- 1School of Natural and Environmental Sciences, Newcastle University, Newcastle upon Tyne, United Kingdom
- 2Centre of Excellence in Marine Biotechnology, Sultan Qaboos University, Al Khoud, Oman
- 3Department of Marine Science and Fisheries, Sultan Qaboos University, Al Khoud, Oman
The vast majority of bacteria present in the natural environment are present in the form of aggregates and/or biofilms. Microbial aggregates are ubiquitous in the marine environment and are inhabited by diverse microbial communities which often express intense extracellular enzymatic activities. However, the secretion of an important group of enzymes, DNases, by bacteria from marine aggregates has not been studied, despite the importance of these aggregates in biogeochemical cycling of nutrients in the oceans. In this work, we therefore, employed both culture-based and bioinformatics approaches to understand the diversity of bacterial DNases in marine bacterioplankton. We found that 34% of 345 strains of attached and non-attached marine bacteria showed extracellular DNase activity. Most of these isolates belong to Proteobacteria (53%) and Firmicutes (34%). Secretion of DNases by bacteria isolated from marine gel particles (MGP) is reported here for the first time. Then, to further understand the wider diversity of the potential to produce DNases, sequences were compared using 2316 whole genome and 42 metagenome datasets. Thirty-nine different taxonomic groups corresponding to 10 bacterial phyla were found to encode genes responsible for DNase secretion. This study highlights the unexpected and widespread presence of DNase secretion in bacteria in general and in MGP more specifically. This has important implications for understanding the dynamics and fate of marine microbial aggregates in the oceans.
Introduction
The marine environment contains an enormous microbial diversity (Salazar and Sunagawa, 2017) and density, where on average, bacteria are present in seawater and can reach concentrations of 106 cells/ml (Azam, 1998; Azam and Malfatti, 2007). Bacteria can live in one of three modes: either as free-living (planktonic), attached to surfaces (biofilm), or in the form of particle aggregates (flocs). In aggregated forms, bacteria live embedded in a hydrated matrix made up of extracellular polymeric substances (EPS) in the form of biofilm or protobiofilm (Verdugo, 2007; Verdugo and Santschi, 2010; Bar-Zeev et al., 2012; Elias and Banin, 2012). Marine gel particles (MGP) is another name for suspended bacterial aggregates found ubiquitously in the oceans and which can also initiate biofilm formation when they become attached to surfaces (Bar-Zeev et al., 2012; Neukermans et al., 2016; Busch et al., 2017).
Marine bacteria can contribute to biogeochemical nutrient cycling processes by secreting extracellular enzymes (ECE) (Azam, 1998; Azam and Malfatti, 2007; Arnosti, 2011; Luo et al., 2017; Ivančić et al., 2018) which play a fundamental role in the breakdown of organic matter by the hydrolysis of high molecular weight material to low molecular weight compounds for bacterial uptake. These enzymes can also affect the integrity and stability of MGP aggregates (Vetter and Deming, 1999; Hoppe et al., 2002; Lechtenfeld et al., 2015; Balmonte et al., 2016; Liu and Liu, 2018). As a result, MGP and their associated bacterial ECE have recently attracted attention due to their important role in the process of sequestration and removal of carbon into the deep ocean during sedimentation of particles (Karner and Herndl, 1992; Grossart et al., 2007; Arnosti et al., 2012; Kellogg and Deming, 2014).
Many hydrolytic enzymes are produced by free-living bacteria and bacteria attached to marine aggregates, for instance, chitinases and proteases (Smith et al., 1992; Baltar et al., 2017). However, information on the presence and diversity of deoxyribonucleases (DNases) in marine bacteria, especially those associated with aggregates, is absent.
DNases catalyze the hydrolysis of deoxyribonucleic acid (DNA) by breaking down phosphodiester bonds (Nishino and Morikawa, 2002). Consequently, DNase is considered to have a pivotal role in DNA utilization and nutrient cycling in the environment (Mulcahy et al., 2010). In addition, extracellular DNA (eDNA) plays an important role in the formation and structure of biofilms (Whitchurch et al., 2002). Indeed, it is now considered a key structural component of the biofilm matrix (Tetz et al., 2009). eDNA plays a critical role in the attachment and stability of the biofilm matrix and DNases are now well recognized as agents which can effectively break up biofilms (Nijland et al., 2010; Jakubovics et al., 2013; Shields et al., 2013; Okshevsky et al., 2015). We were therefore interested in whether eDNA was also an important component of marine flocs, and if so, whether the secretion of DNases by floc-associated bacteria may subsequently affect the structural integrity (and sinking rates) of marine particles. The secretion of extracellular DNase has been reported in several species of marine bacteria such as Bacillus licheniformis (Nijland et al., 2010), Vibrio sp. (Maeda and Taga, 1976), Myroides, Planococcus, Sporosarcina, and Halomonas (Dang et al., 2009) although their precise role remains obscure.
The availability of eDNA as a source of nutrients in the oceans is well recognized (Dell’Anno and Danovaro, 2005) and might also explain the production of DNases. However, little is known about the diversity of DNases produced by marine bacteria generally and in marine aggregates in particular. In this study, we hypothesized that the production of extracellular DNases is common among marine bacteria. Therefore, we investigated the diversity of extracellular DNase production by free-living marine bacteria and by bacteria attached to aggregates.
In addition, to overcome the problem of culturability of environmental bacteria (Vartoukian et al., 2010), and to rapidly assess the presence of DNase genes in a wide variety of microbial species, we carried out analysis of putative DNase genes using a bioinformatics approach (Kennedy et al., 2008). This allows the discovery of enzymes from the dataset of sequences of microbial genomes to include “uncultivable” taxa (Elend et al., 2006). In addition, enzyme discovery using sequence-based databases is often faster than function-based methods (Kennedy et al., 2008) and can deepen our understanding of the diversity of extracellular enzymes in bacteria.
Materials and Methods
Sample Collection
Bacteria were isolated from sediment, seawater, MGP and algae samples. Sediments and seawater were collected in Nalgene bottles (Thermo scientific) from the North Sea, around 15 km off the NE UK coast (55o 07 00 N 01o 20 00 W), on 23/03/2015. Surface sediments from a water depth of 50 m were collected by sediment grab and seawater was collected at 5–10 m depth using a Niskin bottle mounted on a CTD (Conductivity, Temperature, Depth) frame. During R.R.S James Cook JC037, additional sediment samples were collected with a megacore from three stations on the Mid-Atlantic Ridge north and south-east of the Charlie-Gibbs Fracture Zone (CGFZ), 48°–54°N (water depths 2400–2750 m), between 13/07/2007 and 18/08/2007. Only cores with intact surface sediments were selected for use in this work. One core was selected from each station and sectioned at 0–5 mm (surface) and 5–10 mm (subsurface) depth horizons. The sediment subsamples were stored in 50 ml sterile Falcon tubes at -20°C prior to bacterial isolation. North Sea coastal water microbial strains were obtained from the Dove Marine Laboratory (Newcastle University). MGP were collected on 31/10/2016, in 5 m water depth about 2 km off the coast of Northumberland (55° 06 972 N 1°25 600 W). Brown Fucus vesiculosus and red Palmaria palmata algae were collected by hand at low tide on Cullercoat beach (55° 02′ N 1° 25′W) and Boulmer beach (55° 25′N 1° 34′W) in May 2011. Harvested samples were transferred immediately to the laboratory in sterile bags and on ice. The algal surface was washed with filtered seawater and bacterial samples were removed from the surface with a sterile swab. Sections of seaweed from the holdfast, apical tips and growth nodes were cut using a sterile scalpel. Biofilm from the Sea of Oman was sampled during low tide near Muscat at Ras Alhamra 23°37′17″N 58°17′13″E on 06/07/2016. Bacterial samples were removed from rocks at the intertidal zone with sterile swabs. Immediately samples were transferred to the laboratory on ice for further analysis.
Scanning Electron Microscopy of Seaweed
For scanning electron microscopy, the seaweed samples were first fixed with a 2% (v/v) glutaraldehyde solution and kept at 4°C overnight (Dobretsov et al., 2014). Specimens were rinsed twice in 0.2 M phosphate buffer and dehydrated through a series of ethanol washes: 25, 50, 75% (30 min each) and in 100% absolute ethanol (30 min) until completely dried. They were subsequently transferred to Electron Microscopy Research Services at Newcastle University, where they were dried in a critical point dryer (Bal-tec), mounted on aluminum stubs and sputter coated with gold. Finally, biofilms were visualized at ×2000 and ×5000 magnification using a scanning electron microscope (Cambridge Stereoscan 240).
Isolation of Bacterial Strains and Growth Conditions
Sterile swabs (Fisher, United Kingdom) were used to sample filtered particles (MGP 0.4–100 μm), biofilms and the seaweed surfaces. Swabs were streaked onto Difco Marine agar plates (BD DifcoTM Dehydrated Culture Media: Marine Agar 2216, United Kingdom). Sediment samples were diluted (10-1 to 10-8) in sterile phosphate buffered saline (PBS, Sigma, United Kingdom) prior to plating onto marine agar (Difco, BD, United Kingdom). Morphologically distinct isolates were sub-cultured using aseptic technique onto fresh agar plates to obtain pure single colonies. Bacterial cultures were incubated at different temperatures to verify temperature dependent DNase production as well as DNase activity and stability. Psychrophiles were cultivated at 16°C for 5 days, mesophiles at 25 or 37°C for 2 days and thermophiles at 45 or 50°C for 2 days. Isolated bacterial strains were purified and routinely grown in Marine broth (BD Difco, United Kingdom) or LB (Luria-Bertani) (BD Difco, United Kingdom) broth with agitation at 150 rpm using an orbital shaker at room temperature.
The isolation of MGP was carried out by filtration of seawater onto polycarbonate filters (0.4–100 μm) (Alldredge et al., 1993). However, due to the limitations of this method, that generally only allows subsequent analysis of the particles while attached to the filters, rather than as they occur in seawater as suspended particles, we developed a new procedure for concentrating MGPs into small water volumes as suspended particles. Seawater was pre-filtered first through a 100 μm stainless steel sterile sieve and then by gentle vacuum filtration (Glass Vacuum Filtration Device 47/50 mm, Sartorius) through 0.4 μm polycarbonate filters (PC) (Whatman, GE Healthcare Life Sciences). To reduce clogging each PC filter was used only to filter one liter of seawater. The particles remaining on the PC filters were recovered by suspending the filters in 5 ml of 0.2 μm filtered sterile seawater and gently agitating to resuspend the particles. The particles in suspension were then used for bacterial isolation. Five microliters of the particle suspension were spread onto marine agar (BD, United Kingdom) and incubated at room temperature for 1–5 days; isolation was carried out until pure strains were obtained.
