- 1Genomic Sciences Graduate Program, North Carolina State University, Raleigh, NC, United States
- 2Department of Food, Bioprocessing and Nutrition Sciences, North Carolina State University, Raleigh, NC, United States
Lactic acid bacteria (LAB) are frequently harnessed for the delivery of biomolecules to mucosal tissues. Several species of Lactobacillus are commonly employed for this task, of which a subset are known to possess surface-layers (S-layers). S-layers are two-dimensional crystalline arrays of repeating proteinaceous subunits that form the outermost coating of many prokaryotic cell envelopes. Their periodicity and abundance have made them a target for numerous biotechnological applications. In the following review, we examine the multi-faceted S-layer protein (Slp), and its use in both heterologous protein expression systems and mucosal vaccine delivery frameworks, through its diverse genetic components: the strong native promoter, capable of synthesizing as many as 500 Slp subunits per second; the signal peptide that stimulates robust secretion of recombinant proteins; and the structural domains, which can be harnessed for both cell surface display of foreign peptides or adhesion enhancement of a host bacterium. Although numerous studies have established vaccine platforms based on one or more components of the Lactobacillus S-layer, this area of research still remains largely in its infancy, thus this review is meant to not only highlight past works, but also advocate for the future usage of Slps in biotherapeutic research.
Introduction
Lactic acid bacteria (LAB) are Gram-positive, anaerobic or microaerophilic, non-sporulating microorganisms that inhabit diverse environments including milk and plant surfaces, as well as the mouth, gastrointestinal tract, and vaginal tract of humans and animals (Liu et al., 2014). Traditionally, they have been employed for food and dairy fermentations, but have more recently garnered attention for their health-promoting properties with many species used widely as probiotics (Klein et al., 1998; Klaenhammer et al., 2005). Recombinant LABs are frequently harnessed for mucosal delivery of biomolecules such as therapeutic proteins or vaccine antigens (Berlec et al., 2012). In comparison to traditional intravenous or intramuscular vaccine administration, the mucosal route enables immunizations to be performed orally, reducing potential side effects while increasing specificity for chronic illnesses and infections associated with mucosal tissues (Bermudez-Humaran et al., 2011; Wyszynska et al., 2015). Furthermore, many LABs are bile and acid tolerant, act as natural adjuvants, and interact with cells of the immune system (Wells and Mercenier, 2008), making them ideal candidates for antigen carriage.
The LABs most frequently chosen for vaccine delivery are Lactococcus lactis and select species of the Lactobacillus genus (Wells and Mercenier, 2008; Bermudez-Humaran et al., 2011, 2013; Wyszynska et al., 2015). However, unlike L. lactis, several species Lactobacillus have been shown to possess surface-layers (S-layers) (Hynonen and Palva, 2013). S-layers have been detected on both Gram-positive and Gram-negative bacteria and are nearly ubiquitous in archaea (Fagan and Fairweather, 2014). They are defined as two-dimensional crystalline arrays composed of repeating proteinaceous subunits that constitute the outermost layer of a cell envelope (Fagan and Fairweather, 2014). The S-layer proteins (Slps) attach to the underlying peptidoglycan via electrostatic interactions and possess inherent, entropy-driven affinities to self-assemble with each other (Hynonen and Palva, 2013). Thus far, S-layers have been characterized for their role in maintaining cell shape; acting as molecular sieves; serving as binding sites for large molecules, ions, or phages; and mediating surface adhesion (Sleytr et al., 2014). Additionally, Slps are some of the most abundant proteins synthesized by the cell, making them metabolically expensive but also underscoring their importance to the organism (Sara and Sleytr, 2000; Fagan and Fairweather, 2014). Their high expression, periodicity, and self-assembling properties have made them a target for numerous applications in biotechnology and nanotechnology (Avall-Jaaskelainen and Palva, 2005; Hynonen and Palva, 2013; Sleytr et al., 2014).
In the following review, we examine Slp applications in recombinant protein expression and biotherapeutic delivery via their distinct genetic building blocks: the strong native promoter, which can synthesize as many as 500 Slp subunits per second; the signal peptide, that can trigger robust secretion of target molecules; and the structural domains, which can be harnessed for both cell surface display of heterologous proteins or enhancement of host adhesion (Figure 1A). Despite the existence of several recombinant protein expression systems based on one or more components of the Lactobacillus S-layer, this area of research still remains largely underexploited. Thus, the purpose of this review is to not only shed light on past S-layer studies, but also to advocate for future utilization of Slps in mucosal vaccine and biotherapeutic delivery research.
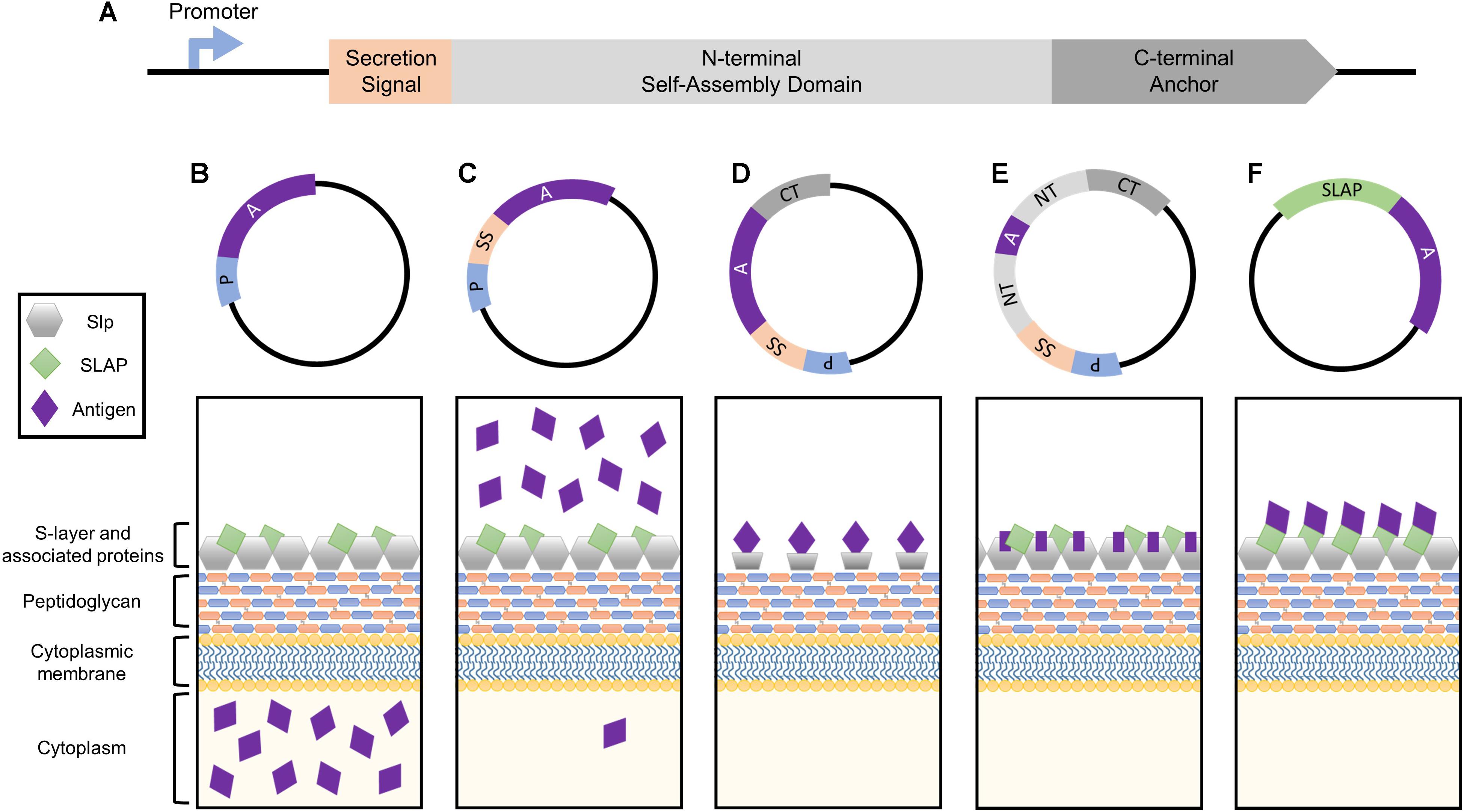
FIGURE 1. Simplified depiction of the genetic components comprising the Slp protein and how they can be harnessed for heterologous protein expression systems. (A) The genetic building blocks of an S-layer gene, including the promoter, secretion signal peptide, N-terminal self-assembly domain, and C-terminal peptidoglycan anchor. Each can be harnessed together or separately for antigen and biotherapeutic delivery applications. (B) The Slp promotor induces the production of high levels of target protein which will accumulate within the cytoplasm of the host bacterium. (C) The Slp secretion signal yields robust secretion of antigen into the extracellular environment. (D) Cell surface display of a foreign peptide can be achieved via fusion to an Slp anchoring domain (N- or C- depending on species). (E) The highest levels of antigen expression and cell surface display can be obtained via direct integration into the Slp, but epitope size is extremely limited in order to maintain array formation. (F) A novel display strategy utilizing S-layer associated protein (SLAP) fusions which theoretically enable the surface display of much larger proteins.
