- 1Infectious Diseases and Tropical Medicine Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Department of Microbiology, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 3Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 4Infectious Diseases Research Center, Golestan University of Medical Sciences, Gorgan, Iran
- 5Department of Microbiology, Faculty of Medicine, Golestan University of Medical Sciences, Gorgan, Iran
- 6Department of Mycobacteriology and Pulmonary Research, Pasteur Institute of Iran, Tehran, Iran
- 7Department of Microbiology, School of Medicine, Babol University of Medical Sciences, Babol, Iran
- 8Microbiology Department, School of Medicine, Gonabad University of Medical Sciences, Gonabad, Iran
- 9Cellular and Molecular Research Center, Qom University of Medical Sciences, Qom, Iran
Stenotrophomonas maltophilia is an environmental Gram-negative bacterium that has rapidly emerged as an important nosocomial pathogen in hospitalized patients. Treatment of S. maltophilia infections is difficult due to increasing resistance to multiple antibacterial agents. The purpose of this study was to determine the phenotypic and genotypic characterization of S. maltophilia isolates recovered from patients referred to several hospitals. A total of 164 clinical isolates of S. maltophilia were collected from hospitals in various regions in Iran between 2016 and 2017. Antibiotic susceptibility testing was performed by disc diffusion method and E-test assay according to the Clinical and Laboratory Standards Institute (CLSI) guideline. The ability of biofilm formation was assessed with crystal violet staining and then, biofilm-associated genes were investigated by PCR-sequencing method. The presence of L1 (a metallo-β-lactamase), L2 (a clavulanic acid-sensitive cephalosporinase), sul1 and sul2 (resistance to Trimethoprim/Sulfamethoxazole), Smqnr (intrinsic resistance to quinolones), and dfrA genes (dihydrofolate reductase enzyme that contributes to trimethoprim resistance) was also examined by PCR-sequencing. Relative gene expression of smeDEF efflux pump was assessed by real-time PCR. Genotyping was performed using the multi-locus sequencing typing (MLST) and repetitive extragenic palindromic-PCR (Rep-PCR). Isolates were resistant to imipenem (100%), meropenem (96%), doripenem (96%), and ceftazidime (36.58%). Notably, 5 (3.04%) isolates showed resistant to trimethoprim-sulfamethoxazole (TMP-SMX), an alarming trend of decreased susceptibility to TMP-SMX in Iran. Minocycline and levofloxacin exhibited the highest susceptibility of 91.46 and 99.39%, respectively. Using the crystal violet staining, 157 (95.73%) isolates had biofilm phenotype: 49 (29.87%), 63 (38.41%), and 45 (27.43%) isolates were categorized as strong-, moderate- and weak-biofilm producer while 7 isolates (4.26%) were identified a non-biofilm producer. Biofilm genes had an overall prevalence of 145 (88.41%), 137 (83.53%), and 164 (100%) of rmlA, rpfF, and spgM, respectively. L1, L2, Smqnr, sul1, and sul2 resistance genes were detected in 145 (88.41%), 156 (96.12%), 103 (62.80%), 89 (54.26%), and 92 (56.09%) isolates, respectively. None of the S. maltophilia isolates were positive for dfrA12, dfrA17, and dfrA27 genes. Gene expression analysis showed that smeD efflux system was overexpressed in two out of the five clinical isolates (40%) that showed resistance to TMP-SMX. Most of the isolates were genetically unrelated. Two new sequence types (ST139 and ST259) were determined. Our results showed that TMP-SMX was still an effective antibiotic against S. maltophilia. The findings of the current study revealed an increasing prevalence of antibiotic resistance and biofilm genes in clinical S. maltophilia isolates in Iran.
Introduction
The genus Stenotrophomonas, together with Xanthomonas, belongs to the γ-β subclass of proteobacteria (Anzai et al., 2000). S. maltophilia isolated in 1943 from pleural effusion of patients was first named as Bacterium bookeri. Later, it was reclassified as a member of the genera Pseudomonas and Xanthomonas in 1961 and 1983, respectively, until it was classified as a new genus, Stenotrophomonas, in 1993 (Al-Anazi and Al-Jasser, 2014).
S. maltophilia is a Gram-negative, non-fermentative, aerobic, motile bacillus that is abundant in the ubiquitous environment with a broad geographical distribution. This organism has emerged as an important opportunistic pathogen in humans worldwide. Although it is considered to have limited pathogenicity (Di Bonaventura et al., 2010), S. maltophilia causes various types of hospital- and community-acquired infections, especially in debilitated or immunocompromised patients, with the mortality rate of 37.5% (Falagas et al., 2009). The bacterium has been increasingly recognized as responsible for a number of clinical syndromes, such as pneumonia, sepsis, bacteremia, endocarditis, septic arthritis, meningitis, endophthalmitis, and urinary infections (Looney et al., 2009; Sumida et al., 2015; Hu et al., 2016).
During the last decade, S. maltophilia has been considered as one of the leading multi-drug resistant (MDR) organisms in hospital settings due to exhibiting high levels of intrinsic and acquired resistance to a broad array of antibacterial agents, including fluoroquinolones, aminoglycosides, and the most common of β-lactam antibiotics (Brooke, 2014). Different types of antimicrobial resistance mechanisms, such as expression of antibiotic hydrolyzing or modifying enzymes, membrane permeability alteration (Hu et al., 2008), and multi-drug efflux systems (Huang et al., 2014) have been identified in S. maltophilia.
This bacterium produces two chromosomal-mediated inducible β-lactamases, known as L1 and L2. The L1 belongs to molecular class B Zn2+-dependent metallo-β-lactamase (MBL), is resistant to clavulanic acid and hydrolyses carbapenems, cephalosporins, and penicillins (Brooke, 2012; Chang et al., 2015). The L2 serine-β-lactamase, an Ambler class A enzyme, is an inducible cephalosporinase that hydrolyses cephalosporins, penicillins, and aztreonam (Flores-Trevino et al., 2014; Mojica et al., 2016). Two mechanisms are associated with resistance to quinolones among S. maltophilia strains, including smeDEF, smeIJK, smeABC, and smeVWX efflux pumps and a novel chromosomal quinolone resistance gene, Smqnr, encoding the pentapeptide repeat protein that protects both topoisomerase IV and gyrase from the quinolones (Sanchez et al., 2009; Chang et al., 2015; Kanamori et al., 2015).
Trimethoprim-sulfamethoxazole (TMP-SMX) is recommended as the first choice for S. maltophilia infections (Abbott et al., 2011; Chong et al., 2017). However, the increasing reports of resistance to TMP-SMX are a matter of concern and have complicated the treatment strategies (Brooke, 2014; Hu et al., 2016; Madi et al., 2016). Resistance to this antibiotic has been recognized due to the presence of sul1 and sul2 genes that are found in class 1 integrons and insertion sequence common region (ISCR) elements, respectively. dfrA gene cassettes are observed in class 1 integrons and encode for the dihydrofolate reductase enzyme, and TolCsm, smeDEF, smeYZ efflux pumps (Hu et al., 2011, 2016; Huang et al., 2013; Lin et al., 2015; Sánchez and Martínez, 2015).
Biofilms are multicellular communities usually held together by extracellular matrix molecules. These extracellular polysaccharides (EPS) produced by the bacteria usually function as highly organized multicellular communities of microorganisms (Bjarnsholt et al., 2009; Irie et al., 2017), appear to be preferred survival strategy of microbes, and confer tolerance to high doses of antimicrobial agents than non-biofilm forming bacteria (Bjarnsholt et al., 2009). In addition, they are increasingly recognized as a contributing factor in the pathogenesis of disease in respiratory diseases often caused by chronic bacterial infections. S. maltophilia strains are well-known biofilm-producing organisms with ability to adhere to biotic and abiotic surfaces (Pompilio et al., 2008). Few genes associated with biofilm formation in S. maltophilia have been experimentally studied (Liu et al., 2017). More recently, the correlation between mutations in rpfF and rmlA genes, encoding enoyl-CoA hydratase and glucose-1-phosphate thymidyltransferase, respectively, and the less extensive biofilm formation have been reported (Huang et al., 2006; Fouhy et al., 2007). In addition, the spgM gene, responsible for the production of phosphoglucomutase (PGM) and phosphomannomutase, could be involved in biofilm-forming ability (McKay et al., 2003; Zhuo et al., 2014).
