- 1School of Microbiology, University College Cork, Cork, Ireland
- 2APC Microbiome Ireland, University College Cork, Cork, Ireland
- 3Laboratory of Probiogenomics, Department of Chemistry, Life Sciences and Environmental Sustainability, University of Parma, Parma, Italy
Plasmids are autonomous, self-replicating, extrachromosomal genetic elements that are typically not essential for growth of their host. They may encode metabolic capabilities, which promote the maintenance of these genetic elements, and may allow adaption to specific ecological niches and consequently enhance survival. Genome sequencing of 16 Lactococcus lactis strains revealed the presence of 83 plasmids, including two megaplasmids. The limitations of Pacific Biosciences SMRT sequencing in detecting the total plasmid complement of lactococcal strains is examined, while a combined Illumina/SMRT sequencing approach is proposed to combat these issues. Comparative genome analysis of these plasmid sequences combined with other publicly available plasmid sequence data allowed the definition of the lactococcal plasmidome, and facilitated an investigation into (bio) technologically important plasmid-encoded traits such as conjugation, bacteriocin production, exopolysaccharide (EPS) production, and (bacterio) phage resistance.
Introduction
Lactococcus lactis is globally applied as a starter culture for dairy-based food fermentations, such as those involved in the production of Cheddar, Colby, Gouda and blue cheeses, and from an economic and (food) biotechnological perspective represents one of the most important bacteria (Ainsworth et al., 2014a). It is widely accepted that L. lactis originated from a plant-associated niche (Price et al., 2012; Wels et al., 2019) and, whilst the majority of sequenced lactococcal representatives are isolated from the dairy environment, this is not representative of the presumed diversity of the taxon. It is evident from genome analyses of L. lactis strains isolated from the dairy niche that genome decay (due to functional redundancy) (Makarova et al., 2006; Goh et al., 2011; Ainsworth et al., 2013; Kelleher et al., 2015, 2017), in parallel with the acquisition of novel plasmid-encoded traits played a significant role in their adaptation to the nutrient-rich environment of milk. Analysis of the plasmid complement has revealed a relatively low abundance of plasmids among lactococcal strains isolated from non-dairy niches (Makarova et al., 2006; Kelly et al., 2010; Ainsworth et al., 2013, 2014c). Since various dairy-associated phenotypes are encoded by plasmids, horizontal acquisition to adapt to the dairy environment is likely to be one of the major drivers of plasmid transfer in L. lactis (Ainsworth et al., 2014c) with dairy strains containing up to twelve plasmids (Van Mastrigt et al., 2018a). Plasmid transfer in L. lactis is believed to be predominantly governed by conjugation and transduction (Ainsworth et al., 2014c), but may also occur as a result of transformation (David et al., 2017; Mulder et al., 2017) Transduction is a process in which DNA transfer is carried out by a (bacterio)phage (i.e., a virus that infects a bacterium) due to unintentional packaging of host DNA, and has previously been observed in L. lactis (Ammann et al., 2008; Wegmann et al., 2012). Conjugation involves the transfer of plasmid material via a conjugative apparatus (Grohmann et al., 2003) and is of particular importance as it represents a natural phenomenon that is suitable for the transfer of genetic traits such as phage resistance systems in food grade processes, bacteriocin production (including nisin), proteinases, and citrate utilization (Neve et al., 1987; Kojic et al., 2005; Mills et al., 2006; Van Mastrigt et al., 2018b). Extensive research into the technological traits of L. lactis has been carried out in the past with a significant focus on lactose utilization (Van Rooijen and De Vos, 1990; Van Rooijen et al., 1992), casein metabolism (Siezen et al., 2005), citrate metabolism (Drider et al., 2004; Van Mastrigt et al., 2018a), flavor formation (McSweeney and Sousa, 2000; McSweeney, 2004), and phage resistance mechanisms (Labrie et al., 2010), all of which represent properties that are commonly plasmid-encoded. Lactose utilization in L. lactis is governed by the lac operon, which provides dairy strains with the ability to rapidly ferment lactose and grow in milk. The L. lactis lac operon, which consists of the genes lacABCDEFGX, is generally plasmid-borne and is regulated by a repressor, encoded by the adjacent lacR gene (Van Rooijen and De Vos, 1990; Van Rooijen et al., 1992). Citrate metabolism is conducted by citrate-positive (Cit+) lactococci and is important as it leads to the production of a number of volatile flavor compounds (McSweeney and Sousa, 2000). Citrate uptake and subsequent diacetyl production is governed by the plasmid-encoded citQRP operon in lactococcal species (Drider et al., 2004). Proteolysis also significantly contributes to flavor production in fermented dairy products, although high levels of proteolysis may cause bitterness in cheese (Broadbent et al., 2002). The plasmid-encoded extracellular cell wall proteinase (lactocepin) has been shown to be directly associated with the bitter flavor defect in Cheddar cheese varieties, specifically involving starters which produce lactocepin of the so-called a, e, or h groups, and its characterization is of particular importance when selecting novel starter cultures (Broadbent et al., 2002).
Lactococcal phages are recognized as the main cause of fermentation problems within the dairy industry with concomitant economic problems. Lactococcal strains possess an arsenal of phage defense mechanisms, such as R-M systems and abortive infection (Abi) systems, many of which are plasmid-encoded. In the current study, we assess the genetic content of lactococcal plasmids, define the current pan-plasmidome of L. lactis, and investigate plasmid-encoded (and technologically relevant) traits.
Materials and Methods
Sequencing
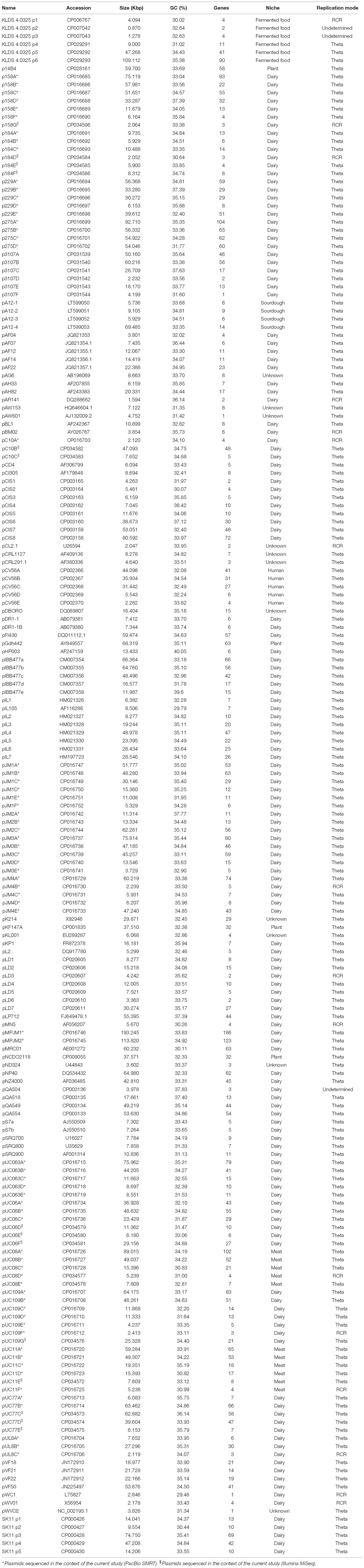
In total, 83 plasmids (81 plasmids and 2 megaplasmids, the latter defined as plasmids that are >100 Kbp in length) were sequenced in the context of this study (Table 1). Sequencing of sixteen lactococcal strains was performed as previously described (Kelleher et al., 2017) utilizing the SMRT sequencing approach on a Pacific Biosciences RS II sequencing platform (executed by GATC Biotech Ltd., Germany). De novo assemblies were performed on the Pacific Biosciences SMRTPortal analysis platform (version 2.3.1), utilizing the RS_HGAP_Assembly.2 protocol. Assemblies were then repeated with a reduced minimum coverage threshold adjusted to 15X to ensure all plasmid-associated contigs had been detected.
In parallel with SMRT sequencing, an Illumina-based approach was applied to the sixteen lactococcal strains to identify strains where plasmids were potentially absent from the completed assemblies. Re-sequencing of genomes was performed on an Illumina MiSeq platform (executed by GenProbio S.R.L., Parma, Italy), to an average coverage of ∼100–125×. Sequences obtained were first quality checked using IlluQC.pl from the NGS QC Toolkit (v2.3) (Patel and Jain, 2012) and assembled with AbySS (v1.9.0) (Simpson et al., 2009). Based on whole genome alignments contigs absent from the SMRT assemblies were identified. Remaining low quality regions and sequence conflicts were then resolved by primer walking and Sanger sequencing of PCR products (performed by Eurofins MWG Operon, Germany).
General Feature Predictions
Annotation of plasmid sequences was performed on both newly sequenced and publically available plasmid sequences using the following protocol. ORF prediction, defined as a continuous stretch of codons without a stop codon was performed with Prodigal v2.5 prediction software1 with a general minimum cut-off of >50 bp and confirmed using BLASTX v2.2.26 alignments (Altschul et al., 1990). ORFs were automatically annotated using BLASTP v2.2.26 (Altschul et al., 1990) analysis against the non-redundant protein databases curated by the National Centre for Biotechnology Information (NCBI)2. Artemis v16 genome browser and annotation tool was used to manually curate identified ORFs3 and for the combination and inspection of ORF results. The final ORF annotations were refined where necessary using additional software tools and database searches, such as Pfam (Bateman et al., 2004), Uniprot/EMBL4 and Bagel3 (Van Heel et al., 2013).
Pan-Plasmidome Analysis
Pan-plasmidome analysis was performed utilizing the PGAP v1.0 pipeline (Zhao et al., 2012) according to Heaps law pan-genome model (Tettelin et al., 2005). The ORF content of each plasmid was organized into functional gene clusters via the Gene Family method. ORFs which produced an alignment with a minimum of 50% sequence identity across 50% of the gene or protein length (both nucleotide and amino acid sequences are applied in parallel) were clustered and a pan-plasmidome profile was subsequently generated (Tettelin et al., 2005).
Comparative Genomics
Tandem Repeats Finder v4.02 (Benson, 1999) was applied to identify nucleotide tandem repeats at a potential plasmid origin of replication. Plasmids were assigned to be employing a Theta mode of replication where the gene encoding replication protein is preceded by 3.5 iterations of a 22 bp tandem repeat with an A/T rich 10 bp direct repeat located further upstream (Kiewiet et al., 1993). Alternatively, plasmids that replicate by rolling circle replication (RCR) can be identified because they rely on a replication protein and a double-stranded origin of replication (dso). Putative dso replication sites were identified based on nucleotide conservation to previously identified dso’s, containing a nic site composed of one or more inverted repeats, and a Rep-binding site consisting of 2–3 direct repeats or an inverted repeat (Del Solar et al., 1993; Mills et al., 2006).
All sequence comparisons at protein level were performed via all-against-all, bi-directional BLAST alignments (Altschul et al., 1990). An alignment cut-off value of >50% amino acid identity across 50% of the sequence length was used (with an associated E-value of <0.0001). For analysis and clustering of these results, the MCL was implemented in the mclblastline pipeline v12-0678 (Enright et al., 2002). TM4 MeV, MultiExperiment Viewer v4.9 was used to view MCL clustering data and conduct hierarchal clustering (HCL)5. The HCL analysis was exported from TM4 MeV in Newick tree format and visualized using ITOL (Interactive Tree of Life) (Letunic and Bork, 2016).
Pulsed Field Gel Electrophoresis (PFGE)
Lactococcus lactis subsp. cremoris strains JM1 and JM2 were cultured in M17 broth (Oxoid) supplemented with 0.5% (w/v) lactose at 30°C without agitation overnight. PFGE plugs were then prepared and restricted with SI nuclease (Thermo Fisher Scientific, Ireland) as previously described (Bottacini et al., 2015).
A 1% (wt/vol) PFGE agarose gel was prepared in 0.5X TBE [89 mM Tris-borate, 2 mM EDTA (pH 8.3)] buffer and the PFGE plugs were melted in and sealed with molten agarose in 0.5X TBE buffer. A CHEF-DR III pulsed-field system (Bio-Rad Laboratories, Hercules, CA, United States) was used to resolve the DNA fragments at 6 V/cm for 18 h in 0.5X TBE running buffer maintained at 14°C with linear increment (interpolation) of pulse time from 3 to 50 s. DNA ladder (Chef DNA lambda) was included in each gel (number 170-3635; Bio-Rad Laboratories). The gels were stained in ethidium bromide (10 mg/ml) (25 μl/500 ml dH2O) for 120 min under light-limited conditions and destained in distilled water for 60 min. Gels were visualized by UV transillumination.
Bacteriocin Assays
Lactococcal strains were cultured in M17 broth (Oxoid) supplemented with 0.5% (w/v) lactose or glucose (strain-dependent) at 30°C without agitation overnight. 3 μl of overnight culture was spotted on M17 agar supplemented with 0.5% (w/v) glucose and left at 30°C overnight. Cells that had grown on the spotted areas were inactivated by exposure to UV light for 30 min. Plates were then overlaid with a semi-solid M17 agar (0.4% agarose) containing indicator strain L. lactis HP. Zones of inhibition were visualized and measured after 24 h.
