- 1Laboratory of Food Chemistry, Wageningen University and Research, Wageningen, Netherlands
- 2Laboratory of Biochemistry, Wageningen University and Research, Wageningen, Netherlands
Lytic polysaccharide monooxygenases (LPMOs) are powerful enzymes that oxidatively cleave glycosidic bonds in polysaccharides. The ability of these copper enzymes to boost the degradation of lignocellulose has greatly stimulated research efforts and biocatalytic applications within the biorefinery field. Initially found as oxidizing recalcitrant substrates, such as chitin and cellulose, it is now clear that LPMOs cleave a broad range of oligo- and poly-saccharides and make use of various electron-donating systems. Herein, substrate specificities and electron-donating systems of fungal LPMOs are summarized. A closer look at LPMOs as part of the fungal enzyme machinery might provide insights into their role in fungal growth and plant-pathogen interactions to further stimulate the search for novel LPMO applications.
Introduction
The discovery of lytic polysaccharide monooxygenases (LPMOs), in particular fungal LPMOs, has contributed to a revised concept of lignocellulose biodegradation. As opposed to the hydrolytic mechanism, LPMOs have been shown to oxidatively cleave the β-(1→4)-linked bonds in polysaccharides and, to a certain degree, in oligosaccharides (Vaaje-Kolstad et al., 2010; Agger et al., 2014; Isaksen et al., 2014). This is not only relevant for second-generation biorefinery approaches, but will also lead to a better understanding of fungal enzyme machineries and plant-pathogen interactions (Beeson et al., 2015; Johansen, 2016).
The degradation of lignocellulosic plant biomass by fungal enzymes is challenging due to the rigid structure and architecture of the plant cell wall (PCW). The PCW, which functions as a protective coat, allows the plant to resist abiotic and biotic stress factors, like wind and pathogenic fungi. The main PCW compounds comprise cellulose, hemicellulose, proteins, and phenolic compounds such as lignin, and together they form a complex network. The complex and rigid PCW structure hampers its efficient degradation through fungal-based glycosyl hydrolases, which has provoked the exploration of novel enzymes, such as lytic polysaccharide monooxygenases (LPMOs). These LPMOs have been shown to improve the activity of hydrolytic enzyme cocktails and are now seen as essential for lignocellulosic degradation (Dotsen et al., 2007; Brown et al., 2008; Harris et al., 2010; Xu et al., 2012; Hu et al., 2014).
Currently, LPMOs are classified based on their amino acid sequence as auxiliary activity (AA) families AA9, AA10, AA11, and AA13 in the Carbohydrate Active enzyme database (CAZy) (Lombard et al., 2014). The number of AA families has been extended by the discovery of family AA14 comprising fungal LPMOs (Couturier et al., 2018). Remarkably, LPMOs have also been isolated from insects and were classified as family AA15 (Sabbadin et al., 2018). LPMOs cleave the β-(1→4)-linked glucan chain via the oxidation of the C1- or C4 atom in the presence of electron-donating reducing agents and molecular oxygen or hydrogen peroxide (Vaaje-Kolstad et al., 2010; Phillips et al., 2011; Bissaro et al., 2017). Among the six AA groups, AA9 LPMOs show the largest variation in substrate specificity. About 37 fungal LPMO members of the AA9 family that act toward various (1→4)-linked PCW polysaccharides have been characterized since 2010 (Table 1).
In this study, we first introduce the main PCW constituents, to facilitate understanding of how and to which extent LPMOs act on these structures according to recent revised concepts. For more details about the structure-function relationships of LPMOs of the different AA families we would like to refer to more specific reviews which have been published recently (Frandsen and Lo Leggio, 2016; Forsberg et al., 2017; Meier et al., 2017; Vaaje-Kolstad et al., 2017). Here, we present an overview of AA9 LPMOs, with the main focus on their reported substrate specificities and regioselectivities. Finally, electron-donors of LPMOs are presented and discussed in relation to their possible physiological role. The substrate- and electron donor acceptance of AA9 LPMOs is intriguingly broad, which is why this will be further discussed in relation to future prospects within the biotechnology field.
Structural and Compositional Features of the Plant Cell Wall
The PCW is composed of a middle lamella, a primary and a secondary cell wall. The middle lamella is the first synthesized layer of the PCW and mainly contains pectins (Reiter, 2002). The next layer is the primary cell wall, which is the major part of the PCW in fruits and vegetables (Fischer and Bennett, 1991). The primary cell wall is classified into Type I and Type II, based on the structure. The Type I cell wall is present in dicots and to a certain degree in monocots as well (Carpita and McCann, 2008). This Type I consists of cellulose microfibrils, which are embedded in a network built of xyloglucans and, to a lesser extent, glucuronoarabinoxylans (GAXs), as well as pectins such as homogalacturonan and rhamnogalacturonan I (Carpita and Gibeaut, 1993; Hoffman et al., 2005). Type II cell walls are present in Poaceae and in related monocots. In these Type II cell walls, cellulose is embedded in a network of GAXs and, to a lower extent, pectins, glucomannans, and xyloglucans (Gordon et al., 1985; Carpita and McCann, 2008).
The third layer is the secondary PCW, which represents the main part of the dry matter of lignocellulosic feedstocks. Examples of the latter are grass-like agricultural by-products or hard- and soft-woods. The secondary cell wall comprises cellulose and hemicellulose, such as (acetylated) GAXs in grasses and hard woods or mannans in soft woods (Carpita and McCann, 2008). In addition, this cell wall layer is further fortified by non-(hemi)cellulose compounds such as lignin (Harris and Stone, 2009).
Cellulose
Cellulose is a homogeneous linear polymer of β-(1→4)-linked glucan chains which aggregate into microfibrils via hydrogen bonds and van der Waals forces (Nishiyama et al., 2002; Figure 1). The structure of cellulose itself is highly polymorph. Alternate glucosyl units, rotated by 180°, are linked to form a flat ribbon. Hydrogen bonds between the O3-H…O5′ tighten this conformation (Jarvis, 2003). Several of these glucan chains are aligned in parallel and form sheets which are stacked on top of each other with a stagger (Fernandes et al., 2011). The type of stagger depends on the crystal form Iα, which is the predominant form found in algae and bacteria, and Iβ that is present in plants (Atalla and Vanderhart, 1984; Nishiyama et al., 2002, 2003). In addition, these crystalline forms vary in their proportion and type of hydrogen bonds between oxygen atoms, dependent on the source. In general, 12–32 glucan chains form one microfibril, which is twisted and forms a diamond or rectangular shape in higher plants, such as wood (Zhang and Lynd, 2004; Fernandes et al., 2011). The microfibrils differ in length because the glucan chains have either a very high degree of polymerization (DP = 14,000–15,000) in the secondary cell wall (e.g., cotton), or a shorter DP (between 500 and 8,000) in the primary cell wall (Mohnen et al., 2008).
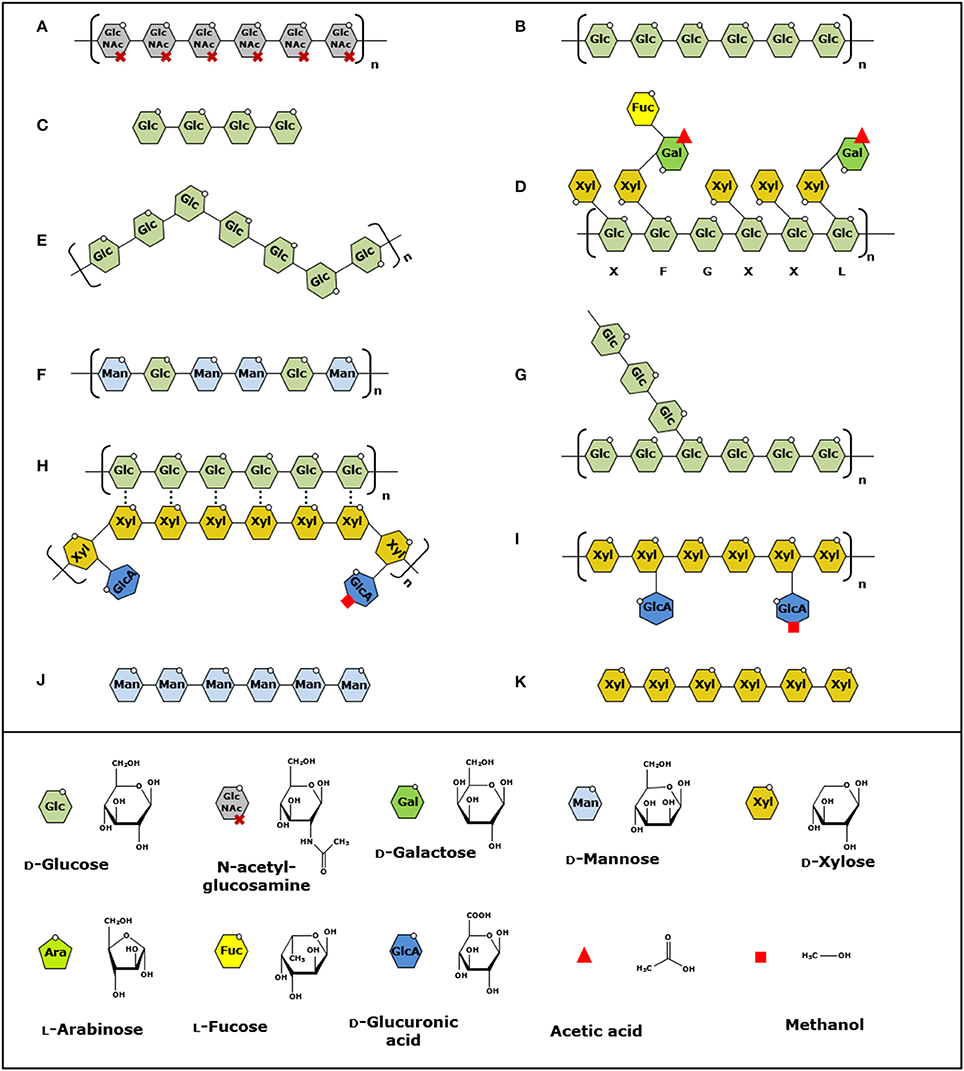
Figure 1. Structural presentation of polysaccharides oxidized by LPMOs. (A–K) Order of the polysaccharides is based on the discovery of the LPMO activity toward these substrates. Polysaccharide bonds cleaved by LPMOs are indicated by linkage type. (A) β-(1→4)-linked chitin (Vaaje-Kolstad et al., 2010), (B) β-(1→4)-linked cellulose (Forsberg et al., 2011), (C) soluble β-(1→4)-linked gluco-oligosaccharides (Isaksen et al., 2014), (D–F), β-(1→4)-linked xyloglucan, mixed β-(1→3, 1→4)-linked glucan and β-(1→4)-linked glucomannan (Agger et al., 2014), (G) α-(1→4)-linked starch (Vu et al., 2014b), (H) β-(1→4)-linked xylan associated to cellulose (Frommhagen et al., 2015), (I–K) β-(1→4)-linked xylan, soluble β-(1→4)-linked manno-oligosaccharides and soluble β-(1→4)-linked xylo-oligosaccharides (Simmons et al., 2017). Structural units are shown in the legend on the bottom. Letters under xyloglucan indicate common side chains according to Fry et al. (1993). Structures are modified and based on Scheller et al. (Scheller and Ulvskov, 2010).
The interaction between the glucan chains and the described packing is not completely coherent within cellulose, which leads to the formation of crystalline and amorphous regions. For example, varying and less strict hydrogen bonds between surface chains of the microfibril allow intermolecular interactions with external molecules in the plant, which were hypothesized to be, for example, hemicelluloses (Viëtor et al., 2002). In addition, pretreatments (e.g., alkaline extraction conditions or heating) can change the structure of cellulose, like its crystallinity (Mohnen et al., 2008; Mittal et al., 2011).
Hemicellulose
Hemicelluloses are heterogeneous polysaccharides that have different backbones and show structural side-chain variations, like different types and distributions of substituents along the backbone (Figure 1). In addition, backbones of hemicelluloses are strongly associated with cellulose via hydrogen bonding, especially the ones with a low degree of substitution or a blockwise distribution of substituents along the backbone (Vincken et al., 1995; Kabel et al., 2007).
Xylans represent the largest group of hemicellulose in the PCW and can account for up to 50% (w/w) (Ebringerová et al., 2005). All xylans consist of a β-(1→4)-linked xylosyl backbone that can be further substituted by arabinosyl, glucuronosyl, or 4-O-methyl-glucuronosyl residues (Darvill et al., 1980; Naran et al., 2008; Gírio et al., 2010). Furthermore, xylans can be covalently bound to lignin via ester and ether linkages through their ferulic and hydrocinnamic acid substitutions (Lam et al., 2001; Hatfield et al., 2008; Kühnel et al., 2012; Figure 1).
Next to xylans, other types of hemicelluloses are found to varying extents in the PCW such as xyloglucans, mixed β-(1→3, 1→4)-linked glucans, mannans, glucomannans, and pectins (Figure 1). Xyloglucans are mostly found in the primary cell wall of higher plants, such as fruit and vegetables or soft- and hard-woods (Park et al., 2004). The backbone of xyloglucan consists of β-(1→4)-linked glycosyl residues that are substituted with xylosyl residues, which are often extended with galactosyl and fucosyl residues (Fry et al., 1993). Mixed β-(1→3, 1→4)-linked glucans are mainly present in the primary cell wall of cereal kernels, such as oat and barley (Scheller and Ulvskov, 2010). In general, three to four β-(1→4)-linked glycosyl units are linked with each other via β-(1→3)-linkages, but longer β-(1→4)-linked segments have also been reported (Bulone et al., 1995; Fry et al., 2008). Mannans and glucomannans mainly consist of β-(1→4)-linked mannosyl units which can be O-2 and O-3 acetyl-esterified. Next to these mannosyl units, the backbone can also contain glycosyl units as reported for glucomannans and galactomannans (Gilbert et al., 2008). Glucomannans are present in soft- and hard-woods, whereas galactomannans are present mainly in softwoods, such as conifers (Popper and Fry, 2003; Ebringerová et al., 2005).
Pectins are highly heterogeneous polymers, which consist of a plethora of different monosaccharides that show a great variability in linkage type. The three backbone structures are homogalacturonan, xylogalacturonan, and rhamnogalacturonan I, whereas the side chain substructures are arabinan, galactan, and rhamnogalacturonan II (Ridley et al., 2001).
Lignin and Aromatic Compounds
Lignin is an aromatic heteropolymer accounting for 10–15% (w/w) of the total dry matter content of grasses and up to 30% of soft- and hard-woods (Rencoret et al., 2009; Del Río et al., 2012). The basic building blocks for the biosynthesis of lignin are the phenylpropane monomers (monolignols) coniferyl, sinapyl, and p-coumaryl alcohol. The resulting guaiacyl (G), syringyl (S) and p-hydroxyphenyl (H) structural units are linked with each other by C-C and C-O bonds and form a complex 3D-network (Constant et al., 2016). Softwoods mostly comprise lignin with G units, whereas hardwoods have both G and S units (Chen and Dixon, 2007). In contrast, grasses comprise all three structural units G, S, and H. In the PCW, lignin is built into a network with hemicellulose via ester and ether linkages. These linkages are formed between lignin and residues of hemicellulose, which comprise glucuronic acid or arabinosyl-ferulic acid substituents (Takahashi and Koshijima, 1988; Jacquet et al., 1995; Lam et al., 2001).
Apart from lignin, plants contain several other aromatic compounds that are mainly non-cell wall components, which can partly be linked to the PCW. Relevant examples are non-esterified aromatic compounds, like p-coumaric acid, ferulic acid, as well as flavonoids such as anthocyanins, which are present in grasses (Cherney et al., 1989). Flavonoids typically consist of two aromatic rings, which are connected via cyclic, aromatic, or aliphatic carbon structures, and have a variety of different substitutions, such as hydroxyl or methoxy groups (Dykes and Rooney, 2007). Other relevant aromatic compounds are tannins, which account for up to 12% (w/w) of the dry matter content in the leaves of some softwoods (Haslam, 2007).
