- 1Department of Food Science and Technology, Advanced Food Safety Research Group, Brain Korea 21 Plus, Chung-Ang University, Anseong, South Korea
- 2Food Safety Research Group, Korea Food Research Institute, Seongnam, South Korea
- 3Food Science and Technology, Oregon State University, Corvallis, OR, United States
The objective of this study was to investigate the virulence factors, genetic relationship, antibiotic resistance profile and the biofilm formation ability of Vibrio parahaemolyticus isolates on shrimp and mussel surfaces at 30∘C. In this study, eight (n = 8) V. parahaemolyticus isolated from mussel were examined. We used the polymerase chain reaction (PCR) to examine the distribution of different genes, and Repetitive Extragenic Palindromic-PCR (REP-PCR) to compare the genetic relationship. Disk diffusion technique was used to assess antibiotic and multiple-antibiotic resistance. The biofilm formation assay, and field emission scanning electron microscopy (FE-SEM) were used to evaluate biofilm formation ability. Transmission Electron Microscope (TEM) was used to observe the morphological structure of bacterial cell. Our results indicated that the biofilm-associated genes, 16S rRNA, toxR, and tdh, were present in all the tested V. parahaemolyticus isolates (n = 8). Approximately, 62.5% (5 isolates among 8 isolates) isolates showed strong multiple-antibiotic resistance index with an average value of 0.56. All isolates (n = 8) showed strong genetic relationship and significant biofilm formation ability on shrimp and mussel surfaces. This study demonstrated that the presence of virulence factors, high multiple antibiotic resistance index (MARI) values, and effective biofilm formation ability of V. parahaemolyticus isolates could be a great threat to human health and economic values in future. It was also suggested that a high resistance rate to antibiotic could be ineffective for treating V. parahaemolyticus infections. The continuous monitoring of V. parahaemolyticus antibiotic, molecular and biofilm characteristics is needed to increase seafood safety.
Introduction
Seafood is recognized as a nutritious and healthy food choice, and is accepted by increasing numbers of consumers worldwide (Hellberg et al., 2012). Every year, above 100.2 million metric tons of seafood are caught and consumed in the world (Cisneros-Montemayor et al., 2016). In 2014, the value of imported seafood in Korea, China, and the United States were 4.16, 1.12 billion, and 222 million, respectively (Mizan et al., 2018). However, the main obstacles in the consumption of seafood are their high perishability and health risk due to contamination by pathogens (Reyhanath and Kutty, 2014). Therefore, seafood safety is considered as necessary to maintain public health and seafood processing (Jahan, 2012; Machado and Gram, 2015).
In the aquaculture industry, black tiger shrimp (Penaeus monodon) plays an important role in the economic aspect and cultured in both inland and marine (Szuster, 2006). Vibrio parahaemolyticus is the most prevalent shrimp pathogen encountered in aquaculture, causes in shrimp Vibriosis with the potential for severe health crisis (Mohammad et al., 2005; Kleter et al., 2009; Sani et al., 2013; Zhang et al., 2014). In China, shrimp contaminated with V. parahaemolyticus has been accompanied with outbreaks of food borne illnesses (Peng et al., 2010). In Australia two outbreaks of gastroenteritis occurred between 1990 and 1992 due to the consumption of V. parahaemolyticus contaminated cooked shrimps imported from Indonesia (Sumner, 2011). The mussel is valued worldwide for its sensory and nutritional qualities. Spain is the main supplier of mussel to the European market, coming 98% of this production from Galicia (Garrido-Maestu et al., 2016). During 1997 to 2010, Global production of mussels has increased up to 1.9 million tons worldwide. This represented 95% of the world mussel production, in comparison to 83% in 1997 (Ferreira et al., 2014). In recent years, Galicia has been recognized as the largest producers of mussels, accounting for the 15 to 25% of the world’s annual mussel production (Miguez et al., 2009; Costas-Rodríguez et al., 2010; Caballero-Miguez et al., 2012). But several studies have demonstrated the presence of pathogenic species of the genus Vibrio in the Galician Rias (Lozano-León et al., 2003; Martinez-Urtaza et al., 2004, 2008; Rodriguez-Castro et al., 2010). As mussel is a good vehicle for Vibrio species, V. parahaemolyticus can survive in mussel with potential contamination (Mannas et al., 2014). Therefore, V. parahaemolyticus constitute a potential risk to consumers for having improperly processed shellfish (FDA BAM, 2004). Several post-harvest processes, including low-temperature pasteurization and irradiation have been developed for reducing Vibrios in aquaculture but they are expensive (Chae et al., 2009).
Vibrio infections occurred due to the presence of virulence factor. The strains of V. parahaemolyticus contain virulence factor, including adhesins (Type I pilus), toxR, biofilm, thermostable direct hemolysin (tdh), TDH-related hemolysin (TRH) encoded by trh gene, VPaI-2, VPaI-3, VPaI-6, type III secretion systems (T3SS), and type VI secretion systems (T6SS) (Chao et al., 2009, 2010; Broberg et al., 2011; Salomon et al., 2013: Letchumanan et al., 2014). In the United States, more than 80% of gastroenteritis and 90% of septicemia infections happened during 1988 to 1997, due to the consumption of oysters (Daniels and Shafaie, 2000). It was also reported in 2006 that V. parahaemolyticus was responsible for 177 cases due to having raw shellfish in the United States (Yoon et al., 2008). Therefore the consumption of V. parahaemolyticus contaminated seafood is one of the greatest source of infection in America as well as in Asia (Hongping et al., 2011).
The other problem associated with V. parahaemolyticus is due to the prevalence of antibiotic resistance in aquaculture. The extensive use of antibiotics for the treatment of infections caused by V. parahaemolyticus, has increased the incidence of antibiotic-resistant strains (Cabello et al., 2013; Yano et al., 2014; Letchumanan et al., 2015; Xie et al., 2017: Yang Y. et al., 2017; Lee et al., 2018). V. parahaemolyticus has shown resistant property against numerous antibiotics including ampicillin, ciprofloxacin, cephazolin, streptomycin, cefotaxime, and cefuroxime sodium (Al-Othrubi et al., 2014; Jiang et al., 2014; Sudha et al., 2014; Yano et al., 2014). The multidrug resistance of V. parahaemolyticus is also increasing gradually due to the excessive use of antibiotics in the fields of agriculture and aquaculture (Kang et al., 2017). Antibiotic-resistant bacteria may represent a potential threat to human health via direct transmission through the food chain or the transfer of resistance genes to other human (Duran and Marshall, 2005; Guglielmetti et al., 2009; Ma et al., 2018). In aquaculture farming, an appropriate policy is essential for using antibiotics (Odeyemi and Stratev, 2016). Moreover, the monitoring of antibiotic resistance patterns of V. parahaemolyticus in seafood is also important (Odeyemi and Stratev, 2016), because it is a high concern for human health (Xie et al., 2017). This emerging incidence of antibiotic resistance in V. parahaemolyticus has generated a growing interest in identifying new strategies for preventing infections related to V. parahaemolyticus biofilms (Su and Liu, 2007; Lopatek et al., 2018; Silva et al., 2018; Jiang et al., 2019).
Bacterial biofilms are architecturally complex assemblies of microorganisms that adhere to biotic or abiotic surfaces and are attached within a matrix of extracellular polymeric substances (Costerton et al., 1999; Hall-Stoodley et al., 2004; Flemming and Wingender, 2010; Mizan et al., 2015). Bacteria in biofilms can be 1,000-fold more resistant to environmental stress than planktonic cells are (Brooun et al., 2000). V. parahaemolyticus is able to produce adherence factors, to adhere to surfaces for biofilm formation (Donlan, 2002). Biofilm cells are more resistant to disinfectants and antibacterial agents than the same bacteria in a free-swimming state, so the environmental survival, infectivity and transmission are enhanced due to strong biofilm formation ability of this pathogen (Kadam et al., 2013; Elexson et al., 2014a). In our knowledge, this is the first study to check the biofilm formation ability of V. parahaemolyticus isolates on mussel surface.
Based on the epidemiological importance of V. parahaemolyticus and the concern of eating undercooked shellfish, the present study was design to verify the genetic relationship, virulence factors, antibiotic resistance profile and biofilm formation ability of V. parahaemolyticus isolated from mussel from the west coast area of Korea.
Materials and Methods
Bacterial Strains, Growth Conditions, and Preparation of Bacterial Suspensions
A total of 10 V. parahaemolyticus strains, were included in this study. Among 10 V. parahaemolyticus strains, two (n = 2) were reference strains (ATCC17802 and ATCC27969), and eight (n = 8) were environmental isolates (Table 1) from sea mussel. These environmental isolates were obtained from the National Institute of Fisheries Science (NIFS), South Korea. V. parahaemolyticus isolates were elementary identified as blue-green colored colonies using thiosulfate citrate bile salts sucrose agar (TCBSA, Difco Laboratories, Sparks, MD, United States). Isolates were transferred onto nutrient agar (Difco Laboratories) and cultivated at 30°C for 24 h. Biochemical tests were directed using a VITEK 2 compact system (bioMerieux, Grenoble, France) to assure the phenotypical identity of the isolates. Prior to each experiment, the strains were activated by transferring from stocks which stored at -80°C to CHROMagar Vibrio plates (CHROMagar, Paris, France) and incubated overnight at 30°C. After 18–24 h incubation a single colony was taken from each plate and inoculated into 5 ml Luria-Bertani (LB) broth (2% NaCl; Difco Laboratories), and then incubated overnight at 30°C in a shaking incubator (Vision Scientific, VS-8480, South Korea) at 220 rpm. Subsequently, the V. parahaemolyticus cultures were centrifuged (11,000 × g at 4°C for 10 min), and then washed and resuspended in peptone water (PW; BD diagnostics, Franklin Lakes, NJ, United States), and made a target concentration (CFU/ml) for the final experiment.
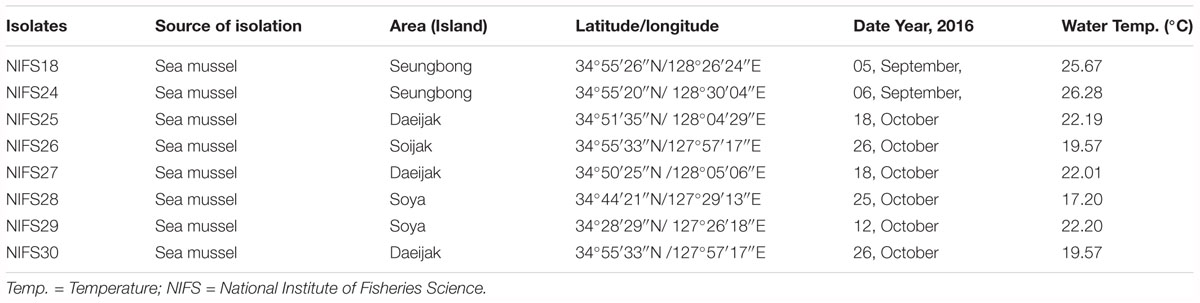
Table 1. The information about regions, season, sources, and water temperature of V. parahaemolyticus isolates used in this study.