Genbank Accession Numbers of Applied Strains
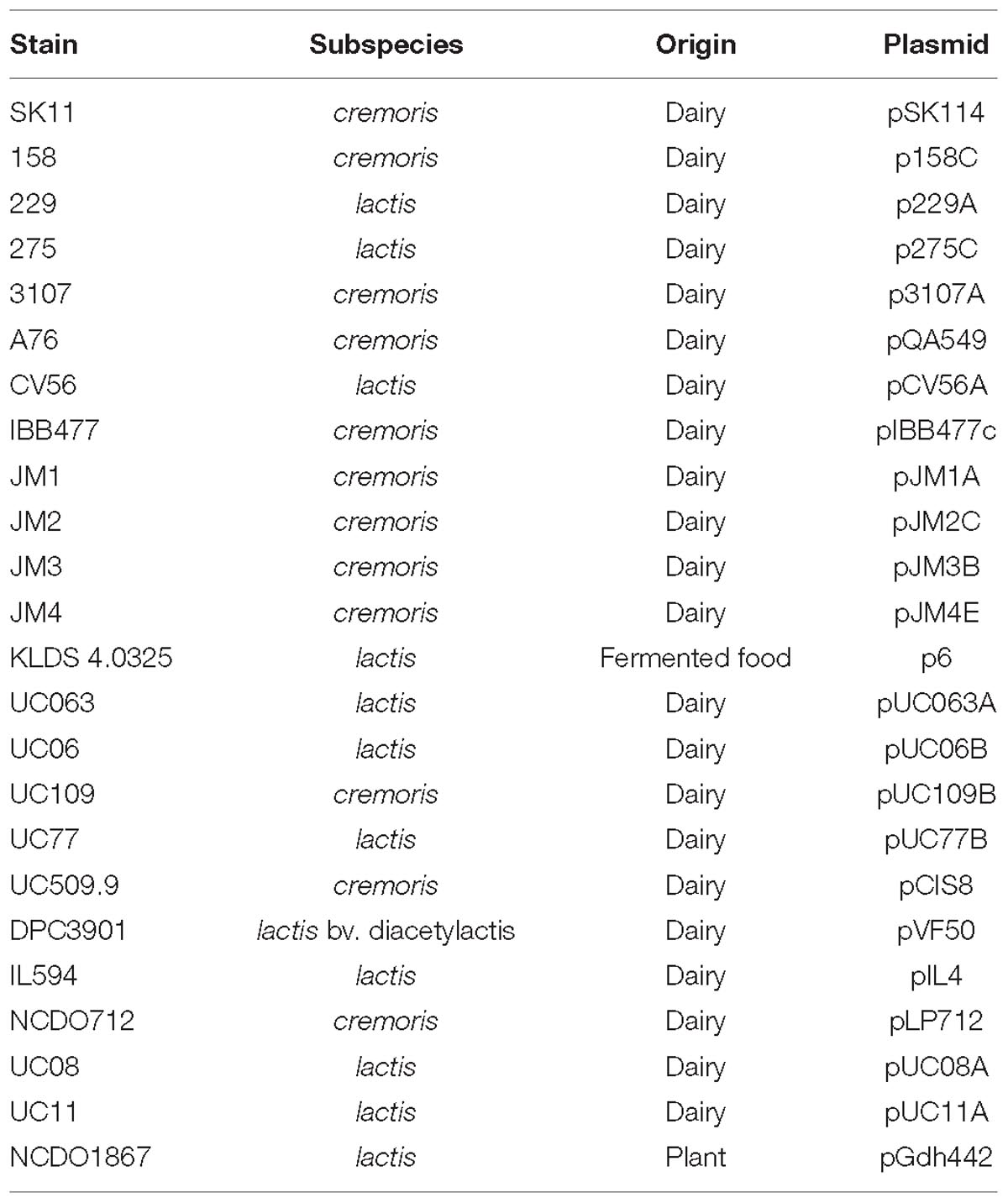
Lactococcus lactis subsp. lactis IL1403: AE005176; L. lactis subsp. lactis IO-1: AP012281; L. lactis subsp. lactis 184: CP015895; L. lactis subsp. lactis 229: CP015896; L. lactis subsp. lactis 275: CP015897; L. lactis subsp. lactis UC06: CP015902; L. lactis subsp. lactis UC08: CP015903; L. lactis subsp. lactis UC11: CP015904; L. lactis subsp. lactis UC063: CP015905; L. lactis subsp. lactis UC77: CP015906; L. lactis subsp. lactis UL8: CP015908; L. lactis subsp. lactis C10: CP015898; L. lactis subsp. cremoris SK11: CP000425; L. lactis subsp. cremoris MG1363: AM406671; L. lactis subsp. cremoris NZ9000: CP002094; L. lactis subsp. cremoris A76: CP003132; L. lactis subsp. cremoris UC509.9: CP003157; L. lactis subsp. cremoris KW2: CP004884; L. lactis subsp. cremoris 158: CP015894; L. lactis subsp. cremoris UC109: CP015907; L. lactis subsp. cremoris JM1: CP015899; L. lactis subsp. cremoris JM2: CP015900; L. lactis subsp. cremoris JM3: CP015901; L. lactis subsp. cremoris JM4: CP015909; L. lactis subsp. cremoris 3107: CP031538; L. lactis subsp. cremoris IBB477: CM007353; L. lactis subsp. lactis A12: LT599049; L. lactis subsp. lactis biovar. diacetylactis FM03: CP020604; L. lactis subsp. lactis 14B4: CP028160; and L. lactis subsp. cremoris HP: JAUH00000000.1.
Results
Plasmid Sequencing
In this study the sequences of 83 plasmids were elucidated utilizing a combined PacBio SMRT sequencing and Illumina MiSeq approach, and represent the detected plasmid complement of 16 lactococcal genomes (Kelleher et al., 2017). Initially 69 plasmids were identified from the SMRT sequencing data by modifying the RS_HGAP_assembly protocol in SMRT portal to a reduced minimum coverage cut-off of 15-fold coverage. To ensure complete coverage of the full plasmid complement the complete genomes of all 16 strains were re-sequenced utilizing an Illumina MiSeq approach which resulted in the eludication of a further 14 plasmids (indicated in Table 1) that had not been detected based on the original SMRT assemblies. These 14 plasmids ranged in size from 6 to 62 Kbp, indicating that their absence from the SMRT dataset was in the majority of cases not associated with exclusion from the library based on their small size. Therefore, it was hypothesized that the absence of some plasmids from the SMRT dataset was either due to a lower plasmid copy number (SMRT library preperation does not incorporate an amplification step) or due to a bias in the DNA extraction protocol. Conversely, no plasmids present in the SMRT assemblies, were absent from the Illumina data, however, Illumina sequencing generated heavily fragmented assemblies (∼100–250 contigs per strain), making eludication of complete plasmid sequences, particular for larger plasmids significantly more challenging if not impossible. The main advantage of SMRT technology is the long read length it achieves. Due to the high frequency of repetitive transposable elements, assembly of lactococcal genomes and plasmids is cumbersome. SMRT sequencing was shown to be very useful in obtaining reliable and accurate assemblies, being particularly beneficial for assembling larger lactococcal plasmids which frequently possess a mosaic type structure and contain multiple identical IS elements (Ainsworth et al., 2014c). Therefore, a combined sequencing approach is suggested as the most effective strategy for the complete sequencing of lactococcal strains.
General Plasmid Features
The sequenced plasmid dataset was combined with a further one hundred and seven plasmids retrieved from the NCBI database (National Centre for Biotechnology Information) (Table 1). In total, the features of one hundred and ninety plasmids derived from fifty three lactococcal strains in addition to seventeen lactococcal plasmids without an assigned strain were investigated. This extra-chromosomal DNA complement amounts to 4,987 Kbp of DNA and is predicted to represent 4,905 CDSs (i.e., ORFs that encode protein products), thus contributing very substantially to the overall genetic content of L. lactis.
The vast majority of currently sequenced plasmids originate from strains that were isolated from the dairy niche (149 out of 190 analyzed plasmids). These dairy lactococci carry between one and twelve plasmids (the latter in L. lactis biovar. diacetylactis FM03P), accounting for up to 355 Kbp of extra-chromosomal DNA in a given strain (as is the case for L. lactis JM1). The size of individual lactococcal plasmids varies widely from the smallest L. lactis KLDS4.0325 plasmid 2, with a size of 0.87 Kbp, to the two megaplasmids, each maintained by L. lactis JM1 and L. lactis JM2, with a size of 193 and 113 Kbp, respectively. The GC content of lactococcal plasmids ranges from ∼30–38%, whilst the average GC content of previously sequenced chromosomes is more constrained (34–36%). Only three lactococcal plasmids deviate from this range; pWC1 29.48, pIL105 29.79, and pHP003 40.05%, where the latter is closer to Streptococcus thermophilus genomic GC-content, which ranges from 39 to 40% (Fernández et al., 2011).
Lactococcal plasmids are known to replicate via either of two alternative methods, RCR or theta-type replication (Mills et al., 2006; Ainsworth et al., 2014c). Based on predicted plasmid replication proteins/origins it appears that the majority of lactococcal plasmids (174 of the current data-set) replicate via the theta-type mechanism, while only a small proportion appears to utilize RCR (sixteen of the current data-set). The relatively small number of plasmids utilizing RCR may be attributed to a number of factors, such as the fact that RCR plasmids can only support a limited replicon size (<10 Kbp), incompatibility with other RCR type plasmids (Leenhouts et al., 1991), and/or intrinsic structural and segregational instability (Ainsworth et al., 2014c). In three instances, the analysis identified plasmids for which the replication mode could not be clearly determined as the origin of replication of these plasmids did not conform to the typical origin of replication associated with RCR or theta replication.
Pan-Plasmidome Calculation
The pan-plasmidome calculation provides an overview of the overall genetic diversity of the L. lactis plasmidome, the latter representing the total plasmid content harbored by (sequenced) members of the L. lactis taxon. To calculate the pan-plasmidome, a pan-genome analysis approach was applied using the PGAP v1.0 pipeline (Zhao et al., 2012). The resultant pan-plasmidome graph (Figure 1) displays an asymptotic curve rising steadily as each of the one hundred and ninety plasmids included in the analysis is added until a total pan-plasmidome size of 1, 315 CDSs was reached. The trend observed in the pan-genome indicates that the pan-plasmidome remains in a fluid or open state, and that, therefore, continued plasmid sequencing efforts will further expand the observed genetic diversity among lactococcal plasmids. The PGAP pipeline was also used to determine the core genome of the lactococcal plasmid sequence data set. Interestingly, no single CDS is conserved across all plasmids resulting in an empty core genome.
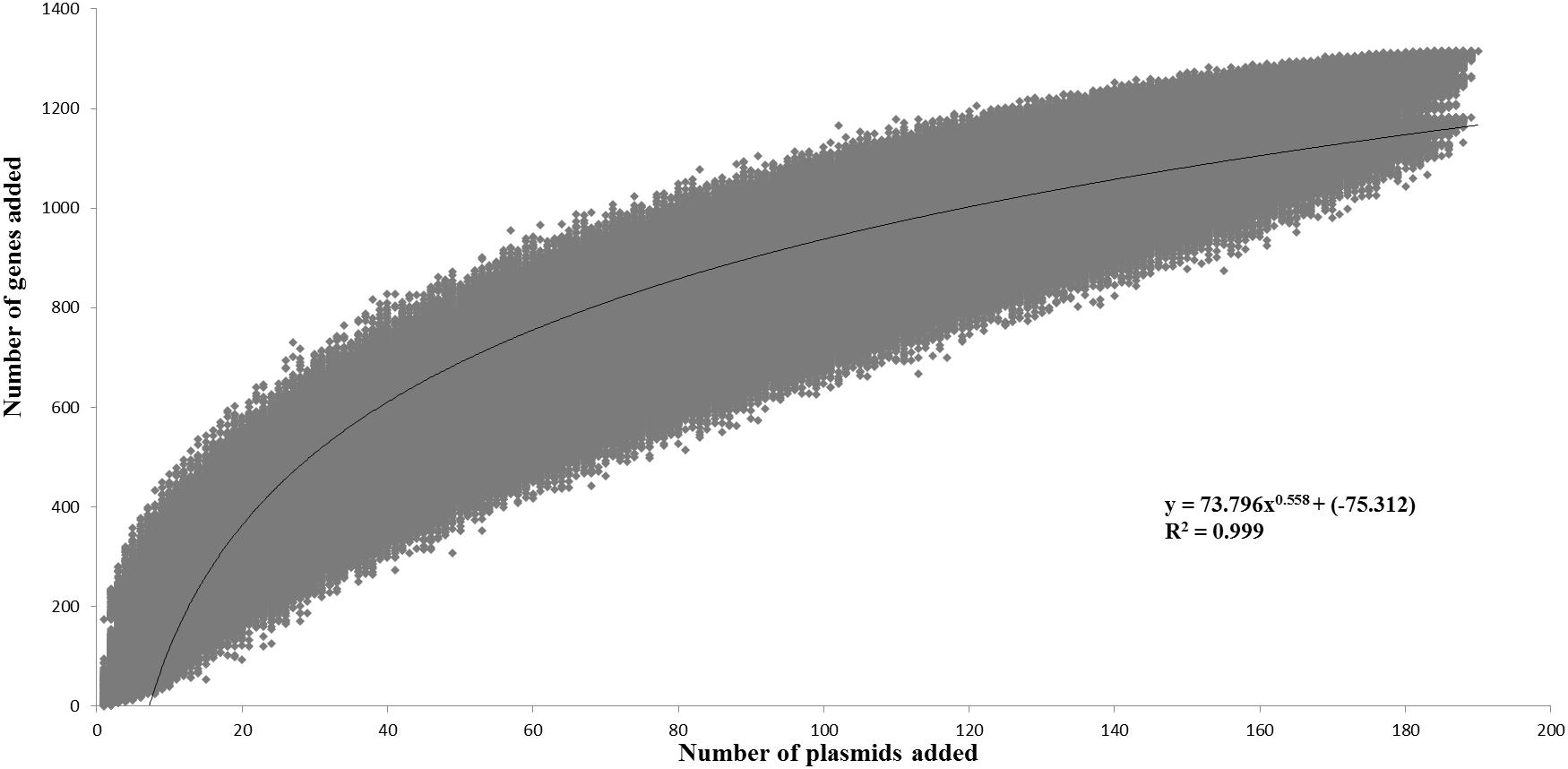
Figure 1. Pan-plasmidome of Lactococcus lactis. This graph represents the accumulated number of new genes in the L. lactis pan-plasmidome plotted against the number of plasmids added. The deduced mathematical equation is also indicated.
The L. lactis pan-genome, based on chromosomal sequences only, has previously been calculated to constitute 5,906 CDSs (Kelleher et al., 2017). When compared with the calculated lactococcal plasmidome (1,315 CDSs), it is obvious that the lactococcal plasmidome contributes very substantially to overall lactococcal genetic diversity.
MCL Analysis of the Pan-Plasmidome
To explore the genetic content of the one hundred and ninety plasmids employed in this study, all-against-all reciprocal BLASTP analysis and MCL (Markov clustering) was conducted (Altschul et al., 1990; Enright et al., 2002). The plasmidome was determined to comprise 885 protein families, of which 413 represented single member protein families, evidence of the divergent nature of the plasmid sequences. Furthermore, 421 of these families constitute hypothetical protein families, being represented by a total of 1,341 individual proteins. These hypothetical proteins encompass 22.7% of the total CDSs in the lactococcal plasmidome.
The second largest constituent of the lactococcal plasmidome is that represented by transposable elements. Transposable elements encompass 825 CDS, or 15.7% of the plasmidome, with members of the IS6, IS30, IS982, and ISL3 insertion families being among the most dominant genetic elements. These mobile elements are responsible for the transfer and recombination of DNA (Nicolas et al., 2007; Machielsen et al., 2011; Alkema et al., 2016) and are likely to contribute to a fluid lactococcal plasmidome.
