- 1State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Yunnan University, Kunming, China
- 2School of Life Sciences, Yunnan University, Kunming, China
- 3Key Laboratory for Microbial Resources of the Ministry of Education, Yunnan University, Kunming, China
The velvet family proteins VosA and VelB are involved in growth regulation and differentiation in the model fungus Aspergillus nidulans and other filamentous fungi. In this study, the orthologs of VosA and VelB, AoVosA, and AoVelB, respectively, were characterized in the nematode-trapping fungus Arthrobotrys oligospora, which captures nematodes by producing trapping devices (traps). Deletion of the AovelB gene resulted in growth defects in different media, and the aerial hyphae from the ΔAovelB mutant lines were fewer in number and their colonies were less dense than those from the wild-type (WT) strain. The ΔAovelB mutants each displayed serious sporulation defects, and the transcripts of several sporulation-related genes (e.g., abaA, flbC, rodA, and vosA) were significantly down-regulated compared to those from the WT strain. Furthermore, the ΔAovelB mutant strains became more sensitive to chemical reagents, including sodium dodecyl sulfate and H2O2. Importantly, the ΔAovelB mutants were unable to produce nematode-capturing traps. Similarly, extracellular proteolytic activity was also lower in the ΔAovelB mutants than in the WT strain. In contrast, the ΔAovosA mutants displayed no obvious differences from the WT strain in these phenotypic traits, whereas conidial germination was lower in the ΔAovosA mutants, which became more sensitive to heat shock stress. Our results demonstrate that the velvet protein AoVelB is essential for conidiation, trap formation, and pathogenicity in A. oligospora, while AoVosA plays a role in the regulation of conidial germination and heat shock stress.
Introduction
Many filamentous fungi naturally produce asexual conidia, and these constitute the main reproductive propagule and infectious particles in these organisms. Conidia formation is a complicated biological process that requires the coordination and cooperation of many genes (Adams et al., 1998; Park and Yu, 2012). Multiple regulatory genes and pathways for conidia production in filamentous fungi have been reported, and many studies have been conducted using Aspergillus as the model fungal species (Park and Yu, 2012, 2016; Chang et al., 2013). The results from previous studies have indicated that conidiophore production is genetically regulated and controlled by multiple activators and repressors in filamentous fungi (Mirabito et al., 1989; Krijgsheld et al., 2013). These regulators govern the coordinated expression of distinct gene sets required for the progression of each stage (Etxebeste et al., 2010; Park and Yu, 2012). In the model fungi Aspergillus nidulans and the pathogenic fungus A. fumigatus, these regulators are divided into three categories: upstream regulators, central regulators, and feedback regulators (Park and Yu, 2012, 2016). The central regulatory pathway contains three key elements, brlA, abaA, and wetA, which coordinate conidiation-specific gene expression, whereas the upstream developmental activators fluG, flbA, flbB, flbC, flbD, and flbE initiate the sporulation process. Velvet family proteins VelB and VosA were identified as the feedback regulators for conidiation (Park and Yu, 2012, 2016).
The velvet regulators VeA, VelB, VelC, and VosA constitute a class of fungi-specific DNA-binding, velvet domain-containing transcription factors (Ahmed et al., 2013; Beyhan et al., 2013). Velvet family proteins share a homologous region of approximately 150 amino acids and were first discovered in A. nidulans (Ni and Yu, 2007; Park and Yu, 2016). In Aspergillus species, the velvet proteins coordinate fungal growth, development, pigmentation, and primary/secondary metabolism (Ni and Yu, 2007; Park et al., 2012a, b, 2014). Velvet proteins, such as VelB/VeA/LaeA, VelB/VosA, and VelB/VelB, can form multimeric velvet complexes that regulate different developmental processes (Bayram et al., 2008; Sarikaya Bayram et al., 2010). Among these complexes the VosA–VelB complex functions as a key functional unit controlling spore maturation, trehalose biosynthesis, and conidial germination (Sarikaya Bayram et al., 2010; Park et al., 2012b). VelB, a primary protein in the composition of velvet complexes, plays a positive role in regulating conidiation in A. nidulans and also plays an important role in its secondary metabolism (Bayram et al., 2008; Sarikaya Bayram et al., 2010). Recent studies have shown that VelB predominantly interacts with VosA in asexual spores and plays an interdependent role in conidial maturation and germination (Park et al., 2012b). These findings suggest that VosA and VelB play crucial roles in the production of asexual spores and conidial maturation.
Nematode-trapping fungi (NTF) are a special group of filamentous fungi by virtue of their innate abilities to capture and digest nematodes by producing traps (trapping devices). Sporulation plays an important role in the growth and reproduction of NTF. Arthrobotrys oligospora is a typical nematode-trapping fungal species that can complete its reproduction asexually by producing abundant conidia (Niu and Zhang, 2011). At present, little is known about the genes involved in conidiation or the related regulatory genes for spore production in A. oligospora and other NTF. In 2011, the genome of the fungus A. oligospora was sequenced (Yang et al., 2011), providing a good opportunity to elucidate the mechanism involved in conidiation in this fungus. Compared with the A. nidulans fungus, many homologous sporulation-related regulators, such as upstream regulators (FluG, FlbC, and FlbD) and feedback regulators (VosA and VelB), were found in A. oligospora. In the present study, we characterized the functions of the homologous VosA (AoVosA) and VelB (AoVelB) proteins in A. oligospora and investigated their roles in the regulation of several important phenotypic traits, including conidiation and cell wall synthesis, using quantitative reverse-transcription PCR (RT-PCR). The results show that AoVelB plays an important role in the growth, conidiation, trap formation, and pathogenicity in A. oligospora.
Materials and Methods
Fungal Strains and Culture Conditions
Arthrobotrys oligospora Fres. (ATCC 24927) and its derived fungal mutants were maintained on potato dextrose agar (PDA) medium at 28°C for standard culturing. Liquid TG (1% tryptone and 1% glucose) medium was used to collect the fungal mycelia. Plasmid pRS426 (a gift from Dr. K. A. Borkovich, University of California, Riverside, CA, United States) was maintained in the Escherichia coli strain DH5α (TaKaRa Bio, Shiga, Japan) and used to construct recombinational plasmids. The PDAS (PDA supplemented with 10 g/L molasses and 0.4 M saccharose) medium was used for protoplast regeneration. TG, TYGA, and CMY media were prepared as previously described (Yang et al., 2018) and were used to analyze the phenotypic traits of A. oligospora and its corresponding mutants. The nematode Caenorhabditis elegans was maintained on oatmeal water medium at 26°C.
Sequence and Phylogenetic Analyses of AoVosA and AoVelB From A. oligospora
The VosA and VelB velvet proteins from A. nidulans were used to search for their homologous sequences in the A. oligospora genome. Their orthologs, AoVosA (AOL_s00054g700) and AoVelB (AOL_s00054g811), were downloaded from GenBank1. The AoVosA and AoVelB homologs from other filamentous fungi identified in GenBank were downloaded and used to construct neighbor-joining trees for VosA and VelB, respectively, using the Mega 7 software package (Kumar et al., 2016).
Deletion of AovosA and AovelB Genes
The cetyltrimethylammonium bromide (CTAB) method was used to isolate genomic DNA from fungal A. oligospora. The replacement fragments from the AovosA and AovelB genes were constructed using a modified yeast cloning procedure (Colot et al., 2006). The 5′ and 3′ flanking sequences of the target genes and the hph cassette (Staben et al., 1989) were cloned from A. oligospora and pCSN44 with paired primers (Supplementary Table S1), respectively. These DNA fragments and the pRS426 plasmid backbone (digested with EcoRI and XhoI) were simultaneously transformed into Saccharomyces cerevisiae strain FY834 by electroporation (Winston et al., 1995). The constructed vectors (pRS426-AoVosA-hph and pRS426-AoVelB-hph) were maintained in E. coli DH5a (TaKaRa Bio) through transformation. Finally, the combined plasmids pRS426-AoVosA-hph and pRS426-AoVelB-hph were individually transformed into the A. oligospora protoplast as previously described (Yang et al., 2018; Zhen et al., 2018, 2019).
The transformants were screened for the presence of the hph resistance gene using hygromycin (200 μg/mL) (Tunlid et al., 1999). The putative transformants were confirmed as authentic by PCR amplification using specific primers (Yf and Yr) and Southern blot analyses with specific probes (Supplementary Table S1). Southern blotting was performed using the North2South Chemiluminescent Hybridization and Detection kit (Pierce, Rockford, IL, United States) according to the manufacturer’s instructions.
Comparison of Mycelial Growth, Conidiation, and Morphology
After incubating the wild-type (WT) strain, ΔAovosA and ΔAovelB mutants on PDA plates at 28°C for 6 days, 7 mm diameter hyphal discs were punched from the edges of plate colonies to compare their growth characteristics in different media, including PDA, TYGA, and TG. After 5–8 days of incubation, the diameter and hyphal morphology of each colony were measured and observed. The WT and mutant strains that were centrally attached to the CMY plates were also cultivated at 28°C for 15 days. The number of conidia and their morphologies were next determined and observed as previously described (Zhen et al., 2018). The mycelia from the WT and mutants were treated according to the methods described previously (Kim et al., 2004). After critical-point drying using a model CPD-030 device (Bal-Tec AG, Balzers, Liechtenstein), the samples were coated with gold using a model SCD 005 sputter coater (Bal-Tec) and observed by scanning electron microscopy (Quanta-200; FEI, Hillsboro, OR, United States).
Stress Tolerance Analysis
The responses of the WT and mutants to different chemical stressors, including osmotic, oxidative, or cell wall-perturbing stressors, were determined by incubating them on TG supplemented with different concentrations of NaCl (0.1, 0.2, and 0.3 M) and sorbitol (0.25, 0.50, and 0.75 M) for osmotic stress, hydrogen peroxide (H2O2; 5, 10, and 15 mM) and menadione (0.01, 0.03, and 0.05 mM) for oxidative stress, or sodium dodecyl sulfate (SDS; 0.01, 0.02, and 0.03%) and Congo red (0.05, 0.07, and 0.09 mg/mL) for cell wall perturbation (Yang et al., 2018; Zhen et al., 2018) at 28°C for 6 days. The diameter and hyphal morphology of each colony were measured and observed.