Cultivation and Identification of Extracellular DNase Producing Bacteria
All isolated bacterial strains were examined for extracellular DNase activity using DNase test agar containing methyl green dye (BD, 263220, United Kingdom). DNase production was evaluated at optimal growth temperatures for each strain tested. The production of extracellular DNase was inferred by the presence of a clear halo on DNase test agar around colonies of the tested strains (Palmer et al., 2012). In order to test whether planktonically grown cells were also able to secrete DNases, strains were also grown in liquid culture in triplicate. Cell-free supernatant (100 μl) of overnight liquid culture (marine broth) was then applied to wells in the DNase test agar, to verify the extracellular activity of the supernatants. Further confirmation of DNase activity was also confirmed by DNA digestion and gel electrophoresis for strains that showed high DNase activity in the filtered supernatant assays.
Measurement of DNase Activity of Seawater
Seawater (100 μl) obtained from the North Sea was used to inoculate 5 mm agar wells of DNase test agar (as above). Any presence of a clear halo was taken to indicate DNA degrading activity. In addition, gel electrophoresis was used to examine the presence of DNA degradation by non-autoclaved and autoclaved seawater.
DNA Extraction and 16S rRNA Gene Sequencing
Total DNA of bacterial isolates was extracted by Invitrogen Purelink® Genomic DNA Mini Kit (according to the manufacturer’s protocol). The 16S rRNA gene was then amplified using the universal 16S rRNA 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-ACG GCT ACC TTG TTA CGA CTT-3′) primers (Eurogentec1) with MyTaqTMRed Mix 2X (Bioline2). PCR cycles used an initial denaturation step at 95°C for 1 min, followed by 35 cycles of 15 s at 95°C, 15 s at 55°C, and 10 s extension at 72°C.
Sequence Analysis and Tentative Identification of Bacterial Isolates With DNase Activity
The extracted DNA from each isolate was amplified and sequenced at Geneius Labs, Newcastle upon Tyne, United Kingdom3. Sequences were edited and aligned using BioEdit4 and submitted to BLAST (NCBI, United States) for identification. Consensus sequences were generated using the DNABASER tool and compared with the sequences in the Ribosomal database 2 (RDB-II). Phylogenetic analysis was performed using MEGA version 6 (Tamura et al., 2013) after obtaining the multiple alignments of data available from public databases by ClustalX 1.83. Phylogenetic trees were generated using a neighbor-joining method with a bootstrap score of 1000 replicates.
Generating Datasets Using Bioinformatics Resources and Sequence Databases
Sequence-based screening was applied for gene discovery of extracellular DNase homologs. A function-specific dataset for the DNase gene(s) of interest was constructed for sequence-based gene analysis. The databases used to generate these datasets are the Enzyme database (Bairoch, 2000), KEGG database (Zhang and Wiemann, 2009), and the UNIPROT protein database (Apweiler et al., 2004). Metagenomic datasets and finished bacterial genomic datasets were available through the IMG/M database (Markowitz et al., 2012). By searching through the list of “genes with extracellular DNases” from every metagenome on the IMG/M database, extracellular DNase homologs (≥99%) were retrieved. The purpose of this was to gain a better insight into the diversity and representation of putative DNases in bacterial metagenomes and genomes. The databases of translated sequences from 43 environmental metagenome projects and 2136 bacterial genomes were used.
Extracellular DNase Diversity and Functional Analysis Using Bioinformatics Tools
Datasets of generated microbial genes which showed characteristics of diverse DNase activity and produced secreted proteins were investigated further through BLAST2GO (Gotz et al., 2008). Each of the retrieved genes was used to perform BLAST-searches and sequences having higher local similarity against the sequences of homologs (if similarity exceeded 99%), were defined as secreted DNases. The taxonomic distribution and diversity analysis was carried out using MEtaGenome ANalyzer, MEGAN 5 (Huson et al., 2011). Within MEGAN, a SEED-based functional analysis of the gene reads was applied to define specific processes associated with the extracellular DNases.
Results
Isolation of Marine Bacteria From Diverse Habitats
In this study, 345 strains of bacteria were isolated from different marine sources and locations: seawater, biofilm, MGP (marine aggregates), seaweed and sediments (Table 1). The majority (45%) of isolates were obtained from Mid-Atlantic Ridge sediment. Diverse epiphytic microorganisms, growing on the surface of the holdfast, apical tip and growth nodes of Fucus vesiculosus and Palmaria palmata, were observed using scanning electron microscopy (Figure 1). The numbers of psychrophilic and thermophilic isolates recovered from deep-sea sediments were higher than from seaweed surfaces, seawater and MGP, while mesophilic bacteria accounted for 70% of the total isolates (Table 1).
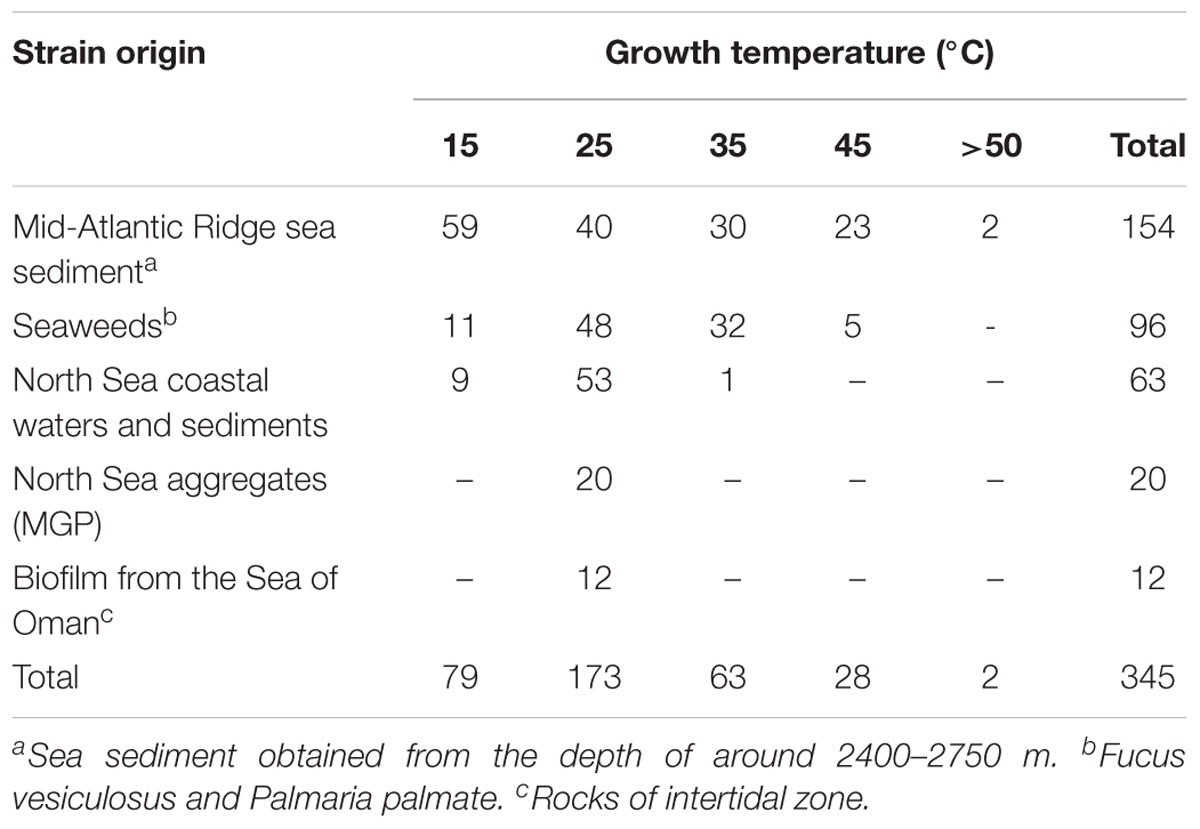
Table 1. Total number of bacterial strains isolated from various environments cultivated at different temperatures.
