- 1School of Food Science and Engineering, South China University of Technology, Guangzhou, China
- 2Institute of Genomics, Huaqiao University, Xiamen, China
- 3State Key Laboratory of Food Safely Technology for Meat Products, Xiamen, China
- 4Institute of Animal Health, Guangdong Academy of Agricultural Sciences, Guangzhou, China
Livestock-associated Staphylococcus aureus colonization and/or infections exist in pigs and people in frequent contact with pigs. In this study, a total of 130 S. aureus isolates obtained from different stages of pork production were subjected to antimicrobial susceptibility, biofilm formation, as well as PCR screening to identify virulence genes, and the accessory gene regulator alleles (agr). Among all 130 S. aureus isolates, 109 (83.8%, 109/130) isolates were positive for agr. All swine farms isolates belonged to agr IV, whereas S. aureus isolated from slaughterhouse and retail indicated diverse agr types. All isolates exhibited biofilm formation ability, and raw meat isolates (belonging to agr I) exhibited a greater ability to form strong biofilms than swine farms isolates (belonging to agr IV). agr-positive isolates were associated with more virulence genes than agr-negative isolates. Most biofilm-producing isolates were positive for microbial surface component recognizing adhesive matrix molecule (MSCRAMM), capsule type and ica group genes. The results illustrate a significant association between the prevalence rate of MSCRAMM, capsule type and ica group genes among isolates producing weak, moderate and strong biofilms. The high prevalence of resistance to ciprofloxacin, gentamicin, tetracycline, clarithromycin, clindamycin, and trimethoprim-sulfamethoxazole were mainly observed in moderate and weak biofilm producers. Our findings indicate that S. aureus isolates from pork production displayed diverse molecular ecology.
Introduction
Staphylococcus aureus is an important zoonotic pathogen that is responsible for a variety of infectious diseases characterized by septicemia and sepsis (Crombe et al., 2013; Song et al., 2015). China is one of the world’s largest pork producers with more than 470 million pigs, accounting for ∼50% of the total numbers in the world (Krishnasamy et al., 2015). Consecutively, several reports suggested transmission between pigs and humans causing livestock-associated S. aureus (LA-SA) colonization in 23–45% of pig-farmers (Voss et al., 2005; Huijsdens et al., 2006; Smith et al., 2008) and 4.6% of pig-care veterinarians (Wulf et al., 2006). The “One Health” concept recognizes that human health or livestock or wildlife health are interconnected and bound to the animal-human-ecosystems in which they (co)exist. Occupational exposure to swine has been associated with increased Staphylococcus aureus carriage, and increased risk of colonization and infections of different hosts (Witte et al., 2007; Graveland et al., 2011; Price et al., 2012; Kock et al., 2013; Ge et al., 2017; Davis et al., 2018). The risk of zoonotic transmission to humans demands our deep understanding of S. aureus contamination and ecology in the swine production.
The rapid development of resistance to multiple antimicrobial agents increases the difficulty treating S. aureus infections and biofilm production facilitate this organism to survive in the presence of antibiotics (Dhanawade et al., 2010; Bhattacharya et al., 2018). Several studies have demonstrated that low doses of certain antibiotics could induce biofilm formation, indicating that biofilm regulation might be involved in the global response to external stresses, including antibiotics (Hoffman et al., 2005; Kaplan, 2011). Previous studies regarding quantitative correlation between biofilm formation and antibiotics resistance have yielded different results. For example, Neopane et al. (2018) concluded that the biofilm-positive strains have a higher tendency to exhibit multidrug resistance and methicillin resistance compared to biofilm-negative strains, while Eyoh et al. (2014) indicated that there was no significant difference in the percentage of multi-drug-resistance (MDR) among biofilm producers and non-biofilm formers for both medical and non-medical personnel.
Staphylococcus aureus produces a wide variety of protein toxins, such as exfoliative toxins, Panton-Valentine leukocidin, hemolysins, enterotoxins, and toxic shock syndrome toxin. Among the large array of S. aureus virulence factors the MSCRAMMs (microbial surface component recognizing adhesive matrix molecules) includes different adhesins, which are essential for initial stages of infection (Magro et al., 2017). MSCRAMMs, which includes fibronectin binding proteins (FnbA and FnbB), fibrinogen binding proteins (ClfA, ClfB and Efb), capsule proteins (Capsule type 5 and 8) and collagen binding proteins (Cna), can bind to a variety of mammalian extracellular proteins and abiotic surfaces (Donlan, 2002). Furthermore, the formation of a highly organized multicellular biofilm is related to the polysaccharide intercellular adhesin (PIA) production, which is controlled by the ica operon (Cramton et al., 1999). Therefore, the numbers and combinations of toxin genes may contribute to the pathogenicity of S. aureus.
While previous studies have documented the prevalence of S. aureus isolates in bovine mastitis (Fluit, 2012; Snel et al., 2015; Artursson et al., 2016; Kot et al., 2016; Magro et al., 2017), there is a lack of data regarding the prevalence and characterization of S. aureus in pork production. A thorough understanding of the correlation between the observed polymorphism in genotype and virulence, and the diversity in production practices is important for targeted mitigation. In this study, an extensive study was conducted involving systematic sampling of three commercial swine farms, a contracted slaughterhouse for the designated farms, and a retail market in Xiamen, China to profile S. aureus isolates along the production, processing and retail chain. The data enabled tracking of the spread of S. aureus from pork production and a better understanding of the evolution of S. aureus.
Materials and Methods
Bacterial Strains and Antibiotic Susceptibility
From September – December 2014, three commercial swine farms with > 5000 pigs, one large slaughterhouse and several terminal markets were selected from Xiamen City, People’s Republic of China, and 501 samples were collected from these places for S. aureus isolation. Pigs were born and raised in these three commercial swine farms with distance for more than 25 km from each other and then were sent to the slaughterhouse. These three swine farms and the slaughterhouse were vertically integrated pork processing plant, meaning pigs originated from these three swine farms contracted to sell hogs exclusively to the slaughterhouse. However, terminal samples from the markets did not totally originate from the slaughterhouse tested in the present study.
Briefly, a total of 501 non-duplicate samples were collected from the pork industry, including three commercial swine farms (sty door and soil, n = 71; nasal swabs, n = 97), one slaughterhouse (pork, n = 173), and terminal markets (pork, n = 160). Isolation and identification of S. aureus were performed according to China’s National Technical Standard GB4789.10-2010 and the special gene nuc was targeted by PCR for identifying S. aureus (Brakstad et al., 1992). Contamination with S. aureus was detected in 26.0% (130/501) of the total samples, and the prevalence of S. aureus was highest in the slaughterhouse (35.8%, 62/173) followed by the market (24.4%, 39/160) and the farm (17.3%, 29/168).