Polymerase Chain Reaction
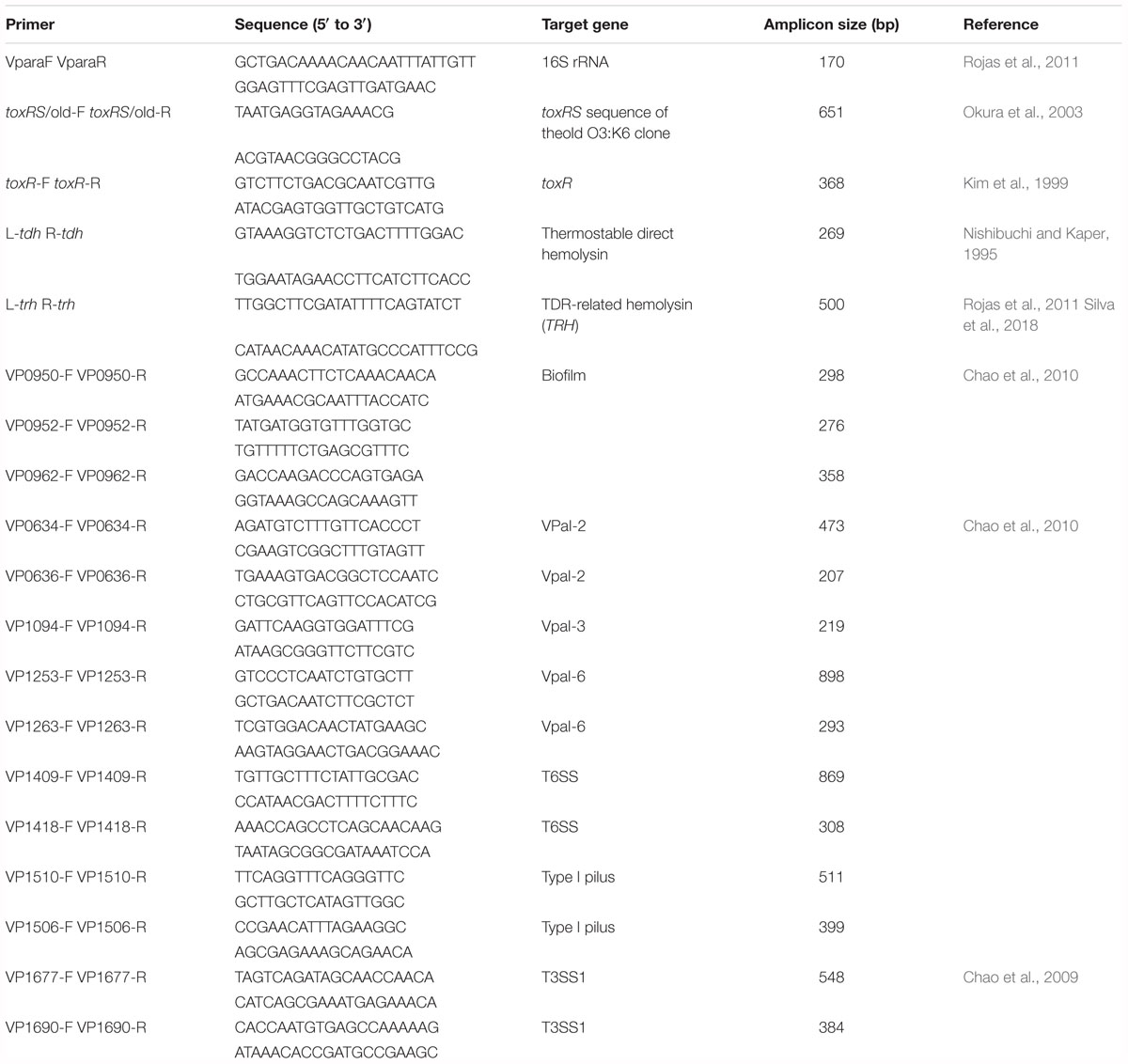
In the present study, a single polymerase chain reaction (PCR) assay was performed to test virulence factor. The nineteen oligonucleotide primer pairs (Table 2) were considered to evaluate the presence and absence of one specific primer for 16S rRNA, two pandemic clone genes, one toxR gene, TDH-related hemolysin trh gene, three biofilm genes, two VPaI-2 open reading frames (ORFs), one VPaI-3 ORFs, two VPaI-6 ORFs, two T6SS genes, two type I pilus genes, and two T3SS1 genes. All of the primers selected in this study were synthesized by the Bioneer Corporation (Daejeon, South Korea). The Tissue Kit and DNeasy Blood (QIAGEN, Venlo, Netherlands) were used to purify total DNA according to the instructions of manufacturer. The PCR reactions (25 μl) contained 14 μl of PCR mix (Solutions for Genetic Technologies, Daejeon, South Korea), 2 μl of each of the primers (10 μM), 2 μl of the DNA template, and milli-Q water. The reactions were as follows: an initial denaturation step at 95°C for 3 min, 40 cycles of 95°C for 30 s, 40–57°C (depending on product annealing temperature) for 30 s, and 72°C for 1 min, followed by a final incubation at 72°C for 5 min. PCR amplification was performed in triplicate for genomic DNA from each of the strains. The products were identified using electrophoresis on a 1.5% agarose gel and Safe View Classic staining (0.008%, v/v) (Applied Biological Materials Inc., Richmond, Canada). A 100-bp ladder (BioFACT, Daejeon, South Korea) was selected as the molecular weight marker.
Repetitive Extragenic Palindromic-PCR (REP-PCR)
REP-PCR, used for chromosomal comparisons of V. parahaemolyticus isolates, was conducted using two primers: REP-1D, 5’-NNN RCG YCG NCA TCM GGC-3’; and REP-2D, 5’-RCG YCT TAT CMG GCC TAC-3’ (where M is A or C, R is A or G, Y is C or T, and N is any nucleotide) as reported previously (Wong and Lin, 2001). The experiment was performed followed by Mizan et al. (2017), and a digital image was captured through a charge coupled device camera (Gel Doc XR system, Bio-Rad). The resulting fingerprints were analyzed using FPQuest software (Bio-Rad Laboratories, Inc., Hercules, CA, United States). Similarities between digitized profiles were counted using Pearson’s correlation, and an average linkage (unweighted pair group method with arithmetic mean, UPGMA) dendrogram was obtained.
Antibiotic Susceptibility Testing
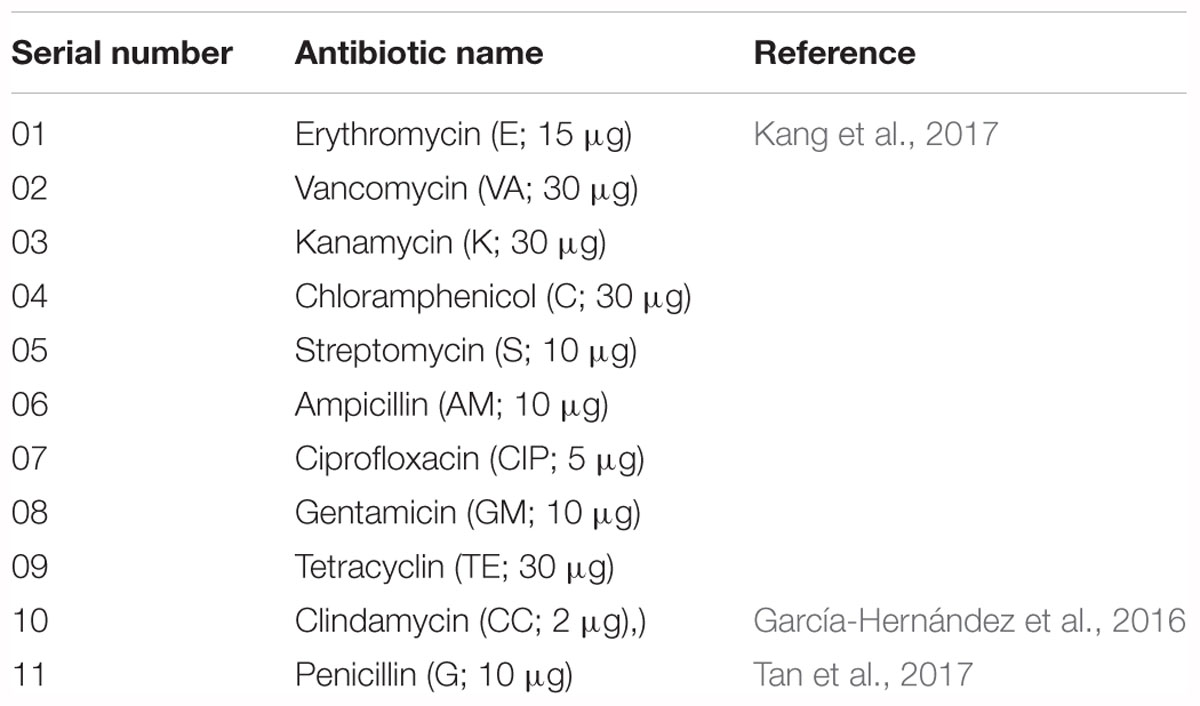
The antibiotic susceptibility of V. parahaemolyticus isolates was determined using the disk diffusion technique (NCCLS, 2003). For testing antibiotic susceptibility, selective media were used with slight modifications as previously described (Temmerman et al., 2003; McLain et al., 2016; Vandeplassche et al., 2017). Eleven antibiotics were tested in this study (Table 3). The 10 V. parahaemolyticus isolates were spread on CHROMagar plates onto which antibiotic disks were then placed. The plates were incubated at 30°C for 18–24 h under aerobic conditions. The zones of inhibition were measured according to the guidelines of Clinical and Laboratory Standards Institute (CLSI, 2010). The multiple-antibiotic resistance (MAR) index of the isolates was defined as x/y, where x represents the number of antibiotics to which a particular isolate was resistant, and y represents the number of antibiotics to which the isolate was susceptible (Krumperman, 1983). Tetracycline and ciprofloxacin are recommended antibiotics to treat V. parahaemolyticus illnesses (Park et al., 2018). Previous reports performed in South Korea showed the antibiotic susceptibility profile of V. parahaemolyticus isolates in seawater samples and found that 3.0–12.2% of isolates were resistant to tetracycline and ciprofloxacin, respectively (Son et al., 2005; Kim et al., 2014). Other studies reported using different antibiotics against V. parahaemolyticus for detecting antibiotic susceptibility (Han et al., 2012; Yang J. H. et al., 2017; Park et al., 2018). Most of the antimicrobials tested in this study are using in agriculture and aquaculture fields (Kim S. et al., 2016; Kang et al., 2017), as well as in the treatment of vibrio infections (Shaw et al., 2014).
Preparation of Inoculum for Food Samples
The cultures in LB containing 2% NaCl were centrifuged (10,000g for 12 min at 4°C) and the pellets were washed with sterile phosphate-buffered saline (PBS, pH 7.2). The pellets were resuspended in the same amount of PBS. These inocula were used to form biofilm on shrimp and mussel surfaces.