Harnessing the Strong Native Promoter
The bacterial S-layer array is composed of an estimated 5 × 105 subunits, representing 10–15% of the total protein content of the bacterium (Sleytr and Messner, 1983; Avall-Jaaskelainen and Palva, 2005). During exponential growth phase, approximately 500 subunits per second must be synthesized, translocated to the cell surface, and incorporated into the existing lattice structure (Sleytr et al., 2014). In order to obtain these high levels of transcription, S-layer genes possess strong, efficient promoters, which can be harnessed for protein production systems (Figure 1B). Within the Lactobacillus genus, this research has predominately been limited to slp promoters of Lactobacillus acidophilus and Lactobacillus brevis.
Several studies have investigated the versatility of Lactobacillus S-layer promoters for driving heterologous protein expression in various LAB hosts. Lactobacilli regularly possess multiple slp genes within the same strain that are not all concurrently active (Hynonen and Palva, 2013). Although SlpA is the major constituent of the L. acidophilus S-layer, it can be moved to an inactive position triggering expression of the once silent SlpB (Boot et al., 1996a; Konstantinov et al., 2008). When the L. acidophilus ATCC 4356 slpA and slpB promoters were evaluated in Lactobacillus casei ATCC 393, only slpA remained active under all tested growth phases (Boot et al., 1996b). However, the same slpA promoter, although highly efficient in L. lactis, was nearly inactive in isolates of Lactobacillus reuteri (Lizier et al., 2010). The L. acidophilus NCFM slpA promoter was used to drive expression of a green fluorescent protein (GFP) in shuttle vectors based on oriV1 and oriV2 replicons (Chen et al., 2014). Similarly, the plasmids exhibited distinct properties based on which strain they were ported into as well as growth-phase-dependent effects. Both plasmids were capable of replicating in strains of L. casei and Lactobacillus delbrueckii, but only the oriV1 plasmid, pEL5.6, could replicate in Lactobacillus paracasei (Chen et al., 2014). The functionality of the L. brevis ATCC 8287 slpA promoter was evaluated in three LAB hosts: L. lactis MG1614, Lactobacillus plantarum NCDO1193 and Lactobacillus gasseri NCK334, via the expression of various reporter genes (Kahala and Palva, 1999). The S-layer promoter was recognized in each strain, but was particularly active in L. lactis and L. plantarum. In fact, aminopeptidase N (PepN) reporter activity within L. plantarum was 30-fold higher compared to the Lactobacillus helveticus PepN native host and composed a staggering 28% of the total cellular protein during late exponential growth phase. In summary, Lactobacillus slp promoters are effective tools for driving recombinant protein expression, but optimization based on host and growth conditions is essential.
The S-layer promoter has also been harnessed for reporter expression in situ. Plasmid instability and antibiotic markers can complicate the use of these constructs when moving into human clinical trials or to market, thus chromosomal insertions have gained immense popularity. For L. acidophilus NCFM, a pORI-based upp counterselective gene replacement system (Goh et al., 2009) has considerably aided this effort (see “Engineering Platforms”). Originally intended for knockout characterizations, it was first employed for a knock-in by cloning a β-glucuronidase (gusA) reporter downstream of the slpA gene (Douglas and Klaenhammer, 2011). The resulting mutant exhibited a three log increase in GusA activity in comparison to the gusA-negative parent, and the study established a framework for the exploitation of highly expressed genomic regions for heterologous protein production (Douglas and Klaenhammer, 2011). Although this particular system was never evaluated within the context of vaccine delivery, the technique was shown effective for expressing antigens using an alternative highly expressed region within the L. acidophilus genome (O’Flaherty and Klaenhammer, 2016).
Moving beyond reporter genes and targeting more specific therapeutics, the L. brevis JCM 1559 slpA promoter was evaluated in an L. casei IGM393 host via incorporation into a mouse interleukin 10 (IL-10) secretion system (Kajikawa et al., 2010). Administration of IL-10 was previously shown to be an effective treatment of murine colitis when delivered via recombinant L. lactis (Steidler et al., 2000). The authors sought to improve upon this design by substituting in a strain hypothesized to be better adapted to mammalian body temperature. The delivery system was successful in that it yielded high levels of IL-10 secretion when cloned into L. casei, but accumulation of the protein varied widely based on pH (Kajikawa et al., 2010). Maximum efficiency occurred at pH 8.0 and dropped drastically as the pH became more acidic. Although the authors attributed low levels of the protein to its physical characteristics (Kajikawa et al., 2010), the selection of a promoter from a free-living species (e.g., L. brevis) over a low pH tolerating, vertebrate-adapted organism (e.g., L. acidophilus), may have also contributed (Duar et al., 2017). Nonetheless, the S-layer promoter has repeatedly demonstrated its utility for driving protein production systems, and thus merits future research with a focus on therapeutic molecule delivery.
Exploitation of the Secretion Signal Peptide
All Lactobacillus S-layer proteins characterized thus far are preceded by a 25–32 amino acid signal peptide sequence indicative of secretion through the general secretory pathway (Hynonen and Palva, 2013). By cloning this short sequence upstream of an antigen or biotherapeutic molecule, one can obtain robust secretion of a target protein (Figure 1C), thus this peptide is frequently harnessed for biotechnological applications.
The L. brevis ATCC 8287 slpA signal sequence, along with the promoter and transcription terminator, were used to drive expression and secretion of the Escherichia coli β-lactamase (bla) reporter gene in L. lactis, L. brevis, L. plantarum, L. gasseri, and L. casei, using a low-copy-number plasmid derived from pGK12 (Savijoki et al., 1997). In all hosts tested, Bla was expressed and released into the culture medium, though highest yields were obtained by recombinant L. lactis and the strain of L. brevis from which the SlpA components were derived. Production of Bla was mainly restricted to exponential growth phase (Savijoki et al., 1997). Since the system was under control of an slpA promoter, it was unsurprising that efficiency was host-specific and growth-phase-dependent (see “Harnessing the strong native promoter”).
The Slp secretion signal has frequently been harnessed for targeted therapeutic applications. In a study seeking to develop a recombinant L. lactis mucosal vaccine against porcine post-weaning diarrhea and edema disease, signal peptides from the L. lactis major extracellular protein, Usp45, and L. brevis SlpA, were used to stimulate the secretion of an E. coli F18 fimbrial adhesion protein (FedF) fused to a proteinase PrtP cell wall anchor (Lindholm et al., 2004). Both expression systems induced secretion of all tested FedF-PrtP fusions, however, the quantity of fusion proteins found in the culture medium was four to sixfold higher in those containing the SlpA signal peptide. This was a compelling find considering that the Usp45 signal sequence, previously shown to be one of the most effective L. lactis secretion signals (Nouaille et al., 2003), was outperformed by the heterologous SlpA signal sequence. A similar result was obtained with the human interferon alpha 2b (hIFNα-2b) gene (Zhang et al., 2010), used worldwide for the treatment of diseases such as hepatitis B and C (Chelbi-Alix and Wietzerbin, 2007). The addition of the L. brevis SlpA signal peptide increased the secretion efficiency threefold in comparison to the lactococcal Usp45 signal sequence (Zhang et al., 2010).