Heat Shock Pressure Tolerance Test
To determine the recuperation of the WT strain and ΔAovosA mutants following heat shock, their growth rates were compared after incubation on PDA plates for 2 days at 28°C, and 8 h at 28, 34, 38, or 40°C followed by incubation at 28°C for 6 days, after which the colony diameter was determined (Zhen et al., 2019). For spore germination analysis under heat shock stress, 150 μL aliquots of conidial suspension (100 conidia) collected from the WT strain and ΔAovosA mutants was incubated in liquid MM medium at 28, 34, or 38°C for 4 and 8 h to assay conidial germination rates (Xie et al., 2012). Additionally, conidial suspension (100 conidia) collected from the WT strain and ΔAovosA mutants was incubated for 2 h at 28, 34, 38, or 42°C, then the spores were inoculated on TYGA plates and incubated at 28°C for 48 h. The surviving colonies were counted (Ni and Yu, 2007).
Extracellular Protease Activity Analysis
WT and mutant strains were incubated on LMZ broth at 28°C for 7 days and the proteolytic activity of the fermentation liquids was assessed on casein plates as previously described (Zhao et al., 2004). Quantitative analysis of protease activity was determined by a caseinolytic method described by Wang et al. (2006). One unit (U) of protease activity was defined as the amount of enzyme that hydrolyzed the substrate and produced 1 μg of tyrosine in 1 min under the assay conditions.
Trap Formation and Pathogenicity of A. oligospora Against Nematodes
WT and mutant strains were incubated on WA (water agar) plates at 28°C for 3–4 days. Approximately 300 nematodes (C. elegans) were added to the middle of each plate to induce trap formation in the fungi. The number of traps and captured nematodes were counted using a light microscope at specific time intervals (Yang et al., 2018).
Quantitative Reverse-Transcription PCR (RT-PCR) Analysis
The WT strain and the ΔAovelB and ΔAovosA mutants were incubated in PD broth at 28°C for 3, 5, and 7 days. Hyphae were collected and stored at −80°C for subsequent RNA extraction. Total RNA from each sample was extracted using an RNA Extraction Kit (AP-MN-MS-RNA, Axygen, Jiangsu, China). The RNAs from the different samples were reverse transcribed into complementary DNAs (cDNAs) using the FastQuant RT Kit (TaKaRa Bio). The cDNAs from all the samples were used as templates to determine the transcriptional levels of the candidate genes (e.g., sporulation and protease-related genes) via RT-PCR with specific paired primers (Supplementary Table S2), and the β-tubulin gene from A. oligospora was used as an internal standard. Additionally, the WT strain was incubated on TYGA at 28°C for 2, 3, 5, or 7 days, and hypha was collected for determining the transcription of genes AovosA and AovelB at different developmental conditions. Similarly, the WT strain was co-cultured with nematodes for 0, 12, 24, or 36 h, and hyphae were collected for determining the transcriptional levels of genes AovosA and AovelB during the trap formation. The relative transcriptional levels (RTLs) of the candidate genes were calculated as the ratio of the transcript in the mutants over that in the WT strain at a given time using the 2–Δ ΔCt method (Livak and Schmittgen, 2001).
Statistical Analyses
One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test was used to differentiate the observations, measurements and estimates. P < 0.05 were considered to be significantly different. GraphPad Prism version 5.00 (GraphPad Software, San Diego, CA, United States) was used for the photographs and the statistical analyses. Every experiment was repeated three times.
Results
Sequence and Phylogenetic Analyses of AoVosA and AoVelB in A. oligospora
An online search through the A. oligospora genome in GenBank using homologous sequences of A. nidulans as the queries identified AovosA (1494 bp) and AovelB (1008 bp). The AovosA gene encodes a polypeptide of 497 amino acids (aa) with a predicted isoelectric point (pI) and molecular weight (MW) of 9.32 and 54.9 kDa, respectively, whereas the AovelB gene encodes a polypeptide of 335 aa with a predicted pI and MW of 7.72 and 36.3 kDa, respectively. The deduced AoVosA and AoVelB proteins contain no signal peptides, and both share two conserved domains in their C-termini: the velvet factor (IPR021740) and the velvet domain (IPR037525).
We also downloaded the AoVosA and AoVelB protein homologs from different fungi in GenBank, and compared their amino acid sequence similarity levels using DNAman software (version 6, Lynnon Biosoft, San Ramon, CA, United States). AoVelB shares 94% identity with VelB from Dactylellina haptotyla, another NTF, and also shares a high degree of sequence similarity (43.8–61.1%) with orthologous VelB from other filamentous fungi (Supplementary Table S3). Similarly, AoVosA shares 89 and 85.6% identity with homologous VosA from the NTF D. haptotyla and Drechslerella stenobrocha, respectively, while also sharing mid-range sequence similarity (24–39.1%) with orthologous VosA from other filamentous fungi (Supplementary Table S3).
We constructed a single phylogenetic tree for VosA and VelB from different fungi based on their amino acid sequences, and the orthologous proteins from VosA and VelB in the various fungi clustered as two groups (A and B). The VelB proteins from the different fungi separated into two sub-groups in this tree (A–I and A–II). Specifically, the VelB proteins from two NTF species, A. oligospora and D. haptotyla, and two species within the Aspergillus genus, A. nidulans, and A. fumigatus, clustered in A–II, whereas the VelB orthologs from other filamentous fungi, including the model fungus N. crassa, the phytopathogenic fungus Magnaporthe oryzae, the entomopathogenic fungi Metarhizium anisopliae and Beauveria bassiana, and the nematode-parasitic fungi Purpureocillium lilacinum, Hirsutella minnesotensis, and Drechmeria coniospora clustered in A–I (Supplementary Figure S1). Similarly, the orthologous VosA proteins from the different fungi separated into two sub-groups (B–I and B–II), with three NTF species and two Aspergillus genus species clustering in B–I, and orthologs of VosA from other filamentous fungi clustering in B–II (Supplementary Figure S1).
Screening and Verifying the ΔAovosA and ΔAovelB Mutants
The pRS426-AoVosA-hph and pRS426-AoVelB-hph plasmids were transformed into the protoplasts of A. oligospora, and their transformants were selected on PDAS plates containing 200 μg/mL of hygromycin B (Tunlid et al., 1999). The genomic DNA from each transformant was isolated using the CTAB method and site-specific insertion was confirmed for each plasmid by PCR. Fragments of 2052 and 1804 bp were amplified from the ΔAovosA and ΔAovelB mutants (Supplementary Figures S2A-b, B-b), respectively, and then compared with the 1356 and 1336 bp fragments from the WT strain using AoVosA-Yf/Yr and AoVelB-Yf/Yr primers (Supplementary Table S1). Finally, three ΔAovosA mutants (1, 2, and 6) and three ΔAovelB mutants (7, 11, and 22) were obtained. These mutants were further confirmed as authentic by southern blot analysis, and single hybridizing bands were observed in the WT, ΔAovosA, and ΔAovelB mutants (Supplementary Figures S2A-c, B-c). Their sizes were consistent with our expectations.
Influence of the AovosA and AovelB Genes on Growth and Conidiation
Compared with the WT strain, the ΔAovelB mutants showed significant growth defects on PDA, TYGA, and TG media, while the ΔAovosA mutants showed no obvious differences in mycelial growth (Figures 1A,B). Colonies produced by the three ΔAovelB mutants were denser than those of the WT strain, and their colony diameters were distinctly smaller than those of the WT strain on the three media (Figure 1B). In addition, the aerial hyphae in the ΔAovelB colonies were also sparser than those of the WT strain (Figure 1A) and the vegetative hyphae in the ΔAovelB colonies were very slender and had many shorter branches than those of the WT strain (Figure 2A). The mycelia were also observed by scanning electron microscopy. While the WT strain had prostrate growth on the surface of the medium, the ΔAovelB mutants grew tightly on the medium and the partial cells became expanded (Figures 2B,C).
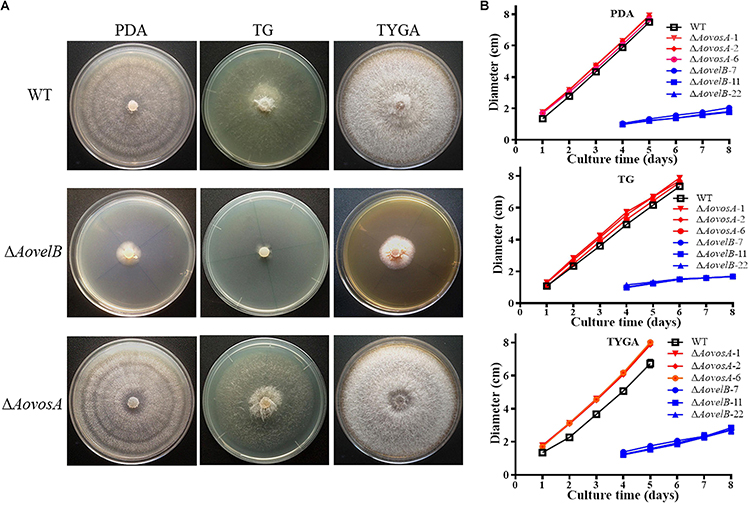
Figure 1. Comparison of colony morphology and mycelial growth between the wild type (WT) and mutant lines. (A) Colonies from the WT and mutant lines were incubated on TYGA media for 5 days at 28°C. (B) Mycelial growth rates of the WT and mutants on PDA, TYGA, and TG plates.
