- 1Beijing Key Laboratory of Environment Friendly Management on Fruit Diseases and Pests in North China, Institute of Plant and Environment Protection, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
- 2Center of Excellence in Fungal Research, Mae Fah Luang University, Mueang Chiang Rai, Thailand
- 3Center for Bioinformatics, School of Life Science and Technology, University of Electronic Science and Technology of China, Chengdu, China
- 4Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Science, Kunming, China
- 5Department of Biology, McMaster University, Hamilton, ON, Canada
- 6College of Plant Protection, China Agricultural University, Beijing, China
Grapevine trunk diseases have become one of the main threats to grape production worldwide, with Diaporthe species as an emerging group of pathogens in China. At present, relatively little is known about the taxonomy and genetic diversity of Chinese Diaporthe populations, including their relationships to other populations worldwide. Here, we conducted an extensive field survey in six provinces in China to identify and characterize Diaporthe species in grape vineyards. Ninety-four isolates were identified and analyzed using multi-locus phylogeny. The isolates belonged to eight species, including three novel taxa, Diaporthe guangxiensis (D. guangxiensis), Diaporthe hubeiensis (D. hubeiensis), Diaporthe viniferae (D. viniferae), and three new host records, Diaporthe gulyae (D. gulyae), Diaporthe pescicola (D. pescicola), and Diaporthe unshiuensis (D. unshiuensis). The most commonly isolated species was Diaporthe eres (D. eres). In addition, high genetic diversity was observed for D. eres in Chinese vineyards. Haplotype network analysis of D. eres isolates from China and Europe showed a close relationship between samples from the two geographical locations and evidence for recombination. In comparative pathogenicity testing, D. gulyae was the most aggressive taxon, whereas D. hubeiensis was the least aggressive. This study provides new insights into the Diaporthe species associated with grapevines in China, and our results can be used to develop effective disease management strategies.
Introduction
In natural ecosystems, plant pathogens play important roles such as regulating host populations and host plant geographic and ecological distributions. Consequently, they can affect the availability of food sources to other living organisms (Lindahl and Grace, 2015). Most microbial pathogens have short generation times and large population sizes, which can result in high genetic variations and rapid adaptations to environmental stresses and to human-mediated factors such as fungicide resistance (Alberts et al., 2002; Lindahl and Grace, 2015). Hence, it is important to understand the genetic diversity and population variation of plant pathogens to develop sustainable control measures.
Grape is one of the most important fruit crops in China. China is the second largest grape-cultivating country and the top producer in the world (OIV, 2016). In 2016, the total grape cultivation area was estimated at 847 kha, and 14.5 million metric tons of fresh grapes were produced in China (OIV, 2016). Therefore, infectious diseases with significant risks to grape production have drawn broad attention from the grapevine industry. Grapevines are affected by several foliar diseases (Gadoury et al., 2012; Zhang et al., 2017), fruit diseases (Daykin and Milholland, 1984; Hong et al., 2008; Greer et al., 2011; Jayawardena et al., 2015), and trunk diseases (Yan et al., 2013; Dissanayake et al., 2015a,b). Grapevine trunk diseases have drawn considerable attention, as these diseases affect the perennial parts of the vine and can limit grape production for many years (Yan et al., 2013, 2015).
The genus Diaporthe Nitschke., belongs to the family Diaporthaceae, and is typified by Diaporthe eres (D. eres) Nitschke (Senanayake et al., 2017). Following the nomenclature rules Rossman et al. (2014) proposed that the genus name Diaporthe over Phomopsis as it was introduced first, represents the majority of species. In earlier species names were given to Diaporthe taxa based on their host specificity. This resulted in over 100 names listed under the genus Diaporthe (http://www.indexfungorum.org/Names/Names.asp and http://www.mycobank.org). With advances in molecular techniques, multi-locus DNA sequence data together with morphological characteristics have been extensively used for the delimitation of Diaporthe species (Udayanga et al., 2011; Gomes et al., 2013; Gao et al., 2017). The internal transcribed spacer (ITS), translation elongation factor-1a (EF-1α), β-tubulin, partial histone H3 (HIS), calmodulin (CAL), genes are the most commonly used gene regions for molecular characterization (Udayanga et al., 2011; Gao et al., 2017; Guarnaccia et al., 2018; Yang et al., 2018). Multiple studies have used different gene combinations to resolve the species boundaries in this genus (Udayanga et al., 2011, 2014a,b; Gao et al., 2017; Marin-Felix et al., 2019). Species belonging to genus Diaporthe are endophytes, pathogenic, and saprobic on wide range of hosts worldwide (Liu et al., 2015; Hyde et al., 2016; Marin-Felix et al., 2019). They are well-known pathogens on economically important crops (Udayanga et al., 2011). Several common disease among those are dieback on forest trees (Yang et al., 2018), leaf spots on tea (Guarnaccia and Crous, 2017), leaf and pod blights and seed decay on soybean (Udayanga et al., 2015), melanose, stem-end rot, and gummosis on Citrus spp. (Mondal et al., 2007; Udayanga et al., 2014a; Guarnaccia and Crous, 2017, 2018) and stem canker on sunflower (Muntañola-Cvetković et al., 1981; Thompson et al., 2011).
Phomopsis cane and leaf spot caused by Diaporthe species on grapevine is one of the most complex grapevine trunk diseases worldwide (Úrbez-Torres et al., 2013; Dissanayake et al., 2015a; Guarnaccia et al., 2018). The disease symptoms of Diaporthe Dieback include shoots breaking off at the base, stunting, dieback, loss of vigor, reduced bunch set, and fruit rot (Pine, 1958, 1959; Pscheidt and Pearson, 1989; Pearson and Goheen, 1994; Wilcox et al., 2015). In woods brown to black necrotic irregular-shaped lesions could be observed. Once clusters are infected rachis necrosis and brown, shriveled berries close to harvest could be observed (Pearson and Goheen, 1994). More than one Diaporthe species is frequently reported as causative agents from one country (Dissanayake et al., 2015a; Guarnaccia et al., 2018). Currently, 27 species have been identified as causal organisms of Diaporthe dieback in grape-producing countries worldwide (Mostert et al., 2001; Van Niekerk et al., 2005; Udayanga et al., 2011, 2014a,b; White et al., 2011; Baumgartner et al., 2013; Úrbez-Torres et al., 2013; Hyde et al., 2014; Dissanayake et al., 2015a; Guarnaccia et al., 2018; Lesuthu et al., 2019). Even though these species characterized under the one disease, disease symptoms, and aggressiveness are varying according to the species. Diaporthe ampelina (D. ampelina) has a long history as the most common and severe pathogenic species together with D. amygdali (Mostert et al., 2001; Van Niekerk et al., 2005). Diaporthe ampelina and Diaporthe kyushuensis (D. kyushuensis) are the causal agent of grapevine swelling arm (Kajitani and Kanematsu, 2000; Van Niekerk et al., 2005). Diaporthe perjuncta (D. perjuncta) and D. ampelina caused cane bleaching (Kuo and Leu, 1998; Kajitani and Kanematsu, 2000; Mostert et al., 2001; Van Niekerk et al., 2005; Rawnsley et al., 2006). Lesuthu et al. (2019) showed that D. ampelina, Diaporthe novem (D. novem), and Diaporthe nebulae (D. nebulae) as the most virulent species of Diaporthe associated with grapevines in South Africa. Diaporthe eres was found as a weak to moderate pathogen in several different studies (Kaliterna et al., 2012; Baumgartner et al., 2013). These results indicate the complexity and high species richness of Diaporthe associated with the grapevines. Up to now in China four Diaporthe species have been reported causing grapevine dieback (Dissanayake et al., 2015a). Those are D. eres, Diaporthe hongkongensis (D. hongkongensis), Diaporthe phaseolorum (D. phaseolorum), and Diaporthe sojae (D. sojae). Their taxonomic placements and pathogenicity under a controlled environment were also studied.
The study conducted by Guarnaccia et al. (2018) showed that species of Diaporthe also associated as endophytes on grapes as well. In that study they observed that Diaporthe bohemiae (D. bohemiae), which was isolated from grape was unable to induce lesions. In addition to grapevines, Diaporthe have been reported on broad range of hosts (Udayanga et al., 2011). However, the most important charter is the ability of endophytic Diaporthe species to be opportunistic pathogens. Huang et al. (2015) observed that some Diaporthe species associated with citrus in China shown to act as opportunistic plant pathogens. Diaporthe foeniculina (D. foeniculina) has been found as both endophyte and opportunistic pathogen on various herbaceous weeds, ornamentals, and fruit trees (Udayanga et al., 2014a; Guarnaccia et al., 2016). So far it is not confirmed the factor that driven into pathogenicity from endophytes either due to environmental changes or the reduction of host's defense. Therefore, further studies are required to understand this in both field level and genomic level.
However, the genetic diversity of Diaporthe spp. associated with Vitis spp., relationships among isolates from different geographical regions, and relationships among isolates from China and those from other countries were not investigated. Therefore, to expand our knowledge on these issues, we performed an extensive field survey to isolate and identify Diaporthe species associated with grapevine dieback in China. We reconstructed a phylogenetic tree for the genus Diaporthe. The present study analyzed the genetic diversity of Diaporthe species associated with grapevines in China and constructed haplotype networks for Diaporthe species from different geographical origins for the first time. Finally, we analyzed the relationship between Diaporthe species from European and Chinese grape vineyards, as Diaporthe dieback is becoming an emerging trunk disease in both regions (Guarnaccia et al., 2018).
Materials and Methods
Sampling and Pathogen Isolation
Field surveys were conducted during 2014 and 2015 in 20 vineyards in the six following provinces in China: Guangxi, Heilongjiang, Hubei, Jilin, Liaoning, and Sichuan (Figure 1). Samples were collected from symptomatic grapevine woody branches that exhibited bark discoloration, shoots breaking off at the base, stunting, wedge-shaped cankers, and light brown streaking of the wood from the following Vitis vinifera (V. vinifera) cultivars: Centennial Seedless, Red Globe, and Summer Black (Figure 2). Symptomatic tissue samples were collected into zip-lock plastic bags that contained wet sterilized tissue papers to maintain humidity. Once the samples were taken into the laboratory, infected trunks or shoots were photographed, and symptoms, location, and other relevant data were documented. The fungal pathogens were isolated using the following procedures. Infected shoots/trunks were cut into small pieces (1–3 mm thick). These pieces were then surface-sterilized by dipping into 70% ethanol for 30 s and then transferred into 1% NaOCl for 1 min. This step was followed by two washes with sterile distilled water. Once the wood pieces were dried, they were placed onto potato dextrose agar (PDA) plates supplemented with ampicillin (0.1 g L−1) and incubated at 25°C. After 5–7 days of incubation, hyphal tips of fungi immerging from wood pieces were transferred onto new PDA plates and incubated until they produce conidia. Once the conidia were developed single spore isolation was done. For the strains do not developed conidia after 4 weeks two-three times hyphal tip isolation was done. All the pure cultures obtained in this study were deposited in the culture collection of Institute of Plant and Environment Protection of Beijing Academy of Agriculture and Forestry Sciences (JZB culture collection) at 4°C.