Discovery of Lytic Polysaccharide Monooxygenases
Due to the PCW complexity a wide range of bacterial and fungal enzymes are involved in the degradation of its PCW polysaccharides. In this respect, enzymes that show hydrolytic activity toward glycosidic bonds are summarized as glycoside hydrolase (GH) families in the CAZy database. Relevant examples are endo- and exoglucanases (GH5, 6, 7, 12, and 45), β-1,4-glucosidases (GH1 and 3) or xylanases (GH10, 11, and 30) (Lombard et al., 2014). GHs interact with their substrates via a pocket, a cleft or a tunnel, which enables them to efficiently degrade hemicelluloses and amorphous regions of cellulose (Davies and Henrissat, 1995; Davies and Williams, 2016). However, in plant biomass, cellulose is mainly present as a crystalline polymer and the surface is hardly accessible for GHs.
Already in 1950, Reese et al. gave first indications of the existence of certain undefined enzymes (“C1”), now considered as LPMOs, that enable cellulolytic bacteria to utilize native cellulose (Reese et al., 1950). In comparison to these cellulolytic prokaryotes, non-cellulolytic bacteria lacked the “C1” enzyme and, therefore, were only able to degrade “shorter linear polyanhydroglucosyl chains.” These results built the foundation of the proposed “C1-Cx-theory” (Reese et al., 1950). A quarter of a century later, Eriksson et al. described an enzyme secreted by Sporotrichum pulverulentum, which is able to oxidize cellulose (Eriksson et al., 1974). In addition, the presence of this oxidative enzyme in a cellulolytic enzyme mixture enhanced the cellulose hydrolysis two-fold compared to the hydrolysis of cellulose by a non-oxidative cellulolytic enzyme mixture. Although not further mentioned in detail, the results of the work conducted within the last decade of the past millennium further paved the way toward the discovery of these unique proteins (Raguz et al., 1992; Schrempf and Walter, 1995; Saloheimo et al., 1997). The first crystal structure of such a bacterial-derived protein from Serratia marcescens was published by Vaaje-Kolstad et al. (2005b) (Figure 2). In 2010, a breakthrough was achieved with the work of Vaaje-Kolstad et al. who first revealed the oxidative mechanism behind the activity of the bacterial AA10 LPMO, which showed activity toward chitin (Vaaje-Kolstad et al., 2010). This discovery accelerated related research in the field and other LPMOs that oxidized cellulose were soon discovered (Harris et al., 2010; Forsberg et al., 2011; Phillips et al., 2011; Quinlan et al., 2011; Westereng et al., 2011; Figure 2). An overview of substrates known being oxidized by AA9 LPMOs is presented in Figure 1.
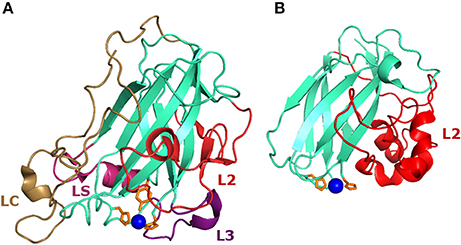
Figure 2. Structure of AA9 and AA10 LPMOs. (A) Structure of the NcLPMO9C (PDB-id: 4D7U) from Neurosspora crassa which has been the first reported LPMO that oxidizes β-(1→4)-linked hemicelluloses, in addition to β-(1→4)-linked cellulose (Borisova et al., 2015) (Table 1). (B) First crystal structure of the chitin-oxidizing CBP21 (SmLPMO19A, PDB-id: 2BEM) from Serratia marcescens (Vaaje-Kolstad et al., 2005b). The loops L2, L3, LC, and LS, which are involved in shaping the substrate binding site, are highlighted in red, purple, magenta, and pale brown, respectively. Histidines coordinating the copper ion (blue) are indicated in orange.
The GH61 classification for cellulose active LPMO-like proteins has been first proposed in 1997 (Saloheimo et al., 1997). In comparison, proteins and protein domains with chitin-binding properties were first classified into several carbohydrate-binding module (CBM) families, such as family 33 (CBM33). The CBM33 family mainly comprises bacterial and viral, as well as some eukaryotic non-catalytic chitin-binding proteins (CBPs) (Henrissat and Davies, 1997; Vaaje-Kolstad et al., 2005b; Aachmann et al., 2012). Later, it was recognized that proteins from both GH61 and CBM33 demand electrons for their oxygen-dependent cleavage of polysaccharides, which led to their classification as (lytic) polysaccharide monooxygenases (Phillips et al., 2011; Horn et al., 2012). In 2013, LPMOs were reclassified as auxiliary activity (AA) families (Levasseur et al., 2013; Lombard et al., 2014) and, based on their amino acid sequence similarities, further categorized into AA families 9, 10, 11, 13 and, more recently, 14 and 15 (Cazy.org; Lombard et al., 2014).
Structural Aspects of Fungal LPMOs
The first crystal structures of AA9 and AA10 LPMOs were determined before the catalytic activity was revealed in 2010 (Vaaje-Kolstad et al., 2005b, 2010; Karkehabadi et al., 2008). Moreover, structures of AA11, AA13, AA14, and AA15 members have also been published (Hemsworth et al., 2014; Vu et al., 2014b; Couturier et al., 2018; Sabbadin et al., 2018). In general, LPMOs feature a conserved immunoglobulin-like β-sheet core and an extended flat face, which can be enlarged by an α-helical loop (Vaaje-Kolstad et al., 2005b; Harris et al., 2010; Li et al., 2012). The planar surface enables LPMOs to bind to the surface of crystalline polysaccharides, which has been demonstrated by using NMR spectroscopy (Aachmann et al., 2012). By using X-ray crystallography, LPMOs have been identified as monocopper enzymes (Quinlan et al., 2011). The copper ion is coordinated by a N-terminal histidine, a second side chain histidine and, depending on the AA class, a third aromatic amino acid such as tyrosine in case of AA9 LPMOs (Karkehabadi et al., 2008). These residues involved in the copper coordination are forming the so-called “histidine brace” (Hemsworth et al., 2013). More details about the structural features of LPMOs are further highlighted in relation to their substrate specificity, C1-/C4-regioselectivity and catalytic mechanism in the following chapters.
Substrate Specificity and C1/C4-Regioselectivity of LPMOs
Substrate Specificity and Structure-Function Relationships
All LPMOs contain multiple extended loops that are involved in polysaccharide recognition and binding via shaping the substrate-binding surface, such as the L2 and L3 loop, the short (LS) and the long C-terminal (LC) loop (Vaaje-Kolstad et al., 2005b; Aachmann et al., 2012; Li et al., 2012; Wu et al., 2013; Borisova et al., 2015; Figure 2). Depending on the substrate specificity, these loops comprise various hydrophilic and aromatic residues that are involved in the substrate binding of LPMOs. For instance, AA9 LPMOs often comprise multiple aromatic amino acids, such as tyrosines, which are expected to take part in the binding of crystalline cellulose (Vaaje-Kolstad et al., 2005b; Forsberg et al., 2011; Li et al., 2012). In contrast to AA9 LPMOs, chitin active LPMOs of the AA10 family feature more hydrophilic amino acids, such as Thr, Gln, or Ser, to bind, compared to cellulose, the more hydrophilic chitin surface (N-acetylglucosamine) via polar interactions (Vaaje-Kolstad et al., 2005b; Aachmann et al., 2012; Li et al., 2012).
Until 2013, AA9 LPMOs were known to oxidize insoluble substrates, like chitin, cellulose, and its derivatives (Figure 1). In 2014, Isaksen et al. reported a fungal LPMO (NcLPMO9C) that oxidizes soluble substrates, in particular, soluble cello-oligosaccharides (Isaksen et al., 2014; Figure 1C). A follow-up study described the activity of NcLPMO9C toward hemicellulose structures such as xyloglucan and mixed β-(1→3, 1→4)-linked glucan (Borisova et al., 2015; Figures 1E–G). On this basis, AA9 LPMOs have been further described to cleave xyloglucan independent of the side chains attached to the β-(1→4)-linked glucan backbone like FgLPMO9A and GtLPMO9A-2, whereas others oxidize xyloglucan more specifically, such as NcLPMO9C and PaLPMO9E (Agger et al., 2014; Bennati-Granier et al., 2015; Kojima et al., 2016; Nekiunaite et al., 2016; Fanuel et al., 2017). Next, we demonstrated that, in the presence of cellulose, MtLPMO9A from M. thermophila C1 is capable of oxidizing xylan associated to cellulose (Frommhagen et al., 2015; Figure 1H). Moreover, it was shown that the CvAA9A from Collariella virescens oxidizes isolated xylan in addition to various hemicelluloses (Simmons et al., 2017) (Figures 1I–K). Recently, the AA14 LPMOs PcAA14A and PcAA14B have been shown to be active toward xylan-coated cellulose fibers, which is indicative that also LPMOs of other AA families may exhibit activity toward was wider range of PCW polysaccharides (Couturier et al., 2018). Similar to xyloglucan, the β-(1→4)-linked xylosyl backbone is also substituted, but it has not been shown yet if the above-mentioned xylan-oxidizing LPMOs show a similar cleavage behavior in the present of side chains as described for xyloglucan-oxidizing LPMOs.
The discovery of LPMOs that act toward soluble polysaccharides, such as NcLPMO9C, PaLPMO9H, Ls(AA9)A, CvAA9A, and MtLPMO9E opened new possibilities to investigate the enzyme-substrate interaction by using X-ray crystallography and NMR spectroscopy (Bennati-Granier et al., 2015; Courtade et al., 2016; Frandsen et al., 2016, 2017a; Frommhagen, 2017; Simmons et al., 2017). One of these studies showed that soluble polysaccharides dock to the histidine brace and an extended flat binding surface, which also involves the L3 and LC loop as determined for NcLPMO9C (Courtade et al., 2016). Interestingly, amino acid residues of structurally investigated LPMOs interact with different sugar moieties of the bound substrates. For example, using X-ray crystallography, Frandsen et al. showed that the +1 sugar is stacked to the methylated His-1 of Ls(AA9)A and that binding ranges from the −4 to +2 sugar (Frandsen et al., 2016). In contrast, based on NMR spectroscopy, Courtrade et al. suggested that the binding range of NcLPMO9C ranges from the −3 to +3 or −2 to +4 sugar (Courtade et al., 2016). Simmons et al. presented a substrate-depending binding mode of LsAA9A and CvAA9A when they investigated enzyme-substrate complexes using different polysaccharides with various substituents (Simmons et al., 2017). The above-mentioned studies clearly showed that different amino acid residues of the binding surface are involved in substrate binding ranging from residues that build up hydrogen bonds [Asn28, His66, and Asn67 for Ls(AA9)A] and aromatic residues, like Tyr204 and Tyr203 in NcLPMO9C and Ls(AA9)A, respectively (Courtade et al., 2016; Frandsen et al., 2016; Simmons et al., 2017).
The phylogenetic relationship between substrate specificity, C1/C4-regioselectivity and amino acid sequence has not been identified so far. A major obstacle to understand the structure-function relationships of LPMOs is the large amino acid sequence variability among these enzymes (Lenfant et al., 2017). Moreover, studies have identified thousands of putative AA9-encoding genes in sequenced genomes among the fungal kingdom and secretomics indicated that a large number of these LPMOs are actually secreted if fungi were grown on lignocellulosic biomass (Mahajan and Master, 2010; Grigoriev et al., 2014; Rytioja et al., 2014a; Kracher et al., 2016; Frommhagen et al., 2017a; Lenfant et al., 2017; Mäkelä et al., 2017). It becomes apparent that more LPMO-substrate interactions, especially involving crystalline polysaccharides, have to be uncovered to enhance the understanding of the structure-function relationships of LPMOs and to revise already implemented sub-classifications of AA9 family members (Li et al., 2012; Hemsworth et al., 2013, 2015; Bennati-Granier et al., 2015). Compared to the widely used global multiple sequence alignment, the recently proposed clustering analysis of fungal AA9 LPMOs, which is based on local pairwise alignments, could be an alternative approach to improve the phylogenetic analysis (Lenfant et al., 2017). Possibly, an enhanced understanding of the structure-function relationships of LPMOs may also help to recognize why the substrate binding affinity and the catalytic performance cannot be easily correlated (Forsberg et al., 2014b).
It is likely that the substrate specificity of AA9 LPMOs is further influenced by CBMs that are appended to LPMOs, like members of the CAZy-CBM family 1, 2, and 33 (Gilkes et al., 1991; Lombard et al., 2014). Appended CBMs have already been described to be involved in the substrate recognition and interaction and, thereby, they can influence the activity of carbohydrate active enzymes (Cantarel et al., 2009). However, the impact of appended CBMs on the activity of LPMOs highly depends on the type of substrate or LPMO used (Forsberg et al., 2014b; Borisova et al., 2015; Crouch et al., 2016; Hansson et al., 2017). It has been reported that truncation or replacement of an appended CBM increased, diminished or did not influence the catalytic performance of the LPMO. Depending on the substrate and LPMO, it is likely that the CBM binding nature influences the positioning of a specific LPMO to which the CBM is appended to, since these modules have already been described to exhibit different binding behaviors and preferences toward polysaccharides (Taylor et al., 2012; King et al., 2015; Arola and Linder, 2016). Hereby, the CBM could impact the mode of action of the LPMO by promoting either a random or more processive substrate oxidation, similar to random and processive glycosyl hydrolases.
Notably, LPMOs are secreted proteins that often contain posttranslational modifications, such as multiple disulfide bonds and N-glycosylation sites. Some of the loops, such as the L2 loop, build disulfide bridges with the conserved β-sheet core (Wu et al., 2013; Figure 2A). Since some AA9 members contain 5–10 monosaccharide units, it was suggested that glycosylations in the planar face could potentially impact the substrate binding, as shown for a CBM1 by using molecular dynamics simulations (Li et al., 2012; Taylor et al., 2012). Fungal AA9 LPMOs contain a τ-N-methylated histidine (His-1), which is involved in coordination of the copper cofactor (Quinlan et al., 2011; Li et al., 2012). In contrast, bacterial AA10 LPMOs and AA9 LPMOs produced in “foreign” hosts, such as Pichia pastoris, do not contain this τ-N-methylation (Li et al., 2012; Hemsworth et al., 2013; Wu et al., 2013; Bennati-Granier et al., 2015). Recently, Frandsen et al. suggested that the methylation of the τ-N-terminal His1 affects the LPMOs stability due to the altered pKa, which makes the imidazole of His1 less likely to be protonated at a lower pH compared to the imidazole of His78 (Frandsen et al., 2017a). Still, the role of this τ-N-methylation remains debatable since some LPMO members either with or without the τ-N-methylation at His1 were active, which led to the conclusion that this posttranslational modification is not strictly necessary to maintain LPMO activity (Westereng et al., 2011; Borisova et al., 2015).
In Vivo and in Vitro Aspects of Substrate Specificity
About 10 AA9 LPMOs have been shown in literature to be active toward hemicellulose or soluble gluco-oligosaccharides (Table 1). Lately, this list has been extended by the discovery of fungal AA14 LPMO that shows activity toward xylan-coated cellulose fibers (Couturier et al., 2018). The discovery of LPMOs, which are active toward hemicellulose structures, led to a paradigm shift in the understanding of PCW degradation. The ability of LPMOs to attack these polysaccharides indicates that these enzymes may have an important function in opening up the lignocellulose matrix to assist subsequent extensive degradation by hydrolytic enzymes. Two examples are xylan and xyloglucan, which are non-covalently associated with cellulose within the PCW (Vincken et al., 1995; Kabel et al., 2007). The planar surface of LPMOs may enable them to oxidize these less accessible regions, which is followed by a disruption of the cellulose and hemicellulose structure and thereby results in an enhanced accessibility for hydrolases.
