- 1Institute of Preventive Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Research Center of Avian Disease, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 3Key Laboratory of Animal Disease and Human Health of Sichuan Province, Chengdu, China
- 4Pasteur Institute, Paris, France
Riemerella anatipestifer is a gram-negative bacterium that causes disease in ducks and other birds. Despite being an important pathogen in poultry, the pathogenesis and drug resistance mechanisms of this bacterium are poorly understood. An analysis of our unpublished RNA-Seq data showed that lptD, a gene encoding one of the lipopolysaccharide transport components, is transcribed at higher levels in strain CH-1 than in strain ATCC11845. In addition, strain CH-1 has been shown to display broader drug resistance than strain ATCC11845. Since LptD is involved in LPS biogenesis and drug resistance, we wondered if lptD is associated with increased R. anatipestifer resistance to glutaraldehyde, a disinfectant used in the production industry. In this study, the minimal inhibitory concentration (MIC) of glutaraldehyde for strain CH-1 was determined to be 0.125% (vol/vol), whereas an MIC of 0.05% (vol/vol) was observed for strain ATCC11845. Furthermore, the level of lptD transcription in strain CH-1 was consistently 2-fold higher than that observed in strain ATCC11845. Moreover, lptD transcription was upregulated in both strains at a subinhibitory concentration of glutaraldehyde. The role of lptD in R. anatipestifer was further assessed by constructing an ATCC11845 mutant strain with low lptD expression, R. anatipestifer ATCC11845 lptD−. The growth of R. anatipestifer ATCC11845 lptD− was severely impaired, and this strain was more susceptible than the wild-type strain to glutaraldehyde. Moreover, compared to the wild-type strain, R. anatipestifer ATCC11845 lptD− exhibited decreased biofilm formation and was more sensitive to duck serum. Finally, low lptD expression led to decreased colonization in ducklings. These results suggest that LptD is involved in R. anatipestifer glutaraldehyde resistance and pathogenicity.
Introduction
The cell envelope of gram-negative bacteria includes an inner membrane (IM), a periplasm, and an outer membrane (OM) (Ruiz et al., 2006). The OM is an asymmetric bilayer with inner and outer leaflets composed of phospholipids and lipopolysaccharide (LPS), respectively (Kamio and Nikaido, 1976). LPS forms a barrier to protect bacteria from hydrophobic antibiotics, dyes and detergents (Nikaido, 2003). Using Escherichia coli as an model system, the biogenesis of LPS has been shown to be a three-step process involving its synthesis in the cytoplasm, transport across the IM to the periplasmic space and insertion into the outer leaf of the OM (Raetz and Whitfield, 2002). The machinery that mediates LPS transports across the IM to the periplasmic space and its insertion into the OM has been well characterized through intense research during the last decade (Putker et al., 2015). Seven LPS transport proteins (Lpt), LptA, LptB, LptC, LptD, LptE, LptF, and LptG, have been reported to be involved in the transport of LPS in E. coli (Sperandeo et al., 2009; Putker et al., 2015). Furthermore, these proteins have been shown to localize to three different regions in the cell envelope. LptB, LptF, and LptG form an ABC transporter in the IM that provides the energy for LPS detachment (not flipping) from the IM and transport across the periplasm (Narita and Tokuda, 2009). LptA and LptC form a periplasmic complex that connects with LptD/E and LptBFG (Freinkman et al., 2012). LptD and LptE form a hetero-oligomeric complex in the OM (Wu et al., 2006) that is responsible for the translocation of LPS to the OM and its final assembly on the cell surface (Chng et al., 2010b). Decreased lptD transcription causes protein extravasation and membrane protein mislocalization in E. coli, suggesting that LptD is essential in this bacterial species and promotes correct cell membrane assembly (Braun and Silhavy, 2002). However, LptD is non-essential in Neisseria meningitidis, as bacteria are viable without LPS (Steeghs et al., 1998). Furthermore, LptD has been shown to be involved in organic solvent tolerance in E. coli and Helicobacter pylori (Ohtsu et al., 2004; Chiu et al., 2009).
Riemerella anatipestifer is a gram-negative bacterium that belongs to the Flavobacteriaceae family and causes septicemic diseases in ducks, geese, turkeys, and other birds (Segers et al., 1993). At present, because significant cross-protection has not been observed for these 21 different serotypes (Pathanasophon et al., 1995, 2002), it is difficult to control this disease in the duck production using vaccines. A number of factors have been reported to be involved in the pathogenesis of R. anatipestifer (Chang et al., 1998; Crasta et al., 2002; Hu et al., 2011; Wang et al., 2017; Yi et al., 2017; Liu et al., 2018). The wide use of antibiotics during poultry feeding has promoted the emergence of R. anatipestifer strains that are resistant to multiple antibiotics (Zhong et al., 2009; Luo et al., 2015, 2018; Huang et al., 2017; Zhang et al., 2017; Zhu et al., 2018). In a previous study, we showed that the strain CH-1 is resistant to many antibiotics, with the strain ATCC11845 being more susceptible to the tested antibiotics than CH-1 (Luo et al., 2015; Xing et al., 2015). The resistance of strain CH-1 and strain ATCC11845 to organic solvents is currently unknown, and glutaraldehyde is a commonly used disinfectant in poultry. According to our unpublished RNA-Seq data, lptD is transcribed at higher levels in strain CH-1 than that in strain ATCC11845 (Supplementary Figure S1 and Supplementary Tables S1, S2). In this study, we investigated whether strain CH-1 is more resistant to glutaraldehyde than strain ATCC11845 and if this phenotype is associated with the level of lptD transcription in these strains, the results of which will be helpful for laying a foundation for studying resistance mechanisms in R. anatipestifer.
Materials and Methods
Bacterial Strains, Primers and Growth Conditions
The bacterial strains and primers used in this study are shown in Table 1. R. anatipestifer was grown in GC broth (GCB) or tryptone soy broth (TSB) medium at 37°C with shaking (Liu et al., 2017). GCB agar plates were prepared by supplementing GCB with 1.5% agar. Alternatively, R. anatipestifer strains were also grown on LB agar supplemented with 5% sheep blood. When required, media were supplemented with erythromycin at a final concentration of 1 μg/ml or with different concentrations of glutaraldehyde or sodium dodecyl sulfate (SDS).
Construction of an R. anatipestifer ATCC11845 Strain Expressing Low Levels of lptD
An R. anatipestifer ATCC11845 strain expressing low levels of lptD was constructed using the natural transformation method as described previously (Liu et al., 2017). Briefly, ∼800-bp fragments upstream and downstream of the start codon of the lptD gene were amplified using the primer pairs lptD Pro-upP1/lptD Pro-upP2 and lptD Pro-downP1/lptD Pro-downP2, respectively (Table 1). A 994-bp erythromycin resistance cassette with a promoter was amplified from strain CH-1 (Luo et al., 2015) using the primers ErmP1/ErmP2 (Table 1). The three PCR fragments were fused by the overlap PCR method (Xiong et al., 2006), purified using a Universal DNA Purification kit (TIANGEN, Beijing, China) and served as donor DNA. Wild-type strain ATCC11845 served as the recipient strain for the fused fragments, which were introduced by natural transformation. Transformants in which the erythromycin resistance cassette with a promoter was inserted upstream of the lptD start codon were selected for on LB plates supplemented with 5% sheep blood and 1 μg/ml erythromycin. A strain expressing low levels of lptD, strain ATCC11845 lptD−, was verified by PCR by amplifying the erythromycin resistance cassette using the primers ErmP1/ErmP2 (Table 1).
