- 1National Research Council Postdoctoral Fellowships, NRC Research Associateship Programs, Washington, DC, United States
- 2United States Naval Research Laboratory, Center for Biomolecular Science and Engineering, Washington, DC, United States
The bacterial quorum sensing phenomenon has been well studied since its discovery and has traditionally been considered to include signaling pathways recognized exclusively within either Gram-positive or Gram-negative bacteria. These groups of bacteria synthesize structurally distinct signaling molecules to mediate quorum sensing, where Gram-positive bacteria traditionally utilize small autoinducing peptides (AIPs) and Gram-negatives use small molecules such as acyl-homoserine lactones (AHLs). The structural differences between the types of signaling molecules have historically implied a lack of cross-talk among Gram-positive and Gram-negative quorum sensing systems. Recent investigations, however, have demonstrated the ability for AIPs and AHLs to be produced by non-canonical organisms, implying quorum sensing systems may be more universally recognized than previously hypothesized. With that in mind, our interests were piqued by the organisms Lactobacillus plantarum, a Gram-positive commensal probiotic known to participate in AIP-mediated quorum sensing, and Pseudomonas aeruginosa, a characterized Gram-negative pathogen whose virulence is in part controlled by AHL-mediated quorum sensing. Both health-related organisms are known to inhabit the human gut in various instances, both are characterized to elicit distinct effects on host immunity, and some studies hint at the putative ability of L. plantarum to degrade AHLs produced by P. aeruginosa. We therefore wanted to determine if L. plantarum cultures would respond to the addition of N-(3-oxododecanoyl)-L-homoserine lactone (3OC12) from P. aeruginosa by analyzing changes on both the transcriptome and proteome over time. Based on the observed upregulation of various two-component systems, response regulators, and native quorum sensing related genes, the resulting data provide evidence of an AHL recognition and response by L. plantarum.
Introduction
Bacteria have been understood to possess a basic level of cell to cell communication for decades (Miller and Bassler, 2001; Waters and Bassler, 2005). The communication phenomenon known as quorum sensing allows organisms to coordinate growth and gene expression efforts, for example, toward a common goal to survive an increasingly harsh environment (Reading and Sperandio, 2006). The traditional model for quorum sensing has been described as the luxR system in multiple Vibrio spp. (Gray et al., 1994; Bassler et al., 1997; Zhu et al., 2002; Hammer and Bassler, 2003; Waters and Bassler, 2006), where a signaling molecule such as an N-acyl homoserine lactone (AHL) is produced and exported into the microenvironment at some undetected basal level. The signaling molecule accumulates over time with proliferation of the producing species until a threshold is reached, whereupon the signal is detected and organisms respond with targeted gene expression. Quorum sensing systems following this model are commonly seen in Gram-negative bacteria such as the aforementioned Vibrio spp. to coordinate luminescent protein production or in pathogens such as Pseudomonas aeruginosa to coordinate virulence and survival (Gray et al., 1994; Storey et al., 1998; Rumbaugh et al., 1999a,b; Winzer et al., 2000; Rampioni et al., 2006, 2007, 2009; Gao et al., 2017; Kariminik et al., 2017). The downstream genetic response to the signal varies among different organisms, but the general model of signal amplification remains consistent and depends on (1) the synthesis and export of the signal molecule into the environment, (2) environmental accumulation of the signal molecule, (3) diffusion of the signal molecule into neighboring cells, and (4) the interaction of the signal molecule with specific transcription factors resulting in the activation of gene expression. Synthesis of the signal molecule is generally upregulated by transcription factor activation as part of a positive feedback loop, and the number of promoters subsequently induced varies widely among organisms and the specific quorum sensing system involved such that induction rarely results in the upregulation of a single gene (Brint and Ohman, 1995; Medina et al., 2003a,b; Jensen et al., 2006).
Gram-positive organisms are capable of quorum sensing by a different mechanism (Miller and Bassler, 2001). Rather than using AHLs with varying acyl chain lengths as signal molecules, Gram-positive organisms employ the use of short peptides known as autoinducer peptides (AIPs), some of which contain unconventional bonding between specific amino acids to produce unique structures. Considering the multicharge potential of peptides, specific proteins are devoted to the export of autoinducers into the microenvironment. These peptides do not typically diffuse through the membranes like their lactone counterparts given their charged characteristics and the peptidoglycan layer of Gram-positive organisms, thus signal recognition occurs by the activation of a two-component system (Kleerebezem et al., 1997; Sturme et al., 2005; Sturme et al., 2007). The agr system in Staphylococcus aureus is a well-studied example of peptide-mediated quorum sensing in Gram-positive organisms (Peng et al., 1988; Janzon and Arvidson, 1990; Booth et al., 1995; Papakyriacou et al., 2000; Jensen et al., 2008; Reyes et al., 2011; Marchand and Collins, 2013, 2016), and homologs have been identified in various Lactic Acid Bacteria such as Lactobacillus sakei, Lactobacillus acidophilus, and Lactobacillus plantarum (Kanehisa and Goto, 2000; Sturme et al., 2005, 2007; Fujii et al., 2008). The Lactic Acid Bacteria are known to produce antimicrobial peptides as one response to specific AIP activity as a way to coordinate a defense or fitness mechanism (Diep et al., 1994; Rekhif et al., 1995; Anderssen et al., 1998; Atrih et al., 2001; Maldonado et al., 2002, 2004a,b), while AIP activity in S. aureus is reported to contribute to virulence (Booth et al., 1995; Papakyriacou et al., 2000) similar to P. aeruginosa. Similar to AHL-mediated sensing, the baseline of genes affected by AIP is unknown given the diversity of organisms that employ this mechanism, and the induction of a single target gene is likely rare.
Quorum sensing traditionally has been split into two classes consisting of AHL- and peptide-mediated signaling assigned to Gram-negative and Gram-positive bacteria, respectively (Miller and Bassler, 2001; Reading and Sperandio, 2006). However, both Gram-negative and Gram-positive bacteria participate in two-component system quorum sensing using autoinducer-2 (AI-2) class molecules, which are structurally unrelated to AHLs and peptides (Reading and Sperandio, 2006). Autoinducer-2 molecules are furanose derivatives of the coenzyme S-adenosyl methionine (SAM) whose final active structures vary expectedly among organisms, with some examples containing the element boron (Schauder et al., 2001; Taga et al., 2001; Semmelhack et al., 2004; Vendeville et al., 2005; Pereira et al., 2013). The synthesis of AI-2 involves the luxS gene detected in a variety of diverse organisms, leading to the theory that AI-2 molecules represent a universal language for bacterial interspecies communication (Vendeville et al., 2005). The emergence and prevalence of the AI-2 systems seemed to offer a more complete picture of the concept of bacterial communication, where the major bacterial groups of Gram-positive and Gram-negative had their own quorum sensing systems, and a third system existed to facilitate interspecies communication. The existence of such an AI-2 system potentially circumvented the curiosity of whether Gram-positive or Gram-negative organisms could respond to each other’s exclusive signaling molecules. Examples that challenge this presumed exclusivity of AHL and AIP quorum sensing, however, have been identified in the last decade, where AHL production has been noted in Cyanobacteria (Sharif et al., 2008), Archaea (Zhang et al., 2012), and a marine Gram-positive organism from the genus Exiguobacterium (Biswa and Doble, 2013). Such discoveries suggest a more universal recognition and utilization of AHLs within the microbial world than previously assumed.
Indeed, AHL recognition by higher order species has been previously observed (Smith et al., 2002a,b; Mathesius et al., 2003; Ritchie et al., 2003, 2005, 2007; Bauer and Mathesius, 2004; Chun et al., 2004; Wagner et al., 2007; Jahoor et al., 2008; Teplitski et al., 2011; Kariminik et al., 2017) with the rationale that the effects imparted by AHLs are due to their structural and functional resemblance to hormones and phytohormones (Teplitski et al., 2011). Within the bacterial kingdom, investigators have previously noted cross-species effects of a Yersinia enterocolitica AHL on enterohemorrhagic Escherichia coli O157:H7 (Nguyen et al., 2013) and S. aureus-derived peptides on both Enterococcus spp. (Firth et al., 1994) and Lactobacillus reuteri (Lubkowicz et al., 2018). While such responses were mostly unexpected and inexplicable, the latter example could be attributed to the close relation of the Lactobacillus and Staphylococcus organisms that diverge at the Class level and the apparent endogenous recognition of the S. aureus AIP-I despite a lack of annotation for agr homologs in L. reuteri (Kanehisa and Goto, 2000). Previous attempts to engineer the agr system into Firmicutes such as Bacillus megaterium were successful and proved the system to be unique to the host (Marchand and Collins, 2013), but the response of L. reuteri to the S. aureus AIP-I in contrast resulted in repression of an exogenous agr promoter rather than stimulation, implying cognate parts of the agr-like signaling system may exist in the Lactobacillus species used for different purposes than in S. aureus. This unexpected result exemplifies the complexity of interspecies quorum sense recognition despite the established genomic characterization of the involved species (Kanehisa and Goto, 2000).
The effects of specific AHLs are presumably localized to cognate and closely related species, at least in terms of an optimized and targeted response based on the number of studies probing the activity of heterologous luxR and lasR systems in E.coli without notable off-target responses (Seed et al., 1995; Latifi et al., 1996; Collins et al., 2005, 2006; Goodson et al., 2015). However, evidence exists of the potential cross-talk, albeit weak, between different AHL-mediated systems based on activity observed in cell-free experiments (Wen et al., 2017; Halleran and Murray, 2018). Considering the above points, it is hard to say universally whether bacteria can sense and respond to any present signaling cues in the microenvironment. In terms of signal fidelity, it makes sense that bacterial species would evolve a unique and uninterceptable signaling regime to coordinate the survival and fitness of itself over others in the community, especially in the context of virulence coordination. P. aeruginosa has been well noted to utilize its lasR-dependent quorum sensing system to establish biofilm formation and anti-host immunity measures as well as cyanide production to solidify colonization in the face of both host responses and microbial competition (Storey et al., 1998; Pessi and Haas, 2000; Winzer and Williams, 2001). Furthermore, L. plantarum has been noted to produce the antimicrobial peptide class of Plantaricins as a result of AIP signaling to similarly diminish bacterial populations in its proximity (Anderssen et al., 1998; Atrih et al., 2001), but this seems to be more of an altruistic endeavor in order to cull the prevalence of organisms potentially harmful to the host (Maldonado et al., 2004b). While the driving force behind the P. aeruginosa signaling system may be attributed to cell density given its occurrence in pure cultures (Valdez et al., 2005), the initiation of the aforementioned response by L. plantarum is unknown, and could be due to either cell density or accumulation of some unrelated environmental cue (Maldonado et al., 2003, 2004a,b).
We therefore set out to investigate the potential for quorum sensing cross-talk between the two organisms P. aeruginosa and L. plantarum considering their activity as quorum sensing bacteria, their contrasting roles in the human gut (Matsumoto et al., 1997; Xia et al., 2011), and the effects they elicit on the host immune system (Schultz et al., 2002; Ko et al., 2007; Puertollano et al., 2008; Peral et al., 2010). Furthermore, there has been nominal evidence that L. plantarum is putatively capable of degrading AHLs produced by P. aeruginosa (Valdez et al., 2005; Peral et al., 2009; Ramos et al., 2012; Ramos et al., 2015), suggesting there could be some mechanism for AHL recognition by L. plantarum. The use of deep analytical techniques involving transcriptomics and proteomics allowed us to gather a detailed global picture of the response of L. plantarum to the presence of the P. aeruginosa AHL N-3-oxo-dodecanoyl homoserine lactone (3OC12). The wealth of resulting data provided us the opportunity to speculate on both the intracellular ripple effect and timeline for the interspecies response to a predominantly Gram-negative signal by a Gram-positive commensal.
Materials and Methods
Reagents
All reagents were obtained from Sigma unless specified otherwise. N-3-oxododecanoyl homoserine lactone (3OC12, Sigma-Aldrich cat# o9139 – manufacturer reported purity of ≤100%) was maintained in 100 mM stocks in molecular biology grade dimethyl sulfoxide (DMSO, purity ≥99.9%, Sigma Aldrich) at -20°C until use.
Culture Methods
Lactobacillus plantarum WCFS1 (BAA-793) was obtained from American Type Culture Collection and maintained in De Man Rogosa Sharp (MRS) media alone or supplemented with 1.5% agar at 37°C in air.
3OC12 Challenge
All experiments carried out in triplicate. Individual colonies of L. plantarum from MRS agar were grown shaking aerobically at 37°C in 5 mL MRS overnight. The following morning, 1 mL overnight cultures were added to 100 mL fresh MRS and grown similarly for approximately 3 h until an OD600 of 0.5, when samples were split into two aliquots of 45 mL of either treated samples with the addition of 3OC12 to a final concentration of 100 μM, or control samples with the addition of 0.1% DMSO (v/v). Samples continued to grow at 37°C shaking for aliquot removal and processing at +1, +4, and +7 h following treatment. Proteomic samples were archived by removing 0.5 mL culture in quadruplicate, flash freezing decanted cell pellets in liquid nitrogen and storing at -80°C before analysis. Transcriptomic samples were archived by pelleting 1.5 mL culture and resuspending in 300 μL RNA Later (Qiagen) and storing at -80°C before library preparation.
