- 1School of Biology and Pharmaceutical Engineering, Wuhan Polytechnic University, Wuhan, China
- 2State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China
Cyclohexylamine (CHAM) is widely used in various industries, but it is harmful to human beings and the environment. Acinetobacter sp. YT-02 can degrade CHAM via cyclohexanone as an intermediate. In this study, the cyclohexylamine oxidase (CHAO) gene from Acinetobacter sp. YT-02 was cloned. Amino acid sequence alignment indicated that the cyclohexylamine oxidase (CHAOYT–02) was 48% identical to its homolog from Brevibacterium oxydans IH-35A (CHAOIH–35). The enzyme was expressed in Escherichia coli BL21 (DE3), and purified to apparent homogeneity by Ni-affinity chromatography. The purified enzyme was proposed to be a dimer of molecular mass of approximately 91 kDa. The enzyme exhibited its maximum activity at 50∘C and at pH 7.0. The enzyme was thermolabile as demonstrated by loss of important percentage of its maximal activity after 30 min incubation at 50∘C. Metal ions Mg2+, Co2+, and K+ had certain inhibitory effect on the enzyme activity. The kinetic parameters Km and Vmax were 0.25 ± 0.02 mM and 4.3 ± 0.083 μM min−1, respectively. The biochemical properties, substrate specificities, and three-dimensional structures of CHAOYT–02 and CHAOIH–35 were compared. Our results are helpful to elucidate the mechanism of microbial degradation of CHAM in the strain YT-02. In addition, CHAOYT–02, as a potential biocatalyst, is promising in controlling CHAM pollution and deracemization of chiral amines.
Introduction
Cyclohexylamine (CHAM), an important fine chemical intermediate, is widely used in industrial manufaction of insecticides, antiseptics and plasticizers. In the process of production and usage, CHAM is released into the atmosphere, water, and soil. Furthermore, It was found that sodium N-cyclohexylsulfamate, which is used as a food additive and produced 100,000 tons annually in China, could be decomposed into CHAM by intestinal bacteria (Collings, 1989). CHAM is classified as a volatile organic compound that can enter human body via inhalation or skin contact. CHAM has drawn increasing attention as a weak carcinogen (Price et al., 1970; Petersen et al., 1972). To eliminate CHAM residual dispersed in the environment, certain measures have to be taken. Microbial degradation has advantages of low cost and less energy consumption and is an effective approach to eliminating CHAM pollution and residues in the environment.
To date, limited studies showed that bacteria utilized CHAM as the only source of carbon and nitrogen. These bacteria included Brevibacterium oxydans IH-35A (Iwaki et al., 1999a). Pseudomonas plecoglossicida NyZ12 (Shen et al., 2008; Yan et al., 2017), and Acinetobacter sp. YT-02 isolated by our group (Yan et al., 2018). Acinetobacter sp. YT-02 is a Gram-negative bacterium isolated from the activated sludge from a sodium N-cyclohexylsulfamate production plant (Yan et al., 2018). The draft genome showed that Acinetobacter sp. YT-02 had poor similarity with cyclohexylamine-degrading bacteria P. plecoglossicida NyZ12 (Yan et al., 2017), indicating that our isolate is a new member of CHAM-degrading bacteria (Yan et al., 2018). These strains can convert CHAM into cyclohexanone which was catalyzed by cyclohexylamine oxidase (CHAO) (E.C.1.4.3.12), a type of monoamine oxidases (MAOs). CHAO catalyzes the oxidative deaminnation of cyclohexylamine to form cyclohexanone, hydrogen peroxide and ammonia, using FAD as cofactor (Iwaki et al., 1999b). To date, only CHAO gene from strain B. oxydans IH-35A has been cloned, and an X-ray structure of CHAO from B. oxydans IH-35A (CHAOIH–35A) was determined (Mirza et al., 2013). Although CHAO derived from Pseudomonas plecoglossicida NyZ12 was confirmed to be involved in the degradation of cyclohexylamine, there were no relevant biochemical properties reported for this enzyme (Yan et al., 2017). The related biochemical properties for CHAO from Pseudomonas sp. (purified enzyme) were investigated, but there is no relevant genetic information (Tokieda et al., 1977). The aim of this study is to clone and heterologously express the CHAO gene from Acinetobacter sp. YT-02. Moreover, the enzymatic properties, kinetic parameters, and three-dimensional structure of Acinetobacter sp. YT-02 (CHAOYT–02) were investigated. The results indicated that CHAOYT–02 could be used for elimination of cyclohexylamine in the environment. In addition, CHAOYT–02, as a potential biocatalyst in the deracemization of chiral amines, is promising in pharmaceutical industry (Alexeeva et al., 2002; Carr et al., 2003; Leisch et al., 2012; Li et al., 2014; Yao et al., 2018).
Materials and Methods
Strains and Plasmids
Acinetobacter sp. YT-02 was cultured in minimal salt medium (Yan et al., 2018) using CHAM–HCl (Sinopharm Chemical Reagent, Shanghai, China) at a concentration of 1 g per liter as the sole carbon and nitrogen sources. Escherichia coli DH5α and pGEM-T Easy vector were used for DNA cloning. E. coli BL21 (DE3) and pET-28b (Novagen, Wisconsin, United States) were used as the host strain and expression vector for gene expression, respectively. When needed, ampicillin and kanamycin were added at final concentrations of 100 and 50 μg/ml, respectively. The restriction enzymes and DNA polymerase were purchased from TaKaRa (Dalian, Liaoning, China). All other chemicals for buffer and medium were of analytical reagent grade.
Gene Cloning
The genomic DNA of Acinetobacter sp. YT-02, extracted using a bacterial DNA kit (OMEGA Bio-Tek, Norcross, United States), was used as the template to amplify the target gene. Primer pair, F1 (5′-TGAATTCGATGAGTGTCAATGACAACCGA-3′) and R1 (5′-CAAGCTTCGAATTGGCCATTGTTTTCT-3′), containing EcoRI and HindIII restriction sites (underlined), respectively, was used for polymerase chain reaction (PCR). PCR amplification was performed as follows: denaturation at 95°C for 5 min, 30 cycles of 95°C for 1 min, 56°C for 1 min, and 72°C for 2 min, and a final extension cycle at 72°C for 8 min. The PCR fragments were cloned into the pGEMT-Easy vector (Promega Corporation, United States) after being added A-tailing using Taq polymerase, and the construct was verified by DNA sequencing. The recombinant plasmid was designated as pGEMT-chao. DNA fragments, encoding CHAO (cleaved down from pGEMT-chao by EcoRI and HindIII), were inserted into pET-28b, which was digested with the same restriction enzymes, generating the recombinant plasmid pET-28b-chao. The correct construct was transformed into E. coli BL21 (DE3) competent cell prepared with CCMB80 solution (Hanahan et al., 1991) for gene expression.
Sequence Analysis
DNA sequence encoding open reading frame (ORFs) of CHAOYT–02 was identified by Glimmer 3.02 (Delcher et al., 2007) and ORF finder1. Promoter regions and regulatory sites were identified based on neural network promoter prediction2. The presence and location of signal peptide were predicted using SignalP server3 (Petersen et al., 2011). The amino acid sequence of CHAOYT–02 (GenBank accession number: PCN59881) was compared with the protein sequences in GenBank database using BLASTp program with default parameters. Amine oxidase homologs with the highest identity in the database, omitting redundant or ambiguous sequences, were selected for amino acid sequence alignment. The alignment was carried out by ClustalW using MEGA6.0 (Tamura et al., 2013). Phylogenetic tree was created based on the amino acid distances of the aligned sequences using the neighbor-joining method with 1,000 bootstrap replications. Amine oxidase from Saccharomyces cerevisiae was used as the outgroup (Landry and Sternglanz, 2003).
