- 1School of Food Science and Engineering, Hefei University of Technology, Hefei, China
- 2State Key Laboratory of Applied Microbiology Southern China, Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, Guangdong Institute of Microbiology, Guangzhou, China
Cronobacter species are linked with life-treating diseases in neonates and show strong tolerances to environmental stress. However, the information about factors involved in oxidative tolerance in Cronobacter remains elusive. Here, factors involved in oxidative tolerance in C. malonaticus were identified using a transposon mutagenesis. Eight mutants were successfully screened based on a comparison of the growth of strains from mutant library (n = 215) and wild type (WT) strain under 1.0 mM H2O2. Mutating sites including thioredoxin 2, glutaredoxin 3, pantothenate kinase, serine/threonine protein kinase, pyruvate kinase, phospholipase A, ferrous iron transport protein A, and alanine racemase 2 were successfully identified by arbitrary PCR and sequencing alignment. Furthermore, the comparison about quantity and structure of biofilms formation among eight mutants and WT was determined using crystal violet staining (CVS), scanning electron microscopy (SEM), and confocal laser scanning microscopy (CLSM). Results showed that the biofilms of eight mutants significantly decreased within 48 h compared to that of WT, suggesting that mutating genes play important roles in biofilm formation under oxidative stress. The findings provide valuable information for deeply understanding molecular mechanism about oxidative tolerance of C. malonaticus.
Introduction
Cronobacter species are important foodborne pathogens causing life-threating infections in infants (Van Acker et al., 2001; Healy et al., 2010). Contaminated powdered infant formula (PIF) is considered to be the major transmission route of Cronobacter infections (Biering et al., 1989; Van Acker et al., 2001; Norberg et al., 2012; Ye et al., 2014). So, the high risks of Cronobacter strains in powdered infant formula on newborn’s health has arouse public concerns.
Cronobacter spp. show unusual abilities to survive under environmental stress (Gurtler et al., 2005). To date, the genus of Cronobacter includes C. sakazakii, C. malonaticus, C. turicensis, C. muytjensii, C. dublinensis, C. universalis, and C. condiment (Iversen et al., 2008). The factors involved in oxidative stress in C. sakazakii have been reported. For example, polymorphisms in RpoS sequence and Significant heterogeneity of stress tolerance including oxidative stress among natural isolates of C. sakazakii has been described (Alvarez-Ordóñez et al., 2012). Johler et al. (2010) demonstrated that genes including crtX, crtE, and crtY involved in yellow pigmenting of C. sakazakii ES5 affected tolerance to oxidative stress. In C. sakazkaii ATCC29544, Hfq, an RNA chaperone, has been found to increase the tolerance to oxidative stress (Kim et al., 2015). C. malonaticus has been implicated in infections in infant and adults (Forsythe et al., 2014; Alsonosi et al., 2015). PIF is the major source of C. malonaticus (Ogrodzki and Forsythe, 2015, 2017). Hydrogen peroxide (H2O2) is a well-studied sanitizer for inactivate foodborne pathogens. In addition, Ye et al. (2018) determined the inhibitory effects of H2O2 on C. malonaticus cells and its biofilm formation. However, information about factors involved in oxidative tolerance in C. malonaticus is largely unknown.
In this study, a transposon mutagenesis approach was applied to reveal the factors involved in resistance to oxidative stress, and the biofilm formation among mutants and WT strains were further detected using crystal violet staining (CVS), scanning electron microscopy (SEM), and confocal laser scanning microscopy (CLSM) to reveal potential relationship between oxidative stress and biofilm formation.
Materials and Methods
The Development of Mutants Library
The procedure of transposon mutagenesis approach was performed as described by Zhang et al. (2018).
Screening of Mutants Tolerance to Oxidative Stress
For screening positive mutants tolerant to oxidative stress, overnight culture (OD600 = 0.8, v/v, 1%) was inoculated into LB broth (Luqiao, Beijing) with 1.0 mM H2O2 at 37°C for 8 h. Growth of mutants (n = 215) and WT strain were measured spectrophotometrically in 96-well culture plates (Corning, New York, NY, United States) by determining the optical density at 600 nm (OD600). Each experiment was independently done in triplicate. Growth of strains were analyzed by the statistical analysis of t-tests using OriginPro 8.5.1 software. A significant difference was defined as a p-value (p < 0.05) between wild-type (WT) and mutants.
Identification of Mutating Sites
The detailed procedure for identification of mutating sites and analysis of inserting sites was performed as described by Zhang et al. (2018). In brief, the mutating genes were amplified by arbitrary PCR, then the fragments were purified for being sequenced and aligned.
Comparison of Biofilm Formation Among Mutants and Wild Type
Under oxidative stress (LB with 1.0 mM H2O2), biofilm formation using CVS was determined ranging from 24 to 72 h described previously by Zhang et al. (2018). In addition, the biofilms on the cell slips at 48 h was detected using SEM (Hitachi, Tokyo, Japan) and CLSM (Zeiss, Berlin, Germany) using LIVE/DEAD BacLight bacterial viability Kit according to instructions (Invitrogen, Carlsbad, CA, United States).
