- 1Division of Immunology and Infectious Diseases, University-Hospital Pediatric Department, Bambino Gesù Children’s Hospital, IRCSS, Rome, Italy
- 2Human Microbiome Unit, Bambino Gesù Children’s Hospital, IRCCS, Rome, Italy
- 3Human Microbiome Unit and Parasitology Unit, Bambino Gesù Children’s Hospital, IRCCS, Rome, Italy
The emergence of antimicrobial resistance (AMR) is of great concern to global public health. Treatment of multi-drug resistant (MDR) infections is a major clinical challenge: the increase in antibiotic resistance leads to a greater risk of therapeutic failure, relapses, longer hospitalizations, and worse clinical outcomes. Currently, there are no validated treatments for many MDR or pandrug-resistant (PDR) infections, and preventing the spread of these pathogens through hospital infection control procedures and antimicrobial stewardship programs is often the only tool available to healthcare providers. Therefore, new solutions to control the colonization of MDR pathogens are urgently needed. In this narrative review, we discuss current knowledge of microbiota-mediated mechanisms of AMR and strategies for MDR colonization control. We focus particularly on fecal microbiota transplantation for MDR intestinal decolonization and report updated literature on its current clinical use.
Introduction
The global emergence and spread of antimicrobial resistance (AMR) are of great concern to public health. The number of deaths associated with multidrug-resistant (MDR) infections each year has been estimated at more than 25,000 in Europe, 38,000 in Thailand, and 23,000 in the United States (Europäisches Zentrum für die Prävention und die Kontrolle von Krankheiten, 2009; Pumart et al., 2012; CDC, 2019). One of the most recent and major threats is the spread of carbapenem-resistant Enterobacteriaceae (CRE), which is becoming a serious healthcare issue in many countries (Munoz-Price et al., 2013; Ivády et al., 2016; van Duin and Paterson, 2016; Del Chierico et al., 2018). Previous studies have reported that CRE infections are associated with a higher frequency of sepsis and an increased early mortality rate (Tumbarello et al., 2012), particularly in vulnerable populations such as pediatric, elderly, hospitalized, transplant recipient, immunosuppressed, and chronically ill patients (Kalpoe et al., 2012; Wang Z. et al., 2018). The emergence and spread of Acinetobacter species resistant to most of the available antimicrobial agents is also of great concern. Acinetobacter infections have high mortality rates; they result in death in up to 65.5% of patients in intensive care unit (ICU) settings (Leão et al., 2016). Resistant gram positive bacterial infections have also increased in hospitals and other environments. Methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococci (VRE) infections are associated with worse clinical outcomes (Schmid et al., 2013), particularly in immunocompromised patients (Prematunge et al., 2016).
Treatment of MDRO (multidrug-resistant organism) infections presents a major clinical challenge, as the increase in antibiotic resistance leads to a greater risk of therapeutic failure, relapses, longer hospitalizations, and worse clinical outcomes (Cosgrove, 2006; Picot-Guéraud et al., 2015). Moreover, the increase in AMR has outpaced the development of new antibiotics. Currently, there are no validated therapeutic strategies for many MDR infections, and preventing the spread of pathogens through hospital infection control procedures and antimicrobial stewardship programs is often the only tool available to healthcare providers (Tacconelli et al., 2014). In this narrative review, we discuss the role of the intestinal microbiota as reservoir of MDRO, and we focus on the use of microbiota transplantation (FMT) for MDR intestinal decolonization, reporting updated literature on its current clinical use.
Gut Microbiota as Reservoir of MDRO
The human gut microbiota is composed of 1014 bacteria organized into microbial communities containing harmless symbionts, commensal bacteria, and opportunistic pathogens, all of which play crucial roles in human health and disease (Sekirov et al., 2010). The gut bacterial community is dominated by Firmicutes and Bacteroidetes in healthy adults, which account for 90% of the total gut bacteria. Small percentages of Actinobacteria, Proteobacteria, and Verrucomicrobia are also present (Qin et al., 2010). Although the microbiota profile of each person is unique, a conserved set of gut bacteria, the core microbiota, is shared amongst most individuals. This suggests that the core microbiota plays a role in maintaining health (Qin et al., 2010). The microbial core aids in several important functions, including digestion of polysaccharides, development of the immune system, biosynthesis of vitamins, and defense against infections (Qin et al., 2010; Sekirov et al., 2010; Cryan and O’Mahony, 2011; Flint et al., 2012; Lynch and Pedersen, 2016). In physiological conditions, the gut microbiota is stable (i.e., eubiotic status), but when perturbative events occur (e.g., dietary changes, stress, antibiotic administration, different hygienic conditions), the equilibrium amongst the bacteria and between the bacteria and the host breaks (i.e., dysbiotic status). This results in adverse health effects and loss of protection against pathogen colonization (Sommer et al., 2017).
The gut is also a large reservoir of opportunistic pathogens. This is particularly relevant for hospitalized patients, especially when antibiotic-based therapies are used that may select MDR bacteria already present in their gut microbiota (Rice, 2008; Biliński et al., 2016). These opportunistic pathogens may translocate across the intestinal barrier or, after fecal contamination of skin and other body sites, cause infections, especially in catheters or intravenous lines (van Schaik, 2015).
The large reservoir of antibiotic-resistance genes that is present in the gut is called the “resistome” (Casals-Pascual et al., 2018). The gut reservoir includes two different resistomes: the resident resistome, which is carried by the commensal bacteria, and the transitory resistome, which is carried by bacteria that are only periodically present in the gut. The latter bacteria can transfer their resistance to the commensal microbiota or become permanent microbiota residents themselves (Baron et al., 2018). Consequently, there is considerable interest in characterizing the antibiotic resistance gene reservoir of the human gut microbiota and to understand to what extent the antibiotic resistance genes can spread among different members of the gut microbiota, particularly between commensals and opportunistic pathogens (Penders et al., 2013).
From Colonization to Antimicrobial Resistance Through Microbiota-Mediated Mechanisms
The exact mechanism by which specific components of the microbiota influence colonization by other pathogens is not yet fully known. Previous studies have found that particular commensal species inhibit the growth of specific MDR bacteria. This has been shown in mice, where VRE did not colonize microbiota containing the obligate anaerobe Barnesiella spp. (Ubeda et al., 2013; Mahieu et al., 2017). Caballero et al. (2017) also demonstrated that administration of a defined bacterial consortium containing Blautia producta and Clostridium bolteae prevented colonization of VRE and cleared persistent VRE in mice. In humans, one study reported that four species of bacteria Desulfovibrio, Oscillospira, Parabacteroides, and Coprococcus, where not present in the microbiota of adult patients colonized by extended-spectrum β-lactamase (ESBL)-producing bacteria, whereas these four species were present in the microbiomes of patients not colonized by ESBL-producing bacteria (Gosalbes et al., 2015).
Little knowledge is available regarding the mechanisms which promote progression from colonization to infection. Martin et al. (2016) and Gorrie et al. (2017) reported that GI carriage of Klebsiella pneumoniae (KP) in hospitalized patients was associated with a greater risk of developing an infection (Martin et al., 2016; Gorrie et al., 2017). Additionally, gut colonization by MDR bacteria has been described as an important risk factor for severity outcomes in both solid organ and hematopoietic stem cell transplantation (HSCT) (Patriarca et al., 2017; Peric et al., 2017; Macesic et al., 2018).
Present Strategies for Fighting Antimicrobial Resistance
Control Measures and Decolonization Regimes
Currently, several strategies have been adopted to fight AMR (Figure 1). In particular, the increase in healthcare-acquired infections has led to a focus on improving control measures and infection prevention. A core component of this strategy is active surveillance for early identification of MDR carriers. MDR transmission usually occurs through person-to-person contact with healthcare providers and through contamination of the hospital environment, especially for pathogens such as Enterobacteriaceae and Acinetobacter baumanii. Depending on the local epidemiology, screening is usually performed for CRE, carbapenem-resistant (CR) Pseudomonas aeruginosa, VRE, and MRSA through rectal and nasal swabs. Early detection tests are usually performed on patients with risk factors for carrying MDR bacteria, including contact with colonized or infected patients, arrival from high incidence regions, and admission to high acuity wards (e.g., ICU, transplant units, onco-hematology units) (Fernando et al., 2017). When MDR carriers are identified, hospitals may have various policies for preventing infections and spreading of colonization, including contact precautions (antiseptic hand hygiene, use of gloves, gowns, single rooms or cohort, and dedicated patient equipment); decolonization of MRSA nasal carriers with mupirocin topic application; intensification of environmental cleaning and disinfection; and education of healthcare providers, patients, and visitors (Ciofi degli Atti et al., 2019). Decolonization of the intestinal tract through bowel preparations or antibiotic treatment has undoubtedly attracted great interest over the past few years. However, most of the available literature on this topic confirms the limited efficacy of using non-absorbable or systemic antibiotics to eradicate MDR bacteria from the intestinal tract (Rieg et al., 2015; Bar-Yoseph et al., 2016). One of the major limits is the limited time of MDR bacterial clearance after the decolonization procedure (Huttner et al., 2013; Katchman et al., 2014). Moreover, it has been observed that decolonization strategies using non-absorbable antibiotics have marked effects on the bacterial population, with resistance increasing during treatment. A considerable “rebound effect” of select antibiotic-resistant organisms in the intestinal tract has been observed after discontinuation of the decolonization regimens (Oostdijk et al., 2010). The latest ESCMID-EUCIC1 clinical guidelines on decolonization of multidrug-resistant Gram negative bacteria carriers (Tacconelli et al., 2019) did not recommend routine decolonization of third-generation cephalosporin-resistant Enterobacteriaceae and CRE carriers and did not find sufficient evidence to provide recommendations for or against any intervention in patients colonized with aminoglycoside-resistant Enterobacteriaceae, colistin-resistant Gram negative organisms, carbapenem-resistant Acinetobacter baumanii, cotrimoxazole-resistant Stenotrophomonas maltophilia, fluoroquinolone-resistant Enterobacteriaceae, pan-drug-resistant Gram negative organisms, or extremely drug-resistant Pseudomonas aeruginosa. Overall, decolonization using antimicrobials is not currently recommended. Conversely, inappropriate use may be responsible for increased dysbiosis and further increases in antibiotic resistance rates (Halaby et al., 2013).