Growth Rate Determination
The in vitro growth rates of the strains were determined as described previously (Wang et al., 2017). Briefly, the bacterial cells were grown overnight on LB plates supplemented with 5% sheep blood, after which a single colony was inoculated into 5 ml of TSB and cultured at 37°C with agitation for 10 h. Subsequently, the cultures were adjusted to an OD600 of 0.05 in 20 ml of fresh and grown at 37°C with shaking at 180 rpm, with OD600 values determined at every 2 h for 16 h.
Determination of the Minimal Inhibitory Concentrations (MICs)
The MICs of glutaraldehyde, SDS and antibiotics (novobiocin, imipenem rifampicin and polymyxin B) for R. anatipestifer were determined in 96-well microtiter plates as described in a previous study (Huang et al., 2017). Briefly, after culturing the strains to the logarithmic growth phase, the turbidity of the cultures was adjusted to 107 colony-forming units (CFU)/ml (100 μl/well). A culture without antibiotics was included as positive control, and a sample of uninoculated broth was used as a negative control. The experiments were repeated three times, with the results determined after a 24 h incubation at 37°C.
Biofilm Formation Assays
The R. anatipestifer strains were for biofilm formation in tubes as described previously with slight modifications (Kita et al., 2016). Cells of the R. anatipestifer strains were collected from LB agar plates supplemented with 5% sheep blood and resuspended in phosphate-buffered saline (PBS). The cells were washed three times with PBS. The bacterial suspensions were adjusted to an OD600 of 1 and then were inoculated into 5 ml of TSB supplemented with 5% serum at an OD600 of 0.1 in glass tubes and cultured at 37°C without shaking. After incubating for 24 h, the OD600 values of the cultures was determined, and the contents of each tube was carefully removed with a pipette. The tubes were washed three times with PBS and stained with 0.1% crystal violet for 30 min at room temperature. After removing the crystal violet solution and washing each tube twice with PBS, the biomass-associated crystal violet was extracted with 3 ml of absolute ethyl alcohol, and the absorbance at OD580 was measured.
Serum Bactericidal Assay
Serum lacking antibodies to R. anatipestifer was obtained from non-immune ducks and filter-sterilized (0.22 μm) for bactericidal assays. Briefly, after adjusting cultures of each bacterial strain tested to an OD600 of 1, the serum was added to the cell cultures at final concentrations of 10 or 20%. Cell cultures without serum were used as a negative control. The samples in group were incubated for 15 and 30 min at 37°C. Subsequently, the cell cultures were serially diluted and plated onto LB agar supplemented with 5% sheep blood. The survival rate was calculated as CFU/ml in pooled serum divided by the CFU/ml of the negative control.
Quantitative Real-Time PCR
Quantitative real-time PCR (qRT-PCR) was performed as described in a previous study with some modifications (Liu et al., 2016). Briefly, the tested strains were cultured in 20 ml of TSB to the exponential growth phase, after which RNA was extracted from the cell cultures at 1 OD600 using an RNAprep pure Cell/Bacteria kit (TIANGENTM, Beijing, China). cDNA was generated using HiScript reverse transcriptase according to the manufacturer’s instructions. qRT-PCR was performed to determine the transcript level of lptD using SYBR Green Master Mix (Vazyme: Q111-01) and the primers qlptD P1/qlptD P2 (Table 1). The gene recA served as an internal reference gene to normalize the level of lptD expression. Three samples and technical replicates were performed, and the fold change was calculated using the ΔΔCt method as previously described (Pfaffl, 2001).
Colonization Assays
Colonization studies were conducted using ten 3-day-old Pekin ducklings per group. The wild-type strain and the mutant expressing low levels of lptD were cultured overnight on sheep blood plates at 37°C. Subsequently, bacterial cells were scraped from the plates, resuspended in TSB medium and cultured to the exponential phase at 37°C with shaking at 180 rpm. After collecting the bacteria by centrifugation at 4°C for 10 min, the cells were washed three times and suspended in PBS. Subsequently, 109 CFU of the bacterial suspensions were intramuscularly injected into the legs of ducklings. The blood, livers and brains of the ducklings were collected at 12 and 18 h postinoculation and homogenized in PBS (0.1 g sample/0.9 ml PBS) using a Nasco WHIRL-PAK (B01245WA, United States) as previously described (Liu et al., 2018). The homogenized contents were serially diluted and spread onto blood agar plates for enumeration.
Ethics Statement
All ducks were handled in strict adherence to the recommendations of the local animal welfare bodies and the Sichuan Agricultural University Ethics Committee (SYXK2014-187). The protocol was approved by the Sichuan Agricultural University Ethics Committee.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 7.0 for Windows (GraphPad Software Inc., San Diego, CA, United States). The significance of the data was ascertained using Student’s t-test, and a value of P < 0.05 was considered significant.
Results
R. anatipestifer ATCC11845 Is More Susceptible to Glutaraldehyde Than Strain CH-1
Glutaraldehyde is both an organic solvent and a hydrophobic drug that is commonly used as a disinfectant (Chiu et al., 2009). To explore whether different R. anatipestifer strains have different tolerances to glutaraldehyde, the MICs of glutaraldehyde for strain CH-1 and strain ATCC11845 were determined. The results showed that the MIC of glutaraldehyde for strain CH-1 and strain ATCC11845 was 0.125% (vol/vol) and 0.05% (vol/vol), respectively (Table 2), revealing that strain ATCC11845 is more susceptible to glutaraldehyde than strain CH-1.
lptD Is Transcribed at Lower Levels in R. anatipestifer ATCC11845 Than Strain CH-1
Bacterial resistance to glutaraldehyde has been reported to be associated with lptD (Chiu et al., 2009). Inprevious study, RNA-Seq data showed that lptD had higher transcription level in strain CH-1, compared to that in strain ATCC11845 (Supplementary Figure S1 and Supplementary Tables S1, S2). To verify whether the tolerance of different R. anatipestifer strains to glutaraldehyde is caused by lptD, the transcription of lptD in strain CH-1 and strain ATCC11845 was assessed by qRT-PCR as described in a previous study (Chiu et al., 2009). As shown in Figure 1, the level of lptD transcription in strain CH-1 was 2-fold higher than that observed in strain ATCC11845. Thus, the different tolerance of the strains to glutaraldehyde was predicted to be associated with the level of lptD transcription in R. anatipestifer.
