Abstract
Usually, 18–48 h are needed for the identification of microbial pathogens causing urinary tract infections (UTIs) by urine culture. Moreover, antimicrobial susceptibility testing (AST) takes an additional 18–24 h. Rapid identification and AST of the pathogens allow fast and precise treatment. The objective of this study was to shorten the time of diagnosis of UTIs by combining pathogen screening through flow cytometry, microbial identification by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry (MALDI-TOF MS), and AST using the VITEK 2 system for the direct analysis of urine samples. We analyzed 1,638 urine samples from patients with suspected UTIs submitted to the microbiology laboratory for culture. Each urine sample had an approximate volume of 30 mL and was divided into three aliquots. Urine processing included differential centrifugation and two washes to enrich the bacterial fraction for direct MALDI-TOF MS and direct AST. From a total of 1,638 urine samples, 307 were found to be positive through UF-1000i screening. Among them, 265 had significant growth of a single-microorganism. Direct identification was obtained in 229 (86.42%) out of these 265 samples, and no pathogens were misidentified. Moreover, species-level identification was obtained in 163 (88.59%) out of the 184 samples with Gram-negative bacteria, and 27 (38.03%) out of the 71 samples with Gram-positive bacteria. VITEK 2 AST was performed for 117 samples with a single-microorganism. Enterobacteriaceae data showed an agreement rate of antimicrobial categories of 94.83% (1,229/1,296), with minor, major, and very major error rates of 4.17% (54/1,296), 0.92% (12/1,296), and 0.08% (1/1,296), respectively. For Enterococcus spp., the overall categorical agreement was 92.94% (158/170), with a minor error rate of 2.94% (5/170) and major error rate of 4.12% (7/170). The turnaround time of this combined protocol to diagnose UTIs was 1 h for pathogen identification and 6–24 h for AST; noteworthily, only 6–8 h are needed for AST of Enterobacteriaceae using the VITEK 2 system. Overall, our findings show that the combination of flow cytometry, MALDI-TOF MS, and VITEK 2 provided a direct, rapid, and reliable identification and AST method for assessing urine samples, especially for Gram-negative bacterial infections.
Introduction
Urinary tract infections (UTIs) are among the most common community- and hospital-associated bacterial infections; affecting about 150 million persons worldwide each year (Flores-Mireles et al., 2015; McLellan and Hunstad, 2016). Uropathogenic Escherichia coli (UPEC) is the most common causative agent of UTIs, it is responsible for more than 80% of the community-acquired UTIs (Terlizzi et al., 2017). Other pathogens include Klebsiella pneumoniae, Proteus mirabilis, Staphylococcus saprophyticus, Enterococcus faecalis, group B Streptococcus (GBS), Pseudomonas aeruginosa, Staphylococcus aureus, and Candida spp., which are particularly relevant as hospital-acquired and catheter-associated infectious agents (Flores-Mireles et al., 2015; McLellan and Hunstad, 2016). Urine culture remains the gold standard method to identify UTI pathogens, but it is time consuming. In conventional laboratory UTI diagnostics, identification of pathogens takes 18–48 h, and antimicrobial susceptibility testing (AST) needs an additional 18–24 h. Moreover, even though traditional screening methods, such as Gram-staining, flow cytometry, and urine dipstick testing, allow the rapid exclusion of negative samples and a primary identification of pathogens in positive samples (Íñigo et al., 2016), information obtained from these screening methods is insufficient for making decisions regarding antibiotic treatment (Davenport et al., 2017). Patients may be empirically treated with antibiotics (sulfamethoxazole/trimethoprim, nitrofurantoin, or ciprofloxacin), and these may be changed if the results from the AST show that it is needed. However, an inappropriate use of antibiotics may delay effective treatment and contribute to the rise of multidrug-resistant organisms. Therefore, a novel methodology is needed for both timely identification of pathogens and AST.
In the past years, matrix-assisted laser desorption ionisation–time of flight mass spectrometry (MALDI-TOF MS) has been considered as a rapid and trustable technique for identifying microorganisms from culture plates, positive blood cultures, and different clinical samples, such as urine (Ferreira et al., 2011; Clark et al., 2013). Previous studies have shown that urine itself can be used for direct identification by MALDI-TOF MS; besides, several studies have been carried out to improve sample processing and MALDI-TOF MS identification capacity (Ferreira et al., 2010; Wang et al., 2013; Burillo et al., 2014; Íñigo et al., 2016; Zboromyrska et al., 2016; Huang et al., 2017; Zboromyrska et al., 2018). However, these studies have focused on direct identification only, while not having addressed direct AST from urine, especially to accurately determine minimum inhibitory concentrations (MICs). Furthermore, AST is the most important step for the determination of a proper antibiotic treatment and a direct AST method could greatly shorten the time to obtain AST results.
In this study, we investigated a whole MALDI-TOF MS-based workflow to shorten the time of pathogenic diagnosis and effective treatment of UTIs by combining flow cytometry for screening, MALDI-TOF MS for microbial identification, and the VITEK 2 system for AST of urine samples directly.
Materials and Methods
Clinical Samples and Study Design
A total of 1,638 consecutive urine samples without any chemical preservatives were submitted for culture to the Clinical Microbiology Laboratory, Department of Clinical Laboratory, Qilu Hospital of Shandong University (Jinan, China) during September and December 2018, from inpatients and outpatients. As shown in Figure 1, each urine sample had an approximate volume of 30 mL, that was divided into three aliquots using 15 mL sterile centrifuge tubes. The first aliquot was used for culture by a Walk Away Specimen Processor (WASP, Copan, Brescia, Italy), and then for a UF-1000i screening (Sysmex, Kobe, Japan). If the result of bacterial counts was ≥5,000 bacteria/μL, the other two aliquots were used to prepare bacterial pellets. The first one was used for direct identification by the MALDI-TOF MS (Bruker Daltonik GmbH, Bremen, Germany). Once with a reliable identification, the second bacterial pellet was used for direct AST by the VITEK 2 system (bioMérieux, Marcy l’Étoile, France).
