |
| ko00230 | Purine metabolism | 18 | 5.5 | 1 | |
| ko00240 | Pyrimidine metabolism | 17 | 5.2 | 1 | |
| ko02020 | Two-component system | 16 | 4.89 | 1 | |
| ko00010 | Glycolysis/gluconeogenesis | 15 | 4.59 | 1 | |
| ko00860 | Porphyrin and chlorophyll metabolism | 15 | 4.59 | 1 | |
| ko00620 | Pyruvate metabolism | 14 | 4.28 | 1 | |
| ko00627 | Aminobenzoate degradation | 14 | 4.28 | 1 | |
| ko00650 | Butanoate metabolism | 12 | 3.67 | 1 | |
| ko00020 | Citrate cycle (TCA cycle) | 12 | 3.67 | 1 | |
| ko00030 | Pentose phosphate pathway | 12 | 3.67 | 0.86 | |
| ko00362 | Benzoate degradation | 12 | 3.67 | 1 | |
| ko00562 | Inositol phosphate metabolism | 10 | 3.06 | 0.29 | |
| ko02060 | Phosphotransferase system | 7 | 2.14 | 0.53 |
The DEG number, proportion, and Q-value in the pathway.
Genetic Organization of ect Gene Cluster in N. gilva YIM 90087T
We analyzed synthesis genes of ectoine and hydroxyectoine in Nocardiopsis. The ectABCD (synthesis genes) was found in Nocardiopsis genome. There are two common organizational types of ectABCD gene cluster on the 18 whole-genome sequences of Nocardiopsis strains (Figure 4). The ectABC were belonging to a gene cluster, and the ectD distributes on other locations of the genome (type A, 15 strains) or the ectABCD belongs to the same gene cluster (type B, 3 strains). Additionally, more than one ectAB or ectD genes were found in N. potens DSM 45234 and N. prasina DSM 43845. However, the organizational types of ect gene cluster from N. gilva YIM 90087T was clearly different from others Nocardiopsis strains. The ectA gene completely separated from ectBCD gene cluster, which deviate from the commonly found genetic organization. Moreover, the ehuABCD, ectoine/hydroxyectoine ABC transporter gene, were situated on the downstream of ectD gene in genome.
FIGURE 4
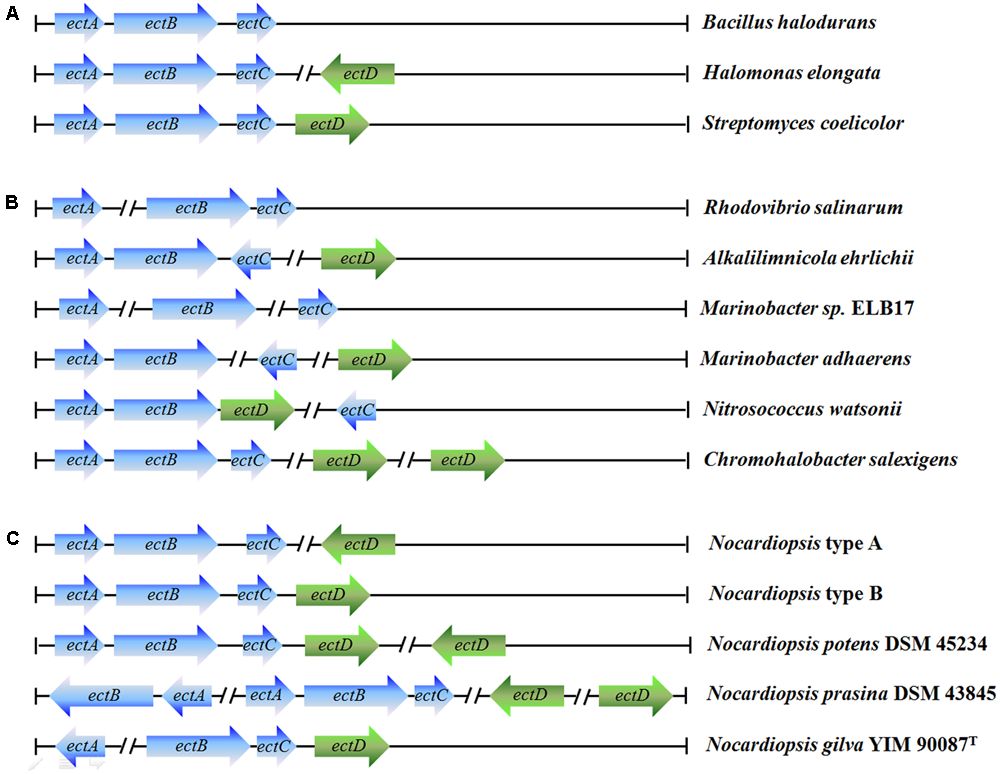
Genetic organization of the ectoine/hydroxyectoine biosynthesis gene clusters. The different types of ectoine and hydroxyectoine biosynthesis gene clusters present in microorganisms: (A) most common organizational types of the ect gene clusters and representative strains; (B) unusual types of the ect gene clusters and representative strains; (C) different types of the ect gene clusters from 21 Nocardiopsis strains.
