- 1Institute of Medical Microbiology, Semmelweis University, Budapest, Hungary
- 2National Public Health Institute, Budapest, Hungary
- 3Institute of Laboratory Medicine, Clinical Microbiology Laboratory, Semmelweis University, Budapest, Hungary
- 4Faculty of Information Technology and Bionics, Pázmány Péter Catholic University, Budapest, Hungary
International high-risk clones of Klebsiella pneumoniae are among the most common nosocomial pathogens. Increased diversity of plasmid-encoded antimicrobial resistance genes facilitates spread of these clones causing significant therapeutic difficulties. The purpose of our study was to investigate fluoroquinolone resistance in extended-spectrum beta-lactamase (ESBL)-producing strains, including four K. pneumoniae and a single K. oxytoca, isolated from blood cultures in Hungary. Whole-genome sequencing and molecular typing including multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) were performed in selected strains. Gene expression of plasmid-mediated quinolone resistance determinants (PMQR) was investigated by quantitative-PCR. MLST revealed that three K. pneumoniae strains belonged to ST11 and one to ST307 whereas K. oxytoca belonged to ST52. The isolates harbored different β-lactamase genes, however, all K. pneumoniae uniformly carried blaCTX-M-15. The K. pneumoniae isolates exhibited resistance to fluoroquinolones and carried various PMQR genes namely, two ST11 strains harbored qnrB4, the ST307 strain harbored qnrB1 and all K. pneumoniae harbored oqxAB efflux pump. Levofloxacin and moxifloxacin MIC values of K. pneumoniae ST11 and ST307 clones correlated with qnr and oqxAB expression levels. The qnrA1 carrying K. oxytoca ST52 exhibited reduced susceptibility to fluoroquinolones. The maintained expression of qnr genes in parallel with chromosomal mutations indicate an additional protective role of Qnr proteins that can support dissemination of high-risk clones. During development of high-level fluoroquinolone resistance, high-risk clones retain fitness thus, enabling them for dissemination in hospital environment. Based on our knowledge this is the first report of ST307 clone in Hungary, that is emerging as a potential high-risk clone worldwide. High-level fluoroquinolone resistance in parallel with upregulated PMQR gene expression are linked to high-risk K. pneumoniae clones.
Introduction
International high-risk clones of Klebsiella pneumoniae are among the most common Gram- negative pathogens. In addition to community-acquired infections, it has been known for decades that due to their ability to spread rapidly in hospital environment, these bacteria can cause several outbreaks. Multi-drug resistant (MDR) K. pneumoniae emerged and dramatically increased prevalence of nosocomial infections while K. oxytoca has been isolated in hospital infections with less frequency (Podschun and Ullmann, 1998; Kang et al., 2006; Zhou et al., 2016).
Multi-drug resistant K. pneumoniae acquires various resistance mechanisms that confer antibiotic resistance to commonly used antibiotics. Among the most frequent resistance mechanisms are extended-spectrum β-lactamases (ESBLs), plasmid-mediated AmpC enzyme (pAmpCs), carbapenemases, plasmid-mediated quinolone resistance (PMQR) genes, aminoglycoside-modifying enzymes (AMEs), as well as exogenously acquired 16S rRNA methyltransferase that have been detected in clinical isolates (Yan et al., 2002; Ko et al., 2010; Cao et al., 2014; Bi et al., 2017). Presence of PMQR genes including qnr determinants, aac(6′)-Ib-cr, qepA and oqxAB efflux pumps confer reduced susceptibility to fluoroquinolones and facilitate selection of fluoroquinolone resistance in Enterobacterales (Rodríguez-Martínez et al., 2011; Carattoli, 2013). High-risk K. pneumoniae clones have acquired these antibiotic resistance determinants, that enabled them to increase their pathogenicity and survival skills. These clones have tenacity and flexibility to accumulate resistance determinants and they have contributed to disseminate global multi-drug resistance (Woodford et al., 2011). Consequently, increased diversity of plasmid-encoded antimicrobial resistance genes facilitates spread of these clones, causing significant therapeutic difficulties.
Multi-drug resistant K. pneumoniae strains mainly belong to certain sequence types (ST) namely, ST11, ST14, ST15, ST37, ST101, ST147, ST258, ST336, ST340, and ST874. These represent high-risk international clones that played major role in dissemination in hospital settings and increased frequency in nosocomial infections (Damjanova et al., 2008; Hrabák et al., 2009; Baquero et al., 2013; Munoz-Price et al., 2013; Rodrigues et al., 2014; Gonçalves et al., 2017). Among these clones ST258 has been reported as a hybrid clone that was created by a large recombination event between ST11 and ST442 (Mathers et al., 2015).
International high-risk K. pneumoniae ST11 has been frequently detected worldwide as a successful pathogen being associated with important co-resistance and virulence factors (Damjanova et al., 2008; Andrade et al., 2014). However, in recent years, new drug-resistant lineages have emerged internationally and among them, KPC-producing K. pneumoniae ST307 has been recognized in the United States which was initially associated with production of CTX-M-15 (Castanheira et al., 2013). Later on, this clone has been reported in several countries including Italy, United Kingdom, Columbia, Pakistan, Morocco, Korea, Tunisia, China, Serbia (Habeeb et al., 2013; Girlich et al., 2014; Gona et al., 2014; Park et al., 2015; Ocampo et al., 2016; Mansour et al., 2017; Novović, 2017; Villa et al., 2017; Xie et al., 2017).
Recent studies related to dissemination and antibiotic resistance of K. pneumoniae clones clearly showed that “fitness cost advantage” associated with high-level resistance to fluoroquinolones contributed to emergence of international high-risk K. pneumoniae clones. In hospital settings where fluoroquinolones are extensively used, international clones are selected out, allowing dominance over other clones (Tóth et al., 2014; Fuzi, 2016; Fuzi et al., 2017). This capacity will provide these clones increased opportunities to spread as well as allow time to acquire antimicrobial drug resistance determinants from other bacteria (Mathers et al., 2015). Whole-genome sequence analysis contributes to detect markers of pathogens, therefore in our study the aim was to investigate high-level fluoroquinolone resistance in K. pneumoniae high-risk clone ST11 and currently emerging ST307.