FIGURE 1
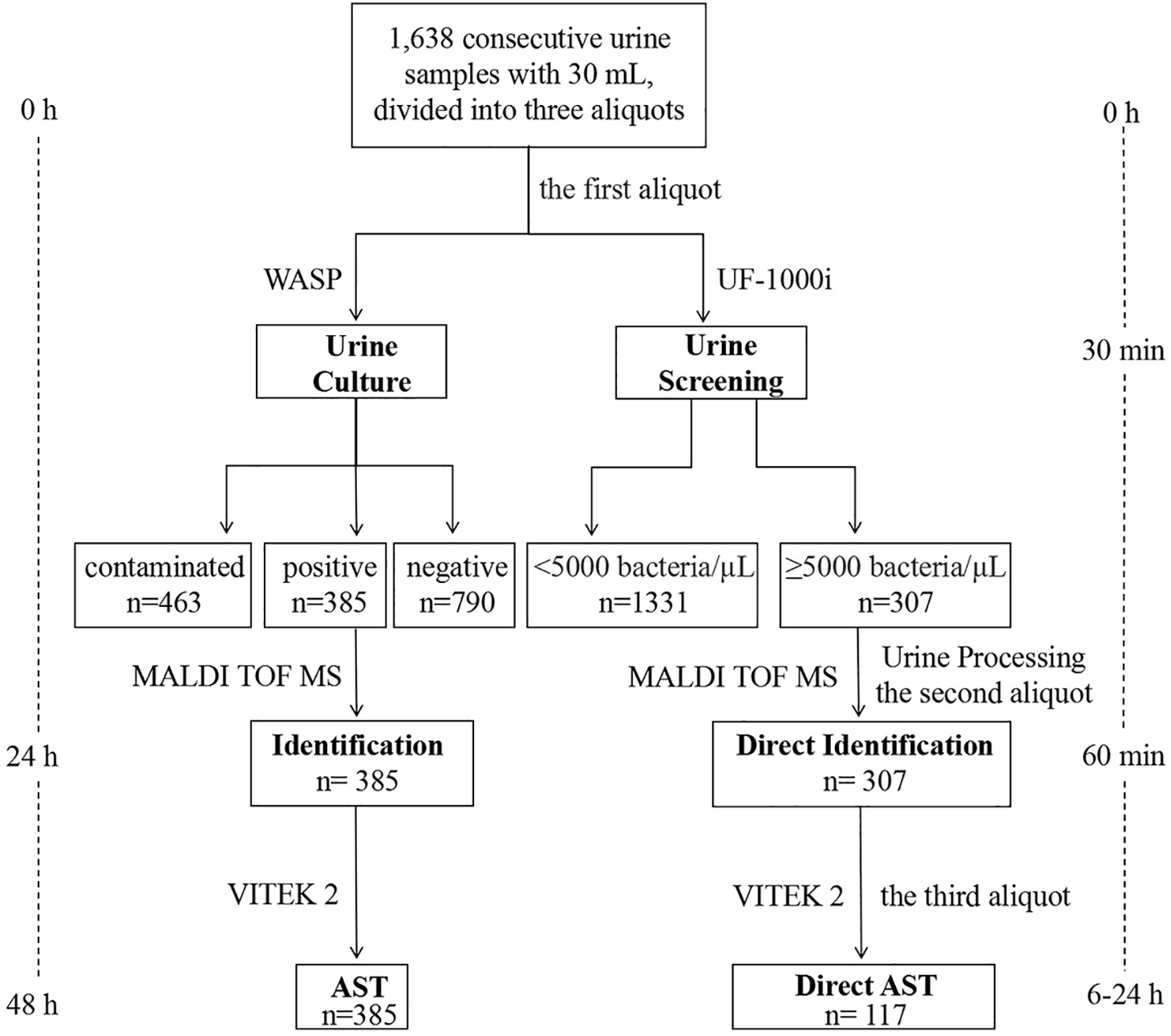
Workflow chart of MALDI-TOF MS-based combined protocol (right) versus conventional culture-dependent protocol (left), showing the turnaround time from arrival of the urine sample in the microbiology laboratory until identification and AST.
Conventional Urine Culture for Pathogen Identification and AST
In our laboratory, all the urine samples submitted for culture were processed by a fully automated plating and streaking instrument, the Copan WASP. The first 10 mL urine aliquot was plated for culturing by the WASP processor using 10 μL sample loops on blood agar and MacConkey agar plates. Plates were incubated aerobically for 24–48 h at 37°C. Urine samples were classified as positive with a UTI pathogen growth >104/mL and a positive leukocyte esterase test result. The recovery of 2 different bacterial species with a similar growth rate, >104/mL, and a positive leukocyte esterase test result was also considered positive. When the bacterial load of one species was >104/mL and lower for the other, the species with a predominant growth was considered clinically significant. However, samples with more than two microorganisms were considered contaminated. Single colonies grown on blood agar plates were counted and the bacterial concentration (CFU/mL) was calculated for each urine sample. Finally, identification was performed by MALDI-TOF MS. Strains with non-reliable identification (NRI) results were confirmed by the VITEK 2 Compact system. The AST was performed on the VITEK 2 Compact system. VITEK cards AST-GN13, AST-GN09, and AST-GP67 were used for Enterobacteriaceae, non-fermenting Gram-negative bacilli, and Staphylococci/Enterococci, respectively. All protocols were performed according to the manufacturers’ instructions. The ATCC strains Escherichia coli 25922, Pseudomonas aeruginosa 27853, Enterococcus faecalis 29212, and Staphylococcus aureus 29213 were used as quality controls of the VITEK 2 Compact System and AST cards to ensure the credibility of AST results.
Flow Cytometry
After WASP processing for culture, the first aliquot was screened for bacteria by using a UF-1000i flow cytometer (Sysmex, Kobe, Japan), according to the manufacturer’s instructions. Samples with bacterial counts of ≥5,000 bacteria/μL were considered as positive for further processing.
Urine Processing and Preparation
The second and third aliquots were vortexed and centrifuged at 2,000 g for 1 min to remove leukocytes, cellular debris and mucus. The resulting supernatant was centrifuged once again at 10,000 g for 5 min, separated from the pellet, and discarded. Bacterial cell pellets were resuspended and washed in 1 mL of 0.45% saline solution (bioMérieux, Marcy l’Étoile, France) and transferred into 1.5 mL microcentrifuge tubes. Once again, bacterial suspensions were pelleted by centrifugation, 10,000 g for 2 min, and washed.