Expression of Genes Involved in Synthesis and Transport of Ectoine and Hydroxyectoine
We analyzed the mRNA expression of the genes involved in ectoine/hydroxyectoine synthesis and transport at different salt concentrations (Table 3). The genes involved in ectoine biosynthesis process presented different levels of up-regulation with the increase of NaCl concentration, particularly in 15% NaCl. Unlike other genes, ectD presented two stepwise up-regulations from 5 to 10% NaCl concentration and from 10 to 15% NaCl concentration. Two potential ABC transporter genes (ehuABCD and proXWV) involved in transport of ectoine were found in N. gilva YIM 90087T. The mRNA expressions of them varied with increase NaCl concentration. The expressions of ehuABCD were positively correlated with NaCl concentration when the NaCl concentration was above 5% in the medium. Yet, the expressions of proXWV had no obvious changes in optimum and mild-tolerated NaCl concentrations. Interestingly, ehuABCD and proXWV were activated strongly at maximum-tolerated NaCl concentrations.
Table 3
| Unigene ID | Genes | Function | Fold change |
||
|---|---|---|---|---|---|
| 5%/0% | 10%/0% | 15%/0% | |||
| GL001695 | ask | Aspartate kinase | 1.29 | 1.51 | 4.45 |
| GL001694 | asd | L-Aspartate-beta-semialdehyde-dehydrogenase | 1.42 | 1.32 | 4.16 |
| GL003065 | ectB | Diaminobutyrate-2-oxoglutarate transaminase | 2.16 | 1.84 | 7.35 |
| GL001325 | ectA | Diaminobutyrate acetyltransferase | 1.33 | 2.23 | 5.26 |
| GL003066 | ectC | Ectoine synthase | 2.58 | 2.22 | 7.59 |
| GL003067 | ectD | Ectoine hydroxylase | 2.60 | 5.91 | 22.94 |
| GL003068 | ehuB | Ectoine/hydroxyectoine ABC transporter substrate-binding protein | 1.17 | 1.73 | 5.85 |
| GL003069 | ehuC | Ectoine/hydroxyectoine ABC transporter permease subunit | 0.88 | 2.67 | 10.48 |
| GL003070 | ehuD | Ectoine/hydroxyectoine ABC transporter permease subunit | 1.19 | 1.95 | 8.48 |
| GL003071 | ehuA | Ectoine/hydroxyectoine ABC transporter ATP-binding protein | 0.85 | 2.262 | 7.18 |
| GL003184 | proX | Glycine betaine/proline transport system substrate-binding protein | 0.80 | 0.90 | 4.07 |
| GL003185 | proX | Glycine betaine/proline transport system substrate-binding protein | 1.00 | 1.28 | 5.31 |
| GL003186 | proW | Glycine betaine/proline transport system permease protein | 0.90 | 1.57 | 9.49 |
| GL003187 | proV | Glycine betaine/proline transport system ATP-binding protein | 0.85 | 1.47 | 10.61 |
The fold changes of the genes involved in ectoine and hydroxyectoine synthesis and transport in response to different salt stress.
Accumulation of Ectoine and Hydroxyectoine in N. gilva YIM 90087T
Ectoine and hydroxyectoine were analyzed by HPLC. The results showed that there were two peaks with the same retention time as the two standards [ectoine (816189, Sigma-Aldrich) and hydroxyectoine (70709, Sigma-Aldrich)] appeared from N. gilva YIM 90087T cell. As shown in Figures 5A,B, the intracellular concentration changes of ectoine and hydroxyectoine were a cumulative progress. Whereas accumulation of ectoine and hydroxyectoine were influenced by NaCl concentration. They were positively correlated with increasing NaCl concentration. Noteworthy, hydroxyectoine were difficult to be synthesized in the medium with 0 or 5% NaCl. After 48 h, hydroxyectoine were detected in 2YT medium with 10 and 15% NaCl. Moreover, the amounts of ectoine and hydroxyectoine were increased with the increasing cultivation time. After 96 h, the highest level of ectoine (33.14 mg/g CDW) and hydroxyectoine (1.17 mg/g CDW) were detected in 15% NaCl concentration. The ectoine concentrations were 42.6 and 28.3 times higher than hydroxyectoine in 10 and 15% NaCl concentration, respectively. Therefore, the results indicates that ectoine was accumulated in N. gilva YIM 90087T cells.
FIGURE 5
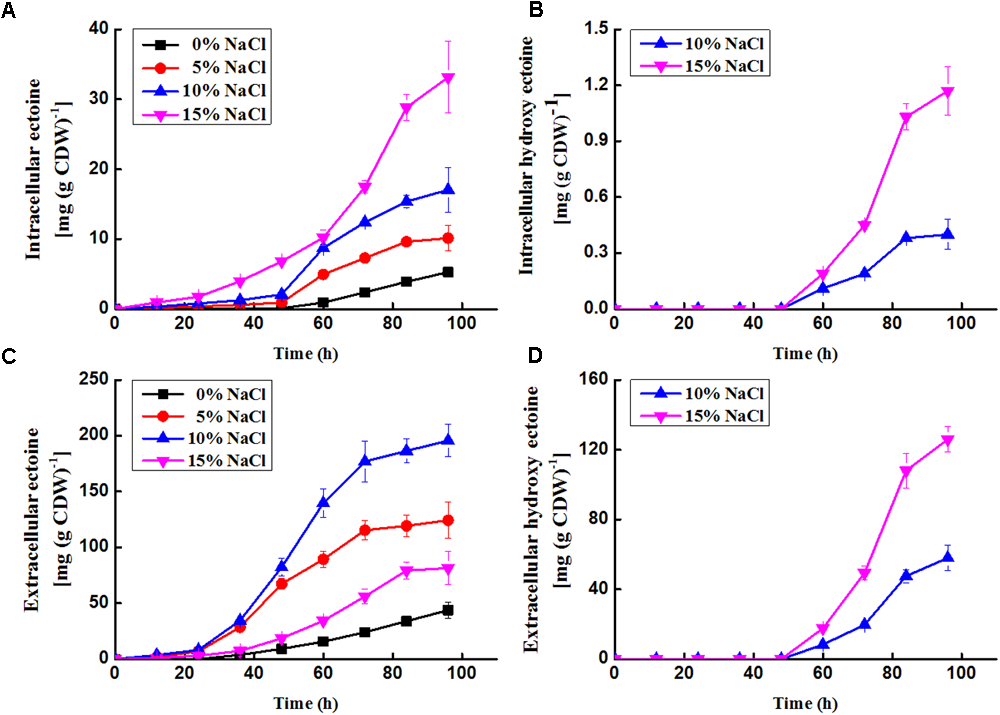
Time profiles of production of ectoine and hydroxyectoine in N. gilva. (A) Intercellular concentration of ectoine; (B) intercellular concentration of hydroxyectoine; (C) extracellular concentration of ectoine; (D) extracellular concentration of hydroxyectoine. 1 ml of the cells (N. gilva YIM 90087T, OD600 = 1) is about 0.00063 g (DCW).