The expectation is that more and more LPMOs will be discovered showing different substrate specificities. Within the fungal kingdom, certain fungal species contain multiple AA9 LPMO-encoding genes, which have been shown to be expressed differently when the fungi are grown on different polysaccharides (Tian et al., 2009; Vanden Wymelenberg et al., 2009; Berka et al., 2011; Mäkelä et al., 2017). Certainly, absolute numbers of putative LPMO-encoding genes have to be interpreted cautiously since not all of these LPMOs have been shown to actually be present in the fungal secretome (Tian et al., 2009; Mahajan and Master, 2010; Rytioja et al., 2014b; Mäkelä et al., 2017). Still, these numbers are indicative of the fact that the discrimination of LPMOs by their substrate specificity is currently not complete. Some fungi even express multiple putative LPMO-encoding genes to utilize only pure cellulose, such as Avicel (Rytioja et al., 2014a). It is likely that LPMOs are exhibiting further modes of action, similar to glycosyl hydrolases, which can range from random to processive hydrolytic cellulose cleavage (Kurašin and Väljamäe, 2011; Jung et al., 2013; Kubicek et al., 2014). Reported AA9 LPMOs oxidize cellulose but recent findings indicate that cellulose-specificity potentially ranges from the preferred oxidation of highly crystalline to completely amorphous regions (Table 1; Eibinger et al., 2014; Villares et al., 2017). Moreover, LPMOs that are active toward crystalline cellulose may further be distinguished by their preference for microfibrils comprising the Iα or Iβ crystal form, which has yet to be investigated (Nishiyama et al., 2002, 2003; Jarvis, 2003). A similar scenario has already been described for CBMs and some members have been shown to be able to bind hardwood cellulose but not bacterial cellulose (Arola and Linder, 2016; Chabbert et al., 2017; Eibinger et al., 2017). Another example is shown by a recent study in which the cleaving behavior of NcLPMO9F on the cellulose surface is revealed by using time-resolved atomic force microscopy (Eibinger et al., 2014). This LPMO oxidized the surface layer of the crystalline cellulose region only, which resulted in the formation of a patch, but amorphous regions were not oxidized. Moreover, a vertical degradation of subjacent layers mediated by NcLPMO9F was hardly determined. In comparison, another study described the activity of PaLPMO9H, which was only active toward amorphous cellulose, but not toward crystalline cellulose fibers (Villares et al., 2017; Table 1).
C1/C4-Regioselectivity
The first C1/C4-regioselectivities have been experimentally determined for two bacterial LPMOs. Both CBP21 (AA10) from S. marcescens and CelS2 (AA10) from Streptomyces coelicolor oxidize the C1-carbon position of chitin and cellulose, respectively (Vaaje-Kolstad et al., 2010; Forsberg et al., 2011). This C1-oxidation of the reducing end sugar of chitin or cellulose leads to the formation of an unstable δ-lactone (−2 Da), which is converted into an aldonic acid in the presence of water (+ 16 Da) (Vaaje-Kolstad et al., 2010; Forsberg et al., 2011; Figure 3). Shortly thereafter, the fungal AA9 LPMO TaGH61 was shown to oxidize cellulose at the C1 as well as at the C4 position (Quinlan et al., 2011; Figure 3). The latter leads to the oxidation of the C4-carbon of the non-reducing end sugar, which results in the formation of a more stable 4-ketoaldose (−2 Da) that tautomerizes into a geminal diol depending on the pH (Quinlan et al., 2011; Isaksen et al., 2014; Westereng et al., 2016; Figure 3). From this point onwards, the number of AA9 and AA10 LPMOs with known C1/C4-regioselectivity grew rapidly (Phillips et al., 2011; Westereng et al., 2011). AA9 LPMOs with experimentally determined C1/C4-regioselectivities are summarized in Table 1. It should be noted that more LPMOs have been expressed and characterized, but their C1-/C4-regioselectivity has not been experimentally determined yet.
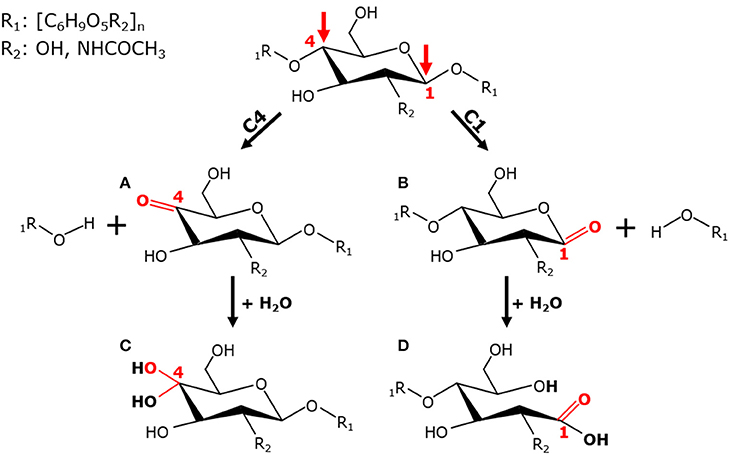
Figure 3. C1/C4-regioselectivity of LPMOs. Oxidation of β-(1→4)-linked glucan at the C1- or C4-carbon position by LPMOs leads to the formation of non-oxidized and C1- or C4-oxidized oligosaccharides. The LPMO-mediated C4-oxidation results in the formation of 4-ketoaldoses (A), which are present as their corresponding hydrates, geminal diols, in aqueous solutions (C). The oxidation at the C1-atom leads to formation of a labile δ-lactone (B), which dissociates in water into an aldonic acid (D).
Until today, AA9 LPMOs have been shown to oxidize either the C1, C4 or both the C1 and C4 position (Table 1). In contrast, bacterial AA10 LPMOs cleave cellulose and chitin by the oxidation of the C1 or both, the C1 and C4 carbon atom, whereas no AA10 member has been published to singly oxidize the C4-position (Forsberg et al., 2011, 2014a, 2017). The described AA11 and AA13 LPMOs, as well as the recently discovered AA14 and AA15 LPMOs, are all C1-oxidizing LPMOs (Hemsworth et al., 2014; Vu et al., 2014b; Lo Leggio et al., 2015; Couturier et al., 2018; Sabbadin et al., 2018).
The reason for the existence of LPMOs with different regioselectivities is not yet fully understood. In fact, it has been shown that a single fungus, such as Neurospora crassa, Myceliophthora thermophila C1, or Podospora anserina, can express LPMOs that oxidize cellulose either at C1 or C4, or at both carbon positions (Table 1). The C1/C4-regioselectivity is likely the result of how the LPMO binds to the substrate, meaning that different LPMOs may bind to differently exposed cellulose-fiber surfaces. As mentioned earlier, cellulose is structurally polymorph and contains of varying microfibril structures, hydrophobic and hydrophilic sides, a different degree of crystallinity and diverse intra- and intermolecular hydrogen bonds and bridges between oxygen atoms (Atalla and Vanderhart, 1984; Nishiyama et al., 2002, 2003; Jarvis, 2003; Fernandes et al., 2011). The thereby established differences in conformation, shape and compactness of the β-(1→4)-linked glucan chains could limit the accessibility of the C1- or C4-carbon atom, which is a possible explanation for the evolution of LPMOs exhibiting different regioselectivities.
So far, published data addressing a possible relation between structural features of LPMOs and C1/C4-regioselectivity is limited (Frandsen and Lo Leggio, 2016; Vaaje-Kolstad et al., 2017). Borisova et al. showed that strictly C1-oxidizing AA9 LPMOs such as TtLPMO9E contain a conserved tyrosine residue (Tyr80 based on NcLPMO9C), which is Ala80 in the C4-oxidizing NcLPMO9C (Borisova et al., 2015). The presence of this residue likely restricts the accessibility to the solvent-facing carbon atom in the axial coordination position (Borisova et al., 2015). Interestingly, Simmons et al. showed that fungal LPMOs seem to feature a substrate-specific C1/C4-regioselectivity. Both of the LPMOs tested, LsAA9A and CvAA9A, oxidized the C4-carbon of shorter oligosaccharides whereas longer polysaccharides were cleaved via the oxidation of the C1- and C4-carbon atom (Simmons et al., 2017). A more recent study demonstrated the involvement of active-site residues, such as the above-mentioned tyrosine, in the C1/C4-regioselectivity of LPMOs (Forsberg et al., 2017). Mutation of active-site residues of the C1/C4-oxidizing MaLPMO10B from Micromonospora aurantiaca let to decrease of the C4-oxidizing activity and loss of chitin specificity, whereas these MaLPMO10B mutants maintained the C1-oxidizing regioselectivity. Moreover, truncation or mutation of residues of the MaLPMO10B-appended CBMII did not alter the C1/C4-regioselectivity of MaLPMO10B (Forsberg et al., 2017). These recent studies illustrate how marginal sequence variations and substrate-binding interactions alter the C1/C4-regioselectivity, which will challenge a further classification of LPMOs based on their experimentally determined substrate specificity and C1/C4-regioselcetivity.
Catalytic Mechanism of LPMOs
Before LPMOs were described as copper-containing oxygenases, isotope labeling (O and 18O2) indicated that one of the two oxygens introduced into the oxidized glucan chain derived from water and the other one from molecular oxygen (Vaaje-Kolstad et al., 2010). In addition, it was shown that this LPMO-mediated reaction demanded electrons, which were provided by reducing agents, such as ascorbic acid or reduced glutathione (Vaaje-Kolstad et al., 2010). In 2011, the first detailed reaction pathway was proposed by Phillips et al. who based the catalytic mechanism on the reaction pathway of copper monooxygenases (Klinman, 2006; Phillips et al., 2011; Solomon et al., 2011).
A key step described the one-electron reduction of Cu(II) to Cu(I) by either an external reducing agent or a cellobiose dehydrogenase (CDH) (Phillips et al., 2011). The Cu(I) binds molecular oxygen and forms a copper superoxo intermediate via an internal electron transfer. Based on the proposed mechanism, the copper superoxo intermediate subtracts a hydrogen atom from the C1-carbon or the C4-carbon atom of the polysaccharide, depending on the C1/C4-regioselectivity of the LPMO. The following reactions include a second reduction step and the recovery of the Cu(II) oxidation state, via formed intermediates such as copper hydroperoxo intermediate and a copper oxo radical, and the release of water (Phillips et al., 2011). Upon the end of this reaction, a substrate radical is formed that reacts further with an additional oxygen atom which destabilizes the β-(1→4)-linkage followed by the elimination reaction and the formation of a non-oxidized and an oxidized glucan chain (Phillips et al., 2011; Beeson et al., 2012). It was also suggested that a direct two electron reduction of oxygen to a Cu-OOH intermediate might be able to abstract a hydrogen (Phillips et al., 2011). However, this reaction pathway was not experimentally underpinned since neither did peroxide enable the reaction nor was the reaction inhibited by catalase addition (Phillips et al., 2011). In the following years, more detailed reaction mechanisms have been proposed (Beeson et al., 2015; Walton and Davies, 2016). These reaction pathways share the idea that Cu(II) is reduced by an external electron to Cu(I) at the start of the reaction. However, for the second step it remains debatable whether the Cu(I)-LPMO first binds oxygen or the substrate (Walton and Davies, 2016). The existence of various intermediate steps, such as the copper-dependent oxygen activation or the binding of oxygen and superoxide, are experimentally and computationally supported by using known LPMO structures (Li et al., 2012; Kjaergaard et al., 2014; Frandsen et al., 2016; Bertini et al., 2017). Intriguingly, new catalytic mechanisms have been proposed, which include H2O2 as a key compound or, dependent on the substrate, facilitate more than one catalytic route (Bissaro et al., 2017; Simmons et al., 2017). Both findings are described below in more detail.
Mechanism of the Electron Transfer
Beside the discussions about the reaction mechanisms, there is an ongoing debate about how LPMOs receive electrons from external reducing agents or enzymes. One major point of discussion is that the close binding of LPMOs to the surface of cellulose or chitin makes the catalytic copper center inaccessible, even for smaller reductants (Courtade et al., 2016; Frandsen et al., 2016; Simmons et al., 2017). Therefore, it was hypothesized that the reduction of the catalytic copper center takes place before the binding of the LPMO to the polysaccharide or, alternatively, electrons are continuously transported to the catalytic copper center through electron tunneling. Li et al. suggested two possible scenarios how fungal AA9 LPMOs could accommodate an internal electron tunneling (Li et al., 2012). In the first scenario, electrons are transferred through an internal conserved ionic network (hydrogen bond network), which connects the copper ligand tyrosine with the surface-exposed Lys under involvement of conserved tyrosine residues. In a second scenario, electrons are provided by aromatic residues from the proximity of the histidines (Li et al., 2012). In addition, computational simulation studies showed that the heme domain of a CDH could potentially bind to a hydrophilic conserved surface patch. The electrons could be transferred to the catalytic copper center by one of the above-proposed scenarios (Li et al., 2012). Interestingly, follow-up research strengthened both of the scenarios by providing more experimental evidence that AA9 LPMOs interact with a reducing agent either via a surface patch centered around the Pro-Gly-Pro triad or a narrow patch around the amino acid residues that coordinate the copper ion (Li et al., 2012; Tan et al., 2015; Courtade et al., 2016).
Catalytic Performance of LPMOs as a Function of the Electron Donor
Interestingly, research has also shown that the reducing agent preference varies among LPMOs (Figure 4). In general, phenolic compounds comprising structural features like the 1,2-dihydroxy (IIa), 1,2-dihydroxy-6-methoxy (IIb), and 1,2,3-trihydroxy moiety (III) efficiently reduce LPMOs (Westereng et al., 2015; Frommhagen et al., 2016; Kracher et al., 2016). In addition, compounds that do not comprise the above-mentioned features have also been shown to reduce LPMOs such as ascorbic acid, glutathione and L-cysteine or the macromolecule lignin (Vaaje-Kolstad et al., 2010; Lo Leggio et al., 2015; Westereng et al., 2015; Figure 4). In comparison, monophenols are less efficient electron donors for LPMOs (Frommhagen et al., 2016). The reducing efficiency of a compound can be determined experimentally by cyclic voltammetry and expressed as the reduction potential. Phenolic compounds comprising a second hydroxyl group attached to the benzene ring have a relatively low reduction potential (≤250 mV), whereas monophenols have a relatively high reduction potential (≥400 mV; Kracher et al., 2016). Kracher et al. (2016) showed that a lower reduction potential of reducing agents results in a higher catalytic performance of LPMOs (Kracher et al., 2016). Hence, it seems possible to predict if a compound is an efficient electron donor for LPMOs, based on its structure and associated reduction potential. But there are exceptions like the ortho-diphenol quercetin, which does not promote LPMO activity for three different AA9 LPMOs tested (Frommhagen et al., 2016).
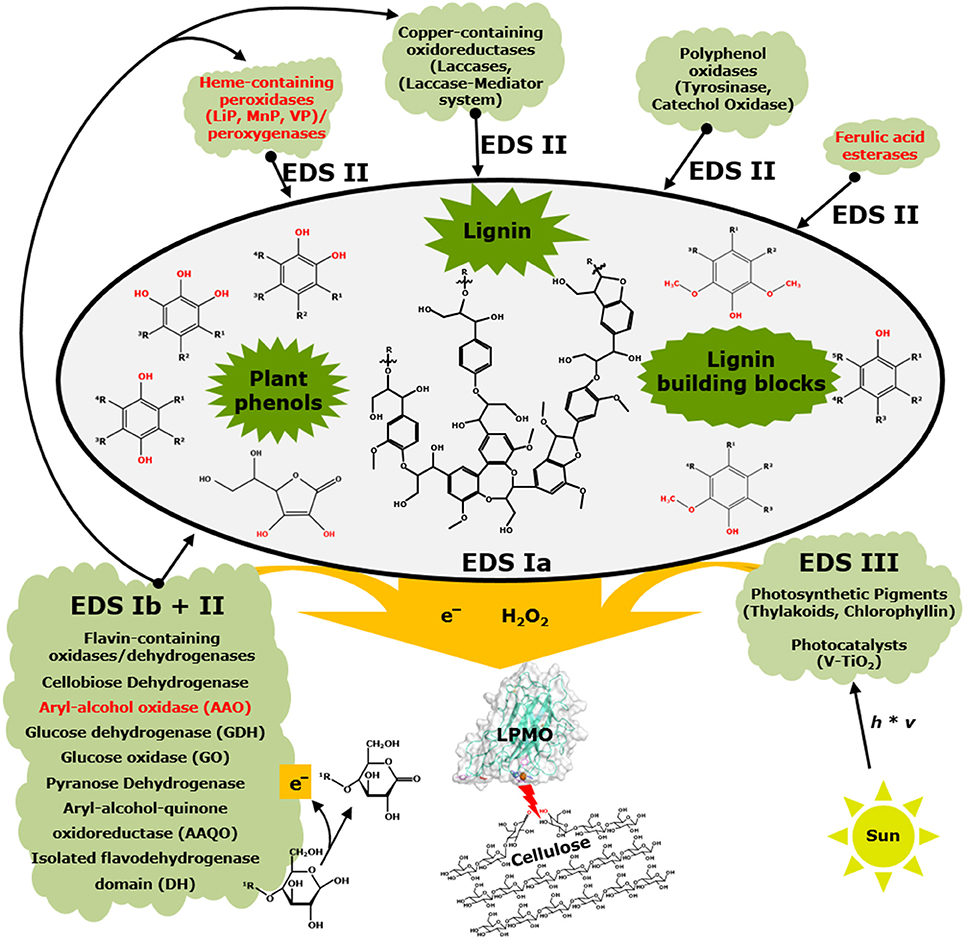
Figure 4. Interactions between non-enzymatic and enzymatic electron-donating systems and their effect on LPMOs. Black fonts represent electron-donating systems Ia and b, II, and III (EDS) that have been published. Red fonts illustrate systems that have not yet been shown to be members of electron-donating systems of LPMOs. This scheme is simplified and co-substrates such as hydrogen peroxide or oxygen as well as enzymes acting on reactive oxygen species (EDS IV) are not included. In addition, aerobic and anaerobic conditions are not further considered. Schematic presentation of lignin is based on Zakzeski et al. (2010). Further details are described in the text.