Preparation of Shrimp and Mussel Surfaces, Biofilm Formation, and Detachment Population
Black tiger shrimp (P. monodon) and mussel (Mytilus coruscus) were purchased from a native grocery store in Anseong, South Korea. Surface preparation, biofilm formation, and detachment procedure were performed followed by Han et al. (2016) with minor modifications. Using a scalpel, the shrimp head surface and mussel cover surface were aseptically cut into 2 × 2 cm2 that were then washed with sterile distilled water to remove the flesh. Prior to inoculation with V. parahaemolyticus, the surfaces were placed in an open sterile petri dish and subjected to ultraviolet-C treatment for 30 min on each side to minimize the background flora. Preliminary experiments confirmed that a UV-C treatment time of 30 min was sufficient to remove the background microbiota below the detection level cultured on Trypticase Soy Agar (TSA) plates (Han et al., 2016; Jahid et al., 2019). The incubated bacterial cultures were centrifuged (10,000 × g at 4°C for 12 min). The resulting pellets were washed three times using PBS, and resuspended into LB to attain the final concentration of bacterial cells (105 CFU/ml), and then used to form biofilm on shrimp and mussel surfaces. Each surface was completely submerged into 10 ml LB in 50-ml Falcon tubes (SPL Life Sciences Co., Ltd., Pocheon, South Korea). Each isolate (105 CFU/ml) was added to a Falcon tube and incubated for 24 h without shaking. After biofilm formation, the shrimp and mussel surfaces were removed from the Falcon tube and washed at least three times with PBS to remove planktonic bacteria, and transferred into a sterile stomacher bag containing 10 ml peptone water (PW; BD Diagnostics, Franklin Lakes, NJ, United States), and processed using a Stomacher (Bag Mixer; Interscience, Saint Nom, France) at the highest speed of 4 for 2 min to release the biofilm-forming bacteria cells from shrimp and mussel surfaces. Enumeration of each V. parahaemolyticus isolate was obtained by serial dilutions and spread onto CHROMagar Vibrio plates. After incubation at 30°C for 24 h, the resulting colonies were counted and expressed as CFU/cm2 for the biofilm populations. V. parahaemolyticus can contaminate both shrimp (Devi et al., 2009; Jun et al., 2012; Saifedden et al., 2016; Ahmed et al., 2018), and mussel (Bauer et al., 2006; Rojas et al., 2011; Jun et al., 2012; Lopatek et al., 2015). Previous studies reported that the V. parahaemolyticus isolates, collected from different sources (shrimp, crab, oysters and mussels) can generate biofilm on other different surfaces (Ahmed et al., 2018; Fang et al., 2018; Rosa et al., 2018), and it was already reported that V. parahaemolyticus can make biofilm on shrimp surface (Mizan et al., 2018). We considered shrimp as a tested surface along with a mussel though strains were isolated from mussel in this study.
Examination of Biofilms via Field Emission Scanning Electron Microscopy (FE-SEM)
The biofilm formation ability of representative isolates (ATCC27969 and NIFS29) were examined by FE-SEM. The surfaces were prepared with some modifications as described previously by Jahid et al. (2013). The surfaces were fixed at room temperature for 4 h with 2.5% glutaraldehyde. The surfaces were then serially treated with ethanol (50% for 15 min, 60% for 15 min, 70% for 15 min, 80% for 15 min, 90% for 15 min, and twice with 100% for 15 min each time) and successively dehydrated by soaking in 33, 50, 66, and 100% hexamethyldisilazane in ethanol for 15 min each time. The dehydrated surfaces were sputter coated with platinum and visualized by FE-SEM (Hitachi/Baltec, S-4700). In this study, NIFS29 was selected as a representative bacterium among all isolates to present our data. NIFS29 was selected based on the higher biofilm formation ability. However, the biofilm formation ability between NIFS28 and NIFS29 were not significantly different.
Transmission Electron Microscope (TEM) Sample Preparation and Imaging
A TEM sample was prepared with minor modifications from the previous study (Golding et al., 2016). The sample was adsorbed for 1 min to a formvar film on a carbon-coated 400-mesh copper grid. It was then washed 3 times in distilled water and negatively contrasted with 2% methylamine tungstate (Nano-W; Nanoprobes, Yaphank, NY, United States). The image was taken at 200 kV using a FEI Tecnai 20 transmission electron microscope (FEI Company, Hillsboro, OR, United States). The magnifications of 3,500× to 19,000× was considered for TEM images.
Statistical Analysis
Each experiment was performed independently in triplicate. The data were expressed as mean ± standard error (SE). Data were analyzed using Microsoft excel and Duncan’s new multiple tests with SAS software (version 9.2, SAS Institute Inc., Cary, NC, United States). P-values < 0.05 were considered significantly different.
Results
Gene Detection of V. parahaemolyticus
The distribution of specific and virulence-associated genes was represented in Table 4. The V. parahaemolyticus isolates shown positive PCR amplification to specific genes: (16S rRNA), marker (toxR), pathogenic gene (L-tdh), biofilm genes VP950 (encoding a lipoprotein-related protein), VP952, and VP962 (encoding a hypothetical protein), type VI secretion T6SS (VP1409), Type I pilus (VP1510), pathogenicity island-2 (VPaI-2), and VPaI-6 (VP1253). Our results also indicated that 87.5% (n = 8) of V. parahaemolyticus isolates harbored the complete type three secretion T3SS (VP1690), tox-RS/Old, and VPaI-6 (VP1263); 50% (n = 8) of isolates harbored the complete Type I pilus (VP1506) and type VI secretion T6SS (VP1418) genes. For trh gene, all isolates (n = 8) shown negative amplification of PCR.

Table 4. Detection of genes involved in biofilm formation as well as the pathogenicity of V. parahaemoliticus isolates.
REP-PCR Fingerprinting
The genetic relationships among the V. parahaemolyticus isolates were analyzed by REP-PCR. All the isolates demonstrated a common band at ∼ 400 bp with amplification ranging from 100 to 2,000 bp. According to the REP-PCR banding pattern, all the isolates were classified into two major clusters, cluster 1 and cluster 2 (Figure 1). Cluster 2 was divided into two sub-clusters (cluster 2a and cluster 2b) having 49.9% similarity. In cluster 2a, two main groups (Cluster 2a1 and Cluster 2a2) were detected for the V. parahaemolyticus isolates. NIFS18, NIFS29, NIFS30, ATCC27969, and ATCC17802 showed 97.4% similarity. The other isolates, NIFS25, NIFS28, NIFS26, NIFS24, and NIFS27, showed 91.7% similarity. ATCC43894 (Escherichia coli) and BB170 (Vibrio harveyi) were used as negative controls. A similarity level of 2.3% was observed between these strains (Figure 1) independently from the V. parahaemolyticus isolates.
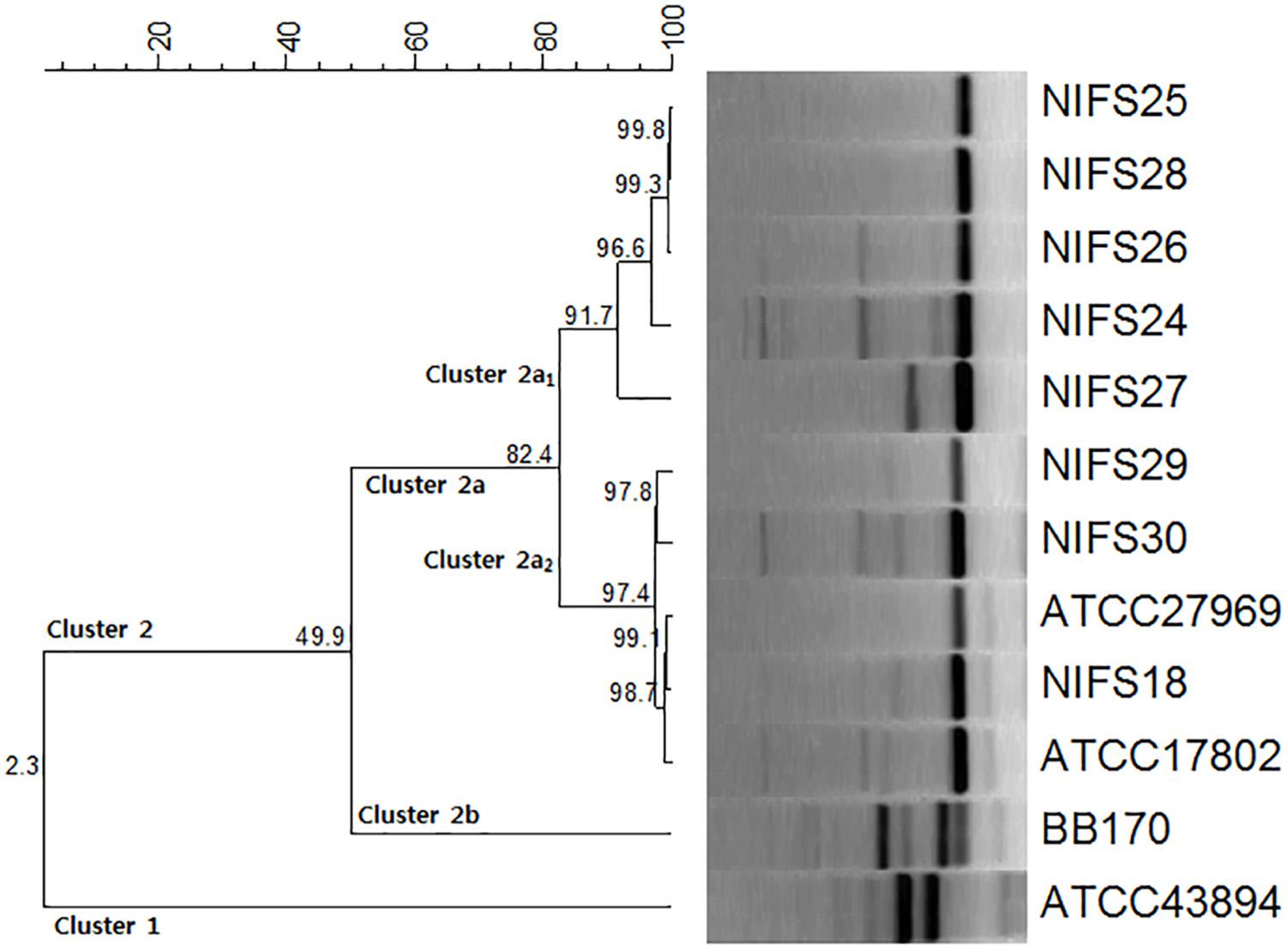
Figure 1. Repetitive extragenic palindromic sequence-based PCR (REP-PCR) results for the 10 Vibrio parahaemolyticus isolates and reference strains Escherichia coli ATCC43894 and Vibrio harveyi BB170.
