- 1Department of Microbiology and Molecular Genetics, University of the Punjab, Lahore, Pakistan
- 2Department of Applied Microbiology, Faculty of Biology, Institute of Microbiology, University of Warsaw, Warsaw, Poland
- 3Provincial TB Control Program, Lahore, Pakistan
Pakistan ranks 5th among the world's highest tuberculosis (TB) burden countries alongside the 6th among countries with the highest burden of drug-resistant TB, including multi-drug resistant (MDR)-TB. Methods for rapid and reliable drug susceptibility testing (DST) are prerequisite for the prompt institution of effective anti-TB treatment. The aim of this study was to evaluate the efficiency of Genotype MTBDRplus and MTBDRsl assays for the detection of MDR and (pre-) extensively drug-resistant (XDR-TB) isolates in Pakistan. The study included 47 pre-XDR and 6 XDR-TB isolates, recovered from 53 patients from Pakistan. Conventional DST was performed using the standard 1% proportion method on the Löwenstein-Jensen medium. For molecular determination of drug resistance, GenoType MTBDRplus and GenoType MTBDRsl assays (Hain Lifescience, Germany) were used. To evaluate discrepancies between conventional and molecular DST results, mutation profiling was performed by amplifying and sequencing seven genetic loci, i.e., katG, inhA, and mabA-inhA promoter, rpoB, gyrA, embB, rrs. The sensitivity of Genotype MTBDRplus was 71.7% for isoniazid (INH) and 79.2% for rifampicin (RIF). Sequence analysis revealed non-synonymous mutations in 93.3 and 27.3% of isolates phenotypically resistant to INH and RIF, respectively, albeit susceptible when tested by GenoType MTBDRplus. GenoType MTBDRsl had a sensitivity of 73.6, 64.7, 20, 25, and 100% for the detection of fluoroquinolones, ethambutol, kanamycin, amikacin, and capreomycin resistance, respectively. Upon sequencing, mutations were detected in 20, 77.8%, and all isolates phenotypically resistant to aminoglycosides, ethambutol, and fluoroquinolones, respectively, yet declared as susceptible with GenoType MTBDRsl. Low sensitivities seriously impede the large-scale application of the Genotype MTBDRplus and MTBDRsl assays. Unless further optimized, the currently available line-probe assays should rather be auxiliary to the conventional, phenotype-based methods in the detection of MDR- and XDR-TB in Pakistan.
Introduction
Tuberculosis (TB) remains an inglorious leader among infectious diseases in mortality, with its annual toll of 1.5 million lives worldwide (Ullah et al., 2016). Although the global TB incidence has been on a downward trend since 2000, the emergence and persistence of drug-resistant (DR) tubercle bacilli strains, particularly those multi-drug resistant (MDR), defined as resistant to at least isoniazid (INH) and rifampicin (RIF), and extensively drug-resistant (XDR), defined as MDR with additional resistance to a fluoroquinolone (FQ) and a second-line injectable drug (SLID), have substantially undermined efforts to control and eliminate the disease. According to a most recent World Health Organization (WHO) report, every seventeenth TB patient expels MDR bacilli. One in sixteen of such patients expel strains of XDR phenotype (World Health Organization, 2017). For almost two decades, Pakistan, with a population of 193 million people, continues to be on the top of the list high TB-burden countries (HBCs), in terms of total TB caseload (TB incidence rate, 268 per 100,000 population) and in terms of DR-TB, including MDR-TB (MDR-TB incidence rate, 14 per 100,000 population), globally (World Health Organization, 2017). Studies on the prevalence of DR-TB, including MDR-TB and XDR-TB in Pakistan are quite scarce and fragmentary (Ali et al., 2011, 2015; Farooqi et al., 2012; Javaid et al., 2016). Also, data collected from national surveys on DR-TB and reported to WHO have to be cautiously treated. The true prevalence of DR-TB is thought to be underestimated, mostly due to a limited number of facilities offering drug susceptibility testing (DST) and poor availability of modern and advanced technologies, allowing for fast and reliable drug resistance profiling (Domínguez et al., 2016).
Conventional DST is laborious, time consuming and requires growth of mycobacteria either on solid or liquid media (Bernard et al., 2015). Identification of the XDR phenotype is particularly protracted as it is a two-step procedure that first involves testing against first-line anti-TB drugs, and then against second-line agents, once multidrug resistance is disclosed (Abebe et al., 2011). Rapid and reliable DST results are prerequisite for the prompt implementation of effective treatment, and reducing the risk of acquired resistance (Hillemann et al., 2009). Over the last decade, diagnosing DR-TB has been greatly improved and expedited with the introduction of various molecular-based DST technologies (Thumamo et al., 2012). They all fall into two major categories that is probe-based and sequence-based methods. The former are represented by Cepheid's GeneXpert MTB/RIF assay (Cepheid, USA) with molecular beacon probes (Ioannidis et al., 2011) and Hain's MTBDRplus and MTBDRsl assays (Hain Life Sciences, Germany) with line probes (Nathavitharana et al., 2017). Whereas molecular beacon-based assays use real-time PCR, dual-labeled probes that form a quenched, stem-loop structure in native state and fluoresce upon hybridization to the target nucleotide sequence (Lawn and Nicol, 2011), line probe assays involve PCR and reverse hybridization with specific oligonucleotide probed fixed to a nitrocellulose strip in parallel lines (World Health Organization, 2008). Sequence-based methods include pyrosequencing (Molina-Moya et al., 2017), Sanger sequencing (Schleusener et al., 2017), and next-generation sequencing (Jagielski et al., 2016).
The implementation of routine sequencing to track drug- resistance profiles has already become a part of a strategy aiming at TB elimination in England, and toward similar plans in other high-income countries (Walker et al., 2017; Satta et al., 2018). However, DNA sequencing-based approaches remains prohibitevely expensive and complex for routine use in low- and middle-income countries, especially those heavily burdened with TB (Votintseva et al., 2017).
In December 2010 WHO approved the GeneXpert MTB/RIF test for detection of DR strains in high-burden, resource-limited countries (World Health Organization, 2013). An important drawback of the system is that it detects resistance to RIF but not to other anti-TB drugs such as INH, FQs, and SLIDs. Furthermore, whereas PCR inhibition or unsuccessful DNA extraction may increase the chances of false-negative results, false positive results might occur in previously treated patients and having mixed TB/non-tuberculous mycobacteria (NTM) infection (Liu et al., 2017).
The Hain probe assays use reverse hybridization technology to detect mutations associated with resistance of tubercle bacilli to both first- and second-line anti-TB drugs. MTBDRplus allows for identification of INH and RIF resistance by disclosing mutations in the katG, inhA, and rpoB genes, while MTBDRsl detects resistance to FQs, ethambutol (EMB), aminoglycosides (kanamycin, KAN; amikacin, AMK; viomycin, VIO), and cyclic peptide (capreomycin, CAP) by finding mutations in three different loci, that is gyrA, embB, and rrs, respectively (Bai et al., 2016).
