- Department of Biology, University of Pennsylvania, Philadelphia, PA, United States
Type IV pili are evolutionarily conserved cell surface filaments that promote surface adhesion and cell aggregation providing bacteria and archaea protection from a variety of stress conditions. In fact, prokaryotic genomes frequently contain several copies of the core biosynthesis genes, pilB and pilC, encoding an ATPase and membrane anchor, respectively. Recent phylogenetic analyses suggest that in haloarchaea, a subset of pilB-C paralogs, such as the Haloferax volcanii pilB1-C1, were gained via horizontal transfer from the crenarchaea, while the co-regulated type IV pilus subunits, the pilins, evolved by duplication, followed by diversification of the ancestral euryarchaeal pilins. Here, we report the identification of an H. volcanii pilB1 transposon mutant that exhibits an adhesion defect in defined media. A similar defect observed in an H. volcanii ∆pilB1-C1 strain can be rescued by expressing pilB1-C1 in trans. However, these proteins cannot rescue the severe adhesion defect of a previously reported ∆pilB3-C3 strain. Conversely, pilB3-C3, which are not predicted to have been laterally transferred, expressed in trans can rescue the adhesion defect of a ∆pilB1-C1 strain. This cross-complementation supports the proposed hybrid origin of the operon containing pilB1-C1 and shows that at least certain euryarchaeal PilB paralogs can work with different pilin sets. Efficient recognition of the euryarchaeal pilins by the crenarchaeal PilB1-C1 may have required some degree of pilin modification, but perhaps the modifications were minor enough that PilB3 recognition of these pilins was not precluded, resulting in modular evolution and an extensive combinatorial diversity that allows for adaptation to a variety of stress conditions and attachment to varied surfaces.
Introduction
Type IV pili are thin filament-like protein complexes that extend from the cell surface and display a great deal of versatility in the roles they play in various cellular processes. As they are found in species that are representative of most bacterial and archaeal phyla, these structures are considered to be evolutionarily ancient (Pohlschroder et al., 2011; Hospenthal et al., 2017; Chaudhury et al., 2018). Among the functional roles that these ancient structures commonly play in species from both prokaryotic domains are promoting adherence to biotic and abiotic surfaces, facilitating close intracellular associations, and mediating the formation and maturation of biofilms; moreover, a type IV pilus-related cell surface structure known as the archaealla drives swimming motility in archaea (Albers and Jarrell, 2018).
Type IV pili are composed of major and minor pilins, with the major pilin comprising the bulk of the filamentous structure. Before these pilins can be assembled into a type IV pilus, a prepilin peptidase, PibD, in archaea, must cleave off their signal peptides, exposing N-terminal hydrophobic domains in the mature pilins. Interactions between these hydrophobic domains then drive the formation of a hydrophobic central core that serves as a scaffold for assembly of the pilus. In addition to the prepilin peptidase, completing the assembly of a functional pilus requires core components that include PilB, an ATPase that provides the energy required for assembly, and PilC, a multispanning transmembrane protein required for anchoring of the surface filament to the cell membrane (Pohlschroder and Esquivel, 2015; Chaudhury et al., 2018; Pohlschroder et al., 2018). Most prokaryotic genomes encode several pilB-C paralogs, many of which are contained in operons that also include pilin genes, indicating that the transcription of paralogs is often co-regulated along with a specific set of pilins (Szabo et al., 2007; Imam et al., 2011; Pohlschroder et al., 2011). Furthermore, many prokaryotic genomes are predicted to encode more than 40 pilins, suggesting that these pilins might be assembled into a variety of type IV pili having a broad range of diverse functional roles to play, likely depending on local environmental conditions. Unfortunately, the specific functional roles played by most of these pilins are not yet known (Szabo et al., 2007; Imam et al., 2011; Losensky et al., 2014; Pohlschroder et al., 2018).
Studies performed in the model euryarchaeon, Haloferax volcanii, which encodes five pil operons (pilB1-C1-pilB5-C5) as well as the archaealla (arl) operon, have revealed still greater complexity in the diversity of type IV pili in archaea (Szabo et al., 2007). In addition to the arl operon, the pilB3-C3 genes, which are in an operon that does not include pilin-encoding genes, have been studied in vivo in some detail (Esquivel et al., 2013, 2016; Esquivel and Pohlschroder, 2014). PilB3-C3 are required for the biosynthesis of pili containing any of the adhesion pilins, PilA[1–6], which are a subset of the 42 pilins encoded by the H. volcanii genome, as predicted by in silico analysis (Esquivel et al., 2013; Tripepi et al., 2013; Esquivel and Pohlschroder, 2014). Each of these predicted pilins contains a completely conserved pilin H-domain. In fact, we have determined that each of the six adhesion pilins can be assembled into a type IV pilus, and that the pili assembled from each of these adhesion pilins appear to play a distinct role during the early stages of biofilm formation (Esquivel and Pohlschroder, 2014). While no significant adhesion can be observed in an H. volcanii ∆pilA[1-6] strain during the first 24 hours of incubation, residual adhesion is present when a ∆pilB3-C3 strain is incubated for that period of time, suggesting that PilB-C paralogs can complement the ∆pilB3-C3 deletion (Esquivel and Pohlschroder, 2014).
To help understand functional and evolutionary relationships between these somewhat disparate biosynthesis systems, Makarova et al. used in silico analyses to phylogenetically group the archaeal PilB ATPases into four clades comprised of clade 1 methanogens, clade 2 euryarchaea, including H. volcanii PilB3, PilB4 and PilB5, clade 3 archaellum, and clade 4 TACK superphylum (Makarova et al., 2016). In these analyses, predicted pilin genes that are associated with pilB-C were also examined. A few haloarchaeal PilB homologs including H. volcanii PilB1 and PilB2 were identified to belong to a subclade of clade 4 (clade 4C). However, it was determined that the genes encoding these PilB homologs are associated with genes encoding pilins belonging to the family of pilins generally associated with clade 2 PilB ATPases, leading to the hypothesis that the PilB and PilC components encoded by some euryarchaeal pilus operons were acquired via horizontal transfer from the crenarchaea, while the pilin genes associated with these pilBC genes are the result of duplication and diversification of ancestral euryarchaeal pilin genes. This proposed hybrid origin led the authors to further propose that the archaeal PilB “can work with different pilin sets resulting in modular evolution and extensive combinatorial diversity” (Makarova et al., 2016).