These isolates were assessed for antimicrobial susceptibility by the Kirby-Bauer disk diffusion method described by the Clinical and Laboratory Standards Institute (CLSI, 2012). The antibiotic disks used (Hangzhou Microbial Reagent Co., Ltd., Hangzhou) included ciprofloxacin (5 μg), penicillin (10 μg), gentamicin (10 μg), tetracycline (30 μg), clarithromycin (15 μg), clindamycin (2 μg), chloramphenicol (30 μg), sulfamethoxazole-trimethoprim (25 μg), nitrofurantoin (30 μg), rifampin (5 μg), cephalothin (30 μg), minocycline (30 μg), cefoxitin (30 μg) and oxacillin (1 μg).
agr Genotyping
Bacterial genomic DNA template was extracted from the isolates by a commercial DNA extraction kit (Biomed, Beijing, China). The agr types (I–IV) were determined by a multiplex PCR assay as described by Gilot et al. (2002). In brief, multiplex PCR was performed with the following primers: Pan (5′-ATG CAC ATG GTG CAC ATG C-3′), agr1 (5′-GTC ACA AGT ACT ATA AGC TGC GAT-3′), agr2 (5′-TAT TAC TAA TTG AAA AGT GGC CAT AGC-3′), agr3 (5′-GTA ATG TAA TAG CTT GTA TAA TAA TAC CCA G-3′) and agr4 (5′-CGA TAA TGC CGT AAT ACC CG-3′). These primers yield a PCR product of 441, 575, 323, or 659 bp corresponding to agr group I, II, III, and IV, respectively. Each assay contained 2 μL of prepared DNA template, 2.5 μL of 10× Easy Taq Buffer [Takara Biomedical Technology (Beijing) Co., Ltd, China], 1 μL of 10 mM deoxynucleotide triphosphate [Takara Biomedical Technology (Beijing) Co., Ltd, China], 1 μL of upstream and downstream primers (10 μM), and 0.125 μL of DNA polymerase (5 U/μL) [Takara Biomedical Technology (Beijing) Co., Ltd, China], and the final system volume was adjusted to 25 μL with sterile ultrapure water. The PCR conditions were as follows: 1 cycle at 94°C for 5 min; 26 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and finally 1 cycle at 72°C for 10 min. All PCR products were analyzed by electrophoresis on a 1.5% (w/v) agarose gel.
Identification of Virulence Determinants
The nucleotide sequences of all PCR primers used in this study and their respective amplified products and specific Tm (°C) are listed in Table 1. All the oligonucleotide primers were synthesized by Sangon Biotech (Shanghai, China). Each assay contained 1 μL of prepared DNA template, 2.5 μL of 10× Easy Taq Buffer [Takara Biomedical Technology (Beijing) Co., Ltd, China], 1 μL of 10 mM deoxynucleotide triphosphate [Takara Biomedical Technology (Beijing) Co., Ltd, China], 1 μL of upstream and downstream primers (10 μM), and 0.125 μL of DNA polymerase (5 U/μL) [Takara Biomedical Technology (Beijing) Co., Ltd, China], and the final system volume was adjusted to 25 μL with sterile ultrapure water. The PCR conditions were as follows: an initial denaturation at 95°C for 5 min; 30 cycles of 95°C for 30 s, specific Tm for 30 s, and 72°C for 40–90 s depending on the PCR product length; and a final extension at 72°C for 10 min. Sequencing of the extracted PCR product was performed by Beijing Genomics Institute (Shenzhen, China) and the data were analyzed with the GenBank database using the BLAST algorithm at the National Center for Biotechnology Information web site1.
Biofilm Formation
Quantification of biofilm formation was performed by spectrophotometry in microplates (Nest Biotechnology Co., Ltd. Wuxi, China) using crystal violet staining as previously described (Pereyra et al., 2016). Briefly, 20 μL of bacterial log phase culture was added to 200 μL of fresh 1% glucose BHI in 96-well flat-bottom microtiter plates. S. aureus ATCC25923 (biofilm-forming) and S. epidermidis ATCC12228 (not biofilm-forming) were used as positive and negative controls, respectively. BHI without bacteria served as the blank. The plates were incubated at 37°C for 24, 48, and 72 h under aerobic conditions. After each sampling time, wells were washed three times with 300 μL of sterile phosphate-buffered saline (PBS; pH 7.2) and drained by inversion. Subsequently, 200 μL of methanol was added to each well and the plates were dried for 15 min. The adherent cells were stained with 150 μL of 0.1% crystal violet solution for 15 min and then washed twice with sterile water. Bound crystal violet was dissolved by treatment with 150 μL of 95% ethanol for 10 min, and OD570 was measured for the stained bacteria and control wells. The experiment was performed in triplicate. An OD570 value of 0.3 was taken as the cutoff point to differentiate between biofilm producers and non-biofilm-producer strains [cut-off value (ODc) = average OD of negative control + 3× standard deviation (SD) of negative control] (Pereyra et al., 2016). The quantitative classification of biofilm production based on ODc and average OD values was carried out, resulting in four categories of strains: strong biofilm producers (OD > 4 × ODc), moderate biofilm producers (4 × ODc > OD > 2 × ODc), weak biofilm producers (2 × ODc > OD > ODc), and no biofilm producers (OD < ODc) (Pereyra et al., 2016).
Growth Rate Analysis
The growth of 12 strong, 12 moderate and 12 weak biofilm formers were measured according to Qi et al. (2016). Briefly, isolates were cultured in BHI agar for 18–24 h and adjusted to 0.5 McFarland units with 0.85% NaCl medium, and diluted 1: 20 in BHI medium. The cultures were incubated for 24 h at 37°C with shaking at 200 rpm and the bacterial growth was monitored by measuring the OD600 values of the culture. All experiments include three independent replicates.
Statistical Analysis
Statistical analysis was performed with SPSS v.22.0 (SPSS Inc., Chicago, IL, United States). Differences groups were compared using the chi-squared test and a p-value of <0.05 was deemed to be significant. Spearman’s rank correlation test was used for comparison of biofilm formation ability and multi-drug-resistance (MDR).
Results
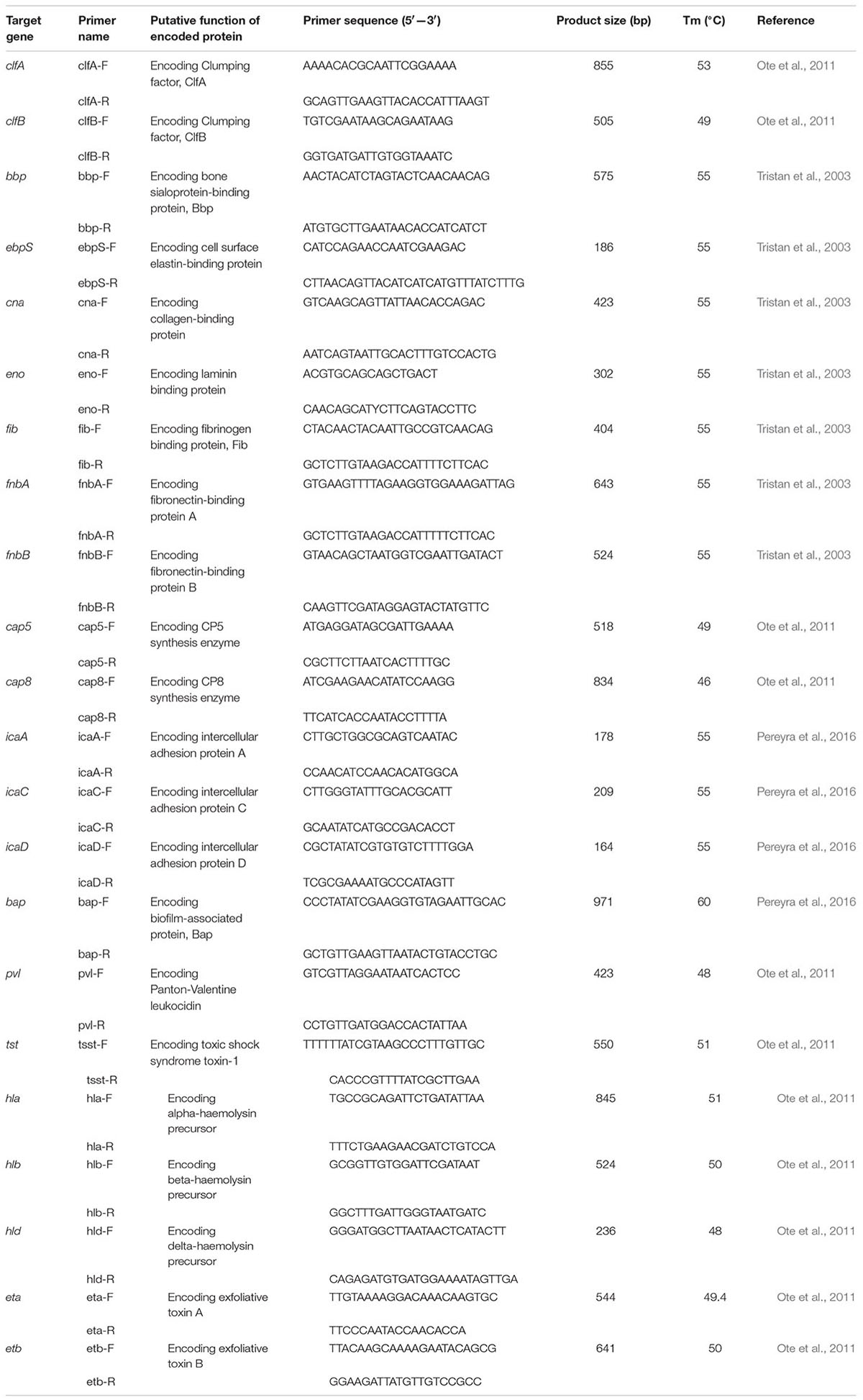
agr Genotyping
By multiplex PCR, the agr types were successfully identified in 109 isolates, and 21 isolates were non-typeable for agr locus. As shown in Table 2, the agr I was most prevalent (39.2%; 51/130), followed by agr IV (32.3%; 42/130), agr II (9.2%; 12/130) and agr III (3.1%; 4/130). All swine farms isolates belonged to agr IV, whereas S. aureus isolated from slaughterhouse and retail indicated diverse agr types.
