- College of Food and Biological Engineering, Anhui Key Laboratory of Intensive Processing of Agricultural Products, Hefei University of Technology, Hefei, China
The development of lignocellulosic bioethanol plays an important role in the substitution of petrochemical energy and high-value utilization of agricultural wastes. The safe and stable expression of cellulase gene sestc was achieved by applying the clustered regularly interspaced short palindromic repeats-Cas9 approach to the integration of sestc expression cassette containing Agaricus biporus glyceraldehyde-3-phosphate-dehydrogenase gene (gpd) promoter in the Saccharomyces cerevisiae chromosome. The target insertion site was found to be located in the S. cerevisiae hexokinase 2 by designing a gRNA expression vector. The recombinant SESTC protein exhibited a size of approximately 44 kDa in the engineered S. cerevisiae. By using orange peel as the fermentation substrate, the filter paper, endo-1,4-β-glucanase, exo-1,4-β-glucanase activities of the transformants were 1.06, 337.42, and 1.36 U/mL, which were 35.3-fold, 23.03-fold, and 17-fold higher than those from wild-type S. cerevisiae, respectively. After 6 h treatment, approximately 20 g/L glucose was obtained. Under anaerobic conditions the highest ethanol concentration reached 7.53 g/L after 48 h fermentation and was 37.7-fold higher than that of wild-type S. cerevisiae (0.2 g/L). The engineered strains may provide a valuable material for the development of lignocellulosic ethanol.
Introduction
Bioethanol, as a promising alternative to petroleum resources, can be produced from lignocellulosic biomass and starch-rich plants (Kou et al., 2017). The development of lignocellulosic ethanol could be more promising in the future than food crop ethanol considering its sustainable, renewable, and environmentally friendly features (Cai et al., 2016). The conversion of lignocellulosic materials requires three key steps, namely, biomass pretreatment, saccharification, and fermentation (Chang et al., 2013). Pretreatment changes the physical and chemical properties of the raw materials. Saccharification produces fermentable sugars from cellulosic materials via enzymatic degradation, acidolysis, and ionic hydrolysis (Zhao et al., 2018). Fermentation converts D-glucose and other monosaccharides into ethanol by using microorganisms. A highly efficient transformation of lignocellulosic materials requires the integration of these three processing technologies (Koppram et al., 2014).
Orange is an important fruit around the world. During the orange processing, orange peel is removed from the flesh and discarded as a waste (Pandiarajan et al., 2018). The wasted orange peel generally decays and severely pollutes the atmosphere, soil, and water. The rational use of wasted orange peel has important value and significance (Rehan et al., 2018). The compositions of dry orange peel (w/w) are 17.5% of cellulose, 8.6% of hemicellulose, 25.4% of pectin, 6.73% of protein, 7.7% of moisture, 4.4% of ash (Miran et al., 2016), and 0.85% of lignin (Kantar et al., 2018). The cellulose can be converted into D-glucose via the hydrolysis of cellulase. Due to its lower lignin content, cellulase is more readily accessible to cellulose (Gao et al., 2014). In this study, orange peel was adopted to produce ethanol using the cellulase-engineering Saccharomyces cerevisiae. By in situ saccharification and fermentation, the production of ethanol from orange peel was investigated. Admittedly, orange peel is a representative of lignocellulosic materials. Other lignocellulosic materials such as corn straw and rice straw would be further investigated for the production of ethanol.
As an engineered host strain, S. cerevisiae can effectively produce ethanol and express heterologous proteins (Romanos et al., 1992; Ruohonen et al., 1997). However, the extremely low activities of cellulase produced by S. cerevisiae severely limit the conversion of lignocellulosic materials into fermentable sugars (Yang et al., 2018). Currently, the high cost of commercial cellulase considerably reduces the competitiveness of bioethanol in the market compared with fossil energy (Aswathy et al., 2010). The construction of engineered S. cerevisiae expressing cellulase is an important approach to degrading lignocellulosic materials (Kroukamp et al., 2018).
Previous reports on the engineered strain of cellulase genes mainly focused on the transformation of expression vectors embracing antibiotic resistance genes (Yang et al., 2016a). These expression vectors are located in the cytoplasm and are easily lost during cell proliferation. The addition of antibiotics raises the production cost, contaminates the broth, and reduces the product purity (Solange et al., 2010). Therefore, the integration of the cellulase gene into the S. cerevisiae genome without the addition of antibiotics should be an economic, safe, and feasible solution to producing bioethanol (Lee et al., 2017). As an RNA-mediated adaptive immune system, clustered regularly interspaced short palindromic repeats-Cas9 (CRISPR-Cas9) has been developed to achieve gene editing through its RNA-guided endonuclease activity (Singh et al., 2017). A segment of DNA is integrated into the host genome by an RNA-mediated approach (Mali et al., 2013). The integrated heterogenous DNA can be sustainably preserved in the engineered strain without antibiotic resistance genes (Jinek et al., 2014). Therefore, CRISPR-Cas9 is an effective approach to construct engineered polyploid industrial yeast and fungal strains (Liu et al., 2017; Lian et al., 2018).