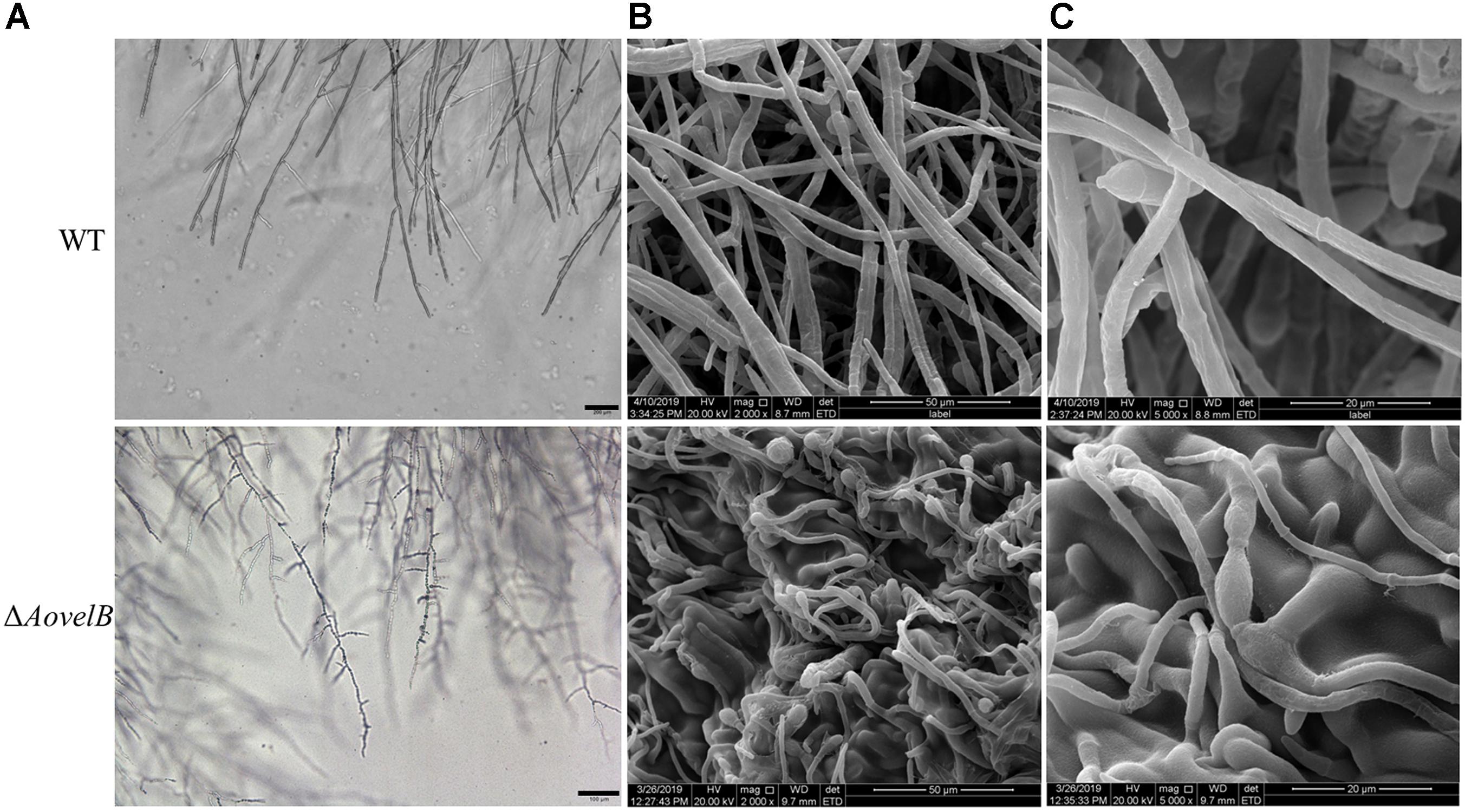
Figure 2. Comparison of mycelial morphology between the wild type (WT) and mutant lines. WT and mutants were cultured for 7 days on CMY plates at 28°C. Mycelial samples were collected and treated as previously described (Kim et al., 2004). (A) Mycelial morphologies of the WT and ΔAovelB mutants as observed by light microscopy. Bar: 100 μm. (B,C) The WT strain and ΔAovelB mutant were coated with gold and observed by scanning electron microscopy.
The conidial yields were determined from the WT and mutants after they were cultured in CMY at 28°C for 15 days. Conidial production in the ΔAovosA mutants (1.05–1.25 × 106 conidia per cm2) were not significantly different compared with the WT strain (1.13 × 106 conidia per cm2). The ΔAovelB mutants lost the ability to produce conidia, although they did produce conidiophores (Figures 3A,B). The transcription of the AovosA and AovelB genes was analyzed in the WT strain during the different developmental conditions, including vegetative growth (day 2) and at different time points of conidition, which included the early stage (day 3), middle stage (day 5), and later stage (day 7). The transcriptional levels of AovosA and AovelB were lower during conidiation than that of vegetative growth. In particular, AovelB was significantly down-regulated during the different stages of conidiation (Figure 3C).
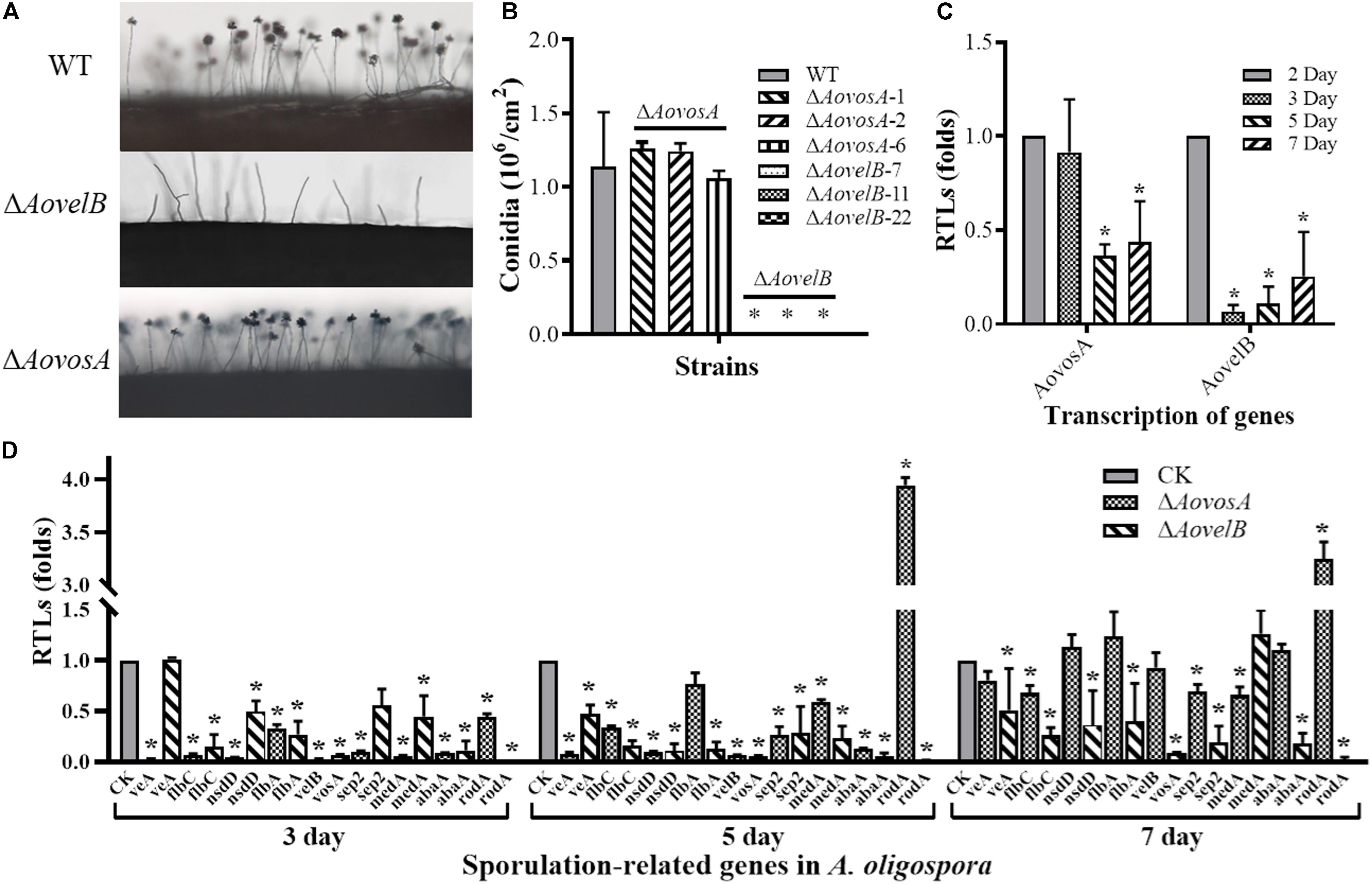
Figure 3. Comparison of conidiation and the transcriptional levels of sporulation-related genes between the wild type (WT) and mutant lines. (A) Conidiophore differentiation in the WT and mutant lines. (B) Sporulation in the WT and mutant lines on CMY medium. (C) The relative transcriptional levels (RTLs) of genes AovosA and AovelB during the vegetative growth and conidiation in A. oligospora. (D) RTLs of sporulation-related genes in the mutants compared with the WT strain at different time points. CK. The level of each gene was computed as a ratio of the transcript level of the gene in the deletion mutant to that in the WT strain under a given condition. An asterisk indicates a significant difference between the mutants and the WT strain (p < 0.05).
The transcriptional levels of 10 sporulation-related genes (veA, flbC, nsdD, flbA, velB, vosA, sep2, medA, abaA, and rodA) retrieved from GenBank based on homologous genes in the model fungus A. nidulans (Park and Yu, 2012; Krijgsheld et al., 2013), were determined in the WT strain and the ΔAovelB and ΔAovosA mutants by RT-PCR after culture on PD broth for 3, 5, and 7 days (Figure 3D). The transcriptional levels of all nine genes changed in the ΔAovelB mutants compared with those of the WT strain. Of these genes, seven (flbC, nsdD, flbA, vosA, sep2, abaA, and rodA) were significantly down-regulated, suggesting a strong interaction between AovelB and sporulation-related genes at the transcriptional level. Moreover, the gene transcript of veA was down-regulated on days 5 and 7, with no obvious change on day 3. The gene transcript of medA was up-regulated on day 7, but was decreased on days 3 and 5 (Figure 3D). Similar to the ΔAovelB mutants, the transcriptional levels of the sporulation-related genes, except rodA, were down-regulated on days 3 and 5 in the ΔAovosA mutant. The transcription of most of the genes was restored to the levels of the WT strain on day 7. The transcription of gene rodA was up-regulated 3.94-, and 3.24-fold in the ΔAovosA mutants compared with the WT strain on days 5 and 7, respectively (Figure 3D).
Contributions of AoVosA to Heat Shock Stress
The recuperation of the WT strain and ΔAovosA mutant was determined, the growth rate of the WT strain and ΔAovosA mutants showed no obvious different at 28, 34, or 38°C, while the mycelial growth rate of the ΔAovosA mutants was significantly lower than that of the WT strain at 40°C (Figures 4A,B). Further, the spore germination rates of the WT strain and ΔAovosA mutants were determined under heat shock stress. The conidia of the WT strain and ΔAovosA mutants germinated at 28 or 34°C, but not at 38°C. Conidia from the WT strain germinated significantly faster than those from the cultures of the ΔAovosA mutants at 28°C, with approximately 55.7 and 95.7% of the conidia from the WT strain germinating at 4 and 8 h, respectively, while only 15.6 and 55.2% of the conidia from the ΔAovosA mutants germinated at the same time points (Figure 4C). Similarly, the spore germination rate of the WT strain was significantly higher than that of the ΔAovosA mutants at 34°C, with approximately 34 and 56% of the conidia from the WT strain germinating at 4 and 8 h, respectively, while only 4.77 and 21.5% of the conidia from the ΔAovosA mutants germinated (Figure 4C). The surviving spores were determined under different heat shock stresses. The spore survival rate of the WT strain and the ΔAovosA mutants showed no obvious differences at 28, 34, or 38°C, while the rates significantly differed at 42°C, with 22.3% survival rate of conidia from the WT strain and essentially none of the spores surviving in the ΔAovosA mutants at 42°C (Figure 4D).