Following MCL analysis, HCL of the pan-plasmidome was used to cluster plasmids based on their genetic content (Figure 2). The high level of diversity within the pan-plasmidome is demonstrated by the observed disparity within the HCL matrix. HCL analysis resulted in thirteen clusters with three outliers; pMPJM1, pWVO2, and pQA504 (Figure 2B). Plasmid pWVO2 encodes a single replication gene, pQA504 contains three CDS (rep gene, mob gene, and hypothetical gene), while pMPJM1 encodes 188 CDS and shares little homology with other lactococcal plasmids. The remaining thirteen clusters did not display subspecies specificity, each cluster containing plasmids from both subsp. lactis and subsp. cremoris hosts.
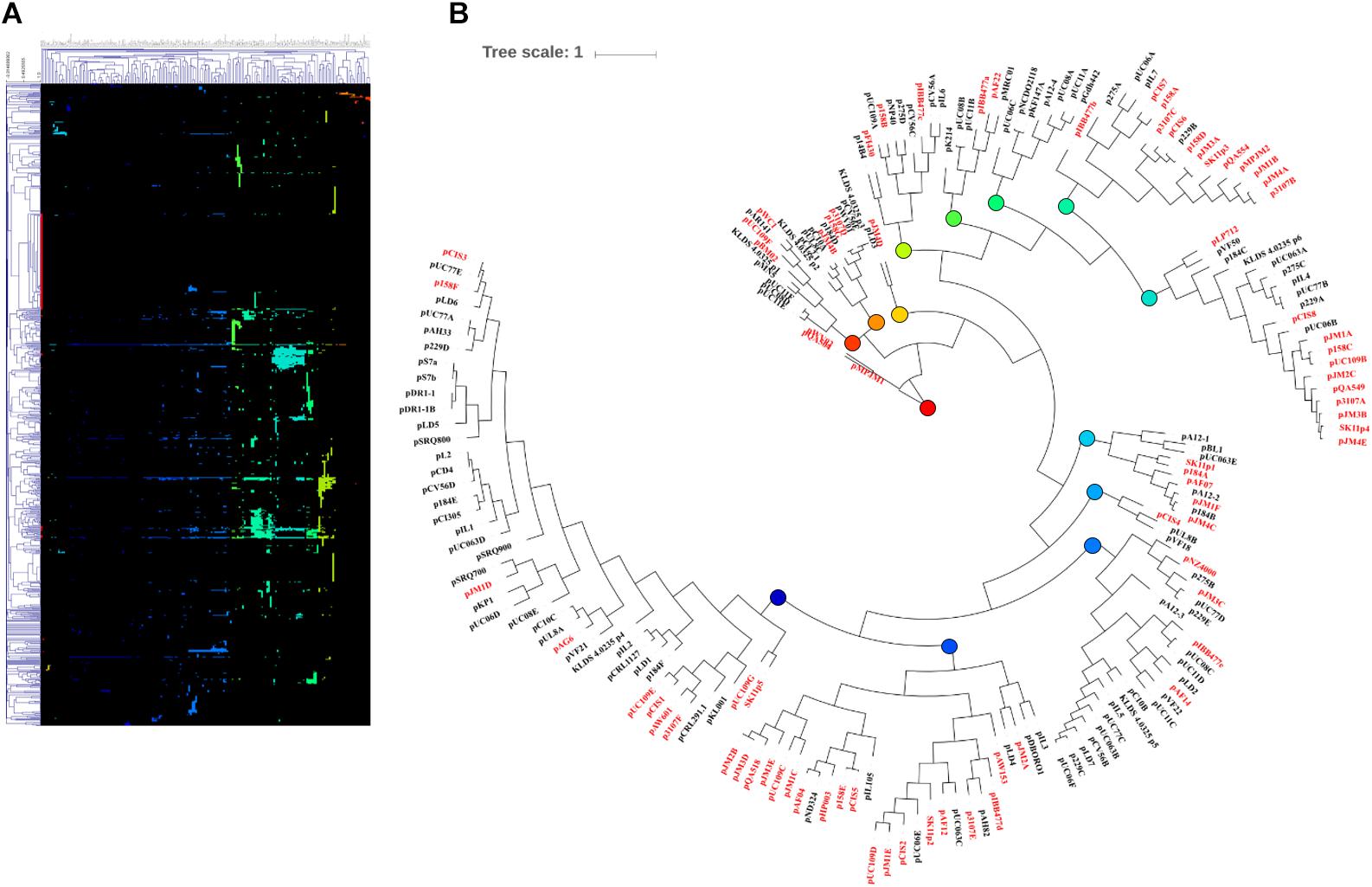
Figure 2. HCL analysis of the lactococcal plasmidome. Hierarchical clustering analysis (A) representing the presence/absence of gene families from 190 lactococcal plasmids. Absence of gene family members is indicated in black and presence of gene family members is indicated in color. (B) Circular tree representation of panel A displaying HCL plasmid groupings. Plasmids from subsp. cremoris strains are labeled in red while plasmids from subsp. Lactis strains are labeled in black. Colored nodes correspond to the presence of genes (colors) in HCL matrix (panel A).
Lactococcal Megaplasmids
Typically, L. lactis plasmids range in size from 1 to 50 Kbp, and, prior to this study, the largest plasmid identified in L. lactis was the self-conjugative mega-plasmid of 155,960 bp in L. lactis subsp. lactis bv. diacetylactis S50 (Kojic et al., 2005). L. lactis S50 p7 represents the first lactococcal megaplasmid and encodes genes for Proteinase PI and lactococcin A and is part of a larger plasmid complement of 7 plasmids totaling 336 Kbp (Kojic et al., 2005). Recently (May 2018) the plasmid complement of L. lactis subsp. lactis KLDS 4.0325 (Yang et al., 2013) has been updated in the public NCBI data base with three additional plasmid sequences, the largest plasmid measuring 109 Kbp (plasmid 6). In the current study, whole genome sequencing efforts resulted in the identification of two plasmids that were larger than 100 Kbp, namely pMPJM1 (193 Kbp) and pMPJM2 (113 Kbp) from L. lactis JM1 and L. lactis JM2, respectively, and owing to their size are defined as megaplasmids (Anton et al., 1995; Barton et al., 1995; Figures 3A,B). Pulsed field gel electrophoresis also identified bands which would be consistent with plasmids of that size, although unambiguous validation will require Southern hybridization analysis (Figure 3C).
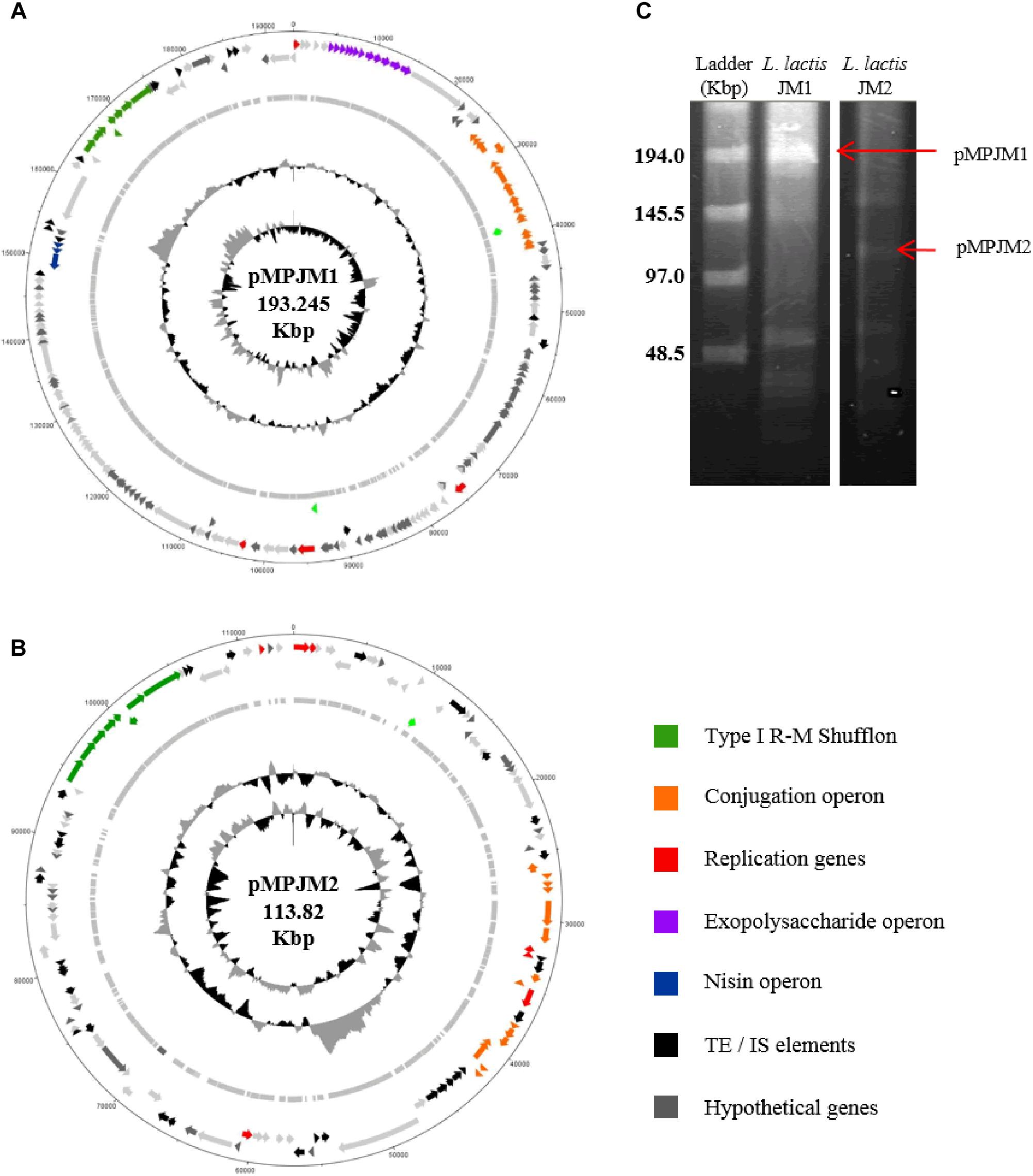
Figure 3. General features of megaplasmids pMPJM1 and pMPJM2. Circular maps of (A) pMPJM1 and (B) pMPJM2. CDS of interest are highlighted in color. (C) PFGE image of pMPJM1 (lane 2) and pMPJM2 (lane 3), the possible position of each of the two megaplasmids is indicated by a red arrow. CHEF lambda (Bio-Rad Laboratories, Hercules, CA, United States) DNA ladder is displayed in lane 1.
The larger of the two megaplasmids, pMPJM1, encompasses 186 CDSs and is presumed to replicate (as expected for such a large replicon) via the theta-type replication mechanism [based on the identification of the origin of replication (ori), comprised of an AT-rich region plus three and a half iterons of 22 bp in length] (Seegers et al., 1994). pMPJM1 encompasses, among others, gene clusters predicted to be responsible for (exo)polysaccharide biosynthesis, conjugation and nisin resistance, while it also specifies an apparently novel type I RM shufflon system (as well as a high proportion of unique/hypothetical CDSs). The overall sequence of the plasmid shows little homology to previously sequenced plasmids in the NCBI databases, however, it shares 24% sequence coverage with 99% nucleotide identity to the other identified megaplasmid pMPJM2, which indicates that they share a common ancestor. pMPJM2 encodes 123 CDSs and BLASTN analysis identified sequence identity to a number of different lactococcal plasmids indicating a mosaic genetic structure commonly seen in large lactococcal plasmids (Ainsworth et al., 2014c). pMPJM2 also encodes a putative conjugation operon and a very close homolog of the type I RM shufflon system of pMPJM1. The third lactococcal megaplasmid KLDS 4.0325 plasmid 6 (109 Kbp) encodes 119 CDSs including the lac operon and associated opp oligopeptide uptake system.
Technological Properties
Strains of L. lactis are commonly used as starter cultures employed by the dairy industry (Beresford et al., 2001), and their dairy adaptations such as citrate metabolism and lactose utilization are frequently plasmid-encoded. In L. lactis, citrate uptake and subsequent diacetyl production is governed by the plasmid-encoded citQRP operon (Drider et al., 2004; Van Mastrigt et al., 2018b). In the current data set, only four plasmids contain the citQRP operon, L. lactis CRL1127 plasmid pCRL1127, L. lactis IL594 plasmid pIL2 (Górecki et al., 2011), L. lactis FM03 plasmid pLD1 and L. lactis 184 plasmid p184F. However, the latter operon in p184F appears to lack citQ which encodes a leader peptide. Lactose metabolism is controlled by the lac operon consisting of the genes lacABCDEFGX and is regulated by a repressor, encoded by the adjacent lacR gene (Cords et al., 1974), both citrate and lactose utilization have previously been described in detail (Cords et al., 1974; Górecki et al., 2011).
The lac operon was found to be present on twenty four plasmids (in 24 different strains) (Table 2). The plasmids analyzed were derived from 53 lactococcal strains in addition to 17 lactococcal plasmids unassigned to a particular strain, and represented the total plasmid complement of 26 such strains. In all cases bar two, the strains were isolated from the dairy environment with the exception of L. lactis NCDO1867 isolated from peas and L. lactis KLDS 4.0325 isolated from fermented food (Table 1). Alternative lactose metabolism methods have previously been observed in L. lactis. For example, L. lactis MG1363 does not harbor the lac operon, yet is capable of growth on lactose-supplemented media due to the activity of a cellobiose-specific phosphotransferase system (PTS), which can act as an alternative lactose utilization pathway (Solopova et al., 2012). Another example of an alternative lactose metabolic pathway is found in the slow lactose fermenter L. lactis NCDO2054, which metabolizes lactose via the Leloir pathway (Bissett and Anderson, 1974). Plasmid integration events have also resulted in the integration of the lac operon in the chromosome of L. lactis SO, where it is located 20 Kbp downstream of an integrated opp operon, sharing significant homology with (the lac operons of) plasmids pCV56B, pSK08, pKF147A, and pNCDO2118 (Kelleher et al., 2017). Due to the lack of sequencing projects that report fully sequenced genomes, defining the true frequency of lactose utilization is challenging. However, of those strains for which complete genome sequencing projects have been described [30 strains in Kelleher et al. (2017)], 22 were found to be capable of metabolizing lactose based on growth in lactose supplemented broth, 19 via plasmid-encoded lac operons, one via a chromosomally encoded lac operon and two by an alternative pathway. This analysis included 12 subsp. cremoris strains, of which all but one possessed genes for a lactose utilization mechanism, the exception being strain KW2, which lacks a plasmid complement.
