- 1Department of Food Science and Technology, Faculty of Veterinary, University Complutense of Madrid, Madrid, Spain
- 2Department of Animal Health, Faculty of Veterinary, University Complutense of Madrid, Madrid, Spain
In food facilities, biofilms or their debris might act as helpers for attracting free floating microorganisms. In this sense, Pseudomonas fluorescens, a dense biofilm producer frequently isolated from food contact surfaces, could be a good candidate for sheltering other microorganisms, such as Listeria monocytogenes. The main objective of this work was to evaluate the ability of L. monocytogenes to colonize pre-established Pseudomonas biofilms. For this, the movement throughout mature Pseudomonas biofilms of a green fluorescent protein (GFP) -tagged strain of L. monocytogenes was tracked for 24 h by confocal laser scanning microscopy (CLSM). Moreover, in order to check the effect of the incorporation of Listeria on the overall matrix production, attached populations of both microorganisms and total biomass (cells + matrix) of the resulting biofilms were measured over time. Planktonic cells of L. monocytogenes efficiently migrated to preformed P. fluorescens biofilms. Moreover, they moved preferentially toward the bottom layers of these structures, suggesting some kind of tropism. When preformed P. fluorescens biofilms were conditioning the surfaces, the L. monocytogenes attached population was on average, 1–2 Log higher than when this organism grew on bare coupons. Furthermore, the arrival of L. monocytogenes to the already established P. fluorescens biofilms led to a matrix over-production. Indeed, biomass values [optical density (OD595 nm)] of the resulting biofilms were double those of the ordinary L. monocytogenes–P. fluorescens mixed biofilms (1.40 vs. 0.6). The fact that L. monocytogenes cells accumulate in the bottom layers of preformed biofilms provides this microorganism an extra protection toward∖physical–chemical damages. This might partly explain why this microorganism can persist in food industry environments.
Introduction
Listeria monocytogenes presence in food processing facilities is a concerning issue for several reasons. Once established in a food processing plant, it can persist there for extended periods that could range from months to years, presumably hosted in and preserved by biofilms (Orgaz et al., 2013; Valderrama and Cutter, 2013). When transferred from contaminated surfaces to food, it might cause listeriosis, a relatively infrequent yet serious human disease, with high morbidity, hospitalization times, and mortality rates among vulnerable individuals (EFSA, 2017). Although many efforts are being done in order to control Listeria’s presence in food facilities, the truth is that in the last 5 years (from 2012 to 2016), there has been an increasing trend of confirmed listeriosis cases in the EU/EEA (EFSA, 2017).
Listeria is known to produce thin biofilms by itself, although very different structures have been described depending on the growing conditions (Rieu et al., 2008; Cherifi et al., 2017; Kocot and Olszewska, 2017). However, in the food industry, as in real environments in general, multispecies biofilms are prevalent (Elias and Banin, 2012; Burmølle et al., 2014; Jahid and Ha, 2014; Giaouris et al., 2015; Sanchez-Vizuete et al., 2015). In many studies investigating the presence of L. monocytogenes on food contact surfaces, the accompanying microbiota is often disregarded (Ortiz et al., 2010). The persistence of L. monocytogenes in food processing plants has been associated with several factors, including its ability to survive under harsh conditions (Carpentier and Cerf, 2011; Orgaz et al., 2013; Ferreira et al., 2014; Puga et al., 2016b; Rychli et al., 2016). However, it is likely that L. monocytogenes has partners or even helpers among the in-house microbiota that contribute to its persistence in certain niches. From an ecological point of view, L. monocytogenes can be considered a cheater, i.e., an organism that does not produce certain goods, but benefits from those produced by others (Cordero et al., 2012; Drescher et al., 2014). Among these goods, the extracellular matrix (ECM) is perhaps the most important. The ECM is the major component of the biofilms and is partly responsible for its resistance to different treatments (Flemming et al., 2016). In this context, the large amount of matrix produced by certain microorganisms can be regarded as a competitive advantage. That is the case when Listeria forms mixed biofilms along with species that produce copious amounts of ECM or extracellular polymeric substances (EPSs), such as Pseudomonas spp. (Huis in’t Veld, 1996; Liao, 2006). There, L. monocytogenes, a poor matrix producer, could find shelter inside the matrix produced by these organisms. Pseudomonas spp. are the most important spoilage microorganisms in many refrigerated products, in which they become the dominant species (Gram, 1993). Several species of this genus have been extensively isolated from dairy, fish, vegetable, and meat processing plants, with P. putida and P. fluorescens being the most prevalent (Chmielewski and Frank, 2003; Dogan and Boor, 2003; Caldera et al., 2016; Langsrud et al., 2016). Moreover, some studies regarding food plant-associated microbiota have frequently co-isolated Pseudomonas spp. and L. monocytogenes from the same food contact surfaces (Rodríguez-López et al., 2015; Langsrud et al., 2016).
A previous study of Puga et al. (2014) described that L. monocytogenes tends to get located in the deepest layers of the mixed biofilms when co-cultivated with P. fluorescens. These positions inside a biofilm, are more restrictive in terms of oxygen concentration and nutrients availability (Stewart and Franklin, 2008) but are tolerable for the facultative anaerobic Listeria. Besides, cells there, though constrained, are less exposed to biofilm damage (Sanchez-Vizuete et al., 2015; Flemming et al., 2016; Puga et al., 2016b).
Most of the studies on the development of multispecies biofilms rely on co-cultivation of different microorganisms in liquid media, but this situation may be not so common in real scenarios. For instance, surfaces conditioned by preformed biofilms that remain unremoved or just partially damaged after defective cleaning, might serve as anchorage points for free floating microorganisms (Castonguay et al., 2006; Klayman et al., 2009). In this case, it is unclear whether the outcome in terms of species distribution inside the biofilm would be the same as in the case of a co-cultivation. Would these new dwellers of the pre-established biofilms remain attached to the upper layers or would they penetrate into the matrix? Would the new comers have an effect, on the matrix production? Answering these questions is useful for the understanding of realistic biofilms, which need to be developed as targets for the improvement of already existing anti-biofilm strategies and for the design of new ones.
In this context, the main objective of this work was to evaluate the effect of a pre-established Pseudomonas biofilm on both the incorporation and positioning of L. monocytogenes in these structures. Confocal laser scanning microscopy (CLSM) was used to follow for 24 h the incorporation of a green fluorescent protein (GFP)-tagged strain of L. monocytogenes into the preformed, DAPI stained, thus blue P. fluorescens biofilms. Moreover, overall attached populations of L. monocytogenes and P. fluorescens and total biomass (cells + matrix) of the resulting biofilms were measured over a 96-h incubation period.
Materials and Methods
Bacterial Strains
Pseudomonas fluorescens ATCC 948TM (isolated from dairy industry waste) and reference strain L. monocytogenes Scott A (serotype 4b, lineage I) were used as biofilm forming microorganisms. They were stored at -20°C in tryptone soya broth (TSB) (Oxoid) with 15% glycerol. Pre-inoculated cultures were incubated overnight while shaking (80 rpm) at 20°C in TSB to attain mid exponential phase. Cells were then harvested by centrifugation at 4000 g for 10 min, washed twice with sterile TSB and their suspension OD600 adjusted to 0.12. The two organisms were inoculated at an initial concentration of 104 CFU mL-1, in both monospecies and dual-species cultures.
Experimental System
Biofilms were developed on commercial 22 mm × 22 mm, thin microscope borosilicate glass coverslips, as described by Orgaz et al. (2011). These coverslips provide single use, cheap, clean, and undamaged smooth surfaces, without scratches or other microtopographic irregularities. They are moderately more hydrophilic than stainless steel, but allow for more reproducible biofilms than reusable metal coupons. Sixteen coverslips held vertically by marginal insertion into the narrow radial slits of a Teflon carousel platform (6.6 cm diameter). The platform and its lid were assembled by an axial metallic rod for handling and placed into a 600-mL beaker (Figure 1). The whole system, i.e., coverslips, carousel, and the covered 600 mL beaker, were heat-sterilized as a unit before aseptically introducing 60 mL of inoculated TSB. For L. monocytogenes–P. fluorescens binary biofilms, both bacterial species were inoculated at the same time with the same initial concentration of 104 CFU mL-1. Incubation was carried out at 20°C for 96 h in a rotating shaker at 80 rpm. Under these conditions, biofilm growth covered approximately 70% of the coverslip’s surface.