Here, we report that a screen of a transposon insertion library has led to the identification of an adhesion mutant that has an insertion in H. volcanii pilB1. We also determined that a ∆pilB1-C1 strain has a similar adhesion defect that can be rescued by the expression of pilB1-C1 in trans. However, we have also shown that a ∆pilB1-C1 ∆pilB3-C3 double mutant does not abolish the residual adhesion observed in the ∆pilB3-C3 strain, nor can the ∆pilB3-C3 adhesion defect be rescued by the expression of pilB1-C1 in trans. Conversely, pilB3-C3 expressed in trans does rescue the adhesion defect of a ∆pilB1-C1 strain, suggesting that the major pilin may have undergone some alterations prior to being efficiently recognized by PilB1-C1 after it was acquired through horizontal transfer while the modification of this pilin was not to such a degree that it could not be recognized by PilB3 anymore.
Materials and Methods
Reagents
For basic molecular biology procedures, reagents were purchased from New England BioLabs, except for the iProof High-Fidelity DNA polymerase, which was purchased from Bio-Rad. For genomic DNA extraction, the Thermo scientific GeneJET Genomic DNA Purification kit was used while for plasmid purification, kits were purchased from Qiagen. Difco agar and Bacto yeast extract were purchased from Becton, Dickinson. Peptone was purchased from Oxoid. 5-fluoroorotic acid (5-FOA) was purchased from Toronto Research Biochemicals. All other chemicals and reagents were purchased from either Thermo Fisher Scientific or Sigma-Aldrich.
Strains and Growth Conditions
The plasmids and strains used in this study are listed in Table 1. H. volcanii strain H53 and its derivatives were grown at 45°C in liquid (orbital shaker at 250 rpm) or on solid casamino acid (CA) medium supplemented with tryptophan and uracil (both at 50 μg ml−1 final concentration). 1.5% agar was used to create solid medium in petri dishes. Strains transformed with pTA963, pGL3, or pRE62 were grown on CA medium supplemented with tryptophan (50 μg ml−1 final concentration). FH37, a variant of the H53 strain expressing a chromosomally encoded tryptophan gene, and GL1 were grown on CA medium supplemented with uracil (50 μg ml−1 final concentration). For selection of the deletion mutant, 5-FOA was added at a final concentration of 150 μg ml−1 in CA medium and uracil was added to a final concentration of 10μgml−1. Escherichia coli strains were grown at 37°C in NZCYM medium supplemented with ampicillin (200 μg ml−1).
Growth Curves
Growth curves were generated using a Biotek PowerWaveX2 microplate spectrophotometer. H. volcanii strains were first incubated in 5-ml liquid cultures in CA medium supplemented with tryptophan and uracil with continuous shaking at 45°C, until suitable OD600 values (0.2–0.5) were reached. Approximately 2 μl of each culture (adjusted for OD600 differences) was then transferred into 198 μl of fresh CA medium supplemented with tryptophan and grown to stationary phase, with OD600 recordings taken every 30 min.
Surface Adhesion Transposon Mutant Screening Assay
Surface adhesion to 96-well plate wells was used to screen transposon mutants and was previously developed, modifying the air-liquid interface (ALI) assay described by O’Toole (O’Toole et al., 1999; Legerme et al., 2016).
To improve consistency, subsequent adhesion assays employing 96-well plates for individual adhesion mutants were modified. Liquid cultures of volume 5 ml were grown to exponential phase in either CA medium supplemented with tryptophan or both tryptophan and uracil. After a second transfer into 5-ml liquid media, 200 μl of this liquid culture was then placed in wells at 45°C with no agitation for 30 h. After the incubation period, the cell adhesion was determined as described above using the Biotek reader.
Cover Slip Adhesion Assay
H. volcanii surface adhesion to a plastic cover slip was assayed using a modified ALI assay as described by Esquivel et al. (2016). Briefly, 3 ml of culture in CA medium supplemented with uracil was grown to an optical density at 600 nm (OD600) of ~0.3 and incubated in each well of a covered 12-well plate. Plastic cover slips (22 mm× 22 mm; 0.19–0.25 mm thick) from Fisher Scientific (Hampton, NH, USA) were inserted into each well at a 90° angle and incubated at 45°C without shaking. To visualize adhering cells, cover slips were removed and fixed with 2% (v/v) acetic acid for 3 min; coverslips were air-dried and then stained in 0.1% (w/v) crystal violet (CV) solution for 10 min. The coverslips were then washed with distilled water, air-dried, and examined using light microscopy.
Identification of Transposon Integration Sites by Next-Generation Sequencing
A Nextera Kit (Illumina; San Diego, CA, USA) was used to prepare the motility and/or adhesion mutant DNA for genome sequencing using a 300-cycle micro v2 MiSeq (Illumina). DNA was purified with the Zymo DNA clean and concentrator Kit (Irvine, CA, USA) and diluted in sterile water to a concentration of 10 ng/μl. Mutant genomic reads were assembled and mapped to the reference transposon sequence using the program, Geneious (Biomatters Ltd; Auckland, New Zealand), and the genomic sequence flanking the transposon was used to identify the transposon-disrupted gene through the National Center for Biotechnology Information’s (NCBI) Basic Local Alignment Search Tool (BLAST) (Altschul et al., 1997). Gene annotations were obtained from NCBI unless otherwise noted.
Generation of Chromosomal Deletions
Chromosomal deletions of hvo_0619 and hvo_0620 were generated in the H53 wild-type strain and RE43 deletion strains using a homologous-recombination-based (pop-in pop-out) method previously described (Allers and Ngo, 2003). Plasmid constructs were generated using overlap PCR with modifications (Tripepi et al., 2010) using primers listed in Table 2. To confirm that the chromosomal replacement event occurred at the proper location on the chromosome, genomic DNA isolated from colonies derived using these techniques was screened by PCR using two primer pairs, one hybridizing at the 5′ end of hvo_0619 and 3′ end of hvo_0620 and the other hybridizing to genomic sequences flanking the deletion (Table 2). The identities of the PCR products were verified by DNA sequencing.
