Abstract
Penicillium chrysogenum (renamed P. rubens) is the most studied member of a family of more than 350 Penicillium species that constitute the genus. Since the discovery of penicillin by Alexander Fleming, this filamentous fungus is used as a commercial β-lactam antibiotic producer. For several decades, P. chrysogenum was subjected to a classical strain improvement (CSI) program to increase penicillin titers. This resulted in a massive increase in the penicillin production capacity, paralleled by the silencing of several other biosynthetic gene clusters (BGCs), causing a reduction in the production of a broad range of BGC encoded natural products (NPs). Several approaches have been used to restore the ability of the penicillin production strains to synthetize the NPs lost during the CSI. Here, we summarize various re-activation mechanisms of BGCs, and how interference with regulation can be used as a strategy to activate or silence BGCs in filamentous fungi. To further emphasize the versatility of P. chrysogenum as a fungal production platform for NPs with potential commercial value, protein engineering of biosynthetic enzymes is discussed as a tool to develop de novo BGC pathways for new NPs.
Introduction
Since the discovery of penicillin by Alexander Fleming produced by the filamentous fungus Penicillium notatum, the genus Penicillium has been deeply studied for its capacity to produce a wide range of natural products (NPs) (secondary metabolites), many of them with biotechnological and pharmaceutical applications. P. chrysogenum (recently renamed as P. rubens) is the most relevant member of more than 354 Penicillium species that constitute the genus (Nielsen et al., 2017). Penicillium is usually found in indoor environments and associated with food spoilage. It is known as an industrial producer of β-lactam antibiotic in particularly penicillin, and current production strains result from several decades of classical strain improvement (CSI) (Gombert et al., 2011; Houbraken et al., 2011). The CSI program began in 1943 with the isolation of P. chrysogenum NRRL 1951 capable of growing in submerged cultures. This strain was subjected to a long serial process of mutations induced by 275 nm ultraviolet and X-ray irradiation, nitrogen mustard gas and nitroso-methyl guanidine exposure, single spore selection and selection for loss of pigments, improved growth in large scale industrial fermenters and enhanced levels of penicillin production. CSI programs were developed in several companies (Barreiro et al., 2012), and this has resulted in an increase of penicillin titers by at least three orders of magnitude (van den Berg, 2010). As consequence, numerous genetic modifications were introduced in P. chrysogenum. Some have been studied in detail, most notably the amplification of the penicillin biosynthetic clusters and DNA inversions in this region (Fierro et al., 1995, 2006; Barreiro et al., 2012). Although the CSI had a major impact on the production of β-lactams by P. chrysogenum, it also affected secondary metabolism in general. Indeed, a proteome analysis performed between P. chrysogenum NRRL 1951 and two derived strains (Wisconsin 54-1255 and AS-P-78) showed reduced levels of proteins related to secondary metabolism in the higher penicillin producer strains (Jami et al., 2010). Genome sequencing of P. chrysogenum Wisconsin 54-1255 revealed the presence of several secondary metabolite encoding biosynthetic gene clusters (BGCs) in addition to the penicillin cluster, most of which have only be poorly studied and remain to be characterized (Figure 1). The products of the BGCs are either nonribosomal peptides (NRPs), polyketides (PKs) or hybrid molecules.
FIGURE 1
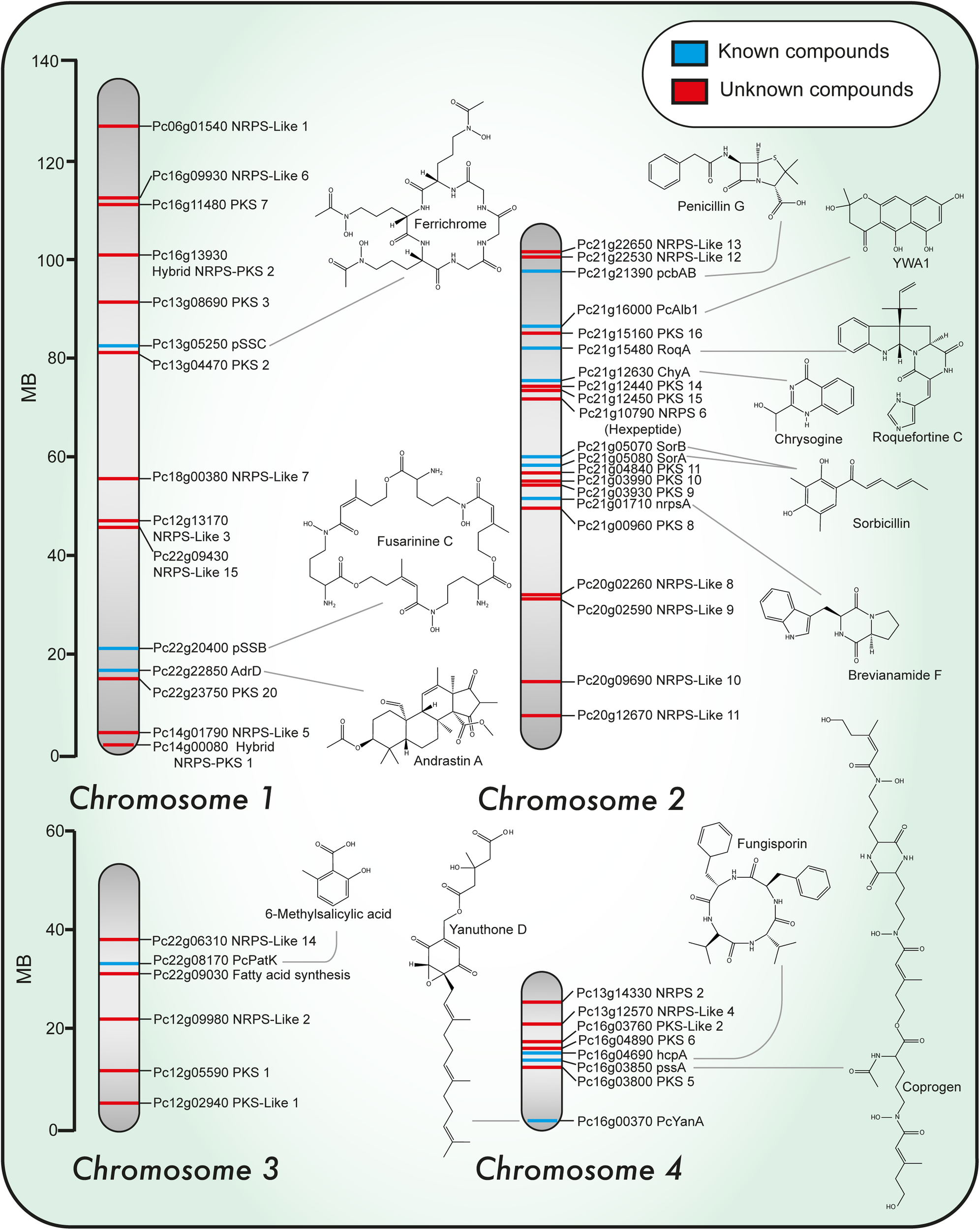
Chromosomal localization of known and predicted PKS and NRPS genes and representative structures of associated secondary metabolites identified in Penicillium chrysogenum. Chromosomal localization of PKS and NRPS genes. Blue and red lines indicate known and unknown associated products so far, respectively.
Penicillium chrysogenum produces a broad range of secondary metabolites such as roquefortines, fungisporin (a cyclic hydrophobic tetrapeptide), siderophores, penitric acid, ω-hydroxyemodin, chrysogenin, chrysogine, sesquiterpene PR-toxin and sorbicillinoids, but likely also possesses the ability to produce other compounds not detected before. For most of the identified compounds, the responsible BGCs are unknown. The development of new bioinformatics tools (SMURF, AntiSMASH) (Khaldi et al., 2010; Weber et al., 2015; Blin et al., 2017) and the increase in the number of fungal genomes sequenced to date has opened the possibility to discover new NPs with novel properties (genome mining). The genes involved in the biosynthesis, regulation and transport of secondary metabolites tend to be arranged in the genome in clusters. Importantly, these gene clusters include the core biosynthetic genes which either encode polyketide synthases (PKSs), nonribosomal peptide synthetases (NRPSs) or terpene synthases genes (Smanski et al., 2016). Recently, a global analysis was performed on 24 genomes of Penicillium species and this identified 1,317 putative BGCs predominated by two classes based on PKS (467) and NRPS (260) (Nielsen et al., 2017). In P. chrysogenum there are 33 core genes in the secondary metabolism that encode 10 NRPS, 20 PKS, 2 hybrid NRPS–PKS, and 1 dimethyl-allyl-tryptophan synthase (van den Berg et al., 2008; Khaldi et al., 2010; Medema et al., 2011; Samol et al., 2016) (Figure 1). A large number of PKS and NRPS enzymes are found also in other Penicillium species but only part of these gene clusters are shared, which suggests an unexplored potential of the secondary metabolome even in a single genus.
Here, we summarize the most recent strategies for engineering filamentous fungi with particular attention to P. chrysogenum, a promising cell factory of novel products with new application spectra. A brief description of the key biosynthetic enzymes involved in biosynthesis of secondary metabolites in fungi is provided.
The Building Enzymes of the Natural Products
Nonribosomal peptide synthetases are large, highly structured and complex enzymatic machineries, closely related to other modular enzymes such as PKSs, NRPS–PKS hybrid synthetases and fatty acid synthetases (FASs). They have certain distinct properties in common, the most striking one being their structural division in domains and modules, which is manifested in their shared evolutionary history (Smith and Sherman, 2008). Every enzyme minimally consists of one module, a functionally distinct unit, which allows for the recruitment and subsequent incorporation of a precursor into a growing product. Domains as well as modules are clearly defined and evolutionary exchangeable structures amongst multi-modular enzymes. In the case of PKS and NRPS, this led to the occurrence of a variety of NRPS–PKS hybrids (Du and Shen, 2001; Shen et al., 2005; Li et al., 2010; Nielsen et al., 2016).
Nonribosomal Peptides (NRPs) and Nonribosomal Peptide Synthetases (NRPSs)
In comparison to most ribosomally derived peptides, NRPs are low molecular weight products. The structural diversity of NRPs is tremendous, mostly due to their chemical complexity. Significantly contributing to this diversity is the fact that NRPS are not only reliant on proteinogenic amino acids, but up until now more than 500 substrates were identified, which serve as NRPS building blocks (Caboche et al., 2008). These molecules are predominantly amino acids, but not exclusively, since fatty acids, carboxylic acids and others substrates have been reported in NRPs (Marahiel et al., 1997). NRP thus represent a diverse group of natural compounds and occur as linear, branched, circular or macrocircular structures (Dang and Sussmuth, 2017; Sussmuth and Mainz, 2017). The natural functions of NRPs are as diverse as their structures. Signaling, communication, metal-ion chelation, host protection are important functions performed by NRPs, though many compounds are not yet fully characterized in this respect. Nevertheless, the characterization of NPs for applied purposes is well developed and led to a vast collection of ground-breaking pharmaceuticals, including antibiotics, anti-fungal agents, immunosuppressants as well as cytostatic drugs (Frisvad et al., 2004; Watanabe et al., 2009; Dang and Sussmuth, 2017).
Structurally, every NRPS module, initiation (1), elongation (n) or termination (1), requires a minimal set of domains (Figure 2A) (Stachelhaus and Marahiel, 1995). The two domains essential to every module are the adenylation domain (A) and the non-catalytic thiolation domain (T). This tandem di-domain enables the specific selection and activation of a given substrate. However, the T-domain must first go through 4′-phosphopantetheinyl transferase (PPTase) and coenzyme A (CoA) dependent activation after expression, by transferring the phosphopantetheine moiety of CoA onto a conserved serine residue, in order to enter the holo state. Also, adenylation domains (A) have accompanying factors, or proteins, called MbtH-like proteins (MLPs) (Quadri et al., 1998; Baltz, 2011). In contrast to PPTases, MLPs are merely interacting with the A-domain, however, they do not have an intrinsic enzymatic activity, but rather a chaperoning function upon binding a distinct part of the A domain (Felnagle et al., 2010; Miller et al., 2016; Schomer and Thomas, 2017). In addition to these domains, any elongation module will require a condensation domain (C), which connects two modules and links up- and downstream activated substrates via a peptide bond. C-domains are stereospecific for both, up- and downstream activated substrates and render the resulting intermediate compound attached to the downstream T-domain. Lastly, the C-terminal termination module essentially requires a thioesterase domain (Te), to catalytically release the covalently bound compound of the NRPS, returning the NRPS complex to the ground state for another reaction cycle. In addition to these essential domains, we can distinguish a series of additional domains, performing epimerization, halogenation, cyclization, macrocyclization, multimerization or methylation (Ansari et al., 2008; Horsman et al., 2016; Bloudoff and Schmeing, 2017).
FIGURE 2
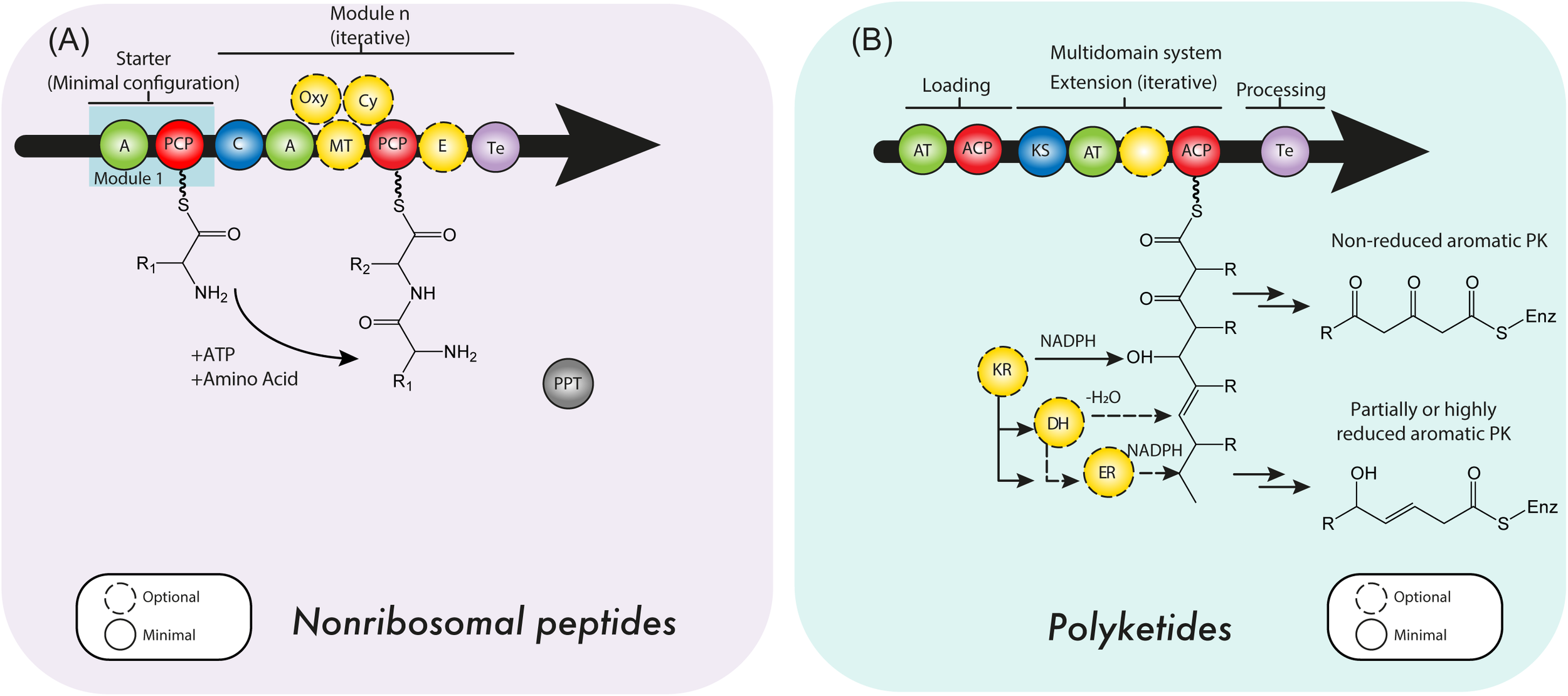
(A) Nonribosomal peptide synthetases (NRPSs) and (B) polyketide synthase (PKS) minimal domain structure. For details see the text. C, condensation domain; PCP, peptidyl carrier protein; A, adenylation domain; E, epimerase; MT, methyltransferase; PPT, 4’-phosphopantetheine transferase domain; Oxy, oxygenation domain; Cy, cyclization domain; ACP, acyl carrier protein; AT, acyltransferase domain; KS, ketosynthase domain; KR, ketoreductase domain; DH, dehydratase domain; ER, enoyl reductase domain; Te, thioesterase domain.
Theses enzymatic machineries can be classified as type I NRPS when the modules are arranged on a single protein, while the type II NRPS are independent proteins in an transient manner during the NRP synthesis (Sattely et al., 2008; Hur et al., 2012). A NRPS can be as simple as a single modular unit containing three domains, although the most complex and largest structure known contains 15 modules with 46 domains (Wang H. et al., 2014; Bode et al., 2015) yielding a 1.8 MDa protein complex (type I NRPS). Although the size of a NRPS, as well as the modular sequence, limits the setup of the resulting NRP, it is common that NRPS cluster and interact with tailoring enzymes in order to produce products of a higher complexity (Yin and Zabriskie, 2006). To enable such specific interactions, NRPS can contain small stretches of up to 30 amino acids at the C- or N-terminus, which form a rather specific recognition point, thus enabling communication (COM-domain) between multiple NRPS of one cluster (type II NRPS) (Hahn and Stachelhaus, 2006; Dowling et al., 2016). To date three types of NRPS system have been described according to their synthesis mode (or strategy of biosynthesis): Type A (linear), type B (iterative) and type C (non-linear) (Figure 3). The type A system harbors the typical domain organization A-T(C-A-T)n-1-Te/C, where n represents the number of amino acids in the peptide. In this linear NRPS, the order and number of modules correlates with the amino acid sequence in the NRP and thus it is possible to predict the product that will be formed. Usually, in fungal NRPSs the cyclisation reaction is performed by a specialized C domain instead of Te domain. Since each module catalyzes one cycle during the chain elongation of the nascent NRP due to its specific activity, this system is considered analogous to type I PKS. In fungi, the most prominent examples of this type of NRPS are ACV synthetases (β-lactams), cyclosporin synthetases (Cyclosporin A) and peptaibol synthetases (peptaboils, a class of antibiotics with a high content of α-aminoisobutyric acid) (Wiest et al., 2002; Tang et al., 2007; Felnagle et al., 2008; Eisfeld, 2009). The type B system is characterized to employ all their modules or domains more than once during the synthesis of a single NRP, which enables the assembling of peptide chains that contain repeated sequences along the structure (Mootz et al., 2002). An example of this mode of synthesis occurs in Fusarium scirpi during the biosynthesis of enniatin (antibiotic), which is achieved through the repeated use of two modules. Other examples of type B NRPSs are the siderophore synthetases, which only contain three A domains that catalyze the biosynthesis of ferrichrome (Mootz et al., 2002; Eisfeld, 2009). In type C system, the non-linear NRPSs have at least one domain conformation that deviate from (C-A-T)n-1 organization contained in linear NRPSs. Likewise, in these synthetases the module arrangement does not correspond to the amino acid sequence in the NRP. Unlike type A NRPS, in type C NRPSs the non-linear peptide is produced by a branch-point synthase and contains uncommon cyclization patterns. Another important difference is that non-linear NRPSs can incorporate small soluble structures, such as amines into the rising NRP through specialized C domains (Tang et al., 2007; Felnagle et al., 2008; Hur et al., 2012). Capreomycin, bleomycin and vibriobactin are examples of NRPs produced by this type of synthetases (Felnagle et al., 2008). In continuation, a brief description of the main NRPS domains features is provided.