Protein Production and Purification
The recombinant E. coli BL21 (DE3) (containing pET-28b-chao) cells were cultivated in 4 ml Luria–Bertani (LB) medium (Sambrook et al., 1989) supplemented with kanamycin (50 μg/ml) overnight at 37°C. The culture was inoculated into 200 ml LB medium containing kanamycin (50 μg/ml) and cultured at 37°C. Gene expression was induced by adding isopropy-β-D-thiogalactoside (IPTG) up to a final concentration of 0.2 mM when the OD600 of the culture reached 0.8–1.0. After the induction at 18°C for 16 h, the cells were collected by centrifugation. The bacterial pellet was washed twice with phosphate buffer (50 mM, pH 7.0), resuspended in the same buffer. The cells were broken by ultrasonic crusher (parameter settings were as follows: work 6 s, pause 3 s, working time 20 min, and power 400 W), and the supernatant was collected by centrifugation at 12,000 rpm for 30 min at 4°C. The mixture was filtered through a 0.45 μm filter and then loaded onto 1 ml HisTrap column (GE Healthcare, United States) using ÄKTA Prime Plus system. The recombinant protein was eluted with gradient imidazole-containing buffer (50 mM phosphate buffer, 300 mM NaCl, 250 mM imidazole, and pH 7.0). To remove the imidazole and other salt ions in the elute, the protein buffer was changed by using a centrifugal ultrafiltration device (Millipore) with a molecular mass cutoff of 10 kDa. The purity of isolated CHAOYT–02 was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) [5% (w/v) stacking gel and 10% (w/v) separating gel] on a vertical mini gel apparatus. The concentration of enzyme was determined by Bradford assay (Bradford, 1976) using bovine serum albumin as a standard.
The molecular mass of CHAOYT–02 in solution was determined by gel filtration on Superose® 6 Increase10/300GL column (GE Healthcare, United States). The column was equilibrated with 10 mM phosphate buffer, pH 7.4, containing NaCl 140 mM. The column was calibrated with apoferritin from equine spleen (443 kDa), alcohol dehydrogenase (150 kDa), bovine albumin (66 kDa), and carbonic anhydrase (29 kDa) (Sigma-Aldrich, United States). A linear relationship was established between the Kd (Kd = Ve/Vo, Ve: elution volume, Vo: void volume) and the logarithm of molecular weight, which was used to calculate the molecular mass of recombinant CHAOYT–02.
Influences of pH and Temperature on the Enzyme Activity and Stability
CHAOYT–02 activity was determined using CHAM as the substrate by a modified method (Li et al., 2013). Enzymatic assays were conducted in triplicate. The effect of pH on CHAOYT–02 activity was investigated at 30°C in buffers of different pH (sodium acetate for pH 4.0–6.0, sodium phosphate for pH 6.0–8.0, and Tris–HCl for pH 8.0–9.0). The effect of temperature on the activity of CHAOYT–02 was measured at pH 7.0. The enzyme was preincubated in buffers of different pH (5.0–9.0) for 30 min at 25°C to determine the pH stability. To determine thermostability, the enzyme was incubated at different temperatures (30–60°C) for 30 min, followed by measurement of the residual activity at 50°C and pH 7.0.
Determination of Kinetic Parameters
Under the optimal condition (sodium phosphate buffer pH 7.0 and 50°C), CHAOYT–02 activity was measured at various CHAM concentrations ranging from 0.1 to 2.5 mM. The Km and Vmax were determined by Lineweaver-Burk double-reciprocal plots (Burk et al., 1934).
Effects of Metal Ions on the Activity of CHAOYT–02
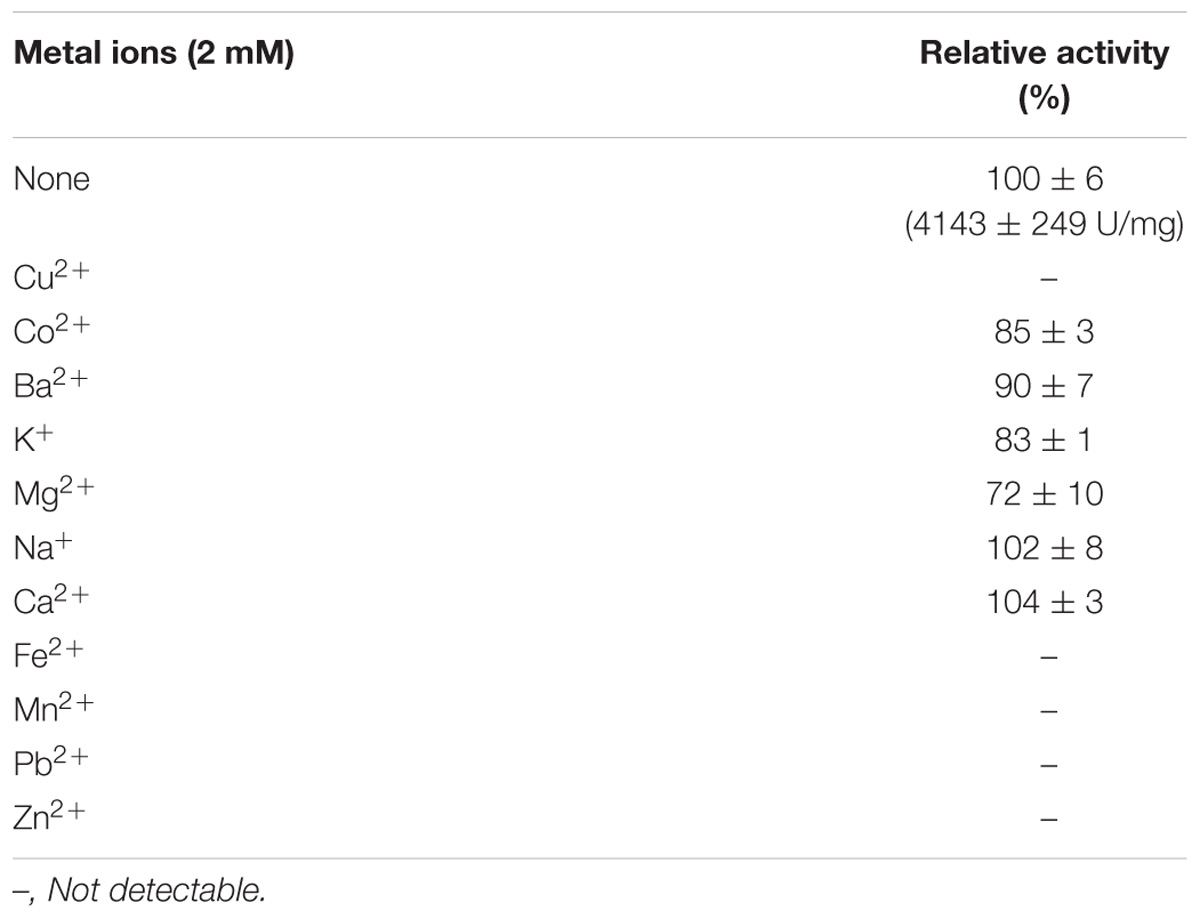
To study the effects of metal ions on the activity of CHAOYT–02, CuCl2, CoCl2, BaCl2, KCl, MgCl2, NaCl, CaCl2, FeCl2, MnCl2, PbCl2, and ZnCl2 were used to prepare the metal ion solutions. The enzyme (1.223 mg/ml) was treated with different metal ions at different concentrations (0.5, 1, and 2 mM). The residual activity was assayed at pH 7.0 and 50°C.