On the other side, we detected ectoine and hydroxyectoine outside the cells (Figures 5C,D). A large proportion of ectoine was released out of the cells. After 96 h, 89.20, 92.46, 91.99, and 71.09% of ectoine were released in the medium with 0, 5, 10, and 15% NaCl, respectively. About 99.31 and 99.08% of hydroxyectoine found out of the cells in medium with 10 and 15% NaCl, respectively. The transfer capacity for ectoine enhanced remarkably in the medium with 15% NaCl. Yet, the capacity for hydroxyectoine had no obvious change as concentration added. In our study, we observed the transformation from ectoine to hydroxyectoine with the increase of NaCl concentration. About 50% ectoine transformed into hydroxyectoine at 15% NaCl concentration. The highest extracellular amount of ectoine and hydroxyectoine were found at 10 and 15% NaCl concentration, respectively.
Effect of Exogenous Ectoine and Hydroxyectoine on Growth of N. gilva YIM 90087T
Exogenous ectoine and hydroxyectoine under salt stress increased the growth rate of N. gilva YIM 90087T. The growth of N. gilva YIM 90087T in absence of NaCl was basically the same, in presence or absence of exogenous ectoine and hydroxyectoine (Figure 6A). However, with the increase of NaCl concentrations, exogenous ectoine, and hydroxyectoine promoted the growth (Figure 6). The addition of ectoine and hydroxyectoine under optimum NaCl concentration of (5%) slightly increased the OD600 values when compared in their absence (Figure 6B). The addition of ectoine and hydroxyectoine at high NaCl concentration (10–15%) drastically increased the OD600 values when compared in their absence (Figures 6C,D). It was also noted that the growth with hydroxyectoine was somewhat better than ectoine at 15% NaCl concentration.
FIGURE 6
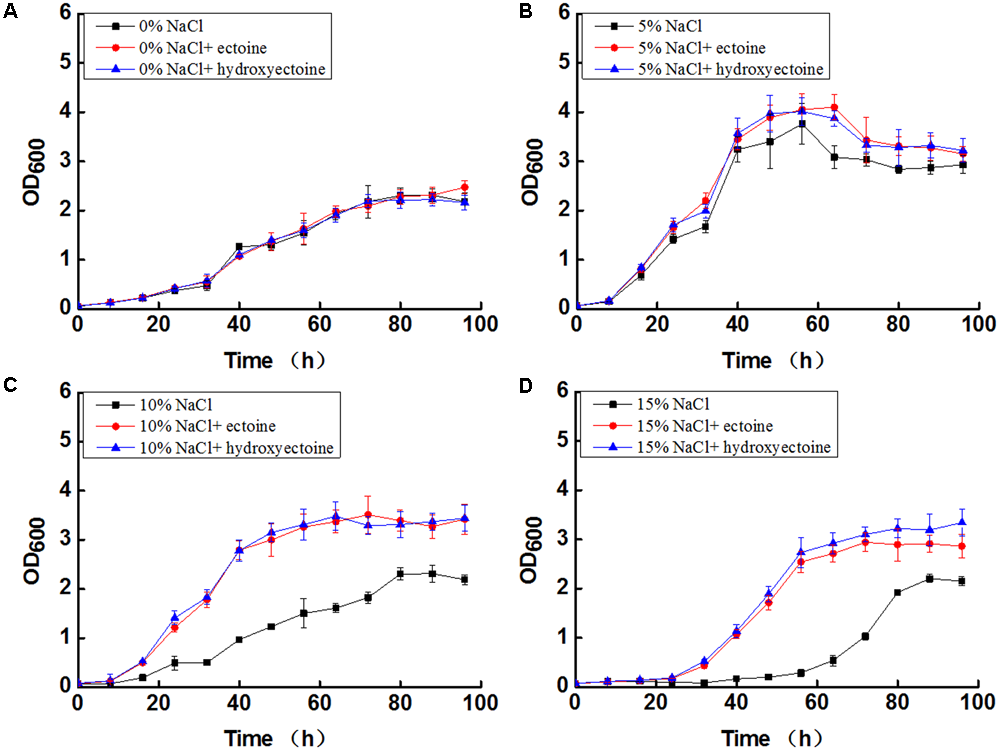
The effect of ectoine and hydroxyectoine on Growth of N. gilva YIM 90087T with varying concentrations of NaCl. (A) Curve of growth in the mediums with 0% NaCl; (B) curve of growth in the mediums with 5% NaCl; (C) curve of growth in the mediums with 10% NaCl; (D) curve of growth in the mediums with 15% NaCl.