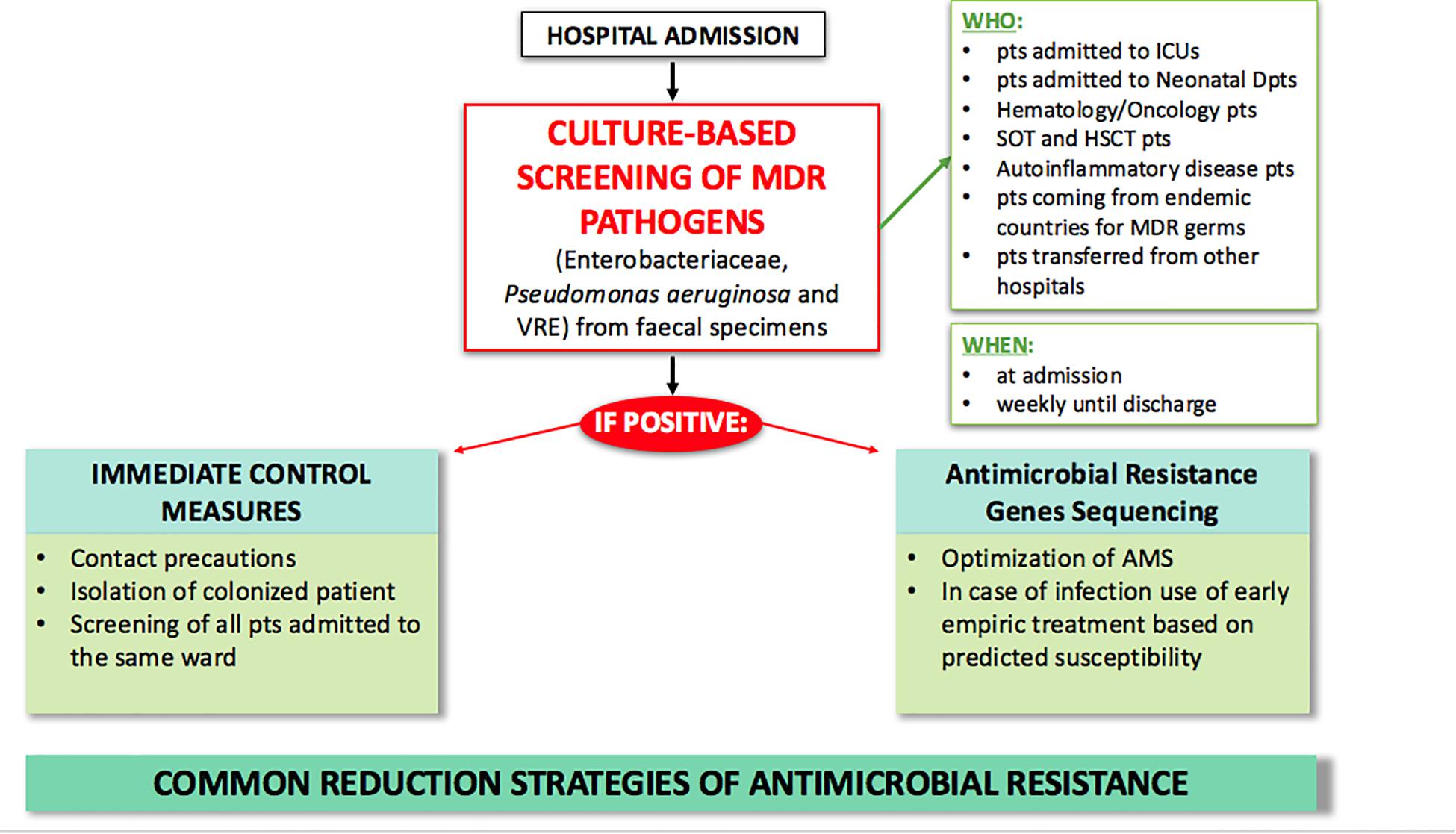
Figure 1. Current strategies for MDR antimicrobial clinical and laboratory stewardship in hospital settings. AMS, antimicrobial stewardship; FMT, fecal microbiota transplantation; HSCT, hematopoietic stem cell transplantation; ICU, intensive care unit; MDR, multi-drug resistant; pts, patients; SOT, solid organ transplant.
Antimicrobial and Diagnostic Stewardship Programs
Antibiotic prescribing policies and implementation of Antimicrobial Stewardship Programs have also played a crucial role in preventing MDR spread and infections. Depending on local infection control committee policies and funding availability, Antimicrobial Stewardship Programs may include a multidisciplinary team, antibiotic prescription guidelines, review of antibiotic appropriateness prescriptions, de-escalation therapy, formulary restriction and pre-authorization requirements, audit of antibiotic use, and monitoring antibiotic consumption with indicators (Septimus, 2018). Antimicrobial stewardship programs are of particular importance, because it is well-known that misuse and overuse of antibiotics accelerate the development of resistance. Therefore, it is necessary to limit the reckless use of antibiotics, especially the newest ones. In the last 40 years, the discovery of new molecules has been limited (World Health Organization, 2017). Recently, new promising antibiotics have been developed, finally reversing the declining trend of antimicrobial development (Talbot et al., 2019). However, resistance to all antibiotics will develop eventually. Hence, the discovery of new antimicrobials, although important, does not seem to be the definitive solution to AMR.
Diagnostic stewardship also plays a crucial role in controlling AMR. Current hospital protocols use culture-based screening as first step for identifying MDR carriers by Matrix Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS), and secondly by sequencing AMR genes for early pre-emptive antimicrobial therapy based on predicted susceptibility (Rodríguez-Sánchez et al., 2014). As technology in microbiologic diagnostics advances, and as outbreaks of MDR infections put 1000s of lives at risk, ad hoc outbreak-specific diagnostics need to be implemented (Willmann and Peter, 2017; Friedrich, 2019). We expect future AMR policies to include microbiome profiling, such as metataxonomic analysis of MDR-related microbiota and use of whole genome sequencing to type MDR strains (Figure 2). These microbiome-based approaches may not only contribute to a better knowledge of MDR pathogens, but also give an additional tool to clinical management of infections.
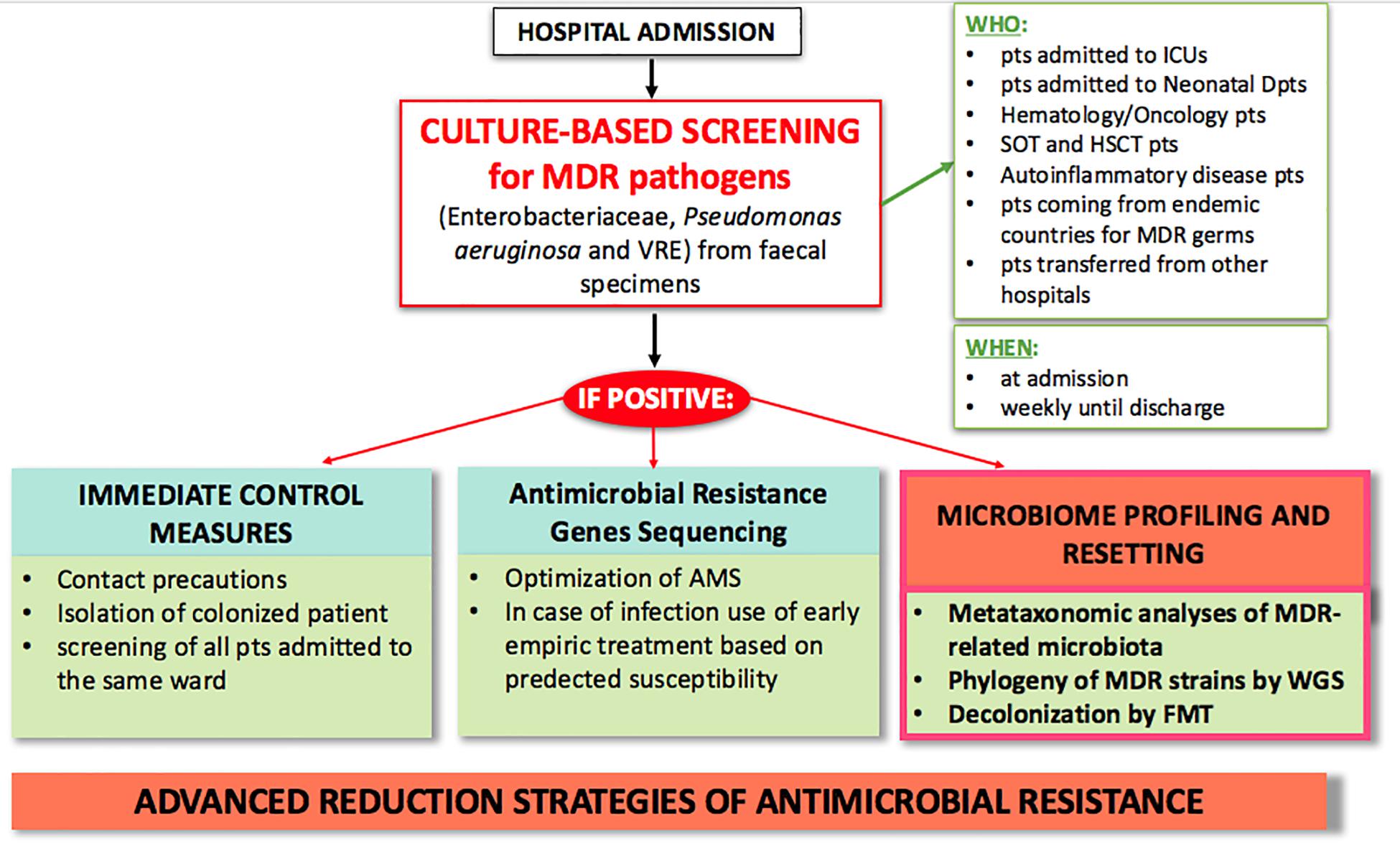
Figure 2. Proposed model of advanced clinical and laboratory stewardship for MDR pathogen screening in hospital settings. AMS, antimicrobial stewardship; FMT, fecal microbiota transplantation; HSCT, hematopoietic stem cell transplantation; ICU, intensive care unit; MDR, multi-drug resistant; pts, patients; SOT, solid organ transplant; WGS, whole genome sequencing.