High genetic diversity was identified among S. maltophilia strains through the use of a variety of molecular biology techniques. Several genotypic profile methods have been used to compare and link clinical isolates to environmental sources, including whole genome sequencing analyses, amplified fragment length polymorphism (AFLP) fingerprinting, PCR-restriction fragment length polymorphism (PCR-RFLP), analysis of the gyrase B gene, PCR-based fingerprinting methods, such as BOX and repetitive extragenic palindromic (rep)-PCR, enterobacterial repetitive intergenic consensus (ERIC)-PCR, pulsed-field gel electrophoresis (PFGE) analysis of XbaI genomic digests, and multi-locus sequence typing (MLST) (Gherardi et al., 2015). Rep-PCR technique is based on the fact that microbial genomes contain a variety of repetitive sequences. Although their function has mostly not been elucidated so far, most rep-PCR-based DNA fingerprinting studies have used short polytrinucleotides, such as (GTG)5 35–40 bp repetitive sequences, and 154 bp BOX element as priming sites for PCR, resulting in amplification of DNA sequences between the repetitive parts (Ishii and Sadowsky, 2009). MLST technique was developed for tracking the source of infections and the distribution of pathogens isolated from hospitalized patients, providing reliable epidemiological data. In addition, because of its accessible related international databases, the results from different laboratories by MLST can be compared (Cho et al., 2012).
The main purpose of this study was to evaluate the antimicrobial resistance patterns and different resistance mechanisms of the clinical S. maltophilia isolated from different regions of Iran. In addition, the ability of biofilm production as well as clonal and genetic diversity of isolates were examined.
Materials and Methods
Ethics Statement
This study was approved by the Ethics Committee of Shahid Beheshti University of Medical Sciences “IR.SBMU. MSP.REC.1397.579.” In order to maintain patients confidentiality participants were anonymous and no personal information was collected or included in the study.
Bacterial Isolation and Species Identification
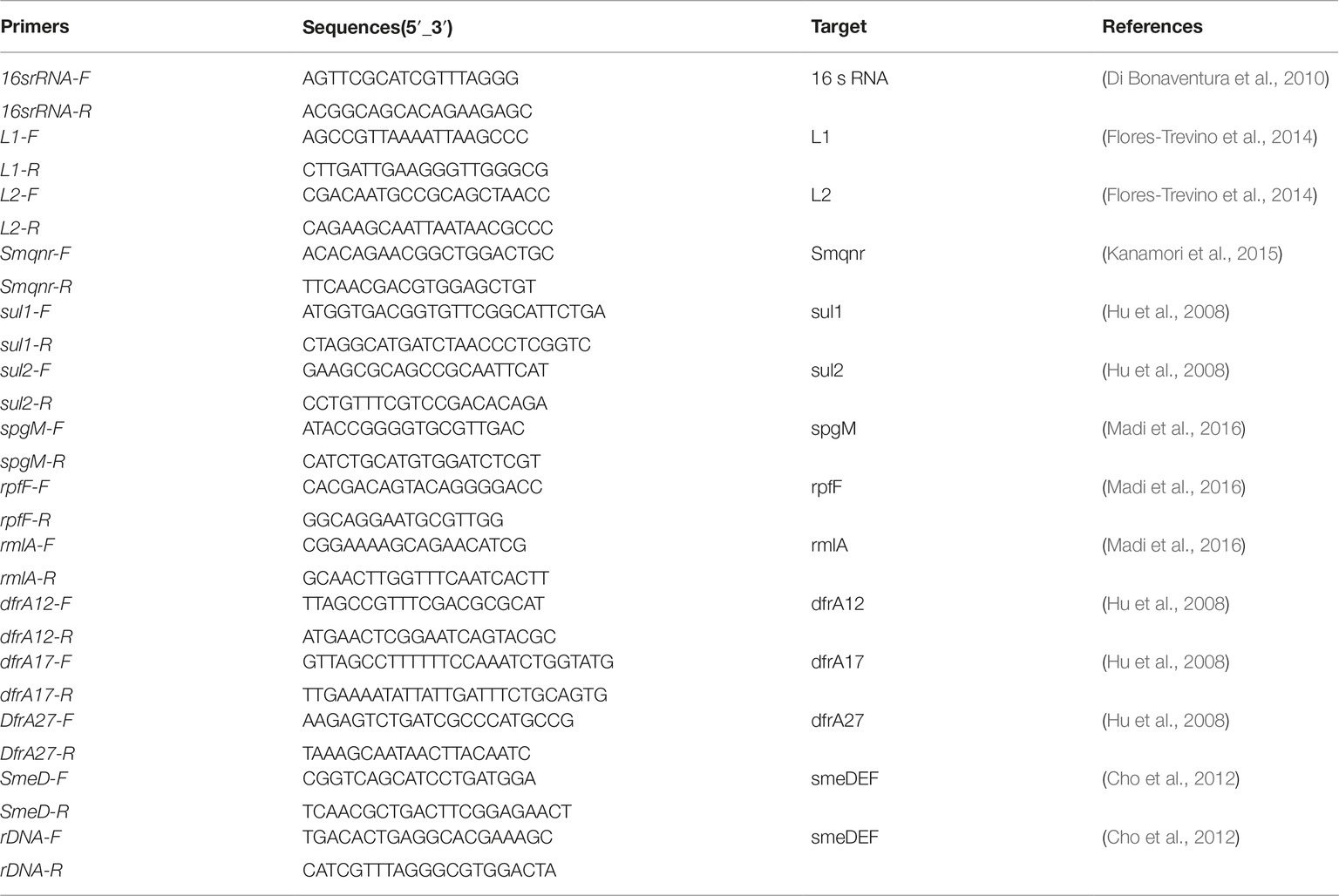
S. maltophilia isolates were collected from different hospitalized patients in selected hospitals in Iran over a 12-months period from May 2016 to May 2017. Laboratory identification of isolates was carried out using the standard biochemical methods, such as oxidase and catalase tests, and reactions in media, including deoxyribonuclease test agar (Merck Cat. No.1.10449.0500), triple sugar iron agar (Merck Cat. No 1.03915.0500), and SIM (Merck Cat. No1.05470.0500). Consequently, isolates were confirmed as S. maltophilia by using the 16S rRNA sequencing with specific primers (Table 1; Kettleson et al., 2013). All isolates were stored in LB with 20% glycerol at −70°C. Escherichia coli ATCC 35218, Pseudomonas aeruginosa ATCC 27853, E. coli ATCC 25922, and S. maltophilia ATCC 13637 were used as the quality control strains.
Antimicrobial Susceptibility Testing
Susceptibility of isolates to different antibiotics was evaluated according to the criteria of the Clinical and Laboratory Standard Institute (Clinical and Laboratory Standards Institute (CLSI) (2016)). Kirby-Bauer disc diffusion method was used for susceptibility testing to imipenem (10 μg), meropenem (10 μg), doripenem (10 μg), levofloxacin (5 μg), minocycline (30 μg), trimethoprim-sulfamethoxazole (1.25/23.75 μg), ceftazidime (30 μg), and tetracycline (30 μg) (Mast, Company). Minimal inhibitory concentration (MIC) was determined by MIC-Test Strip (Liofilchem; Roseto degli Abruzzi, Italy) for four selected antibiotics, including trimethoprim-sulfamethoxazole, chloramphenicol, ceftazidime, and ticarcillin-clavulanate. Quality control was performed using E. coli ATCC 35218 and E. coli ATCC 25922.
DNA Extraction
S. maltophilia isolates were grown on LB for 24 h at 37°C, and genomic DNA was extracted using the high pure PCR Template Preparation Kit (Roche, Germany, and Lot.No.10362400) according to the manufacturer’s guidelines. The total DNA concentration was determined using the Nanodrop instrument (WPA Biowave II Nanospectrophotometer, USA).
PCR-Sequencing Technique
The presence of β-lactamase genes L1 and L2 as well as dfrA12, dfrA17, dfrA27, sul1, sul2, and Smqnr genes were examined using the primers shown in Table 1 (Levesque et al., 1995; Hu et al., 2011, 2016; Liu et al., 2012; Kanamori et al., 2015). As described previously (Hu et al., 2011), PCR was conducted in a final volume of 25 μl containing 1 μl (20 ng) of DNA template and 12.5 μl of 2× Master Mix (SinaClon-Iran, CAT. No., PR901638), including 1× PCR buffer, 0.4 mmol/L dNTPs, 3 mmol/L MgCl2, and 0.08 IU Taq DNA polymerase, 1 μl of 10 pmol of each primer and 9.5 μl of sterile distilled water. Amplification reactions were performed on a thermal cycler (Eppendorf, Master Cycler Gradient, Germany). PCR was initiated by denaturation for 5 min 94°C, followed by 36 cycles of 45 s at 94°C, annealing at 50–59°C, according to the primers for each gene for 45 s, and extension at 72°C for 45 s. PCR products were electrophoresed by 1–1.5% agarose gel, visualized by DNA Safe staining and photographed under UV light. The PCR products were purified using a PCR purification Kit (Bioneer Co., Korea) and then, nucleotide sequencing of amplicons was performed by an ABI PRISM 3700 sequencer (Macrogen Co., Korea). The sequenced data obtained was viewed in Chromas version 1.45 software. In addition, sequence alignment was conducted using the Nucleotide BLAST program1.