Materials and Methods
Bacterial Strains
In our preliminary examination, a total of 54 Klebsiella strains (53 K. pneumoniae and a single K. oxytoca) isolated from bloodstream infections of patients treated at intensive care units of Semmelweis University between 2010 and 2014 were collected. Species identification was done by MALDI-TOF/MS (Bruker Daltonics, Bremen, Germany). Minimum inhibitory concentration determination was performed by microdilution method based on EUCAST recommendation.1 All Klebsiella strains were resistant to third-generation cephalosporins and showed reduced susceptibility or resistance to fluoroquinolones. All strains were tested for presence of PMQR genes and all of them were ESBL producers by phenotypic test. In this study, selection of strains was done based on the following criteria: (1) presence of qnr gene and non-wild type fluoroquinolone MIC values: Kox37 (isolated in 2010); (2) presence of qnr gene and high fluoroquinolone MIC values: Kpn33 (isolated in 2010), Kpn47 (isolated in 2014), Kpn125 (isolated in 2013); (3) multiple PMQR gene carriage together with high fluoroquinolone MIC values: Kpn115 (isolated in 2013) (Domokos et al., 2016).
Multilocus Sequence Typing (MLST)
Genotype of each strain was determined by MLST. The sequences of seven housekeeping genes namely, gapA, infB, mdh, pgi, phoE, rpoB, and tonB were amplified and directly sequenced. Alleles and sequence types were assigned by using the MLST database2 (Diancourt et al., 2005). The distance based relationship between the strains was investigated by BacWGST (Ruan and Feng, 2016) using both the whole-genome MLST and SNP (sequenced based) strategies. Multiple genome analysis was carried out using all the draft genomes of this study and the HS11286_CP003200_ST11 as a reference genome (Figure 1).
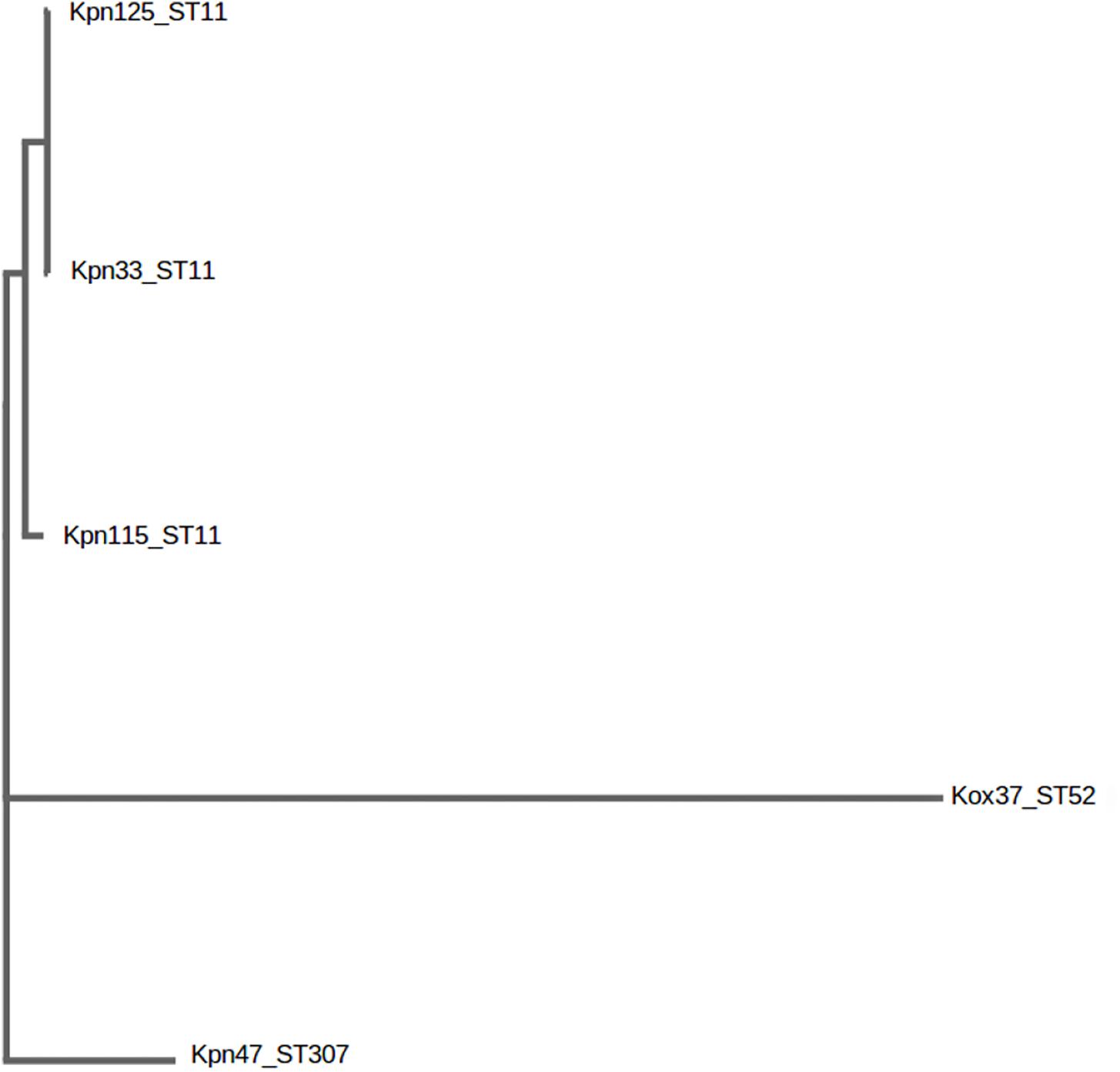
Figure 1. Distance based tree of K. pneumoniae ST11, ST307 and K. oxytoca ST52 after genome based single nucleotide polymorphism (SNP) analysis. BacWGST, Multiple genome analysis http://bacdb.org/BacWGSTdb/Tools.php.
Pulsed Field Gel Electrophoresis (PFGE) Typing
Clonal relatedness of the four K. pneumoniae strains was analyzed by PFGE according to CDC (2000) protocol. Prepared genomic DNA of each strain was digested by XbaI restriction endonuclease (Fermentas, ABI, Germany), and DNA fragments were separated in a PFGE CHEF-DR II system (Bio-Rad Laboratories, Hercules, CA, United States). Banding patterns were analyzed by Fingerprinting II Informatix Software (Bio-Rad). Salmonella enterica serotype Braenderup H9812 was used as a size marker (Hunter et al., 2005).