Conveying the Present to the Future for Fighting Against Antimicrobial Resistance
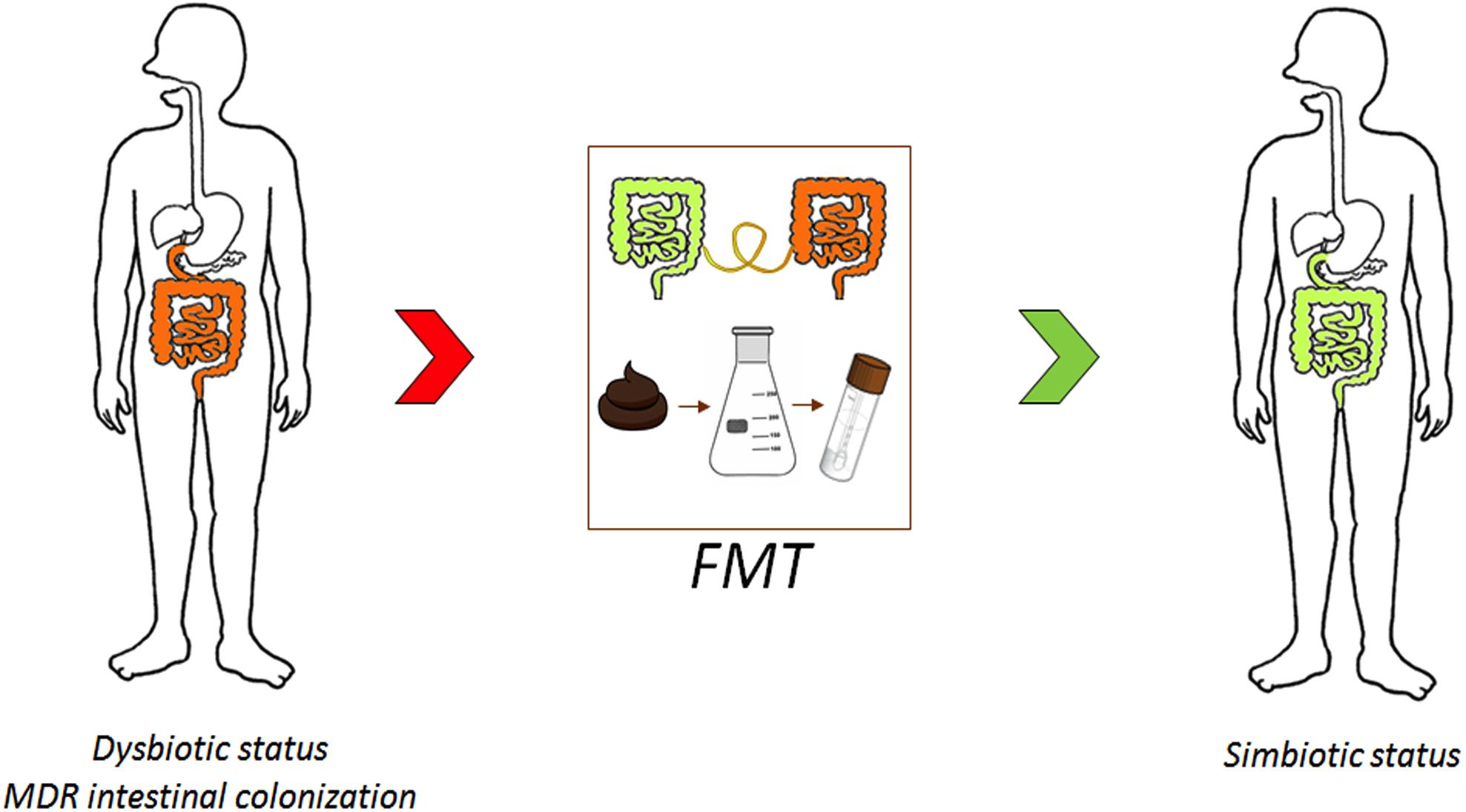
New solutions are urgently required to reduce the prevalence of MDRO and to control colonization. In this section, we discuss FMT as an advanced approach for combating AMR (Figure 3) and other strategies (i.e., probiotics and bacteriophages) as potential alternative antimicrobial stewardship solutions.
FMT: Present Use in Clinical Practice in Adult and Pediatric Populations
In current clinical practice, FMT is being used as a therapeutic option for recurrent infection with toxin-producing Clostridium difficile (rCDI) in adults (van Nood et al., 2013; Cammarota et al., 2015, 2017). As per the latest guidelines of Infectious Diseases Society of America (IDSA) and the Society for Healthcare Epidemiology of America (SHEA)2,3, FMT is recommended for patients with multiple recurrences of CDI when the appropriate antibiotic treatments have failed (strong recommendation, moderate quality of evidence) (McDonald et al., 2018). Use of FMT is also under study for the treatment of primary CDI (Juul et al., 2018). Randomized controlled trials (RCTs), systematic reviews, and meta-analyses have been performed to confirm the effectiveness of FMT, which is reported to be between 77 and 94% with administration via the proximal small bowel, and around 80–100% with instillation in the colon (McDonald et al., 2018). Oral capsules with lyophilized fecal microbiota product have also been found to be equally effective to frozen product given by colonoscopy (Kao et al., 2017) or enema (Jiang et al., 2018).
The safety of FMT has also been established (Kelly et al., 2015; Ianiro et al., 2018), although episodes of bacteremia following FMT have been reported (Baxter and Colville, 2016; Wang S. et al., 2016; Shogbesan et al., 2018). In pediatric patients, an increase in the prevalence of CDI has also been observed over the past decade (Sammons and Toltzis, 2013; Wendt et al., 2014), especially in children given prolonged antibiotic therapy or those who have chronic inflammatory bowel diseases (IBDs), oncological conditions, and who have had recent surgery (Kelsen et al., 2011; Hourigan et al., 2014). An overall increase in recurrences of CDI has also been reported, estimated at around 20–30% (Nicholson et al., 2015). According to the latest joint position paper on FMT for rCDI in children released by the North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition (NASPGHAN) and by the European Society for Pediatric Gastroenterology, Hepatology, and Nutrition (ESPGHAN) in January 2019, 336 FMTs for rCDI have been performed in the pediatric population in the United States alone, with a success rate of 81% after a single delivery and nearly 90% when patients received a second FMT (Davidovics et al., 2019). This is consistent with previous reports demonstrating its effectiveness (between 90 and 100%) and safety (Hourigan and Oliva-Hemker, 2016). In general, FMT-related adverse events in pediatric patients were infrequent (Chen et al., 2017), mainly characterized as minor and transient adverse events of the GI tract. No bacterial translocation or sepsis were reported. Current data suggest that the age of the recipient does not affect efficacy or safety, although definitive data from pediatric RCTs are not yet available. The youngest reported recipient so far was a 0.5-year-old infant (Wang A.Y. et al., 2016), and FMT in other patients less than 2-years-old has also been reported (Kahn et al., 2012; Russell et al., 2014; Walia et al., 2014; Kronman et al., 2015).
In addition to its use for MDR infections, FMT has also been described as a promising therapy for other disorders associated with the alteration of intestinal microbiota, including intestinal inflammatory diseases such as IBD (Wang A.Y. et al., 2016; Paramsothy et al., 2017). Several RCTs, case series, and case reports on FMT in IBD have been published. Clinical remission is seen in 28.8% of patients and a clinical response is seen in 53% during follow-up, according to a recent systematic review and meta-analysis of 459 IBD patients (Fang et al., 2018). In pediatrics, several case series on FMT for IBDs have also been published (De Leon et al., 2013; Barfield et al., 2018); Fang et al. (2018) reported a total of 67 pediatric patients, which will need to be updated with the rising use of FMT in this field. The currently published data show a clinical response rate ranging between 56 and 100% (Kunde et al., 2013; Wang A.Y. et al., 2016; Goyal et al., 2018). This wide variability may be due to the different locations of disease in Crohn’s disease and ulcerative colitis, as well as the different routes of FMT administration used in the published studies (Hourigan and Oliva-Hemker, 2016; Wang A.Y. et al., 2016; Barfield et al., 2018). Side effects in both populations have been confirmed to be modest and transient, involving the GI tract (Hourigan and Oliva-Hemker, 2016; Wang A.Y. et al., 2016). To the best of our knowledge, the youngest reported patient that underwent FMT for very early-onset IBD was 2 years and 3 months old (Yodoshi and Hurt, 2018).
Extensive studies on the microbiome and eventually FMT as a therapeutic strategy have been performed for other diseases, including metabolic diseases (Groen and Nieuwdorp, 2017; Zhang et al., 2017), diabetes mellitus (van Olden et al., 2015), obesity and non-alcoholic liver disease (Abdou et al., 2016; de Clercq et al., 2016; Marotz and Zarrinpar, 2016; Reijnders et al., 2016; Tandon et al., 2017; Nobili et al., 2018), HIV (Vujkovic-Cvijin et al., 2017; Sessa et al., 2019), and autism spectrum disorders (Kang et al., 2017), with initial but promising results in terms of microbiota profiling and clinical outcomes. The use of FMT in the fields of hematology and oncology, specifically HSCT, has also gained interest. The gut microbiota plays a pivotal role in intestinal inflammation and the immune response (Zeiser et al., 2016; Andermann et al., 2018). Indeed, disruption of the host microbiota following prolonged antimicrobial therapy contributes to the pathogenesis of graft-versus-host-disease (GVHD) (Jenq et al., 2012; Holler et al., 2014; Simms-Waldrip et al., 2017), and is an independent predictor of poor outcome (Weber et al., 2017) and overall mortality in allo-HSCT recipients (Taur et al., 2014). Although the initial reports are encouraging (Taur et al., 2018; DeFilipp et al., 2019), RCTs are still ongoing and the results are pending. Depending on these results, FMT may become a useful tool for preventing HSCT-related morbidity and mortality.
FMT: Present Use for Decolonization of MDRO
Based on current clinical practice, and considering the increased threat of AMR, FMT has been considered for the eradication of drug-resistant bacteria from the intestinal reservoir. Studies show that adult patients undergoing prolonged antibiotic therapy for chronic infections, such as rCDI, have a greater amount of antibiotic resistance genes in their intestinal microbiota compared to healthy adults (Jouhten et al., 2016; Millan et al., 2016). Moreover, the number and diversity of antibiotic resistance genes decreased after FMT, especially if repeated FMTs were performed (Jouhten et al., 2016; Millan et al., 2016). However, AMR genes can also be acquired from FMT donor stool, hence healthy stool donor selection is fundamental and needs further standardization (Leung et al., 2018).
