- 1Biomedical Research, The University of Texas Health Science Center, Tyler, TX, United States
- 2Institute of Medical Biology, Polish Academy of Sciences, Lodz, Poland
- 3Department of Pathology and Laboratory Medicine, Rutgers New Jersey Medical School, Newark, NJ, United States
The biological processes regulated by the essential response regulator MtrA and the growth conditions promoting its activation in Mycobacterium tuberculosis, a slow grower and pathogen, are largely unknown. Here, using a gain-of-function mutant, MtrAY 102C, which functions in the absence of the cognate MtrB sensor kinase, we show that the MtrA regulon includes several genes involved in the processes of cell division and cell wall metabolism. The expression of selected MtrA targets and intracellular MtrA levels were compromised under replication arrest induced by genetic manipulation and under stress conditions caused by toxic radicals. The loss of the mtrA gene in M. smegmatis, a rapid grower and non-pathogen, produced filamentous cells with branches and bulges, indicating defects in cell division and cell shape. The ΔmtrA mutant was sensitized to rifampicin and vancomycin and became more resistant to isoniazid, the first line antituberculosis drug. Our data are consistent with the proposal that MtrA controls the optimal cell division, cell wall integrity, and susceptibility to some antimycobacterial drugs.
Introduction
Tuberculosis remains the leading cause of mortality globally and accounts for, on average, 1.5 million deaths a year. Mycobacterium tuberculosis, the causative agent of tuberculosis, is a successful pathogen that employs a host of regulatory networks for its optimal survival upon infection (Cole et al., 1998), one of which is the paired histidine-aspartate two-component regulatory signal transduction system (TCSS) (Hoch, 2000; Stock et al., 2000). This system includes a membrane-bound sensor kinase and a cytosolic response regulator (RR). The sensor kinases respond to specific environmental stimuli, become autophosphorylated and engage in the transphosphorylation of the cognate RR via the phosphorelay signal transduction process. The phosphorylated RR then binds to specific sequences in the promoter regions of several target genes and modulates their expression. The M. tuberculosis genome contains eleven TCSSs and several orphan kinases and response regulators. Among these, MtrAB and PrrAB are essential for M. tuberculosis survival (Zahrt and Deretic, 2000; Haydel et al., 2012). The MtrAB system includes the MtrB (Rv3245c) sensor kinase and the MtrA RR (Rv3246c). Although it was first described nearly 20 years ago, the essential MtrAB system remains poorly characterized.
Earlier studies (Fol et al., 2006) designed to evaluate the roles of MtrA characterized the viability of M. tuberculosis strains overproducing phosphorylation-competent wild-type MtrA+ (WT, Rv78), phosphorylation-defective MtrA (MtrAD56N, Rv129) and MtrB along with MtrA+ (Rv79) upon infection (the uncommon Rv names Rv78, Rv129, Rv79 are the numbers of M. tuberculosis strains from the collection of our laboratory). These studies revealed that the regulation of M. tuberculosis proliferation upon infection is in part dependent upon optimal MtrA levels and MtrA phosphorylation (MtrA∼P) and that MtrB activity is likely involved in regulating MtrA∼P (Fol et al., 2006). These data also indicated that MtrA∼P level is modulated during intracellular growth. Other studies identified that the promoters (P) of the cell wall hydrolase ripA (rv1477), the cell wall mycolyl transferase fbpB (rv1886c) and the replication initiator dnaA (rv0001) are MtrA targets (Rajagopalan et al., 2010; Plocinska et al., 2012). The role of MtrAB two component system was also studied in M. smegmatis cells, a rapid grower and non-pathogen. The MtrB (MSMEG_1875) sensor kinase is not essential in M. smegmatis, and mtrB disruption compromised MtrA-target expression and cell division (Plocinska et al., 2012). The mtrB KO phenotype was reversed by either the production of an intact MtrB or overproduction of MtrA (MSMEG_1874) carrying Y102C mutation which favors MtrA phosphorylation and/or DNA binding even in the absence of MtrB (Plocinska et al., 2012). The overproduction of MtrAY 102C in MtrB KO mutant caused not only the reversal of the ΔmtrB phenotype but also increased the expression levels of MtrA targets (dnaA, ripA fbpB, ftsI and wag31) (Plocinska et al., 2012). Recent studies show MtrB interacts with FtsI (Rv2163c, penicillin-binding protein 3 with transpeptidase activity) and Wag31 (Rv2145c, the cell wall synthesis protein). The expression levels of mtrA, along with MtrA targets dnaA, fbpB and ripA were decreased under FtsI depletion conditions. FtsI, in contrast to Wag31, functions as a positive modulator of MtrB activation and MtrA regulon expression (Plocinska et al., 2014). It was also shown that phosphorylation defective MtrA carrying both the D56N and Y102C mutations was able to bind its DNA targets and reverse, at least partially, mtrB KO phenotypes in M. smegmatis indicating that phosphorylation is not required for the function of MtrAY 102C (Satsangi et al., 2013). More recently it was reported that the access of the wild type MtrA to origin of replication (oriC) in tubercle bacilli depends on its phosphorylation. The abundant oriC binding by phosphorylated MtrA as well as MtrAY 102C reduced the dnaN (rv0002) and dnaA expression, interfered with replication synchrony, and compromised cell division (Purushotham et al., 2015). Moreover, the in vitro study has shown that phosphorylated and non-phosphorylated MtrAY 102C binds oriC with similar affinity (Purushotham et al., 2015).
The high-throughput sequencing (CHIP-seq) analysis was also applied to identify the MtrA targets in two distinct studies (Minch et al., 2015; Chatterjee et al., 2018).
Although the above studies identified a handful of MtrA targets and connected MtrA activity to the cell cycle, several questions remain. For example, how does MtrA activity impact the cell division and cell wall metabolism processes? Which of the genes involved in these such important processes are under MtrA∼P control, and what are the members of the MtrA∼P regulon? Here, we performed chromatin immunoprecipitation of MtrAY 102C phosphorylation competent protein followed by high-throughput sequencing under active and stationary-phase growth conditions to elucidate a comprehensive MtrA∼P regulon. Our CHIP-seq analysis for MtrAY 102C a gain-of-function protein that binds to its targets independent of phosphorylation (Satsangi et al., 2013), identified and moreover expanded new targets, not being identified by previously published CHIP-seq analysis for MtrA, performed by Minch et al. (2015) and Chatterjee et al. (2018). Additionally, we evaluated the growth conditions where the MtrA system is active and, finally, created and characterized a M. smegmatis ΔmtrA mutant strain. Based on transposon mutagenesis studies (TRASH), mtrA is an essential gene for growth and survival of M. tuberculosis (Sassetti et al., 2003). Despite numerous attempts we were not able to construct ΔmtrA mutant in M. tuberculosis cells. These studies revealed that MtrA is a key regulator of optimal cell wall integrity and cell division in replicating cells and its depletion affects susceptibility of M. smegmatis cells to the first line antituberculosis drugs.
Materials and Methods
Bacterial Strains and Proteins
The oligonucleotide primers used in the study are listed in Supplementary Table S1. The description of strains and plasmids is included in Supplementary Table S2. M. tuberculosis (H37Rv) and M. smegmatis (mc2155) strains were grown in Middlebrook 7H9 media supplemented with OADC (oleic acid-albumin-dextrose-catalase) and ADC supplements, respectively. Recombinant maltose binding protein fusions of EnvZ, MtrA, and MtrAY 102C were produced in Escherichia coli as described (Plocinska et al., 2012). Bacterial growth was assayed by measuring changes in absorbance at 600 nm, and viability was determined by assaying colony-forming units per mL. For some experiments, in order to induce stress conditions, the actively growing M. tuberculosis cells were exposed to 0.2% SDS or 100 μM DETA-NO for 2 or 16 h, respectively. Next, cells were harvested and RNA was isolated according to the protocol described previously (Fol et al., 2006).
Cloning and Construction of Plasmids
The plasmids and oligonucleotide primers used in this study are listed in Supplementary Tables S1, S2. The coding regions of various genes were PCR amplified with Phusion DNA polymerase (New England BioLabs Inc., Ipswich, MA, United States) and cloned into various plasmids using standard molecular biology techniques. Cloned regions were confirmed by sequencing. The PCR products corresponding to ChIP-seq peaks were amplified using primer pairs (Supplementary Table S1) and cloned as HindIII-SacI fragments into the plasmid pUC57. FAM-labeled primers targeting the vector sequence flanking the insert were used to generate DNA probes for EMSA.
Phenotype Analysis
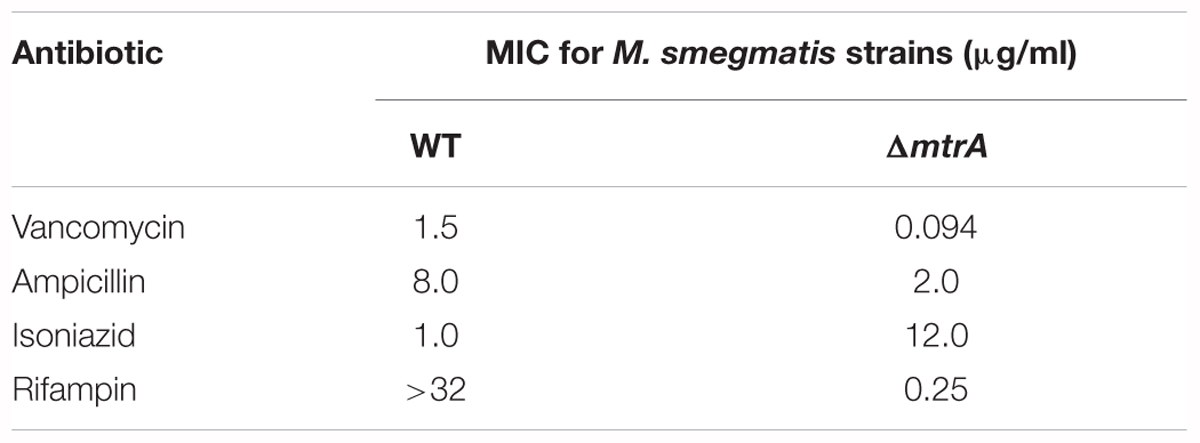
Microscopy: Actively growing M. tuberculosis and M. smegmatis cells were visualized by bright-field and fluorescent microscopy as described (Plocinski et al., 2012). All M. tuberculosis cells were fixed in 4% paraformaldehyde prior to visualization. To evaluate sensitivity to antibiotics, actively growing M. smegmatis cultures were diluted to an OD600 of 0.05 for 6 h, and approximately 1 × 105 cells were spread with sterile cotton-tipped swabs. Next, E-test strips (ampicillin, vancomycin, rifampicin, or isoniazid) were placed on the culture plates and incubated at 37°C for 4 days prior to recording as described (Plocinski et al., 2013; Plocinska et al., 2014). The MIC values were determined following the E-test manufacturer’s recommendations.