Substrate Spectrum of CHAOYT–02
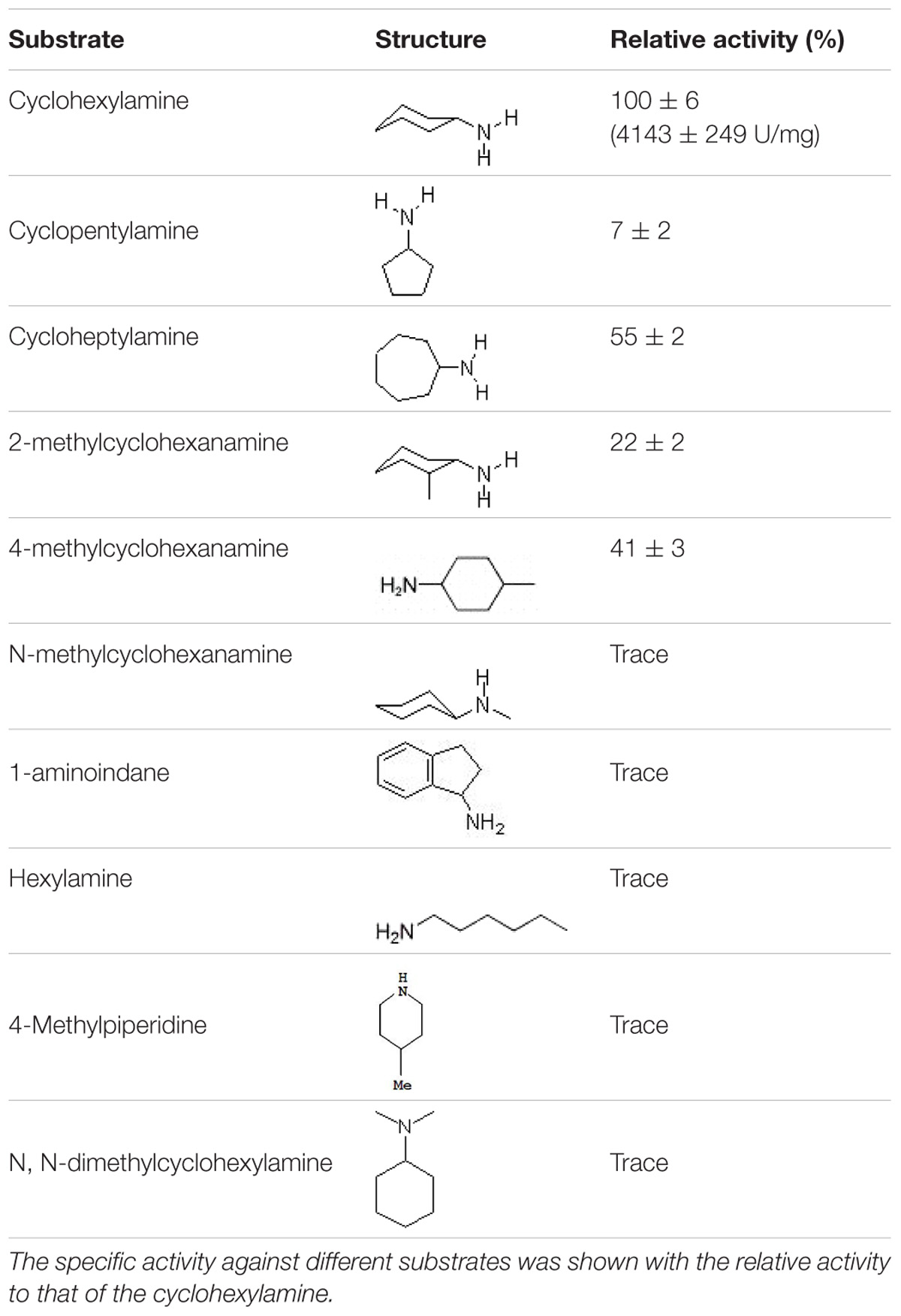
A series of amine substrates (cyclohexylamine, cyclopentylamine, cycloheptylamine, 2-methylcyclohexanamine, 4-methylcyclohexanamine, 1-aminoindane, hexylamine, N-methylcyclohexanamine, 4-methylpiperidine, and N, N-dimethylcyclohexylamine) with diverse structural features were selected to characterize the substrate spectrum of the CHAOYT–02. The activity of CHAOYT–02 toward each substrate was determined individually at a concentration of 10 mM using the purified enzyme (Tokieda et al., 1977).
Structure Model of CHAOYT–02
The three-dimensional structure model of CHAOYT–02 was created by homology modeling using SWISS-MODEL4. The crystal structure of CHAOIH–35A was used as the modeling template (Protein Data Bank entry: 4I58). The overall quality of the model was evaluated by MolProbity (Chen et al., 2010). Structural comparison was performed using DaliLite v. 35 (Holm and Laakso, 2016). Structural figures were prepared using PyMOL (Schrödinger).
Results
Gene Cloning and Sequence∖Bioinformatics Analysis
Gene CF596_10820 in the genome of Acinetobacter sp. YT-02 was predicted to encode CHAOYT–02 (Yan et al., 2018). A putative promoter region consisting of nucleotides 49–54, TTGAAT (the −35 region) and nucleotides 72–77, TATACT (the −10 region) was present before the coding region (Supplementary Figure S1). The gene contained only one long open reading frame, which encoded a protein of 460 amino acids, as suggested by Glimmer and ORF finder. Signal peptide analysis (Petersen et al., 2011) indicated that no signal peptide was present in the polypeptide chain.
The amino-acid sequence of CHAOYT–02 was aligned with amine oxidase homologs selected from a protein database. The phylogenetic neighbor-joining tree indicated that CHAOYT–02 was closest to CHAOIH–35A. These two enzymes shared 48 and 63% amino acid identity and similarity, respectively (Figure 1A). CHAOYT–02 showed relatively low amino acid sequence identity and similarity to CHAONyZ12 (17 and 31%, respectively). The amino acids constituting the active-site cavity of CHAO were largely conserved among CHAOYT–02, CHAOIH–35A, and CHAONyZ12 (Figure 1B). A putative flavin adenine dinucleotide (FAD) binding consensus sequence GXGXXG was present in the N-terminus of CHAOs.