Antibiotic Resistance Profile of V. parahaemolyticus Isolates
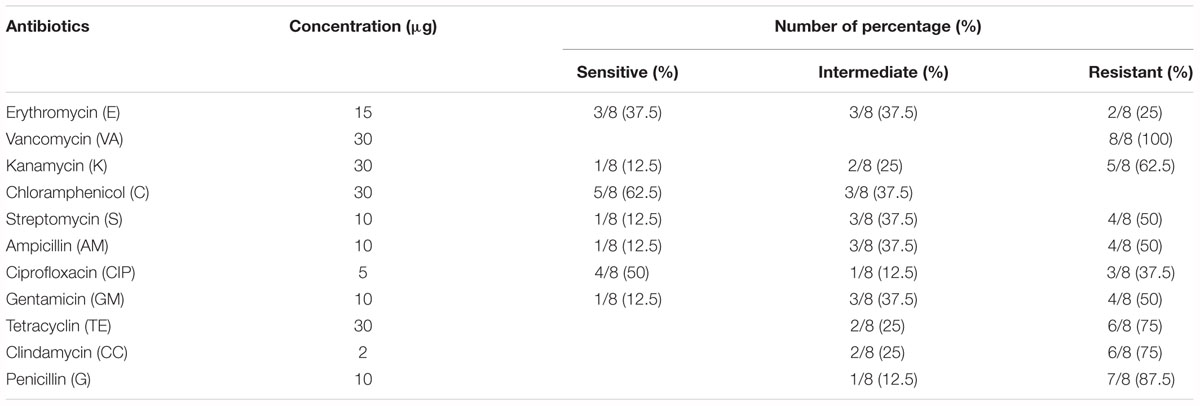
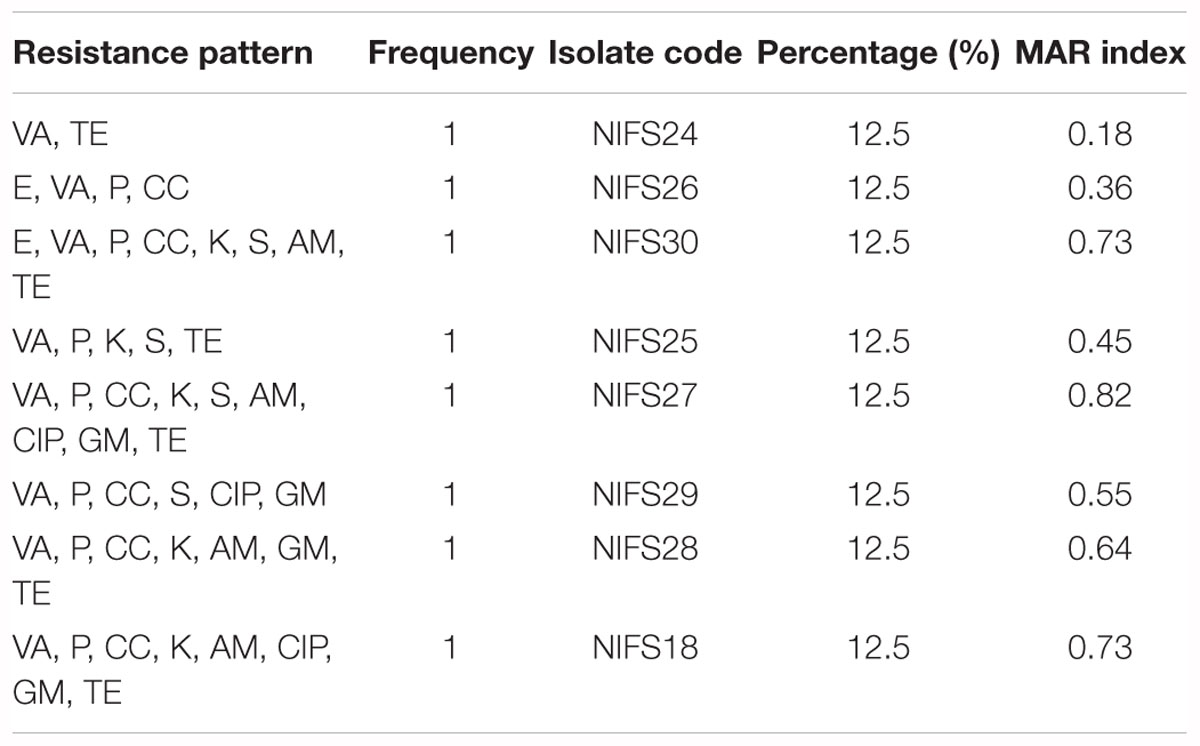
The antibiotic resistance profile of V. parahaemolyticus isolates with respect to 11 antibiotics were shown in Table 5. The majority of isolates were resistant to all the antibiotics. The isolates were highly resistant to vancomycin (100%) and penicillin (87.5%) and showed 75% resistance to clindamycin and tetracycline, and 62.5% resistance to kanamycin. The MAR index values of V. parahaemolyticus isolates were summarized in Table 6. The MAR values ranged from 0.18 to 0.82, with an average of 0.56. One (12.5%) isolate exhibited the highest MAR index value of 0.82, showing resistance to nine antibiotics.
Comparison of Biofilm Formation Ability Between Clinical and Environmental Isolates
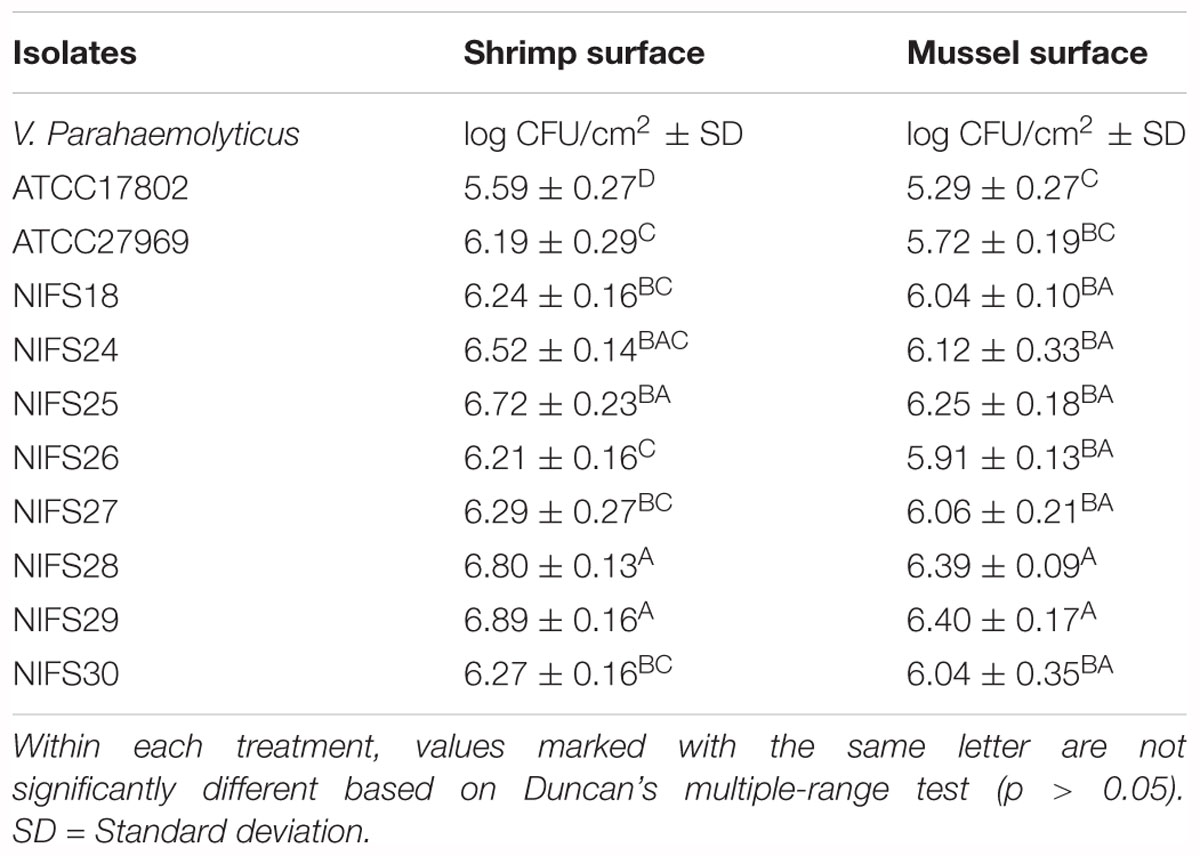
In this study, environmental isolates showed a higher biofilm formation ability than clinical isolates. For shrimp surface, NIFS25, NIFS28 and NIFS29 showed 6.72, 6.80, and 6.89 log CFU/cm2 biofilm formation, respectively (Table 7). For mussel surface, NIFS25, NIFS28 and NIFS29 also shown strong biofilm formation like 6.25, 6.39, and 6.40 log CFU/cm2, respectively (Table 7). On the other hand, clinical isolates ATCC17802 and ATCC27969 showed 5.29 and 5.72 log CFU/cm2 biofilm formation, respectively (Table 7). The biofilm-forming ability of the isolates may vary under different environmental stress and isolation source. This study indicated that environmental isolates show a great ability to contaminate shellfish.
FE-SEM
The FE-SEM analysis of clinical and environmental isolates were shown in Figure 2. The representative isolates for the clinical and environmental were ATCC27969, and NIFS29, respectively. ATCC27969 and NIFS29 were selected based on their higher biofilm formation ability. In the case of both isolates, biofilms were organized in structure with intact cell-to-cell connections. The morphology of the isolates (ATCC27969 and NIFS29) looked smooth with an intact cell membrane (Figure 2). The environmental isolate showed stronger biofilm formation on both surfaces than on the clinical isolate (Figure 2).
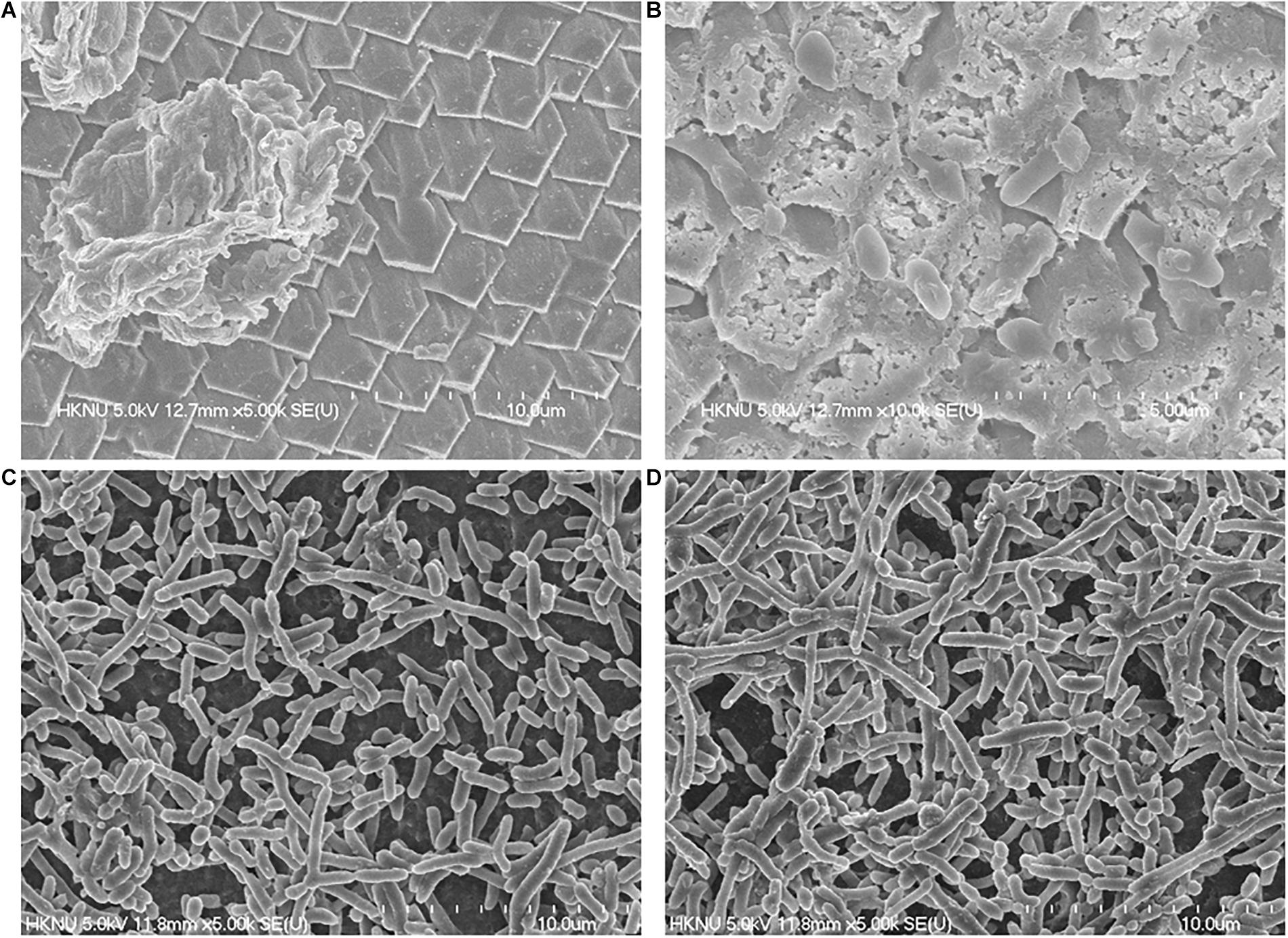
Figure 2. Representative FESEM images of V. parahaemolyticus clinical (ATCC27969) and environmental (NIFS29) isolates biofilm formation on mussel and shrimp surfaces. For mussel surface clinical isolate (A), environmental isolates (B); For shrimp surface clinical isolate (C), environmental isolates (D).