Accumulation of Ectoine in E. coli
Using aspartate as a substrate, glutamate as amino donor and glycerol as an acetyl-CoA donor, ectoine was produced by E. coli pBNABC/BW25113. The changes in the production of ectoine were shown in Figure 7. Ectoine was produced hardly at 15% NaCl buffer, and a few of ectoine was detected in 0% NaCl buffer. The synthetic rate of ectoine dropped significantly after 24 and 48 h at 10 and 5% NaCl buffer, respectively. The maximum values of initial synthetic rate [0.5 mg (g CDW)-1 h-1] and final output [28.85 mg (g CDW)-1] were found in 10 and 5% NaCl buffer, respectively.
FIGURE 7
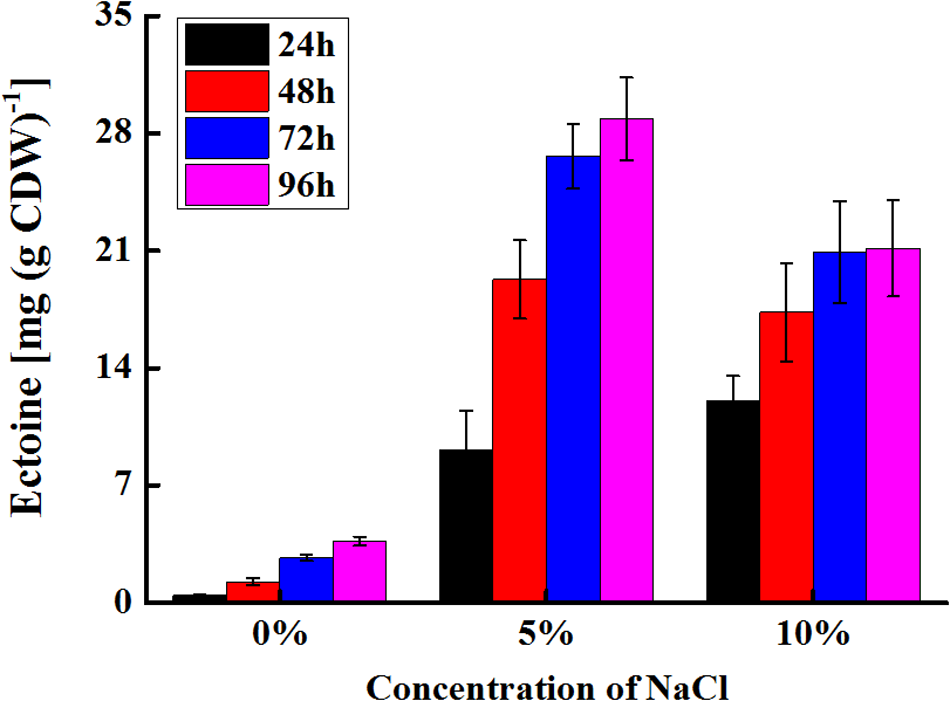
Production changes of ectoine in E. coli. After induced, recombinant E. coli was shaken in ectoine biosynthesis buffer with different NaCl (0–15%). The ectoine productions were measured every 24 h. Ectoine did not detect in the buffer with 15% NaCl. 1 ml of the cells (E. coli pBNABC/BW25113, OD600 = 1) is about 0.00034 g (DCW).
Discussion
Halophilic microorganisms have the ability to adjust saline environments more quickly and efficiently (Liu et al., 2017). Survival and growth of microorganisms in saline environments require numerous morphological ecotypes and adaptations (Kralj Kuncic et al., 2010; Liu et al., 2017). Salt stress causes change in colony morphology especially, colony color hence, in the present study colony color under salt stress was evaluated. The colony color of N. gilva YIM 90087T changed from yellow to white with the increase of salt concentration. Similar result was observed in many halophilic microorganisms when grown in high salt stress (Kralj Kuncic et al., 2010; Liu et al., 2017). Compatible solutes play significant roles to cope with hyperosmotic stress (Poolman and Glaasker, 1998; Ventosa et al., 1998; Roesser and Muller, 2001). These compatible solutes includes sugars (sucrose, trehalose), polyols (glycerol, sorbitol, mannitol, α-glucosylglycerol, mannosyl-glycerol, and mannosyl-glyceramide), N-acetylated diamino acids (N-acetylglutaminylglutamine amide), betaines (glycine betaine and derivatives), amino acids (proline, glutamate, glutamine, and alanine), and ectoine (hydroxyectoine) (Pastor et al., 2010). Halophilic microorganisms can synthesize different compatible solutes according to the external conditions, such as duration of the osmotic stress, level of salinity, availability of substrates, and osmolytes in the surroundings (Roberts, 2005; Aston and Peyton, 2007; Canamas et al., 2007). Nocardiopsis is a special halophilic microorganism, which can synthesize a variety of compatible solutes, such as ectoine, hydroxyectoine, trehalose, glutamate and β-glutamate and so on (Pastor et al., 2010). The related genes involved in compatible solutes synthesis were found in most of the Nocardiopsis species genomes (Li et al., 2013).
In N. gilva YIM 90087T, the transcriptional regulation was influenced by NaCl stress. We found that ABC transports and glycine, serine, and threonine metabolism pathway were involved in essential biological processes in response to salinity (Table 3). The ProP and ProU systems (Aliases of ProXWV) involved in uptake and accumulation of ectoine, proline, and glycine betaine in E. coli (Jebbar et al., 1992). The EhuABCD, whose expression is induced by ectoine, specifically facilitates the movement of ectoine and hydroxyectoine across the membrane in Sinorhizobium meliloti (Jebbar et al., 2005). The increase of expression levels of these proteins with the increase of NaCl concentration were also found in N. xinjiangensis (Zhang et al., 2016a). Glycine betaine and ectoine/hydroxyectoine were two types of significant compatible solutes in glycine, serine, and threonine metabolism pathway. They offered effective protection for cells against salinity, but we found that only expressions of ectoine and hydroxyectoine synthesis genes were positively correlated with increasing of NaCl concentration in N. gilva YIM 90087T.