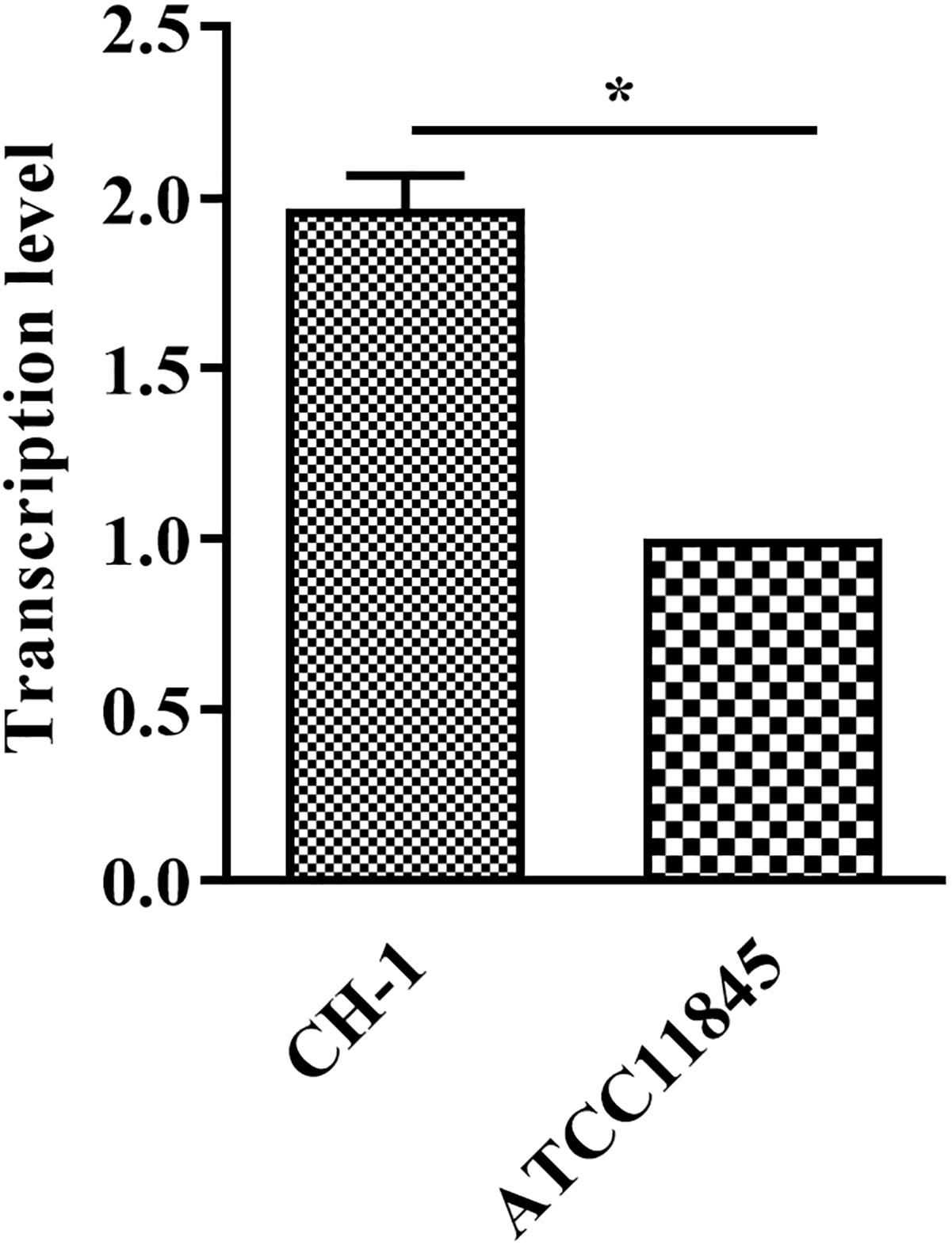
Figure 1. Transcription of lptD in R. anatipestifer CH-1 and ATCC11845. RNA was extracted and performed qRT-PCR. The data were analyzed using Student’s t-test. The error bars represent the standard deviations of three independent experiments, ∗, p < 0.05.
Transcription of the lptD Gene Is Induced by Glutaraldehyde in R. anatipestifer CH-1 and ATCC11845
To further assess whether the tolerance of R. anatipestifer to glutaraldehyde is correlated with lptD expression, strain CH-1 and strain ATCC11845 were treated with a sub-inhibitory concentration of glutaraldehyde [0.01% (vol/vol) and 0.005% (vol/vol) for strain CH-1 and strain ATCC11845, respectively] and assayed for lptD expression by qRT-PCR. The results showed that lptD transcription increased 3- and 10-fold in strain CH-1 and strain ATCC11845 after incubation with glutaraldehyde, respectively (Figure 2), indicating that lptD transcription is induced by glutaraldehyde in both of these strains.
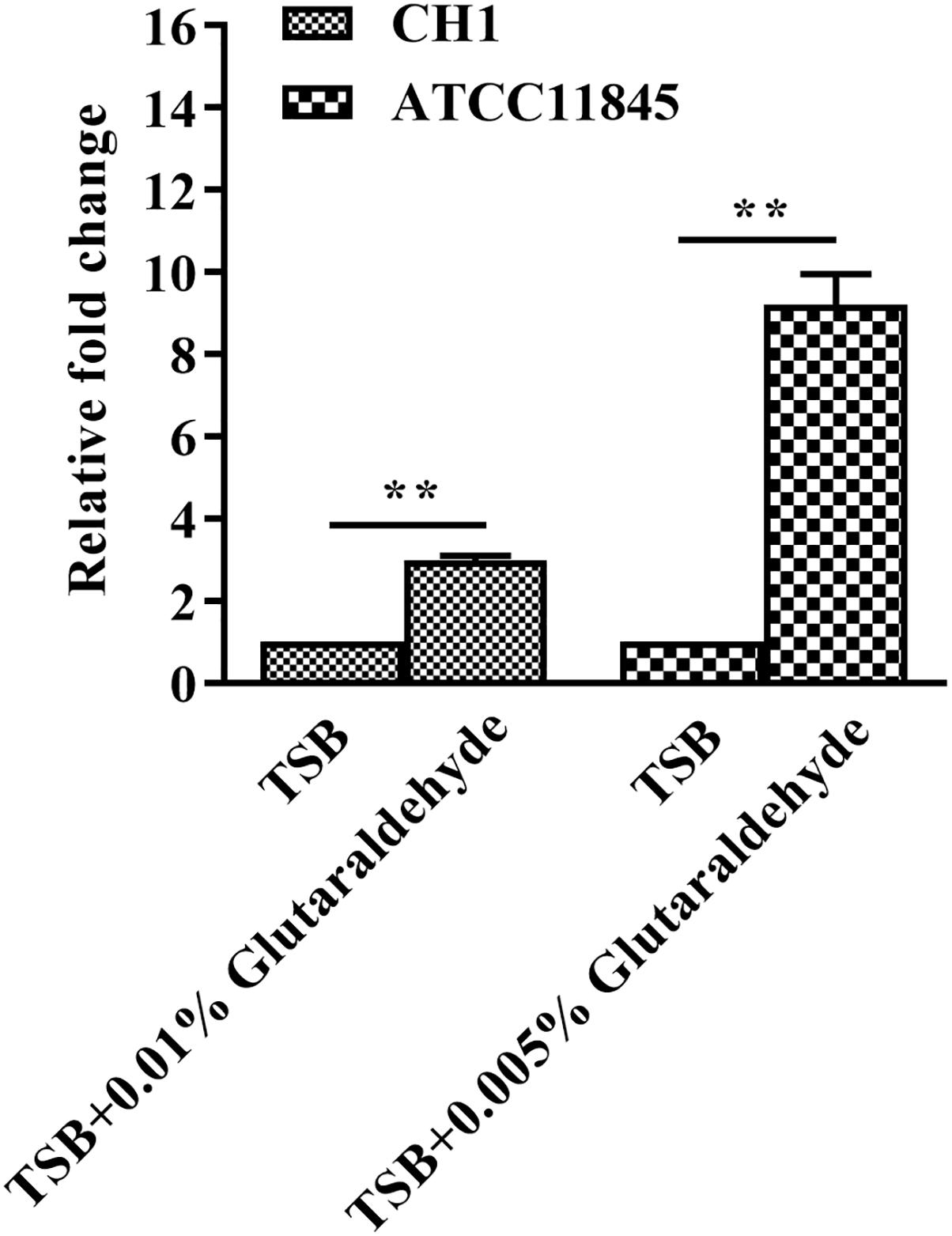
Figure 2. Changes in lptD transcription in R. anatipestifer CH-1 and ATCC11845 in TSB and TSB with glutaraldehyde. qRT-PCR analysis of the relative expression of lptD in strain CH-1 and strain ATCC11845 in TSB and TSB with glutaraldehyde (0.01% (vol/vol) and 0.005% (vol/vol) for strain CH-1 and strain ATCC11845, respectively).The data were analyzed using Student’s t-test. The error bars represent the standard deviations of three independent experiments, ∗∗, p < 0.01.