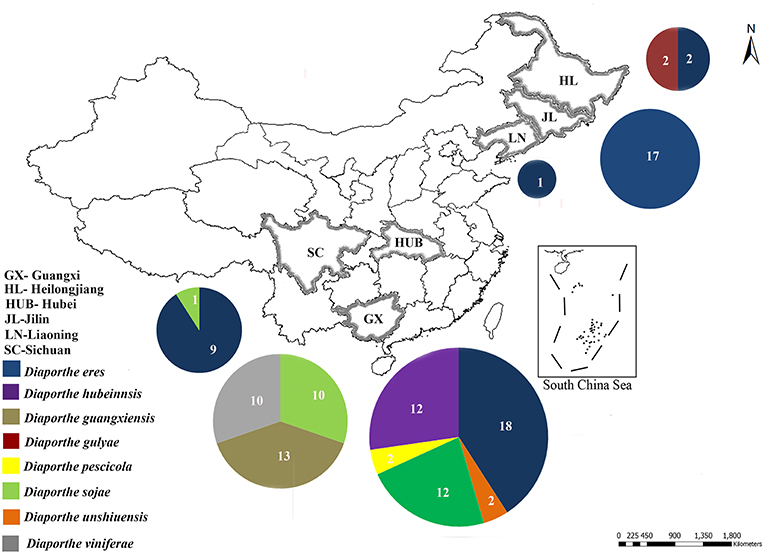
Figure 1. Sample collection sites of Diaporthe dieback in six provinces in China. Circles represent the association frequency of each species in each population sampled, and the number of isolates analyzed in each population is given inside the respective slice.

Figure 2. Symptoms of Diaporthe dieback. (A,B) Field symptoms on trunks and shoots, (C) appearance of fruiting bodies on trunk surface, and (D,E) cross sections of infected trunks.
DNA Extraction, PCR Amplification, and Sequence Assembly
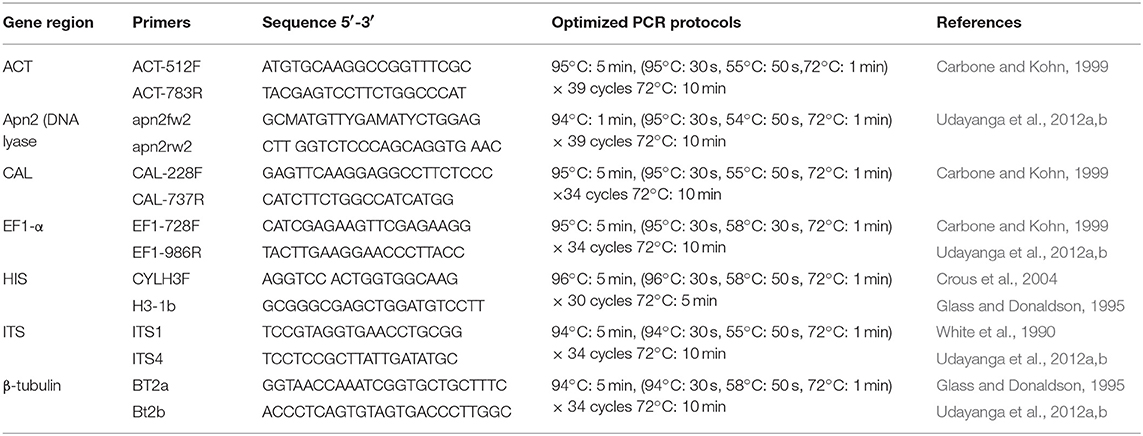
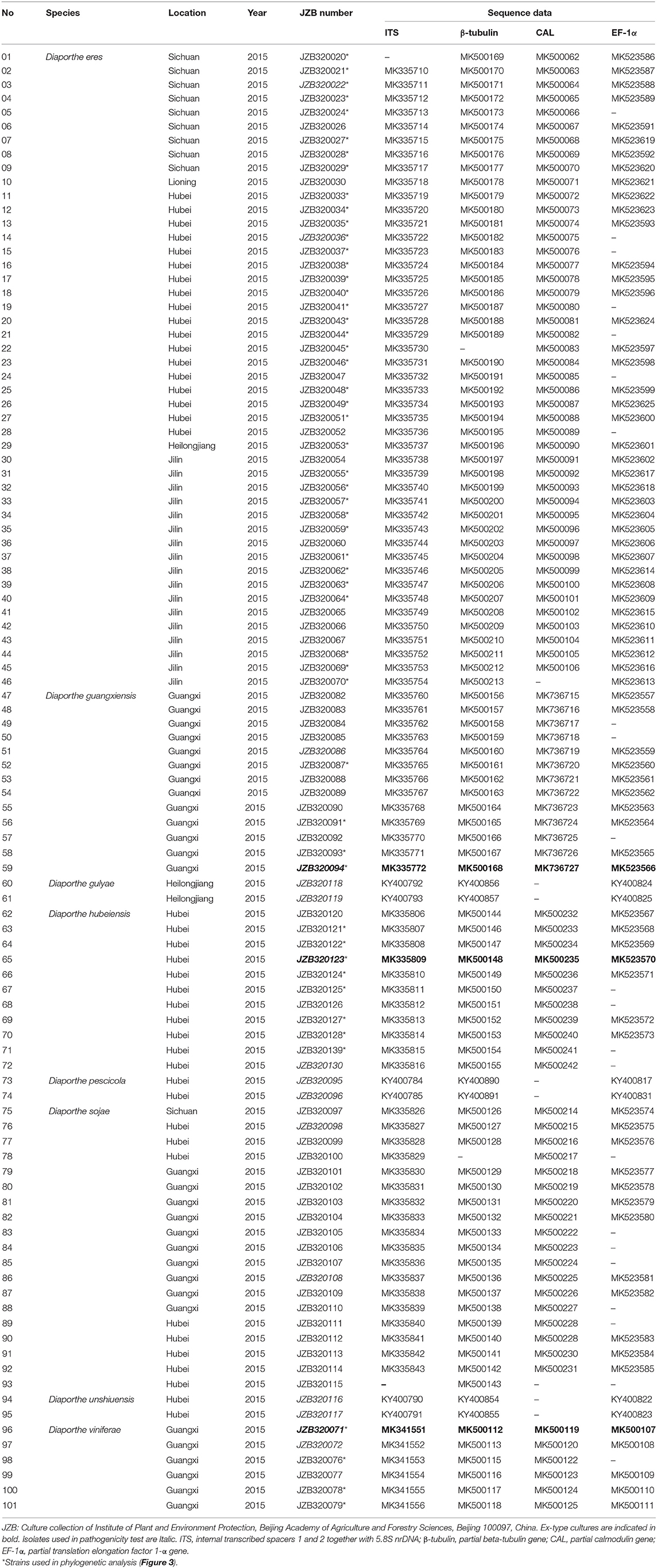
Approximately 10 mg of aerial mycelium was scraped from 5–7 days old isolates grown on PDA (Potato Dextrose Agar) at 25°C. Total genomic DNA was extracted using the DNeasy Plant Mini Kit (QIAGEN GmbH, QIAGEN Strasse 1, 40742 Hilden, Germany). For species confirmation, the internal transcribed spacer (ITS) regions were sequenced for all isolates. The obtained sequences were compared to those in GenBank using the MegaBLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). After isolates were confirmed as belonging to the genus Diaporthe, six additional gene regions, those encoding translation elongation factor-1α (EF-1α), β-tubulin, calmodulin (CAL), partial histone H3 (HIS), partial actin (ACT), and DNA-lyase (Apn2), were sequenced. Table 1 presents the primer pairs with their respective amplification conditions for each of the above gene regions. PCR mixtures of 25 μl total volume consisted of 0.3 μl of TaKaRa Ex-Taq DNA polymerase, 2.5 μl of 10 × Ex-Taq DNA polymerase buffer, 3.0 μl of dNTPs, 2 μl of genomic DNA, 1 μl of each primer, and 15.2 ddH2O. The PCRs were conducted in a Bio-Rad C1000 thermal cycler (Germany). The resulting products were visualized on a 1% agarose gel stained with ethidium bromide under UV light using a Gel DocTM XR Molecular Imager (Bio Rad, USA). All positive amplicons were sequenced by Beijing Biomed Gene Technology Co LTD. The sequence quality was confirmed by checking chromatograms using BioEdit v. 5 (Hall, 2006). Sequences were obtained using both forward and reverse primers, and consensus sequences were generated using DNAStar v. 5.1 (DNASTAR, Inc.). The sequence data generated in the present study have been deposited in GenBank (Table 2).
Phylogenetic Analyses
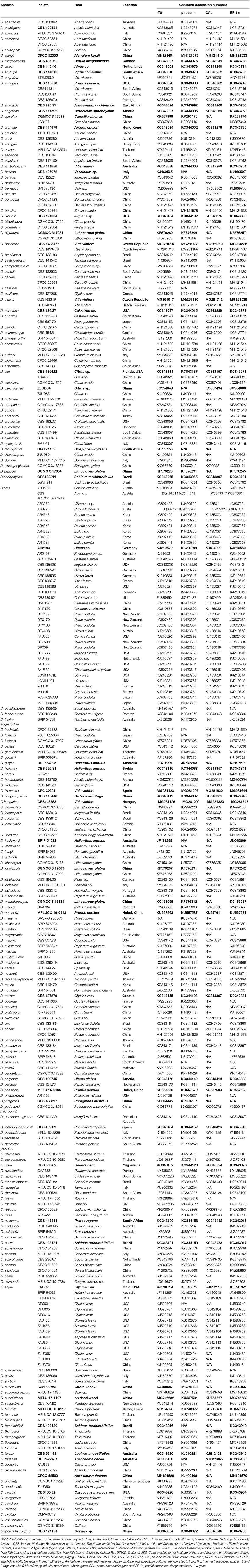
For the phylogenetic analyses, reference sequences representing related taxa in Diaporthe were downloaded from GenBank (Guarnaccia et al., 2018; Yang et al., 2018; Table 3) and aligned with the sequences obtained in this study (Table 2). The sequences were aligned using MAFFT (Katoh and Toh, 2010) (http://www.ebi.ac.uk/Tools/msa/mafft/) and manually adjusted using BioEdit v. 5 (Hall, 2006) whenever necessary. Phylogenetic relationships were inferred using maximum parsimony (MP) implemented in PAUP (v4.0) (Swofford, 2003), maximum likelihood (ML) in RAxML (Silvestro and Michalak, 2010) and Bayesian analyses in MrBayes v. 3.0b4 (Ronquist and Huelsenbeck, 2003). In phylogenetic analysis, single-gene trees were constructed first using ML in RAxML. The phylogenetic tree topologies for different gene fragments were compared for evidence of incongruences with a focus on comparing branches with high bootstrap values. If no conflict was observed, a combined phylogenetic tree was generated.
In PAUP, ambiguous regions in the alignment were excluded for further analyses, and gaps were treated as missing data. The stability of the trees was evaluated by 1000 bootstrap replications. Branches of zero length were collapsed, and all multiple parsimonious trees were saved. Parameters, including tree length (TL), consistency index (CI), retention index (RI), relative consistency index (RC), and homoplasy index (HI) were calculated. Differences between the trees inferred under different optimality criteria were evaluated using Kishino-Hasegawa tests (KHT) (Kishino and Hasegawa, 1989). The evolutionary models for each locus used in Bayesian analysis and ML were selected using MrModeltest v. 2.3 (Nylander, 2004). ML analyses were accomplished using RAxML-HPC2 on XSEDE (8.2.8) (Stamatakis et al., 2008; Stamatakis, 2014) in the CIPRES Science Gateway platform (Miller et al., 2010) using the GTR + I + G model of evolution with 1000 non-parametric bootstrapping iterations. Bayesian analysis was performed in MrBayes v. 3.0b4 (Ronquist and Huelsenbeck, 2003), and posterior probabilities (PPs) were determined by Markov chain Monte Carlo sampling (MCMC). Six simultaneous Markov chains were run for 106 generations, sampling the trees at every 100th generation. From the 10,000 trees obtained, the first 2,000 representing the burn-in phase were discarded. The remaining 8,000 trees were used to calculate PPs in a majority rule consensus tree. Alignment generated in this study is submitted to TreeBASE (https://treebase.org/treebase-web/home.html) under the submission number 24324. Taxonomic novelties were submitted to the Faces of Fungi database (Jayasiri et al., 2015) and Index fungorum (http://www.indexfungorum.org). New species are described following Jeewon and Hyde (2016).