Direct MALDI-TOF MS Identification
The bacterial cell pellet of the second aliquot was dissolved in 50 μL of formic acid 70% by vortexing; then, 50 μL of pure acetonitrile was incorporated into the solution, vortexed, and centrifuged (10,000 g for 2 min) for the following direct MALDI-TOF MS identification assay. In brief, 1 μL supernatant was spotted onto a target plate, and 1 μL of an α-cyano-4-hydroxycinnamic acid (HCCA) matrix solution (Bruker Daltonik GmbH, Bremen, Germany) was added and air dried at room temperature. Identifications were performed by the Bruker microflex MALDI-TOF MS system using the MALDI Biotyper 3.0 RTC database (Bruker Daltonik GmbH, Bremen, Germany). According to the manufacturer, a score of ≥2.0 indicates reliable identification at the species level, while a score between 1.7 and 1.99 indicates a reliable identification at the genus level, and a score below 1.7 indicates a NRI. Thus, these were the criteria used when comparing the results of the direct MALDI-TOF MS identification assay with those from the conventional culture-dependent identification for the evaluation of direct identification performance.
Direct VITEK 2 AST
Once a reliable direct identification by MALDI-TOF MS was achieved, the pellet of the third aliquot was diluted in a saline solution 0.45% and its density adjusted to 0.5 McFarland turbidity standard. Then, it was used for a direct AST by the VITEK-2 Compact System. All protocols were performed according to the manufacturer’s instructions. VITEK cards AST-GN13, AST-GN09, AST-GP67 were used for Enterobacteriaceae, non-fermenting Gram-negative bacilli, and Staphylococci/Enterococci, respectively.
Data Analysis
To compare the results from direct AST with those of conventional culture-dependent AST, the MIC values obtained by both methods were reclassified into clinical categories (susceptible, intermediate, and resistant) according to the Clinical and Laboratory Standards Institute’s (CLSI) clinical breakpoints (Clinical and Laboratory Standards Institute [CLSI], 2018b); the categorical agreement rates as well as the error rates were calculated as recommended by the CLSI M23Ed5 document (Clinical and Laboratory Standards Institute [CLSI], 2018a). For evaluation purposes, we defined the conventional culture-dependent method as our “gold standard” method. We considered that we had a categorical agreement if the results from both AST methods were within the same susceptibility category. The category errors were defined as follows: minor error, bacterial isolates showing an intermediate result by direct AST but either resistant or susceptible by conventional culture-dependent AST, or vice-versa; major error, strains shown to be susceptible by conventional culture-dependent AST but resistant by direct AST (false resistance); very major error, strains were shown to be resistant by conventional culture-dependent AST, but susceptible by direct AST (false susceptibility).
Results
UF-1000i Flow Cytometry Analysis
For this study, 1,638 urine samples submitted to our clinical microbiology laboratory with sufficient volumes were screened for bacteria through a UF-1000i cytometer. Based on a previously established bacterial counts cut-off value of ≥5,000 bacteria/μL (Zboromyrska et al., 2016, 2018), 307 (18.74%) out of 1,638 samples were positive and used for further direct identification. Among these positive samples, 265 had a significant growth of a single microorganism by urine culture, 16 had a significant growth of two microorganisms, 22 were contaminated, and 4 were negative (Table 1). Among the 1,331 urine samples with negative bacterial screening results, 104 were positive by urine culture with a significant growth, 241 were contaminated, and 986 were negative (Table 1). In this study, the omission ratio of urine samples for further direct identification was 6.35% (104/1,638) when the bacterial counts cut-off value was set to ≥5,000 bacteria/μL. From those samples with a concentration <5,000 bacteria/μL, 98 had a significant growth of a single microorganism, including 65 samples with Enterobacteriaceae, 16 with Enterococcus spp., 5 with non-fermenting Gram-negative bacilli, 5 with Staphylococcus spp., and 7 with Candida. Additionally, 6 samples had a significant growth of two microorganisms, including 2 samples with Escherichia coli + Klebsiella pneumoniae, 1 sample with Escherichia coli + Candida glabrata, 1 sample with Candida tropicalis + Enterococcus faecium, 1 sample with Enterococcus avium + Staphylococcus haemolyticus, and 1 sample with Klebsiella pneumoniae + Enterococcus faecalis. Bacterial counts in these 104 urine samples were between 500 and 5,000 bacteria/μL.
Table 1
| UF1000i bacterial count (bacteria/μL) |
||
|---|---|---|
| Culture results | ≥5,000 n (%) | <5,000 n (%) |
| Positive | 281 (91.53) | 104 (7.81) |
| Single-microorganism | 265 (86.32) | 98 (7.36) |
| Two-microorganism | 16 (5.21) | 6 (0.45) |
| Contaminated | 22 (7.17) | 241 (18.11) |
| Negative | 4 (1.3) | 986 (74.08) |
| Total | 307 (100) | 1,331 (100) |
Correlation between UF1000i bacterial count and culture results in 1638 urine samples.
MALDI-TOF MS Direct Identification Results
In this study, 307 out of 1,638 bacterial pellets from urine samples were directly identified by the MALDI-TOF MS. The direct identification results of 265 single-microorganism samples were compared with those from the conventional culture-dependent method. As shown in Table 2, reliable direct identification was obtained in 229 (86.42%) out of 265 single-microorganism samples, while 36 (13.58%) samples were NRI (n = 24) or with no peaks (NP, n = 12). No bacteria were misidentified and species-level identification (score ≥ 2.0) was obtained for 190 out of these 229 samples. Direct identification showed that the dominating microorganisms were Escherichia coli (99/229, 43.23%), Klebsiella pneumoniae (35/229, 15.28%), and Enterococcus spp. (32/229, 13.97%). Among the 184 Gram-negative bacteria samples, 163 (88.59%) showed a score higher than 2, while 17 (9.24%) scored between 1.7 and 2, and 4 (2.17%) had a score below 1.7 or NP (Table 2). Besides, from the 71 Gram-positive bacterial samples, 27 (38.03%) had a score higher than 2, whereas 20 (28.17%) scored between 1.7 and 2, and 24 (33.8%) samples showed a score lower than 1.7 or NP (Table 2). Interestingly, the performance of MALDI-TOF MS direct identification for Staphylococcus spp. was better than that for Enterococcus spp. Reliable direct identification was obtained in 14 out of 15 (93.33%) samples belonging to the genus Staphylococcus, while just 32 out of 51 (62.75%) from the genus Enterococcus were reliably identified. Among the 10 yeast samples found, only 2 were directly identified at the genus level, while 8 samples were NRI (n = 4) or NP (n = 4). As shown in Figure 1, the turn-around time (TAT) of direct MALDI-TOF MS identification was about 1 h.
