- 1Microbial Systems Ecology, Department of Freshwater and Marine Ecology, Institute for Biodiversity and Ecosystem Dynamics, University of Amsterdam, Amsterdam, Netherlands
- 2Department of Food, Environmental and Nutritional Sciences, University of Milan, Milan, Italy
- 3Research Centre of Biotechnology, Winogradsky Institute of Microbiology, Russian Academy of Sciences, Moscow, Russia
- 4Department of Biotechnology, Delft University of Technology, Delft, Netherlands
The genus Thioalkalivibrio includes haloalkaliphilic chemolithoautotrophic sulfur-oxidizing bacteria isolated from various soda lakes worldwide. Some of these lakes possess in addition to their extreme haloalkaline environment also other harsh conditions, to which Thioalkalivibrio needs to adapt. An example is arsenic in soda lakes in eastern California, which is found there in concentrations up to 3000 μM. Arsenic is a widespread element that can be an environmental issue, as it is highly toxic to most organisms. However, resistance mechanisms in the form of detoxification are widespread and some prokaryotes can even use arsenic as an energy source. We first screened the genomes of 76 Thioalkalivibrio strains for the presence of known arsenic oxidoreductases and found 15 putative ArxA (arsenite oxidase) and two putative ArrA (arsenate reductase). Subsequently, we studied the resistance to arsenite in detail in Thioalkalivibrio jannaschii ALM2T, and Thioalkalivibrio thiocyanoxidans ARh2T by comparative genomics and by growing them at different arsenite concentrations followed by arsenic species and transcriptomic analysis. Tv. jannaschii ALM2T, which has been isolated from Mono Lake, an arsenic-rich soda lake, could resist up to 5 mM arsenite, whereas Tv. thiocyanoxidans ARh2T, which was isolated from a Kenyan soda lake, could only grow up to 0.1 mM arsenite. Interestingly, both species oxidized arsenite to arsenate under aerobic conditions, although Tv. thiocyanoxidans ARh2T does not contain any known arsenite oxidases, and in Tv. jannaschii ALM2T, only arxB2 was clearly upregulated. However, we found the expression of a SoeABC-like gene, which we assume might have been involved in arsenite oxidation. Other arsenite stress responses for both strains were the upregulation of the vitamin B12 synthesis pathway, which can be linked to antioxidant activity, and the up- and downregulation of different DsrE/F-like genes whose roles are still unclear. Moreover, Tv. jannaschii ALM2T induced the ars gene operon and the Pst system, and Tv. thiocanoxidans ARh2T upregulated the sox and apr genes as well as different heat shock proteins. Our findings for Thioalkalivibrio confirm previously observed adaptations to arsenic, but also provide new insights into the arsenic stress response and the connection between the arsenic and the sulfur cycle.
Introduction
The genus Thioalkalivibrio comprises a group of metabolically diverse, haloalkaliphilic and chemolithoautotrophic sulfur-oxidizing bacteria thriving under extreme conditions in soda lakes. They are part of the family Ectothiorhodospiraceae within the Gammaproteobacteria (Sorokin et al., 2001a), and include 10 described species and more than 100 isolated strains (Foti et al., 2006; Sorokin et al., 2012). In silico analysis of the genomes of 76 strains classified Thioalkalivibrio in 25 genomic species, indicating a high genomic diversity within this genus (Ahn et al., 2017). Concomitantly, members of this genus are able to use different reduced sulfur compounds as electron donors such as sulfide, polysulfide, thiosulfate, polythionates, and elemental sulfur (Sorokin et al., 2001a, 2002a,b, 2003, 2004, 2012; Banciu et al., 2004). Moreover, the strains Thioalkalivibrio paradoxus ARh1T (Sorokin et al., 2002b), Tv. thiocyanoxidans ARh2T (Sorokin et al., 2002b) and Tv. thiocyanodenitrificans ARhD1T (Sorokin et al., 2004) are also able to oxidize thiocyanate (Sorokin et al., 2001b; Berben et al., 2017), and Tv. denitrificans ALJDT (Sorokin et al., 2001a), Tv. nitratireducens ALEN2T (Sorokin et al., 2003), and Tv. thiocyanodenitrificans ARhD1T (Sorokin et al., 2004) can also grow anaerobically by denitrification. Recently, Andres and Bertin (2016) and Oremland et al. (2017) detected the presence of an arxA gene, which in other bacteria, is responsible for the anaerobic energy-generating oxidation of arsenite [As(III)] to arsenate [As(V)], in the genome of 11 Thioalkalivibrio strains. Furthermore, transcripts of the arxA gene that were highly similar to genes of Thioalkalivibrio were discovered in high abundance in Mono Lake, an arsenic-rich soda lake in eastern California (Edwardson and Hollibaugh, 2017). Soda lakes in this area possess, in addition to their characteristic extreme haloalkaline condition (Jones et al., 1977, 1998), elevated arsenic concentrations that range from 0.8 μM in Crowley Lake, over 200 μM in Mono Lake, to 3000 μM in Searles Lake (Oremland et al., 2004). However, despite the multi-extreme conditions, Thioalkalivibrio are found in abundance in these soda lakes (Stamps et al., 2018).
Numerous microorganisms developed mechanisms to detoxify their cells from arsenic and in some cases to even use it as an energy source. Arsenic is well known to be highly toxic to most organisms. It may contaminate soils and groundwaters that are used for food production or as a drinking water source (Mandal and Suzuki, 2002; Cavalca et al., 2013) posing severe threats to human health (Kapaj et al., 2006). The most common forms in the environment are arsenite [As(III)] and arsenate [As(V)] (Smedley and Kinniburgh, 2002), of which the reduced form is more toxic (Hughes, 2002). This toxicity is due to the fact that As(III) is able to deactivate compounds by binding to sulfhydryl groups, as are present in glutathione (Scott et al., 1993) or in cysteines (Shen et al., 2013). As(V), however, can compete with phosphate in biochemical reactions due to its chemically similar structure and properties (Wolfe-Simon et al., 2009; Tawfik and Viola, 2011). To survive the presence of arsenic, prokaryotes can perform detoxification, which includes the reduction of As(V) to As(III) followed by As(III) methylation (Qin et al., 2006) and/or the active export of As(III) out of the cell (Ben Fekih et al., 2018). In the methylation process, the As(III) S-adenosylmethionine methyltransferase ArsM transforms As(III) into methylated As(III) compounds. By this mechanism the cell forms even more toxic, highly volatile organic arsenic compounds that can escape from the cell (Qin et al., 2006). In the active transport system, bacteria pump arsenic out of the cell using the Ars gene system. It first reduces As(V) to As(III) by the arsenate reductase ArsC (Ji and Silver, 1992; Martin et al., 2001) and subsequently pumps the As(III) out by the efflux pump ArsB or ACR3 (arsenic compounds resistance) (Bobrowicz et al., 1997; Wysocki et al., 1997; Meng et al., 2004). The activity of these pumps can be augmented by an ATPase, the ArsA, which increases the resistance to arsenic even more (Rosen et al., 1988; Dey and Rosen, 1995; Rosen, 2002). ArsD is an As(III) chaperone that transfers As(III) to ArsA (Lin et al., 2006, 2007) and it also possesses a weak activity as transacting regulatory protein (Wu and Rosen, 1993). The main transacting regulatory protein of the Ars cluster is ArsR, which functions as a transcriptional repressor that activates transcription in the presence of As(III) (Wu and Rosen, 1991). In addition to detoxification, there are numerous prokaryotes that can generate energy by the oxidation of As(III) using arsenite oxidases Aio (Anderson et al., 1992) or Arx (Zargar et al., 2010), or by the anaerobic reduction of As(V) by the arsenate respiratory reductase Arr (Saltikov and Newman, 2003). These three proteins belong to the dimethyl sulfoxide (DMSO) reductase family of molybdoenzymes, also known as complex iron-sulfur molybdoenzymes (CISM) (McEwan et al., 2002; Rothery et al., 2008). They are composed by a heterodimer of a large subunit (AioA, ArxA, and ArrA) containing the molybdopterin binding site and a small subunit with an iron-sulfur cluster (AioB, ArxB, and ArrB) (Krafft and Macy, 1998; Ellis et al., 2001; Afkar et al., 2003; Zargar et al., 2010). AioC, ArxC, and ArrC are involved in electron transfer and in the case of the ArxC and the ArrC, are transmembrane proteins anchoring the protein to the periplasmic membrane (Stolz et al., 2006; Zargar et al., 2010, 2012; Van Lis et al., 2012; Kalimuthu et al., 2014; Andres and Bertin, 2016; Oremland et al., 2017; Glasser et al., 2018). Only recently, the clade of the Arx arsenite oxidase was discovered in Alkalilimnicola ehrlichii MLHE-1T (Hoeft et al., 2007; Richey et al., 2009; Zargar et al., 2010) and in Ectothiorhodospira PHS-1 (Zargar et al., 2012), two haloalkaliphilic Gammaproteobacteria isolated from Mono Lake. These bacteria couple oxidation of As(III) as sole electron donor with nitrate reduction (Hoeft et al., 2007; Zargar et al., 2010) or anoxygenic photosynthesis (Kulp et al., 2008; Hernandez-Maldonado et al., 2017), respectively. Interestingly, ArxA is more similar to ArrA than it is to AioA (Richey et al., 2009; Zargar et al., 2010).
The aim of our research was to understand the mechanisms of resistance and adaptation to arsenic within the genus Thioalkalivibrio. We first searched in 76 Thioalkalivibrio genomes for genes that potentially can be involved in arsenic metabolism. Subsequently, we grew two Thioalkalivibrio strains at different As(III) concentrations. For this, we chose Tv. jannaschii ALM2T, which was isolated from Mono Lake (Sorokin et al., 2002a) where arsenic is present at relatively high concentrations (Oremland et al., 2004), and Tv. thiocyanoxidans ARh2T, which was isolated from a Kenyan soda lake (Sorokin et al., 2002b). We measured the As(III) oxidation capacity of the two species and performed RNA-Seq analysis to study their gene expression under arsenite stress. To our knowledge, this is the first transcriptomic work done on the arsenite stress response in chemolithoautotrophic bacteria.
Materials and Methods
Strains and Growth Conditions
Axenic cultures of Tv. jannaschii ALM2T and Tv. thiocyanoxidans ARh2T were grown in 200 ml batch cultures at 30°C on a shaker set at 100 rpm. The medium was composed of 17.5 g/l Na2CO3, 13.9 g/l NaHCO3, 6.1 g/l NaCl, 1 g/l K2HPO4, 0.2 g/l MgCl2, 40 mM Na2S2O3, 5 mM NH4Cl, and 1:1000 trace metals (Pfennig and Lippert, 1966). Sterile solutions of MgCl2, Na2S2O3 and trace elements were added from concentrated stock solutions after autoclaving. The final pH of the culture medium was adjusted to pH 9.8. As(III) as sodium arsenite (NaAsO2) (Sigma Aldrich, United States) was added to the medium just before inoculation of the bacteria. For the growth curves of Tv. jannaschii ALM2T and Tv. thiocyanoxidans ARh2T, the cultures were supplemented with 0.1, 0.5, 5, or 7.5 mM As(III). Cultures without As(III) were used as reference and growth of all cultures was monitored daily by measuring the OD at 600 nm. Tv. thiocyanoxidans ARh2T and Tv. jannaschii ALM2T grew up to a concentration of 0.1 and 5 mM As(III), respectively (Supplementary Figure 1). Therefore, cultures were prepared at 0.1 mM As(III) for Tv. thiocyanoxidans ARh2T, and at 0.1 and 5 mM for Tv. jannaschii ALM2T to study the As(III) resistance mechanisms by transcriptomics. Again, cultures without As(III) were used as reference and their growth was followed by OD measurements at 600 nm. Samples for arsenic species and transcriptomic analysis were taken in the exponential growth phase at an OD600 ∼ 0.1, which corresponded to one [reference; 0 mM As(III)] and two [0.1 mM As(III)] days after inoculation for the Tv. thiocyanoxidans ARh2T cultures, and after one [reference; 0 mM As(III)], one [0.1 mM As(III)], and five [5 mM As(III)] days for the Tv. jannaschii ALM2T cultures. In addition, sterile culture medium was incubated under the same conditions to check for the possibility of chemical As(III) oxidation. To test the growth with As(III) as sole electron donor, Tv. thiocyanoxidans ARh2T was cultivated with 0.1 mM As(III), and Tv. jannaschii ALM2T with 0.1 and 2.5 mM As(III) in culture medium prepared as described above with the exception of containing 0.025 g/l MgSO4 × 7H2O and different Na2S2O3 concentrations depending on the culture (0, 1, 5, 10, and 40 mM). All experiments were done in triplicate.