Whole-Genome Sequencing (WGS)
DNA of each strain was extracted by UltraClean Microbial DNA Isolation Kit (Qiagen GmbH, Hilden, Germany). Libraries were prepared using SureSelect QXT Library Prep Kit (Agilent Technologies, Santa Clara, United States). Sequencing was performed on an Illumina MiSeq system using the MiSeq reagent kit v2 generating 250-bp paired-end reads. Trimmomatic (Bolger et al., 2014) was used for preprocessing the WGS data. If the average quality score was below 20 in a sliding window of 4 the adapter sequences and the leading and trailing bases were removed as well as the first 18 bases. Only the reads longer than 50 nucleotides were used for subsequent analysis. De novo genome assembly was performed with SPAdes Genome Assembler 3.13.0 (Bankevich et al., 2012). Each assembled genome was accepted for further analysis if it met all of the following quality criteria: (i) average coverage > 30 times, (ii) N50 > 15,000 bases, (iii) maximum contig length > 50,000 bases, and (iv) assembled genome size between 5,000,000 and 6,500,000 bases. Assembled genomes were uploaded to the online bioinformatics tools ResFinder (Zankari et al., 2012), PlasmidFinder (Carattoli et al., 2014) (Center for Genomic Epidemiology, Technical University of Denmark, Lyngby, Denmark) to analyse resistome and plasmid replicon types of the isolates.
Quantitative PCR (qPCR)
Total RNA of tested strains was isolated by RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. The qPCR was carried out in a Step One Real-Time PCR System (Applied BioSystems, Thermo Fisher Scientific). Separate expression of qnrA1, qnrB1 qnrB4, oqxA, and oqxB genes were investigated whereas chromosomal rpoB was chosen as housekeeping gene. Set of primers and 6-FAM or VIC labeled probes were designed by Primer Express 3.0 software. All oligonucleotide primers and probes for qPCR are listed in Table 1. Each RNA sample was tested in triplicate. The qPCR was applied in default setting 60°C 30 s; 50°C 5 min; 95°C 10 min; 40 cycles of [95°C 15 s and 60°C 1 min] 60°C 30 s. The CT values of genes of interest were normalized (ΔCT) to the CT values of housekeeping gene rpoB and the relative expression of each gene of interest was calculated as 2−ΔCT = CT (geneofinterest) – CT (rpoB).
Results
In our study, four K. pneumoniae and a single K. oxytoca were investigated by MLST and PFGE. Three different STs were identified, including ST11 (Kpn33, Kpn115, Kpn125), ST307 (Kpn47), and ST52 (Kox37).
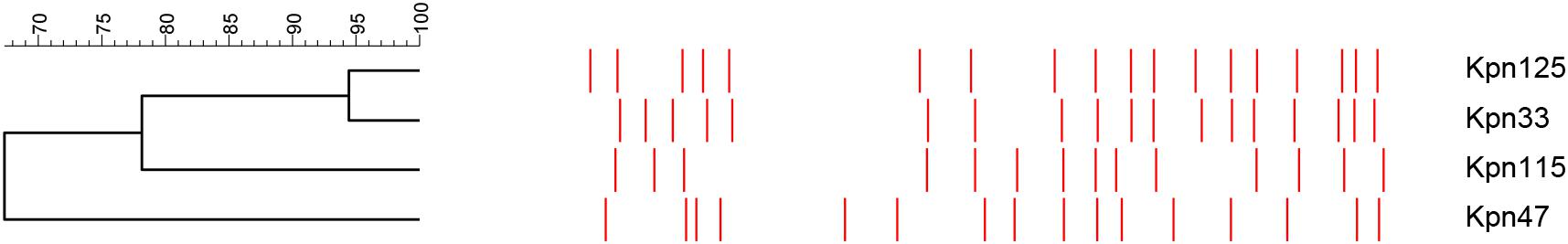