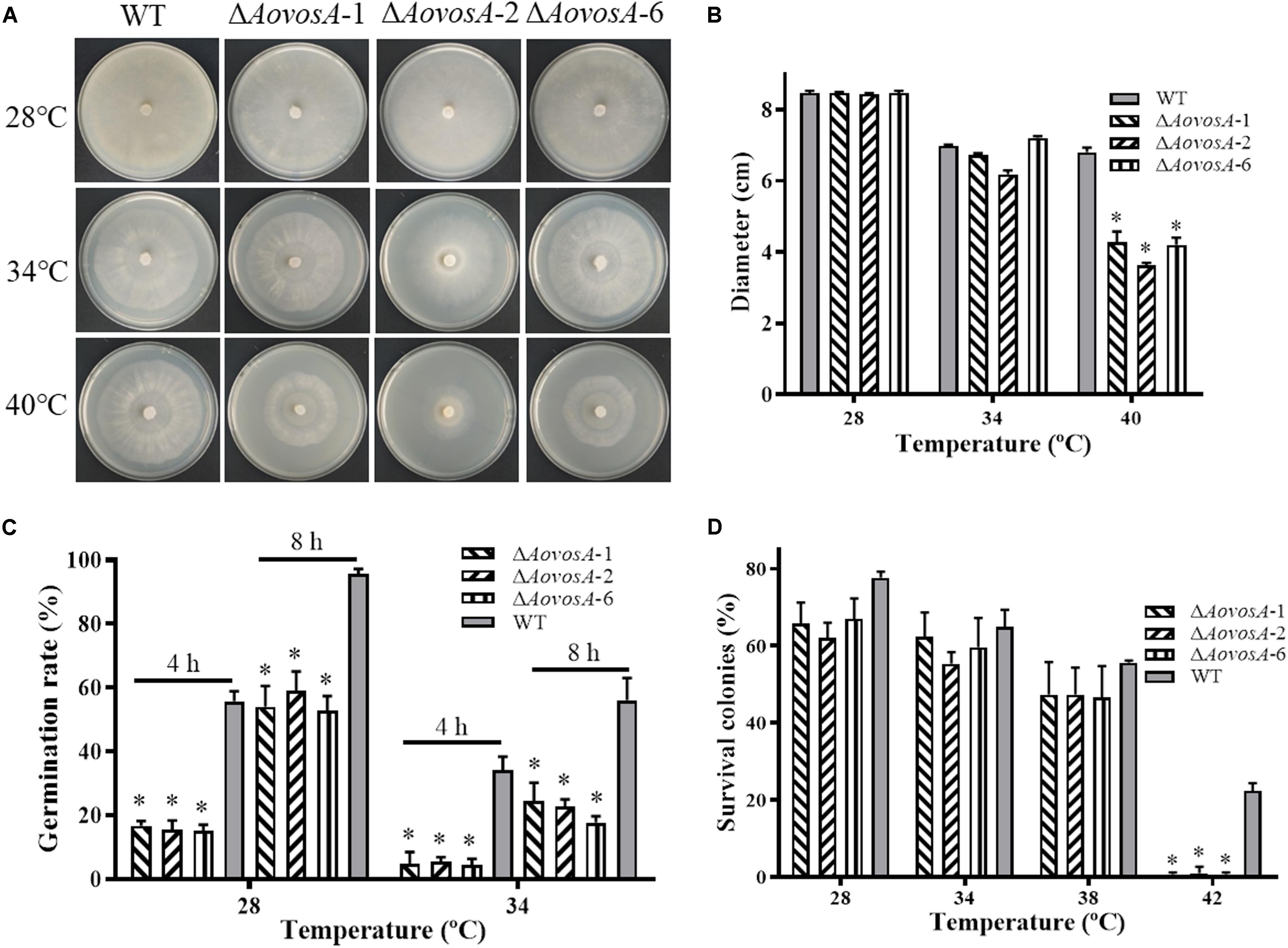
Figure 4. Effect of AoVosA on heat shock stress in A. oligospora. (A) Colonies from the WT strain and ΔAovosA mutants were incubated on PDA for 5 days at 28, 34, and 40°C. (B) Mycelial growth rates of the WT and ΔAovosA mutants on PDA at the three temperatures. (C) Conidia germination rates for the WT strain and ΔAovosA mutants at 28 and 34°C. (D) Spore survival rate of the WT strain and ΔAovosA mutants treated by heat shock at 28, 34, 38, and 42°C. An asterisk indicates a significant difference between the mutants and the WT strain (p < 0.05).
Contributions of AoVosA and AoVelB to Chemical Stress Responses
The WT strain and the mutants were compared for their responses to six types of chemical agents, including oxidants, cell wall perturbing agents, and osmotic agents. The growth of the WT strain and ΔAovosA mutants was significantly inhibited by NaCl and sorbitol. Their colony sizes decreased significantly in response to increasing levels of osmotic agents in the TG plates, unlike the colony sizes of the ΔAovelB mutants where no obvious changes were observed on TG medium supplemented with different concentrations of osmotic agents (Supplementary Figure S3). Similarly, the growth of the WT strain and the ΔAovosA mutants was inhibited by SDS. The ΔAovelB mutants hardly grew on TG plates supplemented with more than 0.02% SDS (Figures 5A,B). In contrast, growth in the WT strain and ΔAovosA mutants was inhibited by Congo red, unlike the ΔAovelB mutants where no obvious changes occurred when they were grown on TG plates supplemented with 0.05, 0.07, and 0.09 mg/mL Congo red (Figures 5A,B). We next selected six putative genes known to be involved in cell wall synthesis and trehalose synthase (trs) (Yang et al., 2018) and measured their transcriptional levels in the WT strain and the ΔAovosA, and ΔAovelB mutants by RT-PCR. They included the genes encoding chitin synthase (chs), glucosamine-fructose-6-phosphate aminotransferase (gfpa), β-glucosidase (glu), 1,3-β-glucan synthase (gls), and chitin synthase G (chsG). With the exception of glu, which was up-regulated at days 3 and 5, the transcriptional levels of the other genes were down-regulated significantly in the ΔAovelB mutants at the tested time points when compared with the WT strain (Figure 5C). Additionally, the transcriptional patterns of these genes in the ΔAovosA mutants were similar to the ΔAovelB mutants, with most genes down-regulated significantly at the tested time points when compared with the WT strain. However, in the ΔAovelB mutant, transcription of glu was down-regulated at days 3 and 5, and up-regulated at day 7 (Figure 5C).
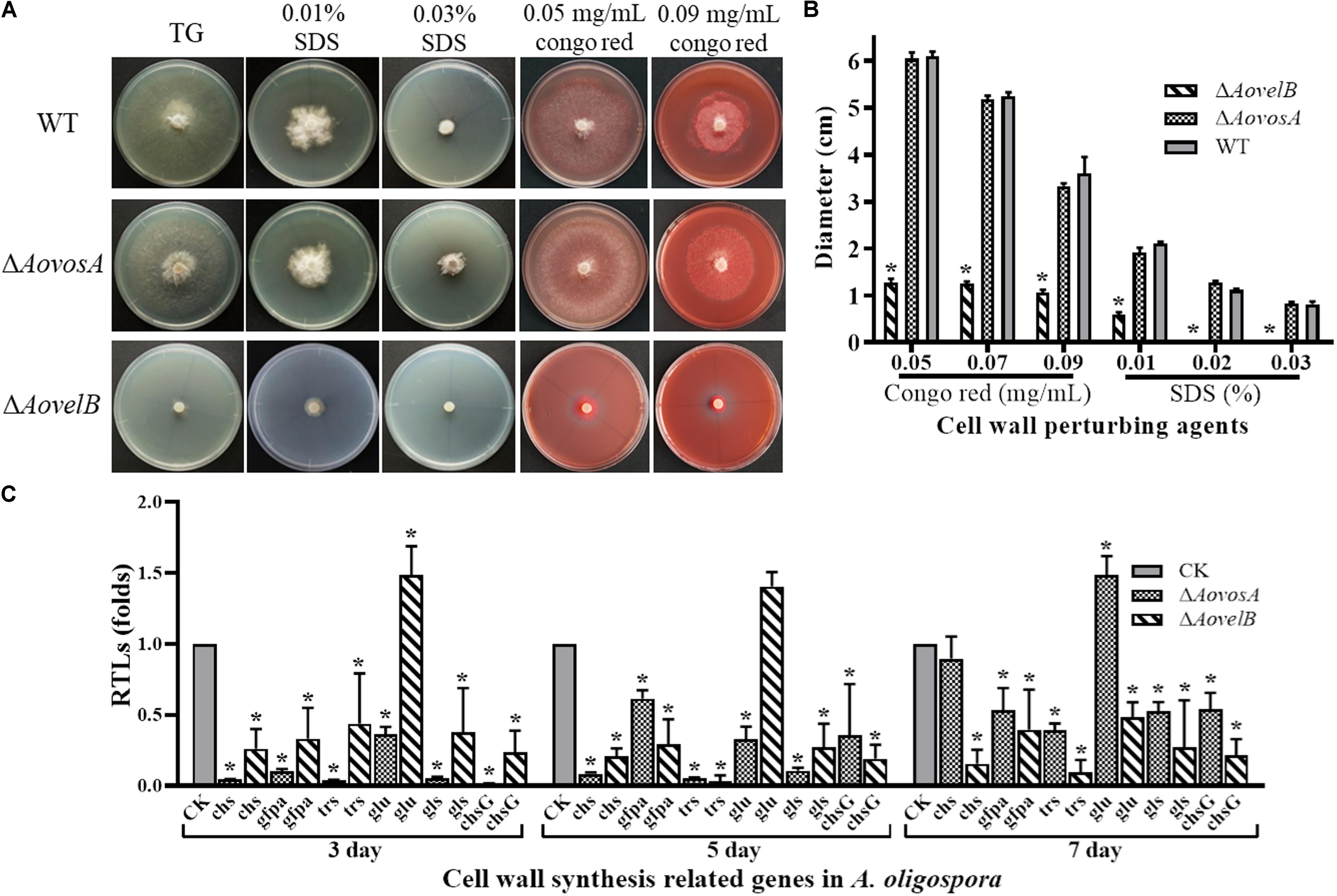
Figure 5. Comparison of stress tolerance to cell wall perturbing agents. (A) Colony morphology of the WT and mutants incubated on TG medium supplemented with sodium dodecyl sulfate (SDS) or Congo red. (B) The colony diameters of the WT and mutants after incubated on TG medium supplemented with 0.01–0.03% SDS or 0.05–0.09 mg/mL Congo red for 5 days. (C) Relative transcriptional levels (RTLs) of cell wall synthesis-related genes in the mutants compared with the WT strain at different time points. CK. The level of each gene was computed as a ratio of the transcript level of the gene in the deletion mutant to that in the WT strain under a given condition. An asterisk indicates a significant difference between the mutants and the WT strain (p < 0.05).