Results and Discussion
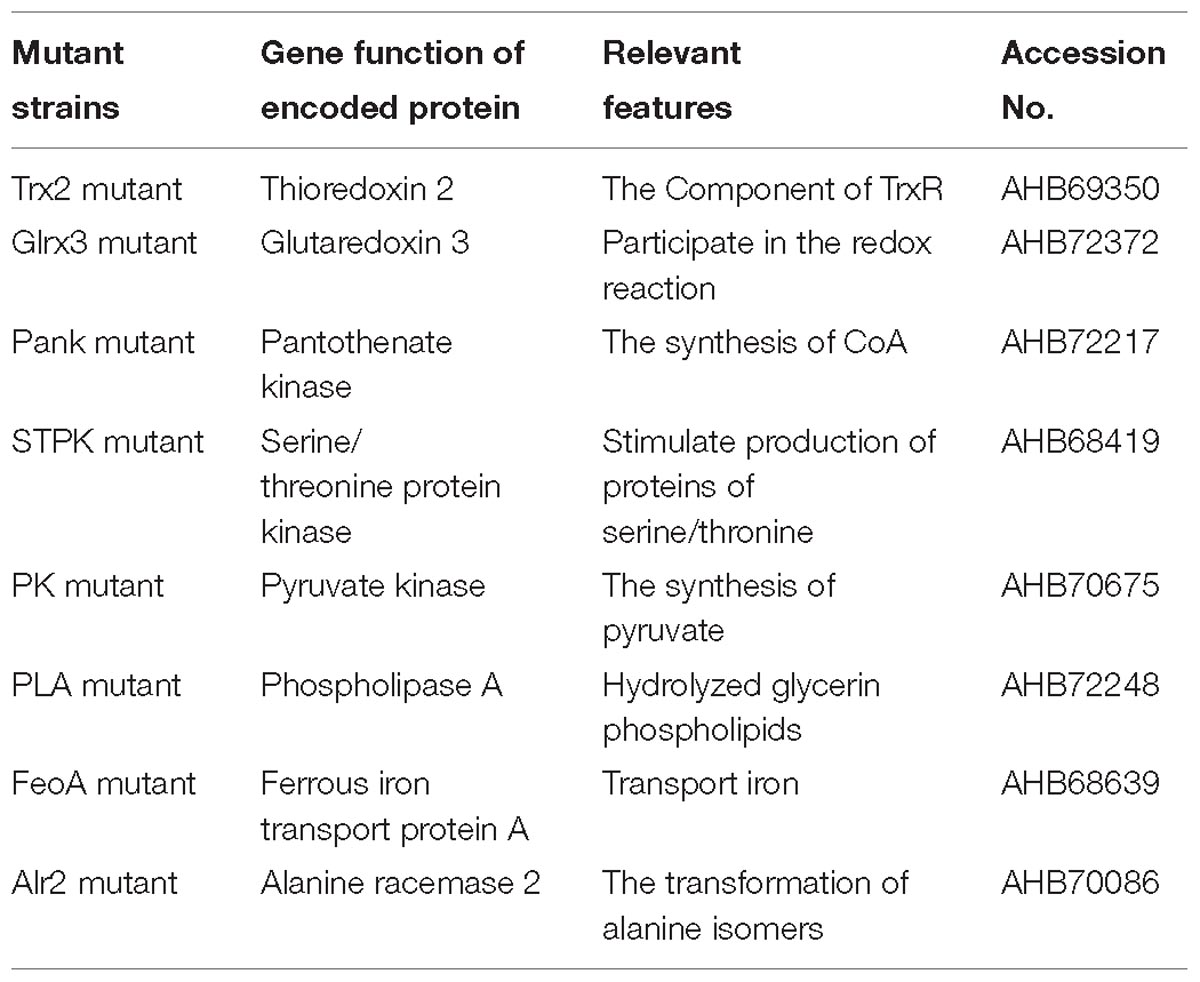
Based on the growth of mutants and WT strain under oxidative stress (1.0 mM H2O2), eight mutants were successfully screened, and the growth of eight mutants under oxidative stress was significantly (p < 0.05) decreased compared with that of WT shown in Figure 1. The mutating genes listed in Table 1 encode thioredoxin 2 (Trx2), glutaredoxin 3 (Grx3), pantothenate kinase (Pank), serine/threonine protein kinase (STPK), pyruvate kinase (PK), phospholipase A (PLA), ferrous iron transport protein A (FeoA), and alanine racemase 2 (Alr2) which contributed to tolerance to oxidative stress in C. malonaticus.
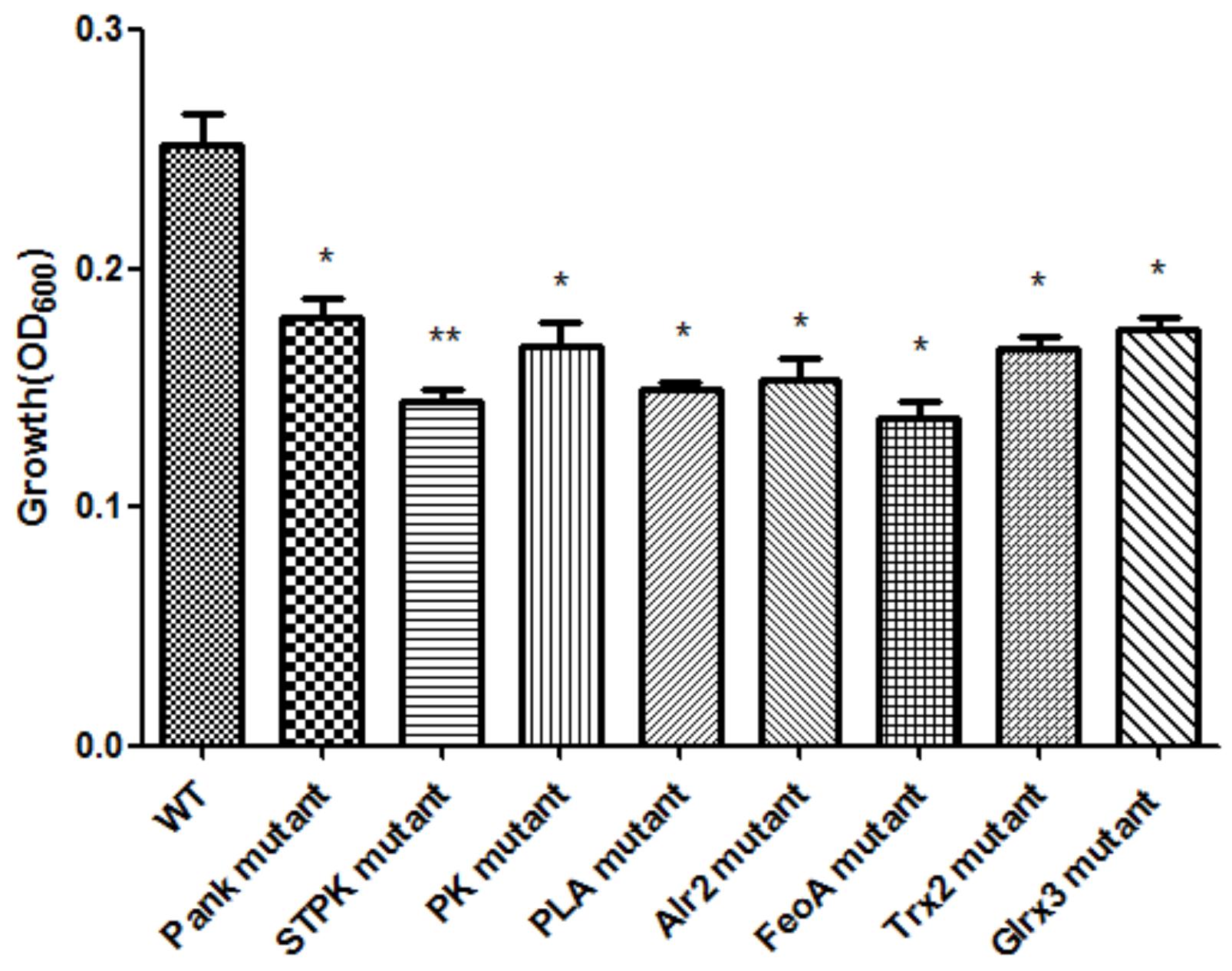
Figure 1. Growth of C. malonaticus wild strain (WT) and mutants under 1.0 mM H2O2 oxidative stress. Bar = mean ± SD. ∗Significant difference about growth under 1.0 mM H202 with WT strain (p < 0.05), ∗∗significant difference about growth under 1.0 mM H202 with WT strain (p < 0.01).