Morphology and Culture Characteristics
Colony morphology and conidial characteristics were examined for Diaporthe species identified by phylogenetic analysis. Colony colors were examined according to Rayner (1970) after 7 days of growth on PDA in the dark at 25°C. Digital images of morphological structures mounted in water were taken using an Axio Imager Z2 photographic microscope (Carl Zeiss Microscopy, Oberkochen, Germany). Measurements were taken using ZEN PRO 2012 (Carl Zeiss Microscopy). Conidial length and width were measured for 40 conidia per isolate, and the mean values were calculated for all measurements. Conidial shape, color, and guttulation were recorded.
Genetic Diversity and Population Structure Analysis
Among the identified species, only one, Diaporthe eres, had a count of >20 individuals. As a result, only D. eres was selected for the analysis of genetic diversity and population relationships. For the D. eres population, diversity indices were calculated for each gene region and the combined sequence dataset. DnaSP v. 6.12 (Librado and Rozas, 2009) was employed to calculate haplotype richness (hR), the total number of haplotypes, Watterson's theta (Θw), and pairwise nucleotide diversity (JI). To overcome the population size effects, hR, Θw and JI were calculated after 1,000 repetitions, and the median estimate was recorded for each parameter. To understand the potential departure from an equilibrium model of evolution, Tajima's D was calculated using DnaSP v. 6.12 with a permutation test of 1,000 replicates. The minimum numbers of recombination events (ZnS) used by Kelly (1997) and the recombination parameters Za and ZZ used by Hudson (1983) were calculated for each gene region and the combined data set. Diaporthe eres haplotype networks were constructed using Network v. 5.0 (Bandelt et al., 1999).
Network Analysis
To understand the relationship among different geographical populations, recombination parameters were calculated, and haplotype networks were constructed. In this analysis, the combined dataset of Diaporthe eres isolates from China alone and Chinese isolates combined with European isolates (Guarnaccia et al., 2018) were used. ZnS, used by Kelly (1997), and the recombination parameters Za and ZZ (Hudson, 1983; Kelly, 1997) were calculated using DnaSP v. 6.12. The haplotype data generated using DnaSP v. 6 were used to construct a median-joining network in Network v. 5.0 (Bandelt et al., 1999).
Pathogenicity Assay
The pathogenicity and aggressiveness of the Diaporthe species were tested using detached green shoots of the V. vinifera cultivar Summer Black. Healthy, 30–50 cm long green shoots (including at least two nodes) were obtained from “Shunyi Xiangyi” vineyard in Beijing, China, where Diaporthe species were not recorded. The cuttings were surface-sterilized with 70% ethanol by wiping with cotton swabs. A shallow wound (5 mm length, 2 mm deep) was made in the center of each shoot using a sterilized scalpel. Mycelial plugs were taken from the growing margin of a 5-day-old culture grown in PDA and inoculated at the wound site. Non-colonized sterile PDA plugs were used for inoculation of shoots as a negative control. To prevent drying, all inoculated areas were covered with Para-film (Bemis, USA). Inoculated shoots were kept in a growth chamber for 21 days at 25°C with a 12 h photoperiod. The experiment was organized with 10 replicates for each isolate. Pathogenicity test was repeated three times with same controlled environment. A total of 16 strains from eight species were tested. The presence of lesions advancing beyond the original 0.5 cm diameter inoculation point was considered indicative of pathogenicity. The experimental design was completely randomized. Data were analyzed with a one-way ANOVA (analysis of variance) using Minitab v. 16.0 (Minitab Inc., Boston, MA, USA), with statistical significance set at the 5% level. The pathogens were re-isolated to confirm their identity.
Results
Initial Species Identification and Phylogenetic Analyses
During our field survey on six grape-growing provinces in China (Figure 1), we collected samples with typical symptoms associated with Diaporthe dieback, such as wedge-shaped cankers, and light brown streaking of the wood (Figure 2). However, these symptoms are sometimes confused with other grape trunk disease symptoms caused by Botryosphaeria dieback, Eupta, and Esca (Mondello et al., 2018). Hence, further confirmation is required by isolating and identifying causal organisms. One hundred and eleven Diaporthe isolates were initially identified by colony characteristics, such as abundant tufted white aerial mycelia on agar medium. The ITS gene regions were sequenced for all fungi isolated from diseased shoots and compared with those in GenBank using the MegaBLAST tool in GenBank. The isolates showed 95–99% similarity to known Diaporthe species in GenBank, and these closely related known species were included in the phylogenetic analysis.
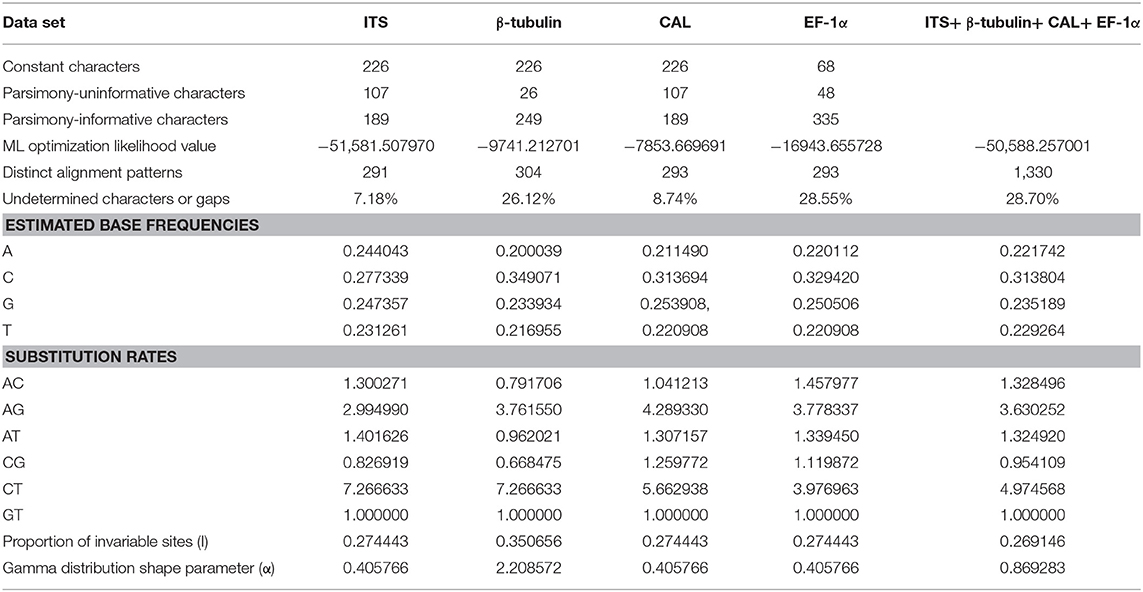
To understand the taxonomic placements of our isolates, additional gene regions, including those encoding EF-1α, β-tubulin, and CAL, were sequenced. Then, phylogenetic trees were constructed for each individual gene region. The concatenated sequence data set consisted of 94 isolates (out of 111, due to sequencing errors) from the current study (Table 3) and 197 isolates originating from GenBank (Table 2), with one outgroup taxon, Diaporthella corylina (CBS 121124). A comparison of maximum likelihood (ML) analysis results for each gene region is given in Table 4. In the ML analysis, the resulting tree of the combined data set of ITS, β-tubulin, CAL, and EF-1α genes had the best resolution of taxa (Figure 3). Therefore, in the present study, we used the combined sequence data to understand the taxonomic placements of the Diaporthe species isolated from grapevines in China. A Bayesian analysis resulted in 10,001 trees after 2,000,000 generations. The first 1,000 trees, representing the burn-in phase of the analyses, were discarded, while the remaining 9,001 trees were used for calculating posterior probabilities (PPs) in the majority-rule consensus tree. The dataset consisted of 1,494 characters with 727 constant characters and 1,006 parsimony-informative and 213 parsimony-uninformative characters. The maximum number of trees generated was 1,000, and the most parsimonious trees had a tree length of 9,862 (CI = 0.249, RI = 0.805, RC = 0.201, HI = 0.751).
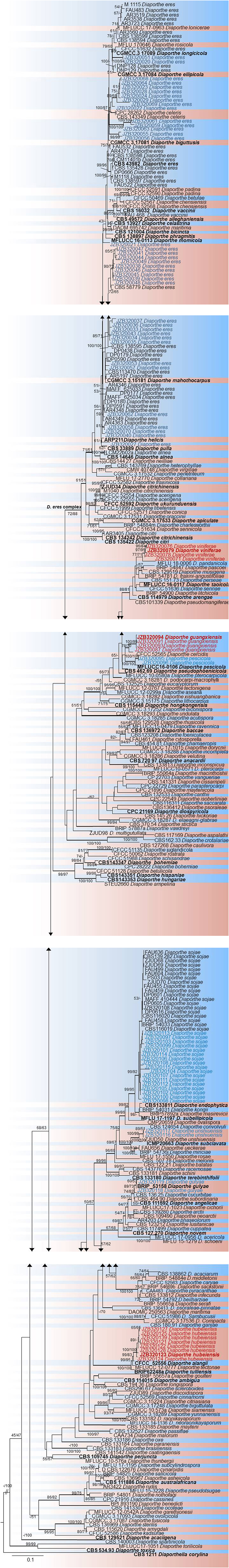
Figure 3. RAxML tree based on analysis of a combined dataset of ITS, β-tubulin, CAL, and EF-1α sequences. Bootstrap support values for ML and MP equal to or >50% are shown as ML/MP above the nodes. The isolates obtained for the present study are shown in blue for already known species, and novel taxa are shown in red. Ex-type strains are indicated in bold. The tree is rooted using Diaporthella corylina. The scale bar represents the expected number of nucleotide substitutions per site.
In the phylogenetic tree generated using the combined data set (Figure 3), 36 isolates from the present study clustered with Diaporthe eres in the D. eres complex. This group represents 37.5% of the total isolates, and these isolates were obtained from five provinces. Sixteen isolates (19.76% of the total isolates) clustered with Diaporthe sojae (D. sojae) species in the D. sojae complex. Two isolates from Heilongjiang province clustered together with Diaporthe gulyae (D. gulyae) (BRIP 54025). In addition, two isolates clustered with Diaporthe unshiuensis (D. unshiuensis) (ZJUD52) from Hubei province, and another two isolates that were also from Hubei province clustered with Diaporthe pescicola (D. pescicola) (MFLUCC 16-0105). The remaining isolates (35 in total) did not cluster with any known Diaporthe species. Thus, these were putatively identified as belonging to three novel species (Figure 3): D. hubeiensis, D. guangxiensis, and D. viniferae. Diaporthe hubeiensis (D. hubeiensis) was isolated from grapevines from Hubei province and represents 12.5% of the total isolates. This species is a sister taxon with Diaporthe alangi (D. alangi) (CFCC52556). The remaining two new taxa were isolated from grapevines from Guangxi Province. Diaporthe guangxiensis (D. guangxiensis) was represented by 11 isolates (13.54%), and it is closely associated with Diaporthe cercidis (D. cercidis) (CFCC5255). Diaporthe viniferae (D. viniferae) was represented by 8 isolates (10.41%), and its closest relative is Diaporthe pandanicola (D. pandanicola) (MFLU 18-0006).
Taxonomic Novelties
Diaporthe guangxiensis (D. guangxiensis) Dissanayake, X.H. Li & K.D. Hyde, sp. nov. (Figure 4).