Based on two recent meta-analyses, including over 50 studies, the overall sensitivities of the two Genotype assays varied widely from 96% for RIF to 44% for KAN, while the specificities ranged from 99% for INH, RIF, AMK, and KAN to 79% for EMB (Feng et al., 2013; Bai et al., 2016). Still, conventional, culture-based DST is considered a gold standard in mycobacteriology.
The aim of the study was to evaluate the efficiency of Genotype MTBDRplus and MTBDRsl assays in the detection of drug resistance in the context of conventional DST profiling and results of PCR-sequencing targeted at selected drug resistance-associated loci.
Materials and Methods
Isolates
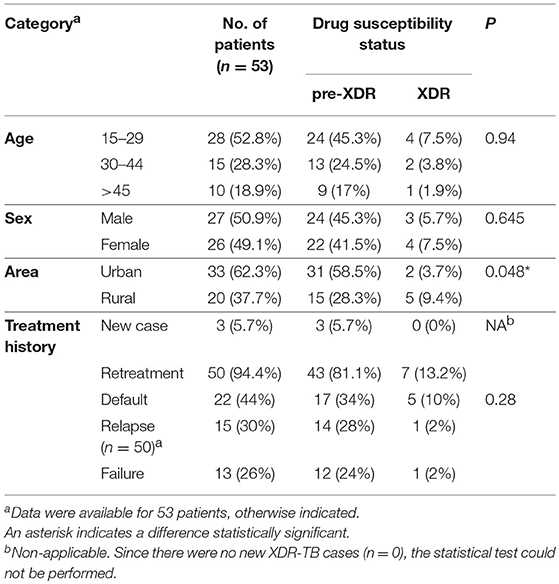
The study sample was selected out of a pool of 3056 Mycobacterium tuberculosis isolates, recovered from as many patients, originally diagnosed as having pulmonary TB and originated from seven major cities of Punjab province of Pakistan from January 2013 to June 2015. MDR-TB, based on the conventional DST results, was initially identified in 362 of the cases (Figure S1). These cases were further reviewed to select those where (pre-)XDR-TB was confirmed. Thereby, the final study group of 53 patients was achieved (Table 1).
pre-XDR phenotype was defined as MDR with either resistance to ofloxacin (OFX) or any of the SLIDs (i.e., AMK, KAN or CAP). XDR-TB was defined as MDR-TB with additional resistance to OFX and one of the SLIDs.
Primary isolation, culturing, and species identification were performed with standard mycobacteriological methods in Provincial TB Reference Laboratory, Institute of Public Health, Lahore. Briefly, clinical samples were decontaminated with 1% N-acetyl-L-cysteine/NaOH and centrifuged. Each sample was then inoculated onto Löwenstein-Jensen and MGIT (Becton Dickinson, Franklin Lakes, NJ, USA) media. Identification was performed using the BACTEC NAP TB Differentiation Test Kit (Becton Dickinson, USA), growth in para-nitrobenzoic acid–containing media, nitrate reduction, and niacin accumulation (Koneman et al., 1983).
This study was carried out in accordance with the recommendations of ethical policy of the University of Punjab.
Drug Susceptibility Testing
Conventional DST was performed using the standard 1% proportion method on the Löwenstein-Jensen (L-J) medium, with the M. tuberculosis H37Rv strain as a quality control, following the WHO recommendations (Van Embden et al., 1993). The critical concentrations for specific drugs were as follows: INH, 0.2 mg/L; RIF, 40 mg/L; EMB, 2 mg/L; STR, 4 mg/L; KAN, 30 mg/L; AMK, 30 mg/L; CAP, 40 mg/L; and OFX, 4 mg/L.
For molecular determination of drug resistance, GenoType MTBDRplus and GenoType MTBDRsl assays (Hain Lifescience, Germany) were used. The principle of these tests was essentially described elsewhere (Anek-vorapong et al., 2010). Both assays were performed and interpreted in accordance with the manufacturer's instructions. A test result was considered valid, only if all control bands appeared correctly. An isolate was declared resistant if at least one wild type probe was absent or if any mutant probe was present. If all wild type probes were present and all mutant probes were absent, an isolate was recognized as susceptible.
DNA Extraction
Genomic DNA from M. tuberculosis isolates, grown on L-J medium, was extracted using the cetyl-trimethyl ammonium bromide (CTAB) method, as described previously (Jagielski et al., 2015).
PCR and Sequencing
To evaluate discrepancies between conventional and molecular (GenoType MTBDRplus and MTBDRsl) DST results, selected fragments of seven genetic loci, containing the hot spots for mutations associated with resistance of tubercle bacilli to INH (katG, inhA, and mabA-inhA promoter), RIF (rpoB), EMB (embB), aminoglycosides (rrs), and FQs (gyrA) were PCR-amplified and sequenced. The oligonucleotide primers used for PCR are described in Table S1. Amplification reactions were set up and performed according to the manufacturer's instructions (TopTaq DNA polymerase, Qiagen, Germany). Purified PCR amplicons (Clean-up kit, A&A Biotechnology, Poland) were sequenced in both directions using the same primers as for PCR amplification. Mutations were detected using Clone Manager software (v. 8.0, Scientific & Educational Software, USA) by comparing the obtained sequences with the M. tuberculosis reference strain H37Rv sequences of respective loci, deposited in the GenBank database (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/). An isolate was declared resistant if at least one non-synonymous mutation was detected in a resistance-associated locus.
Nucleotide bases and codon numbers were reported using either M. tuberculosis (katG, inhA, mabA-inhA, embB, rrs, gyrA) or Escherichia coli (rpoB) numbering system.
Nucelotide Accession Numbers
The sequences with detected mutations were deposited in GenBank (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/) under the following accession numbers: MF145294-MF145302, MF145309, MF145313 for the rpoB gene, MF145303-MF145308, MF145310-MF145312, MF145314-MF145316 for the katG gene, MF145317-MF145331 for the gyrA gene, MF145332-MF145339 for rrs gene, and MF145340- MF145357 for the embB gene.
Statistical Analysis
Data were analyzed with the IBM SPSS software (version 22.0, USA). To analyze differences between categorical variables, χ2-test was used. A value of P < 0.05 was considered statistically significant.