In Escherichia coli, thioredoxin 2 (encoded by trxC) was identified on the basis of sequence similarity (Miranda-Vizuete et al., 1997), but trxC mutants do not show altered sensitivity to H2O2 (Ritz et al., 2000). In addition, inactivity of thioredoxin 1 (encoded by trxA) and thioredoxin reductase (encoded by trxB) caused more sensitive to H2O2 in stationary phase of E. coli (Takemoto et al., 1998). Glutaredoxin (Grx) is a thiol-disulfide oxidoreductase widely distributed from bacteria to higher eukaryotes (Rouhier et al., 2008). In yeast, mutants lacking Grx are sensitive to oxidative stress (Luikenhuis et al., 1998). The OxyR and SoxR in E. coli, and the S. cerevisiae Yap1p transcriptional regulators were modulated by glutathione- and thioredoxin-dependent reduction systems for the adaptive responses to oxidative stress (Carmel-Harel and Storz, 2000). The inactivity of glutaredoxin 2 and glutaredoxin 3 encoded by grxB and grxC, respectively, were found in E. coli strains lacking glutaredoxin 1 and thioredoxin 1 still showed GSH oxidoreductase activity (Aslund et al., 1994). The inactivity of glutaredoxin 2 in E. coli cells were more sensitive to hydrogen peroxide and other oxidants, and the interconnection between catalases and thioredoxin/glutaredoxin pathways in the antioxidant response was observed (Vlamis-Gardikas et al., 2002). Regulators including OxyR, SoxRS, and RpoS in E. coli were associated with the tolerance to oxidative stress (Chiang and Schellhorn, 2012). The redox proteins such as Grx A (Grx1) required for maintaining redox status in bacteria also protect bacteria from oxidative stress (Caldas et al., 2006; Meyer et al., 2009). The pantothenate kinase is required for the biosynthesis of coenzyme A (CoA). In Bacillus anthracis, the type III pantothenate kinase plays important roles in maintenance of cytosolic redox balance and in adaptation to the oxidative stress in B. anthracis (Paige et al., 2008).
Ferrous iron (Fe2+) is one of the essential elements required for growth and virulence of the majority of pathogens (Hayrapetyan et al., 2016). Here, ferrous iron transport contributed to oxidative tolerance in C. malonaticus through the reduction reaction of Fe2+ to attenuate the injuries from oxidation (H2O2). The ferrous iron transport (feo) operon was first discovered in E. coli K12 in 1987 through studies of a series of ferrous iron transport mutants, and the deletion of feo strains cause the failure to taking up ferrous iron (Hantke, 1987). In addition, in the absence of FeoB, H. pylori was unable to colonize the gastric mucosa of mice (Velayudhan et al., 2000). Naikare et al. (2006) found that FeoB is essential for the uptake of ferrous iron, gut colonization and intracellular survival. On the Contrary, feo deletions in V. cholerae do not seem to affect its colonization in the mouse model (Wyckoff et al., 2006).
Through 2-D method combined with MALDI-TOF-MS and database queries, pyruvate kinase was involved in enhancement of oxidative stress in Pichia caribbica (Zhang et al., 2017). In the mitochondrial, pyruvate kinase M2 isoform (PKM2) regulates oxidative stress-induced apoptosis by stabilizing B-cell lymphoma 2 (Bcl2) (Liang et al., 2017). Brien et al. reported that increased placental phospholipase A2 gene expression was implicated in oxidative stress in preeclampsia (Brien et al., 2017). Expression of serine/threonine protein kinase and peroxisomal catalase in P. caribbica were involved in the enhancement of oxidative stress tolerance and biocontrol efficacy of P. caribbica (Zhang et al., 2017). Serine/Threonine kinases activation was induced by oxidative stress in frontotemporal dementia (Palluzzi et al., 2017). S. mutans expresses a eukaryotic serine/threonine type kinase known as STPK which enhances resistance to oxidative stress (Zhu and Kreth, 2010). Likewise, our results also found that inactivity of pantothenate kinase (Pank), serine/threonine protein kinase (STPK), pyruvate kinase (PK) caused sensitive to oxidative stress. To date, roles of Phospholipases (PLs) on tolerance to oxidative stress are not reported in other foodborne pathogens except for C. malonaticus.