Table 2
| No. of samples with MS score by culture method |
No. of samples with MS score by direct method |
|||||||
|---|---|---|---|---|---|---|---|---|
| Microorganisms | ≥2.0 | 1.7–1.99 | <1.7 | ≥2.0 | 1.7–1.99 | <1.7 | NP | |
| Gram-negative bacteria (184) | Escherichia coli | 97 | 4 | 92 | 7 | 1 | 1 | |
| Klebsiella pneumoniae | 34 | 2 | 31 | 4 | 1 | |||
| Klebsiella oxytoca | 4 | 4 | ||||||
| Proteus mirabilis | 7 | 6 | 1 | |||||
| Citrobacter koseri | 4 | 4 | ||||||
| Citrobacter freundii | 2 | 2 | ||||||
| Klebsiella aerogenes | 2 | 1 | 1 | |||||
| Enterobacter asburiae | 2 | 2 | ||||||
| Enterobacter cloacae | 6 | 5 | 1 | |||||
| Morganella morganii | 1 | 1 | ||||||
| Serratia marcescens | 4 | 4 | ||||||
| Pseudomonas aeruginosa | 9 | 1 | 7 | 2 | 1 | |||
| Acinetobacter baumannii | 2 | 2 | ||||||
| Acinetobacter pittii | 2 | 2 | ||||||
| Providencia stuartii | 1 | 1 | ||||||
| Gram-positive bacteria (71) | Enterococcus faecium | 26 | 1 | 10 | 9 | 6 | 2 | |
| Enterococcus faecalis | 23 | 1 | 7 | 6 | 8 | 3 | ||
| Staphylococcus epidermidis | 7 | 5 | 1 | 1 | ||||
| Staphylococcus haemolyticus | 4 | 3 | 1 | |||||
| Staphylococcus aureus | 4 | 2 | 2 | |||||
| Streptococcus agalactiae | 3 | 1 | 1 | 1 | 2 | 2 | ||
| Yeast (10) | Candida albicans | 3 | 1 | 1 | 1 | 2 | 2 | |
| Candida glabrata | 2 | 1 | 1 | 1 | 1 | |||
| Candida tropicalis | 1 | 1 | ||||||
| Candida koseri | 1 | 1 | ||||||
| Total | 251 | 12 | 2 | 190 | 39 | 24 | 12 | |
Results of MALDI-TOF MS identification by conventional culture-dependent method and direct method in 265 single-microorganism samples.
MALDI-TOF MS, matrix-assisted laser desorption ionisation time-of-flight mass spectrometry; NP, No peaks.
A total of 22 out of the 1,638 samples had a significant growth of two microorganisms, including 16 samples with bacterial counts ≥5,000 bacteria/μL, and 6 samples with bacterial counts <5,000 bacteria/μL. The 16 samples with counts above the cut-off value were directly identified by the MALDI-TOF MS. Among them, 9 samples corresponded to combined infections by Candida and other microorganism, and their results from the direct identification assays were NPR or NP. In addition, 5 samples had a combined growth of E. coli + K. pneumoniae, 1 sample had a combined growth of E. coli + Proteus mirabilis, and 1 sample had a combined growth of Enterobacter cloacae + Enterococcus faecalis. In addition to a score value, the MALDI Biotyper also provides a list of top ten microorganisms with the highest scores. Interestingly, 3 out of these 7 “second pathogens” were found in the top ten list with score value >1.7, and even >2.0.
VITEK 2 Direct AST Results
Of the 163 Gram-negative bacterial pellets identified at the species level, the first 90, including 72 Enterobacteriaceae and 18 non-fermenting Gram-negative bacilli, were collected consecutively and used for direct VITEK 2 AST testing. Unfortunately, due to financial reasons, the remaining 73 pellets did not undergo direct VITEK 2 AST testing. Results were compared to those from the conventional culture-dependent VITEK 2 AST method; the distribution of category agreement and error rates between the two methods are shown in Table 3. A total of 1,296 antimicrobial tests were analyzed in the samples of Enterobacteriaceae. There was an overall category agreement rate of 94.83% (1,229/1,296) between the two methods, with minor error rate of 4.17% (54/1,296), major error rate of 0.92% (12/1,296), and very major error rate of 0.08% (1/1,296). The minor errors were mainly found in ampicillin/sulbactam (15.28%), tobramycin (13.89%), cefepime (12.5%), nitrofurantoin (12.5%), imipenem (6.94%), and ceftazidime (5.56%), while the major errors mainly occurred for cefazolin (2.78%), cefepime (2.78%), aztreonam (2.78%), and ertapenem (2.78%). The very major error rate was just found in aztreonam (1.39%). Interestingly, the category agreement rates between the two methods of group U (“urine”) antimicrobial agents, recommended by the CLSI M100 document for UTIs primarily treatment (Clinical and Laboratory Standards Institute [CLSI], 2018b), were very high, 100% for ciprofloxacin, levofloxacin, and sulfamethoxazole/trimethoprim (SMZ/TMP), 97.22% for cefazolin, and 84.72% for nitrofurantoin. A total of 378 antimicrobial tests were analyzed in the samples of the non-fermenting Gram-negative bacilli. There was an overall category agreement of 94.44% (357/378), with rates for minor errors of 2.91% (11/378), major errors of 1.59% (6/378), and very major errors of 1.06% (4/378). The minor errors were mainly found in imipenem (16.67%), meropenem (11.11%), ceftazidime (11.11%), piperacillin (5.56%), cefepime (5.56%), and gentamicin (5.56%), while the major errors mainly occurred for aztreonam (16.67%), piperacillin (5.56%), ceftriaxone (5.56%), and ciprofloxacin (5.56%). The very major errors were found in ceftriaxone (5.56%), imipenem (5.56%), nitrofurantoin (5.56%), and SMZ/TMP (5.56%).