Low lptD Expression Affects R. anatipestifer ATCC11845 Growth in TSB Medium
To elucidate the function of lptD in R. anatipestifer, we attempted to construct an lptD mutant strain; however, this effort failed despite numerous attempts, suggesting that lptD is an essential gene in R. anatipestifer. This result was not unexpected, as lptD has been consistently shown to be essential in E. coli (Sampson et al., 1989; Braun and Silhavy, 2002; Chng et al., 2010a) and Salmonella typhimurium (Dong et al., 2014; Gu et al., 2015). Subsequently, we inserted an erythromycin resistance gene driven by its native promoter upstream of the lptD start codon region to decrease lptD transcription, which was shown 2-fold lower than in the wild-type strain by qRT-PCR (Figure 3A). The strain with low lptD expression was named strain ATCC11845 lptD−. Moreover, strain ATCC11845 lptD− had no significant effect on the transcription of upstream gene RA0C_1120 and downstream gene RA0C_1122, suggesting that it did not cause polar effect to RA0C_1122 (Figure 3A). Later, strain ATCC11845 lptD− was used to evaluate the effect of lptD on the growth of strain ATCC11845. The results showed that wild-type strain ATCC11845 grew well in TSB liquid medium, whereas that of strain ATCC11845 lptD− was severely impaired (Figure 3B).
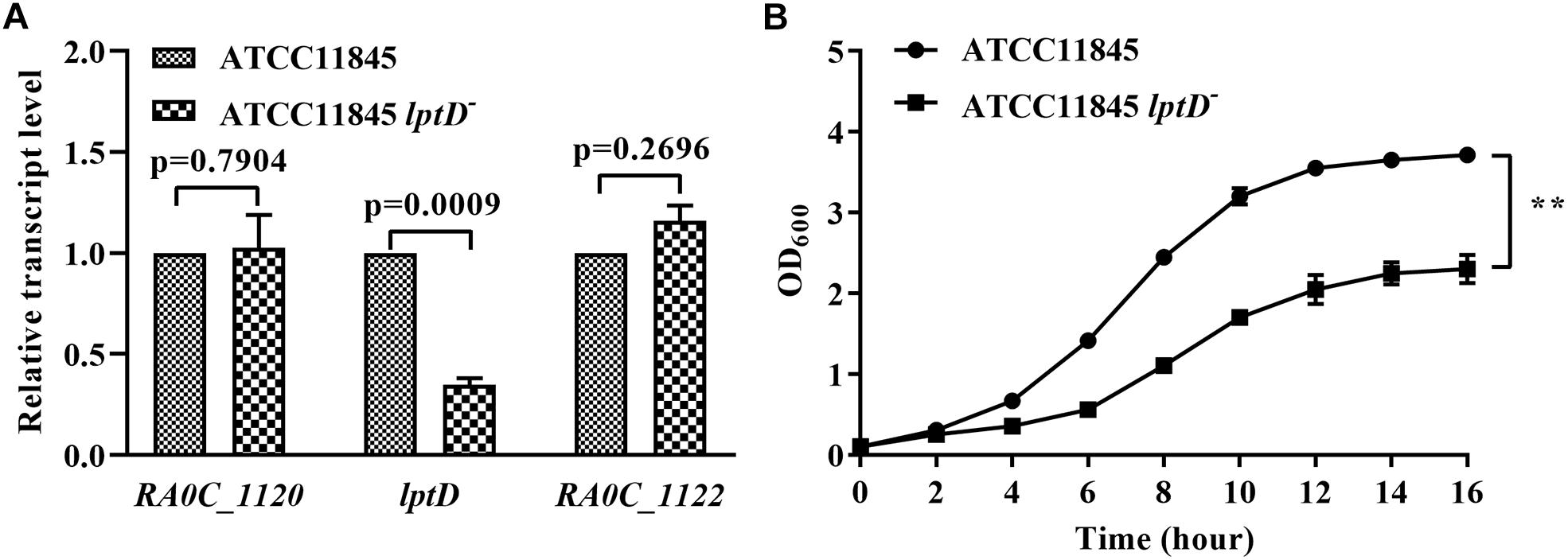
Figure 3. Effect of low lptD expression on the growth of R. anatipestifer ATCC11845 in TSB. (A) The transcription of RA0C_1120, lptD and RA0C_1122 in strain ATCC11845 and ATCC11845 lptD−. qRT-PCR analysis of the transcription of RA0C_1120, lptD and RA0C_1122 in ATCC11845 and ATCC11845 lptD− in TSB. The data were analyzed using a t-test. The error bars represent the standard deviations of three independent experiments. (B) The growth of strain ATCC11845 and ATCC11845 lptD− in TSB. Cells were grown in 20 ml of TSB medium at 37°C with an initial OD600 of 0.05. The OD600 values were subsequently measured every 2 h for 16 h, ∗∗, p < 0.01.
R. anatipestifer ATCC11845 lptD− Is More Susceptible to Glutaraldehyde and Several Antibiotics Than the Wild-Type Strain
To directly assess whether lptD affects the tolerance of R. anatipestifer to glutaraldehyde and several antibiotics, including novobiocin, imipenem, rifampicin and polymyxin B, the MICs of glutaraldehyde and antibiotics for strain ATCC11845 and strain ATCC11845 lptD− were determined. As shown in Table 2, strain ATCC11845 lptD− was more susceptible to glutaraldehyde than strain ATCC11845, with MICs of 0.0125% (vol/vol) and 0.05% (vol/vol) observed for these strains, respectively. The MICs of novobiocin, imipenem, rifampicin and polymyxin B for strain ATCC11845 lptD− were 0.025, 0.125, 0.005, and 250 μg/ml, respectively, whereas the MICs for these antibiotics for strain ATCC11845 were 0.05, 0.5, 0.025, and >500 μg/ml, respectively. These results suggested that the lptD− strain was more susceptible than the wild-type strain to glutaraldehyde, novobiocin, imipenem, rifampicin and polymyxin B. However, compared to that of ATCC11845, the sensitivity of ATCC11845 lptD− to SDS did not have significant change (Table 2).
Decreased lptD Expression Affects R. anatipestifer Biofilm Formation and Resistance to Duck Serum
Previous studies showed that LPS is a primary component of biofilms (Murphy et al., 2014; Alshalchi and Anderson, 2015). Thus, the role of lptD in R. anatipestifer biofilm formation was examined in test tubes. The results showed that strain ATCC11845 lptD− was significantly attenuated in biofilm formation compared to the wild-type strain (Figure 4A). The OD580 values for strain ATCC11845 and strain ATCC11845 lptD− were 1.52 and 0.48, respectively, suggesting that the biofilm formation of strainATCC11845 lptD− was significantly lower than that of the wild-type strain (Figure 4B). These results indicated that the decreased expression of lptD had an effect on R. anatipestifer biofilm formation. Next, a bactericidal assay was performed to determine whether lptD is involved in the resistance of R. anatipestifer to duck serum. As shown in Figure 5, the survival rates of strain ATCC11845 and strain ATCC11845 lptD− in 10% non-inactivated serum for 15 min were 60.1 and 39.9%, respectively. In contrast, when incubated in 20% non-inactivated serum for 15 min, all strain ATCC11845 lptD− bacteria were killed, whereas the survival rate of strain ATCC11845 was 47.6%. When incubated in 10% non-inactivated serum for 30 min, the bacterial survival rates of strain ATCC11845 and strain ATCC11845 lptD− were 26.3 and 15.5%, respectively. When the concentration of non-inactivated serum was increased to 20% for 30 min, all strain ATCC11845 lptD− cells were killed, whereas the survival rate of strain ATCC11845 was 15.9%. Taken together, these results suggested that the decreased expression of lptD in strain ATCC11845 lptD− resulted in significantly greater sensitivity to duck serum than the wild-type strain.