Based on analysis of biofilms using CVS, the strong biofilm-formatting abilities among eight mutants and WT were observed, and biofilms of eight mutants significantly decreased at 48 h compared with that of wild type (WT) shown in Figure 2. Furthermore, the detection of spatial structure of biofilms was confirmed using SEM (Figure 3), and the mature biofilms were formed at 48 h among mutants and WT. From Figure 4, the viable cells and exopolysaccharides (blue) were more predominant at 48 h. Here, inactivity of eight factors caused weak biofilms compared with that of WT under oxidative stress, and a positive relationship between biofilm formation and oxidative tolerance was observed. Hartmann et al. (2010) demonstrated that cellulose and flagella facilitated biofilm formation in C. sakazakii. Using comparative proteomics analysis, genes including LuxS and TolB were found to contribute to biofilm formation in Cronobacter strains (Ye et al., 2016). In addition, the deoB, adh, and nlpD were involved in biofilm formation in C. sakazakii (Du et al., 2012). In addition, environmental conditions such as temperature and pH also greatly affected biofilm formation in C. sakazakii strains (Jung et al., 2013; Ye et al., 2015). In Haemophilus influenzae, expression abundance of peroxiredoxin–glutaredoxin increased in biofilms compared to planktonic cells (Gallaher et al., 2006). Similarly, thioredoxin, peroxidase, and thioredoxin were upregulated in biofilms in Candida albicans (Seneviratne et al., 2008). The biofilm formation in trxB mutant of Neisseria gonorrhoeae on human cervical epithelial cells was greatly reduced compared with wild-type strain (Potter et al., 2009). In S. typhimurium, the Feo system has been found to play important roles in colonization of the mouse intestine (Tsolis et al., 1996). Jiang et al. (2015) found that hydrolase and pantothenate kinase were detected in the Streptococcus mutans 593 biofilm only, indicating that pantothenate kinase was involved in the biofilm formation in S. mutans 593. The high pyruvate kinase activity in S. mutans contributed to the cariogenic biofilm formation in caries patents (Krzyściak et al., 2017). Pyruvate kinase activity in Staphylococcus aureus was regulated by serine/threonine protein kinase, which favors biofilm formation (Vasu et al., 2015). Serine/Threonine kinases (STPKs) have been implicated in biofilm formation of Bacillus subtilis (Madec et al., 2002). Ser/Thr protein kinase PrkC mediates biofilm formation in B. anthracis by regulation of GroEL activity (Arora et al., 2017). Phospholipases (PLs) are considered important factors for C. parapsilosis adherence, tissue penetration, and host invasion (Junior et al., 2011). Meanwhile, the germination, adherence, biofilm formation, phospholipase and proteinase production were considered the virulence factors in Candida albicans (Larkin et al., 2017).
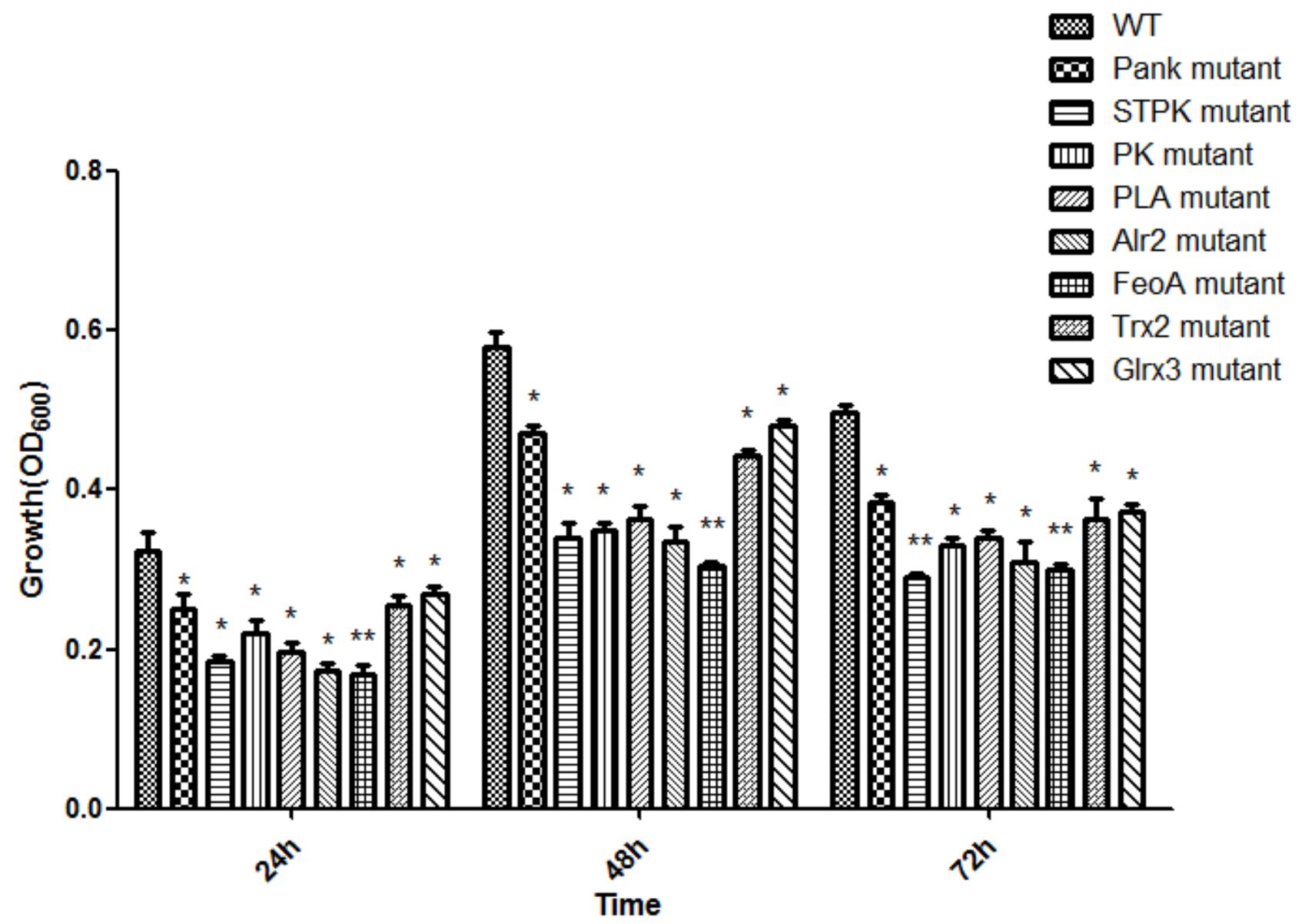
Figure 2. Biofilm formation of C. malonaticus wild strain (WT) and mutants under 1.0 mM H202 oxidative stress. ∗Significant difference about growth under 1.0 mM H202 with WT strain (p < 0.05), ∗∗significant difference about growth under 1.0 mM H202 with WT strain (p < 0.01).