Creation of M. smegmatis mtrA Mutant
A two-step recombination protocol was used to delete the mtrA gene of M. smegmatis. First, the 5′ end of mtrA (125 bp) and the upstream region were PCR amplified using the primers MtrAsmegGR1ScaI and MtrAsmegGR2HindIII and cloned into the p2NIL vector to create pDR45 (Supplementary Tables S1, S2). Next, a 1530-bp fragment including 365 bp from the 3′ end of mtrA and the downstream region was PCR amplified with the primers MtrAsmegGR3HindIII and MtrAsmegGR4PacI and cloned into pDR45 to create pDR47. An 850-bp gentamicin cassette was then cloned into the HindIII site of pDR47 to create pDR49. Finally, a 6000-bp cassette containing the lacZ and sacB genes from pGOAL17 was inserted in pDR49 to create the final suicidal recombination vector pDR51. The plasmid DNA of pDR51 was pre-treated with UV light and electroporated into M. smegmatis competent cells. Prior to double-cross-over (DCO) screening, the plasmid pDR54 expressing the M. tuberculosis mtrA gene under a tetracycline-inducible promoter was transformed into one representative SCO strain. White DCO colonies that were resistant to sucrose and sensitive to kanamycin were further confirmed by PCR and Southern blotting approaches (GE Healthcare). Finally, the pDR54 plasmid was swapped out with the pMV306K vector as described (Chauhan et al., 2006b).
Verification of the Essentiality of mtrB in M. tuberculosis
A two-step recombination protocol was used to verify the essentiality of mtrB in tubercle bacilli. The upstream region of M. tuberculosis mtrB gene (2145 bp) was cloned into suicidal recombination p2Nil vector, followed by the downstream fragment of mtrB sequence (1566 bp), creating pDR56. Next, gentamicin cassette was inserted into HindIII site. Finally, the screening cassette from pGOAL17 vector was cloned and resulting suicide delivery vector pDR58 was used to engineer the direct mtrB mutant as described above and by Dadura et al. (2017). Next, the complementation plasmid expressing mtrB gene under acetamide promoter (pRD102) (Plocinska et al., 2012) was introduced into representative SCO strain, in order to process for DCO screening. Next, the pRD102 was swapped with pKS4 vector expressing mtrB-gfp and kanamycin resistance cassette (Plocinska et al., 2012). Finally, the pKS4 vector was swapped with pDS4, expressing mtrAY 102C and hygromycin cassette. The genotypes of ΔmtrB::mtrAY 102C strain was confirmed by Southern blot hybridization using probe to mtrB gene and following manufacturer instructions (GE Healthcare). Plasmids and primers used for PCR amplification are listed in Supplementary Tables S1, S2.
RNA Extraction and Quantitative Real-Time PCR
Extraction of total RNA and quantitative real-time PCR were performed in a BioRad iCycler iQTM Real-Time PCR detection system using FAM fluorophore-labeled 2X iQ SYBR Supermix (BioRad, Cat# 1708880) as described (Maloney et al., 2009; Rajagopalan et al., 2010). The threshold cycle (Ct) value of each gene of interest was normalized to the Ct value of 16S rRNA, and the fold expression was calculated [fold change = 2-Δ(ΔCt)]. Expression data were obtained from an average of three independent RNA preparations, and each gene of interest was investigated in triplicate. Fold differences of 2 or more were considered significant. Primers used for qRT-PCR are listed in Supplementary Table S1.
Electrophoretic Mobility Shift Assay (EMSA)
Electrophoretic mobility shift assay was carried out to detect MtrA binding to FITC-labeled promoter PrpfB and FAM-labeled promoters: PripA, PsigD, PoxyS, Pwag31, PbetP, Prv3887c, PwhiB3, PdacB1 or MtrAY 102C binding to FAM-labeled promoters: PbetP, Prv2525 and PdacB1 as described (Plocinska et al., 2012; Satsangi et al., 2013; Purushotham et al., 2015). The 200 bp upstream regions of chosen targets were amplified using primers listed in Supplementary Table S1. MtrA/MtrAY102C were phosphorylated by EnvZ (Al Zayer et al., 2011) and incubated at 1, 2, 4, 6, 10 μM concentration with 200 fmols promoter DNA’s in buffer containing 50 mM Tris-HCl pH 7.5, 50 mM NaCl, 10 mM MgCl2, 10 mM CaCl2. Next, 20 pmoles poly dI/dC and shared salmon sperm DNA (1 μg) were added, reactions were incubated at 37°C for 15 min and resolved in 5% polyacrylamide gels. The DNA-protein complexes were visualized using Molecular Imager Fx (BioRad).
Chromatin Immunoprecipitation (ChIP)
Detailed methodology for ChIP-seq sample preparation, processing of libraries, SOLiD sequencing, data analyses and motif prediction are presented in the Supplementary Data. Briefly, M. tuberculosis cultures producing MtrAY 102C growing for 3 (exponential growth) and 8 (stationary growth) days were cross-linked by fixing in 1% formaldehyde and processed for ChIP analysis with anti-MtrA antibodies essentially as described (Fol et al., 2006; Rajagopalan et al., 2010). The protein-DNA complexes obtained following incubation of sheared lysates with anti-MtrA antibodies were recovered using ImmunoPure immobilized Protein G agarose beads (Thermo Fisher Scientific, Rockford, IL, United States), and the cross-links were reversed by incubation at 65°C for 16 h. DNA samples were purified using DNAzol, resuspended in 50 μl of TE buffer, processed and used either for individual target evaluation (Fol et al., 2006; Rajagopalan et al., 2010; Plocinska et al., 2012) or for SOLiD library construction (detailed in the Supplementary Data).
Results
The mtrB of Tubercle Bacilli Is Not Essential in the MtrAY 102C Genetic Background
The high density transposon mutagenesis (TRASH) suggested that in distinction to M. smegmatis the membrane-bound sensor kinase MtrB responsible for the phosphorylation of the response regulator MtrA is essential in M. tuberculosis (Sassetti et al., 2003). Since the MtrAY 102C is believed to be phosphorylation competent and functions in the absence of MtrB, we tested whether mtrB can be inactivated in M. tuberculosis complemented with mtrAY 102C. The gene replacement protocol (Parish and Stoker, 2000; Dziadek et al., 2002b) was used to engineer M. tuberculosis mutants carrying both wild type and ΔmtrB genes (SCO) which were further processed for homologous recombination to select double cross-over mutants (DCO) carrying either wild type or mutated mtrB. We have analyzed over 50 DCO mutants by using PCR screening and in all cases only wild type mtrB gene was detected. Multiple attempts to replace complementing vector pRD102 with empty pMV306 vector confirmed essentiality of MtrB protein in M. tuberculosis cells. On the other hand the native mtrB was replaced with an inactivated copy (ΔmtrB::gmR) when mtrB-gfp or mtrAY 102C were introduced with integration vectors into an attB site. The genotypes of the merodiploid strains (ΔmtrB::gmR-attB::mtrB-gfp or -attB::mtrAY 102C) were confirmed by Southern hybridization (Supplementary Figure S1), confirming that MtrAY 102C is able to replace at least partially the function of phosphorylated MtrA.
The Replacement of Cys With Tyr in Position 102 of MtrA Results in Signal Independent Phosphorylation of MtrA
Based on the crystal structure of MtrA indicating tyrosine at position 102 is at the interdomain interface (Friedland et al., 2007), we have previously engineered M. tuberculosis strain producing MtrAY 102C (RvY102C), which contains cysteine in place of the tyrosine located at the interface of the regulatory and DNA-binding domains (Plocinska et al., 2012). Because MtrAY 102C is phosphorylation competent and functions in the absence of the cognate sensor kinase MtrB (Plocinska et al., 2012; Satsangi et al., 2013), we reasoned that MtrAY 102C would be activated in the absence of the specific exogenous signals that are otherwise necessary for the MtrB autophosphorylation and transphosphorylation activities. The RvY102C mutant initially grew similarly to the WT and MtrA overproducing strain (Rv78) during exponential phase, however, reached the stationary phase much faster (OD600 value for Rv19 – 4,315; Rv78 – 4,144; RvY102C-B – 4,28; RvY102C – 1,812) (Figure 1A). Similar results were also obtained when the plasmid carrying MtrAY 102C was transformed into other M. tuberculosis strain CSU#1, suggesting that the expression of mtrAY 102C is enough to affect the growth in two different genetic background strains (Supplementary Figure S2). Overexpression of mtrB along with mtrAY 102C reversed the observed growth defect associated with MtrAY 102C overproduction (Figure 1A, compare RvY102C-B with RvY102C), corroborating the previous data that MtrB modulates MtrA∼P potential (Fol et al., 2006). Independently, we evaluated the expression levels of the MtrA targets dnaA (Fol et al., 2006) and ftsI [see below, also (Plocinska et al., 2012; Plocinska et al., 2014) in the RvY102C background relative to WT]. Expression of dnaA was decreased, whereas that of the ftsI was increased in RvY102C relative to WT (Figure 1B). No significant differences in the dnaA and ftsI expression were noted in Rv78 (Figure 1B).
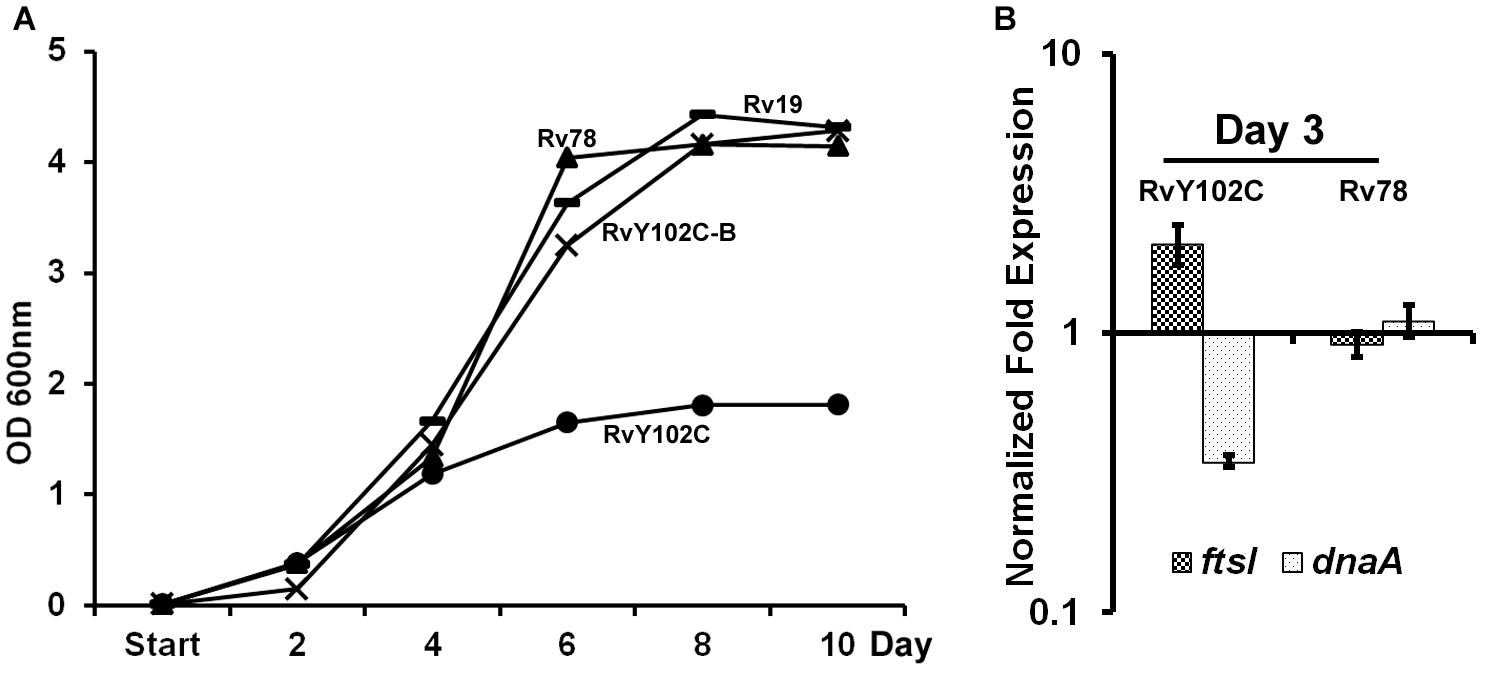
FIGURE 1. Characterization of RvY102C. (A) Growth of M. tuberculosis Rv19, Rv78, RvY102C, RvY102C-B carrying empty vector, mtrA, mtrAY 102C and mtrAY 102C-mtrB plasmids, respectively (see Supplementary Table S2). (B) Expression of dnaA and ftsI in RvY102C (normalized to 16S rRNA) determined by qRT-PCR. Data shown are fold expression in Rv78 and RvY102C relative to Rv19. The total RNA was isolated from three independent biological replicates and the data are represented as the means ± standard errors from three independent experiments.