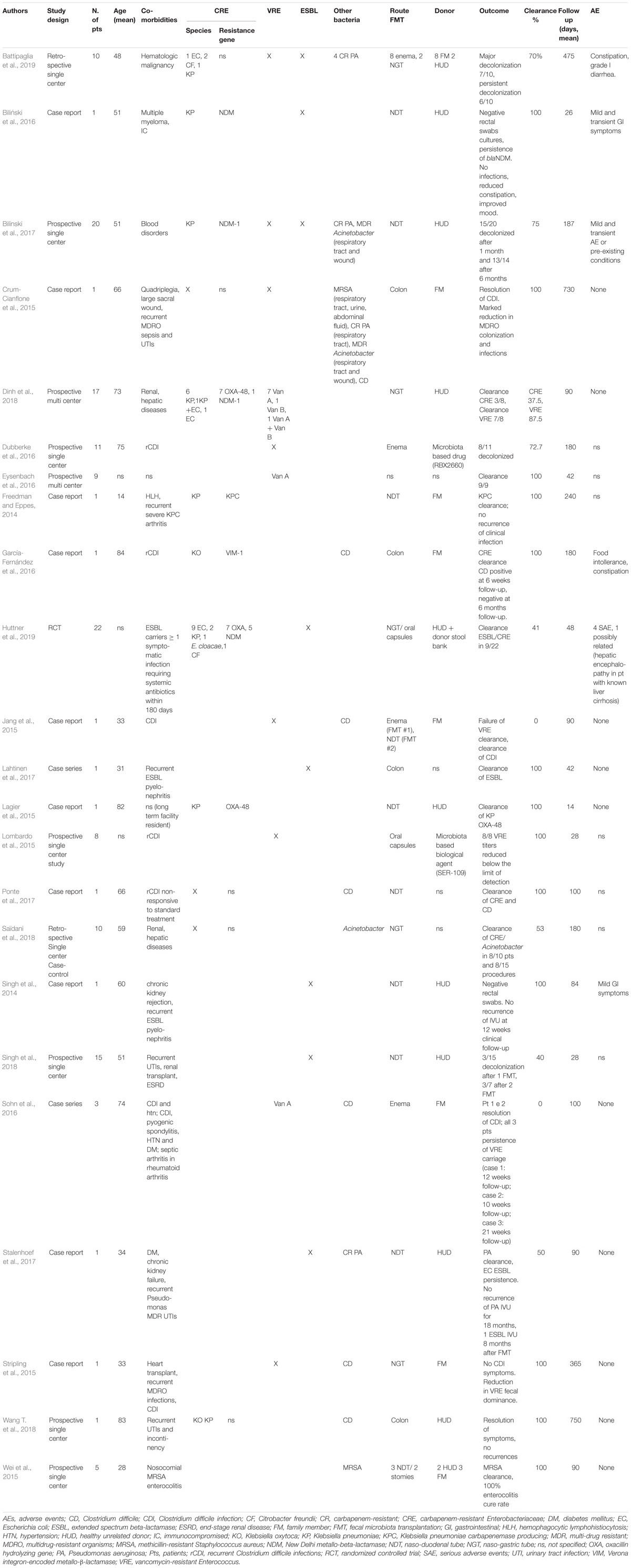
Since 2014, case reports and case series have described the use of FMT for MDR intestinal decolonization (Gopalsamy et al., 2018). Given the rising interest in this new approach, retrospective and prospective single-center and multi-center studies have been successively performed (Table 1). Currently, there are several RCTs evaluating the efficacy of FMT in patients colonized by MDR bacteria (NCT03802461; NCT03786900; NCT03643887; NCT03527056; NCT03391674; NCT03367910; NCT03167398; NCT03063437; NCT03029078; NCT02922816; NCT02906774; NCT02816437; NCT02543866; NCT02472600; NCT02461199; NCT02390622; and NCT023129864). To date, only one RCT is complete (Huttner et al., 2019), which showed a slight decrease of ESBL/CRE carriage compared to controls when using non-absorbable antibiotics followed by FMT. However, the unfavorable results are potentially due to the study design and early trial termination. From the analysis of the literature available so far (Table 1), 142 patients undergoing FMT for MDR intestinal decolonization globally have been described in 23 total reports, with a mean recipient age of 54.8 years (range: 14–89 years). The most commonly isolated pre-FMT MDR intestinal bacteria were CRE and VRE; MDR organisms in other sites, including Pseudomonas aeruginosa, MRSA, and Acinetobacter were also reported. The most commonly used route of administration was via naso-duodenal tube (45/142, 31.6%), followed by naso-gastric tube (36/142, 25.3%), oral capsules (24/142, 16.8%), enema (23/142, 16.1%), colonoscopy (4/142, 2.7%), and through pre-existing stomies (2/142, 1.3%). In 9/142 (6.2%) patients, the route of administration was not specified. Microbiological clearance of MDR bacteria on fecal samples or rectal swabs was achieved on 77.5% of patients. Microbiological follow-up was on average 180.8 days (range: 14–750 days). Donors were chosen among family members of recipients in 19/142 (13.4%) of cases, while most studies used samples from healthy unrelated donors (83/142, 58.5%). The donor was not specified in 21/142 (14.8%) of cases. An experimental drug was used in two studies on 19 patients [RBX2660 (Dubberke et al., 2016); SER-109 (Lombardo et al., 2015)]. Age of donors, where specified, ranged from 6 to 60 years. Preparation of patients for FMT, where reported, included fasting for at least 12 h (21.2%), introduction of treatment with a proton pump inhibitor (PPI) twice daily to neutralize gastric acid (49.3%), discontinuation of antibiotic treatment (55.5%), and bowel lavage with laxative drugs (50%). Administration of oral antibiotics was part of the preparation in one case report (Lagier et al., 2015), in the only RCT completed to date (Huttner et al., 2019), and in a recent retrospective case-control study (Saïdani et al., 2018). Notably, Saïdani et al. (2018) reported a unique preparation protocol that included a 3-day nasopharyngeal decolonization with 0.12% chlorhexidine gluconate 8 days before FMT, two bowel lavages (5 days before FMT and the day before the procedure), 5 days of antimicrobial treatment prior to FMT with an oral non-absorbable bi-antibiotic (mostly colistin and aminoglycoside, replaced by sulfadiazine or fusidic acid in case of resistance), post-FMT environmental decontamination, room transfer, and removal of medical tubes and catheters in order to limit post-FMT recolonization (Saïdani et al., 2018). In the published studies, stool suspension for transplant was prepared using fresh stools for 30 patients (21.1%), frozen samples for 27 patients (19.0%), oral capsules of fecal preparation for 16 patients (11.3%), was not specified in 35.2% of cases, and an experimental drug was used in 19 cases (13.4%). One serious adverse event was reported: in the only completed RCT, a 57-year-old female patient with known liver cirrhosis and recurrent episodes of hepatic encephalopathy was hospitalized 2 weeks after FMT for an episode of encephalopathy. This event was classified as a possible “Suspected Unexpected Serious Adverse Reaction” (Huttner et al., 2019). In the other reports, the adverse events described were minor, transient, and involved the gastro-intestinal tract (loose stools/mild diarrhea, transient mild abdominal discomforts or cramps, food intolerance, constipation).
To the best of our knowledge, only two pediatric cases of FMT for MDR intestinal decolonization have been described so far. The first pediatric case report (Freedman and Eppes, 2014) describes a 14-year-old patient undergoing immunosuppressive treatment for hemophagocytic lymphohistiocytosis, affected by recurrent infections with highly resistant carbapenemase-producing K. pneumoniae. Despite prolonged antibiotic therapy, the patient’s stool samples remained persistently colonized, putting her at risk for further infective relapses. The patient underwent FMT via naso-duodenal tube, with microbiological clearance at an 8-month follow-up, and clinical absence of infections 1.5 years after FMT. The second patient (Battipaglia et al., 2019) has been described in a recent retrospective study among adult hematologic patients. The patient was a 16-year old female with acute myeloid leukemia (AML) that underwent two FMTs for VRE and ESBL-producing bacteria 98 days after HSCT, resulting in decolonization of VRE and persistence of ESBL-producing bacteria, with no reported adverse events.
As reported herein, great variability in terms of the mode of delivery of emulsion, sample preparation, donor selection, and targeted MDRO are seen for MDRO decolonization via FMT. This reflects the lack of knowledge regarding the effects of each variable on the efficacy of FMT. Therefore, further studies and RCTs are needed to determine a standardized methodology and best clinical practice.
Other Possible Strategies for Decolonization of MDR Bacteria
A potential microbial strategy that needs to be considered in the prevention and treatment of MDR is the use of probiotics, which have antimicrobial activity against pathogens. Many studies have been conducted with this purpose and have produced interesting results, especially regarding Gram positive organisms. In fact, clinical trials on intestinal VRE decolonization showed marked efficacy of probiotics in eradicating VRE carriage (Manley et al., 2007; Szachta et al., 2011). Similarly, in a recent clinical trial, MRSA colonization of the GI tract was reduced by daily administration of oral Lactobacillus rhamnosus, although colonization was not reduced in other body sites (Eggers et al., 2018). However, results from studies using probiotics for Gram negative decolonization are not promising. To date, two randomized double-blind placebo-controlled clinical trials showed that probiotics were not effective in decolonizing hospitalized patients or long-term residents harboring MDR Gram negative bacilli (Tannock et al., 2011; Salomão et al., 2016). Ongoing RCTs, such as NCT036449966 on probiotics and MDR urinary tract infections, will definitely give notable contributions to the field. Lactobacilli, Lactococci, and Bifidobacteria are classified as “generally regarded as safe” (Food and Agriculture Organization of the United Nations, and World Health Organization, 2006; Snydman, 2008). However, it has also been reported that RCTs using probiotics, prebiotics, and symbiotics often lack adequate reporting of adverse effects (Bafeta et al., 2018). Although substantial challenges regarding the safety, mechanisms of action, and real-life indications of probiotic use need to be fully addressed, the therapeutic potential of probiotics in reducing and preventing MDR bacterial colonization is promising. RCTs and comprehensive studies are certainly needed to expand this field and to evaluate the potential efficacy of probiotics for the treatment and prevention of MDR infections.
Another potential tool for combating AMR could exploit the use of bacteriophages directed against specific bacterial strains (Wong and Santiago, 2017). Phage therapy could have major advantages over antibiotics, such as host-specificity and consequent low toxicity in humans (Bourdin et al., 2014). Additionally, its efficacy against biofilms could make phage therapy suitable for the treatment of biofilm-associated infections, such as MRSA (Tkhilaishvili et al., 2018) and P. aeruginosa infections (Fong et al., 2017). The safety of all the phage therapy products currently under study is generally excellent. Nevertheless, phage therapy and enzymes can both induce rapid endotoxin release due to rapid cell lysis, as much as bactericidal antibiotics (Maciejewska et al., 2018). Therefore, there are currently no phage therapy products approved for human use in the EU or United States.
Conclusion
Antimicrobial resistance is increasing worldwide and is causing a rise in hospital-acquired infections, deaths, and costs. Microbiota-based strategies need to be considered in the prevention and treatment of MDRO. In particular, FMT is a promising tool, especially in cases where conventional therapies have not proven effective. So far, FMT has been shown to be effective and safe. However, RCTs are needed to standardize the methodology and set regulatory boundaries in order to include FMT in MDR clinical management.
Author Contributions
LG and LP conceived the study. LG, FD, PD’A, and LP wrote, reviewed, and edited the original draft of the manuscript.
Funding
This work was supported by the Ministry of Health, Ricerca corrente 201802G004314: “Clinical analysis of intestinal microbiota profiling of patients colonized by multi-drug resistant germs: preventive and therapeutic actions of FMT,” assigned to LP, Children’s Hospital Bambino Gesù, IRCCS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the experts from BioMed Proofreading LLC for their English revision.