Ectoine and hydroxyectoine are extremely vital compatible solute. Addition of ectoine and hydroxyectoine was helpful for the growth of N. gilva YIM 90087T at maximum-tolerated NaCl concentrations. Ectoine and hydroxyectoine showed similar effect in Virgibacillus pantothenticus in response to high salinity (Kuhlmann et al., 2011). Besides this, ectoine and hydroxyectoine serve as protectants for proteins (Lippert and Galinski, 1992), nucleic acids (Malin et al., 1999), whole cell (Manzanera et al., 2002, 2004a,b), and skin (Heinrich et al., 2007).
Like many halophilic microorganisms (Kuhlmann and Bremer, 2002; Zhang et al., 2009), the synthesis and accumulation of ectoine and hydroxyectoine were response to NaCl stress in N. gilva YIM 90087T. Transcriptional regulation in response to salinity was paramount. The ectABCD genes present different degrees of up-regulation in the range from 5 to 15% NaCl concentration, particularly at 15% NaCl concentration. Ectoine was converted into hydroxyectoine because of higher expression of ectD. Intracellular hydroxyectoine was far lower than ectoine in N. gilva YIM 90087T. It was inclined to use ectoine as compatible solutes in cell. However, in Prauserella alba, a moderately halophile from saline soil (Li et al., 2003), hydroxyectoine replaced ectoine and became main compatible solute in the cell, when the salt concentration reached up to 15% outside the cell (Li et al., 2011).
The EctABC activity was also influenced by NaCl concentration. To study this effect, E. coli pBNABC/BW25113 was constructed. The expression level of EctABC was completely consistent in cell after induction by arabinose. An equal number of the cells were provided in the process of whole-cell catalysis. The aspartate was catalyzed by Ask (aspartate kinase) and Asd (L-aspartate-beta-semialdehyde dehydrogenase) from E. coli, ectB (diaminobutyrate-2-oxoglutarate transaminase), ectA (diaminobutyrate acetyltransferase), and ectC (ectoine synthase) from N. gilva, and finally transformed into ectoine (Figure 8). This way, E. coli could be used to produce ectoine under high-salt conditions, if the related enzymes were not inactive (Lin et al., 2015). Unlike the EctABC from Halomonas elongata (He et al., 2015), EctABC from N. gilva YIM 90087T were activated under the salt stress in E. coli. However, the biosynthesis of ectoine was completely blocked at 15% NaCl concentration. The metabolism of host cell was inhibited in high salt environment (Metris et al., 2014). The enzymes from the host E. coli, such as Ask, Asd, and the enzymes involved in transamination and synthesis of acetyl-CoA, tend to keep the activity at the lower salt environments.
FIGURE 8
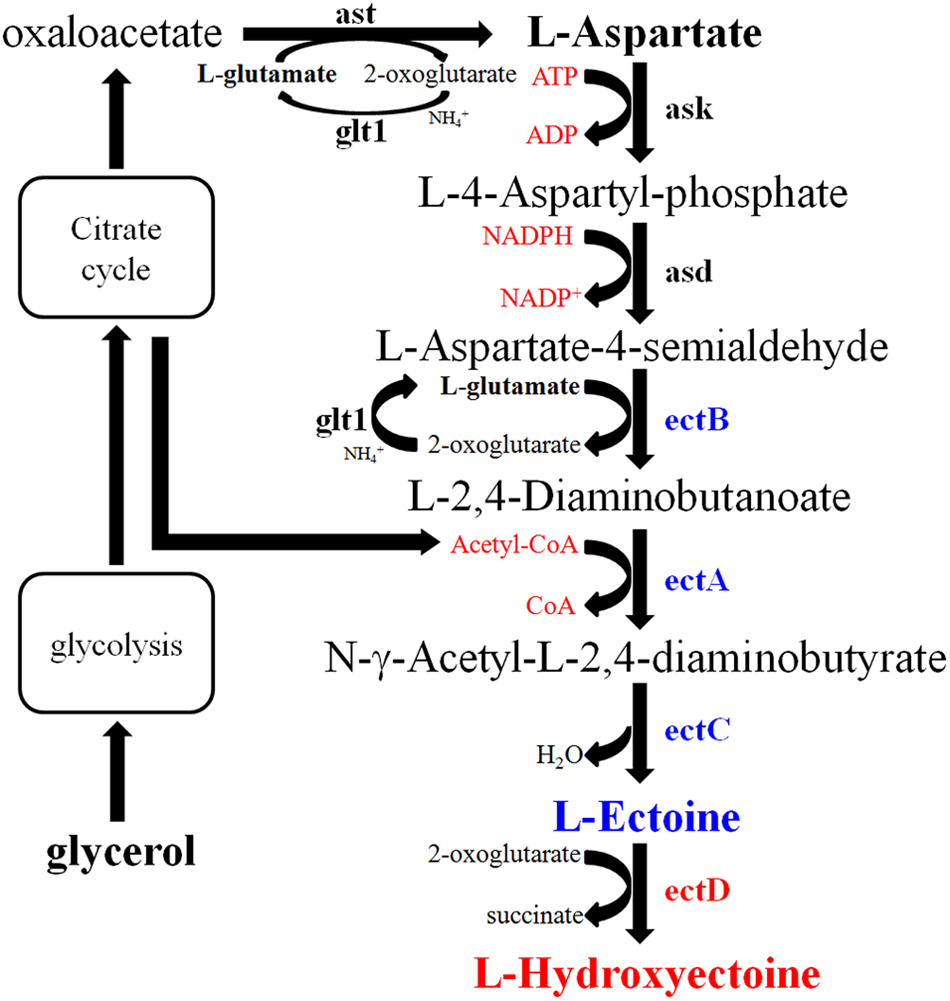
Biosynthetic pathway of ectoine/hydroxyectoine. Ast, aspartate transaminase; Glt1, glutamate synthase; Ask, aspartate kinase; Asd, L-aspartate-beta-semialdehyde-dehydrogenase; ectB, diaminobutyrate-2-oxoglutarate transaminase; ectA, diaminobutyrate acetyltransferase; ectC, ectoine synthase; ectD, ectoine hydroxylase.