The reduction potential of a number of LPMOs has been determined to be around +250 mV (vs. SHE). However, some members show deviating values which are significantly higher (+326 mV vs. SHE) or lower (+155 mV vs. SHE) (Aachmann et al., 2012; Borisova et al., 2015; Garajova et al., 2016; Kracher et al., 2016). These deviating values may result from differences in the protein structure. Thus, LPMOs comprising a low reduction potential (e.g., below +250 mV (vs. SHE)] likely receive electrons from compounds with a relatively low reduction potential. In contrast, LPMOs with a high reduction potential [e.g., above +250 mV (vs. SHE)] can also receive electrons from reducing agents which comprise a reduction potential that is, for example, around +250 mV (vs. SHE).
Still, the catalytic performance in the presence of various efficient electron donors with a low reduction potential (e.g., compounds comprising a 1,2-dihydroxy moiety) differs vastly among LPMOs. These differences are caused by numerous factors, which have to be carefully considered when interpreting obtained results from LPMO activity assays, such as reducing agent stability or operational stability of the LPMO in the presence of different reducing agents (Frommhagen et al., 2016; Bissaro et al., 2017). Furthermore, the amino acid residues present in the surface patch centered around the Pro-Gly-Pro triad or the narrow patch around the copper ion differ in structure and surface charge distribution among LPMOs (Frommhagen et al., 2016). Therefore, it seems plausible that the interaction between these amino acid residues and reducing agents is different for each of the LPMOs. Notably, the above-mentioned properties such as surface charge of the enzyme or reduction potential are largely influenced by incubation conditions like pH, temperature or the type of buffer used (Frommhagen et al., 2017c). High concentrations of reducing agents, in particular the ones with a low reduction potential, have been reported to inactivate LPMOs within minutes by forming reactive oxygen species (ROS) (Bissaro et al., 2017). Alternatively, the catalytic performance of LPMOs might be influenced by the pH due to alterations in the coordination of the copper ion in the active site, as a harsh decrease in the pH may lead to the protonation of the imidazole (Frandsen et al., 2017a). Such pH-dependent rearrangements of active-site histidines could also affect copper and substrate binding (Frandsen et al., 2017b).
New insight in the reaction mechanism of LPMOs might be brought up by an intriguing recent finding of Bissaro et al. (2017), who showed that H2O2 is a co-substrate of LPMOs (Bissaro et al., 2017). These authors proposed that reducing agents are directly involved in the “priming reduction” which describes the reduction of LPMO-Cu(II) to LPMO-Cu(I) (Bissaro et al., 2017). In the reduced stage, the LPMO-Cu(I) reacts with H2O2 and the thereby formed copper-bound oxygen species subtract a hydrogen atom from either the C1- or the C4-carbon atom of the polysaccharide. In a further step, the copper-bound oxygen species reacts with the carbon atom, which leads to the hydroxylation of the substrate and, under molecular rearrangement, to the cleavage of the β-(1→4)-linkage (Bissaro et al., 2017). More recently, quantum mechanical/molecular mechanical (QM/MM) calculations supported the findings of Bissaro and colleagues by demonstrating that LPMOs are able to break the O-O bond of H2O2 (Wang et al., 2018). The subsequently generated Cu(II)-oxyl species oxidizes the C4-H bond and the thereby formed hydroxylated C4-carbon intermediate leads to spontaneous non-enzymatic chain cleavage (Wang et al., 2018). Hangasky et al. (2018) showed that the C4-oxidizing MtLPMO9E utilizes both O2 and H2O2, dependent on the assay condition. Based on their results, MtLPMO9E oxidized cellulose specifically at the C4-position in the presence of O2, whereas H2O2 addition led to a non-specific polysaccharide oxidation (Hangasky et al., 2018). These results will contribute to the ongoing debate about the role of H2O2 on the catalytic mechanism of LPMOs and, furthermore, should be considered regarding the determination of the C1/C4-regioselectivity of LPMOs in the presence of varying assay conditions.
Interestingly, LPMOs of family AA9 and AA10 that exhibit the H2O2-driven peroxygenase-like reaction have a significantly higher catalytic performance compared to LPMOs that are active in the presence of O2 (Bissaro et al., 2017; Kuusk et al., 2017). In addition, Bissaro et al. (2017) have shown that reducing agents, such as ascorbic acid, are also able to stimulate the formation of H2O2 in the absence of AA9 LPMOs (Bissaro et al., 2017). Based on their findings, it is conceivable that more scenarios may impact the reducing agent preference of LPMOs, such as different sensitivities of LPMOs for H2O2 (Bissaro et al., 2017). Still, the proposed priming reduction which describes the activation of the Cu(II)-LPMO via one electron reduction to the Cu(I)-LPMO demands a direct interaction between reducing agent and LPMO. In addition to Bissaro et al. (2017), Simmons et al. (2017) showed evidence that LPMOs may exhibit a substrate-depending catalytic mechanism, which is based on the observation that different oxygen species may be bound to the copper ion if LsAA9 is bound to cello-oligosaccharides compared to xylo-oligosaccharides (Simmons et al., 2017). They observed that the catalytic performance of LsAA9 toward xylan is less dependent on the type of reducing agent present, which corroborates that LPMOs perform more than one catalytic mechanism. Combining the recent findings, it becomes apparent that several factors, which impact the reducing agent preference of LPMOs, are not fully understood so far.
Electron-Donating Systems of LPMOs
In 2010, it became apparent that LPMOs demand electrons from small molecular weight compounds, such as ascorbic acid and gallic acid (Vaaje-Kolstad et al., 2010). Later, other electron-donation systems were discovered, like the co-action of LPMOs with CDH (Phillips et al., 2011). Westereng et al. were the first who reported lignin as a possible electron donor for TtLPMO9E (Westereng et al., 2015). Today, it is generally accepted that LPMOs can receive electrons from multiple types of reducing agents. Therefore, reducing agents published in literature were classified into four different electron-donating systems (EDS I-IV). It should be noted that some members of one electron-donating system (Ib), such as CDH, can also be part of another system (II), and vice versa, dependent on the activity.
Electron-Donating System I
EDS I includes compounds (EDS Ia) and enzymes (EDS Ib) that directly donate electrons to LPMOs. This system includes numerous compounds that are mainly plant phenols and lignin building blocks, such as monophenols and phenolic compounds comprising a 1,2-benzendiol or 1,2,3-benzenetriol moiety, as well as lignin (Westereng et al., 2015; Frommhagen et al., 2016; Kracher et al., 2016; Figure 4). The flavin-containing CDH, which is a member of the GMC (glucose-methanol-choline oxidase/dehydrogenase) oxidoreductase family, was the first enzyme reported to reduce LPMOs directly and, so far, is the most used enzyme system to study the LPMO mechanism in vitro (Phillips et al., 2011; Kittl et al., 2012; Sygmund et al., 2012; Bey et al., 2013; Garajova et al., 2016; Figure 4). Phillips et al. (2011) showed that the deletion of the highly expressed CDH-1 gene from the cellulolytic fungus N. crassa resulted in a decreased cellulose degradation. Moreover, the addition of a purified CDH from M. thermophila to a CDH lacking N. crassa strain improved the cellulose degradation up to two-fold (Phillips et al., 2011). In addition, studies have shown that LPMOs and CDH are co-expressed with various growth substrates, which indicates the relevance of this interaction in nature (Hori et al., 2011; Langston et al., 2011; Yakovlev et al., 2012; Navarro et al., 2014; Couturier et al., 2015).
Next to CDH, other GMC members, such as glucose oxidase (GOx), glucose dehydrogenase (GDH), pyranose dehydrogenase (PDH), aryl-alcohol quinone oxidoreductases (AAQO), aryl-alcohol oxidase (AAO), and an isolated flavodehydrogenase domain (DH), have been shown to either donate electrons directly to LPMOs or regenerate electron-donating plant phenols and lignin building blocks (Garajova et al., 2016; Kracher et al., 2016; Bissaro et al., 2017). In that respect some of the GMC members also belong to “Electron-donating system II.” As an example, a GDH from Glomerella cingulata added to the incubation with NcLPMO9C (LPMO-02916) in the presence of various quinones improved the oxidation of cellulose (Kracher et al., 2016). In addition, sequence analysis showed a positive correlation between the occurrence of GMC oxidoreductase-encoding genes and LPMO-encoding genes among fungi (Kracher et al., 2016). In another study, a GDH from Pycnoporus cinnabarinus was shown to donate electrons to PaLPMO9H (Garajova et al., 2016). In this study two variants of AAQOs (AAQO1 and AAQO2) from P. cinnabarinus were described that can be used to reduce PaLPMO9E. Until now, aryl-alcohol oxidases (AAOs) have not been shown to influence the catalytic performance of LPMOs, such as the AAO from Ustilago maydis, which did not reduce PaLPMO9E (Garajova et al., 2016). Moreover, some GMC family members (e.g., AAO, CDH) are reported to interact either via electron donation or H2O2 formation with other enzymes, such as laccases and peroxidases, which may affect the catalytic performance of LPMO indirectly (Temp and Eggert, 1999; Hildén et al., 2000; Hernández-Ortega et al., 2012; Figure 4). Next to flavin-dependent oxidoreductases, a pyrroloquinoline quinone (PQQ)-dependent pyranose dehydrogenase (PDH) from Coprinopsis cinerea, CcPDH, has been shown to reduce both the C1-oxidizing NcLPMO9F and C4-oxidizing NcLPMO9C from N. crassa (Várnai et al., 2018).
Electron-Donating System II
EDS II comprises enzymes that release and modify reducing agents that influence the activity of LPMOs. Recently, polyphenol oxidases (PPOs), such as MtPPO7 from M. thermophila C1 and AbPPOs from Agaricus bisporus, have been shown to affect the activity of LPMOs indirectly by their monophenolase and diphenolase activity (Frommhagen et al., 2017a; Figure 4). More specifically, the polyphenol oxidase MtPPO7 from M. thermophila C1 converts methoxylated phenolic compounds that are present as lignin-building blocks in the PCW into compounds comprising a 1,2-dihydroxy moiety and, thereby, enhances the catalytic performance of MtLPMO9B. Sequence analysis of genomes of 336 Ascomycota and 208 Basidiomycota revealed a high correlation between genes encoding MtPPO7-like proteins and AA9 LPMOs, which is indicative for the coupled action of PPOs and LPMOs in the concerted degradation of lignocellulosic biomass (Frommhagen et al., 2017a). Peroxidases such as manganese peroxidases (MnPs), lignin peroxidases (LiPs), and versatile peroxidases (VPs) as well as laccases are known to oxidize plant phenols, lignin building blocks and lignin and, therefore, are also likely to effect the electron donation of these compounds toward LPMOs (Hammel et al., 1993; Hofrichter, 2002; Camarero et al., 2005; Pérez-Boada et al., 2005; Figure 4). Moreover, it has been demonstrated that the laccase-mediator system (LMS) can be used to partially break down lignin into small-molecular-weight compounds which are potential electron donors for LPMOs (Brenelli et al., 2018).
Presumably, other reducing agent releasing and/or modifying enzymes, which take part in PCW degradation, will also affect the activity of LPMOs, such as ferulic acid esterases (Figure 4). Ferulic acid esterases have been shown to release coumaric acid and ferulic acid from PCW hemicelluloses, like in grains and grasses (Kühnel et al., 2012; Várnai et al., 2014). As described above, these cinnamic acid derivatives have been shown to be hydroxylated by PPOs into compounds comprising 1,2-dihydroxy moieties, which are efficient electron donors for LPMOs (Frommhagen et al., 2017a).
Electron-Donating System III
EDS III includes light-induced non-enzymatic systems. In 2016, Cannella et al. (2016) reported that light in combination with photosynthetic pigments, such as thylakoids or chlorophillin, are very efficient electron-donating systems for the reduction of LPMOs (Cannella et al., 2016; Figure 4). Next to these pigments, chemical photocatalysts like vanadium-doped titanium dioxide (V-TiO2) can also efficiently reduce LPMOs (Bissaro et al., 2016; Figure 4). Above all, these systems comprise the advantage of a switch-on/off possibility and, therefore, are a useful tool for investigating the catalytic mechanism of LPMOs (Bissaro et al., 2016, 2017; Cannella et al., 2016; Möllers et al., 2017).
Electron-Donating System IV
EDS IV comprises enzymes that alter superoxide or hydrogen peroxide levels to affect the catalytic performance of LPMOs. Catalase and superoxide dismutase can affect the LPMO activity or stability in an indirect way due to altering superoxide or hydrogen peroxide concentrations during the incubation (Scott et al., 2016; Bissaro et al., 2017). As an example, the catalase from Thermoascus aurantiacus (Accession DD046677) reportedly decreases the inactivation of AA9 LPMOs during biomass degradation (Scott et al., 2016). Apart from that, both superoxide dismutase and catalase may also be involved in most of the non-enzymatic and enzymatic electron-donating systems, since superoxide and hydrogen peroxide are released during the oxidation/reduction (redox) reactions (Bissaro et al., 2017). Furthermore, the hydrogen peroxide generating xanthine oxidase (XOD) has been shown to reduce ScLPMO10C, which led to the oxidation of cellulose (Bissaro et al., 2017). The same AA10 LPMO was also active toward cellulose in the presence of chemical superoxide generating systems, such as potassium superoxide (KO2) (Bissaro et al., 2017).
Electron-Donating Systems in Vivo
So far, most LPMO electron-donating systems have been determined in vitro. In general, LPMOs used for the degradation of lignocellulosic biomass are secreted enzymes, which oxidize polysaccharides outside of the fungal cell. The physiological relevance of LPMO electron-donating systems is likely to be determined by the mode of life-style that fungi exhibit.
For instance, saprophytic fungi such as the Ascomycota M. thermophila C1 and N. crassa or the Basidiomycota Phanerochaete chrysosporium, which comprise already multiple characterized AA9 LPMOs (Table 1), mainly feed on dead biomass that often contains high amounts of lignin and other phenolic compounds, next to polysaccharides (Martinez et al., 2004; Phillips et al., 2011; Frommhagen et al., 2017a). For these saprophytic fungi, LPMOs are likely to receive electrons from lignin and lignin-building blocks (Figure 4). Presumably, LPMOs secreted by these fungi benefit from other secreted oxidative enzymes during the concerted degradation of biomass, such as the GMC oxidoreductases, PPOs and, although not shown yet, laccases and peroxidases (Phillips et al., 2011; Kracher et al., 2016; Frommhagen et al., 2017a; Figure 4). It is unlikely that the light-induced non-enzymatic electron-donating system will act as an efficient electron donor for LPMOs, which are secreted by saprophytic fungi. The latter are able to decompose biomass in the absence of light (e.g., in the soil or ground level of a forest) or degrade parts of the plant that do not contain photosynthetic pigments (e.g., the stamp of hardwoods). Also intracellular reducing agents, such as ascorbic acid and reduced glutathione, are not expected to play a major role as reducing agents for LPMOs, since dead biomass will not comprise intact cells that contain these rather instable compounds.