Pulsed-field gel electrophoresis analysis detected three pulsotypes (PT) among K. pneumoniae strains, namely, KP053, S and KP197. Two isolates belonged to KP053 (Kpn33 and Kpn125) and one was detected as S PT (Kpn115). These strains belonged to the ST11 international high-risk clone. By contrast, Kpn47 was classified as KP197 PT (Figure 2).
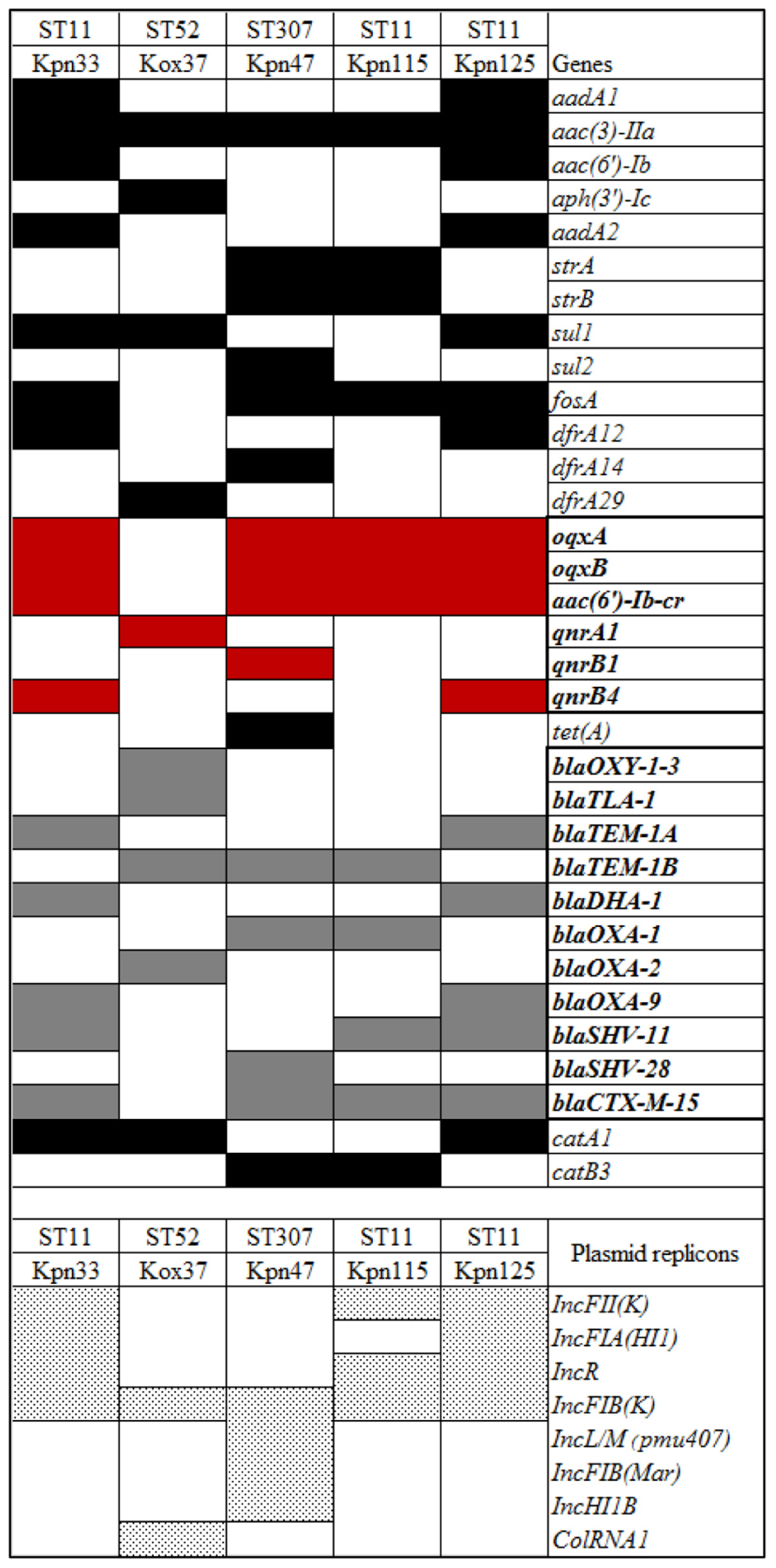
The initial assembled draft genome sequences were 5611026 bp (Kpn33); 6370417 bp (Kox37); 5451744 bp, (Kpn47); 5450412 bp (Kpn115), and 5593358 bp (Kpn125). Seventeen antibiotic resistance genes were found in two ST11 K. pneumoniae strains (Kpn33 and Kpn125), twelve were in the third ST11 strain (Kpn115), sixteen resistant genes were in ST307 strain (Kpn47) and ten resistance genes were detected in Kox37. Sequence analysis revealed that the isolates harbored different β-lactamase genes, including blaDHA–1, blaOXA–1, blaOXA–2, blaOXA–9, blaHV–11, blaHV–28, and blaTEM–1A, blaTEM–1B, blaOXY–1–3, blaTLA–1; and all K. pneumoniae strains carried blaCTX–M–15. Among aminoglycoside resistance genes all isolates were positive for aac(3)-IIa. Only Kpn47 carried a tetracycline resistance (tetA) gene. Except for Kox37, all strains were identified positive for fosA gene nevertheless, sul1 or sul2 and trimethoprim resistance (dfrA12, dfrA14, dfrA29) genes were detected in four strains. PMQR genes were found in each tested strain namely, in Kpn33 qnrB4, in Kox37 qnrA1, in Kpn47 qnrB1, in Kpn125 qnrB4. All K. pneumoniae strains harbored oqxAB efflux pump and aac(6′)-Ib-cr, but one of the ST11 strains (Kpn115) carried no qnr gene. Presence of phenicol resistance gene (catA1 or catB3) was observed in all strains. Chromosomal mutations conferring fluoroquinolone resistance in K. pneumoniae strains were also detected, Ser83Phe and Asp87Ala substitutions were in DNA gyrase subunit A of Kpn115 (ST11), but all other K. pneumoniae strains had only Ser83Ile in gyrase while on the other hand all K. pneumoniae had a Ser80Ile substitution in DNA topoisomerase IV. Based on the sequencing data, IncFIB, IncFII, and IncR replicons were uniformly present in all ST11 strains. In the case of ST307 IncFIB, IncL/M, IncHI1B were detected. The detected resistance genes and plasmid replicons are listed in Table 2 and Figure 3.
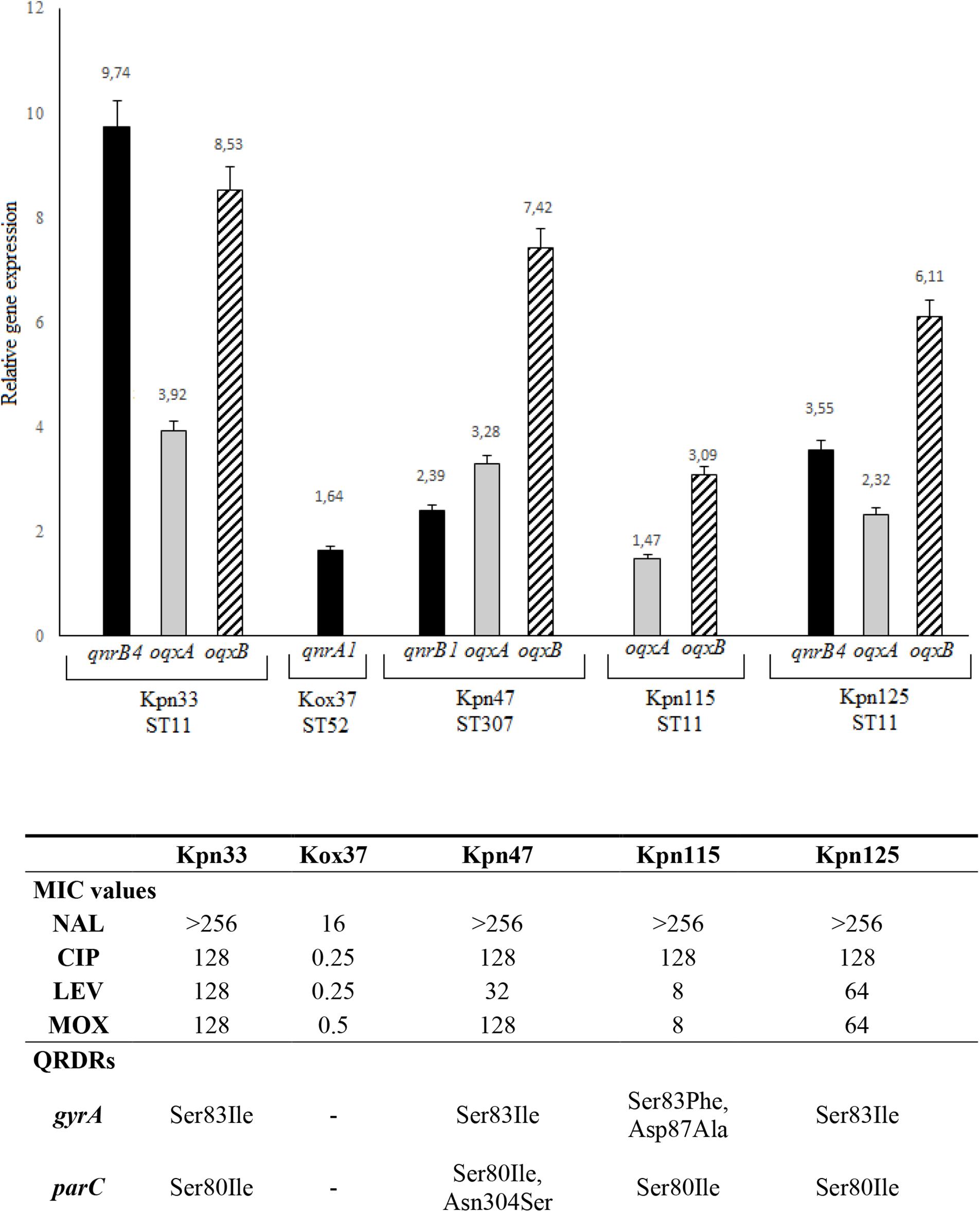
Figure 3. Level of qnrB4 (Kpn33 and Kpn125), qnrA1 (Kox37), and qnrB1, oqxA, and oqxB relative gene expression. QRDRs: quinolone resistance determining regions. All MIC values are in mg/L.