Conjugation
Conjugation and transduction are believed to be the dominant mechanisms of plasmid transfer in L. lactis (Ainsworth et al., 2014c). Particular emphasis has been placed on conjugation as it is considered a naturally occurring DNA transfer process and for this reason may be used in food-grade applications to confer beneficial traits to industrial strains (Mills et al., 2006). Generally, during conjugation the AT-rich, so-called “origin of transfer” or oriT of the conjugative plasmid is nicked by a nickase, and the resulting ssDNA strand is passed to a recipient cell (Grohmann et al., 2003). The tra (transfer) locus is believed to be responsible for the donor-to-recipient DNA transfer process of conjugation, though the precise mechanistic details of the conjugation process in L. lactis has not yet been fully elucidated. Plasmids which do not encode the tra operon, may also be co-transferred by conjugation in instances where a plasmid contains an oriT sequence and at least one mobilization gene (mobA, B, C, or D). Additional genes can also be involved in conjugation in L. lactis; an example of this is cluA, which encodes a cell surface-presented protein, and which is involved in cell aggregation and thought to be essential for high efficiency conjugal transfer (Stentz et al., 2006). Furthermore, a chromosomally associated, so-called sex factor in L. lactis has been shown to facilitate transfer of chromosomal genes during conjugation (Gasson et al., 1995).
The tra locus, which encodes the protein complex responsible for donor-to-recipient DNA transfer has as yet been fully eludicated. Previous studies have identified the role of traF as encoding a membrane-spanning protein involved in channel formation and membrane fusion. In addition, the traE and traG genes have been proposed to encode proteins involved in the formation of the conjugal pilus similar to type IV secretion systems (O’Driscoll et al., 2006; Górecki et al., 2011). Typically, the three tra genes (i.e., traE, traF, and traG) are part of a larger gene cluster (consisting of up to 15 genes; Figure 4), including traA, which encodes a DNA relaxase. In the current data set, 34 genes with homology to traG were identified on 27 plasmids (present in duplicate on seven plasmids) along with five occurrences of traE/F also being present (in the case of plasmids pIBB477A, pUC08B, pUC11B, pAF22, and pMRC01).
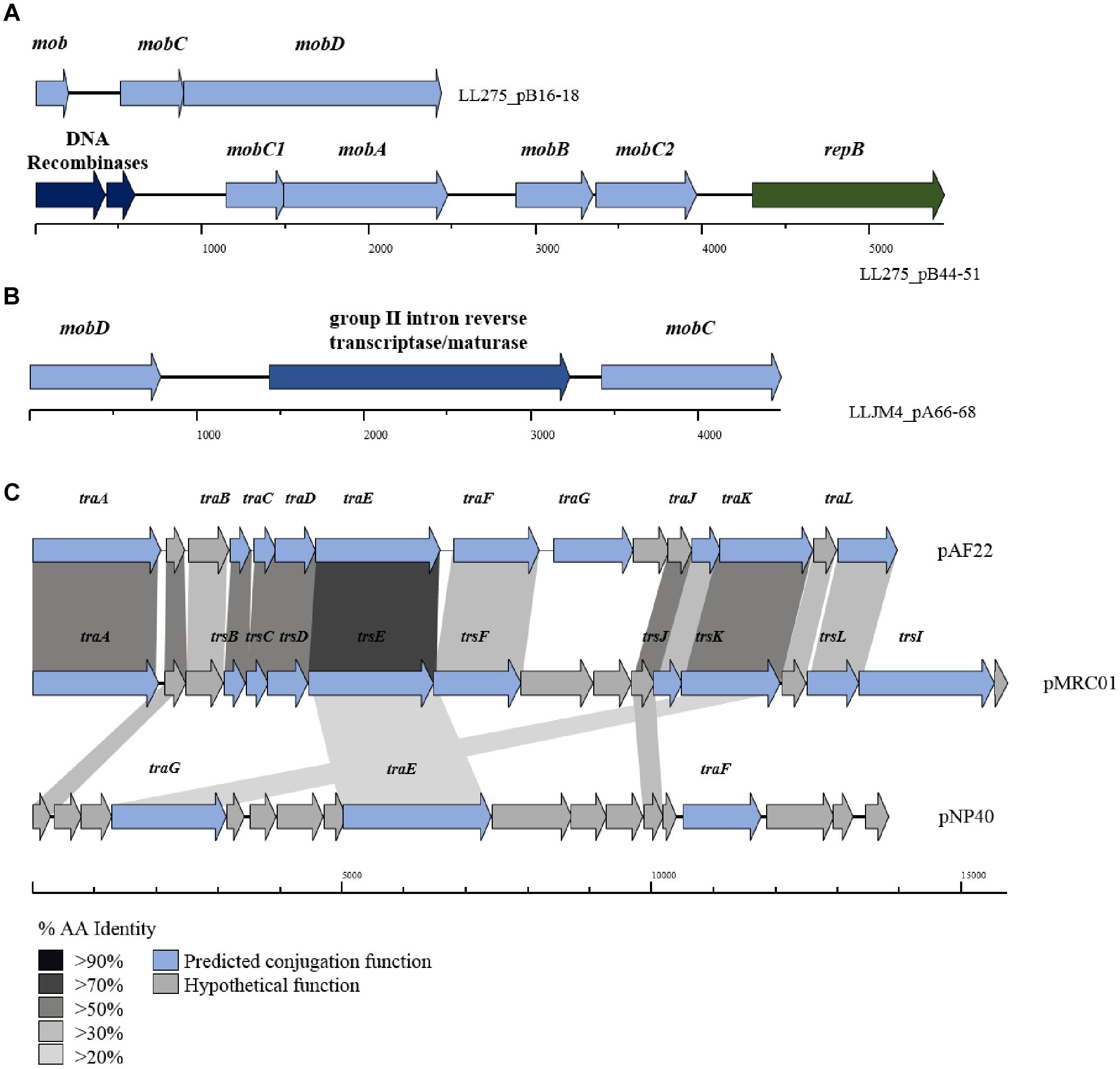
Figure 4. BLAST map of active lactococcal conjugation gene clusters. The image describes the genetic organization of mob genes in (A) p275B, (B) pJM4A, and (C) the conjugation gene clusters from plasmids pAF22, pMRC01, and pNP40. All three plasmids have previously been shown to be self-transmissible by conjugation. Gene synteny is highly conserved between pAF22 and pMRC01, but amino acid identity is not, while pNP40 represents a divergent system. Amino acid identity is indicated by the shaded boxes. Arrows colored blue indicate predicted conjugative function, while arrows shaded gray indicate hypothetical functions.
The precise functions for the remainder of the genes in the tra gene cluster have yet to be elucidated, though additional tra-encoded functions have been predicted in a small number of cases, the majority based on homology to the trs operon in Staphylococcus (Sharma et al., 1994). For example, traJ and traL were identified on plasmids pAF22, pIBB477a and pMRC01, and traB, traC, traD, traF (mating channel formation) and traK (P-loop NTPase) on plasmids pUC08B, pIBB477a, pUC11B, pAF22, and pMRC01. Plasmids pAF22, pMRC01, and pNP40 have all previously been demonstrated to be capable of conjugation (Harrington and Hill, 1991; Coakley et al., 1997; O’Driscoll et al., 2006; Fallico et al., 2012). However, the annotation(s) of the operons involved in conjugation is not well defined and they are currently poorly characterized. This is also amplified by both a lack of sequence conservation and limited synteny within the genes that make up these conjugation-associated genetic clusters (Figure 4).
While the tra operon is thought to be responsible for the formation of conjugal pili, previous studies have identified a number of genes believed to play a role in the mobilization of other (non-self-transmissible) plasmids in L. lactis (Mills et al., 2006; O’Driscoll et al., 2006; Millen et al., 2012); principal among these are the mob (mobilization) genes. Mobilization genes are responsible for nicking the plasmid’s dsDNA at a particular site and forming a relaxosome, which allows the transfer of a single stranded template to a recipient cell. Variants of four main mob genes are distributed throughout the lactococcal plasmidome; mobA and mobD encode nickases, and mobB and mobC, whose protein products are thought to form a relaxosome with an associated nickase (either mobA or mobD) are typically present in the genetic configuration mobABC or mobDC. Comparative analysis identified 422 occurrences of mob genes (any of the afore mentioned mob genes) distributed across the 190 plasmids assessed in this study, including 15 occurrences of a predicted retron-type reverse transcriptase or maturase (located between mobD and mobC) believed to play a role in DNA recombination. The results indicate that 59.5% of plasmids in the lactococcal plasmidome carry at least one or more genes encoding mobilization proteins.
The lactococcal megaplasmids pMPJM1 and pMPJM2 harbor two (16 Kbp) regions putatively involved in conjugation and/or mobilization. In the case of pMPJM2 the predicted region was found to contain homologs of mobC and mobD, encoding a nickase and an associated relaxase near a possible secondary replication origin. However, the presence of five transposase-encoding genes and the lack of predicted tra genes with conserved functions suggest that this plasmid is not capable of autonomous conjugation (though mobilization is possible).
Conversely, analysis of pMPJM1 identified a more divergent system to that typically found in lactococcal plasmids. Three hypothetical proteins were found to contain the PFAM domain usually conserved in conjugation proteins (pfam12846), in addition to a homolog of virB11, whose deduced product acts as a type IV secretory pathway ATPase (pfam00437). Cellular localization analysis of the operon using PsortB was also indicative of a transmembrane complex composed of cytoplasmic, membrane bound, signal and extracellular proteins (Figure 5). The divergence of both operons from typical lactococcal conjugative operons suggests that these two megaplasmids have lost their conjugative ability or may possess a conjugation system with very few identifiable similarities to currently known systems.
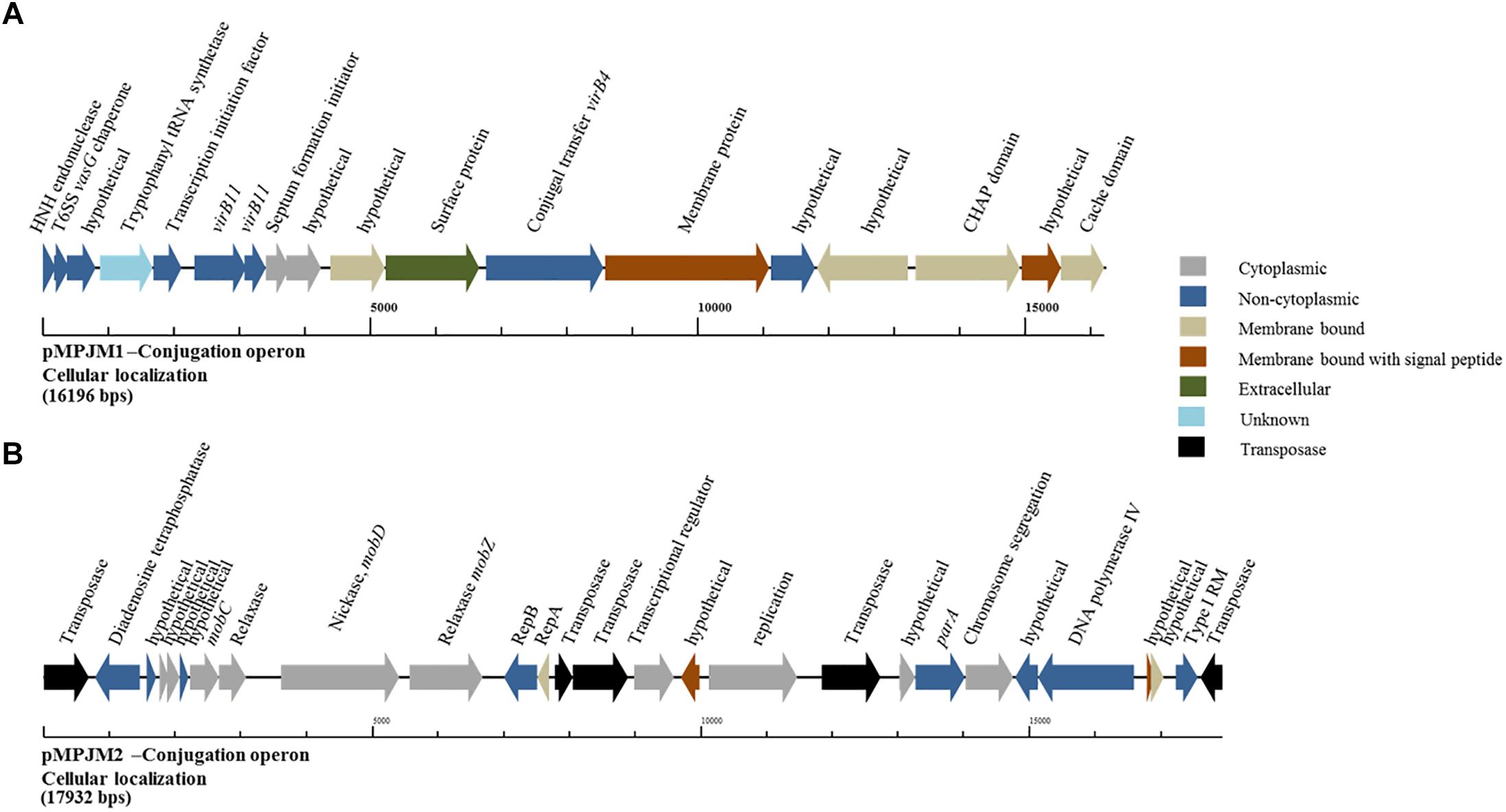
Figure 5. Genetic organization of the putative conjugation gene clusters in pMPJM1 and pMPJM2. (A) Represents the putative conjugation locus in pMPJM1. (B) Represents the putative conjugation locus in pMPJM2. Colors indicate the predicted cellular localization of each product. The system in pMPJM1 appears to encode proteins involved in conjugal transfer, while the cellular localization data is predictive of a transmembrane complex. Conversely, the conjugation locus in pMPJM2 appears to be involved in mobilization rather than conjugation, and the presence of a number of insertion elements suggest it is unlikely to be functional.