In contrast to saprophytes, pathogenic fungi are known to attack the PCW of living cells such as the rice pathogen Magnaporthe grisea or the wheat pathogen Fusarium graminearum, which both comprise multiple putative AA9 LPMO-encoding genes (Dean et al., 2005; Nekiunaite et al., 2016; Frommhagen et al., 2017a). LPMOs secreted by these pathogenic fungi are possibly used to disrupt the cell wall in order to improve the penetration of the fungus into the plant cell. Thereby, LPMOs are likely to get in contact with the intracellular matrix of the plant cell (e.g., in leaves) and endogenous electron donors become available for LPMOs, such as ascorbic acid or reduced glutathione (Figure 4). Moreover, it is likely that photosynthetic pigments, such as chlorophyll, are released from the chloroplast during the decay of the plant cell and donate electrons for LPMOs. Based on the current knowledge, however, it remains debatable to what extent the latter system will contribute to the LPMO-assisted PCW degradation in vivo.
Catalases and superoxide dismutases are secreted by fungi and have been shown to potentially decrease the inactivation of LPMOs via the regulation of the reactive oxygen species concentration (Zintel et al., 2011; Schneider et al., 2016). However, the role of these enzymes during the LPMO-mediated plant biomass degradation is currently challenged due to a not fully understood reaction mechanism, in particular, regarding the role of O2 or H2O2 during the catalytic cleavage of the substrate (Phillips et al., 2011; Scott et al., 2016; Walton and Davies, 2016; Bissaro et al., 2017; Möllers et al., 2017; Simmons et al., 2017; Hangasky et al., 2018).
Future Prospects
Substrate Specificity and C1/C4-Regioselectivity
First described to oxidize crystalline polysaccharides like chitin and cellulose, it became apparent that LPMOs oxidize a much wider range of substrates including various hemicelluloses. At this moment it is not clear which of the above-mentioned substrate specificities of LPMOs are most beneficial to improve the activity of hydrolytic enzyme cocktails toward plant biomass. Presumably, the decomposition of diverse PCW structures demands the presence of LPMOs that comprise diverse substrate specificities like hemicellulose-active LPMOs, but the role of these LPMOs during plant biomass degradation has still to be proven experimentally. Furthermore, we do not know why fungi express LPMOs that oxidize soluble oligosaccharides since hydrolytic enzymes such as cellobiohydrolases can degrade these substrates significantly faster. Certainly, LPMOs that oxidize crystalline polysaccharides are able to enhance the substrate accessibility and improve the activity of hydrolytic enzymes (Vaaje-Kolstad et al., 2005a; Harris et al., 2010). At this moment, it remains enigmatic why chitin-oxidizing LPMOs boost the activity of chitinases toward chitin to a larger extent than cellulose-oxidizing LPMOs added to cellulases in the presence of cellulose (Vaaje-Kolstad et al., 2010; Forsberg et al., 2011, 2014a). Moreover, we suggested that LPMOs may comprise further mode of actions, like processive or random polysaccharide-oxidizing LPMOs, or, as already shown, LPMOs that preferably cleave amorphous cellulose rather than crystalline cellulose (Villares et al., 2017; Table 1). But, how the mode of action of LPMOs affects plant biomass degradation and the activity of hydrolytic enzymes remains to be answered.
Methods like proteomics, in particular, analysis of secretomes or fungi grown on different substrates have a good potential for the identification of LPMOs with promising substrate specificities. These techniques led to the recent discovery of the AA14 family by studying the growth of the white-rot fungus Pycnoporus coccineus in the presence of different types of biomass (Tian et al., 2009; Berka et al., 2011; Couturier et al., 2018). These findings about various substrate specificities and C1/C4-regioselectivities of LPMOs will contribute to the development of more “tailor-made” enzyme cocktails and evoke possibilities for new applications, such as polysaccharide modification to form novel polysaccharides.
Reducing Agent Specificity
Several electron-donating systems of LPMOs have been described so far, but it can only be speculated which system is actually of relevance for different types of fungi that comprise certain modes of life-style, such as saprophytes and plant pathogenic fungi. Further investigation of these systems would potentially provide more answers concerning the role of electron-donating systems during plant biomass degradation, especially when aiming to improve the activity of hydrolytic enzyme cocktails comprising LPMOs (Westereng et al., 2015; Frommhagen et al., 2016). Interestingly, lignin is the most abundant plant polyphenol in biomass. However, only limited amount of work has been done on the interaction between lignin (e.g., high and low molecular weight lignin; H, G, and S units) and LPMOs and its role during LPMO-mediated plant biomass degradation, which merits future investigation (Westereng et al., 2015). In addition, Hu et al. (2014) demonstrated that pretreatments affect the cellulose hydrolysis and thereby also the extent of the LPMO-mediated boosting effect. Therefore, optimization of the plant biomass degradation with LPMO-enriched enzyme cocktails and the simultaneous development of pretreatments should include the focus on the potential of residual lignin as a reducing agent.
Most of the commercial enzyme cocktails that have been recently developed are enriched with LPMOs to varying amounts, like Cellic CTec2 and CTec3 (Chylenski et al., 2017). The increase of the hydrolytic activity that can be achieved by the addition of AA9 LPMOs to these enzyme cocktails varies, depending on the type of the LPMO and the substrate that has been used for the activity assay (Merino and Cherry, 2007; Hu et al., 2014, 2015). Notably, it should be considered that the addition of LPMOs to an enzyme cocktail is limited due to the increased formation of oxidized moieties that are known inhibitors of hydrolases, like β-glucosidases. In addition, these oxidized moieties also inhibit subsequent ethanol fermentation since only a limited amount of these compounds can be metabolized by Saccharomyces cerevisiae species (Peinado et al., 2003, 2006; Cannella et al., 2012).
Catalytic Mechanism and Activity
Vaaje-Kolstad et al. (2010) presented LPMOs as monooxygenases that demand molecular oxygen and external electron donors to oxidize polysaccharides (Vaaje-Kolstad et al., 2010). This widely accepted catalytic mechanism has recently been expanded by findings of Bissaro et al. (2017), which indicate that hydrogen peroxide-fueled LPMOs are able to act under anaerobic conditions (Bissaro et al., 2017). This finding is of high importance for future applications, since biomass degradation at an industrial scale is mainly conducted under oxygen-poor conditions. A changed view on the catalytic mechanism will also improve the understanding of how LPMOs are effected by different electron-donating systems in the presence of different substrates (Simmons et al., 2017). Further investigation should aim at the improved understanding of the structure-function relationships of LPMOs, which would also pave the way for protein engineering methods, such as directed evolution (Forsberg et al., 2017). These methods should initially aim at the improvement of the LPMO's operational stability, rather than the activity of the enzyme (Bissaro et al., 2017). The characterization of LPMOs should be expended by the investigation of the effect of varying incubation conditions on LPMO activity, especially in the presence of different reducing agents. This research is of high importance from an application point of view, since pH and temperature influence the catalytic performance of LPMOs significantly (Frommhagen et al., 2017c).
Concluding Remarks
LPMOs have initially been shown to oxidize hardly accessible substrates, however, it has become apparent that these powerful enzymes cleave a much broader range of PCW polysaccharides in the presence of various electron-donating systems. The discovery of new potential substrates and electron-donating systems of LPMOs is a continuous process, whereas the role of these enzymes during PCW degradation and plant-pathogen interactions has hardly been explored so far. Recent discoveries of new AA families, in particular AA14 and AA15, show the diversity of LPMO sources, ranging from bacteria, fungi, viruses and insects, which illustrate how widely these enzymes are distributed throughout nature. Here, we summarized findings obtained from in vitro studies of LPMO substrate-specificities and electron-donating systems to emphasize their potential role for fungi in vitro. The latter is expected to contribute to the further understanding of these enzymes and, possibly, will promote new possibilities for a better usage of LPMOs within the biotechnology field.
Author Contributions
MF designed and wrote the manuscript. AW and MF built the LPMO models. MK and WvB contributed to the design and performed the critical revision of the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was funded by The Graduate School VLAG (Advanced Studies in Food Technology, Agrobiotechnology, Nutrition and Health Sciences) of Wageningen University & Research (Wageningen, The Netherlands). Part of the content of this manuscript has been published in the Ph.D. thesis of Dr. Matthias Frommhagen (Frommhagen, 2017).
References
Aachmann, F. L., Sorlie, M., Skjak-Braek, G., Eijsink, V. G., and Vaaje-Kolstad, G. (2012). NMR structure of a lytic polysaccharide monooxygenase provides insight into copper binding, protein dynamics, and substrate interactions. Proc. Natl. Acad. Sci. U.S.A. 109, 18779–18784. doi: 10.1073/pnas.1208822109
Agger, J. W., Isaksen, T., Várnai, A., Vidal-Melgosa, S., Willats, W. G., Ludwig, R., et al. (2014). Discovery of LPMO activity on hemicelluloses shows the importance of oxidative processes in plant cell wall degradation. Proc. Natl. Acad. Sci. U.S.A. 111, 6287–6292. doi: 10.1073/pnas.1323629111
Arola, S., and Linder, M. B. (2016). Binding of cellulose binding modules reveal differences between cellulose substrates. Sci. Rep. 6:35358. doi: 10.1038/srep35358
Atalla, R. H., and Vanderhart, D. L. (1984). Native cellulose: a composite of two distinct crystalline forms. Science 223, 283–285. doi: 10.1126/science.223.4633.283
Beeson, W. T., Phillips, C. M., Cate, J. H., and Marletta, M. A. (2012). Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J. Am. Chem. Soc. 134, 890–892. doi: 10.1021/ja210657t
Beeson, W. T., Vu, V. V., Span, E. A., Phillips, C. M., and Marletta, M. A. (2015). Cellulose degradation by polysaccharide monooxygenases. Annu. Rev. Biochem. 84, 923–946. doi: 10.1146/annurev-biochem-060614-034439
Bennati-Granier, C., Garajova, S., Champion, C., Grisel, S., Haon, M., Zhou, S., et al. (2015). Substrate specificity and regioselectivity of fungal AA9 lytic polysaccharide monooxygenases secreted by Podospora anserina. Biotechnol. Biofuels 8:90. doi: 10.1186/s13068-015-0274-3
Berka, R. M., Grigoriev, I. V., Otillar, R., Salamov, A., Grimwood, J., Reid, I., et al. (2011). Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat. Biotechnol. 29, 922–927. doi: 10.1038/nbt.1976
Bertini, L., Breglia, R., Lambrughi, M., Fantucci, P., De Gioia, L., Borsari, M., et al. (2017). Catalytic mechanism of fungal lytic polysaccharide monooxygenases investigated by first-principles calculations. Inorg. Chem. 57, 86–97. doi: 10.1021/acs.inorgchem.7b02005
Bey, M., Zhou, S., Poidevin, L., Henrissat, B., Coutinho, P. M., Berrin, J. G., et al. (2013). Cello-oligosaccharide oxidation reveals differences between two lytic polysaccharide monooxygenases (family GH61) from Podospora anserina. Appl. Environ. Microbiol. 79, 488–496. doi: 10.1128/AEM.02942-12
Bissaro, B., Forsberg, Z., Ni, Y., Hollmann, F., Vaaje-Kolstad, G., and Eijsink, V. G. H. (2016). Fueling biomass-degrading oxidative enzymes by light-driven water oxidation. Green Chem. 18, 5357–5366. doi: 10.1039/C6GC01666A
Bissaro, B., Rohr, A. K., Muller, G., Chylenski, P., Skaugen, M., Forsberg, Z., et al. (2017). Oxidative cleavage of polysaccharides by monocopper enzymes depends on H2O2. Nat. Chem. Biol. 13, 1123–1128. doi: 10.1038/nchembio.2470
Bodenheimer, A. M., O'dell, W. B., Stanley, C. B., and Meilleur, F. (2017). Structural studies of Neurospora crassa LPMO9D and redox partner CDHIIA using neutron crystallography and small-angle scattering. Carbohydr. Res. 448, 200–204. doi: 10.1016/j.carres.2017.03.001
Borisova, A. S., Isaksen, T., Dimarogona, M., Kognole, A. A., Mathiesen, G., Várnai, A., et al. (2015). Structural and functional characterization of a lytic polysaccharide monooxygenase with broad substrate specificity. J. Biol. Chem. 290, 22955–22969. doi: 10.1074/jbc.M115.660183
Brenelli, L., Squina, F. M., Felby, C., and Cannella, D. (2018). Laccase-derived lignin compounds boost cellulose oxidative enzymes AA9. Biotechnol. Biofuels 11:10. doi: 10.1186/s13068-017-0985-8
Breslmayr, E., Hanžek, M., Hanrahan, A., Leitner, C., Kittl, R., Šantek, B., et al. (2018). A fast and sensitive activity assay for lytic polysaccharide monooxygenase. Biotechnol. Biofuels 11:79. doi: 10.1186/s13068-018-1063-6
Brown, K., Harris, P. J., Zaretsky, E., Re, E., Vlasenko, E., McFarland, K. C., et al. (2008). Polypeptides Having Cellulolytic Enhancing Activity and Polynucleotides Encoding Same. U.S. Patent No US20050191736A1. Davies, CA: Novozymes, Inc.