The SlpA signal sequence has also been employed for secretion of chromosomally inserted heterologous proteins. The Bacillus anthracis protective antigen (PA), fused to a dendritic cell (DC)-targeting peptide, was previously shown to induce protective immunity when delivered on a plasmid via L. acidophilus NCFM (Mohamadzadeh et al., 2009) and L. gasseri (Mohamadzadeh et al., 2010). Since a major advantage of using recombinant microbes for vaccine delivery is their ability to express multiple antigens (Wells and Mercenier, 2008), the B. anthracis PA was co-expressed with the Clostridium botulinum Serotype A neurotoxin heavy-chain antigen (O’Flaherty and Klaenhammer, 2016). The C. botulinum vaccine cassette, which utilizes an L. acidophilus SlpA secretion signal, was chromosomally inserted downstream of the highly expressed enolase gene. Western blot analysis and RNA sequencing confirmed expression of the two antigens (O’Flaherty and Klaenhammer, 2016). Although this strain was never evaluated in vivo, a similarly, constructed strain, also utilizing the SlpA secretion signal, but carrying only the C. botulinum antigen (O’Flaherty and Klaenhammer, 2016), was used to vaccinate mice but unable to confer complete protection (Sahay et al., 2018). Rather, vaccine efficacy was enhanced by intradermal injection of the purified immunogenic subunit, which elicited robust memory B cell responses and rendered mice resistant to lethal doses of botulinum neurotoxin serotype A (Sahay et al., 2018).
The Slp signal sequence is capable of stimulating ample secretion of target molecules, prompting its continued use in vaccine research platforms. Although antigen secretion by recombinant LABs has been shown effective for treating disease (Mohamadzadeh et al., 2005, 2009, 2010), exposure to proteolytic enzymes, low pH, and bile salts may encourage protein degradation and therefore decreased functionality (Wells and Mercenier, 2008); consequently cell wall anchoring has become a popular alternative. However, surface display is a balancing act. High exposure of a protein implies optimal host interaction, but also increased susceptibly to degradation. Alternatively, low exposure, via compact protein folding or embedment within the cell wall, confers protection in exchange for diminished efficacy (Michon et al., 2016). Several S-layer-mediated surface display approaches have attempted to resolve this relationship.
The Self-Assembling and Anchoring Domains of the Slp
Cell wall anchoring via fusion to Slp structural domains (Figure 1D) enables direct interaction of target peptides with host mucosal tissues while simultaneously protecting them from degradation (Bermudez-Humaran et al., 2011; Wang et al., 2016). The first S-layer-mediated display of a foreign epitope was generated by fusing the S-layer homology (SLH) domain of B. anthracis to the normally secreted levansucrase of Bacillus subtilis (Mesnage et al., 1999). The surface-exposed levansucrase retained its enzymatic and antigenic properties, prompting a new area of research which exploits the anchoring abilities of the Slp for cell surface presentation. Lactobacillus Slps do not possess SLH domains and are instead composed of two structural domains: a variable terminal for monomer self-assembly (N- or C- depending on the species), and a highly conserved peptidoglycan anchor (Smit et al., 2001; Hynonen and Palva, 2013).
Several studies have used S-layer-mediated anchoring for recombinant protein display on S-layer-deficient bacteria. The cell wall binding domain of the L. crispatus F5.7 Slp (LbsA) in conjunction with the Slp promoter and secretion signal, were used for surface display of a GFP on several Lactobacillus chicken isolates (Mota et al., 2006). The intention was to generate a vaccine delivery framework for the immunization of broilers against infectious diseases, but never evolved past proof of concept. Vaccination via recombinant bacteria is a particularly attractive option for livestock and poultry operations since the lyophilized microorganisms can be blended into feed; a process that is easily scaled up (Wang et al., 2016). For human applications, the complete SlpA from L. acidophilus ATCC 4356 was fused to a GFP reporter for external presentation on a plasmid-cured, lactose-deficient derivative of L. casei ATCC 334 (Qin et al., 2014). The authors were able to develop a food-grade cell surface display vector by substituting lactose metabolism genes in place of antibiotic selection markers and verified gastrointestinal stability via in vitro modeling (Qin et al., 2014). The L. crispatus K2-4-3 SlpB C-terminal domain, LcsB, was employed for the display of not only a GFP reporter (Hu et al., 2011), but also a carcinoembryonic antigen (CEA) (Zhang et al., 2016). Previous studies have shown that CEA is capable of eliciting strong T-cell and humoral immune responses, which can hinder tumor growth (Greiner et al., 2002). Oral administration of recombinant CEA-presenting L. lactis to mice yielded significantly higher levels of CEA-specific secretory IgA and a higher spleen index in comparison to CEA antigen alone or the negative control, demonstrating the potential of L. lactis CEA as a cancer vaccine (Greiner et al., 2002).
Since the S-layer of L. crispatus is capable of binding intestinal extracellular matrices such as collagen and laminin, heterologous expression of Slps has also been used to improve the adhesive capacity of host organisms. For example, the L. crispatus JCM5810 collagen-binding S-layer protein, CbsA (Martinez et al., 2000), as well as its individual domains (Antikainen et al., 2002), were expressed on the surface of L. casei enabling recombinant organisms to bind various extracellular matrices. Similarly, a surface display cassette consisting of the L. brevis ATCC 8287 SlpA receptor-binding domain fused to a PrtP spacer enhanced L. lactis adherence to Intestine 407 cells (Avall-Jaaskelainen et al., 2003). Both approaches were able to significantly increase the adhesive capacity of engineered organisms, but yet to be evaluated is the probable synergistic effect of combining improved adhesion with an S-layer anchored antigen in what could potentially be a potent vaccine design platform.
Unlike much of the work presented in this review, a handful of studies achieved Slp-mediated display of foreign proteins without DNA manipulation in the host by exploiting the inherent ability of Slps to anchor and self-assemble. For instance, the L. acidophilus C-terminal anchor (SAC) was attached to a GFP reporter, then produced and purified in E. coli. The SAC-GFP fusion protein was capable of binding lithium chloride-pretreated surfaces of wild-type L. acidophilus, L. helveticus, and L. crispatus (Smit et al., 2001). Similarly, L. crispatus K2-4-3 LcsB-GFP fusions were able to associate with SDS-pretreated surfaces of various S-layer-forming LABs including L. brevis, L. helveticus, L. crispatus, and Lactobacillus salivarius as well as several non-S-layer-formers, including L. lactis, L. delbrueckii, Lactobacillus johnsonii, and Streptococcus thermophilus (Hu et al., 2011). Interestingly, neither the SAC-GFP fusion nor LcsB-GFP were able to bind the surface of L. casei (Smit et al., 2001; Hu et al., 2011). This approach is unique in that it offers an alternative way to deliver foreign proteins while also circumventing the GMO (genetically modified organism) label, but is limited by its inability to generate additional heterologous protein in vivo and susceptibility to replacement by wild type Slps. However, in general, studies using heterologously expressed Slps and Slp anchors are hindered by inadequate secretion across the cell wall (Hu et al., 2011) or inability to form an array due to irregular folding and/or lack of cell surface exposure (Martinez et al., 2000). Consequently, rather than tease apart the efficient Slp secretion and display system, there is now interest in harnessing it as a whole through the direct insertion of foreign peptides within the context of the protein.
Delivery Via Direct Integration Within the Slp
The extraordinarily high, stable abundance of the Slp, makes it an enticing target for antigen display and delivery via direct integration into its genome sequence (Figure 1E). The presentation of an exogenous protein within the context of the Slp was first achieved in Caulobacter crescentus through the random introduction of a pilin peptide from Pseudomonas aeruginosa strain K. Eleven potential sites of successful insertion were identified, demonstrating for the first time the capacity of the Slp to act as a carrier for foreign epitopes (Bingle et al., 1997).