Arsenic Speciation by ICP-MS Analysis
Culture supernatant was filtered through a 0.2 μm filter and arsenic species were determined according to Kim et al. (2007). To quantify the total As concentration, 5 ml of the filtrate was acidified prior the analysis with 200 μl of 2% (v/v) HNO3. For the determination of inorganic arsenic species As(III) and As(V), 5 ml of the filtrate was added to a Sep-Pak® Plus Acell Plus QMA cartridge (Waters, MA, United States). As(V) remained in the cartridge, whereas As(III) passed through. As(III) was collected and acidified with 200 μl of 2% (v/v) HNO3. The As(V) was then washed off the cartridge with 5 ml 0.16 M HNO3. Total As, As(III), and As(V) concentrations were measured by ICP-MS (Agilent Technologies, United States). Standard solutions ranging from 0 to 1 mg/l of As were prepared from a sodium arsenite (NaAsO2) solution (Sigma Aldrich, United States). All measurements were done in triplicate.
Comparative Sequence Analysis
The phylogenetic tree of ArxA, ArrA, and AioA was constructed based on a multiple alignment of amino acid sequences, which were selected by a BLASTp analysis of 76 Thioalkalivibrio genomes (Ahn et al., 2017) and of reference protein sequences. The selected sequences were aligned with MUSCLE (Edgar, 2004) and the tree was built with the software program MEGA7 (version 7.0.26; Kumar et al., 2016) using the Maximum Likelihood method with 1000 bootstrap replicates, the LG model as substitution model and a discrete gamma distribution (+G) as evolutionary rate differences amongst sites.
The phylogenetic tree of the two SoeA clusters was also built with aligned amino acid sequences found in 76 Thioalkalivibrio genomes and references, which were selected based on a previous BLASTp analysis. The alignment and the tree construction were calculated following the same protocol as described above.
RNA-Sequencing
The biomass was collected in 50 ml Greiner tubes and immediately placed into a centrifuge that was precooled to 4°C. The cells were pelleted by centrifugation at 7,000 × g for 4 min at 4°C. The supernatant was removed until approximately 2 ml, in which the cells were suspended and transferred to a 2 ml Eppendorf tube. The sample was then centrifuged at 15,000 × g for 1 min at 4°C. The supernatant was completely removed, and the cell pellet was immediately frozen in liquid nitrogen and stored at -80°C until further processing.
The frozen cell pellets were homogenized with a mortar and a pestle before being resuspended in QIAzol Lysis Reagent (Qiagen, Germany). Total RNA was extracted and purified with the RNeasy kit (Qiagen) following the manufacturer’s instructions. The purification step comprised a DNase treatment using the RNase-free DNase kit (Qiagen). The concentration was quantified with the NanoDrop ND2000 (Thermo Fisher Scientific, United States) and the integrity of the RNA was checked on the 2200 TapeStation with Agilent RNA ScreenTapes (Agilent Technologies, Netherlands). Ribosomal RNA (rRNA) was removed by the Illumina Ribo-Zero rRNA Removal Kit (Bacteria) (Illumina, United States). Bar-coded RNA libraries were prepared using the Ion Total RNA-Seq kit v2 and the Ion Xpress RNA-Seq barcoding kit according to the supplier’s instructions (Thermo Fisher Scientific). Size distribution and yield were measured on the 2200 TapeStation using Agilent D1000 ScreenTapes (Agilent Technologies). Sequencing templates were prepared on the Ion Chef System with the Ion PI Hi-Q Chef kit (Thermo Fisher Scientific). Samples were sequenced on the Ion Proton platform with an Ion PI Chip v3 (Thermo Fisher Scientific) following the supplier’s instructions.
RNA-Seq Analysis
The genomes of Tv. thiocyanoxidans ARh2T (NZ_ARQK00000000.1) (Berben et al., 2015) and Tv. jannaschii ALM2T (NZ_ARLZ00000000.1) were previously sequenced and annotated. The reference gene and genome sequences of both strains were obtained from the NCBI RefSeq FTP server. The software program kallisto (Bray et al., 2016) (v0.44.0) was used to create index files for the quantification from those references. The quality of the reads was assessed by FastQC (version 0.11.7) and estimated to be sufficient. Therefore, no trimming or filtering was performed. Pseudo-alignments were generated in kallisto by mapping the reads from the fastq RNA-Seq files against the indexed reference and reads were quantified using 100 bootstrap samples. Subsequently, differential expression analysis was performed with the software program sleuth (Pimentel et al., 2017) (0.30.0) using the Wald test. The complete differential expression values are presented in Supplementary Table 1 and consists of the NCBI locus-tag, the b-value (beta-value), the P-value, the q-value, the raw counts and the annotation by NCBI for each gene. The b-value is a biased estimator of the log fold change and is on a natural-log scale (Pimentel et al., 2017).
The sequences were also analyzed with the RNA-Seq analysis module in the software program CLC Genomics Workbench 11.0.1 (QIAGEN). Proton Torrent fastq files were imported and trimmed using the following default settings: (i) removal of low-quality sequences with a limit of 0.05, (ii) removal of ambiguous nucleotides: maximum 2 nucleotides allowed, and (iii) discard reads below a length of 30 nucleotides. Subsequently, the trimmed reads were mapped to the reference genomes. Differential expression data includes the NCBI locus-tag, the max group mean, the log2 (fold change), the fold change, the P-value, the FDR P-value, and the Bonferroni value (Supplementary Table 2).
Results and Discussion
Genomic Features of Arsenic Metabolism and Resistance in Thioalkalivibrio
We searched in Thioalkalivibrio for genes that can be used to grow on arsenic as an energy source. Therefore, a phylogenetic tree was constructed with the putative protein sequences of ArxA, AioA (arsenite oxidases), and ArrA (arsenate reductase) detected in the 76 available genome sequences of different Thioalkalivibrio strains (Figure 1A). In those, a putative ArxA was found in 14 Thioalkalivibrio strains and a putative ArrA in two. Genes coding for AioA were not detected in any of the strains. Tv. nitratireducens ALEN2T was the only strain that contained both ArxA and ArrA. Previously, the presence of ArxA has only been reported in 11 Thioalkalivibrio strains (Andres and Bertin, 2016; Oremland et al., 2017) while the presence of ArrA has been never documented. For the strains used in the cultivation experiment, Tv. jannaschii ALM2T possesses a putative ArxA while Tv. thiocyanoxidans ARh2T lacks any of the known genes to generate energy from inorganic arsenic.
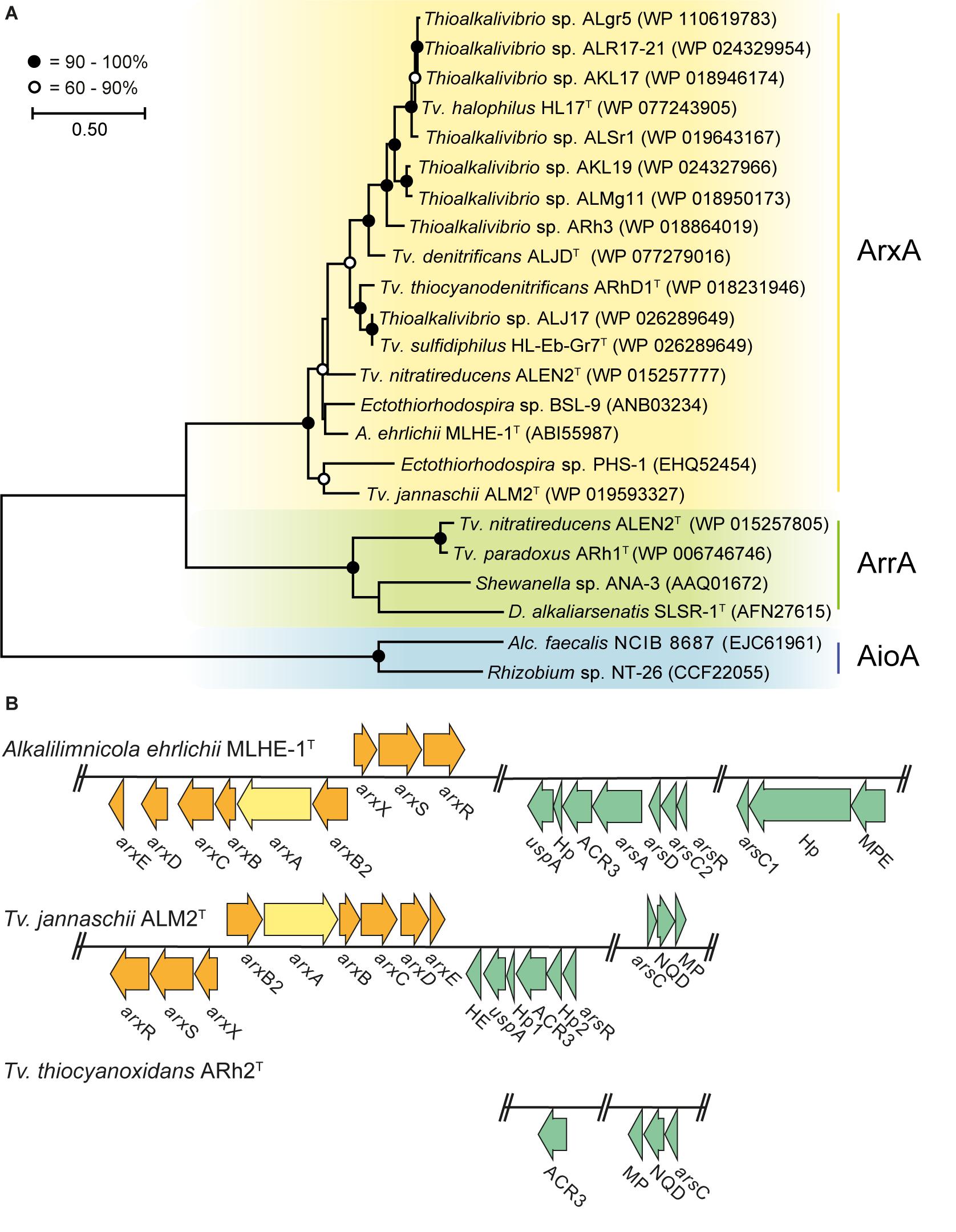
Figure 1. Comparative sequence analysis of arsenic resistance genes in Thioalkalivibrio. Yellow, arsenite oxidase ArxA; green, arsenate reductase ArrA; blue, arsenite oxidase AioA; orange, arsenite oxidase arx gene cluster; turquoise, arsenic resistance ars gene cluster. (A) Phylogenetic tree constructed from ArxA, ArrA, and AioA protein sequences present in the genomes of Thioalkalivibrio strains and other bacteria. Accession number is provided for each sequence in the figure. (B) Arsenic resistance genes in Alkalilimnicola ehrlichii MLHE-1T, Tv. jannaschii ALM2T, and Tv. thiocyanoxidans ARh2T. Hp, hypothetical protein; MPE, metallophosphoesterase; HE, hemerythrin; MP, uncharacterized membrane protein; NQD, NADP(H):quinone dehydrogenase. The locus tags for the genes used in (B) are listed in Supplementary Table 3.