Footnotes
- ^ https://www.escmid.org/research_projects/eucic/
- ^ https://www.idsociety.org/
- ^ https://www.shea-online.org/
- ^ ClinicalTrials.gov
References
Abdou, R. M., Zhu, L., Baker, R. D., and Baker, S. S. (2016). Gut microbiota of nonalcoholic fatty liver disease. Dig. Dis. Sci. 61, 1268–1281. doi: 10.1007/s10620-016-4045-1
Andermann, T. M., Peled, J. U., Ho, C., Reddy, P., Riches, M., Storb, R., et al. (2018). The microbiome and hematopoietic cell transplantation: past, present, and future. Biol. Blood Marrow Transplant. 24, 1322–1340. doi: 10.1016/j.bbmt.2018.02.009
Bafeta, A., Koh, M., Riveros, C., and Ravaud, P. (2018). Harms reporting in randomized controlled trials of interventions aimed at modifying microbiota: a systematic review. Ann. Intern. Med. 169, 240–247. doi: 10.7326/M18-0343
Barfield, E., Small, L., Navallo, L., and Solomon, A. (2018). Going to the bank: fecal microbiota transplantation in pediatrics. Clin. Pediatr. 57, 481–483. doi: 10.1177/0009922817721159
Baron, S. A., Diene, S. M., and Rolain, J.-M. (2018). Human microbiomes and antibiotic resistance. Hum. Microbiome J. 10, 43–52. doi: 10.1016/j.humic.2018.08.005
Bar-Yoseph, H., Hussein, K., Braun, E., and Paul, M. (2016). Natural history and decolonization strategies for ESBL/carbapenem-resistant Enterobacteriaceae carriage: systematic review and meta-analysis. J. Antimicrob. Chemother. 71, 2729–2739. doi: 10.1093/jac/dkw221
Battipaglia, G., Malard, F., Rubio, M. T., Ruggeri, A., Mamez, A. C., Brissot, E., et al. (2019). Fecal microbiota transplantation before or after allogeneic hematopoietic transplantation in patients with hematological malignancies carrying multidrug-resistance bacteria. Haematologica doi: 10.3324/haematol.2018.198549 [Epub ahead of print].
Baxter, M., and Colville, A. (2016). Adverse events in faecal microbiota transplant: a review of the literature. J. Hosp. Infect. 92, 117–127. doi: 10.1016/j.jhin.2015.10.024
Biliński, J., Grzesiowski, P., Muszyński, J., Wróblewska, M., Mądry, K., Robak, K., et al. (2016). Fecal microbiota transplantation inhibits multidrug-resistant gut pathogens: preliminary report performed in an immunocompromised host. Arch. Immunol. Ther. Exp. 64, 255–258. doi: 10.1007/s00005-016-0387-9
Bilinski, J., Grzesiowski, P., Sorensen, N., Madry, K., Muszynski, J., Robak, K., et al. (2017). Fecal microbiota transplantation in patients with blood disorders inhibits gut colonization with antibiotic-resistant bacteria: results of a prospective, single-center study. Clin. Infect. Dis. 65, 364–370. doi: 10.1093/cid/cix252
Bourdin, G., Navarro, A., Sarker, S. A., Pittet, A.-C., Qadri, F., Sultana, S., et al. (2014). Coverage of diarrhoea-associated Escherichia coli isolates from different origins with two types of phage cocktails. Microb. Biotechnol. 7, 165–176. doi: 10.1111/1751-7915.12113
Caballero, S., Kim, S., Carter, R. A., Leiner, I. M., Sušac, B., Miller, L., et al. (2017). Cooperating commensals restore colonization resistance to vancomycin- resistant Enterococcus faecium. Cell Host Microbe 21, 592.e4–602.e4. doi: 10.1016/j.chom.2017.04.002
Cammarota, G., Ianiro, G., Tilg, H., Rajilić-Stojanović, M., Kump, P., Satokari, R., et al. (2017). European consensus conference on faecal microbiota transplantation in clinical practice. Gut 66, 569–580. doi: 10.1136/gutjnl-2016-313017
Cammarota, G., Masucci, L., Ianiro, G., Bibbò, S., Dinoi, G., Costamagna, G., et al. (2015). Randomised clinical trial: faecal microbiota transplantation by colonoscopy vs. vancomycin for the treatment of recurrent Clostridium difficile infection. Aliment. Pharmacol. Ther. 41, 835–843. doi: 10.1111/apt.13144
Casals-Pascual, C., Vergara, A., and Vila, J. (2018). Intestinal microbiota and antibiotic resistance: perspectives and solutions. Hum. Microbiome J. 9, 11–15. doi: 10.1016/j.humic.2018.05.002
CDC (2019). Antibiotic Resistance Threatens Everyone. Atlanta, GA: Centers for Disease Control and Prevention.
Chen, B., Avinashi, V., and Dobson, S. (2017). Fecal microbiota transplantation for recurrent Clostridium difficile infection in children. J. Infect. 74, S120–S127. doi: 10.1016/S0163-4453(17)30202-5
Ciofi degli Atti, M. I., D’Amore, C., and Gagliotti, C. (2019). Strategies to control antibiotic resistance: results from a survey in Italian children’s hospitals. Ann. Ig. 31, 3–12. doi: 10.7416/ai.2019.2253
Cosgrove, S. E. (2006). The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin. Infect. Dis. 42(Suppl. 2), S82–S89. doi: 10.1086/499406
Crum-Cianflone, N. F., Sullivan, E., and Ballon-Landa, G. (2015). Fecal microbiota transplantation and successful resolution of multidrug-resistant-organism colonization. J. Clin. Microbiol. 53, 1986–1989. doi: 10.1128/JCM.00820-15
Cryan, J. F., and O’Mahony, S. M. (2011). The microbiome-gut-brain axis: from bowel to behavior. Neurogastroenterol. Motil. 23, 187–192. doi: 10.1111/j.1365-2982.2010.01664.x
Davidovics, Z. H., Michail, S., Nicholson, M. R., Kociolek, L. K., Pai, N., Hansen, R., et al. (2019). Fecal microbiota transplantation for recurrent Clostridium difficile infection and other conditions in children: a joint position paper from the north american society for pediatric gastroenterology, hepatology, and nutrition and the european society for pediatric gastroenterology, hepatology, and nutrition. J. Pediatr. Gastroenterol. Nutr. 68, 130–143. doi: 10.1097/MPG.0000000000002205
de Clercq, N. C., Groen, A. K., Romijn, J. A., and Nieuwdorp, M. (2016). Gut microbiota in obesity and undernutrition. Adv. Nutr. Int. Rev. J. 7, 1080–1089. doi: 10.3945/an.116.012914
De Leon, L. M., Watson, J. B., and Kelly, C. R. (2013). Transient flare of ulcerative colitis after fecal microbiota transplantation for recurrent Clostridium difficile infection. Clin. Gastroenterol. Hepatol. 11, 1036–1038. doi: 10.1016/j.cgh.2013.04.045
DeFilipp, Z., Hohmann, E., Jenq, R. R., and Chen, Y.-B. (2019). Fecal microbiota transplantation: restoring the injured microbiome after allogeneic hematopoietic cell transplantation. Biol. Blood Marrow Transplant. 25, e17–e22. doi: 10.1016/j.bbmt.2018.10.022
Del Chierico, F., Cardile, S., Pietrobattista, A., Liccardo, D., Russo, A., Candusso, M., et al. (2018). Liver transplantation and gut microbiota profiling in a child colonized by a multi-drug resistant Klebsiella pneumoniae: a new approach to move from antibiotic to “eubiotic” control of microbial resistance. Int. J. Mol. Sci. 19:E1280. doi: 10.3390/ijms19051280
Dinh, A., Fessi, H., Duran, C., Batista, R., Michelon, H., Bouchand, F., et al. (2018). Clearance of carbapenem-resistant Enterobacteriaceae vs vancomycin-resistant enterococci carriage after faecal microbiota transplant: a prospective comparative study. J. Hosp. Infect. 99, 481–486. doi: 10.1016/j.jhin.2018.02.018
Dubberke, E. R., Mullane, K. M., Gerding, D. N., Lee, C. H., Louie, T. J., Guthertz, H., et al. (2016). Clearance of vancomycin-resistant Enterococcus concomitant with administration of a microbiota-based drug targeted at recurrent Clostridium difficile infection. Open Forum Infect. Dis. 3:ofw133. doi: 10.1093/ofid/ofw133
Eggers, S., Barker, A. K., Valentine, S., Hess, T., Duster, M., and Safdar, N. (2018). Effect of Lactobacillus rhamnosus HN001 on carriage of Staphylococcus aureus: results of the impact of probiotics for reducing infections in veterans (IMPROVE) study. BMC Infect. Dis. 18:129. doi: 10.1186/s12879-018-3028-6
Europäisches Zentrum für die Prävention und die Kontrolle von Krankheiten (2009). The Bacterial Challenge, Time to React: a Call to Narrow the Gap Between Multidrug-Resistant Bacteria in the EU and the Development of New Antibacterial Agents. Stockholm: ECDC.
Eysenbach, L., Allegretti, J. R., Aroniadis, O., Brandt, L., Donovan, D., Fischer, M., et al. (2016). Clearance of vancomycin-resistant enterococcus colonization with fecal microbiota transplantation among patients with recurrent Clostridium difficile infection. Open Forum Infect. Dis. 3:2119. doi: 10.1093/ofid/ofw172.1667
Fang, H., Fu, L., and Wang, J. (2018). Protocol for fecal microbiota transplantation in inflammatory bowel disease: a systematic review and meta-analysis. Biomed. Res. Int. 2018:8941340. doi: 10.1155/2018/8941340
Fernando, S. A., Gray, T. J., and Gottlieb, T. (2017). Healthcare-acquired infections: prevention strategies: preventing healthcare-acquired infection. Intern. Med. J. 47, 1341–1351. doi: 10.1111/imj.13642
Flint, H. J., Scott, K. P., Duncan, S. H., Louis, P., and Forano, E. (2012). Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3, 289–306. doi: 10.4161/gmic.19897
Fong, S. A., Drilling, A., Morales, S., Cornet, M. E., Woodworth, B. A., Fokkens, W. J., et al. (2017). Activity of bacteriophages in removing biofilms of Pseudomonas aeruginosa isolates from chronic rhinosinusitis patients. Front. Cell Infect. Microbiol. 7:418. doi: 10.3389/fcimb.2017.00418
Food and Agriculture Organization of the United Nations, and World Health Organization (2006). Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation. Rome: Food and Agriculture Organization of the United Nations.