Diagnostic Performance
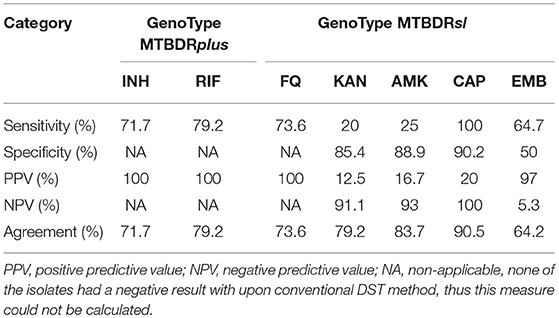
To assess the diagnostic performance of the line-probe assays, sensitivities and positive predictive values (PPV) were calculated for all drugs covered by the GenoType MTBDRplus and MTBDRsl assays. Since the study sample included no INH-, RIF-, FQ-susceptible isolates, specificities and negative predictive values (NPV) were calculated only for KAN, AMK, CAP, and EMB of GenoType MTBDRsl.
Results
Conventional DST
All (53/53) isolates tested were resistant to INH, RIF (MDR) and OFX. Fifty-one (51/53; 96.2%) isolates were resistant to EMB, 39 (39/53; 73.6%) to STR, five (5/53; 9.4%) to KAN, and four (4/53; 7.5%) to AMK. Only one (1/53; 1.9%) isolate was CAP-resistant (Table 2). The most common drug resistance pattern was INH+RIF+OFX+EMB+STR (29/53, 54.7%) followed by INH+RIF+OFX+EMB (12/53, 22.6%). Forty-seven (47/53; 88.7%) isolates were resistant to OFX and not to any other SLIDs, thus meeting the definition of pre-XDR-TB. Six (6/53; 11.3%) isolates were categorized as XDR-TB isolates with additional (to INH+RIF+OFX) resistance to AMK and KAN (2/53; 3.8%), KAN (2/53; 3.8%), AMK (1/53; 1.9%), and AMK, KAN and CAP (1/53; 1.9%) (Table 2).

Table 2. Drug susceptibility profiles detected by conventional DST, GenoType MTBDRplus, GenoType MTBDRsl (LPA), and sequence analysis (SEQ).
Performance of Genotype MTBDRplus
The results of DST with the GenoType MTBDRplus assay are shown in Table 2, while representative patterns obtained with the assay are depicted in Figure 1.
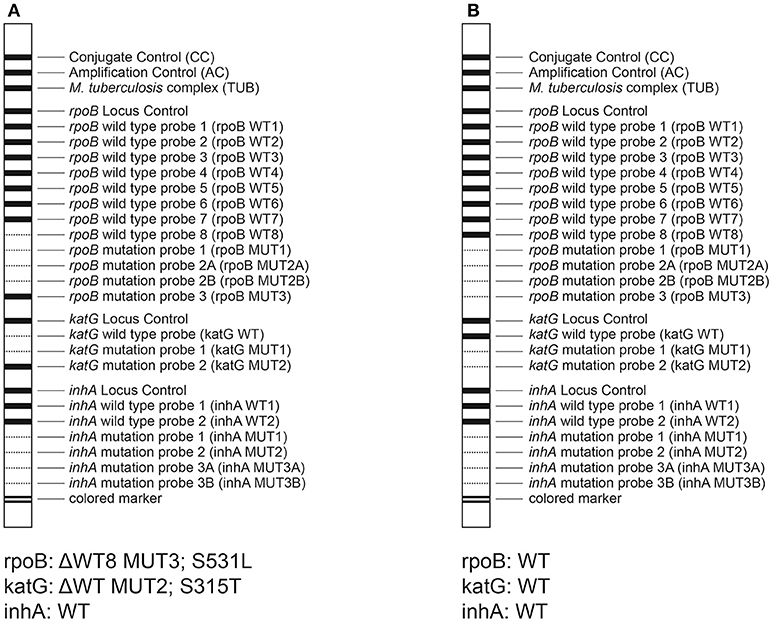
Figure 1. Representative patterns obtained by the MTBDRplus assay. (A) isolate 2A12, RIFr, and INHr; (B) isolate A16, RIFs, and INHs.
Detailed patterns of drug resistance are given in Table S2. Resistance to INH was detected in 38 (38/53; 71.7%) isolates. Of these, all but one (37/53; 69.8%) had S315T (MUT1) alteration in the katG gene along with a WT band present (21/53; 39.6%) or absent ΔWT (16/53; 30.2%). Four (4/53; 7.5%) isolates had mutations in the inhA promoter. Single polymorphism C-15T occurred in three (3/53; 5.6%) isolates (MUT1+ΔWT1, 2/53, 3.8%; MUT1 coupled with WT bands, 1/53, 1.9%). One (1/53; 1.9%) isolate harbored two substitutions, i.e., C-15T and T-8C (MUT1 and MUT3A, with all WT bands present). All but one isolates with mutations in the inhA promoter had their katG alleles unreactive to mutant probes.
GenoType MTBDRplus identified 42 (42/53; 79.2%) isolates as RIF-resistant. The most prevalent mutation pattern was S531L (MUT3) found in 34 (34/53; 64.1%) isolates, coupled with either ΔWT8 (S531L; 32/53; 60.4%), ΔWT3ΔWT4 (D516Y/Δ515; 1/53; 1.9%) or all WT bands (1/53; 1.9%). Five (5/53; 9.4%) and two (2/53; 3.8%) isolates carried mutations in codons 515–516 (ΔWT3ΔWT4, D516Y/Δ515, 3/53, 5.7%; MUT1+ΔWT3ΔWT4, D516V+D516Y/Δ515, 1/53, 1.9%; MUT1+ΔWT7, D516V+H526R/P/W/N/L/S/C, 1/53, 1.9%) and 526 (ΔWT7, H526R/P/W/N/L/S/C), respectively. One (1/53; 1.9%) isolate had S522L/Q (ΔWT5ΔWT6) and L533P/S531Q/W (ΔWT8) mutations.
Concordance between DST results obtained with the MTBDRplus assay and conventional methods is shown in Table 3. The sensitivity of the MTBDRplus assay was calculated at 79.2% and 71.7% for the detection of RIF and INH resistance, respectively.
Performance of Genotype MTBDRsl
The results of DST with the GenoType MTBDRsl are shown in Table 2. Detailed patterns of drug resistance are given in Table S2.
Resistance to FQ was detected in 39 (39/53; 73.6%) isolates. Mutations A90V (MUT1+ΔWT2) and D94G (MUT3C+ΔWT3) in the gyrA gene occurred with equal frequencies, that is in 13 (13/53; 24.5%) isolates, each. The S91P (MUT2+ΔWT2 or MUT2 and all WT bands), D94N/Y (MUT3B+ΔWT3 or MUT3B and all WT bands), D94H (MUT3D+ΔWT3), and D94A (MUT3A+ΔWT3) alterations were detected in 3 (3/53; 5.6%), 3 (3/53; 5.6%), 2 (2/53; 3.8%), and 2 (2/53; 3.8%) isolates, respectively. Three (3/53; 5.6%) isolates carried a double amino acid change; two isolates had A90V and D94G (MUT1+MUT3C, all WT bands) mutations, whereas one had D94N/Y and D94G (MUT3B+MUT3C+ΔWT3) mutations.