Among qnr genes, qnrB4 of two ST11 strains (Kpn33 and Kpn125) showed 9.74 and 3.55 fold expression, respectively. Interestingly, Kpn33 (ST11) was characterized approximately 3-fold higher expression, compared to the genetically similar Kpn125 (ST11). The lowest expression level (1.64) among qnr genes was detected in K. oxytoca, that exhibited reduced susceptibility to ciprofloxacin. In the case of qnrB1 in Kpn47 (ST307), it showed 2.39 fold expression.
Expression of oqxA ranged between 1.47 and 3.92 and that of oqxB from 3.09 to 8.53. The highest oqxA and oqxB expressions were observed in Kpn33 (ST11) and Kpn47 (ST307). These were followed by Kpn125 (ST11) and Kpn115 (ST11). Interestingly, Kpn115 a strain of ST11 high-risk clone carried no qnr gene moreover, it showed the lowest oqxAB expression. It is conspicuous that in every K. pneumoniae strain the oqxB is expressed 2–3 fold higher than oqxA.
Discussion
International high-risk K. pneumoniae ST11 clone has been frequently detected worldwide as a successful pathogen being associated with important virulence (Damjanova et al., 2008; Andrade et al., 2014), and resistance determinants including VIM, NDM and KPC-production (Yan et al., 2002; Kristóf et al., 2010; Qi et al., 2011; Yu et al., 2016; Campana et al., 2017). In our study, all strains of ST11 international high-risk clone carried blaSCTX–M–15 ESBL that correlates well with earlier studies as the most common global ESBLs are the CTX-M type beta-lactamases in Enterobacterales (Nordmann and Poirel, 2014). Recently, in a Bulgarian study among 82 ESBL-producing K. pneumoniae and four K. oxytoca CTX-M-15 (87%) was predominant (Markovska et al., 2017). K. pneumoniae ST11 has been already reported in Hungary, as a widely disseminated clone in all over the country (Damjanova et al., 2008). In Poland, an inter-regional outbreak was reported that was dominated by NDM-1 and CTX-M-15 coproducing K. pneumoniae ST11 clone (Baraniak et al., 2016). A high prevalence (30.2%) of CTX-M-15-producing K. pneumoniae was detected in raw bovine milk too. This finding highlights the spread of CTX-M-15-producing K. pneumoniae also in the food chain (Diab et al., 2017).
In recent years, new drug-resistant international lineages have emerged, among them, KPC-producing K. pneumoniae ST307 has been recognized in several countries (Castanheira et al., 2013; Villa et al., 2017). To the best of our knowledge, our study is the first description of ST307 in Hungary that is has been reported as a potential high-risk clone. High similarity has been found in our ST307 isolate compared to that of detected by Villa et al. (2017).
Three pulsotypes were identified among the investigated K. pneumoniae strains: KP053, S PT, and KP197. Two ST11 isolates belonged to KP053 (Kpn33 and Kpn125) and the third ST11 was detected as S PT (Kpn115) that was earlier reported in Hungary (Damjanova et al., 2008). In a Hungarian study, PFGE typing revealed 12 pulsotypes; of these, KP053 (262/312) and KP070 (38/312) belonged to sequence type ST11 (Kis et al., 2016); these data also prove the spread of KP053/ST11 clone in our country. K. pneumoniae ST307 (Kpn47) was classified as KP197 pulsotype, however, this type was not registered until 2014 by the National Public Health Institute. Since 2015, altogether 30 strains have been identified with this pulsotype in Hungary (unpublished data).
In this study, mutations in gyrase and topoisomerase coding genes and various PMQRs were detected in K. pneumoniae and K. oxytoca. Of the detected PMQRs in this study oqxAB was present in all K. pneumoniae clinical isolates but not in K. oxytoca. This result can be explained by the fact that the oqxAB is a chromosomally-encoded gene in K. pneumoniae (Yuan et al., 2012). The qnrB genes were observed in K. pneumoniae ST11 correlating with the international data (Hidalgo et al., 2013; Jaidane et al., 2018). However, this is the first report of the qnr gene in K. oxytoca ST52. Regarding plasmid replicon types, the most common replicon was IncFIB, that was present in all ST11, ST52, and ST307, which confirms earlier studies (Anes et al., 2017).
Acquisition of qnr determinants can have multiple advantages. In the case of K. oxytoca, the presence and expression of qnrA1 caused reduced susceptibility to quinolones. Levofloxacin and moxifloxacin MIC values of K. pneumoniae ST11 and ST307 clones correlated with qnr and oqxAB expression levels (Figure 3).