Growth of the WT strain and the mutants was sensitive to H2O2. The ΔAovelB mutants barely grew on TG agar supplemented with more than 10 mM H2O2 compared with the WT strain and the ΔAovosA mutants (Figures 6A,B). However, the growth of the WT strain and the mutants were not inhibited on TG agar supplemented with 0.01–0.03 mM menadione. Next, we selected six genes encoding the putative proteins involved in antioxidant enzymes for analysis by RT-PCR in the WT strain and the ΔAovosA and ΔAovelB mutants; these included glutathione reductase (glr), glutathione S-transferase (glt), thioredoxin reductase (thr), peroxidase (per), thioredoxin reductase (gliT), and catalase (cat1). Compared with the WT strain, the transcription of glr, glt, and thr was down-regulated in the ΔAovelB mutants. In contrast, transcription of gliT was up-regulated, cat1 was not changed significantly, and the transcription of per decreased gradually from days 3 to 7 in the ΔAovelB mutants (Figure 6C). Unlike the ΔAovelB mutants, the per and gliT transcripts were down-regulated in the ΔAovosA mutants, and the glt transcript was significantly up-regulated at all tested time points (Figure 6C).
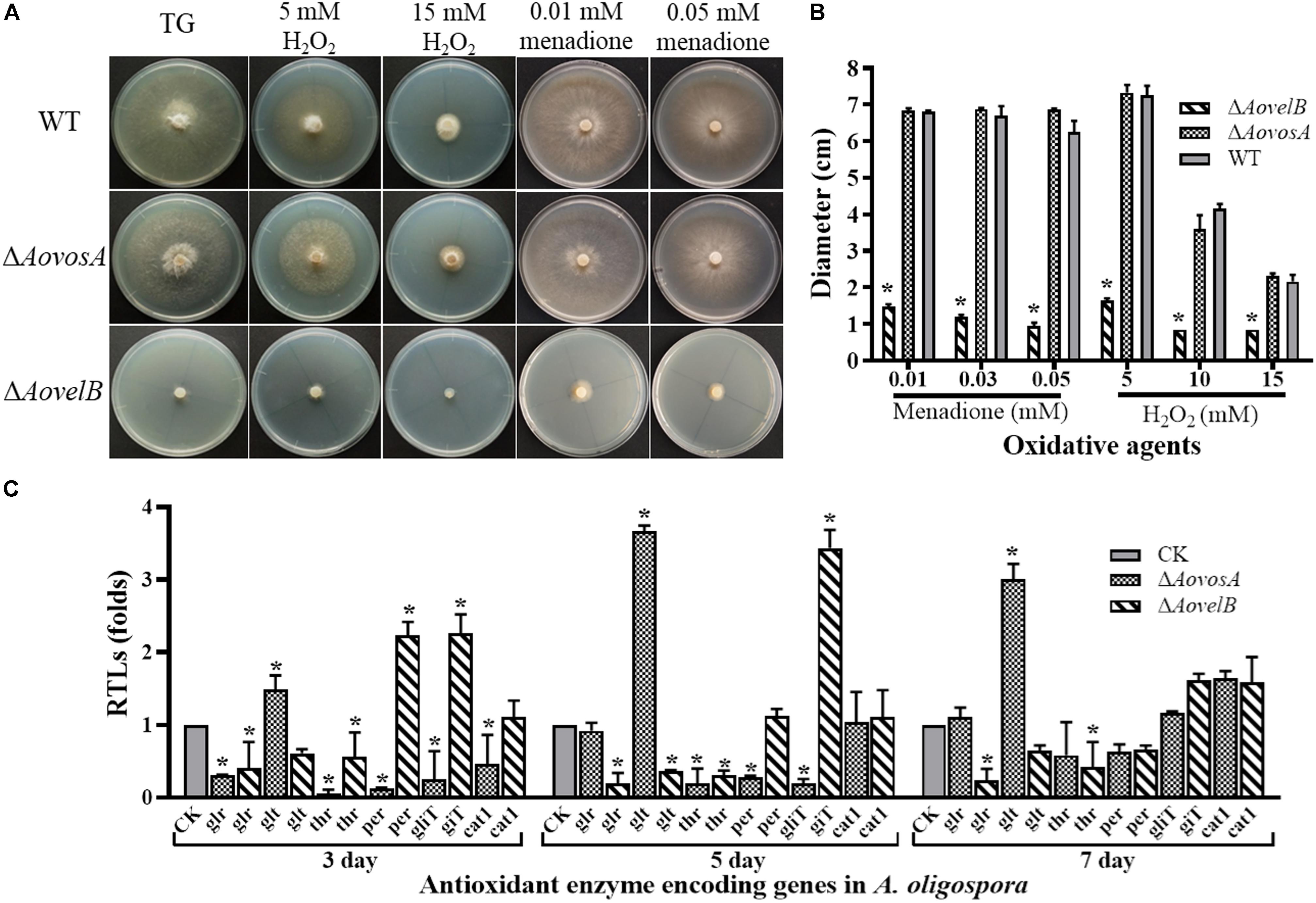
Figure 6. Comparison of stress tolerance to oxidative agents. (A) Colony morphologies of the WT and mutant lines incubated on TG medium supplemented with H2O2 or menadione. (B). Colony diameters of the WT and mutants after incubation on TG medium supplemented with 5–15 mM H2O2 or 0.01–0.05 mM menadione for 5 days. (C) Relative transcriptional levels (RTLs) of oxidation-related genes in the mutants compared with the WT strain at different time points. CK. The level of each gene was computed as a ratio of the transcript level of the gene in the deletion mutant to that in the WT strain under a given condition. An asterisk indicates a significant difference between the mutants and the WT strain (p < 0.05).
AoVelB Plays a Role in Serine Protease Production
Extracellular serine proteases are important virulence factors in NTF. A. oligospora, for example, can produce serine proteases capable of immobilizing nematodes and degrading the proteinaceous components of the nematode cuticle (Tunlid et al., 1994; Yang et al., 2013). Presently, the fermentation broth from the WT strain or from either mutant displayed different proteolytic activities. Compared with the WT strain, disruption of the AovosA gene had little influence on the proteolytic activity of the mutant. In contrast the ΔAovelB mutants all displayed reduced proteolytic activities (50%) (Figures 7A,B). Additionally, the proteolytic activities of the WT strain and mutants were inhibited by 90% in the presence of the serine protease inhibitor phenylmethyl sulfonyl fluoride (Figure 7C). The transcriptional levels of five of the serine proteases genes in A. oligospora were determined by RT-PCR. Of these, 215g702, 78g136 176g95, and 54g992 were down-regulated in the ΔAovelB mutants. In particular, 215g702 and 54g992 were significantly down-regulated at different time points, while the transcriptional level of 188g273 was slightly up-regulated on day 3 and down-regulated on days 5 and 7 (Figure 7C). Unlike the ΔAovelB mutants, the transcriptional levels of all five serine proteases genes were down-regulated on day 3, while the transcription of 215g702 and 176g95 was up-regulated on day 5. An additional gene, 188g273, was also up-regulated on day 7. The transcription of 54g992 was significant at the three tested times, similar to the ΔAovelB mutants (Figure 7C).
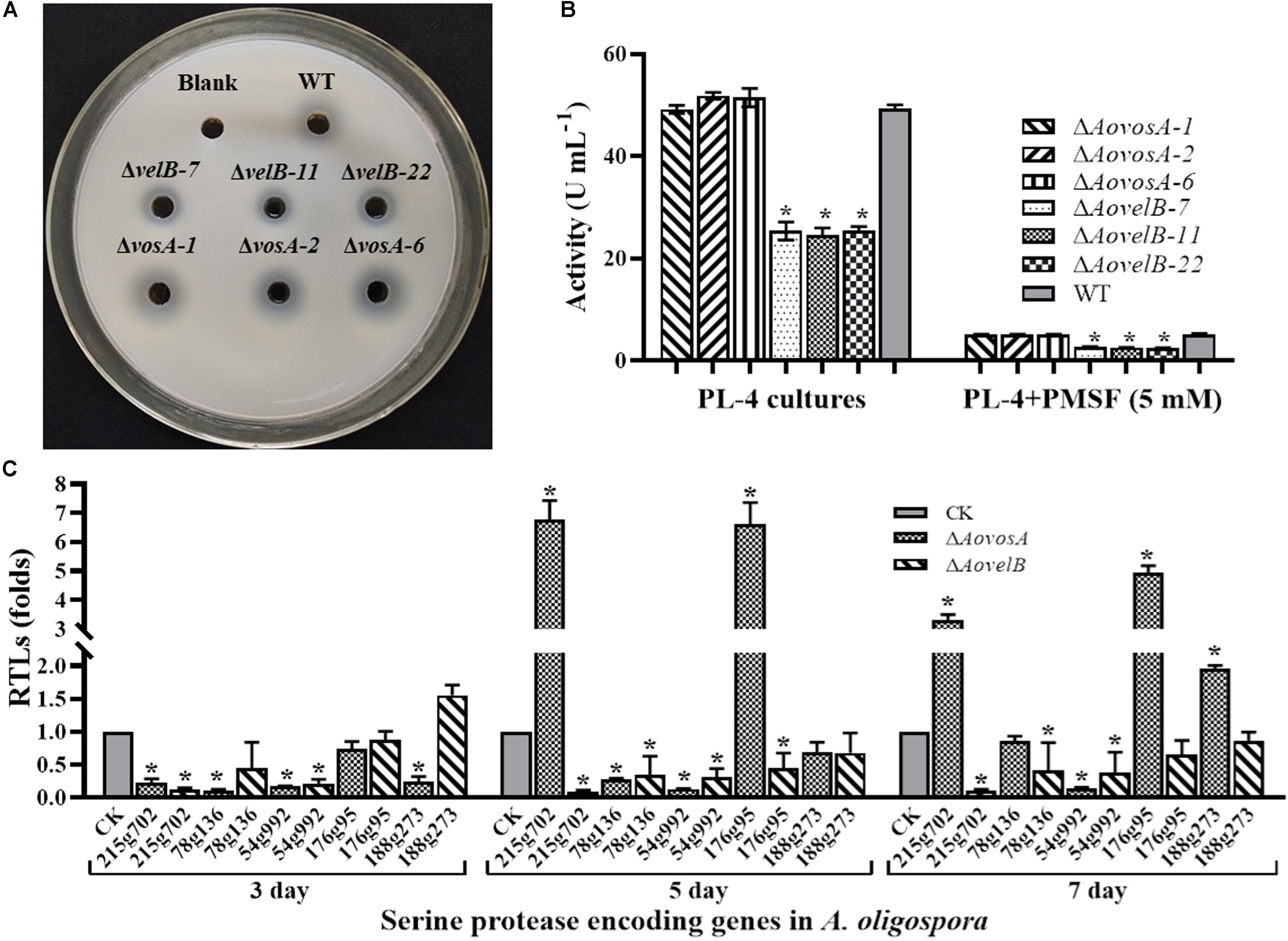
Figure 7. Comparison of extracellular proteolytic activity. (A) Comparison of the extracellular proteolytic activities of the WT and mutant lines on casein plates. (B) Total extracellular protease activity quantified from 7 days old PL-4 cultures. (C) Relative transcriptional levels (RTLs) of serine protease genes in the ΔAovelB andΔAovosA mutants compared with the WT strain at different time points. CK. The level of each gene was computed as a ratio of the transcript level of the gene in the deletion mutant to that in the WT strain under a given condition. An asterisk indicates a significant difference between the mutants and the WT strain (p < 0.05).