The genomes of A. ehrlichii MLHE-1T (another member of the Ectothiorhodospiraceae), Tv. jannaschii ALM2T and Tv. thiocyanoxidans ARh2T encode different gene clusters for the detoxification (ars genes) and for the oxidation (arx genes) of arsenite (Figure 1B). A. ehrlichii MLHE-1T (Zargar et al., 2010, 2012) and Tv. jannaschii ALM2T possess an identical arx gene cluster for arsenite oxidation and a highly similar set of ars genes for arsenic resistance. For the ars genes, A. ehrlichii MLHE-1T possesses the most complete gene cluster including arsADR, ACR3 and two detoxifying arsenate reductases arsC, one glutaredoxin- (arsC1) and one thioredoxin-dependent (arsC2). In Tv. jannaschii ALM2T, a more reduced set including an arsR, a glutaredoxin-dependent arsC and an ACR3 was present. Another annotated ACR3 efflux pump was found in ALM2T outside the shown cluster (Locus-tag: F816_RS0108235) together with three uncharacterized membrane proteins. Interestingly, A. ehrlichii MLHE-1T and Tv. jannaschii ALM2T also encode for a universal stress protein (uspA) in their ars gene cluster. On the contrary, Tv. thiocyanoxidans ARh2T only possesses a truncated ars gene cluster with an ACR3 and a glutaredoxin-dependent arsC, also subdivided in two operons, and without arsR. Outside of the operon, two putative ArsR for Tv. thiocyanoxidans ARh2T were found by BLASTp, but with low identity values, using the ArsR of A. ehrlichii MLHE-1T (Locus-tag: Mlg_2713), and of Tv. jannaschii ALM2T (Locus-tag: F816_RS0102085) (Supplementary Table 4) as subjects. Neither the genome of Tv. thiocyanoxidans ARh2T nor of Tv. jannaschii ALM2T contained the arsM gene necessary for the detoxification of the intracellular As(III) by methylation (Qin et al., 2006). Furthermore, the genomes of Tv. jannaschii ALM2T and Tv. thiocyanoxidans ARh2T were screened for the presence of arsHIJNOPTX via BLASTp. In Tv. jannaschii ALM2T, an ArsI [Locus-tag: F816_RS0102080; query cover of 93% and an identity of 41% with ArsI of Bacillus sp. MD1 (AIA09488)] was found inside the ars gene operon as well as a second putative ArsI positioned directly besides the ars gene operon [Locus-tag: F816_RS14315 (78% query cover and 29% identity to AIA09488)]. In addition, putative sequences for ArsJ were detected in Tv. jannaschii ALM2T [Locus-tag: F816_RS0106725; query cover of 95% and a similarity of 60.3% with ArsJ of Pseudomonas aeruginosa (WP_003109849)] and in Tv. thiocyanoxidans ARh2T (Locus-tag: G372_RS0110690; query cover of 94% and an identity of 59,1% with WP_003109849). The putative ArsJ in Tv. jannaschii ALM2T is encoded together with an annotated glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in the same operon. The combination of these two genes has been described to confer resistance to As(V) (Chen et al., 2016). However, the GAPDH was not found in the operon of the putative ArsJ in Tv. thiocyanoxidans ARh2T. To prove the function of these putative ArsIJ, experimental evidence must follow.
Physiological and Transcriptomic Response to Arsenic Stress
Tv. jannaschii ALM2T and Tv. thiocyanoxidans ARh2T were cultivated in the presence of different concentrations of As(III) to determine their resistance. Tv. jannaschii ALM2T resists much higher As(III) concentrations than Tv. thiocyanoxidans ARh2T (Supplementary Figure 1). Tv. thiocyanoxidans ARh2T was only able to grow until a concentration of 0.1 mM As(III), whereas Tv. jannaschii ALM2T still grew up to 5 mM As(III). All cultures grew aerobically with thiosulfate as an electron donor and they could not grow with As(III) as their sole potential electron donor (Supplementary Figure 2).
To gain deeper insight in their resistance mechanism against arsenic, Tv. thiocyanoxidans ARh2T and Tv. jannaschii ALM2T were both cultivated in absence of As(III) (reference) and at 0.1 mM As(III). Furthermore, Tv. jannaschii ALM2T was also grown at 5 mM As(III). Cultures were harvested in their exponential growth phase to measure the arsenic species composition in the culture fluid and to determine gene expression in both Thioalkalivibrio species.
Arsenic species were measured in the culture medium at the beginning and at the end of the experiment to investigate the potential of both strains to oxidize As(III) to As(V) under aerobic conditions (Figure 2). Additional sterile samples were analyzed to determine the possibility of chemical oxidation of As(III) in the culture medium. Samples inoculated with Tv. jannaschii ALM2T and Tv. thiocyanoxidans ARh2T showed a decrease in As(III) and an increase in As(V) over time. During the same incubation time, As(III) and As(V) concentrations did not change significantly in the sterile samples indicating that As(III) oxidation was biologically induced. Tv. jannaschii ALM2T had a much stronger As(III) oxidizing capacity as compared to Tv. thiocyanoxidans ARh2T. When grown in the presence of 0.1 mM As(III), Tv. jannaschii ALM2T oxidized 57% of the present As(III) in 1 day, whereas Tv. thiocyanoxidans ARh2T only oxidized 26% after 2 days. Most importantly, when grown in the presence of 5 mM As(III), Tv. jannaschii ALM2T was able to oxidize 79% of As(III) within 5 days. These findings resemble previous incubation experiments of Mono Lake surface waters that showed a clear link between aerobic As(III) oxidation capacity and added sulfide or thiosulfate (Fisher et al., 2008). In their research, sulfide-amended lake brines showed the formation of thioarsenates compounds from As(III), which were fairly stable in sterile, oxic surface waters, but which were further oxidized to As(V) in samples containing sulfur-oxidizing bacteria. Molecular analysis of the enrichments identified bacteria closely related to Tv. jannaschii, Tv. versutus and Tv. nitratis. Furthermore, Edwardson et al. (2014) showed that pure cultures of Tv. jannaschii ALM2T were able to oxidize monothioarsenate aerobically, but also that they did not show growth with monothioarsenate as their sole electron donor. In our research, we have now demonstrated growth of Tv. jannaschii ALM2T and Tv. thiocyanoxidans ARh2T with As(III) in combination with thiosulfate. Whether Thioalkalivibrio can gain energy from As(III) or thioarsenate oxidation, or that this oxidation is only used for detoxification purposes, remains an open question. However, it can be excluded that these compounds support growth as a sole electron donor.
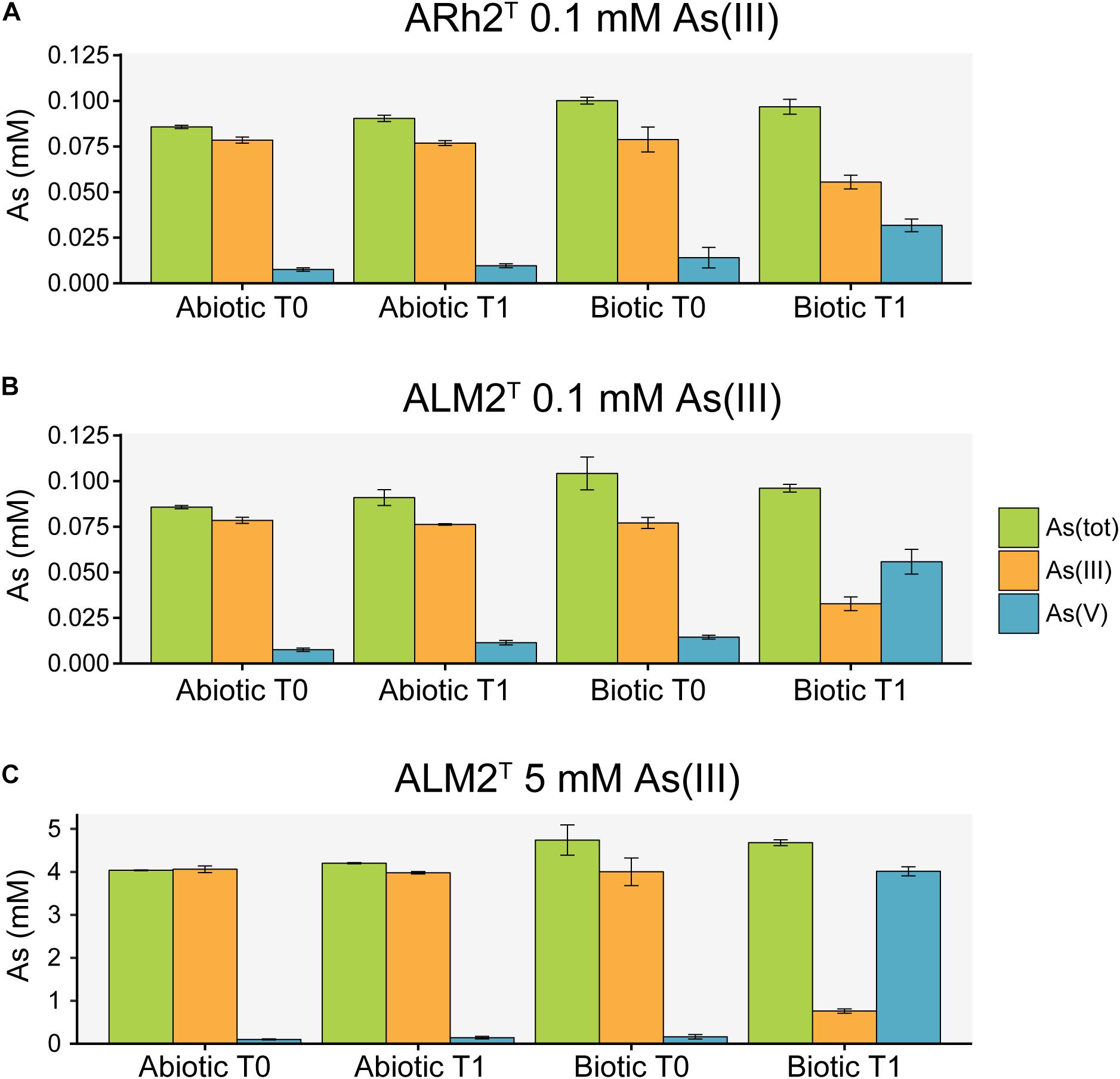
Figure 2. Arsenic species analysis of culture fluid from sterile samples (Abiotic) and samples inoculated with Tv. thiocyanoxidans ARh2T and Tv. jannaschii ALM2T (Biotic) at the inoculation (T0) and sampling time (T1). (A) ARh2T culture incubated with 0.1 mM As(III) for 2 days. (B) ALM2T culture incubated with 0.1 mM As(III) for 1 day. (C) ALM2T culture incubated with 5 mM As(III) for 5 days.