FIGURE 3
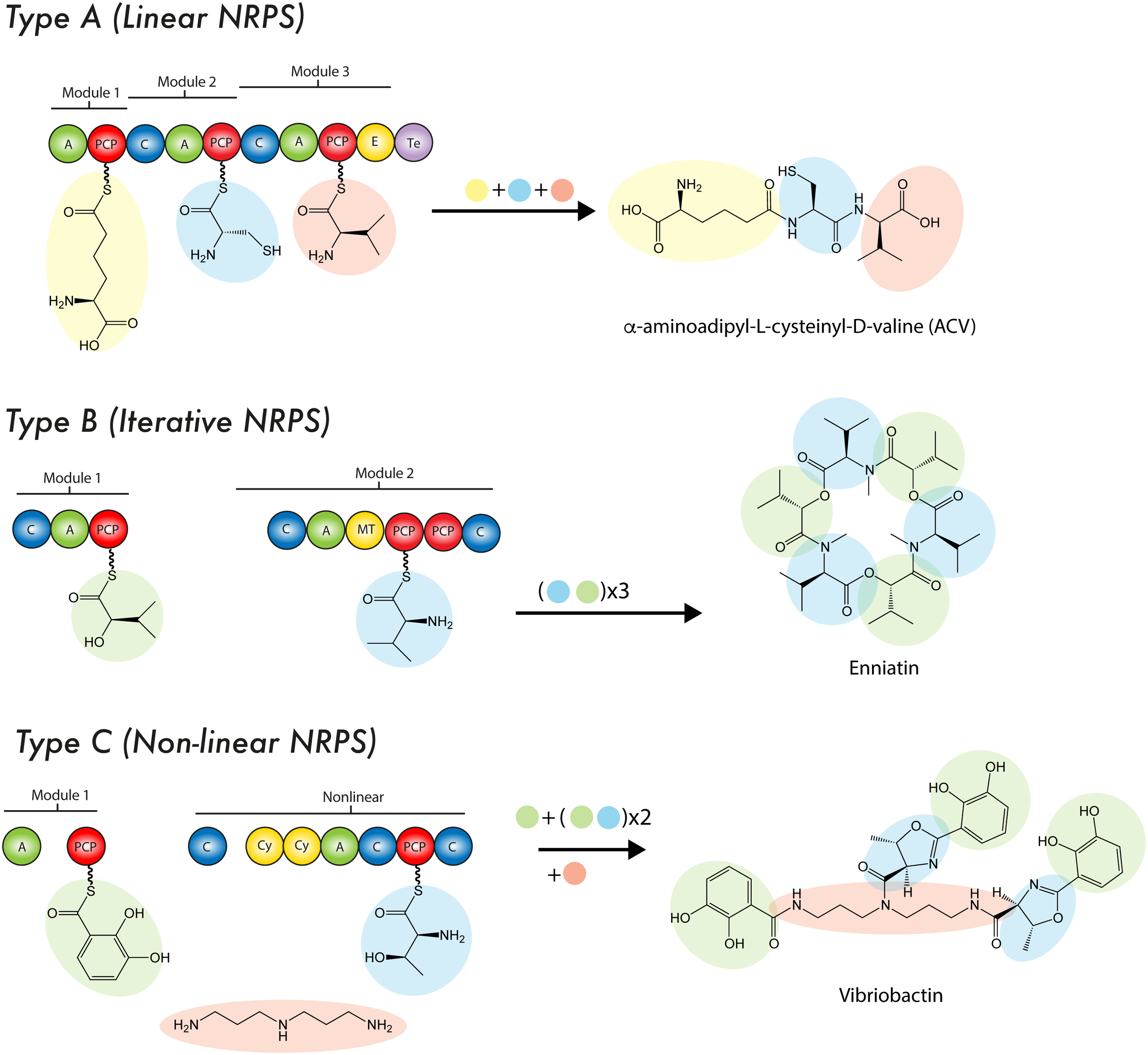
Mode of biosynthesis of nonribosomal peptide synthetases. For details see the text. C, condensation domain; PCP, peptidyl carrier protein; A, adenylation domain; E, epimerase; MT, methyltransferase; Te, thioesterase domain; Cy, cyclization domain.
Adenylation (A) and Thiolation (T) Domains
Any NRPS module minimally consists of an A- and T-domain (or peptidyl carrier protein, PCP), enabling single module functionality and multi-modular functionality upon addition of C domains (Linne and Marahiel, 2000; Bergendahl et al., 2002; Felnagle et al., 2008; Kittilä et al., 2016; Bloudoff and Schmeing, 2017). They are often referred to as “gatekeeper” domains, as there is no subsequent product formation without prior adenylation and thioesterification of a substrate (Sun et al., 2014). The two core functions of the A-domain are characterized first, through the hydrolysis of ATP or adenylation, allowing an AMP-substrate conjugate to be formed, which is subsequently transferred to the free thiol group of the 4′-phosphopantetheinyl-moiety (Ppant), which is anchored to a conserved serine residue in the downstream T-domain (Ku et al., 1997; Weber and Marahiel, 2001; Neville et al., 2005).
Condensation Domains (C)
C-domains are approximately 450 residue NRPS domains, representing a highly versatile class of NRPS domains. Any NRPS composed of more than one module must consequently contain at least one C-domain. However, also single modular NRPS may contain C-domains, especially if they cooperate with other NRPS. Essentially, the primary target of a C-domain is the condensation of the up- and downstream activated substrates through a nucleophilic attack, mainly leading to the formation of an n-peptide linked via a peptide bond. Nonetheless, several residues of the C-domain may have the intrinsic potential to fulfill multiple functions (Balibar et al., 2005; Teruya et al., 2012; Haslinger et al., 2015).
Epimerization Domains (E)
The E-domains are among the most abundant modification domains intrinsic to NRPS. In contrast to the structurally similar C domains they are responsible for the site specific epimerization of a substrate, predominantly performing this function after peptide bond formation has occurred (Bloudoff and Schmeing, 2017).
Thioesterase Domains (Te)
The thiotemplate based enzymatic systems rely on a catalytic activity in order to remove a product or product-scaffold of the primary enzyme. Therefore, most NRPS contain a domain on their C-terminus responsible for precisely this purpose, the thioesterase domain (Te). Te-domains are a common commodity in single and multi-modular NRPS, although, in multi-NRPS systems only the terminal NRPS contains this domain (Horsman et al., 2016). Additionally, this domain harbors a quality control activity (proofreading) to verify the correct configuration of the nascent peptide (Martín and Liras, 2017).
Intrinsic Product Modifying Domains
In addition to the C-domain related epimerization domain, discussed previously, there are cyclization (Cy), oxygenation (Oxy) as well as methyl-transferase (MT). These domains have been characterized to the extent of classifying their functions, although, especially Cy- and Oxy-domains may occur as a singular bi-functional unit or in a serial manner, respectively (Walsh, 2016). Cy- and Oxy-domains, specifically replace the classic function of C-domains, omitting amino acid condensation through peptide bond formation, resulting in thiazoline, oxazoline or methyloxazoline structures (Sundaram and Hertweck, 2016; Walsh, 2016). Those reactions predominantly occur in siderophore producing NRPS and rely on the presence of serine, threonine and cysteine residues (Patel et al., 2003; Kelly et al., 2005). Also MT-domains follow the common di-sub-domain structural patterning, which is also seen in A-, C-, E-, and Cy-domains. Fundamentally, MT-domains, however, are more restricted in their function, which covers the transfer of methyl-groups from S-adenosylmethionine to N (N-MT), C (C-MT), O (O-MT) or for certain residues S (S-MT) atoms resolving around the amino acids Cα carbon (Miller et al., 2003) and in case of S-MT Cβ, respectively (Al-Mestarihi et al., 2014).
In P. chrysogenum 10 NRPS have been identified (Table 1) (van den Berg et al., 2008; Medema et al., 2011; Samol et al., 2016), of which only two have no attributed function. In this fungus, next to fungisporin (Figure 1) which is a cyclic hydrophobic tetrapeptide generated by a singular NRPS, three biosynthetic pathways involving a NRPS have been described in detail: penicillins, roquefortine/meleagrin, fungisporin and chrysogine (Figure 4).
Table 1
| Gene ID | Gene name | Protein | Domain organization | Product/Pathway |
|---|---|---|---|---|
| Pc13g05250 | pssC | Siderophore synthetase | A1TCA2TCTCA3TCTCT | Siderophore |
| Pc13g14330 | – | Tetrapeptide synthetase | CA1TECA2TCA3TCA3TCA4TC | – |
| Pc16g03850 | pssA∗ | Siderophore synthetase | ATCTA | Coprogen |
| Pc16g04690 | hcpA | Cyclic tetrapeptide synthetase | A1TECA2A3TCA4TECTCT | Fungisporin |
| Pc21g01710 | nrpsA | Dipeptide synthetase | A1TCA2T | Brevianamide F |
| Pc21g10790 | – | Hexapeptide synthetase | A1TCA2TCA3TECA4TCA5TCA6TC | – |
| Pc21g12630 | chyA | 2-Aminobenzamide synthetase | A1TCA2TC | Chrysogine |
| Pc21g15480 | roqA | Histidyl-tryptophanyldiketo- piperazine synthetase | A1TCA2TC | Roquefortine/Meleagrin |
| Pc21g21390 | pcbAB | α-Aminoadipyl-cysteinyl-valine synthetase | A1TCA2TCA3TEte | β-lactams |
| Pc22g20400 | pssB | Siderophore synthetase | ATCTC | Fusarinines |
Nonribosomal peptide synthetases (NRPSs) in P. chrysogenum and known associated products.
A, adenylation; T, thiolation; E, epimerization; te, thioesterase; C, condensation. ∗Point mutations present in nrps genes of industrial P. chrysogenum strains subject to CSI program. Modified from Salo et al. (2015), Samol et al. (2016), and Guzmán-Chávez et al. (2018).
FIGURE 4
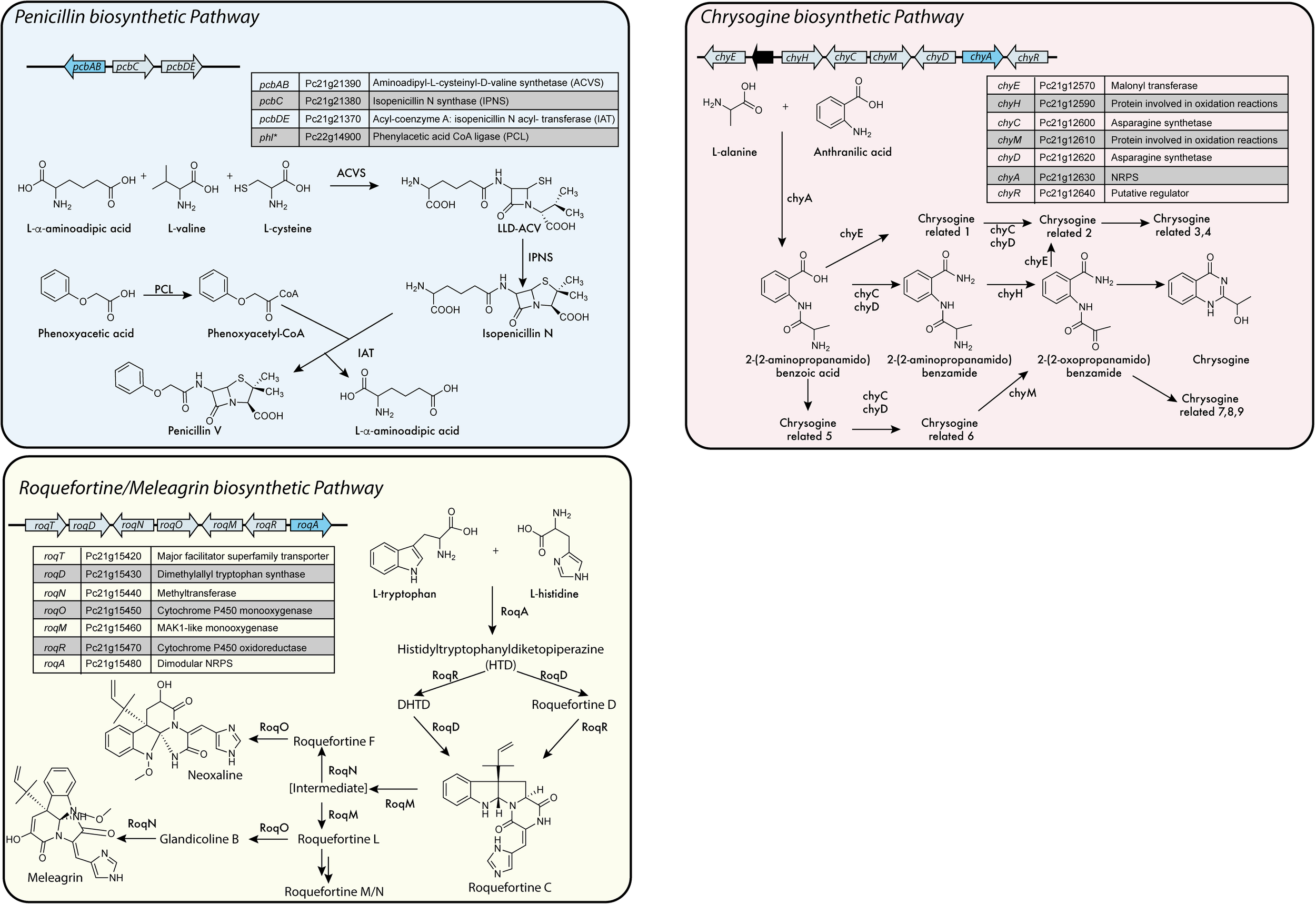
NRPS based biosynthetic pathways in P. chrysogenum.∗Is not part of the biosynthetic gene cluster (BGC). Adapted from Harris et al. (2009), Bartoszewska et al. (2011), García-Estrada et al. (2011), Weber et al. (2012c), Ali et al. (2013), Ozcengiz and Demain (2013), Ries et al. (2013), and Viggiano et al. (2017).
Polyketides and Polyketide Synthase
Polyketides were already discovered in 1883 by James Collie, but the interest in these compounds (enzymes) was revived only as late as the 1950s by the work of Arthur Birch on the aromatic polyketide-6-methyl salicylic acid from P. patulum. These molecules are a class of NPs, that display different types of biological activities such as antibiotic (erythromycin A), antifungal (amphotericin B), immunosuppressant (rapamycin), antitumor (geldanmycin) and hypolipidemic (lovastatin) (Nair et al., 2012; Jenner, 2016; Weissman, 2016). Their assembly process is similar to that in the fatty acid biosynthesis, the main difference is the optional full reduction of the β-carbon in the PK biosynthesis. The group of enzymes that catalyzes the biosynthesis of PKs is referred to as PKSs (Keller et al., 2005; Caffrey, 2012).
In addition to the NRPSs, PKSs are the main enzymes that build the structural scaffold of a wide range of secondary metabolites and NPs in plants, bacteria, insects and fungi (Brakhage, 2012; Nair et al., 2012). Usually, these enzymes are encoded by genes that are grouped into clusters, that also specify genes encoding tailoring enzymes (oxygenases, oxidoreductases, reductases, dehydrogenases, and transferases), that further modify the scaffold produced by the PKS into a final product (Brakhage, 2012; Lim et al., 2012). PKSs are multimodular and multidomain enzymes that use a specific acyl-coenzyme A (acyl-CoA; usually malonyl-CoA or methylmalonyl-CoA) as building block, and subsequently catalyze a decarboxylative Claisen-type condensation of ketide units. The basic structural architecture consists of an acyl carrier protein (ACP), a ketosynthase (KS) and an acyltransferase (AT) domain. These combined domains extent a linear intermediate by two carbon atoms. An optional set of domains (dehydratase (DH), ketoreductase (KR), enoyl reductase (ER) and thioesterase (TE) may provide further modifications of the linear intermediate (Staunton and Weissman, 2001; Brakhage, 2012; Nair et al., 2012; Dutta et al., 2014).
According to their protein architecture and mode of action, PKS enzymes are classified into types I, II, and III (Figure 5). Type I PKSs are mainly found in bacteria and fungi. These multidomain proteins can be further subdivided in two categories: modular and iterative (Nair et al., 2012) Modular type I PKSs or non-iterative PKSs are unique for bacteria and are characterized by presenting a sequence (or set) of modules, each constituted with a set of specific catalytic domains. In consequence, the number of precursors fused in the PK is equivalent to the number of modules which are present (Chan et al., 2009). In contrast, iterative type I PKSs use the same catalytic core domains as modular type I PKSs, but the catalytic reaction is repeated to yield the complete PK backbone. A representative example of this type is LovB, that together with LovC (a enoyl reductase) catalyzes around 35 reactions to produce dihydromonacolin L, an intermediate in the lovastatin biosynthesis (Chan et al., 2009; Campbell and Vederas, 2010). Like iterative type I PKS enzymes, fungal PKSs (Figure 2B) are restricted to a single module and the consecutive domains act in sequential order during the synthesis of the complete PK. They are equipped with basic structural domains typically found in PKS enzymes (ACP-KS-AT domains) but may also contain optional units (KR, DH, ER, and Te domains). Depending on the presence or absence of reducing domains, these enzymes can be divided into highly reducing (HR), non-reducing (NR) and partially reducing (PR) PKS (Figure 2B) (Keller et al., 2005; Crawford and Townsend, 2010; Jenner, 2016).
FIGURE 5
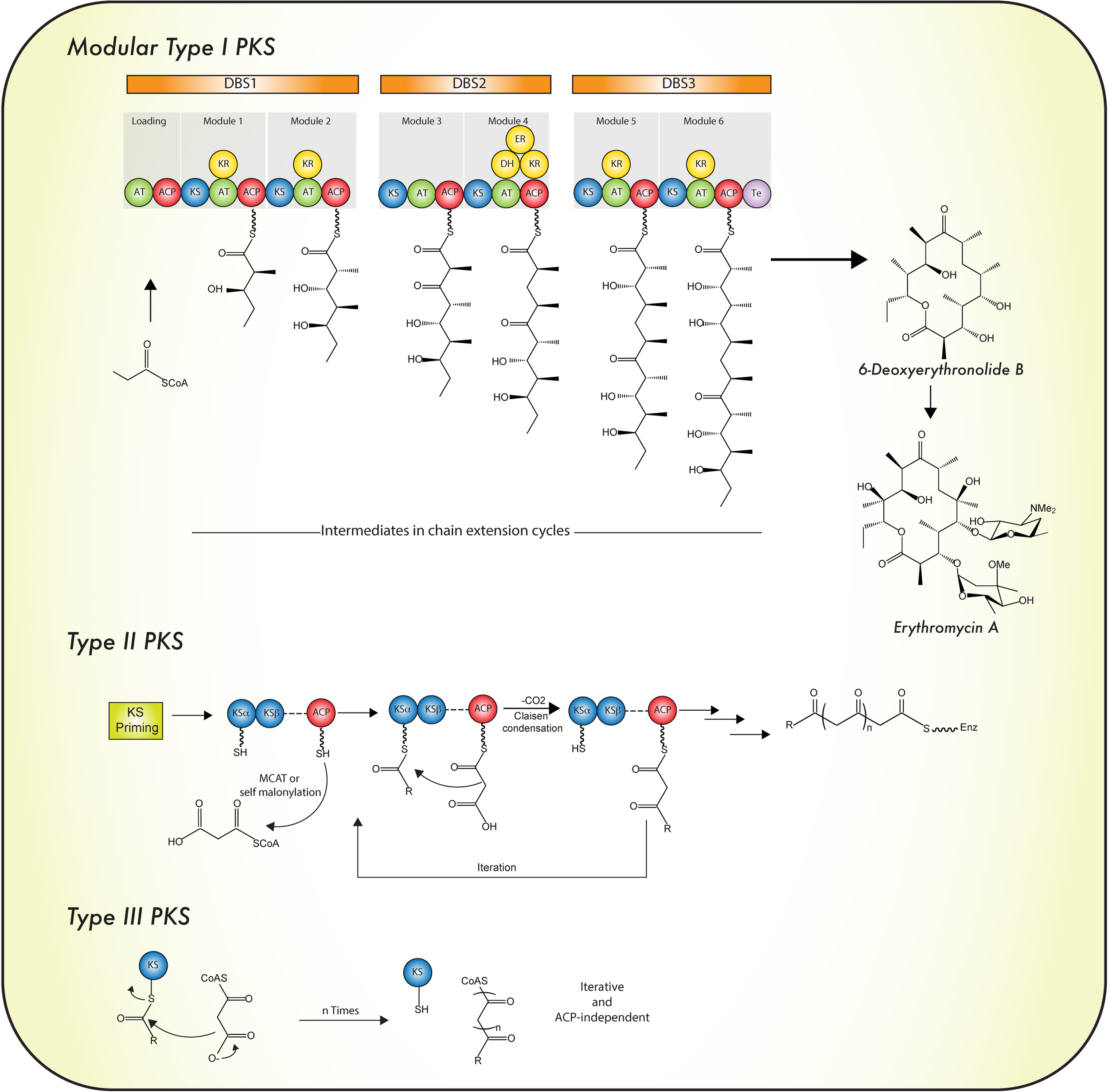
Mode of biosynthesis of polyketide synthases. For details see the text. Abbreviations are as in the legend to Figure 2. Iterative type I PKS is depicted in Figure 2B.