Table 3
| Microorganisms | Antimicrobial agents | No. | Category agreement | Minor error | Major error | Very major error | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Enterobacteriaceae (n = 72) | Ampicillin | 72 | 71 | 98.61% | 1 | 1.39% | ||||
| Ampicillin/Sulbactam | 72 | 61 | 84.72% | 11 | 15.28% | |||||
| Piperacillin/Tazobactam | 72 | 71 | 98.61% | 1 | 1.39% | |||||
| Cefazolin | 72 | 70 | 97.22% | 2 | 2.78% | |||||
| Cefotetan | 72 | 71 | 98.61% | 1 | 1.39% | |||||
| Ceftazidime | 72 | 68 | 94.44% | 4 | 5.56% | |||||
| Ceftriaxone | 72 | 72 | 100% | |||||||
| Cefepime | 72 | 61 | 84.72% | 9 | 12.5% | 2 | 2.78% | |||
| Aztreonam | 72 | 67 | 93.05% | 2 | 2.78% | 2 | 2.78% | 1 | 1.39% | |
| Ertapenem | 72 | 70 | 97.22% | 2 | 2.78% | |||||
| Imipenem | 72 | 67 | 93.05% | 5 | 6.95% | |||||
| Amikacin | 72 | 70 | 97.22% | 2 | 2.78% | |||||
| Gentamicin | 72 | 71 | 98.61% | 1 | 1.39% | |||||
| Tobramycin | 72 | 62 | 86.11% | 10 | 13.89% | |||||
| Ciprofloxacin | 72 | 72 | 100% | |||||||
| Levofloxacin | 72 | 72 | 100% | |||||||
| Nitrofurantoin | 72 | 61 | 84.72% | 9 | 12.5% | 2 | 2.78% | |||
| Sulfamethoxazole/Trimethoprim | 72 | 72 | 100% | |||||||
| Non-fermenting Gram-negative bacilli (n = 18) | Ampicillin | 18 | 18 | 100% | ||||||
| Ampicillin/Sulbactam | 18 | 18 | 100% | |||||||
| Piperacillin | 18 | 16 | 88.89% | 1 | 5.56% | 1 | 5.56% | |||
| Piperacillin/Tazobactam | 18 | 18 | 100% | |||||||
| Cefazolin | 18 | 18 | 100% | |||||||
| Cefuroxime | 18 | 18 | 100% | |||||||
| Cefuroxime Axetil | 18 | 18 | 100% | |||||||
| Cefotetan | 18 | 18 | 100% | |||||||
| Ceftazidime | 18 | 16 | 88.89% | 2 | 11.11% | |||||
| Ceftriaxone | 18 | 16 | 88.89% | 1 | 5.56% | 1 | 5.56% | |||
| Cefepime | 18 | 17 | 94.44% | 1 | 5.56% | |||||
| Aztreonam | 18 | 14 | 77.78% | 1 | 5.56% | 3 | 16.67% | |||
| Imipenem | 18 | 14 | 77.78% | 3 | 16.67% | 1 | 5.56% | |||
| Meropenem | 18 | 16 | 88.89% | 2 | 11.11% | |||||
| Amikacin | 18 | 18 | 100% | |||||||
| Gentamicin | 18 | 17 | 94.44% | 1 | 5.56% | |||||
| Tobramycin | 18 | 18 | 100% | |||||||
| Ciprofloxacin | 18 | 17 | 94.44% | 1 | 5.56% | |||||
| Levofloxacin | 18 | 18 | 100% | |||||||
| Nitrofurantoin | 18 | 17 | 94.44% | 1 | 5.56% | |||||
| Sulfamethoxazole/Trimethoprim | 18 | 17 | 94.44% | 1 | 5.56% | |||||
Categorical agreement and errors for direct AST compared with culture-dependent AST in Gram-negative bacteria.
In addition, 27 Gram-positive bacterial pellets identified at the species level, consisting of 10 Staphylococcus spp. and 17 Enterococcus spp., were used for VITEK 2 direct AST analysis. The distribution of both category agreement and error rates between the direct AST and culture-dependent AST are presented in Table 4. For Staphylococcus spp. with a small sample size, the overall categorical agreement between the two methods was 94.38% (151/160), with rates for minor errors of 3.12% (5/160), major errors of 1.87% (3/160), and very major errors of 0.63% (1/160). High minor error rates were detected for ciprofloxacin (20%), levofloxacin (10%), moxifloxacin (10%), and erythromycin (10%); high major error rates were also observed for oxacillin (10%), ciprofloxacin (10%), and vancomycin (10%), and a high very major error rate was found for moxifloxacin (10%). However, it is important to point out that the category agreement rates of group U antimicrobial agents (nitrofurantoin and SMZ/TMP) between the two methods were of 100%. For Enterococcus spp., the overall categorical agreement between the two methods was 92.94% (158/170), with rates for minor errors of 2.94% (5/170), and for major errors of 4.12% (7/170). The minor errors were mainly found in ciprofloxacin (5.88%), levofloxacin (5.88%), linezolid (5.88%), vancomycin (5.88%), and nitrofurantoin (5.88%), while the major errors were mainly associated to tetracycline (17.65%), ampicillin (5.88%), linezolid (5.88%), vancomycin (5.88%), and nitrofurantoin (5.88%).