Prevalence and Distribution of Virulence Genes
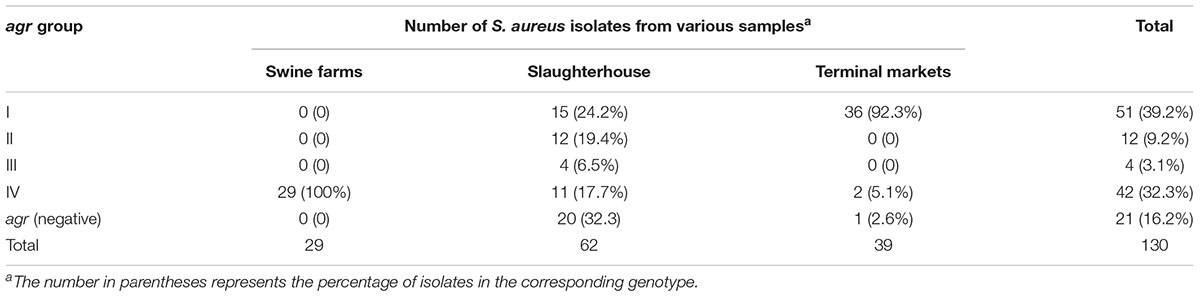
As illustrated in Figure 1, nearly all isolates harbored the hla (95.4%), hlb (100%) and hld (98.5%) genes, encoding alpha-, beta-, and delta-hemolysins respectively. No isolate harbored bap, pvl, or tsst. It was found that the bbp, cna and cap8 genes were detected only in isolates obtained from slaughterhouse and terminal markets. As shown in Table 3, the most frequent numbers of toxin genes per isolate were 11∼14 in all S. aureus isolates (Table 3). Notably, one isolate harbored 16 toxin genes and 5 isolates harbored 15 toxin genes, which were obtained from slaughterhouse (Table 3).
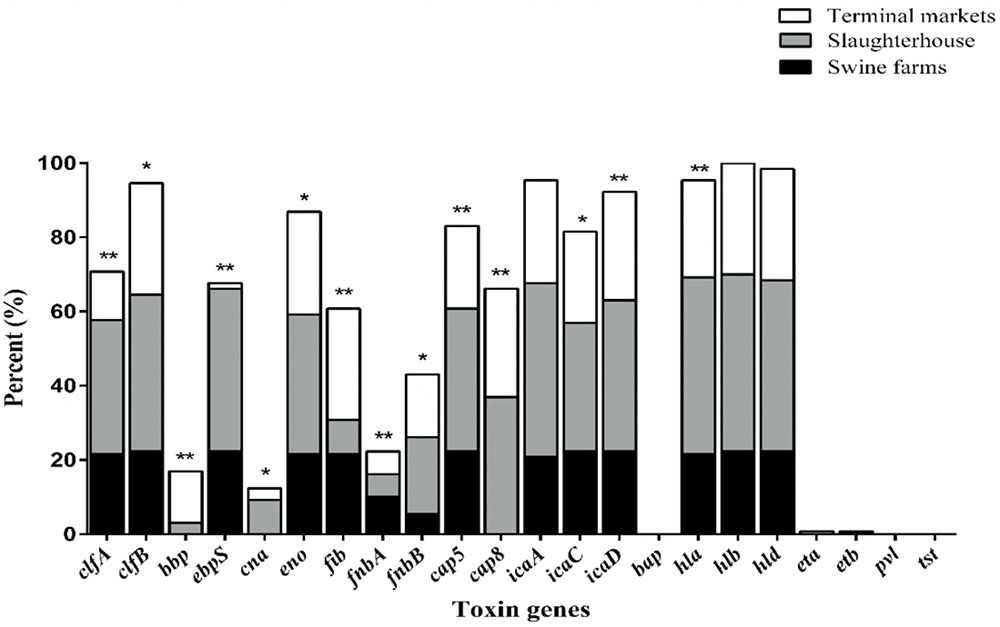
FIGURE 1. The distribution of 22 toxin genes in S. aureus isolates from different stages of pork production. The percentage of strains that were positive for each toxin gene was calculated based on the origin of each sample isolate. Pearson’s chi-square test (two-tailed) was used to test the difference in the virulence determinants distribution among isolates from different sources and run based on multiple group comparison of different sources. ∗Demonstrates that the distribution of toxin genes was statistically significantly different in isolates from different sources (p < 0.05); ∗∗demonstrates that the distribution of toxin genes was more significantly different in isolates from different food sources (p < 0.01).
The average toxin gene number was also examined based on agr genotyping, and a higher average number of toxin genes was found in the agr-positive isolates compared to agr-negative isolates. The agr-positive isolates were associated with a high average number of toxin genes (averaging 13.2 for agr II, 12.6 for agr I, 12.6 for agr IV and 12.0 for agr III), whereas the agr-negative isolates were associated with a lower average number of toxin genes (averaging 9.9) (Figure 2). The distribution of virulence genes differed among the isolates according to the agr genotyping. Among the MSCRAMMs genes, the prevalence of 3 genes was significantly different between the agr-positive and agr-negative isolates: clfA (p < 0.01), clfB (p < 0.01) and fnbA (p < 0.05). The capsule multiple type (carriage of both capsule type 5 and 8) (p < 0.01) and icaC gene (p < 0.01) were positively associated with agr-positive isolates (Figure 3).