Transcriptomic analysis enabled screening for key genes in the metabolism of and in the resistance against arsenic, and it shows differences in gene expression between Tv. jannaschii ALM2T and Tv. thiocyanoxidans ARh2T. General information on the individual RNA-seq samples analyzed by kallisto and sleuth are presented in Supplementary Table 5 and the complete expression data can be found in Supplementary Table 1. In total, 57.4 million sequence reads were produced by the Ion Proton platform ranging from 2.3 million to 4.9 million sequence reads per sample. From those reads, between 57.9% and 72.5% could be assigned to an open reading frame (ORF) depending on the sample analyzed with kallisto. In Tv. jannaschii ALM2T, 2833 ORFs were detected, and 2716 ORFs in Tv. thiocyanoxidans ARh2T. For the analysis performed with sleuth, an ORF is considered differentially expressed if the b-value is greater than 0.7-fold and its P-value is lower than 0.1. The RNA-Seq data analyzed by sleuth gave 101 up- and 84 downregulated genes for Tv. thiocyanoxidans ARh2T at 0.1 mM As(III) [0.1 mM vs. 0 mM As(III)] (Supplementary Table 6), only two up- and one downregulated genes for Tv. jannaschii ALM2T at 0.1 mM As(III) [0.1 mM vs. 0 mM As(III)] (Supplementary Table 7), and 26 up- and 16 downregulated genes for Tv. jannaschii ALM2T at 5 mM As(III) [5 mM vs. 0 mM As(III)] (Supplementary Table 8). As certain pathways could not be completely revealed based on the sleuth results only, we decided to also analyze the RNA-Seq data with CLC Genomics Workbench (Supplementary Table 2). With CLC, between 77.74 and 82.2% of the reads could been allocated to an ORF. Here, an ORF was considered to be differentially expressed if the log2 fold change was higher than 1-fold and its P-value lower than 0.1. With this threshold, CLC found 99 up- and 91 downregulated genes for Tv. thiocyanoxidans ARh2T at 0.1 mM As(III) [0.1 mM vs. 0 mM As(III)], four up-, and five downregulated genes for Tv. jannaschii ALM2T at 0.1 mM As(III) [0.1 mM vs. 0 mM As(III)], and 40 up- and 20 downregulated genes for Tv. jannaschii ALM2T at 5 mM As(III) [5 mM vs. 0 mM As(III)].
The quality of the RNA-Seq data analyzed by kallisto and sleuth was evaluated by principal component analysis and plotted in a graph with the first two principal components as axes (Supplementary Figure 3). On the first principal component, the samples of each condition cluster together and were well separated from the other conditions. Remarkably, the Tv. jannaschii ALM2T samples grown at 0, 0.1, and 5 mM As(III) are not ordered based on the increasing As(III) concentration on the first principle component, but in the order of 0.1, 0, and 5 mM As(III). This phenomenon might be explained by a hormesis reaction, in which an agent, here As(III), at lower level exposes an beneficial effect on the organism and becomes only toxic at higher concentrations (Mattson, 2008).
We summarized the results of the gene expression under As(III) stress in a conceptual model (Figure 3), in which the groups correspond to the subgroups of the discussion: (1) Arsenic influx into the cell, (2) Arsenic metabolism and detoxification, (3) Response to oxidative damage by arsenite, (4) Sulfur metabolism, and (5) Recombination and energy generation.
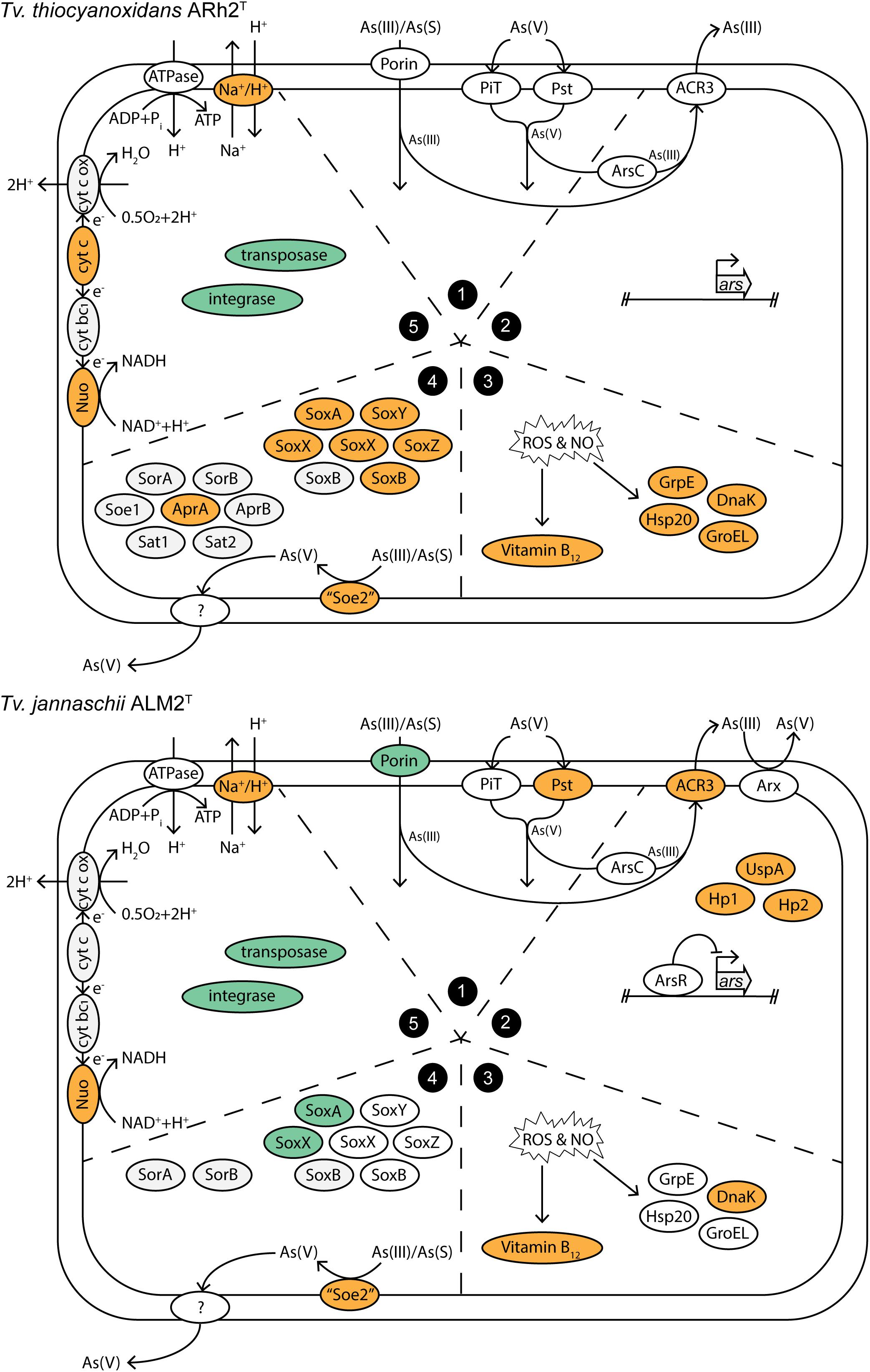
Figure 3. Conceptual model of cellular processes within Tv. jannaschii ALM2T and Tv. thiocyanoxidans ARh2T under As(III) stress. The section numbers correspond to the subgroups in the Results and Discussion section: (1) Arsenic influx into the cell, (2) Arsenic metabolism and detoxification, (3) Response to oxidative damage by arsenite, (4) Sulfur metabolism, and (5) Recombination and energy generation. Upregulated genes are colored in orange and downregulated genes in turquoise. Cyt bc1, cytochrome bc1; cyt c, cytochrome c; cyt c ox, cytochrome c oxidase; As(S), thioarsenate; Hp, hypothetical protein. Locus tags and differential expression values are listed in Supplementary Table 9.
(1) Arsenic Influx Into the Cell
The arsenic species As(III) and As(V) are able to enter the cell by transporters of molecules whose properties they mimic. In E.coli, As(III) can enter the cell by the aquaglyceroporin channel GlpF (Sanders et al., 1997; Meng et al., 2004). These channels normally transport small uncharged molecules such as glycerol (Heller et al., 1980; Borgnia and Agre, 2001), but also As(III) as non-charged As(OH)3 under neutral pH (Ramírez-Solís et al., 2012). However, in environments with a pH higher than its pKa of 9.2, As(III) will be mostly present in its ionic form (Smedley and Kinniburgh, 2002). This is the case for soda lakes whose pH ranges from 9.5 to 11 and, from which most Thioalkalivibrio strains were isolated (Sorokin et al., 2014). Furthermore, thioarsenates are formed in oxic alkaline brines containing sulfide (Stauder et al., 2005; Planer-Friedrich et al., 2007, 2009; Fisher et al., 2008; Härtig and Planer-Friedrich, 2012), conditions present as well in Mono Lake (Hollibaugh et al., 2005). Until now, it is unknown how As(III) or thioarsenates enter the cells under these conditions. A possible porin involved in their influx could be F816_RS0109535 (Supplementary Table 2) in Tv. jannaschii ALM2T as it is highly downregulated at 0.1 and 5 mM As(III). However, no similar protein could be detected in the genome of Tv. thiocyanoxidans ARh2T. The downregulation of a porin responsible for As(III) or thioarsenate influx would keep the intracellular arsenic concentration lower in Tv. jannaschii ALM2T, thus conferring a higher As(III) resistance to the strain.
As(V) possesses a similar chemical structure and properties as phosphate (Wolfe-Simon et al., 2009), and can therefore be taken up by the phosphate uptake systems Pit (inorganic phosphate transporter) and Pst (specific phosphate transporter) (Rosenberg et al., 1977; Willsky and Malamy, 1980a), of which Pst is more specific for phosphate and transports As(V) less efficiently (Willsky and Malamy, 1980b; Elias et al., 2012). The gene for the Pit transporter system did not significantly change in expression in any of the samples. However, when grown with 0.1 mM As(III), Tv. jannaschii ALM2T upregulates the pstA contemporary with the formation of 0.056 mM As(V) (Figure 2). In the presence of 5 mM As(III), the pstABCS and the regulator phoU were upregulated with the simultaneous occurrence of 4.01 mM As(V) in the culture medium (Figure 2). These genes did not change in expression in Tv. thiocyanoxidans ARh2T cultures, which could be explained by the low As(V) concentration of 0.032 mM at the sampling time (Figure 2). Since the Pst transporter is more specific for phosphate than for As(V), many bacteria increase the expression and the production of Pst to increase phosphate uptake (Andres and Bertin, 2016). This would give another advantage for growth in combination with As(III) oxidation for Tv. jannaschii ALM2T. However, as both strains were able to tolerate 30 mM As(V) (data not shown), it is possible that Tv. thiocyanoxidans ARh2T also possesses a similar mechanism, which was not upregulated with the low As(V) concentration deriving from the oxidation process.