Results and Discussion
Identification of pilB1::tn Adhesion Mutant Through Screen of Transposon Insertion Library
The establishment of an H. volcanii transposon insertion library and the modification of the 96-well air-liquid interface (ALI) assay for use with high salt conditions resembling those that haloarchaea inhabit have recently allowed for efficient screening for adhesion mutants in this model haloarchaeon (Kiljunen et al., 2014; Legerme et al., 2016). The fixing and staining of cells in 96-well plates results in observable rings on the well surface at the air-liquid interface, which allows a determination of the relative efficiency with which cells adhere. Cells harboring mutations that result in a significant decrease in the strength of surface adhesion, but do not affect growth, can be readily identified since the rings they form stain less intensely. To help differentiate between normal and deficient adhesion, a wild-type strain and ΔpilA[1-6], an adhesion-deficient mutant that lacks major pilins (Esquivel et al., 2013), were used as standards in these assays. Genome sequencing was then used to identify the transposon insertion location.
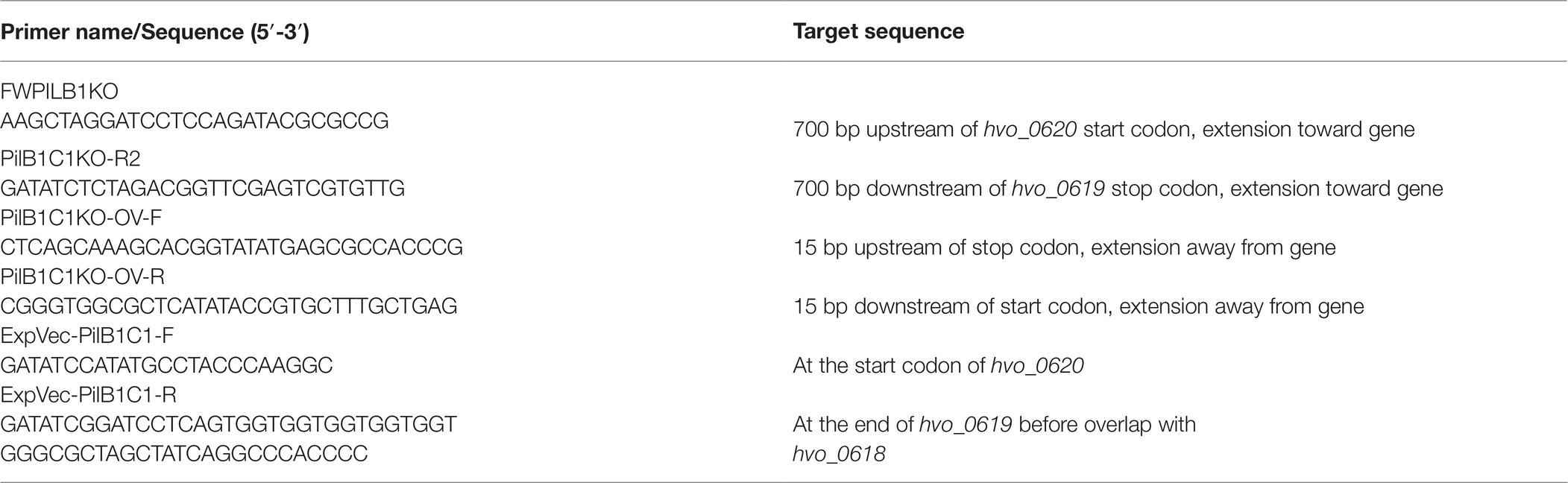
Employing these methods, we were able to identify an adhesion mutant that has a transposon insertion in pilB1, a gene that encodes a paralog of PilB, an ATPase required for pilus assembly (Figure 1A). While the adhesion defect of this transposon mutant is not as severe as that of ΔpilA[1-6], the 96-well plate ALI assay indicates that the strength of adhesion of pilB1::tn cells is less than that of the wild-type (Figure 1B).
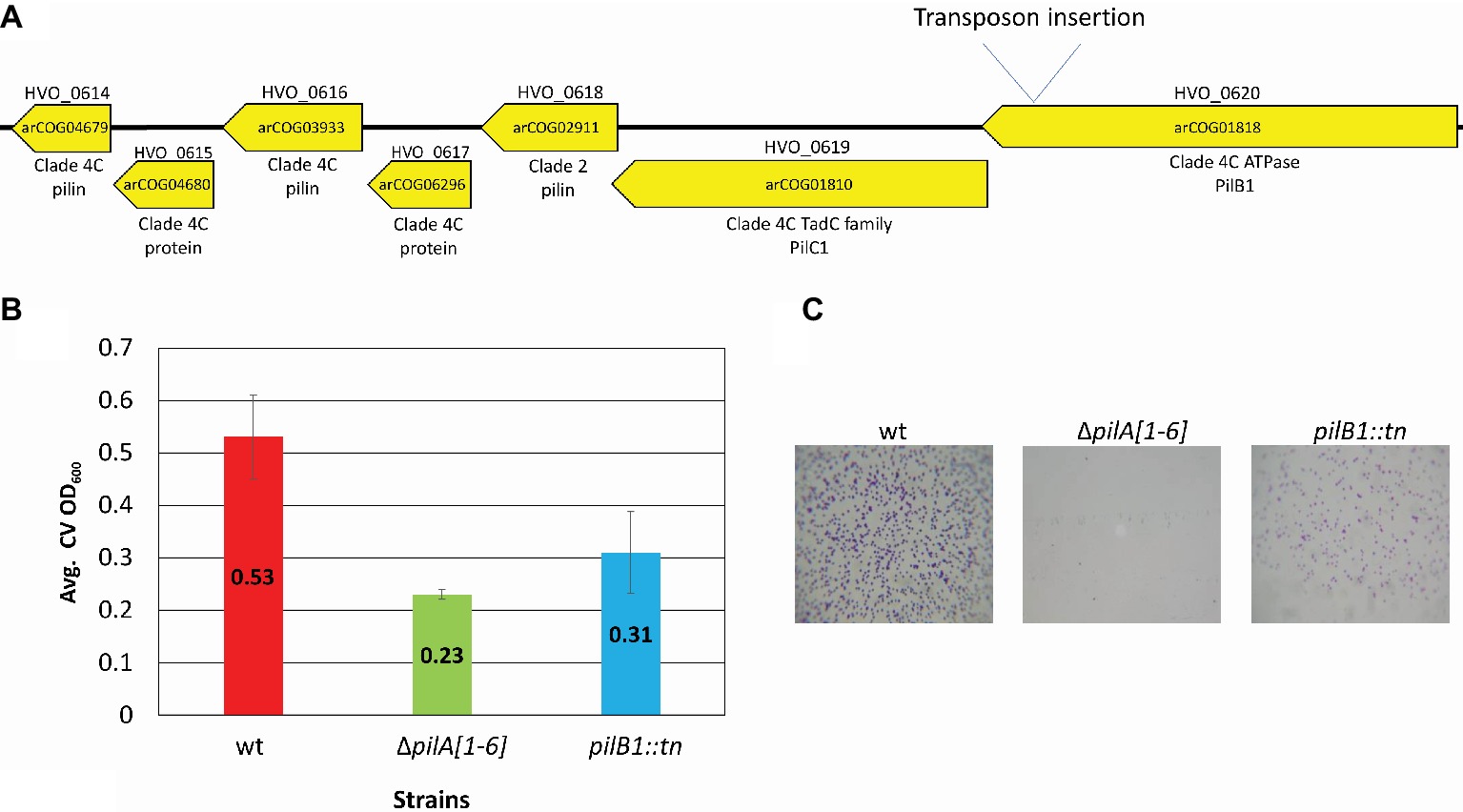
Figure 1. pilB1::tn exhibits a severe adhesion defect. (A) The location of the pilB1::tn insertion between base pair 550,339 and 550,340 was determined by whole genome sequencing. Genes of the pilB1 operon labeled using the H. volcanii locus tag (listed on top of gene), archaeal clusters of orthologous genes (arCOG) (listed within gene), and the clade that they are classified under as determined by Makarova et al. (2016) are listed below the gene. Genes in this operon are either part of clade 4C or clade 2. (B) Using the 96-well surface adhesion assay described in Legerme et al. (2016), the averages ± standard