- 1Department of Microbiology, The University of Tennessee, Knoxville, Knoxville, TN, United States
- 2National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD, United States
- 3Department of Biology, University of Toronto Mississauga, Mississauga, ON, Canada
- 4Department of Biology, James Madison University, Harrisonburg, VA, United States
A Corrigendum on
Genome and Environmental Activity of a Chrysochromulina parva Virus and Its Virophages
by Stough, J. M. A., Yutin, N., Chaban, Y. V., Moniruzzaman, M., Gann, E. R., Pound, H. L., et al. (2019). Front. Microbiol. 10:703. doi: 10.3389/fmicb.2019.00703
In the original article, there was a mistake in Figure 2 as published. In the figure, the tree branch closest to Chrysochromulina parva virus BQ2 labeled as “Phaeocystis globosa virus (group II)” should have been labeled “Phaeocystis globosa virus (group I).” Similarly, the two branches labeled as “Phaeocystis globosa virus (group I)” and “Phaeocystis globosa virus” adjacent to Chrysochromulina parva virus BQ1 should have both been labeled as “Phaeocystis globosa virus (group II).”
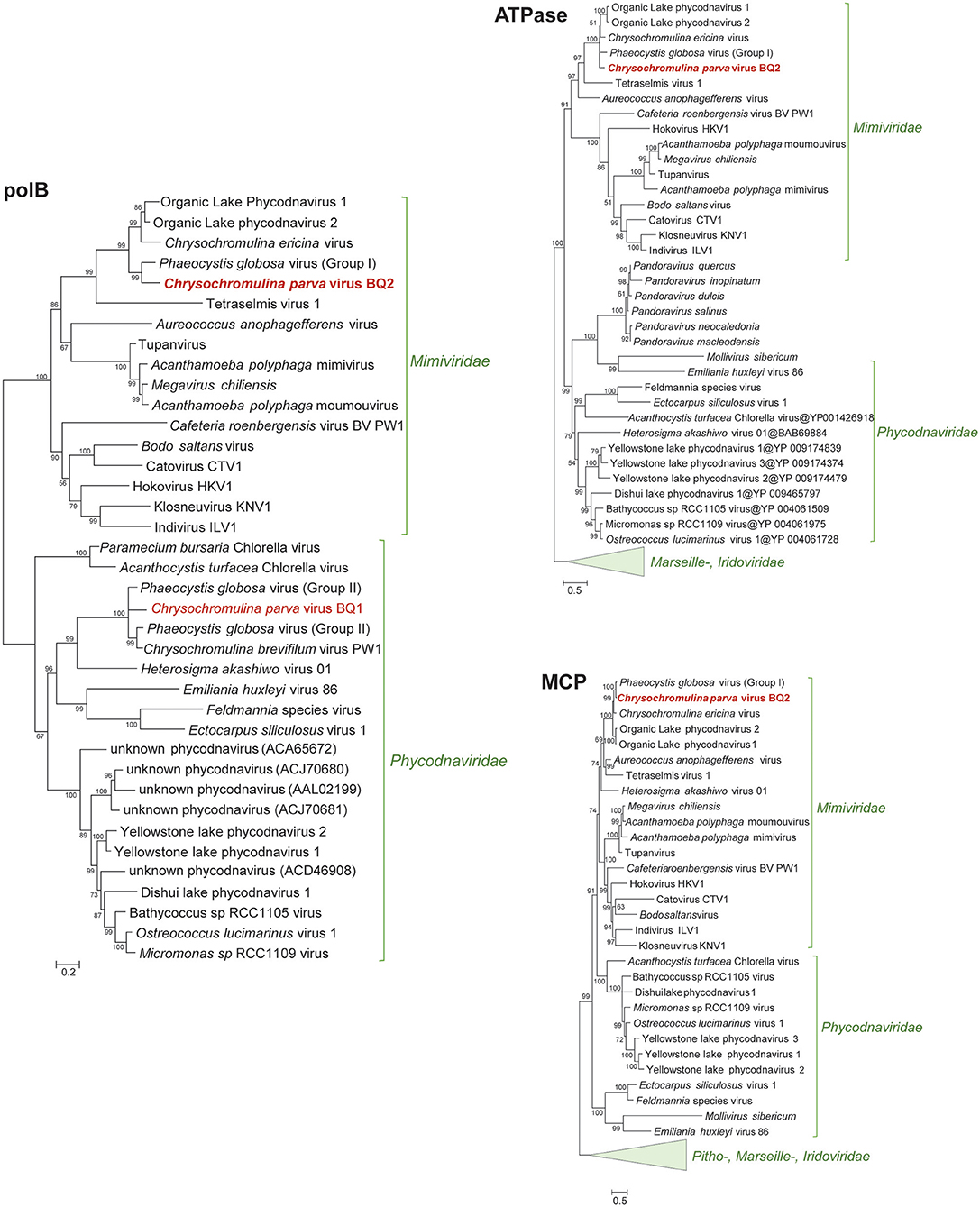
Figure 2. Approximately maximum-likelihood phylogenetic trees of B-family DNA polymerase (polB), A32-like virion packaging ATPase (ATPase), and the major capsid protein (MCP). The polB tree is constructed on protein fragments corresponding to PCR amplicons reported in Mirza et al. (2015). When more than one MCP gene was present in a genome (Mimiviridae), a paralog closest to Phycodnaviridae was chosen for the MCP tree. Node support (aLRT-SH statistic) >50% are shown. Accession numbers for the polB sequences are provided in Supplementary Table 1.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
References
Keywords: giant viruses, algae, NCLDV, freshwater, virophage, genome
Citation: Stough JMA, Yutin N, Chaban YV, Moniruzzaman M, Gann ER, Pound HL, Steffen MM, Black JN, Koonin EV, Wilhelm SW and Short SM (2019) Corrigendum: Genome and Environmental Activity of a Chrysochromulina parva Virus and Its Virophages. Front. Microbiol. 10:907. doi: 10.3389/fmicb.2019.00907
Received: 08 April 2019; Accepted: 09 April 2019;
Published: 24 April 2019.
Approved by:
Frontiers Editorial Office, Frontiers Media SA, SwitzerlandCopyright © 2019 Stough, Yutin, Chaban, Moniruzzaman, Gann, Pound, Steffen, Black, Koonin, Wilhelm and Short. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven W. Wilhelm, d2lsaGVsbUB1dGsuZWR1
Steven M. Short, c3RldmVuLnNob3J0QHV0b3JvbnRvLmNh
 Joshua M. A. Stough
Joshua M. A. Stough Natalya Yutin
Natalya Yutin Yuri V. Chaban
Yuri V. Chaban Mohammed Moniruzzaman
Mohammed Moniruzzaman Eric R. Gann
Eric R. Gann Helena L. Pound
Helena L. Pound Morgan M. Steffen
Morgan M. Steffen Jenna N. Black
Jenna N. Black Eugene V. Koonin
Eugene V. Koonin Steven W. Wilhelm
Steven W. Wilhelm Steven M. Short
Steven M. Short