(2) Arsenic Metabolism and
Detoxification
Transcriptomic analysis of Tv. jannaschii ALM2T grown at 5 mM As(III) showed an upregulation of the arsenite oxidase arxB2 gene, but not of the structural component genes arxABC (Supplementary Tables 1, 2). The Arx protein is only known to work under anaerobic condition coupled to denitrification or anaerobic photosynthesis (Hoeft et al., 2007; Kulp et al., 2008; Zargar et al., 2010; Hernandez-Maldonado et al., 2017). Moreover, as already discussed before, no growth was observed in the tested strains under aerobic condition with As(III) as sole electron donor (Supplementary Figure 2). Previously, Arx has been shown in vitro to function as a bidirectional enzyme able to oxidize As(III) and to reduce As(V) (Richey et al., 2009). Due to the presence of Arx in Tv. jannaschii ALM2T and the incapacity of this strain to perform denitrification, it could be hypothesized that this strain uses Arx to reduce As(V) to As(III) in combination with the oxidation of reduced sulfur compounds under microoxic/anoxic conditions.
Most organisms perform an active extrusion of As(III) as their main arsenic resistance mechanism. This is performed in prokaryotes by the arsenic resistance ars operon, where As(III) produced by the arsenate reductase ArsC is pumped out of the cell by ArsB/ACR3 (Ben Fekih et al., 2018). The ars genes were not differentially expressed in Tv. thiocyanoxidans ARh2T at 0.1 mM As(III) (Supplementary Table 1). For Tv. jannaschii ALM2T at 0.1 mM As(III), only the uspA gene was upregulated. In contrast, the ars gene cluster was highly expressed in Tv. jannaschii ALM2T at 5mM As(III) including the ACR3 efflux pump, two hypothetical proteins and the uspA gene. An exception was the arsC, which explains the high concentration of As(V) observed in the medium at the time of sampling (Figure 2 and Supplementary Tables 1, 2). The uspA gene encodes a uspA that is known to be induced under different stress situations (Kvint et al., 2003), by which it increases the endurance of the cell (Nyström and Neidhardt, 1994). Upregulation of this gene has been shown in bacteria under As(III) stress (Weiss et al., 2009; Cleiss-Arnold et al., 2010; Sacheti et al., 2013). Finally, an operon, which is located next to the arx and ars cluster in Tv. jannaschii ALM2T and which includes a putatively annotated glycosyl transferase involved in the cell wall biosynthesis and a rhodanese-related sulfurtransferase were highly upregulated at 5mM As(III) in ALM2T. However, in Tv. thiocyanoxidans ARh2T, this operon is neither upregulated nor found next to the ars gene cluster.
(3) Response to Oxidative Damage by Arsenite
Arsenic has been shown to induce formation of reactive oxygen species (ROS) and nitric oxide (NO) inside the cell (Andres and Bertin, 2016; Zhang et al., 2016). These radicals can cause damage to nucleic acids, proteins, and lipids (Flora, 2011; Birben et al., 2012; Ray et al., 2012; Espinosa-Diez et al., 2015). To reduce the oxidative damage by arsenic, bacterial cells have developed various responses including the upregulation of Fe- and Mn- superoxide dismutases, thiol peroxidases, thioredoxin reductases, thioredoxins, glutaredoxins, glutathione, organic hydroperoxide resistance proteins, and vitamin B6 (Andres and Bertin, 2016). In our experiments, however, we did not detect changes in expression for the two Thioalkalivibrio strains for any of the genes involved in known antioxidation pathways. However, the As(III) concentrations of 0.1 mM As(III) for Tv. thiocyanoxidans ARh2T and 5 mM As(III) for Tv. jannaschii ALM2T triggered upregulation of the complete vitamin B12 (cobalamin) synthesis pathway (Supplementary Tables 1, 2). Vitamin B12 has been shown to protect eukaryotic cells from oxidative damage by its antioxidant activity (Birch et al., 2009; Suarez-Moreira et al., 2009; Moreira et al., 2011; Alzoubi et al., 2012; Bito et al., 2017) as well as when it is generated by arsenic in hepatic rat cells (Chattopadhyay et al., 2012; Majumdar et al., 2012). In bacterial cells, vitamin B12 has also been shown to be an antioxidant able to protect the cell from oxidative stress in the acidophilic iron-oxidizing bacterium Leptospirillum group II CF-1 (Ferrer et al., 2016). Recently, Qin et al. (2018) discovered that the archaea Nitrosopumilus maritimus SCM1 produces vitamin B12 under Cu2+ stress. Moreover, cobSTU was shown to be expressed by bacteria in an arsenic-rich acid mine drainage, but was only related to the activation of iron oxidation (Bertin et al., 2011). Here, we propose that vitamin B12 is the main antioxidant produced under As(III) stress in Thioalkalivibrio.
In addition, Tv. thiocyanoxidans ARh2T significantly upregulates the expression of the chaperones dnaK and Hsp20, and slightly of groEL, grpE and dnaJ. In contrast, Tv. jannaschii ALM2T does not change the expression of those genes. Chaperones from the Hsp70 (DnaK, DnaJ and GrpE) and Hsp60 (GroEL and GroES) systems have been shown to be commonly induced as an arsenic stress response in bacteria (Andres and Bertin, 2016). These chaperones are essential for the cell viability and the survival under diverse stressful conditions as they facilitate the proper folding of newly translated proteins or maintain it for already translated ones (Houry, 2001; Hayer-Hartl et al., 2016; Hartl, 2017).
(4) Sulfur Metabolism
Thioalkalivibrio strains are sulfur-oxidizing bacteria that possess a high inter-genus diversity of different genes and pathways involved in sulfur oxidation (Berben et al., 2019). Tv. thiocyanoxidans ARh2T and Tv. jannaschii ALM2T differentiate from each other by the fact that Tv. thiocyanoxidans ARh2T possesses the TcDH pathway for thiocyanate oxidation, the Apr-Sat pathway for sulfite oxidation as well as a second homologous copy of Soe for sulfite oxidation, whereas Tv. jannaschii ALM2T does not (Berben et al., 2019). Interestingly, the sequences of the two SoeA in Tv. thiocyanoxidans ARh2T form two distinct clusters, i.e., one cluster grouping around SoeA of Allochromatium vinosum (cluster 1) (Dahl et al., 2013) and a second cluster forming a separate group (cluster 2), hereafter called “Soe-like” gene. Both copies are present together in 41 Thioalkalivibrio strains (Supplementary Figure 4). Various genes in the sulfur oxidation pathway are upregulated in Tv. thiocyanoxidans ARh2T under As(III) stress [soxYZXXAB, aprA and soeABC (cluster 2)], whereas others were not differentially expressed, such as soeABC (cluster 1), sat and sorAB. In Tv. jannaschii ALM2T, only the soeABC (cluster 2) was highly upregulated together with genes necessary for the molybdenum cofactor production of SoeA, the moaA (GTP 3′,8-cyclase) (Mendel and Leimkühler, 2015) and the molybdate ABC transporter.
As(III) was also oxidized in Tv. thiocyanoxidans ARh2T cultures, although this strain does not possess an arsenite oxidase in its genome (Figure 1). Therefore, another as yet unknown enzyme must exist besides the two known arsenite oxidases AioA and ArxA involved in As(III) oxidation under aerobic conditions. Comparing our results with the work of Fisher et al. (2008), we can hypothesize that thioarsenate species have also formed in our cultures, opening new possibilities for enzymatic pathways of As(III) oxidation in the presence of thiosulfate or sulfide. Some enzymes have already been hypothesized to be involved in the oxidation pathway of thioarsenate compounds. Edwardson et al. (2014) proposed the Sox pathway as a potential facilitator of thioarsenate oxidation based on the structural similarity between monothioarsenate and thiosulfate. In this system, Sox enzymes would be able to cleave the thiol group from monothioarsenate. This hypothesis could be supported by the upregulation of the sox cluster in Tv. thiocyanoxidans ARh2T, but is contradicted by the stable or even slight downregulation of these genes in Tv. jannaschii ALM2T where the strongest As(III) oxidation occurred. Furthermore, a sulfide:quinone oxidoreductase and its operon were found upregulated in the presence of sulfide or As(III) in Synechocystis sp. strain PCC6803 (Nagy et al., 2014). As these genes are closely related to genes in Tv. thiocyanodenitrificans ARhD1T, although not found in a single operon in this strain, they proposed that these genes could be involved in thioarsenate oxidation in Thioalkalivibrio. However, no upregulation of these genes was detected in the dataset obtained with Tv. jannaschii ALM2T and Tv. thiocyanoxidans ARh2T growing with As(III). Couture et al. (2012) proposed in a review the involvement of the proteins SelD and SelU coupled to ArxC in thioarsenate production alongside to their normal activity of making selenophosphate and modifying RNA. However, we could not find any homologs to these proteins in the genomes of Tv. jannaschii ALM2T and Tv. thiocyanoxidans ARh2T.
The only genes of the sulfur oxidation pathway that were induced in both strains at their highest respective As(III) concentration were the quinone-dependent sulfite oxidase soeABC (cluster 2). SoeABC is a molybdopterin oxidoreductase of the same family as the arsenic oxidoreductases Aio, Arr, and Arx (Krafft and Macy, 1998; Ellis et al., 2001; McEwan et al., 2002; Zargar et al., 2010; Dahl et al., 2013). Similar to those proteins, SoeA and SoeB form a heterodimer, which is anchored to the cytoplasmic membrane by SoeC. The difference between the SoeA and the arsenic oxidoreductase is that SoeA does not contain a TAT-signal peptide, and therefore it stays in the cytoplasm (Dahl et al., 2013). This TAT-signal peptide also does not exist in the SoeABC-like protein of the cluster 2. Until now, no activity and substrate specificity have been proven for this second Soe-like cluster. Therefore, we propose that this Soe-like protein as a possible candidate for co-oxidation of As(III) and sulfite (SO32-), or oxidation of thioarsenate. Moreover, we hypothesize that the observed oxidation has rather the aim of detoxifying the cell as both strains were unable to grow on As(III) as a sole electron donor (Supplementary Figure 2).
Multiple putative sulfurtransferases annotated as DsrE/F-like genes were found up- or downregulated in the presence of As(III). The sulfurtransferase DsrEFH binds to sulfur via a conserved cysteine of the DsrE and transports it to the DsrC in the reverse Dsr system of elemental sulfur oxidation to sulfite (Stockdreher et al., 2012). The function of a cysteine in an active site is known to be inactivated by the binding of As(III) to the sulfhydryl group (Shen et al., 2013). One possibility for their change in expression could therefore be either their induction to compensate for the inactivation (upregulation) or their reduction to shut down the pathway (downregulation).
(5) Recombination and Energy
Generation
Interestingly, genes for genetic recombination were downregulated. These include different transposases and integrases. This is in contradiction with the findings of Gualco et al. (2004) where a rise in the amount of recombinants in conjugation and transduction, and transposition of the Tn9 was observed when As(III) at a sub-MIC (Minimal Inhibitory Concentration) was added. This is in agreement with the general understanding that stressful environmental conditions induce genetic variation in bacteria via mutations and recombination (Bjedov et al., 2003; Foster, 2005, 2007; Prudhomme et al., 2006; Schuurmans et al., 2014).
In addition, genes are induced that are involved in the various pathways for the electron transfer in oxidative phosphorylation including the NADH:ubiquinone oxidoreductase in both strains, a cytochrome c synthesis gene and a Na+/H+ antiporter subunit in Tv. thiocyanoxidans ARh2T. The transcriptional upregulation of these complexes are commonly observed in bacteria in the presence of arsenic (Andres and Bertin, 2016). One explanation could be that arsenic works as an uncoupler of the membrane potential and as an alternative substrate of ATPase, which could impair NADH and ATP production. To ensuring adequate NADH and ATP production, Tv. thiocyanoxidans ARh2T compensates this effect by the upregulation of the NADH:ubiquinone oxidoreductase, the Na+/H+ antiporter and by increased cytochrome c synthesis.