The cellulase gene used in this study was isolated from the stomach tissue of Ampullaria gigas Spix (Yang et al., 2018). The gene encoding protein possesses three kinds of cellulase activity, namely, endo-1,4-β-glucanase (EG), exo-1,4-β-glucanase (CBH), and β-xylanase, and was named single-enzyme-system-three-cellulase (sestc). In this study, the expression cassette carrying a cellulase gene from A. gigas Spix was integrated into the S. cerevisiae chromosome by using a CRISPR-Cas9-based approach. The novel engineered strains can simultaneously express SESTC cellulase and produce ethanol by consuming the glucose obtained from the SESTC enzymatic hydrolysis. All the processes of SESTC expression, lignocellulosic saccharification, and ethanol production were investigated without the addition of any antibiotics. This study is valuable for developing lignocellulosic ethanol with a lignocellulosic material as the carbon source.
Materials And Methods
Strains, HXK2-gRNA, Cas9-NAT, and sestc Expression Cassette
The S. cerevisiae used in this study was derived from diploid industrial yeast. The Cas9 expression cassette was carried by a yeast single-copy episomal plasmid. The Cas9-NAT plasmid carrying selectable markers nourseothricin and bacterial resistance ampicillin was from Addgene and is reconstructed from the vector backbone pRS414-TEF1p-Cas9-CYC1t (Zhang et al., 2014; Chin et al., 2016). The gRNA expression vector for targeting S. cerevisiae hexokinase 2 gene (HXK2-gRNA) was constructed by amplifying gRNA-trp-HYB, which is preserved in Addgene, and using the primers shown in Table 1.
The sestc expression cassette for cellulase gene expression was isolated as follows. (1) Lentinula edodes glyceraldehyde-3-phosphate dehydrogenase (gpd) gene promoter was isolated from L. edodes genome by using L. edodes gpd promoter primers. The upstream and downstream regions of the L. edodes gpd promoter contained the restriction enzyme cutting sites of Sac I and Spe I. The L. edodes gpd promoter was inserted into the pBluescript II KS(-) plasmid by the double-enzyme digestion of Sac I, Spe I, and ligase. (2) The sestc fragment containing Spe I and BsrG I cutting sites was integrated into the plasmid containing the L. edodes gpd promoter. (3) A pair of primers were designed to amplify a complete sestc cassette embraced by the backbone of pBluescript II KS(-). The isolated sestc cassette contained the L. edodes gpd promoter, sestc, and the terminator.
CRISPR-Cas9 Approach Integrating the sestc Expression Cassette
The sestc expression cassette was integrated into the S. cerevisiae genome by knocking out the hxk2 gene via CRISPR-Cas9-directed gene disruption (Figure 1). A double-strand break was formed at the gRNA target sequence. As a donor DNA, the sestc cassette was inserted into the S. cerevisiae chromosome in the double-strand break. The specific target sequence of the sgRNA (5′-ctcattttggaacaagtcatcgg-3′) for hxk2 deletion was designed using an online software1. The sestc gene cassette was inserted into the S. cerevisiae chromosome by using the two-step CRISPR-Cas9 approach. (1) The Cas9 plasmid was integrated into the S. cerevisiae. The transformation of the Cas9 plasmid containing a nourseothricin resistance gene was performed by the LiAc-PEG method (Schiestl, 2007). Approximately 50 μL of the transformation solution was cultivated on YPD agar plates containing 100 μg/mL nourseothricin at 30°C for 2 days. The true transformants were named S. cerevisiae-Cas9. (2) The HXK2-gRNA expression plasmid containing a hygromycin B resistance gene and sestc expression cassette were synchronously transformed into S. cerevisiae-Cas9 by the LiA-PEG-mediated method (Schiestl, 2007). The transformation solution was cultivated on YPD agar plates containing 80 μg/mL nourseothricin and 200 μg/mL hygromycin B at 30°C for 2 days. The putative transformants were further identified at molecular and protein levels.