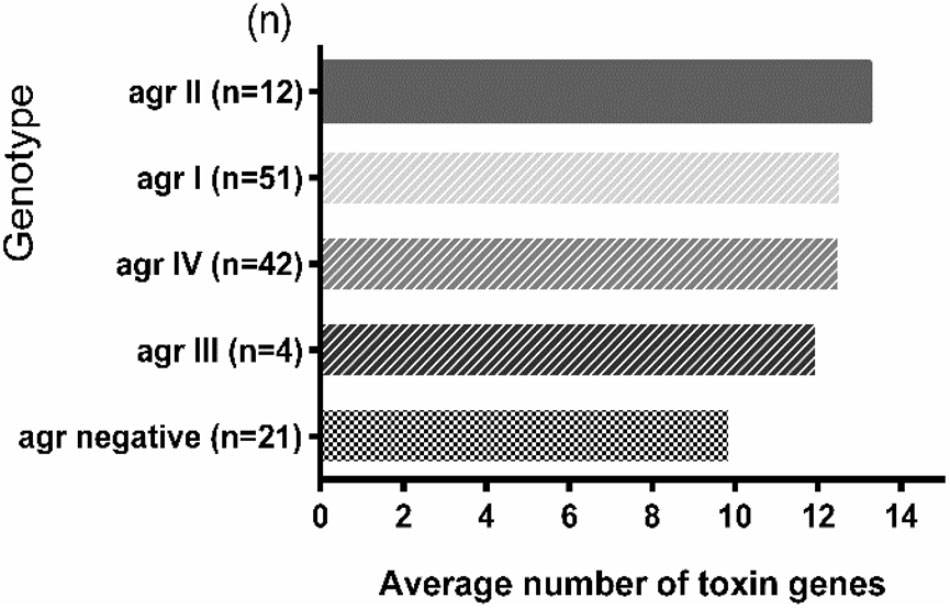
FIGURE 2. The toxin gene content of S. aureus isolates based on agr genotyping. The average number of toxin genes was calculated based on the total number of toxin genes detected in each agr group divided by the total number of isolates in each agr group. The number in parentheses on the Y-axis represents the total number of isolates in the corresponding agr group.
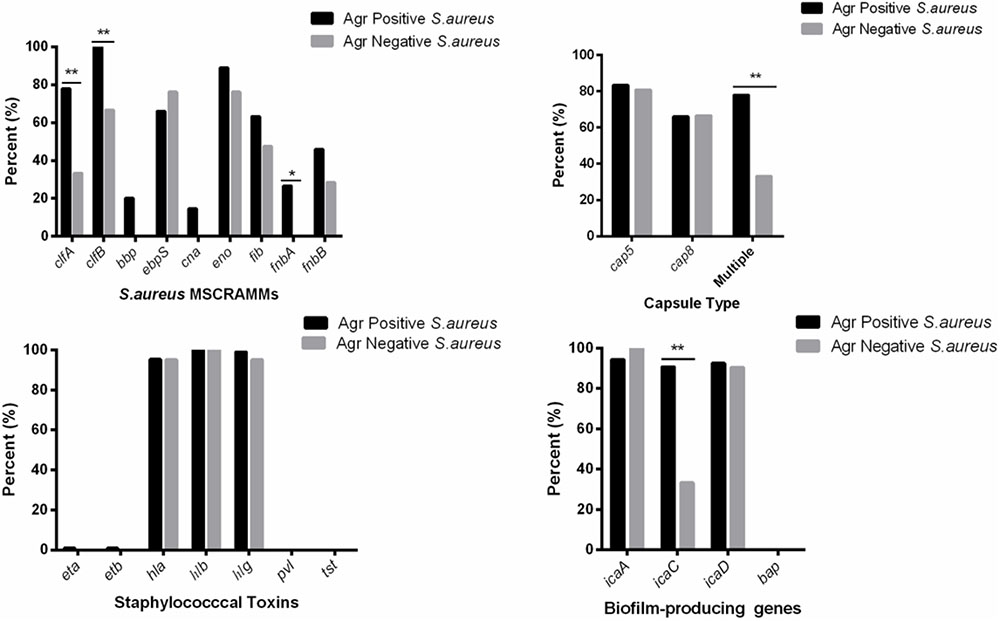
FIGURE 3. Distribution of staphylococcal virulence associated genes between agr-positive and agr-negative S. aureus. Pearson’s chi-square test (two-tailed) was used to test the difference in the virulence determinants distribution between agr-positive isolates and agr-negative isolates. ∗Demonstrates that the distribution of toxin genes was statistically significantly different in isolates from different agr types (p < 0.05); ∗∗Demonstrates that the distribution of toxin genes was more significantly different in isolates from different agr types (p < 0.01).
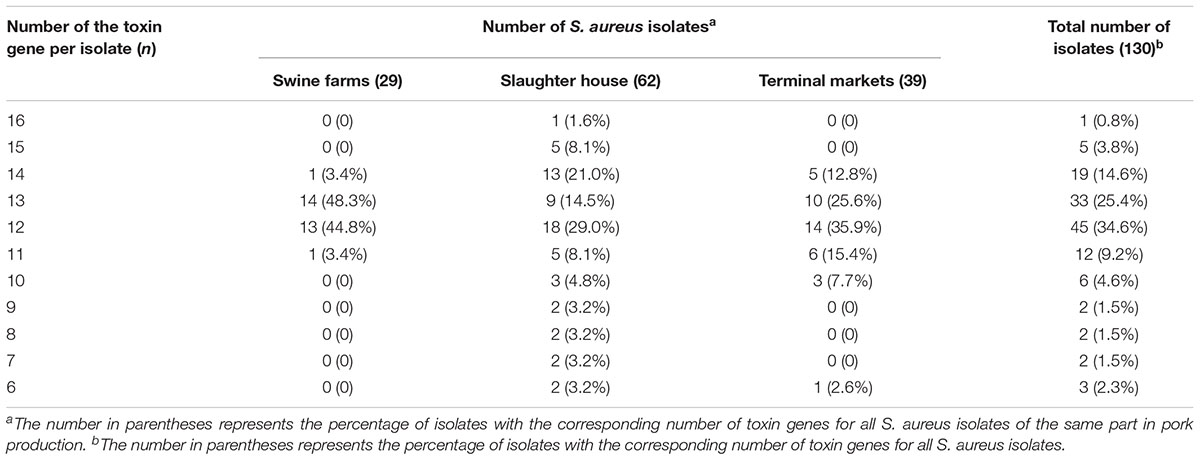
Quantification of Biofilm Biomass and Growth Rate Analysis
Biofilm formation was analyzed, and all the isolates were able to form biofilm. The biomass of biofilms formed by most isolates increased continuously during incubation for 72 h at 37°C (Table 4). Biofilm strong producers are mainly in slaughterhouse and biofilm biomass increase with time. No significant difference in the growth rates of the strong, moderate and weak biofilm formers was observed, indicating that the difference in biofilm formation was not due to the growth rate.
Correlation Between Virulence Genes and Antibiotic Resistance in Biofilm Producing S. aureus
The relationship between prevalence of biofilm-associated genes and biofilm formation ability (incubation for 24 h at 37°C) of S. aureus isolates was further analyzed (Figures 4, 5). Considering the studied gene status, 19 different gene patterns were observed (Table 5). The most prevalent gene pattern was clfA-clfB-ebpS-eno-fib-cap5-icaA-icaC-icaD which was identified in 13 (10.0%) of 130 isolates. However, there was only one strong biofilm producer, nine moderate biofilm producers and three weak biofilm producers in this genes pattern. Conversely, among the genes patterns of clfA-clfB-ebpS-eno-fib-fnbB-cap5-cap8-icaA-icaC-icaD (3.8%,5/130), clfB-eno-fib-fnbB-cap5-cap8-icaA-icaD (1.5%, 2/130), clfB-bbp-eno-fib-cap5-cap8-icaA-icaC-icaD (1.5%, 2/130), clfA-clfB-eno-fib-fnbB-cap5-cap8-icaA-icaD (1.5%, 2/130), and clfB-eno-fib-cap5-cap8-icaA-icaC-icaD (1.5%, 2/130), all isolates showed strong biofilm formation ability (Table 5). A comparison between the strong, moderate, and weak biofilm producers in the isolates showed a significant difference in the prevalence of virulence genes among these isolates.