Conclusion
In this study, we identified the putative potential of arsenic metabolism by the presence of Arx in 14 Thioalkalivibrio strains, and of Arr in two. Furthermore, we investigated the main mechanisms of arsenite resistance for Tv. jannaschii ALM2T and Tv. thiocyanoxidans ARh2T. These strains do not share the same resistance to As(III), which is reflected in their growth response to different As(III) concentrations, in their repertoire of arsenic resistance genes, in their As(III)-oxidizing potential and in their transcriptome. From the gene expression, we discovered an involvement of vitamin B12 as the major player in the protection against arsenic-imposed oxidative stress, as well as the differential expression of DsrE/F-like proteins whose roles need to be elucidated in future research. Moreover, Tv. jannaschii ALM2T induced the transcription of the ars gene operon and the Pst system, and Tv. thiocanoxidans ARh2T increased expression of the sox and apr genes as well as different heat shock proteins. Comparing our results with the work of Fisher et al. (2008), we can postulate the formation of thioarsenates in the Thioalkalivibrio cultures, which were then microbiologically further oxidized by an as yet unknown enzymatic pathway to As(V). We hypothesize that a Soe-like protein is responsible for this oxidation, but evidence must be obtained by future work.
Data Availability
The datasets generated for this study are deposited in the NCBI Sequence Read Archive under SRA accession numbers SRX5567239 to SRX5567253.
Author Contributions
A-CA carried out the cultivation, the comparative sequence analyses, the RNA-Seq data analysis by sleuth, and drafted the manuscript. LC and MC carried out the analysis of the arsenic species. GM carried out the RNA-Seq data analysis by CLC. JS, DS, and GM assisted in the interpretation of the results, and together with LC provided a critical review of the manuscript. All authors read and approved the final version of the manuscript.
Funding
Financial support for A-CA, JS, and GM was provided by the ERC Advanced Grant Parasol (No. 322551). LC and MC were supported by the Cariplo Foundation project 2014-1301. DS was funded by the Russian Foundation for Basic Research (Grant No. 19-04-00401) and the Russian Ministry of Science and Higher Education. The sequencing platform was funded by the NWO Earth and Life Sciences (ALW) project 834.12.003.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Eleanor Spring for the critical proofreading of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01514/full#supplementary-material
References
Afkar, E., Lisak, J., Saltikov, C., Basu, P., Oremland, R. S., and Stolz, J. F. (2003). The respiratory arsenate reductase from Bacillus selenitireducens strain MLS10. FEMS Microbiol. Lett. 226, 107–112. doi: 10.1016/S0378-1097(03)00609-8
Ahn, A. C., Meier-Kolthoff, J. P., Overmars, L., Richter, M., Woyke, T., Sorokin, D. Y., et al. (2017). Genomic diversity within the haloalkaliphilic genus Thioalkalivibrio. PLoS One 12:e0173517. doi: 10.1371/journal.pone.0173517
Alzoubi, K., Khabour, O., Hussain, N., Al-Azzam, S., and Mhaidat, N. (2012). Evaluation of vitamin B12 effects on DNA damage induced by pioglitazone. Mutat. Res. 748, 48–51. doi: 10.1016/j.mrgentox.2012.06.009
Anderson, G. L., Williams, J., and Hille, R. (1992). The purification and characterization of arsenite oxidase from Alcaligenes faecalis, a molybdenum-containing hydroxylase. J. Biol. Chem. 267, 23674–23682.
Andres, J., and Bertin, P. N. (2016). The microbial genomics of arsenic. FEMS Microbiol. Rev. 40, 299–322. doi: 10.1093/femsre/fuv050
Banciu, H., Sorokin, D. Y., Galinski, E. A., Muyzer, G., Kleerebezem, R., and Kuenen, J. G. (2004). Thialkalivibrio halophilus sp. nov., a novel obligately chemolithoautotrophic, facultatively alkaliphilic, and extremely salt-tolerant, sulfur-oxidizing bacterium from a hypersaline alkaline lake. Extremophiles 8, 325–334. doi: 10.1007/s00792-004-0391-6
Ben Fekih, I., Zhang, C., Li, Y. P., Zhao, Y., Alwathnani, H. A., Saquib, Q., et al. (2018). Distribution of arsenic resistance genes in prokaryotes. Front. Microbiol. 9:2473. doi: 10.3389/fmicb.2018.02473
Berben, T., Overmars, L., Sorokin, D. Y., and Muyzer, G. (2017). Comparative genome analysis of three thiocyanate oxidizing Thioalkalivibrio species isolated from soda lakes. Front. Microbiol. 8:254. doi: 10.3389/fmicb.2017.00254
Berben, T., Overmars, L., Sorokin, D. Y., and Muyzer, G. (2019). The diversity and distribution of sulfur oxidation-related genes in Thioalkalivibrio, a genus of chemolithoautotrophic and haloalkaliphilic sulfur-oxidizing bacteria. Front. Microbiol. 10:160. doi: 10.3389/fmicb.2019.00160
Berben, T., Sorokin, D. Y., Ivanova, N., Pati, A., Kyrpides, N., Goodwin, L. A., et al. (2015). Partial genome sequence of the haloalkaliphilic soda lake bacterium Thioalkalivibrio thiocyanoxidans ARh 2T. Stand. Genomic Sci. 10:85. doi: 10.1186/s40793-015-0078-x
Bertin, P. N., Heinrich-Salmeron, A., Pelletier, E., Goulhen-Chollet, F., Arsène-Ploetze, F., Gallien, S., et al. (2011). Metabolic diversity among main microorganisms inside an arsenic-rich ecosystem revealed by meta-and proteo-genomics. ISME J. 5, 1735–1747. doi: 10.1038/ismej.2011.51
Birben, E., Sahiner, U. M., Sackesen, C., Erzurum, S., and Kalayci, O. (2012). Oxidative stress and antioxidant defense. World Allergy Organ. J. 5, 9–19. doi: 10.1097/WOX.0b013e3182439613
Birch, C. S., Brasch, N. E., McCaddon, A., and Williams, J. H. H. (2009). A novel role for vitamin B12: cobalamins are intracellular antioxidants in vitro. Free Radic. Biol. Med. 47, 184–188. doi: 10.1016/j.freeradbiomed.2009.04.023
Bito, T., Misaki, T., Yabuta, Y., Ishikawa, T., Kawano, T., and Watanabe, F. (2017). Vitamin B12 deficiency results in severe oxidative stress, leading to memory retention impairment in Caenorhabditis elegans. Redox Biol. 11, 21–29. doi: 10.1016/j.redox.2016.10.013
Bjedov, I., Tenaillon, O., Gérard, B., Souza, V., Denamur, E., Radman, M., et al. (2003). Stress-induced mutagenesis in bacteria. Science 300, 1404–1409. doi: 10.1126/science.1082240
Bobrowicz, P., Wysocki, R., Owsianik, G., Goffeau, A., and Ułaszewski, S. (1997). Isolation of three contiguous genes, ACR1, ACR2 and ACR3, involved in resistance to arsenic compounds in the yeast Saccharomyces cerevisiae. Yeast 13, 819–828. doi: 10.1002/(sici)1097-0061(199707)13:9<819::aid-yea142>3.0.co;2-y
Borgnia, M. J., and Agre, P. (2001). Reconstitution and functional comparison of purified GlpF and AqpZ, the glycerol and water channels from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 98, 2888–2893. doi: 10.1073/pnas.051628098
Bray, N. L., Pimentel, H., Melsted, P., and Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. doi: 10.1038/nbt.3519
Cavalca, L., Corsini, A., Zaccheo, P., Andreoni, V., and Muyzer, G. (2013). Microbial transformations of arsenic: perspectives for biological removal of arsenic from water. Future Microbiol. 8, 753–768. doi: 10.2217/fmb.13.38
Chattopadhyay, S., Deb, B., and Maiti, S. (2012). Hepatoprotective role of vitamin B12 and folic acid in arsenic intoxicated rats. Drug Chem. Toxicol. 35, 81–88. doi: 10.3109/01480545.2011.589439
Chen, J., Yoshinaga, M., Garbinski, L. D., and Rosen, B. P. (2016). Synergistic interaction of glyceraldehydes-3-phosphate dehydrogenase and ArsJ, a novel organoarsenical efflux permease, confers arsenate resistance. Mol. Microbiol. 100, 945–953. doi: 10.1111/mmi.13371
Cleiss-Arnold, J., Koechler, S., Proux, C., Fardeau, M. L., Dillies, M. A., Coppee, J. Y., et al. (2010). Temporal transcriptomic response during arsenic stress in Herminiimonas arsenicoxydans. BMC Genomics 11:709. doi: 10.1186/1471-2164-11-709
Couture, R. M., Sekowska, A., Fang, G., and Danchin, A. (2012). Linking selenium biogeochemistry to the sulfur-dependent biological detoxification of arsenic. Environ. Microbiol. 14, 1612–1623. doi: 10.1111/j.1462-2920.2012.02758.x
Dahl, C., Franz, B., Hensen, D., Kesselheim, A., and Zigann, R. (2013). Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology 159, 2626–2638. doi: 10.1099/mic.0.071019-0
Dey, S., and Rosen, B. P. (1995). Dual mode of energy coupling by the oxyanion-translocating ArsB protein. J. Bacteriol. 177, 385–389. doi: 10.1128/jb.177.2.385-389.1995
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edwardson, C. F., and Hollibaugh, J. T. (2017). Metatranscriptomic analysis of prokaryotic communities active in sulfur and arsenic cycling in Mono Lake, California, USA. ISME J. 11, 2195–2208. doi: 10.1038/ismej.2017.80
Edwardson, C. F., Planer-Friedrich, B., and Hollibaugh, J. T. (2014). Transformation of monothioarsenate by haloalkaliphilic, anoxygenic photosynthetic purple sulfur bacteria. FEMS Microbiol. Ecol. 90, 858–868. doi: 10.1111/1574-6941.12440
Elias, M., Wellner, A., Goldin-Azulay, K., Chabriere, E., Vorholt, J. A., Erb, T. J., et al. (2012). The molecular basis of phosphate discrimination in arsenate-rich environments. Nature 491, 134–137. doi: 10.1038/nature11517
Ellis, P. J., Conrads, T., Hille, R., and Kuhn, P. (2001). Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 Å and 2.03 Å. Structure 9, 125–132. doi: 10.1016/s0969-2126(01)00566-4
Espinosa-Diez, C., Miguel, V., Mennerich, D., Kietzmann, T., Sánchez-Pérez, P., Cadenas, S., et al. (2015). Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 6, 183–197. doi: 10.1016/j.redox.2015.07.008
Ferrer, A., Rivera, J., Zapata, C., Norambuena, J., Sandoval,Á., Chávez, R., et al. (2016). Cobalamin protection against oxidative stress in the acidophilic iron-oxidizing bacterium Leptospirillum group II CF-1. Front. Microbiol. 7:748. doi: 10.3389/fmicb.2016.00748
Fisher, J. C., Wallschläger, D., Planer-Friedrich, B., and Hollibaugh, J. T. (2008). A new role for sulfur in arsenic cycling. Environ. Sci. Technol. 42, 81–85. doi: 10.1021/es0713936
Flora, S. J. S. (2011). Arsenic-induced oxidative stress and its reversibility. Free Radic. Biol. Med. 51, 257–281. doi: 10.1016/j.freeradbiomed.2011.04.008
Foster, P. L. (2005). Stress responses and genetic variation in bacteria. Mutat. Res. 569, 3–11. doi: 10.1016/j.mrfmmm.2004.07.017
Foster, P. L. (2007). Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42, 373–397. doi: 10.1080/10409230701648494
Foti, M., Ma, S., Sorokin, D. Y., Rademaker, J. L. W., Kuenen, J. G., and Muyzer, G. (2006). Genetic diversity and biogeography of haloalkaliphilic sulphur-oxidizing bacteria belonging to the genus Thioalkalivibrio. FEMS Microbiol. Ecol. 56, 95–101. doi: 10.1111/j.1574-6941.2006.00068.x
Glasser, N. R., Oyala, P. H., Osborne, T. H., Santini, J. M., and Newman, D. K. (2018). Structural and mechanistic analysis of the arsenate respiratory reductase provides insight into environmental arsenic transformations. Proc. Natl. Acad. Sci. U.S.A. 115, E8614–E8623. doi: 10.1073/pnas.1807984115
Gualco, L., Roveta, S., Marchese, A., and Debbia, E. A. (2004). Pleiotropic effect of sodium arsenite on Escherichia coli. Res. Microbiol. 155, 275–282. doi: 10.1016/j.resmic.2004.01.005
Härtig, C., and Planer-Friedrich, B. (2012). Thioarsenate transformation by filamentous microbial mats thriving in an alkaline, sulfidic hot spring. Environ. Sci. Technol. 46, 4348–4356. doi: 10.1021/es204277j
Hartl, F. U. (2017). Unfolding the chaperone story. Mol. Biol. Cell. 28, 2919–2923. doi: 10.1091/mbc.E17-07-0480
Hayer-Hartl, M., Bracher, A., and Hartl, F. U. (2016). The GroEL-GroES chaperonin machine: a nano-cage for protein folding. Trends Biochem. Sci. 41, 62–76. doi: 10.1016/j.tibs.2015.07.009
Heller, K. B., Lin, E. C. C., and Wilson, T. H. (1980). Substrate specificity and transport properties of the glycerol facilitator of Escherichia coli. J. Bacteriol. 144, 274–278.