Conclusion
In this study, we analyzed various response of N. gilva YIM 90087T at different NaCl concentration. Under salt stress, N. gilva YIM 90087T colony color changed. The addition of ectoine and hydroxyectoine provided protection for N. gilva YIM 90087T at high salt environment. Transcriptomic analysis of N. gilva YIM 90087T indicated that, the synthesis and accumulation of ectoine and hydroxyectoine was an effective way to regulate osmotic pressure. Internal and external ectoine and hydroxyectoine analysis showed that synthesis of ectoine and hydroxyectoine raised with increase of NaCl concentration. Ectoine was transformed into hydroxyectoine gradually, however, ectoine was still the dominant compatible solute in the cell. To study the activity, ectABC from N. gilva YIM 90087T transferred into E. coli BW25113. At the same expression level, we study the activity of ectABC with different NaCl concentration, and found that they were activated with a certain NaCl concentrations.
Statements
Author contributions
W-JL, YT, DA, and JH designed the research and project outline. JH, Y-GZ, and OM performed the growth and morphology observation. JH, Y-GZ, LL, and MN performed the transcriptome sample preparation and sequencing. JH, Q-XG, and W-JL provided HPLC and LC–MS analysis of ectoine and hydroxyectoine. JH, MN, MX, WH, YT, and W-JL drafted the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the Xinjiang Uygur Autonomous Region regional coordinated innovation project (Shanghai Cooperation Organization Science and Technology Partnership Program) (Grant No. 2017E01031), the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University, through the Research Groups Program (Grant No. RGP-1438-0004), and China Biodiversity Observation Networks (Sino BON). W-JL is supported by project funded by Guangdong Province Higher Vocational Colleges & Schools Pearl River Scholar Funded Scheme (2014).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Ameur H. Ghoul M. Selvin J. (2011). The osmoprotective effect of some organic solutes on Streptomyces sp. mado2 and Nocardiopsis sp. mado3 growth.Braz. J. Microbiol.42543–553. 10.1590/S1517-838220110002000019
2
Aston J. E. Peyton B. M. (2007). Response of Halomonas campisalis to saline stress: changes in growth kinetics, compatible solute production and membrane phospholipid fatty acid composition.FEMS Microbiol. Lett.274196–203. 10.1111/j.1574-6968.2007.00851.x
3
Barth S. Huhn M. Matthey B. Klimka A. Galinski E. A. Engert A. (2000). Compatible-solute-supported periplasmic expression of functional recombinant proteins under stress conditions.Appl. Environ. Microbiol.661572–1579.
4
Bligh E. G. Dyer W. J. (1959). A rapid method of total lipid extraction and purification.Can. J. Biochem. Physiol.37911–917. 10.1139/o59-099
5
Bouras N. Meklat A. Zitouni A. Mathieu F. Schumann P. Sproer C. et al (2015). Nocardiopsis algeriensis sp. nov., an alkalitolerant actinomycete isolated from Saharan soil.Antonie Van Leeuwenhoek107313–320. 10.1007/s10482-014-0329-7
6
Canamas T. P. Vinas I. Usall J. Magan N. Morello J. R. Teixido N. (2007). Relative importance of amino acids, glycine-betaine and ectoine synthesis in the biocontrol agent Pantoea agglomerans CPA-2 in response to osmotic, acidic and heat stress.Lett. Appl. Microbiol.456–12. 10.1111/j.1472-765X.2007.02156.x
7
Chong L. (2001). Molecular cloning - A laboratory manual, 3rd edition.Science292446–446. 10.1126/science.1060677
8
Galinski E. A. Pfeiffer H. P. Truper H. G. (1985). 1,4,5,6-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid. A novel cyclic amino acid from halophilic phototrophic bacteria of the genus Ectothiorhodospira.Eur. J. Biochem.149135–139. 10.1111/j.1432-1033.1985.tb08903.x
9
Gao G. B. Luo X. X. Xia Z. F. Zhang Y. Wan C. X. Zhang L. L. (2016). Nocardiopsis akesuensis sp. nov., an actinomycete isolated from a salt water beach.Int. J. Syst. Evol. Microbiol.665005–5009. 10.1099/ijsem.0.001460
10
He Y. Z. Gong J. Yu H. Y. Tao Y. Zhang S. Dong Z. Y. (2015). High production of ectoine from aspartate and glycerol by use of whole-cell biocatalysis in recombinant Escherichia coli.Microb. Cell Fact.14:55. 10.1186/s12934-015-0238-0
11
Heinrich U. Garbe B. Tronnier H. (2007). In vivo assessment of ectoin: a randomized, vehicle-controlled clinical trial.Skin Pharmacol. Physiol.20211–218.