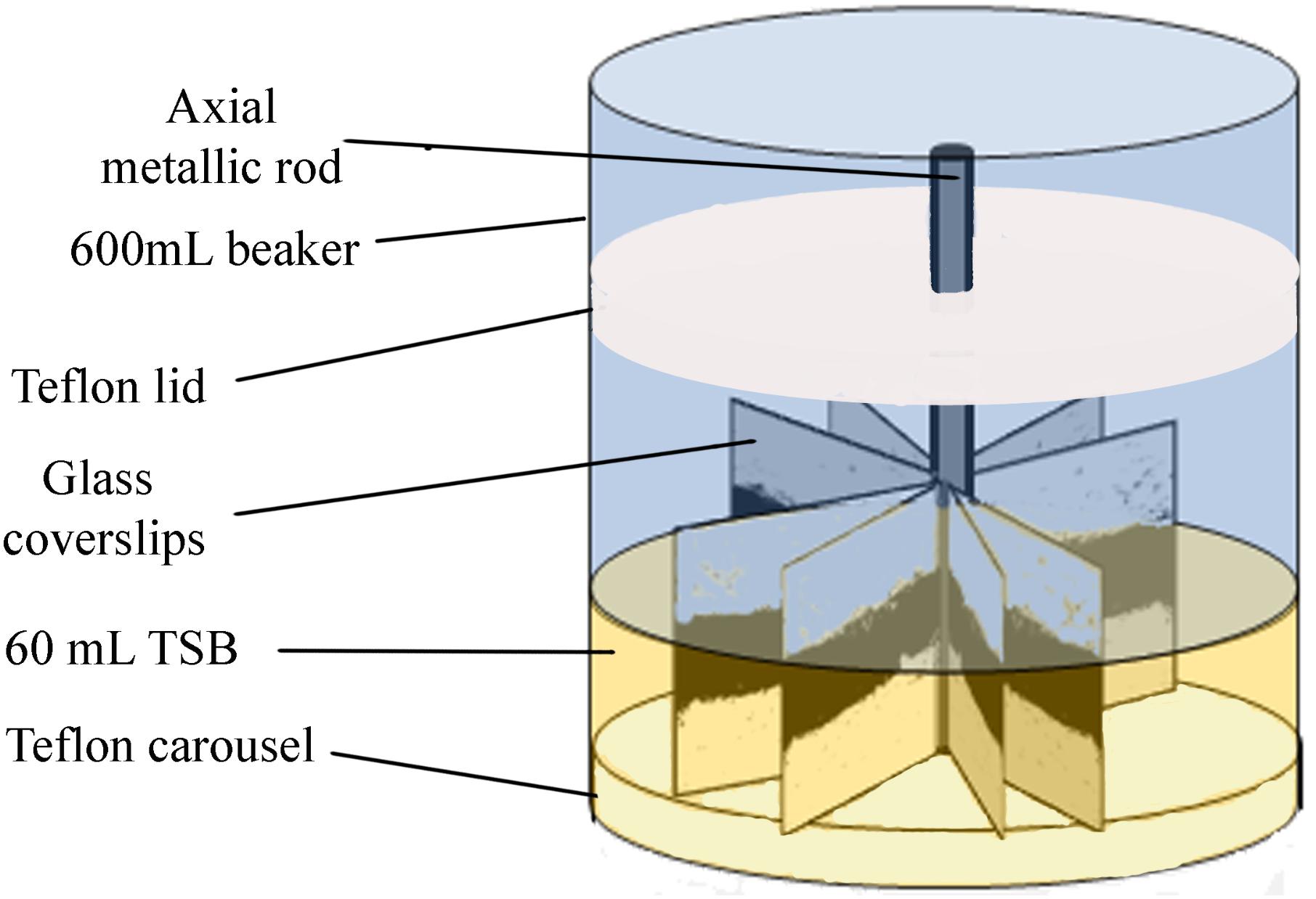
FIGURE 1. Experimental system. Carousel for biofilm development. Darker areas on the coverslips correspond to the more cell dense biofilm zones, near the air/water interface of the culture system. Reprinted from Puga et al. (2016b), with permission from Elsevier.
Surface Conditioning
In order to evaluate the effect of pre-established biofilms on L. monocytogenes incorporation, those formed by P. fluorescens were used as substratum instead of the clean, bare borosilicate glass coverslips. Thus, using the experimental system described above, warm (20°C/48 h) and cold (4°C/10 days) P. fluorescens biofilms were previously developed. The whole carousels bearing them were washed twice in NaCl (0.9%) before being placed into a new beaker containing 60 mL of a L. monocytogenes suspension in TSB (at 104 CFU mL-1). The system was incubated at 20°C for 96 h, under continuous shaking (80 rpm).
Cell Recovery and Counting
For viable cell retrieval and count, attached cells were removed from the surfaces by swabbing both sides of the coverslips. Cells transferred into test tubes with 1.5 mL of peptone water, were vigorously mixed in a vortex stirrer to break up cell aggregates, decimally diluted in peptone water, and pour-plated. P. fluorescens and L. monocytogenes counts were quantified in selective media (Pseudomonas selective agar and PALCAM, respectively, Oxoid) wherein counting was performed after 48 h incubation at 30 and 37°C, respectively. For purity control, plating on Tryptone Soya Agar (TSA, Oxoid) was used to visually spot different colonies. For each type of biofilm, three independent experiments were carried out and two coverslips were taken from each carousel. Data thus correspond to an average of six samples.
Biomass Determination
For biomass (cells plus EPS matrix) quantification, six coverslips of each type of biofilm were first dried and then stained for 2 min with a 1‰ Coomassie Blue (Brilliant Blue R, SIGMA) solution in an acetic acid/methanol/water (1:2.5:6.5) mixture. This step was repeated twice. Afterward, the stained coupons were immersed into 4 mL of the same solvent mixture and the biomass was detached with sterile cell scrapers. After full homogenization of this suspension, optical density (OD) was measured in a spectrophotometer using a wavelength of 595 nm. Bare coupons were stained and used as controls.
Expression of Green Fluorescent Protein (GFP) in Listeria monocytogenes Scott A
Electrocompetent L. monocytogenes cells were prepared as previously described by Monk et al. (2008). The pLSI ROM–GFP plasmid used in this work contains the genes for GFP and resistance to Erythromycin (Fernández de Palencia et al., 2000). Fifty microliter of electro-competent cells were mixed with 2 μL of the plasmid preparation and transferred to a 0.2-cm electroporation cuvette. For electroporation, the electroporation system (Gene Pulser, BioRad) was used with the following settings: Resistance = 400 Ω, Capacitance = 25 μF, and Voltage = 2.5 kV. The average electroporation time was 4.5 s. The electroporation product was then immediately transferred into sterile BHI broth supplemented with 0.5 M sucrose and incubated at 37°C with gentle shaking for 1 h. The suspension was then centrifuged at 8000 g for 2 min. The supernatant was discarded and the pellet was spread on BHI agar containing 5 μg/mL Erythromycin and incubated at 37°C for 48 h. Colonies that grew after 48 h were suspended in sterile BHI broth and visualized under a fluorescent microscope in order to confirm the presence of GFP.
Confocal Laser Scanning Microscopy (CLSM)
Preformed warm (20°C/48 h) and cold (4°C/10 days) P. fluorescens biofilms were used as adhesion substrates for the GFP-tagged L. monocytogenes Scott A strain. Interplay of both species was evaluated by time-series CLSM imaging using a FLUOVIEW® FV 1200 laser scanning microscope (Olympus). Preformed P. fluorescens biofilms were first developed on 27 mm glass bottom culture dishes (cellview) (NuncTM Glass Bottom Dishes, 150686, Thermo Fisher Scientific), vertically held on the carousel platforms, as explained before for coverslips. Preformed P. fluorescens biofilms were rinsed with sterile 0.9% NaCl and stained with DAPI (D9542, Life Technologies), a cell permeable fluorescent probe that binds to DNA. Ten milliliter of 109 CFU mL-1 GFP-tagged L. monocytogenes suspension were added to the glass bottom culture dishes with Pseudomonas biofilms for tracking Listeria movement. Thus, for image analysis, green corresponds to Listeria cells and blue corresponds to Pseudomonas. Most Pseudomonas spp. produce pyoverdin. This fluorescent siderophore is nevertheless produced under iron-deprived conditions (Trapet et al., 2016), which is not the case in our work, in which a rich medium was used for cultivation. Moreover, our parameters for detecting GFP fluorescence were λexcitation = 488 nm and λemission = 520 nm. Pyoverdin fluorescence spectrum shows a maximum Excitation wavelength at 405 nm and a maximum Emission wavelength at 460 nm (Martin et al., 2011). Under these conditions, no residual fluorescence was observed in P. fluorescens biofilms.
Considering zero time the moment at which Listeria suspension was added to the cellview, z-stacks of a representative 0.12 mm × 0.12 mm region of the air–liquid interphase of the biofilm (Figure 1) were acquired every 40 min, for 22 h. An oil immersion objective lens at 60× was selected for image capture. Three-dimensional projections [maximum intensity projection (MIP)] of every time point were reconstructed from z-stacks using the IMARIS® 8.1 software (Bitplane AG, Zurich, Switzerland). To calculate the parameter, here called Biovolume (μm3), the MeasurementPro module of the above mentioned software was used. Each image was segmented into two channels, green and blue, analyzed to estimate the biovolume occupied by Listeria and Pseudomonas cells, respectively. To obtain GFP-Listeria cell distribution along the z-axis of the Pseudomonas biofilm, the Vantage module of IMARIS® 8.1 was used.
Statistical Analysis
At least three independent experiments were performed and two coverslips were sampled each time (n = 6). Data were analyzed using Statgraphics Centurion software (Statistical Graphics Corporation, Rockville, MD, United States). One-way analysis of variance (ANOVA) was carried out to determine whether samples were significantly different at a 95.0% confidence level (P < 0.05).
Results
Confocal Imaging of the Course of Colonization of Pseudomonas Preformed Biofilms by Free Floating L. monocytogenes Cells
Preformed P. fluorescens biofilms developed at 20 and 4°C were used as adhesion substrates for GFP-tagged L. monocytogenes cells. Interplay of both species was monitored by time-series confocal imaging, considering time zero the moment at which Listeria’s suspension was added to the system. z-Stacks of these biofilms were captured every 40 min along a 22-h period, to be reconstructed afterward. Figures 2, 3 display CLSM images of the zenital views (Figures 2A, 3A) and snapshots from 2D cross-sections of biofilms at several time points (Figures 2B, 3B), using warm or cold Pseudomonas biofilms for surface conditioning, respectively.