Further beneficial effect of Qnr proteins can be explained by the toxin-antitoxin effect. Qnr proteins are considered antitoxins, that protect gyrase and topoisomerase IV enzymes from naturally occuring toxins. This theory was described by Ellington and Woodford (2006) and it can be valid also in internationally disseminated high-risk clones (Ellington and Woodford, 2006). During development of fluoroquinolone resistance PMQR determinants play a role in reduced susceptibility, and they maintain low-level fluoroquinolone resistance (Garoff et al., 2018). Later, by chromosomal mutations in QRDRs high-level fluoroquinolone resistance develops, but PMQR expression is maintained thus, indicating further role of PMQRs such as protection of gyrase and topoisomerase IV enzymes (Tran et al., 2005a,b; Redgrave et al., 2014).
It has been also established that the development of fluoroquinolone resistance is diverse among different clones and in the case of international high-risk K. pneumoniae clones the fluoroquinolone resistant strains retain fitness that facilitates their dissemination in hospital environment (Fuzi, 2016). Moreover, Redgrave et al. indicated that fluoroquinolone resistance played a key role in evolutionary success of K. pneumoniae clones (Redgrave et al., 2014).
Emergence and possible dissemination of K. pneumoniae ST307 in hospital settings raises also public health concerns, therefore continous monitoring of high-risk and potential high-risk clones is necessary.
Repository Data
Assembled genomes of all investigated strains were deposited in NCBI Genbank under the following accession numbers. Raw sequence data of each strain in this study was submitted to Sequence Read Archive (SRA)
Kpn 33: Bioproject: PRJNA511518, Biosample SAMN10639440
Kox37: Bioproject PRJNA511522, Biosample: SAMN106 39457
Kpn47: Bioproject: PRJNA511523, Biosample: SAMN10639726
Kpn115 Bioproject: PRJNA511524, BioSamples SAMN10639736
Kpn125: Bioproject: PRJNA511525, BioSamples SAMN10639737.
Author Contributions
JD performed pulsed-field gel electrophoresis, multilocus sequence typing, and handled the manuscript. ID performed pulsed-field gel electrophoresis and whole-genome sequencing. KK identified and handled strains from clinical specimen. BL performed whole-genome sequence analysis. BK performed qPCR, analyzed the data, and handled the manuscript. DS was laboratory chief, contributed to study design, and handled the manuscript.
Funding
This study was financially supported by OTKA Hungarian Research Fund: grant 108481.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Andrade, L. N., Vitali, L., Gaspar, G. G., Bellissimo-Rodrigues, F., Martinez, R., and Darini, A. L. (2014). Expansion and evolution of a virulent, extensively drug-resistant (polymyxin B-resistant), QnrS1-, CTX-M-2-, and KPC-2-producing Klebsiella pneumoniae ST11 international high-risk clone. J. Clin. Microbiol. 52, 2530–2535. doi: 10.1128/JCM.00088-14
Anes, J., Hurley, D., Martins, M., and Fanning, S. (2017). Exploring the genome and phenotype of multi-drug resistant Klebsiella pneumoniae of clinical origin. Front. Microbiol. 8:1913. doi: 10.3389/fmicb.2017.01913
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Baraniak, A., Izdebski, R., Fiett, J., Gawryszewska, I., Bojarska, K., Herda, M., et al. (2016). NDM-producing enterobacteriaceae in poland, 2012–14: inter-regional outbreak of klebsiella pneumoniae ST11 and sporadic cases. J. Antimicrob. Chemother. 71, 85–91. doi: 10.1093/jac/dkv282
Baquero, F., Tedim, A. P., and Coque, T. M. (2013). Antibiotic resistance shaping multi-level population biology of bacteria. Front. Microbiol. 4:15. doi: 10.3389/fmicb.2013.00015
Bi, W., Liu, H., Dunstan, R. A., Li, B., Torres, V. V. L., Cao, J., et al. (2017). Extensively drug-resistant Klebsiella pneumoniae causing nosocomial bloodstream infections in china: molecular investigation of antibiotic resistance determinants, informing therapy, and clinical outcomes. Front. Microbiol. 8:1230. doi: 10.3389/fmicb.2017.01230
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Campana, E. H., Montezzi, L. F., Paschoal, R. P., and Picão, R. C. (2017). NDM-producing Klebsiella pneumoniae ST11 goes to the beach. Int. J. Antimicrob. Agents 49, 119–121. doi: 10.1016/j.ijantimicag.2016.10.006
Cao, X., Xu, X., Zhang, Z., Shen, H., Chen, J., and Zhang, K. (2014). Molecular characterization of clinical multidrug-resistant Klebsiella pneumoniae isolates. Ann. Clin. Microbiol. Antimicrob. 13:16. doi: 10.1186/1476-0711-13-16
Carattoli, A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303, 298–304. doi: 10.1016/j.ijmm.2013.02.001
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Castanheira, M., Farrell, S. E., Wanger, A., Rolston, K. V., Jones, R. N., and Mendes, R. E. (2013). Rapid expansion of KPC-2-producing Klebsiella pneumoniae isolates in two Texas hospitals due to clonal spread of ST258 and ST307 lineages. Microb. Drug Resist. 19, 295–297. doi: 10.1089/mdr.2012.0238
Damjanova, I., Tóth, A., Pászti, J., Hajbel-Vékony, G., Jakab, M., Berta, J., et al. (2008). Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin resistant CTX-M-15-type beta-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005–the new ‘MRSAs’? J. Antimicrob. Chemother. 62, 978–985. doi: 10.1093/jac/dkn287