MtrA Regulon
We reasoned from the above data that the RvY102C background would enable us to elucidate a comprehensive MtrA regulon in the absence of exogenous MtrAB system activation signals. Considering the growth characteristics of RvY102C, samples collected from exponential phase (day 3) and stationary phase (day 8) cultures were processed for ChIP-seq studies to define a full spectrum of the MtrA regulon based on the two different growth states. CHIP-seq analysis were performed using anti-MtrA polyclonal antibodies to detect all MtrA, MtrA-P as well as MtrAY 102C. We identified a total of 216 ChIP-seq peaks that were present in duplicate samples from day 3, extracted the sequences corresponding to ChIP peaks, analyzed the data using the MEME software, and defined the MtrA motif using a Web logo tool, which consists of the a degenerated direct repeat of c-G-T/A-n-A-C/T-c, separated by four nucleobases (Figure 2A). This motif, which begins with either G or T, has a significant sequence similarity to the previously defined MtrA motif based on DNaseI footprinting of oriC and PfbpB (Rajagopalan et al., 2010). However, differences with respect to the weights of the individual bases were noted. The predicted MtrA motif was then used to search for possible matches in the day 8 ChIP-seq peaks with relatively high scores. Using this approach we identified a total of 278 target sites, of which 93 were common to day 3 and day 8, whereas 62 were unique to day 8 only. A comprehensive list of MtrA regulon genes based on both sets of data was generated (Supplementary Table S3 and Supplementary Figure S3A, select peaks at both growth states) and classified into families based on Tuberculist (Supplementary Figure S3B). As could be expected, this list includes new targets not reported previously (Galagan et al., 2013). The identification of the confirmed MtrA targets PfbpB and PripA and the absence of the non-targets PftsZ (rv2150c) and PpfkB (rv2029c) (Rajagopalan et al., 2010; Plocinska et al., 2012) combined with the presence of an MtrA motif in every designated ChIP-seq peak increased the degree of confidence in our analysis. MtrA binding to select targets PsigD (rv3414c), PoxyS (rv0117), Pwag31, PbetP (rv0917), Prv3887, PwhiB3 (rv3416) and MtrAY 102C binding to PbetP, Prv2526 was further validated by EMSA (Supplementary Figure S4). The previously reported MtrA binding to PrpfB (rv1009) (Rajagopalan et al., 2010) and PripA (Plocinska et al., 2012) was a positive control in our experiment. To investigate the possibility of protein-DNA complexes we incubated recombinant MtrA and MtrAY 102C (Plocinska et al., 2012) with FAM-labeled DNA. We identified complexes of MtrA-P as well as MtrA in the absence of phosphorylation with PrpfB, PripA, Prv3887. In case of PsigD and PoxyS we observed better binding to MtrA-P than to MtrA. Only MtrA-P retarded Pwag31 and PwhiB3. Both, MtrA and MtrA-P did not show binding to PbetP.
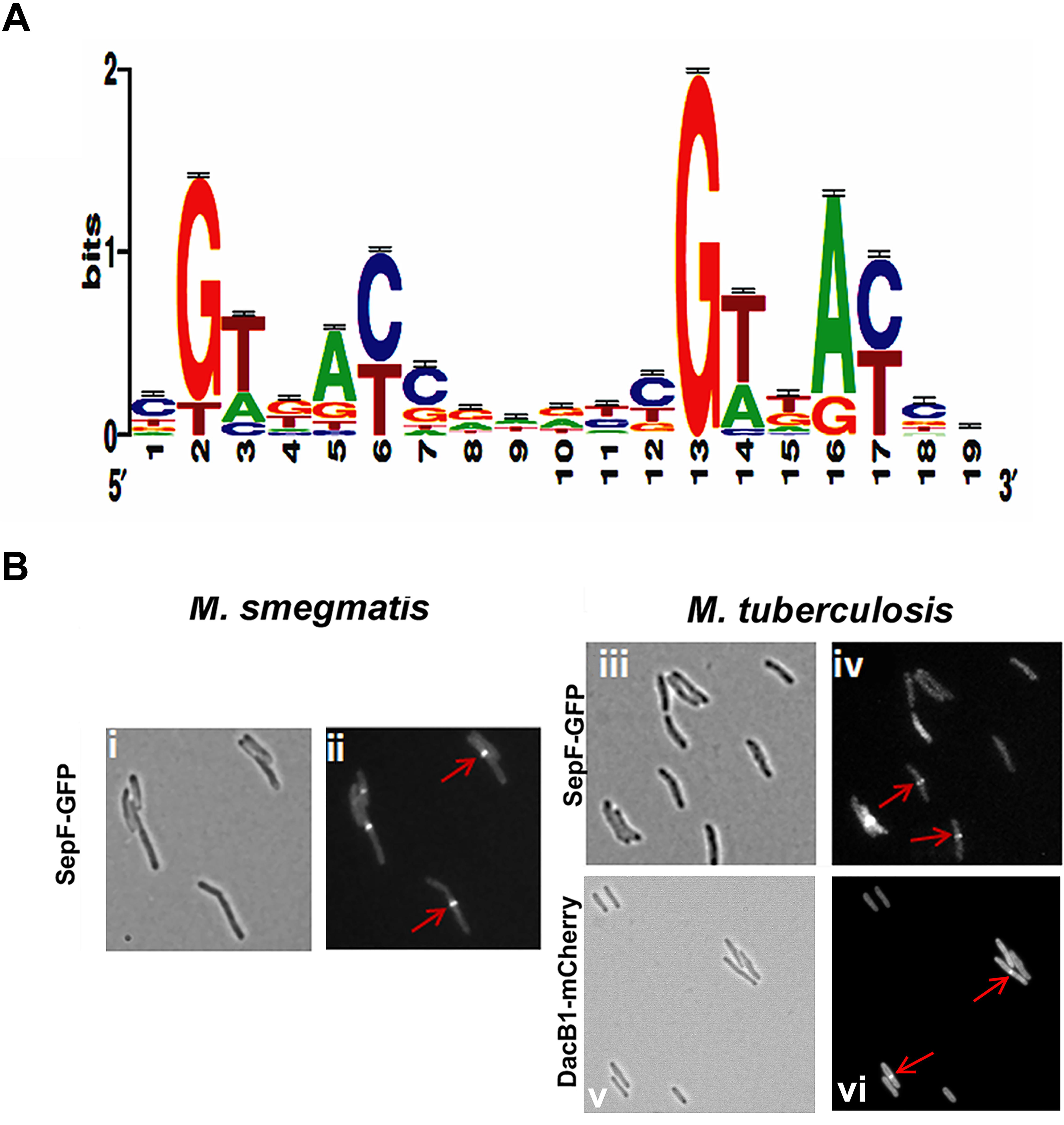
FIGURE 2. MtrA motif and validation of DacB1 and SepF septasomal association: (A) MtrA logo determined by analyzing the MtrA regulon ChIP-Seq peaks. The p-value of the deduced motif is 2.6e-123, and the motif is a direct repeat of 9 nucleotides with a spacing of 2 nucleotides, starting with either G or T and possessing a sequence significantly similar to the previously defined MtrA-motif based on DNase I footprinting of oriC and PfbpB targets (Rajagopalan et al., 2010), as well as to that of the MtrA-motifs of C. glutamicum and TB database (Brocker et al., 2011; Galagan et al., 2013), except for differences with respect to the weightings of the individual bases. (B) Localization of DacB1-mCherry in M. tuberculosis and SepF-GFP in M. smegmatis and M. tuberculosis. Bright-field (i,iii,v) and respective fluorescence (ii,iv,vi) images of and M. tuberculosis producing DacB1-mCherry and M. smegmatis and M. tuberculosis producing SepF-GFP are shown. Arrows denote septal localization.
Cell Division and Cell Wall Metabolism Genes Within MtrA Regulon
Cell division involves the midcell FtsZ assembly (Z-ring formation), followed by successive localization of multiple proteins to a Z-ring, which is also referred to as divisome assembly; septal cell wall synthesis; and cell separation events (Adams and Errington, 2009). The cell division process is interconnected and coordinated with cell wall elongation, which in mycobacteria occurs from the poles (Aldridge et al., 2012; Santi et al., 2013). We noted that the MtrA regulon includes a number of cell division and cell wall metabolism genes, including peptidoglycan hydrolase (ripA), mycolyltransferases (fbpB, fbpC (rv0129c)), penicillin-binding protein involved in peptidoglycan biosynthesis (ftsI, dacB1 (rv3330), FtsZ-interacting protein (sepF) (rv2147c), essential DivIVa family cell division protein (wag31) and resuscitation-promoting factor rpf (A-E) (rv0867c, rv1009, rv1884c, rv2398c, rv2450c, respectively) (Supplementary Table S3). The identification of dacB1 gene by ChIP was positively validated by in vitro study using EMSA assay (Supplementary Figures S5A,Bi,ii). The increasing concentrations of MtrA or MtrAY 102C, phosphorylated by EnvZ and non-phosphorylated were incubated with FAM-labeled promoter of dacB1. EMSA studies showed better binding of MtrA-P than MtrA to PdacB1 (Supplementary Figure S5Bi) and similar binding of MtrAY 102C with and without phosphorylation (Supplementary Figure S5Bii).