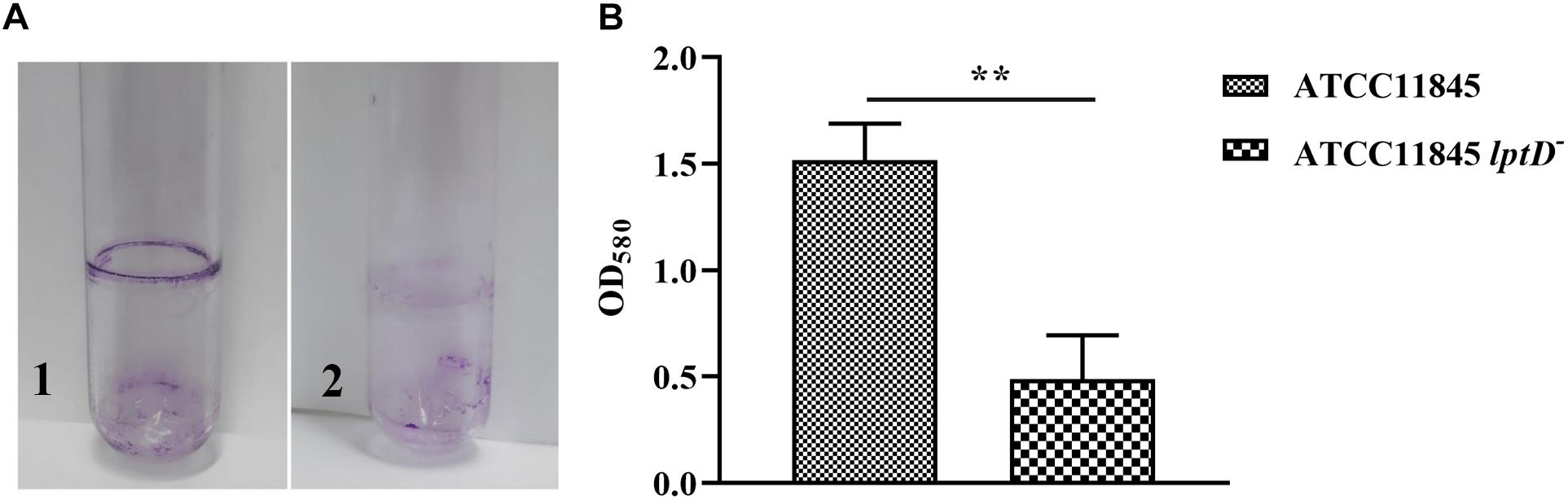
Figure 4. Biofilm formation assay for R. anatipestifer ATCC11845 and ATCC11845 lptD−. (A) Representative images of the results of the crystal violet biofilm formation assay. Strain ATCC11845 (1) and ATCC11845 lptD− (2) were collected from LB agar plates supplemented with 5% sheep blood and resuspended in TSB supplemented with 5% serum at an OD600 of 0.1 in glass tubes and incubated at 37°C without shaking for 24 h. Subsequently, the tubes were washed three times with PBS andstained with 6 ml of 0.1% crystal violet for 30 min at room temperature after carefully removing the bacteria. (B) The absorbance of crystal violet-stained biofilm at OD580, ∗∗, p < 0.01.
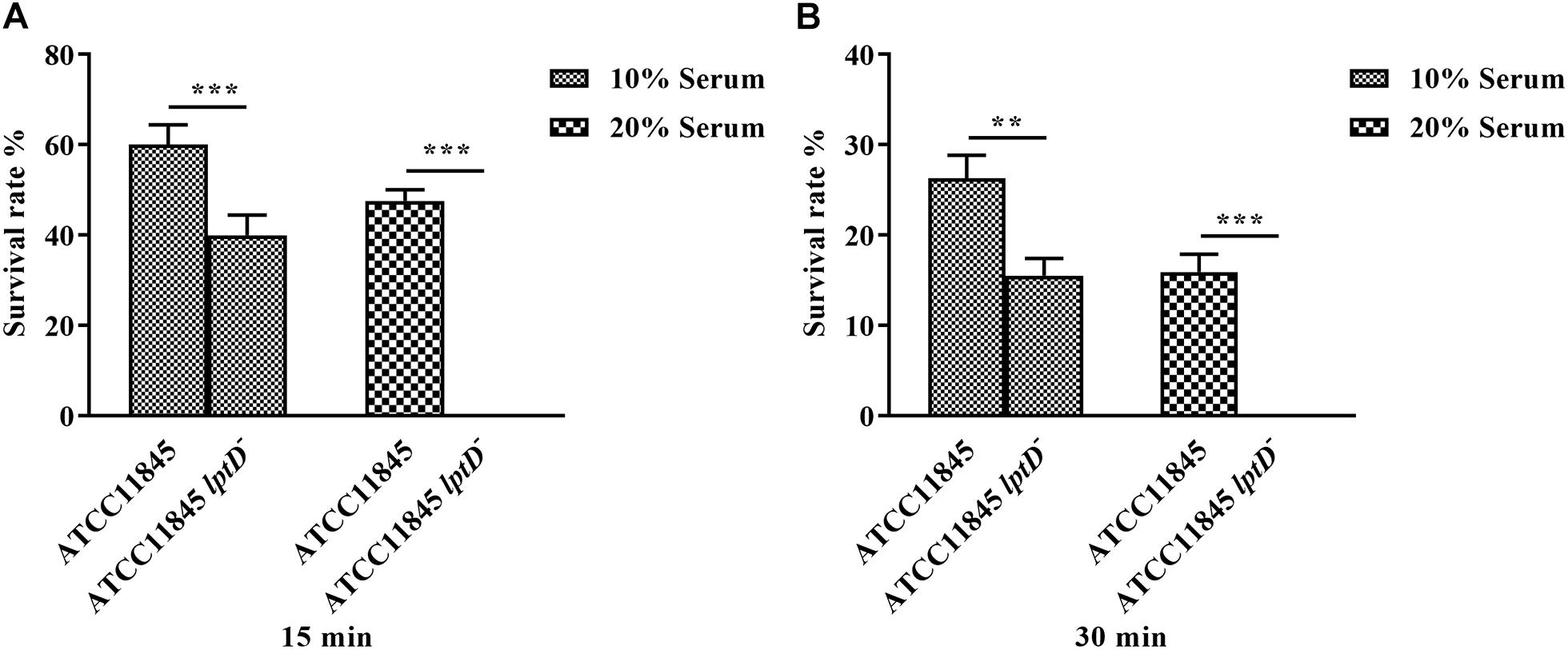
Figure 5. Serum bactericidal assay. Bacteria were incubated with 10 and 20% normal duck serum at 37°C and enumerated after 15 min (A) and 30 min (B) of incubation. The resistance of strain ATCC11845 lptD− to duck serum was significantly reduced compared with that of strain ATCC11845 (∗∗, p < 0.01, ∗∗∗, p < 0.001).The survival rate (%) was calculated as follows: (bacterial CFU with serum treatment/bacterial CFU with PBS treatment) × 100.
Decreased lptD Expression Affects the Colonization of R. anatipestifer ATCC11845 in vivo
To further investigate whether lptD contributes to the colonization dynamics of R. anatipestifer during systemic infection, colonization assay was conducted by infecting 3-day-old ducklings with strain ATCC11845 or strain ATCC11845 lptD− by leg muscle injection. Compared to ducklings infected with strain ATCC11845, at 12 h postinoculation, a notable reduction in the bacterial load was observed in ducklings infected with strain ATCC11845 lptD− in the heart blood (6-fold reduction), liver (4-fold reduction), brain tissue (7-fold reduction) and spleen (2-fold reduction) (Figure 6A). At 18 h postinoculation, compared to strain ATCC11845, significant reductions in the strain ATCC11845 lptD− bacterial loads were still observed in the blood (4-fold reduction), livers (23-fold reduced), brains (42-fold reduction) and spleens (4-fold reduction) (Figure 6B). These results suggest that lptD is involved in the pathogenesis of R. anatipestifer.