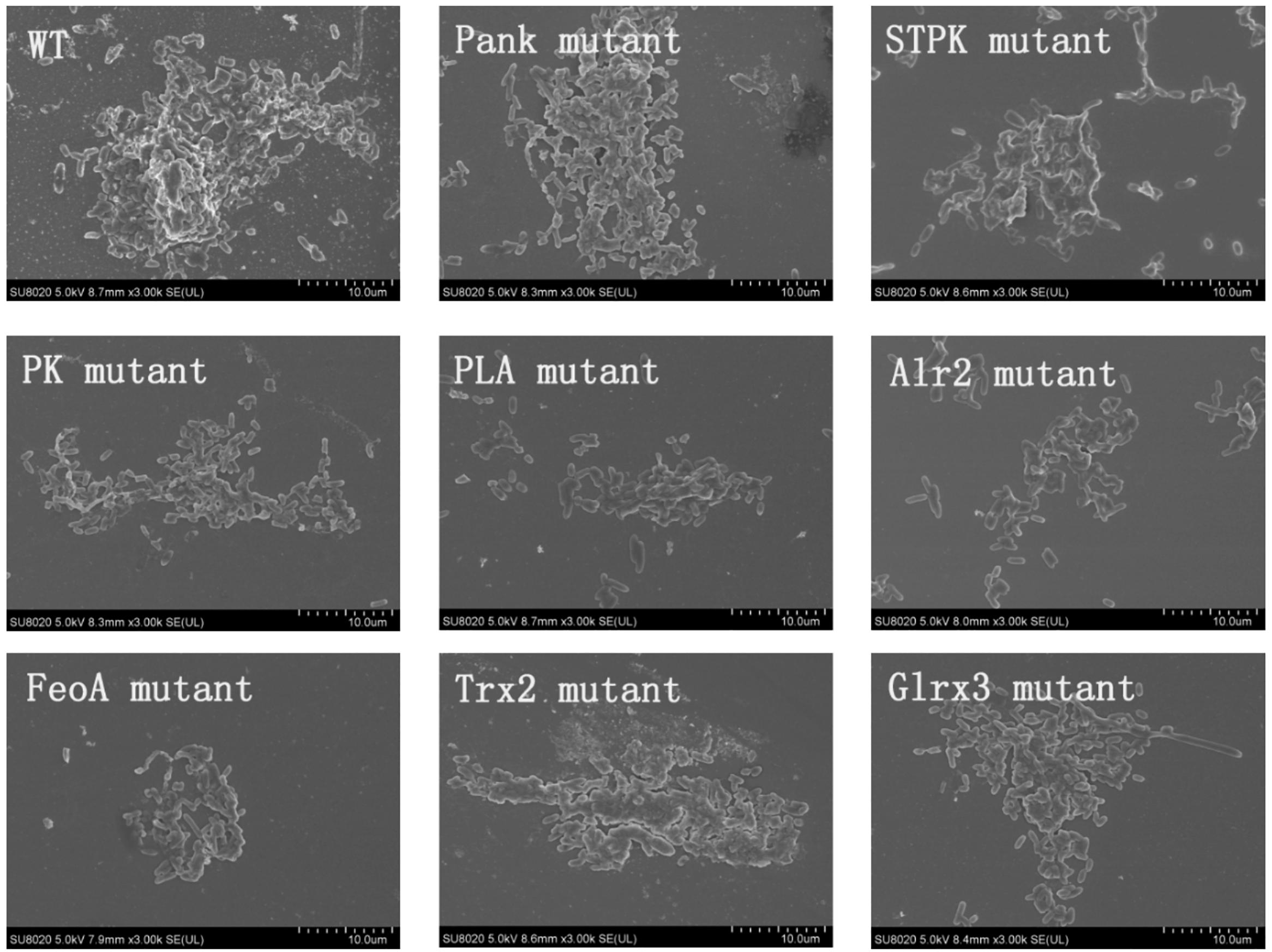
Figure 3. Biofilm formation of C. malonaticus wild strain (WT) and mutants at 48 h under 1.0 mM H202 using SEM.
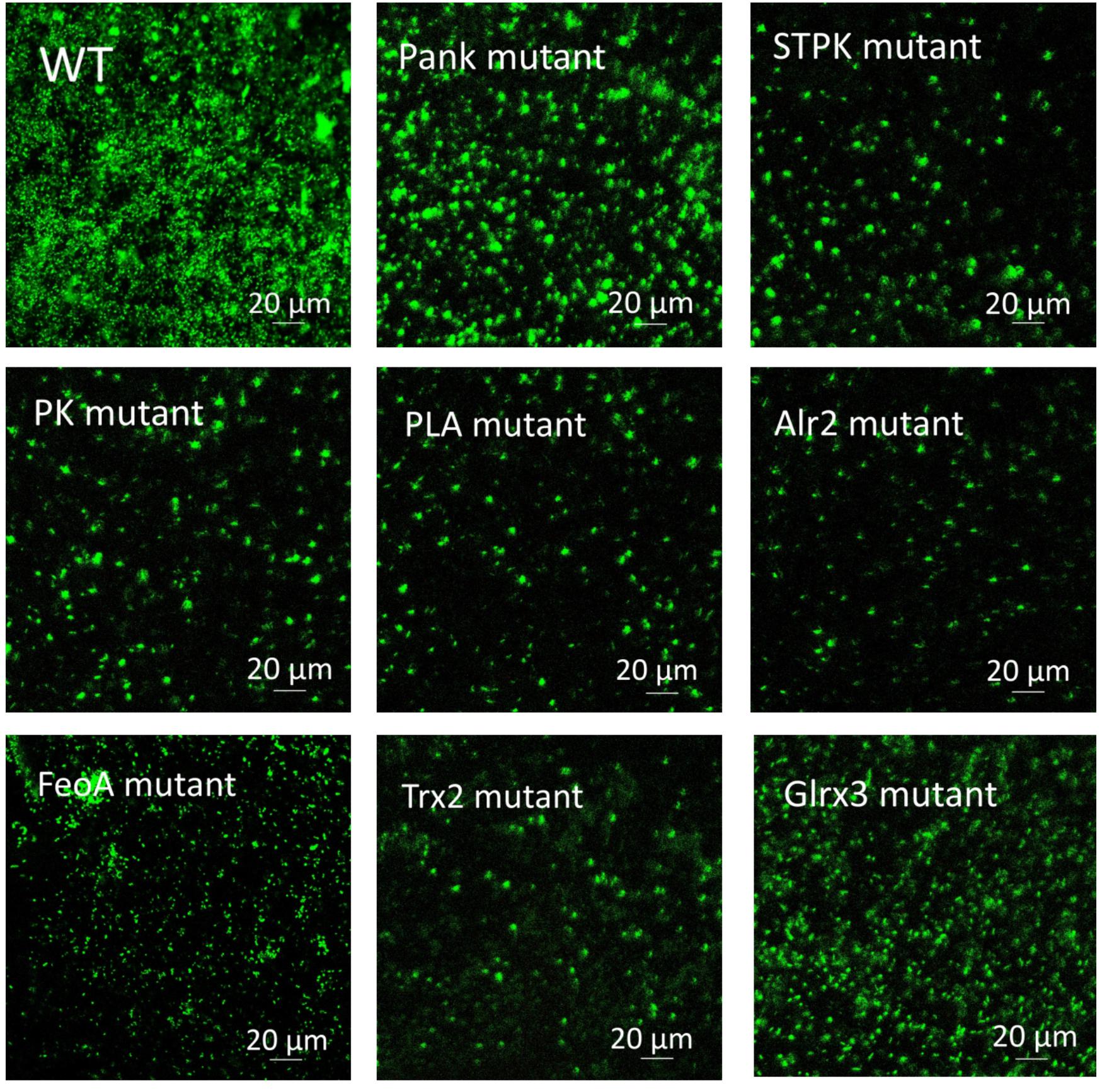
Figure 4. Biofilm formation of C. malonaticus wild strain (WT) and mutants at 48 h under 1.0 mM H202 using CLSM.