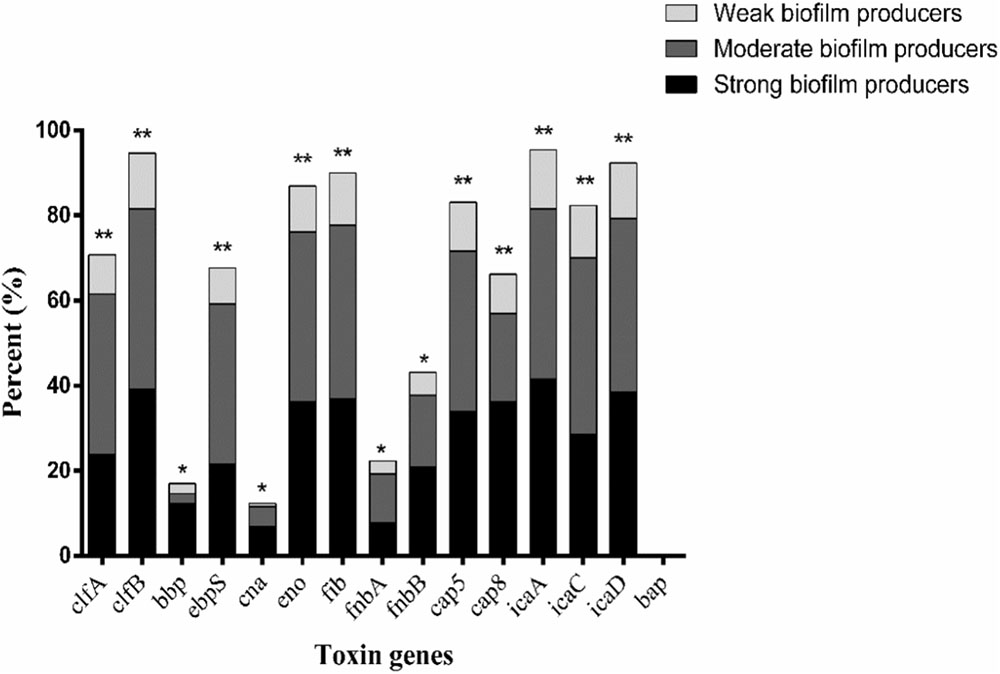
FIGURE 4. Distribution of biofilm-associated genes of S. aureus isolates based on biofilm formation ability. Pearson’s chi-square test (two-tailed) was used to test the difference in the virulence determinants distribution among different ability of biofilm ability. ∗Demonstrates that the distribution of toxin genes was statistically significantly different (p < 0.05); ∗∗demonstrates that the distribution of toxin genes was more significantly different (p < 0.01).
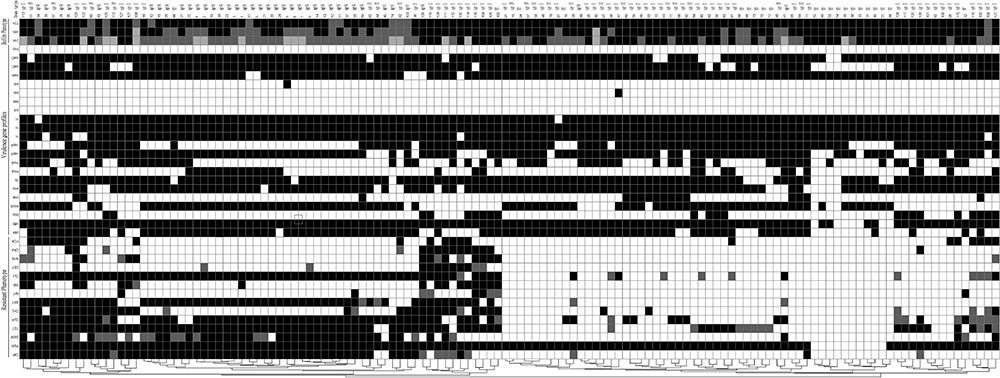
FIGURE 5. Diagram showing the antibiotic resistance pheno- and agr types, virulence genes profiles and biofilm phenotype of S. aureus isolated from different stages of pork production. The diagram was established on the basis of the presence and absence of selected determinants. For antibiotic resistance phenotype, black indicates resistance, gray indicates intermediate, and white indicates susceptible. CIP, ciprofloxacin; PEN, Penicillin; GEM, gentamicin; TET, tetracycline; CLR, clarithromycin; CHL, chloramphenicol; SXT, trimethoprim-sulfamethoxazole; NIT, nitrofurantoin; RIF, rifampicin; CLI, clindamycin; CEF, cephalothin; MIN, minocycline; OXA, oxacillin; FOX, cefoxitin; For virulence genes profiles, black indicates presence and white indicates absence. For biofilm phenotype, black indicates strong, gray indicates moderate, and white indicates weak.
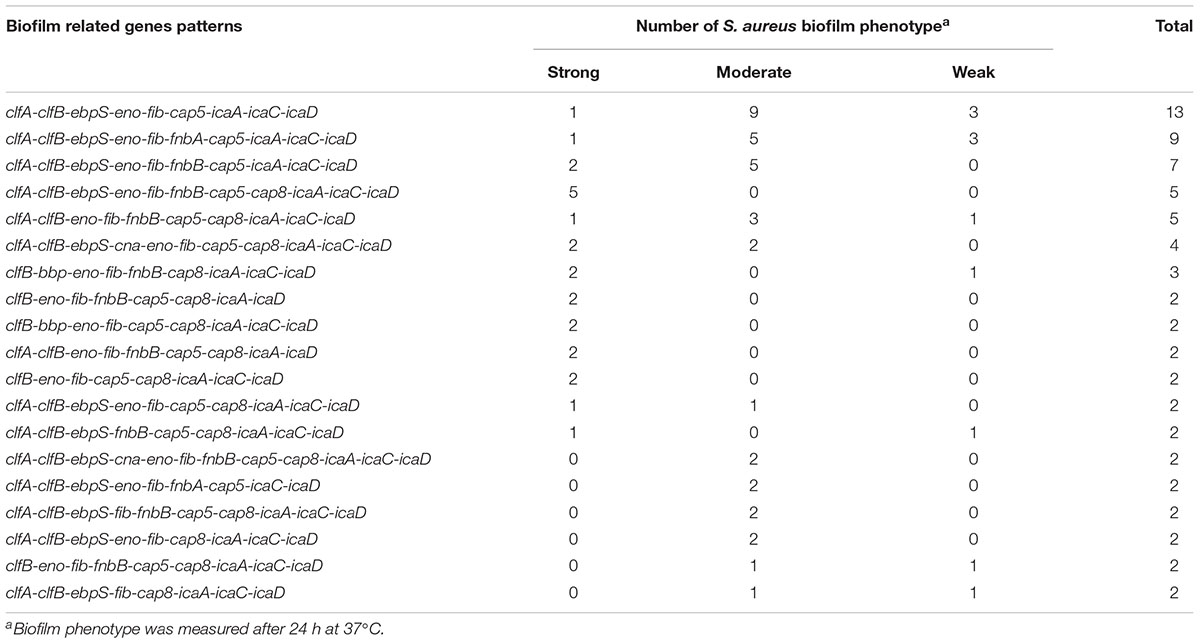
TABLE 5. The prevalence of biofilm related genes pattern and their associations with biofilm production in 130 S. aureus from different stages of pork production.