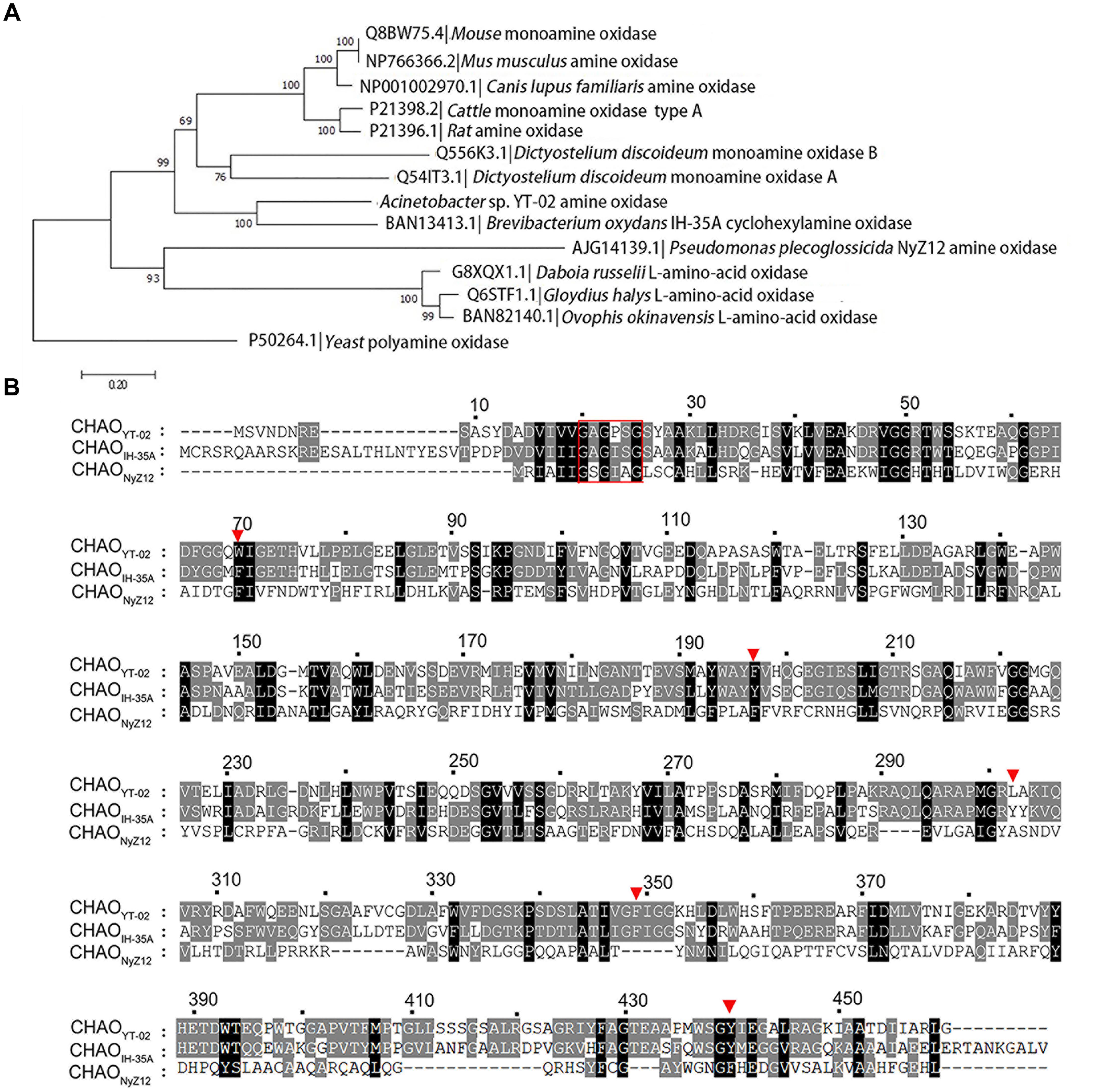
FIGURE 1. Phylogenetic analysis of cyclohexylamine oxidase (CHAO) and other amine oxidases. (A) Phylogenetic tree of CHAO and other amine oxidases. Amino acid sequences were aligned using Clustal W and phylogenetic tree was constructed by using the neighbor joining method using MEGA 6.0. Amine oxidase from Saccharomyces cerevisiae was used as the outgroup. The numbers at each branch of phylogenetic tree represent the bootstrap value (1000 replicates). (B) Amino acid sequence comparison of CHAOYT–02, CHAONyZ12, and CHAOIH–35A. A red rectangular box highlights the putative FAD-binding consensus sequence GXGXXG in the N-terminus of CHAOs. Red triangles indicate amino acids in the active center of the enzymes.
Expression and Purification of Recombinant CHAOYT–02
The recombinant CHAOYT–02 was produced in soluble form in E. coli BL21(DE3) at low temperature (18°C). The enzyme was purified by Ni-affinity chromatography. The purified protein showed the expected molecular size of about 55 kDa in SDS-PAGE (Figure 2), consistent with its theoretical molecular mass of 53,534 Da (including 6xHis tag and translated sequence from pET-28b vector). The oligomeric state of purified CHAO was proposed to be a dimer with a molecular mass of approximately 91 kDa as indicated by gel filtration (Supplementary Figure S2).
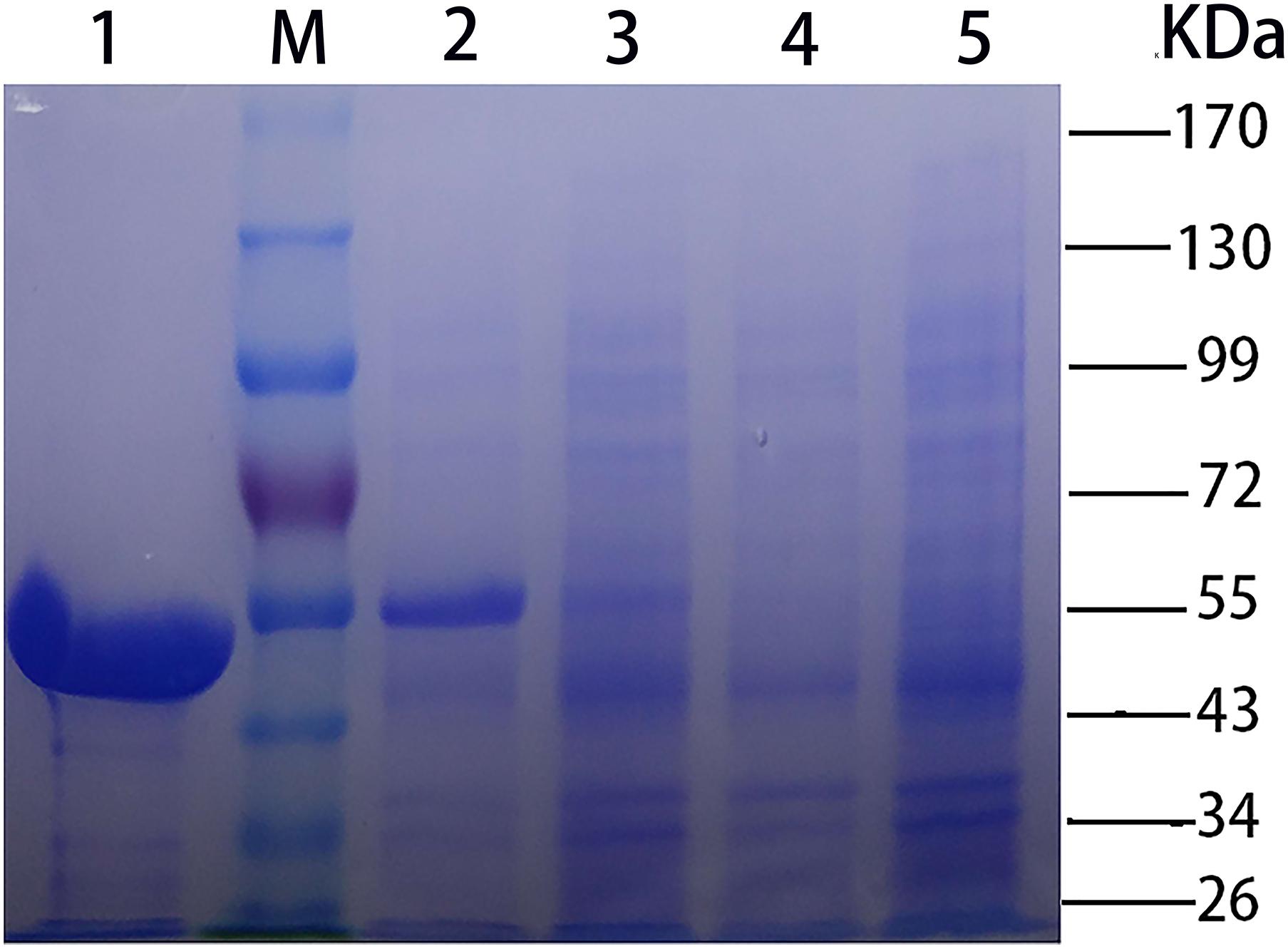
FIGURE 2. SDS-PAGE [stacking gel (5%, w/v) and separating gel (10%, w/v)] analysis of CHAOYT–02. Lanes: M, molecular standard; 1, purified recombinant CHAOYT–02; 2, E. coli BL21 (DE3) harboring pET-28b-chao after induction; 3, E. coli BL21 (DE3) harboring pET-28b-chao without induction; 4, E. coli BL21 (DE3) harboring pET-28b without induction; 5, E. coli BL21 (DE3) harboring pET-28b after induction.