Resistance to EMB was detected in 33 (33/53; 63.3%) isolates. The most prevalent mutation was M306I, found in 18 (18/33; 54.5%) isolates, of which 9 (9/33; 27.3%) had MUT1A+ΔWT and another 9 had ΔWT banding patterns. Fifteen (15/33; 45.5%) isolates harbored M306V (MUT1B+ΔWT) alteration. Of two isolates, identified as EMB-susceptible, upon conventional DST, one was recognized as EMB-resistant with the assay (ΔWT pattern).
Out of six isolates resistant to SLIDs upon conventional DST, only one (1/53; 1.9%) was found resistant (KAN+AMK+CAP) with the MTBDRsl assay. This isolate carried a A1401G (MUT1+ΔWT1) mutation. Seven (7/53; 13.2%) isolates were designated as KAN-, AMK- and/or CAP-resistant, despite being susceptible to those drugs upon culture-based DST. Mutations A1401G (MUT1+ΔWT1 or MUT1 and all WT bands) or G1484T (MUT2+ΔWT2) were found in five (5/7; 71.4%) and two (2/7; 28.6%) of these isolates, respectively.
Overall, GenoType MTBDRsl had sensitivities of 73.6, 64.7, 20, 25, and 100% for the detection of FQ, EMB, KAN, AMK, and CAP resistance. Whereas the specificities ranged from 90.2% for CAP, 88.9% for AMK, 85.4% for KAN to 50% for EMB (Table 2).
Mutation Profiling
Of the 11 (11/53; 20.8%) isolates declared as RIF-resistant but RIF-susceptible with the conventional and GenoType MTBDRplus assay, respectively, three (3/11; 27.3%) harbored non-synonymous alterations S531L, S531W, and I572F upon rpoB gene sequencing. Another seven (7/11; 63.6%) isolates had silent mutations, and one (1/11; 9.1%) isolate had a WT rpoB sequence (Table 2, Table S2).
Among 15 (15/53; 28.3%) isolates in which MTBDRplus assay failed to detect INH resistance, 14 (14/15; 93.3%) had amino acid replacements in the katG gene evidenced by sequencing. The mutation profiles were as follows: R463L (10/15; 66.7%), R463L and S315T (1/15; 6.7%), R463L and S315N (1/15; 6.7%), R463L and P232R (1/15; 6.7%), or R463L and D189Y (1/15; 6.7%). None of the analyzed isolates carried polymorphisms in the inhA promoter and structural gene.
Fourteen (14/53; 26.4%) isolates designated as FQ-resistant with conventional DST, showed FQ susceptibility with the GenoType MTBDRsl assay. Sequence analysis of the gyrA gene revealed that all these isolates carried a S95T substitution, which in case of five (5/14; 35.7%) isolates co-occurred with either A90V, S91P, D94A, D94G, or D94Y polymorphism.
Among 18 isolates for which MTBDRsl assay failed to confirm phenotype-based resistance to EMB, 14 (14/18; 77.8%) had mutations in the embB gene, as shown upon sequence analysis. These mutations translated into the following amino acid alterations: Q497P (2/18; 11.1%), Q497K (2/18; 11.1%), G406D (2/18; 11.1%), Q497R (1/18; 5.5%), G406A (1/18; 5.5%), G406S (1/18; 5.5%), D328I (1/18; 5.5%), D328Y (1/18; 5.5%), M306V (1/18; 5.5%), I284V+C549W (1/18; 5.5%), and D328Y+E378A (1/18; 5.5%). One isolate declared as EMB-susceptible upon conventional DST and as EMB-resistant with GenoType MTBDRsl, carried M306L mutation.
Among five isolates phenotypically resistant to SLIDs yet missed by the GenoType MTBDRsl assay, only one (1/5; 20%) carried a mutation (C517T) in the rrs gene. Two (2/7; 28.6%) out of seven isolates identified as resistant to SLIDs with the GenoType MTBDRsl assay but susceptible with phenotypic DST, carried a single mutation (i.e., G482A or C517T) in their rrs genes.
Discussion
Genotype MTBDRplus and MTBDRsl have become increasingly popular in mycobacteriology laboratories serving as fast molecular assays for detection of drug resistance. However, studies exploring the diagnostic performance of these two assays in HBCs are seriously lacking. This work evaluates the usefulness of Genotype MTBDRplus and MTBDRsl for the detection of drug resistance in M. tuberculosis isolates in Pakistan. It is also the first study from Pakistan investigating drug resistance profiles of tubercle bacilli (MDR-TB isolates) with a three-pronged approach, that is conventional, culture-based method, line-probe, hybridization assays, and PCR sequencing.
The sensitivities of the Genotype MTBDRplus assay evidenced here were lower than those published previously from Pakistan (71.7 vs. 76.3 and 88.8% for INH; 79.2 vs. 90.2 and 92.5% for RIF) (Farooqi et al., 2012; Javaid et al., 2016) and other countries (88–94% for INH; 95–97% for RIF) (Bai et al., 2016).
Most of the isolates had S315T and S531L mutations in the katG and rpoB genes, respectively. This is in line with what was observed among MDR isolates by using not only line-probe assays (69.8 vs. 55.9–90.6% for S315T katG; 64.1 vs. 41.4–67.2% for S531L rpoB) (Farooqi et al., 2012; Shubladze et al., 2013; Sharma et al., 2014; Javaid et al., 2016; Spinato et al., 2016) but also sequencing strategy (66–76% for S315T katG; 46.3–66% for S531L rpoB) (Jou et al., 2005; Ali et al., 2011; Makadia et al., 2012; Jagielski et al., 2015; Unissa et al., 2015). Both these mutations are thought to be low fitness cost mutations, with no adverse effect on transmission capacity (Gagneux, 2009).
Among isolates in which Genotype MTBDRplus assay failed to detect INH resistance, all but one had a missense mutation in codon 463 of the katG gene, as evidenced by PCR-sequencing. This codon is not covered by any of the probe of the Genotype MTBDRplus assay. Mutations at codon 463 of the katG gene have been identified in both INH-resistant (29–59.2%) and INH-susceptible (32–63%) strains, suggesting that they not directly associated with INH resistance (Van Doorn et al., 2001; Arjomandzadegan et al., 2011; Torres et al., 2015). One isolate carried a katG S315N mutation, which had previously been correlated with INH-resistant phenotype (Wei et al., 2003). Since the Genotype MTBDRplus test strip has no probe specific for this mutation, it cannot be detected with the assay. Still, almost a fourth (23.8%) of INH-resistant isolates can harbor this mutation (Jin et al., 2012). Apart from codons 315 and 463, mutations in the katG gene were identified at two other codons i.e., 189 and 232. Only mutation in the latter codon (P232R) had previously been described in one INH-resistant isolate by (Greif et al., 2012), with a frequency of 2.2% among INH-resistant isolates. The precise role of these mutations in the development of INH resistance needs further investigation.