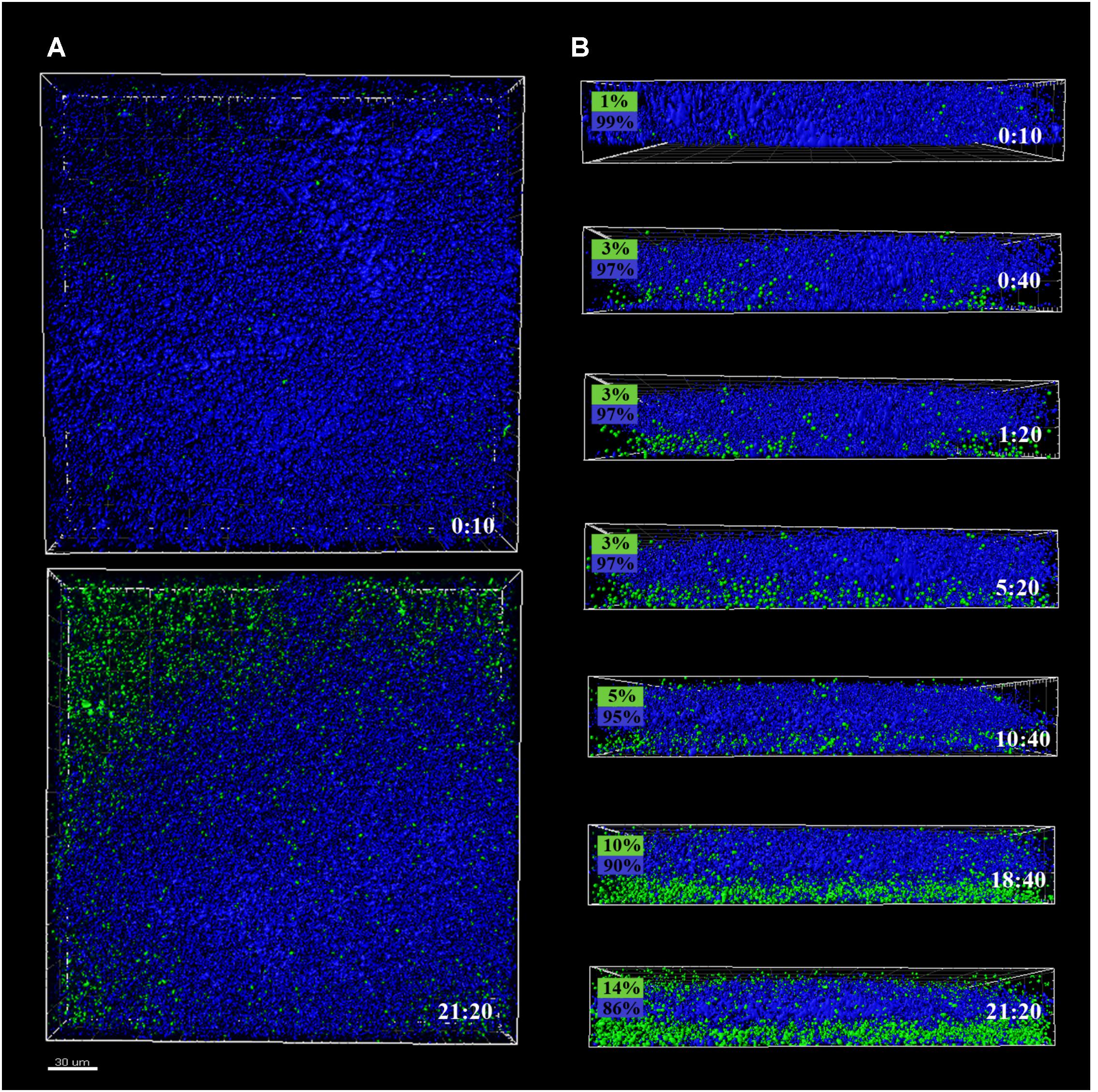
FIGURE 2. CLSM images of different sections of 48 h P. fluorescens biofilms developed at 20°C (warm) pre- and after-incubation with L. monocytogenes Scott A. Pseudomonas cells appear in blue (DAPI) and green cells correspond to the GFP-tagged L. monocytogenes Scott A. (A) Top image corresponds to the zenital 3D view of P. fluorescens biofilm (control) and bottom image corresponds to the same biofilm after 21 h incubation with Listeria. (B) Snapshots from 2D cross-sections of biofilms (35 μm wide) at several time points along the incubation. In boxes on the left is indicated the percentage of biovolume occupied by each microorganism (P. fluorescens in blue and GFP-L. monocytogenes Scott A in green). Scale bar = 20 μm.
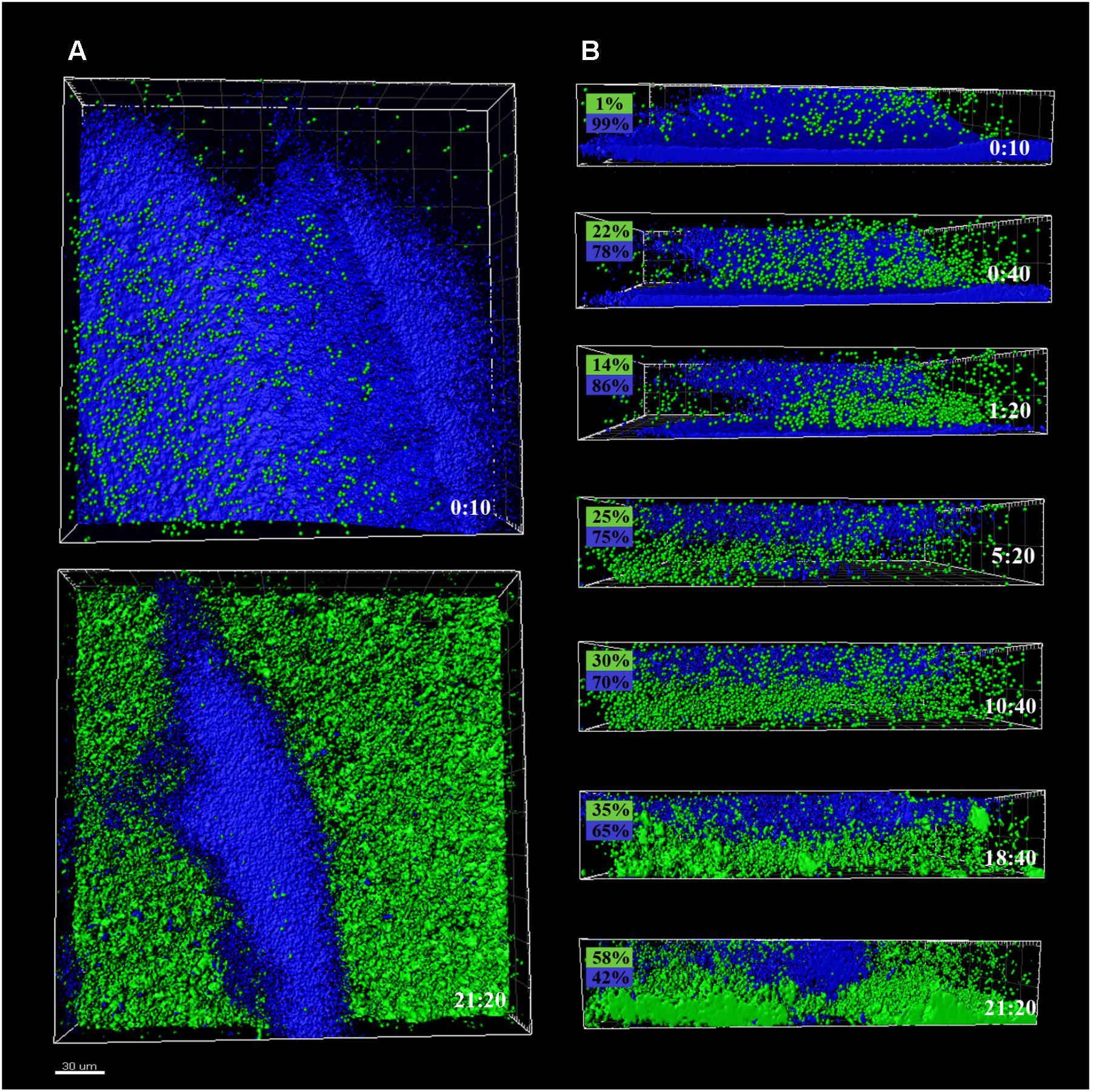
FIGURE 3. CLSM images of different sections of P. fluorescens biofilms developed at 4°C (cold) for 10 days, pre- and after-incubation with L. monocytogenes Scott A. Pseudomonas cells appear in blue (DAPI) and green cells correspond to GFP-tagged L. monocytogenes Scott A. (A) Top image corresponds to the zenital 3D view of P. fluorescens biofilm (control) and bottom image corresponds to the same biofilm after 21 h incubation with Listeria. (B) Snapshots from 2D cross-sections of biofilms (32 μm wide) at several time points along the incubation. In boxes on the left is indicated the percentage of biovolume occupied by each microorganism (P. fluorescens in blue and GFP-L. monocytogenes Scott A in green). Scale bar = 20 μm.
Pseudomonas fluorescens biofilms preformed at 20°C occupied more biovolume than those developed at 4°C (2.5 vs. 1.5 × 105 μm3, respectively) (Figures 2A, 3A). Besides, at 20°C cells had attached more uniformly around the coupon surface. Overall Pseudomonas warm biofilms seemed to be more compact than cold ones.
Colonization had different outcomes in each case. Listeria colonized more efficiently the cold biofilms; after 10 h incubation, almost half of the new structure appeared in green, not too different biovolume values being already occupied by green and blue cells (1 vs. 3.5 × 105 μm3, respectively). After 24 h, the initial Pseudomonas structure was drastically affected by Listeria’s presence. Listeria, in some way, was able to proportionally displace Pseudomonas from the biofilm. Indeed, after 21 h incubation, the biovolume occupied by green cells accounted for 3 × 105 μm3.