Morphological Structure Observation
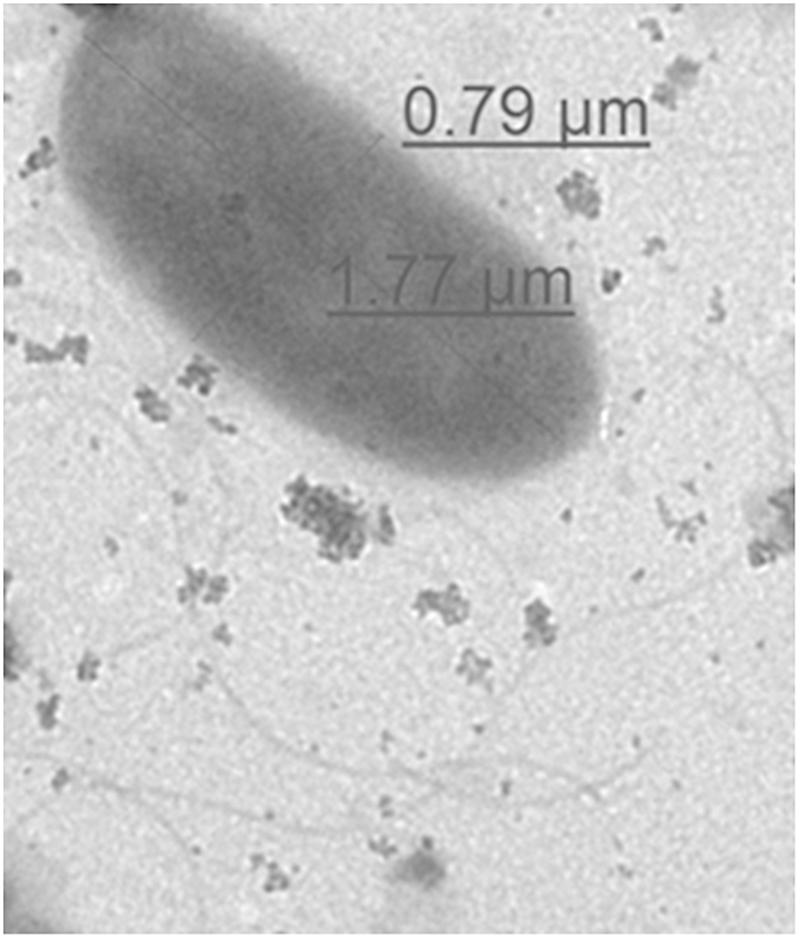
Morphological structure of representative environmental isolate (NIFS29) was observed through TEM. The electron micrographs of NIFS29 cell was displayed in Figure 3. The bacterial cell showed typical character of rod-shaped bacteria. The cell surface was smooth, and the flagella was clear. The diameter of the cell was 0.79 μm in width and 1.77 μm in length (Figure 3), this is within the range of V. parahaemolyticus standard diameter (0.5–0.8 μm in width and 1.4–2.4 μm in length)1.
Discussion
Assays based on molecular PCR are useful in detecting the toxR gene in V. parahaemolyticus isolates (Fabbro et al., 2010). Following detection on the CHROMagar Vibrio plates (data not shown), PCR assays for toxR, and genes associated with biofilm formation and pathogenicity were conducted for molecular characterization of the V. parahaemolyticus isolates. The toxR gene fragment (∼368 bp) specific to V. parahaemolyticus (Zulkifli et al., 2009) and three biofilm-associated genes were effectively amplified from all the isolates (Table 4); similar results were reported in another study (Mizan et al., 2016). It was reported that 82.6% of V. parahaemolyticus isolates were positive for PCR targeting the 16S rRNA, whereas in our study, 100% (Table 4) of V. parahaemolyticus isolates were identified as positive by PCR (Rojas et al., 2011). Our results for VPaI-2, VPaI-3, VPaI-6, Type I pilus, type III secretion systems (T3SS), and type VI secretion systems (T6SS) were consistent with those of other studies (Chao et al., 2009, 2010). The results of our study indicated that 100% of the V. parahaemolyticus isolates harbored the complete L-tdh genes (Table 4). Several studies revealed that 87.4, 93, and 86% of V. parahaemolyticus isolates carried the tdh gene (Bhoopong et al., 2007; Chen et al., 2016; Mizan et al., 2016). In the case of trh gene, all V. parahaemolyticus environmental isolates showed the negative amplification of PCR, and similar results were found in other study (Rojas et al., 2011). Recently, it was demonstrated that the virulence gene trh was absent in all V. parahaemolyticus isolates from water and mollusk (Silva et al., 2018). In another study, among 35 isolates from mussel only 4 isolates showed positive results against trh gene (Ottaviani et al., 2005). It was also reported that only 4 isolates were positive for trh virulence gene among 44 V. parahaemolyticus isolated from oysters (Kang et al., 2017). However, sometimes the presence or absence of virulence genes may depend on the differences in geographical regions, testing methodologies, and sample sources (Raghunath, 2015). For example, in a previous study, the tdh gene was detected in 20.7% of the seafood samples, from southwest coast of India by PCR after 18 h enrichment in ST broth. In the same study, it was isolated tdh carrying V. parahaemolyticus isolates from 19% of seafood samples, by colony hybridization following enrichment using ST broth (Raghunath, 2015), whereas, tdh was detected in 100% of the mussel samples from the west coast area of Korea by PCR after 24 h enrichment in LB broth in this study. It was reported that tdh gene was positive in 55 of environmental (water) samples, by MPN-PCR technique. But no tdh carrying strains were isolated by the conventional MPN-culture procedure (Alam et al., 2002).
In epidemiological research, REP-PCR is an effective and rapid typing method for the comparison and fingerprinting of V. parahaemolyticus isolates (Wong and Lin, 2001; Maluping et al., 2005). An earlier study using REP-PCR explained the intraspecific and interspecific differences between V. parahaemolyticus isolates and other strains (Jun et al., 2012). In our study, we obtained similar results using REP-PCR to compare and differentiating between intraspecific (10 V. parahaemolyticus isolates) and interspecific strains (E. coli ATCC43894, V. harveyi BB170) (Figure 1).
The environmental isolates in our study showed a high level of resistance to vancomycin, tetracycline, kanamycin, ampicillin, and penicillin (Table 5). Our findings were consistent with those of previous reports (Elexson et al., 2014a; Jiang et al., 2014; Xu et al., 2016; Kang et al., 2017; Tan et al., 2017; Ahmed et al., 2018). The high levels of multiple-antibiotic resistance property could be stated by the furthered chance to exchange genetic resistance determinants spotted on the plasmids among microorganisms, due to the extensive use of antibiotics in fishery and for the treatment of different kinds of infections (Ottaviani et al., 2013). It was demonstrated that the low MAR range (0.15) indicated low risk of contamination, whereas the high MAR range (above 0.25) indicated high risk of contamination (Chitanand et al., 2010). In this study, the higher MAR index values were 0.82, 0.73, and 0.64 (Table 6), indicating the high contamination ability of V. parahaemolyticus isolates; this agrees with the results obtained in other studies (Kang et al., 2017; Ahmed et al., 2018). However, appropriate monitoring is essential for developing a new generation of antibiotics and assuring the safety of seafood (Krumperman, 1983; Yu et al., 2016). The variation in the MAR index values may depend on sample sources (Tunung et al., 2010), geographic distribution (Lesley et al., 2011), and different testing methods (Robert-Pillot et al., 2004). It was described that the most possible source was cross-contamination from other products in the sampling location. However, cross-contamination could occur at any stage during the long processing and distribution chain, such as during pre-harvesting or post-harvest stage, contamination might occur through a contaminated container for transporting and improper handling (Chai et al., 2007). It was suggested that geographical locations and selective pressure influence the antibiotic resistance levels as well as multiple antibiotic resistance index. The high multiple antibiotic resistance value of V. parahaemolyticus isolates was 0.94, isolated from local cockles (Anadara granosa) obtained from a harvesting site in Tanjong Karang, Malaysia. The isolates were routinely grown at 35°C in LB broth with addition of 3% (w/v) NaCl (Lesley et al., 2011). In comparison with our study, the high multiple antibiotic resistance value of V. parahaemolyticus isolates was 0.82, isolated from local mussel (M. coruscus) from the west coast area of Korea, and grown at 30°C in LB broth with addition of 2% NaCl. Alternative strategies are urgently needed to overcome the continuous emergence of MDR in V. parahaemolyticus isolates in the environment (Tan et al., 2016).
Clinical strains and V. parahaemolyticus isolated from mussel possesses biofilm-forming abilities as well as pathogenic properties, and the relationship between the virulence genes detected with biofilm formation capabilities of the environmental isolates and clinical strains is variable in this study. The biofilm cell of V. parahaemolyticus has great resistant ability to antibiotics and disinfectants than planktonic cells (Song et al., 2017). The biofilm formation ability of V. parahaemolyticus increase the cells attachment ability to suspended particles, as well as shellfish (Elexson et al., 2014b). In this study, V. parahaemolyticus environmental isolates shown strong biofilm formation ability on both seafood surfaces at 30°C (Table 7). Previous study reported that V. parahaemolyticus environmental isolates produced strong biofilm on abiotic surface at 30°C (Mizan et al., 2016). The temperatures ranging 25–37°C was considered optimum for significant biofilm formation by V. parahaemolyticus isolates (Ahmed et al., 2018). There is limited study about biofilm formation on seafood. This study examined the biofilm formation ability of V. parahaemolyticus isolates on mussel surfaces for the first time. Interestingly, the result indicated that environmental isolates produced strong biofilm on mussel surface as well as shrimp surface (Table 7) than clinical isolates. Various studies described about better biofilm formation ability of environment isolates than clinical isolates (Kim B. R. et al., 2016; Qi et al., 2016). This might be due to differences in structural components such as pili, and fimbriac and adhesive surface proteins (Thompson et al., 2006; Abdallah et al., 2009). It could be also depended on various environmental conditions and bacterial adhesion properties (Wong et al., 2002; Sayen, 2014; Kim B. R. et al., 2016). Previous study reported V. parahaemolyticus biofilm formation ability on shrimp surface (Mizan et al., 2018), but no report was found on mussel surface. Therefore, further studies are needed to confirm the biofilm formation ability of environmental isolates on mussel surface. The SEM images showed the visual biofilm formation ability of both isolates on mussel (Figures 2A,B) and shrimp (Figures 2C,D) surface. Previous studies examined the SEM images of biofilm formation ability of V. parahaemolyticus on shrimp surface with different temperatures (Han et al., 2016). This study indicated that environmental isolates could produce significant biofilm on both surfaces and contaminate seafood, resulting in potential risk to consumer’s health.
Conclusion
In this study, the tested V. parahaemolyticus isolates showed 100% positive amplification to pathogenic gene L-tdh and biofilm genes with strong genetic relationship. The mussel in the west coast area of Korea could be a positive source of resistance genes that may be transmitted to humans through consumption of mussel. The present study demonstrated that V. parahaemolyticus isolates carry the markers of different virulence genes, high antibiotic resistance profile and remarkable biofilm formation ability. These properties could be helpful to influence seafood contamination ability of these isolates significantly. This could be a good source of public health hazard, especially for seafood consumers. Therefore, monitoring of V. parahaemolyticus antibiotic resistance profile and others pathogenic factors are important to protect seafood in the marine environment and improve seafood safety in the seafood industry. The strong point of this study was the comprehensive coverage of genetic relationship, virulence factors as well as biofilm formation ability on seafood. Although only 8 environmental isolates were used in this study, the collected data are expected to be useful to build strong evidence in future research through increasing the number of isolates. This study recommends additional research using V. parahaemolyticus mussel isolates from different countries in the world and making a comprehensive statement about the variation of different virulence factors among the isolates of different countries.