In case of RIF, three out of 11 isolates falsely designated as RIF-susceptible with GenoType MTBDRplus had missense mutations in their rpoB gene sequences i.e., S531L, S531W (in regions covered with the assay), and I572F (region not covered with the assay). I572F mutation had previously been linked with RIF resistance (Siu et al., 2011).
The sensitivities of the Genotype MTBDRsl assay were usually lower than those reported previously, falling in the ranges of 84.5–100% (vs. 73.6% in our study) for FQ, 65.2–70.9% (vs. 64.7%) for EMB, 39.6–89.2% (vs. 20%) for KAN, 77.7–93.8% (vs. 25%) for AMK, and 77.2–91.4% (vs. 100%) for CAP (Feng et al., 2013; Mao et al., 2015; Gardee et al., 2017). When compared with earlier studies from Pakistan, the sensitivities of Genotype MTBDRsl from this study were lower for AMK (56.6 vs. 25%) and EMB (81.8 vs. 64.7%), albeit similar for FQ (72.9 vs. 73.6%).
The most prevalent mutations involved in FQ, EMB, and SLID resistance were D94G (gyrA), M306I (embB), and A1401G (rrs), respectively, with their frequencies ranging from 20 to 35%. This is consistent with previous observations, where mutations D94G and M306I accounted for 32% of ciprofloxacin (CIP)-resistant and 35% of EMB-resistant isolates from Pakistan (Ali et al., 2011, 2015) and 21–32% of FQ-resistant and 22–68% of EMB-resistant isolates from other geographical locales (Bakuła et al., 2013; Avalos et al., 2015; Brossier et al., 2015). The prevalence of A1401G mutants among SLID-resistant isolates from Pakistan was within ranges of 22.2–78.4% (AMK), 22.2–78.4% (KAN), and 20–78.6% (CAP) (Ali et al., 2011, 2015). Globally, the cumulative frequency of that mutation (A1401G), based on a recent meta-analytical study, was 78, 76, and 56% among AMK-, CAP-, and KAN-resistant isolates (Georghiou et al., 2012).
All fourteen isolates falsely designated as FQ-susceptible upon GenoType MTBDRsl carried a S95T gyrA substitution, which is a natural polymorphism, not associated with FQ resistance (Bakuła et al., 2016). Five isolates carried gyrA alterations missed with Genotype MTBDRsl (i.e., A90V, S91P, D94A, D94G, or D94Y). According to the literature, they have all been associated with FQ resistance.
Fourteen out of 18 isolates in which Genotype MTBDRsl failed to detect EMB resistance had a change in the embB gene, as revealed by sequence analysis. The most frequent were amino acid changes in codons 497 and 406. Mutations at both these codons were shown to increase resistance to EMB (Safi et al., 2010). Additionally, mutations in five other codons of the embB gene were identified and four of them (except in codon 284) have been described as conferring resistance to EMB (Ali et al., 2015).
Among five isolates phenotypically resistant to SLIDs but missed by GenoType MTBDRsl, only one, with resistance to KAN, carried a mutation (C517T) in the rrs gene, localized in highly mutable region known as the 530 loop (Jagielski et al., 2014). Mutations in this region are associated with resistance to STR but not to other aminoglycosides (Bakuła et al., 2016).
According to this study, both GenoType MTBDRplus and GenoType MTBDRsl, assays display an important level of inconsistency with conventional DST. When compared to the latter, the line-probe assays were unable to detect 20.7, 28.3, 26.4, 35.3, 80, and 75% of isolates resistant to RIF, INH, FQ, EMB, KAN, and AMK, respectively. These false negative results may be explained by two reasons. First is that mutations not covered by the probes might be more prevalent in Pakistan compared to other geographical locales. Inclusion of probes specific for mutations at two embB codons (Q497 and G406) would considerably increase the sensitivity of the MTBDRsl assay to detect EMB-resistant isolates from our sample (i.e., from 64.7 to 82.3%). Also, addition of other probes specific for mutations conferring resistance to INH (e.g., S315N, KatG) or RMP (e.g., I572F, RpoB) would improve the sensitivity of the MTBDRplus by approximately 2%, each (71.7 vs. 73.6% for INH; 79.2 vs. 81.1% for RMP). Poor outcomes of the GenoType MTBDRplus and MTBDRsl assays due to the lack of probes specific for certain mutations were described in previous studies (Huang et al., 2011; Jin et al., 2012; Maschmann et al., 2013). Second are possible technical errors in phenotypic and molecular assays. Factors such as handling procedures, incubation conditions, and end-point interpretation may influence the outcome of conventional DST (Schön et al., 2017). Whereas, losses on DNA extraction or inhibitors of amplification present in the specimens may affect the efficiency of line probe assays (Mäkinen et al., 2002; Padilla et al., 2004). In this study they missed 10 mutations detected by sequencing of the katG (S315T, one isolate, 1.9%), rpoB (S531L or S531W, two isolates, 3.8%), gyrA (A90V, S91P, D94A, D94G or D94Y, five isolates, 9.4%), and embB (M306V, one isolate, 1.9%) genes. Third, drug resistance may originate from mutations at other genes than those included in the probe-line assays (e.g., in ahpC, kasA, and ndh for INH) (Ferro et al., 2013) and this may also result in false negative results.
Finally, false positive results were noted for 50, 11.1, 14.5, and 9.7% of isolates declared as susceptible to EMB, AMK, KAN, and CAP, respectively, with conventional DST. This can be explained by probe mispriming (due to sample contamination or suboptimal procedure conditions) or the presence of synonymous mutations (Ajileye et al., 2017).
To conclude, for detecting drug resistance in TB cases, especially in high TB incidence countries, such as Pakistan, molecular approaches should still be a complement rather than a replacement to conventional DST. The knowledge on frequencies of drug-resistance conferring mutations in clinically and geographically diverse settings, should guide the inclusion of new specific probes in the test strips in the line-probe assays, such as Genotype MTBDR.