Listeria monocytogenes colonization of Pseudomonas warm biofilms was slower. After 10 h of incubation, the biovolume occupied by green cells was negligible compared to that occupied by blue ones (1 × 104 vs. 3 × 105 μm3). At the end of the incubation period, the Pseudomonas initial structure appeared practically unaltered by the presence of Listeria.
To assess the course of colonization, the distribution of Listeria cells along the z-axis was examined and quantified using the Vantage module of Imaris (Figure 4). The color scale bar shows particle allocation at different biofilm depths. Listeria cells were observed to progressively invade the structure previously formed by P. fluorescens, either by penetration or by basal infiltration, to eventually occupy preferentially the deepest layers of either cold or warm preformed P. fluorescens biofilms.
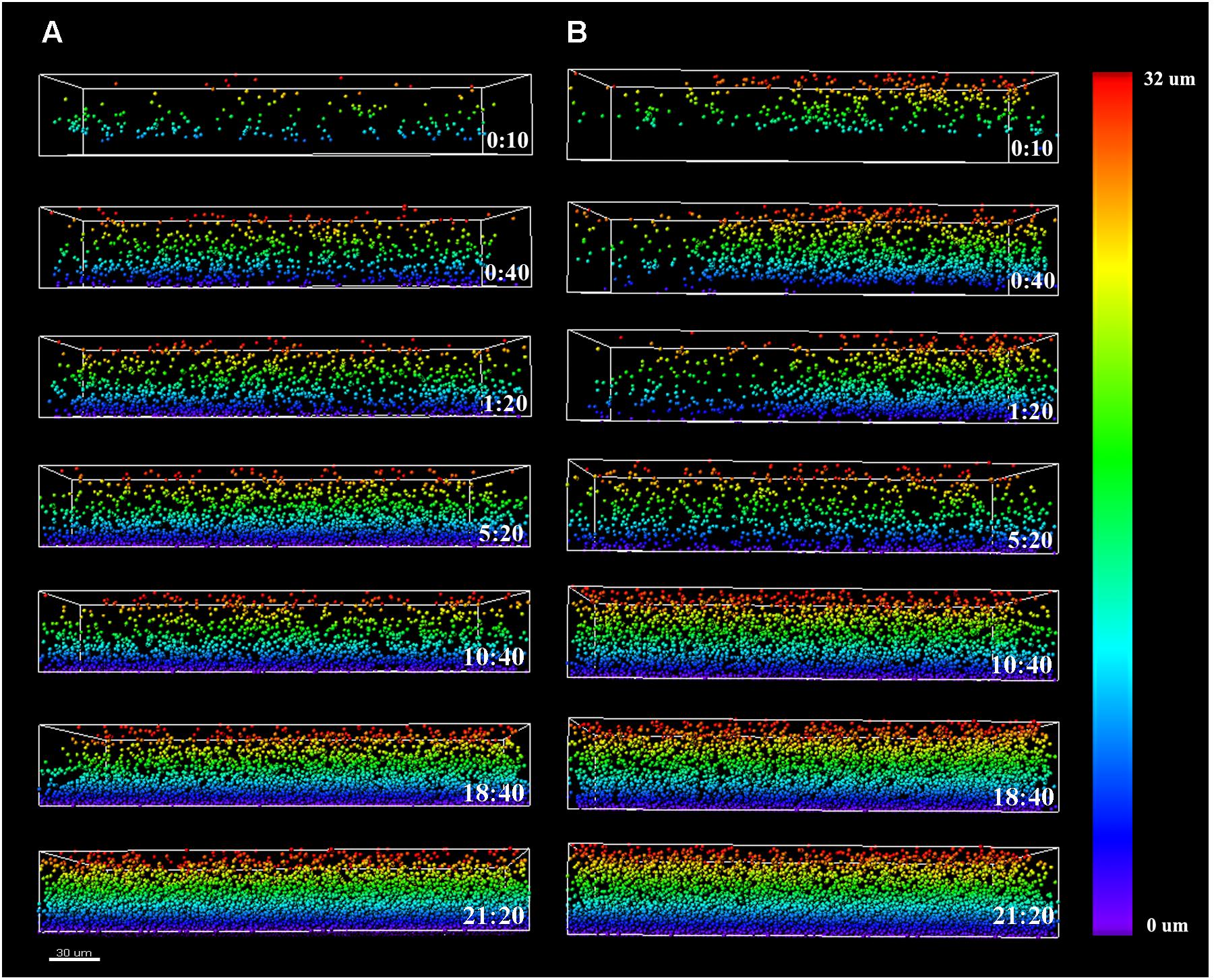
FIGURE 4. Listeria monocytogenes Scott A z-distribution throughout P. fluorescens warm (A) and cold (B) biofilm structures. Color scale bar on the right stands for z-position of Listeria cells over time. Scale bar = 30 μm.
Biofilm Population and Surface Conditioning
To further analyze the advantage effect provided by the already established Pseudomonas biofilms on L. monocytogenes attachment, the involved populations were quantified. Cold and warm Pseudomonas biofilms were first developed, and Listeria suspension was added for further incubation. In parallel, monospecies L. monocytogenes biofilms and L. monocytogenes–P. fluorescens common-start binary biofilms (1:1) were developed. Selective plate counts of each species in biofilms are shown in Figures 5, 6.
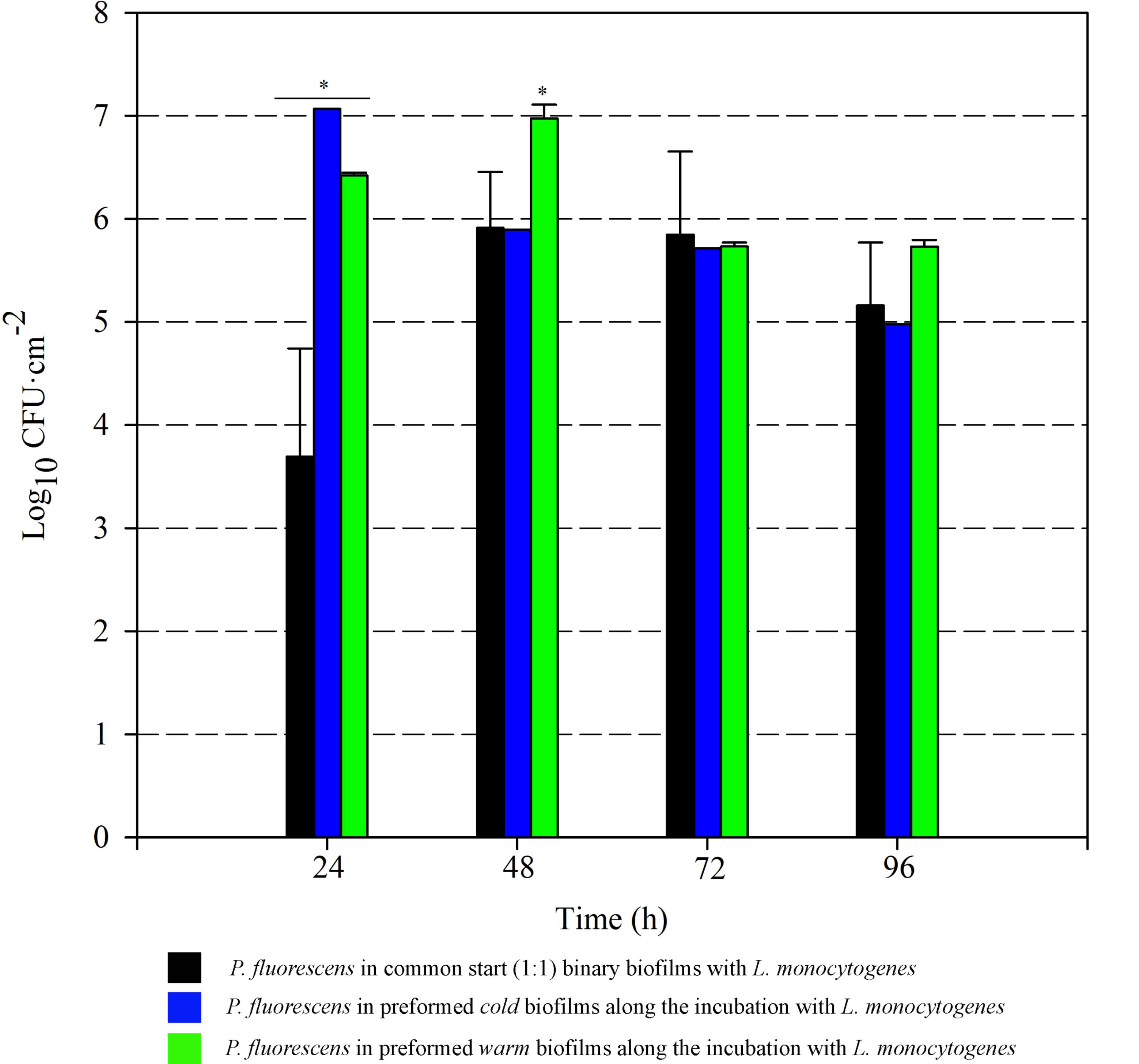
FIGURE 5. Pseudomonas fluorescens attached population over time in different biofilms with L. monocytogenes. Asterisks indicate statistically significant differences (P < 0.01).