With the exception of dacB1, the involvement of the above target in the septum synthesis and cell wall expansion processes in mycobacteria have been established (Belisle et al., 1997; Nguyen et al., 2007; Plocinska et al., 2012; Plocinski et al., 2012; Gupta et al., 2015). In B. subtilis, SepF interacts with FtsZ, promotes FtsZ anchoring to membranes and supports the late steps of septum synthesis (Hamoen et al., 2006; Sauvage et al., 2008). In mycobacteria SepF protein also interacts with FtsZ and regulates the cell division process (Gupta et al., 2015). Through fluorescent protein-fusion experiments, we were able to confirm that M. smegmatis and M. tuberculosis SepF and M. tuberculosis DacB1 are also septasomal components (Figure 2B). We found the SepF-GFP localized to the mid-cell in 42% of M. smegmatis cells (n = 100) and in 36.6% of M. tuberculosis cells (n = 101). DacB1-mCherry localization to the septum was counted in 14.5% of M. tuberculosis cells (n = 103). The GFP expressed alone within the control cells revealed dispersed fluorescence as published (Dziadek et al., 2002a). Thus, the ability of MtrA to target a wide array of cell division and cell wall expansion proteins implies that its activity is crucial for those processes.
The Stress Conditions Down-Regulate MtrA Expression and Its Regulon
The ability of MtrA to affect the cell division and cell wall metabolism processes suggests that its activity is relevant and possibly necessary in replicating cells. To test this possibility, we evaluated MtrA target gene expression under selected growth conditions that compromise proliferation, such as cell envelope stress, DNA and protein damage, and DNA replication inhibition. It is anticipated that M. tuberculosis encounters these types of stresses in the hostile host environment upon infection (Smith, 2003). Accordingly, we exposed actively growing M. tuberculosis to SDS and DETA-NO to induce envelope/membrane stress and nitric oxide (NO) stress, respectively, as described (Chauhan et al., 2006a; He et al., 2006; Pang et al., 2007). SDS stress induces the mprA (rv0981, regulator of TCSS MprAB) and sigE (rv1221) genes and their associated regulons; both MprA and SigE are regulated by each other under SDS stress (He et al., 2006; Pang et al., 2007). As sigE is also a member of the MtrA regulon (this study), we explored the possibility of whether SDS exposure also induces the expression of other MtrA-regulon members. SDS stress led to an induction of mprA and sigE expression as expected (Figure 3A), whereas the expression of mtrA along with mtrB and most of the MtrA targets was decreased by 2- to 3-fold (Figure 3A). Similar to the situation with SDS stress, DETA-NO exposure also compromised the expression of mtrA and its targets while the expression of pfkB, a member of the DosRS regulon and a marker for NO stress, was elevated (Figure 3B). It was previously reported, MtrA-target expression was also compromised in M. smegmatis cells upon exposure to mitomycin C, an agent that damages DNA and interferes with DNA replication (Plocinska et al., 2012). The expression of chiZ (rv2719c), a marker for DNA damage, was elevated in both M. tuberculosis (Chauhan et al., 2006a) and M. smegmatis cells exposed to mitomycin C (Plocinska et al., 2012). It is not readily apparent why the expression of some targets did not change under the above stress conditions; either their expression continues under limiting MtrA∼P conditions or is complex and may perhaps also be impacted by other yet-to-be-identified regulator(s). Together, these data indicate that the replication arrest down-regulate MtrA expression and MtrA regulon. To verify this hypothesis we used temperature-sensitive M. tuberculosis mutant expressing DnaA protein, referred to as dnaATs, that is defective for binding to ATP at 30°C. This strain is cold sensitive for replication initiation at 30°C; however, the non-permissive temperature does not affect the ongoing rounds of replication. Upon a shift to the permissive temperature of 37°C, the mutant resumes replication initiation and DNA synthesis in a synchronous manner (Nair et al., 2009). Accordingly, RNA samples collected from the dnaATs mutant growing at a permissive temperature (37°C) and those incubated at a non-permissive temperature (30°C) for 30 h were processed for mtrA, mtrB, rpfB, dacB1, sepF, ftsI, wag31, sucC and chiZ expression analysis (Please see the schematic of experimental plan in Supplementary Figure S6). Compromised expression of the MtrA targets, with the exception of rpfB and chiZ, at non-permissive temperatures relative to 37°C was noted (Figure 3C). Expression levels were, however, restored upon a shift to 37°C (Purushotham et al., 2015).
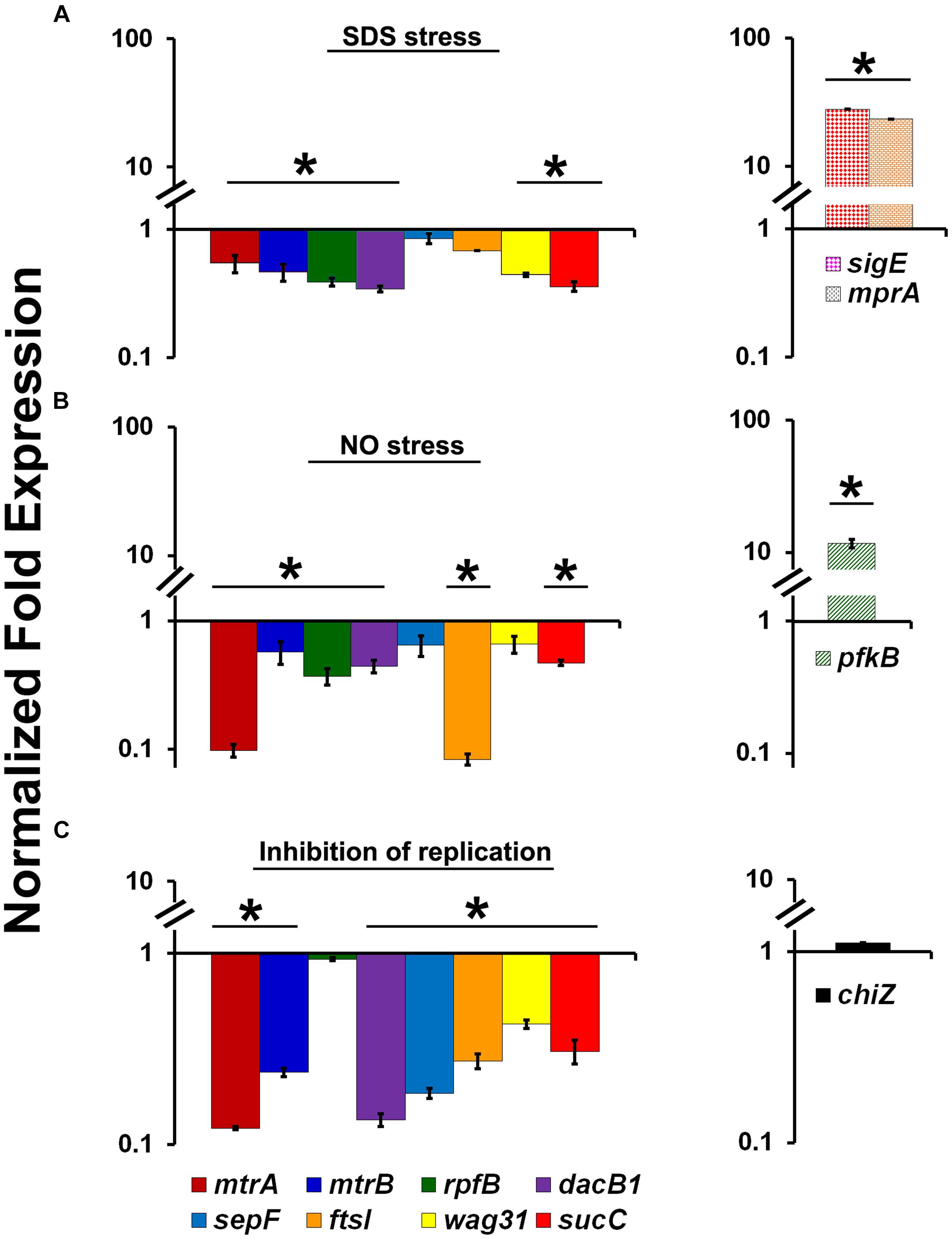
FIGURE 3. Expression profiles of select MtrA target genes (mtrA, mtrB, rpfB, dacB1, sepF, ftsI, wag31, and sucC) under stress and replication-arrest conditions. (A) Total RNA was extracted from actively growing M. tuberculosis cultures exposed to 0.2% SDS for 2 h and the expression levels were analyzed by qRT-PCR as described in Section “Materials and Methods.” (B) Total RNA was extracted from actively growing M. tuberculosis cultures exposed to 100 μM DETA-NO for 16 h and qRT-PCR was performed to analyze gene expression profiles. (C) For evaluating the MtrA-target expression under replication inhibition conditions, M. tuberculosis dnaATs cultures growing at permissive 37°C were shifted to 30°C and incubated for 30 h (non-permissive temperature). RNA samples prepared from cultures growing at both permissive and non-permissive temperatures were processed for evaluation of the target gene expression using qRT-PCR approach. The expression levels of selected genes at non-permissive temperature referred “Inhibition of replication” condition, relative to permissive temperature (37°C) are present on the graph. The qRT-PCR data are normalized to 16S rRNA levels. Mean values and standard deviations from three independent experiments are shown. Expression levels more than twofold was considered as significant upregulation and less than 0.5 fold as the significant reduction in gene expression of each target. Significant expression of each target or group of targets was marked with ‘∗’. The targets showed without ‘∗’ indicates no significant change in expression. The statistical analyses were performed using the Student’s t-test.
We asked whether the compromised expression of the MtrA targets under stress and a replication-arrested state is, in part, due to altered intracellular MtrA levels. Immunoblot analysis of MtrA levels in M. tuberculosis cells, relative to the housekeeping protein SigA, under NO stress revealed a significant reduction in MtrA levels (21 ± 13%) comparing to MtrA levels in wild-type cells, without NO stress (100%) (Figures 4Ai,ii). The NO-damaged proteins are believed to be processed for degradation via the Mpa-dependent proteasome pathway (Darwin et al., 2003), and MtrA is a predicted proteasome target (Festa et al., 2010). We found that MtrA was also degraded, albeit modestly, under NO stress in the mpa mutant (12 ± 5%) in comparison to MtrA levels (100%) in mpa mutant without NO stress (Figure 4Aii). Immunoblot analysis of the MtrA levels in the WT background in the absence of stress revealed 36 ± 5% reduction, assuming the MtrA levels in the mpa mutant as 100% (Figure 4Aiii). MtrA was also degraded in the SDS-exposed cultures, although SigA, DnaA, MtrB, and Wag31 (Figure 4B), were also degraded under these conditions; hence, the data are not normalized. We also found that MtrA levels were decreased by twofold in dnaATs mutant at the non-permissive temperature of 30°C (time point marked as “0” on Figure 4C), but the levels were restored to near WT levels at 24 and 48 h time point upon a shift to permissive conditions (37°C) (time points marked as “24” and “48” on Figure 4C). The diagram representing synchronization plan is shown on Supplementary Figure S6. Together, our results indicate that intracellular MtrA is subject to degradation under stress and in non-replicating cells and that the Mpa proteasome activity is likely important for maintaining MtrA levels.