Characterization of the Optimum pH and Temperature of CHAOYT–02
The optimum pH of CHAOYT–02 activity was evaluated by incubating the enzyme in different pH range of 4.0–9.0. The enzyme showed the maximum activity at pH 7.0 and retained more than 60% residual activity at pH 7.0–9.0 (Figure 3A). The effect of temperature on CHAOYT–02 activity was measured at different temperatures in the range 30—60°C at pH 7.0. The CHAOYT–02 activity increased gradually from 4586 U/mg at 30°C to the maximum of 6724 U/mg at 50°C, and then decreased sharply to 4410 U/mg at 60°C. Therefore, the optimum reaction temperature of CHAOYT–02 is 50°C at pH 7.0 (Figure 3B).
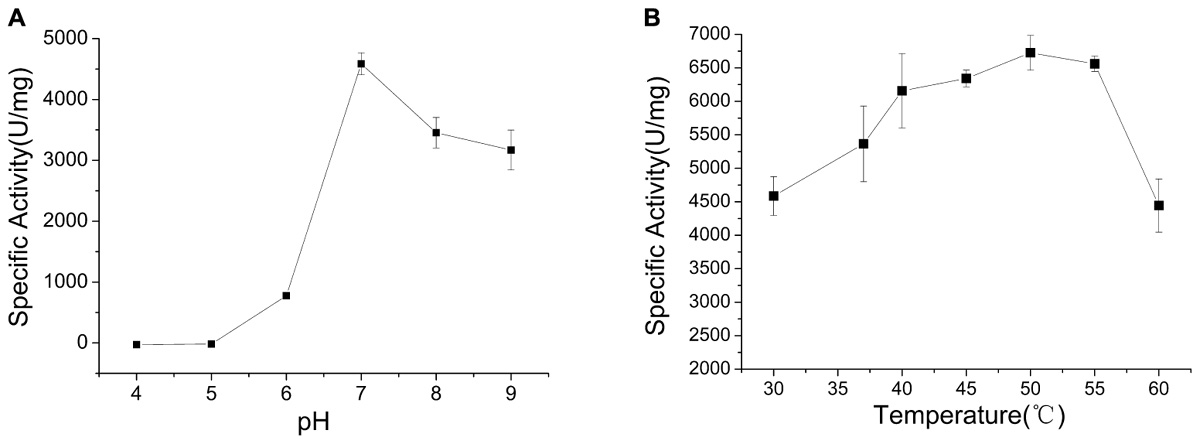
FIGURE 3. Effects of pH and temperature on the activity of CHAOYT–02. (A) Effect of pH on CHAOYT–02 activity at 30°C. (B) Effect of temperature on CHAOYT–02 activity.
Protein Stability
The enzyme was incubated at different pH values (4.0–9.0) for 30 min in the absence of substrate to determine pH stability of CHAOYT–02. CHAOYT–02 was highly unstable under acidic conditions. The enzyme activity was maintained at more than 50% level after incubation for 30 min at pH 7.0–9.0 (Figure 4A).
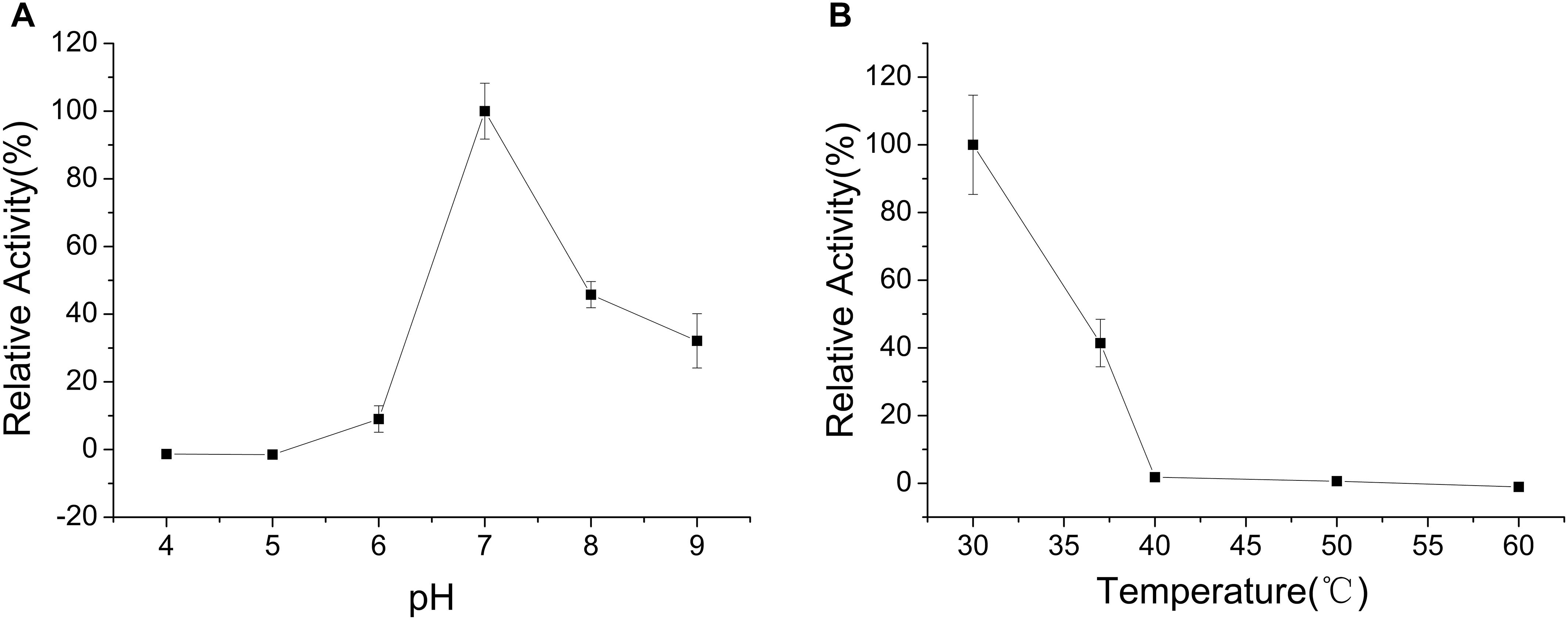
FIGURE 4. Effects of pH and temperature on the stability of CHAOYT–02. (A) Effect of pH on the stability of recombinant CHAOYT–02. The residual activity was determined at 50°C. The value at 50°C is 100%. (B) Effect of temperature on the stability of recombinant CHAOYT–02. The residual activity was determined at pH 7.0 (phosphate buffer, 50 mM) and 50°C. The value at pH 7.0 is 100%.
To investigate the effect of thermal stability of CHAOYT–02, the enzyme was incubated at different temperatures for 30 min in the buffer of pH 7.0. More than 40% activity of the enzyme was retained at 40°C. The activity decreased rapidly when the temperature is higher than 50°C (Figure 4B).