Author Contributions
HJ executed the strain isolation and culturing, analyzed and interpreted the data, and wrote the manuscript. ZB analyzed and interpreted the data, and wrote the manuscript. MP executed the Genotype MTBDRplus and MTBDRsl analysis, and the sequence analysis. HH performed the species-level identification and drug susceptibility testing. ZT executed the administrative, the technical, and the material support. NJ critically revised the manuscript. TJ designed the study and supervised and wrote and critically revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02265/full#supplementary-material
References
Abebe, G., Paasch, F., Apers, L., Rigouts, L., and Colebunders, R. (2011). Tuberculosis drug resistance testing by molecular methods: opportunities and challenges in resource limited settings. J. Microbiol. Methods 84, 155–160. doi: 10.1016/j.mimet.2010.11.014
Ajileye, A., Alvarez, N., Merker, M., Walker, T. M., Akter, S., Brown, K., et al. (2017). Some synonymous and nonsynonymous gyrA mutations in Mycobacterium tuberculosis lead to systematic false-positive fluoroquinolone resistance results with the Hain GenoType MTBDRsl assays. Antimicrob. Agents Chemother. 61:e02169–16. doi: 10.1128/AAC.02169-16
Ali, A., Hasan, R., Jabeen, K., Jabeen, N., Qadeer, E., and Hasan, Z. (2011). Characterization of mutations conferring extensive drug resistance to Mycobacterium tuberculosis isolates in Pakistan. Antimicrobial. Agents Chemother. 55, 5654–5659. doi: 10.1128/AAC.05101-11
Ali, A., Hasan, Z., McNerney, R., Mallard, K., Hill-Cawthorne, G., Coll, F., et al. (2015). Whole genome sequencing based characterization of extensively drug-resistant Mycobacterium tuberculosis isolates from Pakistan. PLoS ONE 10:e0117771. doi: 10.1371/journal.pone.0117771
Anek-vorapong, R., Sinthuwattanawibool, C., Podewils, L. J., McCarthy, K., Ngamlert, K., Promsarin, B., et al. (2010). Validation of the GenoType®; MTBDR plus assay for detection of MDR-TB in a public health laboratory in Thailand. BMC Infect. Dis. 10:123. doi: 10.1186/1471-2334-10-123
Arjomandzadegan, M., Owlia, P., Ranjbar, R., Farazi, A. A., Sofian, M., Sadrnia, M., et al. (2011). Prevalence of mutations at codon 463 of katG gene in MDR and XDR clinical isolates of Mycobacterium tuberculosis in Belarus and application of the method in rapid diagnosis. Acta Microbiol. Immunol. Hung. 58, 51–63. doi: 10.1556/AMicr.58.2011.1.6
Avalos, E., Catanzaro, D., Catanzaro, A., Ganiats, T., Brodine, S., Alcaraz, J., et al. (2015). Frequency and geographic distribution of gyrA and gyrB mutations associated with fluoroquinolone resistance in clinical Mycobacterium tuberculosis isolates: a systematic review. PLoS ONE 10:e0120470. doi: 10.1371/journal.pone.0120470
Bai, Y., Wang, Y., Shao, C., Hao, Y., and Jin, Y. (2016). GenoType MTBDRplus assay for rapid detection of multidrug resistance in Mycobacterium tuberculosis: a meta-analysis. PLoS ONE 11:e015032. doi: 10.1371/journal.pone.0150321
Bakuła, Z., Napiórkowska, A., Bielecki, J., Augustynowicz-Kopeć, E., Zwolska, Z., and Jagielski, T. (2013). Mutations in the embB gene and their association with ethambutol resistance in multidrug-resistant Mycobacterium tuberculosis clinical isolates from Poland. Biomed. Res. Int. 2013:167954. doi: 10.1155/2013/167954
Bakuła, Z., Napiórkowska, A., Kaminski, M., Augustynowicz-Kopeć, E., Zwolska, Z., Bielecki, J., et al. (2016). Second-line anti-tuberculosis drug resistance and its genetic determinants in multidrug-resistant Mycobacterium tuberculosis clinical isolates. J. Microbiol. Immunol. Infect. 49, 439–444. doi: 10.1016/j.jmii.2015.04.003
Bernard, C., Veziris, N., Brossier, F., Sougakoff, W., Jarlier, V., Robert, J., et al. (2015). Molecular diagnosis of fluoroquinolone resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 59, 1519–1524. doi: 10.1128/AAC.04058-14
Brossier, F., Sougakoff, W., Bernard, C., Petrou, M., Adeyema, K., Pham, A., et al. (2015). Molecular analysis of the embCAB locus and embR gene involved in ethambutol resistance in clinical isolates of Mycobacterium tuberculosis in France. Antimicrob. Agents Chemother. 59, 4800–4808. doi: 10.1128/AAC.00150-15
Domínguez, J., Boettger, E., Cirillo, D., Cobelens, F., Eisenach, K., Gagneux, S., et al. (2016). Clinical implications of molecular drug resistance testing for Mycobacterium tuberculosis: a TBNET/RESIST-TB consensus statement. Int. J. Tuberculosis Lung Dis. 20, 24–42. doi: 10.5588/ijtld.15.0221
Farooqi, J. Q., Khan, E., Alam, S. M., Ali, A., Hasan, Z., and Hasan, R. (2012). Line probe assay for detection of rifampicin and isoniazid resistant tuberculosis in Pakistan. J. Pakistan Med. Assoc. 62, 767–772.