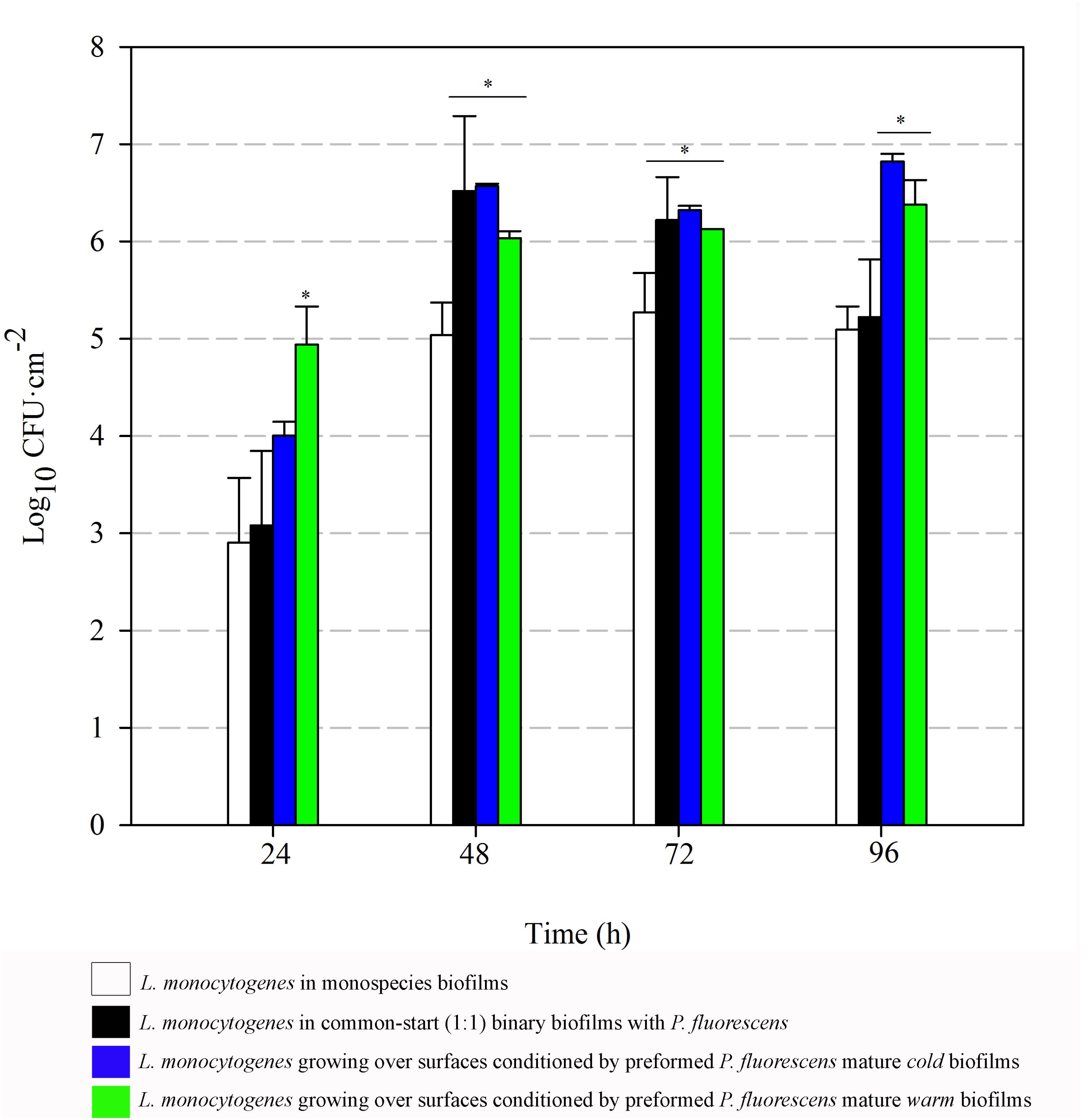
FIGURE 6. Listeria monocytogenes attached population over time in different biofilms. Asterisks indicate statistically significant differences (P < 0.01).
Listeria attached population when preformed P. fluorescens biofilms were used as a conditioned surface, was on average, 1–2 Log higher than when this organism grew on bare coupons in monoculture (Figure 6). A stimulation effect was also observed on the adhesion of Listeria when co-cultivated in a proportion 1:1 with P. fluorescens, that is, when the two species had a common-start. In that case, however, detachment of Listeria cells occurred between 72 and 96 h. When Pseudomonas biofilms were conditioning the substrate surfaces, Listeria counts were higher at those times, reaching values of more than 6 Log (Figure 6). Apparently, L. monocytogenes retention was made possible by the presence of preformed biofilms, either cold or warm.
Biofilm Biomass and Surface Conditioning
In order to complement the results on the retention of Listeria here observed, biofilm matrix volume changes derived from the interaction of the two species were also examined. Optical density values (OD595) after staining biofilms with Coomassie Blue were thus measured. These OD values integrate cellular and non-cellular components of biofilms, as Coomassie Blue binds non-specifically to protein and carbohydrate. Figure 7 shows the data corresponding to both monospecies biofilms, binary biofilms from 1:1 cultures and those of Listeria growing on preformed Pseudomonas biofilms. OD values of monospecies biofilms of L. monocytogenes were particularly low, increased rather poorly and reached a maximum value of 0.11 after 96 h. OD values of the common-start binary biofilms were practically similar to the sum of the parts, i.e., contributions of L. monocytogenes and P. fluorescens, yielding a maximum of 0.62 at 72 h. These data reveal that most of the biofilm matrix volume in this type of consortium is due to the contribution of Pseudomonas, whose production seems quantitatively unaffected by Listeria under these conditions.
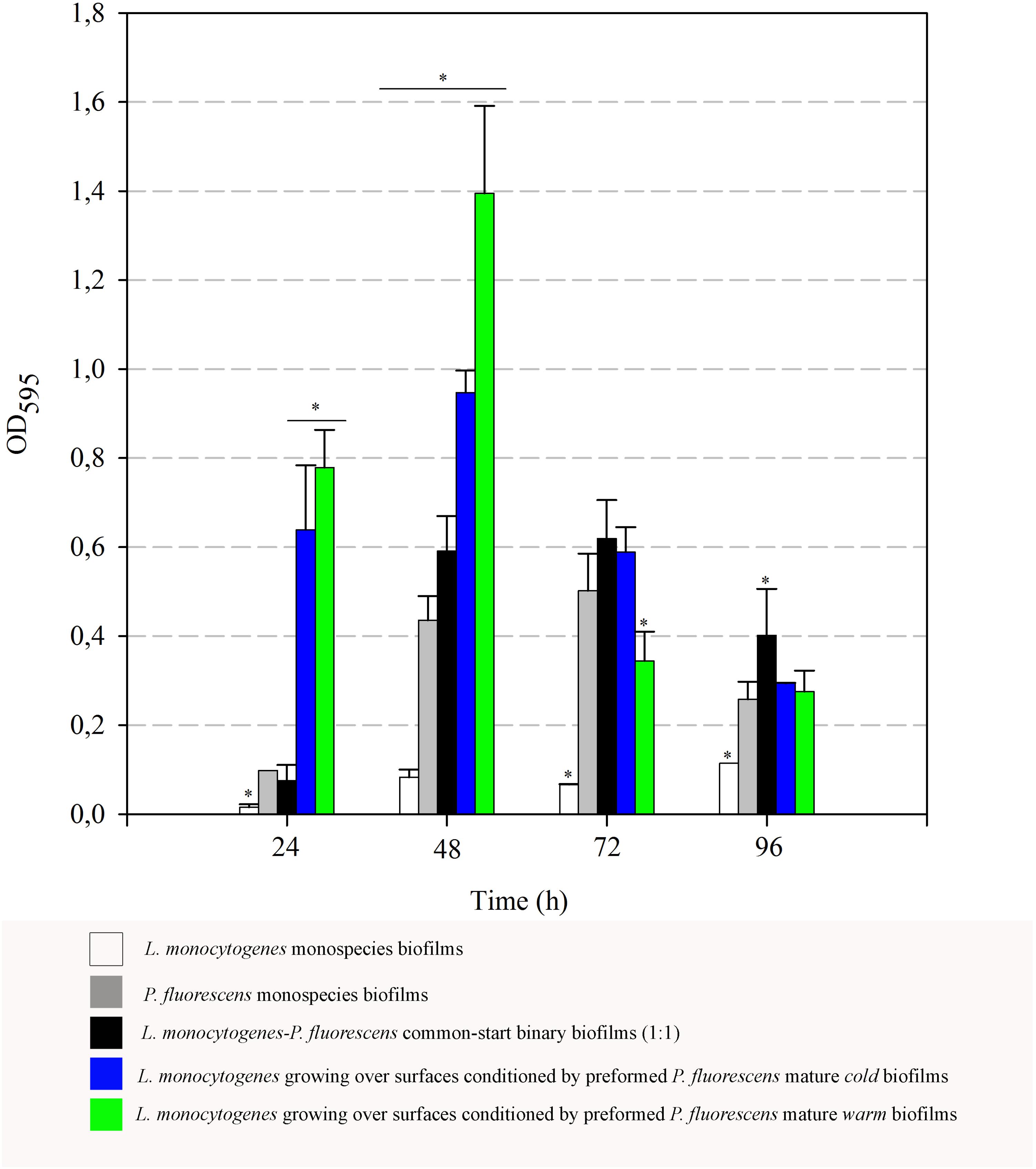
FIGURE 7. Evolution of biomass (OD595) over time. Asterisks indicate statistically significant differences (P < 0.01).
When L. monocytogenes was cultured at 20°C on surfaces conditioned by preformed mature Pseudomonas biofilms, a strong boost of biomass (cells + matrix) values was observed, particularly at the 48-h samples. At that point, maximal OD values of Listeria added to cold preformed biofilms were significantly higher than those of common-start binary biofilms (0.9 vs. 0.6). These differences were even larger when warm preformed biofilms were used as surface conditioners (1.40 vs. 0.6).