Effects of Different Metal Ions on the Activity of CHAOYT–02
The effects of various metal ions on the activity of CHAOYT–02 were determined under standard reaction condition. CHAOYT–02 exhibited robust tolerance to several metal ions. Ba2+, Na+, and Ca2+ slightly affected the enzyme activity. Mg2+, Co2+, and K+ led to slight loss of activity (Table 1). The reaction system turned red when the copper ion was added in the absence of enzyme. This unexpected coloration severely disturbed the measurement. The metal ions such as Pb2+, Zn2+, Fe2+, and Mn2+ produced precipitation in the reaction system, which hindered the measurement of optical density.
Kinetic Parameters of CHAOYT–02
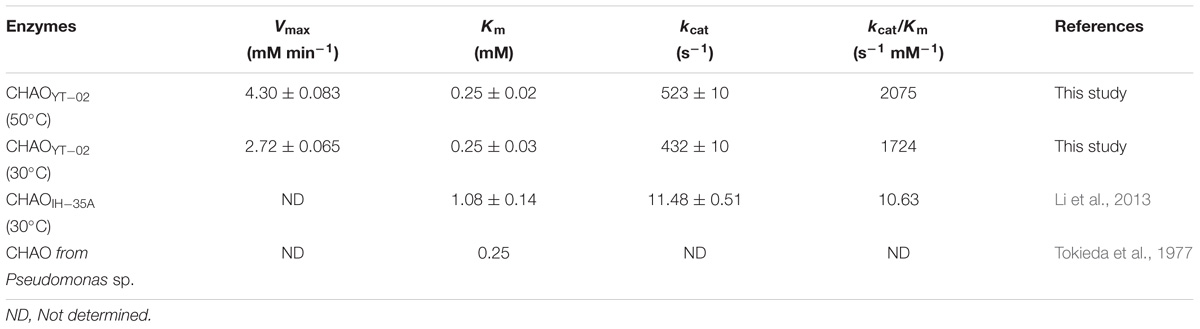
The dependence of the enzyme reaction rate on substrate was investigated under standard conditions at different concentrations (CHAM, 0.1–2.5 mM). The Km and Vmax for the recombinant CHAOYT–02 against CHAM at 50°C, pH 7.0 were 0.25 ± 0.02 mM and 4.3 ± 0.083 mMmin−1, respectively. The kcat and kcat/Km were 523 ± 10 s−1 and 2075 s−1 mM−1 (determined at 50°C), respectively.
Substrate Spectrum of CHAOYT–02
The specific activity of CHAOYT–02 toward amines under the standard condition was investigated as shown in Table 2. CHAOYT–02 exhibited activity toward a wide range of cycloalkyl primary amine. It showed the highest activity toward CHAM, but also had very low activity toward straight-chain, secondary, and tertiary amines.
Structural Model of CHAOYT–02
A structural model of CHAOYT–02 was generated by homology modeling. The overall geometry of the modeled structure displayed a MolProbity of 1.84, thereby indicating good reliability of the theoretical structure. The model of CHAOYT–02 consisted of a cofactor-binding domain and a substrate-binding domain (Figure 5A). It showed considerable similarity to the structure of CHAOIH–35A, with a root mean square deviation (RMSD) of 0.2 Å. The structure alignment between CHAOYT–02 and CHAOIH–35A, indicated that a number of amino-acids involved in the substrate-binding (Trp70, Phe197, and Leu302) and substrate/product-traveling (Ile180, Ile208, and Trp332) in CHAOYT–02 were different from those in CHAOIH–35A (Figure 5B).
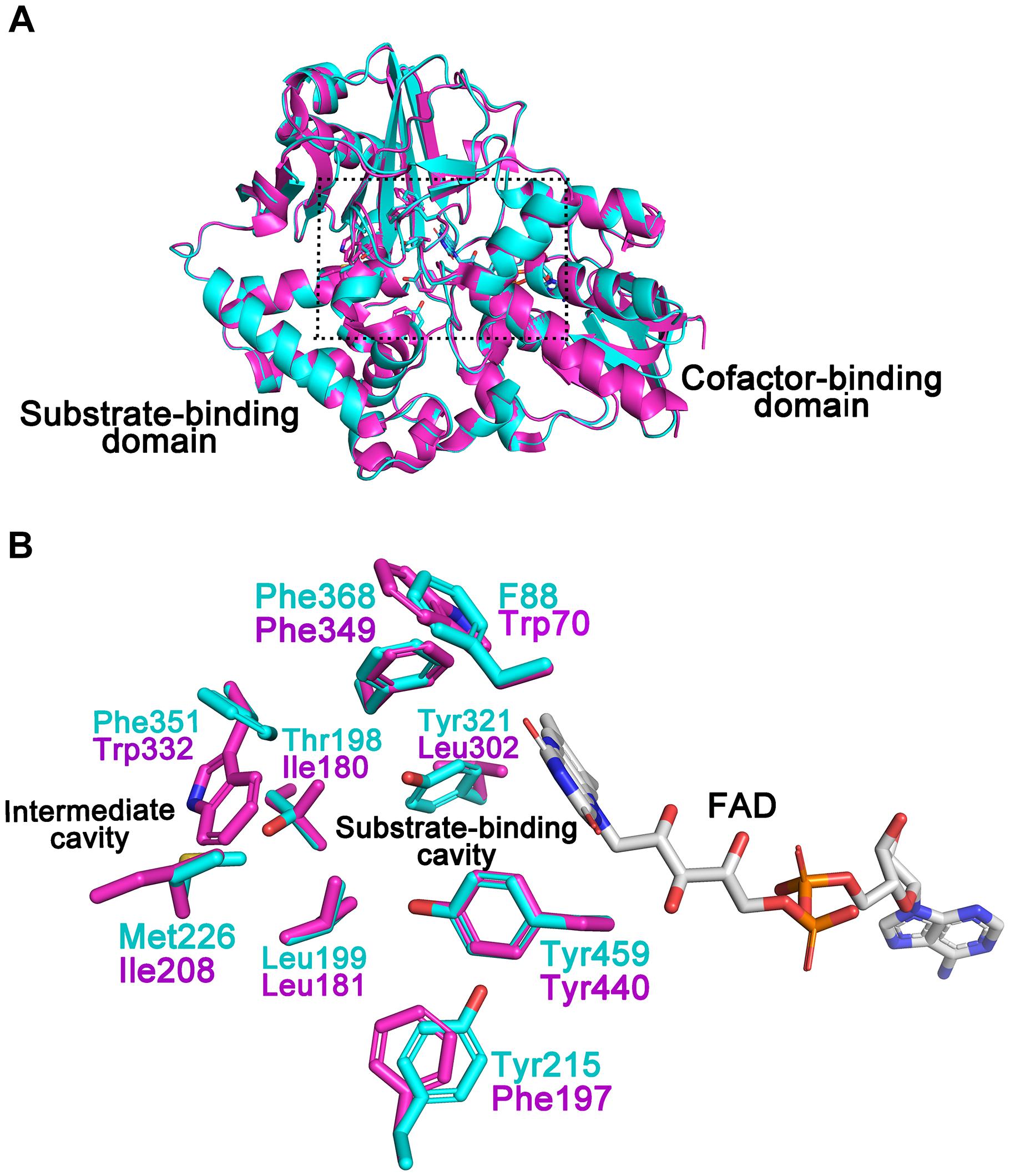
FIGURE 5. Structural model of CHAOYT–02. (A) Superposition of the structural model of CHAOYT–02 (magenta) and crystal structure of CHAOIH–35A (cyan). A rectangular box indicates the putative positions, where the substrate and the cofactor are located. (B) Amino acid compositions of the substrate-binding and intermediate cavities. The carbon atoms in CHAOYT–02 and CHAOIH–35A are shown in magenta and cyan, respectively. The carbon atoms in FAD is presented in gray.