Feng, Y., Liu, S., Wang, Q., Wang, L., Tang, S., Wang, J., et al. (2013). Rapid diagnosis of drug resistance to fluoroquinolones, amikacin, capreomycin, kanamycin and ethambutol using genotype MTBDRsl assay: a meta-analysis. PLoS ONE 8:e55292. doi: 10.1371/journal.pone.0055292
Ferro, B. E., García, P. K., Nieto, L. M., and van Soolingen, D. (2013). Predictive value of molecular drug resistance testing of Mycobacterium tuberculosis isolates in Valle del Cauca, Colombia. J. Clin. Microbiol. 51, 2220–2224. doi: 10.1128/JCM.00429-13
Gagneux, S. (2009). Fitness cost of drug resistance in Mycobacterium tuberculosis. Clin. Microbiol. Infect. 1, 66–68. doi: 10.1111/j.1469-0691.2008.02685.x
Gardee, Y., Dreyer, A. W., Koornhof, H. J., Omar, S. V., da Silva, P., Bhyat, Z., et al. (2017). Evaluation of the genotype MTBDRsl version 2.0 assay for second-line drug resistance detection of Mycobacterium tuberculosis isolates in South Africa. J. Clin. Microbiol. 55, 791–800. doi: 10.1128/JCM.01865-16
Georghiou, S. B., Magana, M., Garfein, R. S., Catanzaro, D. G., Catanzaro, A., and Rodwell, T. C. (2012). Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: a systematic review. PLoS ONE 7:e3327. doi: 10.1371/journal.pone.0033275
Greif, G., Coitinho, C., Rivas, C., van Ingen, J., and Robello, C. (2012). Molecular analysis of isoniazid-resistant Mycobacterium tuberculosis isolates in Uruguay. Int. J. Tuberculosis Lung Dis. 16, 947–949. doi: 10.5588/ijtld.11.0559
Hillemann, D., Rüsch-Gerdes, S., and Richter, E. (2009). Feasibility of the GenoType MTBDRsl assay for fluoroquinolone, amikacin-capreomycin, and ethambutol resistance testing of Mycobacterium tuberculosis strains and clinical specimens. J. Clin. Microbiol. 47, 1767–1772. doi: 10.1128/JCM.00081-09
Huang, W. L., Chi, T. L., Wu, M. H., and Jou, R. (2011). Performance assessment of the GenoType MTBDRsl test and DNA sequencing for detection of second-line and ethambutol drug resistance among patients infected with multidrug-resistant Mycobacterium tuberculosis. J. Clin. Microbiol. 49, 2502–2508. doi: 10.1128/JCM.00197-11
Ioannidis, P., Papaventsis, D., Karabela, S., Nikolaou, S., Panagi, M., Raftopoulou, E., et al. (2011). Cepheid GeneXpert MTB/RIF assay for Mycobacterium tuberculosis detection and rifampin resistance identification in patients with substantial clinical indications of tuberculosis and smear-negative microscopy results. J. Clin. Microbiol. 49, 3068–3070. doi: 10.1128/JCM.00718-11
Jagielski, T., Bakuła, Z., Roeske, K., Kaminski, M., Napiórkowska, A., Augustynowicz-Kopeć, E., et al. (2015). Mutation profiling for detection of isoniazid resistance in Mycobacterium tuberculosis clinical isolates. J. Antimicrob. Chemother. 70, 3214–3212. doi: 10.1093/jac/dkv253
Jagielski, T., Ignatowska, H., Bakuła, Z., Dziewit, L., Napiórkowska, A., Augustynowicz-Kopeć, E., et al. (2014). Screening for streptomycin resistance-conferring mutations in Mycobacterium tuberculosis clinical isolates from Poland. PLoS ONE 9:e100078. doi: 10.1371/journal.pone.0100078
Jagielski, T., Minias, A., Van Ingen, J., Rastogi, N., Brzostek, A., Zaczek, A., et al. (2016). Methodological and clinical aspects of the molecular epidemiology of Mycobacterium tuberculosis and other mycobacteria. Clin. Microbiol. Rev. 29, 239–290. doi: 10.1128/CMR.00055-15
Javaid, M., Ahmed, A., Asif, S., and Raza, A. (2016). Diagnostic plausibility of MTBDRplus and MTBDRsl line probe assays for rapid drug susceptibility testing of drug resistant Mycobacterium tuberculosis Strains in Pakistan. Int. J. Infect. 3:e34903. doi: 10.17795/iji-34903
Jin, J., Zhang, Y., Fan, X., Diao, N., Shao, L., Wang, F., et al. (2012). Evaluation of the GenoType®; MTBDRplus assay and identification of a rare mutation for improving MDR-TB detection. Int. J. Tuberculosis Lung Dis. 16, 521–526. doi: 10.5588/ijtld.11.0269
Jou, R., Chen, H. Y., Chiang, C. Y., and Yu, M. C. (2005). Su genetic diversity of multidrug resistant Mycobacterium tuberculosis isolates and identification of 11 novel rpoB alleles in Taiwan. J. Clin. Microbiol. 43, 1390–1394. doi: 10.1128/JCM.43.3.1390-1394.2005
Koneman, E. W., Dowell, V. R., and Sommers, H. M. (1983). Color Atlas and Text Book of Diagnostic Microbiology. Philadelphia, PA: JB Lippincott Company.
Lawn, S. D., and Nicol, M. P. (2011). Xpert®; MTB/RIF assay: development, evaluation and implementation of a new rapid molecular diagnostic for tuberculosis and rifampicin resistance. Fut. Microbiol. 6, 1067–1082. doi: 10.2217/fmb.11.84
Liu, Q., Li, G.-L., Chen, C., Wang, J.-M., Martinez, L., Lu, W., et al. (2017). Diagnostic performance of the genotype MTBDRplus and MTBDRsl assays to identify tuberculosis drug resistance in eastern china. Chinese Med. J. 130, 1521–0528. doi: 10.4103/0366-6999.208248
Makadia, J. S., Jain, A., Patra, S. K., Sherwal, B. L., and Khanna, A. (2012). Emerging trend of mutation profile of rpoB gene in MDR tuberculosis, North India. Indian J. Clin. Biochem. 27, 370–374. doi: 10.1007/s12291-012-0228-5
Mäkinen, J., Marjamäkia, M., Marttila, H., and Soinia, H. (2002). Evaluation of a novel strip test, genotype Mycobacterium CM/AS, for species identification of mycobacterial cultures. J. Clin. Microbiol. 40, 3478–3481. doi: 10.1128/JCM.40.9.3478-3481.2002
Mao, X., Ke, Z., Shi, X., Liu, S., Tang, B., Wang, J., et al. (2015). Diagnosis of drug resistance to fluoroquinolones, amikacin, capreomycin, kanamycin and ethambutol with genotype MTBDRsl assay: a meta-analysis. Ann. Clin. Lab. Sci. 45, 533–544.