Table 4
| Microorganisms | Antimicrobial agents | No. | Category agreement | Minor error | Major error | Very major error | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cefoxitin screen | 10 | 10 | 100% | |||||||
| Staphylococcus ssp. (n = 10) | Benzylpenicillin | 10 | 10 | 100% | ||||||
| Oxacillin | 10 | 9 | 90% | 1 | 10% | |||||
| Gentamicin | 10 | 10 | 100% | |||||||
| Ciprofloxacin | 10 | 7 | 70% | 2 | 20% | 1 | 10% | |||
| Levofloxacin | 10 | 9 | 90% | 1 | 10% | |||||
| Moxifloxacin | 10 | 8 | 80% | 1 | 10% | 1 | 10% | |||
| Erythromycin | 10 | 9 | 90% | 1 | 10% | |||||
| Clindamycin | 10 | 10 | 100% | |||||||
| Quinupristin-dalfopristin | 10 | 10 | 100% | |||||||
| Linezolid | 10 | 10 | 100% | |||||||
| Vancomycin | 10 | 9 | 90% | 1 | 10% | |||||
| Tetracycline | 10 | 10 | 100% | |||||||
| Nitrofurantoin | 10 | 10 | 100% | |||||||
| Rifampicin | 10 | 10 | 100% | |||||||
| Trimethoprim/Sulfamethoxazole | 10 | 10 | 100% | |||||||
| Enterococcus ssp. (n = 17) | Benzylpenicillin | 17 | 17 | 100% | ||||||
| Ampicillin | 17 | 16 | 94.12% | 1 | 5.88% | |||||
| Ciprofloxacin | 17 | 16 | 94.12% | 1 | 5.88% | |||||
| Levofloxacin | 17 | 16 | 94.12% | 1 | 5.88% | |||||
| Rifampicin | 17 | 17 | 100% | |||||||
| Quinupristin-dalfopristin | 17 | 17 | 100% | |||||||
| Linezolid | 17 | 15 | 88.24% | 1 | 5.88% | 1 | 5.88% | |||
| Vancomycin | 17 | 15 | 88.24% | 1 | 5.88% | 1 | 5.88% | |||
| Tetracycline | 17 | 14 | 82.35% | 3 | 17.65% | |||||
| Nitrofurantoin | 17 | 15 | 88.24% | 1 | 5.88% | 1 | 5.88% | |||
Categorical agreement and error rates for direct AST compared with culture-dependent AST in Gram-positive bacteria.
Discussion
In recent years, MALDI-TOF MS-based combination methods have demonstrated good stability, accuracy, and speed in the direct identification and AST of pathogens present in clinical specimens, such as blood and sterile body fluids (Tian et al., 2016; Di Gaudio et al., 2018; Wu et al., 2019). In our previous study, we developed a fast and easy lysis-centrifugation-wash protocol to prepare bacterial pellets from blood cultures positive for the presence of microbial pathogens, that can be used for direct pathogen identification by MALDI-TOF MS and for direct AST using the VITEK 2 system (Pan et al., 2018). It allows clinically helpful results to be reported from days to hours after a positive blood culture system alarm. However, blood and sterile body fluids need to be injected into blood culture bottles and incubated for several, even dozens of, hours to get a positive alarm, while urine, especially morning urine, is a sample that is naturally incubated inside the bladder. This makes it possible to identify pathogens directly from the urine of patients presenting UTIs, without spending time on incubation.
The purpose of this study was to develop a MALDI-TOF MS-based workflow to shorten the time needed for microbial identification and for obtaining AST results for UTIs. The performance and optimisation of a combination of flow cytometry and MALDI-TOF MS for screening and direct identification of pathogens in urine has been reported previously; several other procedures have been described as well. Initially, Ferreira et al. (2010) established a direct identification method based on flow cytometry screening and MALDI-TOF MS. Sánchez-Juanes et al. (2014) added a pre-treatment step with SDS to this, to lyse cells and release microorganisms, in order to enhance the method’s sensitivity. Veron et al. (2015) performed a comparison between three different methods, and found that the previous 5 h culturing and dual-filtration methods provided the better identification results. Kitagawa et al. (2018) added ultrasonication for 10 min at the beginning of the procedure, followed by centrifugation, to disperse bacterial cell aggregation. Íñigo et al. (2016) established that using a 2 mL volume of urine was optimal for sample manipulation in Eppendorf tubes and centrifuges. While Zboromyrska et al. used 10 mL of urine with bacterial counts of ≥5,000 bacteria/μL for MALDI-TOF MS direct identification and obtained high scores (Zboromyrska et al., 2016). Moreover, these parameters were confirmed by a multicentre study that followed soon after (Zboromyrska et al., 2018).
In this study, we suggest the usage of a differential centrifugation method to prepare bacterial pellets for analysis from a starting urine volume of 30 mL (10 mL for screening, 10 mL for identification, and 10 mL for AST). This is easily achievable as urine is one of the most easily collected clinical samples and 15 mL centrifuge tubes and centrifuges are routinely available in clinical microbiology laboratories. We also confirmed the accuracy of the pre-established cut-off value of ≥5,000 bacteria/μL for bacterial counts as a sample selection criterium for direct identification by MALDI-TOF MS. In this study, flow cytometry analysis results showed that 18.74% (307/1,638) of urine samples were positive for infections and should be used for further direct identification tests, and that the omission ratio of positive culture samples was 6.35% (104/1,638).
Overall, a reliable direct identification was obtained in 86.42% of the single-microorganism samples, in accordance with previous studies (Zboromyrska et al., 2016). Until now, reliable identification of Gram-negative bacteria has proven to be more efficient than that of Gram-positive bacteria, with 97.83% versus 66.2% bacteria identified reliably, respectively. Moreover, the performance of MALDI-TOF MS direct identification for Staphylococcus spp. (93.33%) was better than that for Enterococcus spp. (62.75%). This may be partly explained by the unsuccessful removal of urine cells during sample processing by the differential centrifugation method to prepare pellets of Enterococcus spp. In some urine samples with significant growth of Enterococcus spp., the direct identification results were either NRI or NP, so we prepared a smear using these pellets. After Gram-staining, a certain amount of epithelial cells and leukocytes was found to still be present in the smear (data not shown). Poor performance was also observed in the identification of yeasts; only 2 out of 10 samples were directly identified at the genus-level, in accordance with a study by Burillo and better than results from Íñigo (Burillo et al., 2014; Íñigo et al., 2016). Therefore, direct identification of yeasts from urine samples still constitutes a challenge for achieving quick and efficient diagnosis.
Compared to the conventional culture-dependent VITEK 2 AST, the overall category agreement of the direct AST was 94.56%, with 94.83% for Enterobacteriaceae, 94.44% for the non-fermenting Gram-negative bacilli, 94.38% for Staphylococcus spp., and 92.94% for Enterococcus spp. With respect to Enterobacteriaceae, the most common causative agent of UTIs, a very good concordance was observed between the two AST methods for the group U or first-line antimicrobial agents, which yielded essential agreement rates of 100% for ciprofloxacin, levofloxacin, SMZ/TMP, and ceftriaxone, and 97.22% for cefazolin. However, an unacceptable minor error rate (12.5%) was obtained for nitrofurantoin.