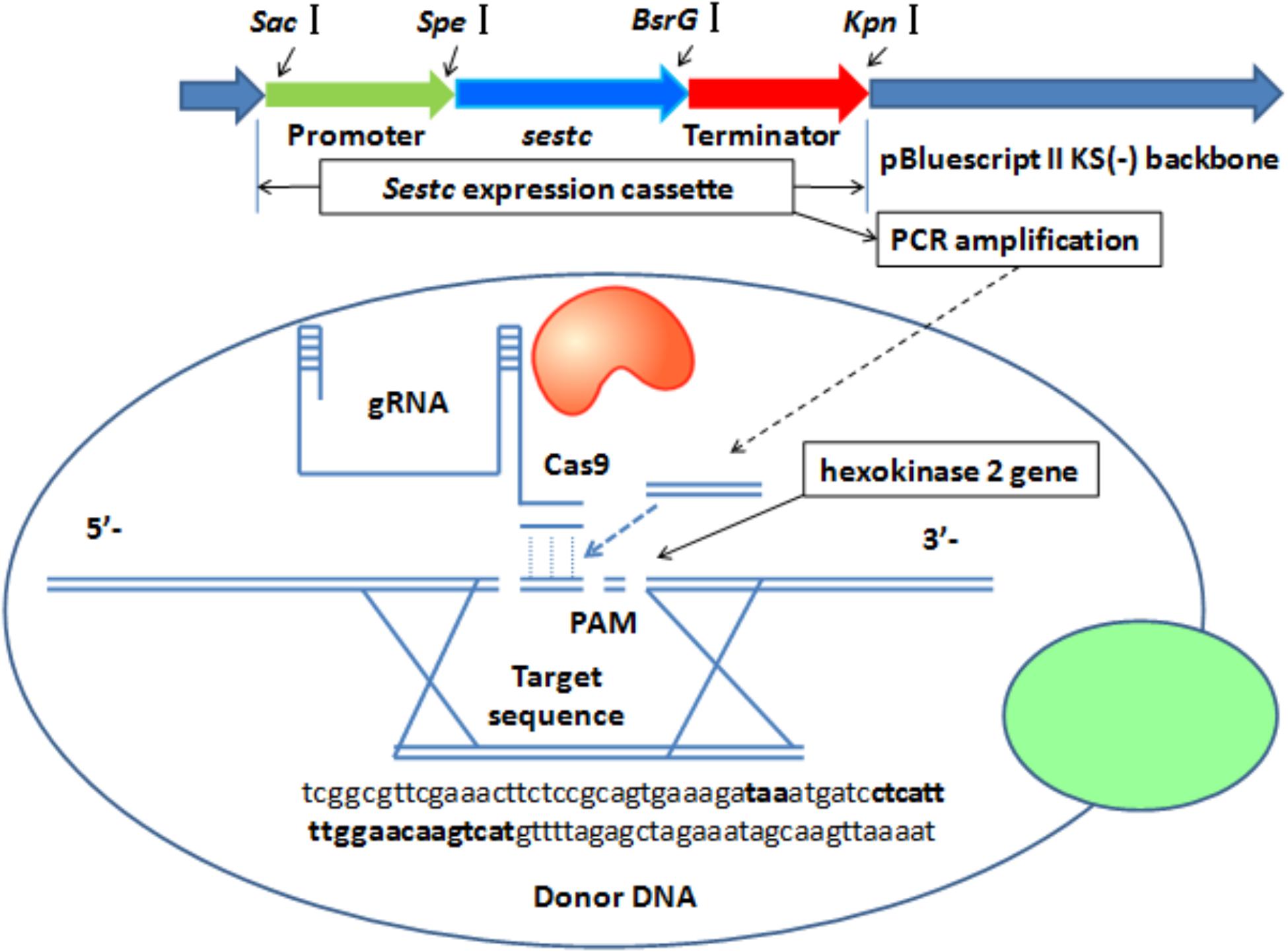
FIGURE 1. CRISPR-Cas-directed gene disruption for the integration of sestc expression cassettes in the Saccharomyces cerevisiae genome. 5′-Tcggcgttcgaaacttctccgcagtgaaagataaatgatcctcattttggaacaagtcatgttttagagctagaaatagcaagttaaaat-3′ were the partial sequences of sgRNA expression vector. Both the left and right sequences, respectively located at bold font taa and 5′-ctcattttggaacaagtcat-3′ are from S. cerevisiae chromosome. 5′-Ctcattttggaacaagtcat-3′ was designed as a 20-nt sgRNA according to S. cerevisiae hexokinase 2 gene. Termination codon taa was designed to stop the gene expression of its upstream sequences and start the synthesis of 20-nt sgRNA. Sestc expression cassette was composed with promoter gene, sestc, and terminator. The cassette amplified via PCR approach was inserted into the locus of S. cerevisiae genome cut by Cas9 Nuclease.
RT-PCR Identification and Protein Analysis of Recombinant S. cerevisiae
Before the identification, the putative transformants were cultured in liquid YPD media at 30°C and a rotational speed of 280 rpm for 3 days. The monoclonal colonies without nourseothricin and hygromycin resistance were selected for further identification. The S. cerevisiae transformant RNA was extracted using a TransZol Plant extraction kit from TransGen Biotech Co., Ltd. EasyScript First-Strand cDNA Synthesis SuperMix from TransGen Biotech Co., Ltd. was used to synthesize the complementary DNA, with the transformant RNA as the template. The designed primers based on the sestc sequences were used to amplify sestc, with wild-type S. cerevisiae as the control. The total proteins of the engineered and wild-type S. cerevisiae were analyzed by the sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) approach.