deviations were determined to compare adhesion of three biological replicates of wt (FH37), ΔpilA[1-6], and pilB1::tn strains grown in casamino acid (CA) media. This showed reduced relative adhesion of pilB1::tn. (C) Representative adhesion to plastic coverslips incubated for 12 h in exponential phase culture in wells of 12-well plates and subsequent staining of attached cells as described previously (Esquivel et al., 2013) confirmed the adhesion defect phenotype observed in (B). (×500 magnification) (n = 3).
Additionally, a surface adhesion assay using immersed coverslips confirmed this phenotype and its strength relative to wild-type and the ΔpilA[1-6] cells (Figure 1C). This result is the first indication of an H. volcanii PilB paralog other than PilB3 being involved in surface adhesion.
However, due to the ambiguities that can result from a transposon insertion, which include the possibility that the insertion disrupts the transcription of the whole operon, a deletion strain to confirm the phenotype observed in the pilB1::tn mutant was generated.
Construction and Characterization of ΔpilB1-C1 Mutant
To confirm the importance of PilB1 for adhesion, a deletion mutant was created. Since pilB1 and pilC1 overlap by four amino acids and are functionally paired in many species, we used homologous recombination to generate an H. volcanii deletion strain lacking both genes (ΔpilB1-C1), except for the last four nucleotides of pilC1, as these nucleotides also overlap with the adjacent gene, the predicted pilin gene hvo_0618, (Allers and Ngo, 2003; Takhar et al., 2013). We confirmed the deletion by PCR using a forward primer homologous to sequences lying just inside pilB1 and a reverse primer homologous to PilC1 sequences starting four nucleotides into pilC1 (Figure 2; Table 1) as done previously for all other deletion strains, such as ΔpilB3-C3 used in this study (Esquivel et al., 2014).
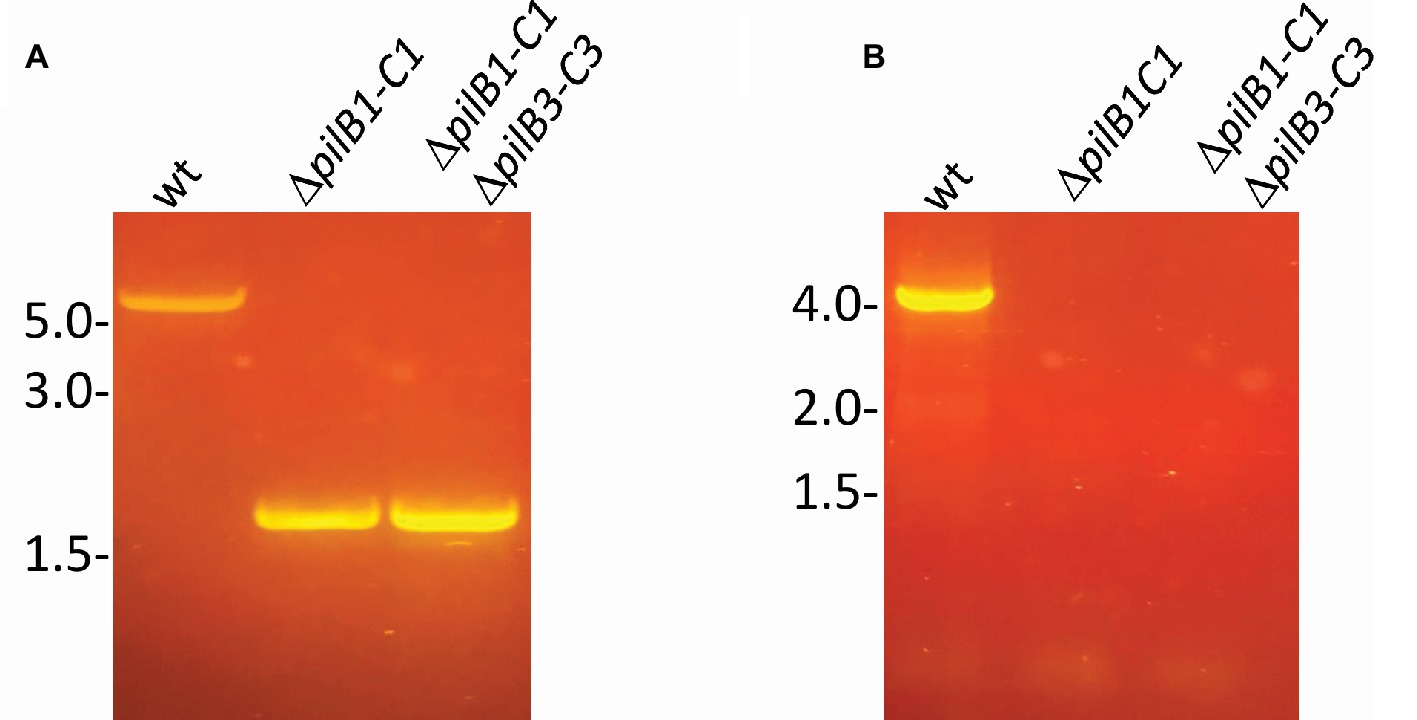
Figure 2. Confirmation of chromosomal pilB1-C1 deletion. PCR amplification was performed using primers (A) against the flanking regions located approximately 700 bp upstream and 700 bp downstream of pilB1-C1 and (B) specific for the pilB1 and pilC1 genes. The template DNA used was isolated from the wild-type or ΔpilB1-C1 strain.