Discussion
Amine oxidases, a large group of enzymes, are widely present among all kind of organisms (Mondovì and Finazzi Agrò, 1982). In higher organisms, they play an important role in polyamine metabolism, whereas in lower eukaryotes and bacteria amine oxidases are used to provide a source of ammonium (Mirza et al., 2013). CHAO is a member of flavin-containing amine oxidases and is highly specific for catalyzing oxidative deamination of CHAM, forming the oxidation product cyclohexanone (Tokieda et al., 1977; Iwaki et al., 1999a,b; Mirza et al., 2013). The enzyme has great potential in eliminating CHAM pollution (Iwaki et al., 1999b; Yan et al., 2017) and synthesis of chiral amines in pharmaceutical industry (Li et al., 2014; Li et al., 2016). Thus far, only a few CHAOs have been cloned and characterized (Iwaki et al., 1999b; Shen et al., 2008; Mirza et al., 2013; Li et al., 2016; Yan et al., 2017, 2018). The limited enzyme resource cannot satisfy the requirement of their biotechnological applications. Therefore, exploring new CHAO becomes very important. In this study, we reported a new CHAO derived from Acinetobacter sp. YT-02. CHAOYT–02 was an acid protein with a molecular mass of approximately 55 kDa. Amino acid sequence alignment revealed that CHAOYT–02 was approximately 48% identical to the well-characterized CHAOIH–35A obtained from Brevibacterium oxydans IH35A.
The optimal temperature for CHAOYT–02 activity was 50°C, which was much higher than that of CHAOIH–35A (30°C). However, both the bacterial CHAOs were thermolabile. CHAOYT–02 lost almost all activities after a short incubation at 50°C. Both CHAOYT–02 and CHAOIH–35A showed their maximum activity at neutral pH (7.0) (Figure 3; Li et al., 2013). However, CHAOYT–02 displayed significant portion of the maximum activity under weak basic condition (pH 7-9) and CHAOIH–35A was only active in a very narrow optimal pH range (7.0–7.5) (Iwaki et al., 1999b). Metal ions Mg2+, Co2+, and K+ had certain inhibitory effect on the enzyme activity. With the CHAM as the substrate and with the same reaction temperature (30°C), CHAOYT–02 had a considerably smaller Km value (0.25 mM) than that of CHAOIH–35A (1.08 mM) (Li et al., 2013). The most notable characteristic of CHAOYT–02 was its much higher turnover number (kcat) (432 s−1) than that of CHAOIH–35A (11 s−1) (Li et al., 2013). Therefore, CHAOYT–02 had a catalytic efficiency 162 times that of CHAOIH–35A (Table 3).
The isolated CHAOYT–02 was yellowish, indicating that oxidized form of cofactor FAD incorporated in active-site of the enzymes, consistent with the presence of putative FAD-binding consensus sequence GXGXXG in its N-terminus (Iwaki et al., 1999b; Figure 1B). In the previous study, the crystal structure of CHAOIH–35A in complex with cyclohexanone revealed a number of residues that were responsible for substrate/product interaction (Mirza et al., 2013). Phe88, Tyr215, Tyr321, Phe368, and Tyr459 constituted the substrate-binding cavity located in the interior of protein. The substrate/product was situated in a so-called “aromatic cage,” which consisted of Tyr321, Phe368, Tyr459, and FAD. A similar architecture was present in the modeled structure of CHAOYT–02 (Figure 5). Amino acid sequence and structure alignments revealed that these residues were invariant in CHAOYT–02, except Tyr321 compared with CHAOIH–35A (Figures 1B, 5). The corresponding residue at the position of Tyr321 in CHAOYT–02 was a leucine (Leu302). The leucine substitution causes broader and more hydrophobic cavity for substrate/product in CHAOYT–02. In addition, molecular dynamic simulations revealed the presence of an intermediate cavity, which connected the substrate-binding cavity with the exterior of protein. The amino acids, which separated the intermediate and substrate-binding cavities, were largely hydrophobic (Mirza et al., 2013). Evidently, these intermediate cavity gating residues in CHAOYT–02 (Ile180, Leu181, Ile208, and Trp332) were more hydrophobic than those in CHAOIH–35A (Thr198, Leu199, Met226, and Phe351) (Figure 5).
Compared with CHAOIH–35A, CHAOYT–02 displayed a relatively narrow substrate spectrum (Table 2). CHAOYT–02 was only active toward primary aliphatic amines with cycloalkane moieties. Similar to CHAOIH–35A, CHAOYT–02 was most active toward CHAM. Compared with CHAOIH–35A, the stronger CHAM-binding affinity (Km value) and faster cyclohexanone-releasing (kcat value) may be attributed to the more hydrophobicity of the entire substrate/product-binding and -releasing path in CHAOYT–02. Site-directed mutagenesis studies showed that mutant T198A displayed enhanced activity relative to wild-type CHAOIH–35A for most (S)-enantiomers of primary amines (Li et al., 2013). The full understanding of enzyme properties and substrate specificity awaits the determination of the crystal structure of CHAOYT–02 and complex structures of CHAOYT–02 and structurally diverse substrates. A structural study on CHAOYT–02 is currently in progress.
Conclusion
In summary, a novel bacterial CHAO gene from Acinetobacter sp. YT-02 was cloned and expressed in E. coli. It is the second CHAO that was cloned and comprehensively characterized to date. The phylogenetic analysis indicated that CHAOYT–02 is a new member of amine oxidases closely related to CHAO from B. oxydans IH-35A. CHAOYT–02 showed higher optimal temperature and catalytic efficiency than the well-characterized CHAOIH–35A suggesting the enzyme is more applicable to be used to eliminate the pollutant CHAM in the environmental treatment. In addition, CHAOYT–02 was only active toward primary aliphatic amines with cycloalkane moieties which implied a potential application of CHAOYT–02 in pharmaceutical industry.
Author Contributions
DY, HZ, ZH, and TF conceived and designed the experiments. HZ, YC, SN, and YG performed the experiments. HZ, ZH, and TF analyzed the data. HZ, ZH, and DY wrote the paper.
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant Nos. 31270112 and 31400114).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02848/full#supplementary-material
FIGURE S1 | Nucleotide sequence of the gene CF596_10820. A putative promoter region was assigned to nucleotides 49–54 (TTGAAT) for the −35 region, and nucleotides 72–77 (TATACT) for the −10 region. The −10 and −35 regions of the promoter are underlined. The translation start codon for the CF596_10820 is shown as the first methionine downstream from T for the transcription start site and GGAGAG for the ribosome binding site (S.D. sequence) (underlined). (A) Nucleotide and deduced amino acid sequence of 5′ flanking regions of cyclohexylamine oxidase gene from Acinetobacter sp. YT-02. (B) Gene sequence chromatogram data for pET-28b-chao (sequencing from T7 promoter primer). (C) Gene sequence chromatogram data for pET-28b-chao (sequencing from T7 terminator primer).
FIGURE S2 | Determination of CHAOYT–02 molecular mass by gel filtration chromatography. The column was equilibrated with 10 mM phosphate buffer, pH 7.4, containing NaCl 140 mM. The column was calibrated with apoferritin from equine spleen (443 kDa), alcohol dehydrogenase (150 kDa), bovine albumin (66 kDa) and carbonic anhydrase (29 kDa) (Sigma-Aldrich, United States). Vo = 8.049 ml, which was determined by the elution volume of blue dextran (2000 KDa).