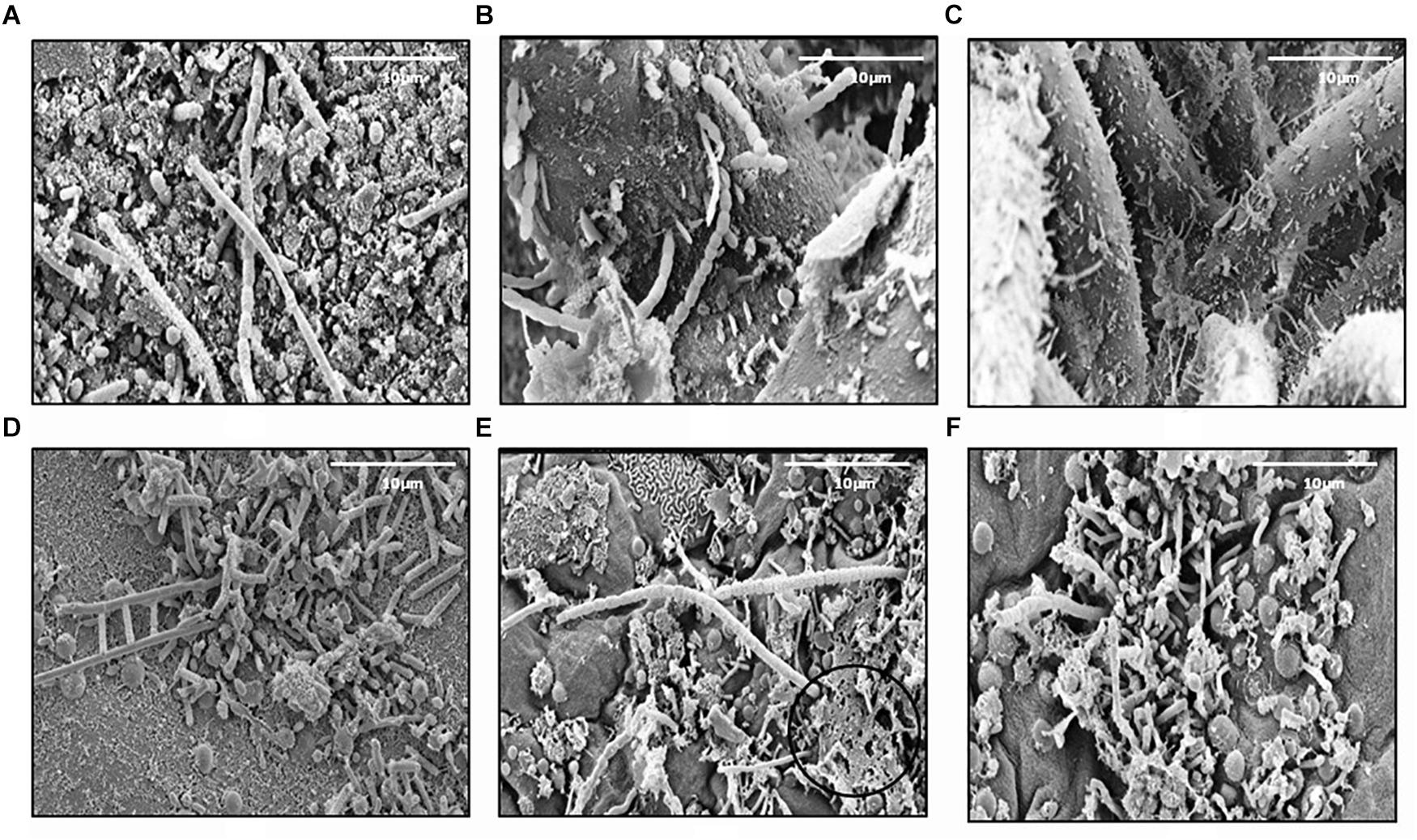
Figure 1. Scanning electron micrographs of the Fucus vesiculosus (A–C) and Palmaria palmata (D–F) seaweed surfaces. Micrographs (A,D) showing holdfast, (B,E) – apical tips, (C,F) – growth nodes. EPS like material can also be observed (E), circled ×100 magnification.
Deoxyribonuclease (DNase) Production by Cultivable Bacterial Isolates
All 345 isolates were examined for their ability to secrete DNase (Table 1). Thirty-five percent of these displayed DNase activity and were subjected to further characterization. This included identification by 1 rRNA gene sequencing. Sequences of the identified strains are available, GenBank Accession numbers (MK599164-MK599196). Most of the identified DNase producing bacteria (Supplementary Table S1) belong to the Proteobacteria (57%) followed by Firmicutes (34%), and Actinobacteria (6%) (Table 2 and Supplementary Figure S2). Most of the isolates belonging to the genera Bacillus (28%), Pseudoalteromonas (19%), and Vibrio (12%) produced DNases. DNase production by the genera Terrabacter, Ochrobactrum, Rhodococcus, Arthrobacter, Planococcus, and Paenisporosarcina is reported for the first time in this study. Furthermore, eight isolates from MGP showed DNase activity: Bacillus cereus, Pseudoalteromonas espejiana, Vibrio atlanticus, Shewanella fidelis, Pseudoalteromonas citrea, Vibrio atlanticus, Cobetia amphilecti, and Pseudoalteromonas issachenkonii.
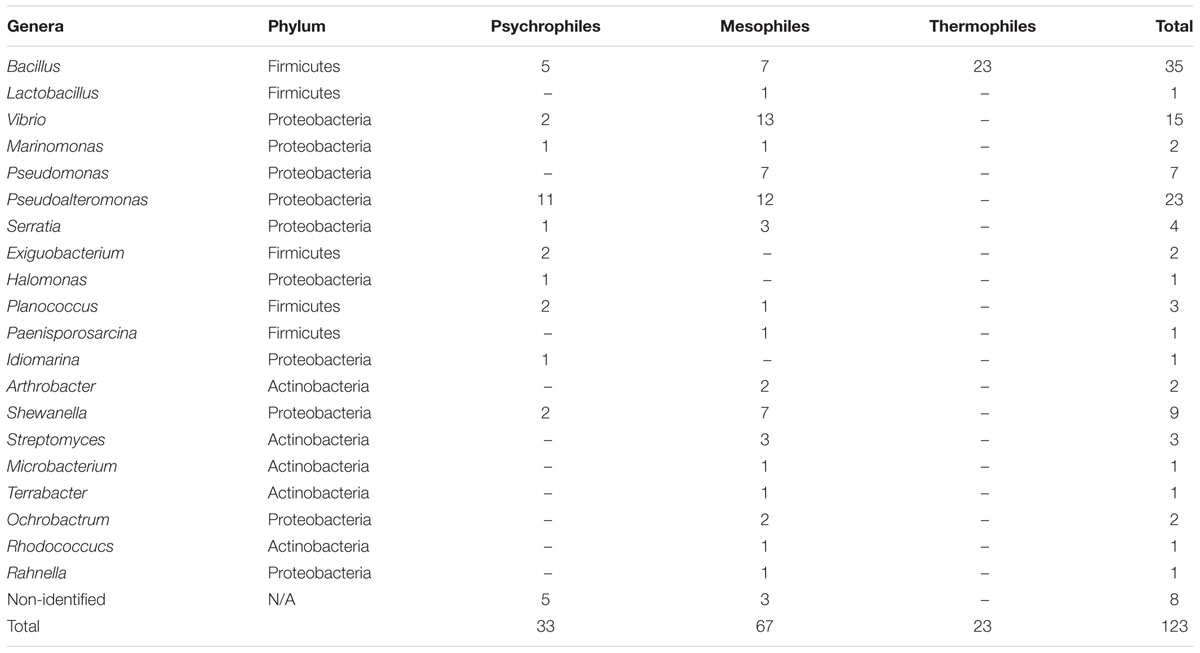
Table 2. Taxonomic classification of isolated bacterial strains based on the abilities to produce DNase enzymes.
The numbers of psychrophiles, mesophiles and thermophiles belonging to each bacterial genera producing DNase are summarized in Table 2. Mesophile strains account for 54% of total DNase producing bacteria. The zone of hydrolysis for DNA degradation of the psychrophilic, mesophilic and thermophilic isolates is shown in Figure 2. Based on quantitative DNase assay, thermophilic and mesophilic bacterial isolates showed higher DNase activity than psychrophilic producers. We also observed no detectable DNAse activity in seawater samples (data not shown).
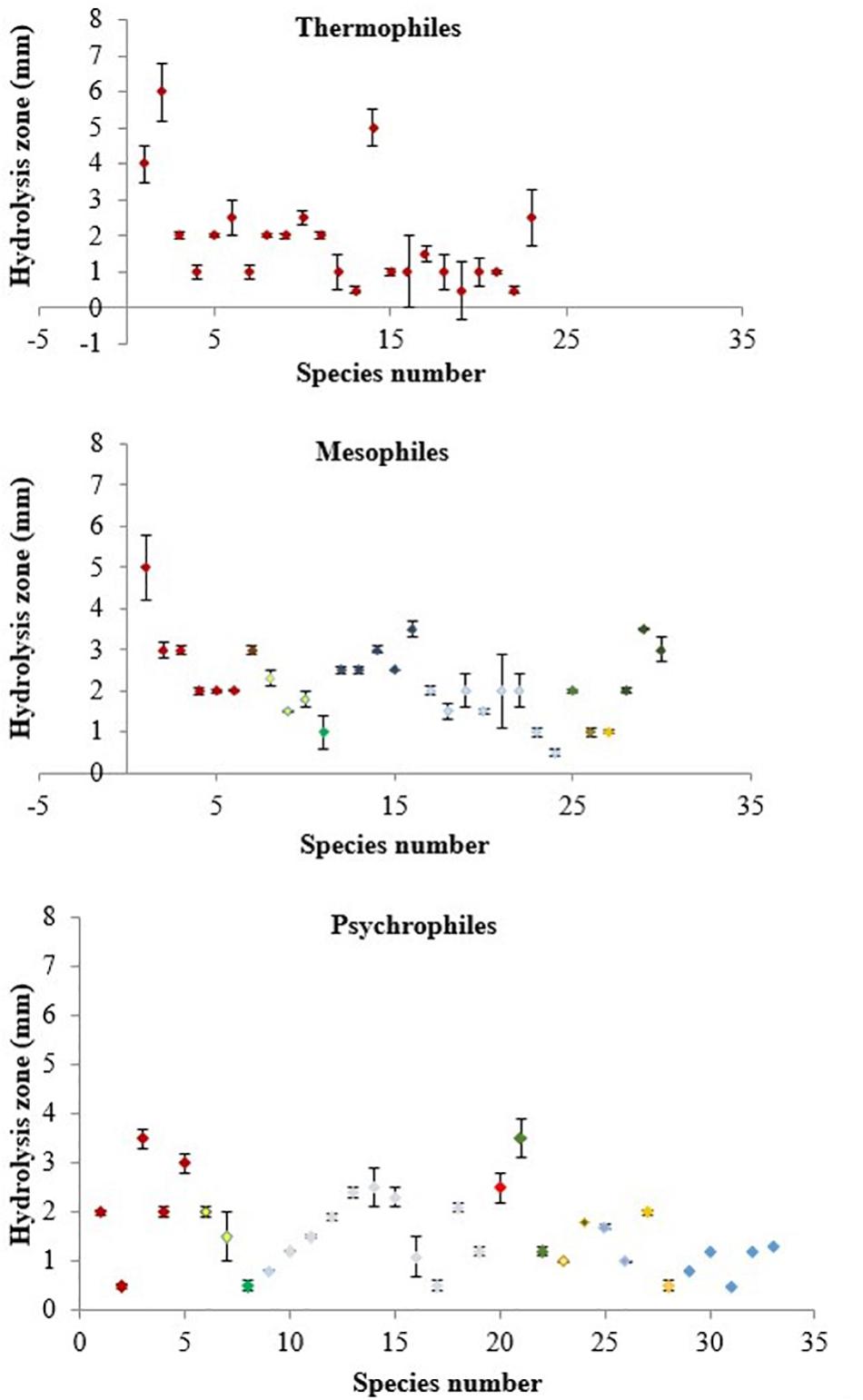
Figure 2. DNase activity in culture supernatants of thermophilic, mesophilic and psychrophilic isolates measured by the DNA hydrolysis zone (mm) produced on DNase Test Agar. Color coding represents each bacterial genera as follows: Bacillus – dark red, Vibrio – yellow, Marinomonas – green, Pseudomonas – dark blue, Pseudoalteromonas – light blue, Planococcus, Planomicrobium, Paenisporosarcina – orange, Serratia – bright red, Arthrobacter – purple, Streptomyces – brown, Halomonas – light purple, Idiomarina – dark purple, Shewanella – light blue, Exiguobacterium – light orange and non-identified – blue.