For Staphylococcus spp. and Enterococcus spp., the error rates of several antimicrobial agents exceeded CLSI recommendations (Clinical and Laboratory Standards Institute [CLSI], 2018a). These results may be due to the small sample size obtained for these bacteria in our study (n = 10 and n = 17, respectively). The small number of Staphylococcus spp. and Enterococcus spp. samples obtained for direct AST analysis may be because of the high bacterial count cut-off value used in this study, which is more frequently achieved by Gram-negative bacteria than by Gram-positive bacteria (Monsen and Rydén, 2015; Zboromyrska et al., 2016). Moreover, it is important to point out that the category agreement rates of nitrofurantoin and SMZ/TMP against Staphylococcus spp. were 100%.
The minor and major errors calculated for vancomycin and linezolid may be due to urine sample contamination with a small amount of Gram-negative bacilli, whose growth on plates may have been covered by the growth of Gram-positive bacteria. The number of these Gram-negative bacilli could have been so small that they were not observed on plates nor directly detected by MALDI-TOF MS. Wang et al. analyzed urine specimens containing different E. coli and Enterococcus faecium proportion ratios by MALDI-TOF MS, and showed that E. coli and Enterococcus faecium were simultaneously detected if the mixture ratio was 1:1 or 1:2, but only the dominant pathogen was detected at mixture ratio of 1:9 (Wang et al., 2013).
Our urine processing methodology includes differential centrifugation and two washes to enrich the bacterial fraction for MALDI-TOF MS and AST assays. The entire process can deliver identification results within 1 h. For the most common pathogens of UTIs, Enterobacteriaceae, it usually takes 6–8 h to yield a final AST report using the VITEK 2 system. Therefore, our MALDI-TOF MS-based combined protocol could provide reliable “same day” reports including identification and ASTs for Enterobacteriaceae, which are important for a quick treatment and to improve clinical outcomes of UTIs. Patients with uncontrolled UTIs are likely to develop secondary blood stream infections, even urosepsis, especially those patients with obstructive uropathy and wearing invasive devices, such as catheters (Dreger et al., 2015). Early identification of pathogens responsible for UTIs and early treatments based on results from fast AST are the best tools for urosepsis prevention (Wagenlehner et al., 2017). The “same day” report achieved through our protocol may result in early pathogenic diagnoses and early treatment of UTIs, reducing the incidence of urinary sepsis.
There are nevertheless some limitations to our method that should be considered for future applications, such as the high starting volume of urine, the need for bacterial counts ≥ 5,000 bacteria/μL, and the exclusivity of application to monomicrobial infections. Additionally, our study has been limited by the reduced number of samples analyzed and the small number of samples found for some bacterial groups, such as Staphylococcus, Enterococcus, and non-fermenting Gram-negative bacilli.
In conclusion, combining flow cytometry, MALDI-TOF MS, and the VITEK 2 system allows the reliable identification and AST of UTI pathogens directly from urine samples, especially in cases of Gram-negative bacterial infections.
Statements
Data availability statement
All datasets generated for this study are included in the manuscript.
Ethics statement
This study was reviewed and approved by the research ethics committee of Qilu Hospital of Shandong University (protocol KYLL-2019-277). The participants provided a written informed consent when their urine samples were included in the study.
Author contributions
CW, YZ, ES, WL, and HP designed the experiments. WL, YW, and HP performed the experiments. YL and CL helped perform the experiments. WL and CW analyzed the data. XZ and LD helped analyze the data. WL and ES wrote the manuscript. All the authors read and approved the final version of the manuscript.
Funding
The work was supported by the National Natural Science Foundation of China (81702066, 81401709), the National Key Research and Development Program of China (2018YFC0114700), the Taishan Scholar Program of Shandong Province, the Key Research and Development Program of Shandong Province (2018YFJH0505), and Natural Science Foundation of Shandong Province (ZR2015PH049).
Acknowledgments
We thank Xiaoli Zhang, Hongxia Zhou and Yue Wu for their help during carrying out the experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Burillo A. Rodríguez-Sánchez B. Ramiro A. Cercenado E. Rodríguez-Créixems M. Bouza E. (2014). Gram-stain plus MALDI-TOF MS (Matrix-assisted laser desorption ionization-time of flight mass spectrometry) for a rapid diagnosis of urinary tract infection.PLoS One9:e86915. 10.1371/journal.pone.0086915
2
Clark A. E. Kaleta E. J. Arora A. Wolk D. M. (2013). Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology.Clin. Microbiol. Rev.26547–603. 10.1128/CMR.00072-12
3
Clinical and Laboratory Standards Institute [CLSI] (2018a). Development of in Vitro Susceptibility Testing Criteria and Quality Control Parameters, 5th Edn.Wayne, PA: CLSI.
4
Clinical and Laboratory Standards Institute [CLSI] (2018b). Performance Standards for Antimicrobial Susceptibility Testing; 28th Informational Supplement M100-S28.Wayne, PA: CLSI.