To determine whether biofilm formation was correlated with resistance to any particular antibiotic(s), we compared the biofilm forming capacities (incubation for 24 h at 37°C) among isolates with different resistance profiles for the 14 antibiotics (Table 6). Resistance to ciprofloxacin, gentamicin, tetracycline, clarithromycin, clindamycin and trimethoprim-sulfamethoxazole were significantly higher in moderate biofilm producers and weak biofilm producers than in strong biofilm producers (Table 6). Notably, resistance to nitrofurantoin was only found in strong biofilm producers (7.1%, 4/56) and moderate biofilm producers (1.8%, 1/56) (Table 6). Resistance to penicillin, cefoxitin and chloramphenicol showed no significant difference among strong biofilm producers, moderate biofilm producers and weak biofilm producers (Table 6). Regarding multidrug resistance, no significant association to strong, moderate or weak biofilm producers was observed (Table 7).
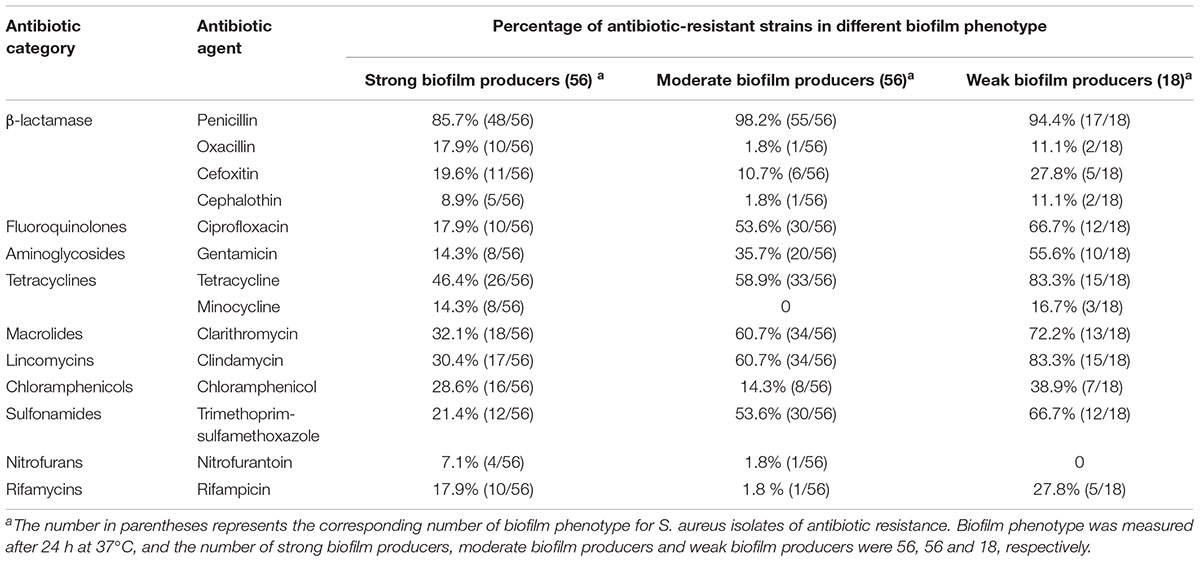
TABLE 6. Biofilm formation and antibiotic resistance pattern of 130 S. aureus isolates from different stages of pork production.
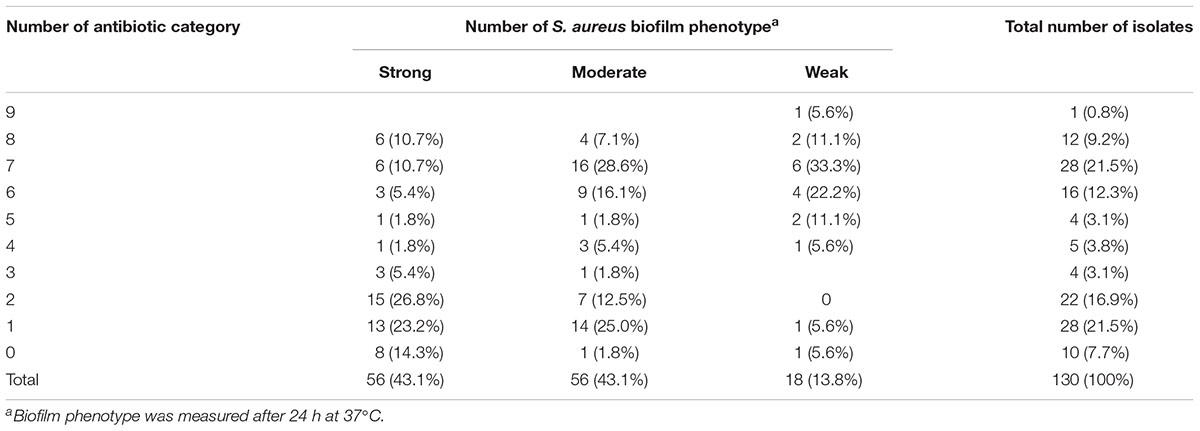
TABLE 7. Occurrence of multidrug resistant pattern and their associations with biofilm phenotype in 130 S. aureus from different stages of pork production.
Discussion
The agr (accessory gene regulator) system is a peptide quorum-sensing system present in all the Staphylococci and a dominant regulator of pathogenesis and biofilm development in S. aureus (Boles and Horswill, 2008; Paharik and Horswill, 2016). All the swine farms isolates were agr type IV, whereas the slaughterhouse and terminal markets isolates indicated diverse agr types. In addition, isolates belonging to agr-positive group had a higher number of toxin genes than those belonging to agr-negative group (p < 0.05), suggesting that agr profiles may be associated with the virulence potential of S. aureus, which is consistent with a previous finding (Cheung et al., 2011). Raw meat isolates (belonging to agr I) exhibited a great ability to form strong biofilms than swine farms isolates (belonging to agr IV). Previous studies have shown that biofilm formation in S. aureus isolated from bovine mastitis with agr I is higher than those with other agr types (Bardiau et al., 2013; Bardiau et al., 2014; Khoramrooz et al., 2016).
The prevalence of virulence genes involved in biofilm formation and staphylococcal toxin genes were investigated. Most biofilm-producing isolates were positive for MSCRAMM, capsule type and ica group genes. The data show a significant association between the prevalence rate of MSCRAMM, capsule type and ica group genes among isolates producing weak, moderate and strong biofilms. Approximately 92.3% (120/130) of all isolates harbored icaA and icaD genes simultaneously, which were similar to those from previous studies (Szweda et al., 2012; Pereyra et al., 2016). Moreover, although both pvl and tst genes were not detected in the tested isolates, hemolysins and enterotoxin-producing genes (data not shown) were found. This suggests that these isolates exhibit pathogenic potential.
In the present study, all S. aureus isolates were biofilm producers. Biofilm formation is influenced by numerous factors, such as sugar content and concentration (glucose versus lactose), proteolytic enzymes and biofilm-associated genes, etc. (Coelho et al., 2008). In this study, biofilm production was higher for raw meat isolates compared to swine farms isolates. There was a difference in the prevalence of several genes involved in adhesion and biofilm production between raw meat and swine farms isolates. However, further studies are required to quantify the expression of relevant genes. Moreover, biofilm biomass increased proportionally as biofilms aged, which is accordance with previous findings (Akinbobola et al., 2017). High variability in biofilm biomass was found among isolates throughout the time course of biofilm formation (24 – 72 h), which is in accordance with previous findings (Marino et al., 2011; Va′zquez-Sa′nchez et al., 2014). Moreover, our study demonstrated the potential association between antibiotic resistance and biofilm-forming ability of S. aureus. Apart from resistance to penicillin, the high prevalence of resistance to ciprofloxacin, gentamicin, tetracycline, clarithromycin, clindamycin and trimethoprim-sulfamethoxazole were mainly observed in moderate and weak biofilm producers. Together, Qi et al. (2016) reported that for Acinetobacter baumannii, there was a statistically negative correlation between antibiotic resistance and biofilm forming capacity, suggesting that biofilm-forming strains are less dependent on antibiotic resistance than no biofilm-forming strains for survival. Previous studies have demonstrated that biofilm resistance to antimicrobials is multifaceted, including reduced penetration of the agent into biofilms due to the presence of extracellular matrix, biofilm heterogeneity and biofilm-specific phenotypes such as expression of efflux pump and persister cells (Stewart and Costerton, 2001; Akinbobola et al., 2017). Moreover biofilm resistance is known to vary from one microorganism to another (Mah and O’Toole, 2001). Thus our further study will focus on the enhancement in resistance of our Staphylococcus aureus after biofilm formation.