Many subsequent studies have focused on mapping the S-layer to gain insight into ideal positioning for novel insertions, including in Lactobacillus. In L. acidophilus ATCC 4356, peptides ranging from 7 to 13 amino acids were randomly introduced into the Slp (Smit et al., 2002). Within the variable N-terminal (SAN), five of these positions maintained paracrystalline structure formation in vitro, while four others resulted in the complete abolishment of any array-forming capacity. Unsurprisingly, an insertion into the cell wall-binding domain had no effect on assembly (Smit et al., 2002). Similarly, the L. brevis SlpA was mapped via cysteine-scanning mutagenesis combined with sulfhydryl modification to identify locations of high surface accessibility and verify that the mutations did not alter self-assembly properties (Vilen et al., 2009). Combined, these works established several stable, surface-accessible insertion sites within the Lactobacillus Slp, yet few researchers have capitalized on this knowledge.
Currently, only two studies have successfully integrated antigens within the context of the Lactobacillus Slp. Through an inducible expression system, the poliovirus VP1 epitope was evaluated in four potential L. brevis ATCC 8287 slpA insertion sites (Avall-Jaaskelainen et al., 2002). The location that demonstrated the best surface expression was then targeted for chromosomal insertion of the c-Myc epitope via direct double-crossover integration (Avall-Jaaskelainen et al., 2002). A uniformly chimeric S-layer was obtained without any effect on array formation. More recently, the membrane proximal external region (MPER) epitope from human immunodeficiency virus type 1 (HIV-1) was inserted into L. acidophilus NCFM SlpA (Kajikawa et al., 2015). L. acidophilus NCFM is regularly employed for mucosal vaccine delivery due in part to its direct interactions with the dendritic cell-specific antigen DC-SIGN (Konstantinov et al., 2008) and adaptation to the harsh conditions associated with gastric transit (Sanders and Klaenhammer, 2001). Vaccination via the recombinant organism, in conjunction with an IL-1β adjuvant, successfully stimulated MPER-specific antibody production in both the serum and mucosal secretions of mice (Kajikawa et al., 2015). This study marks the first and only instance of an Slp-integrated antigen being evaluated in vivo.
The establishment of a uniformly chimeric S-layer translates to approximately 105 instances of epitope display on the surface of a single bacterium (Sleytr and Messner, 1983). Despite these considerable numbers, insert size is exceptionally limited in order to preserve S-layer array formation (Smit et al., 2002). Currently, peptides longer than 19 amino acids are unable to be inserted into SlpA without disrupting the lattice structure (Kajikawa et al., 2015). Therefore, alternative methods exploiting auxiliary proteins associated with the S-layer are now being investigated for the display and delivery of larger antigens, as seen in Slp-mediated anchoring studies (see “The self-assembling and anchoring domains of the Slp”), but at frequencies more akin to direct integration.
S-Layer Associated Proteins
The Lactobacillus S-layer, once thought to be solely composed of repeating monomeric Slp subunits, is actually far more complex (Johnson et al., 2013, 2015). It is now widely accepted that S-layers can act as scaffolds for the external display of numerous auxiliary proteins, termed S-layer associated proteins (SLAPs), which can confer additional physiological functionalities (Johnson et al., 2013, 2015, 2017; Hymes et al., 2016; Johnson and Klaenhammer, 2016; Celebioglu and Svensson, 2017). Recently, the SLAP profile of L. acidophilus NCFM was quantified via multiplexing mass spectrometry (Klotz et al., 2017). Although results revealed significant growth stage-dependent alterations, they also highlighted several proteins with consistent high expression in both logarithmic and stationary growth phases (Klotz et al., 2017). The surface location and abundance of these proteins make them excellent targets for biotherapeutic delivery. Unlike Slp integrants, SLAP fusions are theoretically less limited in epitope size as they are not prone to S-layer array disruption (Figure 1F). Both the native SLAP promoter and secretion signal can be harnessed for this process with the intent to maintain high expression, secretion, and surface localization coupled with the display and delivery of a significantly larger and therefore more potent epitope.
Slp Engineering Platforms
Presently, the two most popular techniques for engineering LABs are the NICE (NIsin Controlled gene Expression) system in L. lactis and the pORI-based upp counterselective gene replacement system in L. acidophilus. There is also growing interest surrounding the use of CRISPR-Cas technology, though its application in bacterial genome editing remains relatively underrepresented (Selle and Barrangou, 2015; Hidalgo-Cantabrana et al., 2017).
The NICE System
The NICE system uses nisin to drive heterologous protein expression in L. lactis (Kuipers et al., 1995). Through the insertion of signal transduction genes from a nisin gene cluster into a nisin-negative L. lactis strain, NZ9000 was created (Kuipers et al., 1998). Subsequently, when a gene of interest is inserted downstream of the inducible nisA promoter, expression of that gene can be obtained by the addition of nisin to the culture medium (Mierau and Kleerebezem, 2005). Since its conception, NICE has become one of the most successful and widely used expression systems in Gram-positive bacteria (Mierau and Kleerebezem, 2005). Indeed, the NICE system was even employed for the production a number of the S-layer fusion proteins mentioned above (Hu et al., 2011; Zhang et al., 2016) and to render the non-adhesive L. lactis NZ9000 adhesive via the addition of an L. brevis SlpA receptor-binding domain (Avall-Jaaskelainen et al., 2003). The availability of an easily engineered, non-S-layer-forming organism, has greatly accelerated not only our understanding of the biological role of an S-layer but also how we can exploit it.
The pORI-Based upp System
The establishment of the pORI-based upp counterselective gene replacement system in S-layer-former L. acidophilus NCFM, first employed for the functional characterization of SlpX (Goh et al., 2009), has since become an invaluable tool for S-layer component engineering. The system uses a upp-encoded uracil phosphoribosyltransferase (UPRTase) as a counterselection marker to positively select for double crossover homologous recombination events. The method has been adapted for numerous Slp studies ranging from reporter integration (Douglas and Klaenhammer, 2011), anchoring/adjuvant assessment (Kajikawa et al., 2011), and targeted antigen delivery systems for disease protection (Kajikawa et al., 2015; O’Flaherty and Klaenhammer, 2016). Similar counterselective systems have also been developed in non-S-layer-formers including L. gasseri ATCC 33323 (Selle et al., 2014) and L. casei ATCC 393 (Song et al., 2014), but have yet to be harnessed for Slp analyses. In general, the technique remains a superior approach for characterizing the functional genetics of lactobacilli without the additional pressures required for plasmid maintenance.
CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) together with CRISPR-associated (Cas) proteins, form the prokaryotic adaptive immune system which provides DNA-encoded, RNA-guided, sequence-specific protection against viral invaders (Barrangou and Doudna, 2016). The current classification system uses two broad classes consisting of six major types (I–VI) which can be allocated into approximately 30 subtypes (Klompe and Sternberg, 2018). The simplest of these is the Type II system which relies on the activity of a singular Cas9 endonuclease and has gained immense popularity for its ability to be repurposed for genome editing (Chylinski et al., 2014; Barrangou and Doudna, 2016; Klompe and Sternberg, 2018). Depending on the organism, this can be done via harnessing of the endogenous system or supplying an exogenous Cas9:single guide RNA (sgRNA) complex. To date, no CRISPR-based genome editing has been performed in S-layer-forming lactobacilli, but it has been conducted in L. casei (Song et al., 2017) and L. lactis (Berlec et al., 2018), frequent hosts for recombinant Slp investigative research. Nevertheless, the development and delivery of a functional CRISPR-Cas9 plasmid remains a critical need, particularly in organisms such as L. acidophilus NCFM (depicted in Figure 2A), which does not possess an active CRISPR system (Crawley et al., 2018), but has proven to be hugely impactful in both probiotic and Slp research.