Cellulase Expression, Saccharification, and Ethanol Production
Exactly 20 mL of 20 OD600 yeast cells/mL was inoculated into a 500 mL Erlenmeyer flask loaded with 200 mL of YP medium and 10 g of oven-dried orange peel powders. After fermentation at 30°C and 180 rpm for 60 h, the cellulase activities were measured using the dinitrosalicylic acid method (Ghose, 1987). The total cellulase activity (filter paper activity, FPA), endo-β-1-4-glucanase (CMCase, EG), and exo-β-1-4-glucanase (cellobiohydrolase, CBH) were measured using Whatman grade 1 filter paper (50 mg, 1 cm × 6 cm), 0.51% CMC–Na (w/v), and 1% microcrystalline cellulose (w/v) as catalytic substrates, respectively. The saccharification process was conducted by incubating the fermentation solution at 37°C for 6 h. Subsequently, the temperature was reduced to 30°C for ethanol production under anaerobic conditions. The ethanol concentration was measured using gas chromatography with the following parameters: Agilent DB-624, FID detector, 50°C column temperature, 250°C detector temperature, 175°C injection temperature, and 1 μL injection volume. High-performance liquid chromatography (HPLC) was used to measure the content of glucose (Zhang et al., 2016).
Results
Cotransformation of Cas9-NAT, HXK2-gRNA, and sestc Expression Cassette
Cas9-NAT, HXK2-gRNA, and the sestc expression cassette were successively transformed into wild-type S. cerevisiae. After the transformation, the solution mixture was cultured on solid YPD media, which contained two antibiotics. The transformation rate of the sestc expression cassette was 37.32 ± 4.23 colonies/μg sestc expression cassettes. No colony was observed on the plates of the two negative controls. The screened colonies were used for further identification.
PCR, RT-PCR, and SDS-PAGE Identification
The colonies that lost both antibiotics were used for PCR identification by using the primers for sestc amplification (Table 1). The sizes of the putative transformant DNA bands were similar to those of the positive control. No band existed in the negative control lane (figure omitted). The positive transformants identified by PCR were further identified by the RT-PCR approach. The sizes of the DNA bands from the transformants were as expected. No DNA band existed in the wild-type S. cerevisiae lane (Figure 2). The SDS-PAGE approach was used to analyze the protein profiles of the identified transformants in comparison with the wild type S. cerevisiae (Figure 3). SESTC was approximately 44 kDa in size. Therefore, the sestc gene was effectively expressed in the engineered S. cerevisiae.
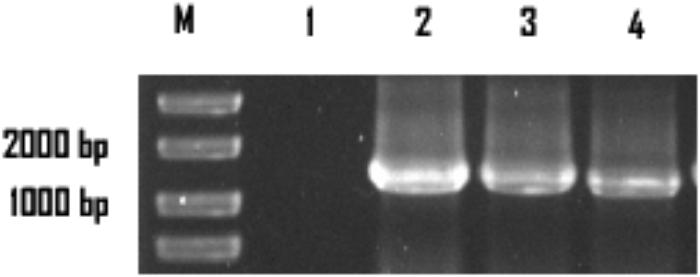
FIGURE 2. RT-PCR approach identifying the sestc S. cerevisiae transformants. Lane M marker; lane 1 wild-type S. cerevisiae; lane 2 the positive control; lane 3,4 the putative transformants.
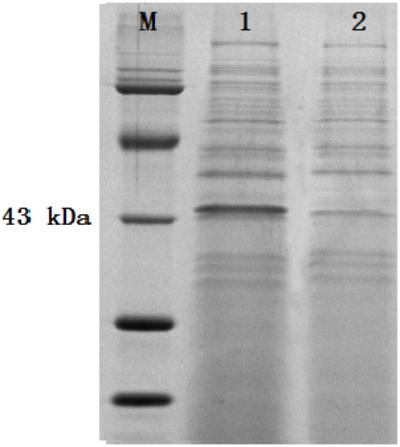
FIGURE 3. SDS-PAGE approach analyzing the total protein profiles of the identified transformants. Lane M protein marker; lane 1 sestc S. cerevisiae transformant; lane 2 wild type S. cerevisiae.