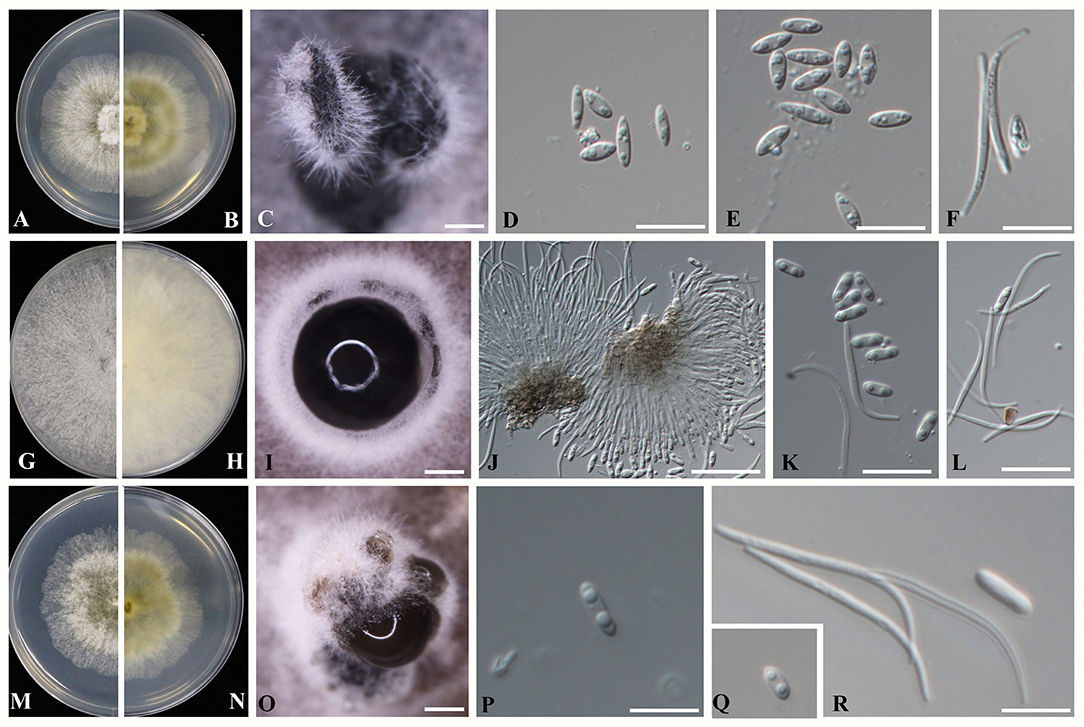
Figure 4. Novel Diaporthe taxa identified in the present study (A–F) Diaporthe guangxiensis (A,B) Culture on PDA after 5 days; (C) Pycnidia on PDA; (D,E) Alpha conidia; and (F) Beta conidia. (G–L) Diaporthe hubeiensis (G,H) Culture on PDA after 5 days; (I) Pycnidia on PDA; (J) Conidiogenous cells for alpha and beta conidia; (K) Alpha conidia, and (L) Beta conidia. (M–R) Diaporthe viniferae (M,N) Culture on PDA after 5 days; (O) Pycnidia on PDA; (P,Q) Alpha conidia; and (R) Beta conidia. Scale bars: (D–F,J–L,P–R) = 1 mm; (C,I,O) = 10 μm.
Index Fungorum number—IF552578, Facesoffungi Number- FoF02725.
Etymology- In reference to the Guangxi Province, from where the fungus was first isolated.
Holotype—JZBH320094.
Description
Sexual morph: efforts were made to initiate sexual morphs, but various methods failed; Asexual morph: pycnidia on PDA 250-1550 μm (x = 1100 μm, n = 20) in diam., superficial, scattered on PDA, dark brown to black, globose, solitary, or clustered in groups of 3–5 pycnidia. Conidiophores aseptate, cylindrical, straight or sinuous, densely aggregated, terminal, slightly tapered toward the apex, 21–35 × 1.5–2.5 μm ( = 27 × 2 μm). Alpha conidia biguttulate, hyaline, fusiform or oval, both ends obtuse 5.3–7.8 × 1.5–3.2 μm ( = 6.8 × 2.5 μm n = 40). Beta conidia aseptate, hyaline, hamate, filiform, guttulate, tapering toward both ends 20–32 × 1–1.5 μm ( = 27 × 1.5 μm, n = 20).
Culture Characteristics
Colonies on PDA reach 70 mm diam. after 7 days at 25°C, producing abundant white aerial mycelia and reverse fuscous black.
Material Examined
CHINA, Guangxi Province, Pingguo County, on diseased trunk of V. vinifera, 3 June 2015, X.H. Li, (JZBH320094, holotype); ex-type living cultures JZB320094).
Notes: Morphological characters such as spores and colony characteristics of D. guangxiensis fit well within the species concept of Diaporthe. DNA sequence analyses of the ITS, CAL, TUB, and EF genes showed a strongly supported monophyletic lineage with 78% ML, 70% MP bootstrap values and 0.95 posterior probabilities (Figure 3). The current species has a particular neighbor relationship with D. cercidis (CFCC52566). Morphologically, D. guangxiensis has larger conidiophores (27 × 2 μm) and smaller conidia (6.8 × 2.5 μm) than D. cercidis (7–17 × 1.4–2.1 μm conidiophores; 8.6 × 3.3 μm conidia) (Yang et al., 2018). In the comparisons of five gene regions between Diaporthe guangxiensis and D. cercidis, 51.5% of 458 nucleotides across the ITS (+5.8S) had base pair differences. In addition, comparisons of the protein-coding genes showed that there were 17.3, 0.66, and 9.06% polymorphic nucleotide sites between the two species for the CAL, β-tubulin and EF-1α genes, respectively.
Diaporthe hubeiensis Dissanayake, X.H. Li & K.D. Hyde, sp. nov. (Figure 4).
Index Fungorum number—IF552579, Facesoffungi Number- FoF 02726.
Etymology- In reference to the Hubei province, from where the fungus was first isolated.
Holotype – JZBH320123.
Description
Sexual morph: efforts were made to initiate sexual morphs, but various methods failed; Asexual morph: pycnidia on PDA varying in size up to 510 μm in diam., subglobose, occurs on PDA and double-autoclaved toothpicks after 3–4 weeks, solitary or forms in groups of stroma with a blackened margin. Ostiolate, up to 100 μm black cylindrical necks. Conidiophores were reduced to conidiogenous cells. Alpha conidia hyaline, smooth, biguttulate, blunt at both ends, ellipsoidal to cylindrical, 5.6–7.1 × 1–3.1 μm ( = 6.1 × 1.8 μm n = 40). Beta conidia filiform, tapering toward both ends, scattered among the alpha conidia 17–27 × 1–1.5 μm ( = 24 × 1.5 μm n = 40).
Culture Characteristics
Colonies on PDA reach 90 mm after 10 days at 25°C (covers total surface), abundant tufted white aerial mycelia, buff, numerous black pycnidia 0.5 mm in diam. occur in the mycelium, typically in the direction of the edge of the colony; reverse buff with concentric lines.
Material Examined
CHINA Hubei Province, Wuhan, on diseased trunk of V. vinifera, 30 June 2015, X. H Li (JZBH320123, holotype); ex-type living cultures JZB320123.
Notes: In phylogenetic analysis, D. hubeiensis was placed in a well-supported clade together with D. alangi (CFCC52556), D. tectonae (MFLUCC 12- 0777) and D. tulliensis (BRIP62248b) with 100% ML, 100% MP bootstrap values and 0.99 posterior probabilities. Diaporthe hubeiensis developed sister clade with D. alangi (CFCC52556) with 99% ML, 83% MP bootstrap values and 0.99 posterior probabilities. Morphologically, Diaporthe hubeiensis has smaller conidiophores and smaller conidia (6.1 × 1.8 μm) than D. alangi (7 × 2 μm), and it has no beta conidia in D. alangi (Yang et al., 2018). Diaporthe hubeiensis differs from D. tectonae by developing wider but shorter conidia (6.1 × 1.8 μm vs 5.5 × 2.6 μm) (Doilom et al., 2017). Compared to D. tulliensis, D. hubeiensis has smaller conidia (6.1 × 1.8 μm vs 5.5–6 μm) (Yang et al., 2018). In the ITS sequence comparison between D. hubeiensis and D. alangi, 44.6% of the 461 nucleotides across the ITS (+5.8S) were different. Of the three protein-coding genes, the two species showed 4.26% and 1.16% and 5.3% polymorphic nucleotide site differences for CAL, β-tubulin and EF-1α genes, respectively.
Diaporthe viniferae Dissanayake, X.H. Li & K.D. Hyde, sp. nov.
Index Fungorum number—IF552002, Facesoffungi Number- FoF 05981.
Etymology- In reference to the host V. vinifera.
Holotype—JZBH320071.
Description
Sexual morph: efforts were made to initiate sexual morphs, but various methods failed; Asexual morph: Pycnidia on PDA 363–937 μm (x = 529 μm, n = 20) in diam., superficial, scattered, dark brown to black, globose, solitary in most. Conidiophores were not observed. Conidiogenous cells were not observed. Alpha conidia biguttulate, hyaline, fusiform or oval, both ends obtuse 5–8.3 × 1.3–2.5 μm ( = 6.4 × 2.1 μm). Beta conidia aseptate, hyaline, hamate, filiform, tapering toward both ends 23–35 × 1–1.5 μm ( = 28 × 1.3 μm n = 40).
Culture Characteristics
Colonies on PDA reach 70 mm diam. after 7 days at 25°C, producing abundant white aerial mycelia and reverse fuscous black.
Material Examined
CHINA, Guangxi Province, Pingguo County, on the diseased trunk of V. vinifera, 3 June 2015, X.H. Li, (JZBH320071 holotype); ex-type living cultures JZB320071).
Notes: In the phylogenetic analysis of D. viniferae, a strongly supported monophyletic lineage with strong 77% ML and 71% MP bootstrap values and 0.95 PP was developed (Figure 3). The current species has a particular close relationship with D. pandanicola (MFLUCC 18-0006). In the original description of D. pandanicola, morphological characteristics were not given (Tibpromma et al., 2018). Therefore, these two species were compared based on only DNA sequence data. ITS sequence comparison between D. viniferae and D. pandanicola revealed that 2.9% of the 478 nucleotide sites across the ITS (+5.8S) regions were different. Similarly, 1.7% of the β-tubulin gene fragment was different.
Genetic Diversity and Population Structure Analysis
Table 5 summarized the genetic diversity data of D. eres associated with grapevines which were estimated using DnaSP V.6. In the analysis, the combined data set of ITS, β-tubulin, HIS, APN, and CAL gene sequences showed 0.16226 segregation sites per sequence and a haplotype diversity of 0.955. A haplotype network was developed for the D. eres species isolated from China using Network v. 5.0 (Figure 5). The resulting network combining ITS, β-tubulin, HIS, EF-1α, and CAL gene sequences gave two main clusters according to geographic origin. In the network, isolates from Hubei province were clustered into two main clades. A single haplotype (H-11) was clustered within the main Jilin clade. Haplotype 7 (from Hubei) and h-13 (from Sichuan Province) were connected with one intermediate haplotype to the two main clusters.