Conclusion
In summary, the factors involved in tolerance to oxidative stress in C. malonaticus were identified including Trx2, Grx3, Pank, STPK, PK, PLA, FeoA, and Alr2. A positive relationship between biofilm-forming ability and oxidative tolerance was also observed, which might indicated that biofilm formation was related with environmental stress. The findings here provide valuable information for deeply understanding molecular mechanism about tolerance to oxidative stress.
Author Contributions
XZ carried out the experiments and analyzed the data. MZ carried out the experiments and analyzed the data. LT analyzed the data and carried out the experiments. DO carried out the partial experiments. YW analyzed the data. JZ modified the manuscript. QW and YY designed and modified the manuscript.
Funding
The financial support of the National Key Research and Development program (2017YFC1601200 and 2017YFC1601202), National Natural Science Foundation of China (31671951), the Anhui provincial Grand Project special of Science and Technology (15czz03109), the Science and Technology Planning Project of Guangdong Province (2016A050502033), and Project of Science and Technology in Guangzhou (201604020036).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge Prof. Xu, M.Y (Guangdong Institute of Microbiology) for presenting the E. coli WM3064.
References
Alsonosi, A., Hariri, S., Kajsik, M., Orieskova, M., Hanulik, V., Roderova, M., et al. (2015). The speciation and genotyping of Cronobacter isolates from hospitalized patients. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1979–1988. doi: 10.1007/s10096-015-2440-8
Alvarez-Ordóñez, A., Begley, M., and Hill, C. (2012). Polymorphisms in rpoS and stress tolerance heterogeneity in natural isolates of Cronobacter sakazakii. Appl. Environ. Microbiol. 78, 3975–3984. doi: 10.1128/AEM.07835-11
Arora, G., Sajid, A., Virmani, R., Singhal, A., Cms, K., Dhasmana, N., et al. (2017). Ser/thr protein kinase PrkC-mediated regulation of GroEL is critical for biofilm formation in Bacillus anthracis. NPJ Biofilms Microb. 3:7. doi: 10.1038/s41522-017-0015-4
Aslund, F., Ehn, B., Miranda-Vizuete, A., Pueyo, C., and Holmgren, A. (1994). Two additional glutaredoxins exist in Escherichia coli: glutaredoxin 3 is a hydrogen donor for ribonucleotide reductase in a thioredoxin/ glutaredoxin 1 double mutant. Proc. Natl. Acad. Sci. U.S.A. 91, 9813–9817. doi: 10.1073/pnas.91.21.9813
Biering, G., Karlsson, S., Clark, N. C., Jónsdóttir, K. E., Lúdvígsson, P., and Steingrímsson, O. (1989). Three cases of neonatal meningitis caused by Enterobacter sakazakii in powdered milk. J. Clin. Microbiol. 27, 2054–2056.
Brien, M., Larose, J., Greffard, K., Julien, P., and Bilodeau, J. F. (2017). Increased placental phospholipase A2 gene expression and free F2-isoprostane levels in response to oxidative stress in preeclampsia. Placenta 55, 54–62. doi: 10.1016/j.placenta.2017.05.004
Caldas, T., Malki, A., Kern, R., Abdallah, J., and Richarme, G. (2006). The Escherichia coli thioredoxin homolog ybbn/trxsc is a chaperone and a weak protein oxidoreductase. Biochem. Biophys. Res. Commun. 343, 780–786. doi: 10.1016/j.bbrc.2006.03.028
Carmel-Harel, O., and Storz, G. (2000). Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54, 439–461. doi: 10.1146/annurev.micro.54.1.439
Chiang, S. M., and Schellhorn, H. E. (2012). Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch. Biochem. Biophys. 525, 161–169. doi: 10.1002/9781119004813.ch60
Du, X.-J., Wang, F., Lu, X. N., Rasco, B. A., and Wang, S. (2012). Biochemical and genetic characteristics of Cronobacter sakazakii biofilm formation. Res. Microbiol. 163, 448–456. doi: 10.1016/j.resmic.2012.06.002
Forsythe, S. J., Dickins, B., and Jolley, K. A. (2014). Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis. BMC Genomics 15:1121. doi: 10.1186/1471-2164-15-1121
Gallaher, T. K., Wu, S., Webster, P., and Aguilera, R. (2006). Identification of biofilm proteins in non-typeable Haemophilus Influenzae. BMC Microbiol. 6:65. doi: 10.1186/1471-2180-6-65
Gurtler, J. B., Kornacki, J. L., and Beuchat, L. R. (2005). Enterobacter sakazakii: a coliform of increased concern to infant health. Int. J. Food Microbiol. 104, 1–34. doi: 10.1016/j.ijfoodmicro.2005.02.013
Hantke, K. (1987). Ferrous iron transport mutants in Escherichia coli K12. FEMS Microbiol. Lett. 44, 53–57. doi: 10.1111/j.1574-6968.1987.tb02241.x
Hartmann, I., Carranza, P., Lehner, A., Stephan, R., Eberl, L., and Riedel, K. (2010). Genes involved in Cronobacter sakazakii biofilm formation. Appl. Environ. Microbiol. 76, 2251–2261. doi: 10.1128/AEM.00930-09
Hayrapetyan, H., Siezen, R., Abee, T., and Nierop, G. M. (2016). Comparative genomics of iron-transporting systems in Bacillus cereus strains and impact of iron sources on growth and biofilm formation. Front. Microbiol. 7:842. doi: 10.3389/fmicb.2016.00842