The ΔpilB1-C1 strain was then characterized using a modified version of the 96-well surface adhesion assay that was used to screen the transposon insertion library. Since the number of analyzed strains was strongly decreased, they could be grown in shaking culture tubes to exponential phase (OD600 0.3) and then be incubated in wells for adhesion, eliminating the need for agitation of the 96-well plate. Although agitation increased aeration and fostered nutrient homogeneity within the media, and thus promoted growth (Juergensmeyer et al., 2007), the turbulence caused by agitation increased shear stress in the wells, reducing adhesion efficiency after initial attachment and perhaps even initial attachment (Moreira et al., 2013). Following static incubation for 30 h, adhesion levels, as determined by crystal violet (CV) absorbance, were more precise, and the differences in adhesion between cultures over time could be determined more effectively, especially given that the initial inoculum was less variable.
Using this surface adhesion assay to characterize strains grown in semi-defined media, it was shown that, like the pilB1::tn strain, ΔpilB1-C1 also has an intermediately reduced adhesion phenotype relative to the wild-type strain. Interestingly, under these conditions, the pilB1::tn strain consistently had an adhesion phenotype that was slightly more deficient than that of ΔpilB1-C1 (Figure 3A), perhaps because the transposon insertion in pilB1 may have affected the transcription of all genes in the operon containing pilB1 as opposed to the effect of completely deleting two specific genes in that operon. The expression of pilB1-C1his in trans under the control of an inducible trp promoter rescues the ΔpilB1-C1 phenotype, underscoring the fact that the adhesion defect of this strain is indeed due to the absence of these pilus biosynthesis genes (Figure 4).
Figure 3. H. volcanii ΔpilB1-C1 has an intermediate adhesion defect similar to that of pilB1::tn; however, PilB1-C1 is not responsible for the residual attachment of ΔpilB3-C3. (A) Using the further modified 96-well surface adhesion assay described in the Materials and Methods with stationary 30-h incubation, H. volcanii wild-type (H53), ΔpilB1-C1, pilB1::tn, ΔpilB3-C3, and ΔpilB1-C1ΔpilB3-C3 strain adhesion was quantified by measuring the OD600 of the stain released in methanol. Three biological replicates with three technical replicates each were completed, and averages (± s.d.), correlating to adhesion, were determined showing similarly reduced adhesion of ΔpilB1-C1 and pilB1::tn. (B) As described in the Materials and Methods, 49-h growth curves were generated for each strain with average growth OD600 calculated and graphed for three biological replicates of each strain, each with three technical replicates showing similar growth patterns.(C) Adhesion profiles of aforementioned strains were determined by incubation of plastic coverslips in exponential phase culture for 24 h in wells of 12-well plates. The images shown are representative of six replicates and are consistent with the results observed in (A).
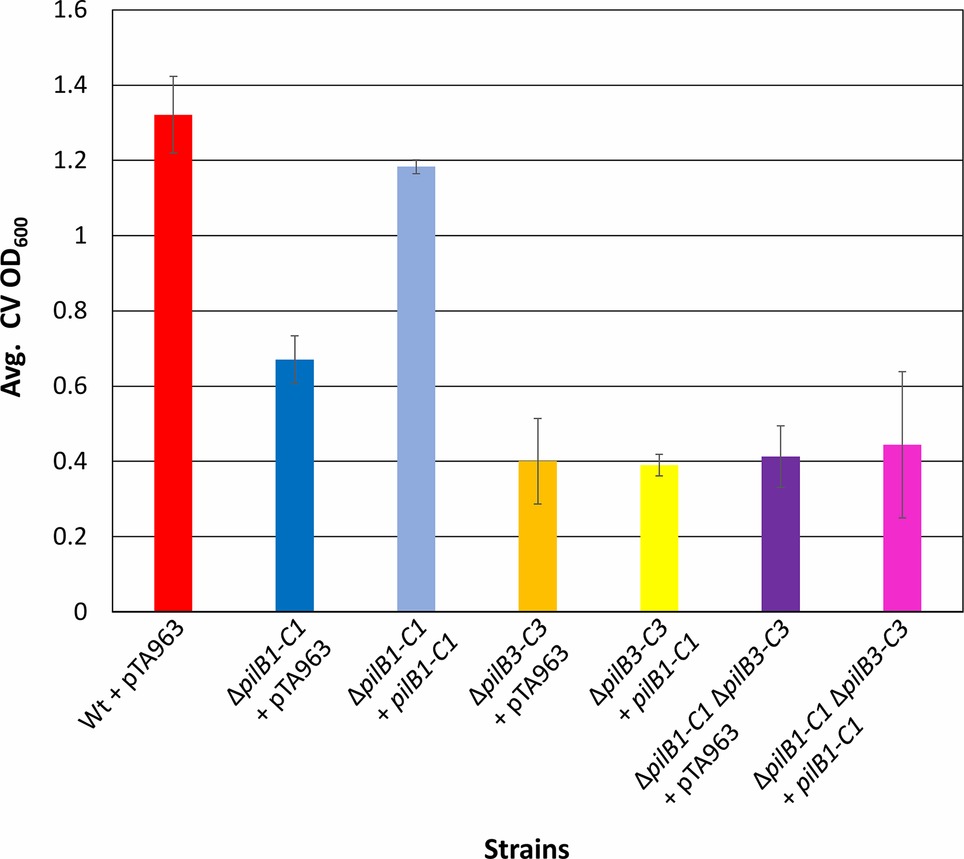
Figure 4. pilB1-C1 rescues adhesion defect of the ΔpilB1-C1 strain to near wild-type adhesion but does not complement ΔpilB3-C3 or ΔpilB1-C1ΔpilB3-C3 adhesion defects. Using the 96-well ALI assay described in Figure 3A, averages (± s.d.) of adhesion quantified by determination of CV OD600 for three biological replicates, with three technical replicates each, of wild-type + pTA963 and ΔpilB1-C1, ΔpilB3-C3, and ΔpilB1-C1ΔpilB3-C3 strains transformed with pTA963 or pTA963 encoding his-tagged pilB1-C1; all transformed strains were grown in CA media. Similar adhesion levels between wild-type + pTA963 and ΔpilB1-C1 + pilB1-C1 are shown but those for ΔpilB3-C3 + pilB1-C1 and ΔpilB1-C1ΔpilB3-C3 + pilB1-C1 are still reduced.