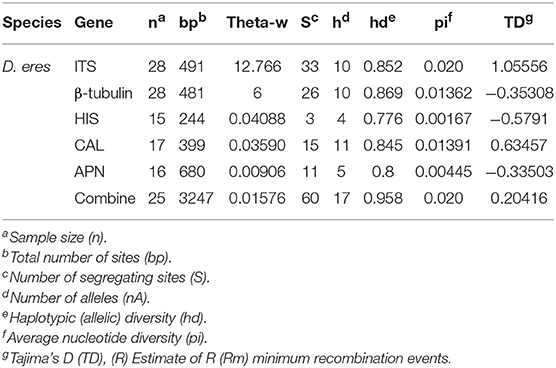
Table 5. Polymorphism and genetic diversity of Diaporthe eres strains associated with Chinese grapevines.
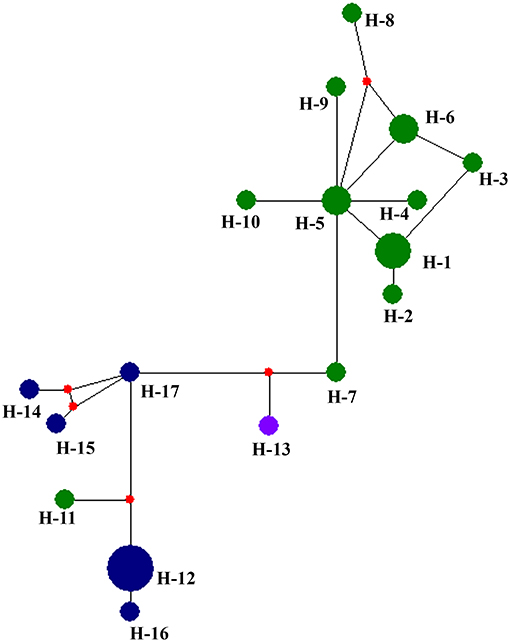
Figure 5. Haplotype network generated for the Diaporthe eres isolates obtained in the present study using Network v 6.0. At each node, sizes are propionate to the number of isolates. Blue, haplotypes from Jilin; Green, haplotypes from Hubei; purple, haplotypes from Sichuan; red, Median vectors.
To understand the relationship between Diaporthe isolates from Chinese vineyards and those from European vineyards, we calculated recombination parameters Z and ZnS. The combined data set consists of 135 sequences with 2203 sites. The estimate of R per gene was 6.6, and the minimum number of recombination events (Rm) was 15. Median-joining networks were constructed using both single-gene data files and a combined data set of ITS, β-tubulin, HIS, EF-1α, and CAL genes. The single-gene networks differed from each other, and the resulting patterns did not give a significant grouping. Therefore, in this study, only the combined network was considered (Figure 6). A total of 33 haplotypes were identified using DnaSP, and the haplotype data file was used to generate the haplotype network. In the resulting network, we found that Chinese haplotypes and Europe haplotypes were not shared and that there was no sharing of haplotypes among different provinces in China. However, the Chinese haplotypes were dispersed in the combined network, with the majority of isolates from Hubei located in two related clusters surrounded by European haplotypes. Similarly, the haplotypes from Sichuan and Jilin provinces were also dispersed in the network and close to both European and Chinese haplotypes.
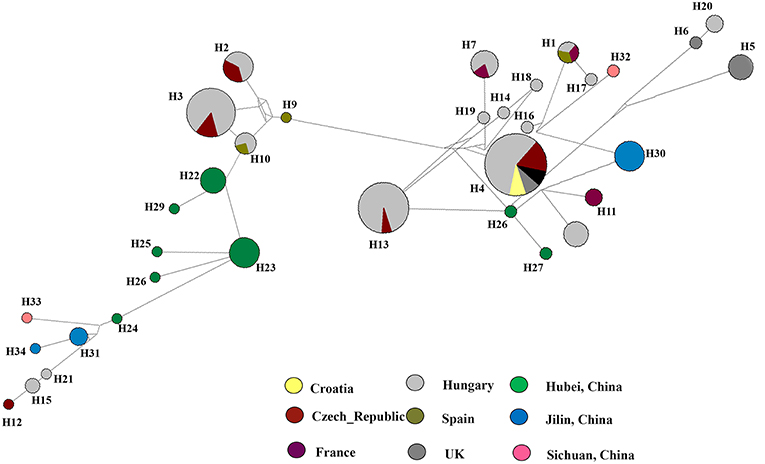
Figure 6. Haplotype network generated for the Diaporthe eres isolates from China and European countries using Network v 6.0. At each node, sizes are proportionate to the number of isolates.
Comparative Aggressiveness Among Diaporthe Species
Pathogenicity and aggressiveness among eight Diaporthe species isolated in our study were compared by inoculating them into the V. vinifera cultivar Summer Black. The inoculated shoots did not show significant lesion development within the first 2 weeks after inoculation. Brown necrotic lesions were detected both on the tissue surface and internally, advancing upwards, and downwards through the inoculation point. Twenty-one days after inoculation, D. gulyae developed the largest lesions (1.23 cm), followed by D. eres (0.94 cm). The remaining species, D. unshiuensis, D. viniferae, D. guangxiensis, D. pescicola, and D. sojae, exhibited similar levels of aggressiveness on grape shoots (Figure 7). Diaporthe hubeiensis was the least aggressive (0.5 cm) among the eight species.
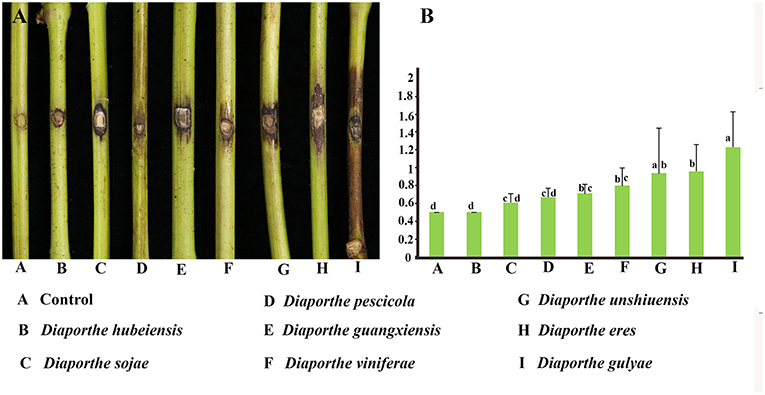
Figure 7. Pathogenicity test results for eight Diaporthe species associated with Chinese grapevines. (A) Variation in the development of lesions. (B) Mean lesion length (cm) at 21 days after inoculation of wounded detached healthy Vitis vinifera (V. vinifera) shoots (n = 10 per species).
Discussion
Grapevine trunk disease has become one of the most devastating grapevine diseases in recent decades. According to data collected worldwide, ~1.5 billion US dollars per year is spent to replace dead grapevines due to these trunk diseases (Hofstetter et al., 2012; Fontaine et al., 2016). This is a great concern among grape-producing countries, as the disease infects perennial parts of the vine and reduces the productive lifespan of vines by several years (Gramaje and Armengol, 2011). The disease ultimately affects the sustainability of the wine industry and table grape production (Fontaine et al., 2016). As the world's top grape-producing country, China has strived to improve the quality and quantity of grapes. Though they are the most important grapevine trunk diseases worldwide, there is no evidence of either the esca complex or Eutypa dieback in China (Fontaine et al., 2016). However, the third most common grapevine trunk disease, caused by the species in Botryosphaeriaceae (Yan et al., 2013, 2018), has been identified as the leading grapevine trunk pathogen complex in China. Unfortunately, over the last few years, diseases caused by Diaporthe species (Dissanayake et al., 2015a, 2017) have become the emerging trunk diseases in China. Understanding the diversity of the causative species and the genetic variation within pathogen populations could help in developing sustainable disease management strategies. In addition, understanding the relationships between European and Chinese isolates can help track disease spread, as both regions share similar disease severity and Diaporthe species that differ from those in North America (Fontaine et al., 2016; Úrbez Torres and O'Gorman, 2019). To achieve these objectives, disease surveys were conducted in six provinces. We isolated and identified 111 Diaporthe strains and showed that they belong to eight species.
In 1958, D. ampelina (= Phomopsis viticola) was identified infecting green shoots of grapevines (Pscheidt and Pearson, 1989). The disease was named “Phomopsis cane and trunk disease.” According to the USDA Fungal—host interaction database, there are 166 records of Diaporthe species associated with grapevines worldwide (https://nt.ars-grin.gov/fungaldatabases/fungushost/fungushost.cfm) (Farr and Rossman, 2019). These records are related to the following 27 Diaporthe species: Diaporthe ambigua (D. ambigua) (Dissanayake et al., 2017), D. ampelina (Úrbez-Torres et al., 2013), Diaporthe amygdali (D. amygdali) (Gomes et al., 2013; Guarnaccia et al., 2018), Diaporthe australafricana (D. australafricana) (Gomes et al., 2013), Diaporthe baccae (D. baccae), D. bohemiae, Diaporthe celeris (D. celeris) (Guarnaccia et al., 2018), Diaporthe chamaeropis (D. chamaeropis) (Lawrence et al., 2015), Diaporthe. Cynaroidis (Lesuthu et al., 2019) Diaporthe cytosporella (D. cytosporella), Diaporthe eres (D. eres), D. foeniculina, Diaporthe helianthi (D. helianthi) (Dissanayake et al., 2017; Guarnaccia et al., 2018; Farr and Rossman, 2019), Diaporthe hispaniae (D. hispaniae), D. hongkongensis (Dissanayake et al., 2017), Diaporthe hungariae (D. hungariae) (Guarnaccia et al., 2018), D. kyushuensis (Kajitani and Kanematsu, 2000), D. nebulae (Lesuthu et al., 2019) Diaporthe neotheicola (D. neotheicola) (Úrbez-Torres et al., 2013), Diaporthe nobilis (D. nobilis) (Dissanayake et al., 2017), D. novem (Lawrence et al., 2015), D. perjuncta (Mostert et al., 2001), Diaporthe perniciosa (D. perniciosa) (Stoykow and Denchev, 2006), D. phaseolorum (Dissanayake et al., 2017), Diaporthe rudis (D. rudis) (Guarnaccia et al., 2018), Diaporthe serafiniae (D. serafiniae) (Lesuthu et al., 2019), and D. sojae (Dissanayake et al., 2017). Among these species D. ampelina is the mostly reported species with 42 records in 12 countries. The present study introduces the three novel taxa D. guangxiensis, D. hubeiensis, and D. viniferae and three new host records: D. gulyae, D. pescicola, and D. unshiuensis.
Diaporthe eres was identified as the most prominent and widespread species associated with grapevine dieback in China (37.5% of total isolates). Other than on grapevines, D. eres has been reported on Aralia elata (A. elata) (Wu et al., 2012), Camellia species (Gao et al., 2016), Citrus species (Huang et al., 2015), peach (Dissanayake et al., 2017), and pear (Bai et al., 2015) plants in China, causing diebacks. Diaporthe eres has been reported in many countries, such as the USA (Úrbez-Torres et al., 2013; Lawrence et al., 2015), Croatia (Kaliterna et al., 2012), Greece (Thomidis and Michailides, 2009), Italy (Cinelli et al., 2016), Latvia (Lombard et al., 2014), Poland (Kowalski et al., 2016), Russia, Serbia (Petrovic et al., 2015), and South Africa (Van Niekerk et al., 2005; Lesuthu et al., 2019) causing diseases on grapevines. These reports reveal that D. eres has a diverse host range and a broad geographical distribution. The second most abundant taxon, D. sojae, has a wide range of hosts as well, including Camptotheca acuminata (C. acuminata) (Chang et al., 2005), Glycine max, Cucumis melo (Lehman, 1923; Santos et al., 2011), Capsicum annuum (C. annuum) (Pennycook, 1989), Stokesia laevis (S. laevis) (Sogonov et al., 2008), and Helianthus annuus (H. annuus) (Thompson et al., 2011). These two Diaporthe species were previously identified and characterized from grapevines in China by Dissanayake et al. (2015a).