Diab, M., Hamze, M., Bonnet, R., Saras, E., Madec, J. Y., and Haenni, M. (2017). OXA-48 and CTX-M-15 extended-spectrum beta-lactamases in raw milk in Lebanon: epidemic spread of dominant Klebsiella pneumoniae clones. J. Med. Microbiol. 66, 1688–1691. doi: 10.1099/jmm.0.000620
Diancourt, L., Passet, V., Verhoef, J., Grimont, P. A., and Brisse, S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 13, 4178–4182. doi: 10.1128/JCM.43.8.4178-4182
Domokos, J., Kristof, K., and Szabo, D. (2016). Plasmid-mediated quinolone resistance among extended-spectrum beta-lactamase producing Enterobacteriaceae from bloodstream infections. Acta Microbiol. Immunol. Hung. 63, 313–323. doi: 10.1556/030.63.2016.002
Ellington, M. J., and Woodford, N. (2006). Fluoroquinolone resistance and plasmid addiction systems: self-imposed selection pressure? J. Antimicrob. Chemother. 57, 1026–1029. doi: 10.1093/jac/dkl110
Fuzi, M. (2016). Dissimilar fitness associated with resistance to fluoroquinolones influences clonal dynamics of various multiresistant bacteria. Front. Microbiol. 7:1017. doi: 10.3389/fmicb.2016.01017
Fuzi, M., Szabo, D., and Csercsik, R. (2017). Double-serine fluoroquinolone resistance mutations advance major international clones and lineages of various multi-drug resistant bacteria. Front. Microbiol. 8:2261. doi: 10.3389/fmicb.2017.02261
Garoff, L., Yadav, K., and Hughes, D. (2018). Increased expression of Qnr is sufficient to confer clinical resistance to ciprofloxacin in Escherichia coli. J. Antimicrob. Chemother. 73, 348–352. doi: 10.1093/jac/dkx375
Girlich, D., Bouihat, N., Poirel, L., Benouda, A., and Nordmann, P. (2014). High rate of faecal carriage of extended-spectrum β-lactamase and OXA-48 carbapenemase-producing Enterobacteriaceae at a university hospital in Morocco. Clin. Microbiol. Infect. 20, 350–354. doi: 10.1111/1469-0691.12325
Gona, F., Barbera, F., Pasquariello, A. C., Grossi, P., Gridelli, B., Mezzatest, M. L., et al. (2014). In vivo multiclonal transfer of blaKPC-3 from Klebsiella pneumoniae to Escherichia coli in surgery patients. Clin. Microbiol. Infect. 20, O633–O635. doi: 10.1111/1469-0691.12577
Gonçalves, G. B., Furlan, J. P. R., Vespero, E. C., Pelisson, M., Stehling, E. G., and Pitondo-Silva, A. (2017). Spread of multidrug-resistant high-risk Klebsiella pneumoniae clones in a tertiary hospital from southern Brazil. Infect. Genet. Evol. 56, 1–7. doi: 10.1016/j.meegid.2017.10.011
Habeeb, M. A., Haque, A., Nematzadeh, S., Iversen, A., and Giske, C. G. (2013). High prevalence of 16S rRNA methylase RmtB among CTX-M extended-spectrum β-lactamase-producing Klebsiella pneumoniae from Islamabad, Pakistan. Int. J. Antimicrob. Agents 41, 524–526. doi: 10.1016/j.ijantimicag
Hidalgo, L., Gutierrez, B., Ovejero, C. M., Carrilero, L., Matrat, S., Saba, C. K., et al. (2013). Klebsiella pneumoniae sequence type 11 from companion animals bearing ArmA methyltransferase, DHA-1 β-lactamase, and QnrB4. Antimicrob. Agents Chemother. 57, 4532–4534. doi: 10.1128/AAC.00491-13
Hrabák, J., Empel, J., Bergerová, T., Fajfrlík, K., Urbásková, P., Kern-Zdanowicz, I., et al. (2009). International clones of Klebsiella pneumoniae and Escherichia coli with extended-spectrum beta-lactamases in a Czech hospital. J. Clin. Microbiol. 47, 3353–3357. doi: 10.1128/JCM.00901-09
Hunter, S. B., Vauterin, P., Lambert-Fair, M. A., Van Duyne, M. S., Kubota, K., Graves, L., et al. (2005). Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43, 1045–1050. doi: 10.1128/JCM.43.3.1045-1050.2005
Jaidane, N., Bonnin, R. A., Mansour, W., Girlich, D., Creton, E., Cotellon, G., et al. (2018). Genomic insights into colistin-resistant Klebsiella pneumoniae from a tunisian teaching hospital. Antimicrob. Agents Chemother. 62:e1601-17. doi: 10.1128/AAC.01601-17
Kang, C. I., Kim, S. H., Bang, J. W., Kim, H. B., Kim, N. J., Kim, E. C., et al. (2006). Community-acquired versus nosocomial Klebsiella pneumoniae bacteremia: clinical features, treatment outcomes, and clinical implication of antimicrobial resistance. J. Korean Med. Sci. 21, 816–822. doi: 10.3346/jkms.2006.21.5.816
Kis, Z., Tóth, Á, Jánvári, L., and Damjanova, I. (2016). Countrywide dissemination of a DHA-1-type plasmid-mediated AmpC β-lactamase-producing Klebsiella pneumoniae ST11 international high-risk clone in hungary, 2009-2013. J. Med. Microbiol. 65, 1020–1027. doi: 10.1099/jmm.0.000302
Ko, K. S., Lee, J. Y., Baek, J. Y., Suh, J. Y., Lee, M. Y., Choi, J. Y., et al. (2010). Predominance of an ST11 extended-spectrum β-lactamase-producing Klebsiella pneumoniae clone causing bacteraemia and urinary tract infections in Korea. J. Med. Microbiol. 59, 822–828. doi: 10.1099/jmm.0.018119-0
Kristóf, K., Tóth, A., Damjanova, I., Jánvári, L., Konkoly-Thege, M., Kocsis, B., et al. (2010). Identification of a blaVIM-4 gene in the internationally successful Klebsiella pneumoniae ST11 clone and in a Klebsiella oxytoca strain in Hungary. J. Antimicrob. Chemother. 65, 1303–1305. doi: 10.1093/jac/dkq133
Mansour, W., Grami, R., Ben Haj Khalifa, A., Dahmen, S., Châtre, P., Haenni, M., et al. (2017). Dissemination of multidrug-resistant blaCTX-M-15/IncFIIk plasmids in Klebsiella pneumoniae isolates from hospital- and community-acquired human infections in Tunisia. Diagn. Microbiol. Infect. Dis. 83, 298–304. doi: 10.1016/j.diagmicrobio.2015.07.023
Markovska, R., Stoeva, T., Boyanova, L., Stankova, P., Pencheva, D., Keuleyan, E., et al. (2017). Dissemination of successful international clone ST15 and clonal complex 17 among bulgarian CTX-M-15 producing K. pneumoniae isolates. Diagn. Microbiol. Infect. Dis. 89, 310–313. doi: 10.1016/j.diagmicrobio.2017.08.012