Additional characterization of these mutant strains included the generation of growth curves to determine whether deletion or disruption of pilB1-C1 had a significant effect on strain growth relative to the wild-type to ensure that reduced adhesion was not a result of diminished growth but rather solely a result of the functional absence of these components of the pilus biosynthesis machinery. Ultimately, we determined that the pilB1::tn and ΔpilB1-C1 strains had growth rates similar to that of the wild-type (Figure 3B). Interestingly, pilB1::tn has slightly delayed growth during exponential phase compared to ΔpilB1-C1 and wild-type, and, although this deficiency is overcome during stationary phase, this could also explain the slight difference in adhesion phenotypes observed in the pilB1::tn and ΔpilB1-C1 strains.
Using the 12-well surface adhesion assay with coverslips, we confirmed the adhesion defects of the mutant strains as the quantity of cells in the adhesion profile of each strain correlated well with the CV quantification results obtained with the 96-well plate assays (Figure 3C). While previous results showed that some pilin mutant strains have phenotypes that include early microcolony formation, it should be noted that neither pilB1::tn nor ΔpilB1-C1 have increased early microcolony formation under the conditions tested (Esquivel et al., 2013).
Analysis of PilB1-C1 Involvement in PilB3-C3-Dependent Adhesion
It was previously shown that, unlike ΔpilA[1-6], which lacks residual attachment, cells lacking the pilB3-C3 biosynthesis genes, which are required for PilA[1–6] pili assembly exhibit residual adhesion (Esquivel and Pohlschroder, 2014). It is possible that PilB1-C1 are responsible for the residual adhesion of the ΔpilB3-C3 strain through the assembly of a subset of PilA[1–6] pili. To test this hypothesis, we deleted pilB1-C1 in the ΔpilB3-C3 strain generating the double mutant, ΔpilB1-C1ΔpilB3-C3. ΔpilB1-C1-ΔpilB3-C3 has the same adhesion phenotype as ΔpilB3-C3 after a 30-h incubation, suggesting that residual attachment of ΔpilB3-C3 is not due to PilB1 and PilC1 driving assembly of PilA[1-6] pili (Figure 3A). Consistent with these results, as noted above, while pilB1-C1his expressed in trans can complement the adhesion defect of ΔpilB1-C1, pilB1-C1his expressed in a ∆pilB3-C3 strain does not affect or recover the adhesion defect of this mutant strain. Therefore, taken together, these results suggest that PilB1-C1 cannot promote PilA[1–6] pilus biosynthesis and that the residual adhesion observed in ΔpilB3-C3 depends upon another mechanism. It is possible that pilins within the membrane are sufficient to mediate the level of adhesion we observed in this strain, as previously suggested (Esquivel and Pohlschroder, 2014). Western blot analyses using anti-his antibodies of protein extracts from H. volcanii ΔpilB3-C3 expressing pilB1-C1his in trans, unfortunately, did not reveal a PilChis band. Therefore, the possibility that it is not stably expressed cannot be ruled out. However, we also failed to detect a PilChis band in protein extracts from the ΔpilB1-C1 background, where adhesion assays suggested that these proteins are expressed (data not shown).
Analysis of PilB3-C3 Involvement in PilB1-C1-Dependent Adhesion
To test the hypothesis put forth by Makarova et al. suggesting that the modular evolution of type IV pili, along with PilB and PilC, proteins required for their assembly, results in combinatorial variability of these protein complexes, we expressed pilB3-C3his, which was previously shown to complement the adhesion defect of a ΔpilB3-C3 strain, in the ΔpilB1-C1 background strains (Makarova et al., 2016). Consistent with this hypothesis, we determined that pilB3-C3his expression strongly complements the adhesion defects of these strains, increasing adhesion of the ΔpilB1-C1 strain to a level similar to that of the pTA963-transformed wild-type strain and ΔpilB1-C1ΔpilB3-C3 adhesion to nearly the same level (Figure 5). To exclude the possibility that PilB3-C3 might promote increased biosynthesis of PilA[1–6] pili, PilB3-C3his was expressed in trans in a wild-type strain, which might then be expected to attach to a surface more effectively. However, that is not the case, as the transformed wild-type strain adheres similarly to the strain transformed with pTA963 (Figure 5). Hence, these results suggest that PilB3-C3 can promote pilus formation of PilB1-C1-dependent pili.