Sequence Driven Identification, Diversity, and Distribution of Microbial Extracellular Nuclease-Like Genes in the Genome and Metagenome Databases
To explore the diversity of putative nucleases a comprehensive bioinformatics search of 43 environmental metagenomes and 2136 bacterial genomes was conducted (Supplementary Figure S1A). As a preliminary study, nuclease-like genes were surveyed across prokaryotic and eukaryotic organisms (Supplementary Table S2). In total, seven microbial genes, which showed distinctive diverse nuclease activity and produced secreted protein, were chosen as a reference for further search. These were: NucB, a sporulation-specific extracellular nuclease from Bacillus subtilis; NucH, a thermonuclease from Staphylococcus aureus; Endo I, a periplasmic nuclease from Escherichia coli; NucA, a DNA/RNA non-specific nuclease from Serratia marcescens; S1P1 nuc, an extracellular nuclease from Penicillium melinii; Aspergillus oryzae, Dns, an extracellular deoxyribonucleases from Aeromonas hydrophila; and End A, a DNA entry nuclease from Streptococcus pneumonia.
This set of seven identified genes were BLAST-searched against databases of draft and completed bacterial genomes and metagenomes, which produced 538 microbial nuclease-like gene reads. An additional search for putative extracellular nucleases identified a further 324 microbial nuclease-like gene reads. Each nuclease-like gene retrieved the 20 highest hits of similar gene sequences through BLAST2GO (Supplementary Table S2).
MEGAN 5 was applied to estimate the microbial diversity and explore the taxonomic content of generated data. This analysis revealed 39 different taxonomic groups (Figure 3), covering 10 bacterial phyla (Supplementary Figure S1B). The highest number of putative nuclease genes was identified in Enterobacteriaceae [26] and Roseiflexus [20], followed by Enterococcus faecalis V583 [19], Vibrio [18], Xanthomonadaceae [16], and Prevotella [15] (Figure 3). The summarized phylum-level analysis (see Supplementary Figure S1A) shows that of a total 862 reads, 87% [751] were assigned to bacterial groups, with a majority of reads assigned to Proteobacteria 33% [248] and Firmicutes 25% [188]. Sequences from Archaea and Eukaryota corresponded only to 4.3 and 0.8% of the total sequences, respectively.
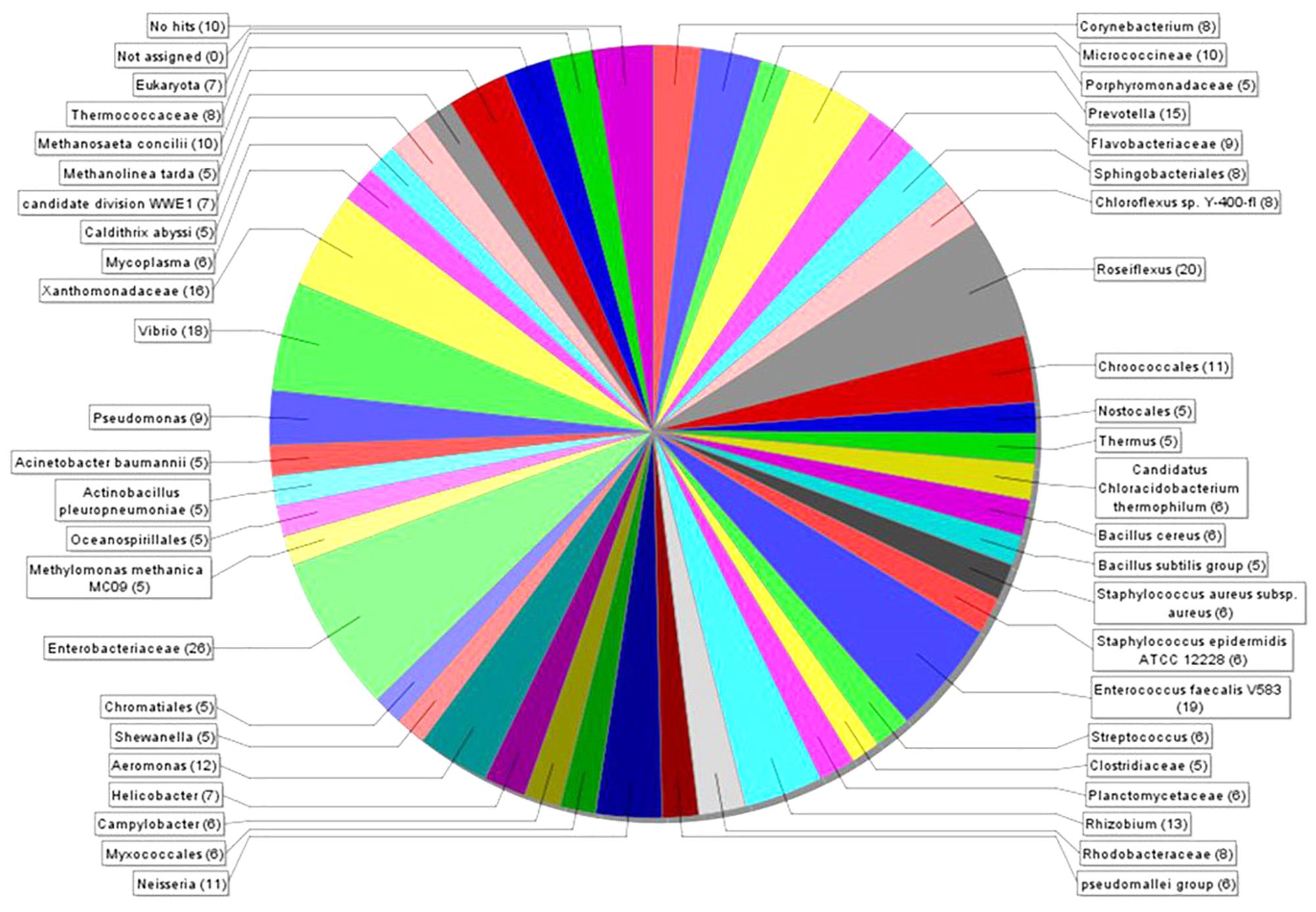
Figure 3. The taxonomic breakdown of microbial diversity analyzed with MEGAN 5. Visualization of the different taxonomic groups encoding 862 DNase-like genes in the bacterial genome and metagenome datasets. Each color represents individual taxonomic groups in the NCBI taxonomy and the number in the brackets denote the number of gene reads assigned to them.
To understand the biological processes involving the genes identified in this study, function-based screening approaches were applied. Within MEGAN, a SEED-based functional analysis of the gene reads defined the specific processes in which extracellular nucleases are implicated (Figure 4). Results revealed that a large portion of functions were expressed for DNA metabolism, phosphorus metabolism and virulence. The biological functions were similar to the functions elucidated by gene mapping performed in BLAST2GO. Noticeably, the nucleic acid phosphodiester bond hydrolysis and sporulation resulting in the formation of a cellular spore showed the greatest annotation score (Supplementary Figure S3). Nevertheless, outcomes (see Supplementary Figure S4) suggest additional important processes involving nucleases, including metabolism of nitrogen, phosphorous, purine, small molecules like tRNA, and response to stress stimulus.
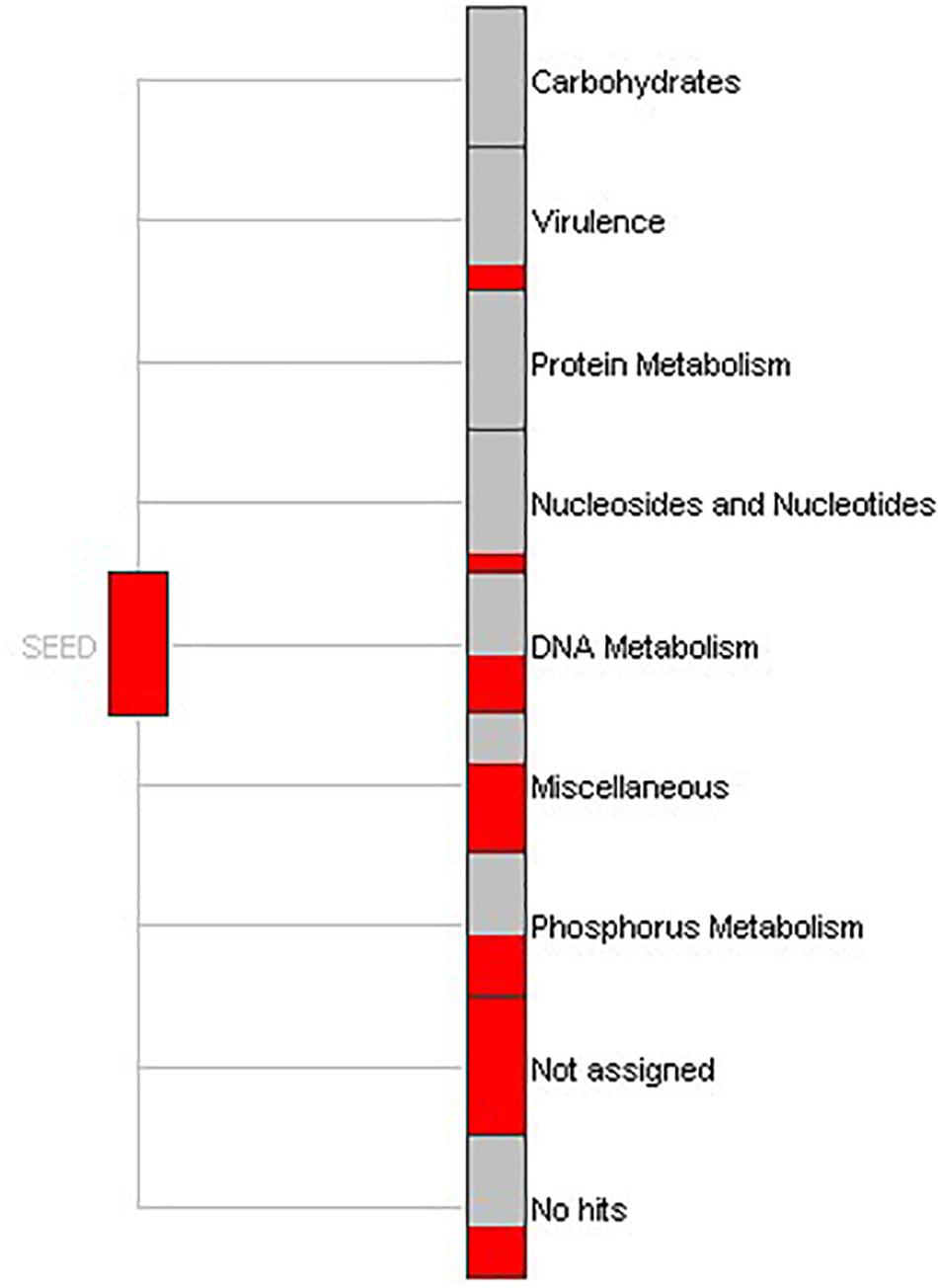
Figure 4. SEED-based functional assignment. Part of a SEED-based functional analysis of 862 DNase-like gene reads obtained from bacterial genome and metagenome. Each item represents a functional role in the SEED and the size of each red colored box is scaled logarithmically to represent the number of reads assigned to this.