Cell Surface Interactions (Adhesion & EPS)
Mucin-binding proteins, i.e., those allowing adhesion to the mucin layer of the gastrointestinal tract, are considered essential for stable and extended gut colonization by LAB (Von Ossowski et al., 2010). While lactococci are typically not associated with the human gut and do not have a growth temperature profile that would be inconsistent with GIT colonization., instances of such proteins encoded by lactococcal plasmids have been reported (Kojic et al., 2011; Lukić et al., 2012; Le et al., 2013). Muco-adhesive proteins are considered of paramount importance for the efficacy of probiotic bacteria (Von Ossowski et al., 2010) and the presence of such elements in L. lactis may have significant commercial impact for their role in functional foods. Analysis of the plasmids assessed in our study identified a number of strains with predicted novel muco-adhesive elements, similar to those found in pKP1 (Kojic et al., 2011). Plasmid pKP1 encodes two proteins, a mucin-binding domain-containing protein and an aggregation-promoting protein AggL, which promotes its binding to colonic mucosa (Lukić et al., 2012). While no direct homolog of AggL was detected, mucus-binding protein-encoding genes were identified on plasmids p14B4, p275A, p275B, pUC08B, and pUC11B perhaps reflecting a potential for gastrointestinal persistence conferred to the strains that carry these plasmids. A number of additional proteins predicted to be host cell surface-associated were detected during the analysis. For example, pUC11C encodes two class C sortases, which are commonly involved in pilus biosynthesis (Von Ossowski et al., 2010; Lebeer et al., 2012), while p275A encodes an LPXTG anchor domain, cell surface-associated protein. Interestingly, each of these strains belongs to subspecies lactis and is capable of growth at 37°C, which would impede growth of their cremoris counterparts, which are generally less thermo-tolerant. L. lactis JM1 is the sole cremoris strain that is predicted to encode proteins directly involved in host cell surface alterations. This plasmid encodes five putative proteins containing a 26-residue repeat domain found in predicted surface proteins (often lipoproteins) and one collagen-binding domain protein.
The plasmid encoded lactococcal cell wall anchored proteinase, PrtP, involved in the breakdown of milk caseins in dairy lactococci, has previously been shown to cause a significant increase in cell adhesion to solid glass and tetrafluoroethylene surfaces (Habimana et al., 2007). More recently, L. lactis subsp. cremoris IBB477 was found to contain two plasmids, pIBB477a and pIBB477b, which encode cell wall-associated peptidases that have been shown to mediate adhesion to bare mucin and fibronectin coated polystyrene and HT29-MTX cells (Radziwill-Bienkowska et al., 2017). Analysis of the current data-set which contains a large number of dairy derived plasmids, identified a further 194 CDS homologous to the cell wall-associated peptidase S8 (PrtP) of IBB477. Whilst extracellular cell wall proteinases have been shown to be directly associated with the bitter flavor defect in Cheddar cheese varieties (Broadbent et al., 2002), a potential role for these peptidases in gut adhesion may present a more positive view of these elements.
Exopolysaccharide production by L. lactis is a characteristic trait of strains isolated from viscous Scandinavian fermented milk products and is widely reported as a plasmid-encoded trait (Vedamuthu and Neville, 1986; Von Wright and Tynkkynen, 1987; Neve et al., 1988; Kranenburg et al., 1997). EPS production by L. lactis strains is of particular importance for functional foods, as the EPS produced by these strains is considered to be a food-grade additive that significantly contributes to properties such as mouth-feel and texture in fermented dairy products (Kleerebezem et al., 1999). The L. lactis EPS biosynthesis gene cluster (eps) contained on pNZ4000 has previously been characterized (Kranenburg et al., 1997) and consists of 14 genes, namely epsRXABCDEFGHIJK. Comparison of the eps gene cluster from pNZ4000 with all sequenced plasmids in the current dataset identified a further four plasmids harboring eps clusters, namely pUC77D, p229E, pJM3C, p275B, and pMPJM1 (Figure 6). In pNZ4000, EPS production is regulated by epsRX, EPS subunit polymerization and export is believed to be executed by the encoded products of epsABIK, while the proteins encoded by epsDEFGH are responsible for the biosynthesis of the EPS subunit (Kranenburg et al., 1997). Homology-based analysis with the five newly identified gene clusters shows that in all cases epsRXABCD are conserved (except in pMPJM1 where epsR is absent), while the remainder of the gene cluster in each case consists of variable genes. These eps gene clusters consist of a highly conserved region at the proximal end of the cluster and a variable distal region, which is not unlike other lactococcal polysaccharide biosynthesis clusters (Mahony et al., 2013; Ainsworth et al., 2014b; Mahony et al., 2015). The conserved epsRX genes are responsible for transcriptional regulation, the products of epsAB are required for EPS export, while the deduced proteins of epsCD are putative glycosyltransferases of which EpsD (priming glycosyltransferase) has previously been demonstrated to be essential for EPS subunit biosynthesis (Kranenburg et al., 1997). The variable region, epsEFGHIJKLP in pNZ4000, encodes predicted or proven functions, such as an acetyltransferase (epsE), glycosyltransferases (epsGHIJ) and a flippase (epsK), together representing the presumed enzymatic machinery responsible for EPS biosynthesis through the addition and export of sugar moieties.
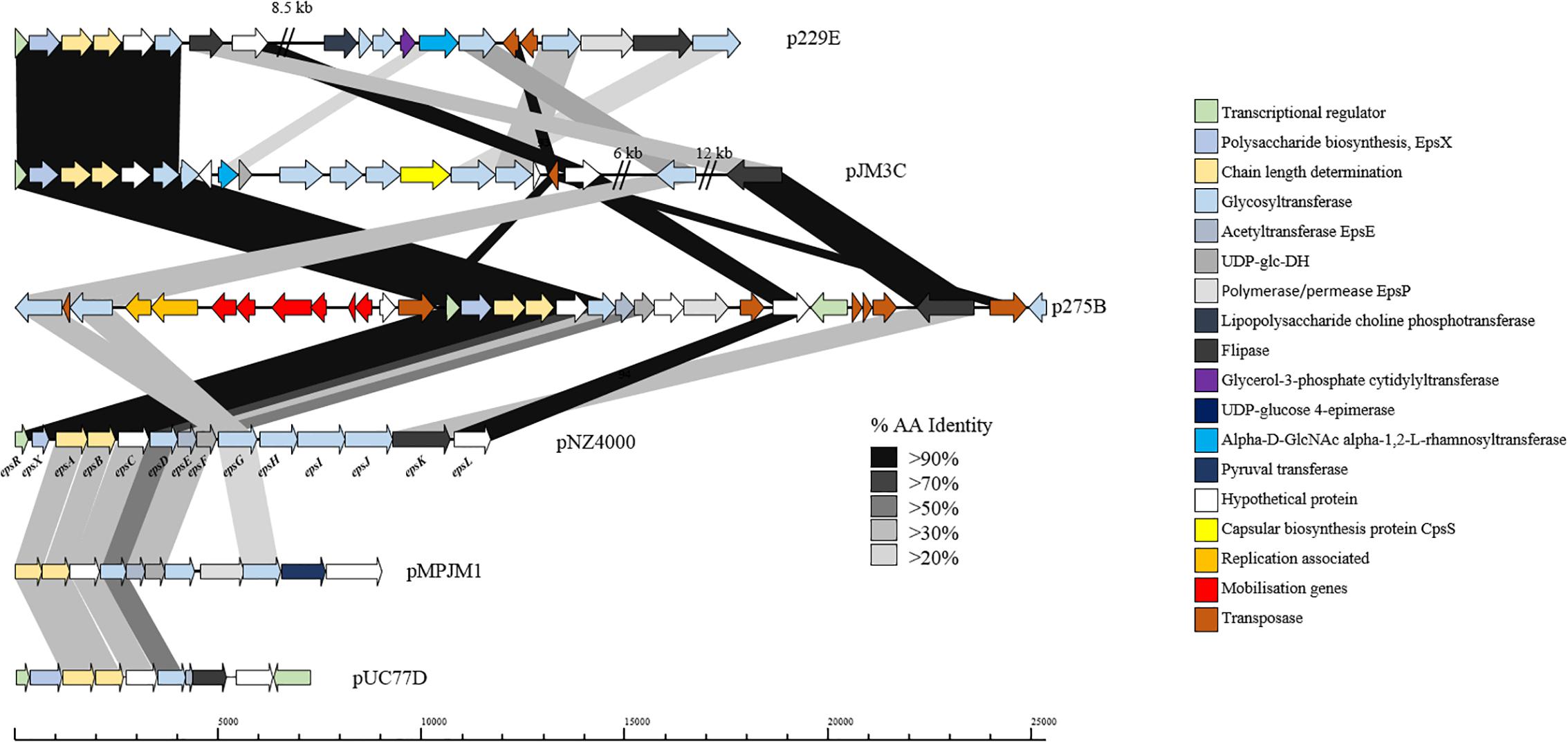
Figure 6. Linear BLAST map of the lactococcal EPS gene clusters. Linear BLAST map of eps gene clusters from (1) p229E, (2) pJM3C, (3) p275B, (4) pNZ4000, (5) pMPJM1, and (6) pUC77D. Arrow color indicates predicted product, while shaded region indicated percentage amino acid identity between BLAST hits. The highly conserved region of the gene cluster is apparent from EpsR to EpsD while the variable region is strain specific.
In the case of p229E, the variable eps region is composed of CDSs predicted to encode products with functions are similar to the chromosomally located cwps gene cluster in strain 229. Plasmid pJM3C contains genes predicted to encode a rhamnosyltransferase, UDP-glucose dehydrogenase, capsular biosynthesis protein and five glycosyltransferases. The p275B variable region is heavily rearranged due to the presence of nine transposase-encoding genes. The megaplasmid pMPJM1 encodes a 9 Kbp predicted EPS region with well conserved functional synteny to that of pNZ4000, although with relatively low homology (Figure 6). Plasmid pUC77D appears to contain the shortest eps gene cluster of 7 Kbp due to the absence of epsFGHIJ genes. Further analysis of these plasmid-borne eps gene clusters revealed that in all cases mob elements are present, indicating that they may be mobilisable via conjugation. To assess if these plasmids have a common lineage, nucleotide homology based analysis was conducted utilizing BLASTN (Altschul et al., 1990). This analysis, however, did not identify significant homology or common hits between the plasmids outside of the conserved region of the EPS gene cluster. Phenotypic analysis of strains L. lactis 275, 229, JM1, JM3, and UC77 indicated a mucoid EPS phenotype in strains 275, 229, and JM3. While strains JM1 and UC77 did not show any EPS production which is probably attributed to the lack of the regulator epsR in strain JM1 and the absence of epsFGHIJ genes in UC77.
Bacteriocins
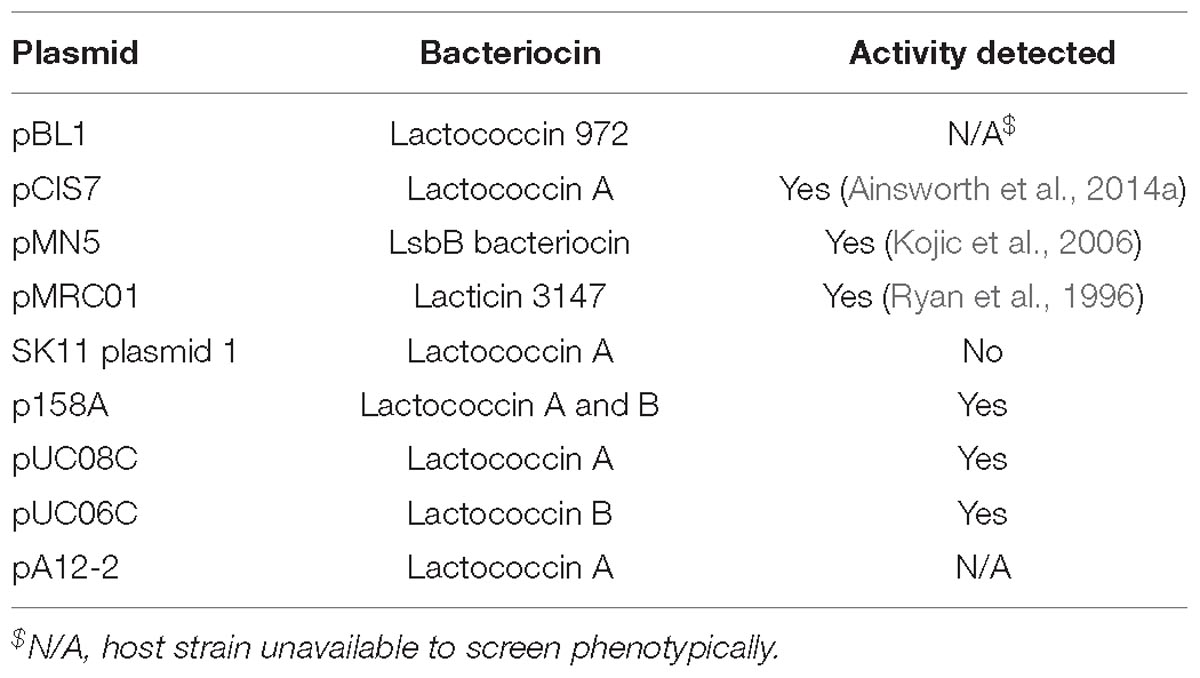
Bacteriocins are a diverse group of ribosomally synthesized bacterial peptides, which when secreted inhibit growth of other bacteria by interfering with cell wall biosynthesis or disrupting membrane integrity (Dobson et al., 2012). The production of bacteriocins by lactococcal strains has been widely reported, including the strain L. lactis subsp. cremoris 9B4 which contains three separate bacteriocin operons, named lactococcins A, B, and M/N are located on one plasmid (Van Belkum et al., 1989, 1991). To investigate bacteriocin production in the lactococcal plasmidome, all the available strains were screened for bacteriocin production against an indicator strain L. lactis subsp. cremoris HP. In total six strains were found to produce clearly defined zones of inhibition, indicating bacteriocin production, namely L. lactis subsp. lactis IO-1, 184, UC06, UC08, UC11, and L. lactis subsp. cremoris 158. Analysis of the plasmid complement of each of these strains indicated that strains 158, UC06 and UC08 each possess a plasmid-borne bacteriocin gene cluster, while IO-1, 184, and UC11 contain a bacteriocin gene cluster of chromosomal origin. In each case, these were identified as lactococcin producers: p158A is predicted to be responsible for lactococcin A and B production, pUC08A for lactococcin A production, and pUC06C for lactococcin B biosynthesis. Lactococcin has a narrow spectrum of activity, targeting predominantly closely related lactococcal species (Geis et al., 1983) and, as such, is an important consideration when selecting strains for application in mixed starter cultures.