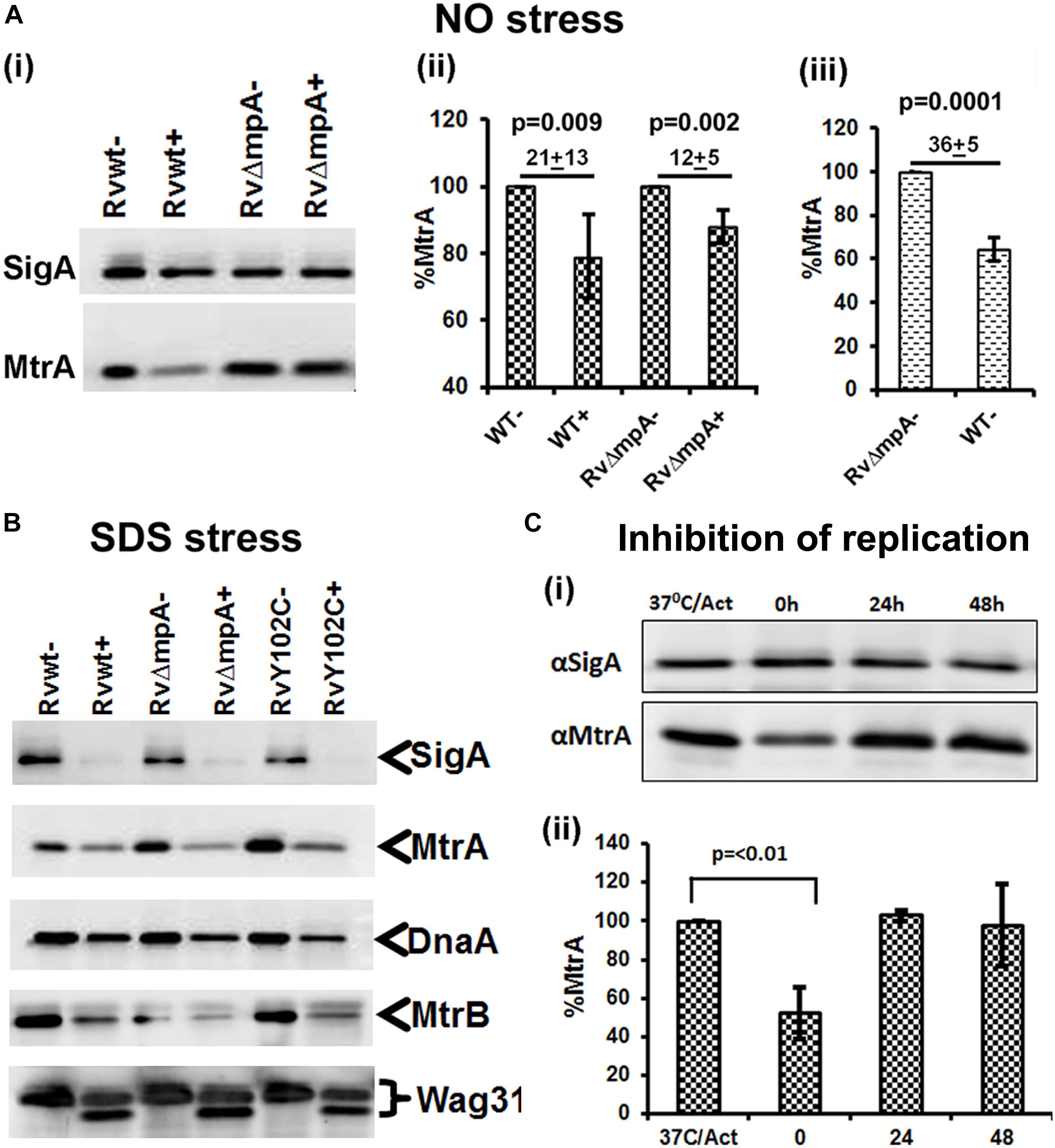
FIGURE 4. MtrA levels decrease under stress and replication-arrest. Two micrograms of total protein lysates from WT and RvΔmpa strains exposed to DETA-NO (A), SDS (B) and isolated from temperature sensitive M. tuberculosis dnaATs mutant strain (Nair et al., 2009) (C) were resolved by SDS-PAGE, transferred to PVDF membranes and immunoblotted with α-MtrA, α-DnaA or α-SigA and quantitated as described (Fol et al., 2006; Rajagopalan et al., 2010). In all cases, untreated cultures are marked with ‘–‘ and treated with ‘+’. (A) Includes 3 sub-panels– (i): immunoblots showing SigA and MtrA proteins. (ii) MtrA and SigA specific bands were quantified using the volumetric analysis tool of the QuantityOne software, the MtrA/SigA ratio was calculated and data presented as percent MtrA level in WT and RvΔmpa mutant strains treated with DATA-NO, treating the MtrA level in WT and mutant strains not treated with DATA-NO as 100%. (iii) Percent of MtrA level in WT strain in comparison to MtrA level in RvΔmpa strain, treated as 100%. (B) Immunoblots of SigA, MtrA, DnaA, MtrB, and Wag31 levels in M. tuberculosis strains. (C) Total protein lysates were isolated from dnaATs cells collected at four different time points of the synchronization plan: 37°C/Act – actively growing cells at 37°C; 0 – cells shifted to 30°C and grown for 30 h; 24 – cells shifted to 37°C and grown for 24 h; 48 – cells shifted to 37°C and grown for 48 h. The schematic of typical dnaATs synchronization plan is shown on Supplementary Figure S6 (i) immunoblots showing MtrA and SigA proteins. (ii) The MtrA/SigA ratio was calculated as described above. The bar graph shows the percent of MtrA protein level at indicated time period: 0, 24, 48 h in synchronized cells, treating the MtrA level in actively growing cells at 37°C (37°C/Act) as 100%. All data shown are averages ± standard error from three independent experiments. The statistical analyses were performed using the Student’s t-test.
The above data support the hypothesis that MtrA activity is intimately associated with replicating cells and is critical for optimal cell division and cell wall metabolism processes. To gain further insights into this issue, we attempted to create and characterize an M. tuberculosis mtrA-defective mutant. Our innumerable attempts to create an mtrA conditional-expression strain were not successful. Given the sequence similarity of the mtrA region (consisting of the mtrA, mtrB, lpqB genes) between M. smegmatis, a non-pathogen, and M. tuberculosis, we attempted to create and characterize the M. smegmatis mtrA mutant as an alternative. The native mtrA gene in M. smegmatis was replaced with a mutant copy wherein a gentamycin cassette replaced the 198-bp internal coding region by homologous recombination (Figure 5A). The genotype of obtained ΔmtrA mutant cells was confirmed by Southern blot hybridization (Figure 5B). The absence of MtrA in the mutant lysates was confirmed by immunoblotting (Figure 5C). These results indicated that the M. smegmatis mtrA gene is not essential, unlike the M. tuberculosis counterpart (Zahrt and Deretic, 2000; Griffin et al., 2011; our unpublished data). M. smegmatisΔmtrA cells were filamentous with branches and buds, indicating cell division and cell shape defects (Figure 5D), and were defective for MtrA-target genes expression (Figure 5E). Expression of the M. tuberculosis mtrA (ΔmtrA::mtrATB) from the amidase (Figure 5D) promoter reversed the cell length and gene expression defects. We also found that production of MtrAY 102C (ΔmtrA::mtrAY 102C) as a sole source for MtrA reversed the filamentous phenotype, except that cells were somewhat wider with an altered cell shape (Figure 5D). Together, these results indicate that the M. tuberculosis mtrA can substitute for the function of the M. smegmatis counterpart and is critical for the regulation of cell division and cell wall metabolism.
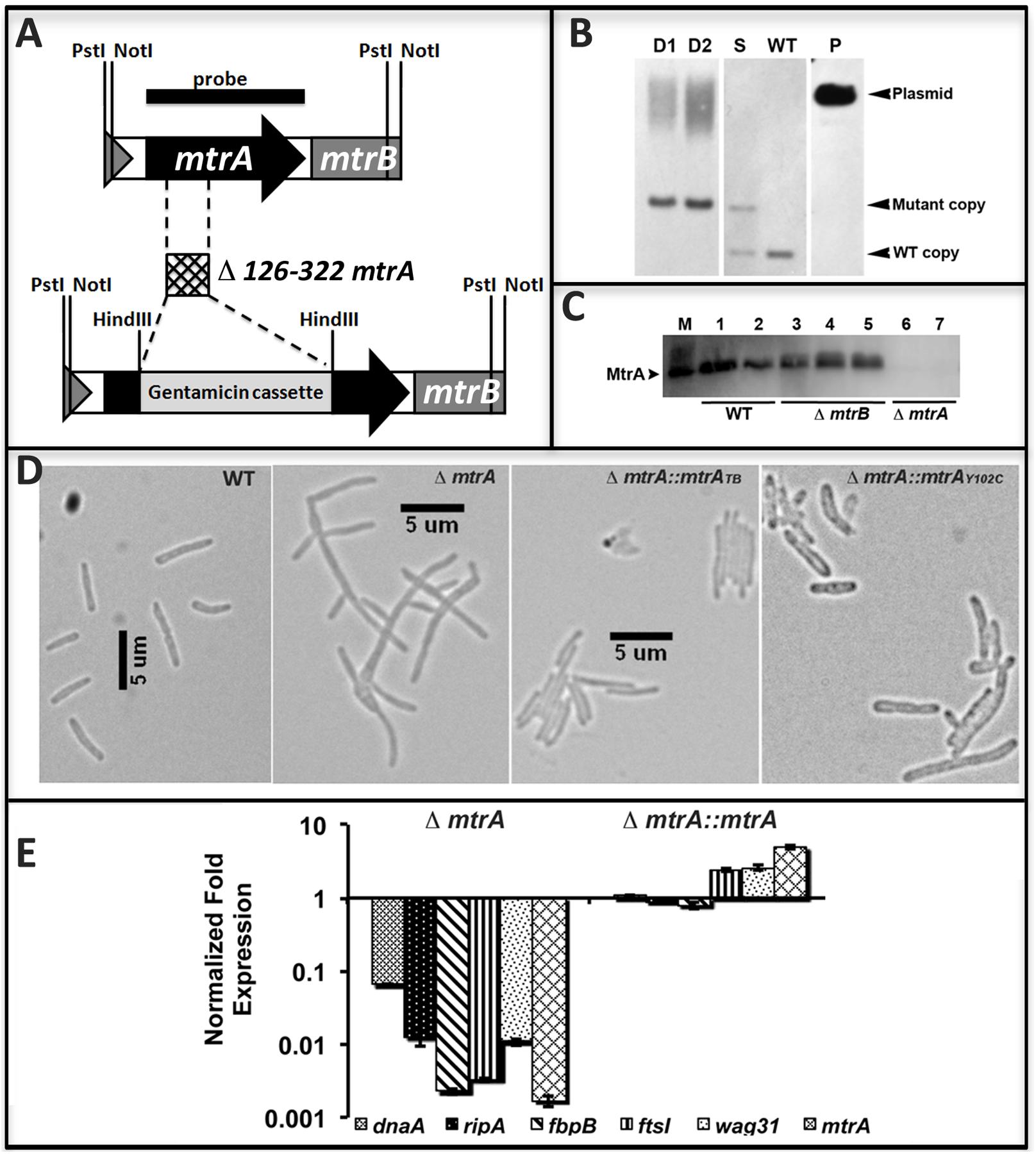
FIGURE 5. Construction and characterization of M. smegmatis mtrA mutant. (A) Schematic of mtrAB region showing the deletion of nucleotides 126–322 followed by insertion of gentamicin cassette in the mtrA coding region. (B) Southern blot confirming the deletion of mtrA. Genomic DNA was isolated from WT M. smegmatis, one single-crossover (S) and two double-crossovers (D1 and D2), digested with NotI, transferred to Hybond-N+ membrane (GE Healthcare) and probed with mtrA fragment (black bar in A). Bands corresponding to the WT (1306-bp) and mutant (1921-bp) copy were seen in single crossover and those corresponding to WT only or mutant only copy were seen in WT and double crossover strains, respectively. Size of mutant band includes 851-bp gentamicin cassette. (C) The loss of MtrA in ΔmtrA strain was confirmed by immunoblotting with MtrA antibodies. (D) Loss of MtrA leads to filamentation and cell shape defects. Exponential cultures of M. smegmatis WT, ΔmtrA, ΔmtrA Pami::mtrA, and ΔmtrA Pami::mtrAY 102C were examined by bright field microscopy and imaged as described in the text. (E) qRT analysis of MtrA targets. Expression levels of select genes was measured, normalized to sigA and data presented as fold expression relative to the WT strain. The results shown are average from three independent experiments.