Contributions of AoVosA and AoVelB to Trap Formation in A. oligospora
Trap formation was induced by adding nematodes to the WA plates. Many traps were observed on the plates with the WT strain and ΔAovosA, but no traps were observed on the plates containing the ΔAovelB mutants after adding the nematodes for 12, 24, and 36 h (Figures 8A,B). At 12 h, the WT strain and the ΔAovosA mutants began to produce immature traps, which only contained one or two circles. Mature traps and three-dimensional nets were formed at 24 and 36 h. Concurrently, approximately 18.3 traps per cm2 were observed with the WT strain at 24 h, while 12.3, 15.5, and 13.6 traps per cm2 were formed by the three ΔAovosA mutants (#1, 2, and 6, respectively) (Figure 8B). The transcription of AovosA and AovelB was determined during the trap formation. Their expression patterns were similarly down-regulated at the tested time points (Figure 8C). The nematicidal activities of the WT strain and its mutants were also calculated at different time points. Thirty two percent and seventy seven percent of the nematodes were captured by the WT strain at 12 and 24 h, respectively, while 20–27% and 71–72% of the nematodes were captured by the ΔAovosA mutants at the same times. By 36 h, almost all of the nematodes were captured, the majority of which were digested by the WT strain and by the ΔAovosA mutants. In contrast, the nematodes remained active and were not captured after they were added to the plates containing the ΔAovelB mutants for 36 h (Figures 8A,D).
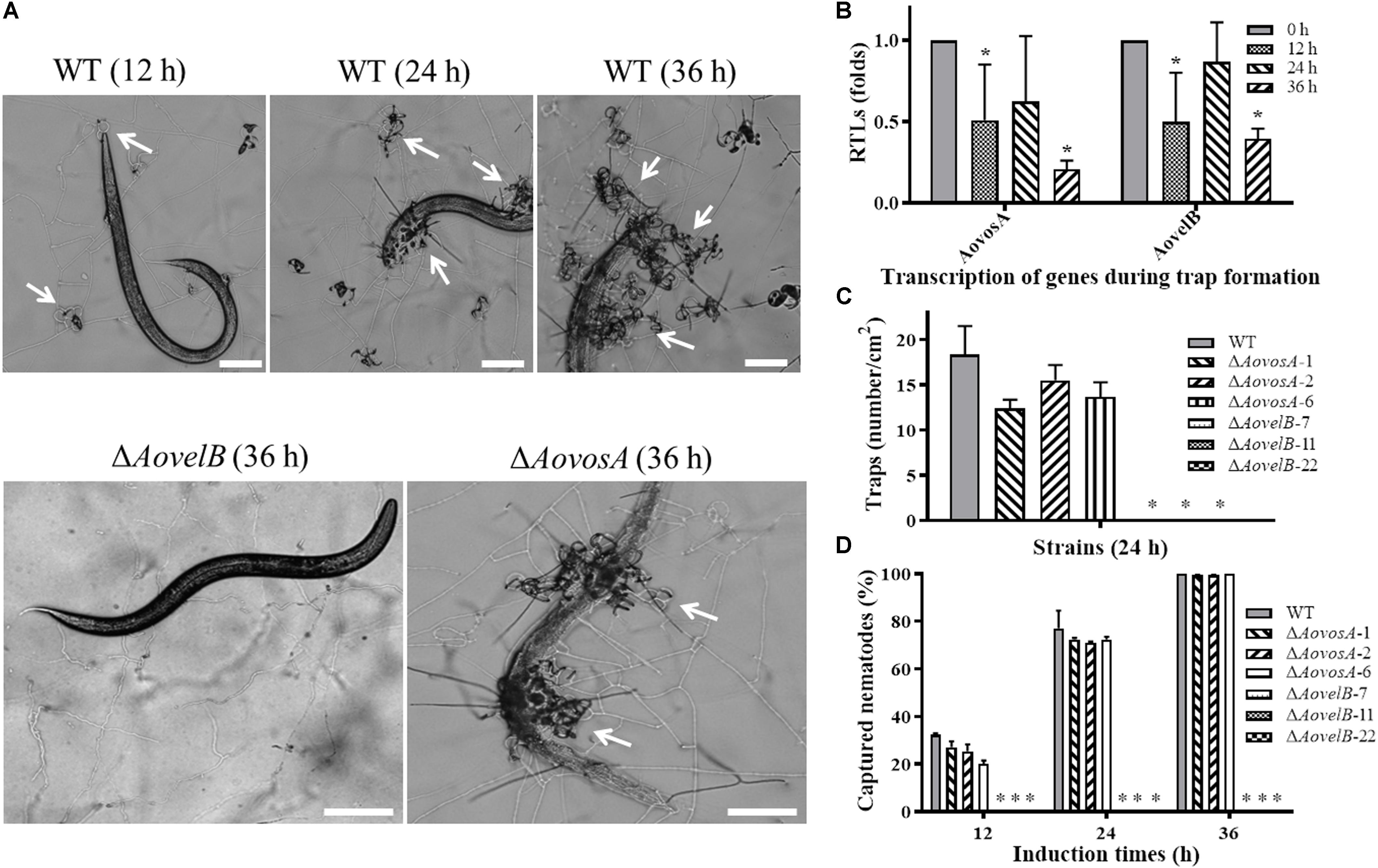
Figure 8. Comparison of trap formation and nematicidal activity in the WT and mutant lines. (A) Trap formation in the WT and mutant lines induced by nematodes at different time points. The arrows show the traps produced by the WT strain and ΔAovosA mutants. Bar: 100 μm. (B) The relative transcriptional levels (RTLs) of genes AovosA and AovelB in A. oligospora during different induction times by nematodes. (C) Nematode-induced trap numbers produced by the WT and mutant lines at 24 h. (D) The percentage values for the nematodes captured by the WT and mutant lines at different time points. An asterisk indicates a significant difference between the mutants and the WT strain (p < 0.05).
Discussion
Previous studies have shown that velvet proteins are fungal-specific, multifunctional regulators that control development and secondary metabolism in various filamentous and dimorphic fungi (Bayram et al., 2008; Bayram and Braus, 2012). In the present study, the homologous velvet proteins AoVelB and AoVosA were characterized in A. oligospora, a typical NTF species, by gene disruption and by phenotypic and RT-PCR analysis. AoVelB significantly impacted mycelial growth, conidiation, trap formation, and pathogenicity in A. oligospora, while AoVosA played limited roles in these biological processes. In addition, we found that the functions of AoVelB and AoVosA in A. oligospora also showed several similarities and differences when compared with their homologous proteins in other filamentous fungi.
VelB is required for mycelial growth, but the drastic severity of hyphal morphology defects attributed to the velB deletion have not been reported in other filamentous fungi. For example, the slow growth rate and reduced aerial hyphae phenotypes are consistent with observations from the plant pathogen Fusarium graminearum (Jiang et al., 2012; Lee et al., 2012). While disruption of the AovelB gene resulted in slow growth, reduced aerial hyphae and more branches, concurrently the mycelia grew closely and tightly to the medium, and the partial cells became swollen (Figure 2). In contrast, growth and mycelial morphology in the ΔAovosA mutants did not differ from those of the WT strain. These results suggest that AoVelB plays an important role in the vegetative growth of A. oligospora.
Apart from defective mycelial growth, AoVelB is evidently essential for sporulation in A. oligospora because its mutants totally lost their conidiation ability (Figures 3A,B). The velB deletion strains reportedly display decreased conidia production in A. nidulans (Park et al., 2012b) and in Aspergillus flavus (Chang et al., 2013), but increased conidial yields coupled with immature or low viability conidia in A. fumigatus (Park et al., 2012a) and F. graminearum (Jiang et al., 2012). However, the conidiation capacity of the ΔAovosA mutants did not differ from that of the WT strain (Figures 3A,B). Previous studies have indicated that VosA functions as a negative regulator of conidiation in A. nidulans (Ni and Yu, 2007; Sarikaya Bayram et al., 2010) and in A. fumigatus (Park et al., 2012a), but is a positive regulator in F. fujikuroi (Wiemann et al., 2010). However, this is not the case for AoVosA in A. oligospora. Furthermore, vosA deletion brought about ascended conidial germination in A. nidulans (Park et al., 2012b) and A. fumigatus (Park et al., 2012a), but decreased conidial germination in B. bassiana (Li et al., 2015). While the conidial morphology of the ΔAovosA mutants was unaltered, their conidial germination decreased by 40 and 40.2% at 4 and 8 h at 28°C, respectively, suggesting that AoVosA plays a particular role in conidial germination in A. oligospora.