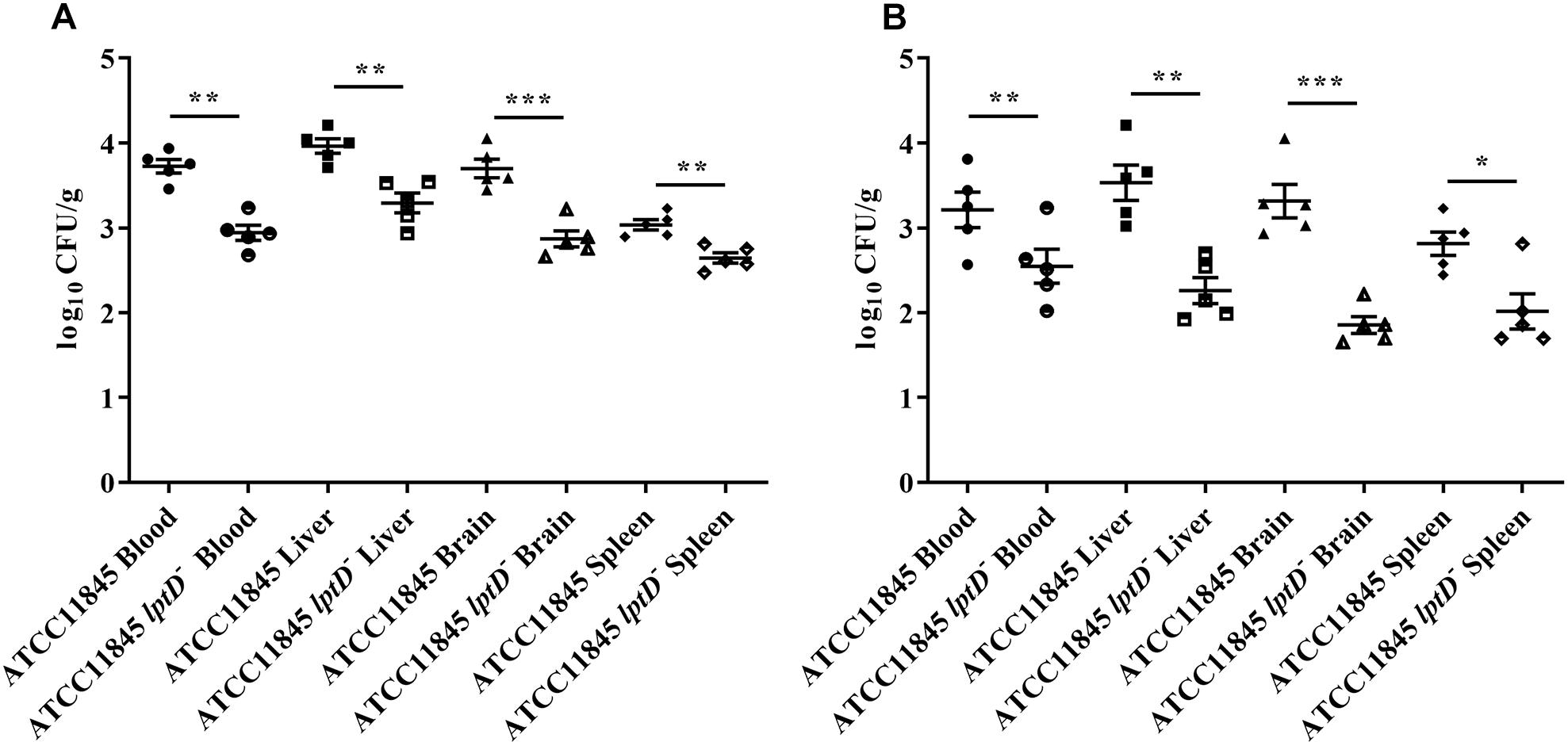
Figure 6. Colonization of R. anatipestifer ATCC11845 and ATCC11845 lptD− in vivo. The strain ATCC11845 (109 CFU) and ATCC11845 lptD− (109 CFU) strains were injected into the leg muscles of 3-day-old ducklings. At 12 h (A) and 18 h (B) postinfection, bacteria were isolated from the livers, brains, spleens, and blood according to the methods described in the “Materials and Methods” section. The data points represent the log10CFU/g of individual animals in the indicated organs, with the bars representing the median values (n = 5), ∗, p < 0.05, ∗∗, p < 0.01, and ∗∗∗, p < 0.001.
Discussion
With the increasing use of antimicrobials, resistance in R. anatipestifer is becoming an important concern. Compared to strain ATCC11845, strain CH-1 is resistant to many antibiotics. At present, it is unknown the resistance to organic solvents in strain CH-1 and strain ATCC11845. Hydrophobic organic solvents are extremely toxic to microorganisms, even at the very low concentration of 0.1% (vol/vol). The first reported organic-solvent-tolerant bacterium was Pseudomonas (Inoue and Horikoshi, 1989). As an organic solvent, glutaraldehyde has been used extensively as a hydrophobic drug. The MIC of glutaraldehyde for strain CH-1 and strain ATCC11845 was assessed to determine if the resistance of these strains to this disinfectant is different. We observed that strain CH-1 and strain ATCC11845 differ in their tolerance to glutaraldehyde, with strain ATCC11845 being more susceptible than strain CH-1.
It was previously reported that imp/ostA (namely, lptD) is involved in glutaraldehyde resistance in a clinical strain of H. pylori (Chiu et al., 2007, 2009). To investigate whether the difference in glutaraldehyde resistance of R. anatipestifer CH-1 and ATCC11845 is associated with lptD, the transcription of this gene was measured in these strains by qRT-PCR. The result showed that the level of lptD transcription in R. anatipestifer CH-1 was 2-fold higher than that observed in strain ATCC11845. After that, when R. anatipestifer was treated with a subinhibitory concentration of glutaraldehyde [0.01% (vol/vol) for strain CH-1, 0.005% (vol/vol) for strain ATCC11845], the transcription levels of lptD were both increased. Altogether, these results suggested that the transcription level of lptD was associated with glutaraldehyde resistance, which is consistent with the results reported for H. pylori (Chiu et al., 2009).
The lpt genes are widely distributed in bacteria that do or do not produce LPS (Putker et al., 2015). LPS transport has been studied extensively in the β- and γ-proteobacteria N. meningitides and E. coli, respectively. Seven Lpt proteins have been shown to be involved in this process. Searching the genome of R. anatipestifer ATCC11845 for homologs of Lpt proteins revealed that LptA, LptB, LptD, LptF, and LptG (RA0C_1913, RA0C_1993, RA0C_1121, RA0C_0335, and RA0C_1496) exhibit 42, 53.78, 48, 19.96, and 30.56% identity to previously identified proteins in E. coli, respectively. Homologs of LptC and LptE were not identified through sequence analysis of the R. anatipestifer ATCC11845 genome. In E. coli, LptD forms a complex with lipoprotein LptE to help LPS transport across outer membrane (Chimalakonda et al., 2011). Overall, it appears that R. anatipestifer transports LPS to the outer membrane via similar Lpt machinery, but there are some differences in the process due to a lack of LptC and LptE. LPS is essential in most gram-negative bacteria, with the notable exception of N. meningitides (Steeghs et al., 1998). The lptD gene has been consistently shown to be an essential gene in E. coli (Sampson et al., 1989; Braun and Silhavy, 2002; Chng et al., 2010a) and S. typhimurium (Putker et al., 2015). Several attempts were made to generate an lptD knockout in R. anatipestifer to study the function of this gene directly. However, the failure to obtain this mutant suggested that lptD is also an essential gene in R. anatipestifer. Subsequently, a R. anatipestifer ATCC11845 lptD− strain was constructed with low lptD expression, which was confirmed by qRT-PCR. The results of the glutaraldehyde sensitivity assay showed that R. anatipestifer ATCC11845 lptD− was more susceptible to glutaraldehyde than strain ATCC11845, suggesting that LPS forms a barrier that protects cells from glutaraldehyde and promotes resistance. Although we constructed a complemented strain using the shuttle plasmid pLMF03, the wild-type phenotype was not restored in this strain. The lack of complementation could be due to problems related with the backbone vector or lptD expression levels. Thus, it was absolutely required to establish a method for conditional mutant to study the function of the essential gene in R. anatipestifer. Furthermore, several attempts to extract LPS from strain ATCC11845 and strain ATCC11845 lptD− failed. In addition, we have analyzed the genetic organization of the lptD in the genome of R. anatipestifer ATCC11845. As shown in Supplementary Figure S2, it was shown that lptD (RA0C_1121) was not located in an operon, however, RA0C_1122 and RA0C_1123 formed an operon through bioinformatic analysis. The direction of transcription of lptD locus and gene RA0C_1120 is reversed. The intergenic region between lptD and RA0C_1120 or RA0C_1122 is 22 bp and 89 bp, respectively. qRT-PCR revealed that strain ATCC11845 lptD− had no significant effect on the transcription level of RA0C_1120 and RA0C_1122.