Phenotypic and Genotypic Detection of Biofilm Formation
Biofilm formation was examined by crystal violet staining as previously described by Stepanović et al. (2007). All experiments were performed in triplicate. An overnight culture of S. maltophilia was adjusted to match the turbidity of a 1.0 McFarland standard. The cultures were then diluted 1:100 in 200 ml tryptic soy broth (TSB) and were transferred into the wells of a flat-bottom polystyrene plate (SPL, Korea). After 24 h incubation at 37°C, plates were washed three times with sterile phosphate buffered saline (PBS with pH 7.3). Adherent biofilms were fixed for 60 min at 65°C, stained for 10 min at room temperature with 250 ml modified crystal violet and then, rinsed with water and allowed to dry. Biofilm samples were destained by treatment with 250 ml 33% glacial acetic acid for 20 min and the optical density (OD) was read at 492 nm (OD492). Grouping of isolates was carried out according to the following criteria: strong-biofilm producer (4 × ODc < OD), moderate-biofilm producer (2 × ODc < OD _ 4 × ODc), weak-biofilm producer (ODc < OD _2 × ODc), and non-biofilm producer (OD _ ODc). In addition, the presence of rpfF, spgM, and rmlA genes was investigated by PCR with specific primers described in Table 1 (Pompilio et al., 2011). Amplicons representing each studied gene was confirmed by sequencing analysis (Macrogen Korea). Obtained sequences were aligned in the NCBI database using BLAST program2.
RNA Preparation and qRT-PCR
TMP-SMX-resistant isolates were assessed for expression of SmeDEF efflux pump. Cell suspensions were prepared and inoculated on LB broth (Cho et al., 2012). After an overnight growth, total RNA was extracted from the cell suspensions by using the RNX-Plus Kit (Cat. No., RN7713C, Sinaclon, Iran) according to the manufacturer’s instructions. The contaminating DNA was removed by RNase-free DNase I (Fermentas, USA). The total RNA concentration was determined using the Nanodrop (WPA Biowave II Nanospectrophotometer, USA). DNase-treated RNA was reverse-transcribed into cDNA using the Takara Kit (Japan). The primers used for real-time PCR are shown in Table 2. Real-time PCR assay was performed on synthesized cDNA using the Power SYBR Green PCR Master Mix (Bioneer, Korea) on a Corbett Rotor-Gene 6000 real-time rotary analyzer (Corbett Life Science, Australia). Each amplification protocol included a first denaturation step of 10 min at 94°C, followed by 40 cycles of 20 s at 94°C and 45 s at 59°C. Samples were run in triplicate. Controls run without reverse transcriptase confirmed the absence of contaminating cDNA in any of the samples. The expression level of smeD gene was normalized using the rDNA housekeeping gene, and was calculated based on 2−ΔΔCT method. Results were obtained as the relative expression of the mRNA compared to that of S. maltophilia ATCC 13637. The parameter Ct was defined as the threshold cycle number at which the first detectable fluorescence generated by the binding of SYBR Green I dye to the minor groove of double-stranded DNA began to increase exponentially. Final results, expressed as n-fold differences in expression of smeD genes, were determined as follows:
Values of n < 1 were considered to indicate overexpression of the Sme efflux system.
Molecular Typing by Multi-Locus Sequence Typing
Multi-Locus Sequence Typing (MLST) technique was performed as the same as described by Kaiser et al. (2009). Briefly, PCR for seven housekeeping genes, including atpD, guaA, gapA, nuoD, ppsA, mutM, and recA was carried out. Amplicons were sequenced according to the PubMLST website recommendations3. Unique sequence (allele) number for each gene was assigned on the basis of the information in the S. maltophilia MLST database4 to determine specific sequence type (ST). A combination of the allelic sequences of the seven genes yielded the allelic profile for each isolate.
Molecular Typing by Repetitive Extragenic Palindromic-Pcr
Rep-PCR analyses were conducted with the single primer BoxA1R (5′-CTA CGG CAA GGC GAC GCT GAC G-3′) according to Versalovic et al. (1994). The PCR reaction mix consisted of 25 μl total volume with 12.5 μl of 2× Master Mix (Genet Bio Cat.No:G-5000) containing 1 unit of Taq polymerase in 2× reaction buffer, 10% dimethyl sulfoxide (DMSO), enzyme stabilizer, sediment, loading dye, 4 mM MgCl2, pH 9.0 and 0.5 mM of each dNTP, 5 μM of primer, and 1 μl of cell extract. Thermal cycling was conducted with an initial denaturation at 94°C for 10 min, followed by 25 cycles of 94°C for 45 s, 50°C for 1.5 min, 65°C for 8 min each, and concluded by a final extension of 65°C for 16 min. To determine phylogenetic relationships, rep-PCR profiles were analyzed by GelCompar II software (Applied Maths, Belgium) using the Pearson’s correlation coefficient with unweighted paired group method using arithmetic averages (UPGMA) as well as at the 80% similarity level (Adamek et al., 2011).
Statistical Analysis
Chi-squared test was performed on the association of TMP-SMX resistance phenotype and resistance genes using SPSS software, 20.0 (SPSS Inc., Chicago, IL, USA). The Pearson’s correlation coefficient was calculated to determine the association between two variables. A significant level of p = 0.05 was considered statistically significant.
Results
Patients and Bacterial Isolates
During 1-year period of study, 164 S. maltophilia isolates were collected from several hospitals in different regions of Iran (Figure 1).
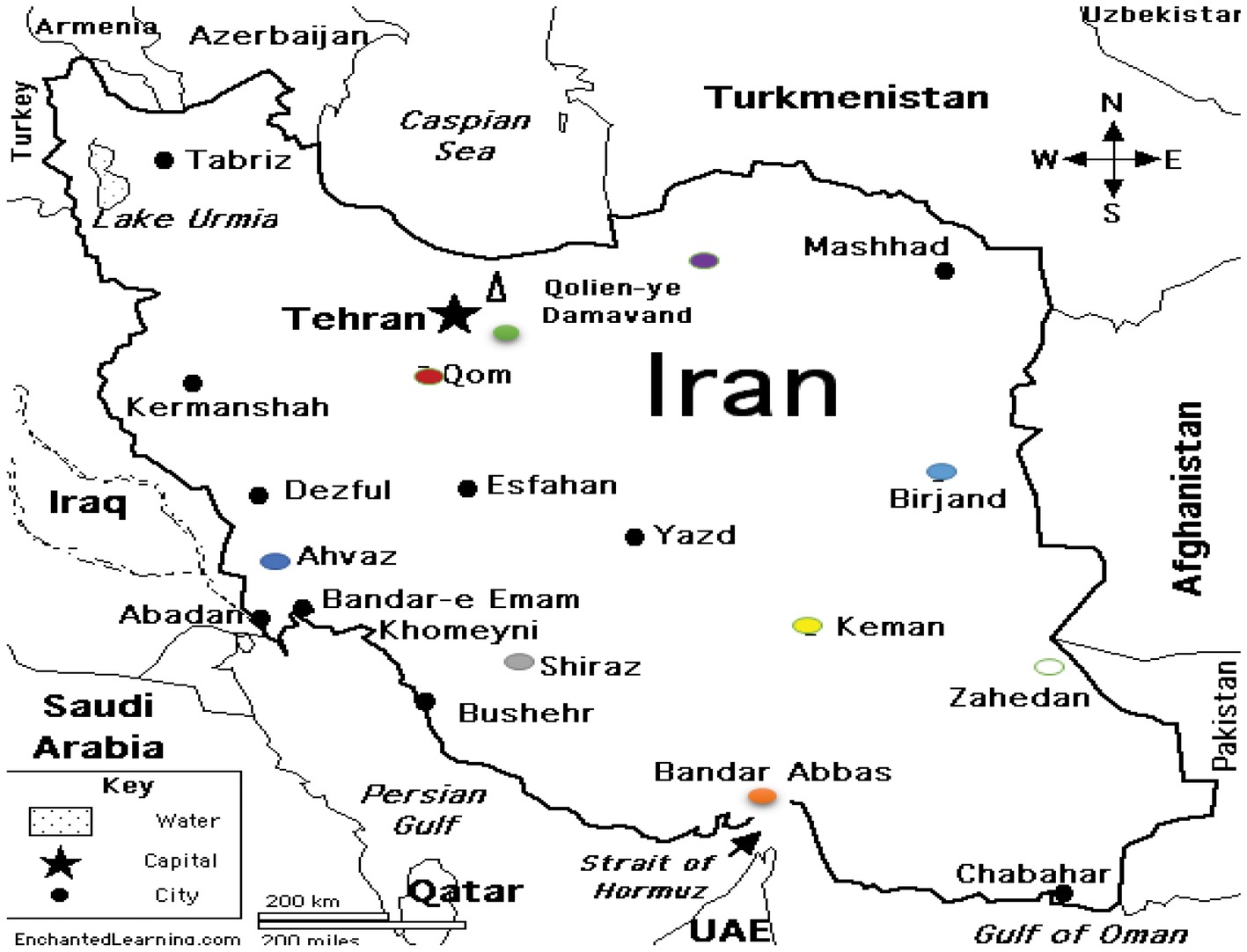
Figure 1. S. maltophilia strains isolated from Iran. 4 isolates from Birjand. 87 isolates from Tehran: Capital of Iran. 32 isolates from Ahwaz. 20 isolates from Shiraz. 14 isolates from Bandar Abbas. 4 isolates from Zahedan. 1 isolate from Kerman. 1 isolate from Gorgan. 1 isolate from Qom.