Bulakhov, A. G., Gusakov, A. V., Chekushina, A. V., Satrutdinov, A. D., Koshelev, A. V., Matys, V. Y., et al. (2016). Isolation of homogeneous polysaccharide monooxygenases from fungal sources and investigation of their synergism with cellulases when acting on cellulose. Biochemistry 81, 530–537. doi: 10.1134/S0006297916050102
Bulakhov, A. G., Volkov, P. V., Rozhkova, A. M., Gusakov, A. V., Nemashkalov, V. A., Satrutdinov, A. D., et al. (2017). Using an inducible promoter of a gene encoding Penicillium verruculosum glucoamylase for production of enzyme preparations with enhanced cellulase performance. PLoS ONE 12:e0170404. doi: 10.1371/journal.pone.0170404
Bulone, V., Fincher, G. B., and Stone, B. A. (1995). In vitro synthesis of a microfibrillar (1→3)-β-glucan by a ryegrass (Lolium multiflorum) endosperm (1→3)-β-glucan synthase enriched by product entrapment. Plant J. 8, 213–225. doi: 10.1046/j.1365-313X.1995.08020213.x
Camarero, S., Ibarra, D., Martínez, M. J., and Martínez, Á. T. (2005). Lignin-derived compounds as efficient laccase mediators for decolorization of different types of recalcitrant dyes. Appl. Environ. Microbiol. 71, 1775–1784. doi: 10.1128/AEM.71.4.1775-1784.2005
Cannella, D., Hsieh, C.-W. C., Felby, C., and Jørgensen, H. (2012). Production and effect of aldonic acids during enzymatic hydrolysis of lignocellulose at high dry matter content. Biotechnol. Biofuels 5:26. doi: 10.1186/1754-6834-5-26
Cannella, D., Möllers, K. B., Frigaard, N. U., Jensen, P. E., Bjerrum, M. J., Johansen, K. S., et al. (2016). Light-driven oxidation of polysaccharides by photosynthetic pigments and a metalloenzyme. Nat. Commun. 7:11134. doi: 10.1038/ncomms11134
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Carpita, N. C., and Gibeaut, D. M. (1993). Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3, 1–30. doi: 10.1111/j.1365-313X.1993.tb00007.x
Carpita, N. C., and McCann, M. C. (2008). Maize and sorghum: genetic resources for bioenergy grasses. Trends Plant Sci. 13, 415–420. doi: 10.1016/j.tplants.2008.06.002
Chabbert, B., Habrant, A., Herbaut, M., Foulon, L., Aguié-Béghin, V., Garajova, S., et al. (2017). Action of lytic polysaccharide monooxygenase on plant tissue is governed by cellular type. Sci. Rep. 7:17792. doi: 10.1038/s41598-017-17938-2
Chen, F., and Dixon, R. A. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25, 759–761. doi: 10.1038/nbt1316
Cherney, J. H., Anliker, K. S., Albrecht, K. A., and Wood, K. V. (1989). Soluble phenolic monomers in forage crops. J. Agric. Food Chem. 37, 345–350. doi: 10.1021/jf00086a016
Chylenski, P., Forsberg, Z., Ståhlberg, J., Várnai, A., Lersch, M., Bengtsson, O., et al. (2017). Development of minimal enzyme cocktails for hydrolysis of sulfite-pulped lignocellulosic biomass. J. Biotechnol. 246, 16–23. doi: 10.1016/j.jbiotec.2017.02.009
Constant, S., Wienk, H. L. J., Frissen, A. E., Peinder, P. D., Boelens, R., Van Es, D. S., et al. (2016). New insights into the structure and composition of technical lignins: a comparative characterisation study. Green Chem. 18, 2651–2665. doi: 10.1039/C5GC03043A
Courtade, G., Wimmer, R., Røhr, Å. K., Preims, M., Felice, A. K. G., Dimarogona, M., et al. (2016). Interactions of a fungal lytic polysaccharide monooxygenase with β-glucan substrates and cellobiose dehydrogenase. Proc. Nat. Acad. Sci. U.S.A. 113, 5922–5927. doi: 10.1073/pnas.1602566113
Couturier, M., Ladevèze, S., Sulzenbacher, G., Ciano, L., Fanuel, M., Moreau, C., et al. (2018). Lytic xylan oxidases from wood-decay fungi unlock biomass degradation. Nat. Chem. Biol. 14, 306–310. doi: 10.1038/nchembio.2558
Couturier, M., Navarro, D., Chevret, D., Henrissat, B., Piumi, F., Ruiz-Dueñas, F. J., et al. (2015). Enhanced degradation of softwood versus hardwood by the white-rot fungus Pycnoporus coccineus. Biotechnol. Biofuels 8:216. doi: 10.1186/s13068-015-0407-8
Crouch, L. I., Labourel, A., Walton, P. H., Davies, G. J., and Gilbert, H. J. (2016). The contribution of non-catalytic carbohydrate binding modules to the activity lytic polysaccharide monooxygenases. J. Biol. Chem. 291, 7439–7449. doi: 10.1074/jbc.M115.702365
Danneels, B., Tanghe, M., Joosten, H.-J., Gundinger, T., Spadiut, O., Stals, I., et al. (2017). A quantitative indicator diagram for lytic polysaccharide monooxygenases reveals the role of aromatic surface residues in HjLPMO9A regioselectivity. PLoS ONE 12:e0178446. doi: 10.1371/journal.pone.0178446
Darvill, J. E., McNeil, M., Darvill, A. G., and Albersheim, P. (1980). Structure of plant cell walls: XI. Glucuronoarabinoxylan, a second hemicellulose in the primary cell wall of suspension-cultured sycamore cells. Plant Physiol. 66, 1135–1139. doi: 10.1104/pp.66.6.1135
Davies, G., and Henrissat, B. (1995). Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859. doi: 10.1016/S0969-2126(01)00220-9
Davies, G. J., and Williams, S. J. (2016). Carbohydrate-active enzymes: sequences, shapes, contortions and cells. Biochem. Soc. Trans. 44, 79–87. doi: 10.1042/BST20150186
Dean, R. A., Talbot, N. J., Ebbole, D. J., Farman, M. L., Mitchell, T. K., Orbach, M. J., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986. doi: 10.1038/nature03449
Del Río, J. C., Rencoret, J., Prinsen, P., Martinez, A. T., Ralph, J., and Gutierrez, A. (2012). Structural characterization of wheat straw lignin as revealed by analytical pyrolysis, 2D-NMR, and reductive cleavage methods. J. Agric. Food Chem. 60, 5922–5935. doi: 10.1021/jf301002n
Dotsen, W. D., Greenier, J., and Ding, H. (2007). Polypeptides Having Cellulolytic Enhancing Activity and Polynucleotides Encoding Same. U.S. Patent No US7271244B2. Davies, CA: Novozymes, Inc.
Dykes, L., and Rooney, L. W. (2007). Phenolic compounds in cereal grains and their health benefits. Cereal Foods World. 52, 105–111. doi: 10.1094/CFW-52-3-0105
Ebringerová, A., Hromádková, Z., and Heinze, T. (2005). Hemicellulose. Adv. Polym. Sci. 186, 1–67. doi: 10.1007/b136816
Eibinger, M., Ganner, T., Bubner, P., Rosker, S., Kracher, D., Haltrich, D., et al. (2014). Cellulose surface degradation by a lytic polysaccharide monooxygenase and its effect on cellulase hydrolytic efficiency. J. Biol. Chem. 289, 35929–35938. doi: 10.1074/jbc.M114.602227
Eibinger, M., Sattelkow, J., Ganner, T., Plank, H., and Nidetzky, B. (2017). Single-molecule study of oxidative enzymatic deconstruction of cellulose. Nat. Commun. 8:894. doi: 10.1038/s41467-017-01028-y
Eriksson, K.-E., Pettersson, B., and Westermark, U. (1974). Oxidation: an important enzyme reaction in fungal degradation of cellulose. FEBS Lett. 49, 282–285. doi: 10.1016/0014-5793(74)80531-4
Fanuel, M., Garajova, S., Ropartz, D., McGregor, N., Brumer, H., Rogniaux, H., et al. (2017). The Podospora anserina lytic polysaccharide monooxygenase PaLPMO9H catalyzes oxidative cleavage of diverse plant cell wall matrix glycans. Biotechnol. Biofuels 10:63. doi: 10.1186/s13068-017-0749-5
Fernandes, A. N., Thomas, L. H., Altaner, C. M., Callow, P., Forsyth, V. T., Apperley, D. C., et al. (2011). Nanostructure of cellulose microfibrils in spruce wood. Proc. Natl. Acad. Sci. U.S.A. 108, E1195–E1203. doi: 10.1073/pnas.1108942108
Fischer, R. L. A., and Bennett, A. B. (1991). Role of cell wall hydrolases in fruit ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42, 675–703. doi: 10.1146/annurev.pp.42.060191.003331
Forsberg, Z., Bissaro, B., Gullesen, J., Dalhus, B., Vaaje-Kolstad, G., and Eijsink, V. G. H. (2017). Structural determinants of bacterial lytic polysaccharide monooxygenase functionality. J. Biol. Chem. 293, 1397–1412. doi: 10.1074/jbc.M117.817130
Forsberg, Z., MacKenzie, A. K., Sørlie, M., Røhr, Å. K., Helland, R., Arvai, A. S., et al. (2014a). Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases. Proc. Natl. Acad. Sci. U.S.A. 111, 8446–8451. doi: 10.1073/pnas.1402771111
Forsberg, Z., Røhr, A. K., Mekasha, S., Andersson, K. K., Eijsink, V. G., Vaaje-Kolstad, G., et al. (2014b). Comparative study of two chitin-active and two cellulose-active AA10-type lytic polysaccharide monooxygenases. Biochemistry 53, 1647–1656. doi: 10.1021/bi5000433
Forsberg, Z., Vaaje-Kolstad, G., Westereng, B., Bunæs, A. C., Stenstrøm, Y., Mackenzie, A., et al. (2011). Cleavage of cellulose by a CBM33 protein. Protein Sci. 20, 1479–1483. doi: 10.1002/pro.689
Frandsen, K. E. H., and Lo Leggio, L. (2016). Lytic polysaccharide monooxygenases: a crystallographer's view on a new class of biomass-degrading enzymes. IUCrJ 3, 448–467. doi: 10.1107/S2052252516014147
Frandsen, K. E. H., Poulsen, J.-C. N., Tandrup, T., and Lo Leggio, L. (2017a). Unliganded and substrate bound structures of the cellooligosaccharide active lytic polysaccharide monooxygenase LsAA9A at low pH. Carbohydr. Res. 448, 187–190. doi: 10.1016/j.carres.2017.03.010
Frandsen, K. E. H., Poulsen, J.-C. N., Tovborg, M., Johansen, K. S., and Lo Leggio, L. (2017b). Learning from oligosaccharide soaks of crystals of an AA13 lytic polysaccharide monooxygenase: crystal packing, ligand binding and active-site disorder. Acta Crystallogr. Sect. D Biol. Crystallogr. 73, 64–76. doi: 10.1107/S2059798316019641
Frandsen, K. E. H., Simmons, T. J., Dupree, P., Poulsen, J.-C. N., Hemsworth, G. R., Ciano, L., et al. (2016). The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases. Nat. Chem. Biol. 12, 298–303. doi: 10.1038/nchembio.2029
Frommhagen, M. (2017). Lytic Polysacchairde Monooxygenases from Myceliophthora thermophila C1. Dissertation, Wageningen University & Research, Wageningen.
Frommhagen, M., Koetsier, M. J., Westphal, A. H., Visser, J., Hinz, S. W. A., Vincken, J.-P., et al. (2016). Lytic polysaccharide monooxygenases from Myceliophthora thermophila C1 differ in substrate preference and reducing agent specificity. Biotechnol. Biofuels 9, 1–17. doi: 10.1186/s13068-016-0594-y
Frommhagen, M., Mutte, S. K., Westphal, A. H., Koetsier, M. J., Hinz, S. W. A., Visser, J., et al. (2017a). Boosting LPMO-driven lignocellulose degradation by polyphenol oxidase-activated lignin building blocks. Biotechnol. Biofuels 10:121. doi: 10.1186/s13068-017-0810-4
Frommhagen, M., Sforza, S., Westphal, A. H., Visser, J., Hinz, S. W. A., Koetsier, M. J., et al. (2015). Discovery of the combined oxidative cleavage of plant xylan and cellulose by a new fungal polysaccharide monooxygenase. Biotechnol. Biofuels 8:101. doi: 10.1186/s13068-015-0284-1
Frommhagen, M., Van Erven, G., Sanders, M., Van Berkel, W. J. H., Kabel, M. A., and Gruppen, H. (2017b). RP-UHPLC-UV-ESI-MS/MS analysis of LPMO generated C4-oxidized gluco-oligosaccharides after non-reductive labeling with 2-aminobenzamide. Carbohydr. Res. 448, 191–199. doi: 10.1016/j.carres.2017.03.006
Frommhagen, M., Westphal, A. H., Hilgers, R., Koetsier, M. J., Hinz, S. W. A., Visser, J., et al. (2017c). Quantification of the catalytic performance of C1-cellulose-specific lytic polysaccharide monooxygenases. Appl. Microbiol. Biotechnol. 102, 1281–1295. doi: 10.1007/s00253-017-8541-9
Fry, S. C., Nesselrode, B. H. W. A., Miller, J. G., and Mewburn, B. R. (2008). Mixed-linkage (1→3,1→4)-β-d-glucan is a major hemicellulose of Equisetum (horsetail) cell walls. New Phytol. 179, 104–115. doi: 10.1111/j.1469-8137.2008.02435.x
Fry, S. C., York, W. S., Albersheim, P., Darvill, A., Hayashi, T., Joseleau, J.-P., et al. (1993). An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol. Plant. 89, 1–3. doi: 10.1111/j.1399-3054.1993.tb01778.x
Garajova, S., Mathieu, Y., Beccia, M. R., Bennati-Granier, C., Biaso, F., Fanuel, M., et al. (2016). Single-domain flavoenzymes trigger lytic polysaccharide monooxygenases for oxidative degradation of cellulose. Sci. Rep. 6:28276. doi: 10.1038/srep28276
Gilbert, H. J., Stålbrand, H., and Brumer, H. (2008). How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Curr. Opin. Plant Biol. 11, 338–348. doi: 10.1016/j.pbi.2008.03.004
Gilkes, N. R., Henrissat, B., Kilburn, D. G., Miller, R. C., and Warren, R. A. (1991). Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol. Rev. 55, 303–315.
Gírio, F. M., Fonseca, C., Carvalheiro, F., Duarte, L. C., Marques, S., and Bogel-Łukasik, R. (2010). Hemicelluloses for fuel ethanol: a review. Bioresour. Technol. 101, 4775–4800. doi: 10.1016/j.biortech.2010.01.088
Gordon, A. H., Lomax, J. A., Dalgarno, K., and Chesson, A. (1985). Preparation and composition of mesophyll, epidermis and fibre cell walls from leaves of perennial ryegrass (lolium perenne) and italian ryegrass (lolium multiflorum). J. Sci. Food Agric. 36, 509–519. doi: 10.1002/jsfa.2740360702
Grigoriev, I. V., Nikitin, R., Haridas, S., Kuo, A., Ohm, R., Otillar, R., et al. (2014). MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42, D699–D704. doi: 10.1093/nar/gkt1183
Gusakov, A. V., Bulakhov, A. G., Demin, I. N., and Sinitsyn, A. P. (2017). Monitoring of reactions catalyzed by lytic polysaccharide monooxygenases using highly-sensitive fluorimetric assay of the oxygen consumption rate. Carbohydr. Res. 452, 156–161. doi: 10.1016/j.carres.2017.10.015
Hammel, K. E., Jensen, K. A., Mozuch, M. D., Landucci, L. L., Tien, M., and Pease, E. A. (1993). Ligninolysis by a purified lignin peroxidase. J. Biol. Chem. 268, 12274–12281.
Hangasky, J. A., Iavarone, A. T., and Marletta, M. A. (2018). Reactivity of O2 versus H2O2 with polysaccharide monooxygenases. Proc. Natl. Acad. Sci. U.S.A. 115, 4915–4920. doi: 10.1073/pnas.1801153115
Hansson, H., Karkehabadi, S., Mikkelsen, N., Douglas, N. R., Kim, S., Lam, A., et al. (2017). High-resolution structure of a lytic polysaccharide monooxygenase from Hypocrea jecorina reveals a predicted linker as an integral part of the catalytic domain. J. Biol. Chem. 292, 19099–19109. doi: 10.1074/jbc.M117.799767
Harris, P. J., and Stone, B. A. (2009). “Chemistry and molecular organization of plant cell walls,” in Biomass Recalcitrance: Deconstructing the Plant Cell Wall for Bioenergy, Vol. 4, ed M. E. Himmel (Oxford: Blackwell Publishing Ltd.), 61–93.