Activity Profiles of Cellulase and Saccharification for Glucose Release
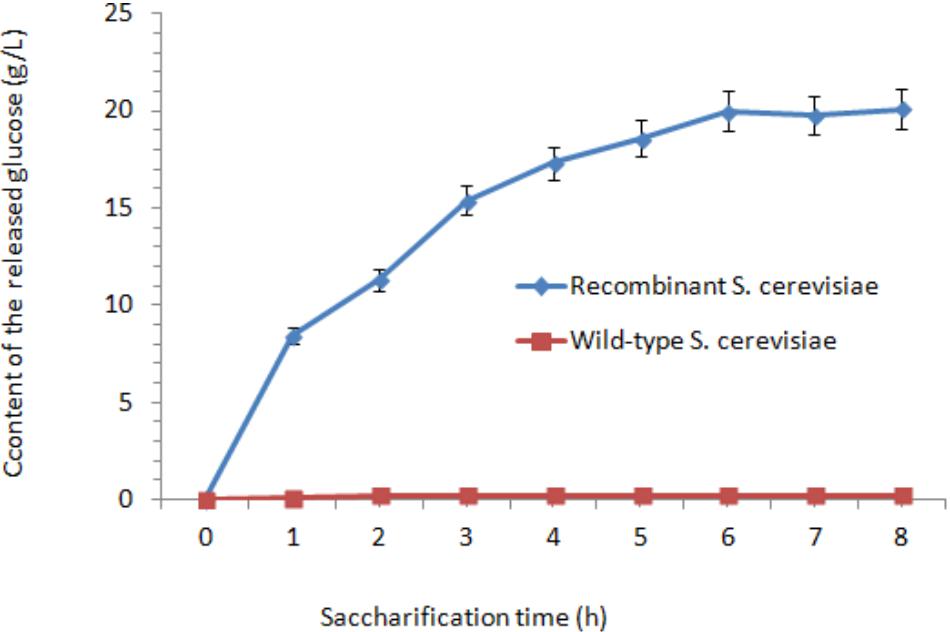
The cellulase activities in wild-type S. cerevisiae were lower than those in the transformants. After fermentation for 48 h, the cellulase activities reached their peaks. The highest FPA, EG, and CBH were 1.06, 337.42, and 1.36 U/mL, which were 35.3-fold, 23.03-fold, and 17-fold higher than those in wild-type S. cerevisiae, respectively. After incubation at 37°C for 18 h, the content of the released glucose was measured. After 6 h treatment, the concentration of glucose reached its peak (20 g/L) (Figure 4). Afterward, the released amount of glucose nearly did not increase when treated for 6–18 h. As the control, nearly no glucose was released from the orange peel, as determined by the HPLC method.
Anaerobic Fermentation for Ethanol Production
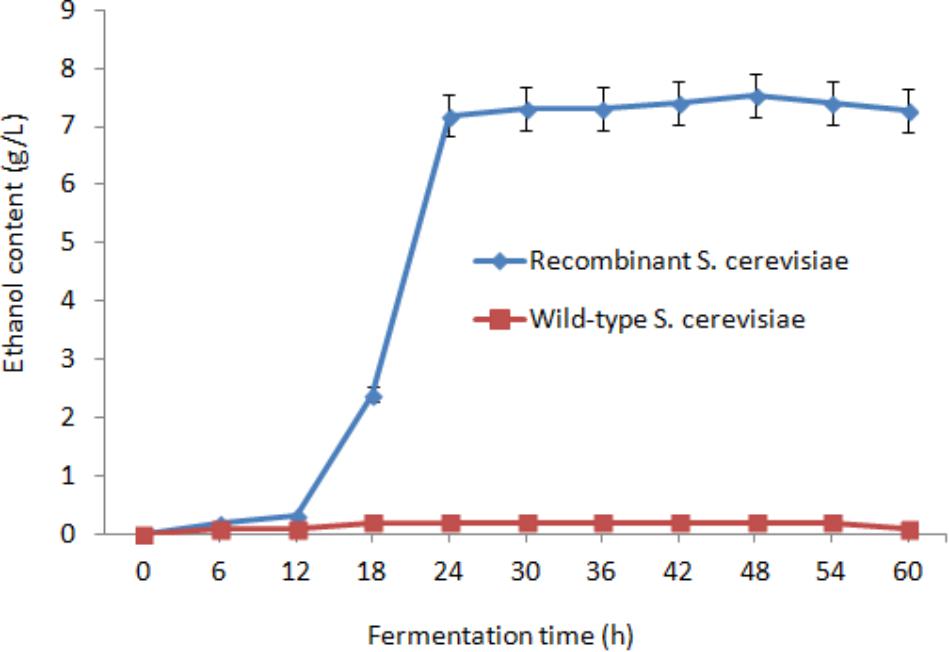
After saccharification, the fermentation mixture containing 15 OD600 of the recombinant S. cerevisiae cells, 20 g/L glucose, residual lignocellulose, and culture medium was treated at 30°C under anaerobic conditions (Figure 5). After 24 h fermentation, the ethanol concentration reached 7.19 g/L. Subsequently, the growth trend of ethanol content gradually decreased. The highest ethanol concentration reached 7.53 g/L after 48 h fermentation and was 37.7-fold higher than that of wild-type S. cerevisiae (0.2 g/L). The conversion rate of ethanol was 0.377 g/g glucose and 0.151 g/g dry orange peel.
Discussion
Lignocellulosic ethanol is an ideal alternative to petrochemical energy. The direct application of lignocellulosic materials can reduce the product cost. The reported materials were mainly from corn stover (Zhao et al., 2017), rice straw (Yu and Li, 2015), spruce (Stenberg et al., 2000), switchgrass (Wu et al., 2018), Miscanthus ×giganteus (Scordia et al., 2013; Table 2). Compared with these lignocellulosic materials, orange peel showed benefits, such as low lignin content, easy decomposition, loose structure, and high sugar content. In addition, orange peel ethanol did not require other treatments, such as steam-, acid-, and alkali-based treatments. The construction of cellulase gene-engineered S. cerevisiae was an effective approach to achieve in situ saccharification and ethanol production. S. cerevisiae coexpressing cellulase/cellodextrin and Trichoderma viride EG3/BGL1 produced ethanol contents of 4.3 g/L (Yamada et al., 2013) and 4.63 g/L (Gong et al., 2014). S. cerevisiae expressing Aspergillus aculeatus β-glucosidase achieved an ethanol content of 15 g/L (Treebupachatsakul et al., 2016). S. cerevisiae expressing Cel3A, Cel7A, and Cel5A achieved an ethanol content of 10% (w/v) (Davison et al., 2016). The current study indicated that the final ethanol concentration reached 7.53 g/L, and the conversion rates of ethanol were 0.377 g/g glucose and 0.151 g/g dry orange peel. These indices approached or exceeded those previously reported (Yamada et al., 2013; Gong et al., 2014; Yu and Li, 2015; Wu et al., 2018).