RNA Preparation and RNAseq
Frozen sample aliquots were thawed at room temperature and pelleted to remove residual RNA Later before treating with 400 μL Lysozyme Solution (1 mg mL-1 lysozyme, 40 mM EDTA, pH 8) for 1 h at 37°C. Lysozyme treated cells were pelleted and resuspended in RNAzol RT (Molecular Research Center) following the manufacturer’s protocol for large RNA isolation. Resulting RNA was resuspended in 20 μL pure water and quantified using a NanoDrop2000 instrument (Thermo Fisher Scientific) and calculations based on absorbance at 260 nm. DNase I (Thermo Fisher Scientific) was added to RNA at 0.5 U μg-1 for 30 min at 37°C followed by ethanol precipitation. RNA was again resuspended in 20 μL pure water, quantified by NanoDrop and stored at -80°C. Approximately 1 μg of RNA was subjected to ribosomal RNA depletion using the Ribo-Zero Bacteria kit (Illumina) following the manufacturer’s protocol. Samples were ethanol precipitated after rRNA depletion using glycogen as a carrier and resuspended in 10 μL pure water, quantified by NanoDrop and stored -80°C. Approximately 100 ng rRNA-depleted RNA was used for Library preparation with the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs), AMPure XP beads (Beckman Coulter, Inc.), and NEBNext Multiplex Oligos for Illumina (New England Biolabs) containing Nextera i7 sequences following the manufacturer’s protocol. Resultant libraries were quantified by the Qubit broad-range protocol (Thermo) and visualized by agarose gel before sequencing by an Illumina MiSeq platform (MiSeq Analyzer v2.5.1.3) using the MiSeq Reagent Kit v3 (Illumina) and the Illumina RNA-seq protocol with paired end reads of 60 bp. Libraries were analyzed by two replicate MiSeq runs generating 47.3 million raw filtered reads.
RNAseq Analysis
Generated MiSeq reads were then analyzed via the RNAseq pipeline described elsewhere (Li et al., 2018). Read quality control was performed where Phred scores of <20 were trimmed using FastQC and Cutadapt as part of Trim Galore (Krueger, 2015). EDGE-pro (Estimated Degree of Gene Expression in PROkaryotes) (Magoc et al., 2013) for paired end reads was used with default settings on the remaining reads for alignment to the L. plantarum WCFS1 genome from NCBI1 generating RPKM files and count tables. Each count table was read into R where DESeq2 (Love et al., 2014) was used for differential expression analysis and generating associated statistics as a function of treatment with 3OC12 and time. The RNA sequence data is available at NCBI GEO accession GSE124050.
General Data Analysis
In-house R scripts were used for data analysis and visualization of both RNAseq and proteomics data, including the addition of KEGG annotation (Cock et al., 2009), the generation of boxplots and bar graphs using ggplot2 (Wickham, 2016), and UpSet plots using UpSetR (Lex et al., 2014).
Proteomics Analysis
Snap frozen pellets were resuspended in 200 μL of 10% n-propanol in 50 mM ammonium bicarbonate, vortexed and sonicated. Cells were lysed in microtubes with 100 μL caps (Pressure Biosciences, Inc., Easton, MA, United States). The tubes were then placed into the HUB-440 Baro-cycler (Pressure Biosciences, Inc., Easton, MA, United States) and lysis was performed by 30 s cycles (20 s ON, 10 s OFF) for 60 cycles at 45 kpsi and 25°C. After lysis, total protein amount in all samples was estimated and adjusted to 12 μg prior to digestion by trypsin in barocycler (60 s cycles – 50 s pressure ON, 10 s pressure OFF, 45 kpsi, 50°C). Digests were dried on speed vac and stored at -20°C prior to LC-MS/MS analysis. Samples were reconstituted in 30 μL of 0.1% formic acid in water and 0.2 μg of total protein was analyzed by LC-MS/MS using U3000 LC coupled to Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific, Waltham, MA, United States). Autosampler loaded sample onto trap column (PepMap 100, C18, 300 μm ID × 5 mm, 5 μm, 100A) via loading pump at 5 μL min-1 flow rate and 2% solvent B. Analytical pump set to 300 nL min-1 was used to elute peptides from the trap onto analytical column (Acclaim PepMap RSLC, 75 μm ID × 150 mm, C18, 2 μm, 100A). A gradient of 2–60% B in 90 min was used for peptide separation. Solvent A was 0.1% formic acid in water and solvent B was 0.1% formic acid in acetonitrile. Mass spectra were acquired on Fusion Lumos Orbitrap equipped with a Nanospray Flex Ion Source in data-dependent acquisition mode with 3 s cycle times. A survey scan range of 400–1,600 Da was acquired on the Orbitrap detector (resolution 120 K). Maximum injection time was 50 ms and AGC target was 400,000. The most intense ions with charges of 2–7 were fragmented using HCD (higher-energy collisional dissociation), and ions were excluded for 30 s from subsequent MS/MS submission. MS/MS detector was IonTrap with 35 ms injection time and AGC target of 10,000. Resulting spectra were extracted, converted into mgf by ProteoWizard software and searched by Mascot (Matrix Science Inc., London, United Kingdom) against a database containing common standards and contaminants, i.e., trypsin, keratin, etc. (190 protein sequences) and database containing all predicted proteins from L. plantarum genome (3,063 sequences). Oxidation of methionine and deamidation of glutamine and asparagine were selected as variable modifications, enzyme was set to trypsin and 3 missed cleavages were allowed. Precursor ion tolerance was set to 100 ppm and fragment ion to 1 Da. Protein identifications were further validated by Scaffold (Proteome Software Inc., Portland, OR, United States). Protein identifications were accepted if they could be established at greater than 90.0% probability and contained at least 2 identified peptides. P Protein probabilities were assigned by the Protein Prophetalgorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Quantitative analysis was done in Scaffold using emPAIs as an input. Only emPAIs satisfying the probability settings were considered for the analysis (lower scoring matches and probabilities <5% were not included). t-test, fold change, and other calculations were performed on emPAIs using Scaffold (Searle, 2010). In order to avoid divide-by-zero errors caused the absence of proteins in fold change calculations, we set missing values to 0.3, as previously described (Bible et al., 2015). The proteomics data is available at ProteomeXchange repository with identifier PDXD012232 and 10.6019/PXD012232.
Results and Discussion
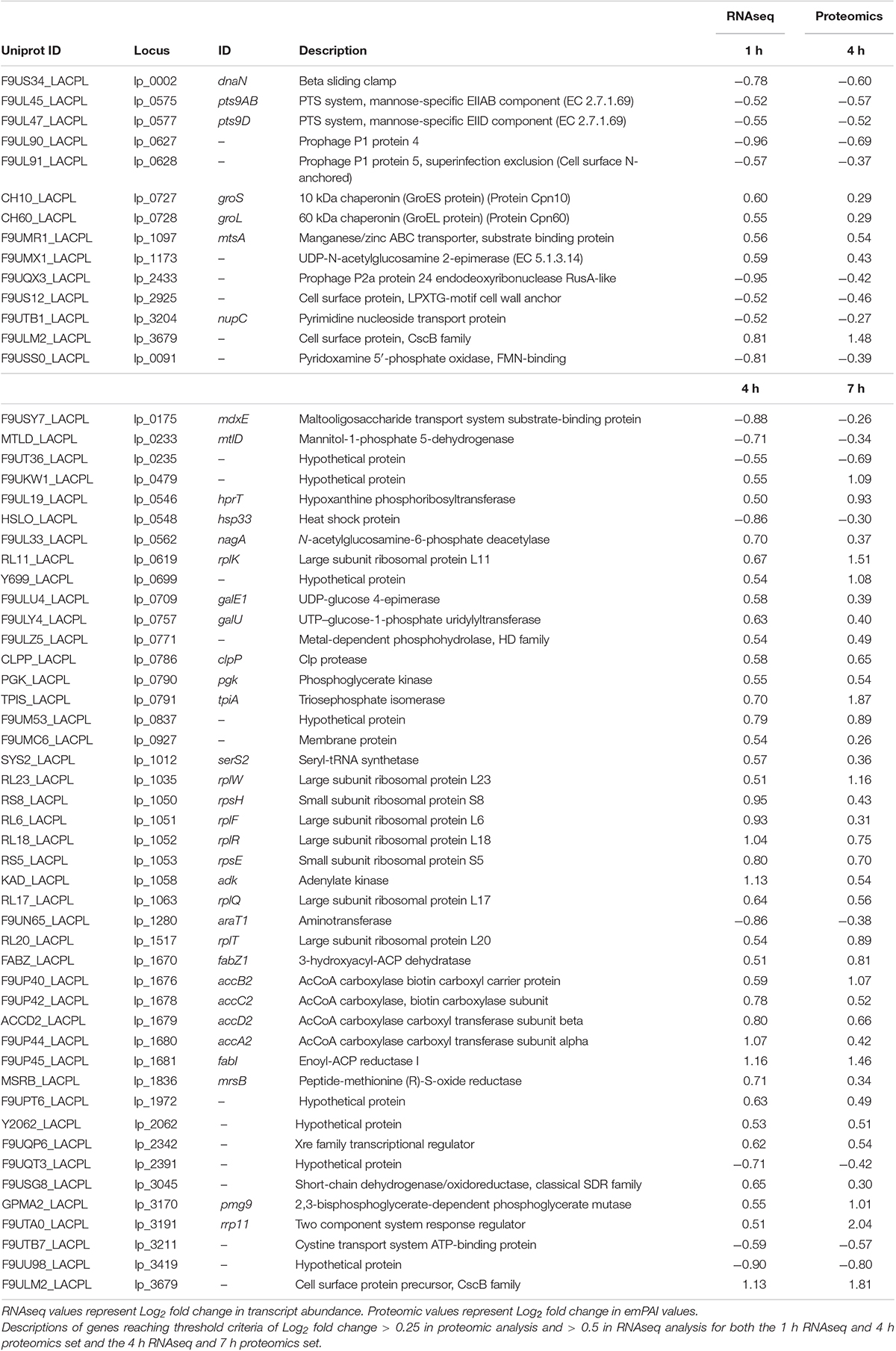
Cultures of L. plantarum WCFS1 were grown from single colonies in overnight cultures of nutrient rich MRS media at 37°C shaking and used the following day to inoculate 100 mL MRS media (1% v/v). The OD600 was monitored throughout growth and when it reached 0.5 (approximately 3 h) cultures were split into 45 mL aliquots for treatments. Cultures continued to be incubated at 37°C and aliquots were removed at 1, 4, and 7 h post-treatment for sample preparation. Transcriptomic data was acquired through replicate RNAseq protocols using Illumina MiSeq and resulted in over 47 million raw filtered reads covering the majority (∼93.6% of the annotated 3,174 genes) of the L. plantarum WCFS1 genome and its three plasmids (Figure 1A). Proteomic analysis resulting from tandem mass spectrometry on the Orbitrap Fusion Lumos (Thermo) were able to corroborate up to 589 of the identified genes from RNAseq at certain time points yielding coverage illustrated in Figure 1B. The resulting datasets from the two techniques were evaluated both independently and as an integrated response in order to dissect the changes displayed by the organism.
Figure 1. Genome coverage. Coverage of L. plantarum genome and its three plasmids (blue region) at 1 h (outermost ring), 4 h (middle ring), and 7 h (innermost ring) by either RNAseq (A) or Proteomic analyses (B). Relative fold changes are mapped as red (upregulated), black (unchanged), or green (downregulated).
Transcriptomic Response
The genomic coverage of RNAseq was relatively consistent throughout all time points (Figure 2A). There were 2,947 genes identified by RNAseq that were common to all three time points, and all genes identified at 1 and 4 h were accounted for at the other times. A large number of the identified genes were determined to be upregulated in comparison to controls (Figure 2B). Of these upregulated genes, 338 were unique to the 1 h samples, 264 were unique to the 4 h samples, and 484 were unique to the 7 h samples, implying that each time point might offer a snapshot of the overall response to 3OC12. In addition to genes identified that were unique to specific times, there were also 372 genes observed to be upregulated at all times. It is interesting that there are sets of genes upregulated consistently as well as those unique to specific timepoints, especially considering that our experiments consisted of a single addition of 3OC12. Based on the hypothesized longevity of AHLs (Yates et al., 2002; Ramos et al., 2010), such an addition might be characterized as an acute environmental change that would elicit an immediate response that then subsides. These initial results, however, implied that this may have consisted of a cascade of genetic changes over the course of the entire experiment where our sampling times were only able to capture small glimpses of the full response. Additionally, the consistent upregulation of one set of genes might indicate a long-term response relative to what might be expected for purportedly transient stimulus.
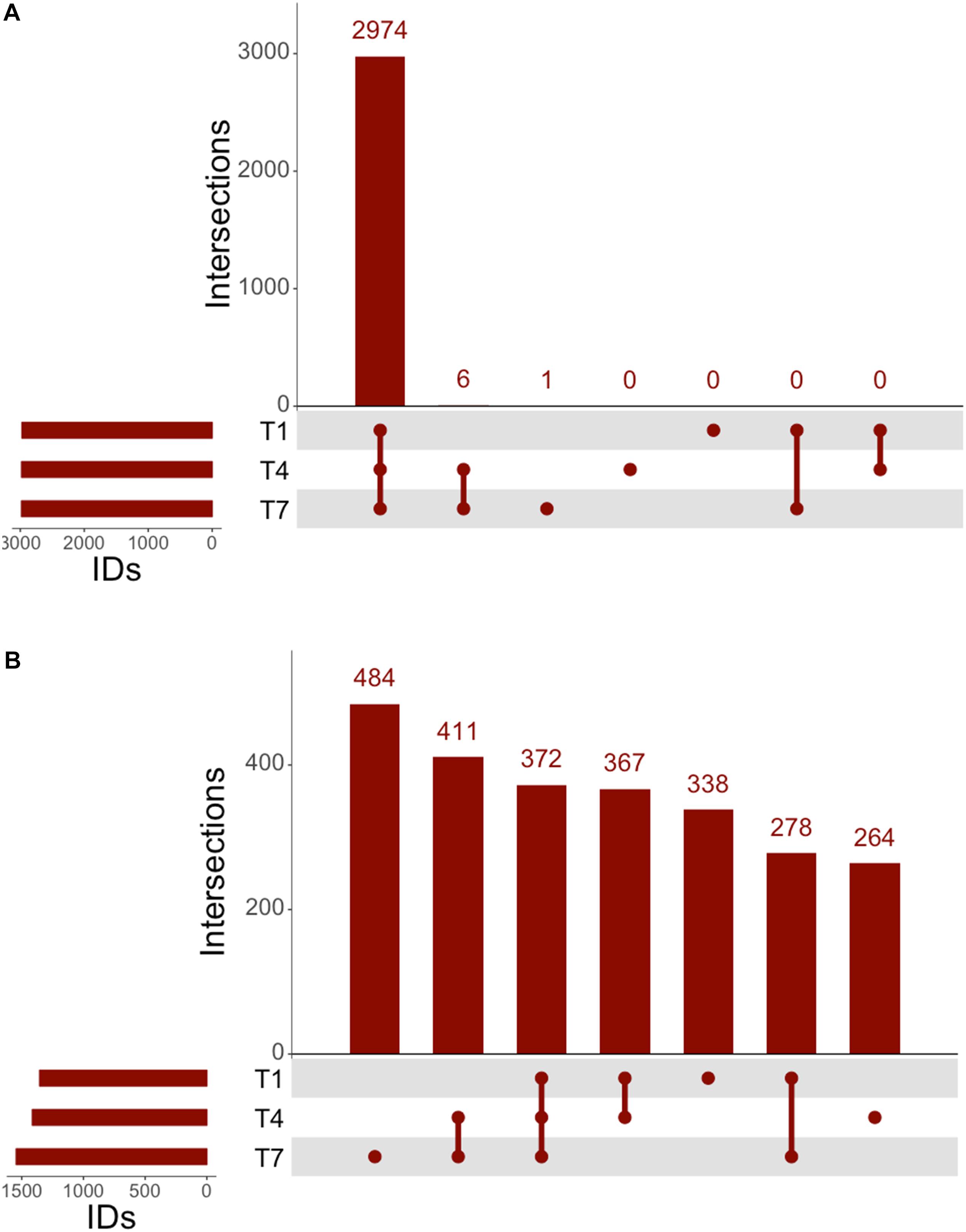
Figure 2. UpSet analysis of RNAseq. (A) Genes identified at each timepoint in RNAseq, consisting of 2,974 genes noted at all timepoints. (B) Upregulated genes identified by RNAseq consisted of 372 consistently upregulated at all times, with 411 genes upregulated after the 1 h timepoint.