Among the 164 isolates obtained, 88 were from males and 76 were from females (male:female ratio = 1.15). The age range of patients was from 1 month to 85 years. The majority of the isolates were originated from blood (83.53%), followed by nose/throat secretions (5.48%), cough swabs (9.75%), sputum (0.6%), and CSF (0.6%).
Antibiotic Susceptibility Profile
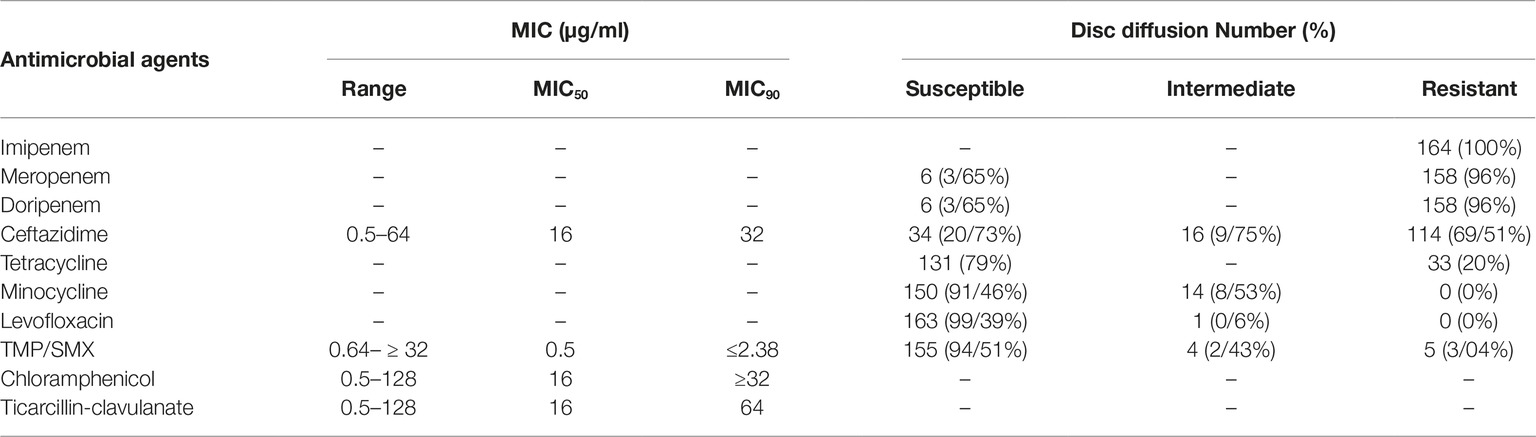
Based on CLSI interpretive criteria (Clinical and Laboratory Standards Institute (CLSI) (2016)), isolates were resistant to imipenem (100%), meropenem (96%), doripenem (96%), and ceftazidime (36.58%). Interestingly, 5 (3.04%) isolates showed resistance to TMP-SMX. Minocycline and levofloxacin exhibited the highest susceptibility of 91.46 and 99.39%, respectively. The MIC ranges, MIC50, MIC90, and the percentages of isolates resistant, intermediate, or susceptible isolates to the six antimicrobial agents are shown in Table 2.
Biofilm Phenotypes and Genotypes
Biofilm phenotypes accounted for 157 out of 164 isolates (95.73%): 49 isolates (29.87%) produced strong biofilm, 63 isolates (38.41%) produced moderate biofilm, and 45 isolates (27.43%) produced weak biofilm; whereas, 7 isolates (4.26%) did not form biofilm (Figure 2). PCR-based typing of biofilm-related genes revealed an overall prevalence of 145 (88.41%), 137 (83.53%), and 164 (100%) of rmlA, rpfF, and spgM, respectively. In addition, the presence of rmlA, rpfF, and spgM had a close relationship with biofilm formation but did not significantly affect the mean amount of biofilm (p ≤ 0.05). Some strong- and weak biofilm-producer phenotypes had mutations within the sequence of each rpfF, spgM, and rmlA genes.
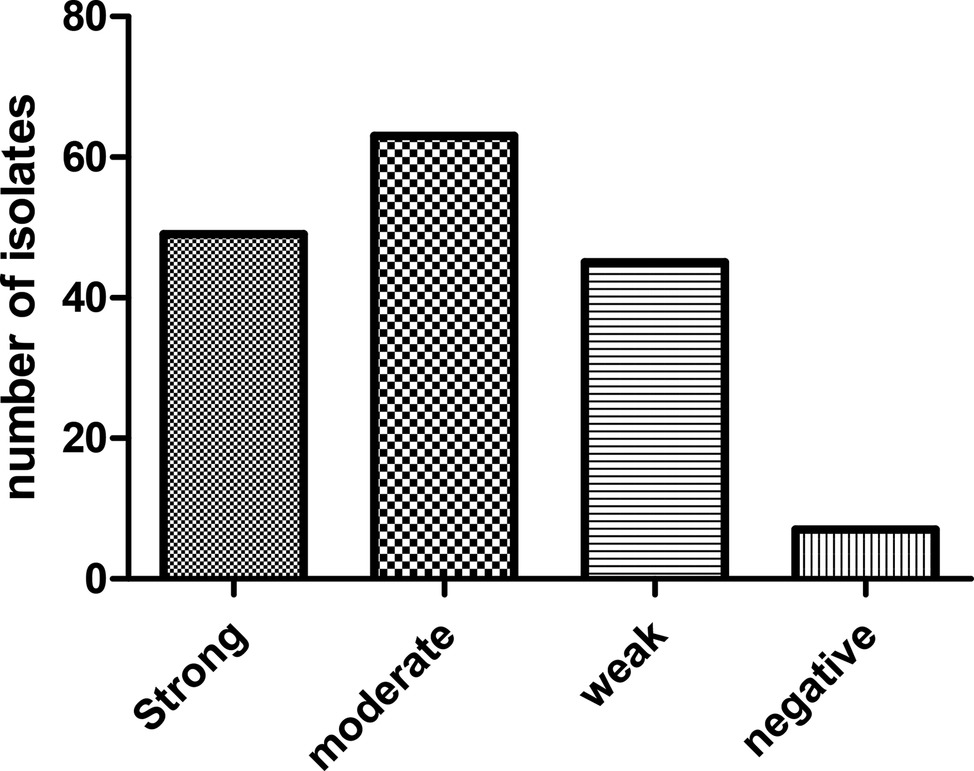
Figure 2. Distribution of S. maltophilia isolates based on the biofilm formation in crystal violet staining assay.
Prevalence of Resistance Genes
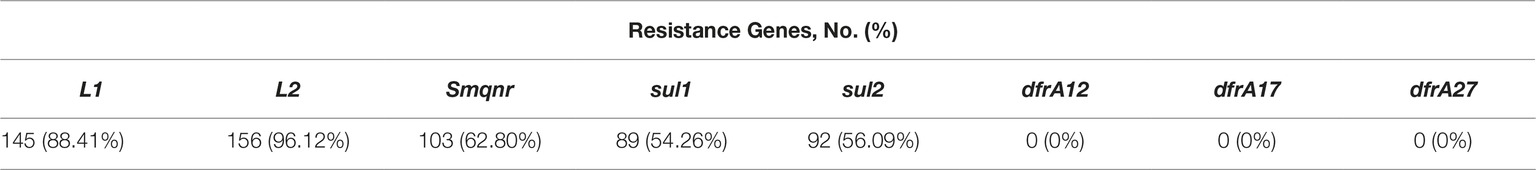
Prevalence of resistance genes among 164 S. maltophilia isolates are shown in Table 3.
Of the 145 isolates that were positive for L1, all 145(100%) and 139(92.3%) showed resistance to imipenem and meropenem, respectively. Amongst 156 isolates carrying the L2 gene, all (100%) were imipenem resistant and 150 (91.1%) were meropenem-resistant (p ≤ 0.001). In addition, 54.19% (89/155) and 58.70% (91/155) TMP-SMX-susceptible isolates and 100% (5/5) and 20% (1/5) TMP-SMX-resistant isolates were detected to contain the sul1, and sul2 genes, respectively.