The present study recorded three Diaporthe species, D. gulyae, D. pescicola, and D. unshiuensis, associated with Vitis dieback for the first time. Diaporthe gulyae was previously reported on H. annuus in Australia (Thompson et al., 2011), Canada, and the United States (Mathew et al., 2015a,b) and on Carthamus lanatus (C. lanatus) in Italy (Andolfi et al., 2015). Diaporthe pescicola was previously described in association with peach shoot dieback in China (Dissanayake et al., 2017). Diaporthe unshiuensis was first described in China in 2015 as an endophyte of a Citrus sp. (Huang et al., 2015).
The identification and characterization of novel taxa and new host records is an indication of the high potential of Diaporthe to evolve rapidly. Host switching is often related to fungal adaptive ability (Bleuven and Landry, 2016). The changing environments and human interference present both challenges and opportunities for fungi, with some capable of switching from endophytic or saprobic lifestyles to pathogenic styles or becoming more aggressive and colonizing new hosts (Manawasinghe et al., 2018). The novel taxa and the new records reported here for grapevine trunk diseases in China might be due to these factors. During the past decade, northern China has become significantly warmer (Piao et al., 2010). The increased temperature could attract new pests and disease agents to the region. On the other hand, human-mediated factors can also influence the development of a new disease (McDonals, 2004). For example, in commercial grape vineyards, significant amounts of chemicals are applied annually in the form of pesticides and fungicides (Úrbez-Torres, 2011). Such applications could lead to the development of resistant strains of the target organism and non-target micro-fungi (Manawasinghe et al., 2018). Over time, strains and species that are more resistant and/or more aggressive could emerge. The recent identification of new species and new host records of Diaporthe in China and in Europe are consistent with the hypothesis. Studying the genetic diversity of pathogens provides clues to how host switches might have occurred and the genetic basis for new pathogen emergence.
The knowledge of the genetic diversity of a particular phytopathogen can be used to develop sustainable management strategies such as resistance breeding and fungicide screening. In this study, D. eres was analyzed, as it had a relatively large number of isolates from which to obtain reasonable estimates of various intraspecific diversity indices. In this study, multi-locus sequences were used as the marker of choice. The use of sequence data as genetic markers facilitated the analysis of genetic variations among isolates within a population. We selected ITS, β-tubulin, HIS, EF-1α, and CAL gene regions, as they were extensively used in phylogenetic analysis of the genus Diaporthe. In addition, ACT and Apn2 genes were selected since those regions provide a large number of polymorphic sites for the Diaporthe eres species complex (Udayanga et al., 2014b). Genetic polymorphisms are required for both phylogenetic and population genetic studies (Xu, 2006). Using these gene regions, we calculated haplotype richness (hR), the total number of haplotypes, Watterson's theta (Θw), and pairwise nucleotide diversity (JI) for Diaporthe eres obtained from Chinese vineyards.
The combined effect of the mutation, recombination, marker ascertainment, and demography of a particular species can be revealed by analyzing and comparing gene genealogies and haplotype diversities within and between genes (Stumpf, 2004; Xu, 2006). The calculated haplotype diversities of Diaporthe eres were higher than 0.5 for Apn2, CAL, HIS, β-tubulin and the combined data, reflecting high genetic diversity. Tajima's D indicates how much population variation can be sustained over time (Tajima, 1989). In the present study, positive D values were observed for coding gene regions (Apn2, CAL, and HIS). This might be due to selective pressure causing a recent population contraction. The selection pressure could have come from the continuous application of fungicides, leading to the loss of certain genotypes. In contrast, Tajima's D for the combined sequences was negative (−0.20416), which indicates a possible recent population expansion of certain multi-locus genotypes (Tajima, 1989). In Hubei, several multi-locus genotypes were over-represented, consistent with this hypothesis.
The Hudson and Kaplan (1985) index for the recombination between Chinese and European isolates was calculated for this study. In our analysis, we calculated the number of recombination events in the history of a sample of sequences (R) and the number of recombination events that can be parsimoniously inferred from a sample of sequences (Rm) (Hudson, 1983; Kelly, 1997). When the rate of recombination equals zero, R gives zero (Hudson, 1983; Hudson and Kaplan, 1985). Since the R is given a value based on the history of the sample, Rm denotes the minimum number of recombination events implied by the data using the four-gamete test. A positive ZZ value, which reflects intragenic recombination, has played an important role in nucleotide variation and a high number of recombination events (Hudson, 1983). Therefore, we can conclude that recent recombination events might have occurred between the Chinese and European isolates. Haplotype networks provide a better understanding of the coexistence of ancestral and derived haplotypes by providing an account for recombination (Huson and Bryant, 2006). Therefore, haplotype networks are intensively used in intraspecific analyses. We used a median-joining network in which the number of mutations separate haplotypes (Castelloe and Templeton, 1994). In each network, the ancestral haplotype was predicted based on rooting probability (Posada and Crandall, 2001). The analyses suggested that the most recent ancestry of the Chinese haplotypes was shared with the Spanish and Hungarian haplotypes. In addition, haplotypes from the UK and Czech Republic shared ancestry with Chinese haplotypes. Overall, the Diaporthe population in China is genetically diverse and might have an admixture population. The current population is likely derived from a combination of endemic D. eres strains and introduced strains from other regions.
Conclusion
Present study provides an account of Diaporthe species associated with Chinese vineyards by their phylogenetic placements. Collectively, in the present study, 111 Diaporthe strains were isolated and characterized into eight species using both morphological and molecular phylogenetic approaches. To identify those taxa, four gene regions were examined. The combination of ITS, CAL, β-tubulin, and EF-1α genes gave the best species delimitation in the genus Diaporthe. The present study introduced three novel taxa and three host records of Diaporthe associated with Chinese grapevines. The most abundant Diaporthe species was D. eres, which was moderately aggressive. D. gulyae was the most aggressive among the eight species on detached green shoots. The Chinese D. eres population was high in nucleotide diversity and haplotype diversity. In haplotype network analysis, the Chinese population was dispersed in the network but showed a certain degree of clustering according to their geographical origins. This result suggests that there is likely geographic structuring of D. eres in China. However, more in-depth analysis is required using more isolates from different provinces. Haplotype networks including Chinese and European isolates suggest a close relationship between the two populations. This is confirmed by the recombination among isolates from these two regions. Our results suggest that the D. eres population in China might be a result of an admixture. The results presented here provide opportunities for several fields, including grapevine breeding for disease-resistant cultivars, screening for new fungicides, and developing appropriate quarantine and management strategies to prevent and control grapevine dieback diseases.
Data Availability
The sequence data generated in this study is deposited in NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank) and the respective accession numbers are given in Table 2. The Alignment generated in the present study available in TreeBASE (https://treebase.org/treebase-web/home.html) under the 24324.
Author Contributions
JY and XL conceived the research. JY, IM, AD, XL, and WZ planned the basic research. ML, YZ, and WSZ provided materials. IM and AD conducted the experiments and prepared manuscript. IM, AD, DW, and JX analyzed data. KH, SB, and JY revised the manuscript. All authors read and approved the final manuscript.
Funding
This project was funded by Beijing Talent Programme for JY, CARS-29 and D171100001617002.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Milan Samarakoon for his guidance in developing phylogenetic trees. We would like to thank the students and non-academic staff of Beijing Key Laboratory of Environment Friendly Management on Fruit Diseases and Pests in North China, for their help with pathogenicity assays. DW would like to thank CAS President's International Fellowship Initiative (PIFI) for funding his postdoctoral research (No. 2019PC0008) as well as the National Science Foundation of China and the Chinese Academy of Sciences for his financial support under the following Grants: 41761144055, 41771063, and Y4ZK111B01.
References
Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., and Walter, P. (2002). “Introduction to Pathogens,” in Molecular Biology of the Cell, 4th Edn (New York, NY: Garland Science). Available online at: https://www.ncbi.nlm.nih.gov/books/NBK26917
Andolfi, A., Boari, A., Evidente, M., Cimmino, A., Vurro, M., Ash, G., et al. (2015). Gulypyrones A and B and phomentrioloxins B and C produced by Diaporthe gulyae, a potential mycoherbicide for saffron thistle (Carthamus lanatus). J. Nat. Prod. 78, 623–629. doi: 10.1021/np500570h
Bai, Q., Zhai, L., Chen, X., Hong, N., Xu, W., and Wang, G. (2015). Biological and molecular characterization of five Phomopsis species associated with pear shoot canker in China. Plant Dis. 99, 1704–1712. doi: 10.1094/PDIS-03-15-0259-RE
Bandelt, H. J., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Baumgartner, K., Fujiyoshi, P. T., Travadon, R., Castlebury, L. A., Wilcox, W. F., and Rolshausen, P. E. (2013). Characterization of species of Diaporthe from wood cankers of grape in eastern North American vineyards. Plant Dis. 97, 912–920. doi: 10.1094/PDIS-04-12-0357-RE
Bleuven, C., and Landry, C. R. (2016). Molecular and cellular bases of adaptation to a changing environment in microorganisms. Proc. R. Soc. 283:20161458. doi: 10.1098/rspb.2016.1458
Carbone, I., and Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91, 553–556. doi: 10.1080/00275514.1999.12061051
Castelloe, J., and Templeton, A. R. (1994) Root probabilities for intraspecific gene trees under neutral coalescent theory. Mol. Phylogenet. Evol. 3, 102–113. doi: 10.1006/mpev.1994.1013
Chang, C., Xi, P., Xiang, M., Jiang, Z., and Chi, P. (2005). New species of Phomopsis on woody plants in hunan province. Mycosystema 2, 145–154. Available online at: http://www.oalib.com/paper/1631068
Cinelli, T., Mondello, V., Marchi, G., Burruano, S., Alves, A., and Mugnai, L. (2016). First report of Diaporthe eres associated with cane blight of grapevine (Vitis vinifera) in Italy. Plant Dis. 100:532. doi: 10.1094/PDIS-08-15-0872-PDN
Crous, P. W., Groenewald, J. Z., Risede, J. M., Simoneau, P., and Hywel-Jones, N. L. (2004). Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Stud. Mycol. 50, 415–430. Available online at: http://edepot.wur.nl/31307
Daykin, M. E., and Milholland, R. D. (1984). Histopathology of ripe rot caused by Colletotrichum gloeosporioides on muscadine grape. J. Phytopathol. 74, 1339–1341. doi: 10.1094/Phyto-74-1339
Dissanayake, A. J., Liu, M., Zhang, W., Chen, Z., Udayanga, D., Chukeatirote, E., et al. (2015a). Morphological and molecular characterization of Diaporthe species associated with grapevine trunk disease in China. Fungal Biol. 119, 283–294. doi: 10.1016/j.funbio.2014.11.003
Dissanayake, A. J., Zhang, W., Liu, M., Hyde, K. D., Zhao, W. S., Li, X. H., et al. (2017). Diaporthe species associated with peach tree dieback in Hubei, China. Mycosphere 8, 512–528. doi: 10.5943/mycosphere/8/5/2
Dissanayake, A. J., Zhang, W., Mei, L., Chukeatirote, E., Yan, J. Y., Li, X., et al. (2015b). Lasiodiplodia pseudotheobromae causes pedicel and peduncle discolouration of grapes in China. Australas. Plant. Dis. 10:21. doi: 10.1007/s13314-015-0170-5
Doilom, M., Dissanayake, A. J., Wanasinghe, D. N., Boonmee, S., Liu, J. K., Bhat, D. J., et al. (2017). Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Divers. 82, 107–182. doi: 10.1007/s13225-016-0368-7
Farr, D. F., and Rossman, A. Y. (2019). Fungal Databases - Fungus-Host Distributions, Systematic Mycology and Microbiology Laboratory, ARS, USDA. Available online at: http://nt.ars-grin.gov/fungaldatabases (accessed March 11, 2019).