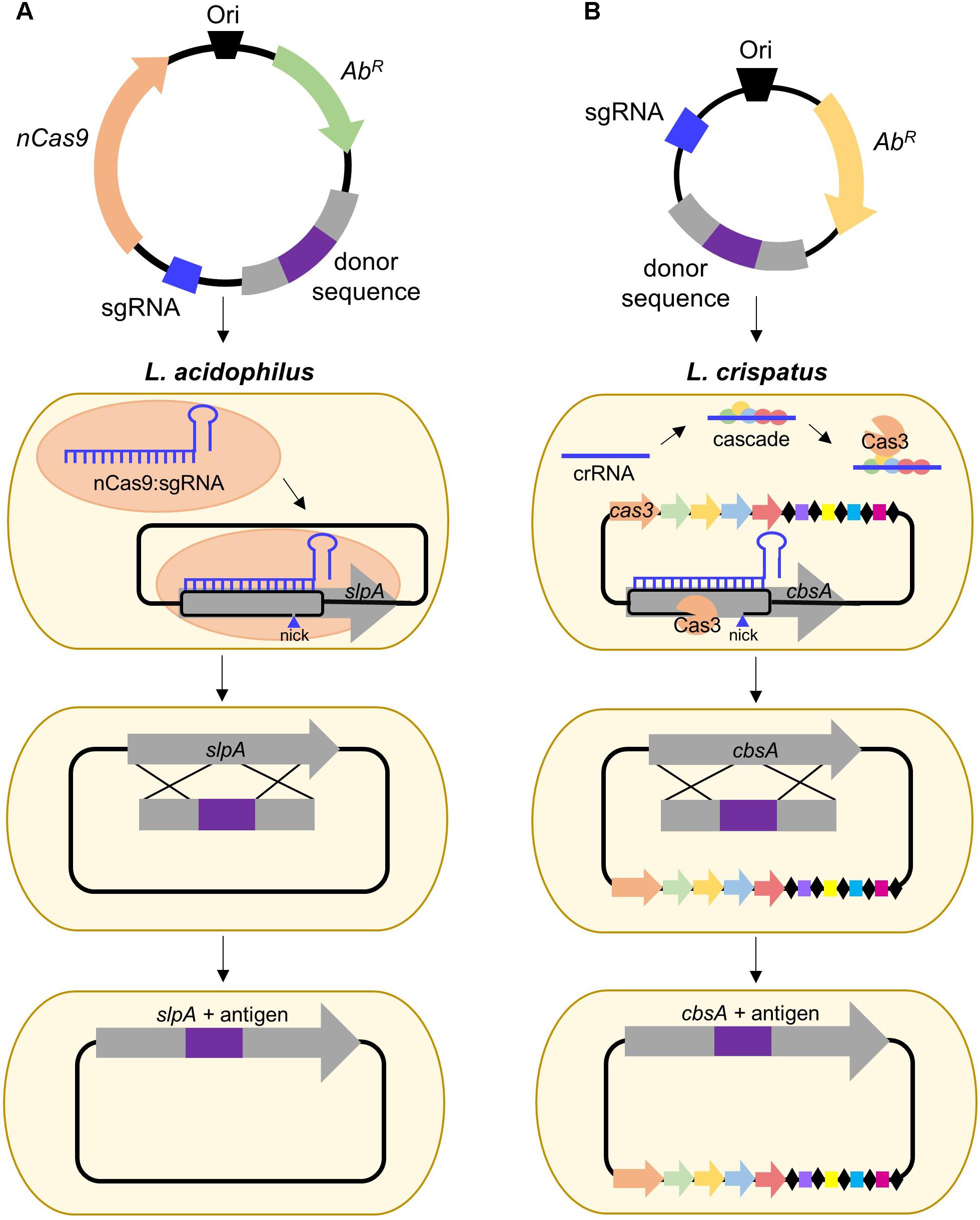
FIGURE 2. Proposed strategies for CRISPR-based genome editing in relevant S-layer-forming lactobacilli. (A) Antigen integration into L. acidophilus SlpA via an exogenous Type II system employing a Cas9 nickase variant which introduces a guide RNA-targeted single-stranded break. Unlike wild-type Cas9 which generates blunt double-stranded breaks (DSB), nickases cut only one strand of the DNA, permitting genome editing in bacteria deficient in DSB repair (Chiang et al., 2016; Song et al., 2017). (B) Antigen integration into L. crispatus CbsA using the endogenous Type I system which consists of the CRISPR-associated complex for antiviral defense (Cascade) and the signature Cas3 nuclease (Barrangou, 2015).
Despite the popularity of Type II engineering, Type I systems have been harnessed for genome editing in Clostridium and archaea (Pyne et al., 2016; Cheng et al., 2017) and transcriptional regulation in E. coli (Luo et al., 2015). S-layer formers, L. crispatus, and L. helveticus, frequently possess Type I systems (Crawley et al., 2018) and are also of interest for vaccine delivery. L. crispatus is a natural inhabitant of the human vaginal tract (Lepargneur, 2016), rendering it suitable for delivery of antigens targeting sexually transmitted diseases, such as HIV, while L. helveticus is predominately associated with dairy (Taverniti and Guglielmetti, 2012), which could be advantageous when considering modes of delivery and stability of the vaccine. A proposed strategy for Type I genome editing in L. crispatus is illustrated in Figure 2B. Harnessing of the endogenous system or delivery of a functional exogenous system both possess the potential to be powerful tools for advancing Slp-mediated biotherapeutic research.
The NICE and pORI-based upp systems have both proven effective for S-layer-mediated biotherapeutic delivery; however, next-generation genome editing tools, such as CRISPR-Cas, hold tremendous potential for bacterial engineering overall (Selle and Barrangou, 2015; Hidalgo-Cantabrana et al., 2017). Many S-layer-forming lactobacilli possess endogenous CRISPR systems, making them promising candidates for future S-layer engineering studies. Alternatively, delivery of a functional CRISPR to strains devoid of a system, will also greatly accelerate the pace at which recombinant organisms can be generated. Ironically, despite CRISPR originating as the bacterial adaptive immune system, CRISPR-based bacterial genome editing still remains relatively underexploited, though recent studies have provided valuable insights for its widespread future implementation (Selle and Barrangou, 2015).
Future Directions
The Slp is a multi-faceted engineering target with both biotherapeutic and biotechnological applications (Sleytr et al., 2014). However, within the Lactobacillus genus, harnessing of this protein remains early in its development (Avall-Jaaskelainen and Palva, 2005; Hynonen and Palva, 2013); nonetheless, the research highlighted above (summarized in Table 1) advocates for its continued pursuit. The slp promoter and signal peptide are undoubtedly adept at driving robust expression and secretion of target proteins, while the structural domains have successfully displayed foreign epitopes and improved the adhesive capacity of host cells. More complex and novel display strategies, such as direct integration into the Slp or SLAP fusions, are innovative approaches for cell surface presentation that also exploit the inherent properties of S-layer-forming lactobacilli. In general, eliciting consistent immune responses via the mucosal route of administration is hindered by rapid elimination or inability to make contact with M cells and other mucosal tissues involved in antigen uptake and processing (Ogra et al., 2001). Thus the ability of recombinant LABs, and the S-layer in particular, to promote antigen uptake and stimulate the adaptive immune response is highly desirable (Konstantinov et al., 2008).