Lipopolysaccharide is a primary component of biofilm in P. aeruginosa (Murphy et al., 2014; Alshalchi and Anderson, 2015). To investigate whether lptD affects biofilm formation, the biofilms of R. anatipestifer ATCC11845 and strain ATCC11845 lptD− were examined. The results showed that biofilms formed by strain ATCC11845 lptD− had decreased biomass compared to those formed by the wild-type strain. Serum bactericidal assays showed that strain ATCC11845 lptD− was significantly more sensitive to duck serum than the wild-type strain. In vivo, the bacterial loads of R. anatipestifer ATCC11845 lptD− were lower than those of the wild-type strain in the blood, livers, brains and spleens of ducklings. Taken together, these results suggested that lptD is involved in glutaraldehyde resistance and bacterial virulence in R. anatipestifer.
Data Availability
The raw data supporting the conclusion of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
ML, AC, and FB conceived and designed the experiments. LH and TM constructed the RA ATCC11845 strain with low lptD expression and assessed the sensitivity of tested strains to glutaraldehyde and SDS. DZ, MW, and YL performed the qRT-PCR to determine the level of lptD transcription. LZ, XC, YY, and JH performed the biofilm formation assay and animal experiments. LP, MR, MW, RJ, SC, and XZ analyzed the data. BT, YW, QY, and SZ contributed to the reagents, materials, and analysis tools. LH, ML, FB, and AC wrote the manuscript. All authors reviewed the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China under Grant Number 31572521, the China Agricultural Research System under Grant Number CARS-42-17, and the Sichuan Veterinary Medicine and Drug Innovation Group of the China Agricultural Research System under Grant Number CARS-SVDIP.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01443/full#supplementary-material
References
Alshalchi, S. A., and Anderson, G. G. (2015). Expression of the lipopolysaccharide biosynthesis gene lpxD affects biofilm formation of Pseudomonas aeruginosa. Arch. Microbiol. 197, 135–145. doi: 10.1007/s00203-014-1030-y
Braun, M., and Silhavy, T. J. (2002). Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol. Microbiol. 45, 1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x
Chang, C. F., Hung, P. E., and Chang, Y. F. (1998). Molecular characterization of a plasmid isolated from Riemerella anatipestifer. Avian Pathol. 27, 339–345. doi: 10.1080/03079459808419349
Chimalakonda, G., Ruiz, N., Chng, S. S., Garner, R. A., Kahne, D., and Silhavy, T. J. (2011). Lipoprotein LptE is required for the assembly of LptD by the beta-barrel assembly machine in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 108, 2492–2497. doi: 10.1073/pnas.1019089108
Chiu, H. C., Lin, T. L., and Wang, J. T. (2007). Identification and characterization of an organic solvent tolerance gene in Helicobacter pylori. Helicobacter 12, 74–81. doi: 10.1111/j.1523-5378.2007.00473.x
Chiu, H. C., Lin, T. L., Yang, J. C., and Wang, J. T. (2009). Synergistic effect of imp/ostA and msbA in hydrophobic drug resistance of Helicobacter pylori. BMC Microbiol. 9:136. doi: 10.1186/1471-2180-9-136
Chng, S. S., Gronenberg, L. S., and Kahne, D. (2010a). Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry 49, 4565–4567. doi: 10.1021/bi100493e
Chng, S. S., Ruiz, N., Chimalakonda, G., Silhavy, T. J., and Kahne, D. (2010b). Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc. Natl. Acad. Sci. U.S.A. 107, 5363–5368. doi: 10.1073/pnas.0912872107
Crasta, K. C., Chua, K. L., Subramaniam, S., Frey, J., Loh, H., and Tan, H. M. (2002). Identification and characterization of CAMP cohemolysin as a potential virulence factor of Riemerella anatipestifer. J. Bacteriol. 184, 1932–1939. doi: 10.1128/jb.184.7.1932-1939.2002
Dong, H., Xiang, Q., Gu, Y., Wang, Z., Paterson, N. G., Stansfeld, P. J., et al. (2014). Structural basis for outer membrane lipopolysaccharide insertion. Nature 511, 52–56. doi: 10.1038/nature13464
Freinkman, E., Okuda, S., Ruiz, N., and Kahne, D. (2012). Regulated assembly of the transenvelope protein complex required for lipopolysaccharide export. Biochemistry 51, 4800–4806. doi: 10.1021/bi300592c
Gu, Y., Stansfeld, P. J., Zeng, Y., Dong, H., Wang, W., and Dong, C. (2015). Lipopolysaccharide is inserted into the outer membrane through an intramembrane hole, a lumen gate, and the lateral opening of LptD. Structure 23, 496–504. doi: 10.1016/j.str.2015.01.001
Hu, Q., Han, X., Zhou, X., Ding, C., Zhu, Y., and Yu, S. (2011). OmpA is a virulence factor of Riemerella anatipestifer. Vet. Microbiol. 150, 278–283. doi: 10.1016/j.vetmic.2011.01.022
Huang, L., Yuan, H., Liu, M. F., Zhao, X. X., Wang, M. S., Jia, R. Y., et al. (2017). Type B chloramphenicol acetyltransferases are responsible for chloramphenicol resistance in Riemerella anatipestifer, China. Front. Microbiol. 8:297. doi: 10.3389/fmicb.2017.00297
Inoue, A., and Horikoshi, K. (1989). A Pseudomonas thrives in high concentrations of toluene. Nature 338, 264–266.