Highly Reducing PKS (HR-PKS)
Highly reducing PKS (HR-PKS) produce the linear or cyclic scaffold of some compounds such as fumonisins, T-toxins, solanapyrone E, squalestatin or/and lovastatin (Chiang et al., 2014; Roberts et al., 2017). Usually, they start with a KS domain, followed by an AT, DH and C-Met domain, although the latter does not always follow the DH domain. The ER domain is an optional unit in HR-PKS enzymes, but when the ER is missing, the corresponding region is filled with a polypeptide domain with an unknown function. Furthermore, these enzymes do not contain a product template domain (PT) or N-terminal SAT domain, whereas these special domains are present in NR-PKS enzymes (Cox and Simpson, 2010).
Partially Reducing PKS (PR-PKS)
Structurally, these enzymes have a domain architecture that is similar to the mammalian FAS: a N-terminal KS-domain followed by an AT-, DH-, and “core”-KR-ACP domain. These enzymes lack an ER domain (Wang L. et al., 2015), and also do not have a Te domain, which suggests an alternative mechanism of product release than hydrolysis. PR-PKS enzymes produce small aromatic molecules such as 6-methylsalicylic acid (MSA), but in most cases the chemical product is unknown (Cox and Simpson, 2009, 2010; Kage et al., 2015).
Non-reducing PKS (NR-PKS; Aromatic PKs)
Non-reducing PKS (NR-PKS; aromatic PKs) typically, consist of six catalytic domains that are covalently associated and arranged in four components: loading (SAT), chain extension (KS-MAT-PT-ACP), cyclisation and processing components (TE-CLC) (Bruegger et al., 2013).
Type II PKSs are unique for bacteria and use a similar iterative mechanism as observed in iterative type I PKSs. However, the different catalytic domains are encoded by independent genes. In general, they often constitute a “minimal PKS,” which comprises of two KS units (KSα and KSβ) and an ACP protein that holds the growing PKS chain. The KSβ domain defines the length of the PK chain. The folding pattern of the poly-β-keto intermediates is determined by optional PKS units such as aromatases, ketoreductases, and cyclases. Other tailoring modifications are performed by oxygenases, methyl and glycosyl transferases. Known metabolites synthetized by type II PKSs are tetracyclines, anthracyclines and aureolic acids (Hertweck et al., 2007; Jenner, 2016).Type III PKSs have originally been discovered in plants but are also present in bacteria and fungi. They consist of a single KS domain that catalyzes a defined number of elongations, usually generating small phenols or naphtol rings. The enzyme transfers the acyl group from the CoA to the active site histidine, which is a highly conserved residue. However, the amino acid sequence of the his motif is not similar to those found in KS domains of type I and II PKS enzymes (Shen, 2003; Chan et al., 2009; Bruegger et al., 2014; Jenner, 2016). Importantly, independent of the mechanistic or structural differences, all the PKs synthetized by PKS enzymes follow the same decarboxylative condensation mechanism of the acyl-CoA precursors. However, these precursors should be activated in prior by the ACP domain, in the case of the type I and II PKS enzymes, whereas type III PKS enzymes act independently of ACP domains (Shen, 2003; Hopwood, 2009). Acridones, pyrenes as well as (and) chalcones are some examples of the compounds produced by type III PKS enzymes (Yu et al., 2012). Below, a brief description is provided on the main catalytic features of PKS domains.
Acyltransferase Domains (AT)
A main unit during PK biosynthesis is the AT domain that selects the starter unit (malonyl-CoA or methylmalonyl-CoA) before it is transferred to the ACP domain for the chain elongation cycle (Dunn et al., 2013). This process involves two steps, i.e., the acylation and the transfer to the ACP (Jenner, 2016).
Acyl Carrier Protein (ACP)
The ACP is an essential cofactor that participates in PK biosynthesis. This protein belongs to a highly conserved carrier family, and consists of 70–100 amino acid residues (Byers and Gong, 2007). To perform the PK biosynthesis, the holo-ACP (active) form is generated by the phosphopantetheinyl transferase enzyme (PPTase) through a post-translational modification of ACP whereby a 4′-phosphopantetheine (4′-PP) moiety from CoA is transferred to the conserved serine (Evans et al., 2008; Kapur et al., 2010; Jenner, 2016) resulting in the formation of the Ppant arm. ACP modulates three important events during PK biosynthesis. First, it allows the condensation during chain elongations since it transfers the starter unit from the AT domain to the KS domain. Second, it shuttles the growing chain between the up and downstream domains, as well as to optional PKS domains, probably involving protein–protein recognition between domains. Third, it prevents premature cyclization and enolization of the PK chain (Yadav et al., 2013).
Ketosynthase Domains (KS)
The KS is a homodimeric condensing domain that catalyzes the extension of the β-ketoacyl intermediate by a decarboxylative Claisen condensation. This domain contains two active sites which are accessible to the ACP through its flexible Ppant arm, which receives the β-carboxyacyl-CoA extender unit from the AT. At that stage, a thioester bond is formed between the active-site cysteines’ thiol group of the KS and the growing PK. Only when both units are covalently attached onto the module, a decarboxylative Claisen condensation occurs, which involves two conserved his residues. Therefore, mechanistically the KS domain acts at three stages: acylation, decarboxylation and condensation (Chen et al., 2006; Caffrey, 2012; Yadav et al., 2013; Jenner, 2016; Robbins et al., 2016).
Ketoreductase Domains (KR)
The KR domain functions as a β-carbon processing unit that belongs to the family of short-chain dehydrogenase/reductases. This domain reduces the β-keto group, that is formed during the condensation process, into a hydroxyl group (a β-hydroxyl intermediate) using NADPH (Keatinge-Clay and Stroud, 2006; Caffrey, 2012). Additionally, some KR domains are equipped with epimerase activity. The epimerizing module has a more open architecture, enabling the catalytic epimerization of methyl groups in acyl-ACP substrates, a reaction that involves the conserved serine and tyrosine residues which are also employed during ketoreduction (Ostrowski et al., 2016c; Bayly and Yadav, 2017).
Dehydratase Domains (DH)
The DH domain is usually coupled to B-type KR domains (B-type). This domain catalyzes water elimination (via syn or anti) at the β-hydroxy acyl chain position thereby producing trans double bonds (α,β-unsaturated moieties) (Caffrey, 2012; Bruegger et al., 2014; Jenner, 2016; Bayly and Yadav, 2017).
Enoyl Reductase Domains (ER)
The ER domain is an optional tailoring unit involved in the final oxidation state of the growing PK. It reduces α,β-enoyl groups and thereby generates saturated α–β bonds. This reaction involves NAD(P)H as hydride donor in a Michael addition type of mechanism. In the enoyl reduction, the products formed during this reaction have a specific stereochemistry (3R,2R) or (3R,2S) due to the β-carbon attack performed by the pro-4R hydride of NADPH, contrasting the KR domain that utilizes the pro-4S hydride (Chen et al., 2006; Bruegger et al., 2014).
Thioesterase Domain (Te)
Termination of PK biosynthesis involves the Te domain, which produces macrolactones via intramolecular cyclization or linear PKs by hydrolysis (Keatinge-Clay, 2012). In both events, an acyl-Te intermediate is formed through the transfer of the PK chain from the last ACP to the active serine on Te domain (Jenner, 2016).
Special Domains
In non-reducing PKS, the ACP transacylase (SAT) domain acts as starter unit that loads the ACP whereupon chain extension is mediated for KS and AT domain. During this process, the malonyl-CoA:ACP transacylase (MAT) domain transfers the extension units from malonyl-CoA to the ACP, while the product template (PT) domain stabilizes the reactive poly-β-keto intermediates. The processing component acts after the initial assembly when the cyclized or PK intermediate is still attached to the ACP. Final cyclization and release is catalyzed by the Te/Claisen cyclase (CLC) domain (Cox and Simpson, 2010; Crawford and Townsend, 2010; Bruegger et al., 2013; Chiang et al., 2014).
In P. chrysogenum, 20 PKS genes have been identified (Table 2) (van den Berg et al., 2008; Medema et al., 2011; Samol et al., 2016), but for only six the products are known. To date, in P. chrysogenum only four PK-related pathways have been described in detail: sorbicillinoids, MSA-6/yanuthones, DHN-melanin and andrastin A (Figure 6).
Table 2
| Gene ID | Gene name | Protein | Domain organization | Product/Pathway |
|---|---|---|---|---|
| Pc12g05590 | pks1 | – | ks-at-dh-mt-kr-acp | – |
| Pc13g04470 | pks2∗ | – | ks-at-dh-mt-er-kr-acp | – |
| Pc13g08690 | pks3 | – | ks-at-dh-mt-er-kr-acp | – |
| Pc16g00370 | yanA | 6-MSA synthase | ks-at-kr-acp | 6-MSA/Yanuthones |
| Pc16g03800 | pks5 | – | ks-at-dh-er-kr-acp | – |
| Pc16g04890 | pks6 | – | ks-at-dh-mt-er-kr-acp | – |
| Pc16g11480 | pks7∗ | – | ks-at-dh-mt-er-kr-acp | – |
| Pc21g00960 | pks8∗ | – | ks-at-dh-mt-er-kr-acp | – |
| Pc21g03930 | pks9 | – | ks-at-dh-mt-er-kr-acp | – |
| Pc21g03990 | pks10 | – | ks-at-dh-er-kr-acp | – |
| Pc21g04840 | pks11 | – | ks-at-dh-er-kr-acp | – |
| Pc21g05070 | sorB∗ | Sorbicillin synthase | ks-at-acp-mt-te/red | Sorbicillinoids |
| Pc21g05080 | sorA∗ | Sorbicillin synthase | ks-at-dh-mt-er-kr-acp | Sorbicillinoids |
| Pc21g12440 | pks14 | – | ks-at-dh-er-kr-acp | – |
| Pc21g12450 | pks15∗ | – | ks-at-acp-te | – |
| Pc21g15160 | pks16 | – | ks-at-dh-mt-er-kr-acp | – |
| Pc21g16000 | alb1∗ | YWA1 synthase | ks-at-acp-acp-te | YWA1/DHN-Melanin |
| Pc22g08170 | patK | 6-MSA synthase | ks-at-kr-acp | 6-MSA |
| Pc22g22850 | adrD | DMOA synthase | ks-at-acp-mt-te/red | DMOA/Andrastin A |
| Pc22g23750 | pks20 | – | ks-at-dh-mt-er-kr-acp | – |
Polyketide synthases in P. chrysogenum and (insofar known) their associated products.
ks, ketosynthase; at, acyltransferase; dh, dehydratase; mt, methyltransferase; er, enoyl reductase; kr, ketoreductase; acp, acyl carrier protein; te/red, thioester reductase. ∗Point mutations present in pks genes of industrial P. chrysogenum strains subject to CSI program. Modified from Salo et al. (2015), Samol et al. (2016), and Guzmán-Chávez et al. (2018).
FIGURE 6
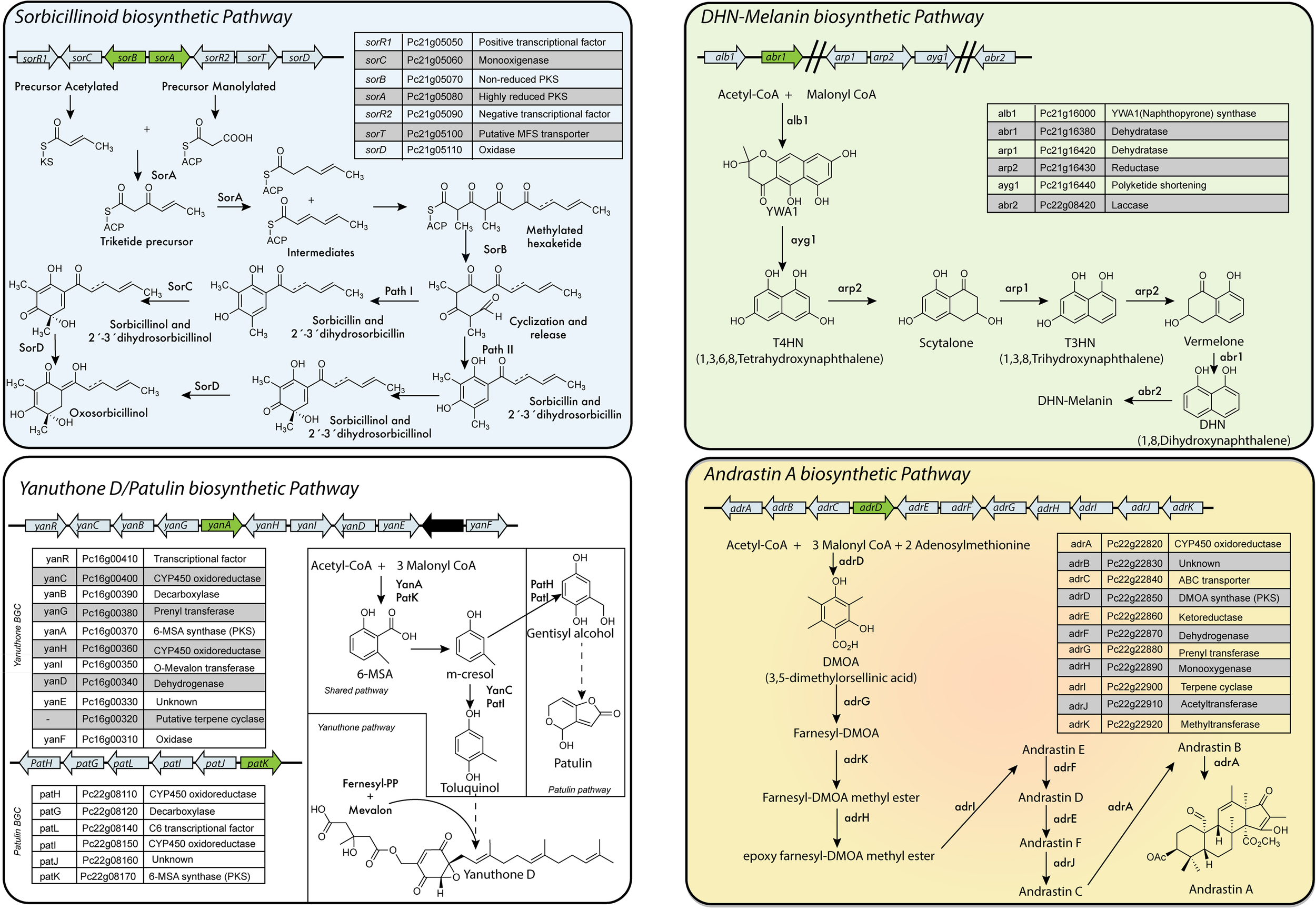
PKS-based biosynthetic pathways in P. chrysogenum. Sorbicillinoids: Despite the fact that this cluster is also present in industrial strains of P. chrysogenum, they do not produce sorbicillinoids due to a point mutation in the ketosynthase domain of SorA. Yanuthones/Patulin: P. chrysogenum only contains a full version of one cluster (yanuthone D BGC), while the second cluster (patulin BGC) is incomplete (Nielsen et al., 2017). The absence of the gene encoding for an isoepoxidon dehydrogenase agrees with the fact that this fungi does not produce patulin (Samol et al., 2016). However, under laboratory conditions, yanuthone D is also not detected in this fungus (Salo, 2016). DHN-Melanin: The genes are only partially clustered in the genome of P. chrysogenum. Andrastin A: P. chrysogenum strains subjected to CSI are not able to produce andrastin A or related compound. Adapted from Staunton and Weissman (2001), Maskey et al. (2005), Cox (2007), Wattanachaisaereekul et al. (2007), Du et al. (2009), Pihet et al. (2009), Crawford and Townsend (2010), Avramovič (2011), Harned and Volp (2011), Gallo et al. (2013), Heinekamp et al. (2013), Matsuda et al. (2013), Salo et al. (2015), Salo et al. (2016), Druzhinina et al. (2016), Meng et al. (2016), Salo (2016), Samol et al. (2016), Guzmán-Chávez et al. (2017), Guzmán-Chávez et al. (2018), Nielsen et al. (2017), and Rojas-Aedo et al. (2017).
Terpenoids Biosynthesis
In addition to NRPs and PKs, terpenoids are another class of NPs that are synthetized by filamentous fungi (Ascomycota) although less abundant as compared to Basidiomycota (Schmidt-Dannert, 2014). Fungal terpenoids or isoprenoids are structurally diverse molecules derived from isoprene units (C5 carbon skeleton): isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), which are synthetized in the mevalonate pathway from acetyl-CoA (Chiang et al., 2014; Soltani, 2016). The head-to-tail condensation of these C5 units is catalyzed by isoprenyl diphosphate synthases (IDSs), producing isoprenyl diphosphates with 10 (geranyl, GPP), 15 (farnesyl, FPP), and 20 (geranylgeranyl, GGPP) carbons. Eventually, these linear chains of different length are further modified by cyclases, terpene synthases (TPs) and prenyl transferases (PTs), yielding different subclasses of terpenoids (Schmidt-Dannert, 2014; Chen et al., 2016). For instance, monoterpenoids, sesquiterpenoids, diterpenoids, sesterterpenoids, and triterpenoids, which harbor two to six isoprene units, respectively (Soltani, 2016). Terpenoids are oxygenated derivatives of terpenes, which are also derived of isoprene (Stashenko and Martinez, 2017).
In filamentous fungi such as Aspergillus, Penicillium, Claviceps, and Neosartorya, ABBA-type PTs are involved in the biosynthesis of a range of toxins (Schmidt-Dannert, 2014). For the synthesis of indole-diterpenoids, IPPS-type PTs transfer GGPP to a indole group, while UbiA-type PTs are involved in the biosynthesis of meroterpenoids, which are hybrid NPs (terpenoids and PKs) (Itoh et al., 2010; Schmidt-Dannert, 2014). In A. nidulans, AusN (UbiA-type TPs) converts the product of a NR-PKS (3,5-dimethylorsellinic acid) as part of an earlier step in the dehydroaustinol/austinol biosynthesis pathway (Lo et al., 2012).
Terpene synthases catalyze cyclization reactions forming the carbocation by substrate ionization (class I) or substrate protonation (class II) (Zhou et al., 2012; Meguro et al., 2014). A relevant example of class I TPs are sesquiterpene synthases, which cyclize the FPP to obtain a sesquiterpene scaffold (C15 backbone) (Quin et al., 2014). Recently, the prx1 to prx4 gene cluster involved in the biosynthesis of PR-toxin in P. roqueforti was cloned and sequenced. This cluster contains the gene prx2 (ari1) that encodes for a aristolochene synthase which forms a sesquiterpene aristolochene derivative (precursor of PR-toxin). Interestingly, an orthologous gene cluster was identified in P. chrysogenum (Pc12g06300 to Pc12g06330), as part of BGC of eleven genes, which is also involved in the biosynthesis of PR-toxin (Hidalgo et al., 2014).
Strategies for Activation of BGCs
Natural products represent a broad range of molecules produced by animals, plants and microorganisms. These molecules may display different biological activities (e.g., antiviral, antimicrobial, anti-tumor, immunosuppressive agents) and it is estimated that the majority of these compounds are derived from filamentous fungal sources and from filamentous bacteria belonging to the genus Streptomyces. With respect to antibiotics, most of the chemical scaffolds used today were discovered during the golden age of antibiotics discovery (1940–1960s). This was followed by four decades during which hardly any new scaffolds from a natural source were developed (Reen et al., 2015; Smanski et al., 2016; Okada and Seyedsayamdost, 2017). However, there is also a current understanding that only a small fraction of the potential possible molecules has been discovered to this date. This follows from genomic studies revealing large numbers of uncharacterized BGCs, while many of these gene clusters are not expressed (silent or sleeping gene clusters) under laboratory conditions (Brakhage and Schroeckh, 2011). Furthermore, metagenomics studies indicate that the majority of microbes present in the environment have not been cultured nor characterized. Thus, there are many challenges that need to be overcome in order to harness the natural diversity of NPs, to cultivate potential strains under laboratory conditions and to activate the BGCs for expression. To achieve the synthesis of new NPs, three main approaches (Figure 7) were used in recent years, which may be successfully applied in P. chrysogenum: manipulation of cultivation conditions, engineering of NRPS and PKS and genetic interference.