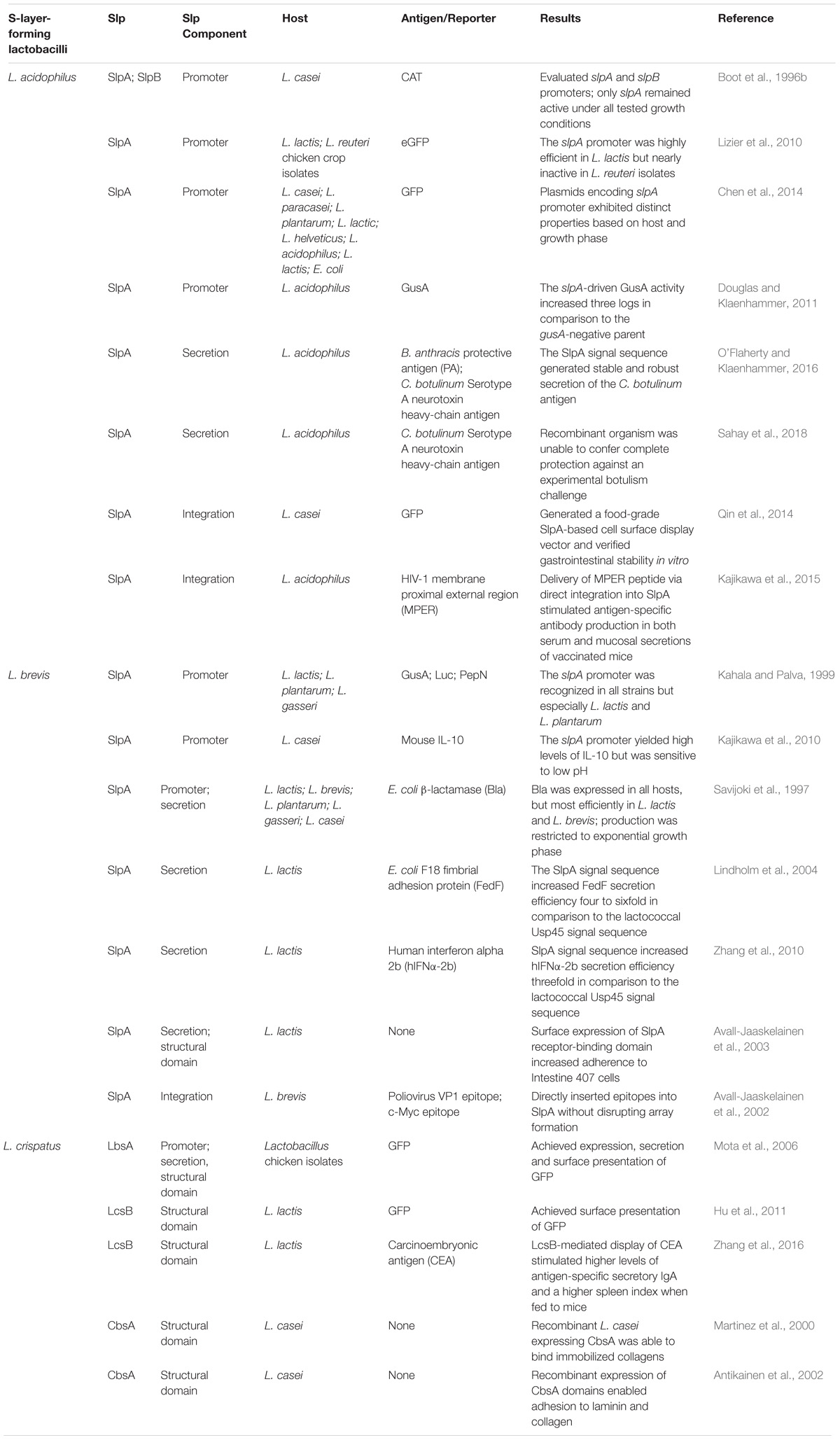
TABLE 1. S-layer protein applications in recombinant protein expression systems and biotherapeutic delivery platforms.
Numerous studies have established vaccine platforms based on one or more components of the Lactobacillus Slp, however, there remains a disconnect between delivery and efficacy. Despite extensive reviews touting the effectiveness of the LAB-based vaccines in vivo (Wells and Mercenier, 2008; LeCureux and Dean, 2018), few S-layer-based delivery frameworks have moved into animal models. Although successful secretion and/or surface display of the reporter/antigen is regularly achieved, only three of the recombinant organisms presented above were tested in mice (Kajikawa et al., 2015; Zhang et al., 2016; Sahay et al., 2018), whereas the remainder were more focused on establishing that antigen production/display was even possible. Thus, an important step moving forward will be connecting the delivery of these antigens with an actual vaccination event, therefore surpassing proof of concept studies and ultimately demonstrating disease protection.
Author Contributions
CK and RB wrote and edited the manuscript.
Funding
This work was supported by DuPont Nutrition and Health and the North Carolina Ag Foundation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Yong Jun Goh, Dr. Sarah O’Flaherty, Dr. Claudio Hidalgo-Cantabrana, and Dr. Alexandra Crawley for their technical expertise and assistance with manuscript preparation.
References
Antikainen, J., Anton, L., Sillanpaa, J., and Korhonen, T. K. (2002). Domains in the S-layer protein CbsA of Lactobacillus crispatus involved in adherence to collagens, laminin and lipoteichoic acids and in self-assembly. Mol. Microbiol. 46, 381–394.
Avall-Jaaskelainen, S., Kyla-Nikkila, K., Kahala, M., Miikkulainen-Lahti, T., and Palva, A. (2002). Surface display of foreign epitopes on the Lactobacillus brevis S-layer. Appl. Environ. Microbiol. 68, 5943–5951.
Avall-Jaaskelainen, S., Lindholm, A., and Palva, A. (2003). Surface display of the receptor-binding region of the Lactobacillus brevis S-layer protein in Lactococcus lactis provides nonadhesive lactococci with the ability to adhere to intestinal epithelial cells. Appl. Environ. Microbiol. 69, 2230–2236.
Avall-Jaaskelainen, S., and Palva, A. (2005). Lactobacillus surface layers and their applications. FEMS Microbiol. Rev. 29, 511–529.
Barrangou, R. (2015). Diversity of CRISPR-Cas immune systems and molecular machines. Genome Biol. 16:247. doi: 10.1186/s13059-015-0816-9
Barrangou, R., and Doudna, J. A. (2016). Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 34, 933–941. doi: 10.1038/nbt.3659
Berlec, A., Ravnikar, M., and Strukelj, B. (2012). Lactic acid bacteria as oral delivery systems for biomolecules. Pharmazie 67, 891–898.
Berlec, A., Skrlec, K., Kocjan, J., Olenic, M., and Strukelj, B. (2018). Single plasmid systems for inducible dual protein expression and for CRISPR-Cas9/CRISPRi gene regulation in lactic acid bacterium Lactococcus lactis. Sci. Rep. 8:1009. doi: 10.1038/s41598-018-19402-1
Bermudez-Humaran, L. G., Aubry, C., Motta, J. P., Deraison, C., Steidler, L., Vergnolle, N., et al. (2013). Engineering lactococci and lactobacilli for human health. Curr. Opin. Microbiol. 16, 278–283. doi: 10.1016/j.mib.2013.06.002
Bermudez-Humaran, L. G., Kharrat, P., Chatel, J. M., and Langella, P. (2011). Lactococci and lactobacilli as mucosal delivery vectors for therapeutic proteins and DNA vaccines. Microbial Cell Factories 10(Suppl. 1)), S4. doi: 10.1186/1475-2859-10-S1-S4
Bingle, W. H., Nomellini, J. F., and Smit, J. (1997). Cell-surface display of a Pseudomonas aeruginosa strain K pilin peptide within the paracrystalline S-layer of Caulobacter crescentus. Mol. Microbiol. 26, 277–288.
Boot, H. J., Kolen, C. P., Andreadaki, F. J., Leer, R. J., and Pouwels, P. H. (1996a). The Lactobacillus acidophilus S-layer protein gene expression site comprises two consensus promoter sequences, one of which directs transcription of stable mRNA. J. Bacteriol. 178, 5388–5394.
Boot, H. J., Kolen, C. P., and Pouwels, P. H. (1996b). Interchange of the active and silent S-layer protein genes of Lactobacillus acidophilus by inversion of the chromosomal slp segment. Mol. Microbiol. 21, 799–809.
Celebioglu, H. U., and Svensson, B. (2017). Exo- and surface proteomes of the probiotic bacterium Lactobacillus acidophilus NCFM. Proteomics 17, 1700019. doi: 10.1002/pmic.201700019
Chelbi-Alix, M. K., and Wietzerbin, J. (2007). Interferon, a growing cytokine family: 50 years of interferon research. Biochimie 89, 713–718.