The high-value utilization of wastes from agricultural products reduces environmental pollution (Tetsuya et al., 2013). Previous studies on cellulase-engineered S. cerevisiae focused on the gene expression in the cytoplasm by using vectors that carry antibiotic resistance genes, such as nourseothricin and hygromycin B resistance genes (Koppram et al., 2014; Yang et al., 2016b). In this study, the cellulase sestc cassette was integrated into the S. cerevisiae genome by using CRISPR-Cas9 technology to achieve the stable expression of the lignocellulolytic enzyme in S. cerevisiae. The sestc gene can be expressed at the level of the S. cerevisiae genome with the gpd promoter by using orange peel as the fermentation substrate. This study provided a valuable reference on cellulase sestc, particularly its expression in cellulase and its role in ethanol production with lignocellulosic materials. However, the cellulase sestc-engineered S. cerevisiae still exhibited problems. The lack of β-glucosidase activity in the sestc-engineered strains caused insufficient substrate hydrolysis. The heterologous expression of β-glucosidase gene in the sestc-engineered strains is a reasonable solution; in addition, the supplement to β-glucosidase in the solution containing SESTC is also an effective strategy.
Conclusion
Crushed orange peel powder was used to produce ethanol via saccharification and fermentation of sestc-engineered S. cerevisiae. The highest ethanol concentration was 7.53 g/L after 48 h fermentation. This value was 37.7-fold higher than that of wild-type S. cerevisiae. The conversion rates of ethanol were 0.377 g/g glucose and 0.151 g/g dry orange peel. This study provided an effective approach to integrate cellulase sestc cassettes into the S. cerevisiae genome via the CRISPR-Cas9 approach. The constructed engineered S. cerevisiae can be applied to biomass ethanol production.
Author Contributions
PY proposed the research and wrote the manuscript. YW performed the experiments. ZZ analyzed the data. XZ made the maps. SJ designed the scheme. DM wrote the discussion section. LC measured the ethanol content.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Aswathy, U., Sukumaran, R., Devi, G., Rajasree, K., Singhania, R., and Pandey, A. (2010). Bio-ethanol from water hyacinth biomass: an evaluation of enzymatic saccharification strategy. Bioresour. Technol. 101, 925–930. doi: 10.1016/j.biortech.2009.08.019
Cai, D., Dong, Z., Wang, Y., Chen, C., Li, P., Qin, P., et al. (2016). Biorefinery of corn cob for microbial lipid and bio-ethanol production: an environmental friendly process. Bioresour. Technol. 211, 677–684. doi: 10.1016/j.biortech.2016.03.159
Chang, J., Ho, F., Ho, C., Wu, Y., Hou, Y., Huang, C., et al. (2013). Assembling a cellulase cocktail and a cellodextrin transporter into a yeast host for CBP ethanol production. Biotechnol. Biofuels 6:19. doi: 10.1186/1754-6834-6-19
Chin, Y., Kang, W., Jang, H., Turner, T., and Kim, H. (2016). CAR1 deletion by CRISPR/Cas9 reduces formation of ethyl carbamate from ethanol fermentation by Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 43, 1517–1525. doi: 10.1007/s10295-016-1831-x
Davison, S., Haan, R., and Zyl, W. (2016). Heterologous expression of cellulase genes in natural Saccharomyces cerevisiae strains. Appl. Microbiol. Biotechnol. 100, 8241–8254. doi: 10.1007/s00253-016-7735-x
Gao, D., Haarmeyer, C., Balan, V., Whitehead, T., Dale, B., and Chundawat, S. (2014). Lignin triggers irreversible cellulase loss during pretreated lignocellulosic biomass saccharification. Biotechnol. Biofuels 7:175. doi: 10.1186/s13068-014-0175-x
Ghose, T. K. (1987). Measurement of cellulase activity. Pureand Appl. Chem. 59, 257–268. doi: 10.1351/pac198759020257
Gong, Y., Tang, G., Wang, M., Li, J., Xiao, W., Lin, J., et al. (2014). Direct fermentation of amorphous cellulose to ethanol by engineered Saccharomyces cerevisiae coexpressing Trichoderma viride EG3 and BGL1. J. Gen. Appl. Microbiol. 60, 198–206. doi: 10.2323/jgam.60.198