All genes identified throughout the experiments were grouped according to KEGG Pathway Categories (Figure 3A) in order to determine where the majority of the transcriptomic activity was occurring. A large portion of identified genes from all samples fell under the Genetic Information Processing group consisting of Transcription, Translation, Folding, Degradation, Replication and Repair. Nearly a quarter of identified genes were assigned to this category at 1 h, and that abundance decreased as the experiments progressed. Samples analyzed at 1 h after 3OC12 addition showed no real changes in Genetic Information Processing, but there was a marked decline in treated samples at 4 h which could indicate the diversion of cellular resources elsewhere for the response. Examples of reductions at 4 h are seen with DNA repair genes such as exoA, recJ, tag2, and mutL, protein export genes such as yidC1, and RNA degradation genes such as recQ2 and rnj. The activity in General Information Processing at 7 h, however, was the lowest of the whole experiment but equivalent in both treated and control samples indicating a restoration to background levels. The response at 1 h showed a decrease in Cellular Growth genes as cells were initially reacting to the AHL stimulus, but by 4 h these genes had returned to the levels observed in the controls. Increases in Membrane Transport and Signal Transduction categories were also observed at 1 h such as upregulation of the transport associated genes mtsABC, metN, and livB and two-component system genes rrp11, hpk1, aad, citCEFX, and pltKR. Increases in Cellular Community genes were also observed at 1 h based on the activity of the known quorum sensing genes lp_0783 (oligopeptide transport) and oroP. Along the same lines of signaling capabilities, there were increases in the gene sip1 that is annotated to function in AIP maturation and export. This might be expected from a cell adjusting to a recently changed environment and initiating signal cascades as a response. Both categories of Membrane Transport and Signal Transduction were returned to control levels after 1 h. The most abundant category of genes noted throughout all time points was that of Global Metabolism, accounting for at least half of all identifications. The amount of RNA devoted to Global Metabolism increased at 4 h in treated samples with increases noted for nagA, argCJ, pts9AB, pgk, tpiA, enoA, galU, pgm, galE1, luxS, adk, and multiple acc and fab genes for example. The amount of total RNA accounted by this category continued at 7 h, but comparisons with control samples showed it to be of background level abundance.
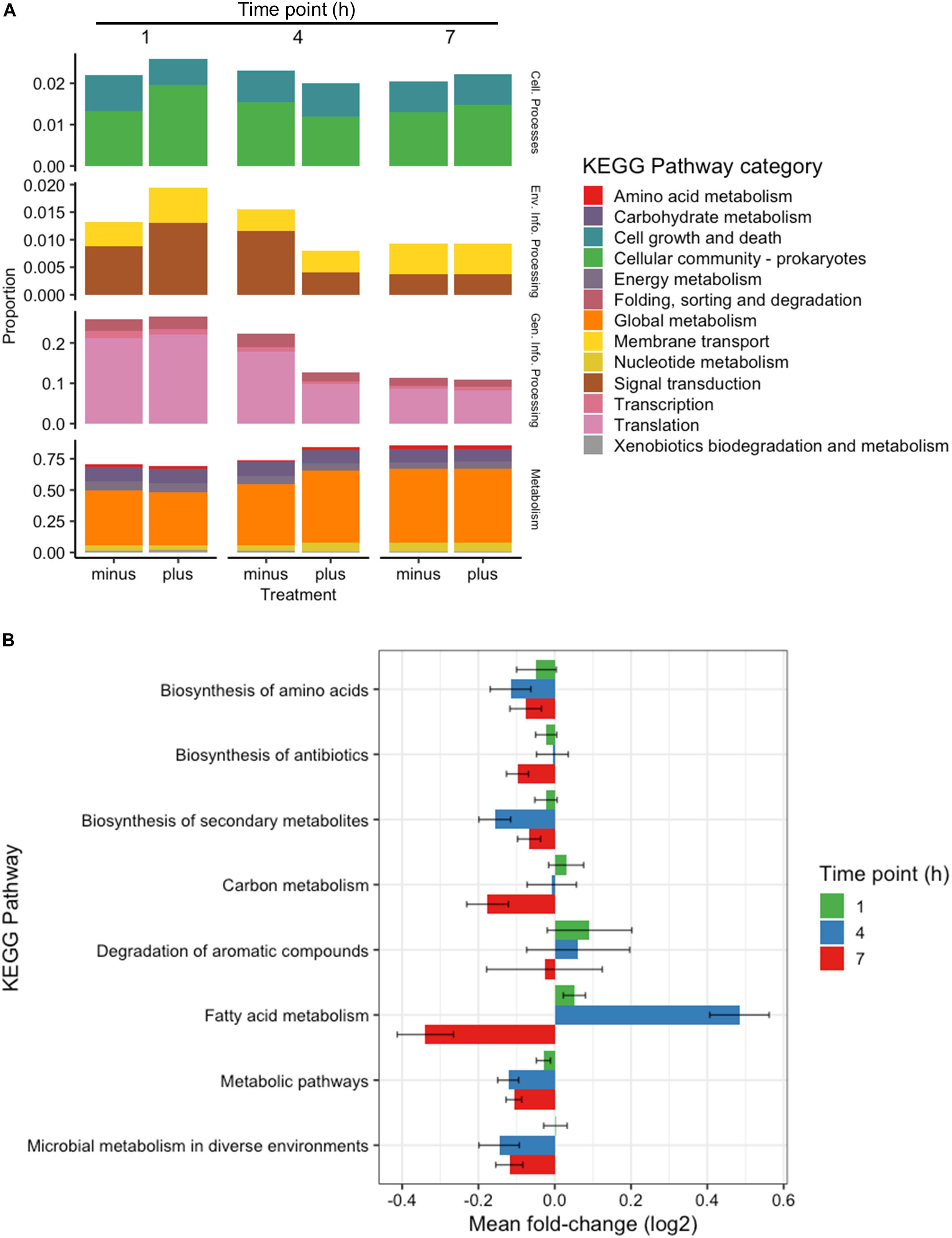
Figure 3. Categorical transcriptomic summary. (A) RNAseq hits mapped to different KEGG Pathway Categories at 1, 4, and 7 h. Control samples are marked as “minus” while treated samples are marked as “plus.” (B) Changes in gene expression falling within the Global Metabolism category at 1, 4, and 7 h plotted as Mean Log2 fold change of 3 biological replicates.
Further investigation of the Global Metabolism response is outlined in Figure 3B. When partitioning the genes of Global Metabolism into 9 further subcategories, it was clear that the increase noted at 4 h was due to activity within Fatty Acid Metabolism. It can be noted here that all subcategories of Global Metabolism revert to a downregulated state at 7 h. Figure 4A shows the further subdivision of Fatty Acid Metabolism into 9 more categories, wherein the Biosynthesis of Unsaturated Fatty Acids and Fatty Acid Biosynthesis groups seem to be the main contributors to the changes noted in Global Metabolism. Both of these groups show upregulation in treated samples at 4 h and downregulation at 7 h. Figure 4B illustrates the components of these two groups and the individual changes noted from treated samples. The incorporated genes make up three groups that represent the conversion from Acetyl-CoA to Malonyl-CoA as a lipid biosynthesis precursor (acc genes), the actual synthesis of fatty acids via acyl-carrier protein-containing genes (fab genes), and the interconversion of Propionyl-CoA and Lipoamide E (pdhD, pflF, and pta). Each of the genes involved show an increase in activity at 4 h and a decrease at 7 h, with the exception of the Propionyl-CoA involved genes pdhD and pflF. Previous investigators have noted that environmental stresses can impart changes on membrane hydrophobicity and adhesion capabilities of Lactobacillus ssp., indicating that these organisms are prone to alter their fatty acid makeup in response to their environment (Haddaji et al., 2017), therefore this comprehensive activation of Fatty Acid Synthesis genes likely follows this reasoning.
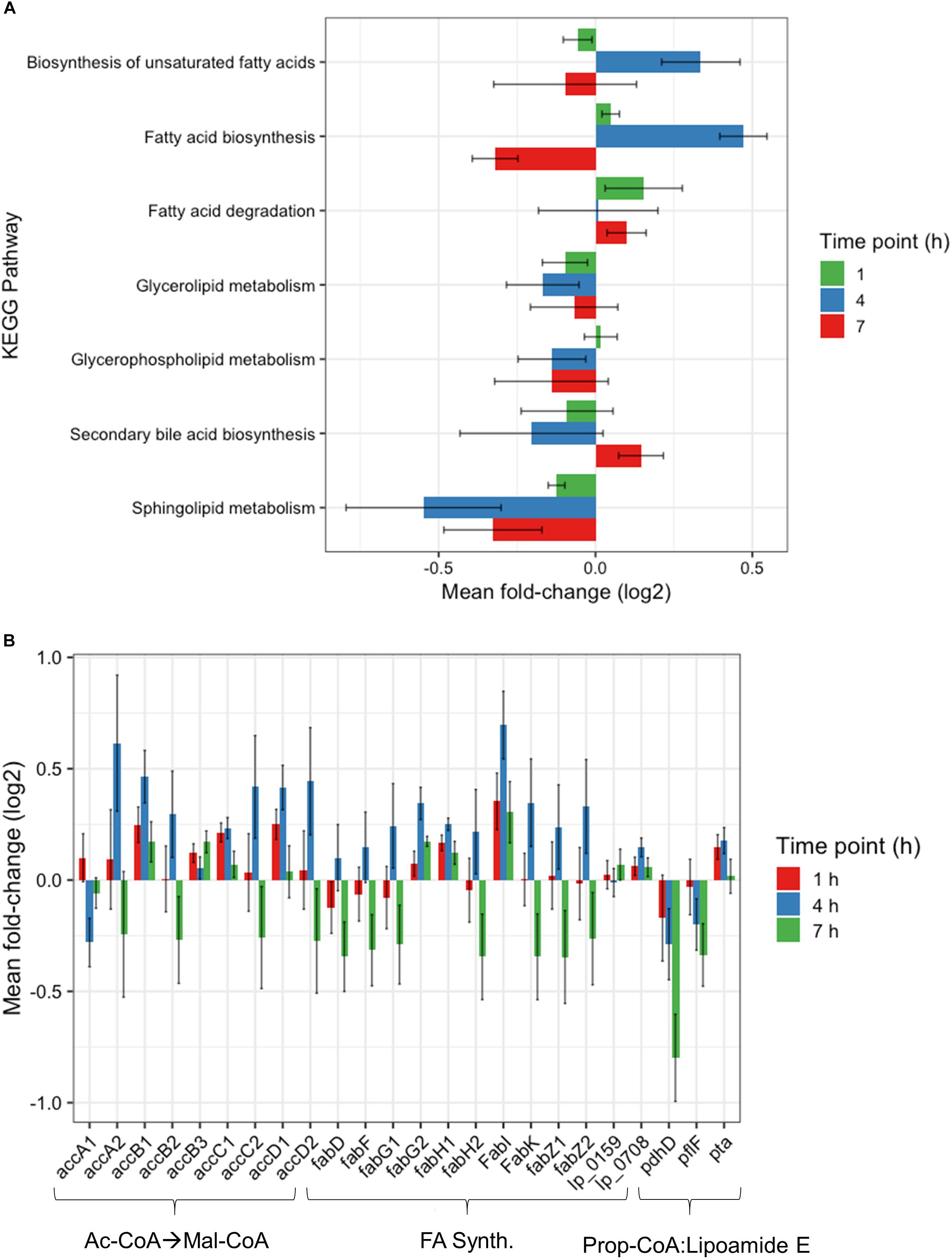
Figure 4. Fatty acid metabolism changes. (A) Changes observed by RNAseq within fatty acid metabolism subcategory at 1, 4, and 7 h plotted as Mean Log2 fold change of 3 biological replicates. (B) RNAseq changes of individual genes involved in Malonyl-CoA synthesis, fatty acid synthesis, and the conversion of propionyl-CoA to Lipoamide E plotted as Mean Log2 fold change of 3 biological replicates at 1, 4, and 7 h.