The severe conidiation defects we observed might contribute to the interactions that occur between AoVelB and other conidiation regulators. For example, deleting the AovelB gene led to the transcriptional depression of almost all of the conidiation regulators. From these regulators, seven genes (flbC, flbA, nsdD, abaA, vosA, and rodA) were significantly down-regulated (Figure 3D). These genes are crucial for A. nidulans conidiation (Park and Yu, 2012). In particular, AbaA is considered to be a central developmental regulator that can activate the expression of vosA and velB in Aspergillus phialides (Park and Yu, 2012). Interestingly, despite having little influence on conidiation, the transcription levels of these sporulation-related genes, except for rodA, were also significantly down-regulated on days 3 and 5 in the ΔAovosA mutants. The transcription of most genes was restored to the level of the WT strain on day 7. The transcription of rodA was significantly different between the ΔAovelB and ΔAovosA mutants on days 5 and 7, being down-regulated in the ΔAovelB mutants and up-regulated in the ΔAovosA mutants (Figure 3D). The rodA gene encodes a hydrophobin and is essential for formation of the rodlet layer and hydrophobicity in conidia. Conidia of the rodA null mutants are hydrophilic due to the absence of the outermost rodlet layer (Thau et al., 1994).
Apart from the abolished conidiation capacity, the sensitivity of the ΔAovelB mutants to oxidative and cell wall perturbation stresses increased under H2O2 and SDS stresses, suggesting that AoVelB is also required to resist such stresses. Previous research findings in A. fumigatus (Park et al., 2012a) are largely consistent with our observations in A. oligospora but are contrary to those in F. graminearum, whose resistance to osmotic stress and cell wall-damaging agents is greater (Jiang et al., 2012). In contrast, the ΔAovosA mutants showed no change in their resistance to chemical agents compared with the WT strain (Figures 5, 6 and Supplementary Figure S3). These phenotypic changes in the ΔAovosA mutants do not coincide with those reported in A. nidulans, whose ΔvosA mutants were highly sensitive to the H2O2 oxidant (Ni and Yu, 2007). Therefore, the AoVosA and AoVelB protein homologs from different fungi might play diverse functions, and their roles likely vary in fungal species.
The spore germination rate of the ΔAovosA mutants was significantly lower than that of the WT strain at 28 or 34°C, the proportion of germinated spores in the ΔAovosA mutants was 28% compared with the WT strain at 28°C (4 h), but the proportion was decreased to 14% at 34°C for 4 h (Figure 4C). Additionally, the recuperation of the ΔAovosA mutants was significantly lower than that of the WT strain at 40°C (Figures 4A,B). Moreover, the spore survival rate of the WT strain was significantly higher than that of the ΔAovosA mutants at 42°C (Figure 4D). These results suggest that the AoVosA protein plays a role in conidial germination and heat shock stress resistance in A. oligospora. Our results coincide with those reported in A. nidulans (Ni and Yu, 2007) and B. bassiana (Li et al., 2015). For example, the deletion of vosA in A. nidulans results in the lack of trehalose in spores; and rapid loss of the cytoplasm, organelles, and viability of spores; and a dramatic reduction in tolerance of conidia to heat (Ni and Yu, 2007).
Presently, A. oligospora began to produce spores on day 4 during incubation on TYGA at 28°C. Spores were abundant by day 6 and peaked in number on day 8. Given that gene expression occurred prior to the corresponding phenotype, the mycelia collected on day 2 were specified as the vegetative growth stage, while mycelia collected on days 3, 5, and 7 were defined as the early stage, middle stage, and later stage of conidiation, respectively. The transcription of AovosA and AovelB during the different developmental conditions was determined by RT-PCR. Both genes were highly expressed in the vegetative growth stage, but their transcription was down-regulated at the early stage of conidiation and increased at the later stage of conidiation (Figure 3C). These results are consistent with the prior finding in A. nidulans, in which the transcript levels of genes vosA and velB were high in conidia and detectable during vegetative growth and increased from 48 h post-developmental light-mediated induction (Eom et al., 2018).
Furthermore, the transcriptional levels of several genes involved in cell wall biosynthesis were down-regulated in the ΔAovelB and ΔAovosA mutants (Figure 5C). Of these, decreased levels of the trs transcript were observed in both the ΔAovelB and ΔAovosA mutants. The gene trs contributes to trehalose synthesis, which is critical for conidia to resist multiple stresses, such as SDS treatment (Ni and Yu, 2007; Zhang and Feng, 2018). Additionally, the transcription pattern of glu was opposite in the ΔAovelB and ΔAovosA mutants, suggesting that glu might be a key gene for cell wall biosynthesis. Analogously, the transcripts from three genes related to antioxidant processes were down-regulated in the ΔAovelB mutants. Previous studies have confirmed that sulfhydryl groups play essential roles in oxidative stress responses (Grant, 2001). Moreover, the transcripts of genes related to antioxidant, with the exception of glt, in the ΔAovosA mutants were down-regulated on days 3 and 5, and no obvious difference was observed between the WT strain and mutants on day 7. In contrast, the transcription of glt was significantly up-regulated at all tested time points (Figure 6C). Glutathione S-transferase encoded by glt is involved in the oxidative stress response in Schizosaccharomyces pombe (Kim et al., 2001).
Pathogenic fungi often secrete a series of virulence factors (e.g., mycotoxins and serine proteases) into the extracellular environment to facilitate infection and allow the pathogen to assimilate essential nutrients from the host (Yang et al., 2013). The extracellular proteolytic activities of the ΔAovelB mutants were reduced when compared with the WT strain, and the transcripts of three serine protease genes (215g702, 78g136, and 54g992) were clearly down-regulated, suggesting that AoVelB is involved in regulating serine protease production. The protein encoded by the 215g702 gene is a cuticle-degrading serine protease capable of degrading the proteinaceous components of nematodes (Yang et al., 2011). The transcripts of serine protease genes were also down-regulated in the ΔAovelB mutants on day 3, while partial genes were up-regulated on days 5 and 7. Especially, the transcripts of 215g702 and 176g95 were significantly up-regulated. Additionally, the proteolytic activities of the WT strain, ΔAovosA and ΔAovelB mutants were significantly inhibited by phenylmethyl sulfonyl fluoride (Figure 7B), suggesting that the main extracellular proteases are serine proteases (Tunlid et al., 1994; Yang et al., 2013).
Notably, deletion of the AovelB gene abolished the ability of A. oligospora to produce the traps required to capture nematodes, suggesting that AoVelB plays a vital role in the biological control potential of A. oligospora. While reduced numbers of traps were produced by the ΔAovosA mutants at 24 h, their nematicidal activities were lower than those of the WT strain at 12 and 24 h, but identical to the WT strain at 36 h (Figures 8C,D). Additionally, the transcripts of AovelB and AovosA showed a similar pattern, in which both were down-regulated at the early stage of trap formation (12 h), increased at the middle stage (24 h), but decreased at the later stage of trap formation (36 h) (Figure 8B). Previous studies revealed that VosA and VelB play important roles in either conidiation or secondary metabolism, which may also affect the pathogenicity of filamentous fungi, such as the entomopathogen B. bassiana whose conidia can infect insects (Li et al., 2015), while F. graminearum (Jiang et al., 2012) and F. fujikuroi (Wiemann et al., 2010) produce mycotoxins that can poison grains. A. oligospora is a typical NTF species, and trap formation by it is considered an important indicator of the A. oligospora lifestyle transition (Su et al., 2017). These observations suggest that trap formation defects result from the absence of AoVelB, which probably causes serious hyphal defects, making it impossible to produce the traps formed by mycelium development and differentiation.
Previous studies have confirmed that VosA and VelB can form a VosA–VelB heterodimer, which plays important roles in spore viability, conidial maturation, and germination (Ni and Yu, 2007; Ahmed et al., 2013; Park et al., 2017). In this study, the transcription of vosA was significantly down-regulated in the ΔAovelB mutants and, in contrast, the transcription of velB was significantly down-regulated in the ΔAovosA mutants. These findings suggest that VosA interacts with VelB at the transcriptional level. At present, no suitable markers are available to construct a double mutant, which hinders studies of the biological function of key genes in A. oligospora, such as AovosA and AovelB. Identification of a novel selectable marker is an important goal.
Our collective results demonstrate that AoVelB is a crucial regulator of multiple biological processes, including mycelial growth, conidiation, trap formation, and serine protease production in A. oligospora, whereas AoVosA plays a role in conidial germination and heat shock stress, with a minor role in trap formation. The data provide the first characterization of AoVelB and AoVosB in a typical NTF species, A. oligospora. Our results provide a basis for further exploration of the mechanism whereby AoVelB regulates conidiation, trap formation, and other phenotypic traits, and will inform investigations of the functions of velvet proteins in NTF to provide a fuller picture of their biological roles.
Conclusion
We identified and characterized the AoVosA and AoVelB velvet proteins from A. oligospora. AoVelB plays a role in conidiation and is important for trap formation, as well as in infection-related morphogenesis in this fungus. AoVosA plays a role in conidial germination and heat shock stress. Our findings enhance current understanding of conidiation, trap formation and the pathogenic mechanisms of NTF.
Data Availability
The datasets generated for this study can be found in GenBank, AOL_s00054g700 and AOL_s00054g811.
Author Contributions
JY and K-QZ conceived and designed the study. GZ, YZ, and JY wrote the manuscript and conducted the experiments. YM, LY, MX, and DZ analyzed the data. JY and XN revised the manuscript. All authors read and approved the final manuscript.