Gene Expression Analysis of smeDEF
Real-time PCR analysis was used to assess the expression of SmeDEF efflux system in TMP-SMX-resistant S. maltophilia isolates (MIC > 4/76 μg/ml). Results showed that smeD gene was overexpressed (5.47–7.87 fold) in two out of five isolates (40%) in comparison to the S. maltophilia ATCC 13637 standard strain.
MLST Analysis
As shown in Table 4, five TMP-SMX-resistant S. maltophilia isolates belonged to two different STs, ST139 and ST259. This is the first report on the detection of ST139 and ST259 from Iran. In addition, ST259 (n = 2) with allelic profile (26, 14, 140, 103, 3, 8, 11) was not previously reported. New allele sequences were deposited at the MLST Database hosted by the Shahid Beheshti University of Medical Science, Tehran, Iran5.

Table 4. Sequence type (ST) of TMP-SMX-resistant S. maltophilia clinical isolates recovered in the present study.
Rep-PCR Fingerprinting
To evaluate the genetic diversity, all 164 S. maltophilia isolates were subjected to rep-PCR fingerprinting. As shown in Figure 3, isolates were divided into 16 common types (CT) containing 2–5 isolates and 114 single types (ST). Among these numerous clones, a dominant one was isolated from Ahwaz and from blood samples. The genotypic pattern of the dominant clone revealed that all isolates harbored sul1 gene.

Figure 3. Dendrogram based on Dice’s coefficient of similarity using UPGMA method applied by the GelComparII program showing relationships between S. maltophilia strains according to BOX-PCR genotyping.
Nucleotide Sequence Accession Numbers
The nucleotide sequence data reported in this study were submitted to the GenBank sequence database and assigned under the accession numbers: MF458984, MF497329, MG601517, MG640120, MG648332, MG597493, MF805867, MG640120, MG560825, MG597494, MG640119, and MG601518 for the L1, L2, sul1, sul2, smqnR, atpD, gapA, guaA, mutM, nuoD, ppsA, and recA genes, respectively.
Discussion
The emergence of S. maltophilia as a nosocomial pathogen in hospitals with intrinsic resistance to multiple antibacterial agents, including carbapenems, aminoglycosides, β-lactams, and quinolones have caused great concern (Farrell et al., 2010). Additionally, some strains have acquired resistance, leading to limited antimicrobial options (Wang et al., 2013; Gholipourmalekabadi et al., 2016). In Iran, decades of misuse of antibiotics resulted in high prevalence of antibiotic resistance in bacteria (Habibzadeh, 2013; Saniee et al., 2018).
Global infectious disease surveillance stipulated that resistance rates for trimethoprim–sulfamethoxazole, ticarcillin-clavulanic acid, levofloxacin, and minocycline in S. maltophilia isolates are less than 4.7, 16.1, 6.5 and 5%, respectively (Sader and Jones, 2005). Among the 164 clinical isolates of S. maltophilia studied in the present study, a significant percentage was resistant to carbapenems (p ≤ 0.001). Resistance to carbapenems in S. maltophilia occurs through several mechanisms, including intrinsic β-lactamase expression. In this study, 145 (88.41%) and 156 (96.12%) isolates harbored L1-and L2- β-lactamase genes, respectively. Also, the results indicate that the susceptibility rate of S. maltophilia isolates against ceftazidime was 20.73%, with the MIC50 and MIC90 of 8 and 32 μg/ml, a figure that was in agreement with previous findings (Nicodemo and Paez, 2007). A study by Jamali et al. showed that susceptibility of S. maltophilia against ceftazidime was 82% with the MIC50 and MIC90 values of 2 and 32 μg/ml, respectively (Jamali et al., 2011). Shahla et al. indicated that among 11 isolate of S. maltophilia, 91.4% were susceptible to ceftazidime (Shahla et al., 2012). In a study by Pfaller, the susceptibility in Canada, United States, and Latin America was respectively 27, 64.7, and 93.3% and Tatmanin Turkey showed the susceptibility of 67% for this drug (Pfaller et al., 1999; Tatman-Otkun et al., 2005). A study by Farrell et al. conducted in North America, Latin America, Europe, and Asian-Pacific reported a susceptibility rate of 27.0–46.1% to ticarcillin-clavulanate among S. maltophilia isolates (Farrell et al., 2010). The present study showed susceptibility rate of 57.92% to ticarcillin-clavulanate. MIC50 and MIC90 for ticarcillin-clavulanate was 12 and 128 μg/ml. A study in a Brazilian hospital showed the susceptibility pattern of S. maltophilia against chloramphenicol differs from 11.5 to 81.4% (Nicodemo and Paez, 2007). In our study, 7.31% of isolates were found to be susceptible to this antibiotic with MIC50 and MIC90 of 24 and 64 μg/ml. This variety in results designate that the susceptibility of S. maltophilia is variable in different countries and even in different hospitals. Other therapeutic alternatives, such as levofloxacin and minocycline, which have been reported as effective agents for treatment of invasive S. maltophilia infections (Wu et al., 2012, 2013; Cho et al., 2014), showed susceptibility rates of 99.39 and 96.41% in our study. Although the prevalence of minocycline and levofloxacin-resistant S. maltophilia is low worldwide, continued surveillance of resistance to such antimicrobials ensures their activity.
Historically, TMP-SMX is considered the first line of defense in S. maltophilia infections (Chung et al., 2015; Kaur et al., 2015). Results from the SENTRY Antimicrobial Surveillance Program in 2004 indicated that 3.8% of S. maltophilia isolates were resistant to TMP-SMX (Fedler et al., 2006). Moreover, the resistance rate reported for Latin America, Argentina, and Malaysia were approximately less than 4.5 and 1% (Barbolla et al., 2004; Farrell et al., 2010; Neela et al., 2012). Resistance rates vary geographically but are commonly less than 10% reported in several studies (Kaur et al., 2015). However, high and different rates of resistance have been reported in patients with cancer and cystic fibrosis (Valenza et al., 2008). In different studies by Shahla et al. (2012), Hu et al. (2016), Tatman-Otkun et al. (2005), Wang et al. (2004), Nicodemo et al. (2004), and Kaur et al. (2015), the susceptibility rates were reported 47.3, 61.3, 95.8, 60, 98.6, and 22.6%, respectively. Jamali et al. showed about 60% susceptibility rate for TMP-SMX and the MIC50 and MIC90 values were 0.5 and 2 μg/ml (Jamali et al., 2011). In our study, based on the CLSI recommended dose of TMP-SMX, the resistance rate of 3.04% and the MIC50% and MIC90% values of 2.38 and 4.76 were found, respectively. We believe that this resistance rate for TMP-SMX, as the treatment of choice for S. maltophilia infection, is sustainable, making necessary the future successive reevaluation of susceptibility to this antibiotic in Iranian hospitals.
The well-known mechanism responsible for TMP-SMX resistance is harboring the sul1, sul2, and/ordfrA resistance genes located either on a chromosome or plasmid (Hu et al., 2011). In this study, sul1 and sul2 genes were detected in both TMP-SMX-resistant and TMP-SMX-susceptible S. maltophilia clinical isolates. Additionally, antimicrobial efflux pump mechanisms have been increasingly recognized as sources of intrinsic and acquired resistance (Song et al., 2010; Hu et al., 2011; Gholami et al., 2015). As reported in other studies, the frequency of sul2 gene in S. maltophilia strains is less than that of sul1 gene (Song et al., 2010; Hu et al., 2011). These reports are contrary to the results of our study, where a higher percentage of sul2 and sul1 (56.9 and 54.26%, respectively) was observed. Furthermore, both sul1 and sul2 genes were found in TMP-SMX -susceptible and –resistant isolates. Similar to our study, Kaur et al. indicated that the percentage of sul1 and sul2 were 50 and 58.3%, respectively (Kaur et al., 2015). In addition, none of the isolates tested were positive for dfrA12, dfrA17, and dfrA27. In contrast, a study showed that 49.1% of TMP-SMX-resistant isolates and 10.3% of TMP-SMX-susceptible isolates were positive for dfrA genes, among them dfrA12 and dfrA17 genes were more prevalent (Hu et al., 2016). Previous reports indicated that overexpression of the SmeDEF efflux system in S. maltophilia plays a significant role in resistance to several antibacterials, including aminoglycosides, β-lactams, and quinolones (Chang et al., 2004; Cho et al., 2012). The results showed overexpression of smeD in 2 (40%) of the 5 TMP-SMX-resistant clinical isolates. Sanchez et al. showed that overexpression of the SmeDEF efflux pump decreases the susceptibility to TMP-SMX (Sánchez and Martínez, 2015).