Healy, B., Cooney, S., Brien, S. O., Iversen, C., Whyte, P., Nally, J., et al. (2010). Cronobacter (Enterobacter sakazakii): an opportunistic foodborne pathogen. Foodborne Pathog. Dis. 7, 339–350. doi: 10.1089/fpd.2009.0379
Iversen, C., Mullane, N. B., Tall, B. D., Lehner, A., Fanning, S., Stephan, R., et al. (2008). Cronobacter gen. nov. a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov. comb. nov. Cronobacter malonaticus sp. nov. Cronobacter turicensis sp. nov. Cronobacter muytjensii sp. nov. Int. J. Syst. Evol. Microbiol. 58, 1442–1447. doi: 10.1099/ijs.0.65577-0
Jiang, S., Huang, X., Zhang, C., Cai, Z., and Zou, T. (2015). Morphological and proteomic analyses of the biofilms generated by Streptococcus mutans isolated from caries-active and caries-free adults. J. Dent. Sci. 10, 206–215. doi: 10.1016/j.jds.2014.09.001
Johler, S., Stephan, R., Hartmann, I., Kuehner, K. A., and Lehner, A. (2010). Genes involved in yellow pigmentation of Cronobacter sakazakii ES5 and influence of pigmentation on persistence and growth under environmental stress. Appl. Environ. Microbiol. 76, 1053–1061. doi: 10.1128/AEM.01420-0
Jung, J. H., Choi, N. Y., and Lee, S. Y. (2013). Biofilm formation and exopolysaccharide (EPS) production by Cronobacter sakazakii depending on environmental conditions. Food Microbiol. 34, 70–80. doi: 10.1016/j.fm.2012.11.008
Junior, A. D. E., Silva, A. F., Rosa, F. C., Monteiro, S. G., Figueiredo, P. M. S., and Monteiro, C. A. (2011). In vitro differential activity of phospholipases and acid proteinases of clinical isolates of Candida. Rev. Soc. Bras. Med. Trop. 44, 334–338. doi: 10.1590/S0037-86822011005000036
Kim, S., Hwang, H., Kim, K.-P., Yoon, H., Kang, D.-H., and Ryu, S. (2015). hfq plays important roles in virulence and stress adaptation in Cronobacter sakazakii ATCC 29544. Infect. Immun. 83, 2089–2098. doi: 10.1128/IAI.03161-14
Krzyściak, W., Papież, M., Jurczak, A., Kościelniak, D., Vyhouskaya, P., Zagórska-Swieży, K., et al. (2017). Relationship between pyruvate kinase activity and cariogenic biofilm formation in Streptococcus mutans biotypes in caries patients. Front. Microbiol. 8:856. doi: 10.3389/fmicb.2017.00856
Larkin, E., Hager, C., Chandra, J., Mukherjee, P. K., Retuerto, M., Salem, I., et al. (2017). The emerging pathogen Candida auris: growth phenotype, virulence factors, activity of antifungals, and effect of scy-078, a novel glucan synthesis inhibitor, on growth morphology and biofilm formation. Antimicrob. Agents Chem. 61:e2396-16. doi: 10.1128/AAC.02396-16
Liang, J., Cao, R., Wang, X., Zhang, Y., Wang, P., Gao, H., et al. (2017). Mitochondrial Pkm2 regulates oxidative stress-induced apoptosis by stabilizing bcl2. Cell Res. 27, 329–351. doi: 10.1038/cr.2016.159
Luikenhuis, S., Perrone, G., Dawes, I. W., and Grant, C. M. (1998). The yeast Saccharomyces cerevisiae contains two glutaredoxin genes that are required for protection against reactive oxygen species. Mol. Biol. Cell 9, 1081–1091. doi: 10.1091/mbc.9.5.1081
Madec, E., Laszkiewicz, A., Iwanicki, A., Obuchowski, M., and Séror, S. (2002). Characterization of a membrane-linked ser/thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol. Microbiol. 46, 571–586. doi: 10.1046/j.1365-2958.2002.03178.x
Meyer, Y., Buchanan, B. B., Vignols, F., and Reichheld, J. P. (2009). Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu. Rev. Genet. 43, 335–367. doi: 10.1146/annurev-genet-102108-134201
Miranda-Vizuete, A., Damdimopoulos, A. E., Gustafsson, J.-A., and Spyrou, G. (1997). Cloning, expression, and characterization of a novel Escherichia coli thioredoxin. J. Biol. Chem. 272, 30841–30847. doi: 10.1074/jbc.272.49.30841
Naikare, H., Palyada, K., Panciera, R., Marlow, D., and Stintzi, A. (2006). Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 74, 5433–5444. doi: 10.1128/iai.00052-06
Norberg, S., Stanton, C., Ross, R. P., Hill, C., Fitzgerald, G. F., and Cotter, P. D. (2012). Cronobacter spp. in powdered infant formula. J. Food Prot. 75, 607–620. doi: 10.4315/0362-028X.JFP-11-285
Ogrodzki, P., and Forsythe, S. (2015). Capsular profiling of the Cronobacter genus and the association of specific Cronobacter sakazakii and C. malonaticus capsule types with neonatal meningitis and necrotizing enterocolitis. BMC Genomics 16, 1–15. doi: 10.1186/s12864-015-1960-z
Ogrodzki, P., and Forsythe, S. J. (2017). DNA-sequence based typing of the Cronobacter genus using MLST, CRISPR-cas array and capsular profiling. Front. Microbiol. 8:1875. doi: 10.3389/fmicb.2017.01875
Paige, C., Reid, S. D., Hanna, P. C., and Claiborne, A. (2008). The type III pantothenate kinase encoded by coax is essential for growth of Bacillus anthracis. J. Bacteriol. 190, 6271–6275. doi: 10.1128/JB.00860-08
Palluzzi, F., Ferrari, R., Graziano, F., Novelli, V., Rossi, G., Galimberti, D., et al. (2017). A novel network analysis approach reveals DNA damage, oxidative stress and calcium/camp homeostasis-associated biomarkers in Frontotemporal dementia. PLoS One 12:e0185797. doi: 10.1371/journal.pone.0185797