Fontaine, F., Gramaje, D., Armengol, J., Smart, R., Nagy, Z. A., Borgo, M., et al. (2016). Grapevine Trunk Diseases A Review, OIV Publications, 1st Edn. Paris.
Gadoury, D. M., Cadle-Davidson, L., Wilcox, W. F., Dry, I. B., Seem, R. C., and Milgroom, M. G. (2012). Grapevine powdery mildew Erysiphe necator: a fascinating system for the study of the biology, ecology and epidemiology of an obligate biotroph. Mol. Plant. Pathol. 13, 1–16. doi: 10.1111/j.1364-3703.2011.00728.x
Gao, Y., Liu, F., Duan, W., Crous, P. W., and Cai, L. (2017). Diaporthe is paraphyletic. Ima Fungus 8, 153–187. doi: 10.5598/imafungus.2017.08.01.11
Gao, Y. H., Liu, F., and Cai, L. (2016). Unravelling Diaporthe species associated with Camellia. Syst. Biodivers. 14, 102–117. doi: 10.1080/14772000.2015.1101027
Glass, N. L., and Donaldson, G. C. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microb. 61, 1323–1330. Available online at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC167388/
Gomes, R. R., Glienke, C., Videira, S. I. R., Lombard, L., Groenewald, J. Z., and Crous, P. W. (2013). Diaporthe: a genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31, 1–41. doi: 10.3767/003158513X666844
Gramaje, D., and Armengol, J. (2011). Fungal trunk pathogens in the grapevine propagation process: potential inoculum sources, detection, identification, and management strategies. Plant Dis. 95, 1040–1055. doi: 10.1094/PDIS-01-11-0025
Greer, L. A., Harper, J. D., Savocchia, S., Samuelian, S. K., and Steel, C. C. (2011). Ripe rot of south-eastern Australian wine grapes is caused by two species of Colletotrichum: C. acutatum and C gloeosporioides with differences in infection and fungicide sensitivity. Aust. J. Grape. Wine. Res. 17, 123–128. doi: 10.1111/j.1755-0238.2011.00143.x
Guarnaccia, V., and Crous, P. W. (2017) Emerging citrus diseases in Europe caused by Diaporthe spp. IMA Fungus 8, 317– 334. doi: 10.5598/imafungus.2017.08.02.07
Guarnaccia, V., and Crous, P. W. (2018). Species of Diaporthe on Camellia and Citrus in the Azores Islands. Phytopathol. Mediter 57, 307–319. doi: 10.14601/Phytopathol_Medi-terr-23254
Guarnaccia, V., Groenewald, J. Z., Woodhall, J., Armengol, J., Cinelli, T., and Eichmeier, A. (2018). Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia 40, 135–153. doi: 10.3767/persoonia.2018.40.06
Guarnaccia, V., Vitale, A., Cirvilleri, G., Aiello, D., Susca, A., Epifani, F., et al. (2016). Characterisation and pathogenicity of fungal species associated with branch cankers and stem-end rot of avocado in Italy. Eur. J. Plant Pathol. 146, 963–976. doi: 10.1007/s10658-016-0973-z
Hall, T. (2006). Bioedit. Department of Microbiology, North Carolina State University Available online at: http://www.mbio.ncsu.edu/BioEdit/page2.html
Hofstetter, V., Buyck, B., Croll, D., Viret, O., Couloux, A., and Gindro, K. (2012). What if esca disease of grapevine were not a fungal disease. Fungal Divers 4, 51–67. doi: 10.1007/s13225-012-0171-z
Hong, S. K., Kim, W. G., Yun, H. K., and Choi, K. J. (2008). Morphological variations, genetic diversity and pathogenicity of Colletotrichum species causing grape ripe rot in Korea. J. Plant Pathol. 24, 269–278. doi: 10.5423/PPJ.2008.24.3.269
Huang, F., Udayanga, D., Wang, X., Hou, X., Mei, X., Fu, Y., et al. (2015). Endophytic Diaporthe associated with Citrus: a phylogenetic reassessment with seven new species from China. Fungal. Biol. 119, 331–347. doi: 10.1016/j.funbio.2015.02.006
Hudson, R. R. (1983). Properties of a neutral allele model with intragenic recombination. Theor. Popul. Biol. 23, 183–201. doi: 10.1016/0040-5809(83)90013-8
Hudson, R. R., and Kaplan, N. L. (1985). Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111, 147–164.
Huson, D. H., and Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. doi: 10.1093/molbev/msj030
Hyde, K. D., Hongsanan, S., Jeewon, R., Bhat, D. J., McKenzie, E. H. C., Jones, E. B. G., et al. (2016). Fungal diversity notes 367-491: taxonomic and phylogenetic contributions to fungal taxa. Fungal. Divers. 80, 1–270. doi: 10.1007/s13225-016-0373-x
Hyde, K. D., Nilsson, R. H., Alias, S. A., Ariyawansa, H. A., Blair, J. E., Cai, L., et al. (2014). One stop shop: backbone trees for important phytopathogenic genera: I. Fungal. Divers. 67, 21–125. doi: 10.1007/s13225-014-0298-1
Jayasiri, S. C., Hyde, K. D., Ariyawansa, H. A., Bhat, J., Buyck, B., Cai, L., et al. (2015). The faces of fungi database: fungal names linked with morphology, molecular and human attributes. Fungal. Divers. 74, 3–18. doi: 10.1007/s13225-015-0351-8
Jayawardena, R. S., Zhang, W., Liu, M., Maharachchikumbura, S. S., Zhou, Y., Huang, J., et al. (2015). Identification and characterization of Pestalotiopsis-like fungi related to grapevine diseases in China. Fungal. Biol. 119, 348–361. doi: 10.1016/j.funbio.2014.11.001
Jeewon, R., and Hyde, K. D. (2016) Establishing species boundaries new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7, 1669–1677. doi: 10.5943/mycosphere/7/11/4
Kajitani, Y., and Kanematsu, S. (2000). Diaporthe kyushuensis, sp. nov. the teleomorph of the causal fungus of grapevine swelling arm in Japan, and its anamorph Phomopsis vitimegaspora. Mycoscience. 41, 111–114. doi: 10.1007/BF02464318
Kaliterna, J., Milicević, T., and Cvjetković, B. (2012). Grapevine trunk diseases associated with fungi from the Diaporthaceae family in Croatian vineyards. Arch. Indust. Hygiene Toxicol. 63, 471–479. doi: 10.2478/10004-1254-63-2012-2226
Katoh, K., and Toh, H. (2010). Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9, 286–298. doi: 10.1093/bib/bbn013
Kelly, J. K. (1997). A test of neutrality based on interlocus associations. Genetics 146, 1197–1206.
Kishino, H., and Hasegawa, M. (1989). Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data. J. Mol. Evol. 29, 170–179. doi: 10.1007/BF02100115
Kowalski, T., Kraj, W., and Bednarz, B. (2016). Fungi on stems and twigs in initial and advanced stages of dieback of European ash Fraxinus excelsior in Poland. Eur. J. For. Res. 135, 565–579. doi: 10.1007/s10342-016-0955-x
Kuo, K. C., and Leu, L. S. (1998) Phomopsis vitimegaspora: a pathogenic Phomopsis from vines. Mycotaxon 66, 497–499.
Lawrence, D. P., Renaud, T., and Kendra, B. (2015). Diversity of Diaporthe species associated with wood cankers of fruit and nut crops in northern California. Mycologia 107, 926–940. doi: 10.3852/14-353
Lehman, S. G. (1923). Pod and stem blight of soybean. Ann. Missouri Botan. Gardens 10, 111–178. doi: 10.2307/2394035
Lesuthu, P., Mostert, L., Spies, C. F., Moyo, P., Regnier, T., and Halleen, F. (2019) Diaporthe nebulae sp. nov. and first report of D. cynaroidis, D. novem, and D. serafiniae on Grapevines in South Africa. Plant Dis. 103, 808–817. doi: 10.1094/PDIS-03-18-0433-RE
Librado, P., and Rozas, J. (2009). DnaSPv5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lindahl, J. F., and Grace, D. (2015). The consequences of human actions on risks for infectious diseases: a review. Infect. Ecol. and Epidemiol. 5:30048. doi: 10.3402/iee.v5.30048
Liu, J. K., Hyde, K. D., Jones, E. B. G., Ariyawansa, H. A., Bhat, D. J., Boonmee, S., et al. (2015). Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal. Divers. 72, 1–197. doi: 10.1007/s13225-015-0324-y
Lombard, L., Van Leeuwen, G. C., Guarnaccia, V., Polizzi, G., Van Rijswick, P. C., Rosendahl, K. C., et al. (2014). Diaporthe species associated with Vaccinium, with specific reference to Europe. Phytopathol. Mediter. 53, 287–299. doi: 10.14601/Phytopathol_Mediterr-14034
Manawasinghe, I. S., Zhang, W., Li, X., Zhao, W., Chethana, K. T., Xu, J., et al. (2018). Novel microsatellite markers reveal multiple origins of Botryosphaeria dothidea causing the Chinese grapevine trunk disease. Fungal. Ecol. 33, 134–142. doi: 10.1016/j.funeco.2018.02.004
Marin-Felix, Y., Hernández-Restrepo, M., Wingfield, M. J., Akulov, A., Carnegie, A. J., Cheewangkoon, R., et al. (2019) Genera of phytopathogenic fungi: GOPHY 2. Stud. Mycol. 92, 47–133. doi: 10.1016/j.simyco.2018.04.002
Mathew, F. M., Alananbeh, K. M., and Jordahl, J. G. (2015b). Phomopsis stem canker: a re-emerging threat to sunflower (Helianthus annuus) in the United States. Phytopathology 105, 990–997. doi: 10.1094/PHYTO-11-14-0336-FI
Mathew, F. M., Rashid, K. Y., and Gulya, T. J. (2015a). First report of Phomopsis stem canker of sunflower (Helianthus annuus) caused by Diaporthe gulyae in Canada. Plant. Dis. 99, 160–160. doi: 10.1094/PDIS-08-14-0858-PDN
McDonals, B. M. (2004). Population Genetics of Plant Pathogens. The Plant Health Instructor. Available online at: https://doi.org/10.1094/PHI-A-2004-0524-01 (assessed Novermber 30, 2018).
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees, in Proceedings of the Gateway Computing Environments Workshop (GCE) 14 Nov 2010. Institute of Electrical and Electronics Engineers, New Orleans, LA,1-8. doi: 10.1109/GCE.2010.5676129
Mondal, S. N., Vincent, A., Reis, R. F., and Timmer, L. W. (2007). Saprophytic colonization of citrus twigs by Diaporthe citri and factors affecting pycnidial production and conidial survival. Plant Dis. 91, 387–392. doi: 10.1094/PDIS-91-4-0387
Mondello, V., Songy, A., Battiston, E., Pinto, C., Coppin, C., Trotel-Aziz, P., et al. (2018). Grapevine trunk diseases: a review of fifteen years of trials for their control with chemicals and biocontrol agents. Plant. Dis. 102, 1189–1217. doi: 10.1094/PDIS-08-17-1181-FE
Mostert, L., Crous, P. W., Kang, J. C., and Phillips, A. J. (2001). Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa: morphological, cultural, molecular and pathological characterization. Mycologia 93, 146–167. doi: 10.1080/00275514.2001.12061286
Muntañola-Cvetković, M., Mihaljčević, M., and Petrov, M. (1981). On the identity of the causative agent of a serious Phomopsis-Diaporthe disease in sunflower plants. Nova Hedwigia 34, 417–435.