12
Huang H. Q. Xing S. S. Yuan W. D. Wang Y. Liu M. Sun Q. G. et al (2015). Nocardiopsis mangrovei sp. nov., isolated from mangrove sediment.Antonie van Leeuwenhoek1071541–1546. 10.1007/s10482-015-0447-x
13
Jebbar M. Sohn-Bosser L. Bremer E. Bernard T. Blanco C. (2005). Ectoine-induced proteins in Sinorhizobium meliloti include an ectoine ABC-type transporter involved in osmoprotection and ectoine catabolism.J. Bacteriol.1871293–1304. 10.1128/JB.187.4.1293-1304.2005
14
Jebbar M. Talibart R. Gloux K. Bernard T. Blanco C. (1992). Osmoprotection of Escherichia coli by ectoine: uptake and accumulation characteristics.J. Bacteriol.1745027–5035. 10.1128/jb.174.15.5027-5035.1992
15
Kralj Kuncic M. Kogej T. Drobne D. Gunde-Cimerman N. (2010). Morphological response of the halophilic fungal genus Wallemia to high salinity.Appl. Environ. Microbiol.76329–337. 10.1128/AEM.02318-09
16
Kuhlmann A. U. Bremer E. (2002). Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp.Appl. Environ. Microb.68772–783. 10.1128/AEM.68.2.772-783.2002
17
Kuhlmann A. U. Hoffmann T. Bursy J. Jebbar M. Bremer E. (2011). Ectoine and hydroxyectoine as protectants against osmotic and cold stress: uptake through the SigB-controlled betaine-choline-carnitine transporter-type carrier EctT from Virgibacillus pantothenticus.J. Bacteriol.1934699–4708. 10.1128/JB.05270-11
18
Li H. W. Zhi X. Y. Yao J. C. Zhou Y. Tang S. K. Klenk H. P. et al (2013). Comparative genomic analysis of the genus Nocardiopsis provides new insights into its genetic mechanisms of environmental adaptability.PLoS One8:e61528. 10.1371/journal.pone.0061528
19
Li W. J. Kroppenstedt R. M. Wang D. Tang S. K. Lee J. C. Park D. J. et al (2006). Five novel species of the genus Nocardiopsis isolated from hypersaline soils and emended description of Nocardiopsis salina Li et al., 2004.Int. J. Syst. Evol. Microbiol.561089–1096. 10.1099/ijs.0.64033-0
20
Li W. J. Xu P. Tang S. K. Xu L. H. Kroppenstedt R. M. Stackebrandt E. et al (2003). Prauserella halophila sp. nov. and Prauserella alba sp. nov., moderately halophilic actinomycetes from saline soil.Int. J. Syst. Evol. Microbiol.531545–1549. 10.1099/ijs.0.02611-0
21
Li Y. Dong L. Wang L. Fang F. He M. Cao Z. et al (2011). Cloning and characterization of gene cluster for biosynthesis of ectoine and 5-hydroxyectoine from extreme halotolerant actinomycete strain Prauserella alba YIM 90005(T).Acta Microbiol. Sin.51603–608.
22
Lin B. Fan K. Zhao J. Ji J. Wu L. Yang K. et al (2015). Reconstitution of TCA cycle with DAOCS to engineer Escherichia coli into an efficient whole cell catalyst of penicillin G.Proc. Natl. Acad. Sci. U.S.A.1129855–9859. 10.1073/pnas.1502866112
23
Lippert K. Galinski E. A. (1992). Enzyme stabilization by ectoine-type compatible solutes - protection against heating. Freezing and Drying.Appl. Microbiol. Biotehnol.3761–65. 10.1007/BF00174204
24
Liu K. H. Ding X. W. Narsing Rao M. P. Zhang B. Zhang Y. G. Liu F. H. (2017). Morphological and transcriptomic analysis reveals the osmoadaptive response of endophytic fungus Aspergillus montevidensis ZYD4 to high salt stress.Front. Microbiol.8:1789. 10.3389/fmicb.2017.01789
25
Malin G. Iakobashvili R. Lapidot A. (1999). Effect of tetrahydropyrimidine derivatives on protein nucleic acids interaction - Type II restriction endonucleases as a model system.J. Biol. Chem.2746920–6929. 10.1074/jbc.274.11.6920
26
Manzanera M. de Castro A. G. Tondervik A. Rayner-Brandes M. Strom A. R. Tunnacliffe A. (2002). Hydroxyectoine is superior to trehalose for anhydrobiotic engineering of Pseudomonas putida KT2440.Appl. Environ. Microb.684328–4333. 10.1128/AEM.68.9.4328-4333.2002
27
Manzanera M. Vilchez S. Tunnacliffe A. (2004a). High survival and stability rates of Escherichia coli dried in hydroxyectoine.FEMS Microbiol. Lett.233347–352. 10.1016/j.femsle.2004.03.005
28
Manzanera M. Vilchez S. Tunnacliffe A. (2004b). Plastic encapsulation of stabilized Escherichia coli and Pseudomonas putida.Appl. Environ. Microb.703143–3145. 10.1128/AEM.70.5.3143-3145.2004
29
Metris A. George S. M. Mulholland F. Carter A. T. Baranyi J. (2014). Metabolic shift of Escherichia coli under salt stress in the presence of glycine betaine.Appl. Environ. Microbiol.804745–4756. 10.1128/AEM.00599-14