Freedman, A., and Eppes, S. (2014). 1805use of stool transplant to clear fecal colonization with carbapenem-resistant Enterobacteraciae (CRE): proof of concept. Open Forum Infect. Dis. 1:S65. doi: 10.1093/ofid/ofu051.177
Friedrich, A. W. (2019). Control of hospital acquired infections and antimicrobial resistance in Europe: the way to go. Wien. Med. Wochenschr. 169, 25–30. doi: 10.1007/s10354-018-0676-5
García-Fernández, S., Morosini, M.-I., Cobo, M., Foruny, J. R., López-Sanromán, A., Cobo, J., et al. (2016). Gut eradication of VIM-1 producing ST9 Klebsiella oxytoca after fecal microbiota transplantation for diarrhea caused by a Clostridium difficile hypervirulent R027 strain. Diagn. Microbiol. Infect. Dis. 86, 470–471. doi: 10.1016/j.diagmicrobio.2016.09.004
Gopalsamy, S. N., Woodworth, M. H., Wang, T., Carpentieri, C. T., Mehta, N., Friedman-Moraco, R. J., et al. (2018). The use of microbiome restoration therapeutics to eliminate intestinal colonization with multidrug-resistant organisms. Am. J. Med. Sci. 356, 433–440. doi: 10.1016/j.amjms.2018.08.015
Gorrie, C. L., Mirceta, M., Wick, R. R., Edwards, D. J., Thomson, N. R., Strugnell, R. A., et al. (2017). Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin. Infect. Dis. 65, 208–215. doi: 10.1093/cid/cix270
Gosalbes, M. J., Vázquez-Castellanos, J. F., Angebault, C., Woerther, P.-L., Ruppé, E., Ferrús, M. L., et al. (2015). Carriage of enterobacteria producing extended-spectrum β-Lactamases and composition of the gut microbiota in an amerindian community. Antimicrob. Agents Chemother. 60, 507–514. doi: 10.1128/AAC.01528-15
Goyal, A., Yeh, A., Bush, B. R., Firek, B. A., Siebold, L. M., Rogers, M. B., et al. (2018). Safety, clinical response, and microbiome findings following fecal microbiota transplant in children with inflammatory bowel disease. Inflamm. Bowel Dis. 24, 410–421. doi: 10.1093/ibd/izx035
Groen, A. K., and Nieuwdorp, M. (2017). An evaluation of the therapeutic potential of fecal microbiota transplantation to treat infectious and metabolic diseases. EMBO Mol. Med. 9, 1–3. doi: 10.15252/emmm.201607035
Halaby, T., Al Naiemi, N., Kluytmans, J., van der Palen, J., and Vandenbroucke-Grauls, C. M. J. E. (2013). Emergence of colistin resistance in Enterobacteriaceae after the introduction of selective digestive tract decontamination in an intensive care unit. Antimicrob. Agents Chemother. 57, 3224–3229. doi: 10.1128/AAC.02634-12
Holler, E., Butzhammer, P., Schmid, K., Hundsrucker, C., Koestler, J., Peter, K., et al. (2014). Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol. Blood Marrow Transplant. 20, 640–645. doi: 10.1016/j.bbmt.2014.01.030
Hourigan, S. K., and Oliva-Hemker, M. (2016). Fecal microbiota transplantation in children: a brief review. Pediatr. Res. 80, 2–6. doi: 10.1038/pr.2016.48
Hourigan, S. K., Oliva-Hemker, M., and Hutfless, S. (2014). The prevalence of Clostridium difficile infection in pediatric and adult patients with inflammatory bowel disease. Dig. Dis. Sci. 59, 2222–2227. doi: 10.1007/s10620-014-3169-4
Huttner, B., Haustein, T., Uçkay, I., Renzi, G., Stewardson, A., Schaerrer, D., et al. (2013). Decolonization of intestinal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae with oral colistin and neomycin: a randomized, double-blind, placebo-controlled trial. J. Antimicrob. Chemother. 68, 2375–2382. doi: 10.1093/jac/dkt174
Huttner, B. D., Lastours, V., de Wassenberg, M., Maharshak, N., Mauris, A., Galperine, T., et al. (2019). A five-day course of oral antibiotics followed by faecal transplantation to eradicate carriage of multidrug-resistant Enterobacteriaceae: a randomized clinical trial. Clin. Microbiol. Infect. 25, 830–838. doi: 10.1016/j.cmi.2018.12.009
Ianiro, G., Maida, M., Burisch, J., Simonelli, C., Hold, G., Ventimiglia, M., et al. (2018). Efficacy of different faecal microbiota transplantation protocols for Clostridium difficile infection: a systematic review and meta-analysis. U. Eur. Gastroenterol. J. 6, 1232–1244. doi: 10.1177/2050640618780762
Ivády, B., Kenesei,É, Tóth-Heyn, P., Kertész, G., Tárkányi, K., Kassa, C., et al. (2016). Factors influencing antimicrobial resistance and outcome of Gram-negative bloodstream infections in children. Infection 44, 309–321. doi: 10.1007/s15010-015-0857-8
Jang, M.-O., An, J. H., Jung, S.-I., and Park, K.-H. (2015). Refractory Clostridium difficile infection cured with fecal microbiota transplantation in vancomycin-resistant enterococcus colonized patient. Intest. Res. 13, 80–84. doi: 10.5217/ir.2015.13.1.80
Jenq, R. R., Ubeda, C., Taur, Y., Menezes, C. C., Khanin, R., Dudakov, J. A., et al. (2012). Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J. Exp. Med. 209, 903–911. doi: 10.1084/jem.20112408
Jiang, Z.-D., Jenq, R. R., Ajami, N. J., Petrosino, J. F., Alexander, A. A., Ke, S., et al. (2018). Safety and preliminary efficacy of orally administered lyophilized fecal microbiota product compared with frozen product given by enema for recurrent Clostridium difficile infection: a randomized clinical trial. PLoS One 13:e0205064. doi: 10.1371/journal.pone.0205064
Jouhten, H., Mattila, E., Arkkila, P., and Satokari, R. (2016). Reduction of antibiotic resistance genes in intestinal microbiota of patients with recurrent Clostridium difficile infection after fecal microbiota transplantation. Clin. Infect. Dis. 63, 710–711. doi: 10.1093/cid/ciw390
Juul, F. E., Garborg, K., Bretthauer, M., Skudal, H., Øines, M. N., Wiig, H., et al. (2018). Fecal microbiota transplantation for primary Clostridium difficile infection. N. Engl. J. Med. 378, 2535–2536. doi: 10.1056/NEJMc1803103
Kahn, S. A., Young, S., and Rubin, D. T. (2012). Colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection in a child. Am. J. Gastroenterol. 107, 1930–1931. doi: 10.1038/ajg.2012.351
Kalpoe, J. S., Sonnenberg, E., Factor, S. H., del Rio Martin, J., Schiano, T., Patel, G., et al. (2012). Mortality associated with carbapenem-resistant Klebsiella pneumoniae infections in liver transplant recipients. Liver Transpl. 18, 468–474. doi: 10.1002/lt.23374
Kang, D.-W., Adams, J. B., Gregory, A. C., Borody, T., Chittick, L., Fasano, A., et al. (2017). Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome 5:10. doi: 10.1186/s40168-016-0225-7
Kao, D., Roach, B., Silva, M., Beck, P., Rioux, K., Kaplan, G. G., et al. (2017). Effect of oral capsule- vs colonoscopy-delivered fecal microbiota transplantation on recurrent Clostridium difficile infection: a randomized clinical trial. JAMA 318, 1985–1993. doi: 10.1001/jama.2017.17077
Katchman, E., Marquez, M., Bazerbachi, F., Grant, D., Cattral, M., Low, C. Y., et al. (2014). A comparative study of the use of selective digestive decontamination prophylaxis in living-donor liver transplant recipients. Transpl. Infect. Dis. 16, 539–547. doi: 10.1111/tid.12235
Kelly, C. R., Kahn, S., Kashyap, P., Laine, L., Rubin, D., Atreja, A., et al. (2015). Update on fecal microbiota transplantation 2015: indications, methodologies, mechanisms, and outlook. Gastroenterology 149, 223–237. doi: 10.1053/j.gastro.2015.05.008
Kelsen, J. R., Kim, J., Latta, D., Smathers, S., McGowan, K. L., Zaoutis, T., et al. (2011). Recurrence rate of Clostridium difficile infection in hospitalized pediatric patients with inflammatory bowel disease. Inflamm. Bowel Dis. 17, 50–55. doi: 10.1002/ibd.21421
Kronman, M. P., Nielson, H. J., Adler, A. L., Giefer, M. J., Wahbeh, G., Singh, N., et al. (2015). Fecal microbiota transplantation via nasogastric tube for recurrent Clostridium difficile infection in pediatric patients. J. Pediatr. Gastroenterol. Nutr. 60, 23–26. doi: 10.1097/MPG.0000000000000545
Kunde, S., Pham, A., Bonczyk, S., Crumb, T., Duba, M., Conrad, H., et al. (2013). Safety, tolerability, and clinical response after fecal transplantation in children and young adults with ulcerative colitis. J. Pediatr. Gastroenterol. Nutr. 56, 597–601. doi: 10.1097/MPG.0b013e318292fa0d
Lagier, J. C., Million, M., Fournier, P. E., Brouqui, P., and Raoult, D. (2015). Faecal microbiota transplantation for stool decolonization of OXA-48 carbapenemase-producing Klebsiella pneumoniae. J. Hosp. Infect. 90, 173–174. doi: 10.1016/j.jhin.2015.02.013
Lahtinen, P., Mattila, E., Anttila, V.-J., Tillonen, J., Teittinen, M., Nevalainen, P., et al. (2017). Faecal microbiota transplantation in patients with Clostridium difficile and significant comorbidities as well as in patients with new indications: a case series. World J. Gastroenterol. 23, 7174–7184. doi: 10.3748/wjg.v23.i39.7174
Leão, A. C. Q., Menezes, P. R., Oliveira, M. S., and Levin, A. S. (2016). Acinetobacter spp. are associated with a higher mortality in intensive care patients with bacteremia: a survival analysis. BMC Infect. Dis. 16:386. doi: 10.1186/s12879-016-1695-8
Leung, V., Vincent, C., Edens, T. J., Miller, M., and Manges, A. R. (2018). Antimicrobial resistance gene acquisition and depletion following fecal microbiota transplantation for recurrent Clostridium difficile infection. Clin. Infect. Dis. 66, 456–457. doi: 10.1093/cid/cix821
Lombardo, M.-J., Vulic, M., Ohsumi, T., Cook, D., Henn, M., Kraft, C., et al. (2015). Vancomycin-resistant enterococcal iters diminish among patients with recurrent Clostridium difficile infection after administration of SER-109, A novel microbiome agent. Open Forum Infect. Dis. 2:757. doi: 10.1093/ofid/ofv133.474
Lynch, S. V., and Pedersen, O. (2016). The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379. doi: 10.1056/NEJMra1600266
Macesic, N., Gomez-Simmonds, A., Sullivan, S. B., Giddins, M. J., Ferguson, S. A., Korakavi, G., et al. (2018). Genomic surveillance reveals diversity of multidrug-resistant organism colonization and infection: a prospective cohort study in liver transplant recipients. Clin. Infect. Dis. 67, 905–912. doi: 10.1093/cid/ciy199
Maciejewska, B., Olszak, T., and Drulis-Kawa, Z. (2018). Applications of bacteriophages versus phage enzymes to combat and cure bacterial infections: an ambitious and also a realistic application? Appl. Microbiol. Biotechnol. 102, 2563–2581. doi: 10.1007/s00253-018-8811-1
Mahieu, R., Cassisa, V., Hilliquin, D., Coron, N., Pailhoriès, H., Kempf, M., et al. (2017). Impact of faecal microbiota transplantation on mouse digestive colonization with two extensively resistant bacteria. J. Infect. 75, 75–77. doi: 10.1016/j.jinf.2017.04.008
Manley, K. J., Fraenkel, M. B., Mayall, B. C., and Power, D. A. (2007). Probiotic treatment of vancomycin-resistant enterococci: a randomised controlled trial. Med. J. Aust. 186, 454–457.