Sequence analysis of the remaining plasmids in the current study (for which strains were not available for phenotypic analysis) identified additional putative bacteriocin-encoding gene clusters (Table 3), which were found to be responsible for the production of lactococcin A or B, and in one case (pMRC01) for the lantibiotic lacticin 3147 (Table 3; Dougherty et al., 1998).
Phage-Resistance Systems
Lactococcal strains typically possess a variety of phage defense mechanisms including superinfection exclusion systems (Sie) (encoded by integrated prophages) (Kelleher et al., 2018), clustered regularly interspaced short palindromic repeats (CRISPR), restriction-modification (R-M), and abortive infection (Abi) systems. Sie systems are a prophage-encoded defense mechanism (Mcgrath et al., 2002; Mahony et al., 2008) and have been reviewed extensively in these strains as part of an investigation into lactococcal prophages (Kelleher et al., 2018). CRISPR and CRISPR-associated (cas) genes specify an acquired adaptive immunity system against invading DNA in bacteria (Horvath and Barrangou, 2010). To date, only one such system has been characterized in Lactococcus on a conjugation-transmissible plasmid, pKLM, which encodes a novel type III CRISPR-Cas system (though it is unable to incorporate new spacers) (Millen et al., 2012). Analysis of plasmid sequences in this study did not detect any further instances of CRISPR systems in lactococci, suggesting CRISPR are not a widespread phenomenon in domesticated lactococci.
Restriction-modification systems are extremely diverse and widespread and are encoded by approximately 90% of all currently available bacterial and archaeal genomes (Roberts et al., 2003). R-M systems are frequently observed in the lactococcal plasmidome and some examples have previously been characterized including the Type II system LlaDCHI from pSRQ700 (Moineau et al., 1995) and LlaJI from pNP40 (O’driscoll et al., 2004). The current dataset holds nine apparently complete Type II systems on plasmids pCV56A, p275D, pJM1D, pUC08B, pUC11B, pNP40, pSRQ700, KLDS 4.0325 plasmid 5, and pAF22; along with multiple orphan methylases and solitary restriction endonucleases. The most commonly encountered R-M systems in lactococcal plasmids are Type I systems. These systems are often incomplete and represented by solitary specificity subunits (77 such orphan specificity subunit-encoding hsdS genes were identified in the current analysis). The high frequency of these systems in lactococcal plasmids is indicative of host adaptation as they predominantly act as a host defense mechanism against phage infection.
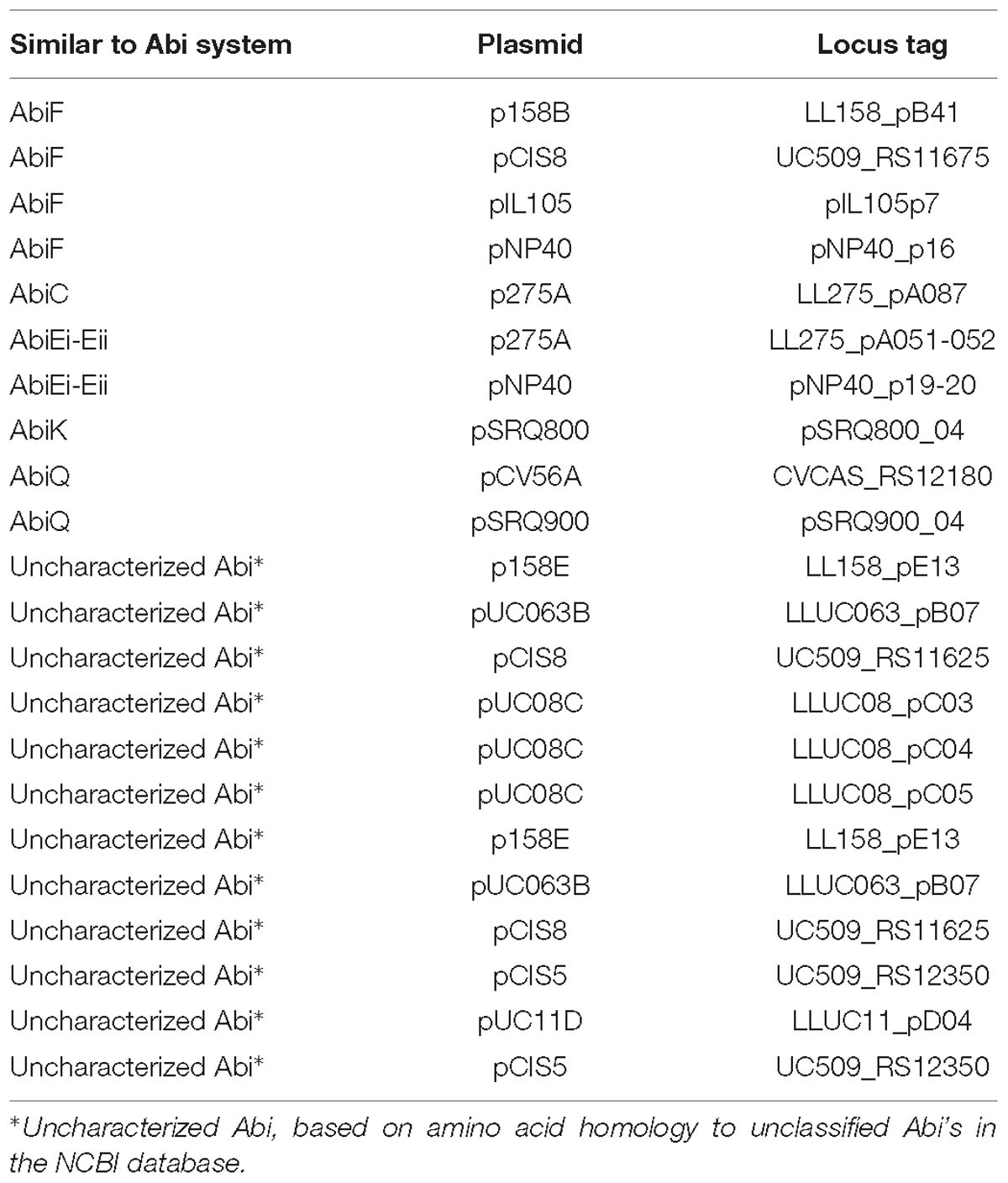
Abortive infection systems represent an abundant phage defense mechanism in L. lactis (Chopin et al., 2005) and are frequently plasmid-encoded (Mills et al., 2006). To date, 23 Abi systems have been identified in L. lactis, of which 21 are plasmid-encoded (Ainsworth et al., 2014c). Most are typically single gene systems, with the exception of three multigene systems, AbiE (Garvey et al., 1995), AbiR (Twomey et al., 2000), and AbiT (Bouchard et al., 2002). Analysis of the plasmids in this study identified eight Abi occurrences based on homology, namely AbiF, AbiC, AbiK, AbiQ, and two occurrences of the two component system AbiEi- AbiEii, alongside twelve predicted uncategorized Abi’s (Table 4), based on amino acid homology to unclassified Abi’s in the NCBI database. The relatively low observed abundance of Abi’s in such a large plasmid dataset is surprising and may be the result of the diversity of Abi’s with the possibility of as yet unidentified systems.
Discussion
The advent of NGS technologies has made genome sequencing much more accessible and has led to a dramatic rise in the number of available genome sequences. In the current study one such technology, SMRT sequencing was applied for the elucidation of 69 novel lactococcal plasmids. However, during the course of the current study some cautionary notes also emerged. These were predominantly related to smaller plasmids and plasmids with lower average consensus coverage, which could potentially be filtered out under standard assembly parameters. It was found that by performing the assembly using a reduced minimum coverage cut-off to 15-fold coverage detection of some of these plasmids was possible. In fact, in order to ensure detection of a given strain’s total plasmid complement we found it necessary to use a combined sequencing approach. This point is strongly supported by the elucidation of a further 14 plasmids from this dataset using an Illumina MiSeq approach which were completely absent from the SMRT assemblies.
The overview of plasmid replication systems presented shows that theta-type replication is the dominant way of replication used in L. lactis. These plasmids are usually viewed as being intrinsically more stable than RCR-type plasmids. However, a recent study of the dynamics of plasmid copy-number in L. lactis FM03-V1 demonstrated that the theta-type replicating plasmid (pLd10) was lost in a retentostat cultivation, while an RCR plasmid was maintained (Van Mastrigt et al., 2018c). During the course of that study, it was found that the reduced copy number of larger theta replicating plasmids increased the likelihood of the loss of these plasmids compared to smaller plasmids regardless of replication type (Van Mastrigt et al., 2018c), while the presence of the partition system (parA and parB) on these plasmids should also be considered as it has been shown to contribute to the stability and maintenance of large plasmids without selection (O’Driscoll et al., 2006). Interestingly, of the 16 plasmids not detected by SMRT sequencing in this study, five were theta replicating plasmids larger than 25 Kbp. This suggests that the lack of an amplification step during library preparation for SMRT sequencing may be a factor in detecting larger plasmids that may have a low copy number.
In the course of this study, the pan-plasmidome of L. lactis was calculated and found to be in a fluid state, making it likely that continued sequencing efforts would expand the diversity of this data set and lead to an increase in the identification of novel plasmid features. At present, the lactococcal plasmidome was found to consist of over ∼5000 Kbp of extra-chromosomal DNA encoding an arsenal of diverse features. Significantly, the current open plasmidome contributes the equivalent of 22.26% of the CDSs contained in the pan-genome of the L. lactis chromosomes that is in a closed state (Kelleher et al., 2017). BLAST-based analysis of these features identified 885 protein families, of which 413 represented unique families, evidence of the divergent nature of the plasmid sequences. There is, however, a skew in the data set toward the dairy niche, which has arisen due to a number of factors. Primarily, the majority of strains sequenced to date have been sequenced due to their commercial value in the production of fermented dairy products. The impact of these strains on the overall data set is further amplified as these strains generally carry a larger plasmid complement than their non-dairy counterparts (Kelleher et al., 2017), since many desirable dairy-associated traits are typically plasmid-encoded (e.g., lac operon). As such, these features account for a large proportion of the plasmidome. However, as efforts to isolate new starter cultures for the dairy industry continue (Cavanagh et al., 2015), screening of more diverse cultures, particularly from the plant niche, is expected to lead to increased novelty and diversity in the lactococcal plasmidome.
Megaplasmids have been found in LAB previously, in particular in members of the Lactobacillus genus (Muriana and Klaenhammer, 1987; Roussel et al., 1993; Claesson et al., 2006; Li et al., 2007; Fang et al., 2008). In the current study, sequencing efforts resulted in the identification of two examples of lactococcal megaplasmids (>100 Kbp), with pMPJM1 (193 Kbp) substantially surpassing the size of the previously largest sequenced plasmid in this taxon L. lactis S50 p7 (155 Kbp) (Kojic et al., 2005), and providing further diversity within the plasmidome. While megaplasmids are not expected to be essential for growth of their host, they can encode additional metabolic capabilities. The lactococcal megaplasmids were also examined for the presence of conjugation machinery. A novel gene cluster encoding a number of conjugation-related proteins located in pMPJM1 suggests that this plasmid is or has been involved in conjugal transfer. Further analysis of mob and tra genes across the plasmidome identified a number of genes predicted to encode proteins involved in conjugal transfer. The frequency (422 mob/tra genes across 190 plasmids) of these genes is indicative of the self-transmissible and/or mobilizable nature of lactococcal plasmids.
There has been limited research performed to date in the area of lactococcal gut adhesion as L. lactis is not commonly associated with the human gut. In this study, the lactococcal plasmidome was shown to contain potential gut adhesion factors, which may allow colonization and/or persistence in the gastrointestinal tract. This trait may offer opportunities for the application of L. lactis as a vector for vaccine and biomolecule delivery (Bermúdez-Humarán, 2009; Bermúdez-Humarán et al., 2013). Further technological properties of L. lactis were investigated including EPS production. Analysis of a large dataset of newly sequenced plasmids facilitated the identification and comparison of a number of novel EPS gene clusters. The major outcome of this work was the definition of “conserved” and “variable” regions within these EPS clusters. The conserved region encodes the transcriptional regulation, export and biosynthesis initiation machinery, while the variable region contains various genes that are predicted to encode glycosyltransferases, which are believed to be responsible for the production of a diverse set of EPS subunits, and thus a polysaccharide with a distinct composition and perhaps different technological properties.
Finally, phage-resistance mechanisms were assessed with particular emphasis on Abi systems. Abi systems confer defense against phage infection and are commonly found in lactococcal strains where they are frequently plasmid-encoded (Mills et al., 2006). Analysis of the plasmid sequences identified 22 plasmid-encoded Abi systems, while further analysis also identified frequent occurrences of these systems within the lactococcal chromosomes (Chopin et al., 2005). The presence of these systems and a range of R-M systems is evidence for the adaptation of these strains toward phage-resistance.
Discovery of the first lactococcal megaplasmids along with a host of novel features is evidence that the diversity of the lactococcal plasmidome represents a significant amount of unexploited genetic diversity, and suggests that continued future sequencing efforts and subsequent functional analysis will increase the observed diversity carried by these elements, potentially leading to new avenues of research, and applications. The current plasmidome contributes the equivalent of 22.26% of the CDSs contained in the pan-genome of the L. lactis chromosomes demonstrating its significant value to this taxon. The importance of which has been built on a long history of use in food fermentations, particularly in the dairy industry. The fact that both the opp and lac operons which have led to this adaptation remain largely plasmid encoded only further demonstrates the fundamental importance of the lactococcal plasmidome in terms of the evolution, adaptation, and application of lactococci.
Data Availability
The datasets generated for this study can be found in NCBI Genbank, CP034577, CP034578, CP034579, CP034580, CP034581, CP034582, CP034583, CP034584, CP034585, and CP034586.
Author Contributions
PK carried out the data analysis with FB. PK performed the experiments. DS and JM provided materials and strains. PK, JM, and DS wrote the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by Science Foundation Ireland (SFI)- grant numbers 15/SIRG/3430 (JM), 12/RC/2273, and 13/IA/1953 (DS). PK was funded by the Department of Agriculture, Food and the Marine under the Food Institutional Research Measure (FIRM) (Ref: 10/RD/TMFRC/704–“CheeseBoard 2015” project).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
BLAST, basic local alignment search tool; CDS, coding sequence; HCL, hierarchical clustering; MCL, Markov clustering algorithm; NGS, next generation sequencing; ORF, open reading frame; PFGE, pulse field gel electrophoresis; qPCR, quantitative polymerase chain reaction; R-M, restriction modification; SMRT, single molecule real time sequencing.