In summary, our study revealed agr type diversity, virulence potential, antibiotic multiresistance and high biofilm formation ability of S. aureus isolated from pork production. All swine farms isolates belonged to agr IV, whereas S. aureus isolated from slaughterhouse and retail indicated diverse agr types. Raw meat isolates (belonging to agr I) exhibited a great ability to form strong biofilms than swine farms isolates (belonging to agr IV). Most biofilm-producing isolates were positive for MSCRAMM, capsule type and ica group genes. The results illustrate a significant association between the prevalence rate of MSCRAMM, capsule type and ica group genes among isolates producing weak, moderate and strong biofilms. Clarifying these mechanisms could provide novel insights that would prevention against S. aureus biofilm-related infections.
Author Contributions
HY and LS participated in the design of this study. RC, DX, LS, and CL provided assistance for concepts, design, literature search, data acquisition, and manuscript preparation. YZ collected important background information, carried out the study, and performed the statistical analysis. HY and YZ drafted the manuscript. HY and DX performed the manuscript review. All the authors have read and approved the content of the manuscript.
Funding
This work was supported by the National Key Basic Research Program (2016YFD0500606), the Science and Technology Planning Project of Guangdong Province, China (2014A020214001 and 2016A020219001), the Fundamental Research Funds for the Central Universities, SCUT (D2170320), and the Central Universities constructs the world first-class university (discipline) and Characteristic Development Guidance Special Fund (K5174960).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnote
References
Akinbobola, A. B., Sherry, L., Mckay, W. G., Ramage, G., and Williams, C. (2017). Tolerance of Pseudomonas aeruginosa in in-vitro biofilms to high-level peracetic acid disinfection. J. Hosp. Infect. 97, 162–168. doi: 10.1016/j.jhin.2017.06.024
Artursson, K., Söderlund, R., Liu, L., Monecke, S., and Schelin, J. (2016). Genotyping of Staphylococcus aureus in bovine mastitis and correlation to phenotypic characteristics. Vet. Microbiol. 193, 156–161. doi: 10.1016/j.vetmic.2016.08.012
Bardiau, M., Detilleux, J., Farnir, F., Mainil, J. G., and Ote, I. (2014). Associations between properties linked with persistence in a collection of Staphylococcus aureus isolates from bovine mastitis. Vet. Microbiol. 169, 74–79. doi: 10.1016/j.vetmic.2013.12.010
Bardiau, M., Yamazaki, K., Duprez, J. N., Taminiau, B., Mainil, J., and Ote, I. (2013). Genotypic and phenotypic characterization of methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk of bovine mastitis. Lett. Appl. Microbiol. 57, 181–186. doi: 10.1111/lam.12099
Bhattacharya, M., Berends, E. T. M., Chan, R., Schwab, E., Roy, S., and Sen, C. K. (2018). Staphylococcus aureus biofilms release leukocidins to elicit extracellular trap formation and evade neutrophil-mediated killing. Proc. Natl. Acad. Sci. U.S.A. 115, 7416–7421. doi: 10.1073/pnas.1721949115
Boles, B. R., and Horswill, A. R. (2008). Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. doi: 10.1371/journal.ppat.1000052
Brakstad, O. G., Aasbakk, K., and Maeland, J. A. (1992). Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 30, 1654–1660.
Cheung, G. Y., Wang, R., Khan, B. A., Sturdevant, D. E., and Otto, M. (2011). Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect. Immun. 79, 1927–1935. doi: 10.1128/IAI.00046-11
CLSI (2012). Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standards Institute.
Coelho, L. R., Souza, R. R., Ferreira, F. A., Guimaraes, M. A., Ferreira-Carvalho, B. T., and Figueiredo, A. M. (2008). Agr RNAIII divergently regulates glucose-induced biofilm formation in clinical isolates of Staphylococcus aureus. Microbiology 154, 3480–3490. doi: 10.1099/mic.0.2007/016014-0
Cramton, S. E., Gerke, C., Schnell, N. F., Nichols, W. W., and Gotz, F. (1999). The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67, 5427–5433.
Crombe, F., Argudin, M. A., Vanderhaeghen, W., Hermans, K., Haesebrouck, F., and Butaye, P. (2013). Transmission dynamics of methicillin-resistant Staphylococcus aureus in pigs. Front. Microbiol. 4:57. doi: 10.3389/fmicb.2013.00057
Davis, M. F., Pisanic, N., Rhodes, S. M., Brown, A., Keller, H., Nadimpalli, M., et al. (2018). Occurrence of Staphylococcus aureus in swine and swine workplace environments on industrial and antibiotic-free hog operations in North Carolina, USA: a one health pilot study. Environ. Res. 163, 88–96. doi: 10.1016/j.envres.2017.12.010
Dhanawade, N. B., Kalorey, D. R., Srinivasan, R., Barbuddhe, S. B., and Kurkure, N. V. (2010). Detection of intercellular adhesion genes and biofilm production in Staphylococcus aureus isolated from bovine subclinical mastitis. Vet. Res. Commun. 34, 81–89. doi: 10.1007/s11259-009-9326-0
Donlan, R. M. (2002). Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8, 881–890. doi: 10.3201/eid0809.020063
Eyoh, A. B., Toukam, M., Atashili, J., Fokunang, C., Gonsu, H., Lyonga, E. E., et al. (2014). Relationship between multiple drug resistance and biofilm formation in Staphylococcus aureus isolated from medical and non-medical personnel in Yaounde, Cameroon. Pan Afr. Med. J. 17:186. doi: 10.11604/pamj.2014.17.186.2363
Fluit, A. C. (2012). Livestock-associated Staphylococcus aureus. Clin. Microbiol. Infect. 18, 735–744. doi: 10.1111/j.1469-0691.2012.03846.x
Ge, B., Mukherjee, S., Hsu, C. H., Davis, J. A., Tran, T., Yang, Q., et al. (2017). MRSA and multidrug-resistant Staphylococcus aureus in U.S. retail meats, 2010-2011. Food Microbiol. 62, 289–297. doi: 10.1016/j.fm.2016.10.029
Gilot, P., Lina, G., Cochard, T., and Poutrel, B. (2002). Analysis of the genetic variability of genes encoding the RNA III-activating components agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J. Clin. Microbiol. 40, 4060–4067. doi: 10.1128/JCM.40.11.4060-4067.2002
Graveland, H., Duim, B., van Duijkeren, E., Heederik, D., and Wagenaar, J. A. (2011). Livestock-associated methicillin-resistant Staphylococcus aureus in animals and humans. Int. J. Med. Microbiol. 301, 630–634. doi: 10.1016/j.ijmm.2011.09.004
Hoffman, L. R., D’Argenio, D. A., MacCoss, M. J., Zhang, Z., Jones, R. A., and Miller, S. I. (2005). Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436, 1171–1175. doi: 10.1038/nature03912