Marotz, C. A., and Zarrinpar, A. (2016). Treating obesity and metabolic syndrome with fecal microbiota transplantation. Yale J. Biol. Med. 89, 383–388.
Martin, R. M., Cao, J., Brisse, S., Passet, V., Wu, W., Zhao, L., et al. (2016). Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere 1:e00261-16. doi: 10.1128/mSphere.00261-16
McDonald, L. C., Gerding, D. N., Johnson, S., Bakken, J. S., Carroll, K. C., Coffin, S. E., et al. (2018). Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the infectious diseases society of america (IDSA) and society for healthcare epidemiology of america (SHEA). Clin. Infect. Dis. 66, e1–e48. doi: 10.1093/cid/cix1085
Millan, B., Park, H., Hotte, N., Mathieu, O., Burguiere, P., Tompkins, T. A., et al. (2016). Fecal microbial transplants reduce antibiotic-resistant genes in patients with recurrent Clostridium difficile infection. Clin. Infect. Dis. 62, 1479–1486. doi: 10.1093/cid/ciw185
Munoz-Price, L. S., Poirel, L., Bonomo, R. A., Schwaber, M. J., Daikos, G. L., Cormican, M., et al. (2013). Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13, 785–796. doi: 10.1016/S1473-3099(13)70190-7
Nicholson, M. R., Thomsen, I. P., Slaughter, J. C., Creech, C. B., and Edwards, K. M. (2015). Novel risk factors for recurrent Clostridium difficile infection in children. J. Pediatr. Gastroenterol. Nutr. 60, 18–22. doi: 10.1097/MPG.0000000000000553
Nobili, V., Putignani, L., Mosca, A., Chierico, F. D., Vernocchi, P., Alisi, A., et al. (2018). Bifidobacteria and lactobacilli in the gut microbiome of children with non-alcoholic fatty liver disease: which strains act as health players? Arch. Med. Sci. 14, 81–87. doi: 10.5114/aoms.2016.62150
Oostdijk, E. A., de Smet, A. M., Blok, H. E., Thieme Groen, E. S., van Asselt, G. J., Benus, R. F., et al. (2010). Ecological effects of selective decontamination on resistant gram-negative bacterial colonization. Am. J. Respir. Crit. Care Med. 181, 452–457. doi: 10.1164/rccm.200908-1210OC
Paramsothy, S., Paramsothy, R., Rubin, D. T., Kamm, M. A., Kaakoush, N. O., Mitchell, H. M., et al. (2017). Faecal microbiota transplantation for inflammatory bowel disease: a systematic review and meta-analysis. J. Crohns Colitis 11, 1180–1199. doi: 10.1093/ecco-jcc/jjx063
Patriarca, F., Cigana, C., Massimo, D., Lazzarotto, D., Geromin, A., Isola, M., et al. (2017). Risk factors and outcomes of infections by multidrug-resistant gram-negative bacteria in patients undergoing hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 23, 333–339. doi: 10.1016/j.bbmt.2016.11.005
Penders, J., Stobberingh, E. E., Savelkoul, P. H. M., and Wolffs, P. F. G. (2013). The human microbiome as a reservoir of antimicrobial resistance. Front. Microbiol. 4:87. doi: 10.3389/fmicb.2013.00087
Peric, Z., Vranjes, V. R., Durakovic, N., Desnica, L., Marekovic, I., Serventi-Seiwerth, R., et al. (2017). Gut colonization by multidrug-resistant gram-negative bacteria is an independent risk factor for development of intestinal acute graft-versus-host disease. Biol. Blood Marrow Transplant. 23, 1221–1222. doi: 10.1016/j.bbmt.2017.03.025
Picot-Guéraud, R., Batailler, P., Caspar, Y., Hennebique, A., and Mallaret, M.-R. (2015). Bacteremia caused by multidrug-resistant bacteria in a French university hospital center: 3 years of collection. Am. J. Infect. Control 43, 960–964. doi: 10.1016/j.ajic.2015.05.004
Ponte, A., Pinho, R., and Mota, M. (2017). Fecal microbiota transplantation: is there a role in the eradication of carbapenem-resistant Klebsiella pneumoniae intestinal carriage? Rev. Esp. Enferm. Dig. 109:392. doi: 10.17235/reed.2017.4425/2016
Prematunge, C., MacDougall, C., Johnstone, J., Adomako, K., Lam, F., Robertson, J., et al. (2016). VRE and VSE bacteremia outcomes in the era of effective VRE therapy: a systematic review and meta-analysis. Infect. Control Hosp. Epidemiol. 37, 26–35. doi: 10.1017/ice.2015.228
Pumart, P., Phodha, T., Thamlikitkul, V., Riewpaiboon, A., Prakongsai, P., and Limwattananon, S. (2012). Health and economic impacts of antimicrobial resistance in Thailand. J. Health Syst. Res. 6, 352–360.
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Reijnders, D., Goossens, G. H., Hermes, G. D. A., Neis, E. P. J. G., van der Beek, C. M., Most, J., et al. (2016). Effects of gut microbiota manipulation by antibiotics on host metabolism in obese humans: a randomized double-blind placebo-controlled trial. Cell Metab. 24, 63–74. doi: 10.1016/j.cmet.2016.06.016
Rice, L. B. (2008). Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 197, 1079–1081. doi: 10.1086/533452
Rieg, S., Küpper, M. F., de With, K., Serr, A., Bohnert, J. A., and Kern, W. V. (2015). Intestinal decolonization of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBL): a retrospective observational study in patients at risk for infection and a brief review of the literature. BMC Infect. Dis. 15:475. doi: 10.1186/s12879-015-1225-0
Rodríguez-Sánchez, B., Sánchez-Carrillo, C., Ruiz, A., Marín, M., Cercenado, E., Rodríguez-Créixems, M., et al. (2014). Direct identification of pathogens from positive blood cultures using matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 20, O421–O427. doi: 10.1111/1469-0691.12455
Russell, G. H., Kaplan, J. L., Youngster, I., Baril-Dore, M., Schindelar, L., Hohmann, E., et al. (2014). Fecal transplant for recurrent Clostridium difficile infection in children with and without inflammatory bowel disease. J. Pediatr. Gastroenterol. Nutr. 58, 588–592. doi: 10.1097/MPG.0000000000000283
Saïdani, N., Lagier, J.-C., Cassir, N., Million, M., Baron, S., Dubourg, G., et al. (2018). Faecal microbiota transplantation shortens the colonisation period and allows re-entry of patients carrying carbapenamase-producing bacteria into medical care facilities. Int. J. Antimicrob. Agents 53, 355–361. doi: 10.1016/j.ijantimicag.2018.11.014
Salomão, M. C., Heluany-Filho, M. A., Menegueti, M. G., Kraker, M. E., Martinez, R., and Bellissimo-Rodrigues, F. (2016). A randomized clinical trial on the effectiveness of a symbiotic product to decolonize patients harboring multidrug-resistant Gram-negative bacilli. Rev. Soc. Bras. Med. Trop. 49, 559–566. doi: 10.1590/0037-8682-0233-2016
Sammons, J. S., and Toltzis, P. (2013). Recent trends in the epidemiology and treatment of C. difficile infection in children. Curr. Opin. Pediatr. 25, 116–121. doi: 10.1097/MOP.0b013e32835bf6c0
Schmid, H., Romanos, A., Schiffl, H., and Lederer, S. R. (2013). Persistent nasal methicillin-resistant staphylococcus aureus carriage in hemodialysis outpatients: a predictor of worse outcome. BMC Nephrol. 14:93. doi: 10.1186/1471-2369-14-93
Sekirov, I., Russell, S. L., Antunes, L. C. M., and Finlay, B. B. (2010). Gut microbiota in health and disease. Physiol. Rev. 90, 859–904. doi: 10.1152/physrev.00045.2009
Septimus, E. J. (2018). Antimicrobial resistance: an antimicrobial/diagnostic stewardship and infection prevention approach. Med. Clin. North Am. 102, 819–829. doi: 10.1016/j.mcna.2018.04.005
Sessa, L., Reddel, S., Manno, E., Quagliariello, A., Cotugno, N., Del Chierico, F., et al. (2019). Distinct gut microbiota profile in ART-treated perinatally HIV-infected patients associated with cardiac and inflammatory biomarkers. AIDS doi: 10.1097/QAD.0000000000002131 [Epub ahead of print].