The Inactivation of mtrA Affects Susceptibility of M. smegmatis to Antimycobacterial Agents
One consequence of the defective cell division and cell wall metabolism of the ΔmtrA mutant could be altered susceptibility to antimycobacterial drugs, notably cell wall targeting antibiotics. When tested, ΔmtrA was nearly 12-fold more resistant to isoniazid than the parent strain and 16-fold more sensitive to vancomycin (Table 1). Intriguingly, the ΔmtrA mutant sensitized to rifampin (Table 1), whereas the parent strain was resistant at the concentrations tested as published (Alexander et al., 2003). The ΔmtrA mutant showed a modest increase in sensitivity to ampicillin (Table 1).
It is pertinent to note that increased susceptibility to the first-line antimycobacterial drug rifampicin with an associated increase in resistance to isoniazid is a characteristic feature of persistent bacilli (see Discussion). The sequencing of katG gene (MSMEG_6380) confirmed the observed high resistance of ΔmtrA M. smegmatis mutant cells to isoniazid is not due to the accumulation of mutation(s) in katG.
Discussion
The response regulators of bacterial two component systems are often required to be phosphorylated by their partnering sensor kinase in order to bind to the target DNA sequences.
The phosphorylation usually occurs under specific, often unidentified conditions and is induced by specific triggers, e.g., change in the pH, osmotic pressure, temperature, concentration of growth limiting substances and ions. The global CHIP-seq analysis is among the most powerful methods currently available for identification of the DNA targets for a given transcription regulator.
This method was previously applied for M. tuberculosis overproducing C-terminal FLAG-MtrA (Minch et al., 2015) and N-terminal His-tagged MtrA (Chatterjee et al., 2018). A total of 114 genes were selected as putative MtrA targets in both studies. Using EMSA approach, Minch et al. (2015) confirmed binding of MtrA to the upstream region of Rv0494. EMSA and q-PCR techniques were used to validate the binding of MtrA to upstream regions of rpfA, rpfC, rpoB, relF, rpfB, and whiB4 genes by Chatterjee et al. (2018). Here, we expressed phosphorylation competent MtrAY 102C mutant protein, able to function in the absence of the cognate sensor kinase MtrB, under the control of acetamide promoter in M. tuberculosis and the CHIP-seq analyses were performed from exponential and stationary phases of growth. The CHIP-seq analysis based on anti-MtrA antibodies recognizing MtrA, MtrA-P as well as MtrAY 102C allowed to identify 278 MtrA targets. Fifty six targets were found to be common to Minch et al. (2015) studies, 27 were common to Chatterjee et al. (2018) and 14 targets were common to both previously published studies. The remaining 195 targets were identified with MtrAY 102C overproduction strain, exclusively. Among the 14 putative MtrA targets identified together with Minch and Chatterjee, are resuscitation promoting factors: rpfA (Rv0867c), rpfB (Rv1009), rpfC (Rv1884c), and PPE family proteins: PPE19 (Rv1361c), PPE35 (Rv1918c), PPE38 (Rv2352c). The EMSA assay was applied to verify previously identified rpfB (Minch et al., 2015; Chatterjee et al., 2018), ripA (Minch et al., 2015), wag31 (Minch et al., 2015) and new targets (sigD, oxyS, wag31, betP, rv3887, whib3) of MtrA. The venn diagram showing comparison of MtrA targets identified in our studies with targets identified by Minch et al. (2015) and Chatterjee et al. (2018) is located in the Supplementary Figure S3C. Common targets are also marked in Supplementary Table S3. The MtrA binding site consensus was determined based on the CHIP-seq datasets. The motif consensus found in our study is consistent with our previous findings and it shows a high level of sequence conservation with the MtrA-binding motifs reported for Corynebacterium glutamicum (Brocker et al., 2011) and Streptomyces coelicolor (Zhang et al., 2017), n-A/G-T-a-A-C-a and n-G-T-n-A-C-c-(c), respectively. Since signals promoting MtrA phosphorylation are unknown, applying CHIP-seq analyses for MtrAY 102C made it possible to identify new targets and expanded the MtrA regulon. On the other hand we have to remember that growth kinetic for RvY102C is affected in stationary phase, reaching it faster than control strains, suggesting that MtrAY 102C, which mimics phosphorylated MtrA, is slowing down the replication and/or cell division, e.g., as a down-regulator of dnaA involved in the initiation of DNA replication (Figure 1B, Fol et al., 2006). On the other hand, we cannot exclude that MtrAY 102C is not fully active and able to replace the function of phosphorylated MtrA for 100%. It may be also associated with partially disturbed regulation of genes involved in the process of cell division. It was previously reported that MtrAY102C is a phosphorylation competent and acts as a gain-of-function (GoF) protein in the absence of MtrB. Its overexpression restores the ΔmtrB phenotype in M. smegmatis cells and increases the expression of dnaA, pfbpB, ripA, ftsI and wag31 – MtrA targets (Plocinska et al., 2012). The Wag31∼P/Wag31 protein levels were reduced in ΔmtrB strain overproducing MtrAY 102C (Plocinska et al., 2014). The MtrA carrying both D56N (phosphorylation-defective) and Y102C (constitutively active protein) mutations was found to be phosporylation defective although bound to its promoters of PfbpB and PripA. Also expression of MtrAD56N-Y 102C reversed ΔmtrB phenotype (Satsangi et al., 2013). The in vivo studies has shown both MtrAY102C∼P/MtrAY102C binds to oriC and mtrA F2, F4, F5 wild-type boxes (Purushotham et al., 2015). In this study, using the qRT-PCR approach, we have shown increased expression of ftsI and decreased expression of dnaA in M. tuberculosis strain producing MtrAY 102C. The CHIP-seq analysis for MtrAY 102C performed under active and stationary phase of growth identified comprehensive MtrA∼P regulon.
Here, we showed that several players involved in the cell division and cell wall metabolism processes are members of the MtrA regulon and that MtrA levels and activity are compromised under stress and replication-arrest conditions. We also showed that the M. smegmatis mtrA is not an essential gene and that MtrA activity contributes to susceptibility to the antimycobacterial agents rifampin and isoniazid. Together, these results are consistent with the proposal that MtrA controls the cell division and cell wall integrity, affecting the susceptibility of bacilli to some antimycobacterial agents in replicating cells.
Despite being one of the two essential two component systems, the activities of the MtrAB TCSSs are unknown. The results presented in this study provide valuable insights into the environmental conditions promoting MtrAB TCSS activation. We observed here the ability of MtrA to target the key players involved in cell division, such as ftsI, dacB1, ripA, fbpB, and fbpC in actively multiplying cells (Supplementary Table S3). Additionally, MtrA protein levels and activity are reduced under replication-arrest and growth-compromising conditions such as exposure to NO and envelope stress (Figures 3, 4). Admittedly, these studies were based on the evaluation of MtrA levels and MtrA-target expression under limited stress and growth-arrest conditions, although it is known that M. tuberculosis encounters multiple stresses upon infection. Although MtrA levels under other environmental stress conditions remain to be evaluated, a consequence of MtrA degradation could be a suboptimal ratio of MtrA∼P/MtrA and, hence, compromised MtrA-regulon expression. This line of thinking leads to a proposal that a growth-arrest state and compromised MtrA activity go hand-in-hand and that replicating cells promote MtrA activation and, hence, are enriched with the optimal MtrA∼P/MtrA levels required for optimal MtrA-target expression. Indeed, MtrAY 102C overproduction which could lead to an increase in the ratio of MtrA∼P/MtrA is associated with an increase in the expression of MtrA-targets (Figure 1B). Because the DosRS and MprAB systems are induced under NO – and envelope-stress conditions (Bretl et al., 2011), it is reasonable to assume that one effective adaptation strategy of M. tuberculosis to cope with NO and envelope stress is the downregulation of the MtrAB with a concomitant upregulation of the MprAB and DosRS, the elements of two component systems. Such a scenario can also be envisioned during intracellular growth and infection. In partial agreement with this line of thought are the reports showing the upregulation of the MtrA targets rpfA-E and ripA, which aid in cell wall processing and the resolution of the septa upon M. tuberculosis reactivation and exit from the dormant and non-replicative persistent states (Mukamolova et al., 2002; Davies et al., 2008; Gupta et al., 2010; Kana and Mizrahi, 2010; Kondratieva et al., 2011).
Our results showing the stabilization of intracellular MtrA levels in the mpa mutant background are consistent with the notion that the intracellular MtrA is reduced via the Mpa-mediated proteasome pathway. Proteins damaged during NO stress are processed for degradation by the Mpa-dependent proteasome pathway, and this type of proteasomal degradation is critical for optimal M. tuberculosis survival upon infection (Darwin et al., 2003; Festa et al., 2010). However, our results also showed that MtrA was degraded, albeit modestly, under envelope and NO stress in the mpa mutant (Figure 4). These results signal the involvement of Mpa-independent pathways for MtrA processing in vivo, although the identity of such pathway(s) is unknown. Unregulated MtrA∼P owing to elevated MtrA production upon infection is detrimental for M. tuberculosis proliferation, and the activity of the MtrB sensor kinase is implicated in maintaining optimal ratios of MtrA∼P to MtrA (Fol et al., 2006). Thus, our results showing the reduction of the intracellular MtrA levels under stress and replication-arrest conditions by Mpa and possibly other proteolytic pathways adds another layer of control for monitoring MtrA∼P potential in replicating cells.