Huijsdens, X. W., van Dijke, B. J., Spalburg, E., van Santen-Verheuvel, M. G., Heck, M. E., Pluister, G. N., et al. (2006). Community-acquired MRSA and pig-farming. Ann. Clin. Microbiol. Antimicrob. 5:26. doi: 10.1186/1476-0711-5-26
Kaplan, J. B. (2011). Antibiotic-induced biofilm formation. Int. J. Artif. Organs 34, 737–751. doi: 10.5301/ijao.5000027
Khoramrooz, S. S., Mansouri, F., Marashifard, M., Malek Hosseini, S. A., Akbarian Chenarestane-Olia, F., Ganavehei, B., et al. (2016). Detection of biofilm related genes, classical enterotoxin genes and agr typing among Staphylococcus aureus isolated from bovine with subclinical mastitis in southwest of Iran. Microb. Pathog. 97, 45–51. doi: 10.1016/j.micpath.2016.05.022
Kock, R., Schaumburg, F., Mellmann, A., Koksal, M., Jurke, A., Becker, K., et al. (2013). Livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) as causes of human infection and colonization in Germany. PLoS One 8:e55040. doi: 10.1371/journal.pone.0055040
Kot, B., Szweda, P., Frankowska-Maciejewska, A., Piechota, M., and Wolska, K. (2016). Virulence gene profiles in Staphylococcus aureus isolated from cows with subclinical mastitis in eastern Poland. J. Dairy Res. 83, 228–235. doi: 10.1017/S002202991600008X
Krishnasamy, V., Otte, J., and Silbergeld, E. (2015). Antimicrobial use in Chinese swine and broiler poultry production. Antimicrob. Resist. Infect. Control 4:17. doi: 10.1186/s13756-015-0050-y
Magro, G., Biffani, S., Minozzi, G., Ehricht, R., Monecke, S., Luini, M., et al. (2017). Virulence genes of S. aureus from dairy cow mastitis and contagiousness risk. Toxins 9:E195. doi: 10.3390/toxins9060195
Mah, T. F., and O’Toole, G. A. (2001). Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9, 34–39. doi: 10.1016/S0966-842X(00)01913-2
Marino, M., Frigo, F., Bartolomeoli, I., and Maifreni, M. (2011). Safety-related properties of staphylococci isolated from food and food environments. J. Appl. Microbiol. 110, 550–561. doi: 10.1111/j.1365-2672.2010.04909.x
Neopane, P., Nepal, H. P., Shrestha, R., Uehara, O., and Abiko, Y. (2018). In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int. J. Gen. Med. 11, 25–32. doi: 10.2147/IJGM.S153268
Ote, I., Taminiau, B., Duprez, J. N., Dizier, I., and Mainil, J. G. (2011). Genotypic characterization by polymerase chain reaction of Staphylococcus aureus isolates associated with bovine mastitis. Vet. Microbiol. 153, 285–292. doi: 10.1016/j.vetmic.2011.05.042
Paharik, A. E., and Horswill, A. R. (2016). The staphylococcal biofilm: adhesins, regulation, and host response. Microbiol. Spectr. 4. doi: 10.1128/microbiolspec.VMBF-0022-2015
Pereyra, E. A., Picech, F., Renna, M. S., Baravalle, C., Andreotti, C. S., Russi, R., et al. (2016). Detection of Staphylococcus aureus adhesion and biofilm-producing genes and their expression during internalization in bovine mammary epithelial cells. Vet. Microbiol. 183, 69–77. doi: 10.1016/j.vetmic.2015.12.002
Price, L. B., Stegger, M., Hasman, H., Aziz, M., Larsen, J., Andersen, P. S., et al. (2012). Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3, e305–e311. doi: 10.1128/mBio.00305-11
Qi, L., Li, H., Zhang, C., Liang, B., Li, J., Wang, L., et al. (2016). Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front. Microbiol. 7:483. doi: 10.3389/fmicb.2016.00483
Smith, T. C., Male, M. J., Harper, A. L., Kroeger, J. S., Tinkler, G. P., Moritz, E. D., et al. (2008). Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS One 4:e4258. doi: 10.1371/journal.pone.0004258
Snel, G. G., Monecke, S., Ehricht, R., and Piccinini, R. (2015). Molecular characteristics of bap-positive Staphylococcus aureus strains from dairy cowmastitis. J. Dairy Res. 82, 312–316. doi: 10.1017/S0022029915000199
Song, M., Bai, Y., Xu, J., Carter, M. Q., Shi, C., and Shi, X. (2015). Genetic diversity and virulence potential of Staphylococcus aureus isolates from raw and processed food commodities in Shanghai. Int. J. Food Microbiol. 195, 1–8. doi: 10.1016/j.ijfoodmicro.2014.11.020
Stewart, P. S., and Costerton, J. W. (2001). Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138. doi: 10.1016/S0140-6736(01)05321-1
Szweda, P., Schielmann, M., Milewski, S., Frankowska, A., and Jakubczak, A. (2012). Biofilm production and presence of ica and bap genes in Staphylococcus aureus strains isolated from cows with mastitis in the eastern Poland. Pol. J. Microbiol. 61, 65–69.
Tristan, A., Ying, L., Bes, M., Etienne, J., Vandenesch, F., and Lina, G. (2003). Use of multiplex PCR to identify Staphylococcus aureus adhesins involved in human hematogenous infections. J. Clin. Microbiol. 41, 4465–4467. doi: 10.1128/JCM.41.9.4465-4467.2003
Va′zquez-Sa′nchez, D., Cabo, M. L., Ibusquiza, P. S., and Rodri′guez-Herrera, J. J. (2014). Biofilm-forming ability and resistance to industrial disinfectants of Staphylococcus aureus isolated from fishery products. Food Control 39, 8–16. doi: 10.1016/j.foodcont.2013.09.029
Voss, A., Loeffen, F., Bakker, J., Klaassen, C., and Wulf, M. (2005). Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11, 1965–1966. doi: 10.3201/eid1112.050428
Witte, W., Strommenger, B., Stanek, C., and Cuny, C. (2007). Methicillin-resistant Staphylococcus aureus ST398 in humans and animals. Central Europe. Emerg. Infect. Dis. 13, 255–258. doi: 10.3201/eid1302.060924
Keywords: Staphylococcus aureus, agr typing, biofilm formation, virulence gene, antibiotic resistance, pork production
Citation: Zhang Y, Xu D, Shi L, Cai R, Li C and Yan H (2018) Association Between agr Type, Virulence Factors, Biofilm Formation and Antibiotic Resistance of Staphylococcus aureus Isolates From Pork Production. Front. Microbiol. 9:1876. doi: 10.3389/fmicb.2018.01876
Received: 08 March 2018; Accepted: 25 July 2018;
Published: 20 August 2018.
Edited by:
Learn-Han Lee, Monash University Malaysia, MalaysiaReviewed by:
Maria José Saavedra, Universidade de Trás-os-Montes e Alto Douro, PortugalFlorence Dubois-Brissonnet, AgroParisTech-Institut des Sciences et Industries du Vivant et de l’Environnement, France
Copyright © 2018 Zhang, Xu, Shi, Cai, Li and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunling Li, bGNsY2xhcmVAMTYzLmNvbQ== He Yan, eWFuaGVAc2N1dC5lZHUuY24=
†These authors have contributed equally to this work
 Yang Zhang
Yang Zhang Dongyang Xu
Dongyang Xu Lei Shi3
Lei Shi3 Rujian Cai
Rujian Cai Chunling Li
Chunling Li He Yan
He Yan