Proteomic Response
The proteomic response of L. plantarum WCFS1 cultures to 3OC12 was assessed by LC-MS/MS of trypsinized samples on the Orbitrap Fusion Lumos, wherein the output spectra were assigned to known annotated proteins using Mascot (Perkins et al., 1999) and normalized with the Exponentially Modified Protein Abundance Index (emPAI) (Ishihama et al., 2005) to determine a relative abundance of protein per sample similar to strategies used in RNAseq methods. The resulting analysis identified 300 proteins common to samples at all times (Figure 5A). In addition, there were 20 proteins detected unique to the 1 h time point, 70 proteins unique to the 4 h time point, and 126 proteins unique to the 7 h time point. Furthermore, there were 160 proteins that were exclusively identified in the two later time points, and only 3 proteins that were registered at 1 and 7 h only. When comparing treated and control samples, a pattern emerged similar to that seen with RNAseq where each time point contained a unique set of proteins that seemed to increase in number as the experiment progressed (Figure 5B). There were 38 unique upregulated proteins identified at 1 h, 67 at 4 h, and 221 by the 7 h time point. Overall, there were 31 proteins that remained upregulated throughout the experiment. This sweeping trend of increasing translational activity similarly seemed to indicate a 3OC12 response.
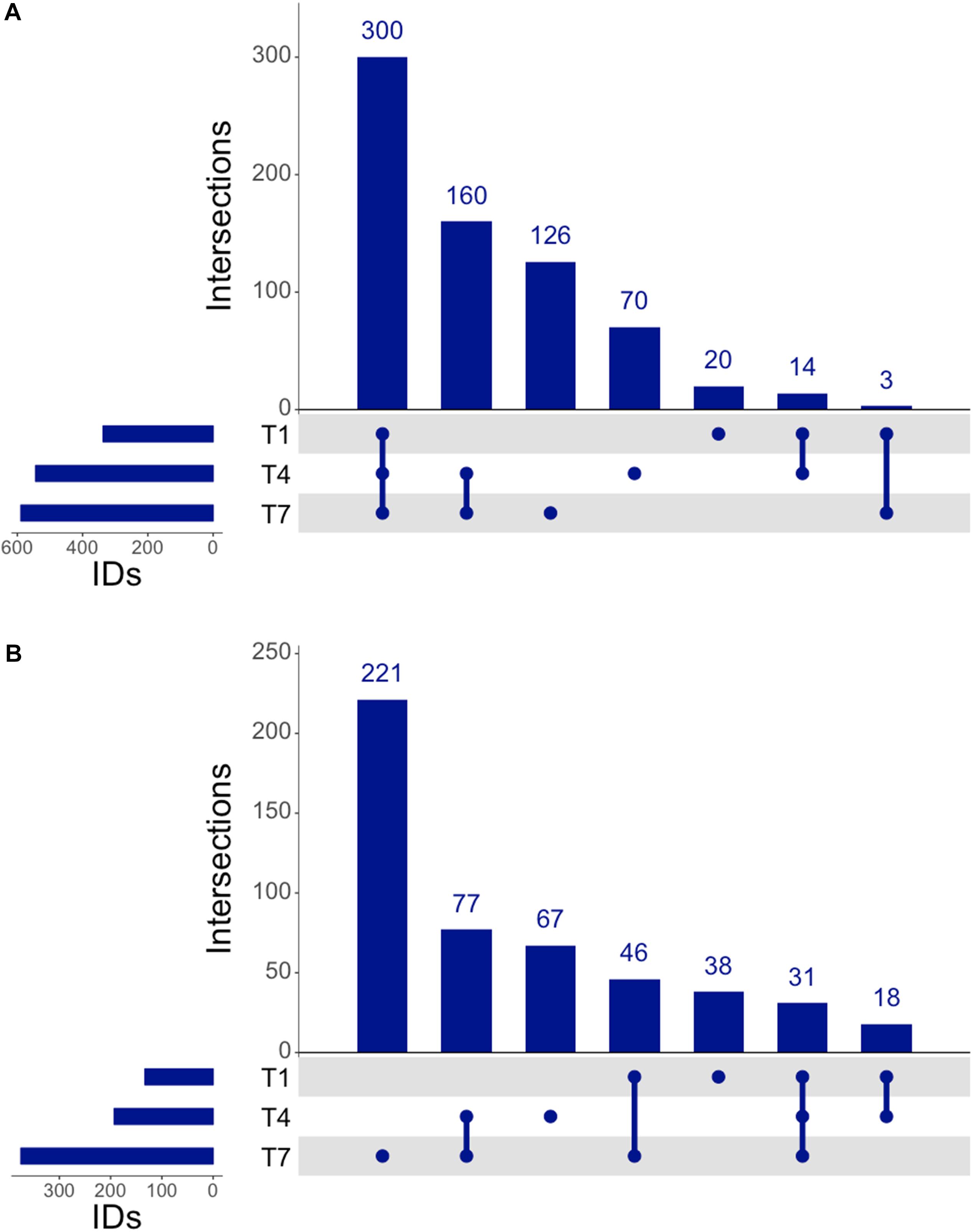
Figure 5. UpSet analysis of proteomics. (A) Genes identified at each time by proteomics, consisting of 300 consistently identified genes throughout experiment and an increasing number of identified proteins at each progressive timepoint. (B) Upregulated genes identified by proteomics showed 31 consistently upregulated genes throughout the experiment, with 77 genes upregulated after the 1 h timepoint.
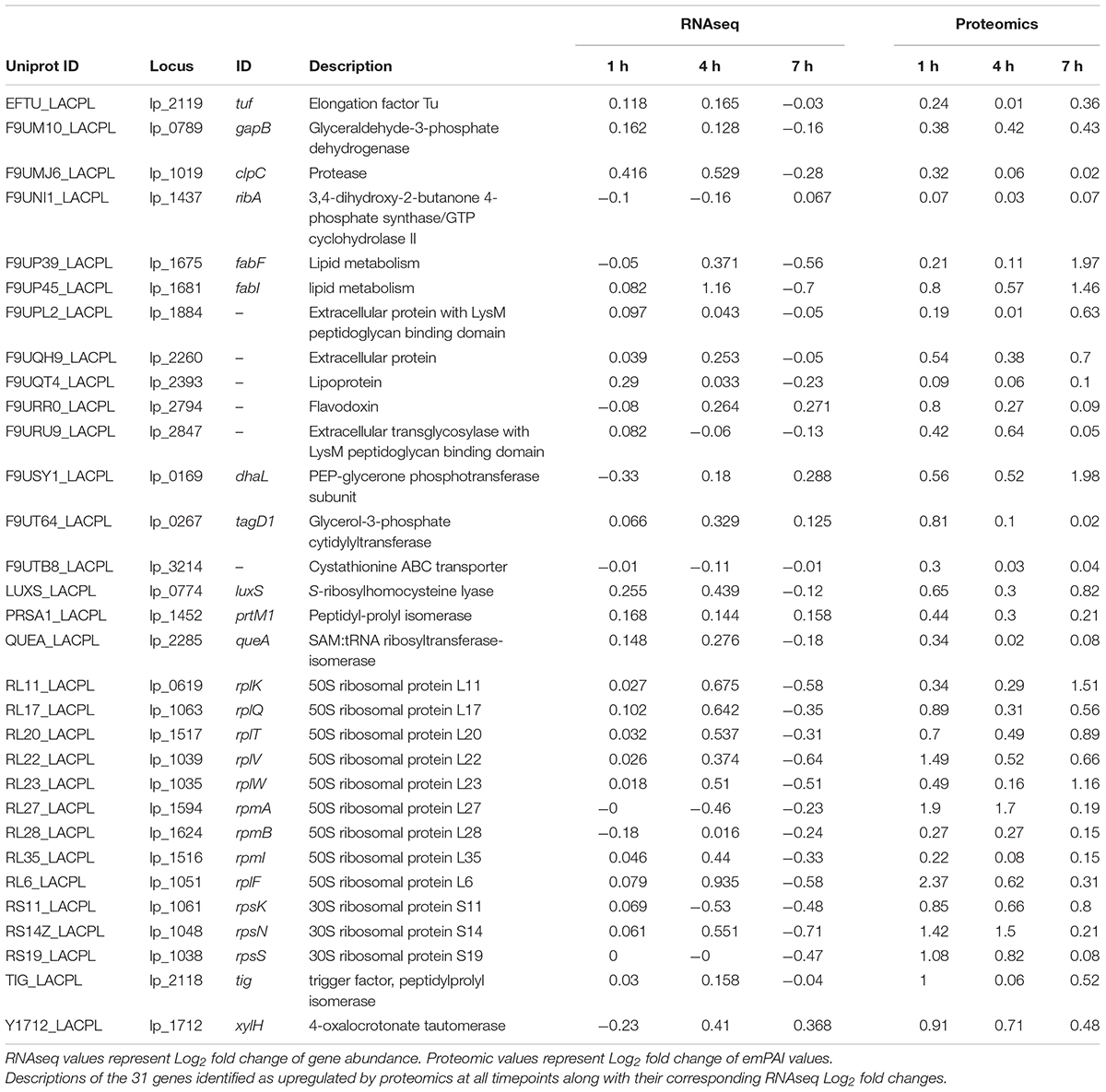
Further investigation into the nature of the identified proteins showed an interesting devotion of resources in both control and treated samples (Figure 6A). Treated samples showed a higher abundance of protein at 1 h contributing to Cellular Processes. In particular, genes relating to processes in the Cellular Community category were higher in treated samples including an increased abundance in Lon protease and the oligopeptide transporter lp_0018, but Cell Growth genes were lower (ClpP, ClpX, FtsZ, and FtsA). At 4 h, however, the Cell Growth genes were comparable in both treated and control samples, and Cellular Community genes in treated samples had reduced to below the levels observed in controls. Detected at a lower abundance than the Cellular Process genes were those related to Environmental Information Processing. A clear increase in this category consisting of Signal Transduction and Membrane Transport was observed for treated samples at 1 h based on the abundance of proteins such as DltD, and greatly diminished thereafter. The second highest category of identified proteins was that of Genetic Information Processing. Treated and control samples had devoted roughly an equivalent amount of resources toward this group at 1 h, but at 4 h the translation in treated cells had diminished to nearly half of that observed in controls. This drop could be attributed to reductions in identifications of chaperones DnaK and Hsp3, as well as repair genes XseAB and MutS2, and large ribosomal proteins RplAMNOBU. Considering the values of top upregulated genes identified at all times (Table 1), however, the abundance of some ribosomal proteins (RplKQTVW, RpmABI, RplF, and RpsKNS) and chaperones (Tig) decreased from 1 h to 4 h but remained above the levels of the control samples, indicating a sort of turnover of proteins that are likely involved in the same process over time.
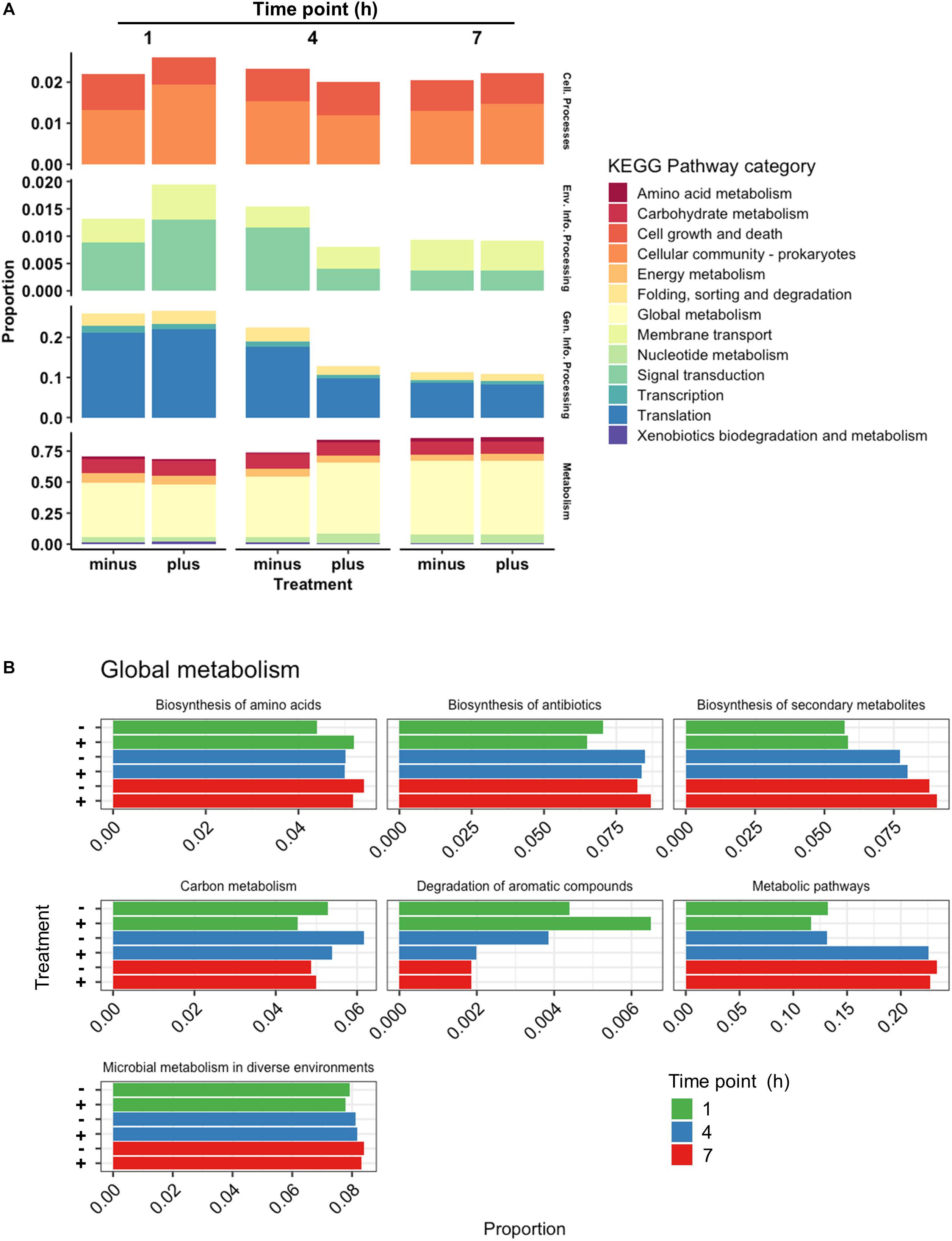
Figure 6. Categorical proteomics summary. (A) Identified proteins were mapped to different KEGG Pathway Categories at 1, 4, and 7 h. (B) Identified proteins mapped to subcategories within Global Metabolism at 1, 4, and 7 h. Control samples are marked as “minus” while treated samples are marked as “plus.”