Hernandez-Maldonado, J., Sanchez-Sedillo, B., Stoneburner, B., Boren, A., Miller, L., McCann, S., et al. (2017). The genetic basis of anoxygenic photosynthetic arsenite oxidation. Environ. Microbiol. 19, 130–141. doi: 10.1111/1462-2920.13509
Hoeft, S. E., Blum, J. S., Stolz, J. F., Tabita, F. R., Witte, B., King, G. M., et al. (2007). Alkalilimnicola ehrlichii sp. nov., a novel, arsenite-oxidizing haloalkaliphilic gammaproteobacterium capable of chemoautotrophic or heterotrophic growth with nitrate or oxygen as the electron acceptor. Int. J. Syst. Evol. Microbiol. 57, 504–512. doi: 10.1099/ijs.0.64576-0
Hollibaugh, J. T., Carini, S., Gürleyük, H., Jellison, R., Joye, S. B., LeCleir, G., et al. (2005). Arsenic speciation in Mono Lake, California: response to seasonal stratification and anoxia. Geochim. Cosmochim. Acta 69, 1925–1937. doi: 10.1016/j.gca.2004.10.011
Houry, W. A. (2001). Chaperone-assisted protein folding in the cell cytoplasm. Curr. Protein Pept. Sci. 2, 227–244. doi: 10.2174/1389203013381134
Hughes, M. F. (2002). Arsenic toxicity and potential mechanisms of action. Toxicol. Lett. 133, 1–16. doi: 10.1016/s0378-4274(02)00084-x
Ji, G., and Silver, S. (1992). Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid p1258. Proc. Natl. Acad. Sci. U.S.A. 89, 9474–9478. doi: 10.1073/pnas.89.20.9474
Jones, B. E., Grant, W. D., Duckworth, A. W., and Owenson, G. G. (1998). Microbial diversity of soda lakes. Extremophiles 2, 191–200. doi: 10.1007/s007920050060
Jones, B. F., Eugster, H. P., and Rettig, S. L. (1977). Hydrochemistry of the lake magadi basin, Kenya. Geochim. Cosmochim. Acta 41, 53–72. doi: 10.1016/0016-7037(77)90186-7
Kalimuthu, P., Heath, M. D., Santini, J. M., Kappler, U., and Bernhardt, P. V. (2014). Electrochemically driven catalysis of Rhizobium sp. NT-26 arsenite oxidase with its native electron acceptor cytochrome c552. Biochim. Biophys. Acta 1837, 112–120. doi: 10.1016/j.bbabio.2013.07.010
Kapaj, S., Peterson, H., Liber, K., and Bhattacharya, P. (2006). Human health effects from chronic arsenic poisoning - a review. J. Environ. Sci. Health A Tox. Hazard Subst. Environ. Eng. 41, 2399–2428. doi: 10.1080/10934520600873571
Kim, Y. T., Yoon, H., Yoon, C., and Woo, N. C. (2007). An assessment of sampling, preservation, and analytical procedures for arsenic speciation in potentially contaminated waters. Environ. Geochem. Health 29, 337–346. doi: 10.1007/s10653-007-9091-3
Krafft, T., and Macy, J. M. (1998). Purification and characterization of the respiratory arsenate reductase of Chrysiogenes arsenatis. Eur. J. Biochem. 255, 647–653. doi: 10.1046/j.1432-1327.1998.2550647.x
Kulp, S. E., Hoeft, M., Asao, M. T., Madigan, J. T., Hollibaugh, J. C., Fisher, J. F., et al. (2008). Arsenic(III) fuels anoxygenic photosynthesis in hot spring biofilms from Mono Lake, California. Science 321, 967–971. doi: 10.1126/science.1160799
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kvint, K., Nachin, L., Diez, A., and Nyström, T. (2003). The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6, 140–145. doi: 10.1016/s1369-5274(03)00025-0
Lin, Y. F., Walmsley, A. R., and Rosen, B. P. (2006). An arsenic metallochaperone for an arsenic detoxification pump. Proc. Natl. Acad. Sci. U.S.A. 103, 15617–15622. doi: 10.1073/pnas.0603974103
Lin, Y. F., Yang, J., and Rosen, B. P. (2007). ArsD: an As(III) metallochaperone for the ArsAB As(III)-translocating ATPase. J. Bioenerg. Biomembr. 39, 453–458. doi: 10.1007/s10863-007-9113-y
Majumdar, S., Maiti, A., Karmakar, S., Das, A. S., Mukherjee, S., Das, D., et al. (2012). Antiapoptotic efficacy of folic acid and vitamin B12 against arsenic-induced toxicity. Environ. Toxicol. 27, 351–363. doi: 10.1002/tox.20648
Mandal, B. K., and Suzuki, K. T. (2002). Arsenic round the world: a review. Talanta 58, 201–235. doi: 10.1016/s0039-9140(02)00268-0
Martin, P., DeMel, S., Shi, J., Gladysheva, T., Gatti, D. L., Rosen, B. P., et al. (2001). Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme. Structure 9, 1071–1081. doi: 10.1016/s0969-2126(01)00672-4
McEwan, A. G., Ridge, J. P., McDevitt, C. A., and Hugenholtz, P. (2002). The DMSO reductase family of microbial molybdenum enzymes; molecular properties and role in the dissimilatory reduction of toxic elements. Geomicrobiol. J. 19, 3–21. doi: 10.1080/014904502317246138
Mendel, R. R., and Leimkühler, S. (2015). The biosynthesis of the molybdenum cofactors. J. Biol. Inorg. Chem. 20, 337–347. doi: 10.1007/s00775-014-1173-y
Meng, Y. L., Liu, Z., and Rosen, B. P. (2004). As(III) and Sb(III) uptake by GlpF and efflux by ArsB in Escherichia coli. J. Biol. Chem. 279, 18334–18341. doi: 10.1074/jbc.M400037200
Moreira, E. S., Brasch, N. E., and Yun, J. (2011). Vitamin B12 protects against superoxide-induced cell injury in human aortic endothelial cells. Free Radic. Biol. Med. 51, 876–883. doi: 10.1016/j.freeradbiomed.2011.05.034
Nagy, C. I., Vass, I., Rákhely, G., Vass, I. Z., Tóth, A., Duzs, Á., et al. (2014). Coregulated genes link sulfide:quinone oxidoreductase and arsenic metabolism in Synechocystis sp. strain PCC6803. J. Bacteriol. 196, 3430–3440. doi: 10.1128/JB.01864-14
Nyström, T., and Neidhardt, F. C. (1994). Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 11, 537–544. doi: 10.1111/j.1365-2958.1994.tb00334.x
Oremland, R. S., Saltikov, C. W., Stolz, J. F., and Hollibaugh, J. T. (2017). Autotrophic microbial arsenotrophy in arsenic-rich soda lakes. FEMS Microbiol. Lett. 364:fnx146. doi: 10.1093/femsle/fnx146
Oremland, R. S., Stolz, J. F., and Hollibaugh, J. T. (2004). The microbial arsenic cycle in Mono Lake, California. FEMS Microbiol. Ecol. 48, 15–27. doi: 10.1016/j.femsec.2003.12.016
Pfennig, N., and Lippert, K. D. (1966). Über das Vitamin B12-bedürfnis phototropher schwefelbakterien. Arch. für Mikrobiol. 55, 245–256. doi: 10.1007/bf00410246
Pimentel, H., Bray, N. L., Puente, S., Melsted, P., and Pachter, L. (2017). Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690. doi: 10.1038/nmeth.4324
Planer-Friedrich, B., Fisher, J. C., Hollibaugh, J. T., Süß, E., and Wallschläger, D. (2009). Oxidative transformation of trithioarsenate along alkaline geothermal drainages-abiotic versus microbially mediated processes. Geomicrobiol. J. 26, 339–350. doi: 10.1080/01490450902755364
Planer-Friedrich, B., London, J., McCleskey, R. B., Nordstrom, D. K., and Wallschläger, D. (2007). Thioarsenates in geothermal waters of Yellowstone national park: determination, preservation, and geochemical importance. Environ. Sci. Technol. 41, 5245–5251. doi: 10.1021/es070273v
Prudhomme, M., Attaiech, L., Sanchez, G., Martin, B., and Claverys, J. P. (2006). Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313, 89–92. doi: 10.1126/science.1127912
Qin, J., Rosen, B. P., Zhang, Y., Wang, G., Franke, S., and Rensing, C. (2006). Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 103, 2075–2080. doi: 10.1073/pnas.0506836103
Qin, W., Amin, S. A., Lundeen, R. A., Heal, K. R., Martens-Habbena, W., Turkarslan, S., et al. (2018). Stress response of a marine ammonia-oxidizing archaeon informs physiological status of environmental populations. ISME J. 12, 508–519. doi: 10.1038/ismej.2017.186
Ramírez-Solís, A., Mukopadhyay, R., Rosen, B. P., and Stemmler, T. L. (2012). Experimental and theoretical characterization of arsenite in water: insights into the coordination environment of As-O. Inorg. Chem. 43, 2954–2959. doi: 10.1021/ic0351592
Ray, P. D., Huang, B. W., and Yoshiaki, T. (2012). Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signalling. Cell. Signal. 24, 981–990. doi: 10.1016/j.cellsig.2012.01.008
Richey, C., Chovanec, P., Hoeft, S. E., Oremland, R. S., Basu, P., and Stolz, J. F. (2009). Respiratory arsenate reductase as a bidirectional enzyme. Biochem. Biophys. Res. Commun. 382, 298–302. doi: 10.1016/j.bbrc.2009.03.045
Rosen, B. P. (2002). Biochemistry of arsenic detoxification. FEBS Lett. 529, 86–92. doi: 10.1016/s0014-5793(02)03186-1
Rosen, B. P., Weigel, U., Karkaria, C., and Gangola, P. (1988). Molecular characterization of an anion pump. The arsA gene product is an arsenite(antimonate)-stimulated ATPase. J. Biol. Chem. 263, 3067–3070.