Author Contributions
MA, MM, HP, K-HB, NL, and SP provided assistance and guidance in throughout the research. MA wrote the manuscript. MM assisted the manuscript checking. All authors checked the manuscript and submitted final version.
Funding
This research was supported by the Chung-Ang University excellent freshman scholarship grants (2017–2018). The Basic Science Research Program and National Research Foundation of Korea (NRF) were also supported this research through funded by Ministry of Science, ICT and Future Planning (2016R1A2B4007960).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^ https://www.msdsonline.com/resources/sds-resources/free-safety-data-sheet-index/Vibrio-parahaemolyticus/
References
Abdallah, F. B., Chaieb, K., Zmantar, T., Kallel, H., and Bakhrouf, A. (2009). Adherence assays and slime production of Vibrio alginolyticus and Vibrio parahaemolyticus. Brazil. J. Microbiol. 40, 394–398. doi: 10.1590/S1517-838220090002000033
Ahmed, H. A., El Bayomi, R. M., Hussein, M. A., Khedr, M. H., Remela, E. M. A., and El-Ashram, A. M. (2018). Molecular characterization, antibiotic resistance pattern and biofilm formation of Vibrio parahaemolyticus and V. cholerae isolated from crustaceans and humans. Int. J. Food Microbiol. 274, 31–37. doi: 10.1016/j.ijfoodmicro.2018.03.013
Alam, M. J., Tomochika, K. I., Miyoshi, S. I., and Shinoda, S. (2002). Environmental investigation of potentially pathogenic Vibrio parahaemolyticus in the Seto-Inland Sea, Japan. FEMS Microbiol. Lett. 208, 83–87. doi: 10.1111/j.1574-6968.2002.tb11064.x
Al-Othrubi, S., Kqueen, C., Mirhosseini, H., Hadi, Y., and Radu, S. (2014). Antibiotic resistance of Vibrio parahaemolyticus isolated from cockles and shrimp sea food marketed in Selangor, Malaysia. Clin. Microbiol. 3, 148–154.
Bauer, A., Østensvik, Ø, Florvåg, M., Ørmen, Ø, and Rørvik, L. M. (2006). Occurrence of Vibrio parahaemolyticus, V. cholerae, and V. vulnificus in Norwegian Blue Mussels (Mytilus edulis). Appl. Environ. Microbiol. 72, 3058–3061. doi: 10.1128/AEM.72.4.3058-3061.2006
Bhoopong, P., Palittapongarnpim, P., Pomwised, R., Kiatkittipong, A., Kamruzzaman, M., Nakaguchi, Y., et al. (2007). Variability of properties of Vibrio parahaemolyticus strains isolated from individual patients. J. Clin. Microbiol. 45, 1544–1550. doi: 10.1128/JCM.02371-06
Broberg, C. A., Calder, T. J., and Orth, K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 13, 992–1001. doi: 10.1016/j.micinf.2011.06.013
Brooun, A., Liu, S., and Lewis, K. (2000). A dose-response study of antibiotic resistance inPseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 44, 640–646. doi: 10.1128/AAC.44.3.640-646.2000
Caballero-Miguez, G., Garza-Gil, M. D., and Varela-Lafuente, M. M. (2012). Legal change, property rights system and institutional stability: the case of the floating raft culture in the Galician mussel sector. Ocean Coast. Manag. 55, 84–93. doi: 10.1016/j.ocecoaman.2011.10.002
Cabello, F. C., Godfrey, H. P., Tomova, A., Ivanova, L., Dölz, H., Millanao, A., et al. (2013). Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 15, 1917–1942. doi: 10.1111/1462-2920.12134
Chae, M., Cheney, D., and Su, Y. C. (2009). Temperature effects on the depuration of Vibrio parahaemolyticus and Vibrio vulnificus from the American oyster (Crassostrea virginica). J. Food Sci. 74, M62–M66. doi: 10.1111/j.1750-3841.2008.01031.x
Chai, L. C., Robin, T., Ragavan, U. M., Gunsalam, J. W., Bakar, F. A., Ghazali, F. M., et al. (2007). Thermophilic Campylobacter spp. in salad vegetables in Malaysia. Int. J. Food Microbiol. 117, 106–111. doi: 10.1016/j.ijfoodmicro.2007.02.014
Chao, G., Jiao, X., Zhou, X., Wang, F., Yang, Z., Huang, J., et al. (2010). Distribution of genes encoding four pathogenicity islands (VPaIs), T6SS, biofilm, and type I pilus in food and clinical strains of Vibrio parahaemolyticus in China. Foodborne Pathog. Dis. 7, 649–658. doi: 10.1089/fpd.2009.0441
Chao, G., Jiao, X., Zhou, X., Yang, Z., Pan, Z., Huang, J., et al. (2009). Systematic functional pandemic strain–specific genes, three genomic islands, two t3sss in foodborne, and clinical Vibrio parahaemolyticus isolates in China. Foodborne Pathog. Dis. 6, 689–698. doi: 10.1089/fpd.2009.0274
Chen, Y., Chen, X., Yu, F., Wu, M., Wang, R., Zheng, S., et al. (2016). Serology, virulence, antimicrobial susceptibility and molecular characteristics of clinical Vibrio parahaemolyticus strains circulating in southeastern China from 2009 to 2013. Clin. Microbiol. Infect. 22:258.e9-16. doi: 10.1016/j.cmi.2015.11.003
Chitanand, M., Kadam, T., Gyananath, G., Totewad, N., and Balhal, D. (2010). Multiple antibiotic resistance indexing of coliforms to identify high risk contamination sites in aquatic environment. Indian J. Microbiol. 50, 216–220. doi: 10.1007/s12088-010-0042-9
Cisneros-Montemayor, A. M., Pauly, D., Weatherdon, L. V., and Ota, Y. (2016). A global estimate of seafood consumption by coastal indigenous peoples. PLoS One 11:e0166681. doi: 10.1371/journal.pone.0166681
CLSI (2010). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria; Approved Guideline, 2nd Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Costas-Rodríguez, M., Lavilla, I., and Bendicho, C. (2010). Classification of cultivated mussels from Galicia (Northwest Spain) with european protected designation of Origin using trace element fingerprint and chemometric analysis. Anal. Chim. Acta 664, 121–128. doi: 10.1016/j.aca.2010.03.003
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Daniels, N. A., and Shafaie, A. (2000). A review of pathogenic Vibrio infections for clinicians. Infect. Med. 17, 665–685.
Devi, R., Surendran, P. K., and Chakraborty, K. (2009). Antibiotic resistance and plasmid profiling of Vibrio parahaemolyticus isolated from shrimp farms along the southwest coast of India. World J. Microbiol. Biotechnol. 25, 2005–2012. doi: 10.1007/s11274-009-0101-8
Donlan, R. M. (2002). Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8, 881–890. doi: 10.3201/eid0809.020063
Duran, G. M., and Marshall, D. L. (2005). Ready-to-eat shrimp as an international vehicle of antibiotic-resistant bacteria. J. Food Prot. 68, 2395–2401. doi: 10.4315/0362-028X-68.11.2395
Elexson, N., Afsah-Hejri, L., Rukayadi, Y., Soopna, P., Lee, H., Zainazor, T. T., et al. (2014a). Effect of detergents as antibacterial agents on biofilm of antibiotics-resistant Vibrio parahaemolyticus isolates. Food Control 35, 378–385. doi: 10.1016/j.foodcont.2013.07.020
Elexson, N., Yaya, R., Nor, A., Kantilal, H., Ubong, A., Nishibuchi, M., et al. (2014b). Biofilm assessment of Vibrio parahaemolyticus from seafood using random amplified polymorphism DNA-PCR. Int. Food Res. J. 21, 59–65.
Fabbro, C., Cataletto, B., and Del Negro, P. (2010). Detection of pathogenic Vibrio parahaemolyticus through biochemical and molecular-based methodologies in coastal waters of the Gulf of Trieste (North Adriatic Sea). FEMS Microbiol. Lett. 307, 158–164. doi: 10.1111/j.1574-6968.2010.01969.x
Fang, Z., Sun, D., Li, C., Sun, L., Wang, Y., Guo, M., et al. (2018). Regulatory effects of Shewanella putrefaciens isolated from shrimp Penaeus orientalis on the virulence factors of Vibrio parahaemolyticus and evaluation of the role of quorum sensing in virulence factors regulation. FEMS Microbiol. Ecol. 94:fiy097. doi: 10.1093/femsec/fiy097
FDA BAM (2004). Bacteriological Analytical Manual Chapter 9: Vibrio. Washington DC: Food & Drug Administration.
Ferreira, M., Lago, J., Vieites, J. M., and Cabado, A. G. (2014). “World production of bivalve mollusks and socioeconomic facts related to the impact of marine biotoxins,” in Seafood and Freshwater Toxins, ed. L. M. Botana (Boca Raton, FL: CRC Press), 291–311.
Flemming, H.-C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
García-Hernández, Y., Pérez-Sánchez, T., Boucourt, R., Balcázar, J. L., Nicoli, J. R., Moreira-Silva, J., et al. (2016). Isolation, characterization and evaluation of probiotic lactic acid bacteria for potential use in animal production. Res. Vet. Sci. 108, 125–132. doi: 10.1016/j.rvsc.2016.08.009
Garrido-Maestu, A., Lozano-León, A., Rodríguez-Souto, R. R., Vieites-Maneiro, R., Chapela, M.-J., and Cabado, A. G. (2016). Presence of pathogenic Vibrio species in fresh mussels harvested in the southern Rias of Galicia (NW Spain). Food Control 59, 759–765. doi: 10.1016/j.foodcont.2015.06.054
Golding, C. G., Lamboo, L. L., Beniac, D. R., and Booth, T. F. (2016). The scanning electron microscope in microbiology and diagnosis of infectious disease. Sci. Rep. 6:26516. doi: 10.1038/srep26516
Guglielmetti, E., Korhonen, J. M., Heikkinen, J., Morelli, L., and Von Wright, A. (2009). Transfer of plasmid-mediated resistance to tetracycline in pathogenic bacteria from fish and aquaculture environments. FEMS Microbiol. Lett. 293, 28–34. doi: 10.1111/j.1574-6968.2009.01512.x
Hall-Stoodley, L., Costerton, J. W., and Stoodley, P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108. doi: 10.1038/nrmicro821
Han, A., Yoon, Y. J., and Kim, J. W. (2012). Antibiotic resistance and plasmid profile of Vibrio parahaemolyticus strains isolated from kyunggi-incheon coastal area. Korean J. Microbiol. 48, 22–28. doi: 10.7845/kjm.2012.48.1.022
Han, N., Mizan, M. F. R., Jahid, I. K., and Ha, S. D. (2016). Biofilm formation by Vibrio parahaemolyticus on food and food contact surfaces increases with rise in temperature. Food Control 70, 161–166. doi: 10.1016/j.foodcont.2016.05.054
Hellberg, R. S., Dewitt, C. A. M., and Morrissey, M. T. (2012). Risk-benefit analysis of seafood consumption: a review. Compr. Rev.Food Sci. Food Safety 11, 490–517. doi: 10.1111/j.1541-4337.2012.00200.x
Hongping, W., Jilun, Z., Ting, J., Yixi, B., and Xiaoming, Z. (2011). Insufficiency of the Kanagawa hemolytic test for detecting pathogenic Vibrio parahaemolyticus in Shanghai, China. Diagn. Microbiol. Infect. Dis. 69, 7–11. doi: 10.1016/j.diagmicrobio.2010.08.016
Jahan, S. (2012). “Epidemiology of foodborne illness,” in Scientific, Health and Social Aspects of the Food Industry, ed. B. Valdez (London: Intech Open).