FIGURE 7
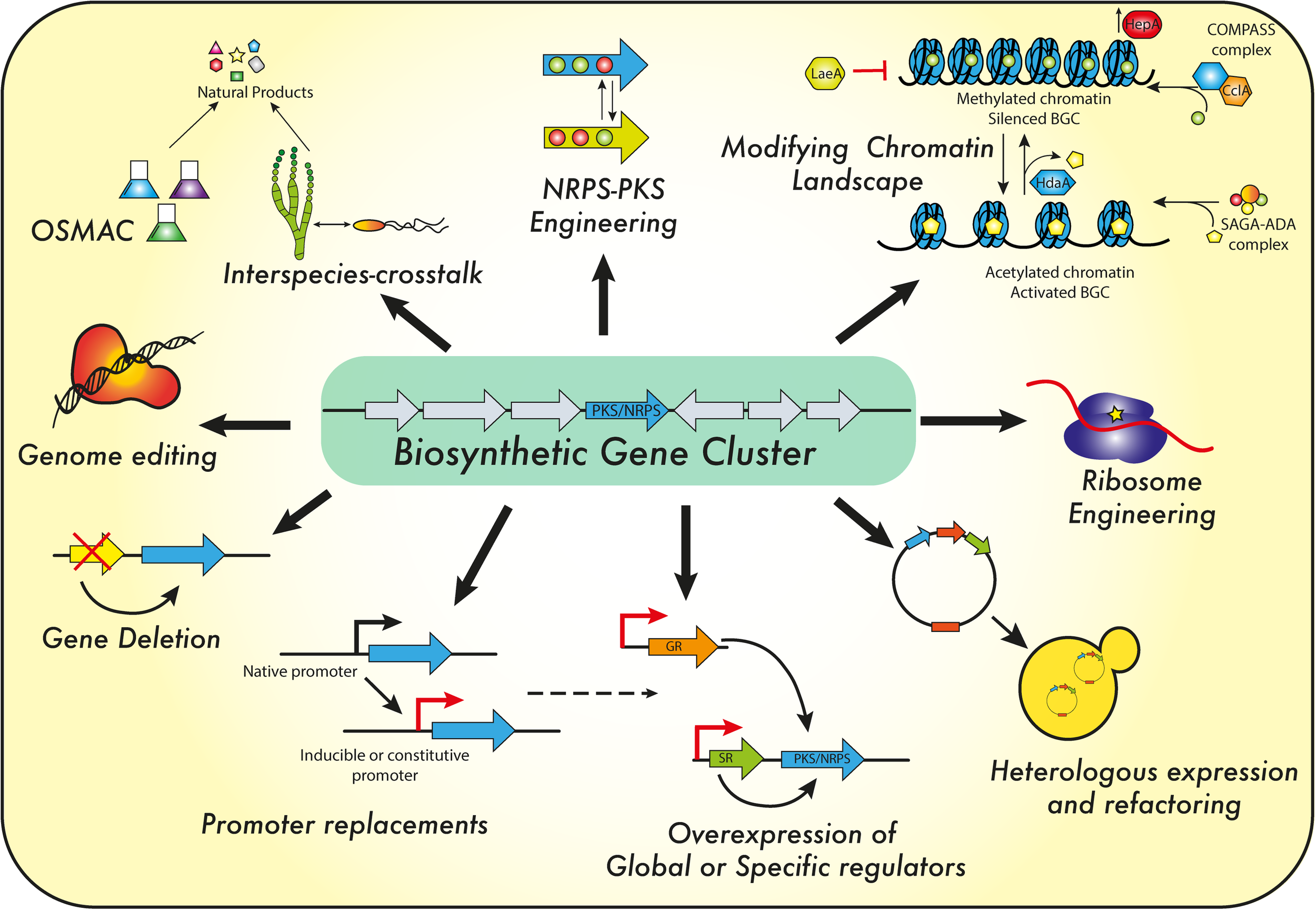
Regulatory targets and strategies to engineer P. chrysogenum and other filamentous fungi for secondary metabolite formation. For details see the text.
Manipulation of Cultivation Conditions
Under natural conditions, fungi face a variety of biotic and abiotic conditions to survive. The cellular response to the environment involves complex regulatory networks that respond to stimuli such as light, pH, availability of carbon and nitrogen sources, reactive oxygen species, thermal stress, and interspecies-crosstalk (Brakhage, 2012; Reen et al., 2015).
OSMAC (One Strain Many Compounds) Approach
This strategy is derived from the observation that changes in the metabolic output of microorganisms can be achieved by alternating the medium composition and other cultivation parameters. It is well known that glucose, ammonium, or phosphate at high concentrations act as repressors of secondary metabolism, whereas iron starvation and nitrogen limitation can stimulate secondary metabolite production. The latter is for instance exploited for the production of terrain by A. terreus (Bode et al., 2002; Brakhage and Schroeckh, 2011; Gressler et al., 2015). This strategy can readily be implemented using high-throughput methods, where an array of culture conditions can be screened for new metabolite profiles (Spraker and Keller, 2014). In combination with bioinformatics tools, this strategy can be a powerful tool to investigate the production of new molecules, as exemplified by the discovery of aspoquinolones A–D in A. nidulans (Scherlach and Hertweck, 2006). However, despite the fact that the OSMAC approach has led to the discovery of increased numbers of new molecules with antimicrobial activity, some chemical and physical conditions are still missing under the laboratory tested conditions as the activation often concerns a limited number of BGCs (Chiang et al., 2009).
Interspecies-Crosstalk
The production of secondary metabolites is a natural strategy that microorganisms have developed to cope with specific environmental conditions and challenges. They serve as intermediary agents to establish a symbiotic association between species or as a weapon against other organism to compete for nutrients and space. These conditions, that are not present in axenic cultures, boost the production of molecules that are constitutively present and/or that are cryptic and normally are not synthetized due to silencing of the respective BGCs (Demain and Fang, 2000; Marmann et al., 2014). The strategy in which different organisms are cultivated together is called “co-culture,” which has been successful in several cases yielding new metabolites. A. fumigatus produces fumiformamide when co-cultivated with Streptomyces peucetius, while co-cultivation of this fungi with S. rapamycinicus results in the production of fumicyclines A and B, two novel PKs with antibacterial activity, are examples of the use of this strategy (Netzker et al., 2015; Adnani et al., 2017). Interestingly, the association of two marine organisms, Emericella sp. and Salinispora arenicola, results in the biosynthesis of emericellamides A and B which are equipped with antibacterial activity (Oh et al., 2007). Also, the interactions between fungi and insects result in the production of volatile secondary metabolites (Rohlfs and Churchill, 2011).
Engineering of NRPS and PKS
Nonribosomal peptide synthetase and PKS are highly structured and multi-facetted enzymes, containing a tremendous potential for the exploitation of their product scaffold structure for the generation of novel, bioactive compounds. However, due to the complexity of all interactions within these mega enzymes, the elucidation and implementation of engineering strategies is an extremely challenging task. Several strategies have been developed and applied with different degrees of success, though the overall approaches can be grouped as module, domain, sub-domain or site directed, respectively. Owing to their large size, utilization of a random mutagenesis approach proved to be difficult, but other more directed strategies are met with a great success. Nevertheless, all of these strategies have their inherent difficulties, advantages and disadvantages in respect to the complexity and success rate of NRPS/PKS engineering efforts.
Subunit, Module, and Domain Swapping
Extensive efforts targeting the active site of A-domains has been a major focus in NRPS engineering. Multiple studies confirmed that the substrate specificity of a NRPS A-domain can be successfully altered, however, at the cost of substantially lowered catalytic velocity (Thirlway et al., 2012; Zhang et al., 2013). Similar successes and limitations were observed when domains were swapped or replaced by synthetic versions (Beer et al., 2014). The most challenging way of obtaining novel NRPS, however, is the swapping or combining of entire modules (Kries, 2016). Domain swapping overall created not only functional parts or domains, but also complete NRPS though with limited success (Beer et al., 2014).
Due to the strict arrangement of NRPS in domains and modules, the possibility of exchanging a unit appears to be the most straight forward approach for altering its intrinsic properties. A series of studies targeting the enzymes linked to the production of daptomycin (Nguyen et al., 2006; Baltz, 2014) elucidated the possibilities and borders of a combinatorial swapping strategy in context of novel compound production. The daptomycin biosynthetic cluster comprises three NRPS containing a total of 13 modules for the incorporation of an equal number of substrates. Different levels of domain and module swap approaches were followed, starting with the exchange of modules 8 and 11 (C-A-T), representing an internal module exchange. The resulting NRPS exhibited the production of novel daptomycin compounds with an inverted amino acid composition at the predicted sites at a near native rate (Nguyen et al., 2006). A similar combinatorial approach has been chosen for altering the PK stereochemistry. The exchange of a R domain with a TE domain in a NR-PKS from A. niger produced two alternative NR-PKS that harbor carboxylic acids instead of the aldehydes present in the original products (Yeh et al., 2013; Weissman, 2016). In Aspergillus, this rational domain swap has also been used to diversify the native substrates that NR-PKS takes as starter unit to produce new products. This involved exchanging the starter unit ACP transacylase domain in the PKS (Liu et al., 2014). Likewise, an analogous approach was used to produce new hybrids (PK–NRPs) in A. nidulans via module swapping of the two PKS–NRPS natural hybrids involved in the syn2 and cytochalasin E pathways from Magnaporthe oryzae and A. clavatus respectively (Nielsen et al., 2016). Despite the successful use of this strategy in some filamentous fungi, the engineering of NRPS and PKS in P. chrysogenum remains unexplored.
Genetic Interference
Another mechanism to stimulate the expression of silent BGCs in P. chrysogenum is by genetic interference, for instance by direct manipulation of the regulatory network related to BGCs expression. The regulation of BGCs is effected at many levels, through specific (or local) and global regulators up to epigenetic regulation involving the modification of the chromatin landscape (Lim et al., 2012; Spraker and Keller, 2014).
Global and Specific Regulators
Global regulator-based regulation
Pleiotropic transcriptional regulators or global regulators are proteins that respond to environmental signals such as pH, temperature, and N- and C-sources. They provide the link between the production of secondary metabolites and external cues. In fungi, these proteins control the regulation of BGCs that do not contain other regulatory factors. Up to 40% of the known clusters do not encode a local and specific regulator (or obvious regulatory genes). Additionally, global regulators also act on genes that do not belong to secondary metabolism (Brakhage, 2012; Rutledge and Challis, 2015; Fischer et al., 2016). Global regulators that have been reported as key players in the biosynthesis of secondary metabolites are featured below.
Velvet complex
This heterotrimeric complex is a conserved regulator present in most of the fungi, except yeast. It consists of at least three proteins: VeA, VelB, and LaeA. Likewise, this complex provides a link between sexual development and secondary metabolism through light regulation (Yin and Keller, 2011; Deepika et al., 2016), since light has an inhibitory effect on VeA expression. The formation of the velvet complex takes place in the nucleus, where the complex VeA–VelB via the α-importin KapA meets the methyltransferase LaeA. It has been hypothesized that the velvet complex acts as a transcriptional factor as it contains a DNA binding fold that resembles the corresponding region of the NF-κB transcription factor of mammals (Sarikaya-Bayram et al., 2015). The role of the velvet complex in secondary metabolism mostly follows from the control that the LaeA protein executes on several BGCs in filamentous fungi. LaeA (loss of aflR expression-A) was identified in 2004 as a global regulator in Aspergillus. Deletion of this gene results in the repression of many BGC, such as the one responsible for the production of penicillin, lovastatin, and sterigmatocystin. Overexpression of LaeA causes an opposite phenotype. Interestingly, LaeA is negatively regulated by AflR (Zn2Cy6 transcriptional factor) in a feed loop mechanism (Bok and Keller, 2004). It has been hypothesized that LaeA acts at different levels, i.e., as a methyltransferase, epigenetically and as a direct member of the velvex complex. Structurally, LaeA has a S-adenosyl methionine (SAM)-binding site with a novel S-methylmethionine auto-methylation activity, although this activity does not seem to be essential for its function. LaeA is not a DNA-binding protein, but it does affect chromatin modifications. In an A. nidulans ΔlaeA strain, high levels of the heterochromatin protein 1 (HepA) are detected and an increase in trimethylation of the H3K9 in the sterigmatocystin cluster. When LaeA is present, the levels of HepA, ClrD (H3K9 methyltransferase) and H3K9me3 decrease while the sterigmatocystin levels are raised. The heterochromatic marks stay until the sterigmatocystin cluster is activated, and apparently LaeA influences the offset of these marks in this particular cluster (Reyes-Dominguez et al., 2010; Brakhage, 2012; Jain and Keller, 2013; Sarikaya-Bayram et al., 2015; Bok and Keller, 2016). Orthologs of LaeA have been discovered in many other filamentous fungi as Penicillium, Fusarium, Trichoderma, Monascus spp. and LaeA exhibits positive and negative effects on the synthesis of NPs. For instance, LaeA1 of F. fujikuroi positively regulates the production of fusarin C, fumonisins and gibberellins, and represses bikaverin biosynthesis. In P. chrysogenum, LaeA controls the biosynthesis of penicillin, pigmentation and sporulation (Keller et al., 2005; Kosalková et al., 2009; Jain and Keller, 2013). In Trichoderma reesei, Lae1 positively modulates the expression of cellulases, xylanases, β-glucosidases. Interestingly the stimulation of these genes was not directly influenced by the methylation of H3K4 or H3K9 (Wiemann et al., 2010; Yin and Keller, 2011; Lim et al., 2012; Seiboth et al., 2012; Jain and Keller, 2013).
LaeA is not the only member of the velvet complex that has influence on the regulation of secondary metabolite production. VeA of A. parasiticus is necessary for the expression of two transcriptional factors of the aflatoxin cluster (AflR and AflJ), which regulate the pathway. In A. fumigatus, veA regulates 12e BGCs (Dhingra et al., 2013). This study also revealed that veA modulates the biosynthesis of fumagillin via the regulation of FumR, a transcriptional factor of the fumagillin cluster, which in turn is also regulated by LaeA. Similarly, a transcriptome analysis in A. flavus revealed that 28 of 56 BGCs are dependent on veA, in particular the aflavarin cluster which is differentially expressed. Likewise, orthologs of veA are also present in other fungi such as in P. chrysogenum, F. oxysporum, Botrytis cinerea, F. verticillioides (Yin and Keller, 2011; Dhingra et al., 2013; Jain and Keller, 2013; Cary et al., 2015). Despite the clear interaction between veA and LaeA in the velvet complex and its influence on secondary metabolism, it is thought that veA may be acting as molecular scaffold of the velvet complex, since it interacts also with three other methyl transferases [LaeA-like methyltransferase F (LlmF), velvet interacting protein C (VipC), and VipC associated protein B (VapB)]. This suggests that veA functions in a supercomplex or in dynamic network control. Taken together, modulation of the velvet complex is a useful tool to activate BGCs (Sarikaya-Bayram et al., 2015) but results are difficult to predict.
bZIP transcription factors
Basic leucine zipper (bZIP) transcription factors are highly conserved in the eukaryotes. The dimeric bZIP transcriptional factors play an important role in the cellular responses to the environment. Regarding their structure, they contain a conserved leucine zipper domain and a basic region, which controls the dimerization of the protein and establishes sequence-specific DNA-binding, respectively. Once dimeric, bZIPs target palindromic DNA sequences by two mechanisms: redox and phosphorylation (Amoutzias et al., 2006; Knox and Keller, 2015). In fungi, bZIP proteins have been implicated in multiple metabolic processes, such as in the regulation of development, morphology and in stress responses. Several orthologs of the Yap family bZIPs, which were first described in yeast, have been characterized in Aspergillus spp. (AtfA, NapA, Afyap1, Aoyap1, and Apyap1) and these regulators have recently been associated with the production of secondary metabolites in filamentous fungi. In A. nidulans, overexpression of RsmA (restorer of the secondary metabolism A, Yap-like bZIP) has a compensatory effect on secondary metabolism in a strain in which LaeA and veA are missing. However, these transcription factors also display negative regulation. For instance, an increase in the biosynthesis of aflatoxin and chratoxin has been observed when yap1 is deleted in A. parasiticus and A. ochraceus (Yin et al., 2013; Knox and Keller, 2015; Wang X. et al., 2015). MeaB is another bZIP transcriptional factor which was discovered in A. nidulans. Its function is associated in nitrogen regulation and has a negative effect on the biosynthesis of aflatoxin in A. flavus and bikaverin production in F. fujikuroi (Wagner et al., 2010; Amaike et al., 2013).
Other global regulators
AreA is a highly conserved transcriptional factor in fungi that belongs to the GATA family and it is characterized by Cys2His2 zinc finger DNA binding domains. Likewise, it is involved in the repression of nitrogen metabolism when ammonium or glutamine are present. Recently, this transcription factor and its orthologs have been shown to influence secondary metabolism. For instance, areA deletion strains of F. verticillioides are not able to produce fumonisins on mature maize kernels. In Acremonium chrysogenum, the deletion of areA resulted in the reduction of cephalosporin because of a reduced expression of the enzymes involved in cephalosporin biosynthesis. Additionally, AreA is a positive regulator of the production of gibberellins, trichothecene deoxynivalenol (DON), fusarielin H, beauvericin and zearalenone (Li et al., 2013; Tudzynski, 2014; Knox and Keller, 2015 and Keller, 2015). The carbon catalytic repressor CreA also influences secondary metabolism. CreA is a Cys2His2 zinc finger transcription factor that is involved in the repression of genes associated with the use of carbon sources other than glucose (Knox and Keller, 2015). This transcription factor acts by direct competition with activator proteins for specific binding sites (5′-SYGGRG-3′) and by direct interaction with activators (Janus et al., 2008). In P. chrysogenum CreA represses penicillin biosynthesis and causes a reduced expression of the pcbAB gene that encodes a NRPS involved in this pathway. Mutations in the putative CreA binding site in the pcbAB promoter result in enhanced enzyme expression when cells are grown in the presence of glucose (Cepeda-García et al., 2014). In contrast, mutations in the CreA binding sites of the ipnA promoter (pcbC in other species) of A. nidulans revealed that in this organism repression of penicillin biosynthesis by glucose is independent of CreA (Knox and Keller, 2015). CreA has been implicated in the variable metabolite profiles when fungi are grown in the presence of different carbon sources (Yu and Keller, 2005). Recently, the xylanase promoter binding protein (Xpp1) of Trichoderma reesei was used as a reporter to fulfill a dual role in the regulation of primary and secondary metabolism. Xpp1 is an activator of primary metabolism, while its deletion boosts the production of secondary metabolites, including sorbicillinoids (Derntl et al., 2017b). Another Cys2His2 zinc finger transcription factor conserved in fungi is PacC, which is involved in pH dependent regulation. Deletion of the ortholog of this gene (BbpacC) in Beauveria bassiana resulted in a loss of dipicolinic acid (insecticide compound) and oxalic acid production, compounds that reduce the pH of the medium. However, also production of a yellow pigment was noted. When A. nidulans is grown at alkaline pH, PacC modulates the expression of the acvA (pcbAB) and ipnA of the penicillin BGC, while it acts negatively on the expression of the sterigmatocystin BGC (Deepika et al., 2016; Luo et al., 2017). In filamentous fungi, another global regulatory element is the CCAAT-binding complex (CBC). This complex consists of three proteins (HapB, HapC, and HapE) that respond to redox stimuli and an additional unit HapX, a bZIP protein that interacts with the complex for modulating the iron levels. In A. nidulans this complex binds to CAATT motifs, which are present in the penicillin BGC stimulating the expression of the ipnA and aatA (penDE) genes (Bayram and Braus, 2012; Brakhage, 2012). Whereas in F. verticillioides the ortholog core of this complex (FvHAP2, FvHAP3, and FvHAP5) is deleted, cells show an altered hyphal morphology, reduction of growth, reduced pathogenesis and a deregulation of secondary metabolism (Ridenour and Bluhm, 2014).