Footnotes
- ^ http://compbio.ornl.gov/prodigal/
- ^ https://www.ncbi.nlm.nih.gov/
- ^ http://www.sanger.ac.uk/science/tools/artemis
- ^ http://www.uniprot.org/
- ^ http://www.tm4.org/mev.html
References
Ainsworth, S., Mahony, J., and Van Sinderen, D. (2014a). The plasmid complement of Lactococcus lactis UC509. 9 encodes multiple bacteriophage resistance systems. Appl. Environ. Microbiol. 80, 4341–4349. doi: 10.1128/AEM.01070-14
Ainsworth, S., Sadovskaya, I., Vinogradov, E., Courtin, P., Guerardel, Y., Mahony, J., et al. (2014b). Differences in lactococcal cell wall polysaccharide structure are major determining factors in bacteriophage sensitivity. MBio 5:e880-14. doi: 10.1128/mBio.00880-14
Ainsworth, S., Stockdale, S., Bottacini, F., Mahony, J., and Van Sinderen, D. (2014c). The Lactococcus lactis plasmidome: much learnt, yet still lots to discover. FEMS Microbiol. Rev. 38, 1066–1088. doi: 10.1111/1574-6976.12074
Ainsworth, S., Zomer, A., De Jager, V., Bottacini, F., Van Hijum, S. A., Mahony, J., et al. (2013). Complete genome of Lactococcus lactis subsp. cremoris UC509. 9, host for a model lactococcal P335 bacteriophage. Genome Announc. 1:e119-12. doi: 10.1128/genomeA.00119-12
Alkema, W., Boekhorst, J., Wels, M., and Van Hijum, S. A. (2016). Microbial bioinformatics for food safety and production. Brief. Bioinformatics 17, 283–292. doi: 10.1093/bib/bbv034
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Ammann, A., Neve, H., Geis, A., and Heller, K. J. (2008). Plasmid transfer via transduction from Streptococcus thermophilus to Lactococcus lactis. J. Bacteriol. 190, 3083–3087. doi: 10.1128/JB.01448-07
Anton, J., Amilsz, R., Smith, C. L., and Lopez-Garcia, P. (1995). Comparative restriction maps of the archaeal megaplasmid Phm300 in different Haloferax mediterranei strains. Syst. Appl. Microbiol. 18, 439–447. doi: 10.1016/S0723-2020(11)80436-4
Barton, B. M., Harding, G. P., and Zuccarelli, A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226, 235–240. doi: 10.1006/abio.1995.1220
Bateman, A., Coin, L., Durbin, R., Finn, R. D., Hollich, V., Griffiths-Jones, S., et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32, D138–D141. doi: 10.1093/nar/gkh121
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580. doi: 10.1093/nar/27.2.573
Beresford, T. P., Fitzsimons, N. A., Brennan, N. L., and Cogan, T. M. (2001). Recent advances in cheese microbiology. Int. Dairy J. 11, 259–274. doi: 10.1016/S0958-6946(01)00056-5
Bermúdez-Humarán, L. G. (2009). Lactococcus lactis as a live vector for mucosal delivery of therapeutic proteins. Hum. Vaccin. 5, 264–267. doi: 10.4161/hv.5.4.7553
Bermúdez-Humarán, L. G., Aubry, C., Motta, J.-P., Deraison, C., Steidler, L., Vergnolle, N., et al. (2013). Engineering lactococci and lactobacilli for human health. Curr. Opin. Microbiol. 16, 278–283. doi: 10.1016/j.mib.2013.06.002
Bissett, D. L., and Anderson, R. L. (1974). Lactose and d-galactose metabolism in group N streptococci: presence of enzymes for both the d-Galactose 1-phosphate and d-tagatose 6-phosphate pathways1. J. Bacteriol. 117:318.
Bottacini, F., O’connell Motherway, M., Casey, E., Mcdonnell, B., Mahony, J., Ventura, M., et al. (2015). Discovery of a Conjugative megaplasmid in Bifidobacterium breve. Appl. Environ. Microbiol. 81, 166–176. doi: 10.1128/AEM.02871-14
Bouchard, J. D., Dion, E., Bissonnette, F., and Moineau, S. (2002). Characterization of the two-component abortive phage infection mechanism AbiT from Lactococcus lactis. J. Bacteriol. 184, 6325–6332. doi: 10.1128/JB.184.22.6325-6332.2002
Broadbent, J. R., Barnes, M., Brennand, C., Strickland, M., Houck, K., Johnson, M. E., et al. (2002). Contribution of Lactococcus lactis cell envelope proteinase specificity to peptide accumulation and bitterness in reduced-fat Cheddar cheese. Appl. Environ. Microbiol. 68, 1778–1785. doi: 10.1128/AEM.68.4.1778-1785.2002
Cavanagh, D., Casey, A., Altermann, E., Cotter, P. D., Fitzgerald, G. F., and Mcauliffe, O. (2015). Evaluation of Lactococcus lactis isolates from nondairy sources with potential dairy applications reveals extensive phenotype-genotype disparity and implications for a revised species. Appl. Environ. Microbiol. 81, 3961–3972. doi: 10.1128/AEM.04092-14
Chopin, M.-C., Chopin, A., and Bidnenko, E. (2005). Phage abortive infection in lactococci: variations on a theme. Curr. Opin. Microbiol. 8, 473–479. doi: 10.1016/j.mib.2005.06.006
Claesson, M. J., Li, Y., Leahy, S., Canchaya, C., Van Pijkeren, J. P., Cerdeño-Tárraga, A. M., et al. (2006). Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. U.S.A. 103, 6718–6723. doi: 10.1073/pnas.0511060103
Coakley, M., Fitzgerald, G., and Ros, R. (1997). Application and evaluation of the phage resistance-and bacteriocin-encoding plasmid pMRC01 for the improvement of dairy starter cultures. Appl. Environ. Microbiol. 63, 1434–1440.
Cords, B. R., Mckay, L. L., and Guerry, P. (1974). Extrachromosomal elements in group N streptococci. J. Bacteriol. 117, 1149–1152.
David, B., Radziejwoski, A., Toussaint, F., Fontaine, L., Henry De Frahan, M., Patout, C., et al. (2017). Natural DNA transformation is functional in Lactococcus lactis ssp. cremoris KW2. Appl. Environ. Microbiol. doi: 10.1128/AEM.01074-17 [Epub ahead of print].
Del Solar, G., Moscoso, M., and Espinosa, M. (1993). In vivo definition of the functional origin of replication (ori +) of the promiscuous plasmid pLS1. Mol. Gen. Genet. 237, 65–72. doi: 10.1007/BF00282785
Dobson, A., Cotter, P. D., Ross, R. P., and Hill, C. (2012). Bacteriocin production: a probiotic trait? Appl. Environ. Microbiol. 78, 1–6. doi: 10.1128/AEM.05576-11
Dougherty, B. A., Hill, C., Weidman, J. F., Richardson, D. R., Venter, J. C., and Ross, R. P. (1998). Sequence and analysis of the 60 kb conjugative, bacteriocin-producing plasmid pmrc01 from Lactococcus lactis DPC3147. Mol. Microbiol. 29, 1029–1038. doi: 10.1046/j.1365-2958.1998.00988.x
Drider, D., Bekal, S., and Prévost, H. (2004). Genetic organization and expression of citrate permease in lactic acid bacteria. Genet. Mol. Res. 3, 271–281.
Enright, A. J., Van Dongen, S., and Ouzounis, C. A. (2002). An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584. doi: 10.1093/nar/30.7.1575
Fallico, V., Ross, R., Fitzgerald, G., and Mcauliffe, O. (2012). Novel conjugative plasmids from the natural isolate Lactococcus lactis subspecies cremoris DPC3758: a repository of genes for the potential improvement of dairy starters. J. Dairy Sci. 95, 3593–3608. doi: 10.3168/jds.2011-5255
Fang, F., Flynn, S., Li, Y., Claesson, M. J., Van Pijkeren, J.-P., Collins, J. K., et al. (2008). Characterization of endogenous plasmids from Lactobacillus salivarius UCC118. Appl. Environ. Microbiol. 74, 3216–3228. doi: 10.1128/AEM.02631-07
Fernández, E., Alegría, Á, Delgado, S., Martín, M. C., and Mayo, B. (2011). Comparative phenotypic and molecular genetic profiling of wild Lactococcus lactis subsp. lactis strains of the L. lactis subsp. lactis and L. lactis subsp. cremoris genotypes, isolated from starter-free cheeses made of raw milk. Appl. Environ. Microbiol. 77, 5324–5335. doi: 10.1128/AEM.02991-10
Garvey, P., Fitzgerald, G. F., and Hill, C. (1995). Cloning and DNA sequence analysis of two abortive infection phage resistance determinants from the lactococcal plasmid PNP40. Appl. Environ. Microbiol. 61, 4321–4328.
Gasson, M., Godon, J., Pillidge, C., Eaton, T., Jury, K., and Shearman, C. (1995). Characterization and exploitation of conjugation in Lactococcus lactis. Int. Dairy J. 5, 757–762. doi: 10.1016/0958-6946(95)00030-5
Geis, A., Singh, J., and Teuber, M. (1983). Potential of lactic streptococci to produce bacteriocin. Appl. Environ. Microbiol. 45, 205–211.
Goh, Y. J., Goin, C., O’flaherty, S., Altermann, E., and Hutkins, R. (2011). Specialized adaptation of a lactic acid bacterium to the milk environment: the comparative genomics of Streptococcus thermophilus LMD-9. Microb. Cell Fact 10(Suppl. 1):S22. doi: 10.1186/1475-2859-10-S1-S22
Górecki, R. K., Koryszewska-Bagińska, A., Gołȩbiewski, M., Żylińska, J., Grynberg, M., and Bardowski, J. K. (2011). Adaptative potential of the Lactococcus lactis IL594 strain encoded in its 7 plasmids. PLoS One 6:e22238. doi: 10.1371/journal.pone.0022238
Grohmann, E., Muth, G., and Espinosa, M. (2003). Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67, 277–301. doi: 10.1128/MMBR.67.2.277-301.2003
Habimana, O., Le Goff, C., Juillard, V., Bellon-Fontaine, M. N., Buist, G., Kulakauskas, S., et al. (2007). Positive role of cell wall anchored proteinase PrtP in adhesion of lactococci. BMC Microbiol. 7:36. doi: 10.1186/1471-2180-7-36
Harrington, A., and Hill, C. (1991). Construction of a bacteriophage-resistant derivative of Lactococcus lactis subsp. lactis 425A by using the conjugal plasmid PNP40. Appl. Environ. Microbiol. 57, 3405–3409.
Horvath, P., and Barrangou, R. (2010). Crispr/Cas, the immune system of bacteria and archaea. Science 327, 167–170. doi: 10.1126/science.1179555
Kelleher, P., Bottacini, F., Mahony, J., Kilcawley, K. N., and Van Sinderen, D. (2017). Comparative and functional genomics of the Lactococcus lactis taxon; insights into evolution and niche adaptation. BMC Genomics 18:267. doi: 10.1186/s12864-017-3650-5
Kelleher, P., Mahony, J., Schweinlin, K., Neve, H., Franz, C. M., and Van Sinderen, D. (2018). Assessing the functionality and genetic diversity of lactococcal prophages. Int. J. Food Microbiol. 272, 29–40. doi: 10.1016/j.ijfoodmicro.2018.02.024
Kelleher, P., Murphy, J., Mahony, J., and Van Sinderen, D. (2015). Next-generation sequencing as an approach to dairy starter selection. Dairy Sci. Technol. 95, 545–568. doi: 10.1007/s13594-015-0227-4
Kelly, W. J., Ward, L. J. H., and Leahy, S. C. (2010). Chromosomal diversity in Lactococcus lactis and the origin of dairy starter cultures. Genome Biol. Evol. 2, 729–744. doi: 10.1093/gbe/evq056
Kiewiet, R., Bron, S., De Jonge, K., Venema, G., and Seegers, J. F. (1993). Theta replication of the lactococcal plasmid pwvo2. Mol. Microbiol. 10, 319–327. doi: 10.1111/j.1365-2958.1993.tb01958.x
Kleerebezem, M., Van Kranenburg, R., Tuinier, R., Boels, I. C., Zoon, P., Looijesteijn, E., et al. (1999). “Exopolysaccharides produced by Lactococcus lactis: from genetic engineering to improved rheological properties?,” in Lactic Acid Bacteria: Genetics, Metabolism and Applications, eds W. Konings, O. P. Kuipers, and J. H. J. Huis in ’t Veld (Berlin: Springer).