Discussion
Bacterial extracellular enzymes are ubiquitous in the ocean and play an important role in the cycling and fate of organic matter. However, knowledge regarding DNase diversity in marine bacteria in general, and in marine aggregates more specifically, remains rudimentary. DNase production has been reported in several bacterial species, including Serratia marcescens (Nestle and Roberts, 1969), marine isolates of Vibrio sp. (Maeda and Taga, 1976), Pseudomonas aeruginosa (Mulcahy et al., 2010), Myroides, Planococcus, Sporosarcina and Halomonas (Dang et al., 2009), and Bacillus licheniformis (Nijland et al., 2010). In addition, there are some studies on DNases produced by the human pathogen Streptococcus (Porschen and Sonntag, 1974). However, most studies have focused only on individual cultivable species. There are no studies on the overall diversity of DNases produced by bacteria in general or by marine aggregate associated bacteria. In this study we used a culture based approach and bioinformatics tools together, to highlight the diversity of marine bacterial production of nucleases and DNases.
Deoxyribonuclease Production by Cultivable Marine Bacteria
In our current study, bacteria were isolated from various environments (seaweed surfaces, North Sea waters, mid-Atlantic Ridge sediments and coastal waters of Oman) (Table 1) and are of different types (free-living or attached) and from a range of temperatures (15–45oC). For seaweed associated bacteria, an EPS matrix was visualized on the seaweed surface by ESM circled in Figure 1E. This layer is presumably secreted by the algae and the commensal epibiotic bacteria on the seaweed surface to reduce fouling, and protection against harsh conditions and predation (Armstrong et al., 2001; Wiese et al., 2009; Goecke et al., 2010; Wahl et al., 2012). Subsequently, these macroalgae may release EPS materials into the water column, that are then colonized by bacteria and would contribute to the MGP pool in the ocean. These particles may aggregate further to form marine snow and can settle into the deep ocean through sedimentation. Exopolymer release by algae and diatoms is well evidenced (Thornton, 2004, 2014; Chen and Thornton, 2015; Li et al., 2015) but the chemical composition of these particles as they evolve and sink is not well studied. In particular, the presence of DNA in these particles is unknown. It is thought that DNA is abundant in higher concentrations in sediments compared to seawater and that 90% of the DNA found in sediments is extracellular DNA (Dell’Anno et al., 2002; Nielsen et al., 2007; Torti et al., 2015). This suggests that this DNA is transported into the deep sea by sinking particles.
In this work, 35% of all the cultivable bacteria obtained produced extracellular DNase. The production of DNase seems to be common among a diverse array of marine bacterial isolates and is greater in mesophilic bacteria (54%) than in psychrophilic or thermophilic as our results demonstrated (Table 2 and Figure 2). A possible explanation is that marine bacterial DNase may be more active at mesophile conditions of 20–45oC (Ahrenholtz et al., 1994). Most of the cultivable bacteria that produced DNase in this study belong to 20 genera, affiliated to Proteobacteria (53%) and Firmicutes (34%) (Supplementary Figure S2). It has been observed that Proteobacteria employ nuclease based bacteriocins (NBs) as a defense mechanism (Parret and De Mot, 2002; Sharp et al., 2017).
Most of the isolates that were able to produce DNase belonged to the genus Bacillus (28%) and all were identified as thermophiles. Bacillus are well-known enzyme producers (Ferrero et al., 1996; Lei et al., 2017; Parab et al., 2017; Anjum et al., 2018) including nuclease production, for example, NucB production by Bacillus licheniformis (Nijland et al., 2010). The next highest cultivable genus was Pseudoalteromonas (18%), which species of which are known for their production of bioactive compounds and enzymes (Tutino et al., 2002; Zeng et al., 2006; de Pascale et al., 2008; Bian et al., 2012; Bhattacharya et al., 2018). A recent study of extracellular enzymes from marine cultivable bacteria from The New Britain Trench showed that Pseudoalteromonas produced various extracellular enzymes, mostly proteases and chitinases (García-Fraga et al., 2015; Liu et al., 2018). Another study on cultivable bacteria in Laizhou Bay, China, showed that Pseudoalteromonas was the most common cultivable genus producing extracellular proteases (Li et al., 2017). Moreover, Pseudoalteromonas spp. have also been reported to produce nuclease baceriocins (Desriac et al., 2010).
Here, bacteria associated with marine aggregates were investigated for nuclease production for the first time. Our results revealed that eight out of 20 isolates from MGP were able to secrete nucleases. These isolates belong to the families Pseudoalteromonadacea (Azam and Malfatti, 2007), Vibrionaceae (Azam, 1998), Bacillaceae (Salazar and Sunagawa, 2017), Shewanellaceae (Salazar and Sunagawa, 2017), and Halomonadaceae (Salazar and Sunagawa, 2017) most of which have been reported previously (Dang et al., 2009; Balabanova et al., 2017). Although extracellular enzymes from Vibrio (Bunpa et al., 2016) and extracellular agents and enzymes from Pseudoalteromonas citrea have been reported previously (Holmström and Kjelleberg, 1999; Roca et al., 2016; Yamada et al., 2016; Imbs et al., 2018), extracellular DNases from bacteria isolated specifically from MGPs has not been described previously. In addition, there is evidence that Pseudomonas and Vibrio produce extracellular deoxyribonucleases to utilize DNA as a nutrient source as well as synthesizing DNases for horizontal gene transfer of DNA as part of natural transformation (Blokesch and Schoolnik, 2008; Mulcahy et al., 2010). However, the observation that Vibrio atlanticus and Psudoalteromonas citrea isolated from MGP can produce DNase, is interesting as it may implicate DNase in the dispersal of MGP, based on our hypothesis that eDNA is a key component of MGPs and plays a role in their physical integrity.
Extracellular enzymes produced by heterotrophic bacteria are key players in organic matter cycling in the ocean (Azam and Malfatti, 2007; Rier et al., 2014; Balmonte et al., 2016; Burns et al., 2016) and bacterial extracellular enzyme activity in aggregates is reported to be two orders of magnitude higher than in free-living bacteria (Smith et al., 1992; Grossart et al., 2006; Ziervogel et al., 2010; Kellogg and Deming, 2014). These extracellular enzymes are thought to be involved in the dissolution of MGPs and marine snow (D’ambrosio et al., 2014; Balmonte et al., 2016; Luo et al., 2017; Baltar, 2018; Ivančić et al., 2018). However, the effect of nucleases, specifically has hitherto been overlooked, with regard to the implications for MGP and marine snow dispersal. We were also unable to detect DNase activity in seawater samples, indicating that the level of enzyme activity may be much lower than concentrations which might be expected to occur inside MGPs.
Additional studies are required to study the production of nucleases in situ. However, results with cultivable isolates revealed that DNase is expressed by various bacteria. The versatile ability of marine isolates to produce DNase suggests that they play an important ecological role in extracellular DNA degradation in the marine environment. The present study provides data on DNase production by marine cultivable bacteria from aggregates for the first time and hence provides further insights into the diversity of extracellular enzyme production by marine bacteria.
DNase Gene Diversity a Bioinformatics Study
The MetaBIOME database was employed previously to explore and identify commercially applicable enzymes from metagenomes (Sharma et al., 2010; Sharma and Taylor, 2015) and as an advanced tool for exploring novel genes involved in marine microbial metabolism (Kennedy et al., 2008, Kennedy et al., 2010). Similarly, in this study, the IMG/M data management and analysis system was explored for nucleases. The IMG/M database comprises both microbial whole genomes and metagenomes. As a simple, yet comprehensive analysis tool to search for currently available genes of interest, it has advantages over other applied databases, for example, GenDB/JCoast (Richter et al., 2008) and Magnifying Genome/MicroScope (Vallenet et al., 2013).
A bioinformatics survey of 43 environmental metagenomes and 2136 bacterial genomes, together with taxonomic analysis, revealed a high diversity of microbial extracellular nuclease-like genes. Interestingly, nuclease-like enzyme positive genotypes were found in 39 different taxonomic groups, representing one-third of bacterial phyla. Taxonomic classification indicated that the dominant bacterial phyla were Proteobacteria and Firmicutes, accounting for 33 and 25%, respectively, of the total bacterial reads, respectively (Figure 3). This reinforces our experimental data where 53% of the cultured nuclease enzyme producing bacteria belonged to the phylum Proteobacteria and 34% to Firmicutes. This concurrence in the results of the culture-based methods and the bioinformatics tools suggest that marine Proteobacteria and Firmicutes are important producers of nucleases. Furthermore, data presented in Table 2 are also in agreement with a recent study that developed a bioinformatics pipeline to identify nuclease bacteriocins (NBs) in bacteria that revealed that NBs are found commonly in gamma-proteobacteria (Sharp et al., 2017).