Specific regulator-based regulation
In addition to the global regulators, the expression of BGCs can be also modulated by specific regulatory elements, which most of the times are encoded by genes that are part of the same cluster that they regulate. In some cases, such regulators also influence the expression of other BGCs. It is estimated that about 60% of the fungal BGCs contain a gene encoding a potential regulator amidst the gene cluster. With PKS containing BGCs mostly containing a regulator that belongs to the Zn2Cys6 binuclear cluster domain family (around 90%). With NRPS containing BGCs, the putative transcription factors are more diverse. The Zn2Cys6 family of transcription factors contain a DNA binding domain (DBD) that has two zinc atoms coordinated by six cysteines. There are three sub regions: a linker, a zinc finger and a dimerization domain. Additional to a DBD, these proteins contain two further functional domains, the acidic region and the regulatory domain. These transcription factors can act as monomers, hetero- and homodimers. They recognize single or multiple trinucleotide sequences, commonly CCG triples, in a symmetric or asymmetrical format. The affinity of the DBD for a given DNA stretch is also determined by the nucleotides surrounding this triplet. The transcriptional activity of these proteins is regulated by phosphorylation, exposing the activation and DNA binding domains for DNA binding (MacPherson et al., 2006; Brakhage, 2012). Some of these regulators have been shown to control the expression of BGCs. For instance, in F. verticillioides the disruption of FUM21 gene, that encodes a Zn2Cys6 protein, reduces fumonisin production as a result of a downregulation of the BGC (Brown et al., 2007). Interestingly, fumonisin production is also regulated by another Zn2Cys6 protein that is encoded by a gene located outside of the fumonisin cluster (Flaherty and Woloshuk, 2004). Mlcr is another example of a positive regulator that controls compactin production in P. citrum (Abe et al., 2002). AflR is a Zn2Cys6 protein that regulates the biosynthesis of aflatoxin/sterigmatocystin through binding to a palindromic sequence (5′-TCG(N5)GCA) that is found in most of the promoters of this BGC, albeit a second binding sequence has been reported that is associated with the autoregulation mechanism of the expression of AflR. The disruption of AlfR abolishes the production of aflatoxin/sterigmatocystin. Likewise, some BGCs encode multiple regulatory proteins. Next to the aflR gene in the aflatoxin cluster resides the aflS (formerly aflJ) gene. The corresponding transcription factor binds to AflR to enhance the transcription of early and mid-biosynthetic genes in the aflatoxin pathway (Georgianna and Payne, 2009; Yin and Keller, 2011). In P. chrysogenum and Trichoderma reesei, the sorbicillin BGC is regulated by two transcriptional factors through a coordinated action (Derntl et al., 2016, 2017a; Guzmán-Chávez et al., 2017). Also, regulation of BGCs via crosstalk has been observed in filamentous fungi. For instance, the alcohol dehydrogenase promoter has been used to induce the expression of putative pathway-specific regulatory gene (scpR) in A. nidulans, which controls the expression of two pathway associated NRPS genes (inpA and inpB). Surprisingly, two PKS genes (afoE and afoG) and one transcriptional activator (afoA) belonging to the asperfuranone BGC are also upregulated by ScpR, allowing the production of asperfuranone (Bergmann et al., 2010). For some regulators, no clear phenotype is observed. For instance deletion of the chyR gene of the chrysogine BGC in P. chrysogenum, has no effect on the expression of the corresponding BGC (Viggiano et al., 2017).
Manipulation of regulatory elements as strategies for the activation of BGCs
Gene deletion
It is a classical strategy that consists of the abolishment of the expression of a certain gene by its elimination whereupon the impact on the metabolite profile is examined by HPLC or LC-MS. A major limitation of this approach is that it can only be used in BGCs that are not totally silenced under laboratory conditions. Using this strategy, it was possible to elucidate the highly branched biosynthetic pathway for the synthesis of roquefortine as well as the biosynthetic pathways of sorbicillinoids and chrysogine in P. chrysogenum (García-Estrada et al., 2011; Ali et al., 2013; Ries et al., 2013; Deepika et al., 2016; Guzmán-Chávez et al., 2017; Viggiano et al., 2017). Likewise, this approach can be used to remove transcriptional repressor genes, as in the case of TetR-like pathway-specific repressor proteins, whose deletion induced the production of gaburedins in Streptomyces venezuelae (Rutledge and Challis, 2015). Global regulators, such as LaeA have also been targeted using this strategy (Chiang et al., 2009).
Promoter replacement
Another method concerns the replacement of the endogenous promoter of the gene(s) in a BGC by a strong constitutive or inducible promoter. For instance in A. nidulans replacement of the native promotor of the scpR gene (secondary metabolism cross-pathway regulator) for the inducible promoter of alcohol dehydrogenase AlcA induced the expression of a silent cluster that contained two NRPS genes (inpA and inpB) and scpR itself. Additionally, it also led to the expression of the asperfuranone BGC, which is normally silent (Bergmann et al., 2010; Yin and Keller, 2011; Lim et al., 2012). Recently in P. chrysogenum a promising promoter toolbox for bioengineering purposes was developed. This included the analysis of four constitutive promoters from P. chrysogenum and six from A. niger, which were evaluated using a reporter system and assorted by promoter strength (Polli et al., 2016).
Overexpression of a specific or global regulator
This approach is one of the most used strategies to turn on cryptic BGCs, since a change in expression level of a regulator may boost the expression of a whole cluster. Usually, this strategy is applied in combination with the promoter replacement approach. Using this strategy, i.e., overexpression of the transcription activator ApdR under control of the alcohol dehydrogenase promoter alcAp, it has been possible to induce the expression of a hybrid PKS-NRPS BGC in A. nidulans. This resulted in the production of aspyridones A and B (Bergmann et al., 2007). Similarly when the global regulator FfSge, which is associated with vegetative growth of F. fujikuroi, is overexpressed, some BGCs are forced to express under these unfavorable conditions (low nitrogen concentrations) leading to the identification of the corresponding products (Michielse et al., 2015).
Chromatin-Mediated Regulation
In fungal cells, chromosomal DNA is wrapped in a complex of DNA, histone proteins and RNA called chromatin. This chromatin structure consists of a basic unit called nucleosome, which consists of superhelical DNA (147 base pairs) that binds an octamer of four different core histone proteins (two each of H2A, H2B, H3, and H4) in 1.75 turns. It has been shown that modifications of the chromatin structure (boosts or alters) changes gene expression, amongst other genes involved in the biosynthesis of secondary metabolites. Structurally, chromatin represents an obstacle that complicates access of DNA-binding factors to their corresponding binding regions. According to the compaction level, chromatin can be in a dense (heterochromatin) or relaxed (euchromatin) state. These compaction levels are regulated by post-translational modification of the histone proteins by acetylation, methylation, ubiquitination, ethylation, propylation, butylation, and phosphorylation events. Regions that display low transcriptional activity have been associated with the heterochromatic conformation. In contrast, the euchromatic conformation is present in regions with abundant coding sequences and is usually highly active during transcription. Such regions are also linked with hyper-acetylated nucleosomal histones. Likewise, it has been reported that methylation of H3K9, H3K27, and H4K20 are typical markers of the heterochromatin, while in euchromatin methylation occurs at H3K4 (Brosch et al., 2008; Strauss and Reyes-Dominguez, 2011; Gacek and Strauss, 2012; Spraker and Keller, 2014; Rutledge and Challis, 2015).
Histone methylation, acetylation, and sumoylation
As mentioned above, LaeA influences secondary metabolite production through chromatin modification. The methylation state of H3K9 has been correlated with the heterochromatin protein A (HepA), since this protein needs the di- and tri- methylation of H3K9 for binding to chromatin and to form heterochromatin. Deletion of LaeA allows the unobstructed binding of HepA to the AlfR promoter, thereby affecting the expression of the sterigmatocystin pathway. The deletion of the methyltransferase encoding clrD and ezhB genes in Epichloe festucae, that act on H3K9 and H3K27, respectively (in axenic culture), results in the activation of the ergot alkaloids and lolitrem BGCs. These compounds are necessary to establish a symbiotic association with the plant Lolium perenne. Compass (complex of proteins associated with Set1) which methylates H3K4 in yeast, also impacts secondary metabolism in filamentous fungi. The deletion of one of its components (cclA) in A. nidulans allowed the activation of a cryptic BGC and the production of emodin (Palmer and Keller, 2010; Gacek and Strauss, 2012; Chujo and Scott, 2014; Netzker et al., 2015; Deepika et al., 2016). Likewise, in F. fujikuroi and F. graminearum, the deletion of cclA caused the overproduction of secondary metabolites derived from BGCs close to the telomeres, but this seems to relate to a H3K4 methylation independent mechanism (Studt et al., 2017). Other types of histone modification may alter the chromatin landscape, such as acetylation which is a reversible process governed by two antagonist enzymes: histone acetyltransferases (HATs) and deacetylases (HDACs). Active transcription is usually associated with histone acetylation, although recently the deacetylation of histones has been shown to cause activation of genes (Brosch et al., 2008). Usually, histones are acetylated by several complexes with acyltransferase activity, such as Saga/Ada and NuA4. In A. nidulans a chromatin immunoprecipitation (ChIP) analysis revealed that GcnE and AdaB, the catalytic subunits of the complex Saga/Ada, are needed for acetylation of histone H3 (Deepika et al., 2016). Indeed, the interaction between A. nidulans and Streptomyces rapamycinicus can be linked to a GcnE dependent increase in the acetylation of H3K14 that shields the promoters of the orsellinic acid BGC. The Saga/Ada complex is a key player in the induction of the penicillin, terrequinone and sterigmatocystin BGCs (Nutzmann et al., 2011; Brakhage, 2012). In contrast, deletion of hdaA (encoding a HDAC) in A. nidulans resulted in major changes in the metabolite profile (Rutledge and Challis, 2015). HdaA is a class 2 histone deacetylase involved in the regulation of BGCs that are located near the telomeres, such as the penicillin and sterigmatocystin clusters in A. nidulans. Indeed, deletion of the hdaA gene results in the increased and early gene expression of these two BGCs, and the production of the corresponding secondary metabolites. In A. fumigatus, the hdaA gene is involved in growth and production of secondary metabolites, and the deletion of this gene increases the production of many secondary metabolites while it causes a reduction of gliotoxin production. In contrast, HdaA overexpression shows the opposite effect (Shwab et al., 2007; Lee et al., 2009). In P. chrysogenum was demonstrated that HdaA (histone deacetylase) mediates the transcriptional crosstalk among sorbicillinoids biosynthesis and other BGCs, since a new compound as detected only under conditions of sorbicillinoids production (Guzmán-Chávez et al., 2018).
Histone deacetylases are ubiquitously distributed in filamentous fungi, and therefore HDAC inhibitors can be used to improve the synthesis of NPs by epigenome manipulation (Shwab et al., 2007; Lee et al., 2009). For instance, the metabolite profile of Cladosporium cladosporioides and A. niger underwent a significant change when these strains were exposed to suberoylanilide hydroxamic acid (SAHA), a HDAC inhibitor, allowing the detection of two new compounds, cladochrome and nygerone A, respectively (Rutledge and Challis, 2015). An exploratory analysis performed in 12 fungi treated with different types of DNA methyltransferase and histone deacetylase inhibitors, revealed the production of new secondary metabolites but also the elevated amounts of known compounds (Williams et al., 2008). In this respect, the chromatin state can directly influence the binding of transcription factors, and thereby modulate expression (Palmer and Keller, 2010; Macheleidt et al., 2016). It has been hypothesized that histone sumoylation may modulate secondary metabolite production. This process is mediated by a small protein termed SUMO (small ubiquitin-like modifier) that shares structural similarity to the ubiquitin protein. In A. nidulans, deletion of the sumO gene enhanced the production of asperthecin, whereas synthesis of austinol, dehydroaustinol, and sterigmatocystin was reduced. Although the molecular mechanism still needs to be elucidated, it is thought that sumoylation acts at several levels, such as on epigenetic regulators (COMPASS, Clr4, SAGA/ADA and HDACs) or at the level of transcriptional regulators (Brakhage and Schroeckh, 2011; Spraker and Keller, 2014; Wu and Yu, 2015).
Modification of the chromatin landscape to activate BGCs
Many fungal BGCs are located in distal regions of the chromosomes. In these heterochromatin rich regions, transcription of the BGCs can be activated by epigenetic regulation. Therefore, the encoding genes of proteins that influence histone modification are prime targets, although these modifications can also be achieved by chemical treatment (Williams et al., 2008; Brakhage, 2012). A recent study in P. chrysogenum showed that the expression of a set of PKS and NRPS encoding genes is induced when an ortholog of a class 2 histone deacetylase (HdaA) is deleted. This allowed for the overproduction of sorbicillinoids, the reduction of chrysogine related metabolites and the detection of a new compound whose origin still unknown (Guzmán-Chávez et al., 2018).
Other Targets for Regulation
Secondary metabolites produced by fungi can be toxic to the producer organisms, and often fungi are equipped with detoxification mechanisms. One of these mechanisms is toxin excretion by transporters, which are membrane proteins whose genes often localize to the BGCs. Transporters may belong to different protein families but the major facilitator superfamily (MFS) and ABC superfamily are most commonly encoded by BGCs (Keller, 2015). Since biosynthesis of secondary metabolites may take place in different cell compartments, also intracellular transport may be evident (Kistler and Broz, 2015). Despite their assumed biological importance, the deletion of transporter genes from the BGCs often does not impact secondary metabolite production. For instance, deletion of the A. parasiticus aflT gene, that encodes a MFS transporter, does not result in reduced aflatoxin excretion, despite the fact that aflT belongs to the aflatoxin BGC and its expression is regulated by a specific transcription factor, AflR, of the pathway. Probably, this protein is redundant, and other transporters may participate in excretion, detoxification or self-defense. In A. fumigatus, GliA facilitates the excretion of gliotoxin. Similarly, the tri12 gene contained in the trichothecene BGC encodes for a membrane protein required for the biosynthesis of trichothecene and virulence of F. graminearum on wheat crops (Chang et al., 2004; Menke et al., 2012; Wang D.N. et al., 2014; Keller, 2015). Often, however, the deletion of the transporter gene in BGCs has no effect on production. Possibly, these metabolites are also recognized by other promiscuous transporters, or transporters that are not part of the BGC (Keller, 2015). For example, ZRA1 of Gibberella zae, whose gene is not localized to the zearalenona BGC, impacts zearalenone production. However, the expression of the zra1 gene is regulated by the transcriptional factor ZEB2, whose gene localizes to the corresponding BGC (Lee et al., 2011). Also, the penicillin BGC of P. chrysogenum lacks a transporter gene whereas export of penicillin occurs against the concentration gradient, probably through the activity of multiple transporter proteins (van den Berg et al., 2008; Kistler and Broz, 2015). Furthermore, compartmentalization of the biosynthesis of penicillin is well documented requiring transport of penicillin precursors across the membrane of intracellular organelles (Weber et al., 2012a,b).
Other Genetic Engineering Strategies for the Activation of BGCs
Several approaches have been used to activate the expression of cryptic BGCs in a targeted manner. Usually, this is achieved by manipulation of pathway-specific regulatory genes, or by replacing endogenous promoters for inducible systems or strong promoters (Rutledge and Challis, 2015). The various approaches are summarized in Figure 7.
Manipulating biosynthetic pathways by genome editing
Due to the increasing number of sequenced filamentous fungi, it is necessary to make use of efficient genome editing tools to explore new potential sources of secondary metabolites. For many years, the unique strategy available for the genome edition of P. chrysogenum was based on the use of ku70/80 disrupted strains to improve the homologous recombination instead of the Non-Homologous End Joining (NHEJ) pathway (Weber et al., 2012a). This strategy allowed for the generation P. chrysogenum strains with high copy numbers of the penicillin cluster, the identification of a biosynthetic branch of the roquefortine cluster and the reactivation of the sorbicillinoid gene cluster (Nijland et al., 2010; García-Estrada et al., 2011; Ali et al., 2013; Ries et al., 2013; Salo et al., 2016; Guzmán-Chávez et al., 2017). Recently, a CRISPR/Cas9 based system was developed for genome modifications in P. chrysogenum (Pohl et al., 2016, 2018). This study demonstrated that the deletion of full gene clusters is feasible with minimal cloning efforts, which opens the possibilities to engineer new synthetic pathways and the re-factoring P. chrysogenum as platform organism.
Ribosome engineering
This approach has been applied for activating silent or poorly expressed BGCs (Ochi and Hosaka, 2013). Basically, this concept is derived from the activation of the actinorhodin BGC in S. lividans due to a point mutation in the rpsL gene, which encodes for the ribosomal S12 protein (Shima et al., 1996). Another successful examples in the BGCs activation have been reported by modifying the transcription and translation pathways via targeting different ribosomal proteins, RNA polymerases (RNAP) and translation factors (Ochi and Hosaka, 2013). In P. purpurogenum G59, a marine derived strain, the insertion of gentamicin resistance after treatment with high concentrations of this antibiotic, altered ribosomal functions of this fungus which allowed for the activation of dormant secondary metabolite gene clusters (Chai et al., 2012).
Heterologous expression and refactoring
Due to the broad range of molecular tools available to express heterologous pathways in yeast, several attempts have been undertaken to express NRPS and PKS genes with the remainder of the pathway in yeast (Rutledge and Challis, 2015). A recent study demonstrated that the baker’s yeast Saccharomyces cerevisiae can be used as a platform to produce and secrete penicillin when the biosynthetic genes are expressed in this organism (Awan et al., 2016). Although the first step was performed when the acetyl-CoA:isopenicillin N acyltransferase (IAT), which catalyzes the last step in the penicillin biosynthesis was amplified from the P. chrysogenum penDE gene and expressed in Hansenula polymorpha (Lutz et al., 2005). However, most of the times the main obstacle is the large size (>40 kbp) of the DNA fragment that needs to be cloned, the effective activation/maturation of the expressed enzymes, and the toxicity of the produced compounds (Rutledge and Challis, 2015). Alternatively, fungi may be used as platform organism, as it was for instance demonstrated with the reconstruction of the citrinin gene cluster of Monascus purpurea in A. oryzae (Spraker and Keller, 2014). Likewise, the in vivo assembly of genetic elements has been successfully applied in P. chrysogenum through the overlapping of bi-partite fragments that reconstituted a functional amdS gene (marker), which eventually is integrated in the genome of this fungus proving the uncharacterized potential of P. chrysogenum as heterologous host (Polli et al., 2016). The potential of this approach follows a recent study employing A. nidulans as a host for the plasmid based expression of a diverse range of BGCs from other filamentous fungi (Clevenger et al., 2017).
The introduction of revolutionary new genetic tools, such as CRISPR/Cas9 offers more effective solutions to express specific BGCs. Such methods can contribute to product identification but also to the production of unique compounds by introduction of specific tailoring enzymes. These are the main strategies that are used for the activation of silent BGCs or for the modification/redirection of known biosynthetic pathways in order to increase NP diversification (Smanski et al., 2016). Specifically, this involves the expression of pathways from a plasmid in a suitable production host and a screen for product formation.
Concluding Remarks
For many years P. chrysogenum has been used as one of the main industrial strains to produce penicillins (β-lactams). Its genome sequence revealed an unexplored potential of P. chrysogenum as a source of NPs (van den Berg et al., 2008). Despite the development of bioinformatics tools for genome mining of BGCs to identify novel molecules (Blin et al., 2017), the experimental validation of product structure and identity is still necessary. However, most of the secondary metabolite associated genes in P. chrysogenum are silent or poorly expressed. Given the urgent need for new molecules based on novel chemical scaffolds for the use in the medical and biotechnological fields (e.g., antibiotics, anti-cancer agents, antivirals, nutraceuticals, pigments, surfactants and many more), the use of organisms that have been genetically domesticated offers a promising target solution for NP discovery due to the availability of molecular tools for their genetic modification. Here, we have summarized the main approaches that have been applied for P. chrysogenum and other filamentous fungi to bioengineer secondary metabolite BGC pathways which have led to a greater understanding of the main obstacles to be overcome to use this fungus as a generic cell factory. We discussed the main characteristics of the building enzymes (PKS and NRPS) in filamentous fungi. Despite the apparent modular organization, the complexity of these mega enzymes and the inherent interactions between the various domains within their structures has not allowed a straight forward approach for the PKS/NRPS engineering (Thirlway et al., 2012; Zhang et al., 2013; Weissman, 2016). However, the combinatorial swapping strategy of structural elements such as recognition regions has increased the perspectives for designing de novo biosynthetic pathways. Further research needs to be focused around PKS/NRPS engineering in filamentous fungi to facilitate the rational design of biosynthetic enzymes to produce another generation of novel compounds. To mine the secondary metabolome of filamentous fungi, general methods such as manipulating cultivation conditions have been used that can also be implemented as a high-throughput strategy. Another avenue is interfering with the genetic regulatory systems, either through the manipulation of specific or global regulators. This strategy also revealed crosstalk between certain BGCs and an important role of chromatin remodeling in BGC expression. Because of its pleiotropic effect that leads to the activation or silencing of biosynthetic pathways, chromatin remodeling might be used as a more general strategy to explore the production of new metabolites in filamentous fungi (Guzmán-Chávez et al., 2018).