Nylander, J. A. A. (2004). MrModeltest 2.0. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
OIV (2016). (International Organization of vine and wine Intergovernmental Organization) World congress of vine and Wine. Statistical report on world vitiviniculture Bento Gonçalves Brazil. 28th October 2016.
Pearson, R. C., and Goheen, C. (1994). “Phomopsis cane and leaf spot,” in Compendium of Grape Diseases, eds W. B. Hewitt and R. C. Pearson (St Paul, MI: APS Press, 17–18.
Pennycook, S. R. (1989). Plant Diseases Recorded in New Zealand, Vol. 3, Plant Disease Division Auckland: DSIR.
Petrovic, K., Vidic, M., and Riccioni, L. (2015). First report of Diaporthe eres species complex causing seed decay of soybean in Serbia. Plant. Dis. 99, 1186–1186. doi: 10.1094/PDIS-01-15-0056-PDN
Piao, S., Ciais, P., Huang, Y., Shen, Z., Peng, S., Li, J., et al. (2010). The impacts of climate change on water resources and agriculture in China. Nature 467, 43–51. doi: 10.1038/nature09364
Posada, D., and Crandall, K. A. (2001). Intraspecific gene genealogies: trees grafting into networks. TREE 16, 37–45. doi: 10.1016/S0169-5347(00)02026-7
Pscheidt, J. W., and Pearson, R. C. (1989). Time of infection and control of Phomopsis fruit rot of Grape. Plant. Dis. 73, 829–833 doi: 10.1094/PD-73-0829
Rawnsley, B., Wicks, T. J., Stummer, B. E., and Scott, E. S. (2006). The role of Diaporthe perjuncta, in delayed budburst and death of grapevine buds. Australas. Plant. Pathol. 35, 265–273.
Rayner, R. W. (1970). A Mycological Colour Chart. Commonwealth Mycological Institute (Great Britain), & British Mycological Society.
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Rossman, A., Udayanga, D., Castlebury, L. A., and Hyde, K. D. (2014). (2304) Proposal to conserve the name Diaporthe eres against twenty-one competing names (Ascomycota: Diaporthales: Diaporthaceae). Taxon 63, 934–935. doi: 10.12705/634.23
Santos, J. M., Vrandečić, K., Cosić, J., Duvnjak, T., and Phillips, A. J. (2011). Resolving the Diaporthe species occurring on soybean in Croatia. Persoonia 27, 9–19. doi: 10.3767/003158511X603719
Senanayake, I. C., Crous, P. W., Groenewald, J. Z., Maharachchikumbura, S. S., Jeewon, R., Phillips, A. J. L., et al. (2017). Families of diaporthales based on morphological and phylogenetic evidence. Stud. Mycol. 86, 217–296. doi: 10.1016/j.simyco.2017.07.003
Silvestro, D., and Michalak, I. (2010). RaxmlGUI: a Graphical Front-End for RAxML. Available online at: http://sourceforgenet/projects/raxmlgui/ (accessed February 25, 2019).
Sogonov, M. V., Castlebury, L. A., Rossman, A. Y., Mejía, L. C., and White, J. F. (2008). Leaf-inhabiting genera of the gnomoniaceae, diaporthales. Stud. Mycol. 62, 1–79. doi: 10.3114/sim.2008.62.01
Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033.
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57,758–771. doi: 10.1080/10635150802429642
Stoykow, D. Y., and Denchev, C. M. (2006). Current knowledge of Diaporthales (Ascomycota) in Bulgaria. Mycol. Balcan. 3, 179–185. doi: 10.5281/zenodo.2547618
Stumpf, M. P. (2004). Haplotype diversity and SNP frequency dependence in the description of genetic variation. Eur. J. Hum. Genet. 12, 469–477. doi: 10.1038/sj.ejhg.5201179
Swofford, D. L. (2003). PAUP: Phylogenetic Analysis Using Parsimony and Other Methods, Version 4 Sinauer Associates, Sunderland, Massachusetts.
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595.
Thomidis, T., and Michailides, T. J. (2009). Studies on Diaporthe eres as a new pathogen of peach trees in Greece. Plant. Dis. 93, 1293–1297. doi: 10.1094/PDIS-93-12-1293
Thompson, S. M., Tan, Y. P., Young, A. J., Neate, S. M., Aitken, E. A. B., and Shivas, R. G. (2011). Stem cankers on sunflower Helianthus annuus in Australia reveal a complex of pathogenic Diaporthe (Phomopsis) species. Persoonia 27, 80–89. doi: 10.3767/003158511X617110
Tibpromma, S., Hyde, K. D., Bhat, J. D., Mortimer, P. E., Xu, J., Promputtha, I., et al. (2018). Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from southern Thailand. MycoKeys 33, 25–67. doi: 10.3897/mycokeys.33.23670
Udayanga, D., Castlebury, L. A., Rossman, A. Y., Chukeatirote, E., and Hyde, K. D. (2014b). Insights into the genus Diaporthe: phylogenetic species delimitation in the D. eres species complex. Fungal Div. 67, 203–229. doi: 10.1007/s13225-014-0297-2
Udayanga, D., Castlebury, L. A., Rossman, A. Y., Chukeatirote, E., and Hyde, K. D. (2015). The Diaporthe sojae species complex: phylogenetic re-assessment of pathogens associated with soybean, cucurbits and other field crops. Fungal. Biol. 119, 383–407. doi: 10.1016/j.funbio.2014.10.009
Udayanga, D., Castlebury, L. A., Rossman, A. Y., and Hyde, K. D. (2014a). Species limits in Diaporthe: molecular re-assessment of D citri, D cytosporella, D foeniculina and D rudis. Persoonia 32, 83–101. doi: 10.3767/003158514X679984
Udayanga, D., Liu, X., McKenzie, E. H., Chukeatirote, E., Bahkali, A. H., and Hyde, K. D. (2011). The genus Phomopsis: biology, applications, species concepts and names of common pathogens. Fungal. Divers. 50, 189–225. doi: 10.1007/s13225-011-0126-9
Udayanga, D., Liu, X. X., Crous, P. W., McKenzie, E. H. C., Chukeatirote, E., and Hyde, K. D. (2012b). Multilocus phylogeny of Diaporthe reveals three new cryptic species from Thailand. Cryptogam. Mycol. 33, 295–309. doi: 10.7872/crym.v33.iss3.2012.295
Udayanga, D., Liu, X. Z., Crous, P. W., McKenzie, E. H. C., Chukeatirote, E., and Hyde, K. D. (2012a). A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal. Divers. 56,157–171. doi: 10.1007/s13225-012-0190-9
Úrbez Torres, J. R., and O'Gorman, D. T. (2019). Grapevine Trunk Diseases in British Columbia: From Identification to Control. Summerland Research and Development Centre, Agriculture and Agri-Food Canada.
Úrbez-Torres, J. R. (2011). The status of Botryosphaeriaceae species infecting grapevines. Phytopathol. Mediterr. 50, 5–45. doi: 10.14601/Phytopathol_Mediterr-9316
Úrbez-Torres, J. R., Peduto, F., Smith, R. J., and Gubler, W. D. (2013) Phomopsis Dieback: a grapevine trunk disease caused by Phomopsis viticola in California. Plant. Dis. 97, 1571–1579. doi: 10.1094/PDIS-11-12-1072-RE
Van Niekerk, J. M., Groenewald, J. Z., Farr, D. F., Fourie, P. H., Halleer, F., and Crous, P. W. (2005). Re-assessment of Phomopsis species on grapevines. Austr. Plant. Pathol. 34, 27–39. doi: 10.1071/AP04072
White, C., Halleen, F., Fischer, M., and Mostert, L. (2011). Characterisation of the fungi associated with esca diseased grapevines in South Africa. Phytopathol. Mediterr. 50, 204–223.
White, T. J., Bruns, T., Lee, S. J. W. T., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols 18, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wilcox, W. A., Ellis, M. A., Ramsley, B., Rossaman, A., and Pscheidt, J. (2015). “Phomopsis cane and leaf spot,” in Compendium of Grape Diseases, Disorders, and Pests, 2nd Edn, eds W. F. Wilcox, W. D. Gubler, and J. K. Uyemoto (S. Paul, MN: APS Press), 46–51. doi: 10.1094/9780890544815
Wu, H., Yang, H., You, X., and Li, Y. (2012). Isolation and characterization of saponin-producing fungal endophytes from Aralia elata in Northeast China. Int. J. Mol. Sci. 13, 16255–16266. doi: 10.3390/ijms131216255
Xu, J. (2006). Fundamentals of fungal molecular population genetic analyses. Curr. Issues. Mol. Boil. 8, 75–90. doi: 10.21775/cimb.008.075
Yan, J. Y., Jayawardena, M. M. R. S., Goonasekara, I. D., Wang, Y., Zhang, W., Liu, M., et al. (2015). Diverse species of Colletotrichum associated with grapevine anthracnose in China. Fungal. Divers. 71, 233–246. doi: 10.1007/s13225-014-0310-9
Yan, J. Y., Xie, Y., Zhang, W., Wang, Y., Liu, J. K., Hyde, K. D., et al. (2013). Species of Botryosphaeriaceae involved in grapevine dieback in China. Fungal. Divers. 61, 221–236. doi: 10.1007/s13225-013-0251-8
Yan, J. Y., Zhao, W. S., Chen, Z., Xing, Q. K., Zhang, W., Chethana, K. W. T., et al. (2018). Comparative genome and transcriptome analyses reveal adaptations to opportunistic infections in woody plant degrading pathogens of Botryosphaeriaceae. DNA Res. 25, 87–102. doi: 10.1093/dnares/dsx040
Yang, Q., Fan, X. L., Guarnaccia, V., and Tian, C. M. (2018). High diversity of Diaporthe species associated with twelve new species described. MycoKeys 39, 97–149. doi: 10.3897/mycokeys.39.26914
Keywords: novel species, new host record, network analysis, phylogeography, phomopsis
Citation: Manawasinghe IS, Dissanayake AJ, Li X, Liu M, Wanasinghe DN, Xu J, Zhao W, Zhang W, Zhou Y, Hyde KD, Brooks S and Yan J (2019) High Genetic Diversity and Species Complexity of Diaporthe Associated With Grapevine Dieback in China. Front. Microbiol. 10:1936. doi: 10.3389/fmicb.2019.01936
Received: 21 May 2019; Accepted: 06 August 2019;
Published: 02 September 2019.
Edited by:
Giorgio Gambino, Institute for Sustainable Plant Protection (CNR), ItalyReviewed by:
Vladimiro Guarnaccia, University of Turin, ItalyFrancois Halleen, Agricultural Research Council of South Africa (ARC-SA), South Africa
Copyright © 2019 Manawasinghe, Dissanayake, Li, Liu, Wanasinghe, Xu, Zhao, Zhang, Zhou, Hyde, Brooks and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiye Yan, aml5ZXlhbkB2aXAuMTYzLmNvbQ==
†These authors have contributed equally to this work
 Ishara S. Manawasinghe
Ishara S. Manawasinghe Asha J. Dissanayake
Asha J. Dissanayake Xinghong Li
Xinghong Li Mei Liu
Mei Liu Dhanushka N. Wanasinghe
Dhanushka N. Wanasinghe Jianping Xu
Jianping Xu Wensheng Zhao
Wensheng Zhao Wei Zhang
Wei Zhang Yueyan Zhou
Yueyan Zhou Kevin D. Hyde
Kevin D. Hyde Siraprapa Brooks
Siraprapa Brooks Jiye Yan
Jiye Yan