Jahid, I. K., Lee, N. Y., Kim, A., and Ha, S. D. (2013). Influence of glucose concentrations on biofilm formation, motility, exoprotease production, and quorum sensing in Aeromonas hydrophila. J. Food Prot. 76, 239–247. doi: 10.4315/0362-028X.JFP-12-321
Jahid, I. K., Mizan, M. F. R., Myoung, J., and Ha, S. D. (2019). Aeromonas hydrophila biofilm, exoprotease, and quorum sensing responses to co-cultivation with diverse foodborne pathogens and food spoilage bacteria on crab surfaces. Biofouling doi: 10.1080/08927014.2018.1519069 [Epub ahead of print].
Jiang, Y., Chu, Y., Xie, G., Li, F., Wang, L., Huang, J., et al. (2019). Antimicrobial resistance, virulence and genetic relationship of Vibrio parahaemolyticus in seafood from coasts of Bohai Sea and Yellow Sea, China. Int. J. Food Microbiol. 290, 116–124. doi: 10.1016/j.ijfoodmicro.2018.10.005
Jiang, Y., Yao, L., Li, F., Tan, Z., Zhai, Y., and Wang, L. (2014). Characterization of antimicrobial resistance of Vibrio parahaemolyticus from cultured sea cucumbers (Apostichopus japonicas). Lett. Appl. Microbiol. 59, 147–154. doi: 10.1111/lam.12258
Jun, J. W., Kim, J. H., Choresca, C. H. Jr., Shin, S. P., Han, J. E., Han, S. Y., et al. (2012). Isolation, molecular characterization, and antibiotic susceptibility of Vibrio parahaemolyticus in Korean seafood. Foodborne Pathog. Dis. 9, 224–231. doi: 10.1089/fpd.2011.1018
Kadam, S. R., Den Besten, H. M., Van Der Veen, S., Zwietering, M. H., Moezelaar, R., and Abee, T. (2013). Diversity assessment of Listeria monocytogenes biofilm formation: impact of growth condition, serotype and strain origin. Int. J. Food Microbiol. 165, 259–264. doi: 10.1016/j.ijfoodmicro.2013.05.025
Kang, C. H., Shin, Y., Jang, S., Yu, H., Kim, S., An, S., et al. (2017). Characterization of Vibrio parahaemolyticus isolated from oysters in Korea: resistance to various antibiotics and prevalence of virulence genes. Mar. Pollut. Bull. 118, 261–266. doi: 10.1016/j.marpolbul.2017.02.070
Kim, B. R., Bae, Y. M., and Lee, S. Y. (2016). Effect of environmental conditions on biofilm formation and related characteristics of Staphylococcus aureus. J. Food Safety 36, 412–422. doi: 10.1111/jfs.12263
Kim, S., An, S., Park, B., Oh, E. G., Song, K. C., Kim, J. W., et al. (2016). Virulence factors and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from the oyster Crassostrea gigas. Korean J. Fish. Aquat. Sci. 49, 116–123. doi: 10.5657/KFAS.2016.0116
Kim, T. O., Eum, I. S., Jo, S. M., Seo, J. K., Lee, N. Y., Lim, S. Y., et al. (2014). Antimicrobial-Resistance Profiles and Virulence Genes of Vibrio parahaemolyticus Isolated from Seawater in Wando Area. Korean Soc. Fish. Aquat. Sci. 47, 220–226. doi: 10.5657/KFAS.2014.0220
Kim, Y. B., Okuda, J., Matsumoto, C., Takahashi, N., Hashimoto, S., and Nishibuchi, M. (1999). Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J. Clin. Microbiol. 37, 1173–1177.
Kleter, G. A., Groot, M., Poelman, M., Kok, E., and Marvin, H. (2009). Timely awareness and prevention of emerging chemical and biochemical risks in foods: proposal for a strategy based on experience with recent cases. Food Chem. Toxicol. 47, 992–1008. doi: 10.1016/j.fct.2008.08.021
Krumperman, P. H. (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46, 165–170.
Lee, L. H., Ab Mutalib, N. S., Law, J. W. F., Wong, S. H., and Letchumanan, V. (2018). Discovery on antibiotic resistance patterns of Vibrio parahaemolyticus in Selangor reveals carbapenemase producing Vibrio parahaemolyticus in marine and freshwater fish. Front. Microbiol. 9:2513. doi: 10.3389/fmicb.2018.02513
Lesley, M., Velnetti, L., Cheah, Y., Son, R., Kasing, A., Samuel, L., et al. (2011). Antibiotic resistance and plasmid profiling of Vibrio parahaemolyticus isolated from cockles (Anadara granosa) at Tanjung Karang, Kuala Selangor. Int. Food Res. J. 18, 1183–1188.
Letchumanan, V., Chan, K. G., and Lee, L. H. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Letchumanan, V., Yin, W.-F., Lee, L.-H., and Chan, K.-G. (2015). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6:33. doi: 10.3389/fmicb.2015.00033
Lopatek, M., Wieczorek, K., and Osek, J. (2015). Prevalence and antimicrobial resistance of Vibrio parahaemolyticus isolated from raw shellfish in Poland. J. Food Prot. 78, 1029–1033. doi: 10.4315/0362-028X.JFP-14-437
Lopatek, M., Wieczorek, K., and Osek, J. (2018). Characterization and genetic diversity of Vibrio parahaemolyticus isolated from seafoods. Appl. Environ. Microbiol. 84:AEM-00537-18.
Lozano-León, A., Torres, J., Osorio, C. R., and MartıìNez-Urtaza, J. (2003). Identification of tdh-positive Vibrio parahaemolyticus from an outbreak associated with raw oyster consumption in Spain. FEMS Microbiol. Lett. 226, 281–284. doi: 10.1016/S0378-1097(03)00604-9
Ma, Z., Ginn, A., Kang, M., Galvao, K., and Jeong, K. C. (2018). Genomic and virulence characterization of intrauterine pathogenic Escherichia coli with multi-drug resistance isolated from cow uteri with metritis. Front. Microbiol. 9:3137. doi: 10.3389/fmicb.2018.03137
Machado, H., and Gram, L. (2015). The fur gene as a new phylogenetic marker for Vibrionaceae species identification. Appl. Environ. Microbiol. 81, 2745–2752. doi: 10.1128/AEM.00058-15
Maluping, R., Ravelo, C., Lavilla-Pitogo, C., Krovacek, K., and Romalde, J. (2005). Molecular typing of Vibrio parahaemolyticus strains isolated from the Philippines by PCR-based methods. J. Appl. Microbiol. 99, 383–391. doi: 10.1111/j.1365-2672.2005.02571.x
Mannas, H., Mimouni, R., Chaouqy, N., Hamadi, F., and Martinez-Urtaza, J. (2014). Occurrence of Vibrio and Salmonella species in mussels (Mytilus galloprovincialis) collected along the Moroccan Atlantic coast. Springerplus 3:265. doi: 10.1186/2193-1801-3-265
Martinez-Urtaza, J., Lozano-Leon, A., Depaola, A., Ishibashi, M., Shimada, K., Nishibuchi, M., et al. (2004). Characterization of pathogenic Vibrio parahaemolyticus isolates from clinical sources in Spain and comparison with Asian and North American pandemic isolates. J. Clin. Microbiol. 42, 4672–4678. doi: 10.1128/JCM.42.10.4672-4678.2004
Martinez-Urtaza, J., Lozano-Leon, A., Varela-Pet, J., Trinanes, J., Pazos, Y., and Garcia-Martin, O. (2008). Environmental determinants of the occurrence and distribution of Vibrio parahaemolyticus in the rias of Galicia, Spain. Appl. Environ. Microbiol. 74, 265–274. doi: 10.1128/AEM.01307-07
McLain, J. E., Cytryn, E., Durso, L. M., and Young, S. (2016). Culture-based methods for detection of antibiotic resistance in agroecosystems: advantages, challenges, and gaps in knowledge. J. Environ. Qual. 45, 432–440. doi: 10.2134/jeq2015.06.0317
Miguez, G. C., Gil, M. D. G., and Lafuente, M. M. V. (2009). The institutional foundations of economic performance of mussel production: the Spanish case of the Galician floating raft culture. Mar. Policy 33, 288–296. doi: 10.1016/j.marpol.2008.07.008
Mizan, M. F., Jahid, I. K., Kim, M., Lee, K. H., Kim, T. J., and Ha, S. D. (2016). Variability in biofilm formation correlates with hydrophobicity and quorum sensing among Vibrio parahaemolyticus isolates from food contact surfaces and the distribution of the genes involved in biofilm formation. Biofouling 32, 497–509. doi: 10.1080/08927014.2016.1149571
Mizan, M. F. R., Ashrafudoulla, M., Sadekuzzaman, M., Kang, I., and Ha, S.-D. (2018). Effects of NaCl, glucose, and their combinations on biofilm formation on black tiger shrimp (Penaeus monodon) surfaces by Vibrio parahaemolyticus. Food Control 89, 203–209. doi: 10.1016/j.foodcont.2017.12.004
Mizan, M. F. R., Bang, H.-J., Sadekuzzaman, M., Lee, N., Kim, T.-J., and Ha, S.-D. (2017). Molecular characteristics, biofilm-forming abilities, and quorum sensing molecules in Vibrio parahaemolyticus strains isolated from marine and clinical environments in Korea. Biofouling 33, 369–378. doi: 10.1080/08927014.2017.1316840
Mizan, M. F. R., Jahid, I. K., and Ha, S.-D. (2015). Microbial biofilms in seafood: a food-hygiene challenge. Food Microbiol. 49, 41–55. doi: 10.1016/j.fm.2015.01.009
Mohammad, A., Hashim, J., Gunasalam, J., and Radu, S. (2005). Microbiological Risk Assessment: Risk Assessment of Vibrio parahaemolyticus in Black Tiger Prawn (Penaeus monodon). Technical Report. Putrajaya: Ministry of Health Malaysia.
NCCLS (2003). Performance Standards for Antimicrobial Disc Susceptibility Tests: Approved Standard. Pennsylvania: National Committee for Clinical Laboratory Standard of Antimicrobial Susceptibility.
Nishibuchi, M., and Kaper, J. B. (1995). Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect. Immun. 63:2093.
Odeyemi, O., and Stratev, D. (2016). Occurrence of antimicrobial resistant or pathogenic Vibrio parahaemolyticus in seafood. A mini review. Revue de Médecine Vétérinaire 67, 93–98.
Okura, M., Osawa, R., Iguchi, A., Arakawa, E., Terajima, J., and Watanabe, H. (2003). Genotypic analyses of Vibrio parahaemolyticus and development of a pandemic group-specific multiplex PCR assay. J. Clin. Microbiol. 41, 4676–4682. doi: 10.1128/JCM.41.10.4676-4682.2003
Ottaviani, D., Leoni, F., Talevi, G., Masini, L., Santarelli, S., Rocchegiani, E., et al. (2013). Extensive investigation of antimicrobial resistance in Vibrio parahaemolyticus from shellfish and clinical sources, Italy. Int. J. Antimicrob. Agents 42, 191–193. doi: 10.1016/j.ijantimicag.2013.05.003
Ottaviani, D., Santarelli, S., Bacchiocchi, S., Masini, L., Ghittino, C., and Bacchiocchi, I. (2005). Presence of pathogenic Vibrio parahaemolyticus strains in mussels from the Adriatic Sea, Italy. Food Microbiol. 22, 585–590. doi: 10.1016/j.fm.2005.01.005
Park, K., Mok, J. S., Kwon, J. Y., Ryu, A. R., Kim, S. H., and Lee, H. J. (2018). Food-borne outbreaks, distributions, virulence, and antibiotic resistance profiles of Vibrio parahaemolyticus in Korea from 2003 to 2016: a review. Fish. Aquat. Sci. 21:3. doi: 10.1186/s41240-018-0081-4
Peng, F., Jiang, D., Ruan, H., Liu, H., and Zhou, L. (2010). Pathogenic investigation on a food poisoning induced by Vibrio parahaemolyticus. Prev. Med. Trib. 16, 746–747.