An important feature of S. maltophilia is its ability to form biofilms on hospital surfaces as well as on human tissues; biofilms have been related to 65% of hospital-acquired infections (Zhuo et al., 2014). In this study, the majority of isolates were biofilm-producer as well as biofilm-related gene (rpfF, rmlA and spgM) carrier. In a study by Flores-Trevino et al., they showed that all S. maltophilia isolates were able to form biofilm and 47.9, 38.7, and 13.4% of the isolates were weak-, moderate-, and strong-biofilm producers, respectively (Flores-Trevino et al., 2014). Zhou et al. showed that the results of a biofilm formation assay on polystyrene was strong in 49 (29.87%) strains, moderate in 63 strains (38.41%), and weak in 45 (27.43%) strains, while nine strains (4.26%) were non-biofilm former. Furthermore, the presence of rpfF and spgM was significantly correlated to biofilm formation. Pompilio et al. reported that spgM gene played a significant role in formation of strong biofilm among S. maltophilia isolates (Zhuo et al., 2014). Similarly, the presence of rmlA, rpfF, and spgM genes in the present study improved significantly biofilm formation by S. maltophilia isolates tested (p ≤ 0.05). Indeed, the isolates with rpfF+/spgM+/rmlA+ genotype were associated with production of moderate or strong biofilm. In addition, amino acid substitution in genes encoding SpgM, RpfF and RmlA were found among some strains (Corlouer et al., 2017). However, it is still unclear which gene mutation results to change in biofilm formation.
High genetic diversity among S. maltophilia isolates has been described worldwide. Although occurrences of outbreaks within hospital settings have also been reported (Flores-Trevino et al., 2014). Recently, molecular epidemiologic studies, like MLST is developed for S. maltophilia strain-typing that focuses on conserved housekeeping genes (Corlouer et al., 2017). In the present study, MLST analysis was performed for determining genetic diversity of five TMP-SMX-resistant isolates. The results revealed two STs (ST139 and ST259), of which ST259 was identified for the first time in this study. Similarly, studies in Spain in 2004, and Korea in 2010, a high rate of genetic diversity among S. maltophilia isolates despite their source in a single hospital (Valdezate et al., 2004; Cho et al., 2012; Corlouer et al., 2017). These findings indicate that S. maltophilia has a high potential for environmental distribution, although database analysis shows that there are noticeably fewer STs for S. maltophilia isolates than other bacterial isolates. Rep-PCR fingerprinting is a method with lower cost and the best time efficiency. According to the cluster analysis of S. maltophilia strains, this study detected high clonal diversity among the isolates. The only exception is the dominant common type including strains isolated from blood culture of patients hospitalized in Ahwaz. In addition, all these isolates harbored sul1 gene. As a result, the presence and spread of these strains with resistance gene could be a significant threat.
Conclusions
This multi-institutional study revealed that S. maltophilia is an emerging MDR opportunistic pathogen in hospital settings, especially among immunocompromised patients. TMP-SMX remains the most effective antibacterial agent against S. maltophilia. So, a significant effort is required to maintain antibacterial properties of this antibiotic. Due to the low prevalence of resistance to two antibiotics levofloxacin and minocycline, clinical usage of these agents can be continued. The findings of this study showed an increasing presence of antibacterial resistance-and biofilm genes among the clinical isolates of S. maltophilia strains in Iran. Clinicians must consider that S. maltophilia as a co-pathogen or co-colonizer in polymicrobial infections can have negative effect on the success rate of antibacterial treatment and clinical outcome. In our opinion, this is significant medical problem, which should be of great concern.
Author Contributions
NB, ZG, FF, and AH conceived and designed the experiments. NB, ZG, AH, AY, JM, AA, SS, ST, AA, FF, and AP performed the experiments and analyzed the data. NB, AA, and AH wrote the paper.
Funding
The present study was financially supported by the Infectious Diseases and Tropical Medicine Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran (grant No 11533).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the curator of MLST database, for assigning our new alleles and STs to Stenotrophomonas maltophilia MLST database.
Footnotes
1. http://www.ncbi.nlm.nih.gov/nucleotide/
2. http://www.ncbi.nlm.nih.gov/nucleotide/
3. http://pubmlst.org/smaltophilia/
4. http://pubmlst.org/smaltophilia/
5. http://pubmlst.org/perl/bigsdb/bigsdb.pl?db=pubmlst_smaltophilia_isolates&page=query
References
Abbott, I. J., Slavin, M. A., Turnidge, J. D., Thursky, K. A., and Worth, L. J. (2011). Stenotrophomonas maltophilia: emerging disease patterns and challenges for treatment. Expert Rev. Anti-Infect. Ther. 9, 471–488. doi: 10.1586/eri.11.24
Adamek, M., Overhage, J., Bathe, S., Winter, J., Fischer, R., and Schwartz, T. (2011). Genotyping of environmental and clinical Stenotrophomonas maltophilia isolates and their pathogenic potential. PLoS One 6:e27615. doi: 10.1371/journal.pone.0027615
Al-Anazi, K. A., and Al-Jasser, A. M. (2014). Infections caused by Stenotrophomonas maltophilia in recipients of hematopoietic stem cell transplantation. Front. Oncol. 4, 1–11. doi: 10.3389/fonc.2014.00311
Anzai, Y., Kim, H., Park, J.-Y., Wakabayashi, H., and Oyaizu, H. (2000). Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 50, 1563–1589. doi: 10.1099/00207713-50-4-1563
Barbolla, R., Catalano, M., Orman, B. E., Famiglietti, A., Vay, C., Smayevsky, J., et al. (2004). Class 1 integrons increase trimethoprim-sulfamethoxazole MICs against epidemiologically unrelated Stenotrophomonas maltophilia isolates. Antimicrob. Agents Chemother. 48, 666–669. doi: 10.1128/AAC.48.2.666-669.2004
Bjarnsholt, T., Jensen, P. Ø., Fiandaca, M. J., Pedersen, J., Hansen, C. R., Andersen, C. B., et al. (2009). Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44, 547–558. doi: 10.1002/ppul.21011
Brooke, J. S. (2012). Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin. Microbiol. Rev. 25, 2–41. doi: 10.1128/CMR.00019-11
Brooke, J.S. (2014). New strategies against Stenotrophomonas maltophilia: a serious worldwide intrinsically drug-resistant opportunistic pathogen. 12, 1–4. doi: 10.1586/14787210.2014.864553
Chang, L.-L., Chen, H.-F., Chang, C.-Y., Lee, T.-M., and Wu, W.-J. (2004). Contribution of integrons, and Sme ABC and SmeDEF efflux pumps to multidrug resistance in clinical isolates of Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 53, 518–521. doi: 10.1093/jac/dkh094
Chang, Y.-T., Lin, C.-Y., Chen, Y.-H., and Hsueh, P.-R. (2015). Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front. Microbiol. 6:893. doi: 10.3389/fmicb.2015.00893
Cho, S. Y., Kang, C.-I., Kim, J., Ha, Y. E., Chung, D. R., Lee, N. Y., et al. (2014). Can levofloxacin be a useful alternative to trimethoprim-sulfamethoxazole for treating Stenotrophomonas maltophilia bacteremia? Antimicrob. Agents Chemother. 58, 581–583. doi: 10.1128/AAC.01682-13
Cho, H. H., Sung, J. Y., Kwon, K. C., and Koo, S. H. (2012). Expression of Sme efflux pumps and multilocus sequence typing in clinical isolates of Stenotrophomonas maltophilia. Ann. Lab. Med. 32, 38–43. doi: 10.3343/alm.2012.32.1.38
Chong, S. Y., Lee, K., Chung, H.-S., Hong, S. G., Suh, Y., and Chong, Y. (2017). Levofloxacin efflux and smeD in clinical isolates of Stenotrophomonas maltophilia. Microb. Drug Resist. 23, 163–168. doi: 10.1089/mdr.2015.0228
Chung, H.-S., Kim, K., Hong, S. S., Hong, S. G., Lee, K., and Chong, Y. (2015). The sul1 gene in Stenotrophomonas maltophilia with high-level resistance to trimethoprim/sulfamethoxazole. Ann. Lab. Med. 35, 246–249. doi: 10.3343/alm.2015.35.2.246
Clinical and Laboratory Standards Institute (CLSI) (2016). Performance standards for antimicrobial susceptibility testing; Twenty-sixth informational supplement. CLSI document M100-S26 (Wayne, PA: CLSI).