Microbial diversity of extracellular enzymes using an openly available sequence-based approach has been used successfully for esterases (Elend et al., 2006). However, information regarding the variety of secreted nucleases is still scant. The sequence driven analysis suggests that microbial communities are able to produce diverse extracellular nuclease-like enzymes. However, the taxonomy-based function predictions are limited, as they depend on databases that are constructed from sequences of cultured bacteria, which restricts the understanding of natural community functions.
BLAST2GO gene mapping and SEED-based functional analysis underlined the fact that DNases are also centrally important in DNA metabolism and involved in a number of intracellular DNA processing steps (Figure 4). Because bacteria can produce DNases to utilize extracellular DNA as a source of carbon, nitrogen and phosphorus we also surveyed intracellular putative DNase enzyme genes in this work. DNases also play a role in virulence, degradation of neutrophil DNA extracellular traps (NETs) and in biofilm degradation (Dubnau, 1999; Thomas and Nielsen, 2005; Vorkapic et al., 2016; Veening and Blokesch, 2017). Bacteria employ DNases to also compete with other bacteria for space or resources and thus these enzymes have an important ecological function in survival (Cao et al., 2017). Nucleases digest DNA that can be used as the sole nutrient source by biofilm-forming bacteria Pseudomonas aeruginosa and Shewanella oneidensis (Mulcahy et al., 2010; Godeke et al., 2011). Another study reported that the balance of extracellular DNA in the marine environment is largely regulated by DNase, which is important for the functioning of deep-sea ecosystems (Dell’Anno and Danovaro, 2005). Our findings regarding the role of DNases in cellular metabolism of nitrogen and phosphorus, suggest that DNase secretion may also play an important role in the fate of extracellular DNA in the natural environment, particularly with regard to the dynamics and stability of marine aggregates.
Culture-based and bioinformatics tools used to investigate the diversity of DNases allowed us to gain insight into the presence and expression of DNase in cultivable and non-cultivable marine bacteria. Additionally diverse bacterial genera that were recovered were able to produce DNases, and the function based investigation of the sequence reads facilitated an appreciation of the ecological importance of extracellular DNases. The present study provides new insights into the abundance, hidden diversity, and environmentally important functions of microbes carrying DNase-like genes.
The most notable finding to emerge from this study is that the bacterial community associated with MGPs is able to secrete extracellular DNases. This is an important element that could contribute to our understanding of extracellular enzymes present within gel particles and marine snow in the ocean. In addition, it might provide answers to the key questions surrounding what proportion of particulate organic matter sinks into the deep ocean and what regulates this process.
There is a need to further understand the diversity of MGP associated bacteria and their ability to produce extracellular DNases in the ocean. We are now investigating the effects of extracellular DNases on MGP dissolution. This can expand our knowledge of bacterial DNases and the role they play in the dynamics of MGP and consequently carbon cycling in the oceans.
Author Contributions
JB contributed to the conception and design of the work. NR, EL, and AA-W has contributed in the acquisition, analysis and interpretation of the data. AA-W and NR wrote and drafted the manuscript. JB, SD, and RU-G critically revised the manuscript.
Funding
Part of the project was funded by the Ministry of Higher Education in the Sultanate of Oman.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the support of the NERC ECOMAR programme (NE/C512961/1) and the officers and crew of RRS James Cook for the provision of data and samples made available for this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00969/full#supplementary-material
FIGURE S1 | Phylogenetic diversity of the putative DNase-like enzyme producing taxonomic groups as calculated by MEGAN 5. (A) Phylum-level summary of MEGAN analysis. (B) A low level view of the MEGAN analysis. Each node represents a taxon in the NCBI taxonomy, and the size of the node is scaled logarithmically to represent the number of reads assigned directly to the taxon.
FIGURE S2 | Phylogenetic diversity at phylum level of the cultured DNase enzyme producing bacteria where 53% belong to the phylum Proteobacteria, 34% Firmicutes, and 6% Actinobacteria.
FIGURE S3 | A Directed Acyclic Graph (DAG) visualizing the hierarchical structure of the Gene Ontology (GO) in biological processes involving DNases resulting in sporulation. The darker the color of the node, the greater the number of BLAST hits and the higher the score values. All nodes contain the hit annotation scores in numbers.
FIGURE S4 | A Directed Acyclic Graph (DAG) visualizing the biological process involving DNAses resulting in metabolism of nitrogen, phosphorous, purine, small molecules such as tRNA and response to stress stimulus. The darker the color of the node the greater the number of BLAST hits and the higher the score values. All nodes contain the hit annotation scores in numbers.
TABLE S1 | List of strains used.
TABLE S2 | The dataset of the BLAST2GO results of 832 DNase-like gene reads.
Footnotes
- ^ https://secure.eurogentec.com/eu-home.html
- ^ https://www.bioline.com/mytaq-red-mix.html
- ^ http://www.geneiuslabs.co.uk/
- ^ http://www.mbio.ncsu.edu/BioEdit/bioedit.html
References
Ahrenholtz, I., Lorenz, M. G., and Wackernagel, W. (1994). The extracellular nuclease of Serratia marcescens: studies on the activity in vitro and effect on transforming DNA in a groundwater aquifer microcosm. Arch. Microbiol. 161, 176–183. doi: 10.1007/s002030050039
Alldredge, A. L., Passow, U., and Logan, B. E. (1993). The abundance and significance of a class of large, transparent organic particles in the ocean. Deep Sea Res. Part I Oceanogr. Res. Pap. 40, 1131–1140. doi: 10.1016/0967-0637(93)90129-q
Anjum, K., Bi, H., Chai, W., Lian, X. Y., and Zhang, Z. (2018). Antiglioma pseurotin A from marine Bacillus sp. FS8D regulating tumour metabolic enzymes. Nat. Prod. Res. 32, 1353–1356. doi: 10.1080/14786419.2017.1343329
Apweiler, R., Bairoch, A., and Wu, C. H. (2004). Protein sequence databases. Curr. Opin. Chem. Biol. 8, 76–80.
Armstrong, E., Yan, L., Boyd, K. G., Wright, P. C., and Burgess, J. G. (2001). The symbiotic role of marine microbes on living surfaces. Hydrobiologia 461, 37–40.
Arnosti, C. (2011). Microbial extracellular enzymes and the marine carbon cycle. Ann. Rev. Mar. Sci. 3, 401–425. doi: 10.1146/annurev-marine-120709-142731
Arnosti, C., Fuchs, B., Amann, R., and Passow, U. (2012). Contrasting extracellular enzyme activities of particle-associated bacteria from distinct provinces of the North Atlantic Ocean. Front. Microbiol. 3:425. doi: 10.3389/fmicb.2012.00425
Azam, F. (1998). Microbial control of oceanic carbon flux: the plot thickens. Science 280, 694–696. doi: 10.1126/science.280.5364.694
Azam, F., and Malfatti, F. (2007). Microbial structuring of marine ecosystems. Nat. Rev. Micro. 5, 782–791. doi: 10.1038/nrmicro1747
Bairoch, A. (2000). The ENZYME database in 2000. Nucleic Acids Res. 28, 304–305. doi: 10.1093/nar/28.1.304
Balabanova, L., Podvolotskaya, A., Slepchenko, L., Eliseikina, M., Noskova, Y., Nedashkovskaya, O., et al. (2017). Nucleolytic enzymes from the marine bacterium Cobetia amphilecti KMM 296 with antibiofilm activity and biopreservative effect on meat products. Food Control. 78, 270–278. doi: 10.1016/j.foodcont.2017.02.029
Balmonte, J., Teske, A., and Arnosti, C., eds (2016). Composition and Extracellular Enzymatic Function of Pelagic, Particle-Associated, and Benthic Bacterial Communities in the Central Arctic Ocean. Washington, DC: American Geophysical Union.
Baltar, F. (2018). Watch out for the “living dead”: cell-free enzymes and their fate. Front. Microbiol. 8:2438. doi: 10.3389/fmicb.2017.02438
Baltar, F., Morán, X. A. G., and Lønborg, C. (2017). Warming and organic matter sources impact the proportion of dissolved to total activities in marine extracellular enzymatic rates. Biogeochemistry 133, 307–316. doi: 10.1007/s10533-017-0334-9
Bar-Zeev, E., Berman-Frank, I., Girshevitz, O., and Berman, T. (2012). Revised paradigm of aquatic biofilm formation facilitated by microgel transparent exopolymer particles. Proc. Natl. Acad. Sci. U.S.A. 109, 9119–9124. doi: 10.1073/pnas.1203708109
Bhattacharya, S., Choudhury, J. D., Gachhui, R., and Mukherjee, J. (2018). A new collagenase enzyme of the marine sponge pathogen Pseudoalteromonas agarivorans NW4327 is uniquely linked with a TonB dependent receptor. Int. J. Biol. Macromol. 109, 1140–1146. doi: 10.1016/j.ijbiomac.2017.11.106
Bian, F., Xie, B. B., Qin, Q. L., Shu, Y. L., Zhang, X. Y., Yu, Y., et al. (2012). Genome sequences of six Pseudoalteromonas strains isolated from arctic sea ice. J. Bacteriol. 194, 908–909. doi: 10.1128/JB.06427-11
Blokesch, M., and Schoolnik, G. K. (2008). The extracellular nuclease Dns and its role in natural transformation of Vibrio cholerae. J. Bacteriol. 190, 7232–7240. doi: 10.1128/JB.00959-08
Bunpa, S., Sermwittayawong, N., and Vuddhakul, V. (2016). Extracellular enzymes produced by Vibrio alginolyticus isolated from environments and diseased aquatic animals. Proced. Chem. 18, 12–17. doi: 10.1016/j.proche.2016.01.002
Burns, W., Marchetti, A., White, B., Prairie, J., and Ziervogel, K., eds (2016). Algal-Bacteria Interactions and the Effects on Organic Matter Flux and Carbon Remineralization in the Ocean. American Geophysical Union, Washington, DC.