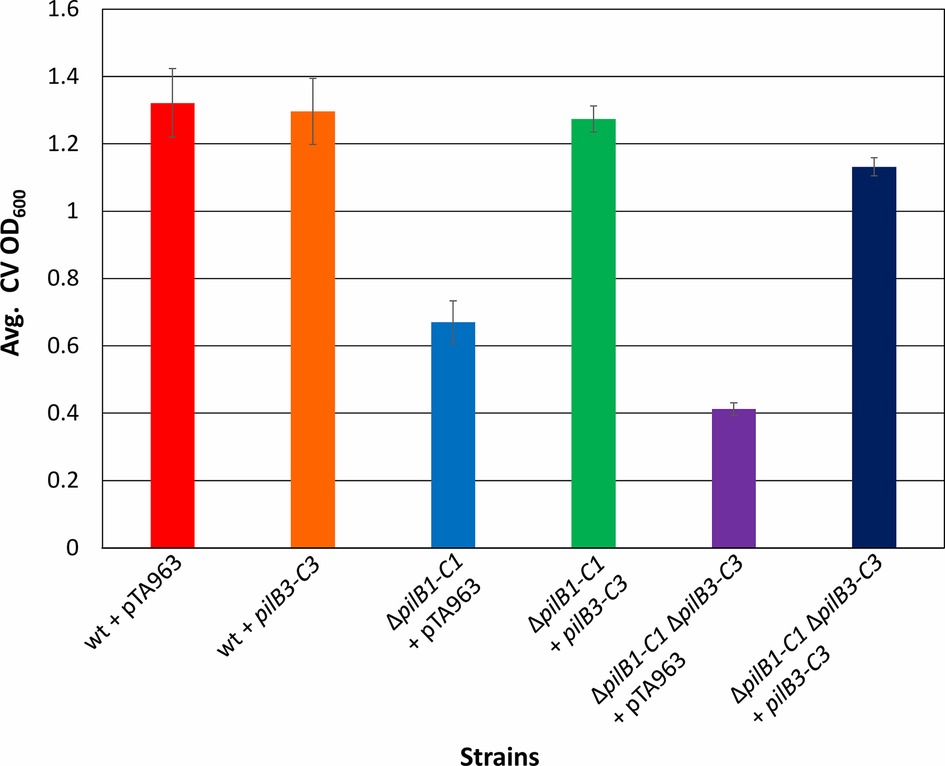
Figure 5. pilB3-C3 expression in trans complements the ΔpilB1-C1 and ΔpilB1-C1 ΔpilB3-C3 adhesion defects. Using the 96-well ALI assay, as described in Figure 3A, averages (± s.d.) of adhesion quantified through OD600 measurements of CV for three biological replicates with three technical replicates each, of wild-type, ΔpilB1-C1, and ΔpilB1-C1 ΔpilB3-C3 strains transformed with pTA963 and pTA963 encoding his-tagged pilB3-C3 grown in CA media. It is shown that ΔpilB1-C1 + pilB3-C3 has similar adhesion levels to wild-type + pTA963 unlike ΔpilB1-C1 + pTA963.
Concluding Remarks
The results of the deletion and complementation studies of pilB and pilC paralogs in the model archaeon, H. volcanii, presented in this study support the hypothesis posited by Makarova et al. that pilB1-C1, which phylogenetically clusters with type IV pilus biosynthesis genes of clade 4C, were acquired through lateral transfer from a crenarchaeal species, but are involved in assembling type IV pili composed, at least in part, of euryarchaeal type IV pilins belonging to clade 2. This was determined, at least in part, based on previous in vivo studies in H. volcanii that identified the major pilins of this clade demonstrating a complementary synergism of in silico and in vivo approaches for facilitating biological discoveries (Esquivel and Pohlschroder, 2014). Furthermore, these combined results might suggest future experiments for determining the mechanisms used by a distinct PilB-C paralog to recognize and assemble specific pilins and exclude others in the generation of a pilus perhaps allowing us to better understand the complexities involved in the biosynthesis of this universally conserved surface structure. While the adhesion defect of ∆pilB1-C1 revealed by the transposon insertion mutant was invaluable for these studies, further testing may well reveal stronger, or weaker, phenotypes for this mutant under other conditions, underscoring the evolutionary advantage of the acquisition of an additional paralog by H. volcanii through lateral transfer. When expressed under certain conditions, these acquired paralogs might facilitate adhesion to additional abiotic surfaces, or to other cells. For example, in Sulfolobus acidocaldarius, UV-induced type IV pili promote cell aggregation leading to increased DNA exchange in this hyperthermophilic archaeon (Frols et al., 2008; Ajon et al., 2011). At least under the growth conditions used and with the abiotic surfaces tested, pilB3-C3, when constitutively expressed, can complement the phenotypic defects caused by a pilB1-C1 deletion. Undoubtedly, the accumulation of additional transcriptomic and proteomic data for H. volcanii, as well as other euryarchaea containing these clades of type IV pili and pilins, will lead to a better understanding of the specific roles these PilB-C paralogs play in the cell surface biology of the euryarchaea.
Author Contributions
GL and MP conceived and designed the experiments. GL performed the experiments involving screening, characterization of adhesion mutants, and adhesion assays. GL and MP analyzed the data. GL and MP wrote the manuscript.
Funding
GL and MP were supported by the National Science Foundation Grant MCB-1413158. GL also received funding through the National Aeronautics and Space Administration MIRS Program.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Pohlschroder lab for helpful discussions, editing, and guidance.