Corlouer, C., Lamy, B., Desroches, M., Ramos-Vivas, J., Mehiri-Zghal, E., Lemenand, O., et al. (2017). Stenotrophomonas maltophilia healthcare-associated infections: identification of two main pathogenic genetic backgrounds. J. Hosp. Infect. 96, 183–188. doi: 10.1016/j.jhin.2017.02.003
Di Bonaventura, G., Pompilio, A., Zappacosta, R., Petrucci, F., Fiscarelli, E., Rossi, C., et al. (2010). Role of excessive inflammatory response to Stenotrophomonas maltophilia lung infection in DBA/2 mice and implications for cystic fibrosis. Infect. Immun. 78, 2466–2476. doi: 10.1128/IAI.01391-09
Falagas, M. E., Kastoris, A. C., Vouloumanou, E. K., Rafailidis, P. I., Kapaskelis, A. M., and Dimopoulos, G. (2009). Attributable mortality of Stenotrophomonas maltophilia infections: a systematic review of the literature. Future Microbiol 4, 1103–1109. doi: 10.2217/fmb.09.84
Farrell, D. J., Sader, H. S., and Jones, R. N. (2010). Antimicrobial susceptibilities of a worldwide collection of Stenotrophomonas maltophilia isolates tested against tigecycline and agents commonly used for S. maltophilia infections. Antimicrob. Agents Chemother. 54, 2735–2737. doi: 10.1128/AAC.01774-09
Fedler, K. A., Biedenbach, D. J., and Jones, R. N. (2006). Assessment of pathogen frequency and resistance patterns among pediatric patient isolates: report from the 2004 SENTRY Antimicrobial Surveillance Program on 3 continents. Diagn. Microbiol. Infect. Dis. 56, 427–436. doi: 10.1016/j.diagmicrobio.2006.07.003
Flores-Trevino, S., Gutierrez-Ferman, J. L., Morfín-Otero, R., Rodríguez-Noriega, E., Estrada-Rivadeneyra, D., Rivas-Morales, C., et al. (2014). Stenotrophomonas maltophilia in Mexico: antimicrobial resistance, biofilm formation and clonal diversity. J. Med. Microbiol. 63, 1524–1530. doi: 10.1099/jmm.0.074385-0
Fouhy, Y., Scanlon, K., Schouest, K., Spillane, C., Crossman, L., Avison, M. B., et al. (2007). Diffusible signal factor-dependent cell-cell signaling and virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J. Bacteriol. 189, 4964–4968. doi: 10.1128/JB.00310-07
Gherardi, G., Creti, R., Pompilio, A., and Di Bonaventura, G. (2015). An overview of various typing methods for clinical epidemiology of the emerging pathogen Stenotrophomonas maltophilia. Diagn. Microbiol. Infect. Dis. 81, 219–226. doi: 10.1016/j.diagmicrobio.2014.11.005
Gholami, M., Hashemi, A., Hakemi-Vala, M., Goudarzi, H., and Hallajzadeh, M. (2015). Efflux pump inhibitor phenylalanine-arginine Β-naphthylamide effect on the minimum inhibitory concentration of imipenem in Acinetobacter baumannii strains isolated from hospitalized patients in Shahid Motahari burn hospital, Tehran, Iran. Jundishapur J. Microbiol. 8:e19048. doi: 10.5812/jjm.19048
Gholipourmalekabadi, M., Sameni, M., Hashemi, A., Zamani, F., Rostami, A., and Mozafari, M. (2016). Silver-and fluoride-containing mesoporous bioactive glasses versus commonly used antibiotics: activity against multidrug-resistant bacterial strains isolated from patients with burns. Burns 42, 131–140. doi: 10.1016/j.burns.2015.09.010
Hu, L.-F., Chang, X., Ye, Y., Wang, Z.-X., Shao, Y.-B., Shi, W., et al. (2011). Stenotrophomonas maltophilia resistance to trimethoprim/sulfamethoxazole mediated by acquisition of sul and dfrA genes in a plasmid-mediated class 1 integron. Int. J. Antimicrob. Agents 37, 230–234. doi: 10.1016/j.ijantimicag.2010.10.025
Hu, L.-F., Chen, G.-S., Kong, Q.-X., Gao, L.-P., Chen, X., Ye, Y., et al. (2016). Increase in the prevalence of resistance determinants to trimethoprim/sulfamethoxazole in clinical Stenotrophomonas maltophilia isolates in China. PLoS One 11:e0157693. doi: 10.1371/journal.pone.0167792
Hu, R.-M., Huang, K.-J., Wu, L.-T., Hsiao, Y.-J., and Yang, T.-C. (2008). Induction of L1 and L2 β-lactamases of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 52, 1198–1200. doi: 10.1128/AAC.00682-07
Huang, Y.-W., Hu, R.-M., and Yang, T.-C. (2013). Role of the pcm-tolCsm operon in the multidrug resistance of Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 68, 1987–1993. doi: 10.1093/jac/dkt148
Huang, Y.-W., Liou, R.-S., Lin, Y.-T., Huang, H.-H., and Yang, T.-C. (2014). A linkage between SmeIJK efflux pump, cell envelope integrity, and σE-mediated envelope stress response in Stenotrophomonas maltophilia. PLoS One 9:e111784. doi: 10.1371/journal.pone.0115743
Huang, T.-P., Somers, E. B., and Wong, A. C. L. (2006). Differential biofilm formation and motility associated with lipopolysaccharide/exopolysaccharide-coupled biosynthetic genes in Stenotrophomonas maltophilia. J. Bacteriol. 188, 3116–3120. doi: 10.1128/JB.188.8.3116-3120.2006
Irie, Y., Roberts, A., Kragh, K., Gordon, V., Hutchison, J., Allen, R., et al. (2017). The Pseudomonas aeruginosa PSL polysaccharide is a social but noncheatable trait in biofilms. mBio 8, pii:e00374-17. doi: 10.1128/mBio.00374-17
Ishii, S., and Sadowsky, M. J. (2009). Applications of the rep-PCR DNA fingerprinting technique to study microbial diversity, ecology and evolution. Environ. Microbiol. 11, 733–740. doi: 10.1111/j.1462-2920.2008.01856.x
Jamali, F., Boroumand, M. A., Yazdani, F., Anvari, M. S., Pourgholi, L., Mahfouzi, S., et al. (2011). Minimal inhibitory concentration of ceftazidime and co-trimoxazole for Stenotrophomonas maltophilia using E-test. J. Global Infect. Dis. 3, 254–258. doi: 10.4103/0974-777X.83531
Kaiser, S., Biehler, K., and Jonas, D. (2009). A Stenotrophomonas maltophilia multilocus sequence typing scheme for inferring population structure. J. Bacteriol. 191, 2934–2943. doi: 10.1128/JB.00892-08
Kanamori, H., Yano, H., Tanouchi, A., Kakuta, R., Endo, S., Ichimura, S., et al. (2015). Prevalence of Smqnr and plasmid-mediated quinolone resistance determinants in clinical isolates of Stenotrophomonas maltophilia from Japan: novel variants of Smqnr. New Microbes New Infect. 7, 8–14. doi: 10.1016/j.nmni.2015.04.009
Kaur, P., Gautam, V., and Tewari, R. (2015). Distribution of class 1 integrons, sul1 and sul2 genes among clinical isolates of Stenotrophomonas maltophilia from a tertiary care hospital in North India. Microb. Drug Resist. 21, 380–385. doi: 10.1089/mdr.2014.0176
Kettleson, E., Kumar, S., Reponen, T., Vesper, S., Méheust, D., Grinshpun, S. A., et al. (2013). Stenotrophomonas, Mycobacterium, and Streptomyces in home dust and air: associations with moldiness and other home/family characteristics. Indoor Air 23, 387–396. doi: 10.1111/ina.12035
Levesque, C., Piche, L., Larose, C., and Roy, P. H. (1995). PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39, 185–191. doi: 10.1128/AAC.39.1.185
Lin, Y.-T., Huang, Y.-W., Chen, S.-J., Chang, C.-W., and Yang, T.-C. (2015). The SmeYZ efflux pump of Stenotrophomonas maltophilia contributes to drug resistance, virulence-related characteristics, and virulence in mice. Antimicrob. Agents Chemother. 59, 4067–4073. doi: 10.1128/AAC.00372-15
Liu, W., Tian, X.-Q., Wei, J.-W., Ding, L.-L., Qian, W., Liu, Z., et al. (2017). BsmR degrades c-di-GMP to modulate biofilm formation of nosocomial pathogen Stenotrophomonas maltophilia. Sci. Rep. 7:4665. doi: 10.1038/s41598-017-18286-x
Liu, W., Zou, D., Wang, X., Li, X., Zhu, L., Yin, Z., et al. (2012). Proteomic analysis of clinical isolate of Stenotrophomonas maltophilia with bla NDM-1, bla L1 and bla L2 β-lactamase genes under imipenem treatment. J. Proteome Res. 11, 4024–4033. doi: 10.1021/pr300062v
Looney, W. J., Narita, M., and Mühlemann, K. (2009). Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect. Dis. 9, 312–323. doi: 10.1016/S1473-3099(09)70083-0