Busch, K., Endres, S., Iversen, M. H., Michels, J., Nöthig, E. M., and Engel, A. (2017). Bacterial colonization and vertical distribution of marine gel particles (TEP and CSP) in the Arctic fram strait. Front. Mar. Sci. 4:166. doi: 10.3389/fmars.2017.00166
Cao, Z., Casabona, M. G., Kneuper, H., Chalmers, J. D., and Palmer, T. (2017). The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nature Microbiol. 2:16183. doi: 10.1038/nmicrobiol.2016.183
Chen, J., and Thornton, D. C. O. (2015). Transparent exopolymer particle production and aggregation by a marine planktonic diatom (Thalassiosira weissflogii) at different growth rates. J. Phycol. 51, 381–393. doi: 10.1111/jpy.12285
D’ambrosio, L., Ziervogel, K., MacGregor, B., Teske, A., and Arnosti, C. (2014). Composition and enzymatic function of particle-associated and free-living bacteria: a coastal/offshore comparison. ISME J. 8:2167. doi: 10.1038/ismej.2014.67
Dang, H., Zhu, H., Wang, J., and Li, T. (2009). Extracellular hydrolytic enzyme screening of culturable heterotrophic bacteria from deep-sea sediments of the Southern Okinawa Trough. World J. Microbiol. Biotechnol. 25, 71–79. doi: 10.1007/s11274-008-9865-5
de Pascale, D., Cusano, A. M., Autore, F., Parrilli, E., Di Prisco, G., Marino, G., et al. (2008). The cold-active Lip1 lipase from the antarctic bacterium Pseudoalteromonas haloplanktis TAC125 is a member of a new bacterial lipolytic enzyme family. Extremophiles 12, 311–323. doi: 10.1007/s00792-008-0163-9
Dell’Anno, A., and Danovaro, R. (2005). Extracellular DNA plays a key role in deep-sea ecosystem functioning. Science 309:2179. doi: 10.1126/science.1117475
Dell’Anno, A., Stefano, B., and Danovaro, R. (2002). Quantification, base composition, and fate of extracellular DNA in marine sediments. Limnol. Oceanogr. 47, 899–905. doi: 10.4319/lo.2002.47.3.0899
Desriac, F., Defer, D., Bourgougnon, N., Brillet, B., Le Chevalier, P., and Fleury, Y. (2010). Bacteriocin as weapons in the marine animal-associated bacteria warfare: inventory and potential applications as an aquaculture probiotic. Mar. Drugs 8, 1153–1177. doi: 10.3390/md8041153
Dobretsov, S., Thomason, J. C., and Williams, D. N. (2014). Biofouling Methods. Hoboken: John Wiley & Sons, Inc., 291–314. doi: 10.1002/9781118336144
Dubnau, D. (1999). DNA uptake in bacteria. Annu. Rev. Microbiol. 53, 217–244. doi: 10.1146/annurev.micro.53.1.217
Elend, C., Schmeisser, C., Leggewie, C., Babiak, P., Carballeira, J. D., Steele, H. L., et al. (2006). Isolation and biochemical characterization of two novel metagenome-derived esterases. Appl. Environ. Microb. 72, 3637–3645. doi: 10.1128/aem.72.5.3637-3645.2006
Elias, S., and Banin, E. (2012). Multi-species biofilms: living with friendly neighbors. FEMS Microbiol. Rev. 36, 990–1004. doi: 10.1111/j.1574-6976.2012.00325.x
Ferrero, M., Castro, G., Abate, C., Baigori, M., and Sineriz, F. (1996). Thermostable alkaline proteases of Bacillus licheniformis MIR 29: isolation, production and characterization. Appl. Microbiol. Biotechnol. 45, 327–332. doi: 10.1007/s002530050691
García-Fraga, B., da Silva, A. F., López-Seijas, J., and Sieiro, C. (2015). A novel family 19 chitinase from the marine-derived Pseudoalteromonas tunicata CCUG 44952T: heterologous expression, characterization and antifungal activity. Biochem. Eng. J. 93, 84–93. doi: 10.1016/j.bej.2014.09.014
Godeke, J., Heun, M., Bubendorfer, S., Paul, K., and Thormann, K. M. (2011). Roles of two shewanella oneidensis MR-1 extracellular endonucleases. Appl. Environ. Microb. 77, 5342–5351. doi: 10.1128/AEM.00643-11
Goecke, F., Labes, A., Wiese, J., and Imhoff, J. F. (2010). Chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Progr. Ser. 409, 267–299. doi: 10.3354/meps08607
Gotz, S., Garcia-Gomez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Grossart, H. P., Tang, K. W., Kiørboe, T., and Ploug, H. (2006). Comparison of cell-specific activity between free-living and attached bacteria using isolates and natural assemblages. FEMS Microbiol. Lett. 266, 194–200. doi: 10.1111/j.1574-6968.2006.00520.x
Grossart, H. P., Tang, K. W., Kiørboe, T., and Ploug, H. (2007). Comparison of cell-specific activity between free-living and attached bacteria using isolates and natural assemblages. FEMS Microbiol. Lett. 266, 194–200. doi: 10.1111/j.1574-6968.2006.00520.x
Holmström, C., and Kjelleberg, S. (1999). Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol. Ecol. 30, 285–293. doi: 10.1016/s0168-6496(99)00063-x
Hoppe, H. G., Arnosti, C., and Herndl, G. J. (2002). Ecological significance of bacterial enzymes in the marine environment. Enzym. Environ. Activity Ecol. Appl. 2002, 73–107.
Huson, D. H., Mitra, S., Ruscheweyh, H. J., Weber, N., and Schuster, S. C. (2011). Integrative analysis of environmental sequences using MEGAN4. Genome Res. 21, 1552–1560. doi: 10.1101/gr.120618.111
Imbs, T. I., Silchenko, A. S., Fedoreev, S. A., Isakov, V. V., Ermakova, S. P., and Zvyagintseva, T. N. (2018). Fucoidanase inhibitory activity of phlorotannins from brown algae. Algal Res. 32, 54–59. doi: 10.1016/j.algal.2018.03.009
Ivančić, I., Paliaga, P., Pfannkuchen, M., Djakovac, T., Najdek, M., Steiner, P., et al. (2018). Seasonal variation of extracellular enzymatic activity in marine snow-associated microbial communities and their impact on the surrounding water. FEMS Microbiol. Ecol. 94:fiy198
Jakubovics, N. S., Shields, R. C., Rajarajan, N., and Burgess, J. G. (2013). Life after death: the critical role of extracellular DNA in microbial biofilms. Lett. Appl. Microbiol. 57, 467–475. doi: 10.1111/lam.12134
Karner, M., and Herndl, G. J. (1992). Extracellular enzymatic activity and secondary production in free-living and marine-snow-associated bacteria. Mar. Biol. 113, 341–347.
Kellogg, C. T., and Deming, J. W. (2014). Particle-associated extracellular enzyme activity and bacterial community composition across the Canadian Arctic Ocean. FEMS Microbiol. Ecol. 89, 360–375. doi: 10.1111/1574-6941.12330
Kennedy, J., Flemer, B., Jackson, S. A., Lejon, D. P., Morrissey, J. P., O’gara F., et al. (2010). Marine metagenomics: new tools for the study and exploitation of marine microbial metabolism. Mar. Drugs 8, 608–628. doi: 10.3390/md8030608
Kennedy, J., Marchesi, J. R., and Dobson, A. D. W. (2008). Marine metagenomics: strategies for the discovery of novel enzymes with biotechnological applications from marine environments. Microb. Cell Fact. 7:1. doi: 10.1016/bs.afnr.2016.05.001
Lechtenfeld, O. J., Hertkorn, N., Shen, Y., Witt, M., and Benner, R. (2015). Marine sequestration of carbon in bacterial metabolites. Nat. Commun. 6:6711. doi: 10.1038/ncomms7711
Lei, F., Zhao, Q., Sun-Waterhouse, D., and Zhao, M. (2017). Characterization of a salt-tolerant aminopeptidase from marine Bacillus licheniformis SWJS33 that improves hydrolysis and debittering efficiency for soy protein isolate. Food Chem. 214, 347–353. doi: 10.1016/j.foodchem.2016.07.028
Li, S., Winters, H., Villacorte, L. O., Ekowati, Y., Emwas, A. H., Kennedy, M. D., et al. (2015). Compositional similarities and differences between transparent exopolymer particles (TEPs) from two marine bacteria and two marine algae: significance to surface biofouling. Mar. Chem. 174, 131–140. doi: 10.1016/j.marchem.2015.06.009
Li, Y., Wu, C., Zhou, M., Wang, E. T., Zhang, Z., Liu, W., et al. (2017). Diversity of cultivable protease-producing bacteria in laizhou bay sediments, Bohai Sea, China. Front. Microbiol. 8:405. doi: 10.3389/fmicb.2017.00405
Liu, Q., Fang, J., Li, J., Zhang, L., Xie, B. B., Chen, X. L., et al. (2018). Depth-resolved variations of cultivable bacteria and their extracellular enzymes in the water column of the new britain trench. Front. Microbiol. 9:135. doi: 10.3389/fmicb.2018.00135
Liu, S., and Liu, Z. (2018). Free extracellular enzymes dominate initial peptide hydrolysis in coastal seawater. Mar. Chem. 199, 37–43. doi: 10.1016/j.marchem.2018.01.005
Luo, L., Meng, H., and Gu, J. D. (2017). Microbial extracellular enzymes in biogeochemical cycling of ecosystems. J. Environ. Manage. 197, 539–549. doi: 10.1016/j.jenvman.2017.04.023
Maeda, M., and Taga, N. (1976). Extracellular nuclease produced by a marine bacterium. II. Purification and properties of extracellular nuclease from a marine Vibrio sp. Can. J. Microbiol. 22, 1443–1452. doi: 10.1139/m76-214
Markowitz, V. M., Chen, I. M. A., Chu, K., Szeto, E., Palaniappan, K., Grechkin, Y., et al. (2012). IMG/M: the integrated metagenome data management and comparative analysis system. Nucleic Acids Res. 40:D123-D9.
Mulcahy, H., Charron-Mazenod, L., and Lewenza, S. (2010). Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ. Microbiol. 12, 1621–1629. doi: 10.1111/j.1462-2920.2010.02208.x
Nestle, M., and Roberts, W. (1969). An extracellular nuclease from Serratia marcescens II. Specificity of the enzyme. J. Biol. Chem. 244, 5219–5225.