Kojic, M., Jovcic, B., Strahinic, I., Begovic, J., Lozo, J., Veljovic, K., et al. (2011). Cloning and expression of a novel lactococcal aggregation factor from Lactococcus lactis subsp. lactis BGKP1. BMC Microbiol. 11:265. doi: 10.1186/1471-2180-11-265
Kojic, M., Strahinic, I., Fira, D., Jovcic, B., and Topisirovic, L. (2006). Plasmid content and bacteriocin production by five strains of Lactococcus lactis isolated from semi-hard homemade cheese. Can. J. Microbiol. 52, 1110–1120. doi: 10.1139/w06-072
Kojic, M., Strahinic, I., and Topisirovic, L. (2005). Proteinase Pi and lactococcin A genes are located on the largest plasmid in Lactococcus lactis subsp. lactis bv. diacetylactis S50. Can. J. Microbiol. 51, 305–314. doi: 10.1139/w05-009
Kranenburg, R. V., Marugg, J. D., Van Swam, I. I., Willem, N. J., and De Vos, W. M. (1997). Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24, 387–397. doi: 10.1046/j.1365-2958.1997.3521720.x
Labrie, S. J., Samson, J. E., and Moineau, S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317. doi: 10.1038/nrmicro2315
Le, D. T., Tran, T. L., Duviau, M. P., Meyrand, M., Guerardel, Y., Castelain, M., et al. (2013). Unraveling the role of surface mucus-binding protein and pili in muco-adhesion of Lactococcus lactis. PLoS One 8:e79850. doi: 10.1371/journal.pone.0079850
Lebeer, S., Claes, I., Tytgat, H. L. P., Verhoeven, T. L. A., Marien, E., Von Ossowski, I., et al. (2012). Functional analysis of Lactobacillus rhamnosus GG Pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 78, 185–193. doi: 10.1128/AEM.06192-11
Leenhouts, K. J., Tolner, B., Bron, S., Kok, J., Venema, G., and Seegers, J. F. (1991). Nucleotide sequence and characterization of the broad-host-range lactococcal plasmid PWVO1. Plasmid 26, 55–66. doi: 10.1016/0147-619X(91)90036-V
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, Y., Canchaya, C., Fang, F., Raftis, E., Ryan, K. A., Van Pijkeren, J.-P., et al. (2007). Distribution of megaplasmids in Lactobacillus salivarius and other lactobacilli. J. Bacteriol. 189, 6128–6139. doi: 10.1128/JB.00447-07
Lukić, J., Strahinić, I., Jovčić, B., Filipić, B., Topisirović, L., Kojić, M., et al. (2012). Different roles for lactococcal aggregation factor and mucin binding protein in adhesion to gastrointestinal mucosa. Appl. Environ. Microbiol. 78, 7993–8000. doi: 10.1128/AEM.02141-12
Machielsen, R., Siezen, R. J., Van Hijum, S. A., and Van Hylckama Vlieg, J. E. (2011). Molecular description and industrial potential of Tn6098 conjugative transfer conferring alpha-galactoside metabolism in Lactococcus lactis. Appl. Environ. Microbiol. 77, 555–563. doi: 10.1128/AEM.02283-10
Mahony, J., Kot, W., Murphy, J., Ainsworth, S., Neve, H., Hansen, L. H., et al. (2013). Investigation of the relationship between lactococcal host cell wall polysaccharide genotype and 936 phage receptor binding protein phylogeny. Appl. Environ. Microbiol. 79, 4385–4392. doi: 10.1128/AEM.00653-13
Mahony, J., Mcgrath, S., Fitzgerald, G. F., and Van Sinderen, D. (2008). Identification and characterization of lactococcal-prophage-carried superinfection exclusion genes. Appl. Environ. Microbiol. 74, 6206–6215. doi: 10.1128/AEM.01053-08
Mahony, J., Randazzo, W., Neve, H., Settanni, L., and Van Sinderen, D. (2015). Lactococcal 949 group phages recognize a carbohydrate receptor on the host cell surface. Appl. Environ. Microbiol. 81, 3299–3305. doi: 10.1128/AEM.00143-15
Makarova, K., Slesarev, A., Wolf, Y., Sorokin, A., Mirkin, B., Koonin, E., et al. (2006). Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 15611–15616. doi: 10.1073/pnas.0607117103
Mcgrath, S., Fitzgerald, G. F., and Sinderen, D. V. (2002). Identification and characterization of phage-resistance genes in temperate lactococcal bacteriophages. Mol. Microbiol. 43, 509–520. doi: 10.1046/j.1365-2958.2002.02763.x
McSweeney, P. L. H. (2004). Biochemistry of cheese ripening. Int. J. Dairy Technol. 57, 127–144. doi: 10.1111/j.1471-0307.2004.00147.x
McSweeney, P. L. H., and Sousa, M. J. (2000). Biochemical pathways for the production of flavour compounds in cheeses during ripening: a review. Lait 80, 293–324. doi: 10.1051/lait:2000127
Millen, A. M., Horvath, P., Boyaval, P., and Romero, D. A. (2012). Mobile crispr/cas-mediated bacteriophage resistance in Lactococcus lactis. PLoS One 7:e51663. doi: 10.1371/journal.pone.0051663
Mills, S., Mcauliffe, O. E., Coffey, A., Fitzgerald, G. F., and Ross, R. P. (2006). Plasmids of lactococci – genetic accessories or genetic necessities? FEMS Microbiol. Rev. 30, 243–273.
Moineau, S., Walker, S. A., Vedamuthu, E. R., and Vandenbergh, P. A. (1995). Cloning and sequencing of Lladchi restriction/modification genes from Lactococcus lactis and relatedness of this system to the Streptococcus pneumoniae Dpnii system. Appl. Environ. Microbiol. 61, 2193–2202.
Mulder, J., Wels, M., Kuipers, O. P., Kleerebezem, M., and Bron, P. A. (2017). Unleashing natural competence in Lactococcus lactis by induction of the competence regulator ComX. Appl. Environ. Microbiol. doi: 10.1128/AEM.01320-17 [Epub ahead of print].
Muriana, P. M., and Klaenhammer, T. R. (1987). Conjugal transfer of plasmid-encoded determinants for bacteriocin production and immunity in Lactobacillus acidophilus 88. Appl. Environm. Microbiol. 53, 553–560.
Neve, H., Geis, A., and Teuber, M. (1987). Conjugation, a common plasmid transfer mechanism in lactic acid streptococci of dairy starter cultures. Syst. Appl. Microbiol. 9, 151–157. doi: 10.1016/S0723-2020(87)80070-X
Neve, H., Geis, A., and Teuber, M. (1988). Plasmid-encoded functions of ropy lactic acid streptococcal strains from Scandinavian fermented milk. Biochimie 70, 437–442. doi: 10.1016/0300-9084(88)90218-0
Nicolas, P., Bessières, P., Ehrlich, S. D., Maguin, E., and Van De Guchte, M. (2007). Extensive horizontal transfer of core genome genes between two Lactobacillus species found in the gastrointestinal tract. BMC Evol. Biol. 7:1. doi: 10.1186/1471-2148-7-141
O’driscoll, J., Glynn, F., Cahalane, O., O’connell-Motherway, M., Fitzgerald, G. F., and Van Sinderen, D. (2004). Lactococcal plasmid PNP40 encodes a novel, temperature-sensitive restriction-modification system. Appl. Environ. Microbiol. 70, 5546–5556. doi: 10.1128/AEM.70.9.5546-5556.2004
O’Driscoll, J., Glynn, F., Fitzgerald, G. F., and Sinderen, D. V. (2006). Sequence analysis of the lactococcal plasmid PNP40: a mobile replicon for coping with environmental hazards. J. Bacteriol. 188, 6629–6639. doi: 10.1128/JB.00672-06
Patel, R. K., and Jain, M. (2012). NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. doi: 10.1371/journal.pone.0030619
Price, C. E., Zeyniyev, A., Kuipers, O. P., and Kok, J. (2012). From meadows to milk to mucosa – adaptation of Streptococcus and Lactococcus species to their nutritional environments. FEMS Microbiol. Rev. 36, 949–971. doi: 10.1111/j.1574-6976.2011.00323.x
Radziwill-Bienkowska, J. M., Robert, V., Drabot, K., Chain, F., Cherbuy, C., Langella, P., et al. (2017). Contribution of plasmid-encoded peptidase S8 (PrtP) to adhesion and transit in the gut of Lactococcus lactis Ibb477 strain. Appl. Microbiol. Biotechnol. 101, 5709–5721. doi: 10.1007/s00253-017-8334-1
Roberts, R. J., Belfort, M., Bestor, T., Bhagwat, A. S., Bickle, T. A., Bitinaite, J., et al. (2003). A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 31, 1805–1812. doi: 10.1093/nar/gkg274
Roussel, Y., Colmin, C., Simonet, J. M., and Decaris, B. (1993). Strain characterization, genome size and plasmid content in the Lactobacillus acidophilus group (Hansen and Mocquot). J. Appl. Bacteriol. 74, 549–556.
Ryan, M. P., Rea, M. C., Hill, C., and Ross, R. P. (1996). An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl. Environ. Microbiol. 62, 612–619.
Seegers, J. F., Bron, S., Franke, C. M., Venema, G., and Kiewiet, R. (1994). The majority of lactococcal plasmids carry a highly related replicon. Microbiology 140(Pt 6), 1291–1300. doi: 10.1099/00221287-140-6-1291
Sharma, V. K., Johnston, J. L., Morton, T. M., and Archer, G. L. (1994). Transcriptional regulation by TrsN of conjugative transfer genes on staphylococcal plasmid PGO1. J. Bacteriol. 176, 3445–3454. doi: 10.1128/jb.176.12.3445-3454.1994
Siezen, R. J., Renckens, B., Van Swam, I., Peters, S., Van Kranenburg, R., Kleerebezem, M., et al. (2005). Complete sequences of four plasmids of Lactococcus lactis subsp. cremoris SK11 reveal extensive adaptation to the dairy environment. Appl. Environ. Microbiol. 71, 8371–8382. doi: 10.1128/AEM.71.12.8371-8382.2005
Simpson, J. T., Wong, K., Jackman, S. D., Schein, J. E., Jones, S. J., and Birol, I. (2009). ABYSS: a parallel assembler for short read sequence data. Genome Res. 19, 1117–1123. doi: 10.1101/gr.089532.108
Solopova, A., Bachmann, H., Teusink, B., Kok, J., Neves, A. R., and Kuipers, O. P. (2012). A specific mutation in the promoter region of the silent cel cluster accounts for the appearance of lactose-utilizing Lactococcus lactis MG1363. Appl. Environ. Microbiol. 78, 5612–5621. doi: 10.1128/AEM.00455-12
Stentz, R., Gasson, M., and Shearman, C. (2006). The Tra domain of the lactococcal CluA surface protein is a unique domain that contributes to sex factor DNA transfer. J. Bacteriol. 188, 2106–2114. doi: 10.1128/JB.188.6.2106-2114.2006
Tettelin, H., Masignani, V., Cieslewicz, M. J., Donati, C., Medini, D., Ward, N. L., et al. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U.S.A. 102, 13950–13955. doi: 10.1073/pnas.0506758102
Twomey, D. P., De Urraza, P. J., Mckay, L. L., and O’sullivan, D. J. (2000). Characterization of AbiR, a novel multicomponent abortive infection mechanism encoded by plasmid PKR223 of Lactococcus lactis subsp. lactis KR2. Appl. Environ. Microbiol. 66, 2647–2651. doi: 10.1128/AEM.66.6.2647-2651.2000
Van Belkum, M. J., Hayema, B. J., Geis, A., Kok, J., and Venema, G. (1989). Cloning of two bacteriocin genes from a lactococcal bacteriocin plasmid. Appl. Environ. Microbiol. 55, 1187–1191.
Van Belkum, M. J., Hayema, B. J., Geis, A., Kok, J., and Venema, G. (1991). Organization and nucleotide sequences of two lactococcal bacteriocin operons. Appl. Environ. Microbiol. 57, 492–498.
Van Heel, A. J., De Jong, A., Montalban-Lopez, M., Kok, J., and Kuipers, O. P. (2013). BAGEL3: automated identification of genes encoding bacteriocins and (non-) bactericidal posttranslationally modified peptides. Nucleic Acids Res. 41, W448–W453. doi: 10.1093/nar/gkt391
Van Mastrigt, O., Di Stefano, E., Hartono, S., Abee, T., and Smid, E. J. (2018a). Large plasmidome of dairy Lactococcus lactis subsp. lactis biovar diacetylactis FM03P encodes technological functions and appears highly unstable. BMC Genomics 19:620. doi: 10.1186/s12864-018-5005-2
Van Mastrigt, O., Lommers, M. M., De Vries, Y. C., Abee, T., and Smid, E. J. (2018b). Citrate, low pH and amino acid limitation induce citrate utilization in Lactococcus lactis biovar diacetylactis. Microbial. Biotechnol. 11, 369–380. doi: 10.1111/1751-7915.13031
Van Mastrigt, O., Lommers, M. M., De Vries, Y. C., Abee, T., and Smid, E. J. (2018c). Dynamics in copy numbers of five plasmids of a dairy Lactococcus lactis in dairy-related conditions including near-zero growth rates. Appl. Environ. Microbiol. 84, AEM.00314–AEM.318. doi: 10.1128/AEM.00314-18
Van Rooijen, R., Gasson, M., and De Vos, W. (1992). Characterization of the Lactococcus lactis lactose operon promoter: contribution of flanking sequences and LacR repressor to promoter activity. J. Bacteriol. 174, 2273–2280. doi: 10.1128/jb.174.7.2273-2280.1992
Van Rooijen, R. J., and De Vos, W. (1990). Molecular cloning, transcriptional analysis, and nucleotide sequence of lacR, a gene encoding the repressor of the lactose phosphotransferase system of Lactococcus lactis. J. Biol. Chem. 265, 18499–18503.
Vedamuthu, E. R., and Neville, J. M. (1986). Involvement of a plasmid in production of ropiness (mucoidness) in milk cultures by Streptococcus cremoris MS. Appl. Environ. Microbiol. 51, 677–682.
Von Ossowski, I., Reunanen, J., Satokari, R., Vesterlund, S., Kankainen, M., Huhtinen, H., et al. (2010). Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG spacba and SpaFED pilin subunits. Appl. Environ. Microbiol. 76, 2049–2057. doi: 10.1128/AEM.01958-09
Von Wright, A., and Tynkkynen, S. (1987). Construction of Streptococcus lactis subsp. lactis strains with a single plasmid associated with mucoid phenotype. Appl. Environ. Microbiol. 53, 1385–1386.
Wegmann, U., Overweg, K., Jeanson, S., Gasson, M., and Shearman, C. (2012). Molecular characterization and structural instability of the industrially important composite metabolic plasmid plp712. Microbiology 158, 2936–2945. doi: 10.1099/mic.0.062554-0
Wels, M., Siezen, R., Van Hijum, S., Kelly, W., and Bachmann, H. (2019). Comparative genome analysis of Lactococcus lactis indicates niche adaptation and resolves genotype/phenotype disparity. Front. Microbiol. 10:4. doi: 10.3389/fmicb.2019.00004
Yang, X., Wang, Y., and Huo, G. (2013). Complete Genome Sequence of Lactococcus lactis subsp. lactis KLDS4. 0325. Genome Announc. 1:e962-13. doi: 10.1128/genomeA.00962-13
Keywords: lactococcal, plasmid, SMRT sequencing, dairy fermentation, conjugation, lactic acid bacteria
Citation: Kelleher P, Mahony J, Bottacini F, Lugli GA, Ventura M and van Sinderen D (2019) The Lactococcus lactis Pan-Plasmidome. Front. Microbiol. 10:707. doi: 10.3389/fmicb.2019.00707
Received: 17 January 2019; Accepted: 20 March 2019;
Published: 04 April 2019.
Edited by:
Kimberly Kline, Nanyang Technological University, SingaporeReviewed by:
Milan Kojic, University of Belgrade, SerbiaMaría de Toro, Centro de Investigación Biomédica de La Rioja, Spain
Copyright © 2019 Kelleher, Mahony, Bottacini, Lugli, Ventura and van Sinderen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Douwe van Sinderen, ZC52YW5zaW5kZXJlbkB1Y2MuaWU=
 Philip Kelleher
Philip Kelleher Jennifer Mahony
Jennifer Mahony Francesca Bottacini
Francesca Bottacini Gabriele A. Lugli
Gabriele A. Lugli Marco Ventura
Marco Ventura Douwe van Sinderen
Douwe van Sinderen