Jinek, M., Jiang, F., Taylor, D., Sternberg, S., Kaya, E., Ma, E., et al. (2014). Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343, 1247997–1247997. doi: 10.1126/science.1247997
Kantar, S. E., Boussetta, N., Rajha, H. N., Maroun, R. G., Louka, N., and Vorobiev, E. (2018). High voltage electrical discharges combined with enzymatic hydrolysis for extraction of polyphenols and fermentable sugars from orange peels. Food Res. Int. 107, 755–762. doi: 10.1016/j.foodres.2018.01.070
Koppram, R., Elia, P., and Xiros, C. (2014). Lignocellulosic ethanol production at high-gravity: challenges and perspectives. Trends Biotechnol. 32, 46–53. doi: 10.1016/j.tibtech.2013.10.003
Kou, L., Song, Y., Zhang, X., and Tan, T. (2017). Comparison of four types of energy grasses as lignocellulosic feedstock for the production of bio-ethanol. Bioresour. Technol. 241, 424–429. doi: 10.1016/j.biortech.2017.04.078
Kroukamp, H., Haan, R., Zyl, J., and Zyl, W. (2018). Rational strain engineering interventions to enhance cellulase secretion by Saccharomyces cerevisiae. Biofuels Bioprod. Biorefin. 12, 108–124. doi: 10.1002/bbb.1824
Lee, Y., Jin, Y., Cha, Y., and Seo, J. (2017). Bioethanol production from cellulosic hydrolysates by engineered industrial Saccharomyces cerevisiae. Bioresour. Technol. 228, 355–361. doi: 10.1016/j.biortech.2016.12.042
Lian, J., Bao, Z., and Zhao, H. (2018). Engineered CRISPR/Cas9 system for multiplex genome engineering of polyploid industrial yeast strains. Biotechnol. Bioeng. 115, 1630–1635. doi: 10.1002/bit.26569
Liu, Q., Gao, R., Li, J., Zhao, J., Sun, W., and Tian, C. (2017). Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol. Biofuels 10:1. doi: 10.1186/s13068-016-0693-9
Mali, P., Yang, L., Esvelt, M., Aach, J., Guell, M., Dicarlo, J., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. doi: 10.1126/science.1232033
Miran, W., Nawaz, M., Jang, J., and Lee, D. (2016). Conversion of orange peel waste biomass to bioelectricity using a mediator-less microbial fuel cell. Sci. Total Environ. 547, 197–205. doi: 10.1016/j.scitotenv.2016.01.004
Pandiarajan, A., Kamaraj, R., Vasudevan, S., and Vasudevan, S. (2018). OPAC (orange peel activated carbon) derived from waste orange peel for the adsorption of chlorophenoxyacetic acid herbicides from water: adsorption isotherm, kinetic modelling and thermodynamic studies. Bioresour. Technol. 261, 329–334. doi: 10.1016/j.biortech.2018.04.005
Rehan, M., Abdel-Wahed, N. A. M., Farouk, A., and El-Zawahry, M. M. (2018). Extraction of valuable compounds from orange peel waste for advanced functionalization of cellulosic surfaces. ACS Sustain. Chem. Eng. 6, 5911–5928. doi: 10.1021/acssuschemeng.7b04302
Romanos, M., Scorer, C., and Clarke, J. (1992). Foreign gene expression in yeast: a review. Yeast 8, 423–488. doi: 10.1002/yea.320080602
Ruohonen, L., Toikkanen, J., Tieaho, V., Outola, M., Soderlund, H., and Keranen, S. (1997). Enhancement of protein secretion in Saccharomyces cerevisiae by overproduction of Sso protein, a late-acting component of the secretory pathway. Yeast 13, 337–351. doi: 10.1002/(SICI)1097-0061(19970330)13:4<337::AID-YEA98>3.0.CO;2-K
Schiestl, R. (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34. doi: 10.1038/nprot.2007.13
Scordia, D., Cosentino, S., and Jeffries, T. (2013). Effectiveness of dilute oxalic acid pretreatment of Miscanthus x giganteus biomass for ethanol production. Biomass Bioenergy 59, 540–548. doi: 10.1016/j.biombioe.2013.09.011
Singh, V., Braddick, D., and Dhar, P. (2017). Exploring the potential of genome editing CRISPR-Cas9 technology. Gene 599, 1–18. doi: 10.1016/j.gene.2016.11.008
Solange, M., Giuliano, D., Pedro, M., Guimarães, J., Lívia, M., Inês, C., et al. (2010). Technological trends, global market, and challenges of bio-ethanol production. Biotechnol. Adv. 28, 817–830. doi: 10.1016/j.biotechadv.2010.07.001
Stenberg, K., Bollok, M., Reczey, K., Galbe, M., and Zacchi, G. (2000). Effect of substrate and cellulase concentration on simultaneous saccharification and fermentation of steam-pretreated softwood for ethanol production. Biotechnol. Bioeng. 68, 204–210. doi: 10.1002/(SICI)1097-0290(20000420)68:2<204::AID-BIT9>3.0.CO;2-4