Neukermans, G., Reynolds, R. A., and Stramski, D. (2016). Optical classification and characterization of marine particle assemblages within the western Arctic Ocean. Limnol. Oceanogr. 61, 1472–1494. doi: 10.1002/lno.10316
Nielsen, K. M., Johnsen, P. J., Bensasson, D., and Daffonchio, D. (2007). Release and persistence of extracellular DNA in the environment. Environ. Biosaf. Res. 6, 37–53. doi: 10.1051/ebr:2007031
Nijland, R., Hall, M. J., and Burgess, J. G. (2010). Dispersal of biofilms by secreted, matrix degrading, bacterial DNase. PLoS One 5:e15668. doi: 10.1371/journal.pone.0015668
Nishino, T., and Morikawa, K. (2002). Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors. Oncogene 21, 9022–9032. doi: 10.1038/sj.onc.1206135
Okshevsky, M., Regina, V. R., and Meyer, R. L. (2015). Extracellular DNA as a target for biofilm control. Curr. Opin. Biotechnol. 33, 73–80. doi: 10.1016/j.copbio.2014.12.002
Palmer, L., Chapple, I., Wright, H., Roberts, A., and Cooper, P. (2012). Extracellular deoxyribonuclease production by periodontal bacteria. J. Periodont. Res. 47, 439–445. doi: 10.1111/j.1600-0765.2011.01451.x
Parab, P., Khandeparker, R., Amberkar, U., and Khodse, V. (2017). Enzymatic saccharification of seaweeds into fermentable sugars by xylanase from marine Bacillus sp. strain BT21. 3 Biotech. 7:296.
Parret, A. H., and De Mot, R. (2002). Bacteria killing their own kind: novel bacteriocins of Pseudomonas and other γ-proteobacteria. Trends Microbiol. 10, 107–112. doi: 10.1016/s0966-842x(02)02307-7
Porschen, R. K., and Sonntag, S. (1974). Extracellular deoxyribonuclease production by anaerobic bacteria. Appl. Microbiol. 27, 1031–1033.
Richter, M., Lombardot, T., Kostadinov, I., Kottmann, R., Duhaime, M. B., Peplies, J., et al. (2008). JCoast - a biologist-centric software tool for data mining and comparison of prokaryotic (meta)genomes. BMC Bioinformatics 9:177. doi: 10.1186/1471-2105-9-177
Rier, S. T., Shirvinski, J. M., and Kinek, K. C. (2014). In situ light and phosphorus manipulations reveal potential role of biofilm algae in enhancing enzyme-mediated decomposition of organic matter in streams. Freshw. Biol. 59, 1039–1051. doi: 10.1111/fwb.12327
Roca, C., Lehmann, M., Torres, C. A., Baptista, S., Gaudêncio, S. P., and Freitas, F., et al. (2016). Exopolysaccharide production by a marine Pseudoalteromonas sp. strain isolated from Madeira Archipelago ocean sediments. New Biotechnol. 33, 460–466. doi: 10.1016/j.nbt.2016.02.005
Salazar, G., and Sunagawa, S. (2017). Marine microbial diversity. Curr. Biol. 27, R489–R494. doi: 10.1016/j.cub.2017.01.017
Sharma, V. K., Kumar, N., Prakash, T., and Taylor, T. D. (2010). MetaBioME: a database to explore commercially useful enzymes in metagenomic datasets. Nucleic Acids Res. 38, D468–D472. doi: 10.1093/nar/gkp1001
Sharma, V. K., and Taylor, T. D. (2015). “MetaBioME: computational tool for mining metagenomic datasets to discover novel biocatalysts by using a homology-based approach,” in Encyclopedia of Metagenomics, ed. K. E. Nelson (Berlin: Springer), 372–383.
Sharp, C., Bray, J., Housden, N. G., Maiden, M. C., and Kleanthous, C. (2017). Diversity and distribution of nuclease bacteriocins in bacterial genomes revealed using hidden markov models. PLoS Comput. Biol. 13:e1005652. doi: 10.1371/journal.pcbi.1005652
Shields, R. C., Mokhtar, N., Ford, M., Hall, M. J., Burgess, J. G., ElBadawey, M. R., et al. (2013). Efficacy of a marine bacterial nuclease against biofilm forming microorganisms isolated from chronic rhinosinusitis. PLoS One 8:e55339. doi: 10.1371/journal.pone.0055339.
Smith, D. C., Simon, M., Alldredge, A. L., and Azam, F. (1992). Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359, 139–142. doi: 10.1038/359139a0
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tetz, G. V., Artemenko, N. K., and Tetz, V. V. (2009). Effect of DNase and antibiotics on biofilm characteristics. Antimicrob. Agents Chemother. 53, 1204–1209. doi: 10.1128/AAC.00471-08
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711. doi: 10.1038/nrmicro1234
Thornton, D. C. (2004). Formation of transparent exopolymeric particles (TEP) from macroalgal detritus. Mar. Ecol. Progr. Ser. 282, 1–12. doi: 10.3354/meps282001
Thornton, D. C. (2014). Dissolved organic matter (DOM) release by phytoplankton in the contemporary and future ocean. Eur. J. Phycol. 49, 20–46. doi: 10.1080/09670262.2013.875596
Torti, A., Lever, M. A., and Jørgensen, B. B. (2015). Origin, dynamics, and implications of extracellular DNA pools in marine sediments. Mar. Genomics 24, 185–196. doi: 10.1016/j.margen.2015.08.007
Tutino, M. L., Parrilli, E., Giaquinto, L., Duilio, A., Sannia, G., Feller, G., et al. (2002). Secretion of alpha-amylase from Pseudoalteromonas haloplanktis TAB23: two different pathways in different hosts. J. Bacteriol. 184, 5814–5817. doi: 10.1128/jb.184.20.5814-5817.2002
Vallenet, D., Belda, E., Calteau, A., Cruveiller, S., Engelen, S., Lajus, A., et al. (2013). MicroScope-an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 41:E636–E47.
Vartoukian, S. R., Palmer, R. M., and Wade, W. G. (2010). Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol. Lett. 309, 1–7. doi: 10.1111/j.1574-6968.2010.02000.x
Veening, J. W., and Blokesch, M. (2017). Interbacterial predation as a strategy for DNA acquisition in naturally competent bacteria. Nat. Rev. Microbiol. 15:621. doi: 10.1038/nrmicro.2017.66
Verdugo, P. (2007). Dynamics of marine biopolymer networks. Polym. Bull. 58, 139–143. doi: 10.1007/s00289-006-0615-2
Verdugo, P., and Santschi, P. H. (2010). Polymer dynamics of DOC networks and gel formation in seawater. Deep Sea Res. Part II Top. Stud. Oceanogr. 57, 1486–1493. doi: 10.1016/j.dsr2.2010.03.002
Vetter, Y. A., and Deming, J. W. (1999). Growth rates of marine bacterial isolates on particulate organic substrates solubilized by freely released extracellular enzymes. Microb. Ecol. 37, 86–94. doi: 10.1007/s002489900133
Vorkapic, D., Pressler, K., and Schild, S. (2016). Multifaceted roles of extracellular DNA in bacterial physiology. Curr. Genet. 62, 71–79. doi: 10.1007/s00294-015-0514-x
Wahl, M., Goecke, F., Labes, A., Dobretsov, S., and Weinberger, F. (2012). The second skin: ecological role of epibiotic biofilms on marine organisms. Front. Microbiol. 3:292. doi: 10.3389/fmicb.2012.00292
Whitchurch, C. B., Tolker-Nielsen, T., Ragas, P. C., and Mattick, J. S. (2002). Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487
Wiese, J., Thiel, V., Nagel, K., Staufenberger, T., and Imhoff, J. F. (2009). Diversity of antibiotic-active bacteria associated with the brown alga Laminaria saccharina from the Baltic Sea. Mar. Biotechnol. 11, 287–300. doi: 10.1007/s10126-008-9143-4
Yamada, Y., Fukuda, H., Tada, Y., Kogure, K., and Nagata, T. (2016). Bacterial enhancement of gel particle coagulation in seawater. Aquat. Microb. Ecol. 77, 11–22. doi: 10.3354/ame01784
Zeng, R. Y., Xiong, P. J., and Wen, J. J. (2006). Characterization and gene cloning of a cold-active cellulase from a deep-sea psychrotrophic bacterium Pseudoalteromonas sp DY3. Extremophiles 10, 79–82. doi: 10.1007/s00792-005-0475-y
Zhang, J. D., and Wiemann, S. (2009). KEGGgraph: a graph approach to KEGG PATHWAY in R and bioconductor. Bioinformatics 25, 1470–1471. doi: 10.1093/bioinformatics/btp167
Keywords: marine gel particle aggregates, bacterial diversity in the ocean, biofilm, extracellularDNA (eDNA), extracellular bacterial nuclease, protobiofilm, extracellular DNA (eDNA), marine enzymes
Citation: Al-Wahaibi ASM, Lapinska E, Rajarajan N, Dobretsov S, Upstill-Goddard R and Burgess JG (2019) Secretion of DNases by Marine Bacteria: A Culture Based and Bioinformatics Approach. Front. Microbiol. 10:969. doi: 10.3389/fmicb.2019.00969
Received: 27 November 2018; Accepted: 16 April 2019;
Published: 07 May 2019.
Edited by:
Hongyue Dang, Xiamen University, ChinaReviewed by:
Verdugo Pedro, University of Washington, United StatesXiu-Lan Chen, Shandong University, China
Copyright © 2019 Al-Wahaibi, Lapinska, Rajarajan, Dobretsov, Upstill-Goddard and Burgess. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: J. Grant Burgess, Z3JhbnQuYnVyZ2Vzc0BuY2wuYWMudWs=
†Present address: Nithyalakshmy Rajarajan, Fujifilm Diosynth Biotechnologies, Billingham, United Kingdom
 Aisha S. M. Al-Wahaibi
Aisha S. M. Al-Wahaibi Emilia Lapinska
Emilia Lapinska Nithyalakshmy Rajarajan
Nithyalakshmy Rajarajan Sergey Dobretsov
Sergey Dobretsov Robert Upstill-Goddard
Robert Upstill-Goddard J. Grant Burgess
J. Grant Burgess