The most abundant proteins at all times belonged to the Metabolism category, specifically Global Metabolism, accounting for at least half of the proteins identified similar to what was seen with RNAseq data. While approximately equivalent in abundance at 1 h, proteins identified in this category from treated cells increased notably at 4 h before finally returning to levels similar to controls at 7 h. The activity noted in the Global Metabolism category appeared to come from genes functioning in Metabolic Pathways (Figure 6B), where at 4 h the abundance of proteins in treated samples were near double those identified from controls. Proteins encoded from genes such as xylH (putative tautomerase EC 5.3.2.6, lp_1712), dltC1 (D-alanyl carrier protein 1, lp_2017), iolE (inosine dehydratase, lp_3607), fabI (enoyl-ACP reductase, lp_1681), and dak2 (dihydroxyacetone phosphotransferase, lp_0169) are examples of contributors to this increase in abundance. While FabI and Dak2 can both be attributed to different aspects of lipid metabolism, XylH is an enzyme participating in Xylene Degradation, and the lack of other enzymes that L. plantarum possesses in this pathway suggests it may be a participant of a community effort to degrade aromatic compounds. The presence of IolE is also curious considering its role in inositol phosphate metabolism, a similarly incomplete pathway within L. plantarum itself. DltC1, however, poses an interesting observation given its role in cationic antimicrobial peptide resistance. The expression of dltC1 is controlled by a two-component system along with other dlt genes all found in close proximity to each other on the chromosome, and showed a similar upregulation profile as it’s cohort dltD. Indeed the scope of two-component systems is vast and diverse among studied bacteria and have been known to commonly participate in cross-talk (Procaccini et al., 2011), implying an activation of a two-component system by some facet of 3OC12 treatment whose downstream cascade may resemble that of cationic antimicrobial peptide resistance.
The top identified proteins throughout the experiment outlined in Table 1 are a more detailed reflection of the summarized categorical changes above. Very few of the proteins identified with the criteria of having an emPAI Log2 fold change > 0.25 at one or more time points belong to an Energy Metabolism pathway. The only protein that meets these criteria is GapB, which seemed to steadily increase in abundance throughout the experiment. While there were a number of ribosomal proteins detected at all time points, they do not all maintain similar expression trends based on observed spectra. The most abundant group of proteins identified either belonged to Lipid Metabolism or were involved in cell membrane architecture. Previous investigators have noted L. plantarum to undergo changes in membrane composition as a response to environmental stressors (Haddaji et al., 2017), which was interesting based on the presence of LuxS and ClpC in this list of identified proteins. While the latter is a characterized member of the CtsR regulated stress response pathway (Fiocco et al., 2010), the former is a member of various pathways of interest. The LuxS enzyme is a well-characterized S-ribosylhomocysteine lyase that plays a part in various amino acid syntheses, but is also responsible for the synthesis of the bacterial interspecies signaling molecule autoinducer-2 (AI-2). Recently LuxS has been linked with the acid stress response in certain strains of L. plantarum as well as the increased adherence capabilities of cells (Jia et al., 2018) which has been characterized as a basic bacterial survival mechanism (Hunt et al., 2004). It is intriguing, however, to consider the potential that L. plantarum is initiating a quorum sensing event in response to sensing the pathogen-associated 3OC12. Based on recent studies, it is not farfetched to deduce that if LuxS is used in the face of environmental stress to increase the fitness of L. plantarum, and 3OC12 is causing the upregulation of luxS in our cultures, then L. plantarum possesses some way of recognizing the AHL as an indication of environmental stress.
Integrated Transcriptomic and Proteomic Response
The integration of both proteomic and RNAseq datasets resulted in the reinforcement of previous hypotheses generated from independent analyses. The cross-referencing of both datasets allowed the verification by proteomics of 337 genes previously detected by RNAseq at 1 h, 544 at 4 h, and 589 detected at 7 h (Figures 7A,C,E). The threshold values considered in our analyses were Log2-fold change > | 0.25| for proteomic data and > | 0.5| for transcriptomic data in order to gain a broader view of the response using both methods. Correlation plots at specific time points show the number of genes satisfying both threshold criteria at given timepoints (Figures 7B,D,F and Table 1), where the biggest overlap of identifications from proteomics and transcriptomics occurs at 4 h (Figure 7D). When analyzing the data produced across all timepoints by both analyses, we noticed a trend of genes that show upregulation via RNAseq that later appear as protein identifications via proteomics in the next timepoint. We therefore decided to additionally check correlation plots of identifications that met our threshold criteria with a 3 h time offset in mind (Figure 8). As a result of analyzing the correlation of changes in RNAseq at 1 h and proteomics at 4 h (Figure 8A), we identified 13 genes satisfying our previously described criteria wherein all RNAseq results (except those for lp_2433 and lp_0091) met the threshold of p < 0.05 (Wald test, Table 2). Using a similar approach to analyze identifications by RNAseq at 4 h and proteomics at 7 h (Figure 8B), we noted 45 genes of interest, of which 71% similarly showed p < 0.05 (Wald test, Table 2).
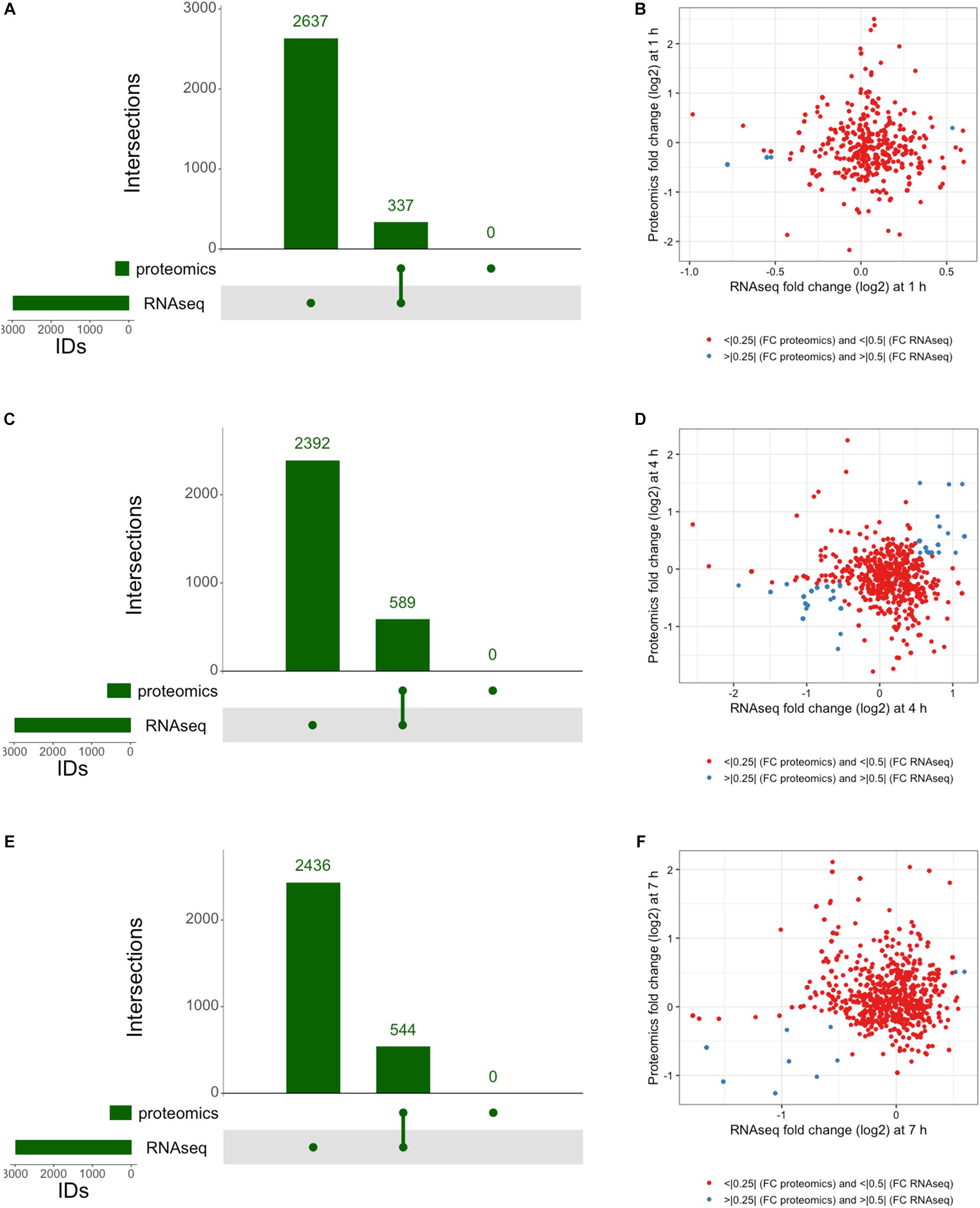
Figure 7. Integrated response analysis. UpSet plot of genes identified by both RNAseq and proteomics at (A) 1 h showing 337 genes in common, (C) 4 h with 544 genes in common, and (E) 7 h with 589 genes in common. Correlation plots of genes identified by RNAseq and Proteomics using threshold cutoffs for genes with Log2 fold change > 0.25 for proteomics and > 0.5 for RNAseq (blue dots) at (B) 1 h, (D) 4 h, and (F) 7 h.
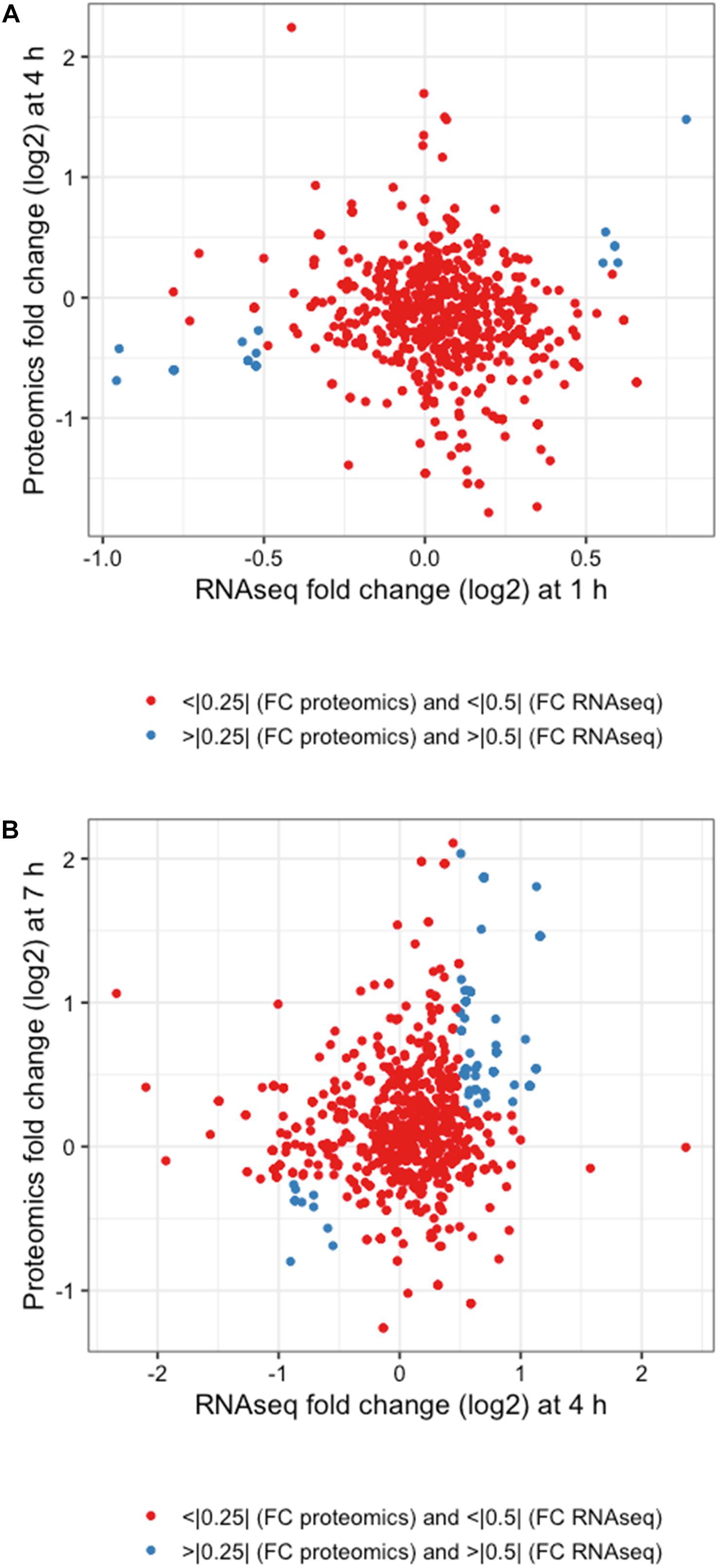
Figure 8. Integrated response analysis with time offset. Correlation plots for (A) 1 h RNAseq with 4 h proteomics and (B) 4 h RNAseq and 7 h proteomics using threshold cutoffs of Log2 fold changes of >0.25 for proteomics and >0.5 for RNAseq (blue dots).