5
Davenport M. Mach K. E. Shortliffe L. M. D. Banaei N. Wang T. H. Liao J. C. (2017). New and developing diagnostic technologies for urinary tract infections.Nat. Rev. Urol.14296–310. 10.1038/nrurol.2017.20
6
Di Gaudio F. Indelicato S. Indelicato S. Tricoli M. R. Stampone G. Bongiorno D. (2018). Improvement of a rapid direct blood culture microbial identification protocol using MALDI-TOF MS and performance comparison with SepsiTyper kit.J. Microbiol Methods.1551–7. 10.1016/j.mimet.2018.10.015
7
Dreger N. M. Degener S. Ahmad-Nejad P. Wöbker G. Roth S. (2015). Urosepsis–etiology, diagnosis, and treatment.Dtsch. Arztebl. Int.112837–847. 10.3238/arztebl.2015.0837
8
Ferreira L. Sánchez-Juanes F. González-Avila M. Cembrero-Fuciños D. Herrero-Hernández A. González-Buitrago J. M. et al (2010). Direct identification of urinary tract pathogens from urine samples by matrix-assisted laser desorption ionization-time of flight mass spectrometry.J. Clin. Microbiol.482110–2115. 10.1128/JCM.02215-09
9
Ferreira L. Sánchez-Juanes F. Muñoz-Bellido J. L. González-Buitrago J. M. (2011). Rapid method for direct identification of bacteria in urine and blood culture samples by matrix-assisted laser desorption ionization time-of-flight mass spectrometry: intact cell vs. extraction method.Clin. Microbiol. Infect.171007–1012. 10.1111/j.1469-0691.2010.03339.x
10
Flores-Mireles A. L. Walker J. N. Caparon M. Hultgren S. J. (2015). Urinary tract infections: epidemiology, mechanisms of infection and treatment options.Nat. Rev. Microbiol.13269–284. 10.1038/nrmicro3432
11
Huang B. Zhang L. Zhang W. Liao K. Zhang S. Zhang Z. et al (2017). Direct detection and identification of bacterial pathogens from urine with optimized specimen processing and enhanced testing algorithm.J. Clin. Microbiol.551488–1495. 10.1128/JCM.02549-16
12
Íñigo M. Coello A. Fernández-Rivas G. Rivaya B. Hidalgo J. Quesada M. D. et al (2016). Direct identification of urinary tract pathogens from urine samples, combining urine screening methods and matrix-assisted laser desorption ionization-time of flight mass spectrometry.J. Clin. Microbiol.54988–993. 10.1128/JCM.02832-15
13
Kitagawa K. Shigemura K. Onuma K. I. (2018). Improved bacterial identification directly from urine samples with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.J. Clin. Lab. Anal.32:e22301. 10.1002/jcla.22301
14
McLellan L. K. Hunstad D. A. (2016). Urinary tract infection: pathogenesis and outlook.Trends Mol. Med.22946–957. 10.1016/j.molmed.2016.09.003
15
Monsen T. Rydén P. (2015). Flow cytometry analysis using sysmex UF-1000i classifies uropathogens based on bacterial, leukocyte, and erythrocyte counts in urine specimens among patients with urinary tract infections.J. Clin. Microbiol.53539–545. 10.1128/JCM.01974-14
16
Pan H. W. Li W. Li R. G. Li Y. Zhang Y. Sun E. H. (2018). Simple sample preparation method for direct microbial identification and susceptibility testing from positive blood cultures.Front. Microbiol.9:481. 10.3389/fmicb.2018.00481
17
Sánchez-Juanes F. Siller Ruiz M. Moreno Obregón F. Criado González M. Hernández Egido S. de Frutos Serna M. et al (2014). Pretreatment of urine samples with SDS improves direct identification of urinary tract pathogens with matrix-assisted laser desorption ionization-time of flight mass spectrometry.J. Clin. Microbiol.52335–338. 10.1128/JCM.01881-13
18
Terlizzi M. E. Gribaudo G. Maffei M. E. (2017). UroPathogenic Escherichia coli (UPEC) infections: virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies.Front. Microbiol.8:1566. 10.3389/fmicb.2017.01566
19
Tian Y. Zheng B. Wang B. Lin Y. Li M. (2016). Rapid identification and multiple susceptibility testing of pathogens from positive-culture sterile body fluids by a combined MALDI-TOF mass spectrometry and vitek susceptibility system.Front. Microbiol.7:523. 10.3389/fmicb.2016.00523
20
Veron L. Mailler S. Girard V. Muller B. H. L’Hostis G. Ducruix C. et al (2015). Rapid urine preparation prior to identification of uropathogens by MALDI-TOF MS.Eur. J. Clin. Microbiol. Infect. Dis.341787–1795. 10.1007/s10096-015-2413-y
21
Wagenlehner F. M. Tandogdu Z. Bjerklund Johansen T. E. (2017). An update on classification and management of urosepsis.Curr. Opin. Urol.27133–137. 10.1097/MOU.0000000000000364
22
Wang X. H. Zhang G. Fan Y. Y. Yang X. Sui W. J. Lu X. X. (2013). Direct identification of bacteria causing urinary tract infections by combining matrix-assisted laser desorption ionization-time of flight mass spectrometry with UF-1000i urine flow cytometry.J. Microbiol. Methods92231–235. 10.1016/j.mimet.2012.12.016
23
Wu S. Xu J. Qiu C. Xu L. Chen Q. Wang X. (2019). Direct antimicrobial susceptibility tests of bacteria and yeasts from positive blood cultures by using serum separator gel tubes and MALDI-TOF MS.J. Microbiol. Methods15716–20. 10.1016/j.mimet.2018.12.011
24
Zboromyrska Y. Bosch J. Aramburu J. Cuadros J. García-Riestra C. Guzmán-Puche J. et al (2018). A multicentre study investigating parameters which influence direct bacterial identification from urine.PLoS One13:e0207822. 10.1371/journal.pone.0207822
25
Zboromyrska Y. Rubio E. Alejo I. Vergara A. Mons A. Campo I. et al (2016). Development of a new protocol for rapid bacterial identification and susceptibility testing directly from urine samples.Clin. Microbiol. Infect.22561.e1–561.e6. 10.1016/j.cmi.2016.01.025
Summary
Keywords
bacterial identification, antimicrobial susceptibility, MALDI-TOF MS, urinary tract infections, rapid diagnosis, urine
Citation
Li W, Sun E, Wang Y, Pan H, Zhang Y, Li Y, Zhang X, Li C, Du L and Wang C (2019) Rapid Identification and Antimicrobial Susceptibility Testing for Urinary Tract Pathogens by Direct Analysis of Urine Samples Using a MALDI-TOF MS-Based Combined Protocol. Front. Microbiol. 10:1182. doi: 10.3389/fmicb.2019.01182
Received
03 March 2019
Accepted
09 May 2019
Published
05 June 2019
Volume
10 - 2019
Edited by
Bing Gu, Affiliated Hospital of Xuzhou Medical University, China
Reviewed by
Omai Garner, University of California, Los Angeles, United States; Giovanni Gherardi, Campus Bio-Medico University, Italy
Updates
Copyright
© 2019 Li, Sun, Wang, Pan, Zhang, Li, Zhang, Li, Du and Wang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuanxin Wang, cxwang@sdu.edu.cn
This article was submitted to Infectious Diseases, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.