Chen, Z. J., Lin, J. Z., Ma, C. J., Zhao, S. M., She, Q. X., and Liang, Y. X. (2014). Characterization of pMC11, a plasmid with dual origins of replication isolated from Lactobacillus casei MCJ and construction of shuttle vectors with each replicon. Appl. Microbiol. Biotechnol. 98, 5977–5989. doi: 10.1007/s00253-014-5649-z
Cheng, F., Gong, L., Zhao, D., Yang, H., Zhou, J., Li, M., et al. (2017). Harnessing the native type I-B CRISPR-Cas for genome editing in a polyploid archaeon. J Genet Genomics 44, 541–548. doi: 10.1016/j.jgg.2017.09.010
Chiang, T. W., Le Sage, C., Larrieu, D., Demir, M., and Jackson, S. P. (2016). CRISPR-Cas9(D10A) nickase-based genotypic and phenotypic screening to enhance genome editing. Sci Rep 6, 24356. doi: 10.1038/srep24356
Chylinski, K., Makarova, K. S., Charpentier, E., and Koonin, E. V. (2014). Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 42, 6091–6105. doi: 10.1093/nar/gku241
Crawley, A., Henriksen, E., Stout, E., Brandt, K., and Barrangou, R. (2018). Characterizing the activity of abundant, diverse and active CRISPR-Cas systems in lactobacilli. Sci Rep 8, 11544. doi: 10.1038/s41598-018-29746-3
Douglas, G. L., and Klaenhammer, T. R. (2011). Directed chromosomal integration and expression of the reporter gene gusA3 in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 77, 7365–7371. doi: 10.1128/AEM.06028-11
Duar, R. M., Lin, X. X. B., Zheng, J. S., Martino, M. E., Grenier, T., Perez-Munoz, M. E., et al. (2017). Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 41, S27–S48. doi: 10.1093/femsre/fux030
Fagan, R. P., and Fairweather, N. F. (2014). Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol. 12, 211–222. doi: 10.1038/nrmicro3213
Goh, Y. J., Azcarate-Peril, M. A., O’Flaherty, S., Durmaz, E., Valence, F., Jardin, J., et al. (2009). Development and application of a upp-based counterselective gene replacement system for the study of the S-layer protein SlpX of Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 75, 3093–3105. doi: 10.1128/AEM.02502-08
Greiner, J. W., Zeytin, H., Anver, M. R., and Schlom, J. (2002). Vaccine-based therapy directed against carcinoembryonic antigen demonstrates antitumor activity on spontaneous intestinal tumors in the absence of autoimmunity. Cancer Res. 62, 6944–6951.
Hidalgo-Cantabrana, C., O’Flaherty, S., and Barrangou, R. (2017). CRISPR-based engineering of next-generation lactic acid bacteria. Curr. Opin. Microbiol. 37, 79–87. doi: 10.1016/j.mib.2017.05.015
Hu, S., Kong, J., Sun, Z., Han, L., Kong, W., and Yang, P. (2011). Heterologous protein display on the cell surface of lactic acid bacteria mediated by the s-layer protein. Microb Cell Fact 10, 86. doi: 10.1186/1475-2859-10-86
Hymes, J. P., Johnson, B. R., Barrangou, R., and Klaenhammer, T. R. (2016). Functional analysis of an S-layer-associated fibronectin-binding protein in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 82, 2676–2685. doi: 10.1128/AEM.00024-16
Hynonen, U., and Palva, A. (2013). Lactobacillus surface layer proteins: structure, function and applications. Appl. Microbiol. Biotechnol. 97, 5225–5243.
Johnson, B., Selle, K., O’Flaherty, S., Goh, Y. J., and Klaenhammer, T. (2013). Identification of extracellular surface-layer associated proteins in Lactobacillus acidophilus NCFM. Microbiology 159, 2269–2282. doi: 10.1099/mic.0.070755-0
Johnson, B. R., Hymes, J., Sanozky-Dawes, R., Henriksen, E. D., Barrangou, R., and Klaenhammer, T. R. (2015). Conserved S-layer-associated proteins revealed by exoproteomic survey of S-layer-forming Lactobacilli. Appl. Environ. Microbiol. 82, 134–145. doi: 10.1128/AEM.01968-15
Johnson, B. R., and Klaenhammer, T. R. (2016). AcmB Is an S-Layer-associated beta-N-acetylglucosaminidase and functional autolysin in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 82, 5687–5697. doi: 10.1128/AEM.02025-16
Johnson, B. R., O’Flaherty, S., Goh, Y. J., Carroll, I., Barrangou, R., and Klaenhammer, T. R. (2017). The S-layer associated serine protease homolog PrtX impacts cell surface-mediated microbe-host interactions of Lactobacillus acidophilus NCFM. Front. Microbiol. 8:1185. doi: 10.3389/fmicb.2017.01185
Kahala, M., and Palva, A. (1999). The expression signals of the Lactobacillus brevis slpA gene direct efficient heterologous protein production in lactic acid bacteria. Appl. Microbiol. Biotechnol. 51, 71–78.
Kajikawa, A., Ichikawa, E., and Igimi, S. (2010). Development of a highly efficient protein-secreting system in recombinant Lactobacillus casei. J. Microbiol. Biotechnol. 20, 375–382.
Kajikawa, A., Nordone, S. K., Zhang, L., Stoeker, L. L., Lavoy, A. S., Klaenhammer, T. R., et al. (2011). Dissimilar properties of two recombinant Lactobacillus acidophilus strains displaying Salmonella FliC with different anchoring motifs. Appl. Environ. Microbiol. 77, 6587–6596. doi: 10.1128/AEM.05153-11
Kajikawa, A., Zhang, L., Lavoy, A., Bumgardner, S., Klaenhammer, T. R., and Dean, G. A. (2015). Mucosal immunogenicity of genetically modified Lactobacillus acidophilus expressing an HIV-1 epitope within the surface layer protein. PLoS One 10:e0141713. doi: 10.1371/journal.pone.0141713
Klaenhammer, T. R., Barrangou, R., Buck, B. L., Azcarate-Peril, M. A., and Altermann, E. (2005). Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiol. Rev. 29, 393–409.
Klein, G., Pack, A., Bonaparte, C., and Reuter, G. (1998). Taxonomy and physiology of probiotic lactic acid bacteria. Int. J. Food Microbiol. 41, 103–125.
Klompe, S., and Sternberg, S. (2018). Harnessing a billion years of experimentation: the ongoing exploration and exploitation of CRISPR–cas immune systems. CRISPR J. 1, 141–158.
Klotz, C., O’Flaherty, S., Goh, Y. J., and Barrangou, R. (2017). Investigating the effect of growth phase on the surface-layer associated proteome of Lactobacillus acidophilus using quantitative proteomics. Front. Microbiol. 8:2174. doi: 10.3389/fmicb.2017.02174
Konstantinov, S. R., Smidt, H., De Vos, W. M., Bruijns, S. C., Singh, S. K., Valence, F., et al. (2008). S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. U.S.A. 105, 19474–19479. doi: 10.1073/pnas.0810305105
Kuipers, O. P., Beerthuyzen, M. M., De Ruyter, P. G., Luesink, E. J., and De Vos, W. M. (1995). Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270, 27299–27304.
Kuipers, O. P., De Ruyter, P. G. G. A., Kleerebezem, M., and De Vos, W. M. (1998). Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64, 15–21.
LeCureux, J. S., and Dean, G. A. (2018). Lactobacillus mucosal vaccine vectors: immune responses against bacterial and viral antigens. mSphere 3:e00061-e18. doi: 10.1128/mSphere.00061-18
Lepargneur, J. P. (2016). Lactobacillus crispatus as biomarker of the healthy vaginal tract. Ann. Biol. Clin. 74, 421–427. doi: 10.1684/abc.2016.1169
Lindholm, A., Smeds, A., and Palva, A. (2004). Receptor binding domain of Escherichia coli F18 fimbrial adhesin FedF can be both efficiently secreted and surface displayed in a functional form in Lactococcus lactis. Appl. Environ. Microbiol. 70, 2061–2071.
Liu, W., Pang, H., Zhang, H., and Cai, Y. (2014). “Biodiversity of Lactic Acid Bacteria,” in Lactic Acid Bacteria: Fundamentals and Practice, eds H. Zhang and Y. Cai (Dordrecht: Springer Netherlands), 103–203.
Lizier, M., Sarra, P. G., Cauda, R., and Lucchini, F. (2010). Comparison of expression vectors in Lactobacillus reuteri strains. FEMS Microbiol. Lett. 308, 8–15. doi: 10.1111/j.1574-6968.2010.01978.x
Luo, M. L., Mullis, A. S., Leenay, R. T., and Beisel, C. L. (2015). Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 43, 674–681. doi: 10.1093/nar/gku971
Martinez, B., Sillanpaa, J., Smit, E., Korhonen, T. K., and Pouwels, P. H. (2000). Expression of cbsA encoding the collagen-binding S-protein of Lactobacillus crispatus JCM5810 in Lactobacillus casei ATCC 393(T). J. Bacteriol. 182, 6857–6861.
Mesnage, S., Tosi-Couture, E., and Fouet, A. (1999). Production and cell surface anchoring of functional fusions between the SLH motifs of the Bacillus anthracis S-layer proteins and the Bacillus subtilis levansucrase. Mol. Microbiol. 31, 927–936.