Qi, L., Li, H., Zhang, C., Liang, B., Li, J., Wang, L., et al. (2016). Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front. Microbiol. 7:483. doi: 10.3389/fmicb.2016.00483
Raghunath, P. (2015). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 5:805. doi: 10.3389/fmicb.2014.00805
Reyhanath, P. V., and Kutty, R. (2014). Incidence of multidrug resistant Vibrio parahaemolyticus isolated from Ponnani, South India. Iran. J. Microbiol. 6:60.
Robert-Pillot, A., Guénolé, A., Lesne, J., Delesmont, R., Fournier, J.-M., and Quilici, M.-L. (2004). Occurrence of the tdh and trh genes in Vibrio parahaemolyticus isolates from waters and raw shellfish collected in two French coastal areas and from seafood imported into France. Int. J. Food Microbiol. 91, 319–325. doi: 10.1016/j.ijfoodmicro.2003.07.006
Rodriguez-Castro, A., Ansede-Bermejo, J., Blanco-Abad, V., Varela-Pet, J., Garcia-Martin, O., and Martinez-Urtaza, J. (2010). Prevalence and genetic diversity of pathogenic populations of Vibrio parahaemolyticus in coastal waters of Galicia, Spain. Environ. Microbiol. Rep. 2, 58–66. doi: 10.1111/j.1758-2229.2009.00064.x
Rojas, M. V. R., Matté, M. H., Dropa, M., Silva, M. L. D., and Matté, G. R. (2011). Characterization of Vibrio parahaemolyticus isolated from oysters and mussels in São Paulo, Brazil. Revista do Instituto de Medicina Tropical de São Paulo 53, 201–205. doi: 10.1590/S0036-46652011000400005
Rosa, J. V. D., Conceição, N. V. D., Conceição, R. D. C. D. S., and Timm, C. D. (2018). Biofilm formation by Vibrio parahaemolyticus on different surfaces and its resistance to sodium hypochlorite. Ciência Rural 48:e20180612. doi: 10.1590/0103-8478cr20180612
Saifedden, G., Farinazleen, G., Nor-Khaizura, A., Kayali, A. Y., Nakaguchi, Y., Nishibuchi, M., et al. (2016). Antibiotic Susceptibility profile of Vibrio parahaemolyticus isolated from shrimp in Selangor, Malaysia. Int. Food Res. J. 23, 2732–2736.
Salomon, D., Gonzalez, H., Updegraff, B. L., and Orth, K. (2013). Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS One 8:e61086. doi: 10.1371/journal.pone.0061086
Sani, N. A., Ariyawansa, S., Babji, A. S., and Hashim, J. K. (2013). The risk assessment of Vibrio parahaemolyticus in cooked black tiger shrimps (Penaeus monodon) in Malaysia. Food Control 31, 546–552. doi: 10.1016/j.foodcont.2012.10.018
Sayen, S. A. (2014). Biofilm Control and Antimicrobial Agents, 1st Edn. Cambridge, MA: Apple Academic Press. doi: 10.1201/b16593
Shaw, K. S., Goldstein, R. E. R., He, X., Jacobs, J. M., Crump, B. C., and Sapkota, A. R. (2014). Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of chesapeake bay and maryland coastal bays. PLoS One 9:e89616. doi: 10.1371/journal.pone.0089616
Silva, I. P., de Souza Carneiro, C., Saraiva, M. A. F., de Oliveira, T. A. S., de Sousa, O. V., and Evangelista-Barreto, N. S. (2018). Antimicrobial resistance and potential virulence of Vibrio parahaemolyticus isolated from water and bivalve mollusks from Bahia, Brazil. Mar. Pollut. Bull. 131, 757–762. doi: 10.1016/j.marpolbul.2018.05.007
Son, K. T., Oh, E. G., Lee, T. S., Lee, H. J., Kim, P. H., and Kim, J. H. (2005). Antimicrobial susceptibility of Vibrio parahaemolyticus and Vibrio alginolyticus from fish farms on the southern coast of Korea. Korean J. Fish. Aquat. Sci. 38, 365–371.
Song, X., Ma, Y., Fu, J., Zhao, A., Guo, Z., Malakar, P. K., et al. (2017). Effect of temperature on pathogenic and non-pathogenic Vibrio parahaemolyticus bio-film formation. Food Control 73, 485–491. doi: 10.1016/j.foodcont.2016.08.041
Su, Y.-C., and Liu, C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24, 549–558. doi: 10.1016/j.fm.2007.01.005
Sudha, S., Mridula, C., Silvester, R., and Hatha, A. (2014). Prevalence and antibiotic resistance of pathogenic Vibrios in shellfishes from Cochin market. Indian J. Mar. Sci. 43, 815–824.
Sumner, J. (2011). Food Safety Risks Associated with Prawns Consumed in Australia. Available at: http://australianwildprawns.com.au/wp-content/uploads/2017/02/2009-787-Food-safety-risks-for-prawns.pdf
Szuster, B. (2006). Coastal Shrimp Farming in Thailand: Searching for Sustainability. Environment and Livelihoods in Tropical Coastal Zones: Managing Agriculture-Fishery-Aquaculture Conflicts. Wallingford: CAB International, 86–97. doi: 10.1079/9781845931070.0086
Tan, C. W., Malcolm, T. T., Kuan, C. H., Thung, T. Y., Chang, W. S., Loo, Y. Y., et al. (2017). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from short mackerels (Rastrelliger brachysoma) in Malaysia. Front. Microbiol. 8:1087. doi: 10.3389/fmicb.2017.01087
Tan, L. T.-H., Chan, K.-G., Lee, L.-H., and Goh, B.-H. (2016). Streptomyces bacteria as potential probiotics in aquaculture. Front. Microbiol. 7:79. doi: 10.3389/fmicb.2016.00079
Temmerman, R., Pot, B., Huys, G., and Swings, J. (2003). Identification and antibiotic susceptibility of bacterial isolates from probiotic products. Int. J. Food Microbiol. 81, 1–10. doi: 10.1016/S0168-1605(02)00162-9
Thompson, F. L., Klose, K. E., and Group, A. (2006). Vibrio 2005: the first international conference on the biology of vibrios. J. Bacteriol. 188, 4592–4596. doi: 10.1128/JB.00141-06
Tunung, R., Margaret, S., Jeyaletchumi, P., Chai, L., Tuan Zainazor, T., Ghazali, F., et al. (2010). Prevalence and quantification of Vibrio parahaemolyticus in raw salad vegetables at retail level. J. Microbiol. Biotechnol. 20, 391–396.
Vandeplassche, E., Coenye, T., and Crabbé, A. (2017). Developing selective media for quantification of multispecies biofilms following antibiotic treatment. PLoS One 12:e0187540. doi: 10.1371/journal.pone.0187540
Wong, H.-C., Chung, Y.-C., and Yu, J.-A. (2002). Attachment and inactivation of Vibrio parahaemolyticus on stainless steel and glass surface. Food Microbiol. 19, 341–350. doi: 10.1006/fmic.2002.0478
Wong, H.-C., and Lin, C.-H. (2001). Evaluation of typing of Vibrio parahaemolyticus by three PCR methods using specific primers. J. Clin. Microbiol. 39, 4233–4240. doi: 10.1128/JCM.39.12.4233-4240.2001
Xie, T., Wu, Q., Zhang, J., Xu, X., and Cheng, J. (2017). Comparison of Vibrio parahaemolyticus isolates from aquatic products and clinical by antibiotic susceptibility, virulence, and molecular characterisation. Food Control 71, 315–321. doi: 10.1016/j.foodcont.2016.06.046
Xu, X., Cheng, J., Wu, Q., Zhang, J., and Xie, T. (2016). Prevalence, characterization, and antibiotic susceptibility of Vibrio parahaemolyticus isolated from retail aquatic products in North China. BMC Microbiol. 16:32. doi: 10.1186/s12866-016-0650-6
Yang, J. H., Mok, J. S., Jung, Y. J., Lee, K. J., Kwon, J. Y., Park, K., et al. (2017). Distribution and antimicrobial susceptibility of Vibrio species associated with zooplankton in coastal area of Korea. Mar. Pollut. Bull. 125, 39–44. doi: 10.1016/j.marpolbul.2017.07.054
Yang, Y., Xie, J., Li, H., Tan, S., Chen, Y., and Yu, H. (2017). Prevalence, antibiotic susceptibility and diversity of Vibrio parahaemolyticus isolates in seafood from South China. Front. Microbiol. 8:2566. doi: 10.3389/fmicb.2017.02566
Yano, Y., Hamano, K., Satomi, M., Tsutsui, I., Ban, M., and Aue-Umneoy, D. (2014). Prevalence and antimicrobial susceptibility of Vibrio species related to food safety isolated from shrimp cultured at inland ponds in Thailand. Food control 38, 30–36. doi: 10.1016/j.foodcont.2013.09.019
Yoon, K., Min, K., Jung, Y., Kwon, K., Lee, J., and Oh, S. (2008). A model of the effect of temperature on the growth of pathogenic and nonpathogenic Vibrio parahaemolyticus isolated from oysters in Korea. Food Microbiol. 25, 635–641. doi: 10.1016/j.fm.2008.04.007
Yu, Q., Niu, M., Yu, M., Liu, Y., Wang, D., and Shi, X. (2016). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shellfish in Shanghai. Food control 60, 263–268. doi: 10.1016/j.foodcont.2015.08.005
Zhang, X.-J., Yan, B.-L., Bai, X.-S., Bi, K.-R., Gao, H., and Qin, G.-M. (2014). Isolation and characterization of Vibrio parahaemolyticus and Vibrio rotiferianus associated with mass mortality of Chinese shrimp (Fenneropenaeus chinensis). J. Shellfish Res. 33, 61–68. doi: 10.2983/035.033.0108
Keywords: Vibrio parahaemolyticus, seafood, genetic relationship, antibiotic resistance, biofilm
Citation: Ashrafudoulla M, Mizan MFR, Park H, Byun K-H, Lee N, Park SH and Ha S-D (2019) Genetic Relationship, Virulence Factors, Drug Resistance Profile and Biofilm Formation Ability of Vibrio parahaemolyticus Isolated From Mussel. Front. Microbiol. 10:513. doi: 10.3389/fmicb.2019.00513
Received: 02 January 2019; Accepted: 27 February 2019;
Published: 20 March 2019.
Edited by:
Kwangcheol Casey Jeong, University of Florida, United StatesReviewed by:
Sunae Kim, Ewha Womans University, South KoreaLearn-Han Lee, Monash University Malaysia, Malaysia
Copyright © 2019 Ashrafudoulla, Mizan, Park, Byun, Lee, Park and Ha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang-Do Ha, c2FuZ2RvaGFAY2F1LmFjLmty
†These authors have contributed equally to this work
 Md. Ashrafudoulla
Md. Ashrafudoulla Md. Furkanur Rahaman Mizan
Md. Furkanur Rahaman Mizan Heedae Park
Heedae Park Kye-Hwan Byun
Kye-Hwan Byun Nari Lee
Nari Lee Si Hong Park
Si Hong Park Sang-Do Ha
Sang-Do Ha