Mathers, A. J., Peirano, G., and Pitout, J. D. (2015). The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28, 565–591. doi: 10.1128/CMR.00116-14
Munoz-Price, L. S., Poirel, L., Bonomo, R. A., Schwaber, M. J., Daikos, G. L., Cormican, M., et al. (2013). Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13, 785–796. doi: 10.1016/S1473-3099(13)70190-7
Nordmann, P., and Poirel, L. (2014). The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 20, 821–830. doi: 10.1111/1469-0691.12719
Novović, K., Trudić, A., Brkić, S., Vasiljević, Z., Kojić, M., Medić, D., et al. (2017). Molecular epidemiology of colistin-resistant, carbapenemase-producing Klebsiella pneumoniae in Serbia from 2013 to 2016. Antimicrob. Agents Chemother. 61:e2550-16. doi: 10.1128/AAC.02550-16
Ocampo, A. M., Chen, L., Cienfuegos, A. V., Roncancio, G., Chavda, K. D., Kreiswirth, B. N., et al. (2016). A two-year surveillance in five Colombian tertiary care hospitals reveals high frequency of non-CG258 clones of carbapenem-resistant Klebsiella pneumoniae with distinct clinical characteristics. Antimicrob. Agents Chemother. 60, 332–342. doi: 10.1128/AAC.01775-15
Park, D. J., Yu, J. K., Park, K. G., and Park, Y. J. (2015). Genotypes of ciprofloxacin-resistant Klebsiella pneumoniae in Korea and their characteristics according to the genetic lineages. Microb. Drug. Resist. 21, 622–630. doi: 10.1089/mdr.2015.0001
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603. doi: 10.1128/CMR.11.4.589
Qi, Y., Wei, Z., Ji, S., Du, X., Shen, P., and Yu, Y. (2011). ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 66, 307–312. doi: 10.1093/jac/dkq431
Redgrave, L. S., Sutton, S. B., Webber, M. A., and Piddock, L. J. (2014). Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 22, 438–445. doi: 10.1016/j.tim.2014.04.007
Rodrigues, C., Machadoa, E., Ramos, H., Peixea, L., and Novais, Â (2014). Expansion of ESBL-producing Klebsiella pneumoniae in hospitalized patients: a successful story of international clones (ST15, ST147, ST336) and epidemic plasmids (IncR, IncFIIK). Int. J. Med. Microbiol. 304, 1100–1108. doi: 10.1016/j.ijmm.2014.08.003
Rodríguez-Martínez, J. M., Cano, M. E., Velasco, C., Martínez-Martínez, L., and Pascual, A. (2011). Plasmid-mediated quinolone resistance: an update. J. Infect. Chemother. 17, 149–182. doi: 10.1007/s10156-010-0120-2
Ruan, Z., and Feng, Y. (2016). BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 44, D682–D687. doi: 10.1093/nar/gkv1004
Tóth, A., Kocsis, B., Damjanova, I., Kristóf, K., Jánvári, L., Pászti, J., et al. (2014). Fitness cost associated with resistance to fluoroquinolones is diverse across clones of Klebsiella pneumoniae and may select for CTX-M-15 type extended-spectrum β-lactamase. Eur. J. Clin. Microbiol. Infect. Dis. 33, 837–843. doi: 10.1007/s10096-013-2022-6
Tran, J. H., Jacoby, G. A., and Hooper, D. C. (2005a). Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 49, 118–125. doi: 10.1128/AAC.49.1.118-125.2005
Tran, J. H., Jacoby, G. A., and Hooper, D. C. (2005b). Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob. Agents Chemother. 49, 3050–3052. doi: 10.1128/AAC.49.7.3050-3052.2005
Villa, L., Feudi, C., Fortini, D., Brisse, S., Passet, V., Bonura, C., et al. (2017). Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb. Genom. 3:e000110. doi: 10.1099/mgen.0.000110
Woodford, N., Turton, J. F., and Livermore, D. M. (2011). Multiresistant gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35, 736–755. doi: 10.1111/j.1574-6976.2011.00268.x
Xie, L., Dou, Y., Zhou, K., Chen, Y., Han, L., Guo, X., et al. (2017). Coexistence of blaOXA-48 and truncated blaNDM-1 on different plasmids in a Klebsiella pneumoniae isolate in China. Front. Microbiol. 8:133. doi: 10.3389/fmicb.2017.00133
Yan, J. J., Ko, W. C., Jung, Y. C., Chuang, C. L., and Wu, J. J. (2002). Emergence of Klebsiella pneumoniae isolates producing inducible DHA-1 β-lactamase in a university hospital in Taiwan. J. Clin. Microbiol. 40, 3121–3126. doi: 10.1128/JCM.40.9.3121-3126.2002
Yu, J., Tan, K., Rong, Z., Wang, Y., Chen, Z., Zhu, X., et al. (2016). Nosocomial outbreak of KPC-2- and NDM-1-producing Klebsiella pneumoniae in a neonatal ward: a retrospective study. BMC Infect. Dis. 16:563. doi: 10.1186/s12879-016-1870-y
Yuan, J., Xu, X., Guo, Q., Zhao, X., Ye, X., Guo, Y., et al. (2012). Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates. J. Antimicrob. Chemother. 67, 1655–1659. doi: 10.1093/jac/dks086
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: international clones, multi-drug resistance, whole genome sequence analysis, gene expression, plasmid-mediated quinolone resistance
Citation: Domokos J, Damjanova I, Kristof K, Ligeti B, Kocsis B and Szabo D (2019) Multiple Benefits of Plasmid-Mediated Quinolone Resistance Determinants in Klebsiella pneumoniae ST11 High-Risk Clone and Recently Emerging ST307 Clone. Front. Microbiol. 10:157. doi: 10.3389/fmicb.2019.00157
Received: 15 December 2017; Accepted: 22 January 2019;
Published: 12 February 2019.
Edited by:
Gilberto Igrejas, University of Trás-os-Montes and Alto Douro, PortugalCopyright © 2019 Domokos, Damjanova, Kristof, Ligeti, Kocsis and Szabo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bela Kocsis, a29jc2lzLmJlbGFAbWVkLnNlbW1lbHdlaXMtdW5pdi5odQ==
 Judit Domokos1
Judit Domokos1 Bela Kocsis
Bela Kocsis Dora Szabo
Dora Szabo