Kamio, Y., and Nikaido, H. (1976). Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry 15, 2561–2570. doi: 10.1021/bi00657a012
Kita, D., Shibata, S., Kikuchi, Y., Kokubu, E., Nakayama, K., Saito, A., et al. (2016). Involvement of the Type IX secretion system in Capnocytophaga ochracea gliding motility and biofilm formation. Appl. Environ. Microbiol. 82, 1756–1766. doi: 10.1128/AEM.03452-15
Liu, M., Huang, M., Shui, Y., Biville, F., Zhu, D., Wang, M., et al. (2018). Roles of B739_1343 in iron acquisition and pathogenesis in Riemerella anatipestifer CH-1 and evaluation of the RA-CH-1DeltaB739_1343 mutant as an attenuated vaccine. PLoS One 13:e0197310. doi: 10.1371/journal.pone.0197310
Liu, M., Wang, M., Zhu, D., Wang, M., Jia, R., Chen, S., et al. (2016). Investigation of TbfA in Riemerella anatipestifer using plasmid-based methods for gene over-expression and knockdown. Sci. Rep. 6:37159. doi: 10.1038/srep37159
Liu, M., Zhang, L., Huang, L., Biville, F., Zhu, D., Wang, M., et al. (2017). Use of natural transformation to establish an easy knockout method in Riemerella anatipestifer. Appl. Environ. Microbiol. 83:e00127-17. doi: 10.1128/AEM.00127-17
Luo, H., Liu, M., Wang, L., Zhou, W., Wang, M., Cheng, A., et al. (2015). Identification of ribosomal RNA methyltransferase gene ermF in Riemerella anatipestifer. Avian Pathol. 44, 162–168. doi: 10.1080/03079457.2015.1019828
Luo, H. Y., Liu, M. F., Wang, M. S., Zhao, X. X., Jia, R. Y., Chen, S., et al. (2018). A novel resistance gene, lnu(H), conferring resistance to lincosamides in Riemerella anatipestifer CH-2. Int. J. Antimicrob. Agents 51, 136–139. doi: 10.1016/j.ijantimicag.2017.08.022
Murphy, K., Park, A. J., Hao, Y., Brewer, D., Lam, J. S., and Khursigara, C. M. (2014). Influence of O polysaccharides on biofilm development and outer membrane vesicle biogenesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 196, 1306–1317. doi: 10.1128/JB.01463-13
Narita, S., and Tokuda, H. (2009). Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 583, 2160–2164. doi: 10.1016/j.febslet.2009.05.051
Nikaido, H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656. doi: 10.1128/mmbr.67.4.593-656.2003
Ohtsu, I., Kakuda, N., Tsukagoshi, N., Dokyu, N., Takagi, H., Wachi, M., et al. (2004). Transcriptional analysis of the ostA/imp gene involved in organic solvent sensitivity in Escherichia coli. Biosci. Biotechnol. Biochem. 68, 458–461. doi: 10.1271/bbb.68.458
Pathanasophon, P., Phuektes, P., Tanticharoenyos, T., Narongsak, W., and Sawada, T. (2002). A potential new serotype of Riemerella anatipestifer isolated from ducks in Thailand. Avian Pathol. 31, 267–270. doi: 10.1080/03079450220136576
Pathanasophon, P., Sawada, T., and Tanticharoenyos, T. (1995). New serotypes of Riemerella anatipestifer isolated from ducks in Thailand. Avian Pathol. 24, 195–199. doi: 10.1080/03079459508419059
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45.
Putker, F., Bos, M. P., and Tommassen, J. (2015). Transport of lipopolysaccharide to the gram-negative bacterial cell surface. FEMS Microbiol. Rev. 39, 985–1002. doi: 10.1093/femsre/fuv026
Raetz, C. R. H., and Whitfield, C. (2002). Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700. doi: 10.1146/annurev.biochem.71.110601.135414
Ruiz, N., Kahne, D., and Silhavy, T. J. (2006). Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 4, 57–66. doi: 10.1038/nrmicro1322
Sampson, B. A., Misra, R., and Benson, S. A. (1989). Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics 122, 491–501.
Segers, P., Mannheim, W., Vancanneyt, M., Brandt, K. D., Hinz, K. H., Kersters, K., et al. (1993). Riemerella anatipestifer gen. nov., comb. nov., the causative agent of septicemia anserum exudativa, and its phylogenic affiliation within the Flavobacterium-Cytophaga rRNA homology group. Int. J. Syst. Bacteriol. 43, 768–776. doi: 10.1099/00207713-43-4-768
Sperandeo, P., Deho, G., and Polissi, A. (2009). The lipopolysaccharide transport system of gram-negative bacteria. Biochim. Biophys. Acta 1791, 594–602. doi: 10.1016/j.bbalip.2009.01.011
Steeghs, L., den Hartog, R., den Boer, A., Zomer, B., Roholl, P., and van der Ley, P. (1998). Meningitis bacterium is viable without endotoxin. Nature 392, 449–450. doi: 10.1038/33046
Wang, M., Zhang, P., Zhu, D., Wang, M., Jia, R., Chen, S., et al. (2017). Identification of the ferric iron utilization gene B739_1208 and its role in the virulence of R. anatipestifer CH-1. Vet. Microbiol. 201, 162–169. doi: 10.1016/j.vetmic.2017.01.027
Wu, T., McCandlish, A. C., Gronenberg, L. S., Chng, S. S., Silhavy, T. J., and Kahne, D. (2006). Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 103, 11754–11759. doi: 10.1073/pnas.0604744103
Xing, L., Yu, H., Qi, J., Jiang, P., Sun, B., Cui, J., et al. (2015). ErmF and ereD are responsible for erythromycin resistance in Riemerella anatipestifer. PLoS One 10:e0131078. doi: 10.1371/journal.pone.0131078
Xiong, A. S., Yao, Q. H., Peng, R. H., Duan, H., Li, X., Fan, H. Q., et al. (2006). PCR-based accurate synthesis of long DNA sequences. Nat. Protoc. 1, 791–797. doi: 10.1038/nprot.2006.103
Yi, H., Yuan, B., Liu, J., Zhu, D., Wu, Y., Wang, M., et al. (2017). Identification of a wza-like gene involved in capsule biosynthesis, pathogenicity and biofilm formation in Riemerella anatipestifer. Microb. Pathog. 107, 442–450. doi: 10.1016/j.micpath.2017.04.023
Zhang, X., Wang, M. S., Liu, M. F., Zhu, D. K., Biville, F., Jia, R. Y., et al. (2017). Contribution of RaeB, a putative RND-type transporter to aminoglycoside and detergent resistance in Riemerella anatipestifer. Front. Microbiol. 8:2435. doi: 10.3389/fmicb.2017.02435
Zhong, C. Y., Cheng, A. C., Wang, M. S., Zhu de, K., Luo, Q. H., Zhong, C. D., et al. (2009). Antibiotic susceptibility of Riemerella anatipestifer field isolates. Avian Dis. 53, 601–607. doi: 10.1637/8552-120408-ResNote.1
Keywords: Riemerella anatipestifer, lipopolysaccharide, LptD, hydrophobic drug resistance, membrane permeability
Citation: Huang L, Wang M, Mo T, Liu M, Biville F, Zhu D, Jia R, Chen S, Zhao X, Yang Q, Wu Y, Zhang S, Huang J, Tian B, Liu Y, Zhang L, Yu Y, Pan L, Rehman MU, Chen X and Cheng A (2019) Role of LptD in Resistance to Glutaraldehyde and Pathogenicity in Riemerella anatipestifer. Front. Microbiol. 10:1443. doi: 10.3389/fmicb.2019.01443
Received: 10 March 2019; Accepted: 07 June 2019;
Published: 21 June 2019.
Edited by:
Xian-Zhi Li, Health Canada, CanadaReviewed by:
Paola Sperandeo, University of Milan, ItalyFrancesco Imperi, Roma Tre University, Italy
Copyright © 2019 Huang, Wang, Mo, Liu, Biville, Zhu, Jia, Chen, Zhao, Yang, Wu, Zhang, Huang, Tian, Liu, Zhang, Yu, Pan, Rehman, Chen and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anchun Cheng, Y2hlbmdhbmNodW5AdmlwLjE2My5jb20=
†These authors have contributed equally to this work
 Li Huang
Li Huang Mingshu Wang
Mingshu Wang Ting Mo1,2,3†
Ting Mo1,2,3† Mafeng Liu
Mafeng Liu Dekang Zhu
Dekang Zhu Shun Chen
Shun Chen Bin Tian
Bin Tian Mujeeb Ur Rehman
Mujeeb Ur Rehman Xiaoyue Chen
Xiaoyue Chen Anchun Cheng
Anchun Cheng