A further major advance is the development of genome-editing tools that allow for efficient genetic engineering of complex fungal cell factories (Jakoèiunas et al., 2016). In P. chrysogenum, improved methods for homologous recombination and CRISPR/Cas9 as genome editing tool now facilitates more advanced engineering of this fungus (Pohl et al., 2016, 2018). This is further stimulated by the development of a synthetic biology toolbox using promoters and terminators as building blocks and more complex regulatory devices to control the expression of genes. Importantly, the in vivo assembly of genetic elements in P. chrysogenum offers a promising tool to build entire pathways from scratch with reducing cloning efforts at minimal costs (Polli et al., 2016). In particular the low cost of DNA synthesis will allow rapid progress using such approaches as exemplified by a study using A. nidulans as a host where a diverse set of BGCs was expressed from an extra-chromosomal vector, the AMA plasmid (Clevenger et al., 2017). A further challenge is the generation of a platform strain in which endogenous BGCs have been removed to allow for more optimal carbon and nitrogen flow toward the production of the compounds of interest. Herein, the CRISPR/Cas9 based methods are instrumental (Pohl et al., 2016, 2018). Such a Penicillium platform might be used as a heterologous host to express a vast arsenal of BGCs from others filamentous fungi and represents a good alternative to yeast as expression host, an organism that does not naturally produce NRP and PK. The use of a such industrial strains to rapidly achieve the high level production of a novel metabolite has proven to be successful for pravastatin production (McLean et al., 2015) but a further step is to make use of secondary metabolite deficient industrial strains.
Despite the progress in genetic engineering and bioinformatics tools to identify BGCs, the main bottleneck to identify potentially interesting compounds has not yet been solved. Bioinformatic tools perform poorly in the prediction of the structures formed, and therefore future discovery programs will mostly dependent on high throughput methods to express foreign pathways and then use advanced metabolomics to identify the novel products. Since such approaches depend on high throughput, further efforts are needed to implement high throughput cloning methods to P. chrysogenum which will enable further studies to harness the enormous untapped source for NPs hidden in fungal (meta-)genomes.
Statements
Author contributions
FG-C and RZ wrote the manuscript. AD supervised, conceived, and designed the manuscript. RB co-supervised the manuscript.
Funding
FG-C was supported by Consejo Nacional de Ciencia y Tecnología (CONACyT, Mexico) and Becas Complemento SEP (Mexico). RZ was supported by the BE-Basic Foundation, an international public–private partnership.
Conflict of interest
RB is an employee of DSM Biotechnology, Delft. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
AbeY.OnoC.HosobuchiM.YoshikawaH. (2002). Functional analysis of mlcR, a regulatory gene for ML-236B (compactin) biosynthesis in Penicillium citrinum.Mol. Genet. Genomics268352–361. 10.1007/s00438-002-0755-5
2
AdnaniN.RajskiS. R.BugniT. S. (2017). Symbiosis-inspired approaches to antibiotic discovery.Nat. Prod. Rep.34784–814. 10.1039/C7NP00009J
3
AliH.RiesM. I.NijlandJ. G.LankhorstP. P.HankemeierT.BovenbergR. A. L.et al (2013). A branched biosynthetic pathway is involved in production of roquefortine and related compounds in Penicillium chrysogenum.PLoS One8:e65328. 10.1371/journal.pone.0065328
4
Al-MestarihiA. H.VillamizarG.FernandezJ.ZolovaO. E.LomboF.Garneau-TsodikovaS. (2014). Adenylation and S-methylation of cysteine by the bifunctional enzyme TioN in thiocoraline biosynthesis.J. Am. Chem. Soc.13617350–17354. 10.1021/ja510489j
5
AmaikeS.AffeldtK. J.YinW.-B.FrankeS.ChoithaniA.KellerN. P. (2013). The bZIP protein MeaB mediates virulence attributes in Aspergillus flavus.PLoS One8:e74030. 10.1371/journal.pone.0074030
6
AmoutziasG.Bornberg-BauerE.OliverS.RobertsonD. (2006). Reduction/oxidation-phosphorylation control of DNA binding in the bZIP dimerization network.BMC Genomics7:107. 10.1186/1471-2164-7-107
7
AnsariM. Z.SharmaJ.GokhaleR. S.MohantyD. (2008). In silico analysis of methyltransferase domains involved in biosynthesis of secondary metabolites.BMC Bioinformatics9:454. 10.1186/1471-2105-9-454
8
AvramovičM. (2011). Analysis of the Genetic Potential of the Sponge- Derived Fungus Penicillium chrysogenum E01- 10 / 3 for Polyketide Production.Doctoral dissertation, Bonn, Rheinischen Friedrich-Wilhelms-Universität.
9
AwanA. R.ShawW. M.EllisT. (2016). Biosynthesis of therapeutic natural products using synthetic biology.Adv. Drug Deliv. Rev.105(Pt A), 96–106. 10.1016/j.addr.2016.04.010
10
BalibarC. J.VaillancourtF. H.WalshC. T. (2005). Generation of D amino acid residues in assembly of arthrofactin by dual condensation/epimerization domains.Chem. Biol.121189–1200. 10.1016/j.chembiol.2005.08.010
11
BaltzR. H. (2011). Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery.J. Ind. Microbiol. Biotechnol.381747–1760. 10.1007/s10295-011-1022-8
12
BaltzR. H. (2014). Combinatorial biosynthesis of cyclic lipopeptide antibiotics: a model for synthetic biology to accelerate the evolution of secondary metabolite biosynthetic pathways.ACS Synth. Biol.3748–758. 10.1021/sb3000673
13
BarreiroC.MartínJ. F.García-EstradaC. (2012). Proteomics shows new faces for the old penicillin producer Penicillium chrysogenum.J. Biomed. Biotechnol.20121–15. 10.1155/2012/105109
14
BartoszewskaM.OpaliŁ;skiŁ.VeenhuisM.van der KleiI. J. (2011). The significance of peroxisomes in secondary metabolite biosynthesis in filamentous fungi.Biotechnol. Lett.331921–1931. 10.1007/s10529-011-0664-y
15
BaylyC.YadavV. (2017). Towards precision engineering of canonical polyketide synthase domains: recent advances and future prospects.Molecules22:E235. 10.3390/molecules22020235
16
BayramÖ.BrausG. H. (2012). Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins.FEMS Microbiol. Rev.361–24. 10.1111/j.1574-6976.2011.00285.x
17
BeerR.HerbstK.IgnatiadisN.KatsI.AdlungL.MeyerH.et al (2014). Creating functional engineered variants of the single-module non-ribosomal peptide synthetase IndC by T domain exchange.Mol. Biosyst.101709–1718. 10.1039/c3mb70594c
18
BergendahlV.LinneU.MarahielM. A. (2002). Mutational analysis of the C-domain in nonribosomal peptide synthesis.Eur. J. Biochem.269620–629. 10.1046/j.0014-2956.2001.02691.x
19
BergmannS.FunkA. N.ScherlachK.SchroeckhV.ShelestE.HornU.et al (2010). Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide.Appl. Environ. Microbiol.768143–8149. 10.1128/AEM.00683-10
20
BergmannS.SchümannJ.ScherlachK.LangeC.BrakhageA. A.HertweckC. (2007). Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans.Nat. Chem. Biol.3213–217. 10.1038/nchembio869
21
BlinK.MedemaM. H.KottmannR.LeeS. Y.WeberT. (2017). The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters.Nucleic Acids Res.45D555–D559. 10.1093/nar/gkw960
22
BloudoffK.SchmeingT. M. (2017). Structural and functional aspects of the nonribosomal peptide synthetase condensation domain superfamily: discovery, dissection and diversity.Biochim. Biophys. Acta18651587–1604. 10.1016/j.bbapap.2017.05.010
23
BodeH. B.BetheB.HöfsR.ZeeckA. (2002). Big effects from small changes: possible ways to explore nature’s chemical diversity.ChemBioChem3619–627. 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9
24
BodeH. B.BrachmannA. O.JadhavK. B.SeyfarthL.DauthC.FuchsS. W.et al (2015). Structure elucidation and activity of kolossin A, the D-/L-pentadecapeptide product of a giant nonribosomal peptide synthetase.Angew. Chem.5410352–10355. 10.1002/anie.201502835
25
BokJ. W.KellerN. P. (2004). LaeA, a regulator of secondary metabolism in Aspergillus spp.Eukaryot. Cell3527–535. 10.1128/EC.3.2.527-535.2004
26
BokJ. W.KellerN. P. (2016). “Insight into fungal secondary metabolism from ten years of LaeA research,” inBiochemistry and Molecular Biology, ed.HoffmeisterD. (Cham: Springer International Publishing), 21–29. 10.1007/978-3-319-27790-5_2
27
BrakhageA. A. (2012). Regulation of fungal secondary metabolism.Nat. Rev. Microbiol.1121–32. 10.1038/nrmicro2916
28
BrakhageA. A.SchroeckhV. (2011). Fungal secondary metabolites – Strategies to activate silent gene clusters.Fungal Genet. Biol.4815–22. 10.1016/j.fgb.2010.04.004
29
BroschG.LoidlP.GraessleS. (2008). Histone modifications and chromatin dynamics: a focus on filamentous fungi.FEMS Microbiol. Rev.32409–439. 10.1111/j.1574-6976.2007.00100.x
30
BrownD. W.ButchkoR. A. E.BusmanM.ProctorR. H. (2007). The Fusarium verticillioides FUM gene cluster encodes a Zn(II)2Cys6 protein that affects FUM gene expression and fumonisin production.Eukaryot. Cell61210–1218. 10.1128/EC.00400-06
31
BrueggerJ.CaldaraG.BeldJ.BurkartM. D.TsaiS. S. (2014). “Polyketide synthase: sequence, structure, and function,” inNatural Products, edsOsbournA.GossR. J.CarterG. T. (Hoboken, NJ: John Wiley & Sons, Inc.), 219–243. 10.1002/9781118794623.ch12
32
BrueggerJ.HaushalterB.VagstadA.ShakyaG.MihN.TownsendC. A.et al (2013). Probing the selectivity and protein⋅protein interactions of a nonreducing fungal polyketide synthase using mechanism-based crosslinkers.Chem. Biol.201135–1146. 10.1016/j.chembiol.2013.07.012
33
ByersD. M.GongH. (2007). Acyl carrier protein: structure–function relationships in a conserved multifunctional protein family.Biochem. Cell Biol.85649–662. 10.1139/O07-109
34
CabocheS.PupinM.LeclèreV.FontaineA.JacquesP.KucherovG. (2008). NORINE: a database of nonribosomal peptides.Nucleic Acids Res.36D326–D331. 10.1093/nar/gkm792
35
CaffreyP. (2012). Dissecting complex polyketide biosynthesis.Comput. Struct. Biotechnol. J.3:e201210010. 10.5936/csbj.201210010
36
CampbellC. D.VederasJ. C. (2010). Biosynthesis of lovastatin and related metabolites formed by fungal iterative PKS enzymes.Biopolymers93755–763. 10.1002/bip.21428
37
CaryJ. W.HanZ.YinY.LohmarJ. M.ShantappaS.Harris-CowardP. Y.et al (2015). Transcriptome analysis of Aspergillus flavus reveals veA -dependent regulation of secondary metabolite gene clusters, including the novel aflavarin cluster.Eukaryot. Cell14983–997. 10.1128/EC.00092-15
38
Cepeda-GarcíaC.Domínguez-SantosR.García-RicoR. O.García-EstradaC.CajiaoA.FierroF.et al (2014). Direct involvement of the CreA transcription factor in penicillin biosynthesis and expression of the pcbAB gene in Penicillium chrysogenum.Appl. Microbiol. Biotechnol.987113–7124. 10.1007/s00253-014-5760-1
39
ChaiY. J.CuiC. B.LiC. W.WuC. J.TianC. K.HuaW. (2012). Activation of the dormant secondary metabolite production by introducing gentamicin-resistance in a marine-derived Penicillium purpurogenum G59.Mar. Drugs10559–582. 10.3390/md10030559
40
ChanY. A.PodevelsA. M.KevanyB. M.ThomasM. G. (2009). Biosynthesis of polyketide synthase extender units.Nat. Prod. Rep.2690–114. 10.1039/B801658P
41
ChangP.-K.YuJ.YuJ.-H. (2004). aflT, a MFS transporter-encoding gene located in the aflatoxin gene cluster, does not have a significant role in aflatoxin secretion.Fungal Genet. Biol.41911–920. 10.1016/j.fgb.2004.06.007
42
ChenA. Y.SchnarrN. A.KimC.-Y.CaneD. E.KhoslaC. (2006). Extender unit and Acyl carrier protein specificity of ketosynthase domains of the 6-deoxyerythronolide B synthase.J. Am. Chem. Soc.1283067–3074. 10.1021/ja058093d
43
ChenX.KöllnerT. G.JiaQ.NorrisA.SanthanamB.RabeP.et al (2016). Terpene synthase genes in eukaryotes beyond plants and fungi: occurrence in social amoebae.Proc. Natl. Acad. Sci. U.S.A.11312132–12137. 10.1073/pnas.1610379113
44
ChiangY. M.LeeK. H.SanchezJ. F.KellerN. P.WangC. C. (2009). Unlocking cryptic fungal natural product clusters.Nat. Prod. Commun.41505–1510. 10.1055/s-0033-1348488
45
ChiangY.-M.WangC. C. C.OakleyB. R. (2014). “Analyzing fungal secondary metabolite genes and gene clusters,” inNatural Products, edsOsbournA.GossR. J.CarterG. T. (Hoboken, NJ: John Wiley & Sons, Inc.), 171–193. 10.1002/9781118794623.ch10
46
ChujoT.ScottB. (2014). Histone H3K9 and H3K27 methylation regulates fungal alkaloid biosynthesis in a fungal endophyte-plant symbiosis.Mol. Microbiol.92413–434. 10.1111/mmi.12567
47
ClevengerK. D.BokJ. W.YeR.MileyG. P.VerdanM. H.VelkT.et al (2017). A scalable platform to identify fungal secondary metabolites and their gene clusters.Nat. Chem. Biol.13895–901. 10.1038/nchembio.2408
48
CoxR. J. (2007). Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets.Org. Biomol. Chem.52010–2016. 10.1039/b704420h
49
CoxR. J.SimpsonT. J. (2009). “Fungal type I polyketide synthases,” inMethods in Enzymology, ed.HopwoodD. A. (Norwich: Academic Press), 49–78. 10.1016/S0076-6879(09)04603-5
50
CoxR. J.SimpsonT. J. (2010). “Fungal type I polyketides,” inComprehensive Natural Products II, edsTownsendC. A.EbizukaT. (Amsterdam: Elsevier), 347–383. 10.1016/B978-008045382-8.00017-4
51
CrawfordJ. M.TownsendC. A. (2010). New insights into the formation of fungal aromatic polyketides.Nat. Rev. Microbiol.8879–889. 10.1038/nrmicro2465
52
DangT.SussmuthR. D. (2017). Bioactive peptide natural products as lead structures for medicinal use.Acc. Chem. Res.501566–1576. 10.1021/acs.accounts.7b00159
53
DeepikaV. B.MuraliT. S.SatyamoorthyK. (2016). Modulation of genetic clusters for synthesis of bioactive molecules in fungal endophytes: a review.Microbiol. Res.182125–140. 10.1016/j.micres.2015.10.009
54
DemainA. L.FangA. (2000). The natural functions of secondary metabolites.Adv. Biochem. Eng. Biotechnol.691–39.10.1007/3-540-44964-7_1
55
DerntlC.Guzmán-ChávezF.Mello-de-SousaT. M.BusseH.-J.DriessenA. J. M.MachR. L.et al (2017a). In vivo study of the sorbicillinoid gene cluster in Trichoderma reesei.Front. Microbiol.8:2037. 10.3389/fmicb.2017.02037
56
DerntlC.KlugerB.BueschlC.SchuhmacherR.MachR. L.Mach-AignerA. R. (2017b). Transcription factor Xpp1 is a switch between primary and secondary fungal metabolism.Proc. Natl. Acad. Sci. U.S.A.114E560–E569. 10.1073/pnas.1609348114
57
DerntlC.RassingerA.SrebotnikE.MachR. L.Mach-AignerA. R. (2016). Identification of the main regulator responsible for synthesis of the typical yellow pigment produced by Trichoderma reesei.Appl. Environ. Microbiol.826247–6257. 10.1128/AEM.01408-16
58
DhingraS.LindA. L.LinH. -C.TangY.RokasA.CalvoA. M. (2013). The fumagillin gene cluster, an example of hundreds of genes under veA control in Aspergillus fumigatus.PLoS One8:e77147. 10.1371/journal.pone.0077147
59
DowlingD. P.KungY.CroftA. K.TaghizadehK.KellyW. L.WalshC. T.et al (2016). Structural elements of an NRPS cyclization domain and its intermodule docking domain.Proc. Natl. Acad. Sci. U.S.A.11312432–12437. 10.1073/pnas.1608615113
60
DruzhininaI. S.KubicekE. M.KubicekC. P. (2016). Several steps of lateral gene transfer followed by events of ‘birth-and-death’ evolution shaped a fungal sorbicillinoid biosynthetic gene cluster.BMC Evol. Biol.16:269. 10.1186/s12862-016-0834-6
61
DuL.ShenB. (2001). Biosynthesis of hybrid peptide-polyketide natural products.Curr. Opin. Drug Discov. Dev.4215–228.
62
DuL.ZhuT.LiL.CaiS.ZhaoB.GuQ. (2009). Cytotoxic sorbicillinoids and bisorbicillinoids from a marine-derived fungus Trichoderma sp.Chem. Pharm. Bull.57220–223. 10.1248/cpb.57.220
63
DunnB. J.CaneD. E.KhoslaC. (2013). Mechanism and specificity of an acyltransferase domain from a modular polyketide synthase.Biochemistry521839–1841. 10.1021/bi400185v
64
DuttaS.WhicherJ. R.HansenD. A.HaleW. A.ChemlerJ. A.CongdonG. R.et al (2014). Structure of a modular polyketide synthase.Nature510512–520. 10.1038/nature13423
65
EisfeldK. (2009). “Non-ribosomal peptide synthetases of fungi,” inPhysiology and Genetics: Selected Basic and Applied Aspects,edsAnkeT.Weber BerlinD. (Heidelberg: Springer). 10.1007/978-3-642-00286-1
66
EvansS. E.WilliamsC.ArthurC. J.BurstonS. G.SimpsonT. J.CrosbyJ.et al (2008). An ACP structural switch: conformational differences between the Apo and Holo forms of the actinorhodin polyketide synthase Acyl carrier protein.ChemBioChem92424–2432. 10.1002/cbic.200800180
67
FelnagleE. A.BarkeiJ. J.ParkH.PodevelsA. M.McMahonM. D.DrottD. W.et al (2010). MbtH-like proteins as integral components of bacterial nonribosomal peptide synthetases.Biochemistry498815–8817. 10.1021/bi1012854
68
FelnagleE. A.JacksonE. E.ChanY. A.PodevelsA. M.BertiA. D.McMahonM. D.et al (2008). Nonribosomal peptide synthetases involved in the production of medically relevant natural products.Mol. Pharm.5191–211. 10.1021/mp700137g
69
FierroF.BarredoJ. L.DiezB.GutierrezS.FernandezF. J.MartinJ. F. (1995). The penicillin gene cluster is amplified in tandem repeats linked by conserved hexanucleotide sequences.Proc. Natl. Acad. Sci. U.S.A.926200–6204. 10.1073/pnas.92.13.6200
70
FierroF.García-EstradaC.CastilloN. I.RodríguezR.Velasco-CondeT.MartínJ.-F. (2006). Transcriptional and bioinformatic analysis of the 56.8kb DNA region amplified in tandem repeats containing the penicillin gene cluster in Penicillium chrysogenum.Fungal Genet. Biol.43618–629. 10.1016/j.fgb.2006.03.001
71
FischerJ.SchroeckhV.BrakhageA. A. (2016). “Awakening of fungal secondary metabolite gene clusters,” inGene Expression Systems in Fungi: Advancements and Applications Fungal Biology, edsSchmollM.DattenböckC. (Cham: Springer International Publishing), 253–273. 10.1007/978-3-319-27951-0
72
FlahertyJ. E.WoloshukC. P. (2004). Regulation of fumonisin biosynthesis in.Society702653–2659. 10.1128/AEM.70.5.2653
73
FrisvadJ. C.SmedsgaardJ.LarsenT. O.SamsonR. A. (2004). Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium.Stud. Mycol.49201–241.