30
Meyer J. (1976). Nocardiopsis, a new genus of the order Actinomycetales. Int. J. Syst. Bacteriol.26487–493. 10.1099/00207713-26-4-487
31
Muangham S. Suksaard P. Mingma R. Matsumoto A. Takahashi Y. Duangmal K. (2016). Nocardiopsissediminis sp. nov., isolated from mangrove sediment.Int. J. Syst. Evol. Microbiol.663835–3840. 10.1099/ijsem.0.001273
32
Pan H. Q. Zhang D. F. Li L. Jiang Z. Cheng J. Zhang Y. G. et al (2015). Nocardiopsis oceani sp. nov. and Nocardiopsis nanhaiensis sp. nov., actinomycetes isolated from marine sediment of the South China Sea.Int. J. Syst. Evol. Microbiol.653384–3391. 10.1099/ijsem.0.000425
33
Pastor J. M. Salvador M. Argandona M. Bernal V. Reina-Bueno M. Csonka L. N. et al (2010). Ectoines in cell stress protection: uses and biotechnological production.Biotechnol. Adv.28782–801. 10.1016/j.biotechadv.2010.06.005
34
Poolman B. Glaasker E. (1998). Regulation of compatible solute accumulation in bacteria.Mol. Microbiol.29397–407. 10.1046/j.1365-2958.1998.00875.x
35
Roberts M. F. (2005). Organic compatible solutes of halotolerant and halophilic microorganisms.Saline Syst.1:5. 10.1186/1746-1448-1-5
36
Roesser M. Muller V. (2001). Osmoadaptation in bacteria and archaea: common principles and differences.Environ. Microbiol.3743–754. 10.1046/j.1462-2920.2001.00252.x
37
Sharma A. K. Singh S. P. (2016). Effect of amino acids on the repression of alkaline protease synthesis in haloalkaliphilic Nocardiopsis dassonvillei.Biotechnol. Rep.1240–51. 10.1016/j.btre.2016.10.004
38
Sun M. W. Zhang X. M. Bi H. L. Li W. J. Lu C. H. (2017). Two new sesquiterpenoids produced by halophilic Nocardiopsis chromatogenes YIM 90109.Nat. Prod. Res.3177–83. 10.1080/14786419.2016.1214831
39
Sun M. W. Zhang X. M. Hao H. L. Li W. J. Lu C. H. (2015). Nocarbenzoxazoles A-G, benzoxazoles produced by halophilic Nocardiopsis lucentensis DSM 44048.J. Nat. Prod.782123–2127. 10.1021/acs.jnatprod.5b00031
40
Tsujibo H. Kubota T. Yamamoto M. Miyamoto K. Inamori Y. (2003). Characterization of chitinase genes from an alkaliphilic actinomycete. Nocardiopsis prasina OPC-131.Appl. Environ. Microbiol.69894–900. 10.1128/AEM.69.2.894-900.2003
41
Ventosa A. Nieto J. J. Oren A. (1998). Biology of moderately halophilic aerobic bacteria.Microbiol. Mol. Biol. Rev.62504–544.
42
Zhang L. H. Lang Y. J. Nagata S. (2009). Efficient production of ectoine using ectoine-excreting strain.Extremophiles13717–724. 10.1007/s00792-009-0262-2
43
Zhang Y. Li Y. Zhang Y. Wang Z. Zhao M. Su N. et al (2016a). Quantitative proteomics reveals membrane protein-mediated hypersaline sensitivity and adaptation in halophilic Nocardiopsis xinjiangensis.J. Proteome Res.1568–85. 10.1021/acs.jproteome.5b00526
44
Zhang Y. G. Liu Q. Wang H. F. Park D. J. Guo J. W. Kim C. J. et al (2016b). Nocardiopsis ansamitocini sp. nov., a new producer of ansamitocin P-3 of the genus Nocardiopsis.Int. J. Syst. Evol. Microbiol.66230–235. 10.1099/ijsem.0.000703
45
Zhang Y. G. Lu X. H. Ding Y. B. Zhou X. K. Wang H. F. Guo J. W. et al (2016c). Nocardiopsis rhizosphaerae sp. nov., isolated from rhizosphere soil of Halocnermum strobilaceum (Pall.) Bieb.Int. J. Syst. Evol. Microbiol.665129–5133. 10.1099/ijsem.0.001483
Summary
Keywords
Nocardiopsis gilva , halotolerant, ectoine, hydroxyectoine, transcriptomic analysis, whole-cell catalysis
Citation
Han J, Gao Q-X, Zhang Y-G, Li L, Mohamad OAA, Rao MPN, Xiao M, Hozzein WN, Alkhalifah DHM, Tao Y and Li W-J (2018) Transcriptomic and Ectoine Analysis of Halotolerant Nocardiopsis gilva YIM 90087T Under Salt Stress. Front. Microbiol. 9:618. doi: 10.3389/fmicb.2018.00618
Received
15 December 2017
Accepted
16 March 2018
Published
29 March 2018
Volume
9 - 2018
Edited by
Masahiro Ito, Toyo University, Japan
Reviewed by
Masahiro Kamekura, Halophiles Research Institute, Japan; Rie Yatsunami, Tokyo Institute of Technology, Japan
Updates
Copyright
© 2018 Han, Gao, Zhang, Li, Mohamad, Narsing Rao, Xiao, Hozzein, Alkhalifah, Tao and Li.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dalal H. M. Alkhalifah, dhalkalifah@pnu.edu.sa Yong Tao, taoyong@im.ac.cn Wen-Jun Li, liwenjun3@mail.sysu.edu.cn
This article was submitted to Extreme Microbiology, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.