Rosenberg, H., Gerdes, R. G., and Chegwidden, K. (1977). Two systems for the uptake of phosphate in Escherichia coli. J. Bacteriol. 131, 505–511.
Rothery, R. A., Workun, G. J., and Weiner, J. H. (2008). The prokaryotic complex iron-sulfur molybdoenzyme family. Biochim. Biophys. Acta 1778, 1897–1929. doi: 10.1016/j.bbamem.2007.09.002
Sacheti, P., Bhonsle, H., Patil, R., Kulkarni, M. J., Srikanth, R., and Gade, W. (2013). Arsenomics of Exiguobacterium sp. PS (NCIM 5463). RSC Adv. 3, 9705–9713. doi: 10.1039/C3RA40897C
Saltikov, C. W., and Newman, D. K. (2003). Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. U.S.A. 100, 10983–10988. doi: 10.1073/pnas.1834303100
Sanders, O. I., Rensing, C., Kuroda, M., Mitra, B., and Rosen, B. P. (1997). Antimonite is accumulated by the glycerol facilitator GlpF in Escherichia coli. J. Bacteriol. 179, 3365–3367. doi: 10.1128/jb.179.10.3365-3367.1997
Schuurmans, J. M., Van Hijum, S. A. F. T., Piet, J. R., Händel, N., Smelt, J., Brul, S., et al. (2014). Effect of growth rate and selection pressure on rates of transfer of an antibiotic resistance plasmid between E. coli strains. Plasmid 72, 1–8. doi: 10.1016/j.plasmid.2014.01.002
Scott, N., Hatlelid, K. M., MacKenzie, N. E., and Carter, D. E. (1993). Reactions of arsenic (III) and arsenic (V) species with glutathione. Chem. Res. Toxicol. 6, 102–106. doi: 10.1021/tx00031a016
Shen, S., Li, X. F., Cullen, W. R., Weinfeld, M., and Le, X. C. (2013). Arsenic binding to proteins. Chem. Rev. 113, 7769–7792. doi: 10.1021/cr300015c
Smedley, P. L., and Kinniburgh, D. G. (2002). A review of the source, behaviour and distribution of arsenic in natural waters. Chem. Rev. 17, 517–568. doi: 10.1016/s0883-2927(02)00018-5
Sorokin, D. Y., Berben, T., Melton, E. D., Overmars, L., Vavourakis, C. D., and Muyzer, G. (2014). Microbial diversity and biogeochemical cycling in soda lakes. Extremophiles 18, 791–809. doi: 10.1007/s00792-014-0670-9
Sorokin, D. Y., Gorlenko, V. M., Tourova, T. P., Tsapin, A. I., Nealson, K. H., and Kuenen, G. J. (2002a). Thioalkalimicrobium cyclium sp. nov. and Thioalkalivibrio jannaschii sp. nov., novel species of haloalkaliphilic, obligately chemolithoautotrophic sulfur-oxidizing bacteria from hypersaline alkaline Mono Lake (California). Int. J. Syst. Evol. Microbiol. 52, 913–920. doi: 10.1099/00207713-52-3-913
Sorokin, D. Y., Tourova, T. P., Lysenko, A. M., Mityushina, L. L., and Kuenen, J. G. (2002b). Thioalkalivibrio thiocyanoxidans sp. nov. and thioalkalivibrio paradoxus sp. nov., novel alkaliphilic, obligately autotrophic, sulfur-oxidzing bacteria capable of growth on thiocyanate, from soda lakes. Int. J. Syst. Evol. Microbiol. 52, 657–664. doi: 10.1099/00207713-52-2-657
Sorokin, D. Y., Lysenko, A. M., Mityushina, L. L., Tourova, T. P., Jones, B. E., Rainey, F. A., et al. (2001a). Thioalkalimicrobium aerophilum gen. nov., sp. nov. and Thioalkalimicrobium sibericum sp. nov., and Thioalkalivibrio versutus gen. nov., sp. nov., Thioalkalivibrio nitratis sp. nov. and Thioalkalivibrio denitrificans sp. nov., novel obligately alkaliphilic. Int. J. Syst. Evol. Microbiol. 51, 565–580. doi: 10.1099/00207713-51-2-565
Sorokin, D. Y., Tourova, T. P., Lysenko, A. M., and Kuenen, J. G. (2001b). Microbial thiocyanate utilization under highly alkaline conditions. Appl. Environ. Microbiol. 67, 528–538. doi: 10.1128/AEM.67.2.528-538.2001
Sorokin, D. Y., Muntyan, M. S., Panteleeva, A. N., and Muyzer, G. (2012). Thioalkalivibrio sulfidiphilus sp. nov., a haloalkaliphilic, sulfur-oxidizing gammaproteobacterium from alkaline habitats. Int. J. Syst. Evol. Microbiol. 62, 1884–1889. doi: 10.1099/ijs.0.034504-0
Sorokin, D. Y., Tourova, T. P., Antipov, A. N., Muyzer, G., and Kuenen, J. G. (2004). Anaerobic growth of the haloalkaliphilic denitrifying sulfur-oxidizing bacterium Thialkalivibrio thiocyanodenitrificans sp. nov. with thiocyanate. Microbiology 150, 2435–2442. doi: 10.1099/mic.0.27015-0
Sorokin, D. Y., Tourova, T. P., Sjollema, K. A., and Kuenen, J. G. (2003). Thialkalivibrio nitratireducens sp. nov., a nitrate-reducing member of an autotrophic denitrifying consortium from a soda lake. Int. J. Syst. Evol. Microbiol. 53, 1779–1783. doi: 10.1099/ijs.0.02615-0
Stamps, B. W., Nunn, H. S., Petryshyn, V. A., Oremland, R. S., Miller, L. G., Rosen, M. R., et al. (2018). Metabolic capability and phylogenetic diversity of Mono Lake during a bloom of the eukaryotic phototroph Picocystis sp. strain ML. Appl. Environ. Microbiol. 84, 1171–1189. doi: 10.1128/AEM.01171-18
Stauder, S., Raue, B., and Sacher, F. (2005). Thioarsenates in sulfidic waters. Environ. Sci. Technol. 39, 5933–5939. doi: 10.1021/es048034k
Stockdreher, Y., Venceslau, S. S., Josten, M., Sahl, H. G., Pereira, I. A. C., and Dahl, C. (2012). Cytoplasmic sulfurtransferases in the purple sulfur bacterium Allochromatium vinosum: evidence for sulfur transfer from DsrEFH to DsrC. PLoS One 7:e40785. doi: 10.1371/journal.pone.0040785
Stolz, J. F., Basu, P., Santini, J. M., and Oremland, R. S. (2006). Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 60, 107–130. doi: 10.1146/annurev.micro.60.080805.142053
Suarez-Moreira, E., Yun, J., Birch, C. S., Williams, J. H. H., McCaddon, A., and Brasch, N. E. (2009). Vitamin B12 and redox homeostasis: cob(II)alamin reacts with superoxide at rates approaching superoxide dismutase (SOD). J. Am. Chem. Soc. 131, 15078–15079. doi: 10.1021/ja904670x
Tawfik, D. S., and Viola, R. E. (2011). Arsenate replacing phosphate - alternative life chemistries and ion promiscuity. Biochemistry 50, 1128–1134. doi: 10.1021/bi200002a
Van Lis, R., Nitschke, W., Warelow, T. P., Capowiez, L., Santini, J. M., and Schoepp-Cothenet, B. (2012). Heterologously expressed arsenite oxidase: a system to study biogenesis and structure/function relationships of the enzyme family. Biochim. Biophys. Acta 1817, 1701–1708. doi: 10.1016/j.bbabio.2012.06.001
Weiss, S., Carapito, C., Cleiss, J., Koechler, S., Turlin, E., Coppee, J. Y., et al. (2009). Enhanced structural and functional genome elucidation of the arsenite-oxidizing strain Herminiimonas arsenicoxydans by proteomics data. Biochimie 91, 192–203. doi: 10.1016/j.biochi.2008.07.013
Willsky, G. R., and Malamy, M. H. (1980a). Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J. Bacteriol. 144, 356–365.
Willsky, G. R., and Malamy, M. H. (1980b). Effect of arsenate on inorganic phosphate transport in Escherichia coli. J. Bacteriol. 144, 366–374.
Wolfe-Simon, F., Davies, P. C. W., and Anbar, A. D. (2009). Did nature also choose arsenic? Int. J. Astrobiol. 8, 69–74. doi: 10.1017/S1473550408004394
Wu, J., and Rosen, B. P. (1991). The ArsR protein is a trans-acting regulatory protein. Mol. Microbiol. 5, 1331–1336. doi: 10.1111/j.1365-2958.1991.tb00779.x
Wu, J., and Rosen, B. P. (1993). The arsD gene encodes a second trans-acting regulatory protein of the plasmid-encoded arsenical resistance operon. Mol. Microbiol. 8, 615–623. doi: 10.1111/j.1365-2958.1993.tb01605.x
Wysocki, R., Bobrowicz, P., and Ułaszewski, S. (1997). The Saccharomyces cerevisiae ACR3 gene encodes a putative membrane protein involved in arsenite transport. J. Biol. Chem. 272, 30061–30066. doi: 10.1074/jbc.272.48.30061
Zargar, K., Conrad, A., Bernick, D. L., Lowe, T. M., Stolc, V., Hoeft, S., et al. (2012). ArxA, a new clade of arsenite oxidase within the DMSO reductase family of molybdenum oxidoreductases. Environ. Microbiol. 14, 1635–1645. doi: 10.1111/j.1462-2920.2012.02722.x
Zargar, K., Hoeft, S., Oremland, R., and Saltikov, C. W. (2010). Identification of a novel arsenite oxidase gene, arxA, in the haloalkaliphilic, arsenite-oxidizing bacterium Alkalilimnicola ehrlichii strain MLHE-1. J. Bacteriol. 192, 3755–3762. doi: 10.1128/JB.00244-10
Keywords: RNA-Seq, arsenic, resistance, adaptation, sulfur-oxidizing bacteria, soda lake, soeABC
Citation: Ahn A-C, Cavalca L, Colombo M, Schuurmans JM, Sorokin DY and Muyzer G (2019) Transcriptomic Analysis of Two Thioalkalivibrio Species Under Arsenite Stress Revealed a Potential Candidate Gene for an Alternative Arsenite Oxidation Pathway. Front. Microbiol. 10:1514. doi: 10.3389/fmicb.2019.01514
Received: 05 April 2019; Accepted: 17 June 2019;
Published: 04 July 2019.
Edited by:
Masahiro Ito, Toyo University, JapanReviewed by:
Ronald Oremland, United States Geological Survey, United StatesJohn Stolz, Duquesne University, United States
Copyright © 2019 Ahn, Cavalca, Colombo, Schuurmans, Sorokin and Muyzer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerard Muyzer, Zy5tdWlqemVyQHV2YS5ubA==
 Anne-Catherine Ahn
Anne-Catherine Ahn Lucia Cavalca
Lucia Cavalca Milena Colombo2
Milena Colombo2 J. Merijn Schuurmans
J. Merijn Schuurmans Dimitry Y. Sorokin
Dimitry Y. Sorokin Gerard Muyzer
Gerard Muyzer