The question then arises of what signals in the replicating cells promote MtrA activation. One possibility is that cell wall synthetic and hydrolytic products, which accumulate as a consequence of cell wall metabolism, promote MtrA-regulon activation. Earlier studies indicated that the association between MtrB and the septum promotes MtrA∼P and MtrA-regulon expression (Plocinska et al., 2012). Thus, nascent septa and or cell wall metabolic products could be two of the many triggers that promote MtrA activation and, hence, the output of the MtrAB two component system. Further studies are required to address these issues.
Elucidation of the MtrA regulon and the identification of cell division and cell wall metabolism players as the regulon members combined with the filamentation and cell shape-defect phenotypes of the M. smegmatis ΔmtrA mutant (Figure 5 and Supplementary Table S3) support the notion that MtrA plays a vital role in the regulation of cell division and cell shape maintenance in mycobacteria. Interestingly, ftsZ, whose gene product is critical for the initiation of the septal Z-ring and cell division, is not an MtrA target (Fol et al., 2006; Rajagopalan et al., 2010). These data imply that FtsZ rings can be formed independently of MtrA and that MtrA activity impacts cell division and cell wall metabolism at steps subsequent to the FtsZ-ring assembly. It is pertinent to note that response regulators affecting cell division in other bacteria are known. Analyses of their activities lead to a proposal that the MtrA system has departed significantly from the known eubacterial response regulators affecting cell division and cell wall metabolism processes. For example, the essential response regulator CtrA of Caulobacter crescentus promotes the cell cycle-dependent ordered expression of ftsZ, ftsQ, and ftsA, the core genes required for the initiation and progression of cell division. CtrA∼P has been shown to act as a transcriptional repressor of ftsZ and an activator of ftsQ and ftsA (Kelly et al., 1998; Wortinger et al., 2000). Similarly, the essential response regulator YycF in B. subtilis affects the expression levels of ftsZ and ftsA (Fukuchi et al., 2000) and those genes are involved in cell wall metabolism such as autolytic enzymes and hydrolases (Bisicchia et al., 2010; Fukushima et al., 2010). The ftsA-like gene is absent in the mycobacterial genomes; MtrA also distinguishes itself from CtrA and YycF in that it specifically targets the players involved in septum and cell wall synthesis, i.e., ftsI, dacB1, sepF, wag31, and cell wall expansion and remodeling, i.e., ripA, rpfA-E, fbpB, fbpC (Figures 3, 4, Plocinska et al., 2014). Thus, unlike in the other organism, MtrA activity targets septum synthesis and cell wall expansion, the late steps of the cell division process. This leaves open a question how the regulation and stabilization of FtsZ-mediated Z-ring assembly in mycobacteria is accomplished.
Antibiotic susceptibility experiments with M. smegmatisΔmtrA mutant clearly showed that MtrA impacts tolerance to antimycobacterial drugs. The ΔmtrA mutant is 16-fold more resistant to isoniazid than the parent strain but still significantly sensitive to rifampin (Table 1). Isoniazid is a first-line mycobactericidal drug that rapidly kills actively dividing bacilli, whereas rifampin is a sterilizing drug that kills both replicating and persistent bacilli (Tomasz et al., 1970; Dickinson et al., 1977). It is pertinent to note that increased tolerance to isoniazid with continued susceptibility to rifampin is a characteristic feature of persistent bacilli and a clinical feature of latent tuberculosis infection (Mc, 1959). Although the antibiotic susceptibility experiments were based on M. smegmatisΔmtrA, given the conserved organization of the MtrA region and the players involved in cell division and cell wall metabolism in M. tuberculosis and M. smegmatis, it is logical to speculate that MtrA activity is reduced during persistence, thereby impacting antibiotic tolerance.
The MtrAB TCSS is conserved in high G+C rich actinobacteria such as Corynebacterium glutamicum, C. diphtheria and Streptomyces sps (Hoskisson and Hutchings, 2006); mtrAB genes can be deleted in C. glutamicum (Moker et al., 2004) much like the situation with the MtrAB system of M. smegmatis (this study and Plocinska et al., 2012). Also, the C. glutamicum mtrAB mutant is sensitive to vancomycin and ampicillin (Moker et al., 2004), like the M. smegmatis counterpart.
On the other hand, in the contradiction to our observations, Li and colleagues reported that M. smegmatis construct expressing antisense mtrA RNA is sensitive to isoniazid and did not show susceptibility to rifampicin (Li et al., 2010). Although the levels of MtrA protein are unknown, the mtrA expression is reported to be reduced 0.38 fold under antisense mtrA condition (Li et al., 2010). Our studies were carried out in a clean mtrA mutant background, hence cannot directly be comparable with the reported mtrA antisense studies.
Author Contributions
PG, RP, KS, AS, EP, and RD conceived and designed the experiments. PG, RP, KS, AS, EP, and RD performed the experiments. PG, RP, KS, AS, EP, RD, JD, MR, and MM analyzed the data. MR, MM, JD, PG, and RP wrote the paper.
Funding
This work was supported, in part, by National Institutes of Health grants (United States)– AI084374 (to MM), MI48417 (to MR); Ministry of Science and Higher Education grant (Poland): IP2011 042571 (to RP); The National Science Centre (Poland), SONATA 6 – 2013/11/D/NZ6/02888 (to RP).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02839/full#supplementary-material
References
Adams, D. W., and Errington, J. (2009). Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7, 642–653. doi: 10.1038/nrmicro2198
Al Zayer, M., Stankowska, D., Dziedzic, R., Sarva, K., Madiraju, M. V., and Rajagopalan, M. (2011). Mycobacterium tuberculosis mtrA merodiploid strains with point mutations in the signal-receiving domain of MtrA exhibit growth defects in nutrient broth. Plasmid 65, 210–218. doi: 10.1016/j.plasmid.2011.01.002
Aldridge, B. B., Fernandez-Suarez, M., Heller, D., Ambravaneswaran, V., Irimia, D., Toner, M., et al. (2012). Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 335, 100–104. doi: 10.1126/science.1216166
Alexander, D. C., Jones, J. R., and Liu, J. (2003). A rifampin-hypersensitive mutant reveals differences between strains of Mycobacterium smegmatis and presence of a novel transposon, IS1623. Antimicrob. Agents Chemother. 47, 3208–3213. doi: 10.1128/AAC.47.10.3208-3213.2003
Belisle, J. T., Vissa, V. D., Sievert, T., Takayama, K., Brennan, P. J., and Besra, G. S. (1997). Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276, 1420–1422. doi: 10.1126/science.276.5317.1420
Bisicchia, P., Lioliou, E., Noone, D., Salzberg, L. I., Botella, E., Hubner, S., et al. (2010). Peptidoglycan metabolism is controlled by the WalRK (YycFG) and PhoPR two-component systems in phosphate-limited Bacillus subtilis cells. Mol. Microbiol. 75, 972–989. doi: 10.1111/j.1365-2958.2009.07036.x
Bretl, D. J., Demetriadou, C., and Zahrt, T. C. (2011). Adaptation to environmental stimuli within the host: two-component signal transduction systems of Mycobacterium tuberculosis. Microbiol. Mol. Biol. Rev. 75, 566–582. doi: 10.1128/MMBR.05004-11
Brocker, M., Mack, C., and Bott, M. (2011). Target genes, consensus binding site, and role of phosphorylation for the response regulator MtrA of Corynebacterium glutamicum. J. Bacteriol. 193, 1237–1249. doi: 10.1128/JB.01032-10
Chatterjee, A., Sharma, A. K., Mahatha, A. C., Banerjee, S. K., Kumar, M., Saha, S., et al. (2018). Global mapping of MtrA-binding sites links MtrA to regulation of its targets in Mycobacterium tuberculosis. Microbiology 164, 99–110. doi: 10.1099/mic.0.000585
Chauhan, A., Lofton, H., Maloney, E., Moore, J., Fol, M., Madiraju, M. V., et al. (2006a). Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol. Microbiol. 62, 132–147.
Chauhan, A., Madiraju, M. V., Fol, M., Lofton, H., Maloney, E., Reynolds, R., et al. (2006b). Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J. Bacteriol. 188, 1856–1865.
Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. doi: 10.1038/31159
Dadura, K., Plocinska, R., Rumijowska-Galewicz, A., Plocinski, P., Zaczek, A., Dziadek, B., et al. (2017). PdtaS deficiency affects resistance of mycobacteria to ribosome targeting antibiotics. Front. Microbiol. 8:2145. doi: 10.3389/fmicb.2017.02145
Darwin, K. H., Ehrt, S., Gutierrez-Ramos, J. C., Weich, N., and Nathan, C. F. (2003). The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 302, 1963–1966. doi: 10.1126/science.1091176
Davies, A. P., Dhillon, A. P., Young, M., Henderson, B., Mchugh, T. D., and Gillespie, S. H. (2008). Resuscitation-promoting factors are expressed in Mycobacterium tuberculosis-infected human tissue. Tuberculosis 88, 462–468. doi: 10.1016/j.tube.2008.01.007
Dickinson, J. M., Aber, V. R., and Mitchison, D. A. (1977). Bactericidal activity of streptomycin, isoniazid, rifampin, ethambutol, and pyrazinamide alone and in combination against Mycobacterium Tuberculosis. Am. Rev. Respir. Dis. 116, 627–635. doi: 10.1164/arrd.1977.116.4.627
Dziadek, J., Madiraju, M. V. V. S., Rutherford, S. A., Atkinson, M. A. L., and Rajagopalan, M. (2002a). Physiological consequences associated with overproduction of Mycobacterium tuberculosis FtsZ in mycobacterial hosts. Microbiology 148, 961–971.
Dziadek, J., Rajagopalan, M., Parish, T., Kurepina, N., Greendyke, R., Kreiswirth, B. N., et al. (2002b). Mutations in the CCGTTCACA DnaA box of Mycobacterium tuberculosis oriC that abolish replication of oriC plasmids are tolerated on the chromosome. J. Bacteriol. 184, 3848–3855.