Harris, P. V., Welner, D., McFarland, K. C., Re, E., Navarro Poulsen, J. C., Brown, K., et al. (2010). Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry 49, 3305–3316. doi: 10.1021/bi100009p
Haslam, E. (2007). Vegetable tannins – Lessons of a phytochemical lifetime. Phytochemistry 68, 2713–2721. doi: 10.1016/j.phytochem.2007.09.009
Hatfield, R. D., Marita, J. M., and Frost, K. (2008). Characterization of p-coumarate accumulation, p-coumaroyl transferase, and cell wall changes during the development of corn stems. J. Sci. Food Agric. 88, 2529–2537. doi: 10.1002/jsfa.3376
Hemsworth, G. R., Davies, G. J., and Walton, P. H. (2013). Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr. Opin. Struct. Biol. 23, 660–668. doi: 10.1016/j.sbi.2013.05.006
Hemsworth, G. R., Henrissat, B., Davies, G. J., and Walton, P. H. (2014). Discovery and characterization of a new family of lytic polysaccharide mono-oxygenases. Nat. Chem. Biol. 10, 122–126. doi: 10.1038/nchembio.1417
Hemsworth, G. R., Johnston, E. M., Davies, G. J., and Walton, P. H. (2015). Lytic polysaccharide monooxygenases in biomass conversion. Trends Biotechnol. 33, 747–761. doi: 10.1016/j.tibtech.2015.09.006
Henrissat, B., and Davies, G. (1997). Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7, 637–644. doi: 10.1016/S0959-440X(97)80072-3
Hernández-Ortega, A., Ferreira, P., and Martínez, A. T. (2012). Fungal aryl-alcohol oxidase: a peroxide-producing flavoenzyme involved in lignin degradation. Appl. Microbiol. Biotechnol. 93, 1395–1410. doi: 10.1007/s00253-011-3836-8
Hildén, L., Johansson, G., Pettersson, G., Li, J., Ljungquist, P., and Henriksson, G. (2000). Do the extracellular enzymes cellobiose dehydrogenase and manganese peroxidase form a pathway in lignin biodegradation? FEBS Lett. 477, 79–83. doi: 10.1016/S0014-5793(00)01757-9
Hoffman, M., Jia, Z., Peña, M. J., Cash, M., Harper, A., Blackburn Ii, A. R., et al. (2005). Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr. Res. 340, 1826–1840. doi: 10.1016/j.carres.2005.04.016
Hofrichter, M. (2002). Review: lignin conversion by manganese peroxidase (MnP). Enzyme Microb. Technol. 30, 454–466. doi: 10.1016/S0141-0229(01)00528-2
Hori, C., Igarashi, K., Katayama, A., and Samejima, M. (2011). Effects of xylan and starch on secretome of the basidiomycete Phanerochaete chrysosporium grown on cellulose. FEMS Microbiol. Lett. 321, 14–23. doi: 10.1111/j.1574-6968.2011.02307.x
Horn, S. J., Vaaje-Kolstad, G., Westereng, B., and Eijsink, V. (2012). Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 5:45. doi: 10.1186/1754-6834-5-45
Hu, J., Arantes, V., Pribowo, A., Gourlay, K., and Saddler, J. N. (2014). Substrate factors that influence the synergistic interaction of AA9 and cellulases during the enzymatic hydrolysis of biomass. Energy Environ. Sci. 7, 2308–2315. doi: 10.1039/C4EE00891J
Hu, J., Chandra, R., Arantes, V., Gourlay, K., Susan Van Dyk, J., and Saddler, J. N. (2015). The addition of accessory enzymes enhances the hydrolytic performance of cellulase enzymes at high solid loadings. Biores. Technol. 186, 149–153. doi: 10.1016/j.biortech.2015.03.055
Isaksen, T., Westereng, B., Aachmann, F. L., Agger, J. W., Kracher, D., Kittl, R., et al. (2014). A C4-oxidizing lytic polysaccharide monooxygenase cleaving both cellulose and cello-oligosaccharides. J. Biol. Chem. 289, 2632–2642. doi: 10.1074/jbc.M113.530196
Jacquet, G., Pollet, B., Lapierre, C., Mhamdi, F., and Rolando, C. (1995). New ether-linked ferulic acid-coniferyl alcohol dimers identified in grass straws. J. Agric. Food Chem. 43, 2746–2751. doi: 10.1021/jf00058a037
Jagadeeswaran, G., Gainey, L., Prade, R., and Mort, A. J. (2016). A family of AA9 lytic polysaccharide monooxygenases in Aspergillus nidulans is differentially regulated by multiple substrates and at least one is active on cellulose and xyloglucan. Appl. Microbiol. Biotechnol. 100, 4535–4547. doi: 10.1007/s00253-016-7505-9
Johansen, K. S. (2016). Discovery and industrial applications of lytic polysaccharide mono-oxygenases. Biochem. Soc. Trans. 44, 143–149. doi: 10.1042/BST20150204
Jung, J., Sethi, A., Gaiotto, T., Han, J. J., Jeoh, T., Gnanakaran, S., et al. (2013). Binding and movement of individual Cel7A cellobiohydrolases on crystalline cellulose surfaces revealed by single-molecule fluorescence imaging. J. Biol. Chem. 288, 24164–24172. doi: 10.1074/jbc.M113.455758
Kabel, M. A., Van Den Borne, H., Vincken, J.-P., Voragen, A. G. J., and Schols, H. A. (2007). Structural differences of xylans affect their interaction with cellulose. Carbohydr. Polym. 69, 94–105. doi: 10.1016/j.carbpol.2006.09.006
Karkehabadi, S., Hansson, H., Kim, S., Piens, K., Mitchinson, C., and Sandgren, M. (2008). The first structure of a glycoside hydrolase family 61 member, Cel61B from Hypocrea jecorina, at 1.6 A resolution. J. Mol. Biol. 383, 144–154. doi: 10.1016/j.jmb.2008.08.016
Karnaouri, A., Muraleedharan, M. N., Dimarogona, M., Topakas, E., Rova, U., Sandgren, M., et al. (2017). Recombinant expression of thermostable processive MtEG5 endoglucanase and its synergism with MtLPMO from Myceliophthora thermophila during the hydrolysis of lignocellulosic substrates. Biotechnol. Biofuels 10:126. doi: 10.1186/s13068-017-0813-1
Kim, I. J., Seo, N., An, H. J., Kim, J.-H., Harris, P. V., and Kim, K. H. (2017). Type-dependent action modes of TtAA9E and TaAA9A acting on cellulose and differently pretreated lignocellulosic substrates. Biotechnol. Biofuels 10:46. doi: 10.1186/s13068-017-0721-4
King, J. R., Bowers, C. M., and Toone, E. J. (2015). Specific binding at the cellulose binding module–cellulose interface observed by force spectroscopy. Langmuir 31, 3431–3440. doi: 10.1021/la504836u
Kittl, R., Kracher, D., Burgstaller, D., Haltrich, D., and Ludwig, R. (2012). Production of four Neurospora crassa lytic polysaccharide monooxygenases in Pichia pastoris monitored by a fluorimetric assay. Biotechnol. Biofuels 5:79. doi: 10.1186/1754-6834-5-79
Kjaergaard, C. H., Qayyum, M. F., Wong, S. D., Xu, F., Hemsworth, G. R., Walton, D. J., et al. (2014). Spectroscopic and computational insight into the activation of O2 by the mononuclear Cu center in polysaccharide monooxygenases. Proc. Natl. Acad. Sci. U.S.A. 111, 8797–8802. doi: 10.1073/pnas.1408115111
Klinman, J. P. (2006). The copper-enzyme family of dopamine β-monooxygenase and peptidylglycine α-hydroxylating monooxygenase: resolving the chemical pathway for substrate hydroxylation. J. Biol. Chem. 281, 3013–3016. doi: 10.1074/jbc.R500011200
Kojima, Y., Várnai, A., Ishida, T., Sunagawa, N., Petrovic, D. M., Igarashi, K., et al. (2016). A lytic polysaccharide monooxygenase with broad xyloglucan specificity from the brown-rot fungus Gloeophyllum trabeum and its action on cellulose-xyloglucan complexes. Appl. Environ. Microbiol. 82, 6557–6572. doi: 10.1128/AEM.01768-16
Kracher, D., Andlar, M., Furtmüller, P. G., and Ludwig, R. (2017). Active-site copper reduction promotes substrate binding of fungal lytic polysaccharide monooxygenase and reduces stability. J. Biol. Chem. 293, 1676–1687. doi: 10.1074/jbc.RA117.000109
Kracher, D., Scheiblbrandner, S., Felice, A. K. G., Breslmayr, E., Preims, M., Ludwicka, K., et al. (2016). Extracellular electron transfer systems fuel cellulose oxidative degradation. Science 352, 1098–1101 doi: 10.1126/science.aaf3165
Kubicek, C. P., Starr, T. L., and Glass, N. L. (2014). Plant cell wall–degrading enzymes and their secretion in plant-pathogenic fungi. Ann. Rev. Phytopathol. 52, 427–451. doi: 10.1146/annurev-phyto-102313-045831
Kühnel, S., Pouvreau, L., Appeldoorn, M. M., Hinz, S. W. A., Schols, H. A., and Gruppen, H. (2012). The ferulic acid esterases of Chrysosporium lucknowense C1: purification, characterization and their potential application in biorefinery. Enzyme Microb. Technol. 50, 77–85. doi: 10.1016/j.enzmictec.2011.09.008
Kurašin, M., and Väljamäe, P. (2011). Processivity of cellobiohydrolases is limited by the substrate. J. Biol. Chem. 286, 169–177. doi: 10.1074/jbc.M110.161059
Kuusk, S., Bissaro, B., Kuusk, P., Forsberg, Z., Eijsink, V. G. H., Sørlie, M., et al. (2017). Kinetics of H2O2-driven degradation of chitin by a bacterial lytic polysaccharide monooxygenase. J. Biol. Chem. 293, 523–531. doi: 10.1074/jbc.M117.817593
Ladevèze, S., Haon, M., Villares, A., Cathala, B., Grisel, S., Herpoël-Gimbert, I., et al. (2017). The yeast Geotrichum candidum encodes functional lytic polysaccharide monooxygenases. Biotechnol. Biofuels 10:215. doi: 10.1186/s13068-017-0903-0
Lam, T. B. T., Kadoya, K., and Iiyama, K. (2001). Bonding of hydroxycinnamic acids to lignin: ferulic and p-coumaric acids are predominantly linked at the benzyl position of lignin, not the β-position, in grass cell walls. Phytochemistry 57, 987–992. doi: 10.1016/S0031-9422(01)00052-8
Langston, J. A., Shaghasi, T., Abbate, E., Xu, F., Vlasenko, E., and Sweeney, M. D. (2011). Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl. Environ. Microbiol. 77, 7007–7015. doi: 10.1128/AEM.05815-11
Lenfant, N., Hainaut, M., Terrapon, N., Drula, E., Lombard, V., and Henrissat, B. (2017). A bioinformatics analysis of 3400 lytic polysaccharide oxidases from family AA9. Carbohydr. Res. 448, 166–174. doi: 10.1016/j.carres.2017.04.012
Levasseur, A., Drula, E., Lombard, V., Coutinho, P. M., and Henrissat, B. (2013). Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 6:41. doi: 10.1186/1754-6834-6-41
Li, X., Beeson, I., William, T, Phillips, C. M., Marletta, M. A., and Cate, J. H. D. (2012). Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure 20, 1051–1061. doi: 10.1016/j.str.2012.04.002
Liu, B., Kognole, A. A., Wu, M., Westereng, B., Crowley, M. F., Kim, S., et al. (2018). Structural and molecular dynamics studies of a C1-oxidizing lytic polysaccharide monooxygenase from Heterobasidion irregulare reveal amino acids important for substrate recognition. FEBS J. doi: 10.1111/febs.14472. [Epub ahead of print].
Liu, B., Olson, Å., Wu, M., Broberg, A., and Sandgren, M. (2017). Biochemical studies of two lytic polysaccharide monooxygenases from the white-rot fungus Heterobasidion irregulare and their roles in lignocellulose degradation. PLoS ONE 12:e0189479. doi: 10.1371/journal.pone.0189479
Lo Leggio, L., Simmons, T. J., Poulsen, J.-C. N., Frandsen, K. E. H., Hemsworth, G. R., Stringer, M. A., et al. (2015). Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nat. Commun. 6:5961. doi: 10.1038/ncomms6961
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Mahajan, S., and Master, E. R. (2010). Proteomic characterization of lignocellulose-degrading enzymes secreted by Phanerochaete carnosa grown on spruce and microcrystalline cellulose. Appl. Microbiol. Biotechnol. 86, 1903–1914. doi: 10.1007/s00253-010-2516-4
Mäkelä, M. R., Bouzid, O., Robl, D., Post, H., Peng, M., Heck, A., et al. (2017). Cultivation of Podospora anserina on soybean hulls results in an efficient enzyme cocktail for plant biomass hydrolysis. N. Biotechnol. 37, 162–171. doi: 10.1016/j.nbt.2017.02.002
Martinez, D., Larrondo, L. F., Putnam, N., Gelpke, M. D. S., Huang, K., Chapman, J., et al. (2004). Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22, 695–700. doi: 10.1038/nbt967
Meier, K. K., Jones, S. M., Kaper, T., Hansson, H., Koetsier, M. J., Karkehabadi, S., et al. (2017). Oxygen activation by Cu LPMOs in recalcitrant carbohydrate polysaccharide conversion to monomer sugars. Chem. Rev. 118, 2593–2635. doi: 10.1021/acs.chemrev.7b00421
Merino, S. T., and Cherry, J. (2007). “Progress and challenges in enzyme development for biomass utilization,” in Biofuels, ed L. Olsson (Berlin; Heidelberg: Springer), 95–120.
Mittal, A., Katahira, R., Himmel, M. E., and Johnson, D. K. (2011). Effects of alkaline or liquid-ammonia treatment on crystalline cellulose: changes in crystalline structure and effects on enzymatic digestibility. Biotechnol. Biofuels 4, 41–41. doi: 10.1186/1754-6834-4-41
Mohnen, D., Bar-Peled, M., and Somerville, C. (2008). “Biosynthesis of plant cell walls,” in Biomass Recalcitrance, ed M. Himmel (Oxford: Blackwell Publishing), 94–187.
Möllers, K. B., Mikkelsen, H., Simonsen, T. I., Cannella, D., Johansen, K. S., Bjerrum, M. J., et al. (2017). On the formation and role of reactive oxygen species in light-driven LPMO oxidation of phosphoric acid swollen cellulose. Carbohydr. Res. 448, 182–186. doi: 10.1016/j.carres.2017.03.013
Müller, G., Várnai, A., Johansen, K. S., Eijsink, V. G. H., and Horn, S. J. (2015). Harnessing the potential of LPMO-containing cellulase cocktails poses new demands on processing conditions. Biotechnol. Biofuels 8, 1–9. doi: 10.1186/s13068-015-0376-y
Naran, R., Chen, G., and Carpita, N. C. (2008). Novel rhamnogalacturonan I and arabinoxylan polysaccharides of flax seed mucilage. Plant Physiol. 148, 132–141. doi: 10.1104/pp.108.123513
Navarro, D., Rosso, M.-N., Haon, M., Olivé, C., Bonnin, E., Lesage-Meessen, L., et al. (2014). Fast solubilization of recalcitrant cellulosic biomass by the basidiomycete fungus Laetisaria arvalis involves successive secretion of oxidative and hydrolytic enzymes. Biotechnol. Biofuels 7, 1–14. doi: 10.1186/s13068-014-0143-5
Nekiunaite, L., Petrovi,ć, D. M., Westereng, B., Vaaje-Kolstad, G., Abou Hachem, M., Várnai, A., et al. (2016). FgLPMO9A from Fusarium graminearum cleaves xyloglucan independently of the backbone substitution pattern. FEBS Lett. 590, 3346–3356. doi: 10.1002/1873-3468.12385
Nishiyama, Y., Langan, P., and Chanzy, H. (2002). Crystal structure and hydrogen-bonding system in cellulose Iβ from synchrotron X-ray and neutron fiber diffraction. J. Am. Chem. Soc. 124, 9074–9082. doi: 10.1021/ja0257319
Nishiyama, Y., Sugiyama, J., Chanzy, H., and Langan, P. (2003). Crystal structure and hydrogen bonding system in cellulose Iα from synchrotron X-ray and neutron fiber diffraction. J. Am. Chem. Soc. 125, 14300–14306. doi: 10.1021/ja037055w
Park, Y. W., Baba, K. I., Furuta, Y., Iida, I., Sameshima, K., Arai, M., et al. (2004). Enhancement of growth and cellulose accumulation by overexpression of xyloglucanase in poplar. FEBS Lett. 564, 183–187. doi: 10.1016/S0014-5793(04)00346-1
Patel, I., Kracher, D., Ma, S., Garajova, S., Haon, M., Faulds, C. B., et al. (2016). Salt-responsive lytic polysaccharide monooxygenases from the mangrove fungus Pestalotiopsis sp. NCi6. Biotechnol. Biofuels 9, 1–12. doi: 10.1186/s13068-016-0520-3
Peinado, R. A., Moreno, J. J., Ortega, J. M., and Mauricio, J. C. (2003). Effect of gluconic acid consumption during simulation of biological aging of sherry wines by a flor yeast strain on the final volatile compounds. J. Agri. Food Chem. 51, 6198–6203. doi: 10.1021/jf034512j
Peinado, R. A., Mauricio, J. C., and Moreno, J. (2006). Aromatic series in sherry wines with gluconic acid subjected to different biological aging conditions by Saccharomyces cerevisiae var. capensis. Food Chem. 94, 232–239. doi: 10.1016/j.foodchem.2004.11.010
Pérez-Boada, M., Ruiz-Dueñas, F. J., Pogni, R., Basosi, R., Choinowski, T., Martínez, M. J., et al. (2005). Versatile peroxidase oxidation of high redox potential aromatic compounds: site-directed mutagenesis, spectroscopic and cystallographic investigation of three long-range electron transfer pathways. J. Mol. Biol. 354, 385–402. doi: 10.1016/j.jmb.2005.09.047
Phillips, C. M., Beeson, W. T., Cate, J. H., and Marletta, M. A. (2011). Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 6, 1399–1406. doi: 10.1021/cb200351y
Pierce, B. C., Agger, J. W., Wichmann, J., and Meyer, A. S. (2017a). Oxidative cleavage and hydrolytic boosting of cellulose in soybean spent flakes by Trichoderma reesei Cel61A lytic polysaccharide monooxygenase. Enzyme Microb. Technol. 98, 58–66. doi: 10.1016/j.enzmictec.2016.12.007
Pierce, B. C., Agger, J. W., Zhang, Z., Wichmann, J., and Meyer, A. S. (2017b). A comparative study on the activity of fungal lytic polysaccharide monooxygenases for the depolymerization of cellulose in soybean spent flakes. Carbohydr. Res. 449, 85–94. doi: 10.1016/j.carres.2017.07.004
Popper, Z. A., and Fry, S. C. (2003). Primary cell wall composition of Bryophytes and Charophytes. Ann. Bot. 91, 1–12. doi: 10.1093/aob/mcg013
Quinlan, R. J., Sweeney, M. D., Lo Leggio, L., Otten, H., Poulsen, J. C., Johansen, K. S., et al. (2011). Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. U.S.A. 108, 15079–15084. doi: 10.1073/pnas.1105776108
Raguz, S., Yagüe, E., Wood, D. A., and Thurston, C. F. (1992). Isolation and characterization of a cellulose-growth-specific gene from Agaricus bisporus. Gene 119, 183–190. doi: 10.1016/0378-1119(92)90270-Y
Reese, E. T., Siu, R. G. H., and Levinson, H. S. (1950). The biological degradation or soluble cellulose derivatives and its relationship to the mechanism of cellulose hydrolysis. J. Bacteriol. 59, 485–497.