The genes identified satisfying Log2 fold change thresholds for both RNAseq and proteomics with offset times are summarized in Table 2. Changes in RNAseq that were verified with proteomics consisted of upregulation of membrane-associated proteins encoded by lp_3679 (CscB family cell surface protein) and mtsA (metal transporter), chaperones GroES and GroEL, and lp_1173 (UDP-N-acetylglucosamine 2-epimerase) that could be involved in membrane architecture. The transcriptional changes at 4 h that were corroborated with a similar protein response at 7 h consisted of a variety of processes. There seemed to be an emphasis on ATP generation and conservation among the observed changes in abundance. Genes such as mtlD involved in fructose/mannose metabolism were downregulated, as were genes such as mdxE responsible for maltooligosaccharide transport and lp_3211 involved in cystine transport. ABC transporters similar to these have been shown to be downregulated during the onset of stationary phase-related environmental stressors (Cohen et al., 2006) based on the assumption that they require ATP, which at that time is a precious commodity. When checking the rest of our data for this trend of ATP conservation we found that of the 93 ABC transporters detected by RNAseq at 4 h, all but 16 were downregulated compared to controls, and 11 of the 14 detected by proteomics at 7 h were also downregulated. While L. plantarum cells remained in log phase of growth throughout these experiments, it’s possible a similar environmental pressure was asserted on the cells causing them to reprioritize their use of ATP. The genes pgk, tpiA and pmg9 involved in glycolysis were upregulated, presumably to facilitate an influx of metabolites into this pathway to more readily produce ATP. Also noted was an increase in the abundance of adk. This gene encodes the adenylate kinase enzyme responsible for generating ADP and dADP as substrates for ndk to convert into ATP. Increases were also noted in the levels of hprT, which is responsible for the generation of GMP from guanine as a precursor to GTP synthesis. In addition to energy conservation, genes involved in nucleotide sugar metabolism such as nagA, galE1, and galU were all observed to be upregulated during this portion of the response as well. UDP-glucose 4-epimerase, encoded by galE1, is also a participant in the Leloir pathway and has been observed to be upregulated during late log phase of growth as cells alter their membrane structure to deal with the changing environment (Cohen et al., 2006). Like what was observed with the ABC transporters, cells did not enter stationary phase but it is possible they are adapting to environmental stressors by altering their membrane composition (Haddaji et al., 2017). This hypothesis is reinforced by the upregulation of a number of genes involved in the conversion of Acetyl-CoA to Malonyl CoA (accA2, accB2, accC2, and accD2) and the subsequent conversion of Malonyl-CoA to different fatty acids (fabZ1, fabI, and lp_3045).
Stress Response
Stress responses consist of a variety of differentially expressed genes and can originate from a number of different stimuli as mentioned above. While there have been a number of studies on Lactobacillus ssp. responding to various stresses such as oxidative stress (Serrano et al., 2007), general acid stress (Heunis et al., 2014; Seme et al., 2015), phenolic acid stress (Gury et al., 2009), lactic acid stress (Pieterse et al., 2005), alkaline stress (Lee et al., 2011), metal stress (Tong et al., 2017), growth phase transition stress (Cohen et al., 2006), and the general Classes I and III stress responses (Van Bokhorst-van de Veen et al., 2013), most of these studies indicate a large number of participating genes with no apparent timeframe on induction of the response or the restoration to the pre-stimulus state presumably due to the nature of the individual stress tested. Although 3OC12 is not traditionally considered a stressor, we observed similarities in gene and protein activity with stress responses characterized in similar organisms when we analyzed our cultures after exposure to this AHL.
One of the top identified genes in both RNAseq and proteomics was that of mrsB (Table 2). The enzyme peptide-methionine (R)-S-oxide reductase encoded by this gene is responsible for reducing oxidized methionine caused by reactive oxygen species. Aside from the specific activity of MrsB, thioredoxin is a more general oxidative-stress response protein whose activity has been established in L. plantarum (Serrano et al., 2007). While the investigators emphasized trxB1 to be the key player involved in the stress response, our data showed significant upregulation of the genes trxB, trxA2, and trxH at 1 and 4 h in transcriptomics. The collective upregulations of trxB, trxA2, trxH, and mrsB indicate the cells to be responding similarly as they might in oxidative stress conditions.
The universal stress protein (Usp) family involves nucleotide-binding proteins that function in various non-specific stress conditions. Research has been done in particular on their involvement with phenolic acid stress in L. plantarum (Gury et al., 2009) focusing on the activity of Usp1. There are 10 uncharacterized universal stress proteins in L. plantarum WCFS1, and all but 1 show upregulation to some extent at 1 h by RNAseq. Of those 9, 4 are upregulated still at 4 h while 3 are downregulated, and 5 are upregulated at 7 h while only 1 is downregulated. Without further elucidation of the roles of these individual universal stress proteins, no conclusions can be drawn about the nature of the stress response displayed. The only thing that can be concluded is that it does not follow the pattern observed from stress brought on by the presence of organic acids, as the Usp immediately downstream of the PadR repressor studied (Gury et al., 2009) displayed changes opposite to those that would be expected from such a response.
Studies on the acid stress response in L. plantarum strains (Heunis et al., 2014) have shown increasing activity in a number of seemingly unrelated genes, characterizing a profile for the adaptation to the acidic environment that includes changes in energy metabolism, for example. Of the 18 genes previously observed as upregulated in response to acid stress (Heunis et al., 2014), only 3 are upregulated in our RNAseq data at 1 h (pmi or lp_2384, ldhL1 or lp_0537, and pta or lp_0807), 2 at 4 h (nagB or lp_0226, and ldhL2 or lp_1101), and 1 at 7 h (acdH or lp_0329). The genes upregulated at 1 h and 4 h are involved in carbohydrate metabolism, while acdH encodes a putative acetaldehyde dehydrogenase. Furthermore, the ldhL gene observed as upregulated at multiple times encodes the enzyme lactate dehydrogenase responsible for the reduction of pyruvate to lactate, which is the hallmark of the Lactic Acid Bacteria as a method of regenerating NAD+. Another acid-induced stress marker in L. plantarum has been shown to be LuxS (Jia et al., 2018). While it didn’t reach the thresholds set above (Log2 fold change > 0.5), RNAseq was able to identify the slight but significant luxS upregulation occurring at 1 h (Log2 fold change = 0.25, or +19% increase; p = 2 × 10-5) and at 4 h (Log2 fold change = 0.44, or +37% increase; p = 0.002). Proteomic analysis was further able to confirm LuxS upregulation throughout all treated samples. While previous studies have shown LuxS to be involved in acid response, we did not observe any other significant markers for such stress. Based on the function of LuxS, however, it is highly probable that other factors could be responsible for its induction.
Studies on transcriptomic activity during growth phase transitions in L. plantarum provide a long list of differentially expressed genes (Cohen et al., 2006). While cultures in our experiments never entered stationary phase of growth, there were some similar changes in gene expression observed in our experiments to those noted in late-log phase and stationary phase. Examples include dacA1, hpk11, and rrp11 which all were noted to increase at 1 and 4 h in RNAse q. The Rrp11 protein functions as a two-component system response regulator that was also identified by proteomics at 7 h, further reinforcing its upregulation over time. These three genes have been hypothesized to be involved in a two-component system involved in cell wall maintenance, adding to the list of similarly tasked genes that have been previously discussed above. Cohen et al. (2006) also noted the high abundance of Plantaricin genes throughout growth that decreased with the onset of stationary phase. Our RNAseq data showed that of the 21 annotated pln genes, all were downregulated at 1 h, 5 were upregulated at 4 h, and by 7 h there were 13 that were upregulated. As all time points in our experiments represent log phase of growth, the increasing expression of Pln genes over time does not disagree with previous observations of their phase-specific expression patterns. Their upregulation in comparison to controls, however, makes the activity of these Plantaricin genes interesting. Considering previous instances when L. plantarum has employed the use of Plantaricins such as when grown in co-culture with other bacteria (Maldonado et al., 2003, 2004a,b), the idea of 3OC12 stimulating a Plantaricin response is an intriguing example of a stress-induced defense mechanism. Cohen et al. (2006) also noted a small group of stress-related proteins that peak during stationary phase, specifically dps1, grpE, clpP, and kat, all of which are upregulated at 1 h in our experiments. With exception of clpP, all of these genes are also upregulated at 4 h as well.
General stress responses can be grouped into classes based on their regulators. The causes for such responses are traditionally attributed to heat shock or general stress conditions (Derre et al., 1999). The Class I and III stress response regulons have recently been characterized in L. plantarum by a transcriptomic analysis of single- and double-mutants of their respective regulators HrcA and CtsR (Van Bokhorst-van de Veen et al., 2013). The Class I response regulon governed by the HrcA repressor showed the involvement of genes such as the chaperones groS and groL, the hsp1 small heat shock protein, and three putative genes annotated as an integrase/recombinase (lp_1268), a CAAX family membrane-bound protease (lp_0726), and an uncharacterized protein (lp_1880). Each of these Class I regulon members with the exception of the protease and the recombinase were observed to be upregulated via RNAseq at 1 and 4 h. Proteomics further confirmed increased numbers of GroES and GroEL at 4 h and increased Hsp1 at 7 h. The Class III response regulon that is controlled by the repressor CtsR has been studied a couple of times in L. plantarum (Fiocco et al., 2009; Fiocco et al., 2010; Van Bokhorst-van de Veen et al., 2013), and consists mainly of the Clp proteases and Clp ATPases. The existing clp genes in the L. plantarum WCFS1 genome (clpPCEBXL) were all determined to be upregulated by RNAseq at 1 h (with the exception of clpL) and at 4 h (with the exception of clpE), and all were downregulated compared to controls at 7 h in our experiments. Proteomic analysis was further able to confirm the upregulation of ClpP at 7 h. Aside from clp genes, CtsR has also been shown in L. plantarum to control the small heat shock protein encoded by hsp1, the protease subunits encoded by hslU and hslV, a tyrosine recombinase xerC, and an annotated aldose-1-epimerase (lp_1843) (Van Bokhorst-van de Veen et al., 2013). The putative aldose 1-epimerase (lp_1843), the two protease subunits (lp_1845 and lp_1846) and the tyrosine recombinase (lp_1847) all fall within the same region of the chromosome and are likely regulated by a single promoter. As such, our RNAseq data show these four genes are upregulated at 1 h, but only the distal lp_1843 remained upregulated by 4 h. Proteomics, however, were only able to confirm the upregulation of the protease subunits HslU and HslV.
Despite the abundance of genes described as part of various stress responses, the responses we observed from our cultures did not perfectly align with any established stress response. Although 3OC12 is not traditionally considered a stressor, and the genes upregulated in response to its addition to cultures of L. plantarum are do not traditionally follow any single previously defined stress response pathway, we consider this AHL sensing to be a form of a stress response based on our observations of downregulated cellular growth genes in addition to upregulation of previously characterized stress response genes compared to untreated controls. In particular, this AHL response shares similarities with a number of different characterized stress responses, namely oxidative stress responses, general Classes I and III stress responses, and late growth stage transition responses. Based on the nature of our experiments we can conclude that our cultures were free of any traditional initiator of these set responses, and therefore these findings represent a unique response that may borrow facets of other commonly used response pathways to achieve the most desirable survival phenotype.
Conclusion
Here we have shown that L. plantarum WCFS1 is capable of sensing the Gram-negative quorum sensing molecule 3OC12 from P. aeruginosa. Transcriptomic and proteomic analyses indicate a number genes specifically upregulated as a result of 3OC12 titration into pure cultures of L. plantarum. The majority of genes identified by both methods fall within the category of Global Metabolism with an emphasis on Fatty Acid Synthesis, although a number of identified genes also hint at the organism’s alteration of energy metabolism in order to conserve ATP similar to what might occur in a transition to stationary phase of growth. Further changes registered by both methods include genes consistent with both a Classes I and III stress response traditionally caused by general environmental stress. While no stress response profiles could be perfectly matched to previous omic investigations of stress responses, our data showed similarities with cellular responses to oxidative stress and those associated with growth phase transitions, among others, such as thioredoxin activity, fatty acid synthesis, and membrane maintenance. Based on the genes identified we can conclude that the cell is responding to environmental stress unlike other previously established responses. The upregulation of the AI-2 synthesizing enzyme LuxS is an intriguing occurrence that implicates the attempt of L. plantarum to externalize a cell signaling event as a response to AHL addition. The upregulation of the Plantaricin genes is a similar event that when taken together with the luxS activity paints the picture of a probiotic organism initiating a defense mechanism in response to a pathogen-associated small molecule. We have therefore provided evidence that L. plantarum responds to the presence of 3OC12 by initiating multiple quorum sensing systems of its own given the luxS activity and the upregulation of Plantaricin that results from AIP signaling. In addition, the induction of two-component systems and of multiple putative transcription factors further hints at the complex cell signaling cascade initiated by this AHL.
Author Contributions
JS, SD, DL, and SW contributed to the experimental work described in this manuscript and the preparation of the manuscript. JS and SD cultured bacteria and prepared samples for transcriptomics and proteomics. JS performed library preparation and transcriptomics studies. DL was responsible for all proteomics work. SD performed data compiling and preliminary analysis. SW directed experimental design and analysis.
Funding
The authors were funds of the Naval Research Laboratory (MA041-06-41) and funds associated with the Office of the Secretary of Defense Applied Research for the Advancement of S&T Priorities (ARAP) Synthetic Biology for Military-Relevant Environments (SBME) program.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^RefSeq NC_004567.2, NC_006375.1, NC_006376.1, NC_006377.1.
References
Anderssen, E. L., Diep, D. B., Nes, I. F., Eijsink, V. G., and Nissen-Meyer, J. (1998). Antagonistic activity of Lactobacillus plantarum C11: two new two-peptide bacteriocins, plantaricins EF and JK, and the induction factor plantaricin A. Appl. Environ. Microbiol. 64, 2269–2272.
Atrih, A., Rekhif, N., Moir, A. J., Lebrihi, A., and Lefebvre, G. (2001). Mode of action, purification and amino acid sequence of plantaricin C19, an anti-Listeria bacteriocin produced by Lactobacillus plantarum C19. Int. J. Food Microbiol. 68, 93–104. doi: 10.1016/S0168-1605(01)00482-2
Bassler, B. L., Greenberg, E. P., and Stevens, A. M. (1997). Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179, 4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997
Bauer, W. D., and Mathesius, U. (2004). Plant responses to bacterial quorum sensing signals. Curr. Opin. Plant Biol. 7, 429–433. doi: 10.1016/j.pbi.2004.05.008
Bible, A. N., Khalsa-Moyers, G. K., Mukherjee, T., Green, C. S., Mishra, P., Purcell, A., et al. (2015). Metabolic adaptations of Azospirillum brasilense to oxygen stress by cell-to-cell clumping and flocculation. Appl. Environ. Microbiol. 81, 8346–8357. doi: 10.1128/AEM.02782-15
Biswa, P., and Doble, M. (2013). Production of acylated homoserine lactone by gram-positive bacteria isolated from marine water. FEMS Microbiol. Lett. 343, 34–41. doi: 10.1111/1574-6968.12123
Booth, M. C., Atkuri, R. V., Nanda, S. K., Iandolo, J. J., and Gilmore, M. S. (1995). Accessory gene regulator controls Staphylococcus aureus virulence in endophthalmitis. Invest. Ophthalmol. Vis. Sci. 36, 1828–1836.