Shogbesan, O., Poudel, D. R., Victor, S., Jehangir, A., Fadahunsi, O., Shogbesan, G., et al. (2018). A systematic review of the efficacy and safety of fecal microbiota transplant for Clostridium difficile infection in immunocompromised patients. Can. J. Gastroenterol. Hepatol. 2018:1394379. doi: 10.1155/2018/1394379
Simms-Waldrip, T. R., Sunkersett, G., Coughlin, L. A., Savani, M. R., Arana, C., Kim, J., et al. (2017). Antibiotic-induced depletion of anti-inflammatory clostridia is associated with the development of graft-versus-host disease in pediatric stem cell transplantation patients. Biol. Blood Marrow Transpl. 23, 820–829. doi: 10.1016/j.bbmt.2017.02.004
Singh, R., de Groot, P. F., Geerlings, S. E., Hodiamont, C. J., Belzer, C., Berge, I. J. M. T., et al. (2018). Fecal microbiota transplantation against intestinal colonization by extended spectrum beta-lactamase producing Enterobacteriaceae: a proof of principle study. BMC Res. Notes 11:190. doi: 10.1186/s13104-018-3293-x
Singh, R., van Nood, E., Nieuwdorp, M., van Dam, B., ten Berge, I. J. M., Geerlings, S. E., et al. (2014). Donor feces infusion for eradication of extended spectrum beta-lactamase producing Escherichia coli in a patient with end stage renal disease. Clin. Microbiol. Infect. 20, O977–O978. doi: 10.1111/1469-0691.12683
Snydman, D. R. (2008). The safety of probiotics. Clin. Infect. Dis. 46(Suppl. 2), S104–S111. doi: 10.1086/523331
Sohn, K. M., Cheon, S., and Kim, Y.-S. (2016). Can fecal microbiota transplantation (FMT) eradicate fecal colonization with vancomycin-resistant Enterococci (VRE)? Infect. Control Hosp. Epidemiol. 37, 1519–1521. doi: 10.1017/ice.2016.229
Sommer, F., Anderson, J. M., Bharti, R., Raes, J., and Rosenstiel, P. (2017). The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 15, 630–638. doi: 10.1038/nrmicro.2017.58
Stalenhoef, J. E., Terveer, E. M., Knetsch, C. W., Van’t Hof, P. J., Vlasveld, I. N., Keller, J. J., et al. (2017). Fecal microbiota transfer for multidrug-resistant gram-negatives: a clinical success combined with microbiological failure. Open Forum Infect. Dis. 4:ofx047. doi: 10.1093/ofid/ofx047
Stripling, J., Kumar, R., Baddley, J. W., Nellore, A., Dixon, P., Howard, D., et al. (2015). Loss of vancomycin-resistant enterococcus fecal dominance in an organ transplant patient with Clostridium difficile colitis after fecal microbiota transplant. Open Forum Infect. Dis. 2:ofv078. doi: 10.1093/ofid/ofv078
Szachta, P., Ignyś, I., and Cichy, W. (2011). An evaluation of the ability of the probiotic strain Lactobacillus rhamnosus GG to eliminate the gastrointestinal carrier state of vancomycin-resistant enterococci in colonized children. J. Clin. Gastroenterol. 45, 872–877. doi: 10.1097/MCG.0b013e318227439f
Tacconelli, E., Cataldo, M. A., Dancer, S. J., De Angelis, G., Falcone, M., Frank, U., et al. (2014). ESCMID guidelines for the management of the infection control measures to reduce transmission of multidrug-resistant Gram-negative bacteria in hospitalized patients. Clin. Microbiol. Infect. 20(Suppl. 1), 1–55. doi: 10.1111/1469-0691.12427
Tacconelli, E., Mazzaferri, F., de Smet, A. M., Bragantini, D., Eggimann, P., Huttner, B. D., et al. (2019). ESCMID-EUCIC clinical guidelines on decolonization of multidrug-resistant Gram-negative bacteria carriers. Clin. Microbiol. Infect. 25, 807–817. doi: 10.1016/j.cmi.2019.01.005
Talbot, G. H., Jezek, A., Murray, B. E., Jones, R. N., Ebright, R. H., Nau, G. J., et al. (2019). The infectious diseases society of America’s 10 × ’20 initiative (ten new systemic antibacterial agents FDA-approved by 2020): is 20 × ’20 a possibility? Clin. Infect. Dis. 69, 1–11. doi: 10.1093/cid/ciz089
Tandon, P., Madsen, K., and Kao, D. (2017). Fecal microbiota transplantation for hepatic encephalopathy: ready for prime time? Hepatology 66, 1713–1715. doi: 10.1002/hep.29396
Tannock, G. W., Tiong, I. S., Priest, P., Munro, K., Taylor, C., Richardson, A., et al. (2011). Testing probiotic strain Escherichia coli Nissle 1917 (Mutaflor) for its ability to reduce carriage of multidrug-resistant E. coli by elderly residents in long-term care facilities. J. Med. Microbiol. 60, 366–370. doi: 10.1099/jmm.0.025874-0
Taur, Y., Coyte, K., Schluter, J., Robilotti, E., Figueroa, C., Gjonbalaj, M., et al. (2018). Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci. Transl. Med. 10:eaa9489. doi: 10.1126/scitranslmed.aap9489
Taur, Y., Jenq, R. R., Perales, M.-A., Littmann, E. R., Morjaria, S., Ling, L., et al. (2014). The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 124, 1174–1182. doi: 10.1182/blood-2014-02-554725
Tkhilaishvili, T., Lombardi, L., Klatt, A.-B., Trampuz, A., and Di Luca, M. (2018). Evaluation of Sb-1 bacteriophage activity in enhancing antibiotic efficacy against biofilm, degrading the exopolysaccharide matrix and targeting persister cells of Staphylococcus aureus. bioRxiv
Tumbarello, M., Viale, P., Viscoli, C., Trecarichi, E. M., Tumietto, F., Marchese, A., et al. (2012). Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin. Infect. Dis. 55, 943–950. doi: 10.1093/cid/cis588
Ubeda, C., Bucci, V., Caballero, S., Djukovic, A., Toussaint, N. C., Equinda, M., et al. (2013). Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect. Immun. 81, 965–973. doi: 10.1128/IAI.01197-12
van Duin, D., and Paterson, D. (2016). Multidrug resistant bacteria in the community: trends and lessons learned. Infect. Dis. Clin. North Am. 30, 377–390. doi: 10.1016/j.idc.2016.02.004
van Nood, E., Vrieze, A., Nieuwdorp, M., Fuentes, S., Zoetendal, E. G., de Vos, W. M., et al. (2013). Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415. doi: 10.1056/NEJMoa1205037
van Olden, C., Groen, A. K., and Nieuwdorp, M. (2015). Role of intestinal microbiome in lipid and glucose metabolism in diabetes mellitus. Clin. Ther. 37, 1172–1177. doi: 10.1016/j.clinthera.2015.03.008
van Schaik, W. (2015). The human gut resistome. Philos. Trans. R. Soc. B Biol. Sci. 370:20140087. doi: 10.1098/rstb.2014.0087
Vujkovic-Cvijin, I., Rutishauser, R. L., Pao, M., Hunt, P. W., Lynch, S. V., McCune, J. M., et al. (2017). Limited engraftment of donor microbiome via one-time fecal microbial transplantation in treated HIV-infected individuals. Gut Microbes 8, 440–450. doi: 10.1080/19490976.2017.1334034
Walia, R., Garg, S., Song, Y., Girotra, M., Cuffari, C., Fricke, W. F., et al. (2014). Efficacy of fecal microbiota transplantation in 2 children with recurrent Clostridium difficile infection and its impact on their growth and gut microbiome. J. Pediatr. Gastroenterol. Nutr. 59, 565–570. doi: 10.1097/MPG.0000000000000495
Wang, A. Y., Popov, J., and Pai, N. (2016). Fecal microbial transplant for the treatment of pediatric inflammatory bowel disease. World J. Gastroenterol. 22, 10304–10315. doi: 10.3748/wjg.v22.i47.10304
Wang, S., Xu, M., Wang, W., Cao, X., Piao, M., Khan, S., et al. (2016). Systematic review: adverse events of fecal microbiota transplantation. PLoS One 11:e0161174. doi: 10.1371/journal.pone.0161174
Wang, T., Kraft, C. S., Woodworth, M. H., Dhere, T., and Eaton, M. E. (2018). Fecal microbiota transplant for refractory Clostridium difficile infection interrupts 25-year history of recurrent urinary tract infections. Open Forum Infect. Dis. 5:ofy016. doi: 10.1093/ofid/ofy016
Wang, Z., Qin, R.-R., Huang, L., and Sun, L.-Y. (2018). Risk factors for carbapenem-resistant Klebsiella pneumoniae infection and mortality of Klebsiella pneumoniae infection. Chin. Med. J. 131, 56–62. doi: 10.4103/0366-6999.221267
Weber, D., Jenq, R. R., Peled, J. U., Taur, Y., Hiergeist, A., Koestler, J., et al. (2017). Microbiota disruption induced by early use of broad-spectrum antibiotics is an independent risk factor of outcome after allogeneic stem cell transplantation. Biol. Blood Marrow Transplant. 23, 845–852. doi: 10.1016/j.bbmt.2017.02.006
Wei, Y., Gong, J., Zhu, W., Guo, D., Gu, L., Li, N., et al. (2015). Fecal microbiota transplantation restores dysbiosis in patients with methicillin resistant Staphylococcus aureus enterocolitis. BMC Infect. Dis. 15:265. doi: 10.1186/s12879-015-0973-1
Wendt, J. M., Cohen, J. A., Mu, Y., Dumyati, G. K., Dunn, J. R., Holzbauer, S. M., et al. (2014). Clostridium difficile infection among children across diverse US geographic locations. Pediatrics 133, 651–658. doi: 10.1542/peds.2013-3049
Willmann, M., and Peter, S. (2017). Translational metagenomics and the human resistome: confronting the menace of the new millennium. J. Mol. Med. 95, 41–51. doi: 10.1007/s00109-016-1478-0
Wong, W. F., and Santiago, M. (2017). Microbial approaches for targeting antibiotic-resistant bacteria. Microb. Biotechnol. 10, 1047–1053. doi: 10.1111/1751-7915.12783
World Health Organization (2017). Antibacterial Agents in Clinical Development: an Analysis of the Antibacterial Clinical Development Pipeline, Including Tuberculosis. Geneva: World Health Organization.
Yodoshi, T., and Hurt, T. L. (2018). Fecal microbiota transplantation to patients with refractory very early onset ulcerative colitis. Pediatr. Gastroenterol. Hepatol. Nutr. 21, 355–360. doi: 10.5223/pghn.2018.21.4.355
Zeiser, R., Socié, G., and Blazar, B. R. (2016). Pathogenesis of acute graft-versus-host disease: from intestinal microbiota alterations to donor T cell activation. Br. J. Haematol. 175, 191–207. doi: 10.1111/bjh.14295
Keywords: antimicrobial resistance, clinical and laboratory advanced stewardship, microbiota profiling, fecal microbiota transplantation (FMT), antimicrobial stewardship (AMS), antimicrobial stewardship program (ASP), multidrug resistance (MDR) bacteria, microbiota modulation strategies
Citation: Gargiullo L, Del Chierico F, D’Argenio P and Putignani L (2019) Gut Microbiota Modulation for Multidrug-Resistant Organism Decolonization: Present and Future Perspectives. Front. Microbiol. 10:1704. doi: 10.3389/fmicb.2019.01704
Received: 01 April 2019; Accepted: 10 July 2019;
Published: 25 July 2019.
Edited by:
Peter Mullany, University College London, United KingdomReviewed by:
Markus M. Heimesaat, Charité Medical University of Berlin, GermanySrishti Saha, Mayo Clinic, United States
Copyright © 2019 Gargiullo, Del Chierico, D’Argenio and Putignani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lorenza Putignani, bG9yZW56YS5wdXRpZ25hbmlAb3BiZy5uZXQ=
 Livia Gargiullo
Livia Gargiullo Federica Del Chierico
Federica Del Chierico Patrizia D’Argenio
Patrizia D’Argenio Lorenza Putignani
Lorenza Putignani