74
GacekA.StraussJ. (2012). The chromatin code of fungal secondary metabolite gene clusters.Appl. Microbiol. Biotechnol.951389–1404. 10.1007/s00253-012-4208-8
75
GalloA.FerraraM.PerroneG. (2013). Phylogenetic study of polyketide synthases and nonribosomal peptide synthetases involved in the biosynthesis of mycotoxins.Toxins5717–742. 10.3390/toxins5040717
76
García-EstradaC.UllánR. V.AlbillosS. M.Fernández-BodegaM. Á.DurekP.von DöhrenH.et al (2011). A single cluster of coregulated genes encodes the biosynthesis of the mycotoxins roquefortine C and meleagrin in Penicillium chrysogenum.Chem. Biol.181499–1512. 10.1016/j.chembiol.2011.08.012
77
GeorgiannaD. R.PayneG. A. (2009). Genetic regulation of aflatoxin biosynthesis: from gene to genome.Fungal Genet. Biol.46113–125. 10.1016/j.fgb.2008.10.011
78
GombertA. K.VeigaT.Puig-MartinezM.LambooF.NijlandJ. G.DriessenA. J. M.et al (2011). Functional characterization of the oxaloacetase encoding gene and elimination of oxalate formation in the β-lactam producer Penicillium chrysogenum.Fungal Genet. Biol.48831–839. 10.1016/j.fgb.2011.04.007
79
GresslerM.MeyerF.HeineD.HortschanskyP.HertweckC.BrockM. (2015). Phytotoxin production in Aspergillus terreus is regulated by independent environmental signals.eLife4:e07861. 10.7554/eLife.07861
80
Guzmán-ChávezF.SaloO.NygårdY.LankhorstP. P.BovenbergR. A. L.DriessenA. J. M. (2017). Mechanism and regulation of sorbicillin biosynthesis by Penicillium chrysogenum.Microb. Biotechnol.10958–968. 10.1111/1751-7915.12736
81
Guzmán-ChávezF.SaloO.SamolM.RiesM.KuipersJ.BovenbergR. A. L. L.et al (2018). Deregulation of secondary metabolism in a histone deacetylase mutant of Penicillium chrysogenum.Microbiologyopen7:e00598. 10.1002/mbo3.598
82
HahnM.StachelhausT. (2006). Harnessing the potential of communication-mediating domains for the biocombinatorial synthesis of nonribosomal peptides.Proc. Natl. Acad. Sci. U.S.A.103275–280. 10.1073/pnas.0508409103
83
HarnedA. M.VolpK. A. (2011). The sorbicillinoid family of natural products: isolation, biosynthesis, and synthetic studies.Nat. Prod. Rep.281790–1810. 10.1039/c1np00039j
84
HarrisD. M.WesterlakenI.SchipperD.van der KrogtZ. A.GombertA. K.SutherlandJ.et al (2009). Engineering of Penicillium chrysogenum for fermentative production of a novel carbamoylated cephem antibiotic precursor.Metab. Eng.11125–137. 10.1016/j.ymben.2008.12.003
85
HaslingerK.PeschkeM.BriekeC.MaximowitschE.CryleM. J. (2015). X-domain of peptide synthetases recruits oxygenases crucial for glycopeptide biosynthesis.Nature521105–109. 10.1038/nature14141
86
HeinekampT.ThywißenA.MacheleidtJ.KellerS.ValianteV.BrakhageA. A. (2013). Aspergillus fumigatus melanins: interference with the host endocytosis pathway and impact on virulence.Front. Microbiol.3:440. 10.3389/fmicb.2012.00440
87
HertweckC.LuzhetskyyA.RebetsY.BechtholdA. (2007). Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork.Nat. Prod. Rep.24162–190. 10.1039/B507395M
88
HidalgoP. I.UllánR. V.AlbillosS. M.MonteroO.Fernández-BodegaM. Á.García-EstradaC.et al (2014). Molecular characterization of the PR-toxin gene cluster in Penicillium roqueforti and Penicillium chrysogenum: cross talk of secondary metabolite pathways.Fungal Genet. Biol.6211–24. 10.1016/j.fgb.2013.10.009
89
HopwoodD. A. (2009). Complex Enzymes in Microbial Natural Product Biosynthesis. Part B, Polyketides, Aminocoumarins, and Carbohydrates.Amsterdam: Elsevier, 581.
90
HorsmanM. E.HariT. P. A.BoddyC. N. (2016). Polyketide synthase and non-ribosomal peptide synthetase thioesterase selectivity: logic gate or a victim of fate?Nat. Prod. Rep.33183–202. 10.1039/c4np00148f
91
HoubrakenJ.FrisvadJ. C.Samson’R. A. (2011). Fleming’s penicillin producing strain is not Penicillium chrysogenum butP. rubens. IMA Fungus287–95. 10.5598/imafungus.2011.02.01.12
92
HurG. H.VickeryC. R.BurkartM. D. (2012). Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology.Nat. Prod. Rep.291074–1098. 10.1039/c2np20025b
93
ItohT.TokunagaK.MatsudaY.FujiiI.AbeI.EbizukaY.et al (2010). Reconstitution of a fungal meroterpenoid biosynthesis reveals the involvement of a novel family of terpene cyclases.Nat. Chem.2858–864. 10.1038/nchem.764
94
JainS.KellerN. (2013). Insights to fungal biology through LaeA sleuthing.Fungal Biol. Rev.2751–59. 10.1016/j.fbr.2013.05.004
95
JakoèiunasT.JensenM. K.KeaslingJ. D. (2016). CRISPR/Cas9 advances engineering of microbial cell factories.Metab. Eng.3444–59. 10.1016/j.ymben.2015.12.003
96
JamiM.-S.BarreiroC.García-EstradaC.MartínJ.-F. (2010). Proteome analysis of the penicillin producer Penicillium chrysogenum.Mol. Cell. Prot.91182–1198. 10.1074/mcp.M900327-MCP200
97
JanusD.HortschanskyP.KückU. (2008). Identification of a minimal cre1 promoter sequence promoting glucose-dependent gene expression in the β-lactam producer Acremonium chrysogenum.Curr. Genet.5335–48. 10.1007/s00294-007-0164-8
98
JennerM. (2016). Using Mass Spectrometry for Biochemical Studies on Enzymatic Domains from Polyketide Synthases.Cham: Springer International Publishing. 10.1007/978-3-319-32723-5
99
KageH.RivaE.ParascandoloJ. S.KreutzerM. F.TosinM.NettM. (2015). Chemical chain termination resolves the timing of ketoreduction in a partially reducing iterative type I polyketide synthase.Org. Biomol. Chem.1311414–11417. 10.1039/C5OB02009C
100
KapurS.ChenA. Y.CaneD. E.KhoslaC. (2010). Molecular recognition between ketosynthase and acyl carrier protein domains of the 6-deoxyerythronolide B synthase.Proc. Natl. Acad. Sci. U.S.A.10722066–22071. 10.1073/pnas.1014081107
101
Keatinge-ClayA. T. (2012). The structures of type I polyketide synthases.Nat. Prod. Rep.291050–1073. 10.1039/c2np20019h
102
Keatinge-ClayA. T.StroudR. M. (2006). The structure of a ketoreductase determines the organization of the β-carbon processing enzymes of modular polyketide synthases.Structure14737–748. 10.1016/j.str.2006.01.009
103
KellerN. P. (2015). Translating biosynthetic gene clusters into fungal armor and weaponry.Nat. Chem. Biol.11671–677. 10.1038/nchembio.1897
104
KellerN. P.TurnerG.BennettJ. W. (2005). Fungal secondary metabolism - from biochemistry to genomics.Nat. Rev. Microbiol.3937–947. 10.1038/nrmicro1286
105
KellyW. L.HillsonN. J.WalshC. T. (2005). Excision of the epothilone synthetase B cyclization domain and demonstration of in trans condensation/cyclodehydration activity.Biochemistry4413385–13393. 10.1021/bi051124x
106
KhaldiN.SeifuddinF. T.TurnerG.HaftD.NiermanW. C.WolfeK. H.et al (2010). SMURF: genomic mapping of fungal secondary metabolite clusters.Fungal Genet. Biol.47736–741. 10.1016/j.fgb.2010.06.003
107
KistlerH. C.BrozK. (2015). Cellular compartmentalization of secondary metabolism.Front. Microbiol.6:68. 10.3389/fmicb.2015.00068
108
KittiläT.MolloA.CharkoudianL. K.CryleM. J. (2016). New structural data reveal the motion of carrier proteins in nonribosomal peptide synthesis.Angew. Chem. Int. Ed.559834–9840. 10.1002/anie.201602614
109
KnoxB. P.KellerN. P. (2015). “Key players in the regulation of fungal secondary metabolism,” inBiosynthesis and Molecular Genetics of Fungal Secondary Metabolites Fungal Biology., edsZeilingerS.MartínJ.-F.García-EstradaC. (New York, NY: Springer), 13–28. 10.1007/978-1-4939-2531-5_2
110
KosalkováK.García-EstradaC.UllánR. V.GodioR. P.FeltrerR.TeijeiraF.et al (2009). The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine C synthesis in Penicillium chrysogenum.Biochimie91214–225. 10.1016/j.biochi.2008.09.004
111
KriesH. (2016). Biosynthetic engineering of nonribosomal peptide synthetases.J. Pept. Sci.22564–570. 10.1002/psc.2907
112
KuJ.MirmiraR. G.LiuL.SantiD. V. (1997). Expression of a functional non-ribosomal peptide synthetase module in Escherichia coli by coexpression with a phosphopantetheinyl transferase.Chem. Biol.4203–207. 10.1016/S1074-5521(97)90289-1
113
LeeI.OhJ.-H.Keats ShwabE.DagenaisT. R. T.AndesD.KellerN. P. (2009). HdaA, a class 2 histone deacetylase of Aspergillus fumigatus, affects germination and secondary metabolite production.Fungal Genet. Biol.46782–790. 10.1016/j.fgb.2009.06.007
114
LeeS.SonH.LeeJ.LeeY. -R.LeeY.-W. (2011). A putative ABC transporter gene, ZRA1, is required for zearalenone production in Gibberella zeae.Curr. Genet.57343–351. 10.1007/s00294-011-0352-4
115
LiJ.PanY.LiuG. (2013). Disruption of the nitrogen regulatory gene AcareA in Acremonium chrysogenum leads to reduction of cephalosporin production and repression of nitrogen metabolism.Fungal Genet. Biol.6169–79. 10.1016/j.fgb.2013.10.006
116
LiY.WeissmanK. J.MullerR. (2010). Insights into multienzyme docking in hybrid PKS-NRPS megasynthetases revealed by heterologous expression and genetic engineering.Chembiochem111069–1075. 10.1002/cbic.201000103
117
LimF. Y.SanchezJ. F.WangC. C.KellerN. P. (2012). “Toward awakening cryptic secondary metabolite gene clusters in filamentous fungi,” in,Methods in enzymologyed.HopwoodD. A. (Amsterdam: Elsevier Inc.), 303–324. 10.1016/B978-0-12-404634-4.00015-2
118
LinneU.MarahielM. A. (2000). Control of directionality in nonribosomal peptide synthesis: role of the condensation domain in preventing misinitiation and timing of epimerization.Biochemistry3910439–10447. 10.1021/bi000768w
119
LiuT.SanchezJ. F.ChiangY. M.OakleyB. R.WangC. C. C. (2014). Rational domain swaps reveal insights about chain length control by ketosynthase domains in fungal nonreducing polyketide synthases.Org. Lett.161676–1679. 10.1021/ol5003384
120
LoH. C.EntwistleR.GuoC. J.AhujaM.SzewczykE.HungJ. H.et al (2012). Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans.J. Am. Chem. Soc.1344709–4720. 10.1021/ja209809t
121
LuoZ.RenH.MousaJ. J.RangelD. E. N.ZhangY.BrunerS. D.et al (2017). The PacC transcription factor regulates secondary metabolite production and stress response, but has only minor effects on virulence in the insect pathogenic fungus Beauveria bassiana.Environ. Microbiol.19788–802. 10.1111/1462-2920.13648
122
LutzM. V.BovenbergR. A. L.van der KleiI. J.VeenhuisM. (2005). Synthesis of acetyl-CoA:isopenicillin acyltransferase in: first step towards the introduction of a new metabolic pathway.FEMS Yeast Res.51063–1067. 10.1016/j.femsyr.2005.07.002
123
MacheleidtJ.MatternD. J.FischerJ.NetzkerT.WeberJ.SchroeckhV.et al (2016). Regulation and role of fungal secondary metabolites.Annu. Rev. Genet.50371–392. 10.1146/annurev-genet-120215-035203
124
MacPhersonS.LarochelleM.TurcotteB. (2006). A fungal family of transcriptional regulators: the zinc cluster proteins.Microbiol. Mol. Biol. Rev.70583–604. 10.1128/MMBR.00015-06
125
MarahielM. A.StachelhausT.MootzH. D. (1997). Modular peptide synthetases involved in nonribosomal peptide synthesis.Chem. Rev.972651–2674. 10.1021/cr960029e
126
MarmannA.AlyA.LinW.WangB.ProkschP. (2014). Co-cultivation—a powerful emerging tool for enhancing the chemical diversity of microorganisms.Mar. Drugs121043–1065. 10.3390/md12021043
127
MartínJ. F.LirasP. (2017). “Insights into the structure and molecular mechanisms of β-lactam synthesizing enzymes in fungi,” inBiotechnology of Microbial Enzymes, edsBrahmachariG.DemainA. L.AdrioJ. L. (New York, NY: Elsevier), 215–241. 10.1016/B978-0-12-803725-6.00009-1
128
MaskeyR. P.Grün-WollnyI.LaatschH. (2005). Sorbicillin analogues and related dimeric compounds from Penicillium notatum.J. Nat. Prod.68865–870. 10.1021/np040137t
129
MatsudaY.AwakawaT.AbeI. (2013). Reconstituted biosynthesis of fungal meroterpenoid andrastin A.Tetrahedron698199–8204. 10.1016/j.tet.2013.07.029
130
McLeanK. J.HansM.MeijrinkB.van ScheppingenW. B.VollebregtA.TeeK. L.et al (2015). Single-step fermentative production of the cholesterol-lowering drug pravastatin via reprogramming of Penicillium chrysogenum.Proc. Natl. Acad. Sci. U.S.A.1122847–2852. 10.1073/pnas.1419028112
131
MedemaM. H.BlinK.CimermancicP.de JagerV.ZakrzewskiP.FischbachM. A.et al (2011). antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences.Nucleic Acids Res.39W339–W346. 10.1093/nar/gkr466
132
MeguroA.TomitaT.NishiyamaM.KuzuyamaT. (2014). Identification and characterization of bacterial diterpene cyclases that synthesize the cembrane skeleton.ChemBioChem15913–913. 10.1002/cbic.201402114
133
MengJ.WangX.XuD.FuX.ZhangX.LaiD.et al (2016). Sorbicillinoids from fungi and their bioactivities.Molecules211–19. 10.3390/molecules21060715
134
MenkeJ.DongY.KistlerH. C. (2012). Fusarium graminearum Tri12p influences virulence to wheat and trichothecene accumulation.Mol. Plant Microbe Interact.251408–1418. 10.1094/MPMI-04-12-0081-R
135
MichielseC. B.StudtL.JanevskaS.SieberC. M. K.ArndtB.EspinoJ. J.et al (2015). The global regulator FfSge1 is required for expression of secondary metabolite gene clusters but not for pathogenicity in F usarium fujikuroi.Environ. Microbiol.172690–2708. 10.1111/1462-2920.12592
136
MillerB. R.DrakeE. J.ShiC.AldrichC. C.GulickA. M. (2016). Structures of a nonribosomal peptide synthetase module bound to MbtH-like proteins support a highly dynamic domain architecture.J. Biol. Chem.29122559–2257110.1074/jbc.M116.746297
137
MillerD. J.OuelletteN.EvdokimovaE.SavchenkoA.EdwardsA.AndersonW. F. (2003). Crystal complexes of a predicted S-adenosylmethionine-dependent methyltransferase reveal a typical AdoMet binding domain and a substrate recognition domain.Protein Sci.121432–1442. 10.1110/ps.0302403
138
MootzH. D.SchwarzerD.MarahielM. A. (2002). Ways of assembling complex natural products on modular nonribosomal peptide synthetases a list of abbreviations can be found at the end of the text.ChemBioChem3490–504. 10.1002/1439-7633(20020603)3:6<490::AID-CBIC490>3.0.CO;2-N
139
NairD. R.AnandS.VermaP.MohantyD.GokhaleR. S. (2012). Genetic, biosynthetic and functional versatility of polyketide synthases.Curr. Sci.102277–287.