References
Abdul Halim, M. F., Karch, K. R., Zhou, Y., Haft, D. H., Garcia, B. A., and Pohlschroder, M. (2015). Permuting the PGF signature motif blocks both archaeosortase-dependent C-terminal cleavage and prenyl lipid attachment for the Haloferax volcanii S-layer glycoprotein. J. Bacteriol. 198, 808–815. doi: 10.1128/JB.00849-15
Ajon, M., Frols, S., van Wolferen, M., Stoecker, K., Teichmann, D., Driessen, A. J., et al. (2011). UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili. Mol. Microbiol. 82, 807–817. doi: 10.1111/j.1365-2958.2011.07861.x
Albers, S. V., and Jarrell, K. F. (2018). The archaellum: an update on the unique archaeal motility structure. Trends Microbiol. 26, 351–362. doi: 10.1016/j.tim.2018.01.004
Allers, T., Barak, S., Liddell, S., Wardell, K., and Mevarech, M. (2010). Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl. Environ. Microbiol. 76, 1759–1769. doi: 10.1128/AEM.02670-09
Allers, T., and Ngo, H. P. (2003). Genetic analysis of homologous recombination in archaea: Haloferax volcanii as a model organism. Biochem. Soc. Trans. 31, 706–710. doi: 10.1042/bst0310706
Allers, T., Ngo, H. P., Mevarech, M., and Lloyd, R. G. (2004). Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70, 943–953. doi: 10.1128/AEM.70.2.943-953.2004
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Blyn, L. B., Braaten, B. A., and Low, D. A. (1990). Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J. 9, 4045–4054. doi: 10.1002/j.1460-2075.1990.tb07626.x
Chaudhury, P., Quax, T. E. F., and Albers, S. V. (2018). Versatile cell surface structures of archaea. Mol. Microbiol. 107, 298–311. doi: 10.1111/mmi.13889
Esquivel, R. N., and Pohlschroder, M. (2014). A conserved type IV pilin signal peptide H-domain is critical for the post-translational regulation of flagella-dependent motility. Mol. Microbiol. 93, 494–504. doi: 10.1111/mmi.12673
Esquivel, R. N., Schulze, S., Xu, R., Hippler, M., and Pohlschroder, M. (2016). Identification of Haloferax volcanii pilin N-glycans with diverse roles in pilus biosynthesis, adhesion, and microcolony formation. J. Biol. Chem. 291, 10602–10614. doi: 10.1074/jbc.M115.693556
Esquivel, R. N., Xu, R., and Pohlschroder, M. (2013). Novel archaeal adhesion pilins with a conserved N terminus. J. Bacteriol. 195, 3808–3818. doi: 10.1128/JB.00572-13
Frols, S., Ajon, M., Wagner, M., Teichmann, D., Zolghadr, B., Folea, M., et al. (2008). UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol. Microbiol. 70, 938–952. doi: 10.1111/j.1365-2958.2008.06459.x
Hospenthal, M. K., Costa, T. R. D., and Waksman, G. (2017). A comprehensive guide to pilus biogenesis in gram-negative bacteria. Nat. Rev. Microbiol. 15, 365–379. doi: 10.1038/nrmicro.2017.40
Imam, S., Chen, Z., Roos, D. S., and Pohlschroder, M. (2011). Identification of surprisingly diverse type IV pili, across a broad range of gram-positive bacteria. PLoS One 6:e28919. doi: 10.1371/journal.pone.0028919
Juergensmeyer, M. A., Nelson, E. S., and Juergensmeyer, E. A. (2007). Shaking alone, without concurrent aeration, affects the growth characteristics of Escherichia coli. Lett. Appl. Microbiol. 45, 179–183. doi: 10.1111/j.1472-765X.2007.02172.x
Kiljunen, S., Pajunen, M. I., Dilks, K., Storf, S., Pohlschroder, M., and Savilahti, H. (2014). Generation of comprehensive transposon insertion mutant library for the model archaeon, Haloferax volcanii, and its use for gene discovery. BMC Biol. 12, 103. doi: 10.1186/s12915-014-0103-3
Legerme, G., Yang, E., Esquivel, R., Kiljunen, S., Savilahti, H., and Pohlschroder, M. (2016). Screening of a Haloferax volcanii transposon library reveals novel motility and adhesion mutants. Lifestyles 6, 1–14. doi: 10.3390/life6040041
Losensky, G., Vidakovic, L., Klingl, A., Pfeifer, F., and Frols, S. (2014). Novel pili-like surface structures of Halobacterium salinarum strain R1 are crucial for surface adhesion. Front. Microbiol. 5:755. doi: 10.3389/fmicb.2014.00755
Makarova, K. S., Koonin, E. V., and Albers, S. V. (2016). Diversity and evolution of type IV pili systems in archaea. Front. Microbiol. 7:667. doi: 10.3389/fmicb.2016.00667
Moreira, J. M., Teodosio, J. S., Silva, F. C., Simoes, M., Melo, L. F., and Mergulhao, F. J. (2013). Influence of flow rate variation on the development of Escherichia coli biofilms. Bioprocess Biosyst. Eng. 36, 1787–1796. doi: 10.1007/s00449-013-0954-y
O’Toole, G. A., Pratt, L. A., Watnick, P. I., Newman, D. K., Weaver, V. B., and Kolter, R. (1999). Genetic approaches to study of biofilms. Methods Enzymol. 310, 91–109.
Pohlschroder, M., and Esquivel, R. N. (2015). Archaeal type IV pili and their involvement in biofilm formation. Front. Microbiol. 6:190. doi: 10.3389/fmicb.2015.00190
Pohlschroder, M., Ghosh, A., Tripepi, M., and Albers, S. V. (2011). Archaeal type IV pilus-like structures–evolutionarily conserved prokaryotic surface organelles. Curr. Opin. Microbiol. 14, 357–363. doi: 10.1016/j.mib.2011.03.002
Pohlschroder, M., Pfeiffer, F., Schulze, S., and Halim, M. F. A. (2018). Archaeal cell surface biogenesis. FEMS Microbiol. Rev. 42, 694–717. doi: 10.1093/femsre/fuy027
Szabo, Z., Stahl, A. O., Albers, S. V., Kissinger, J. C., Driessen, A. J., and Pohlschroder, M. (2007). Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J. Bacteriol. 189, 772–778. doi: 10.1128/JB.01547-06
Takhar, H. K., Kevin, K., Melissa, K., Howell, P. L., and Burrows, L. L. (2013). The platform protein is essential for type IV pilus biogenesis. J. Biol. Chem. 288, 9721–9728. doi: 10.1074/jbc.m113.453506
Tripepi, M., Esquivel, R. N., Wirth, R., and Pohlschroder, M. (2013). Haloferax volcanii cells lacking the flagellin FlgA2 are hypermotile. Microbiology 159, 2249–2258. doi: 10.1099/mic.0.069617-0
Keywords: Haloferax volcanii, pilus biosynthesis, pilin, archaea, type IV pili, adhesion, biofilm formation
Citation: Legerme G and Pohlschroder M (2019) Limited Cross-Complementation Between Haloferax volcanii PilB1-C1 and PilB3-C3 Paralogs. Front. Microbiol. 10:700. doi: 10.3389/fmicb.2019.00700
Edited by:
Gloria Paz Levicán, Universidad de Santiago de Chile, ChileReviewed by:
James A. Coker, University of Maryland University College, United StatesKelly Bidle, Rider University, United States
Copyright © 2019 Legerme and Pohlschroder. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mechthild Pohlschroder, cG9obHNjaHJAc2FzLnVwZW5uLmVkdQ==
 Georgio Legerme
Georgio Legerme Mechthild Pohlschroder
Mechthild Pohlschroder