Maschmann, R. A., Spies, F. S., Nunes, L. S., Ribeiro, A. W., Machado, T. M., and Zaha, A. (2013). Performance of the genotype MTBDRplus assay directly on sputum specimens from Brazilian patients with tuberculosis treatment failure or relapse. J, Clin. Microbiol. 51, 1606–1608. doi: 10.1128/JCM.00364-13
Molina-Moya, B., Lacoma, A., García-Sierra, N., Blanco, S., Haba, L., Samper, S., et al. (2017). PyroTyping, a novel pyrosequencing-based assay for Mycobacterium tuberculosis genotyping. Sci. Rep. 7:6777. doi: 10.1038/s41598-017-06760-5
Nathavitharana, R. R., Cudahy, P. G., Schumacher, S. G., Steingart, K. R., Pai, M., and Denkinger, C. M. (2017). Accuracy of line probe assays for the diagnosis of pulmonary and multidrug-resistant tuberculosis: a systematic review and meta-analysis. Eur. Respir. J. 49:1601075. doi: 10.1183/13993003.01075-2016
Padilla, E., González, V., Manterola, J. M., Pérez, A., Quesada, M. D., Gordillo, S., et al. (2004). Comparative evaluation of the new version of the INNO-LiPA Mycobacteria and genotype Mycobacterium assays for identification of Mycobacterium species from MB/BacT liquid cultures artificially inoculated with Mycobacterial strains. J. Clin. Microbiol. 42, 3083–3088. doi: 10.1128/JCM.42.7.3083-3088.2004
Safi, H., Fleischmann, R. D., Peterson, S. N., Jones, M. B., Jarrahi, B., and Alland, D. (2010). Allelic exchange and mutant selection demonstrate that common clinical embCAB gene mutations only modestly increase resistance to ethambutol in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 54, 103–108. doi: 10.1128/AAC.01288-09
Satta, G., Atzeni, A., and McHugh, T. D. (2018). Mycobacterium tuberculosis and whole genome sequencing: a practical guide and online tools available for the clinical microbiologist. Clin. Microbiol. Infect. 23, 69–72. doi: 10.1016/j.cmi.2016.09.005
Schleusener, V., Köser, C. U., Beckert, P., Niemann, S., and Feuerriegel, S. (2017). Mycobacterium tuberculosis resistance prediction and lineage classification from genome sequencing: comparison of automated analysis tools. Sci. Rep. 7:46327. doi: 10.1038/srep46327
Schön, T., Miotto, P., Köser, C. U., Viveiros, M., Böttger, E., and Cambau, E. (2017). Mycobacterium tuberculosis drug-resistance testing: challenges, recent developments and perspectives. Clin. Microbiol. Infect. 23, 154–160. doi: 10.1016/j.cmi.2016.10.022
Sharma, S., Madan, M., Agrawal, C., and Asthana, A. K. (2014). Genotype MTBDR plus assay for molecular detection of rifampicin and isoniazid resistance in Mycobacterium tuberculosis. Indian J. Pathol. Microbiol. 57,423–426. doi: 10.4103/0377-4929.138738
Shubladze, N., Tadumadze, N., and Bablishvili, N. (2013). Molecular patterns of multidrug resistance of Mycobacterium tuberculosis in Georgia. Int. J. Mycobacteriol. 2, 73–78. doi: 10.1016/j.ijmyco.2013.02.002
Siu, G. K., Zhang, Y., Lau, T. C., Lau, R. W., Ho, P. L., Yew, W. W., et al. (2011). Mutations outside the rifampicin resistance-determining region associated with rifampicin resistance in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 66, 730–733. doi: 10.1093/jac/dkq519
Spinato, J., Boivin, É., Bélanger-Trudelle, É., Fauchon, H., Tremblay, C., and Soualhine, H. (2016). Genotypic characterization of drug resistant Mycobacterium tuberculosis in Quebec, 2002-2012. BMC Microbiol.16:164. doi: 10.1186/s12866-016-0786-4
Thumamo, B. P., Asuquo, A. E., Abia-Bassey, L. N., Lawson, L., Hill, V., Zozio, T., et al. (2012). Molecular epidemiology and genetic diversity of Mycobacterium tuberculosis complex in the Cross River State, Nigeria. Infect. Genetics Evol. 12, 671–677. doi: 10.1016/j.meegid.2011.08.011
Torres, J. N., Paul, L. V., Rodwell, T. C., Victor, T., Amallraja, AM1, Elghraoui, A., Goodmanson, A. P., et al. (2015). Novel katG mutations causing isoniazid resistance in clinical Mycobacterium tuberculosis isolates. Emerg. Microbes Infect. 4:e42. doi: 10.1038/emi.2015.42
Ullah, I., Javaid, A., Tahir, Z., Ullah, O., Shah, A. A., Hasan, F., et al. (2016). Pattern of drug resistance and risk factors associated with development of drug resistant Mycobacterium tuberculosis in Pakistan. PLoS ONE 11:e0147529. doi: 10.1371/journal.pone.0147529
Unissa, A. N., Selvakumar, N., Narayanan, S., Suganthi, C., and Hanna, L. E. (2015). Investigation of Ser315 substitutions within katG gene in isoniazid-resistant clinical isolates of Mycobacterium tuberculosis from south India. Biomed. Res. Int. 2015:257983. doi: 10.1155/2015/257983
Van Doorn, H. R., Kuijper, E. J., van der Ende, A., Welten, A. G., van Soolingen, D., de Haas, P. E., et al. (2001). The susceptibility of Mycobacterium tuberculosis to isoniazid and the Arg → Leu mutation at codon 463 of katG are not associated. J. Clin. Microbiol. 39, 1591–1594. doi: 10.1128/JCM.39.4.1591-1594.2001
Van Embden, J., Cave, M. D., Crawford, J. T., Dale, J., Eisenach, K., Gicquel, B., et al. (1993). Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31, 406–409.
Votintseva, A. A., Bradley, P., Pankhurst, L., Del Ojo Elias, C., Loose, M., Nilgiriwala, K., et al. (2017). Same-day diagnostic and surveillance data for tuberculosis via Whole-Genome Sequencing of direct respiratory samples. J. Clin. Microbiol. 55, 1285–1298. doi: 10.1128/JCM.02483-16
Walker, T. M., Merker, M., Kohl, T. A., Crook, D. W., Nieman, S., and Peto, T. E. A. (2017). Whole genome sequencing for M/XDR tuberculosis surveillance and for resistance testing. Clin. Microbiol. Infect. 23, 161–166. doi: 10.1016/j.cmi.2016.10.014
Wei, C. J., Lei, B., Musser, J. M., and Tu, S. C. (2003). Isoniazid activation defects in recombinant Mycobacterium tuberculosis catalase-peroxidase (KatG) mutants evident in InhA inhibitor production. Antimicrob. Agents Chemother. 47, 670–675. doi: 10.1128/AAC.47.2.670-675.2003
World Health Organization (2008). Molecular Line Probe Assays for Rapid Screening of Patients at Risk of Multidrug-Resistant Tuberculosis (MDR-TB). Geneva: World Health Organization.
Keywords: Genotype MTBDRplus, Genotype MTBDRsl, line probe assay, Mycobacterium tuberculosis, drug resistance
Citation: Javed H, Bakuła Z, Pleń M, Hashmi HJ, Tahir Z, Jamil N and Jagielski T (2018) Evaluation of Genotype MTBDRplus and MTBDRsl Assays for Rapid Detection of Drug Resistance in Extensively Drug-Resistant Mycobacterium tuberculosis Isolates in Pakistan. Front. Microbiol. 9:2265. doi: 10.3389/fmicb.2018.02265
Received: 26 February 2018; Accepted: 05 September 2018;
Published: 26 September 2018.
Edited by:
Xiao-Yong Fan, Fudan University, ChinaReviewed by:
Jason Sahl, Northern Arizona University, United StatesGurvinder Kaur, All India Institute of Medical Sciences, India
Jianping Xie, Southwest University, China
Copyright © 2018 Javed, Bakuła, Pleń, Hashmi, Tahir, Jamil and Jagielski. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomasz Jagielski, dC5qYWdpZWxza2lAYmlvbC51dy5lZHUucGw=
 Hasnain Javed1
Hasnain Javed1 Zofia Bakuła
Zofia Bakuła Małgorzata Pleń
Małgorzata Pleń Tomasz Jagielski
Tomasz Jagielski