Michon, C., Langella, P., Eijsink, V. G. H., Mathiesen, G., and Chatel, J. M. (2016). Display of recombinant proteins at the surface of lactic acid bacteria: strategies and applications. Microb. Cell Fact. 15:70. doi: 10.1186/s12934-016-0468-9
Mierau, I., and Kleerebezem, M. (2005). 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68, 705–717.
Mohamadzadeh, M., Duong, T., Sandwick, S. J., Hoover, T., and Klaenhammer, T. R. (2009). Dendritic cell targeting of Bacillus anthracis protective antigen expressed by Lactobacillus acidophilus protects mice from lethal challenge. Proc. Natl. Acad. Sci. U.S.A. 106, 4331–4336. doi: 10.1073/pnas.0900029106
Mohamadzadeh, M., Durmaz, E., Zadeh, M., Pakanati, K. C., Gramarossa, M., Cohran, V., et al. (2010). Targeted expression of anthrax protective antigen by Lactobacillus gasseri as an anthrax vaccine. Future Microbiol 5, 1289–1296. doi: 10.2217/fmb.10.78
Mohamadzadeh, M., Olson, S., Kalina, W. V., Ruthel, G., Demmin, G. L., Warfield, K. L., et al. (2005). Lactobacilli activate human dendritic cells that skew T cells toward T helper 1 polarization. Proc. Natl. Acad. Sci. U.S.A. 102, 2880–2885.
Mota, R. M., Moreira, J. L., Souza, M. R., Horta, M. F., Teixeira, S. M., Neumann, E., et al. (2006). Genetic transformation of novel isolates of chicken Lactobacillus bearing probiotic features for expression of heterologous proteins: a tool to develop live oral vaccines. BMC Biotechnol. 6:2. doi: 10.1186/1472-6750-6-2
Nouaille, S., Ribeiro, L. A., Miyoshi, A., Pontes, D., Le Loir, Y., Oliveira, S. C., et al. (2003). Heterologous protein production and delivery systems for Lactococcus lactis. Genet. Mol. Res. 2, 102–111.
O’Flaherty, S., and Klaenhammer, T. R. (2016). Multivalent chromosomal expression of the Clostridium botulinum serotype a neurotoxin heavy-chain antigen and the Bacillus anthracis protective antigen in Lactobacillus acidophilus. Appl. Environ. Microbiol. 82, 6091–6101.
Ogra, P. L., Faden, H., and Welliver, R. C. (2001). Vaccination strategies for mucosal immune responses. Clin. Microbiol. Rev 14, 430–445.
Pyne, M. E., Bruder, M. R., Moo-Young, M., Chung, D. A., and Chou, C. P. (2016). Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium. Sci. Rep. 6:25666. doi: 10.1038/srep25666
Qin, J., Wang, X., Kong, J., Ma, C., and Xu, P. (2014). Construction of a food-grade cell surface display system for Lactobacillus casei. Microbiol. Res. 169, 733–740. doi: 10.1016/j.micres.2014.02.001
Sahay, B., Colliou, N., Zadeh, M., Ge, Y., Gong, M., Owen, J. L., et al. (2018). Dual-route targeted vaccine protects efficiently against botulinum neurotoxin A complex. Vaccine 36, 155–164. doi: 10.1016/j.vaccine.2017.11.008
Sanders, M. E., and Klaenhammer, T. R. (2001). Invited review: the scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic. J. Dairy Sci. 84, 319–331.
Savijoki, K., Kahala, M., and Palva, A. (1997). High level heterologous protein production in Lactococcus and Lactobacillus using a new secretion system based on the Lactobacillus brevis S-layer signals. Gene 186, 255–262.
Selle, K., and Barrangou, R. (2015). Harnessing CRISPR-Cas systems for bacterial genome editing. Trends Microbiol 23, 225–232. doi: 10.1016/j.tim.2015.01.008
Selle, K., Goh, Y. J., O’Flaherty, S., and Klaenhammer, T. R. (2014). Development of an integration mutagenesis system in Lactobacillus gasseri. Gut Microbes 5, 326–332. doi: 10.4161/gmic.29101
Sleytr, U. B., and Messner, P. (1983). Crystalline surface layers on bacteria. Annu. Rev. Microbiol. 37, 311–339.
Sleytr, U. B., Schuster, B., Egelseer, E. M., and Pum, D. (2014). S-layers: principles and applications. FEMS Microbiol. Rev. 38, 823–864. doi: 10.1111/1574-6976.12063
Smit, E., Jager, D., Martinez, B., Tielen, F. J., and Pouwels, P. H. (2002). Structural and functional analysis of the S-layer protein crystallisation domain of Lactobacillus acidophilus ATCC 4356: evidence for protein-protein interaction of two subdomains. J. Mol. Biol. 324, 953–964.
Smit, E., Oling, F., Demel, R., Martinez, B., and Pouwels, P. H. (2001). The S-layer protein of Lactobacillus acidophilus ATCC 4356: identification and characterisation of domains responsible for S-protein assembly and cell wall binding. J. Mol. Biol. 305, 245–257.
Song, B. F., Ju, L. Z., Li, Y. J., and Tang, L. J. (2014). Chromosomal insertions in the Lactobacillus casei upp gene that are useful for vaccine expression. Appl. Environ. Microbiol. 80, 3321–3326. doi: 10.1128/AEM.00175-14
Song, X., Huang, H., Xiong, Z., Ai, L., and Yang, S. (2017). CRISPR-Cas9(D10A) nickase-assisted genome editing in Lactobacillus casei. Appl. Environ. Microbiol 83, e01259-17. doi: 10.1128/AEM.01259-17
Steidler, L., Hans, W., Schotte, L., Neirynck, S., Obermeier, F., Falk, W., et al. (2000). Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289, 1352–1355.
Taverniti, V., and Guglielmetti, S. (2012). Health-promoting properties of Lactobacillus helveticus. Front. Microbiol. 3:392. doi: 10.3389/fmicb.2012.00392
Vilen, H., Hynonen, U., Badelt-Lichtblau, H., Ilk, N., Jaaskelainen, P., Torkkeli, M., et al. (2009). Surface Location of Individual Residues of SlpA Provides Insight into the Lactobacillus brevis S-Layer. J. Bacteriol. 191, 3339–3349. doi: 10.1128/JB.01782-08
Wang, M., Gao, Z., Zhang, Y., and Pan, L. (2016). Lactic acid bacteria as mucosal delivery vehicles: a realistic therapeutic option. Appl. Microbiol. Biotechnol. 100, 5691–5701. doi: 10.1007/s00253-016-7557-x
Wells, J. M., and Mercenier, A. (2008). Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat. Rev. Microbiol. 6 349–362. doi: 10.1038/nrmicro1840
Wyszynska, A., Kobierecka, P., Bardowski, J., and Jagusztyn-Krynicka, E. K. (2015). Lactic acid bacteria–20 years exploring their potential as live vectors for mucosal vaccination. Appl. Microbiol. Biotechnol. 99, 2967–2977. doi: 10.1007/s00253-015-6498-0
Zhang, Q., Zhong, J., Liang, X., Liu, W., and Huan, L. (2010). Improvement of human interferon alpha secretion by Lactococcus lactis. Biotechnol. Lett. 32, 1271–1277. doi: 10.1007/s10529-010-0285-x
Keywords: Lactobacillus, probiotic, S-layer, biotherapeutic, mucosal vaccine, CRISPR
Citation: Klotz C and Barrangou R (2018) Engineering Components of the Lactobacillus S-Layer for Biotherapeutic Applications. Front. Microbiol. 9:2264. doi: 10.3389/fmicb.2018.02264
Received: 05 June 2018; Accepted: 05 September 2018;
Published: 02 October 2018.
Edited by:
Aleš Berlec, Jožef Stefan Institute, SloveniaReviewed by:
Akinobu Kajikawa, Tokyo University of Agriculture, JapanOlivia Cano Garrido, Centre for Research in Agricultural Genomics (CRAG), Spain
Copyright © 2018 Klotz and Barrangou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rodolphe Barrangou, cmJhcnJhbkBuY3N1LmVkdQ==
 Courtney Klotz
Courtney Klotz Rodolphe Barrangou
Rodolphe Barrangou