There was a general biomass downshift at late incubation stages. That was larger when preformed P. fluorescens biofilms were involved. Minor changes in viable cell count (Figures 5, 6) in these old biofilms suggest a specific decline in matrix volume.
Discussion
Pseudomonas fluorescens is frequently isolated from food processing plants and is commonly found on the same surfaces with L. monocytogenes (Rodríguez-López et al., 2015). Moreover, Langsrud et al. (2016) have recently found that Pseudomonas spp. is the dominant genus in the microbiota surviving biofilm sanitation, where the Listeria population also represented 0.1–0.01%. The ecological relationships between Pseudomonas spp. and L. monocytogenes have been studied for many years, both in planktonic environments and in biofilms. Positive, neutral, and negative interactions have been described between these two species, their outcome depending on particular strains and experimental conditions (Breen et al., 1982; Farrag and Marth, 1989; Gram, 1993; Carpentier and Chassaing, 2004; Heir et al., 2018).
The main and more recurrently proposed factor for Pseudomonas and Listeria cooperation in biofilms has been the ability of Pseudomonas to produce large amounts of EPS that would physically engulf Listeria cells, or would condition the surfaces as a “first colonizer” before Listeria arrives (Sasahara and Zottola, 1993; Hassan et al., 2004). In a previous work, we found that P. fluorescens adhered faster than L. monocytogenes, although the latter was occupying the deeper layers in mixed biofilms of these species (Puga et al., 2014). This position within the biofilm structure provides an extra protection from physical–chemical damage. Most of the studies involving mixed biofilms use similar levels of microorganisms at the beginning of the incubation, but in real environments, some of them that are free floating might found a pre-established biofilm. Under this scenario, the outcome of the resulting biofilm structure could be rather different. In this work, we wanted to know what happened when planktonic Listeria cells found mature Pseudomonas biofilms already conditioning a surface.
Listeria monocytogenes Penetration of Pseudomonas fluorescens Biofilms
Green fluorescent protein-tagged L. monocytogenes planktonic cells did penetrate both types of Pseudomonas biofilms, but this penetration was faster in cold ones, suggesting planktonic cells found less physical impediment for movement across these cold structures. Structurally, Pseudomonas warm biofilms were initially denser than cold ones (Figures 2, 3). In a previous work, we observed that in dual biofilms, biomass values changed in a temperature-depending manner, being much lower in biofilms developed at 4°C (Puga et al., 2014). As cell numbers were rather similar, this was attributed to the fact that matrix production is affected by cold stress. In this work, OD values for warm biofilms were almost double those of cold ones at 48 h (0.44 vs. 0.23) (Figure 7). As a material, a biofilm exhibits viscoelastic behavior and is considered a gel-like structure, where cells are dispersed into a heterogeneous polymeric matrix (Stewart, 2014). Generally, the gel’s viscosity increases in proportion to the matrix concentration, leading to movement constraints of cells across the biofilm matrix (Stewart, 1998). Matsui et al. (2005) found, using a very different system, that the ability of leukocytes to penetrate the mucus of the lung was restricted in patients with cystic fibrosis whose mucus viscosity is increased by Pseudomonas aeruginosa EPS. Houry et al. (2012) demonstrated that several strains of motile bacilli were able to swim across Staphylococcus aureus biofilms at a relatively quicker pace. Nevertheless, when old biofilms (72 h) were used as substratum, the swimmers’ movement across the biofilm matrix was slower. All the changes in matrix mechanical properties that lead to more viscous and copious matrices are supposed to impede, to a certain extent, the movement of planktonic microorganisms.
Independently of the state of the conditioning Pseudomonas biofilm (warm or cold), we noted that by the end of the cultivation period, Listeria cells preferentially occupied the bottom layers (Figure 4). The structural pattern in which layers of cells from one microorganism buried those of other species has been previously observed in several consortia (An et al., 2006; Almeida et al., 2011; Habimana et al., 2011; Puga et al., 2014). In co-culture biofilms, this phenomenon, known as blanketing, is generally attributed to the fast rate of growing exhibited by of one of the species in the consortium (An et al., 2006). Nevertheless, while using a pre-established Pseudomonas biofilm, this pattern was somehow unexpected, as one might think incoming Listeria cells would incorporate at the top layers. Instead, as shown by the particle distribution along the z-axis, and the sagittal profile of the CLSM images over time (Figures 2–4), Listeria cells seemed to drill through Pseudomonas biofilms in order to reach the bottom layers of the structure. This “addressed” migration could be due to signaling compounds concentrated in the Pseudomonas biofilm matrix. Charlton et al. (2000) found that the biofilm concentration of longer acyl side-chain homoserin lactones (3-oxo-C12 and 3-oxo-C14) was about 4.5 orders of magnitude higher than that measured in open systems (i.e., effluents). Horswill et al. (2007) developed a mathematical model to measure the effect of a hydrodynamic environment on the movement of signaling molecules from the biofilm to the bulk fluid. According to this model, in a closed system, signals produced by a biofilm may induce a quorum sensing response in neighboring bacteria that are not part of such biofilm, whereas in open systems, signals might be continuously washed away. Taking that into consideration, it might be hypothesized that once Listeria planktonic cells establish contact with Pseudomonas biofilms, they would be somewhat attracted by Pseudomonas signaling molecules concentrated at the bottom of the biofilm structure. Nevertheless, we cannot discard the possibility that direct cell contact is also necessary for this observation.
Flagellum-mediated motility is critical for L. monocytogenes biofilm formation (Lemon et al., 2007). Previous studies have shown that biosynthesis of flagella in this microorganism is temperature dependent, being motile at 30°C and below (O’Neil and Marquis, 2006). Therefore, flagella expression could have been increased under the experimental conditions used in this work for biofilm formation, acting as the driving force for L. monocytogenes to reach the bottom layer in the biofilm matrix. Moreover, L. monocytogenes produces certain enzymes, such as chitinases, that might have helped to reach these deep locations inside the biofilm. Chitinases are expressed during the stationary phase so that chitin could be used as a carbon source, although chitin-like structures present in other substrates may serve when the former is not present (Chaudhuri et al., 2010; Paspaliari et al., 2015). The Pseudomonas matrix is rich in acetylated polysaccharides, which somehow resemble those of chitin backbone (Kives et al., 2006), a fact that could explain the type of interaction that is taking place between these species within the biofilm.
Established Pseudomonas Biofilms Entrap Listeria Cells
Pseudomonas stimulated Listeria adhesion in both 1:1 co-cultures and cultures with previously attached Pseudomonas biofilms. One of the reasons frequently suggested to account for positive effects on the growth of L. monocytogenes is Pseudomonas’ ability to produce extracellular proteinases that could mobilize essential amino acids, particularly in rich media. Extracellular enzymes, such as proteinases, lipases, and other hydrolases, are very often produced by Pseudomonas spp. (Sorhaug and Stepaniak, 1997) and could contribute to nutrient commensalism. Nevertheless, we observed the retention of Listeria cells increased if preformed Pseudomonas biofilms were already present (Figure 6). Moreover, Listeria populations remained almost unaltered along the incubation period when coupons were conditioned (Figure 6). On the contrary, Pseudomonas dispersal was registered (between 1 and 2 Log) after having achieved a threshold cellular density (approximately 7 Log CFU cm-2) (Figure 5). As the biofilm ages, dispersal of the cells located at the top layers of the structure occurs, while those buried in the biofilm remain practically unaltered (An et al., 2006; Petrova and Sauer, 2016; Puga et al., 2016a). In our case, these top layers were mainly occupied by Pseudomonas, as shown CLSM images (Figures 2, 3), partly explaining why its cells detach first whereas those of Listeria were unaffected by biofilm aging.
Since co-cultivation of Listeria and Pseudomonas did not attain the same outcomes, it seems that, apart from Pseudomonas cells, other components present in the biofilm matrix could be playing an important role in attracting Listeria cells to these structures and trapping them more efficiently. In 1:1 co-cultures, both species have to compete at first for nutrients and space. Matrix production is obviously costly, so the process of constructing a solid matrix takes time. Indeed, in 1:1 co-cultures, the highest rate of matrix production took place between 24 and 48 h incubation (Figure 7). On the other hand, while incubating Listeria with preformed biofilms, this organism found an already stablished biofilm with high cellular density and a copious amount of matrix (P. fluorescens 48 h biofilm). This structure was quickly penetrated by Listeria as shown in Figure 4. In this scenario, the Pseudomonas matrix could have become a constraint for Listeria movement afterward, explaining why its counts did not change along the incubation period (Figure 5). Coufort et al. (2007) found that the basal layer of the biofilm was very cohesive and could resist shear stresses up to 13 Pa. Similarly, Ahimou et al. (2007) demonstrated that cohesive energy increased with biofilm depth. Accordingly, once Listeria cells reached these deep layers, most of them will probably become immobilized within the structure, as stiffness also increases with biofilm width (Safari et al., 2015).
Interestingly, the presence of Pseudomonas preformed biofilms led to a drastic increase in OD values at 48 h (Figure 7). As changes in cell population were negligible, this data suggests that matrix production is somehow over-stimulated by Listeria’s arrival to the already established biofilms. Furthermore, this stimulation leads later on to a drastic disaggregation phenomenon, suggesting once the matrix amount reaches a threshold, biofilm dispersal is rapidly induced. This most probably happens by secreting degradative enzymes that are necessary for breaking down polymeric matrices (Li et al., 2014). In our static system, there was no extra nutrient supply, so cellular dispersal phenomena could have been stimulated for this reason. Some authors have reported that, in systems where nutrients are periodically renewed, longer incubation periods are required in order to detect robust biofilms produced by L. monocytogenes (Papaioannou et al., 2018; Ripolles-Avila et al., 2018).