Reiter, W.-D. (2002). Biosynthesis and properties of the plant cell wall. Curr. Opin. Plant Biol. 5, 536–542. doi: 10.1016/S1369-5266(02)00306-0
Rencoret, J., Marques, G., Gutiérrez, A., Nieto, L., Santos, J. I., Jiménez-Barbero, J., et al. (2009). HSQC-NMR analysis of lignin in woody (Eucalyptus globulus and Picea abies) and non-woody (Agave sisalana) ball-milled plant materials at the gel state 10th EWLP, Stockholm, Sweden, August 25–28, 2008. Holzforschung 63:6. doi: 10.1515/HF.2009.070
Ridley, B. L., O'Neill, M. A., and Mohnen, D. (2001). Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57, 929–967. doi: 10.1016/S0031-9422(01)00113-3
Rytioja, J., Hildén, K., Hatakka, A., and Mäkel,ä, M. R. (2014a). Transcriptional analysis of selected cellulose-acting enzymes encoding genes of the white-rot fungus Dichomitus squalens on spruce wood and microcrystalline cellulose. Fungal Genet. Biol. 72, 91–98. doi: 10.1016/j.fgb.2013.12.008
Rytioja, J., Hildén, K., Yuzon, J., Hatakka, A., De Vries, R. P., and Mäkel,ä, M. R. (2014b). Plant-polysaccharide-degrading enzymes from basidiomycetes. Microbiol. Mol. Biol. Rev. 78, 614–649. doi: 10.1128/MMBR.00035-14
Sabbadin, F., Hemsworth, G. R., Ciano, L., Henrissat, B., Dupree, P., Tryfona, T., et al. (2018). An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion. Nat. Commun. 9:756. doi: 10.1038/s41467-018-03142-x
Saloheimo, M., Nakari-Setäl,ä, T., Tenkanen, M., and Penttilä, M. (1997). cDNA Cloning of a Trichoderma reesei cellulase and demonstration of endoglucanase activity by expression in yeast. Eur. J. Biochem. 249, 584–591. doi: 10.1111/j.1432-1033.1997.00584.x
Scheller, H., and Ulvskov, P. (2010). Hemicelluloses. Ann. Rev. Plant Biol. 61, 263–289. doi: 10.1146/annurev-arplant-042809-112315
Schneider, W. D. H., Gonçalves, T. A., Uchima, C. A., Couger, M. B., Prade, R., Squina, F. M., et al. (2016). Penicillium echinulatum secretome analysis reveals the fungi potential for degradation of lignocellulosic biomass. Biotechnol. Biofuels 9:66. doi: 10.1186/s13068-016-0476-3
Schrempf, H., and Walter, S. (1995). The cellulolytic system of Streptomyces reticuli. Int. J. Biol. Macromol. 17, 353–355. doi: 10.1016/0141-8130(96)81845-9
Scott, B. R., Huang, H. Z., Frickman, J., Halvorsen, R., and Johansen, K. S. (2016). Catalase improves saccharification of lignocellulose by reducing lytic polysaccharide monooxygenase-associated enzyme inactivation. Biotechnol. Lett. 38, 425–434. doi: 10.1007/s10529-015-1989-8
Simmons, T. J., Frandsen, K. E. H., Ciano, L., Tryfona, T., Lenfant, N., Poulsen, J. C., et al. (2017). Structural and electronic determinants of lytic polysaccharide monooxygenase reactivity on polysaccharide substrates. Nat. Commun. 8:1064. doi: 10.1038/s41467-017-01247-3
Solomon, E. I., Ginsbach, J. W., Heppner, D. E., Kieber-Emmons, M. T., Kjaergaard, C. H., Smeets, P. J., et al. (2011). Copper dioxygen (bio)inorganic chemistry. Faraday Discuss. 148, 11–108. doi: 10.1039/C005500J
Song, B., Li, B., Wang, X., Shen, W., Park, S., Collings, C., et al. (2018). Real-time imaging reveals that lytic polysaccharide monooxygenase promotes cellulase activity by increasing cellulose accessibility. Biotechnol. Biofuels 11:41. doi: 10.1186/s13068-018-1023-1
Span, E. A., Suess, D. L. M., Deller, M. C., Britt, R. D., and Marletta, M. A. (2017). The role of the secondary coordination sphere in a fungal polysaccharide monooxygenase. ACS Chem. Biol. 12, 1095–1103. doi: 10.1021/acschembio.7b00016
Sygmund, C., Kracher, D., Scheiblbrandner, S., Zahma, K., Felice, A. K. G., and Harreither, W. (2012). Characterization of the two Neurospora crassa cellobiose dehydrogenases and their connection to oxidative cellulose degradation. Appl. Environ. Microbiol. 78, 6161–6171. doi: 10.1128/AEM.01503-12
Takahashi, N., and Koshijima, T. (1988). Ester linkages between lignin and glucuronoxylan in a lignin-carbohydrate complex from beech (Fagus crenata) wood. Wood Sci. Technol. 22, 231–241. doi: 10.1007/BF00386018
Tan, T.-C., Kracher, D., Gandini, R., Sygmund, C., Kittl, R., Haltrich, D., et al. (2015). Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation. Nat. Commun. 6:7542. doi: 10.1038/ncomms8542
Tanghe, M., Danneels, B., Camattari, A., Glieder, A., Vandenberghe, I., Devreese, B., et al. (2015). Recombinant expression of Trichoderma reesei Cel61A in Pichia pastoris: optimizing yield and N-terminal processing. Mol. Biotechnol. 57, 1010–1017. doi: 10.1007/s12033-015-9887-9
Taylor, C. B., Talib, M. F., McCabe, C., Bu, L., Adney, W. S., Himmel, M. E., et al. (2012). Computational investigation of glycosylation effects on a family 1 carbohydrate-binding module. J. Biol. Chem. 287, 3147–3155. doi: 10.1074/jbc.M111.270389
Temp, U., and Eggert, C. (1999). Novel interaction between laccase and cellobiose dehydrogenase during pigment synthesis in the white rot fungus Pycnoporus cinnabarinus. Appl. Environ. Microbiol. 65, 389–395.
Tian, C., Beeson, W. T., Iavarone, A. T., Sun, J., Marletta, M. A., Cate, J. H. D., et al. (2009). Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc. Natl. Acad. Sci. U.S.A. 106, 22157–22162. doi: 10.1073/pnas.0906810106
Vaaje-Kolstad, G., Forsberg, Z., Loose, J. S. M., Bissaro, B., and Eijsink, V. G. H. (2017). Structural diversity of lytic polysaccharide monooxygenases. Curr. Opin. Struct. Biol. 44, 67–76. doi: 10.1016/j.sbi.2016.12.012
Vaaje-Kolstad, G., Horn, S. J., Van Aalten, D. M., Synstad, B., and Eijsink, V. G. H. (2005a). The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J. Biol. Chem. 280, 28492–28497. doi: 10.1074/jbc.M504468200
Vaaje-Kolstad, G., Houston, D. R., Riemen, A. H., Eijsink, V. G. H., and Van Aalten, D. M. (2005b). Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J. Biol. Chem. 280, 11313–11319. doi: 10.1074/jbc.M407175200
Vaaje-Kolstad, G., Westereng, B., Horn, S. J., Liu, Z., Zhai, H., Sorlie, M., et al. (2010). An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222. doi: 10.1126/science.1192231
Vanden Wymelenberg, A., Gaskell, J., Mozuch, M., Kersten, P., Sabat, G., Martinez, D., et al. (2009). Transcriptome and secretome analyses of Phanerochaete chrysosporium reveal complex patterns of gene expression. Appl. Environ. Microbiol. 75, 4058–4068. doi: 10.1128/AEM.00314-09
Várnai, A., Costa, T. H. F., Faulds, C. B., Milagres, A. M. F., Siika-Aho, M., and Ferraz, A. (2014). Effects of enzymatic removal of plant cell wall acylation (acetylation, p-coumaroylation, and feruloylation) on accessibility of cellulose and xylan in natural (non-pretreated) sugar cane fractions. Biotechnol. Biofuels 7:153. doi: 10.1186/s13068-014-0153-3
Várnai, A., Umezawa, K., Yoshida, M., and Eijsink, V. G. H. (2018). The pyrroloquinoline-quinone dependent pyranose dehydrogenase from Coprinopsis cinerea (CcPDH) drives lytic polysaccharide monooxygenase (LPMO) action. Appl. Environ. Microbiol. doi: 10.1128/AEM.00156-18. [Epub ahead of print].
Viëtor, R. J., Newman, R. H., Ha, M.-A., Apperley, D. C., and Jarvis, M. C. (2002). Conformational features of crystal-surface cellulose from higher plants. Plant J. 30, 721–731. doi: 10.1046/j.1365-313X.2002.01327.x
Villares, A., Moreau, C., Bennati-Granier, C., Garajova, S., Foucat, L., Falourd, X., et al. (2017). Lytic polysaccharide monooxygenases disrupt the cellulose fibers structure. Sci. Rep. 7:40262. doi: 10.1038/srep40262
Vincken, J. P., De Keizer, A., Beldman, G., and Voragen, A. G. J. (1995). Fractionation of xyloglucan fragments and their interaction with cellulose. Plant Physiol. 108, 1579–1585. doi: 10.1104/pp.108.4.1579
Vu, V. V., Beeson, W. T., Phillips, C. M., Cate, J. H., and Marletta, M. A. (2014a). Determinants of regioselective hydroxylation in the fungal polysaccharide monooxygenases. J. Am. Chem. Soc. 136, 562–565. doi: 10.1021/ja409384b
Vu, V. V., Beeson, W. T., Span, E. A., Farquhar, E. R., and Marletta, M. A. (2014b). A family of starch-active polysaccharide monooxygenases. Proc. Natl. Acad. Sci. U.S.A. 111, 13822–13827. doi: 10.1073/pnas.1408090111
Vuong, T. V., Liu, B., Sandgren, M., and Master, E. R. (2017). Microplate-based detection of lytic polysaccharide monooxygenase activity by fluorescence-labeling of insoluble oxidized products. Biomacromolecules 18, 610–616. doi: 10.1021/acs.biomac.6b01790
Walton, P. H., and Davies, G. J. (2016). On the catalytic mechanisms of lytic polysaccharide monooxygenases. Curr. Opin. Chem. Biol. 31, 195–207. doi: 10.1016/j.cbpa.2016.04.001
Wang, B., Johnston, E. M., Li, P., Shaik, S., Davies, G. J., Walton, P. H. H., et al. (2018). QM/MM studies into the H2O2-dependent activity of lytic polysaccharide monooxygenases: evidence for the formation of a caged hydroxyl radical intermediate. ACS Catal. 8, 1346–1351. doi: 10.1021/acscatal.7b03888
Westereng, B., Agger, J. W., Horn, S. J., Vaaje-Kolstad, G., Aachmann, F. L., Stenstrom, Y. H., et al. (2013). Efficient separation of oxidized cello-oligosaccharides generated by cellulose degrading lytic polysaccharide monooxygenases. J. Chromatogr. A 1271, 144–152. doi: 10.1016/j.chroma.2012.11.048
Westereng, B., Arntzen, M. Ø., Aachmann, F. L., Várnai, A., Eijsink, V. G. H., and Agger, J. W. (2016). Simultaneous analysis of C1 and C4 oxidized oligosaccharides, the products of lytic polysaccharide monooxygenases acting on cellulose. J. Chromatogr. A 1445, 46–54. doi: 10.1016/j.chroma.2016.03.064
Westereng, B., Cannella, D., Wittrup Agger, J., Jørgensen, H., Larsen Andersen, M., Eijsink, V. G. H., et al. (2015). Enzymatic cellulose oxidation is linked to lignin by long-range electron transfer. Sci. Rep. 5:18561. doi: 10.1038/srep18561
Westereng, B., Ishida, T., Vaaje-Kolstad, G., Wu, M., Eijsink, V. G. H., Igarashi, K., et al. (2011). The putative endoglucanase PcGH61D from Phanerochaete chrysosporium is a metal-dependent oxidative enzyme that cleaves cellulose. PLoS ONE 6:e27807. doi: 10.1371/journal.pone.0027807
Wu, M., Beckham, G. T., Larsson, A. M., Ishida, T., Kim, S., Payne, C. M., et al. (2013). Crystal structure and computational characterization of the lytic polysaccharide monooxygenase GH61D from the Basidiomycota fungus Phanerochaete chrysosporium. J. Biol. Chem. 288, 12828–12839. doi: 10.1074/jbc.M113.459396
Xu, F., Sweeney, M. D., Quinlan, R. J., and Johansen, K. S. (2012). Compositions Comprising a Polypeptide Having Cellulolytic Enhancing Activity and an Organic Compound and Uses Thereof. U.S. Patent No WO2012021396A1. Davies, CA: Novozymes, Inc.
Yakovlev, I., Vaaje-Kolstad, G., Hietala, A. M., Stefanczyk, E., Solheim, H., and Fossdal, C. G. (2012). Substrate-specific transcription of the enigmatic GH61 family of the pathogenic white-rot fungus Heterobasidion irregulare during growth on lignocellulose. Appl. Microbiol. Biotechnol. 95, 979–990. doi: 10.1007/s00253-012-4206-x
Zakzeski, J., Bruijnincx, P. C. A., Jongerius, A. L., and Weckhuysen, B. M. (2010). The catalytic valorization of lignin for the production of renewable chemicals. Chem. Rev. 110, 3552–3599. doi: 10.1021/cr900354u
Zhang, Y.-H. P., and Lynd, L. R. (2004). Toward an aggregated understanding of enzymatic hydrolysis of cellulose: noncomplexed cellulase systems. Biotechnol. Bioeng. 88, 797–824. doi: 10.1002/bit.20282
Keywords: LPMO, lytic polysaccharide monooxygenase, substrate specificity, C1/C4-oxidation, electron donor, reducing agent, oxygen, hydrogen peroxide
Citation: Frommhagen M, Westphal AH, van Berkel WJH and Kabel MA (2018) Distinct Substrate Specificities and Electron-Donating Systems of Fungal Lytic Polysaccharide Monooxygenases. Front. Microbiol. 9:1080. doi: 10.3389/fmicb.2018.01080
Received: 31 January 2018; Accepted: 07 May 2018;
Published: 29 May 2018.
Edited by:
Selin Kara, Technische Universität Hamburg, GermanyReviewed by:
Jean-Guy Berrin, Institut National de la Recherche Agronomique (INRA), FranceAlberto A. Iglesias, National University of the Littoral, Argentina
Copyright © 2018 Frommhagen, Westphal, van Berkel and Kabel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Willem J. H. van Berkel, d2lsbGVtLnZhbmJlcmtlbEB3dXIubmw=
 Matthias Frommhagen
Matthias Frommhagen Adrie H. Westphal
Adrie H. Westphal Willem J. H. van Berkel
Willem J. H. van Berkel Mirjam A. Kabel
Mirjam A. Kabel