Brint, J. M., and Ohman, D. E. (1995). Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177, 7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995
Chun, C. K., Ozer, E. A., Welsh, M. J., Zabner, J., and Greenberg, E. P. (2004). Inactivation of a Pseudomonas aeruginosa quorum-sensing signal by human airway epithelia. Proc. Natl. Acad. Sci. U.S.A. 101, 3587–3590. doi: 10.1073/pnas.0308750101
Cock, P. J., Antao, T., Chang, J. T., Chapman, B. A., Cox, C. J., Dalke, A., et al. (2009). Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423. doi: 10.1093/bioinformatics/btp163
Cohen, D. P., Renes, J., Bouwman, F. G., Zoetendal, E. G., Mariman, E., De Vos, W. M., et al. (2006). Proteomic analysis of log to stationary growth phase Lactobacillus plantarum cells and a 2-DE database. Proteomics 6, 6485–6493. doi: 10.1002/pmic.200600361
Collins, C. H., Arnold, F. H., and Leadbetter, J. R. (2005). Directed evolution of vibrio fischeri LuxR for increased sensitivity to a broad spectrum of acyl-homoserine lactones. Mol. Microbiol. 55, 712–723. doi: 10.1111/j.1365-2958.2004.04437.x
Collins, C. H., Leadbetter, J. R., and Arnold, F. H. (2006). Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nat. Biotechnol. 24, 708–712. doi: 10.1038/nbt1209
Derre, I., Rapoport, G., and Msadek, T. (1999). CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31, 117–131. doi: 10.1046/j.1365-2958.1999.01152.x
Diep, D. B., Havarstein, L. S., Nissen-Meyer, J., and Nes, I. F. (1994). The gene encoding plantaricin A, a bacteriocin from Lactobacillus plantarum C11, is located on the same transcription unit as an agr-like regulatory system. Appl. Environ. Microbiol. 60, 160–166.
Fiocco, D., Capozzi, V., Collins, M., Gallone, A., Hols, P., Guzzo, J., et al. (2010). Characterization of the CtsR stress response regulon in Lactobacillus plantarum. J. Bacteriol. 192, 896–900. doi: 10.1128/JB.01122-09
Fiocco, D., Collins, M., Muscariello, L., Hols, P., Kleerebezem, M., Msadek, T., et al. (2009). The Lactobacillus plantarum ftsH gene is a novel member of the CtsR stress response regulon. J. Bacteriol. 191, 1688–1694. doi: 10.1128/JB.01551-08
Firth, N., Fink, P. D., Johnson, L., and Skurray, R. A. (1994). A lipoprotein signal peptide encoded by the staphylococcal conjugative plasmid pSK41 exhibits an activity resembling that of Enterococcus faecalis pheromone cAD1. J. Bacteriol. 176, 5871–5873. doi: 10.1128/jb.176.18.5871-5873.1994
Fujii, T., Ingham, C., Nakayama, J., Beerthuyzen, M., Kunuki, R., Molenaar, D., et al. (2008). Two homologous Agr-like quorum-sensing systems cooperatively control adherence, cell morphology, and cell viability properties in Lactobacillus plantarum WCFS1. J. Bacteriol. 190, 7655–7665. doi: 10.1128/JB.01489-07
Gao, Y., Duan, J., Geng, X., Zhang, Z., Zhang, R., Li, X., et al. (2017). Deficiency of quorum sensing system inhibits the resistance selection of Pseudomonas aeruginosa to ciprofloxacin and levofloxacin in vitro. J. Glob. Antimicrob. Resist. 10, 113–119. doi: 10.1016/j.jgar.2017.04.008
Goodson, M. S., Harbaugh, S. V., Chushak, Y. G., and Kelley-Loughnane, N. (2015). Integrating and amplifying signal from riboswitch biosensors. Methods Enzymol. 550, 73–91. doi: 10.1016/bs.mie.2014.10.032
Gray, K. M., Passador, L., Iglewski, B. H., and Greenberg, E. P. (1994). Interchangeability and specificity of components from the quorum-sensing regulatory systems of Vibrio fischeri and Pseudomonas aeruginosa. J. Bacteriol. 176, 3076–3080. doi: 10.1128/jb.176.10.3076-3080.1994
Gury, J., Seraut, H., Tran, N. P., Barthelmebs, L., Weidmann, S., Gervais, P., et al. (2009). Inactivation of PadR, the repressor of the phenolic acid stress response, by molecular interaction with Usp1, a universal stress protein from Lactobacillus plantarum, in Escherichia coli. Appl. Environ. Microbiol. 75, 5273–5283. doi: 10.1128/AEM.00774-09
Haddaji, N., Mahdhi, A. K., Ismaiil, M. B., and Bakhrouf, A. (2017). Effect of environmental stress on cell surface and membrane fatty acids of Lactobacillus plantarum. Arch. Microbiol. 199, 1243–1250. doi: 10.1007/s00203-017-1395-9
Halleran, A. D., and Murray, R. M. (2018). Cell-free and in vivo characterization of Lux, Las, and Rpa quorum activation systems in E. coli. ACS Synth. Biol. 7, 752–755. doi: 10.1021/acssynbio.7b00376
Hammer, B. K., and Bassler, B. L. (2003). Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50, 101–104. doi: 10.1046/j.1365-2958.2003.03688.x
Heunis, T., Deane, S., Smit, S., and Dicks, L. M. (2014). Proteomic profiling of the acid stress response in Lactobacillus plantarum 423. J. Proteome Res. 13, 4028–4039. doi: 10.1021/pr500353x
Hunt, S. M., Werner, E. M., Huang, B., Hamilton, M. A., and Stewart, P. S. (2004). Hypothesis for the role of nutrient starvation in biofilm detachment. Appl. Environ. Microbiol. 70, 7418–7425. doi: 10.1128/AEM.70.12.7418-7425.2004
Ishihama, Y., Oda, Y., Tabata, T., Sato, T., Nagasu, T., Rappsilber, J., et al. (2005). Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell Proteomics 4, 1265–1272. doi: 10.1074/mcp.M500061-MCP200
Jahoor, A., Patel, R., Bryan, A., Do, C., Krier, J., Watters, C., et al. (2008). Peroxisome proliferator-activated receptors mediate host cell proinflammatory responses to Pseudomonas aeruginosa autoinducer. J. Bacteriol. 190, 4408–4415. doi: 10.1128/JB.01444-07
Janzon, L., and Arvidson, S. (1990). The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 9, 1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x
Jensen, R. O., Winzer, K., Clarke, S. R., Chan, W. C., and Williams, P. (2008). Differential recognition of Staphylococcus aureus quorum-sensing signals depends on both extracellular loops 1 and 2 of the transmembrane sensor AgrC. J. Mol. Biol. 381, 300–309. doi: 10.1016/j.jmb.2008.06.018
Jensen, V., Lons, D., Zaoui, C., Bredenbruch, F., Meissner, A., Dieterich, G., et al. (2006). RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and -independent pathways. J. Bacteriol. 188, 8601–8606. doi: 10.1128/JB.01378-06
Jia, F. F., Zheng, H. Q., Sun, S. R., Pang, X. H., Liang, Y., Shang, J. C., et al. (2018). Role of luxS in Stress Tolerance and Adhesion Ability in Lactobacillus plantarum KLDS1.0391. Biomed. Res. Int. 2018:4506829. doi: 10.1155/2018/4506829
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kariminik, A., Baseri-Salehi, M., and Kheirkhah, B. (2017). Pseudomonas aeruginosa quorum sensing modulates immune responses: an updated review article. Immunol. Lett. 190, 1–6. doi: 10.1016/j.imlet.2017.07.002
Kleerebezem, M., Quadri, L. E., Kuipers, O. P., and De Vos, W. M. (1997). Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24, 895–904. doi: 10.1046/j.1365-2958.1997.4251782.x
Ko, J. S., Yang, H. R., Chang, J. Y., and Seo, J. K. (2007). Lactobacillus plantarum inhibits epithelial barrier dysfunction and interleukin-8 secretion induced by tumor necrosis factor-alpha. World J. Gastroenterol. 13, 1962–1965. doi: 10.3748/wjg.v13.i13.1962
Krueger, F. (2015). Trim Galore: A Wrapper Tool Around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files. Available at: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed January 12, 2017).
Latifi, A., Foglino, M., Tanaka, K., Williams, P., and Lazdunski, A. (1996). A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21, 1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x
Lee, K., Rho, B. S., Pi, K., Kim, H. J., and Choi, Y. J. (2011). Proteomic analysis of protein expression in Lactobacillus plantarum in response to alkaline stress. J. Biotechnol. 153, 1–7. doi: 10.1016/j.jbiotec.2011.02.008
Lex, A., Gehlenborg, N., Strobelt, H., Vuillemot, R., and Pfister, H. (2014). UpSet: visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph. 20, 1983–1992. doi: 10.1109/TVCG.2014.2346248
Li, L., Tetu, S. G., Paulsen, I. T., and Hassan, K. A. (2018). “A transcriptomic approach to identify novel drug efflux pumps in bacteria,” in Bacterial Multidrug Exporters, eds A. Yamaguchi and K. Nishino (New York, NY: Humana Press), 221–235.
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lubkowicz, D., Ho, C. L., Hwang, I. Y., Yew, W. S., Lee, Y. S., and Chang, M. W. (2018). Reprogramming probiotic Lactobacillus reuteri as a biosensor for Staphylococcus aureus derived AIP-I detection. ACS Synth. Biol. 7, 1229–1237. doi: 10.1021/acssynbio.8b00063
Magoc, T., Wood, D., and Salzberg, S. L. (2013). EDGE-pro: estimated degree of gene expression in prokaryotic genomes. Evol. Bioinform. Online 9, 127–136. doi: 10.4137/EBO.S11250
Maldonado, A., Jimenez-Diaz, R., and Ruiz-Barba, J. L. (2004a). Induction of plantaricin production in Lactobacillus plantarum NC8 after coculture with specific gram-positive bacteria is mediated by an autoinduction mechanism. J. Bacteriol. 186, 1556–1564.
Maldonado, A., Ruiz-Barba, J. L., and Jimenez-Diaz, R. (2004b). Production of plantaricin NC8 by Lactobacillus plantarum NC8 is induced in the presence of different types of gram-positive bacteria. Arch. Microbiol. 181, 8–16.