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/orffinder/
- ^ http://www.fruitfly.org/seq_tools/promoter.html
- ^ http://www.cbs.dtu.dk/services/SignalP/
- ^ http://swissmodel.expasy.org/
- ^ http://ekhidna2.biocenter.helsinki.fi/dali/oldstyle.html
References
Alexeeva, M., Enright, A., Dawson, M. J., Mahmoudian, M., and Turner, N. J. (2002). Deracemization of a-methylbenzylamine using an enzyme obtained by in vitro evolution. Angew. Chem. Int. Ed. Engl. 41, 3177–3180. doi: 10.1002/1521-3773(20020902)41:17<3177::AID-ANIE3177>3.0.CO;2-P
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microorganism quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Burk, D., Lineweaver, H., and Horner, C. K. (1934). The specific influence of acidity on the mechanism of nitrogen fixation by Azotobacter. J. Bacteriol. 27, 325–340.
Carr, R., Alexeeva, M., Enright, A., Eve, T. S. C., Dawson, M. J., and Turner, N. J. (2003). Directed evolution of an amine oxidase possessing both broad substrate specificity and high enantioselectivity. Angew. Chem. Int. Ed. Engl. 42, 4807–4810. doi: 10.1002/anie.200352100
Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., et al. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21. doi: 10.1107/S0907444909042073
Collings, A. J. (1989). Metabolism of cyclamate and its conversion to cyclohexylamine. Diabetes Care 12, 50–55. doi: 10.2337/diacare.12.1.50
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Hanahan, D., Jessee, J., and Bloom, F. R. (1991). Plasmid transformation of E. coli and other bacteria. Methods Enzymol. 204, 63–113. doi: 10.1016/0076-6879(91)04006-A
Holm, L., and Laakso, L. M. (2016). Dali server update. Nucleic Acids Res. 44, 351–355. doi: 10.1093/nar/gkw357
Iwaki, H., Shimizu, M., Tokuyama, T., and Hasegawa, Y. (1999a). Biodegradation of cyclohexylamine by Brevibacterium oxydans IH-35A. Appl. Environ. Microbiol. 65, 2232–2234.
Iwaki, H., Shimizu, M., Tokuyama, T., and Hasegawa, Y. (1999b). Purification and characterization of a novel cyclohexylamine oxidase from the cyclohexylamine-degrading Brevibacterium oxydans IH-35A. J. Biosci. Bioeng. 88, 264–268. doi: 10.1016/S1389-1723(00)80007-9
Landry, J., and Sternglanz, R. (2003). Yeast Fms1 is a FAD-utilizing polyamine oxidase. Biochem. Biophys. Res. Commun. 303, 771–776. doi: 10.1016/S0006-291X(03)00416-9
Leisch, H., Grosse, S., Iwaki, H., Hasegawa, Y., and Lau, P. C. K. (2012). Cyclohexylamine oxidase as a useful biocatalyst for the kinetic resolution and dereacemization of amines. Can. J. Chem. 90, 39–45. doi: 10.1139/v11-086
Li, G. Y., Ren, J., Iwaki, H., Zhang, D. L., Hasegawa, Y., Wu, Q. Q., et al. (2013). Substrate profiling of cyclohexylamine oxidase and its mutants reveals new biocatalytic potential in deracemization of racemic amines. Appl. Microbiol. Biotechnol. 98, 1681–1689. doi: 10.1007/s00253-013-5028-1
Li, G. Y., Ren, J., Yao, P. Y., Duan, Y. T., Zhang, H. L., Wu, Q. Q., et al. (2014). Deracemization of 2-Methyl-1,2,3,4-tetrahydroquinoline using mutant cyclohexylamine oxidase obtained by iterative saturation mutagenesis. ACS Catal. 4, 903–908. doi: 10.1021/cs401065n
Li, G. Y., Yao, P. Y., Cong, P. Q., Ren, J., Wang, L., Feng, J. H., et al. (2016). New recombinant cyclohexylamine oxidase variants for deracemization of secondary amines by orthogonally assaying designed mutants with structurally diverse substrates. Sci. Rep. 6:24973. doi: 10.1038/srep24973
Mirza, I. A., Burk, D. L., Xiong, B., Iwaki, H., Hasegawa, Y., Grosse, S., et al. (2013). Structural analysis of a novel cyclohexylamine oxidase from Brevibacterium oxydans IH-35A. PLoS One 8:e60072. doi: 10.1371/journal.pone.0060072
Mondovì, B., and Finazzi Agrò, A. (1982). Structure and function of amine oxidase. Adv. Exp. Med. Biol. 148, 141–153. doi: 10.1201/9781351076951
Petersen, K. W., Legator, M. S., and Figge, F. H. (1972). Dominant-lethal effects of cyclohexylamine in C57 B1-Femice. Mutat. Res. 14, 126–129. doi: 10.1016/0027-5107(72)90116-9
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Price, J. M., Biava, C. G., Oser, B. L., Vogin, E. E., Steinfeld, J., and Ley, H. L. (1970). Bladder tumors in rats fed cyclohexylamine or high doses of a mixture of cyclamate and saccharin. Science 167, 1131–1132. doi: 10.1126/science.167.3921.1131
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor, NY: C.S.H.Laboratory.
Shen, Y., Yan, D. Z., Chi, X. Q., Yang, Y. Y., Leak, D. J., and Zhou, N. Y. (2008). Degradation of cyclohexylamine by a new isolate of Pseudomonas plecoglossicida. World. J. Microbiol. Biotechnol. 24, 1623–1625. doi: 10.1007/s11274-007-9651-9
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30,2725–2729. doi: 10.1093/molbev/mst197
Tokieda, T., Niimura, T., Takamura, F., and Yamaha, T. (1977). Purification and some properties of cyclohexylamine oxidase from a Pseudomonas sp. J. Biochem. 81, 851–858. doi: 10.1093/oxfordjournals.jbchem.a131549
Yan, D. Z., Gan, Y. T., Zhou, H., Liu, J., and Li, X. (2018). Draft genome sequence of cyclohexylamine-degrading strain Acinetobacter sp. YT-02 isolated. Curr. Microbiol. 75, 284–287. doi: 10.1007/s00284-017-1377-9
Yan, D. Z., Li, X., Li, C. Z., Mao, L. Q., Chi, X. Q., Zhou, N. Y., et al. (2017). Genome-wide identification and characterization of genes encoding cyclohexylamine degradation in a novel cyclohexylamine-degrading bacterial strain of Pseudomonas plecoglossicida NyZ12. J. Biotechnol. 251, 166–173. doi: 10.1016/j.jbiotec.2017.04.036
Keywords: cyclohexylamine oxidase, Acinetobacter sp. YT-02, cyclohexylamine, cyclohexanone, biodegradation
Citation: Zhou H, Han Z-G, Fang T, Chen Y-Y, Ning S-B, Gan Y-T and Yan D-Z (2018) Characterization of a New Cyclohexylamine Oxidase From Acinetobacter sp. YT-02. Front. Microbiol. 9:2848. doi: 10.3389/fmicb.2018.02848
Received: 26 August 2018; Accepted: 06 November 2018;
Published: 22 November 2018.
Edited by:
Mariusz Cycoń, Medical University of Silesia, PolandReviewed by:
Jun-Jie Zhang, Indiana University School of Medicine, United StatesChandradipa Ghosh, Vidyasagar University, India
Xinjiong Fan, Anhui Medical University, China
Gabriela Briceño, Universidad de La Frontera, Chile
Copyright © 2018 Zhou, Han, Fang, Chen, Ning, Gan and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Da-zhong Yan, eWFuZHo2ODA4QDE2My5jb20=
†These authors have contributed equally to this work
 Hui Zhou
Hui Zhou Zheng-gang Han
Zheng-gang Han Ti Fang
Ti Fang Yuan-yuan Chen
Yuan-yuan Chen Shang-bo Ning
Shang-bo Ning Ya-ting Gan
Ya-ting Gan Da-zhong Yan
Da-zhong Yan