140
NetzkerT.FischerJ.WeberJ.MatternD. J.KönigC. C.ValianteV.et al (2015). Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters.Front. Microbiol.6:299. 10.3389/fmicb.2015.00299
141
NevilleC.MurphyA.KavanaghK.DoyleS. (2005). A 4′-phosphopantetheinyl transferase mediates non-ribosomal peptide synthetase activation in Aspergillus fumigatus.Chembiochem6679–685. 10.1002/cbic.200400147
142
NguyenK. T.RitzD.GuJ.-Q.AlexanderD.ChuM.MiaoV.et al (2006). Combinatorial biosynthesis of novel antibiotics related to daptomycin.Proc. Natl. Acad. Sci. U.S.A.10317462–17467. 10.1073/pnas.0608589103
143
NielsenJ. C.GrijseelsS.PrigentS.JiB.DainatJ.NielsenK. F.et al (2017). Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species.Nat. Microbiol.21–9. 10.1038/nmicrobiol.2017.44
144
NielsenM. L.IsbrandtT.PetersenL. M.MortensenU. H.AndersenM. R. R.HoofJ. B. B.et al (2016). Linker flexibility facilitates module exchange in fungal hybrid PKS-NRPS engineering.PLoS One11:e0161199. 10.1371/journal.pone.0161199
145
NijlandJ. G.EbbendorfB.WoszczynskaM.BoerR.BovenbergR. A. L.DriessenA. J. M. (2010). Nonlinear biosynthetic gene cluster dose effect on penicillin production by Penicillium chrysogenum.Appl. Environ. Microbiol.767109–7115. 10.1128/AEM.01702-10
146
NutzmannH.-W.Reyes-DominguezY.ScherlachK.SchroeckhV.HornF.GacekA.et al (2011). Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation.Proc. Natl. Acad. Sci. U.S.A.10814282–14287. 10.1073/pnas.1103523108
147
OchiK.HosakaT. (2013). New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters.Appl. Microbiol. Biotechnol.9787–98. 10.1007/s00253-012-4551-9
148
OhD.-C.KauffmanC. A.JensenP. R.FenicalW. (2007). Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture.J. Nat. Prod.70515–520. 10.1021/np060381f
149
OkadaB. K.SeyedsayamdostM. R. (2017). Antibiotic dialogues: induction of silent biosynthetic gene clusters by exogenous small molecules.FEMS Microbiol. Rev.4119–33. 10.1093/femsre/fuw035
150
OstrowskiM. P.CaneD. E.KhoslaC. (2016). Recognition of acyl carrier proteins by ketoreductases in assembly line polyketide synthases.J. Antibiot.69507–510. 10.1038/ja.2016.41
151
OzcengizG.DemainA. L. (2013). Recent advances in the biosynthesis of penicillins, cephalosporins and clavams and its regulation.Biotechnol. Adv.31287–311. 10.1016/j.biotechadv.2012.12.001
152
PalmerJ. M.KellerN. P. (2010). Secondary metabolism in fungi: does chromosomal location matter?Curr. Opin. Microbiol.13431–436. 10.1016/j.mib.2010.04.008
153
PatelH. M.TaoJ.WalshC. T. (2003). Epimerization of an L-cysteinyl to a D-cysteinyl residue during thiazoline ring formation in siderophore chain elongation by pyochelin synthetase from Pseudomonas aeruginosa.Biochemistry4210514–10527. 10.1021/bi034840c
154
PihetM.VandeputteP.TronchinG.RenierG.SaulnierP.GeorgeaultS.et al (2009). Melanin is an essential component for the integrity of the cell wall of Aspergillus fumigatus conidia.BMC Microbiol.9:177. 10.1186/1471-2180-9-177
155
PohlC.KielJ. A. K. W.DriessenA. J. M.BovenbergR. A. L.NygardY. (2016). CRISPR/Cas9 based genome editing of Penicillium chrysogenum.ACS Synth. Biol.5754–764. 10.1021/acssynbio.6b00082
156
PohlC.MózsikL.DriessenA. J. M.BovenbergR. A. L.NygårdY. I. (2018). “Genome editing in Penicillium chrysogenum using Cas9 ribonucleoprotein particles,” in Synthetic Biology: Methods and Protocols, ed.BramanJ. C. (Clifton, NJ: Springer Protocols), 213–232. 10.1007/978-1-4939-7795-6_12
157
PolliF.MeijrinkB.BovenbergR. A. L.DriessenA. J. M. (2016). New promoters for strain engineering of Penicillium chrysogenum.Fungal Genet. Biol.8962–71. 10.1016/j.fgb.2015.12.003
158
QuadriL. E.SelloJ.KeatingT. A.WeinrebP. H.WalshC. T. (1998). Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin.Chem. Biol.5631–645. 10.1016/S1074-5521(98)90291-5
159
QuinM. B.FlynnC. M.Schmidt-DannertC. (2014). Traversing the fungal terpenome.Nat. Prod. Rep.311449–1473. 10.1039/C4NP00075G
160
ReenF.RomanoS.DobsonA.O’GaraF. (2015). The sound of silence: activating silent biosynthetic gene clusters in marine microorganisms.Mar. Drugs134754–4783. 10.3390/md13084754
161
Reyes-DominguezY.BokJ. W.BergerH.ShwabE. K.BasheerA.GallmetzerA.et al (2010). Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans.Mol. Microbiol.761376–1386. 10.1111/j.1365-2958.2010.07051.x
162
RidenourJ. B.BluhmB. H. (2014). The HAP complex in Fusarium verticillioides is a key regulator of growth, morphogenesis, secondary metabolism, and pathogenesis.Fungal Genet. Biol.6952–64. 10.1016/j.fgb.2014.05.003
163
RiesM. I.AliH.LankhorstP. P.HankemeierT.BovenbergR. A. L.DriessenA. J. M.et al (2013). Novel key metabolites reveal further branching of the roquefortine/meleagrin biosynthetic pathway.J. Biol. Chem.28837289–37295. 10.1074/jbc.M113.512665
164
RobbinsT.KapilivskyJ.CaneD. E.KhoslaC. (2016). Roles of conserved active site residues in the ketosynthase domain of an assembly line polyketide synthase.Biochemistry554476–4484. 10.1021/acs.biochem.6b00639
165
RobertsD. M.BartelC.ScottA.IvisonD.SimpsonT. J.CoxR. J. (2017). Substrate selectivity of an isolated enoyl reductase catalytic domain from an iterative highly reducing fungal polyketide synthase reveals key components of programming.Chem. Sci.81116–1126. 10.1039/C6SC03496A
166
RohlfsM.ChurchillA. C. L. (2011). Fungal secondary metabolites as modulators of interactions with insects and other arthropods.Fungal Genet. Biol.4823–34. 10.1016/j.fgb.2010.08.008
167
Rojas-AedoJ. F.Gil-DuránC.Del-CidA.ValdésN.ÁlamosP.VacaI.et al (2017). The biosynthetic gene cluster for andrastin A in Penicillium roqueforti.Front. Microbiol.8:813. 10.3389/fmicb.2017.00813
168
RutledgeP. J.ChallisG. L. (2015). Discovery of microbial natural products by activation of silent biosynthetic gene clusters.Nat. Rev. Microbiol.13509–523. 10.1038/nrmicro3496
169
SaloO. V.RiesM.MedemaM. H.LankhorstP. P.VreekenR. J.BovenbergR. A. L.et al (2015). Genomic mutational analysis of the impact of the classical strain improvement program on β – lactam producing Penicillium chrysogenum.BMC Genomics16:937. 10.1186/s12864-015-2154-4
170
SaloO. (2016). Secondary Metabolism by Industrially Improved Penicillium chrysogenum Strains.Doctoral dissertation, Groningen, University of Groningen.
171
SaloO.Guzmán-ChávezF.RiesM. I.LankhorstP. P.BovenbergR. A. L.VreekenR. J.et al (2016). Identification of a polyketide synthase involved in sorbicillin biosynthesis by Penicillium chrysogenum.Appl. Environ. Microbiol.823971–3978. 10.1128/AEM.00350-16.Editor
172
SamolM. M.SaloO.LankhorstP.BovenbergR. A. L.DriessenA. J. M. (2016). “Secondary metabolite formation by the filamentous fungus Penicillium chrysogenum in the post-genomic era,” inAspergillus and Penicillium in the Post-Genomic Era, edsde VriesR. P.GelberI. B.AndersenM. R. (Norfolk: Caister Academic Press), 145–172. 10.21775/9781910190395.09
173
Sarikaya-BayramÃ.PalmerJ. M.KellerN.BrausG. H.BayramÃ. (2015). One Juliet and four Romeos: VeA and its methyltransferases.Front. Microbiol.6:1. 10.3389/fmicb.2015.00001
174
SattelyE. S.FischbachM. A.WalshC. T. (2008). Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal peptide pathways.Nat. Prod. Rep.25757–793. 10.1039/b801747f
175
ScherlachK.HertweckC. (2006). Discovery of aspoquinolones A–D, prenylated quinoline-2-one alkaloids from Aspergillus nidulans, motivated by genome mining.Org. Biomol. Chem.43517–3520. 10.1039/B607011F
176
Schmidt-DannertC. (2014). “Biosynthesis of terpenoid natural products in fungi,” inBiotechnology of Isoprenoids. Advances in Biochemical Engineering/Biotechnology, edsSchraderJ.BohlmannJ. (Cham: Springer), 19–61. 10.1007/10_2014_283
177
SchomerR. A.ThomasM. G. (2017). Characterization of the functional variance in MbtH-like protein interactions with a nonribosomal peptide synthetase.Biochemistry565380–5390. 10.1021/acs.biochem.7b00517
178
SeibothB.KarimiR. A.PhataleP. A.LinkeR.HartlL.SauerD. G.et al (2012). The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei.Mol. Microbiol.841150–1164. 10.1111/j.1365-2958.2012.08083.x
179
ShenB. (2003). Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms.Curr. Opin. Chem. Biol.7285–295. 10.1016/S1367-5931(03)00020-6
180
ShenB.ChenM.ChengY.DuL.EdwardsD. J.GeorgeN. P.et al (2005). Prerequisites for combinatorial biosynthesis: evolution of hybrid NRPS/PKS gene clusters.Ernst Schering Res. Found. Workshop51107–126. 10.1007/3-540-27055-8_5
181
ShimaJ.HeskethA.OkamotoS.KawamotoS.OchiK. (1996). Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2).J. Bacteriol.1787276–7284. 10.1128/JB.178.24.7276-7284.1996
182
ShwabE. K.BokJ. W.TribusM.GalehrJ.GraessleS.KellerN. P. (2007). Histone deacetylase activity regulates chemical diversity in Aspergillus.Eukaryot. Cell61656–1664. 10.1128/EC.00186-07
183
SmanskiM. J.ZhouH.ClaesenJ.ShenB.FischbachM. A.VoigtC. A. (2016). Synthetic biology to access and expand nature’s chemical diversity.Nat. Rev. Microbiol.14135–149. 10.1038/nrmicro.2015.24
184
SmithJ. L.ShermanD. H. (2008). Biochemistry. An enzyme assembly line.Science3211304–1305. 10.1126/science.1163785
185
SoltaniJ. (2016). “Secondary metabolite diversity of the genus Aspergillus: recent advances,” inNew and Future Developments in Microbial Biotechnology and Bioengineering: Aspergillus System Properties and Applications, ed.GuptaV. K. (Amsterdam: Elsevier B.V.), 275–292. 10.1016/B978-0-444-63505-1.00035-X
186
SprakerJ.KellerN. (2014). “Waking sleeping pathways in filamentous fungi,” inNatural Products, edsOsbournA.GossR. J.CarterG. T. (Hoboken, NJ: John Wiley & Sons, Inc.), 277–292. 10.1002/9781118794623.ch15
187
StachelhausT.MarahielM. A. (1995). Modular structure of peptide synthetases revealed by dissection of the multifunctional enzyme GrsA.J. Biol. Chem.2706163–6169. 10.1074/jbc.270.11.6163
188
StashenkoE. E.MartinezJ. R. (2017). “Identification of essential oil components,” inEssential Oils in Food Processing, edsHashemiS. M. B.KhaneghahA. M.de Souza Sant’AnaA. (Chichester: John Wiley & Sons, Ltd.), 57–117. 10.1002/9781119149392.ch3
189
StauntonJ.WeissmanK. J. (2001). Polyketide biosynthesis: a millennium review.Nat. Prod. Rep.18380–416. 10.1039/a909079g
190
StraussJ.Reyes-DominguezY. (2011). Regulation of secondary metabolism by chromatin structure and epigenetic codes.Fungal Genet. Biol.4862–69. 10.1016/j.fgb.2010.07.009
191
StudtL.JanevskaS.ArndtB.BoediS.SulyokM.HumpfH. -U.et al (2017). Lack of the COMPASS component Ccl1 reduces H3K4 trimethylation levels and affects transcription of secondary metabolite genes in two plant–pathogenic Fusarium species.Front. Microbiol.7:2144. 10.3389/fmicb.2016.02144
192
SunX.LiH.AlfermannJ.MootzH. D.YangH. (2014). Kinetics profiling of gramicidin S synthetase A, a member of nonribosomal peptide synthetases.Biochemistry537983–7989. 10.1021/bi501156m
193
SundaramS.HertweckC. (2016). On-line enzymatic tailoring of polyketides and peptides in thiotemplate systems.Curr. Opin. Chem. Biol.3182–94. 10.1016/j.cbpa.2016.01.012
194
SussmuthR. D.MainzA. (2017). Nonribosomal peptide synthesis-principles and prospects.Angew. Chem.563770–3821. 10.1002/anie.201609079
195
TangG.-L.ChengY.-Q.ShenB. (2007). Chain initiation in the leinamycin-producing hybrid nonribosomal peptide/polyketide synthetase from Streptomyces atroolivaceus S-140.J. Biol. Chem.28220273–20282. 10.1074/jbc.M702814200
196
TeruyaK.TanakaT.KawakamiT.AkajiK.AimotoS. (2012). Epimerization in peptide thioester condensation.J. Pept. Sci.18669–677. 10.1002/psc.2452
197
ThirlwayJ.LewisR.NunnsL.Al NakeebM.StylesM.StruckA. W.et al (2012). Introduction of a non-natural amino acid into a nonribosomal peptide antibiotic by modification of adenylation domain specificity.Angew. Chem. Int. Ed.517181–7184. 10.1002/anie.201202043
198
TudzynskiB. (2014). Nitrogen regulation of fungal secondary metabolism in fungi.Front. Microbiol.5:656. 10.3389/fmicb.2014.00656
199
van den BergM. A. (2010). Functional characterisation of penicillin production strains.Fungal Biol. Rev.2473–78. 10.1016/j.fbr.2010.03.006
200
van den BergM. A.AlbangR.AlbermannK.BadgerJ. H.DaranJ. -M.DriessenA. J. M.et al (2008). Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum.Nat. Biotechnol.261161–1168. 10.1038/nbt.1498
201
ViggianoA.SaloO.AliH.SzymanskiW.LankhorstP.NygårdY.et al (2017). Elucidation of the biosynthetic pathway for the production of the pigment chrysogine by Penicillium chrysogenum.Appl. Environ. Microbiol.84;e02246–17. 10.1128/AEM.02246-17
202
WagnerD.SchmeinckA.MosM.MorozovI. Y.CaddickM. X.TudzynskiB. (2010). The bZIP transcription factor MeaB mediates nitrogen metabolite repression at specific loci.Eukaryot. Cell91588–1601. 10.1128/EC.00146-10
203
WalshC. T. (2016). Insights into the chemical logic and enzymatic machinery of NRPS assembly lines.Nat. Prod. Rep.33127–135. 10.1039/C5NP00035A
204
WangD.-N.ToyotomeT.MuraosaY.WatanabeA.WurenT.BunsupaS.et al (2014). GliA in Aspergillus fumigatus is required for its tolerance to gliotoxin and affects the amount of extracellular and intracellular gliotoxin.Med. Mycol.52506–518. 10.1093/mmy/myu007
205
WangH.FewerD. P.HolmL.RouhiainenL.SivonenK. (2014). Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes.Proc. Natl. Acad. Sci. U.S.A.1119259–9264. 10.1073/pnas.1401734111
206
WangL.WangY.WangQ.LiuF.SelvarajJ. N.LiuL.et al (2015). Functional characterization of new polyketide synthase genes involved in ochratoxin A biosynthesis in Aspergillus ochraceus fc-1.Toxins72723–2738. 10.3390/toxins7082723
207
WangX.WuF.LiuL.LiuX.CheY.KellerN. P.et al (2015). The bZIP transcription factor PfZipA regulates secondary metabolism and oxidative stress response in the plant endophytic fungus Pestalotiopsis fici.Fungal Genet. Biol.81221–228. 10.1016/j.fgb.2015.03.010
208
WatanabeK.OguriH.OikawaH. (2009). Diversification of echinomycin molecular structure by way of chemoenzymatic synthesis and heterologous expression of the engineered echinomycin biosynthetic pathway.Curr. Opin. Chem. Biol.13189–196. 10.1016/j.cbpa.2009.02.012
209
WattanachaisaereekulS.LantzA. E.NielsenM. L.AndressonO. S.NielsenJ. (2007). Optimization of heterologous production of the polyketide 6-MSA in Saccharomyces cerevisiae.Biotechnol. Bioeng.97893–900. 10.1002/bit.21286
210
WeberS. S.BovenbergR. A. L.DriessenA. J. M. (2012a). Biosynthetic concepts for the production of β -lactam antibiotics in Penicillium chrysogenum.Biotechnol. J.7225–236. 10.1002/biot.201100065
211
WeberS. S.KovalchukA.BovenbergR. A. L.DriessenA. J. M. (2012b). The ABC transporter ABC40 encodes a phenylacetic acid export system in Penicillium chrysogenum.Fungal Genet. Biol.49915–921. 10.1016/j.fgb.2012.09.003
212
WeberS. S.PolliF.BoerR.BovenbergR. A. L.DriessenA. J. M. (2012c). Increased penicillin production in Penicillium chrysogenum production strains via balanced overexpression of isopenicillin N acyltransferase.Appl. Environ. Microbiol.787107–7113. 10.1128/AEM.01529-12
213
WeberT.BlinK.DuddelaS.KrugD.KimH. U.BruccoleriR.et al (2015). antiSMASH 3.0–a comprehensive resource for the genome mining of biosynthetic gene clusters.Nucleic Acids Res.43W237–W243. 10.1093/nar/gkv437
214
WeberT.MarahielM. A. (2001). Exploring the domain structure of modular nonribosomal peptide synthetases.Structure9R3–R9. 10.1016/S0969-2126(00)00560-8
215
WeissmanK. J. (2016). Genetic engineering of modular PKSs: from combinatorial biosynthesis to synthetic biology.Nat. Prod. Rep.33203–230. 10.1039/C5NP00109A
216
WiemannP.BrownD. W.KleigreweK.BokJ. W.KellerN. P.HumpfH.-U.et al (2010). FfVel1 and FfLae1, components of a velvet-like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism and virulence.Mol. Microbiol.77972–994. 10.1111/j.1365-2958.2010.07263.x
217
WiestA.GrzegorskiD.XuB. -W.GoulardC.RebuffatS.EbboleD. J.et al (2002). Identification of peptaibols from Trichoderma virens and cloning of a peptaibol synthetase.Journal of Biological Chemistry27720862–20868. 10.1074/jbc.M201654200
218
WilliamsR. B.HenriksonJ. C.HooverA. R.LeeA. E.CichewiczR. H. (2008). Epigenetic remodeling of the fungal secondary metabolome.Org. Biomol. Chem.61895–1897. 10.1039/b804701d
219
WuM.-Y.YuJ.-H. (2015). “Epigenetics of fungal secondary metabolism related genes,” inBiosynthesis and Molecular Genetics of Fungal Secondary Metabolites, edsZeilingerS.MartínJ.-F.García-EstradaC. (New York, NY: Springer), 29–42. 10.1007/978-1-4939-2531-5_3
220
YadavG.AnandS.MohantyD. (2013). Prediction of inter domain interactions in modular polyketide synthases by docking and correlated mutation analysis.J. Biomol. Struct. Dyn.3117–29. 10.1080/07391102.2012.691342
221
YehH. H.ChangS. L.ChiangY. M.BrunoK. S.OakleyB. R.WuT. K.et al (2013). Engineering fungal nonreducing polyketide synthase by heterologous expression and domain swapping.Org. Lett.15756–759. 10.1021/ol303328t
222
YinW.KellerN. P. (2011). Transcriptional regulatory elements in fungal secondary metabolism.J. Microbiol.49329–339. 10.1007/s12275-011-1009-1
223
YinW.-B.ReinkeA. W.SzilagyiM.EmriT.ChiangY.-M.KeatingA. E.et al (2013). bZIP transcription factors affecting secondary metabolism, sexual development and stress responses in Aspergillus nidulans.Microbiology15977–88. 10.1099/mic.0.063370-0
224
YinX.ZabriskieT. M. (2006). The enduracidin biosynthetic gene cluster from Streptomyces fungicidicus.Microbiology1522969–2983. 10.1099/mic.0.29043-0
225
YuD.XuF.ZengJ.ZhanJ. (2012). Type III polyketide synthases in natural product biosynthesis.IUBMB Life64285–295. 10.1002/iub.1005
226
YuJ. -H.KellerN. (2005). Regulation of secondary metabolism in filamentous fungi.Annu. Rev. Phytopathol.43437–458. 10.1146/annurev.phyto.43.040204.140214
227
ZhangK.NelsonK. M.BhuripanyoK.GrimesK. D.ZhaoB.AldrichC. C.et al (2013). Engineering the substrate specificity of the dhbe adenylation domain by yeast cell surface display.Chem. Biol.2092–101. 10.1016/j.chembiol.2012.10.020
228
ZhouK.GaoY.HoyJ. A.MannF. M.HonzatkoR. B.PetersR. J. (2012). Insights into diterpene cyclization from structure of bifunctional abietadiene synthase from Abies grandis.J. Biol. Chem.2876840–6850. 10.1074/jbc.M111.337592
Summary
Keywords
Penicillium chrysogenum, natural products, nonribosomal peptides, polyketides, gene activation, biosynthetic gene clusters, cell factory
Citation
Guzmán-Chávez F, Zwahlen RD, Bovenberg RAL and Driessen AJM (2018) Engineering of the Filamentous Fungus Penicillium chrysogenum as Cell Factory for Natural Products. Front. Microbiol. 9:2768. doi: 10.3389/fmicb.2018.02768
Received
19 June 2018
Accepted
29 October 2018
Published
15 November 2018
Volume
9 - 2018
Edited by
Eung-Soo Kim, Inha University, South Korea
Reviewed by
Gang Liu, Institute of Microbiology (CAS), China; Juan F. Martin, Universidad de León, Spain
Updates
Copyright
© 2018 Guzmán-Chávez, Zwahlen, Bovenberg and Driessen.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arnold J. M. Driessen, a.j.m.driessen@rug.nl
†Present address: Fernando Guzmán-Chávez, Synthetic Biology and Reprogramming of Plant Systems, Department of Plant Sciences, University of Cambridge, Cambridge, United Kingdom
This article was submitted to Microbial Physiology and Metabolism, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.