Funding
This research was jointly supported by the NSFC-Yunnan Joint Fund (U1402265 and U1502262), the National Basic Research Program of China (2013CB127503), the National Natural Science Foundation of China (31272093), and co-supported by the Yunnan University’s Research Innovation Fund for Graduate Students (YDY2018190).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01917/full#supplementary-material
Footnotes
References
Adams, T. H., Wieser, J. K., and Yu, J. H. (1998). Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62, 35–54. doi: 10.1128/AAC.45.11.3009-3013.2001
Ahmed, Y. L., Gerke, J., Park, H. S., Bayram, Ö., Neumann, P., Ni, M., et al. (2013). The velvet family of fungal regulators contains a DNA-binding domain structurally similar to NF-κB. PLoS Biol. 11:e1001750. doi: 10.1371/journal.pbio.1001750
Bayram, Ö., and Braus, G. H. (2012). Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol. Rev. 36, 1–24. doi: 10.1111/j.1574-6976.2011.00285.x
Bayram, Ö., Krappmann, S., Ni, M., Bok, J. W., Helmstaedt, K., Valerius, O., et al. (2008). Velb/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320, 1504–1506. doi: 10.1126/science.1155888
Beyhan, S., Gutierrez, M., Voorhies, M., and Sil, A. (2013). A temperature-responsive network links cell shape and virulence traits in a primary fungal pathogen. PLoS Biol. 11:e1001614. doi: 10.1371/journal.pbio.1001614
Chang, P. K., Scharfenstein, L. L., Li, P., and Ehrlich, K. C. (2013). Aspergillus flavus VelB acts distinctly from VeA in conidiation and may coordinate with FluG to modulate sclerotial production. Fungal Genet. Biol. 58, 71–79. doi: 10.1016/j.fgb.2013.08.009
Colot, H. V., Park, G., Turner, G. E., Ringelberg, C., Crew, C. M., Litvinkova, L., et al. (2006). A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U.S.A. 103, 10352–10357. doi: 10.1073/pnas.0601456103
Eom, T. J., Moon, H., Yu, J. H., and Park, H. S. (2018). Characterization of the velvet regulators in Aspergillus flavus. J. Microbiol. 56, 893–901. doi: 10.1007/s12275-018-8417-4
Etxebeste, O., Garzia, A., Espeso, E. A., and Ugalde, U. (2010). Aspergillus nidulans asexual development: making the most of cellular modules. Trends Microbiol. 18, 569–576. doi: 10.1016/j.tim.2010.09.007
Grant, C. M. (2001). Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol. Microbiol. 39, 533–541. doi: 10.1046/j.1365-2958.2001.02283.x
Jiang, J. H., Yun, Y. Z., Liu, Y., and Ma, Z. H. (2012). FgVELB is associated with vegetative differentiation, secondary metabolism and virulence in Fusarium graminearum. Fungal Genet. Biol. 49, 653–662. doi: 10.1016/j.fgb.2012.06.005
Kim, H. G., Park, K. N., Cho, Y. W., Park, E. H., Fuchs, J. A., and Lim, C. J. (2001). Characterization and regulation of glutathione S-transferase gene from Schizosaccharomyces pombe. Biochim. Biophys. Acta. 1520, 179–185. doi: 10.1016/s0167-4781(01)00265-2
Kim, M. J., Park, S. M., Kim, Y. H., Cha, B. J., Yang, M. S., and Kim, D. H. (2004). Deletion of a hypoviral-regulated cppk1 gene in a chestnut blight fungus, Cryphonectria parasitica, results in microcolonies. Fungal Genet. Biol. 41, 482–492. doi: 10.1016/j.fgb.2003.12.006
Krijgsheld, P., Bleichrodt, R. V., Van Veluw, G. J., Wang, F., Müller, W. H., Dijksterhuis, J., et al. (2013). Development in Aspergillus. Stud. Mycol. 74, 1–29. doi: 10.3114/sim0006
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, J. K., Myong, K., Kim, J. E., Kim, H. K., Yun, S. H., and Lee, Y. W. (2012). FgVelB globally regulates sexual reproduction, mycotoxin production and pathogenicity in the cereal pathogen Fusarium graminearum. Microbiology 158, 1723–1733. doi: 10.1099/mic.0.059188-0
Li, F., Shi, H. Q., Ying, S. H., and Feng, M. G. (2015). WetA and VosA are distinct regulators of conidiation capacity, conidial quality, and biological control potential of a fungal insect pathogen. Appl. Microbiol. Biotechnol. 99, 10069–10081. doi: 10.1007/s00253-015-6823-7
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mirabito, P. M., Adams, T. H., and Timberlake, W. E. (1989). Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell 57, 859–868. doi: 10.1016/0092-8674(89)90800-3
Ni, M., and Yu, J. H. (2007). A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans. PLoS One 2:e970. doi: 10.1371/journal.pone.0000970
Niu, X. M., and Zhang, K. Q. (2011). Arthrobotrys oligospora: a model organism for understanding the interaction between fungi and nematodes. Mycology 2, 59–78. doi: 10.1080/21501203.2011.562559
Park, H. S., Bayram, Ö., Braus, G. H., Kim, S. C., and Yu, J. H. (2012a). Characterization of the velvet regulators in Aspergillus fumigatus. Mol. Microbiol. 86, 937–953. doi: 10.1111/mmi.12032
Park, H. S., Ni, M., Jeong, K. C., Kim, Y. H., and Yu, J. H. (2012b). The role, interaction and regulation of the velvet regulator VelB in Aspergillus nidulans. PLoS One 7:e45935. doi: 10.1371/journal.pone.0045935
Park, H. S., Lee, M. K., Kim, S. C., and Yu, J. H. (2017). The role of VosA/VelB-activated developmental gene vadA in Aspergillus nidulans. PLoS One 12:e0177099. doi: 10.1371/journal.pone.0177099
Park, H. S., Nam, T. Y., Han, K. H., Kim, S. C., and Yu, J. H. (2014). VelC positively controls sexual development in Aspergillus nidulans. PLoS One 9:e89883. doi: 10.1371/journal.pone.0089883
Park, H. S., and Yu, J. H. (2012). Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 15, 669–677. doi: 10.1016/j.mib.2012.09.006
Park, H. S., and Yu, J. H. (2016). Developmental regulators in Aspergillus fumigatus. J. Microbiol. 54, 223–231. doi: 10.1007/s12275-016-5619-5
Sarikaya Bayram, Ö., Bayram, Ö., Valerius, O., Park, H. S., Irniger, S., Gerke, J., et al. (2010). LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet. 6:e1001226. doi: 10.1371/journal.pgen.1001226
Staben, C., Jensen, B., Singer, M., Pollock, J., Schechtman, M., Kinsey, J., et al. (1989). Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet. Rep. 36, 79–81. doi: 10.4148/1941-4765.1519
Su, H., Zhao, Y., Zhou, J., Feng, H., Jiang, D., Zhang, K. Q., et al. (2017). Trapping devices of nematode-trapping fungi: formation, evolution, and genomic perspectives. Biol. Rev. 92, 357–368. doi: 10.1111/brv.12233
Thau, N., Monod, M., Crestani, B., Rolland, C., Tronchin, G., Latge, J. P., et al. (1994). Rodletless mutants of Aspergillus fumigatus. Infect. Immun. 62, 4380–4388. doi: 10.1016/0167-5699(94)90199-6
Tunlid, A., Ahman, J., and Oliver, R. P. (1999). Transformation of the nematode-trapping fungus Arthrobotrys oligospora. FEMS Microbiol. Lett. 173, 111–116. doi: 10.1111/j.1574-6968.1999.tb13491.x
Tunlid, A., Rosen, S. E., Ek, B., and Rask, L. (1994). Purification and characterization of an extracellular serine protease from the nematode-trapping fungus Arthrobotrys oligospora. Microbiology 140, 1687–1695. doi: 10.1099/13500872-140-7-1687
Wang, M., Yang, J., and Zhang, K. Q. (2006). Characterization of an extracellular protease and its cDNA from the nematode-trapping fungus Monacrosporium microscaphoides. Can. J. Microbiol. 52, 130–139. doi: 10.1139/w05-110
Wiemann, P., Brown, D. W., Kleigrewe, K., Bok, J. W., Keller, N. P., Humpf, H. U., et al. (2010). FfVel1 and FfLae1, components of a velvet-like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism and virulence. Mol. Microbiol. 77, 972–994. doi: 10.1111/j.1365-2958.2010.07263.x
Winston, F., Dollard, C., and Ricupero-Hovasse, S. L. (1995). Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11, 53–55. doi: 10.1002/yea.320110107
Xie, X. Q., Li, F., Ying, S. H., and Feng, M. G. (2012). Additive contributions of two manganese-cored superoxide dismutases (MnSODs) to antioxidation, UV tolerance and virulence of Beauveria bassiana. PLoS One 7:e30298. doi: 10.1371/journal.pone.0030298
Yang, J. K., Liang, L. M., Li, J., and Zhang, K. Q. (2013). Nematicidal enzymes from microorganisms and their applications. Appl. Microbiol. Biotechnol. 97, 7081–7095. doi: 10.1007/s00253-013-5045-0
Yang, J. K., Wang, L., Ji, X. L., Feng, Y., Li, X. M., Zou, C. G., et al. (2011). Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 7:e1002179. doi: 10.1371/journal.ppat.1002179
Yang, X. W., Ma, N., Yang, L., Zheng, Y. Q., Zhen, Z. Y., Li, Q., et al. (2018). Two Rab GTPases play different roles in conidiation, trap formation, stress resistance, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 102, 4601–4613. doi: 10.1007/s00253-018-8929-1
Zhang, L. B., and Feng, M. G. (2018). Antioxidant enzymes and their contributions to biological control potential of fungal insect pathogens. Appl. Microbiol. Biotechnol. 102, 4995–5004. doi: 10.1007/s00253-018-9033-2
Zhao, M. L., Mo, M. H., and Zhang, K. Q. (2004). Characterization of a neutral serine protease and its full-length cDNA from the nematode-trapping fungus Arthrobotrys oligospora. Mycologia 96, 16–22. doi: 10.1080/15572536.2005.11832991
Zhen, Z. Y., Xing, X. J., Xie, M. H., Yang, L., Yang, X. W., Zheng, Y. Q., et al. (2018). MAP kinase Slt2 orthologs play similar roles in conidiation, trap formation, and pathogenicity in two nematode-trapping fungi. Fungal Genet. Biol. 116, 42–50. doi: 10.1016/j.fgb.2018.04.011
Zhen, Z. Y., Zhang, G. S., Yang, L., Ma, N., Li, Q., Ma, Y. X., et al. (2019). Characterization and functional analysis of the calcium/calmodulin-dependent protein kinases (CaMKs) in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 103, 819–832. doi: 10.1007/s00253-018-9504-5
Keywords: Arthrobotrys oligospora, velvet proteins, mutants, conidiation, trap formation, pathogenicity
Citation: Zhang G, Zheng Y, Ma Y, Yang L, Xie M, Zhou D, Niu X, Zhang K-Q and Yang J (2019) The Velvet Proteins VosA and VelB Play Different Roles in Conidiation, Trap Formation, and Pathogenicity in the Nematode-Trapping Fungus Arthrobotrys oligospora. Front. Microbiol. 10:1917. doi: 10.3389/fmicb.2019.01917
Received: 26 April 2019; Accepted: 05 August 2019;
Published: 20 August 2019.
Edited by:
Jae-Hyuk Yu, University of Wisconsin-Madison, United StatesReviewed by:
Hee-Soo Park, Kyungpook National University, South KoreaIstván Pócsi, University of Debrecen, Hungary
Hokyoung Son, Seoul National University, South Korea
Copyright © 2019 Zhang, Zheng, Ma, Yang, Xie, Zhou, Niu, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinkui Yang, amlua3VpOTYwQHludS5lZHUuY24=
†These authors have contributed equally to this work
 Guosheng Zhang
Guosheng Zhang Yaqing Zheng
Yaqing Zheng Yuxin Ma
Yuxin Ma Le Yang
Le Yang Meihua Xie1,2,3
Meihua Xie1,2,3 Xuemei Niu
Xuemei Niu Ke-Qin Zhang
Ke-Qin Zhang Jinkui Yang
Jinkui Yang