Madi, H., Lukić, J., Vasiljević, Z., Biočanin, M., Kojić, M., Jovčić, B., et al. (2016). Genotypic and phenotypic characterization of Stenotrophomonas maltophilia strains from a Pediatric Tertiary Care Hospital in Serbia. PLoS One 11:e0165660. doi: 10.1371/journal.pone.0165660
McKay, G. A., Woods, D. E., Macdonald, K. L., and Poole, K. (2003). Role of phosphoglucomutase of Stenotrophomonas maltophilia in lipopolysaccharide biosynthesis, virulence, and antibiotic resistance. Infect. Immun. 71, 3068–3075. doi: 10.1128/IAI.71.6.3068-3075.2003
Mojica, M. F., Ouellette, C. P., Leber, A., Becknell, M. B., Ardura, M. I., Perez, F., et al. (2016). Successful treatment of bloodstream infection due to metallo-β-lactamase-producing Stenotrophomonas maltophilia in a renal transplant patient. Antimicrob. Agents Chemother. 60, 5130–5134. doi: 10.1128/AAC.00264-16
Neela, V., Rankouhi, S. Z. R., Van Belkum, A., Goering, R. V., and Awang, R. (2012). Stenotrophomonas maltophilia in Malaysia: molecular epidemiology and trimethoprim–sulfamethoxazole resistance. Int. J. Infect. Dis. 16, e603–e607. doi: 10.1016/j.ijid.2012.04.004
Nicodemo, A., Araujo, M., Ruiz, A., and Gales, A. C. (2004). In vitro susceptibility of Stenotrophomonas maltophilia isolates: comparison of disc diffusion, Etest and agar dilution methods. J. Antimicrob. Chemother. 53, 604–608. doi: 10.1093/jac/dkh128
Nicodemo, A., and Paez, J. G. (2007). Antimicrobial therapy for Stenotrophomonas maltophilia infections. Eur. J. Clin. Microbiol. Infect. Dis. 26, 229–237. doi: 10.1007/s10096-007-0279-3
Pfaller, M., Jones, R., Doern, G., Sader, H., Kugler, K., Beach, M., et al. (1999). Survey of blood stream infections attributable to gram-positive cocci: frequency of occurrence and antimicrobial susceptibility of isolates collected in 1997 in the United States, Canada, and Latin America from the SENTRY Antimicrobial Surveillance Program. Diag. Microbiol. Infect. Dis. 33, 283–297. doi: 10.1016/S0732-8893(98)00149-7
Pompilio, A., Piccolomini, R., Picciani, C., D’antonio, D., Savini, V., and Di Bonaventura, G. (2008). Factors associated with adherence to and biofilm formation on polystyrene by Stenotrophomonas maltophilia: the role of cell surface hydrophobicity and motility. FEMS Microbiol. Lett. 287, 41–47. doi: 10.1111/j.1574-6968.2008.01292.x
Pompilio, A., Pomponio, S., Crocetta, V., Gherardi, G., Verginelli, F., Fiscarelli, E., et al. (2011). Phenotypic and genotypic characterization of Stenotrophomonas maltophilia isolates from patients with cystic fibrosis: genome diversity, biofilm formation, and virulence. BMC Microbiol. 11:159. doi: 10.1186/1471-2180-11-159
Sader, H. S., and Jones, R. N. (2005). Antimicrobial susceptibility of uncommonly isolated non-enteric Gram-negative bacilli. Int. J. Antimicrob. Agents 25, 95–109. doi: 10.1016/j.ijantimicag.2004.10.002
Sanchez, M. B., Hernandez, A., and Martinez, J. L. (2009). Stenotrophomonas maltophilia drug resistance. Future Microbiol 4, 655–660. doi: 10.2217/fmb.09.45
Sánchez, M. B., and Martínez, J. L. (2015). The efflux pump SmeDEF contributes to trimethoprim-sulfamethoxazole resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 59, 4347–4348. doi: 10.1128/AAC.00714-15
Saniee, P., Hosseini, F., Kadkhodaei, S., Siavoshi, F., and Khalili-Samani, S. (2018). Helicobacter pylori multidrug resistance due to misuse of antibiotics in Iran. Arch. Iran. Med. 21, 283–288.
Shahla, M., Mozhdeh, R., Fatemeh, N., and Sasan, G. N. (2012). Prevalence of β-Lactamase production and antimicrobial susceptibility of multidrug resistant clinical isolates of non-fermenting Gram negative bacteria from hospitalized patients in Kerman/Iran. Jundishapur J. Microbiol. 2012, 405–410. doi: 10.5812/jjm.3399
Song, J. H., Sung, J. Y., Kwon, K. C., Park, J. W., Cho, H. H., Shin, S. Y., et al. (2010). Analysis of acquired resistance genes in Stenotrophomonas maltophilia. Korean J. Lab. Med. 30, 295–300. doi: 10.3343/kjlm.2010.30.3.295
Stepanović, S., Vuković, D., Hola, V., Bonaventura, G. D., Djukić, S., Ćirković, I., et al. (2007). Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115, 891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x
Sumida, K., Chong, Y., Miyake, N., Akahoshi, T., Yasuda, M., Shimono, N., et al. (2015). Risk factors associated with Stenotrophomonas maltophilia bacteremia: a matched case-control study. PLoS One 10:e0133731. doi: 10.1371/journal.pone.0133731
Tatman-Otkun, M., Gürcan, Ş., Özer, B., Aydoslu, B., and Bukavaz, Ş. (2005). The antimicrobial susceptibility of Stenotrophomonas maltophilia isolates using three different methods and their genetic relatedness. BMC Microbiol. 5:24. doi: 10.1186/1471-2180-5-24
Valdezate, S., Vindel, A., Martín-Dávila, P., Del Saz, B. S., Baquero, F., and Cantón, R. (2004). High genetic diversity among Stenotrophomonas maltophilia strains despite their originating at a single hospital. J. Clin. Microbiol. 42, 693–699. doi: 10.1128/JCM.42.2.693-699.2003
Valenza, G., Tappe, D., Turnwald, D., Frosch, M., König, C., Hebestreit, H., et al. (2008). Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J. Cyst. Fibros. 7, 123–127. doi: 10.1016/j.jcf.2007.06.006
Versalovic, J., Schneider, M., De Bruijn, F., and Lupski, J. R. (1994). Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5, 25–40.
Wang, W.-S., Liu, C.-P., Lee, C.-M., and Huang, F.-Y. (2004). Stenotrophomonas maltophilia bacteremia in adults: four years’ experience in a medical center in northern Taiwan. J. Microbiol. Immunol. Infect. 37, 359–365.
Wang, Y. L., Scipione, M. R., Dubrovskaya, Y., and Papadopoulos, J. (2013). Monotherapy with fluoroquinolone or trimethoprim-sulfamethoxazole for the treatment of Stenotrophomonas maltophilia infections. Antimicrob. Agents Chemother. 58, 01324–01313. doi: 10.1128/AAC.01324-13
Wu, H., Wang, J.-T., Shiau, Y.-R., Wang, H.-Y., Lauderdale, T.-L. Y., and Chang, S.-C. (2012). A multicenter surveillance of antimicrobial resistance on Stenotrophomonas maltophilia in Taiwan. J. Microbiol. Immunol. Infect. 45, 120–126. doi: 10.1016/j.jmii.2011.09.028
Wu, K., Yau, Y. C., Matukas, L., and Waters, V. (2013). Biofilm compared to conventional antimicrobial susceptibility of Stenotrophomonas maltophilia isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 57, 1546–1548. doi: 10.1128/AAC.02215-12
Keywords: antibiotic resistance genes, biofilm, efflux pump, sequence type, Stenotrophomonas maltophilia, trimethoprim-sulfamethoxazole
Citation: Bostanghadiri N, Ghalavand Z, Fallah F, Yadegar A, Ardebili A, Tarashi S, Pournajaf A, Mardaneh J, Shams S and Hashemi A (2019) Characterization of Phenotypic and Genotypic Diversity of Stenotrophomonas maltophilia Strains Isolated From Selected Hospitals in Iran. Front. Microbiol. 10:1191. doi: 10.3389/fmicb.2019.01191
Edited by:
Gilberto Igrejas, University of Trás-os-Montes and Alto Douro, PortugalReviewed by:
Xu Jia, Chengde Medical College, ChinaJason Sahl, Northern Arizona University, United States
Leila Vali, Kuwait University, Kuwait
Copyright © 2019 Bostanghadiri, Ghalavand, Fallah, Yadegar, Ardebili, Tarashi, Pournajaf, Mardaneh, Shams and Hashemi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ali Hashemi, YWxpLmhhc2hlbWlAc2JtdS5hYy5pcg==; aGFzaGVtaTEzODhAeWFob28uY29t
 Narjess Bostanghadiri1
Narjess Bostanghadiri1 Abbas Yadegar
Abbas Yadegar Samira Tarashi
Samira Tarashi Ali Hashemi
Ali Hashemi