Maldonado, A., Ruiz-Barba, J. L., Floriano, B., and Jimenez-Diaz, R. (2002). The locus responsible for production of plantaricin S, a class IIb bacteriocin produced by Lactobacillus plantarum LPCO10, is widely distributed among wild-type Lact. plantarum strains isolated from olive fermentations. Int. J. Food Microbiol. 77, 117–124. doi: 10.1016/S0168-1605(02)00049-1
Maldonado, A., Ruiz-Barba, J. L., and Jimenez-Diaz, R. (2003). Purification and genetic characterization of plantaricin NC8, a novel coculture-inducible two-peptide bacteriocin from Lactobacillus plantarum NC8. Appl. Environ. Microbiol. 69, 383–389. doi: 10.1128/AEM.69.1.383-389.2003
Marchand, N., and Collins, C. H. (2013). Peptide-based communication system enables Escherichia coli to Bacillus megaterium interspecies signaling. Biotechnol. Bioeng. 110, 3003–3012. doi: 10.1002/bit.24975
Marchand, N., and Collins, C. H. (2016). Synthetic quorum sensing and cell-cell communication in gram-positive Bacillus megaterium. ACS Synth. Biol. 5, 597–606. doi: 10.1021/acssynbio.5b00099
Mathesius, U., Mulders, S., Gao, M., Teplitski, M., Caetano-Anolles, G., Rolfe, B. G., et al. (2003). Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc. Natl. Acad. Sci. U.S.A. 100, 1444–1449. doi: 10.1073/pnas.262672599
Matsumoto, T., Tateda, K., Miyazaki, S., Furuya, N., Ohno, A., Ishii, Y., et al. (1997). Immunomodulating effect of fosfomycin on gut-derived sepsis caused by Pseudomonas aeruginosa in mice. Antimicrob. Agents Chemother. 41, 308–313. doi: 10.1128/AAC.41.2.308
Medina, G., Juarez, K., Diaz, R., and Soberon-Chavez, G. (2003a). Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiology 149, 3073–3081. doi: 10.1099/mic.0.26282-0
Medina, G., Juarez, K., Valderrama, B., and Soberon-Chavez, G. (2003b). Mechanism of Pseudomonas aeruginosa RhlR transcriptional regulation of the rhlAB promoter. J. Bacteriol. 185, 5976–5983. doi: 10.1128/JB.185.20.5976-5983.2003
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Nesvizhskii, A. I., Keller, A., Kolker, E., and Aebersold, R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658. doi: 10.1021/ac0341261
Nguyen, Y. N., Sheng, H., Dakarapu, R., Falck, J. R., Hovde, C. J., and Sperandio, V. (2013). The acyl-homoserine lactone synthase YenI from Yersinia enterocolitica modulates virulence gene expression in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 81, 4192–4199. doi: 10.1128/IAI.00889-13
Papakyriacou, H., Vaz, D., Simor, A., Louie, M., and Mcgavin, M. J. (2000). Molecular analysis of the accessory gene regulator (agr) locus and balance of virulence factor expression in epidemic methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 181, 990–1000. doi: 10.1086/315342
Peng, H. L., Novick, R. P., Kreiswirth, B., Kornblum, J., and Schlievert, P. (1988). Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170, 4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988
Peral, M. C., Martinez, M. A., and Valdez, J. C. (2009). Bacteriotherapy with Lactobacillus plantarum in burns. Int. Wound J. 6, 73–81. doi: 10.1111/j.1742-481X.2008.00577.x
Peral, M. C., Rachid, M. M., Gobbato, N. M., Huaman Martinez, M. A., and Valdez, J. C. (2010). Interleukin-8 production by polymorphonuclear leukocytes from patients with chronic infected leg ulcers treated with Lactobacillus plantarum. Clin. Microbiol. Infect. 16, 281–286. doi: 10.1111/j.1469-0691.2009.02793.x
Pereira, C. S., Thompson, J. A., and Xavier, K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37, 156–181. doi: 10.1111/j.1574-6976.2012.00345.x
Perkins, D. N., Pappin, D. J., Creasy, D. M., and Cottrell, J. S. (1999). Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2
Pessi, G., and Haas, D. (2000). Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 182, 6940–6949. doi: 10.1128/JB.182.24.6940-6949.2000
Pieterse, B., Leer, R. J., Schuren, F. H., and Van Der Werf, M. J. (2005). Unravelling the multiple effects of lactic acid stress on Lactobacillus plantarum by transcription profiling. Microbiology 151, 3881–3894. doi: 10.1099/mic.0.28304-0
Procaccini, A., Lunt, B., Szurmant, H., Hwa, T., and Weigt, M. (2011). Dissecting the specificity of protein-protein interaction in bacterial two-component signaling: orphans and crosstalks. PLoS One 6:e19729. doi: 10.1371/journal.pone.0019729
Puertollano, E., Puertollano, M. A., Cruz-Chamorro, L., Alvarez De Cienfuegos, G., Ruiz-Bravo, A., and De Pablo, M. A. (2008). Orally administered Lactobacillus plantarum reduces pro-inflammatory interleukin secretion in sera from Listeria monocytogenes infected mice. Br. J. Nutr. 99, 819–825. doi: 10.1017/S0007114507832533
Ramos, A. N., Cabral, M. E., Noseda, D., Bosch, A., Yantorno, O. M., and Valdez, J. C. (2012). Antipathogenic properties of Lactobacillus plantarum on Pseudomonas aeruginosa: the potential use of its supernatants in the treatment of infected chronic wounds. Wound Repair Regen. 20, 552–562. doi: 10.1111/j.1524-475X.2012.00798.x
Ramos, A. N., Gobbato, N., Rachid, M., Gonzalez, L., Yantorno, O., and Valdez, J. C. (2010). Effect of Lactobacillus plantarum and Pseudomonas aeruginosa culture supernatants on polymorphonuclear damage and inflammatory response. Int. Immunopharmacol. 10, 247–251. doi: 10.1016/j.intimp.2009.11.007
Ramos, A. N., Sesto Cabral, M. E., Arena, M. E., Arrighi, C. F., Arroyo Aguilar, A. A., and Valdez, J. C. (2015). Compounds from Lactobacillus plantarum culture supernatants with potential pro-healing and anti-pathogenic properties in skin chronic wounds. Pharm. Biol. 53, 350–358. doi: 10.3109/13880209.2014.920037
Rampioni, G., Bertani, I., Zennaro, E., Polticelli, F., Venturi, V., and Leoni, L. (2006). The quorum-sensing negative regulator RsaL of Pseudomonas aeruginosa binds to the lasI promoter. J. Bacteriol. 188, 815–819. doi: 10.1128/JB.188.2.815-819.2006
Rampioni, G., Schuster, M., Greenberg, E. P., Bertani, I., Grasso, M., Venturi, V., et al. (2007). RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 66, 1557–1565. doi: 10.1111/j.1365-2958.2007.06029.x
Rampioni, G., Schuster, M., Greenberg, E. P., Zennaro, E., and Leoni, L. (2009). Contribution of the RsaL global regulator to Pseudomonas aeruginosa virulence and biofilm formation. FEMS Microbiol. Lett. 301, 210–217. doi: 10.1111/j.1574-6968.2009.01817.x
Reading, N. C., and Sperandio, V. (2006). Quorum sensing: the many languages of bacteria. FEMS Microbiol. Lett. 254, 1–11. doi: 10.1111/j.1574-6968.2005.00001.x
Rekhif, N., Atrih, A., and Lefebvre, G. (1995). Activity of plantaricin SA6, a bacteriocin produced by Lactobacillus plantarum SA6 isolated from fermented sausage. J. Appl. Bacteriol. 78, 349–358. doi: 10.1111/j.1365-2672.1995.tb03417.x
Reyes, D., Andrey, D. O., Monod, A., Kelley, W. L., Zhang, G., and Cheung, A. L. (2011). Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J. Bacteriol. 193, 6020–6031. doi: 10.1128/JB.05436-11
Ritchie, A. J., Jansson, A., Stallberg, J., Nilsson, P., Lysaght, P., and Cooley, M. A. (2005). The Pseudomonas aeruginosa quorum-sensing molecule N-3-(oxododecanoyl)-L-homoserine lactone inhibits T-cell differentiation and cytokine production by a mechanism involving an early step in T-cell activation. Infect. Immun. 73, 1648–1655. doi: 10.1128/IAI.73.3.1648-1655.2005
Ritchie, A. J., Whittall, C., Lazenby, J. J., Chhabra, S. R., Pritchard, D. I., and Cooley, M. A. (2007). The immunomodulatory Pseudomonas aeruginosa signalling molecule N-(3-oxododecanoyl)-L-homoserine lactone enters mammalian cells in an unregulated fashion. Immunol. Cell Biol. 85, 596–602. doi: 10.1038/sj.icb.7100090
Ritchie, A. J., Yam, A. O., Tanabe, K. M., Rice, S. A., and Cooley, M. A. (2003). Modification of in vivo and in vitro T- and B-cell-mediated immune responses by the Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)-L-homoserine lactone. Infect. Immun. 71, 4421–4431. doi: 10.1128/IAI.71.8.4421-4431.2003
Rumbaugh, K. P., Griswold, J. A., and Hamood, A. N. (1999a). Contribution of the regulatory gene lasR to the pathogenesis of Pseudomonas aeruginosa infection of burned mice. J. Burn. Care Rehabil. 20, 42–49.
Rumbaugh, K. P., Griswold, J. A., Iglewski, B. H., and Hamood, A. N. (1999b). Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect. Immun. 67, 5854–5862.
Schauder, S., Shokat, K., Surette, M. G., and Bassler, B. L. (2001). The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41, 463–476. doi: 10.1046/j.1365-2958.2001.02532.x
Schultz, M., Veltkamp, C., Dieleman, L. A., Grenther, W. B., Wyrick, P. B., Tonkonogy, S. L., et al. (2002). Lactobacillus plantarum 299V in the treatment and prevention of spontaneous colitis in interleukin-10-deficient mice. Inflamm. Bowel Dis. 8, 71–80. doi: 10.1097/00054725-200203000-00001
Searle, B. C. (2010). Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 10, 1265–1269. doi: 10.1002/pmic.200900437
Seed, P. C., Passador, L., and Iglewski, B. H. (1995). Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J. Bacteriol. 177, 654–659. doi: 10.1128/jb.177.3.654-659.1995
Seme, H., Gjuracic, K., Kos, B., Fujs, S., Stempelj, M., Petkovic, H., et al. (2015). Acid resistance and response to pH-induced stress in two Lactobacillus plantarum strains with probiotic potential. Benef. Microbes 6, 369–379. doi: 10.3920/BM2014.0069
Semmelhack, M. F., Campagna, S. R., Hwa, C., Federle, M. J., and Bassler, B. L. (2004). Boron binding with the quorum sensing signal AI-2 and analogues. Org. Lett. 6, 2635–2637. doi: 10.1021/ol048976u
Serrano, L. M., Molenaar, D., Wels, M., Teusink, B., Bron, P. A., De Vos, W. M., et al. (2007). Thioredoxin reductase is a key factor in the oxidative stress response of Lactobacillus plantarum WCFS1. Microb. Cell Fact 6:29. doi: 10.1186/1475-2859-6-29
Sharif, D. I., Gallon, J., Smith, C. J., and Dudley, E. (2008). Quorum sensing in Cyanobacteria: N-octanoyl-homoserine lactone release and response, by the epilithic colonial cyanobacterium Gloeothece PCC6909. ISME J. 2, 1171–1182. doi: 10.1038/ismej.2008.68
Smith, R. S., Harris, S. G., Phipps, R., and Iglewski, B. (2002a). The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J. Bacteriol. 184, 1132–1139.
Smith, R. S., Kelly, R., Iglewski, B. H., and Phipps, R. P. (2002b). The Pseudomonas autoinducer N-(3-oxododecanoyl) homoserine lactone induces cyclooxygenase-2 and prostaglandin E2 production in human lung fibroblasts: implications for inflammation. J. Immunol. 169, 2636–2642.
Storey, D. G., Ujack, E. E., Rabin, H. R., and Mitchell, I. (1998). Pseudomonas aeruginosa lasR transcription correlates with the transcription of lasA, lasB, and toxA in chronic lung infections associated with cystic fibrosis. Infect. Immun. 66, 2521–2528.
Sturme, M. H., Francke, C., Siezen, R. J., De Vos, W. M., and Kleerebezem, M. (2007). Making sense of quorum sensing in lactobacilli: a special focus on Lactobacillus plantarum WCFS1. Microbiology 153, 3939–3947. doi: 10.1099/mic.0.2007/012831-0
Sturme, M. H., Nakayama, J., Molenaar, D., Murakami, Y., Kunugi, R., Fujii, T., et al. (2005). An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J. Bacteriol. 187, 5224–5235. doi: 10.1128/JB.187.15.5224-5235.2005
Taga, M. E., Semmelhack, J. L., and Bassler, B. L. (2001). The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42, 777–793. doi: 10.1046/j.1365-2958.2001.02669.x
Teplitski, M., Mathesius, U., and Rumbaugh, K. P. (2011). Perception and degradation of N-acyl homoserine lactone quorum sensing signals by mammalian and plant cells. Chem. Rev. 111, 100–116. doi: 10.1021/cr100045m
Tong, Y., Zhai, Q., Lu, W., Tian, F., Zhao, J., Zhang, H., et al. (2017). New insights in integrated response mechanism of Lactobacillus plantarum under excessive manganese stress. Food Res. Int. 102, 323–332. doi: 10.1016/j.foodres.2017.10.014
Valdez, J. C., Peral, M. C., Rachid, M., Santana, M., and Perdigon, G. (2005). Interference of Lactobacillus plantarum with Pseudomonas aeruginosa in vitro and in infected burns: the potential use of probiotics in wound treatment. Clin. Microbiol. Infect. 11, 472–479. doi: 10.1111/j.1469-0691.2005.01142.x
Van Bokhorst-van de Veen, H., Bongers, R. S., Wels, M., Bron, P. A., and Kleerebezem, M. (2013). Transcriptome signatures of class I and III stress response deregulation in Lactobacillus plantarum reveal pleiotropic adaptation. Microb. Cell Fact 12:112. doi: 10.1186/1475-2859-12-112
Vendeville, A., Winzer, K., Heurlier, K., Tang, C. M., and Hardie, K. R. (2005). Making ’sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3, 383–396. doi: 10.1038/nrmicro1146
Wagner, C., Zimmermann, S., Brenner-Weiss, G., Hug, F., Prior, B., Obst, U., et al. (2007). The quorum-sensing molecule N-3-oxododecanoyl homoserine lactone (3OC12-HSL) enhances the host defence by activating human polymorphonuclear neutrophils (PMN). Anal. Bioanal. Chem. 387, 481–487. doi: 10.1007/s00216-006-0698-5
Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Waters, C. M., and Bassler, B. L. (2006). The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 20, 2754–2767. doi: 10.1101/gad.1466506
Wen, K. Y., Cameron, L., Chappell, J., Jensen, K., Bell, D. J., Kelwick, R., et al. (2017). A Cell-Free Biosensor for Detecting Quorum Sensing Molecules in P. aeruginosa-Infected Respiratory Samples. ACS Synth. Biol. 6, 2293–2301. doi: 10.1021/acssynbio.7b00219
Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis. Berlin: Springer. doi: 10.1007/978-3-319-24277-4
Winzer, K., Falconer, C., Garber, N. C., Diggle, S. P., Camara, M., and Williams, P. (2000). The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J. Bacteriol. 182, 6401–6411. doi: 10.1128/JB.182.22.6401-6411.2000
Winzer, K., and Williams, P. (2001). Quorum sensing and the regulation of virulence gene expression in pathogenic bacteria. Int. J. Med. Microbiol. 291, 131–143. doi: 10.1078/1438-4221-00110
Xia, Y., Chen, H. Q., Zhang, M., Jiang, Y. Q., Hang, X. M., and Qin, H. L. (2011). Effect of Lactobacillus plantarum LP-Onlly on gut flora and colitis in interleukin-10 knockout mice. J. Gastroenterol. Hepatol. 26, 405–411. doi: 10.1111/j.1440-1746.2010.06498.x
Yates, E. A., Philipp, B., Buckley, C., Atkinson, S., Chhabra, S. R., Sockett, R. E., et al. (2002). N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 70, 5635–5646. doi: 10.1128/IAI.70.10.5635-5646.2002
Zhang, G., Zhang, F., Ding, G., Li, J., Guo, X., Zhu, J., et al. (2012). Acyl homoserine lactone-based quorum sensing in a methanogenic archaeon. ISME J. 6, 1336–1344. doi: 10.1038/ismej.2011.203
Keywords: Lactobacillus plantarum, quorum sense, Pseudomonas aeruginosa, transcriptomics, proteomics, homoserine lactone
Citation: Spangler JR, Dean SN, Leary DH and Walper SA (2019) Response of Lactobacillus plantarum WCFS1 to the Gram-Negative Pathogen-Associated Quorum Sensing Molecule N-3-Oxododecanoyl Homoserine Lactone. Front. Microbiol. 10:715. doi: 10.3389/fmicb.2019.00715
Received: 16 January 2019; Accepted: 21 March 2019;
Published: 05 April 2019.
Edited by:
Konstantinos Papadimitriou, Agricultural University of Athens, GreeceReviewed by:
Mansel William Griffiths, University of Guelph, CanadaSang Jun Lee, Chung-Ang University, South Korea
Copyright © 2019 Spangler, Dean, Leary and Walper. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Scott A. Walper, c2NvdHQud2FscGVyQG5ybC5uYXZ5Lm1pbA==
 Joseph R. Spangler
Joseph R. Spangler Scott N. Dean
Scott N. Dean Dagmar H. Leary
Dagmar H. Leary Scott A. Walper
Scott A. Walper