Festa, R. A., Mcallister, F., Pearce, M. J., Mintseris, J., Burns, K. E., Gygi, S. P., et al. (2010). Prokaryotic ubiquitin-like protein (Pup) proteome of Mycobacterium tuberculosis [corrected]. PLoS One 5:e8589. doi: 10.1371/journal.pone.0008589
Fol, M., Chauhan, A., Nair, N. K., Maloney, E., Moomey, M., Jagannath, C., et al. (2006). Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol. Microbiol. 60, 643–657. doi: 10.1111/j.1365-2958.2006.05137.x
Friedland, N., Mack, T. R., Yu, M., Hung, L. W., Terwilliger, T. C., Waldo, G. S., et al. (2007). Domain orientation in the inactive response regulator Mycobacterium tuberculosis MtrA provides a barrier to activation. Biochemistry 46, 6733–6743. doi: 10.1021/bi602546q
Fukuchi, K., Kasahara, Y., Asai, K., Kobayashi, K., Moriya, S., and Ogasawara, N. (2000). The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146(Pt 7), 1573–1583. doi: 10.1099/00221287-146-7-1573
Fukushima, T., Furihata, I., Emmins, R., Daniel, R. A., Hoch, J. A., and Szurmant, H. (2010). A role for the essential YycG sensor histidine kinase in sensing cell division. Mol. Microbiol. 79, 503–522. doi: 10.1111/j.1365-2958.2010.07464.x
Galagan, J. E., Minch, K., Peterson, M., Lyubetskaya, A., Azizi, E., Sweet, L., et al. (2013). The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499, 178–183. doi: 10.1038/nature12337
Griffin, J. E., Gawronski, J. D., Dejesus, M. A., Ioerger, T. R., Akerley, B. J., and Sassetti, C. M. (2011). High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7:e1002251. doi: 10.1371/journal.ppat.1002251
Gupta, R. K., Srivastava, B. S., and Srivastava, R. (2010). Comparative expression analysis of rpf-like genes of Mycobacterium tuberculosis H37Rv under different physiological stress and growth conditions. Microbiology 156, 2714–2722. doi: 10.1099/mic.0.037622-0
Gupta, S., Banerjee, S. K., Chatterjee, A., Sharma, A. K., Kundu, M., and Basu, J. (2015). Essential protein SepF of mycobacteria interacts with FtsZ and MurG to regulate cell growth and division. Microbiology 161, 1627–1638. doi: 10.1099/mic.0.000108
Hamoen, L. W., Meile, J. C., De Jong, W., Noirot, P., and Errington, J. (2006). SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol. Microbiol. 59, 989–999. doi: 10.1111/j.1365-2958.2005.04987.x
Haydel, S. E., Malhotra, V., Cornelison, G. L., and Clark-Curtiss, J. E. (2012). The prrAB two-component system is essential for Mycobacterium tuberculosis viability and is induced under nitrogen-limiting conditions. J. Bacteriol. 194, 354–361. doi: 10.1128/JB.06258-11
He, H., Hovey, R., Kane, J., Singh, V., and Zahrt, T. C. (2006). MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J. Bacteriol. 188, 2134–2143. doi: 10.1128/JB.188.6.2134-2143.2006
Hoch, J. A. (2000). Two component and phosphorely signal transduction. Curr. opin. Microbiol. 3, 165–170. doi: 10.1016/S1369-5274(00)00070-9
Hoskisson, P. A., and Hutchings, M. I. (2006). MtrAB-LpqB: a conserved three-component system in actinobacteria? Trends Microbiol. 14, 444–449.
Kana, B. D., and Mizrahi, V. (2010). Resuscitation-promoting factors as lytic enzymes for bacterial growth and signaling. FEMS Immunol. Med. Microbiol. 58, 39–50. doi: 10.1111/j.1574-695X.2009.00606.x
Kelly, A. J., Sackett, M. J., Din, N., Quardokus, E., and Brun, Y. V. (1998). Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 12, 880–893. doi: 10.1101/gad.12.6.880
Kondratieva, T., Rubakova, E., Kana, B. D., Biketov, S., Potapov, V., Kaprelyants, A., et al. (2011). Mycobacterium tuberculosis attenuated by multiple deletions of rpf genes effectively protects mice against TB infection. Tuberculosis 91, 219–223. doi: 10.1016/j.tube.2011.01.005
Li, Y., Zeng, J., Zhang, H., and He, Z. G. (2010). The characterization of conserved binding motifs and potential target genes for M. tuberculosis MtrAB reveals a link between the two-component system and the drug resistance of M. smegmatis. BMC Microbiol. 10:242. doi: 10.1186/1471-2180-10-242
Maloney, E., Stankowska, D., Zhang, J., Fol, M., Cheng, Q. J., Lun, S., et al. (2009). The two-domain LysX protein of Mycobacterium tuberculosis is required for production of lysinylated phosphatidylglycerol and resistance to cationic antimicrobial peptides. PLoS Pathog. 5:e1000534. doi: 10.1371/journal.ppat.1000534
Mc, D. W. (1959). Inapparent infection: relation of latent and dormant infections to microbial persistence. Public Health Rep. 74, 485–499. doi: 10.2307/4590490
Minch, K. J., Rustad, T. R., Peterson, E. J. R., Winkler, J., Reiss, D. J., Ma, S. Y., et al. (2015). The DNA-binding network of Mycobacterium tuberculosis. Nat. Commun. 6:5829. doi: 10.1038/ncomms6829
Moker, N., Brocker, M., Schaffer, S., Kramer, R., Morbach, S., and Bott, M. (2004). Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 54, 420–438. doi: 10.1111/j.1365-2958.2004.04249.x
Mukamolova, G. V., Turapov, O. A., Young, D. I., Kaprelyants, A. S., Kell, D. B., and Young, M. (2002). A family of autocrine growth factors in Mycobacterium tuberculosis. Mol. Microbiol. 46, 623–635. doi: 10.1046/j.1365-2958.2002.03184.x
Nair, N., Dziedzic, R., Greendyke, R., Muniruzzaman, S., Rajagopalan, M., and Madiraju, M. V. (2009). Synchronous replication initiation in novel Mycobacterium tuberculosis dnaA cold-sensitive mutants. Mol. Microbiol. 71, 291–304. doi: 10.1111/j.1365-2958.2008.06523.x
Nguyen, L., Scherr, N., Gatfield, J., Walburger, A., Pieters, J., and Thompson, C. J. (2007). Antigen 84, an effector of pleiomorphism in Mycobacterium smegmatis. J. Bacteriol. 189, 7896–7910. doi: 10.1128/JB.00726-07
Pang, X., Vu, P., Byrd, T. F., Ghanny, S., Soteropoulos, P., Mukamolova, G. V., et al. (2007). Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology 153, 1229–1242. doi: 10.1099/mic.0.29281-0
Parish, T., and Stoker, N. G. (2000). Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146, 1969–1975. doi: 10.1099/00221287-146-8-1969
Plocinska, R., Martinez, L., Gorla, P., Pandeeti, E., Sarva, K., Blaszczyk, E., et al. (2014). Mycobacterium tuberculosis MtrB sensor kinase interactions with FtsI and Wag31 proteins reveal a role for MtrB distinct from that regulating MtrA activities. J. Bacteriol. 196, 4120–4129. doi: 10.1128/JB.01795-14
Plocinska, R., Purushotham, G., Sarva, K., Vadrevu, I. S., Pandeeti, E. V., Arora, N., et al. (2012). Septal localization of the Mycobacterium tuberculosis MtrB sensor kinase promotes MtrA regulon expression. J. Biol. Chem. 287, 23887–23899. doi: 10.1074/jbc.M112.346544
Plocinski, P., Arora, N., Sarva, K., Blaszczyk, E., Qin, H., Das, N., et al. (2012). Mycobacterium tuberculosis CwsA interacts with CrgA and Wag31 and the CrgA-CwsA complex is involved in peptidoglycan synthesis and cell shape determination. J. Bacteriol. 194, 6398–6409. doi: 10.1128/JB.01005-12
Plocinski, P., Martinez, L., Sarva, K., Plocinska, R., Madiraju, M., and Rajagopalan, M. (2013). Mycobacterium tuberculosis CwsA overproduction modulates cell division and cell wall synthesis. Tuberculosis 93(Suppl.), S21–S27. doi: 10.1016/S1472-9792(13)70006-4
Purushotham, G., Sarva, K. B., Blaszczyk, E., Rajagopalan, M., and Madiraju, M. V. (2015). Mycobacterium tuberculosis oriC sequestration by MtrA response regulator. Mol. Microbiol. 98, 586–604. doi: 10.1111/mmi.13144
Rajagopalan, M., Dziedzic, R., Al Zayer, M., Stankowska, D., Ouimet, M. C., Bastedo, D. P., et al. (2010). Mycobacterium tuberculosis origin of replication and the promoter for immunodominant secreted antigen 85B are the targets of MtrA, the essential response regulator. J. Biol. Chem. 285, 15816–15827. doi: 10.1074/jbc.M109.040097
Santi, I., Dhar, N., Bousbaine, D., Wakamoto, Y., and Mckinney, J. D. (2013). Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria. Nat. Commun. 4:2470. doi: 10.1038/ncomms3470
Sassetti, C. M., Boyd, D. H., and Rubin, E. J. (2003). Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84. doi: 10.1046/j.1365-2958.2003.03425.x
Satsangi, A. T., Pandeeti, E. P., Sarva, K., Rajagopalan, M., and Madiraju, M. V. (2013). Mycobacterium tuberculosis MtrAY102C is a gain-of-function mutant that potentially acts as a constitutively active protein. Tuberculosis 93(Suppl.), S28–S32. doi: 10.1016/S1472-9792(13)70007-6
Sauvage, E., Kerff, F., Terrak, M., Ayala, J. A., and Charlier, P. (2008). The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 234–258. doi: 10.1111/j.1574-6976.2008.00105.x
Smith, I. (2003). Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 16, 463–496. doi: 10.1128/CMR.16.3.463-496.2003
Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215. doi: 10.1146/annurev.biochem.69.1.183
Tomasz, A., Albino, A., and Zanati, E. (1970). Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature 227, 138–140. doi: 10.1038/227138a0
Wortinger, M., Sackett, M. J., and Brun, Y. V. (2000). CtrA mediates a DNA replication checkpoint that prevents cell division in Caulobacter crescentus. EMBO J. 19, 4503–4512. doi: 10.1093/emboj/19.17.4503
Zahrt, T. C., and Deretic, V. (2000). An essential two-component signal transduction system in Mycobacterium tuberculosis. J. Bacteriol. 182, 3832–3838. doi: 10.1128/JB.182.13.3832-3838.2000
Keywords: Mycobacterium, two component regulatory system, MtrA-CHIPSeq, MtrA-cell division, MtrA-response regulator
Citation: Gorla P, Plocinska R, Sarva K, Satsangi AT, Pandeeti E, Donnelly R, Dziadek J, Rajagopalan M and Madiraju MV (2018) MtrA Response Regulator Controls Cell Division and Cell Wall Metabolism and Affects Susceptibility of Mycobacteria to the First Line Antituberculosis Drugs. Front. Microbiol. 9:2839. doi: 10.3389/fmicb.2018.02839
Received: 13 September 2018; Accepted: 05 November 2018;
Published: 23 November 2018.
Edited by:
Marie-Joelle Virolle, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Jean-François Cavalier, UMR7255 Laboratoire d’Ingénierie des Systèmes Macromoléculaires (LISM), FranceXiuhua Pang, Shandong University, China
Copyright © 2018 Gorla, Plocinska, Sarva, Satsangi, Pandeeti, Donnelly, Dziadek, Rajagopalan and Madiraju. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Renata Plocinska, cnBsb2NpbnNrYUBjYm0ucGFuLnBs Malini Rajagopalan, bWFsaW5pcjFAZ21haWwuY29t Murty V. Madiraju, bW1hZGlyYWp1QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Purushotham Gorla
Purushotham Gorla Renata Plocinska
Renata Plocinska Krishna Sarva
Krishna Sarva Akash T. Satsangi
Akash T. Satsangi Emmanuel Pandeeti1
Emmanuel Pandeeti1 Robert Donnelly
Robert Donnelly Jaroslaw Dziadek
Jaroslaw Dziadek Malini Rajagopalan
Malini Rajagopalan Murty V. Madiraju
Murty V. Madiraju