Potter, A. J., Kidd, S. P., Edwards, J. L., Falsetta, M. L., Apicella, M. A., Jennings, M. P., et al. (2009). Thioredoxin reductase is essential for protection of Neisseria gonorrhoeae against killing by nitric oxide and for bacterial growth during interaction with cervical epithelial cells. J. Infect. Dis. 199, 227–235. doi: 10.1086/595737
Ritz, D., Patel, H., Doan, B., Zheng, M., Aslund, F., Storz, G., et al. (2000). Thioredoxin 2 is involved in the oxidative stress response in Escherichia coli. J. Biol. Chem. 275, 2505–2512. doi: 10.1074/jbc.275.4.2505
Rouhier, N., Lemaire, S. D., and Jacquot, J. P. (2008). The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 59, 143–166. doi: 10.1146/annurev.arplant.59.032607.092811
Seneviratne, C. J., Wang, Y., Jin, L., Abiko, Y., and Samaranayake, L. P. (2008). Candida albicans biofilm formation is associated with increased anti-oxidative capacities. Proteomics 8, 2936–2947. doi: 10.1002/pmic.200701197
Takemoto, T., Zhang, Q.-M., and Yonei, S. (1998). Different mechanisms of thioredoxin in its reduced and oxidized forms in defense against hydrogen peroxide in Escherichia coli. Free Radic. Biol. Med. 24, 556–562. doi: 10.1016/S0891-5849(97)00287-6
Tsolis, R. M., Baumler, A. J., Heffron, F., and Stojiljkovic, I. (1996). Contribution of TonBand Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect. Immun. 64, 4549–4556. doi: 10.1016/S1380-2933(96)00052-8
Van Acker, J., Smet, F. D., Muyldermans, G., Bougatef, A., Naessens, A., and Lauwers, S. (2001). Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J. Clin. Microbiol. 39, 293–297. doi: 10.1128/jcm.39.1.293-297.2001
Vasu, D., Sunitha, M. M., Srikanth, L., Swarupa, V., Prasad, U. V., Sireesha, K., et al. (2015). In Staphylococcus aureus, the regulation of pyruvate kinase activity by serine/threonine protein kinase favors biofilm formation. 3 Biotech 5, 505–512. doi: 10.1007/s13205-014-0248-3
Velayudhan, J., Hughes, N. J., McColm, A. A., Bagshaw, J., Clayton, C. L., Andrews, S. C., et al. (2000). Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37, 274–286. doi: 10.1046/j.1365-2958.2000.01987.x
Vlamis-Gardikas, A., Potamitou, A., Zarivach, R., Hochman, A., and Holmgren, A. (2002). Characterization of Escherichia coli null mutants for glutaredoxin 2. J. Biol. Chem. 277, 10861–10868. doi: 10.1074/jbc.m111024200
Wyckoff, E. E., Mey, A. R., Leimbach, A., Fisher, C. F., and Payne, S. M. (2006). Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J. Bacteriol. 188, 6515–6523. doi: 10.1128/JB.00626-06
Ye, Y., Jiao, R., Gao, J., Li, H., Ling, N., Wu, Q. P., et al. (2016). Proteins involved in responses to biofilm and planktonic modes in Cronobacter sakazakii. LWT Food Sci. Technol. 65, 1093–1099. doi: 10.1016/j.lwt.2015.09.039
Ye, Y., Li, H., Wu, Q., Zhang, J., and Lu, Y. (2014). The Cronobacter sp. in milk and dairy products: detection and typing. Int. J. Dairy Technol. 67, 167–175. doi: 10.1111/1471-0307.12111
Ye, Y., Ling, N., Jiao, R., Wu, Q., Han, Y., and Gao, J. (2015). Effects of culture conditions on the biofilm formation of Cronobacter sakazakii strains and distribution of genes involved in biofilm formation. LWT Food Sci. Technol. 62, 1–6. doi: 10.1016/j.lwt.2015.01.035
Ye, Y., Zhang, M., Jiao, R., Ling, N., Zhang, X., Tong, L., et al. (2018). Inactivation of Cronobacter malonaticus cells and inhibition of its biofilm formation exposed to hydrogen peroxide stress. J. Dairy Sci. 101, 66–74. doi: 10.3168/jds.2017-13463
Zhang, M., Zhang, X., Tong, L., Wang, Y., Ou, D., Zhang, J., et al. (2018). Genes involved in tolerance to osmotic stress by random mutagenesis in Cronobacter malonaticus. J. Dairy Sci. 101, 3851–3858. doi: 10.3168/jds.2017-13995
Zhang, X., Zhang, G., Li, P., Yang, Q., Chen, K., Zhao, L., et al. (2017). Mechanisms of glycine betaine enhancing oxidative stress tolerance and biocontrol efficacy of Pichia caribbica against blue mold on apples. Biol. Control 108, 55–63. doi: 10.1016/j.biocontrol.2017.02.011
Keywords: Cronobacter malonaticus, random mutagenesis, arbitrary PCR, oxidative stress, biofilm formation
Citation: Zhang M, Zhang X, Tong L, Ou D, Wang Y, Zhang J, Wu Q and Ye Y (2019) Random Mutagenesis Applied to Reveal Factors Involved in Oxidative Tolerance and Biofilm Formation in Foodborne Cronobacter malonaticus. Front. Microbiol. 10:877. doi: 10.3389/fmicb.2019.00877
Received: 19 January 2019; Accepted: 05 April 2019;
Published: 01 May 2019.
Edited by:
Learn-Han Lee, Monash University Malaysia, MalaysiaReviewed by:
Chaoxin Man, Northeast Agricultural University, ChinaPeng Fei, Henan University of Science and Technology, China
Copyright © 2019 Zhang, Zhang, Tong, Ou, Wang, Zhang, Wu and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingping Wu, d3VxcDIwM0AxNjMuY29t Yingwang Ye, eWV5dzA0QG1haWxzLmd1Y2FzLmFjLmNu
†These authors have contributed equally to this work
 Maofeng Zhang1,2†
Maofeng Zhang1,2† Jumei Zhang
Jumei Zhang Yingwang Ye
Yingwang Ye