In summary, the fast dispersal phenomenon here observed could be somehow frequent in real settings, such as food processing plants. In these environments, biofilm fragments might persist after cleaning and disinfection procedures, and some of them could eventually be colonized by free floating microorganisms. This would in turn led to the formation of the type of biofilms we proposed in this work, in which dispersal mechanisms are really effective. Considering that dispersed cells exhibited a distinct phenotype from both planktonic and biofilm cells (Sauer et al., 2002; Li et al., 2014), the organisms disseminated from these structures would be a persistent source of contamination. Moreover, the capacity of L. monocytogenes to penetrate deeper into preformed biofilms could be an added feature to explain its persistence in food processing plants. More studies including persistent strains of L. monocytogenes will be necessary to confirm whether the pattern observed here is widespread among this species.
Author Contributions
CP and BO designed the study, performed experiments and data analysis, interpreted the analyzed results, and prepared the manuscript for submission. ED designed and prepared the GFP-tagged strain of L. monocytogenes. CS coordinated research and critically revised the data. All authors listed have made a substantial, valuable, direct, and intellectual contribution to the work, and approved it for publication.
Funding
This work was funded by the Spanish Ministry of Economy and Competition (Project No. AGL2010-22212-C02-01) and by the Santander-University Complutense of Madrid financial actions (PR26/16-10B-2).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the Cytometry and Fluorescence Microscopy Center of the University Complutense of Madrid for its skillful assistance.
References
Ahimou, F., Michael, J. S., Greg, H., and Paige, J. N. (2007). Effect of protein, polysaccharide, and oxygen concentration profiles on biofilm cohesiveness. Appl. Environ. Microbiol. 73, 2905–2910. doi: 10.1128/AEM.02420-06
Almeida, C., Azevedo, N. F., Santos, S., Keevil, C. W., and Vieira, M. J. (2011). Discriminating multi-species populations in biofilms with peptide nucleic acid fluorescence in situ hybridization (PNA FISH). PLoS One 6:e14786. doi: 10.1371/journal.pone.0014786
An, D., Danhorn, T., Fuqua, C., and Parsek, M. R. (2006). Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc. Natl. Acad. Sci. U.S.A. 103, 3828–3833. doi: 10.1073/pnas.0511323103
Breen, J., Barnes, D., and Ganguly, R. (1982). Protective effect of pseudomonas slime polysaccharide against listeria monocytogenes. Int. J. Immunopharmacol. 4:262.
Burmølle, M., Ren, D., Bjarnsholt, T., and Sørensen, S. J. (2014). Interactions in multispecies biofilms: Do they actually matter? Trends Microbiol. 22, 84–91. doi: 10.1016/j.tim.2013.12.004
Caldera, L., Franzetti, L., Van Coillie, E., De Vos, P., Stragier, P., De Block, J., et al. (2016). Identification, enzymatic spoilage characterization and proteolytic activity quantification of Pseudomonas spp. isolated from different foods. Food Microbiol. 54, 142–153. doi: 10.1016/j.fm.2015.10.004
Carpentier, B., and Cerf, O. (2011). Review — persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 145, 1–8. doi: 10.1016/j.ijfoodmicro.2011.01.005
Carpentier, B., and Chassaing, D. (2004). Interactions in biofilms between Listeria monocytogenes and resident microorganisms from food industry premises. Int. J. Food Microbiol. 97, 111–122. doi: 10.1016/j.ijfoodmicro.2004.03.031
Castonguay, M. H., Van der Schaaf, S., Koester, W., Krooneman, J., Van der Meer, W., Harmsen, H., et al. (2006). Biofilm formation by Escherichia coli is stimulated by synergistic interactions and co-adhesion mechanisms with adherence-proficient bacteria. Res. Microbiol. 157, 471–478. doi: 10.1016/j.resmic.2005.10.003
Charlton, T. S., de Nys, R., Netting, A., Kumar, N., Hentzer, M., Givskov, M., et al. (2000). A novel and sensitive method for the quantification of N-3-oxoacyl homoserine lactones using gas chromatography–mass spectrometry: application to a model bacterial biofilm. Environ. Microbiol. 2, 530–541. doi: 10.1046/j.1462-2920.2000.00136.x
Chaudhuri, S., Bruno, J. C., Alonzo, F., Xayarath, B., Cianciotto, N. P., and Freitag, N. E. (2010). Contribution of chitinases to Listeria monocytogenes pathogenesis. Appl. Environ. Microbiol. 76, 7302–7305. doi: 10.1128/AEM.01338-10
Cherifi, T., Jacques, M., Quessy, S., and Fravalo, P. (2017). Impact of nutrient restriction on the structure of Listeria monocytogenes biofilm grown in a microfluidic system. Front. Microbiol. 8:864. doi: 10.3389/fmicb.2017.00864
Chmielewski, R. A. N., and Frank, J. F. (2003). Biofilm formation and control in food processing facilities. Compr. Rev. Food Sci. Food Saf. 2, 22–32. doi: 10.1111/j.1541-4337.2003.tb00012.x
Cordero, O. X., Ventouras, L. A., DeLong, E. F., and Polz, M. F. (2012). Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc. Natl. Acad. Sci. U.S.A. 109, 20059–20064. doi: 10.1073/pnas.1213344109
Coufort, C., Derlon, N., Ochoa-Chaves, J., Liné, A., and Paul, E. (2007). Cohesion and detachment in biofilm systems for different electron acceptor and donors. Water Sci. Technol. 55, 421–428. doi: 10.2166/wst.2007.286
Dogan, B., and Boor, K. J. (2003). Genetic diversity and spoilage potentials among Pseudomonas spp. isolated from fluid milk products and dairy processing plants. Appl. Environ. Microbiol. 69, 130–138. doi: 10.1128/AEM.69.1.130-138.2003
Drescher, K., Nadell, C. D., Stone, H. A., Wingreen, N. S., and Bassler, B. L. (2014). Solutions to the public goods dilemma in bacterial biofilms. Curr. Biol. 24, 50–55. doi: 10.1016/j.cub.2013.10.030
EFSA (2017). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 15:5077.
Elias, S., and Banin, E. (2012). Multi-species biofilms: living with friendly neighbors. FEMS Microbiol. Rev. 36, 990–1004. doi: 10.1111/j.1574-6976.2012.00325.x
Farrag, S. A., and Marth, E. H. (1989). Growth of Listeria monocytogenes in the presence of Pseudomonas fluorescens at 7-degrees-c or 13-degrees-c in skim milk. J. Food Prot. 52, 852–855. doi: 10.4315/0362-028X-52.12.852
Fernández de Palencia, P., Nieto, C., Acebo, P., Espinosa, M., and López, P. (2000). Expression of green fluorescent protein in Lactococcus lactis. FEMS Microbiol. Lett. 183, 229–234. doi: 10.1111/j.1574-6968.2000.tb08963.x
Ferreira, V., Wiedmann, M., Teixeira, P., and Stasiewicz, M. J. (2014). Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J. Food Prot. 77, 150–170. doi: 10.4315/0362-028X.JFP-13-150
Flemming, H. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Giaouris, E., Heir, E., Desvaux, M., Hébraud, M., Møretrø, T., Langsrud, S., et al. (2015). Intra-and inter-species interactions within biofilms of important foodborne bacterial pathogens. Front. Microbiol. 6:841. doi: 10.3389/fmicb.2015.00841
Gram, L. (1993). Inhibitory effect against pathogenic and spoilage bacteria of Pseudomonas strains isolated from spoiled and fresh fish. Appl. Environ. Microbiol. 59, 2197–2203.
Habimana, O., Guillier, L., Kulakauskas, S., and Briandet, R. (2011). Spatial competition with Lactococcus lactis in mixed-species continuous-flow biofilms inhibits Listeria monocytogenes growth. Biofouling 27, 1065–1072. doi: 10.1080/08927014.2011.626124
Hassan, A. N., Birt, D. M., and Frank, J. F. (2004). Behavior of Listeria monocytogenes in a Pseudomonas putida biofilm on a condensate-forming surface. J. Food Prot. 67, 322–327. doi: 10.1371/journal.pone.0077276
Heir, E., Møretrø, T., Simensen, A., and Langsrud, S. (2018). Listeria monocytogenes strains show large variations in competitive growth in mixed culture biofilms and suspensions with bacteria from food processing environments. Int. J. Food Microbiol. 275, 46–55. doi: 10.1016/j.ijfoodmicro.2018.03.026
Horswill, A. R., Stoodley, P., Stewart, P. S., and Parsek, M. R. (2007). The effect of the chemical, biological, and physical environment on quorum sensing in structured microbial communities. Anal. Bioanal. Chem. 387, 371–380. doi: 10.1007/s00216-006-0720-y
Houry, A., Gohar, M., Deschamps, J., Tischenko, E., Aymerich, S., Gruss, A., et al. (2012). Bacterial swimmers that infiltrate and take over the biofilm matrix. Proc. Natl. Acad. Sci. U.S.A. 109, 13088–13093. doi: 10.1073/pnas.1200791109
Huis in’t Veld, J. H. (1996). Microbial and biochemical spoilage of foods: an overview. Int. J. Food Microbiol. 33, 1–18. doi: 10.1016/0168-1605(96)01139-7
Jahid, I. K., and Ha, S. D. (2014). The paradox of mixed-species biofilms in the context of food safety. Compr. Rev. Food Sci. Food Saf. 13, 990–1011. doi: 10.1111/1541-4337.12087
Kives, J., Orgaz, B., and SanJosé, C. (2006). Polysaccharide differences between planktonic and biofilm-associated EPS from Pseudomonas fluorescens B52. Colloids Surf. B Biointerfaces 52, 123–127. doi: 10.1016/j.colsurfb.2006.04.018
Klayman, B. J., Volden, P. A., Stewart, P. S., and Camper, A. K. (2009). Escherichia coli O157: H7 requires colonizing partner to adhere and persist in a capillary flow cell. Environ. Sci. Technol. 43, 2105–2111. doi: 10.1021/es802218q
Kocot, A. M., and Olszewska, M. A. (2017). Biofilm formation and microscopic analysis of biofilms formed by Listeria monocytogenes in a food processing context. LWT Food Sci. Technol. 84, 47–57. doi: 10.1016/j.lwt.2017.05.042
Langsrud, S., Moen, B., Møretrø, T., Løype, M., and Heir, E. (2016). Microbial dynamics in mixed culture biofilms of bacteria surviving sanitation of conveyor belts in salmon-processing plants. J. Appl. Microbiol. 120, 366–378. doi: 10.1111/jam.13013
Lemon, K. P., Higgins, D. E., and Kolter, R. (2007). Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 189, 4418–4424. doi: 10.1128/JB.01967-06
Li, Y., Petrova, O. E., Su, S., Lau, G. W., Panmanee, W., Na, R., et al. (2014). BdlA, DipA and induced dispersion contribute to acute virulence and chronic persistence of Pseudomonas aeruginosa. PLoS Pathog. 10:e1004168. doi: 10.1371/journal.ppat.1004168
Liao, C. H. (2006). “Food spoilage microorganisms,” in Pseudomonas and Related Genera, ed. C. W. Blackburn (Cambridge: Woodhead Publishing Ltd.).