Tang, H., Hou, J., Shen, Y., Xu, L., Yang, H., Fang, X., et al. (2013). High beta-glucosidase secretion in Saccharomyces cerevisiae improves the efficiency of cellulase hydrolysis and ethanol production in simultaneous saccharification and fermentation. J. Microbiol. Biotechnol. 23, 1577–1585. doi: 10.4014/jmb.1305.05011
Tetsuya, G., Kanako, N., Masaharu, T., Hiroyuki, I., and Shinichi, Y. (2013). Ethanol fermentation from xylose by metabolically engineered strains of Kluyveromyces marxianus. J. Biosci. Bioeng. 116, 551–554. doi: 10.1016/j.jbiosc.2013.05.010
Treebupachatsakul, T., Nakazawa, H., Shinbo, H., Fujikawa, H., Nagaiwa, A., Ochiai, N., et al. (2016). Heterologously expressed Aspergillus aculeatus beta-glucosidase in Saccharomyces cerevisiae is a cost-effective alternative to commercial supplementation of beta-glucosidase in industrial ethanol production using Trichoderma reesei cellulases. J. Biosci. Bioeng. 121, 27–35. doi: 10.1016/j.jbiosc.2015.05.002
Wu, W., Rondon, V., Weeks, K., Pullammanappallil, P., Ingram, L., and Shanmugam, K. (2018). Phosphoric acid based pretreatment of switchgrass and fermentation of entire slurry to ethanol using a simplified process. Bioresour. Technol. 251, 171–180. doi: 10.1016/j.biortech.2017.12.041
Yamada, R., Nakatani, Y., Ogino, C., and Kondo, A. (2013). Efficient direct ethanol production from cellulose by cellulase- and cellodextrin transporter-co-expressing Saccharomyces cerevisiae. AMB Express 3:34. doi: 10.1186/2191-0855-3-34
Yang, P., Zhang, H., Cao, L., Zheng, Z., and Jiang, S. (2016a). Construction of Aspergillus niger integrated with cellulase gene from Ampullaria gigas spix for improved enzyme production and saccharification of alkaline-pretreated rice straw. 3 Biotech 6, 1–10. doi: 10.1007/s13205-016-0545-0
Yang, P., Zhang, H., and Jiang, S. (2016b). Construction of recombinant sestc Saccharomyces cerevisiae for consolidated bioprocessing, cellulase characterization, and ethanol production by in situ fermentation. 3 Biotech 6:192. doi: 10.1007/s13205-016-0512-9
Yang, P., Zhang, H., Cao, L., Zheng, Z., Mu, D., Jiang, S., et al. (2018). Combining sestc engineered A. niger with sestc engineered S. cerevisiae to produce rice straw ethanol via step-by-step and in situ saccharification and fermentation. 3 Biotech 8, 1–8. doi: 10.1007/s13205-017-1021-1
Yu, H., and Li, X. (2015). Alkali-stable cellulase from a halophilic isolate. Biomass Bioenergy 81, 19–25. doi: 10.1007/s10529-011-0698-1
Zhang, G., Kong, I., Kim, H., Liu, J., Cate, J., and Jin, Y. (2014). Construction of a quadruple auxotrophic mutant of an industrial polyploid saccharomyces cerevisiae strain by using RNA-guided Cas9 nuclease. Appl. Environ. Microbiol. 80, 7694–7701. doi: 10.1128/AEM.02310-14
Zhang, G., Kong, I., Wei, N., Peng, D., Turner, T., Sung, B., et al. (2016). Optimization of an acetate reduction pathway for producing cellulosic ethanol by engineered yeast. Biotechnol. Bioeng. 113, 2587–2596. doi: 10.1002/bit.26021
Zhao, C., Zou, Z., Li, J., Jia, H., Liesche, J., and Chen, S. (2018). Efficient bioethanol production from sodium hydroxide pretreated corn stover and rice straw in the context of on-site cellulase production. Renew. Energy 118, 14–24. doi: 10.1016/j.renene.2017.11.001
Keywords: biomass ethanol, cellulase, Saccharomyces cerevisiae, CRISPR-Cas9, orange peel, sestc
Citation: Yang P, Wu Y, Zheng Z, Cao L, Zhu X, Mu D and Jiang S (2018) CRISPR-Cas9 Approach Constructing Cellulase sestc-Engineered Saccharomyces cerevisiae for the Production of Orange Peel Ethanol. Front. Microbiol. 9:2436. doi: 10.3389/fmicb.2018.02436
Received: 04 July 2018; Accepted: 24 September 2018;
Published: 10 October 2018.
Edited by:
Fernando Segato, Universidade de São Paulo, BrazilReviewed by:
Tikam Chand Dakal, Manipal University Jaipur, IndiaFabiano Jares Contesini, Universidade Estadual de Campinas, Brazil
Copyright © 2018 Yang, Wu, Zheng, Cao, Zhu, Mu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peizhou Yang, eWFuZ3BlaXpob3VAMTYzLmNvbQ==; eWFuZ3BlaXpob3VAaGZ1dC5lZHUuY24=
 Peizhou Yang
Peizhou Yang Yun Wu
Yun Wu