Martin, L. W., Reid, D. W., Sharples, K. J., and Lamont, I. L. (2011). Pseudomonas siderophores in the sputum of patients with cystic fibrosis. Biometals 24, 1059–1067. doi: 10.1007/s10534-011-9464-z
Matsui, H., Verghese, M. W., Kesimer, M., Schwab, U. E., Randell, S. H., Sheehan, J. K., et al. (2005). Reduced three-dimensional motility in dehydrated airway mucus prevents neutrophil capture and killing bacteria on airway epithelial surfaces. J. Immunol. 175, 1090–1099. doi: 10.4049/jimmunol.175.2.1090
Monk, I., Gahan, C. G. M., and Hill, C. (2008). Tools for functional postgenomic analysis of Listeria monocytogenes. Appl. Environ. Microbiol. 74, 3921–3934. doi: 10.1128/AEM.00314-08
O’Neil, H. S., and Marquis, H. (2006). Listeria monocytogenes flagella are used for motility, not as adhesins, to increase host cell invasion. Infect. Immun. 74, 6675–6681. doi: 10.1128/IAI.00886-06
Orgaz, B., Lobete, M. M., Puga, C. H., and Sanjose, C. (2011). Effectiveness of chitosan against mature biofilms formed by food related bacteria. Int. J. Mol. Sci. 12, 817–828. doi: 10.3390/ijms12010817
Orgaz, B., Puga, C. H., Martínez-Suárez, J. V., and SanJose, C. (2013). Biofilm recovery from chitosan action: a possible clue to understand Listeria monocytogenes persistence in food plants. Food Control 32, 484–489. doi: 10.1016/j.foodcont.2013.01.024
Ortiz, S., López, V., Villatoro, D., López, P., Dávila, J. C., and Martínez-Suárez, J. V. (2010). A 3-year surveillance of the genetic diversity and persistence of Listeria monocytogenes in an Iberian pig slaughterhouse and processing plant. Foodborne Pathog. Dis. 7, 1177–1184. doi: 10.1089/fpd.2010.0535
Papaioannou, E., Giaouris, E. D., Berillis, P., and Boziaris, I. S. (2018). Dynamics of biofilm formation by Listeria monocytogenes on stainless steel under mono-species and mixed-culture simulated fish processing conditions and chemical disinfection challenges. Int. J. Food Microbiol. 267, 9–19. doi: 10.1016/j.ijfoodmicro.2017.12.020
Paspaliari, D. K., Loose, J. S. M., Larsen, M. H., and Vaaje-Kolstadet, G. (2015). Listeria monocytogenes has a functional chitinolytic system and an active lytic polysaccharide monooxygenase. FEBS J. 282, 921–936. doi: 10.1111/febs.13191
Petrova, O. E., and Sauer, K. (2016). Escaping the biofilm in more than one way: desorption, detachment or dispersion. Curr. Opin. Microb. 30, 67–78. doi: 10.1016/j.mib.2016.01.004
Puga, C. H., Orgaz, B., and SanJose, C. (2016a). Listeria monocytogenes impact on mature or old Pseudomonas fluorescens biofilms during growth at 4 and 20°C. Front. Microbiol. 7:134. doi: 10.3389/fmicb.2016.00134
Puga, C. H., Sanjose, C., and Orgaz, B. (2014). “Spatial distribution of Listeria monocytogenes and Pseudomonas fluorescens in mixed biofilms,” in Listeria monocytogenes: Food Sources, Prevalence and Management Strategies, ed. E. C. Hambrick (New York, NY: Nova Publisher), 115–131.
Puga, C. H., Sanjose, C., and Orgaz, B. (2016b). Biofilm development at low temperatures enhances Listeria monocytogenes resistance to chitosan. Food Control 65, 143–151. doi: 10.1016/j.foodcont.2016.01.012
Rieu, A., Briandet, R., Habimana, O., Garmyn, D., Guzzo, J., and Piveteau, P. (2008). Listeria monocytogenes EGD-e biofilms: no mushrooms but a network of knitted chains. Appl. Environ. Microbiol. 74, 4491–4497. doi: 10.1128/AEM.00255-08
Ripolles-Avila, C., Hascoët, A. S., Guerrero-Navarro, A. E., and Rodríguez-Jerez, J. J. (2018). Establishment of incubation conditions to optimize the in vitro formation of mature Listeria monocytogenes biofilms on food-contact surfaces. Food Control 92, 240–248. doi: 10.1016/j.foodcont.2018.04.054
Rodríguez-López, P., Saá-Ibusquiza, P., Mosquera-Fernández, M., and López-Cabo, M. (2015). Listeria monocytogenes-carrying consortia in food industry. Composition, subtyping and numerical characterisation of mono-species biofilm dynamics on stainless steel. Int. J. Food Microbiol. 206, 84–95. doi: 10.1016/j.ijfoodmicro.2015.05.003
Rychli, K., Grunert, T., Ciolacu, L., Zaiser, A., Razzazi-Fazeli, E., Schmitz-Esser, S., et al. (2016). Exoproteome analysis reveals higher abundance of proteins linked to alkaline stress in persistent Listeria monocytogenes strains. Int. J. Food Microbiol. 218, 17–26. doi: 10.1016/j.ijfoodmicro.2015.11.002
Safari, A., Tukovic, Z., Walter, M., Casey, E., and Ivankovic, A. (2015). Mechanical properties of a mature biofilm from a wastewater system: from microscale to macroscale level. Biofouling 31, 651–664. doi: 10.1080/08927014.2015.1075981
Sanchez-Vizuete, P., Orgaz, B., Aymerich, S., Le Coq, D., and Briandet, R. (2015). Pathogens protection against the action of disinfectants in multispecies biofilms. Front. Microbiol. 6:705. doi: 10.3389/fmicb.2015.00705
Sasahara, K. C., and Zottola, E. A. (1993). Biofilm formation by Listeria monocytogenes utilizes a primary colonizing microorganism in flowing systems. J. Food Prot. 56, 1022–1028. doi: 10.4315/0362-028X-56.12.1022
Sauer, K., Camper, A. K., Ehrlich, G. D., Costerton, J. W., and Davies, D. G. (2002). Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184, 1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002
Sorhaug, T., and Stepaniak, L. (1997). Psychrotrophs and their enzymes in milk and dairy products: quality aspects. Trends Food Sci. Technol. 8, 35–41. doi: 10.1017/S0022029916000728
Stewart, P. S. (1998). A review of experimental measurements of effective diffusive permeabilities and effective diffusion coefficients in biofilms. Biotechnol. Bioeng. 59, 261–272. doi: 10.1002/(SICI)1097-0290(19980805)59:3<261::AID-BIT1>3.0.CO;2-9
Stewart, P. S., and Franklin, M. J. (2008). Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6, 199–210. doi: 10.1038/nrmicro1838
Trapet, P., Avoscan, L., Klinguer, A., Pateyron, S., Citerne, S., Chervin, C., et al. (2016). Pyoverdine impact on plant growth/defense balance. Plant Physiol. Prev. 171, 675–693. doi: 10.1104/pp.15.01537
Keywords: Listeria monocytogenes, biofilms, food industry, Pseudomonas, persistence, CLSM
Citation: Puga CH, Dahdouh E, SanJose C and Orgaz B (2018) Listeria monocytogenes Colonizes Pseudomonas fluorescens Biofilms and Induces Matrix Over-Production. Front. Microbiol. 9:1706. doi: 10.3389/fmicb.2018.01706
Received: 25 April 2018; Accepted: 09 July 2018;
Published: 31 July 2018.
Edited by:
Jennifer Ronholm, McGill University, CanadaReviewed by:
Beatrix Stessl, Veterinärmedizinische Universität Wien, AustriaGonçalo Nieto Almeida, Instituto Nacional de Investigação Agrária e Veterinária, Portugal
Arun K. Bhunia, Purdue University, United States
Copyright © 2018 Puga, Dahdouh, SanJose and Orgaz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Belen Orgaz, YmVsZW5AdmV0LnVjbS5lcw==
 Carmen H. Puga
Carmen H. Puga Elias Dahdouh
Elias Dahdouh Carmen SanJose1
Carmen SanJose1 Belen Orgaz
Belen Orgaz